Abstract
Cartilage cannot self-repair and thus regeneration is a promising approach to its repair. Here we developed new electrospun nanofibers, made of poly (ε-caprolactone)/polytetrahydrofuran (PCL-PTHF urethane) and collagen I from calf skin (termed PC), to trigger the chondrogenic differentiation of mesenchymal stem cells (MSCs) and the cartilage regeneration in vivo. We found that the PC nanofibers had a modulus (4.3 Mpa) lower than the PCL-PTHF urethane nanofibers without collagen I from calf skin (termed P) (6.8 Mpa) although both values are within the range of the modulus of natural cartilage (1–10 MPa). Both P and PC nanofibers did not show obvious difference in the morphology and size. Surprisingly, in the absence of the additional chondrogenesis inducers, the softer PC nanofibers could induce the chondrogenic differentiation in vitro and cartilage regeneration in vivo more efficiently than the stiffer P nanofibers. Using mRNA-sequence analysis, we found that the PC nanofibers outperformed P nanofibers in inducing chondrogenesis by specifically blocking the NF-kappa B signaling pathway to suppress inflammation. Our work shows that the PC nanofibers can serve as building blocks of new scaffolds for cartilage regeneration and provides new insights on the effect of the mechanical properties of the nanofibers on the cartilage regeneration.
Keywords: Poly(ε-caprolactone)/polytetrahydrofuran, (PCL-PTHF) nanofibers, Collagen type I, Stiffness, Mesenchymal stem cells (MSCs), Cartilage tissue engineering
1. Introduction
Cartilage lacks the ability to self-repair after significant lesions or disease. This problem has challenged orthopedic surgeons for decades [1]. As mesenchymal stem cells (MSCs) are multipotent with low immunogenicity and self-renewal capability, and can differentiate into a variety of cells such as chondrocytes, osteoblasts, cardiomyocytes and etc., they have been widely used in cartilage tissue engineering [2,3]. Although stem cells hold promise in cartilage regeneration, directed differentiation through cell-scaffold interaction is seldom reported. A better understanding of the interaction between MSCs and the matrices is of fundamental importance to stem cell-based therapy of cartilage defects.
It is known that mechanical signals from materials have a profound impact on the lineage specification of MSCs [4–6]. The matrix stiffness is one of the most important physical attributes of the solid extracellular matrix (ECM) and directs the lineage specification of naive MSCs [7,8]. For instance, matrix with intermediate stiffness resembling the elasticity of muscle tissue induce myogenic commitment, while relatively hard matrix mimicking collagenous bone cause commitment to an osteogenic lineage specification [7,9]. For chondrogenic differentiation in the presence of growth factors, less stiff substrates, including natural, synthetic or hybrid materials, are preferable [10]. MSCs on soft substrates have a greater expression of chondrogenic markers COL2A1/ACAN and a lower expression level of non-chondrogenic marker COL1A1 [11–13]. However, these scaffolds induced chondrogenic differentiation in the presence of growth factors. Currently, the mechanical properties of most scaffolds can hardly be tuned to match the stiffness of natural cartilage (1–10 MPa) [14,15]. Appropriate substrate that can trigger chondrogenesis in the absence of cytokines and growth factors has not been reported yet, not to mention the corresponding signaling pathways, probably because the substrates do not have cartilage-like mechanical property.
Poly (ε-caprolactone) (PCL) is one of the most exclusively investigated polymers that has been engineered into electrospun nanofibers for cartilage tissue engineering applications. PCL is a biodegradable aliphatic linear polyester approved by FDA. It can be made into nanofibers by electrospinning [16]. Electrospinning is an established processing method for producing nanofibrous structures that mimic the native extra cellular matrix. Compared to other tissue engineering scaffolds, electrospun nanofibers exhibit several advantages, such as high surface area and high porosity. By adjusting the solution parameters and spinning conditions, electrospinning can easily control the fiber properties, including the diameter, structure, surface and mechanical properties of the fibers for constructing scaffolds to facilitate the differentiation of stem cells into different types of cells including osteoblasts [17], neuronal cells [18], chondrocytes [19], and cardiomyocytes [20]. PCL can be easily synthesized with polytetrahydrofuran (PTHF) to form PCL-PTHF copolymers [21]. Other components such as 1,4-butanediol (1,4-BD) [22] and 2-ureido-4 [1H]-pyrimidinone (UPy) [23] were incorporated into PCL-PTHF. One of the most important features of most PCL-PTHF-based fibers is that they are soft and elastic with the Young’s modulus [22,23] almost within the range of natural cartilage (1–10 MPa) [3,14,24]. However, PCL-PTHF has never been applied to chondrogenesis and cartilage tissue engineering probably because PCL-PTHF lacks bioactivity, which is unfavorable for cell differentiation.
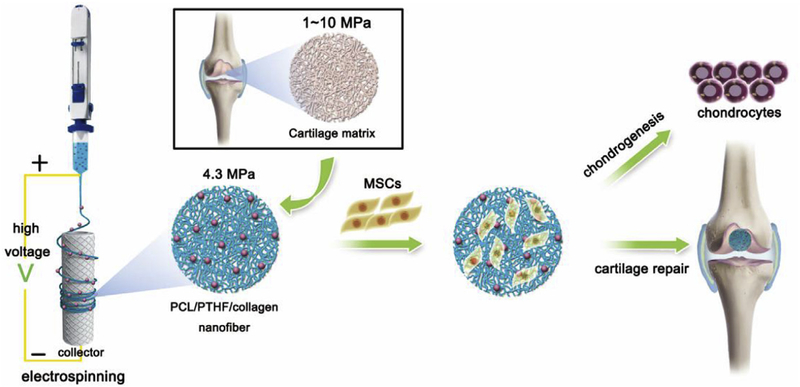
To fill this gap, here we synthesized novel soft and elastic electrospun nanofiber membranes made of PCL-PTHF polyurethane and calf skin derived type I collagen. The resultant nanofibers exhibited a stiffness similar to natural cartilage. We then investigated their potential in chondrogenesis and cartilage repair (Fig. 1). The PCL-PTHF polyurethane without and with type I collagen is termed as PCL-PTHF (P) and PCL-PTHF/collagen (PC), respectively. We found that P and PC nanofiber membranes, especially PC, induced chondrogenic differentiation and promoted cartilage regeneration efficiently. We also explored the signaling mechanisms by which the matrix stiffness induces chondrogenesis.
Fig. 1.
Schematic description of cartilage-mimicking electrospun PC nanofiber membranes for inducing chondrogenic differentiation and cartilage regeneration.
2. Materials and methods
2.1. Synthesis and characterization of poly(PCL-PTHF urethane)
All reagents were obtained from Sigma-Aldrich (Billerica, MA, USA) without further purification. Mn of the poly (ε-caprolactone)-diol (PCL-diol) and poly (tetrahydrofuran carbonate) diol (PTHF-diol) was ca. 2000. The nuclear magnetic resonance (NMR) and differential scanning calorimetry (DSC) analysis of PCL-diol and PTHF-diol are described in Figure S1 and Table S1. Poly (PCL-PTHF urethane) was synthesized from PCL-diol and PTHF-diol using hexamethylenediisocyanate (HDI, 98%) as a coupling reagent and a 1/1 ratio of HDI and the reactive OH groups. The synthesis method is generally similar to those reported by our group [25,26]. The yield was greater than 80% after isolation and purification. The obtained poly (PCL-PTHF urethane) was characterized by 1H NMR and gel permeation chromatography (GPC).
2.2. Electrospinning to form nanofibers
Poly (PCL-PTHF urethane) and calf skin derived collagen type I (Advanced Biomatrix, CA, USA) were mixed in 1,1,1,3,3,3-hexafluoro-2-propanol at a mass ratio of 90:10 between poly (PCL-PTHF urethane) and collagen type I. This mass ratio was chosen because the poly (PCL-PTHF) already displayed a desired mechanical property and only a small amount of collagen was needed to improve its bioactivity. The total concentration of the solution was 5% (w/v). After 72 h of stirring, the solution was transferred into a 10 ml syringe. Electrospinning was conducted at a pumping ratio of 1 ml/h and a voltage of 12 kV. The resultant nanofibers (termed PC nanofibers) were collected onto a collector wrapped with an aluminum foil (15 cm away from the needle tip) and dried overnight. Neat poly (PCL-PTHF urethane) nanofibers, termed P nanofibers, were prepared in the absence of collagen using the same parameters.
2.3. Nanofibers characterization
The surface topographies of the electrospun fibers were characterized using scanning electron microscopy (SEM, JSM6700F, JEOL, Japan). The mechanical properties, including tensile strength (σTS), Young’s modulus (Е) and elongation at break (εb), were measured using a uniaxial tensile testing technique with a 10-N load capacity at a rate of 10 mm/min. Thermogravimetric analysis (TGA) and DSC were used to determine the thermal properties, including enthalpy change (ΔHm) and crystallinity (Xc).
2.4. Cell culture and cell seeding
MSCs were derived from the femoral marrow of 7-day-old Sprague Dawley rats. The newborn Sprague Dawley rats were sacrificed by an overdose injection of pentobarbital sodium salt (Solarbio, Beijing, China). Bilateral femurs were harvested with the proximal and distal ends snipped off. Then, the femoral marrow cavities were washed with alpha-modified Eagle’s medium (α-MEM, Gibco, Beijing, China) and 1% penicillin-streptomycin (Solarbio, Beijing, China). The extracted MSCs were cultured in the culture media containing α-MEM, 10% fetal bovine serum, and 1% penicillin/streptomycin. The medium was changed every 2 or 3 days.
Passage 3 MSCs were seeded on the P and PC nanofiber membranes that were placed in 24-well plates (Corning, NY, USA) at a density of 2 × 104 cells/cm2 respectively. The cells cultured on glass coverslips (GC) at the same cellular density were used as controls. The cell-scaffold composites were cultured in the normal culture medium containing α-MEM, 10% FBS and 1% penicillin-streptomycin. Samples were harvested after 7, 14 and 21 days, respectively, for subsequent analyses.
2.5. Scanning electron microscope (SEM) detection
Cells on the scaffolds were observed using SEM. After 7, 14 and 21 days of culturing, the samples were fixed using 2.5% glutaraldehyde at 4 °C for 2 h after being washed three times using PBS. Then, the fixed samples were gradually dehydrated, and freezedried. After being sprayed with gold, the samples were observe dusing JEOL (Tokyo, Japan) JSM-6300V at 10 kV.
2.6. Cell viability and proliferation assay
After 7, 14 and 21 days of culturing, the cell viability was tested using a LIVE/DEAD® Viability/Cytotoxicity Kit (Thermo Fisher, MA, USA). Cell proliferation was assessed using a standard DNA content analysis [27].
2.7. Measurement of glycosaminoglycan (GAG) secretion
GAG was quantified using a 1,9-dimethylmethyleneblue (DMMB) spectrophotometric assay. Briefly, cells cultured on the scaffolds were suspended in 1 ml PBS, which contained proteinase K (0.2 μg), and incubated at 60 °C for 10 h followed by enzymolysis with 0.25% trypsin after 7, 14 and 21 days, respectively. Then, the cell pellets were incubated with 16 mg/L DMMB (Sigma-Aldrich, Billerica, MA, USA). Finally, a microplate reader was used to measure the absorbance at 525 nm [28]. The GAG content was normalized to the DNA content of the cells.
2.8. Immunofluorescence staining
After 21 days of culture, collagen type II expression was verified by immunohistochemistry using a primary antibody against collagen II (Boster, Wuhan, China, 1:100). The samples were washed and a secondary antibody (Goat Anti-Rabbit IgG H&L). Cell nuclei was stained using DAPI (Solarbio, Beijing, China). Fluorescence images were captured using an Olympus fluorescence microscope (BX53, Olympus, Japan).
2.9. Animal procedure
A total of sixty Sprague Dawley rats (200–300 g, aged 8–10 weeks) were used in this study. All animal procedures were approved by the Animal Ethics and Welfare Committee of Guangxi Medical University. The animals were housed in a controlled environmental (25 ± 3 °C, 40–60% relative humidity) and provided with food and water normally.
Animals were anesthetized by intraperitoneal injection with sodium pentobarbital. The patellar tendon was detached, allowing the patellar groove to be exposed on the chondral surface. A defect (2 mm × 1.5 mm) at the center of the patella groove was pared on the surface using a scalpel. The cartilage defects were covered with the following materials: (1) non-treated (Control); (2) P nanofiber membranes (2 mm × 1.5 mm) seeded with MSCs (1 × 105 cells/mm2); (3) PC nanofiber membranes (2 mm × 1.5 mm) seeded with MSCs (1 × 105 cells/mm2). After surgery, the patella was repositioned again, followed by suturing of the incisions. Intramuscular antibiotics (Cephalexin, 50 mg/kg × 3 for 24 h) were administered postoperatively.
2.10. Gross observation and grading
The animals were sacrificed on week 4 and 8 after the surgery. The repaired articular cartilage was harvested. The morphological evaluation was based on the ICRS [29].
2.11. Histological staining and analysis
After macroscopic observation, the engineered cartilages were fixed in 4% paraformaldehyde for 48 h and then decalcified using EDTA. The tissues were then characterized by standard hematoxylin-eosin staining (HE), Masson’s trichrome, Safranin O/Fast green staining, and immunohistochemistry. Images were taken using a microscope (BX53, Olympus, Japan). ICRS Visual Histological Assessment Scale was adopted to grade the repaired tissues [30].
2.12. Transcriptome sequencing (mRNA-seq) and analysis
Total RNA from MSCs after 21 days of culture on the nanofiber scaffolds and glass coverslips was purified and then analyzed by Chi Biotech (completed by Chi Biotech, Shenzhen, China). Then, 2 μg of RNA was taken to establish database according to VAHTSTM mRNA-seq Kit protocol. Briefly, the mRNA separation/fragmentation reaction was performed at 94 °C for 5 min, and then a 200–300bp fragment was inserted using the hold fragmentation program at 4 °C. After the synthesis and purification of double stranded cDNA, the DNA termini were repaired and connected to each other. To perform library amplification, the total DNA was screened to isolate 320–420 bp strands after further purification. Finally, fragments were amplified through polymerase chain reaction (PCR).
Data quality control was performed using Fastqc. Quantitative mapping was performed using fanse2 algorithm (parameter: -E 5%, -I 0, -S 13) with Rat_rn6_refMrnaas a reference library. The edge R (Version: 3.12.0) software was used to screen out the different genes. The screening threshold was 2 times the difference multiple. p < 0.05 was considered significant. Finally, the KEGG pathway analysis was performed using David (https://david.ncifcrf.gov/).
2.13. Real-time fluorescent quantitative polymerase chain reaction (RT-qPCR)
In each PCR reaction, 1 μL cDNA was used to PCR by mixing with 10 μL of Mastermix (Life Technologies, USA), 1.4 μL of forward primer, 1.4 μL of reverse primer and 6.2 μL of DNase/RNase-free H2O in a total volume of 20 μL at the annealing temperature. β-actin (Actb) was chosen as an internal reference. The primers included the following genes (Table S2): SRY (sex determining region Y)-box 9 (Sox9), collagen type II (Col2a1), collagen type I (Col1a1), aggrecan (Acan), lymphotoxin beta (Ltb), tumor necrosis factor receptor associated factor 1 (Traf1) vascular cell adhesion molecule 1(Vcam1), matrix metallopeptidase (Mmp)-13 and Actb.
2.14. Aomic force microscope (AFM) characterization
AFM characterization was performed on a BL-AC40TS model (Asylum Research, Oxford Instruments Company, Santa Barbara, CA, USA). According to the manufacturer’s instruction, the cantilevers have a Force Constant of 0.05–1.2 N/m, a Resonant Frequency of 50–350 kHz and a Radius of Curvature of 9 nm. Cantilevers were pre-incubated in integrin β1 (Sino Biological Inc., Shanghai, China) at a concentration of 10 μg/ml 10 μM PF-573228 (MedChem Express, NJ, USA) was used for focal adhesion blocking. Young’s modulus was detected and calculated.
2.15. Statistical analysis
The results are expressed as the mean ± standard deviation. The data were evaluated by a one-way ANOVA and Tukey’s t-test using SPSS 16.0 (IBM, USA) with p < 0.05 as a significance level.
3. Results and discussion
3.1. Fabrication of the electrospun nanofibers
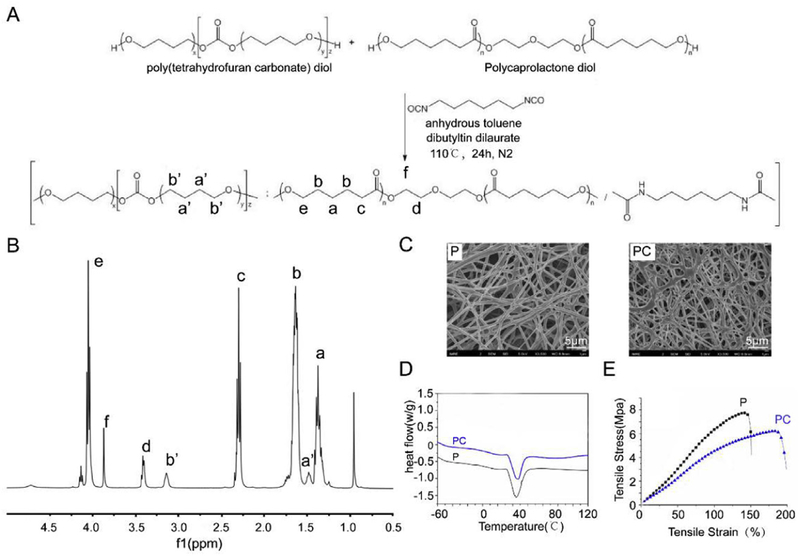
The synthesis route of poly (PCL-PTHF urethane) is shown in Fig. 2A. The successful synthesis of the copolymer was confirmed using 1H NMR. As shown in Fig. 2B, the appearance of a strong signal was detected at 1.35 ppm (Fig. 2B–a), 1.62 ppm (Fig. 2B–b), 2.30 ppm (Fig. 2B–c) and 4.05 ppm (Fig. 2B–e) corresponding to the α-, β-, γ-, and ε-methylene protons in PCL chains, respectively. The characteristic chemical shifts of PTHF were observed at 1.50 ppm (Fig. 2B-a’) and 3.41 ppm (Fig. 2B–d) corresponding to the methylene protons. Based on the integrated areas of peak a’ and b’, the poly (PCL-PTHF urethane) was made of PCL (91.4%) and PTHF (8.6%). The Mn and Mw of the polymer were determined to be 60.6 kDa and 195.0 kDa, respectively, by gel permeation chromatography (GPC).
Fig. 2.
Synthesis and characterization of the electrospun nanofibers. (A)The synthesis route of poly (PCL/PTHF urethane). (B) 1H NMR spectrum of poly (PCL/PTHF urethane). (C) SEM of P and PC nanofibers. Scale bar: 5 μm. (D) Heat flow of P and PC nanofibers. (E) Tensile test of P and PC nanofibers.
The solution concentration and electrospinning parameters were optimized to obtain bead-free nanofibers. As shown in Fig. 2C and Table 1, 5% poly (PCL-PTHF urethane) in HPF generated randomly oriented P nanofibers with a fiber diameter of 523 ± 129 nm while the addition of collagen resulted in the formation of PC nanofibers with a diameter 444 ± 67 nm. Small particles were observed on the PC nanofibers probably due to the deposition of collagen. Such collagen particles could increase the surface roughness and favor cell attachment. More importantly, collagen could prevent MSCs transformation and mimicked triplehelices similar to hyaline cartilage and therefore promote MSCs differentiation into chondrocytes [31,32].
Table 1.
Composition, fiber diameter, thermal and mechanical properties of electrospun nanofibers.
| Fiber code | Biomolecules | Fiber diameter (nm) | Tma (ºC) | ΔHma (J/g) | Xcb (%) | σTSc(MPa) | Еd (MPa) | εbd (%) |
|---|---|---|---|---|---|---|---|---|
| P | N.A. | 523 ± 129 | 34 | 36.1 | 28.8 | 7.4 ± 1.1 | 6.8 ± 1.5 | 138 ± 18 |
| PC | collagen I from calf skin | 444 ± 67 | 35 | 27.5 | 24.3 | 5.8 ± 0.9 | 4.3 ± 0.7 | 184 ± 24 |
Tm (melting temperature) and ΔHm (enthalpy change) determinated by the first heating run of DSC curves.
Crystallinity (Xc) determinated by the equation: where is 139.5 J/g for 100% crystalline PCL.
σTS = Tensile strength, εb = elongation and Е = Young’s modulus determined by tensile test. Each experiment was done independently for 4 times.
The DSC curves (Table 1 and Fig. 2D) showed that P nanofibers had a Tm of 34 °C and Xc of 28.8%, and the addition of collagen to form PC did not alter the Tm but decreased their Xc. The PC nanofibers had the lower Xc of 24.3%. The results indicated that the incorporation of collagen interfered the crystallization of PCL segments. Collagen is a structural protein composed of polypeptide chains and self-organized in a unique 3D architecture (triple helix). Unlike PCL-PTHF, collagen does not have a specific melting temperature but only displays a denaturing temperature of ~40 °C. The polypeptide chains in the triple helix of collagen are rigid and hard to move. Therefore the addition of such biopolymer reduced the ability of molecular chains to move in the fiber system. Also collagen is a big biomolecule with a relatively high molecular weight, which may decrease the nucleation ability of PCL segments.
The tensile test (Table 1 and Fig. 2E) showed that the P nanofibers presented a typical stress-strain curve of an elastic material with σTS of 7.4 ± 1.1 MPa, Е of 6.8 ± 1.5 MPa and εb of 138 ± 18%. Comparing to P nanofibers, the PC nanofibers had a lower value of σTS (5.8 ± 0.9 MPa) and Е (4.3 ± 0.7 MPa). Namely, PC nanofibers are softer than the P nanofibers and both have a modulus within the range of natural cartilage. The degree of polymer’s crystallinity is highly related to its mechanical properties. A highly-crystalized polymer always exhibits a high stiffness and strength, while a material with a low degree of crystallinity is commonly soft and weak. The reason is that the crystallinity reduces the degree of freedom for the molecular chains to move. Here, the addition of collagen reduced the movement capability of the molecular chains and decreased the crystallinity degree of the system, which weakened the mechanical strength of PC nanofibers. In addition, PCL-PTHF and collagen have a low binding energy between each other. Under the deformation, cracks tended to form at the interface, therefore leading to the softer behavior of PC nanofibers.
3.2. Regulating chondrogenesis in vitro by P and PC nanofiber membranes
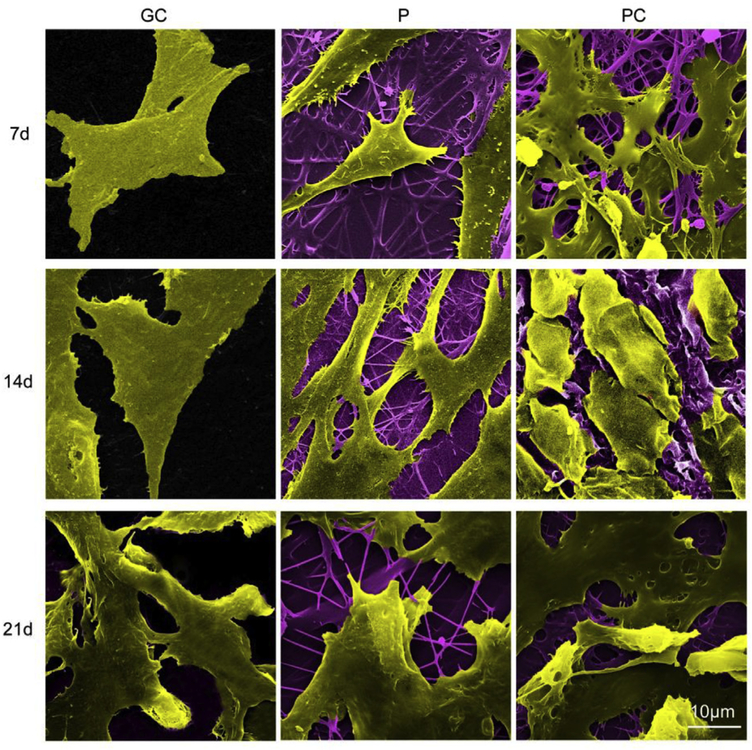
The morphology of MSCs on the scaffolds was analyzed using SEM (Fig. 3). On the glass coverslips (GC) control, cells exhibited a stretched, long, spindle-shaped, fibroblast-like morphology during the culture period. On Day 21, a portion of the cells on P nanofiber membranes became round on day 21, while they were flat and spindle-like on day 7 and 14. However, on the PC nanofiber membranes, the MSCs gradually became more spherical and displayed chondrocyte-like morphology on day 21. The results indicate that the topography of P and PC nanofibers, especially PC, may better support cell aggregation and thus promote chondrogenic phenotype of MSCs [33].
Fig. 3.
The morphology of MSCs on the electrospun P and PC nanofiber membranes for different periods of time was analyzed using SEM. GC is used as control. Scale bar: 10 μm.
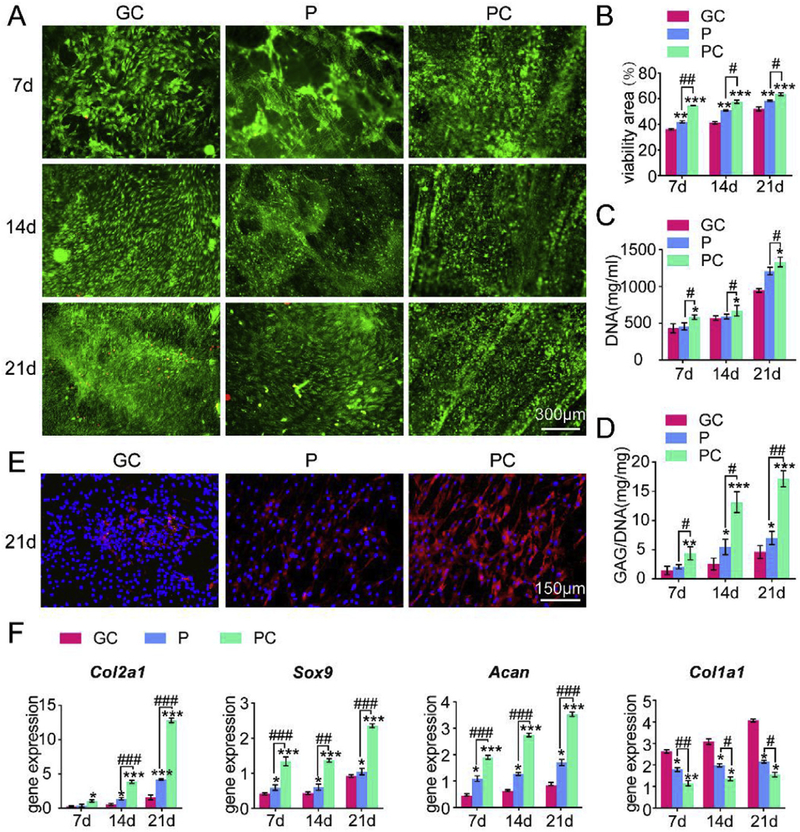
Viable and dead cells were assessed by a calcium AM/PI staining kit (Fig. 4A) and quantified (Fig. 4B). In each group, cells proliferated over time. The results demonstrated that PC nanofiber membranes supported the survival of more MSCs than the P nanofibers mem-branes and GC because more live cells and fewer dead cells were found on the PC nanofiber membranes. Clumps of cells were also observed on day 14 and 21 on the PC nanofiber membranes. PC nanofibers enhanced MSC proliferation, which may be due to the increase of cell-adhesive properties by collagen that contains various amino acids and forms triple-helices [32]. As shown in Fig. 4C, DNA contents increased in a time-dependent manner among all the groups. At each time point, the DNA content was decreased in the order of PC, P and GC. Among all the groups, the DNA content was the greatest in the PC groups. The glycosaminoglycan (GAG) content was assessed to quantify cartilaginous matrix production by MSCs on the scaffolds. The GAG expression increased significantly over time in all groups (Fig. 4D). Among the groups, GAG content was increased in the order of GC, P and PC at each time point. The increase on the PC nanofiber membranes was 201.82%, 412.03% and 271.36% compared with GC on day 7, 14 and 21, respectively.
Fig. 4.
P and PC nanofiber membranes induced chondrogenesis of MSCs in vitro. (A) Cell viability of MSCs cultured on the scaffolds. (B) Quantification of cell viability of (A), n = 3. (C) DNA content of MSCs cultured on the scaffolds. (D) GAG content of MSCs cultured on the scaffolds. (E) Immunofluorescent staining for collagen type II in MSCs cultured on the scaffolds. Scale bar: 150 μm (F) mRNA expression of Acan, Col2a1, Sox9 and Col1a1 of MSCs cultured on the scaffolds. Mean ± SD; n = 3; *,# indicate p < 0.05, **, ## indicate p < 0.01, ***, ### indicate p < 0.001.
Immunofluorescent staining (Fig. 4E) for collagen type II, which is specific for cartilage, also showed a significantly increased accumulation of collagen type II on the P and PC nanofiber mem-branes after 21 days of culture with PC showing more accumulation than P. Namely, among all the groups, MSCs on PC produced the most abundant extracellular matrix that was rich in collagen type II.
Expressions of Acan, Col2a1, Sox9 and Col1a1 gene were measured using RT-qPCR to assess the chondrogenic differentiation of MSCs on the scaffolds. As shown in Fig. 4F, the P and PC nanofiber membranes promoted the expression of cartilage specific genes (Acan, Col2a1 and Sox9) during the culture time compared with GC. For each time point, the expression of these genes was increased in the order of GC, P and PC, suggesting that PC nanofibers increased the expression of cartilage-specific genes the most prominently among all groups over the time. In addition, the expression of Col1a1, which is a fibrocartilage marker, was downregulated in contrast to the expression of cartilage specific genes by the nanofibers, with the PC nanofibers downregulated the expression of the fibrocartilage marker most. The results indicate that the cell aggregation induced by a soft matrix similar to the cartilage matrix may recapitulate the in vivo scenario during the chondrogenesis. This confirmed the earlier hypothesis that a cell fate towards the chondrogenic lineage is more favored on the softer substrates [34].
3.3. P and PC nanofiber membranes promoted cartilage regeneration in vivo
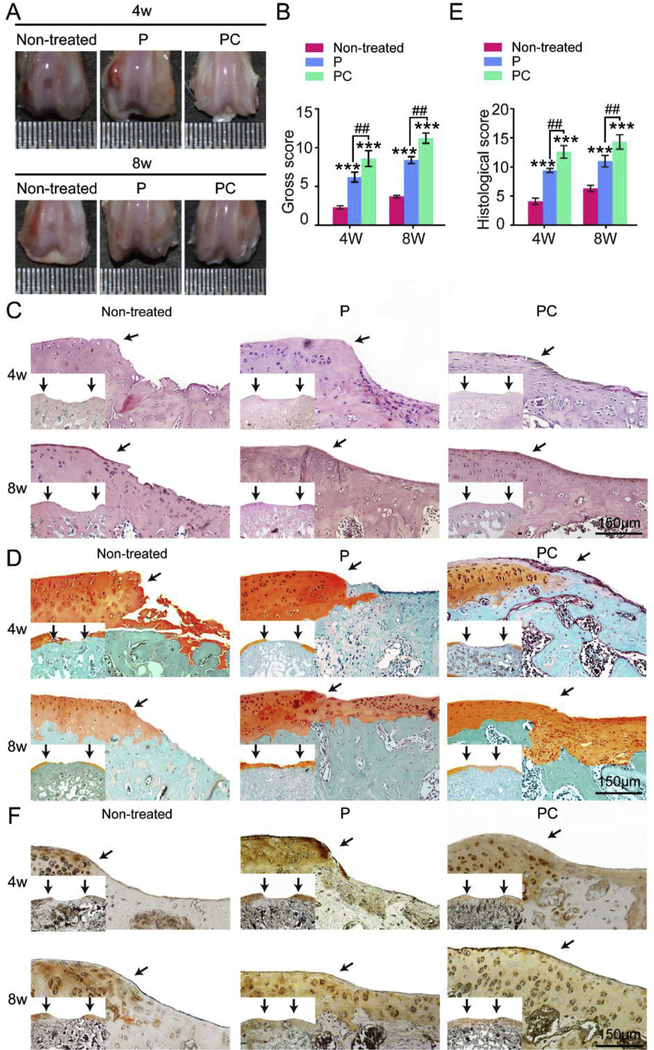
The above results show that PC nanofiber membranes might better support cartilage repair than the P nanofibers membranes. To evaluate the cartilage repair capacity in vivo, P and PC nanofiber membranes were used to repair 2 mm × 1.5 μm defects on the surface of the patellar groove of rat femurs. No inflammation and synovial hyperplasia were detected after 4 and 8 weeks by gross examination of knee joints (Fig. 5A). After 4 weeks, the non-treated group exhibited only a few neo-tissues in the defect, and these tissues had a distinct boundary with the surrounding cartilage. The defects in the P groups contained neo-tissue that was connected with the surrounding tissue. In contrast, repaired tissues that were glossy white were almost connected with the neighboring tissue in the PC group. However, only small fissures were found in the defect center.
Fig. 5.
P and PC nanofiber membranes with seeded MSCs for repairing cartilage defects in vivo. (A) Gross graphics of engineered cartilage. (B) Gross score of the engineered cartilage. (C) HE staining of the engineered cartilage. (D) Safranin O/Fast Green staining of the engineered cartilage. (E) Histological score of the engineered cartilage. Scale bar: 150 μm. (F) Immunohistochemical staining for collagen type II of the engineered cartilage. Scale bar: 150 μm. The black arrows in (C) (D) and (E) indicate the interface between the repaired tissues and the original cartilage. Mean ± SD; n = 5; * and # indicate p < 0.05, ** and ## indicate p < 0.01, *** and ### indicate p < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
After the scaffolds were transplanted for eight weeks, we still could find defects in the center with a loose boundary in the control untreated group. White neo-tissue was connected with the neighboring tissue in the P group. Moreover, the defects in the PC group were improved by filling of cartilage-like tissue. We deter-mined the International Cartilage Repair Society (ICRS) scores from macroscopic observations. The PC group presented the highest overall scores, 8.67 ± 0.57 and 11.3 ± 0.58 at 4 weeks and 8 weeks, respectively. The scores were decreased in the order of PC, P and non-treated group (Fig. 5B).
Histological evaluation was performed 4 and 8 weeks after the surgery (Fig. 5C-E). After 4 weeks of therapy, severe erosion was observed in the non-treated group (Fig. 5CeD and Figure S1). In contrast, newly formed tissues were present in the both P and PC groups with minor inflammatory cells indicating the favorable biocompatibility of the scaffolds. The newly formed tissues were primarily composed of fibrous tissue with a loose and detached interface in the P group. In the PC group, the tissue was fibrocartilage-like and was integrated with the surrounding tissue more tightly than other groups. After 8 weeks, the neo-tissue was primarily composed of fibrous tissue with a loose and detached interface in the untreated group. However, the tissues repaired using P and PC nanofiber membranes were hyaline cartilage with round cells in the lacuna and were well integrated with the sur-rounding tissues. Specifically, the PC engineered cartilage exhibited a more uniform and compact tissue with more round cells in the lacuna than the other groups.
These results were also confirmed by the histological scores (Fig. 5E). At postoperative week 4 and 8, the mean score of the defect treated using the PC nanofiber membranes was greater than that treated using the P nanofiber scaffolds, which was greater than when the defect was left untreated. The mean scores in the PC group were determined to be 11.33 ± 1.53 and 14.33 ± 0.58 on week 4 and 8, respectively (Fig. 5E). Immunohistochemistry assays were performed to evaluate the protein levels of collagen type II (Fig. 5F).
The results showed almost negative staining in the repaired tissue of the untreated groups. Weakly positive staining of the repaired tissues was present in the P group after 4 weeks of therapy. How-ever, after 8 weeks, strong positive staining was shown in P group. Stronger positive immunohistochemical staining of collagen (type could be noted in the tissue repaired using the PC nanofiber membranes after 4 and 8 weeks than the other groups. The results indicated that PC nanofiber membranes with moderate stiffness and favorable biocompatibility are ideal scaffolds for cartilage regeneration.
3.4. P and PC nanofiber membranes directed chondrogenesis through a series of signaling pathways associated with stem cell differentiation
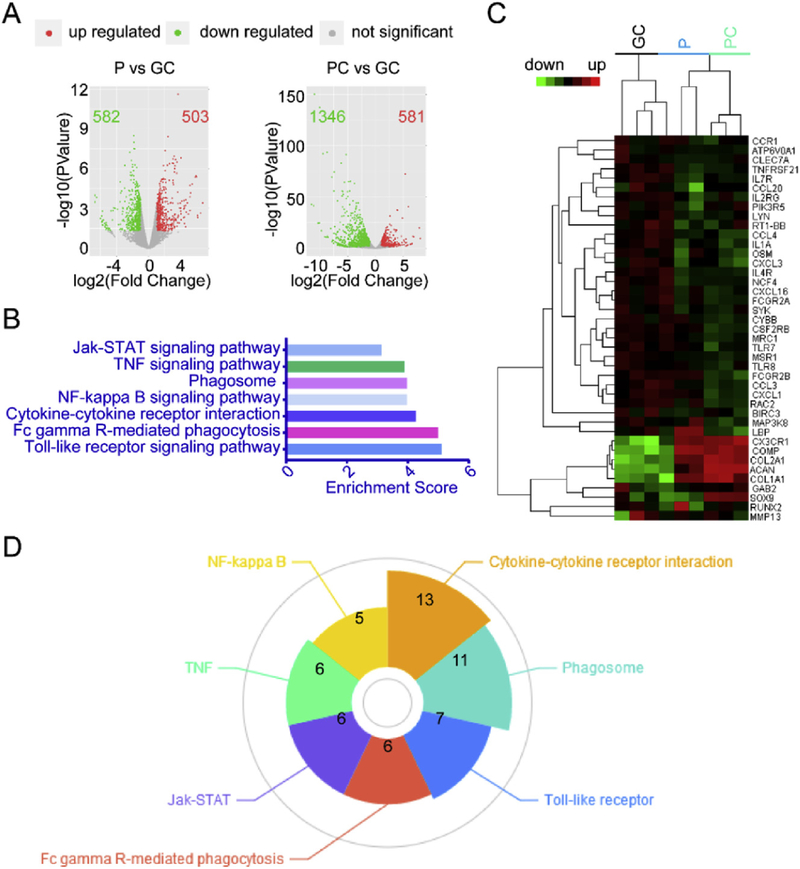
An mRNA-seq analysis was performed after 21 days of culture in vitro. As shown in Fig. 6A, there were 1085 and 1927 differentially expressed mRNAs in the P and PC groups, respectively, whereas the GC group had 104 intersectional genes. We carried out the analysis of the KEGG pathways where the intersectional target genes were involved. The analysis was done by the DAVID tool using 2 gene counts as a threshold under a p-value less than 0.05. By this approach, we found 7 high-degreed signaling pathways associated with chondrogenesis induced by P and PC membranes (Table 2, Fig. 6B) including cytokine-cytokine receptor, NF-kappa B, Toll-like receptor, TNF, Jak-STAT, Fc gamma R-mediated phagocytosis and phagosome. These pathways are involved with stem cell differentiation [35–37]. A heat map of the predicted target genes that were highly enriched in the 7 signaling pathways is shown in Fig. 6C. As shown in Fig. 6D, among the 7 signaling pathways, the most genes were found in the cytokine-cytokine receptor pathway, indicating its pivotal role in chondrogenesis induced by P and PC membranes. The results indicate that P and PC nanofibers may play a role similar as growth factors in triggering the signaling cascades from cell-to-cell interactions during chondrogenic differentiation.
Fig. 6.
Transcriptome profiles of chondrogenic differentiation of MSCs driven by P or PC nanofiber membranes. (A) Volcano plots of MSCs on P or PC membranes vs. MSCs on GC. (B) KEGG Enrichment Score of MSCs on P or PC membranes vs. MSCs on GC. (C) Heatmap of MSCs on P or PC membranes vs. MSCs on GC. (D) Gene counts of the 7 signaling pathways associated with P and PC membranes.
Table 2.
Signaling pathways associated with chondrogenesis induced by P and PC nanofiber membranes.
| KEGG Pathway | Gene count | Genes |
|---|---|---|
| Cytokine-cytokine receptor interaction | 13 | Osm, Tnfrsf21, Ccl3, Ccl20, Cxcl16, Ccr1, Il4r, Cx3cr1, Csf2rb, Il2rg, Il7r, Ccl4, Il1a |
| Phagosome | 11 | Marco, Mrc1, Cybb, Msr1, Fcgr2b, Ncf4, Comp, Atp6v0a1, Fcgr2a, Clec7a, Rt1-Bb |
| Toll-like receptor | 7 | Ccl3, Map3k8, Pik3r5, Lbp, Ccl4, Tlr7, Tlr8 |
| Fc gamma R-mediated phagocytosis | 6 | Rac2, Gab2, Lyn, Fcgr2b, Pik3r5, Syk |
| Jak-STAT | 6 | Osm, Il4r, Csf2rb, Il2rg, Pik3r5, Il7r |
| TNF | 6 | Cxcl1, Ccl20, Cxcl3, Map3k8, Pik3r5, Birc3 |
| NF-kappa B | 5 | Lyn, Lbp, Birc3, Ccl4, Syk |
3.5. PC nanofiber membranes specifically suppressed NK-kappa B signaling pathway to promote chondrogenic differentiation
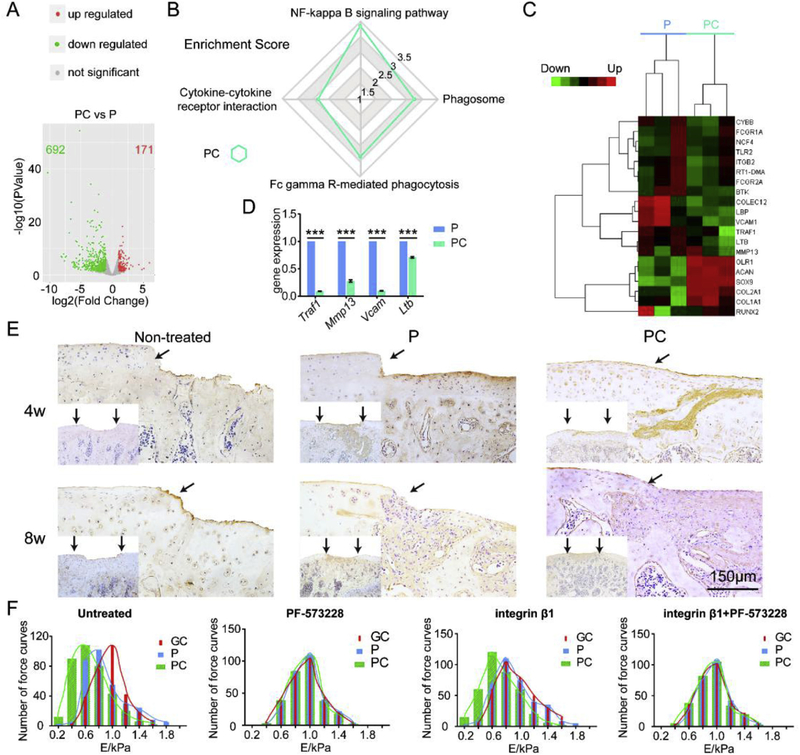
Further mRNA-seq analysis was performed to investigate the difference between the P and PC nanofiber membranes. As shown in Fig. 7A, 863 differentially expressed mRNAs were present in the MSCs cultured on PC vs. P. There were 4 signaling pathways that were highly enriched in the PC group (Fig. 7B). Specifically, the enrichment score of the NK-kappa B signaling pathway were 3.25 in the PC group. The associated molecules in these pathways were analyzed using a cluster assay (Fig. 7C). Most of the genes in the heatmap were enriched in the NK-kappa B signaling pathway. To verify the results of mRNA-seq analysis, the mRNA levels of Ltb, Traf1, Vcam1 and Mmp13 were validated using RT-qPCR. As shown in Fig. 7D, expression of Ltb, Traf1, Vcam1 and Mmp13 was significantly lower in the PC group than in the P group. The down-regulation of the non-specific genes correlates with the upregulation of the cartilage-specific genes Col2a1, Acan and Sox9 on day 21 (Fig. 4D).
Fig. 7.
Enhanced chondrogenic differentiation of MSCs by PC nanofiber membranes. (A) Volcano plots of gene expression significantly influenced by PC on day 21. (B) KEGG Enrichment Score of PC vs. P on day 21. (C) Heatmap of PC vs. P on day 21. (D) mRNA expressions of Ltb, Traf1, Vcam1 and Mmp13 in both P and PC groups on day 21 using RT-PCR analysis. Mean ± SD; n = 3; *** indicate p < 0.001. (E) Immunohistochemical staining for Il-17 of the engineered cartilage. Scale bar: 150 μm. The black arrows indicate the interface between the repaired tissues and the original cartilage. (F) The histograms of Young’s modulus obtained from AFM characterization of (a) untreated MSCs, (b) PF-573228 blocked MSCs after 6 h, (c) MSCs with AFM cantilever was functionalized by integrin β1, and (d) PF-573228 blocked MSCs after 6 h and with AFM cantilever was functionalized by integrin β1. The cells were cultured on the GC, P and PC nanofibers.
The suppression of chondrogenesis by IL-1 beta and TNF alpha has been reported to be associated with a marked activation of NF-kappa B, which was suppressed by the delivery of NF-kappa B inhibitor [38]. NF-kappa B can block the expression of Sox9 [39] and down regulate the expression of the TGF-β receptor II, both of which are essential for chondrogenesis [40]. In the NF-kappa B signaling pathway, lymphotoxin beta (Ltb), tumor necrosis factor receptor associated factor 1 (Traf1), vascular cell adhesion molecule 1 (Vcam1), and matrix metallopeptidase (Mmp)-13 are down-regulated while cartilage specific markers Sox9, Col2a1 and Acan are upregulated by PC compared with P. It has been reported that Ltb is an inducer of the inflammatory response system, which induces the expression of Vcam1 [41] or Traf1 [42] or Mmp13, [43] contributing to inflammation and prevention of Sox9-based chondrogenesis [44]. Findings from Anghelina et al. [45] and Futosi et al. [46] support our explanation and suggest that a physiologically low level of mechanical loading has anti-inflammatory effects through the downregulation of Ltb, Traf1, Vcam1 and Mmp13 and simultaneously enhances matrix synthesis (Acan and Col2a1) in the cartilage/chondrocyte microenvironment. Il-17 is reported as a downstream molecule of Traf1, and both are pro-inflammatory mediators [47]. As shown in Fig. 7E, a lower amount of Il-17 is expressed in the engineered cartilage of PC group than in that of P group, demonstrating that PC enhanced chondrogenesis by suppressing pro-inflammatory mediators.
As indicated by the AFM studies (Fig. 7F), the cells cultured on both the P and PC membranes were stimulated by integrin β1 modified cantilevers, similar to normal chondrocytes [48]. Specifically, the stiffness of the cells on the PC nanofibers was closer to normal chondrocytes than that on the P nanofibers. However, those non-stimulated cells were shown on GC, close to normal MSCs [49]. After the cells cultured on the GC, P and PC nanofibers were blocked by a Focal adhesion inhibitor (PF-573228), their Young’s modulus did not present significant differences determined using cantilevers with or without integrin β1 modified. The results indicated that PC nanofibers promoted the chondrogenic differentiation by activating the mechanotransduction via integrin β1-FAK pathway to suppress inflammation (Fig. 8).
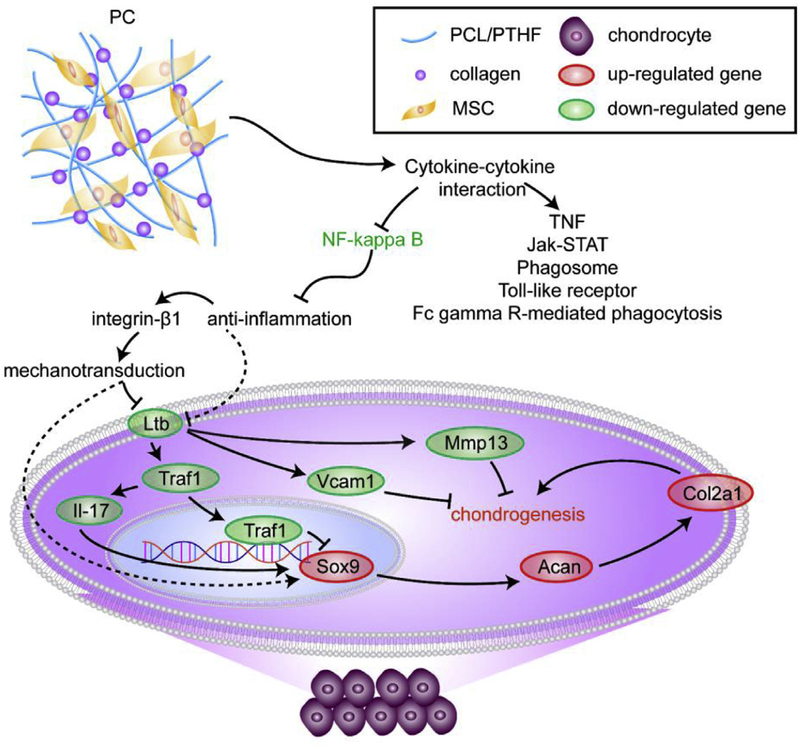
Fig. 8.
The schematic graphic of the mechanism underlying chondrogenesis driven by PC nanofiber membranes. PC induced chondrogenic differentiation through several possible pathways, including cytokine-cytokine receptor interaction (dominant), Toll-like receptor, TNF, Jak-STAT, Fc gamma R-mediated phagocytosis signaling and phagosome. Particularly, inhibition of the NF–kappa B signaling pathway to suppress inflammation is specific for PC induced chondrogenesis.
4. Conclusion
In conclusion, our study provides compelling evidence that cartilage-mimicking electrospun PC nanofiber membranes have the potential to induce the chondrogenic differentiation of MSCs and are favorable scaffolds for cartilage resurfacing. We also identified the differential expression transcript profiles during chondrogenic differentiation. Specifically, the softer PC nanofibers show the better chondrogenic potential than the stiffer P nanofibers because of the suppression of the NF–kappa B signaling pathway to suppress inflammation. Our studies provide a reference for under-standing the precise role and mechanism of the mechanical cues controlling differentiation of MSCs.
Supplementary Material
Acknowledgements
This study was financially supported by National key research and development program of China (2016YFB0700804 and 2016YFA0100900), National Natural Science Fund of China (81760326 and 51673168), the Guangxi Science and Technology Major Project (Guike AA17204085), High level innovation teams and outstanding scholars in Guangxi Universities (The third batch), the Distinguished Young Scholars Program of Guangxi Medical University. YZ and CM would also like to thank the financial support from National Institutes of Health (CA195607 and EB021339).
Footnotes
Conflicts of interest
No conflict of interests exists in this study.
Data availability statement
The data is available upon request.
References
- [1].Gomoll AH, Minas T, The quality of healing: articular cartilage, official publication of the Wound Healing Society [and] the European Tissue Repair Society, Wound Repair Regen 22 (Suppl 1) (2014) 30–38. [DOI] [PubMed] [Google Scholar]
- [2].Chanda D, Kumar S, Ponnazhagan S, Therapeutic potential of adult bone marrow-derived mesenchymal stem cells in diseases of the skeleton, J. Cell. Biochem 111 (2010) 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jiang T, Xu G, Wang Q, Yang L, Zheng L, Zhao J, et al. , In vitro expansion impaired the stemness of early passage mesenchymal stem cells for treatment of cartilage defects, Cell Death Dis 8 (2017), e2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Reilly GC, Engler AJ, Intrinsic extracellular matrix properties regulate stem cell differentiation, J. Biomech 43 (2010) 55–62. [DOI] [PubMed] [Google Scholar]
- [5].Kaur G, Wang C, Sun J, Wang Q, The synergistic effects of multivalent ligand display and nanotopography on osteogenic differentiation of rat bone marrow stem cells, Biomaterials 31 (2010) 5813–5824. [DOI] [PubMed] [Google Scholar]
- [6].Dhand C, Ong ST, Dwivedi N, Diaz SM, Venugopal JR, Navaneethan B, et al. , Bio-inspired in situ crosslinking and mineralization of electrospun collagen scaffolds for bone tissue engineering, Biomaterials 104 (2016) 323–338. [DOI] [PubMed] [Google Scholar]
- [7].Engler AJ, Sen S, Sweeney HL, Discher DE, Matrix elasticity directs stem cell lineage specification, Cell 126 (2006) 677–689. [DOI] [PubMed] [Google Scholar]
- [8].McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS, Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment, Dev. Cell 6 (2004) 483–495. [DOI] [PubMed] [Google Scholar]
- [9].Huebsch N, Arany PR, Mao AS, Shvartsman D, Ali OA, Bencherif SA, et al. , Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate, Nat. Mater 9 (2010) 518–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Elder BD, Athanasiou KA, Hydrostatic pressure in articular cartilage tissue engineering: from chondrocytes to tissue regeneration, Tissue Eng. B Rev 15 (2009) 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wang T, Lai JH, Han LH, Tong X, Yang F, Chondrogenic differentiation of adipose-derived stromal cells in combinatorial hydrogels containing cartilage matrix proteins with decoupled mechanical stiffness, Tissue Eng 20 (2014) 2131–2139. [DOI] [PubMed] [Google Scholar]
- [12].Park JS, Chu JS, Tsou AD, Diop R, Tang Z, Wang A, et al. , The effect of matrix stiffness on the differentiation of mesenchymal stem cells in response to TGF-beta, Biomaterials 32 (2011) 3921–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kwon HJ, Yasuda K, Chondrogenesis on sulfonate-coated hydrogels is regulated by their mechanical properties, J. Mech. Behav. Biomed. Mater 17 (2013) 337–346. [DOI] [PubMed] [Google Scholar]
- [14].Zhang Y, Chen S, Pei M, Biomechanical signals guiding stem cell cartilage engineering: from molecular adaption to tissue functionality, Eur. Cell. Mater 31 (2016) 59–78. [DOI] [PubMed] [Google Scholar]
- [15].Schuh E, Kramer J, Rohwedel J, Notbohm H, Muller R, Gutsmann T, et al. , Effect of matrix elasticity on the maintenance of the chondrogenic phenotype, Tissue Eng 16 (2010) 1281–1290. [DOI] [PubMed] [Google Scholar]
- [16].Cipitria A, Skelton A, Dargaville TR, Dalton PD, Hutmacher DW, Design, fabrication and characterization of PCL electrospun scaffolds-a review, J. Mater. Chem 21 (2011) 9419–9453. [Google Scholar]
- [17].Yao QQ, Cosme JGL, Xu T, Miszuk JM, Picciani PHS, Fong H, et al. , Three dimensional electrospun PCL/PLA blend nanofibrous scaffolds with significantly improved stem cells osteogenic differentiation and cranial bone for-mation, Biomaterials 115 (2017) 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bahrami N, Bayat M, Mohamadnia A, Khakbiz M, Yazdankhah M, Ai J, et al. , Purmorphamine as a Shh signaling activator small molecule promotes motor neuron differentiation of mesenchymal stem cells cultured on nanofibrous PCL scaffold, Mol. Neurobiol 54 (2017) 5668–5675. [DOI] [PubMed] [Google Scholar]
- [19].Lu Z, Lei D, Jiang T, Yang L, Zheng L, Zhao J, Nerve growth factor from Chinese cobra venom stimulates chondrogenic differentiation of mesen-chymal stem cells, Cell Death Dis 8 (2017), e2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Han J, Kim B, Shin JY, Ryu S, Noh M, Woo J, et al. , Iron oxide nanoparticle-mediated development of cellular gap junction crosstalk to improve mesenchymal stem cells’ therapeutic efficacy for myocardial infarction, ACS Nano 9 (2015) 2805–2819. [DOI] [PubMed] [Google Scholar]
- [21].Tanaka T, Tsutsui A, Tanaka K, Yamamoto K, Kadokawa JI, Evaluation of Stability of amylose inclusion complexes depending on guest polymers and their application to supramolecular polymeric materials, Biomolecules 7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rueda-Larraz L, d’Arlas BF, Tercjak A, Ribes A, Mondragon I, Eceiza A, Synthesis and microstructureemechanical property relationships of segmented polyurethanes based on a PCLePTHFePCL block copolymer as soft segment, Eur. Polym. J 45 (2009) 2096–2109. [Google Scholar]
- [23].Shokrollahi P, Mirzadeh H, Huck WTS, Scherman OA, Effect of self-complementary motifs on phase compatibility and material properties in blends of supramolecular polymers, Polymer 51 (2010) 6303–6312. [Google Scholar]
- [24].Afoke NY, Byers PD, Hutton WC, Contact pressures in the human hip joint, J. Bone Joint Surg. Br. Vol 69 (1987) 536–541. [DOI] [PubMed] [Google Scholar]
- [25].Gao Y, Lim J, Han Y, Wang L, Chong MS, Teoh SH, et al. , Cryomilling for the fabrication of doxorubicin-containing silica-nanoparticle/polycaprolactone nanocomposite films, Nanoscale 8 (2016) 2568–2574. [DOI] [PubMed] [Google Scholar]
- [26].Kai D, Tan MJ, Prabhakaran MP, Chan BQY, Liow SS, Ramakrishna S, et al. , Biocompatible electrically conductive nanofibers from inorganic-organic shape memory polymers, Colloids Surfaces B Biointerfaces 148 (2016) 557–565. [DOI] [PubMed] [Google Scholar]
- [27].Cesarone CF, Bolognesi C, Santi L, Improved microfluorometric DNA deter-mination in biological material using 33258 Hoechst, Anal. Biochem 100 (1979) 188–197. [DOI] [PubMed] [Google Scholar]
- [28].Farndale RW, Buttle DJ, Barrett AJ, Improved quantitation and discrimi-nation of sulphated glycosaminoglycans by use of dimethylmethylene blue, Biochim. Biophys. Acta 883 (1986) 173e177. [DOI] [PubMed] [Google Scholar]
- [29].Betz VM, Keller A, Foehr P, Thirion C, Salomon M, Rammelt S, et al. , BMP-2 gene activated muscle tissue fragments for osteochondral defect regeneration in the rabbit knee, J. Gene Med 19 (2017), e2972. [DOI] [PubMed] [Google Scholar]
- [30].Niederauer GG, Slivka MA, Leatherbury NC, Korvick DL, Harroff HH, Ehler WC, et al. , Evaluation of multiphase implants for repair of focal osteochondral defects in goats, Biomaterials 21 (2000) 2561–2574. [DOI] [PubMed] [Google Scholar]
- [31].Yuan T, Zhang L, Li K, Fan H, Fan Y, Liang J, et al. , Collagen hydrogel as an immunomodulatory scaffold in cartilage tissue engineering, J. Biomed. Mater. Res. B Appl. Biomater 102 (2014) 337–344. [DOI] [PubMed] [Google Scholar]
- [32].Jiang T, Liu J, Ouyang Y, Wu H, Zheng L, Zhao J, et al. , Intra-hydrogel culture prevents transformation of mesenchymal stem cells induced by monolayer expansion, Biomater. Sci 6 (2018) 1168–1176. [DOI] [PubMed] [Google Scholar]
- [33].Mahapatra C, Jin G-Z, Kim H-W, Alginate-hyaluronic acid-collagen composite hydrogel favorable for the culture of chondrocytes and their phenotype maintenance, Tissue Eng. Regen. Med 13 (2016) 538–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].VT L, Vg R, DN P, Effect of stiffness of chitosan-hyaluronic acid dialdehyde hydrogels on the viability and growth of encapsulated chondrocytes, Int. J. Biol. Macromol 104 (2017) 1925–1935. [DOI] [PubMed] [Google Scholar]
- [35].Binger T, Stich S, Andreas K, Kaps C, Sezer O, Notter M, et al. , Migration potential and gene expression profile of human mesenchymal stem cells induced by CCL25, Exp. Cell Res 315 (2009) 1468–1479. [DOI] [PubMed] [Google Scholar]
- [36].Roson-Burgo B, Sanchez-Guijo F, Del Canizo C, De Las Rivas J, Insights into the human mesenchymal stromal/stem cell identity through integrative transcriptomic profiling, BMC Genom 17 (2016) 944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhang W, Dong R, Diao S, Du J, Fan Z, Wang F, Differential long noncoding RNA/mRNA expression profiling and functional network analysis during osteogenic differentiation of human bone marrow mesenchymal stem cells, Stem Cell Res. Ther 8 (2017) 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wehling N, Palmer GD, Pilapil C, Liu F, Wells JW, Muller PE, et al. , Interleukin-1beta and tumor necrosis factor alpha inhibit chondrogenesis by human mesenchymal stem cells through NF-kappaB-dependent pathways, Arthritis Rheum 60 (2009) 801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sitcheran R, Cogswell PC, Baldwin AS Jr., NF-kappaB mediates inhibition of mesenchymal cell differentiation through a posttranscriptional gene silencing mechanism, Genes Dev 17 (2003) 2368–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bauge C, Legendre F, Leclercq S, Elissalde JM, Pujol JP, Galera P, et al. , Interleukin-1beta impairment of transforming growth factor beta1 signaling by down-regulation of transforming growth factor beta receptor type II and up-regulation of Smad7 in human articular chondrocytes, Arthritis Rheum 56 (2007) 3020–3032. [DOI] [PubMed] [Google Scholar]
- [41].Madge LA, Kluger MS, Orange JS, May MJ, Lymphotoxin-alpha 1 beta 2 and LIGHT induce classical and noncanonical NF-kappa B-dependent proinflammatory gene expression in vascular endothelial cells, J. Immunol 180 (2008) 3467–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Abdul-Sater AA, Edilova MI, Clouthier DL, Mbanwi A, Kremmer E, Watts TH, The signaling adaptor TRAF1 negatively regulates Toll-like receptor signaling and this underlies its role in rheumatic disease, Nat. Immunol 18 (2017) 26–35. [DOI] [PubMed] [Google Scholar]
- [43].Fehrenbacher A, Steck E, Rickert M, Roth W, Richter W, Rapid regulation of collagen but not metalloproteinase 1, 3, 13, 14 and tissue inhibitor of metalloproteinase 1, 2, 3 expression in response to mechanical loading of cartilage explants in vitro, Arch. Biochem. Biophys 410 (2003) 39–47. [DOI] [PubMed] [Google Scholar]
- [44].Sai J, Raman D, Liu Y, Wikswo J, Richmond A, Parallel phosphatidylinositol 3-kinase (PI3K)-dependent and Src-dependent pathways lead to CXCL8-mediated Rac2 activation and chemotaxis, J. Biol. Chem 283 (2008) 26538–26547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Anghelina M, Sjostrom D, Perera P, Nam J, Knobloch T, Agarwal S, Regulation of biomechanical signals by NF-kappaB transcription factors in chondrocytes, Biorheology 45 (2008) 245–256. [PMC free article] [PubMed] [Google Scholar]
- [46].Futosi K, Fodor S, Mocsai A, Neutrophil cell surface receptors and their intracellular signal transduction pathways, Int. Immunopharm 17 (2013) 638–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Bonaventura P, Lamboux A, Albarede F, Miossec P, Differential effects of TNF-alpha and IL-1beta on the control of metal metabolism and cadmium-induced cell death in chronic inflammation, PLoS One 13 (2018), e0196285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zhang Q, Yu Y, Zhao H, The effect of matrix stiffness on biomechanical properties of chondrocytes, Acta Biochim. Biophys. Sin 48 (2016) 958–965. [DOI] [PubMed] [Google Scholar]
- [49].Wang X, Yang Y, Hu X, Kawazoe N, Yang Y, Chen G, Morphological and mechanical properties of osteosarcoma microenvironment cells explored by atomic Force microscopy, Anal. Sci.: Int. J. Japan Soc. Anal. Chem 32 (2016) 1177–1182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.