Abstract
In the life sciences, researchers increasingly want to access multiple databases in an integrated way. However, different databases currently use different formats and vocabularies, hindering the proper integration of heterogeneous life science data. Adopting the Resource Description Framework (RDF) has the potential to address such issues by improving database interoperability, leading to advances in automatic data processing. Based on this idea, we have advised many Japanese database development groups to expose their databases in RDF. To further promote such activities, we have developed an RDF-based life science dataset repository called the National Bioscience Database Center (NBDC) RDF portal. All the datasets in this repository have been reviewed by the NBDC to ensure interoperability and queryability. As of July 2018, the service includes 21 RDF datasets, comprising over 45.5 billion triples. It provides SPARQL endpoints for all datasets, useful metadata and the ability to download RDF files. The NBDC RDF portal can be accessed at https://integbio.jp/rdf/.
Introduction
In the life sciences, enormous amounts of diverse data are continually being produced and numerous databases have been made available on the Internet (1). It is becoming increasingly important to unify and integrate these databases in order to study complex biological phenomena (2), but these independently developed databases use a variety of different data formats, vocabularies and identifiers, making it extremely difficult to use multiple databases in an integrated way (3). However, the Semantic Web (SW) is attracting attention as a promising approach to address these issues (4, 5).
The SW is a set of technologies that aims to create a web of data, consisting of interlinked machine-readable data on the web. It includes the following core technologies: the Resource Description Framework (RDF) to describe the data, SPARQL to query RDF datasets, RDF Schema (RDFS) to provide a vocabulary for modeling RDF data and the Web Ontology Language to describe the properties and classes needed to develop ontologies. The RDF is a framework for representing information about resources on the web in the form of subject–predicate–object triples. The subject and predicate are described using Uniform Resource Identifiers (URIs) that act as global identifiers, while the object can be described using either a URI or a literal. Objects represented by URIs can become the subject of another triple thus connecting them and resulting in RDF datasets forming graph structures.
Life science data are currently being provided in a wide variety of formats, such as flat files and dump files from relational database management systems (RDBMSs) as well as in JavaScript Object Notation, Extensible Markup Language and comma-separated values (CSV) formats. It is often extremely time-consuming for users to extract the necessary data from these diverse sources and construct a dataset for use in their research. In fact, according to the first National Institutes of Health Strategic Plan for Data Science (https://datascience.nih.gov/sites/default/files/NIH_Strategic_Plan_for_Data_Science_Final_508.pdf), data scientists in a wide array of fields are reported to spend ∼80% of their work time obtaining existing datasets and organizing data. In order to load the gathered data into a local RDBMS, it is also necessary to normalize the data and design a database schema. In contrast, with RDF, it is possible to load several different RDF databases into an RDF store without any additional processing, avoiding the work that would otherwise be required. In addition, since RDF data are described using global URIs, there is no need to consider issues such as the same identifiers being assigned to different entities in different databases. Several attempts have been made to utilize such SW technology features that enhance data interoperability in the life sciences (6–9). In addition, fundamental databases, such as UniProt (10), PDB (11), PubChem (12) and Ensembl (13), are already available in RDF.
The National Bioscience Database Center (NBDC) in Japan aims to promote the development of life science databases. Since its foundation, the NBDC has recognized the potential of SW technologies to integrate diverse databases. To achieve that goal, the NBDC and the Database Center for Life Science (DBCLS) have organized the BioHackathon series (8, 9, 14, 15) that is designed to encourage discussions about applying the SW to life science databases and facilitate the development of RDF datasets and tools.
The NBDC has also funded the development of various life science databases and advised the groups involved to release them in RDF. This has led to a variety of databases becoming available in RDF, produced by both funded groups and other domestic research groups. Initially, each research group was left to decide how to publish their RDF datasets. However, it has proved difficult to provide SPARQL endpoints for all groups and it has become apparent that there is a need for a service that allows people to list, download and query RDF datasets. Given this, we began developing the NBDC RDF portal to meet these needs.
The NBDC RDF portal has the following two features. First, it is an RDF dataset repository, hosting datasets developed by Japanese research groups in a wide variety of research fields. Second, each submitted dataset is reviewed by the NBDC and only those that ultimately pass this review are accepted. We have compiled a set of guidelines for converting databases into RDF and utilize these to review the quality of each dataset in terms of interoperability and queryability.
This article describes our new RDF repository service, the NBDC RDF portal, in detail.
RDF portal guidelines and review policy
Background to creating the guidelines
All datasets provided by the RDF portal have been reviewed by the NBDC to assess their conformance to the guidelines below. In 2018, we also began using an automatic verification tool prior to the manual review. Before discussing the guidelines themselves, however, we first describe the background to creating them and the associated review policy.
The DBCLS hosts a monthly hackathon event, called SPARQLthon, that aims to promote SW applications in the life sciences and technical information sharing among developers. Based on experience and knowledge gathered from these events, we have compiled a set of useful practices known as the ‘DBCLS guidelines for RDFizing databases’ (https://github.com/dbcls/rdfizing-db-guidelines).
Several useful guidelines have already been published, such as a collection of patterns for modeling linked data (Linked Data Patterns, http://patterns.dataincubator.org/book/) and instructions on how to represent data in RDF for exposure in Open PHACTS (http://www.openphacts.org/specs/2013/WD-rdfguide-20131007/) or select bio-ontologies (16). By combining these, our guidelines aim to answer some of the questions that life science database developers with little SW experience may have when creating datasets in RDF.
From these guidelines, we then selected topics that could be used to objectively evaluate such datasets, compiling a guideline subset designed for the RDF portal (called the RDF portal guidelines from now on). Before being included in the RDF portal, all datasets are first reviewed according to these guidelines to ensure a certain level of interoperability.
RDF portal guidelines
Now, we summarize the RDF portal guidelines. The qualified name (QNames: https://www.w3.org/TR/REC-xml-names/#ns-qualnames) prefixes used in this article are shown in Table 1.
Table 1.
QName prefixes used in this article
Primary resources should be instances of an ontology class
Life science databases usually cover either one or a few subjects and their content is organized by subject. For example, UniProt (10) is a database of protein sequences, each represented as an instance of the up:Protein class in the UniProt RDF. As another example, ChEMBL (17) is a database on the bioactivity of chemical compounds, and its entries are instances of classes such as cco:Assay, cco:Activity or cco:Substance. URIs that represent such subjects (called primary resources from now on) should be defined as instances of an ontology class. This helps to reduce the search spaces of SPARQL queries.
Primary resources should have human-readable labels
Even though RDF is primarily intended to make data more machine-readable, providing natural-language labels for resources can be useful, especially when writing SPARQL queries or displaying application results. Linked Data Patterns, the previously mentioned online design pattern catalog for linked data development, advises us to ‘Ensure that every resource in a dataset has an rdfs:label property.’ Our guidelines also recommend adding labels to as many URIs as possible but at minimum all primary URIs must be labeled using the rdfs:label property. When multiple labels are needed, we recommend using the skos:altLabel property.
Some of the datasets in the RDF portal contain labels written in Japanese, partly because they were developed in Japan. For resources with multiple labels in different languages, each label should have a language tag so that labels in a specific language can be selected. On the other hand, language-independent literals, such as numerical values and database entry IDs, should not have language tags.
Primary resources should provide their local database IDs
The local database ID is generally placed after the last slash at the end of each primary URI. However, when printing search results and showing them in an application’s user interface, users often find it easier to work with local database IDs rather than full URIs and local IDs can also be convenient when writing SPARQL queries, for example. To enable this, the primary URI should have a dcterms:identifier property whose value is a literal containing the local ID.
Links to external resources should be provided in the specified format
With the SW, it is essential that both users and machines can explore the RDF-based web of data. Life science databases often provide abundant crosslinks to external database entries, but there are often several different URIs referring to the same database entry, and no general rules as to which URI to use when linking to external databases. Therefore, just converting such databases into RDF may not enhance the web of data because these different URIs, even if they are ultimately redirected to the same Internet URI, are regarded as different RDF resources.
To address this problem, we require all external resources to be referred to using the URIs provided by identifiers.org (18) and the rdfs:seeAlso property. This ensures that the same URI will always be used to refer to the same resource in different RDF datasets. One exception to this is that references to the primary resources within an RDF dataset officially released by the database provider must use the URIs defined in the dataset because datasets do not usually use identifiers.org URIs to describe their own resources. In such cases, redundant links must therefore be included to both the canonical and identifers.org URIs. The canonical URIs used for the main RDF datasets are listed in Table 2.
Table 2.
Canonical URIs used in the main RDF datasets
| RDF dataset | A representative class of primary resources | Prefix of canonical URL |
|---|---|---|
| UniProt | core:Protein | http://purl.uniprot.org/uniprot/ |
| Ensembl | obo:SO_0001217 | http://rdf.ebi.ac.uk/resource/ensembl/ |
| ChEMBL | cco:Substance | http://rdf.ebi.ac.uk/resource/chembl/molecule/ |
| ExpressionAtlas | atlas:BaseLineExpressionValue | http://rdf.ebi.ac.uk/resource/expressionatlas/ |
| atlas:DifferentialExpressionRatio | http://rdf.ebi.ac.uk/resource/expressionatlas/ | |
| Reactome | biopax3:Pathway | http://identifiers.org/reactome/ |
| BioModels | sbmlrdf:SBMLModel | http://identifiers.org/biomodels.vocabulary# |
| BioSamples | biosd-terms:Sample | http://rdf.ebi.ac.uk/resource/biosamples/sample |
| PubChem | compound | http://rdf.ncbi.nlm.nih.gov/pubchem/compound/ |
| substance | http://rdf.ncbi.nlm.nih.gov/pubchem/substance/ | |
| MESH | meshv:TopicalDescriptor | http://id.nlm.nih.gov/mesh/ |
| wwPDB | PDBo:datablock | http://rdf.wwpdb.org/pdb/ |
These URIs are used to represent primary resources in each officially released RDF datasets.
There are two other exceptions to this rule for external resources. References to articles or books should use the relevant PubMed URI or digital object identifier (DOI) with the dcterms:references property, and images should use the foaf:depiction property.
Metadata should be provided
Dataset submitters should provide the following metadata: the dataset providers’ and creators’ names, the version, the date issued, the license and the NBDC database classification tags. It is particularly important that license information is provided, so users can determine how the dataset can be used. This is also a condition for the dataset to be findable, accessible, interoperable and reusable (FAIR) (19). The RDF portal only accepts datasets provided with some type of open license. Currently, most datasets are available under the Creative Commons license.
Existing ontologies should be used where possible
Using common ontologies for different datasets is one of the most important ways of enhancing the interoperability of RDF datasets. Although the semantics of individual RDF datasets are left to their developers, we encourage the use of existing ontologies where possible. The DBCLS guidelines for RDFizing databases therefore list the ontologies we recommend.
The domain and range of each user-defined property should be explicitly defined
When converting a database into RDF, it may be necessary to define new properties, particularly to express relationships between concepts. When doing so, each property’s domain and range should be defined as explicitly as possible. This helps to make queries more efficient and create applications that build SPARQL queries automatically.
A schema diagram should be provided
When writing SPARQL queries for an RDF dataset, it is a great help to have a schema diagram available. Such a diagram should therefore be provided.
Sample queries should be provided
It is very helpful to see examples of typical queries when querying RDF datasets using SPARQL. At least one example query should therefore be provided.
DNA and protein sequence coordinates should be described using FALDO
Many life science databases provide structural and functional annotations to genome or protein sequences. The Feature Annotation Location Description Ontology (FALDO) ontology (20) should be used to specify the point in a sequence to be annotated. This is already used in various RDF datasets, such as UniProt, Ensembl and DDBJ (21), and using common sequence coordinates will enable us to achieve highly interoperable annotations.
Structured values should be used for values with units
Structured values should be used to describe numerical values with units by using the Semanticscience Integrated Ontology (SIO) (22) and giving at least an sio:SIO_000300 property (i.e. sio:has-value) for each value and an sio:SIO_000221 property (i.e. sio:has-unit) for each unit, as in the example below. Structured values should be typed using an appropriate ontology class, included as an sio:SIO_000216 property (i.e. sio:has-measurement-value). The Units of Measurement Ontology (UO) (http://bioportal.bioontology.org/ontologies/UO) should be used to express units where possible but other ontology can be used for units not included in the UO. The following example shows a resource (ex:m1) representing a measurement that the amount of fibrinogen (cmo:CMO_0000209) in a subject’s blood was 21.5 mg/m2 (uo:UO_0000309).
ex:m1 sio:SIO_000216 [
rdf:type cmo:CMO_0000209;
sio:SIO_000300 21.5;
sio:SIO_000221 uo:UO_0000309
].
Review policy
With RDF, any type of information can be described explicitly on the Internet. However, current specifications provide no clues as to how to model particular knowledge or what type of ontology should be used to represent data or knowledge using an RDF. Different ontologies and models can be used to describe the same information, so just exposing databases in RDF will not necessarily improve interoperability from a semantic viewpoint without guidelines or agreement about the semantics. In order to achieve maximum interoperability, it is clearly essential for different communities to agree on common ontologies and models, but, at present, coming to such an agreement appears to be extremely difficult.
With regard to semantics in the life sciences, our policy is essentially to respect the original description in each submitted RDF because we assume that the developers working in each field fully understand these semantics. On the other hand, for general statements that appear in all research areas, such as linking to other database entries, labeling resources, mapping onto genome coordinates and describing numerical values with units, we require the use of specific ontologies and models to increase interoperability among different RDF datasets. Developers can thus retain their original statements, except where they are required to use vocabularies defined in the RDF portal guidelines, due to RDF allowing redundant statements, an advantage that comes from the flexibility of its graph structure.
In the following simple example, resource ex:r1 cites document pubmed:12345 as providing an authoritative description:
(i) ex:r1 cito:citesAsAuthority pubmed:12345.
However, the guidelines require the dcterms:references property to be used when referring to the literature:
(ii) ex:r1 dcterms:references pubmed:12345.
Although statement (i) has more detailed citation semantics than statement (ii), using the same property in all datasets makes it easier to search across datasets. We would therefore instruct the submitter to add statement (ii) to their dataset, leaving it to them to decide whether or not to include statement (i) as well. The SW also offers another solution that satisfies the need both to represent detailed meaning and to use a common property for increased interoperability, namely defining a user-defined property, representing the detailed semantics, as a sub-property of dcterms:references:
(iii) ex2:newCitesAsAuthority rdfs:subClassOf dcterms:references.
However, with regard to the RDF portal guidelines, we ask submitters to add statement (i), even if it is redundant. This is because doing otherwise would unnecessarily complicate writing queries and making inferences on huge life science datasets. With the current RDF store, it would also be generally impractical in terms of performance.
Implementation
The RDF portal currently uses OpenLink Virtuoso version 7.2.4 as its RDF store, running on a Unix server with 48 cores and 1.2 TB memory. The user interface of the site is implemented in JavaScript with several libraries: CodeMirror 5.0, D3.js 4.13.0, JQuery v2.1.4, JQuery UI 1.11.4, jQuery Cookie Plugin 1.4.1, jQuery Easing 1.3 and webcomponents 0.5.5. The SPARQL endpoint uses the SPARQL-proxy software (https://github.com/dbcls/sparql-proxy) for its front end, which enables query verification, scheduling large numbers of queries and improving response time by caching.
Although it would be desirable, from a usability standpoint, to store all the datasets in one RDF store instance, we have created separate virtuoso instances for particularly large datasets because, in our experience, a single virtuoso instance can only handle roughly 20 billion triples without problems in our environment. Currently, the DDBJ and DBKERO RDFs (23, 24) are each stored in their own instances. The metadata is always stored in the primary instance, for all datasets. Figure 1 shows an overview of the system architecture.
Figure 1.
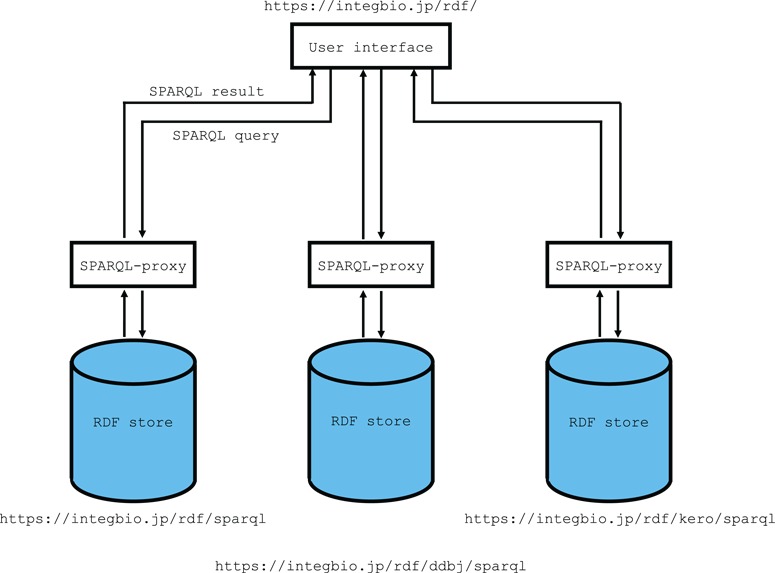
Overview of the system architecture. The RDF portal uses OpenLink Virtuoso as its RDF store. The SPARQL endpoint uses the SPARQL-proxy software for its front end. Currently, there are three virtuoso instances for the primary instance, the DDBJ RDF and the DBKERO RDF.
Persistent Uniform Resource Locators
The use of Cool (i.e. persistent) URIs is recommended for all SW URIs (https://www.w3.org/TR/cooluris) but designing them is not easy. In addition, it is sometimes necessary to use existing (non-Cool) URIs. For example, Cool URIs should not change, but if (for example) a research institute closes, its domain may also become unavailable. Persistent Uniform Resource Locators (PURLs) can address this problem to some extent by redirecting a fixed URL to the current actual web address. To support RDF development, we have created the purl.jp PURL service, which can be used to create new URLs when converting datasets to RDF. It is intended as a general-purpose service, not limited to the life sciences, and issues new URLs for life science applications under http://purl.jp/bio/.
Results
Current status
The NBDC RDF portal (https://integbio.jp/rdf/) was launched in November 2015. As of July 2018, it contains 21 RDF datasets submitted by Japanese research groups, comprising over 45.5 billion triples (Table 3). An up-to-date list and other statistics are available at https://integbio.jp/rdf/?view=matrix. It includes datasets from a wide variety of research areas, such as protein orthology, cancer genomics, glycobiology, transcriptomes and toxicogenomics. At present, most datasets are only accessible as SPARQL endpoints from this site. We rely on developers to provide dataset updates, but we regularly update the datasets as far as possible at their request. For example, we currently update the Worldwide Protein Data Bank (wwPDB)/RDF and the Biological Magnetic Resonance Data Bank (BMRB)/RDF every 3 months and Integbio Database Catalog/RDF every week.
Table 3.
RDF datasets available via the NBDC RDF portal
| RDF dataset | The number of triples |
|---|---|
| DDBJ | 20 067 185 022 |
| DBKERO RDF | 11 017 998 412 |
| Open TG-GATEs | 6 800 384 609 |
| wwPDB/RDF | 4 481 680 698 |
| MBGD RDF | 1 609 018 143 |
| Linked ICGC dataset | 577 082 774 |
| NBDC KikkajiRDF | 333 968 051 |
| MBRB/RDF | 281 996 472 |
| RefEx RDF | 123 447 370 |
| Quanto | 107 782 639 |
| jPOST database RDF | 99 128 038 |
| FAMSBASE GPCR | 21 297 786 |
| PGDBj Ortholog Database RDF | 13 652 175 |
| Dataset of WURCS-RDF | 6 213 789 |
| GlyTouCan | 1 749 648 |
| Integbio Database Catalog/RDF | 92 875 |
| PAConto | 81 785 |
| SSBD: meta-information of quantitative data and microscopy images | 40 300 |
| GGDonto | 39 439 |
| GlycoEpitope | 27 796 |
| Metadata of JCM resources | 8 896 |
| Total number of triples | 45 542 876 717 |
As of July 2018, 21 RDF datasets, comprising over 45.5 billion triples, are available.
Each dataset has its own page; the page for RefEx (25) is shown in Figure 2. These pages contain the dataset’s metadata, the number of out-links and other statistics, RDF model schema diagrams, sample SPARQL queries (linked to the SPARQL endpoint) and links to download the submitted RDF files. The RDF model schema for RefEx RDF is shown in Figure 3.
Figure 2.
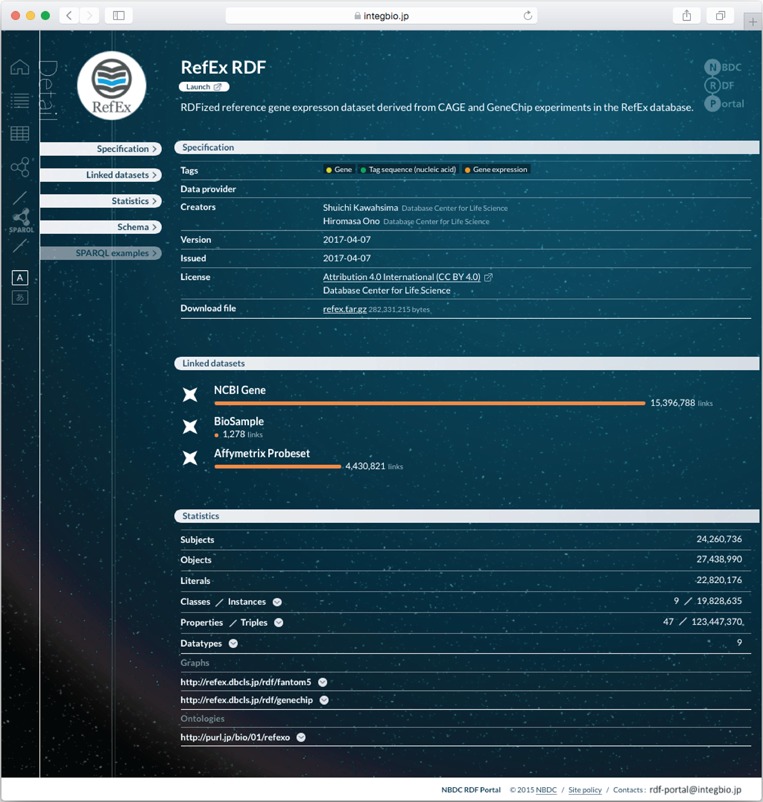
Example dataset page from the NBDC RDF portal. Each RDF dataset has its own page that provides metadata, statistics, links to the RDF files, SPARQL query samples and a link to the SPARQL endpoint.
Figure 3.
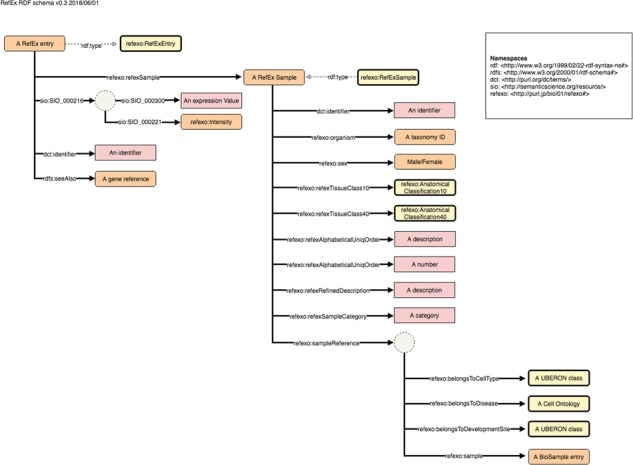
Example schema diagram from the NBDC RDF portal. This example schema diagram is taken from the RefEx RDF. The orange, yellow and pink rectangles represent instances, ontology classes and literals, respectively; the solid and dashed arrows represent properties and rdf:type relationships; the dotted circles represent blank nodes.
When loading an RDF dataset, the number of triples representing out-links (complying with guideline 4) is counted and used to automatically generate a network view (Figure 4). This shows that the site’s datasets complement the main existing RDF datasets and contribute to enriching linked open data in the life sciences.
Figure 4.
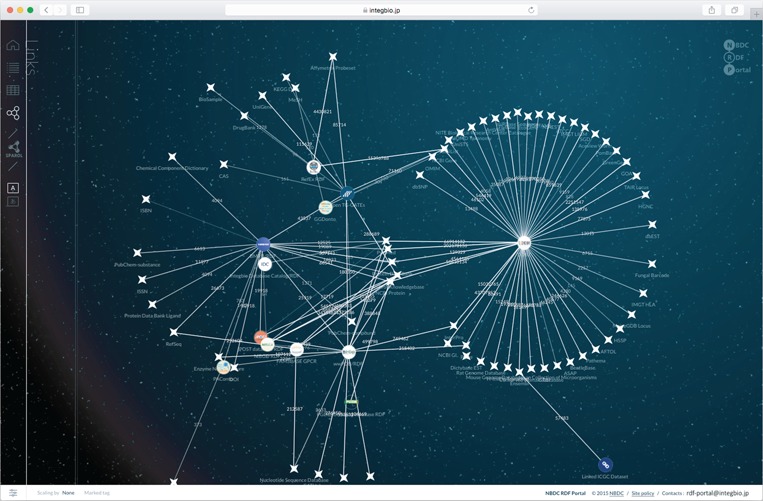
Network view of the NBDC RDF portal. This network view dynamically shows how the datasets are connected. The circles represent datasets registered with the RDF portal, while the stars represent external datasets. When two datasets are linked, they are connected by a straight line, and the number on the line represents the number of links.
Querying multiple datasets
One consequence of the review process is that it enables us to efficiently query multiple datasets. For example, Figure 5 shows a SPARQL query that counts the number of PubMed document citations in each dataset; the results are shown in Table 4. Initially, we encountered cases where rdfs:seeAlso, dcterms:references and other user-defined properties were used in the literature citations. In addition, six different URIs were used to refer to the same PubMed resource (Table 5). Adding statements that used common vocabularies and specifying URIs according to the guidelines therefore enabled us to increase the accuracy of queries across multiple datasets.
Figure 5.

SPARQL query that counts the references in each RDF graph. According to guideline 4, all datasets refer to the PubMed literature using the dcterms:references property.
Table 4.
Results of the SPQRQL query in Figure 5
| Dataset | Graph | The number of references |
|---|---|---|
| wwPDB | http://rdf.integbio.jp/dataset/pdbj | 57546 |
| BMRB | http://bmrbpub.protein.osaka-u.ac.jp/rdf/bmr | 14679 |
| MBGD | http://mbgd.genome.ad.jp/rdf/resource/organism | 2690 |
| GlycoEpitope | http://rdf.glycoinfo.org/glycoepitope | 2354 |
| Integbio Database Catalog | http://rdf.integbio.jp/dataset/dbcatalog/main | 1380 |
| PACONTO | http://jcggdb.jp/rdf/diseases/paconto | 214 |
| SSBD | http://metadb.riken.jp/db/SSBD | 46 |
| GGDONTO | http://jcggdb.jp/rdf/diseases/ggdonto | 15 |
| INSDC ontology | http://integbio.jp/rdf/ontology/nucleotide | 13 |
| BMRB | http://bmrbpub.protein.osaka-u.ac.jp/rdf/bms | 7 |
| JPOST | http://jpost.org/graph/database | 4 |
From left to right are the dataset name, graph name of the dataset in the RDF portal and number of triples referring PubMed URIs.
Table 5.
Six different URIs that refer to the same PubMed resource
In this way, the same resource may be referenced from different URIs, which is one of the reasons that interfere with RDF dataset interoperability.
Next, Figure 6 shows an example SPARQL query against RefEx (25) and Open TG-GATEs (26), which store transcriptomic data. RefEx provides reference transcriptome datasets from 40 normal human, mouse and rat tissues and cells, while Open TG-GATEs is a large-scale toxicogenomics database that includes transcriptome data for human samples exposed to various drugs. The query returns the expression values for probe 210049_at and the chemical compounds the human liver samples were exposed to from Open TG-GATEs, together with reference expression values for the same probe from RefEx; partial query results are shown in Table 6. Both databases include gene expression data measured using the same GeneChip technology, refer to organs in the samples using the Uberon ontology (27) and use the common RDF model to describe measured numerical data, enabling us to integrate them using a single SPARQL query. In addition to the two examples given here, we provide some examples of SPARQL queries that query multiple datasets in the documents section of the RDF portal.
Figure 6.
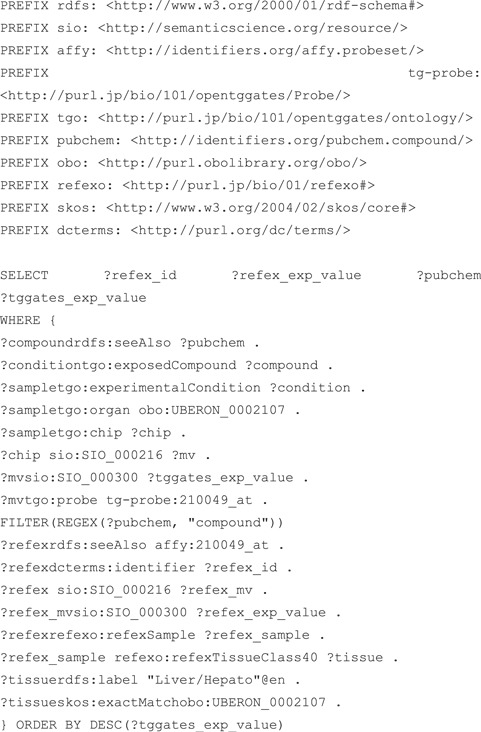
SPARQL query that performs an integrated search of the RefEx and KERO RDFs. Both RefEx and Open TG-GATEs RDF include transcriptome data measured using the same GeneChip technology and use the RDF model defined in guideline 11 to describe measured numerical data.
Table 6.
Partial results of the SPQRQL query in Figure 6
| refex_id | refex_exp_value | PubChem | tggates_exp_value |
|---|---|---|---|
| RFX0016058250 | 12.3 | http://identifiers.org/pubchem.compound/4449 | 319.3662702 |
| RFX0016058250 | 12.3 | http://bio2rdf.org/pubchem.compound:4449 | 319.3662702 |
| RFX0016058250 | 12.3 | http://identifiers.org/pubchem.compound/31703 | 314.3898251 |
| RFX0016058250 | 12.3 | http://bio2rdf.org/pubchem.compound:31703 | 314.3898251 |
| RFX0016058250 | 12.3 | http://identifiers.org/pubchem.compound/31703 | 310.6747304 |
| RFX0016058250 | 12.3 | http://bio2rdf.org/pubchem.compound:31703 | 310.6747304 |
| RFX0016058250 | 12.3 | http://identifiers.org/pubchem.compound/31703 | 306.8218267 |
| RFX0016058250 | 12.3 | http://bio2rdf.org/pubchem.compound:31703 | 306.8218267 |
| RFX0016058250 | 12.3 | http://identifiers.org/pubchem.compound/4449 | 297.3405856 |
| RFX0016058250 | 12.3 | http://bio2rdf.org/pubchem.compound:4449 | 297.3405856 |
From left to right are the RefEx ID, expression value of the probe 210049_at in RefEx, URI of the compound exposed to the sample of Open TG-GATEs and expression value of the probe 210049_at in Open TG-GATEs
Discussion
It is unrealistic to expect that independently created RDF datasets will be highly interoperable. The European Bioinformatics Institute (EBI) RDF platform succeeded in generating interoperable datasets by providing URI design guidelines and using common ontologies and RDF models as far as possible (13). This was largely because they had the advantage that the groups developing the databases and the RDFs belonged to the same institute. Although we could not participate in developing each RDF, we were able to achieve reasonable interoperability by reviewing the RDFs when they were submitted.
Although we want all datasets to comply with all the guidelines, we have been willing to accept non-compliance with some guidelines if there is a sound reason. For example, wwPDB/RDF includes over 1000 classes and 5000 properties in its ontology, making it difficult to draw an appropriately sized schema diagram, so it does not provide schema diagrams. Currently, the guidelines only require the use of certain limited property types; however, to further facilitate the semantic integration of life science data, we plan to ask developers to use more common properties and classes in the future. For example, we are asking developers to represent bio-sample resources as instances of sio:SIO_001050 (sio:sample).
With regard to the system’s operational aspects, we faced the problem of being unable to include all the datasets in a single virtuoso instance due to their enormous combined size. To deal with this, we have set up separate instances to host large datasets, such as DDBJ. However, this means we need to write federated SPARQL queries to query across instances and these generally have performance issues, as well as not always returning answers to more complex queries. That said, we expect to improve the RDF store’s performance in this area in the future.
Acknowledgements
We gratefully acknowledge Katsuhiko Ohkubo, Takehiro Kato, Akio Nagano, Keita Urashima and Yoji Shidara for developing the software and maintaining the National Bioscience Database Center Adopting the Resource Description Framework portal. We would also like to thank all those who participated in the SPARQLthon events for fruitful discussions.
Database URL: https://integbio.jp/rdf/
Funding
Life Science Database Integration Program from National Bioscience Database Center, Japan Science and Technology Agency [18-181023821].
Conflict of interest. None declared.
References
- 1. Rigden,D.J. and Fernández,X.M. (2018) The 2018 Nucleic Acids Research database issue and the online molecular biology database collection. Nucleic Acids Res., 46, D1–D7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stein,L.D. (2003) Integrating biological databases. Nat. Rev. Genet., 4, 337–345. [DOI] [PubMed] [Google Scholar]
- 3. Slater,T., Bouton,C. and Huang,E.S. (2008) Beyond data integration. Drug Discov. Today, 13, 584–589. [DOI] [PubMed] [Google Scholar]
- 4. Antezana,E., Kuiper,M. and Mironov,V. (2009) Biological knowledge management: the emerging role of the Semantic Web technologies. Brief. Bioinform., 10, 392–407. [DOI] [PubMed] [Google Scholar]
- 5. Chen,H., Yu,T. and Chen,J.Y. (2013) Semantic Web meets integrative biology: a survey. Brief. Bioinform., 14, 109–125. [DOI] [PubMed] [Google Scholar]
- 6. Belleau,F., Nolin,M.A., Tourigny,N.et al. (2008) Bio2RDF: towards a mashup to build bioinformatics knowledge systems. J. Biomed. Inform., 41, 706–716. [DOI] [PubMed] [Google Scholar]
- 7. Marshall,M.S., Boyce,R., Deus,H.F.et al. (2012) Emerging practices for mapping and linking life sciences data using RDF—a case series. Web Semant., 14, 2–13. [Google Scholar]
- 8. Katayama,T., Wilkinson,M.D., Micklem,G.et al. (2013) The 3rd DBCLS BioHackathon: improving life science data integration with Semantic Web technologies. J. Biomed. Semantics, 4, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Katayama,T., Wilkinson,M.D., Aoki-Kinoshita,K.F.et al. (2014) BioHackathon series in 2011 and 2012: penetration of ontology and linked data in life science domains. J. Biomed. Semantics, 5, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. UniProt Consortium (2015) UniProt: a hub for protein information. Nucleic Acids Res., 43, D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kinjo,A.R., Bekker,G.J., Suzuki,H.et al. (2016) Protein Data Bank Japan (PDBj): updated user interfaces, resource description framework, analysis tools for large structures. Nucleic Acids Res., 45, 282–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fu,G., Batchelor,C., Dumontier,M.et al. (2014) PubChemRDF: towards the semantic annotation of PubChem compound and substance databases. J. Cheminform., 7, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jupp,S., Malone,J., Bolleman,J.et al. (2014) The EBI RDF platform: linked open data for the life sciences. Bioinformatics, 30, 1338–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Katayama,T., Arakawa,K., Nakao,M.et al. (2010) The DBCLS BioHackathon: standardization and interoperability for bioinformatics web services and workflows. J. Biomed. Semantics, 1, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Katayama,T., Wilkinson,M.D., Vos,R.et al. (2011) The 2nd DBCLS BioHackathon: interoperable bioinformatics web services for integrated applications. J. Biomed. Semantics, 2, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Malone,J., Stevens,R., Jupp,S.et al. (2016) Ten simple rules for selecting a bio-ontology. PLoS Comput. Biol., 12, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Willighagen,E.L., Waagmeester,A., Spjuth,O.et al. (2013) The ChEMBL database as linked open data. J. Cheminform., 5, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Juty,N., Le Nover̀e,N. and Laibe,C. (2012) Identifiers.org and MIRIAM Registry: community resources to provide persistent identification. Nucleic Acids Res., 40, 580–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wilkinson,M.D., Dumontier,M., Aalbersberg,I.J.et al. (2016) The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data., 3, 160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bolleman,J.T., Mungall,C.J., Strozzi,F.et al. (2014) FALDO: a semantic standard for describing the location of nucleotide and protein feature annotation. J. Biomed. Semantics, 7, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kodama,Y., Mashima,J., Kosuge,T.et al. (2015) The DDBJ Japanese genotype–phenotype archive for genetic and phenotypic human data. Nucleic Acids Res., 43, D18–D22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dumontier,M., Baker,C.J.O., Baran,J.et al. (2014) The Semanticscience Integrated Ontology (SIO) for biomedical research and knowledge discovery. J. Biomed. Semantics, 5, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mashima,J., Kodama,Y., Fujisawa,T.et al. (2017) DNA Data Bank of Japan. Nucleic Acids Res., 45, D25–D31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Suzuki,A., Kawano,S., Mitsuyama,T.et al. (2018) DBTSS/DBKERO for integrated analysis of transcriptional regulation. Nucleic Acids Res., 46, D229–D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ono,H., Ogasawara,O., Okubo,K.et al. (2017) RefEx, a reference gene expression dataset as a web tool for the functional analysis of genes. Sci. Data., 4, 170105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Igarashi,Y., Nakatsu,N., Yamashita,T.et al. (2014) Open TG-GATEs: a large-scale toxicogenomics database. Nucleic Acids Res., 43, D921–D927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mungall,C.J., Torniai,C., Gkoutos,G.V.et al. (2012) Uberon, an integrative multi-species anatomy ontology. Genome Biol., 13, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]


