ABSTRACT
Rab1a, a member RAS oncogene family, has been reported playing important role in tumor proliferation and migration. However, the role of Rab1a in intrahepatic cholangiocarcinoma (ICC) is not clear. In this study, we found Rab1a was overexpressed in ICC tissues both in mRNA and protein level. Kaplan-meier analysis showed that high expression of Rab1a was associated with poor prognosis of ICC patients. Suppression of Rab1a led to lower proliferation rate and migration ability both in vitro and in vivo by inhibiting process of cell cycle and Epithelial-Mesenchymal Transition (EMT). Further study showed that Rab1a was targeting regulated by miR-212-3p.In addition, expression of Rab1a was increased while miR-212-3p was decreased under hypoxia condition. In conclusion, these findings extend our understanding of Rab1a in progression of ICC, and we found hypoxia/miR-212-3p/Rab1a pathway played important role for progression of ICC. This newly identified pathway should promote the development of novel therapeutic biomarker for ICC.
KEYWORD: Rab1a, Hypoxia, HIF1a, Intrahepatic Cholangiocarcinoma (ICC), MiR-212-3p; Cell cycle, Epithelial-MesenchymalTransition (EMT)
Introduction
Cholangiocarcinoma (CCA), belongs to all gastrointestinal cancers with 3%, is the second most common primary hepatic tumor after hepatocellular carcinoma.1,2 Early lymphatic and metastatic spread is main characteristic of CCA, thus patients with CCA always have poor prognosis and only few of them are eligible for surgical operation.3,4 Even though patients have chance for curative surgery, only 20–30% of them can survive 5 ‘y’ for the high rate of recurrence after surgery and no effective chemotherapy drugs.5,6 In a word, more studies are needed to understand initiation and progression of CCA and find effective biomarker for disease monitoring.
Rab1a, a member RAS oncogene family, is first known for its function of controlling vesicle traffic from the endoplasmic reticulum to the Golgi apparatus.7,8 Further studies showed that Rab1a play important role in progression of many tumors, including tongue cancer,9 prostate cancer,10 large B cell lymphoma,11 hepatocellular carcinoma,12,13 colorectal cancer,14 lung cancer15 and breast cancer.16 In addition, Thomas JD and his colleges found Rab1a is an mTORC1 activator and can enhance oncogenesis through hyperactive Amino acid signaling.14 Others showed that oncogene Rab1a is downregulated by miR-634 and miR-15b-5p in hepatocellular carcinoma.12,17 However, the function of Rab1a in CCA is unclear, especially in intrahepatic Cholangiocarcinoma. Thus we suppose whether Rab1a plays vital role in the progression of ICC.
In our study, we found Rab1a was upregulated in ICC tissues and overexpression of Rab1a was associated with poor prognosis of ICC patients. Further study showed that suppression of Rab1a could inhibit proliferation and migration capacity of CCA cells. In addition, we found Rab1a was targeting regulated by miR-212-3p, and expression of Rab1a was increased while miR-212-3p was decrease under hypoxia condition. In conclusion, our study demonstrated that hypoxia/miR-212-3p/Rab1a axis played important role in proliferation and migration of ICC.
Materials and methods
Patients, tissue samples and follow-up
All cancer tissue samples and neighbor tissue samples were obtained from 69 Cholangiocarcinoma patients who underwent surgery at Henan Provicial people's Hospital. All patients were diagnosed CCA by Imaging and Serological examination and accepted operation without Radiation and chemotherapy. Fresh cancer tissues and adjacent matched normal tissues were collected and made for paraffin blocks. Two pathologists examined the cancer tissues and the matched normal tissue. The Medical Ethics Committee of The Henan Provicial people's Hospital approved the protocol, and written informed consent was obtained from all participants. Clinical data of all these patients including age, gender, size of tumor, number of tumors, Vessel invasion and tumor grade were collected. Two investigators were responsible for the follow-up of these patients every three months. All these patients were followed until the follow-up termination date (Sep1, 2016) or death. Overall survival was calculated in months from the diagnosis until the date of death, last known to be alive, or the study closing date.
Cell lines and cell culture
RBE, CCLP and 293T cell lines were purchased from the Shanghai Cell Bank at the Chinese Academy of Sciences (Shanghai, China). Such cell lines were cultured in RPMI-1640 medium (Gibco, GrandIsland, NY) supplemented with 10% fetal bovine serum (Hyclone, Shanghai, China) at 37°C in a humidified incubator containing 5% CO2. To induce hypoxia, cells were cultured in the atmosphere of 1% O2, 5% CO2 and 94% N2.
Transfection of interference RNA
Human-specific Rab1a and HIF1a interference RNA (siRNA) were designed, constructed and purified by Jima Biotech Co (Shanghai, China). Rab1a: 5′-CAATCACCTCCAGTTATTA-3′.HIF1-a:5-GGAAATGAGAGAAATGCTTAC-3. The transfection was conducted using lipofectamine 2000 (Invitrogen) according to manufacturer's construction. And Rab1a shRNA was used the sequence of siRNA, the lentivirus was purchased from Ji Kai corporation.
Total RNA Extraction and RT–PCR
The RNA extraction kit and Rever Tre Ace-a-reverse transcription kit were purchased from Invitrogen Co. All procedures were conducted as the instructions from the kits. Primers were synthesized by Shanghai Sangon Biological Engineering Technology Services Co., Ltd. The nucleotide sequences of the primers were as follows: GAPDH, 5′-ATGGGGAAGGTGAAGGTCG-3′and 5′-GGGGTCATTGATGGCAACAATA-3; Rab1a, 5′-TATGGGACACAGCAGGCCAGG-3′and 5′-ACGGAATTCCAAGGGAATCAGC-3′;. Each sample was taken in triplicate. GAPDH was used as an internal reference, and the 2−ΔΔCT method was used to analyze PCR results.
Western blot analysis
All the western blot analysis was based on RBE and CCLP cell lysis. Total proteins were extracted from harvested cells by protein extract kit (Beyontime, Jiangsu, China).All the proteins were denatured by boiling, separated by SDS-PAGE and transferred onto PVDF membranes. Then, membranes were blocked by 5% milk. Next, the membranes were incubated by first antibodies overnight at 4oC. Then washing third times by TBST, the membranes were incubated by second antibody (goat anti-rabbit) for 2 h under 37oC incubator. Finally, all the membranes were visualized by a chemiluminescence kit (Beyontime, Jiangsu, China) on a Bio-Rad imaging system. Primary antibodies used in this study are as follows: CDK2, CDK4 and CyclinB1(1:1000 dilution, Proteintech, USA) HIF1-a (1:1,000 dilution, Abcam Biotechnology, Cambridge, UK), Rab1a (1:1,000 dilution, Abcam Biotechnology, Cambridge, UK) E-cadherin and N-cadherin (1:1000 dilution (EMT) Antibody Sampler Kit #9782,Cell signaling technology), β-actin (1:3000 dilution, Abcam Biotechnology, Cambridge, UK) and GAPDH(1:3000 dilution, proteintech, USA).
Immunohistochemistry
The 69 paired paraffin-embedded caner tissues and neighbor tissues of CCA patients were cut at 3 um, deparaffinized in xylene, and rehydrated in a series of graded alcohol dilutions. Then antigen retrieval were performed by heating the slides in citrate buffer (pH 6.2) with microwave oven for 20 min and washed in PBS 3 times before staining with immunoperoxidase. Slides were incubated in a humidified chamber with blocking agent (5% FBS) for 60 min at room temperature. Next, slides were incubated overnight with anti-Rab1a and anti-HIF1a antibodies in 5% FBS overnight at 4oC. After PBS wash, they were incubated with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin for 40 min at room temperature. Results were visualized by reaction with diaminobenzidine (DAB, 3, 30-diaminobenzidine tetrahydrochloride) and counterstained with hematoxylin. Then the tissues were scored according to the intensity of the dye color and the number of positive cells. The method for IHC score was as follows: 0, negative; 1, <30% positive tumor cells; 2, 30–50% positive tumor cells; 3, 50%-70% positive tumor cells; and 4, >70% positive tumor cells. The intensity of the dye color was graded as 0 (no color), 1 (light yellow), 2 (light brown), or 3 (brown). The staining index was calculated as follows: staining index = staining intensity × tumor cell staining grade. High Rab1a expression was defined as a staining index score ≥6, while low expression was defined as a staining index <6.
Cell viability assay
To detect the relative cell viability, RBE and CCLP cell lines were seeded into 96-well microplates at a density of 5,000 cells per well. Then cell counting kit-8 (CCK-8) assay (Dojindo, Kumamoto, Japan) was then performed according to the manufacturer's instructions in 24h, 48h and 72h. Briefly, 10 μL of CCK-8 working solution per 100 μL of medium was added into the microplates and the cells were incubated for 2 h. The OD450 value was determined by using a MRX II microplate reader (Dynex, Chantilly,VA, USA).
Cell cycle analysis
RBE and CCLP cells with Rab1a siRNA, and negative control were trypsinized and washed 3 times with pre-chilled PBS buffer, and resuspended in 100 μL PBS at 1 × 106cells/ml after 48 h of transfection. For cell cycle analysis, samples of cells were fixed in 75% ethanol overnight, and DNA was stained for cell cycle analysis. RNA was removed by mixing 0.5 ml DNA Prep Stain (Coulter DNA-Prep Reagents kit, Beckman Coulter) in dark at room temperature for 30 min. The DNA content of stained cells was measured with BD LSR II (BD Biosciences). Each histogram was constructed with data from at least 10,000 events. The percentage of a cell population in each phase of the experimental data was calculated using ModFit LT software.
Cell migration assay
Migration assays were performed using Transwell plates (Corning, NY). After 48 h of transfection, RBE and CCLP cells transfected with Rab1a siRNA, and negative control were trypsinized and collected. Then 1 × 105 cells were cultured in a serum-free 1640 on an insert coated without matrigel (migration assay; BD Bioscience, Bedford, MA) for 24 h. In the lower compartment, medium was changed with 1640 complete medium. After fixation and staining, cells on the bottom surface that invaded across the membranes were counted and photographed. All experiments were performed in triplicate.
Immunofluorescence
Immunofluorescence was used to observe E-cadherin and N-cadherin expression. CCA cells were seeded into confocal dishes containing a sterile coverslip on the bottom. After transfected with Rab1a for 48h, cells were washed with PBS, fixed using 2% paraformaldehyde for 5 min, permeabilised using Triton X-100 0.1% for 10 min, and blocked using 10% FBS for 1 h at room temperature. Next, the cells were incubated with a 1:200 dilution of anti-E-cadherin monoclonal antibody or anti-N-cadherin monoclonal antibody overnight at 4°C. All samples were washed twice in PBS and incubated with goat-anti-rabbit CY3 at 1:200 dilution as the secondary antibody (Beyotime Institute of Biotechnology, China). Finally, the samples were incubated with DAPI for 5min, then washed twice in PBS and examined by confocal microscopy (Olympus, Tokyo, Japan) to produce a merged image.
Colony-forming assay
To assay the effect of Rab1a on colony formation, Rab1a shRNA and negative control clonies were dissociated with trypsin, resuspended in MEM complete medium with 10% FBS. Then, they were inoculated into a 6-well plate at a density of 2000 cells/well. After 16 ‘d’, colonies were washed twice with PBS, dyed with crystal violet and photographed. All experiments were independently repeated at least 3 times under identical condition.
In vivo tumorigenesis assay and lung metastasis assay
All experimental procedures involving animals were in accordance with the Guide for the Care and Use of Laboratory Animals and were performed according to the institutional ethical guidelines for animal experiment. The study protocol was also approved by the Committee on the Use of Live Animals in Teaching and Research, Zheng Zhou University. Cells were washed in PBS, counted and injected into the right axilla of each mouse (5 × 106). Tumor size was measured by the following formula: volume = (L × W2) /2, where L and W are the longest and shortest diameters, respectively. Mice were sacrificed about five weeks after injection. And the nude mice were injected with CCA cells who were transfected with Rab1a-shRNA or Rab1a-NC (1 × 106) via the tail vein respectively. Mice were sacrificed 40 ‘d’ after the tail vein injection, and lung was embedded in paraffin and sliced for HE staining to examine for metastasis.
Luciferase reporter assay
293T cells were seeded in 24-well plates and then the cells were co-transfected with miR-212-3p and control and 100 ng either wild-type or mutant-type 3’UTR of Rab1a firefly luciferase reporter plasmid. After incubation for 48 h, firefly and renilla luciferase activities were measured by a dual-luciferase reporter assay (promega, E2920).
Statistical Analysis
Data are presented as a mean ± standard deviation (SD). For comparisons, the Student's t-test, paired-samples t-test, and Fisher's exact test were performed as appropriate. Cumulative recurrence and survival probabilities were evaluated using the Kaplan-Meier method, and differences were assessed using the log-rank test. All analyses were performed with SPSS 18.0 software (SPSS Inc., Chicago, IL). Differences were considered significant at P < 0.05.
Results
Rab1a was overexpressed in ICC tissues and associated with poor prognosis
To investigate whether Rab1a played important role in the progression of ICC, we collected 69 paired ICC tissues and neighbor tissues, and detected expression of Rab1a by immunohistochemistry (IHC).As shown in (Fig. 1A–B), we found Rab1a was overexpressed in 49 paired ICC tissues, other 14 paired were no change, only 6 paired were low expressed. Then we compared mRNA level of Rab1a in 22 paired ICC tissues and neighbor tissues, and found cancer tissues were higher than neighbor tissues (Fig. 1C). Then we used Kaplan-meier analysis to investigate whether expression of Rab1a was associated with the prognosis of ICC patients. As expected, we found patients with high expression of Rab1a always had a poor prognosis (Fig. 1D).In addition, we used single-factor analysis and found expression of Rab1a was correlated with tumor size, vessel invasion and tumor grade (Table 1).
Figure 1.
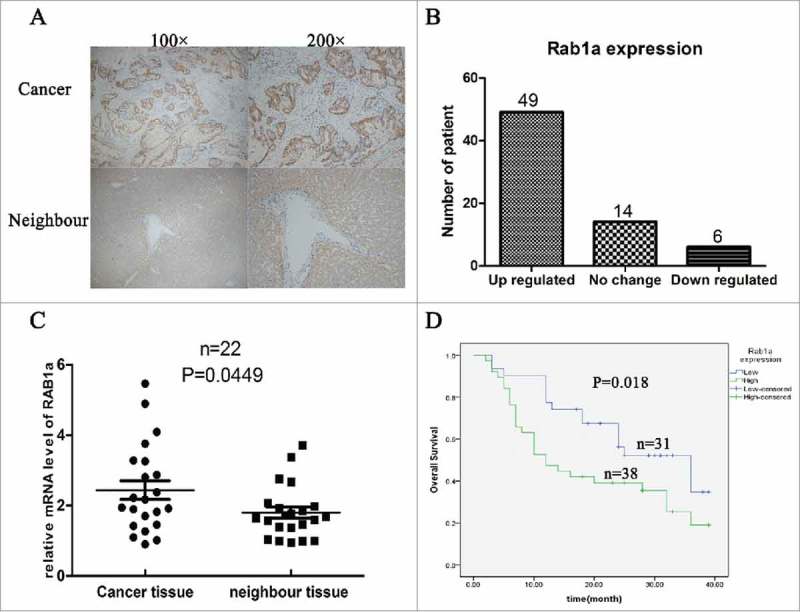
Rab1a was overexpressed in ICC and associated with poor prognosis (A) Shown the IHC results of Rab1a expression in CCA tissues, 100* and 200* mean the magnification (B) The patients number of different expression of Rab1a in ICC tissues were shown with a histogram (C) The result of mRNA level of Rab1a between cancer tissue and neighbor tissue were showed and statistically analyzed (D) Overall survival of 69 ICC patients were analyzed with Kaplan-meier method.
Table 1.
Relationship between Rab1a expression and clinicopathologic features.
| All patients | Rab1a expression |
|||
|---|---|---|---|---|
| Variables | (N = 69) | Low* | High* | P value* |
| Age(years) | ||||
| ≦55 | 22 | 10 | 12 | 0.998 |
| >55 | 47 | 21 | 26 | |
| Gender | ||||
| Male | 23 | 9 | 14 | 0.610 |
| Female | 46 | 22 | 24 | |
| Size of tumor(cm) | ||||
| ≤5 | 29 | 18 | 11 | 0.027 |
| >5 | 40 | 13 | 27 | |
| Number of tumor | ||||
| Single | 49 | 22 | 27 | 0.994 |
| Multiple | 20 | 9 | 11 | |
| Vessel invasion | ||||
| Negative | 40 | 22 | 18 | |
| Positive | 29 | 9 | 20 | 0.041 |
| Grade | ||||
| Well+Moderate | 28 | 17 | 11 | 0.048 |
| Poor | 41 | 14 | 27 | |
The median expression level was used as the cut-off. Low expression of Rab1a in 31 patients was classified as values below the 50th percentile. High Rab1a expression in 38 patients was classified as values at or above the 50th percentile.
For analysis of correlation between Rab1a expression and clinical features, chi-square tests were used. Results were considered statistically significant at P < 0 .05.
Suppression of Rab1a can inhibit the proliferation rate of CCA cells by inducing cell cycle G1-phase arrest
To study function of Rab1a in progression of ICC, Rab1a siRNA and negative control were successfully transfected into RBE and CCLP cell lines, and the efficiency of transfection was verified by western blot (Fig. 2A). Then we cell viability of RBE and CCLP cell lines were detected by CCK-8 assay and showed suppression of Rab1a leaded to lower cell viability than negative control both in two cell lines (Fig 2B). To investigate whether Rab1a can regulate progression of cell cycle, we used flow cytometry to detect cell cycle of RBE and CCLP under treatment of Rab1a siRNA and negative control, and found both these two cell lines reflected high G1 phase proportion in siRNA group than control group (Fig. 2C). And such results were quantified and statistical analyzed (Fig. 2D). In addition, the protein level of CDK2,CDK4 and CyclinB1 were decreased under condition of Rab1a suppression (Fig. 2E–F).
Figure 2.

Suppression of Rab1a can inhibit proliferation rate of CCA cells by inducing cell cycle G1-phase arrest (A) The transfection efficiency of Rab1a siRNA in RBE and CCLP cells were verified by western blot (B) The results of CCK-8 analysis in RBE and CCLP cells under condition of Rab1a siRNA and negative control were showed,**means P < 0.001 (C) Showed the results of cell cycle analysis in RBE and CCLP cells under condition of Rab1a siRNA and negative control (D) the cell cycle results were quantified,*means P < 0.01 (E-F) the protein level of CDK2,CDK4 and CyclinB1 in RBE and CCLP cells were detected under condition of Rab1a siRNA and negative control, GAPDH was the internal control, and the results were quantified and statistically analyzed.
Suppression of Rab1a inhibits migration ability of CCA cell lines by hindering progression of EMT
To study whether Rab1a can regulate migration of CCA cell lines, we used transwell assay to detect cell migration ability and found knockdown of Rab1a in RBE and CCLP cell lines lead to lower migration ability than negative control. And the results were quantified and statistical analyzed (Fig. 3A). Considering EMT process plays important role in cell migration, we used immunofluorescence to detect expression change of E-cadherin and Vimentin under treatment of siRNA and negative control, and found E-cadherin was upregulated and Vimentin was downregulated with suppression of Rab1a comparing with negative control. Such phenomenon was proofed by western blot (Fig. 3B).
Figure 3.

Suppression of Rab1a inhibits migration ability of CCA cell lines by hindering progression of EMT (A) showed results of transwell assay in RBE-Rab1a-siRNA, RBE-Rab1a-NC,CCLP-Rab1a-siRNA and CCLP-Rab1a-NC cells, and these results were quantified respectively,***means P < 0.0001 (B) Expression change of E-cadherin and Vimentin in RBE and CCLP cells under condition of Rab1a siRNA and negative were detected by immunofluorescence and western blot, GAPDH was as the internal control.
Suppression of Rab1a inhibits proliferation and migration of CCA cells in vivo
To verified function of Rab1a in vitro and in vivo, firstly we conducted the stably Rab1a-shRNA transfected cells, and the efficiency was verified both in mRNA and protein levels (Fig. 4A). Then tumorigenesis assay with cells of CCLP-Rab1a-shRNA and CCLP-Rab1a-NC in vitro and vivo were performed, We found the number of colonies and the volume of tumors in Rab1a-NC group were more and larger than the Rab1a-shRNA group (Fig. 4B–C). Then we conducted lung metastatic assay to prove the function of Rab1a in migration of CCA cells in vivo. After 40 ‘d’, we sacrificed the mice and collected the lungs for HE analysis. Interestingly, even though the two groups both had the metastatic tumor, the Rab1a-NC group displayed larger tumor and more numbers than the Rab1a-shRNA group (Fig. 4D). Thus we concluded that Rab1a could play important role in the proliferation and migration of CCA cells.
Figure 4.

Suppression of Rab1a inhibits proliferation and migration of CCA cells in vivo (A) Both mRNA and protein levels were detected for verifying the transfect efficiency (B) Colony formation assay was conducted with Rab1a-shRNA and negative control cells (C) The effect of Rab1a suppression on tumorigenic capacity of CCLP cells was investigated in vivo through subcutaneously injection. The mice were sacrificed after 5 weeks, and subcutaneous tumors were shown. (D) The effect of Rab1a suppression on tumor migration of CCLP cells was investigated in vivo through tail vein injection. The mice were sacrificed after 40 ‘d’, and HE staining of mouse lung tissue containing metastasis was shown, the result was quantified and statistically analyzed.
Rab1a was targeting regulated by miR-212-3p in CCA cells
To investigate upstream regulatory mechanism of Rab1a in CCA cells, we used TargetScan database to predict associated miRNAs and found miR-212-3p may be the candidate miRNA. Then we constructed wild type and mutant type of Rab1a 3’UTR according to sequence of miR-212-3p and designed luciferase reported system to validate our supposition (Fig. 5A). The results of luciferase report experiment showed that miR-212-3p could be targeting combination with 3’UTR of Rab1a mRNA and decrease expression of Rab1a mRNA (Fig. 5B). Then we used miR-212-3p mimics to verify such finding, and the results showed that mRNA level of Rab1a was significantly decreased under condition of miR-212-3p mimics (Fig. 5C). In addition, we detected protein level of Rab1a under condition of miR-212-3p mimics,miR-212-3p inhibitor or negative control, and the results showed that expression of Rab1a in miR-212-3p inhibitor and negative control group was significantly higher than miR-212-3p mimics group (Fig. 5D). Taken together, we found that Rab1a was negative targeting regulated by miR-212-3p in CCA cells.
Figure 5.

Rab1a was targeting regulated by miR-212-3p (A) the complementary paired sequences between 3’UTR of Rab1a and miR-212-3p were showed, and the wild type and mutant type of 3’UTR of Rab1a were designed. (B) The 293T cells were co-transfected with miR-212-3p mimics or NC and 100 ng firefly luciferase reporter plasmid containing wild type or mutant type 3’UTR ofRab1a. After incubation for 48 h, the firefly luciferase activity of each sample was detected and normalized to the renilla luciferase activity, ***p < 0.0005.(C) The mRNA level of Rab1a was compared between miR-212-3p mimics group or negative control group in CCA cells,**p < 0.005.(D) the protein level of Rab1a was detected by western blot under treatment of miR-212-3p mimics,miR-212-3p inhibitor or negative control. GAPDH was as the internal control.
Hypoxia induced expression of Rab1a via inhibiting miR-212-3p
As hypoxia was a microenvironment occurred in tumors and played a vital role in the progression of tumor, we supposed whether Rab1a was regulated under hypoxia condition. We first detected expression of Rab1a both in mRNA and protein levels under normoxia and hypoxia condition, and found Rab1a was increased under hypoxia condition both in mRNA and protein levels (Fig. 6A–B). While we compared expression level of miR-212-3p between normoxia and hypoxia condition and found that miR-212-3p was significantly decreased under hypoxia condition (Fig. 6C). Considering HIF1a was important transcription factor under hypoxia condition, thus we used HIF1a siRNA to suppress expression of HIF1a and compared expression of miR-212-3p and Rab1a among three groups(normoxia group, hypoxia group and hypoxia+HIF1a siRNA group).Then we found that expression of miR-212-3p was higher while Rab1a was lower in normoxia and hypoxia+HIF1a siRNA groups than hypoxia group (Fig. 6D–E). Therefore, these findings showed that hypoxia condition could increase expression of Rab1a through decreasing miR-212-3p level.
Figure 6.

Hypoxia could induce expression of Rab1a through inhibiting miR-212-3p (A-B) the mRNA and protein levels of Rab1a in CCA cells were detected by RT-PCR and western blot under normoxia and hypoxia condition,***p < 0.0005.(C) the expression of miR-212-3p in CCA cells under normoxia and hypoxia condition was detected by RT-PCR,**p < 0.005.(D-E) the expression of miR-212-3p and Rab1a were compared among three different groups, including normoxia condition, hypoxia condition and hypoxia+HIF1a siRNA condition,*p < 0.05,**p < 0.005.
Discussion
Among all kinds of malignant tumors in Hepatobiliary system, the prognosis of CCA was worse than HCC, while correlated study for CCA was less than HCC. Thus more studies are needed for investigating the initial and progression of CCA. Since CCA was stratified according to surgical (extra-hepatic, intrahepatic, hilar, etc), our study mainly investigated the function of Rab1a on progression of intrahepatic cholangiocarcinoma (ICC).
In this study, we first found Rab1a was overexpressed in ICC tissues both in protein and mRNA level. Then we used Kaplan-meier analysis found that those ICC patients who had high expression of Rab1a always leaded to poor prognosis. Further analysis of the relationship between expression of Rab1a and clinicopathologic features showed that high expression of Rab1a always associated with large tumor size, vessel invasion and poor tumor grade. Through loss of function experiments, we demonstrated that suppression of Rab1a led to lower proliferation rate and migration capacity in CCA cell lines. Further study found that hypoxia condition could increase expression of Rab1a via inhibiting miR-212-3p level.
Rab1a, a member RAS oncogene family, played important role in controlling vesicle traffic from the endoplasmic reticulum to the Golgi apparatus.7,8 And further studies showed that Rab1a played vital role in tumor progression.9-15 As X-Z Yang concluded that Rab1 had key roles in regulating mTORC1, Notch and integrin cell signaling pathways, as well as autophagy and localization of cell-surface receptors.18 Our study found Rab1a also could regulate cell cycle and EMT progression. We first used CCK-8 and EDU assay found suppression of Rab1a can affect cell proliferation of RBE and CCLP cell lines. As cell cycle phase was important for cell proliferating, each phase had its own checkpoint, and only passing through the detection of checkpoint the cell can proliferate for the next generation.19,20 So we used flow cytometer to detect change of cell phase under condition of suppression of Rab1a in RBE and CCLP cell lines. Interestingly, the percentage of G1 phase extremely increased and the expression of CDK2, CDK4 and CyclinB1 those G1 phase proteins were deregulated with knockdown of Rab1a, thus we can know that Rab1a was associated with regulating the cell cycle. We also used transwell assay found suppression of Rab1a can inhibit the migration ability of RBE and CCLP cell lines. Epithelial-mesenchymal transition (EMT) which is a reversible dynamic process during which epithelial cells gradually adopt structural and functional characteristics of mesenchymal cells, has gained a lot of attention regarding metastatic dissemination.21,22 Considering this, we used IF and western blot to detect expression change of E-cadherin and N-cadherin under condition of Rab1a suppression. As expected, we found suppression of Rab1a can lead to increasing of E-cadherin and decreasing of N-cadherin. So we verified that Rab1a can regulate the process of EMT in CCA cells.
According to the above findings, we could know that Rab1a was important molecule on progression of ICC. However, there was no reports about how was Rab1a regulated in the CCA cells. As miRNAs was a class of highly conserved short RNAs of 21–23 nucleotides, which inducing degradation of mRNAs via binding to the 3’untranslated region (3’UTR) of their targeted mRNAs.23 And recent studies had proved that Rab1a was targeting regulated by miR-15b-5p,12 miR-63417 and miR-22110 in different solid tumors, we supposed that Rab1a may regulated by miRNAs in CCA cells. By TargetScan database prediction and luciferase reporter experiments, we demonstrated that Rab1a was targeting regulated by miR-212-3p in CCA cells.
Hypoxia was the condition existing in all kinds of tumors, which involved in cancer proliferation, drug resistance, migration and invasion, Immune escape and so on, and HIF1-a was the critical role in the function of hypoxia. And we found Rab20 and Rab22A were reported regulated by HIF1a under hypoxia condition.24,25 So we supposed that whether Rab1a was regulated by HIF1a.Our study found that expression of Rab1a was increased while miR-212-3p was decreased under hypoxia condition. And when knockdown expression of HIF1a under hypoxia condition, the expression of Rab1a was decreased while miR-212-3p was increased. Thus, we demonstrated that hypoxia condition could inhibit transcription of miR-212-3p, such phenomenon led to high expression of Rab1a.
In conclusion, our study found that Rab1a was overexpressed in ICC tissues and associated with poor prognosis of ICC patients. And we found hypoxia/miR-212-3p/Rab1a axis enhanced proliferation and migration of CCA cells through promoting progression of cell cycle and EMT process (Fig. 7). These findings may give new biomarker for developing therapeutic target of ICC. However, further studies were needed to illuminate deep mechanism of Rab1a function on progression of ICC.
Figure 7.

Schematic representation of the relationship between Rab1a and the HIF-1a/miR-212-3p/Rab1a regulatory pathway in HCC proliferation and migration.
Funding Statement
A crucial plan of medical science and technology in henan province, the province department project (No. 201301009).
Ethical approval
The Henan Provicial people's Hospital Ethics Committee approved this study. Informed consent was obtained from all individual participants included in the study. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Author contributions
Panzhang Hou contributes to data collection, experiment conduction and manuscript writing; Yi Kang contributes to fund support and manuscript revision; Jianchao Luo contributes to experiments design, fund support and manuscript revision.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
This study was supported by grants from a crucial plan of medical science and technology in Henan province, the province department project (target gene therapy kill HBVccc DNA in liver cells) (No. 201301009).
References
- 1.Ebata T, Ercolani G, Alvaro D, Ribero D, Di Tommaso L, Valle JW. Current Status on Cholangiocarcinoma and Gallbladder Cancer. Liver Cancer. 2016;6(1):59–65. doi: 10.1159/000449493. PMID:27995089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Global Burden of Disease Cancer C The Global Burden of Cancer 2013. JAMA Oncol. 2015;1(4):505–27. doi: 10.1001/jamaoncol.2015.0735. PMID:26181261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banales JM, Cardinale V, Carpino G, Marzioni M, Andersen JB, Invernizzi P, Lind GE, Folseraas T, Forbes SJ, Fouassier L, et al.. Expert consensus document: Cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA). Nat Rev Gastroenterol Hepatol. 2016;13(5):261–80. doi: 10.1038/nrgastro.2016.51. PMID:27095655. [DOI] [PubMed] [Google Scholar]
- 4.Chong DQ, Zhu AX. The landscape of targeted therapies for cholangiocarcinoma: current status and emerging targets. Oncotarget. 2016;7(29):46750–67. doi: 10.18632/oncotarget.8775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lazaridis KN, Gores GJ. Cholangiocarcinoma. Gastroenterology. 2005;128(6):1655–67. doi: 10.1053/j.gastro.2005.03.040. PMID:15887157. [DOI] [PubMed] [Google Scholar]
- 6.Rizvi S, Gores GJ. Pathogenesis, Diagnosis, and Management of Cholangiocarcinoma. Gastroenterology. 2013;145(6):1215–29. doi: 10.1053/j.gastro.2013.10.013. PMID:24140396.1525821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seabra MC, Brown MS, Slaughter CA, Südhof TC, Goldstein JL. Purification of component A of Rab geranylgeranyl transferase: Possible identity with the choroideremia gene product. Cell. 1992;70(6):1049–57. doi: 10.1016/0092-8674(92)90253-9. PMID:1525821. [DOI] [PubMed] [Google Scholar]
- 8.Tisdale EJ BJ, Khosravi-Far R, Der CJ, Balch WE. GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J Cell Biol. 1992;119(4):749–61. doi: 10.1083/jcb.119.4.749. PMID:1429835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimada K, Uzawa K, Kato M, Endo Y, Shiiba M, Bukawa H, Yokoe H, Seki N, Tanzawa H. Aberrant expression of RAB1A in human tongue cancer. Br J Cancer.. 2005;92(10):1915–21. doi: 10.1038/sj.bjc.6602594. PMID:15870709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun T, Wang X, He HH, Sweeney CJ, Liu SX, Brown M, Balk S, Lee GS, Kantoff PW. MiR-221 promotes the development of androgen independence in prostate cancer cells via downregulation of HECTD2 and RAB1A. Oncogene. 2014;33(21):2790–800. doi: 10.1038/onc.2013.230. PMID:23770851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim SJ, Sohn I, Do I-G, Jung SH, Ko YH, Yoo HY, Paik S, Kim WS. Gene expression profiles for the prediction of progression-free survival in diffuse large B cell lymphoma: Results of a DASL assay. Ann Hematol. 2014;93(3):437–47. doi: 10.1007/s00277-013-1884-0. PMID:23975159. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y, Hou N, Wang X, Wang L, Chang Se, He K, Zhao Z, Zhao X, Song T, Huang C. miR-15b-5p induces endoplasmic reticulum stress and apoptosis in human hepatocellular carcinoma, both in vitro and in vivo, by suppressing Rab1A. Oncotarget. 2015;6(18):16227–38. doi: 10.18632/oncotarget.3970. PMID:26023735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu B-H, Li X-X, Yang Y, Zhang M-Y, Rao H-L, Wang H-Y, Zheng XF. Aberrant amino acid signaling promotes growth and metastasis of hepatocellular carcinomas through Rab1A-dependent activation of mTORC1 by Rab1A. Oncotarget. 2015;6(25):20813–28. doi: 10.18632/oncotarget.5175. PMID:26308575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas JD, Zhang Y, Wei Y, Cho J-H, Morris LE, Wang H-Y, Zheng XF. Rab1A Is an mTORC1 Activator and a Colorectal Oncogene. Cancer Cell. 2014;26(5):754–69. doi: 10.1016/j.ccell.2014.09.008. PMID:25446900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Liu F, Qin X, Huang T, Huang B, Zhang Y, Jiang B. Expression of Rab1A is upregulated in human lung cancer and associated with tumor size and T stage. Aging (Albany NY). 2016;8(11):2790–8. doi: 10.18632/aging.101087. PMID:27902464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu H, Qian M, Zhao B, Wu C, Maskey N, Song H, Li D, Song J, Hua K, Fang L. Inhibition of RAB1A suppresses epithelial-mesenchymal transition and proliferation of triple-negative breast cancer cells. Oncology Rep. 2017;37(3):1619–26. doi: 10.3892/or.2017.5404. [DOI] [PubMed] [Google Scholar]
- 17.Zhang CZ, Cao Y, Fu J, Yun J-P, Zhang M-F. miRߚ634 exhibits antiߚtumor activities toward hepatocellular carcinoma via Rab1A and DHX33. Mol Oncol. 2016;10(10):1532–41. doi: 10.1016/j.molonc.2016.09.001. PMID:27693040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang XZ, Li XX, Zhang YJ, Rodriguez-Rodriguez L, Xiang MQ, Wang HY, Zheng XF. Rab1 in cell signaling, cancer and other diseases. Oncogene. 2016;35(44):5699–704. doi: 10.1038/onc.2016.81. PMID:27041585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murray AW. Recycling the Cell Cycle: Cyclins Revisited. Cell. 2004;116(2):221–34. doi: 10.1016/S0092-8674(03)01080-8. PMID:14744433. [DOI] [PubMed] [Google Scholar]
- 20.Sears RC, Nevins JR. Signaling Networks That Link Cell Proliferation and Cell Fate. J Biol Chem. 2002;277(14):11617–20. doi: 10.1074/jbc.R100063200. PMID:11805123. [DOI] [PubMed] [Google Scholar]
- 21.Nieto MA, Cano A. The epithelial–mesenchymal transition under control: Global programs to regulate epithelial plasticity. Semin Cancer Biol. 2012;22(5–6):361–8. doi: 10.1016/j.semcancer.2012.05.003. PMID:22613485. [DOI] [PubMed] [Google Scholar]
- 22.Guarino M, Rubino B, Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 2007;39(3):305–18. doi: 10.1080/00313020701329914. PMID:17558857. [DOI] [PubMed] [Google Scholar]
- 23.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522. doi: 10.1038/nrg1379. PMID:15211354. [DOI] [PubMed] [Google Scholar]
- 24.Hackenbeck T, Huber R, Schietke R, Knaup KX, Monti J, Wu X, Klanke B, Frey B, Gaipl U, Wullich B, et al.. The GTPase RAB20 is a HIF target with mitochondrial localization mediating apoptosis in hypoxia. Biochim Biophys Acta (BBA)–Mol Cell Res. 2011;1813(1):1–13. doi: 10.1016/j.bbamcr.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 25.Wang T, Gilkes DM, Takano N, Xiang L, Luo W, Bishop CJ, Chaturvedi P, Green JJ, Semenza GL. Hypoxia-inducible factors and RAB22A mediate formation of microvesicles that stimulate breast cancer invasion and metastasis. Proc Natl Acad Sci U S A. 2014;111(31):E3234–E42. doi: 10.1073/pnas.1410041111. PMID:24938788. [DOI] [PMC free article] [PubMed] [Google Scholar]


