Abstract
Acid-sensing ion channels (ASICs) belong to the DEG/ENaC gene family. While ASIC1a, ASIC1b and ASIC3 are activated by extracellular protons, ASIC4 and the closely related bile acid-sensitive ion channel (BASIC or ASIC5) are orphan receptors. Neuropeptides are important modulators of ASICs. Moreover, related DEG/ENaCs are directly activated by neuropeptides, rendering neuropeptides interesting ligands of ASICs. Here, we performed an unbiased screen of 109 short neuropeptides (<20 amino acids) on five homomeric ASICs: ASIC1a, ASIC1b, ASIC3, ASIC4 and BASIC. This screen revealed no direct agonist of any ASIC but three modulators. First, dynorphin A as a modulator of ASIC1a, which increased currents of partially desensitized channels; second, YFMRFamide as a modulator of ASIC1b and ASIC3, which decreased currents of ASIC1b and slowed desensitization of ASIC1b and ASIC3; and, third, endomorphin-1 as a modulator of ASIC3, which also slowed desensitization. With the exception of YFMRFamide, which, however, is not a mammalian neuropeptide, we identified no new modulator of ASICs. In summary, our screen confirmed some known peptide modulators of ASICs but identified no new peptide ligands of ASICs, suggesting that most short peptides acting as ligands of ASICs are already known.
Introduction
Acid-sensing ion channels form a small family of proton-gated ion channels that belongs to the degenerin/epithelial Na+ channel (DEG/ENaC) gene family1. There are four genes coding for ASICs, ACCN1-ACCN4, plus a closely related gene coding for an ion channel that is sensitive to bile acids and has therefore been named bile acid-sensitive ion channel (BASIC)2; alternatively, BASIC is called ASIC5. ASICs are trimers3,4. Homomeric ASIC1a and ASIC3 have a high proton sensitivity (EC50 ~pH 6.5); homomeric ASIC1b, a splice variant of ASIC1a, has a lower proton sensitivity (EC50 ~pH 5.9) and homomeric ASIC2a has a very low proton sensitivity (EC50 < pH 4.5)5–7. Homomeric ASIC2b, a splice variant of ASIC2a, is not activated by low pH, but contributes to heteromeric channels8. The physiological stimuli of ASIC4 and of ASIC5 are unknown9,10.
Some neuropeptides bind to ASICs and modulate their function11. For example, FMRFamide and several FMRFamide-related peptides, including the mammalian neuropeptides FF (NPFF; FLFQPQRFamide) and AF (NPAF; AGEGLSSPFWSLAAPQRFamide)12 and the conorfamide RPRFamide13, probably bind to a cavity in the lower palm domain14 and slow desensitization of ASIC312,13. Other examples are some endogenous opioid peptides, like endomorphin-1 (YPWFamide) and endomorphin-2 (YPFFamide), which slow desensitization of ASIC315, and big dynorphin, which limits steady-state desensitization of ASIC1a16.
Interestingly, other channels in the DEG/ENaC gene family are directly activated by neuropeptides. The first peptide-gated channels to be characterized, the FMRFamide-activated Na+ channel (FaNaC) from snails, and the Hydra Na+ channels (HyNaCs) from Hydra, are activated by RFamide neuropeptides17–19. The recent discovery of a DEG/ENaC activated by a Wamide, the myoinhibitory peptide gated ion channel (MGIC), which is closely related to FaNaC, however, demonstrated that peptides activating DEG/ENaCs are heterogenous20. Modulation of ASICs by neuropeptides and direct activation of peptide-gated DEG/ENaCs by neuropeptides render peptides candidate agonists also for ASIC4 and BASIC.
In this study, we performed an unbiased screen of 109 neuropeptides that contain <20 amino acids. We assessed, first, whether any of the 109 peptides could directly activate ASICs, in particular ASIC4 and BASIC, and, second, whether any of the 109 peptides modulated gating of homomeric ASIC1a, homomeric ASIC1b or homomeric ASIC3.
Results
We selected 109 neuropeptides from the online database NeuroPep (http://isyslab.info/NeuroPep/)21 that met the following criteria: first, organism: Rattus, second, sequence length: below 20 amino acids. To search for potential modulatory effects of neuropeptides on ASIC1a, ASIC1b and ASIC3, we pooled the 109 neuropeptides in 13 mixes as indicated in Table 1. We designed an experimental protocol which captures effects of neuropeptides on both activation and steady-state-desensitization: ASICs were preconditioned with a pH that desensitizes 50% of the channels and were then activated with a pH that opens 50% of the remaining channels, which were not desensitized. Thus, 50% of the ASICs were desensitized, 25% were opened and 25% stayed closed during activation by low pH. We alternately applied preconditioning solutions with or without peptides and looked for changes in current amplitude and desensitization kinetics in the presence of the peptides. Pre-application of peptides would also reveal a direct activation of the channels.
Table 1.
Composition of the 13 peptide mixes screened in this study.
| Name | Amino acid sequence | Name | Amino acid sequence |
|---|---|---|---|
| Mix 1 | Mix 7 | ||
| Macrophage inhibitory factor | TKP | 7B2 (SCG5; Secretogranin-V) C-terminal peptide | SVPHFSEEEKEPE |
| Thyrotropin-releasing hormone precursor | QHP-NH2 | Neurotensin | pQLYENKPRRPYIL |
| Thyrotropin-releasing hormone | pQHP-NH2 | Apelin-13 | QRPRLSHKGPMPF |
| N-Tyr-Melanocyte-inhibiting factor (MIF-1) | YPLG-NH2 | Rimorphin | YGGFLRRQFKVVT |
| Angiotensin I/II(1–4) | DRVY | DynorphinA(1–13) | YGGFLRRIRPKLK |
| Endomorphin-1 | YPWF-NH2 | α-Melanocyte-stimulating hormone | SYSMEHFRWGKPV-NH2 |
| Leu-enkephalin | YGGFL | Pro-Melanin-concentrating hormone(131–144) | EIGDEENSAKFPIG |
| Met-enkephalin | YGGFM | Propeptide21–45(1–14) | QPVVPVEAVDPMEQ |
| long proCART(55–59) | IPIYE | Secretogranin I(186–199) | HTEESGEKHNAFSN |
| Angiotensin I/II(1–5) | DRVYI | Secretogranin II(599–612) | LNQEQAEQGREHLA |
| Mix 2 | Mix 8 | ||
| Tyr-FMRFamide | YFMRF-NH2 | WE-14 | WSRMDQLAKELTAE |
| Neuromedin-N | KIPYIL | Urotensin 2 | QHGTAPECFWKYCI |
| NT-2 (fragment analogue of neurotensin) | RKPWLL | Somatostatin 14 | AGCKNFFWKTFTSC |
| Angiotensin IV | VYIHPF | Secretogranin I(437–451) | DEGHDPVHESPVDTA |
| ORG2766 | (M-O)EHF(D-K)F | VGF(180–194) | pQQETAAAETETRTHT |
| Selank | TKPRPGP | Thymosin beta-10(30–44) | PTKETIEQEKRSEIS |
| KEP | ARPVKEP | Secretogranin II(598–612) | YLNQEQAEQGREHLA |
| ACTH(4–10) | MEHFRWG | [des-Ser1]-cerebellin | GSAKVAFSAIRSTNH |
| Angiotensin I/II(1–7) | DRVYIHP | BigLEN | LENSSPQAPARRLLPP |
| Propeptide 219–229(1–7) | VGRPEWW | VGF(220–235) | VPERAPLPPSVPSQFQ |
| Mix 3 | Mix 9 | ||
| Angiotensin III | RVYIHPF | Secretogranin III(38–53) | ELSAERPLNEQIAEAE |
| Met-enkephalin-RGL | YGGFMRGL | Cerebellin | SGSAKVAFSAIRSTNH |
| Substance P-like | RPQQFGLM -NH2 | Neuropeptide AF-like | AGEGLSSPFWSLAAPQR-NH2 |
| Dynorphin A(1–8) | YGGFLRRI | Thymosin beta-4(28–44) | PLPSKETIEQEKQAGES |
| Neuropeptide FF | FLFQPQRF-NH2 | Chromogranin A(377–393) | LEGEDDPDRSMKLSFRA |
| Angiotensin II | DRVYIHPF | Neuropeptide2 (NocII) | FSEFMRQYLVLSMQSSQ |
| Oxytocin | CYIQNCPLG-NH2 | Nociceptin | FGGFTGARKSARKLANQ |
| 7B2 (SCG5; Secretogranin-V) C-terminal peptide(5–13) | FSEEEKEPE | Dynorphin A(1–17) | YGGFLRRIRPKLKWDNQ |
| Secretogranin I(454–462) | YPQSKWQEQ | LittleSAAS | SLSAASAPLAETSTPLRL |
| Bradykinin | RPPGFSPFR | Tubulin beta-4B chain(168–185) | SVVPSPKVSDTVVEPYNA |
| Mix 4 | Mix 10 | ||
| Gonadotropin-releasing hormone-1-like | HWSYGLRPG | Octadecaneuropeptide | QATVGDVNTDRPGLLDL(K-Ac) |
| β-neoendorphin | YGGFLRKYP | Neuropeptide NPVF | ANMEAGTMSHFPSLPQRF-NH2 |
| Arg-vasopressin | CYFQNCPRG-NH2 | β-Melanocyte-stimulating hormone | ADGPYRVEHFRWGNPPKD |
| Angiotensin I(1–9) | DRVYIHPFH | Neuroendocrine regulatory peptide-4 | NAPPEPVPPPRAAPAPTHV |
| LittleLEN | LENSSPQAPA | Neuropeptide-GE | GPAVFPAENGVQNTESTQE |
| Secretogranin I(371–380) | SEESQEKEYP | long proCART(10–28) | ALDIYSAVDDASHEKELPR |
| HFHH-10 | HFHHALPPAR | LQEQ-19 | LQEQEELENYIEHVLLHRP |
| Antrin | APSDPRLRQF | ||
| Gonadotropin-releasing hormone-1 | pQHWSYGLRPG-NH2 | ||
| Substance P-like | RPKPQQFGLM-NH2 | ||
| Mix 5 | Mix 11 | ||
| α-neoendorphin | YGGFLRKYPK | VGF(211–217) | ASWGEFQ |
| Kisspeptin-10 | YNWNSFGLRY-NH2 | Cholecystokinin-8 | DYMGWMDF-NH2 |
| [Ser2]-Neuromedin-C | GSHWAVGHLM-NH2 | Neurokinin-B | DMHDFFVGLM-NH2 |
| Secretogranin-I (585–594) | SFAKAPHLDL | Proenkephalin 114–133(7–20) | VEPEEEANGGEILA |
| Neurokinin-A | HKTDSFVGLM-NH2 | Chromogranin A(377–391) | LEGEDDPDRSMKLSF |
| Angiotensin I | DRVYIHPFHL | β-Preprotachykinin | ALNSVAYERSAMQNYE |
| Chromogranin A derivative | AYGFRDPGPQL | Gastrin | pQRPPMEEEEEAYGWMDF |
| TLQP-11 (VGF-derived peptide) | TLQPPASSRRR | ||
| Neuropeptide AF-like (C-terminal part of AF) | EFWSLAAPQRF-NH2 | ||
| T-kinin | ISRPPGFSPFR | ||
| Mix 6 | Mix 12 | ||
| Substance P | RPKPQQFFGLM-NH2 | Tail peptide (potential) | ASYYY |
| Neuropeptide FF precursor | NPAFLFQPQRF-NH2 | β-casomorphin-7 | YPFPGP |
| γ-Melanocyte-stimulating hormone-like | YVMGHFRWDRF-NH2 | ||
| VGF(496–507) | PPPRAAPAPTHV | Mix 13 | |
| Somatostatin-28(1–12) | SANSNPAMAPRE | Met-enkephalin-RF | YGGFMRF |
| Cholecystokinin-12 | ISDRDYMGWMDF-NH2 | Urotensin-2B | ACFWKYCV |
| Neurotensin-like | LYENKPRRPYIL | Melanin-concentrating hormone | DFDMLRCMLGRVYRPCWQV |
| RFamide-related peptide-1 | VPHSAANLPLRF-NH2 | ||
| Obestatin-13(C-terminal fragment) | LSGAQYQQHGRAL-NH2 | ||
| Neuropeptide-EI | EIGDEENSAKFPI-NH2 | ||
Sequences of peptides are given in the one-letter code. ACTH, Adrenocorticotropic hormone; CART, Cocaine and amphetamine regulated transcript; pQ, pyro-glutamic acid; (M-O), methionine-sulfoxide; (D-K), D-lysine; (K-Ac), acetylated lysine. Sulfo- or phosphotyrosin modifications were not incorporated into peptides (peptides affected: Cholecystokinin-8, Kisspeptin-10, Cholecystokinin-12 and Gastrin). Incidentally, gastrin was not amidated at the C-terminus. Disulfide bridges were introduced in case of the following peptides: Oxytocin, Arg-Vasopressin, Urotensin-2 and Somatostatin-14.
Modulatory effects of neuropeptides on ASIC1a
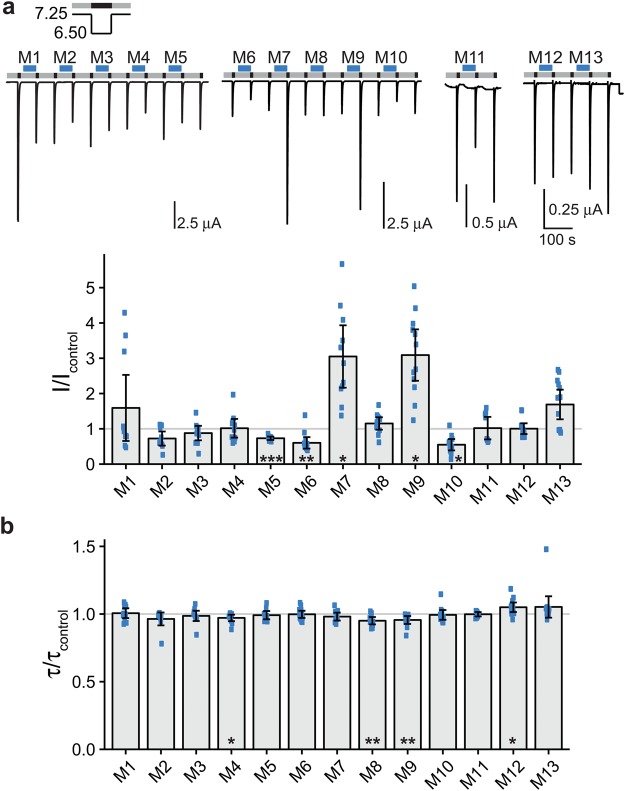
For ASIC1a, we used a preconditioning pH of 7.25 and an activation pH of 6.5. Under these conditions, current amplitude was somewhat variable also without application of peptides (Fig. 1a). While pre-application of peptide mixes 5, 6 or 10 slightly decreased the ASIC1a current amplitude (mix 5: I/Icontrol = 0.73 ± 0.06, mean ± SD, n = 10, p < 0.001; mix 6: I/Icontrol = 0.60 ± 0.26, n = 13, p < 0.01; mix 10: I/Icontrol = 0.55 ± 0.24, n = 12, p < 0.05; Fig. 1a), pre-application of mixes 7 and 9 increased current amplitude more than 3-fold (mix 7: I/Icontrol = 3.05 ± 1.25, n = 11, p < 0.05; mix 9: I/Icontrol = 3.09 ± 1.10, n = 12, p < 0.05; Fig. 1a). To confirm the decrease in current amplitude by mixes 5, 6 and 10, we split each mix in two submixes, the first submix consisting of the first five peptides and the second of the last five peptides of the original mix. Submixes 5.1, 5.2, 6.1, 6.2, 10.1 and 10.2, however, did not significantly decrease the current amplitude of ASIC1a; mixes 5.2, 6.2, 10.1 and 10.2 even slightly increased current amplitude (Supplementary Fig. 1). We conclude that the change in current amplitude was not very robust and perhaps unspecific and did not investigate further the effect of mixes 5, 6 and 10 on ASIC1a current amplitude.
Figure 1.
Effects of peptide mixes on ASIC1a current amplitude and desensitization. (a) Top, representative current traces of ASIC1a expressing oocytes conditioned with pH 7.25 (grey bars) and activated with pH 6.5 (black bars). Peptide mixes 1–13 (M1-M13) were present in the conditioning solution as indicated by the blue bars with a concentration of 20 μM per individual peptide. Bottom, scatter plot showing ratios of the peak currents after pre-application of peptides (I) to the mean of the two flanking control peak currents (Icontrol). Bars show the mean and error bars the SD. (b) Scatter plot showing ratios of time constants of desensitization after pre-application of peptides (τ) to the time constants of desensitization of the two flanking control activations (τcontrol). *p < 0.05, **p < 0.01, ***p < 0.001 (paired Student’s t-test).
Pre-application of dynorphin A(1–13) and dynorphin A(1–17) increased ASIC1a currents
Mixes 7 and 9 contained fragments of dynorphin A (Table 1) which is known to shift the pH dependence of the steady-state desensitization of ASIC1a to more acidic values16. Such a shift is expected to increase current amplitude by retaining more channels in the closed state (and thus available for activation) at conditioning pH 7.25. We tested if the presence of dynorphin A fragments could explain the increased current amplitude after pre-application of mix 7 or 9 by removing dynorphin A(1–13) and dynorphin A(1–17) from these mixes. We then individually applied dynorphin A(1–13) (mix 7) and dynorphin A(1–17) (mix 9) and mixes 7 and 9 without dynorphins (mix 7′ and mix 9′) (Fig. 2). As expected, dynorphin A(1–13) and dynorphin A(1–17) robustly increased current amplitude of ASIC1a (dynorphin A(1–13): I/Icontrol = 2.51 ± 0.53, p < 0.01; dynorphin A(1–17): I/Icontrol = 2.58 ± 0.65, p < 0.001; n = 8) while mixes 7′ and 9′ did not (mix 7′: I/Icontrol = 1.09 ± 0.25; mix 9′: I/Icontrol = 1.35 ± 0.31; n = 8; p > 0.05; Fig. 2). We conclude that dynorphin A was responsible for the increased current amplitude after pre-application of mixes 7 and 9. In summary, except for fragments of dynorphin A, we did not identify another peptide that affected ASIC1a current amplitude when pre-applied. We estimated the power of our screen for different effect sizes (see Methods): it had a high power (0.97) to reveal a change of 50% in current amplitude, but a relatively low power (0.49) to detect a small change of 25% (Table 2). Thus, we might have missed peptides that slightly changed current amplitudes.
Figure 2.
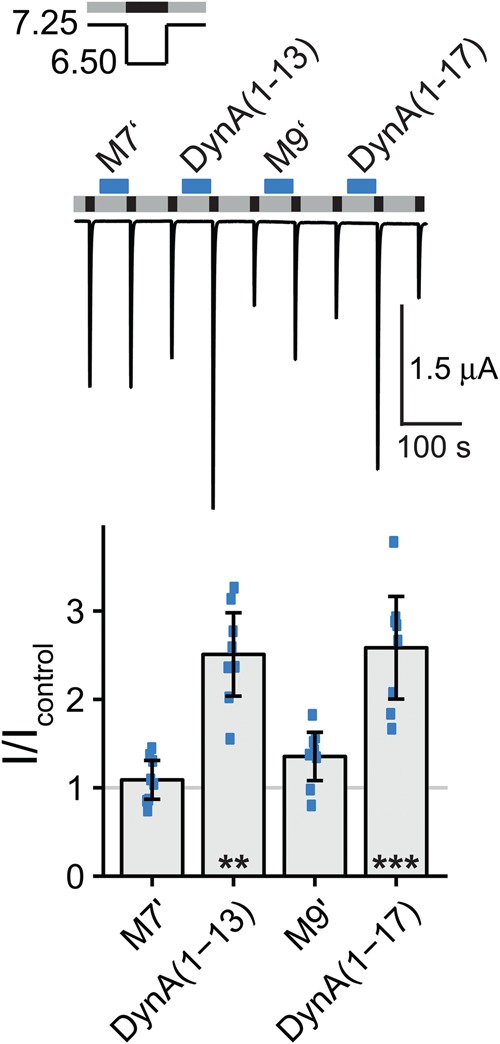
Dynorphin A(1–13) and dynorphin A(1–17) increase ASIC1a current amplitude. Top, representative current trace of an ASIC1a expressing oocyte conditioned with pH 7.25 (grey bars) and activated with pH 6.5 (black bars). Peptide mixes 7′ and 9′ (M7′ and M9′), dynorphin A(1–13) (DynA(1–13)) or dynorphin A(1–17) (DynA(1–17)) were present in the conditioning solution as indicated by the blue bars with a concentration of 20 μM per individual peptide. Mix 7′ contained all peptides of mix 7 except for dynorphin A(1–13), mix 9′ contained all peptides of mix 9 except for dynorphin A(1–17) (See Table 1). Bottom, scatter plot showing ratios of I to Icontrol. Bars show the mean and error bars the SD. **p < 0.01, ***p < 0.001 (paired Student’s t-test).
Table 2.
Calculation of the statistical power of our screen.
| Channel | Condition | Mean | SD | n | 50% reduction | 25% reduction | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Iexp/τexp | d | Power | Iexp/τexp | d | Power | |||||
| ASIC1a | Amplitude (μA) | −1.02 | 0.32 | 8 | −0.51 | −1.59 | 0.97 | −0.25 | −0.79 | 0.49 |
| τdes (msec) | 1376 | 85 | 8 | 688 | 8.09 | >0.99 | 344 | 4.05 | >0.99 | |
| ASIC1b | Amplitude (μA) | −0.42 | 0.05 | 8 | −0.21 | −4.58 | >0.99 | −0.11 | −2.29 | >0.99 |
| τdes (msec) | 732 | 55 | 8 | 366 | 6.65 | >0.99 | 183 | 3.32 | 0.99 | |
| ASIC3 | Amplitude (μA) | −0.24 | 0.12 | 8 | −0.12 | −0.99 | 0.67 | −0.06 | −0.50 | 0.23 |
| τdes (msec) | 436 | 87 | 8 | 218 | 2.50 | 0.86 | 109 | 1.25 | 0.17 | |
Power was calculated for a 50% reduction of the current amplitude and of τdes, and for a 25% reduction, respectively. Power for a 50% or a 25% increase would be identical. See Methods for details.
We analyzed effects on desensitization by determining time constants of desensitization (τdes). Four mixes (mix 4, 8, 9 and 12) changed τdes with a p-value p < 0.05, but produced small effect sizes (mix 4: τ/τcontrol = 0.97 ± 0.03, n = 11, p < 0.05; mix 8: τ/τcontrol = 0.95 ± 0.04, n = 12, p < 0.01; mix 9: τ/τcontrol = 0.96 ± 0.04, n = 12, p < 0.01; mix 12: τ/τcontrol = 1.05 ± 0.06, n = 13, p < 0.05; Fig. 1b). Therefore, we decided to use a stricter significance level (α = 0.001) to decide if a mix should be considered as a relevant modulator of current desensitization of ASICs. For ASIC1a, none of the submixes reached this level of significance; the estimated power of our screen to detect changes in τdes was high (>0.99; Table 2).
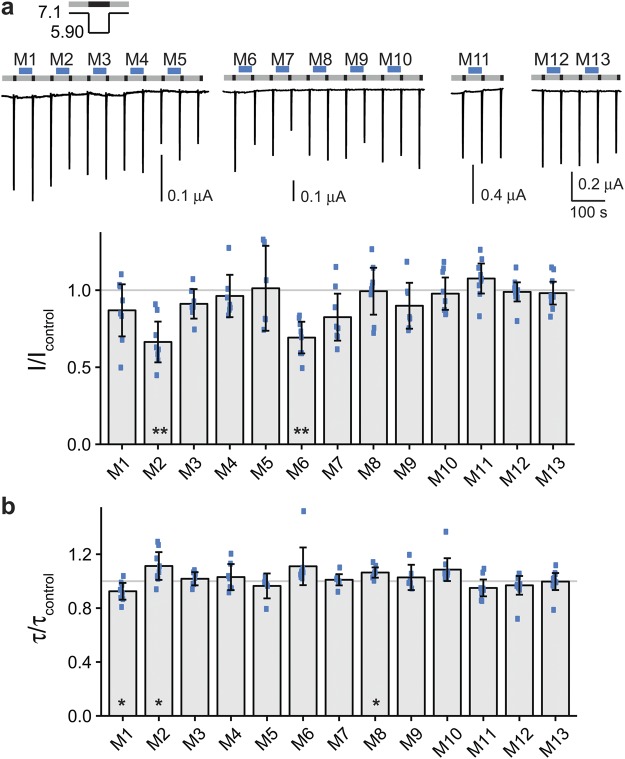
Modulatory effects of neuropeptides on ASIC1b
For ASIC1b, we used pH 7.1 for preconditioning and pH 5.9 for activation. Pre-application of mix 2 and of mix 6 significantly decreased current amplitudes of ASIC1b (mix 2: I/Icontrol = 0.66 ± 0.15; mix 6: I/Icontrol = 0.69 ± 0.12; n = 8; p < 0.01; Fig. 3a). Therefore, we split each of these mixes in half (submixes 2.1, 2.2, 6.1 and 6.2). Only mix 2.1 decreased the current amplitude significantly (I/Icontrol = 0.82 ± 0.04; n = 10; p < 0.05) whereas mixes 2.2, 6.1 and 6.2 did not (Supplementary Fig. 2).
Figure 3.
Effect of peptide mixes on ASIC1b current amplitude and desensitization. (a) Top, representative current traces of ASIC1b expressing oocytes conditioned with pH 7.1 (grey bars) and activated with pH 5.9 (black bars). Peptide mixes 1–13 (M1-M13) were present in the conditioning solution as indicated by the blue bars with a concentration of 20 μM per individual peptide. Bottom, scatter plot showing ratios of I to Icontrol. Bars show the mean and error bars the SD. (b) Scatter plot showing ratios of τ to τcontrol. *p < 0.05, **p < 0.01 (paired Student’s t-test).
Pre-application of YFMRFamide reduced current amplitude and slowed desensitization of ASIC1b
Next, we individually pre-applied the five neuropeptides of mix 2.1 (YFMRFamide, neuromedin N, NT-2 (fragment analogue of neurotensin), angiotensin IV and ORG2766). Only pre-application of YFMRFamide significantly decreased the current amplitude (I/Icontrol = 0.58 ± 0.04; n = 7; p < 0.01; Fig. 4a); additionally, it slightly slowed the desensitization of ASIC1b (τ/τcontrol = 1.13 ± 0.04; n = 7; p < 0.001; Fig. 4b). In contrast, none of the peptide mixes changed τdes of ASIC1b with a high level of significance (p < 0.001) (Fig. 3b). The estimated power of our screen to detect changes in current amplitude and in τdes was high (0.99 or higher; Table 2). We conclude that among the 109 neuropeptides only YFMRFamide modulated ASIC1b. YFMRFamide is related to FMRFamide, which does not strongly modulate ASIC1b22. But YFMRFamide is not a mammalian neuropeptide23 and therefore no endogenous modulator of ASIC1b.
Figure 4.
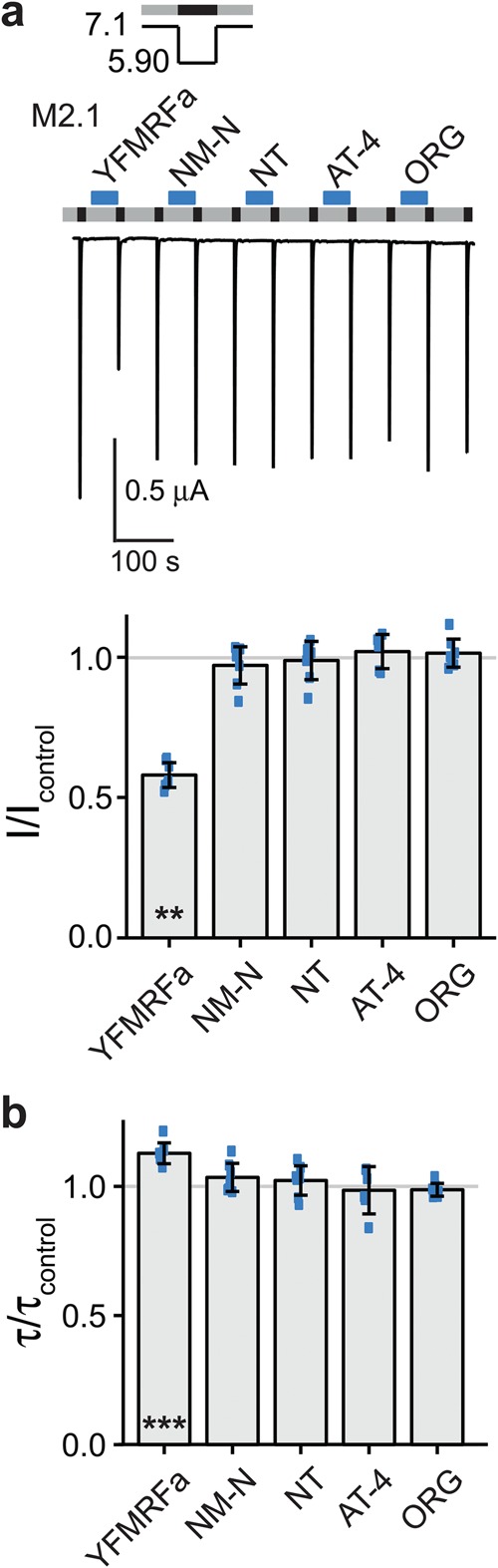
YFMRFamide decreases peak currents and slows desensitization of ASIC1b. (a) Top, representative current trace of an ASIC1b expressing oocyte conditioned with pH 7.1 (grey bars) and activated with pH 5.9 (black bars). Individual peptides of mix 2.1 (M2.1) were applied as indicated by the blue bars with a concentration of 20 μM each: YFMRFamide (YFMRFa), neuromedin N (NM-N), NT-2 (fragment analogue of neurotensin) (NT), angiotensin IV (AT-4) and ORG2766 (ORG). Bottom, scatter plot showing ratios of I to Icontrol. Bars show the mean and error bars the SD. (b) Scatter plot showing ratios of τ to τcontrol. **p < 0.01, ***p < 0.001 (paired Student’s t-test).
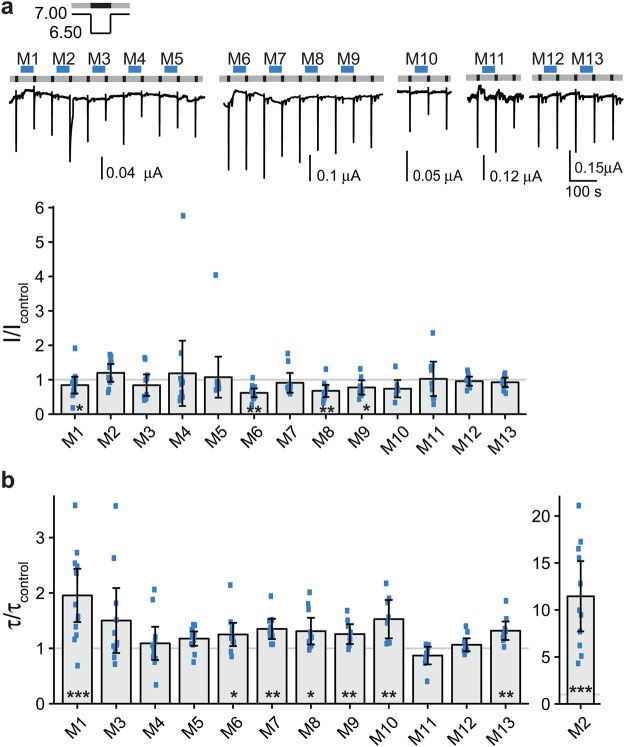
Modulatory effects of neuropeptides on ASIC3
For ASIC3, we used a conditioning pH of 7.0 and an activation pH of 6.5. Pre-application of mixes 1, 6, 8 and 9 slightly but significantly decreased peak current amplitudes (mix 1: I/Icontrol = 0.84 ± 0.39, n = 13, p < 0.05; mix 6: I/Icontrol = 0.62 ± 0.19, n = 12, p < 0.01; mix 8: I/Icontrol = 0.68 ± 0.25, n = 11, p < 0.01; mix 9: I/Icontrol = 0.77 ± 0.25, n = 9, p < 0.05; Fig. 5a); mix 1 also slowed desensitization kinetics highly significantly (τ/τcontrol = 1.96 ± 0.77; n = 13; p < 0.001; Fig. 5b). To confirm these effects and to identify individual neuropeptides, we split each mix in half (submixes 1.1, 1.2, 6.1, 6.2, 8.1, 8.2, 9.1 and 9.2). Only mix 1.2 decreased current amplitude significantly (I/Icontrol = 0.80 ± 0.09; n = 8; p < 0.05; Supplementary Fig. 3a). In addition, mix 1.2 also slowed desensitization significantly (τ/τcontrol = 1.21 ± 0.08; n = 8; p < 0.001; Supplementary Fig. 3b).
Figure 5.
Effect of peptide mixes on ASIC3 current amplitude and desensitization. (a) Top, representative current traces of ASIC3 expressing oocytes conditioned with pH 7.0 (grey bars) and activated with pH 6.5 (black bars). Peptide mixes 1–13 (M1-M13) were present in the conditioning solution as indicated by the blue bars with a concentration of 20 μM per individual peptide. Bottom, scatter plot showing ratios of I to Icontrol. Bars show the mean and error bars the SD. (b) Scatter plot showing ratios of τ to τ control. *p < 0.05, **p < 0.01, ***p < 0.001 (paired Student’s t-test).
Pre-application of endomorphin-1 and YFMRFa slowed desensitization of ASIC3
We next asked which individual neuropeptide decreased current amplitude and slowed desensitization and pre-applied the individual peptides of mix 1.2 (endomorphin-1, leu-enkephalin, met-enkephalin, long proCART(55–59) and angiotensin I/II(1–5)) and identified endomorphin-1 to slow desensitization of ASIC3 (τ/τcontrol = 1.92 ± 0.34; n = 7; p = 0.001; Fig. 6b). In contrast, none of the individual peptides significantly decreased current amplitude (Fig. 6a). A slowing of ASIC3 desensitization by endomorphin-1 is in line with a recently published article15 and can fully account for the slowed desensitization by mix 1 (Fig. 5b).
Figure 6.
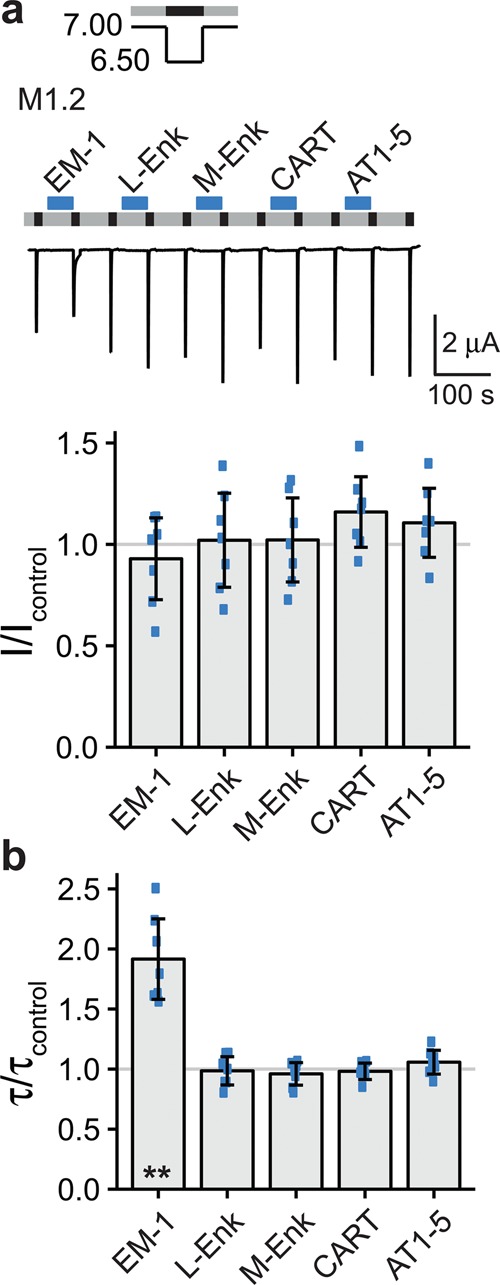
Endomorphin-1 slows the desensitization of ASIC3. (a) Top, representative current trace of an ASIC3 expressing oocyte conditioned with pH 7.0 (grey bars) and activated with pH 6.5 (black bars). Individual peptides of mix 1.2 (M1.2) were applied as indicated by the blue bars with a concentration of 20 μM each: endomorphin-1 (EM-1), leu-enkephalin (L-Enk), met-enkephalin (M-Enk), long proCART(55–59) (CART) and angiotensin I/II(1–5) (AT1–5). Bottom, scatter plot showing ratios of I to Icontrol. Bars show the mean and error bars the SD. (b) Scatter plot showing ratios of τ to τcontrol. **p < 0.01 (paired Student’s t-test).
Like mix 1, mix 2 also significantly slowed desensitization of ASIC3 (mix 2: τ/τcontrol = 11.45 ± 5.31, n = 11; p < 0.001; Fig. 5b). Mix 2 contained the RFamide YFMRFa, which is a variant of FMRFamide (FMRFa), a peptide known to slow desensitization of ASIC312. This suggested that YFMRFa might be the active compound in mix 2. To confirm this hypothesis, we applied YFMRFa individually and compared its effect to the remaining nine peptides of mix 2 (mix 2′; Fig. 7a). YFMRFa did indeed significantly slow the desensitization of ASIC3 (τ/τcontrol = 12.12 ± 9.40; n = 10; p < 0.01; Fig. 7b) whereas, surprisingly, the nine remaining peptides decreased the current amplitude significantly (I/Icontrol = 0.78 ± 0.09; n = 10; p < 0.01; Fig. 7a); YFMRFa had no effect on the current amplitude. To find a potentially active peptide within the nine peptides of mix 2′, whose effect on current amplitude might have been masked by YFMRFa, we split mix 2′ in two submixes (mix 2.1′ contained the first four and mix 2.2′ the remaining five peptides; Supplementary Fig. 4). No significant effect on current amplitude could, however, be observed for the two submixes 2.1′ and 2.2′ (Supplementary Fig. 4). We conclude that mix 2 did not contain a peptide, in addition to YFMRFa, that robustly modulated ASIC3. In summary, among the 109 neuropeptides, our screen identified endomorphin-1 and YFMRFa as modulators of ASIC3. The estimated power of our screen to detect changes in current amplitude was relatively low (0.67 for a change of 50%) and to detect changes in τdes it was modest (0.86 for a 50% change; Table 2). Thus, our screen could only identify peptides with a strong effect on current amplitude or on τdes of ASIC3 (change of at least 50%).
Figure 7.
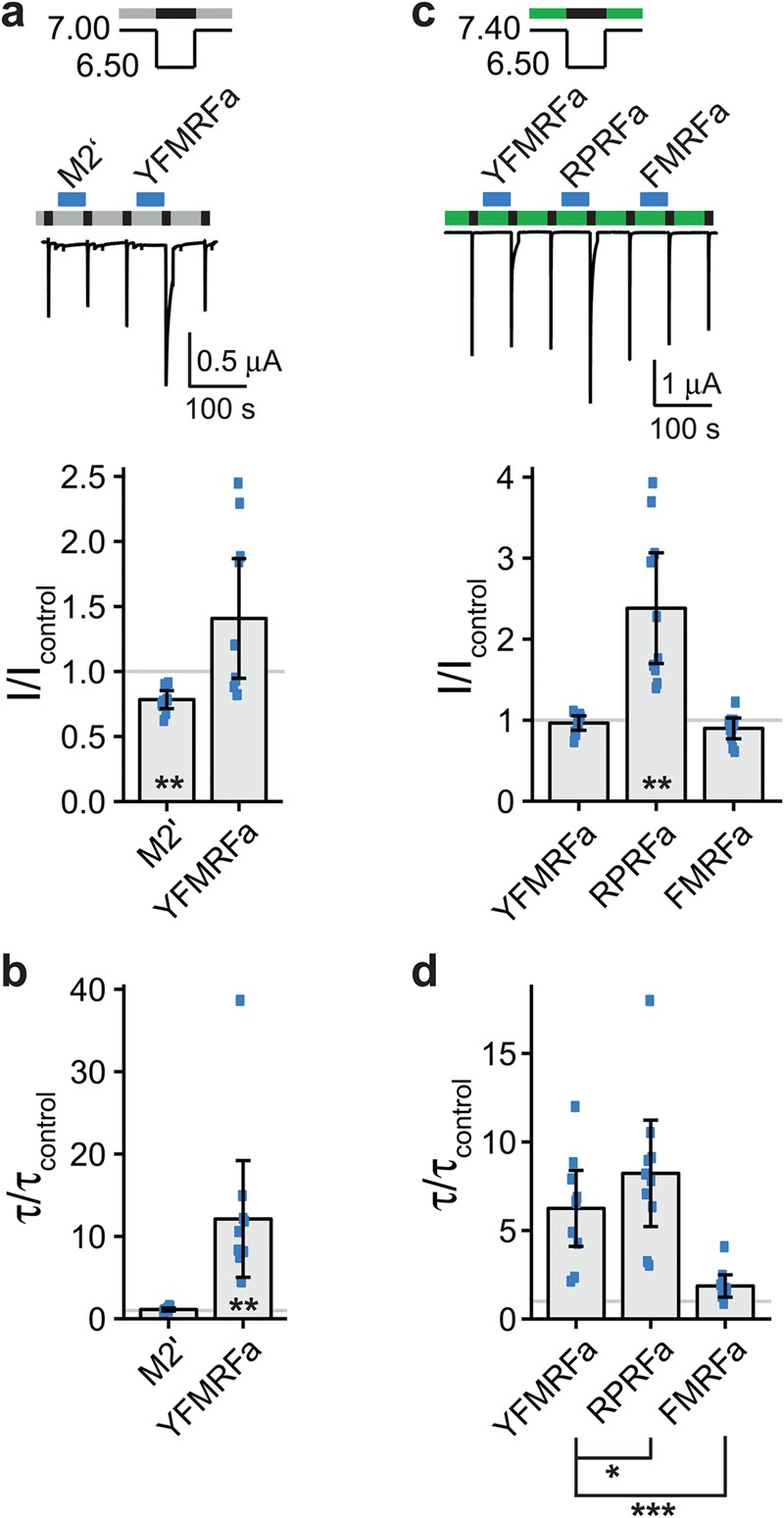
YFMRFamide slows the desensitization of ASIC3. (a) Top, representative current trace of an ASIC3 expressing oocyte conditioned with pH 7.0 (grey bars) and activated with pH 6.5 (black bars). Peptide mix 2′ (M2′) and YFMRFamide (YFMRFa) were present in the conditioning solution as indicated by the blue bars with a concentration of 20 μM per individual peptide. Mix 2′ contained all peptides of mix 2 except for YFMRFa. (See Table 1). Bottom, scatter plot showing ratios of I to Icontrol. Bars show the mean and error bars the SD. (b) Scatter plot showing ratios of τ to τcontrol. (c) Top, representative current trace of an ASIC3 expressing oocyte conditioned with pH 7.4 (green bars) and activated with pH 6.5 (black bars). YFMRFamide (YFMRFa), RPRFamide (RPRFa) and FMRFamide (FMRFa) were present in the conditioning solution as indicated by the blue bars with a concentration of 20 μM per individual peptide. Bottom, scatter plot showing ratios of I to Icontrol. Bars show the mean and error bars the SD. **p < 0.01 (paired Student’s t-test). (d) Scatter plot showing ratios of τ to τcontrol. *p < 0.05, **p < 0.01, ***p < 0.001 (paired Student’s t-test followed by Bonferroni correction).
Since modulation of ASIC3 by YFMRFa has not been previously reported, we compared its effect on ASIC3 to two other RFamides that are known to slow desensitization of ASIC3: the conorfamide RPRFamide (RPRFa) and FMRFa13. To evaluate modulation, we used preconditioning pH 7.4 and activation pH 6.5; RFamides were pre-applied individually. RPRFa significantly increased the peak current (I/Icontrol = 2.38 ± 0.91; n = 10; p < 0.01), whereas YFMRFa and FMRFa did not (Fig. 7c). Concerning desensitization, RPRFa had a stronger effect on ASIC3 than YFMRFa (RPRFa: τ/τcontrol = 8.23 ± 3.99; YFMRFa: τ/τcontrol = 6.25 ± 2.85; n = 10; p = 0.04; Fig. 7d) but YFMRFa had a stronger effect than FMRFa (FMRFa: τ/τcontrol = 1.86 ± 0.84; n = 10; p < 0.001; Fig. 7d). Thus, the three peptides slowed desensitization with the following rank order RPRFa > YFMRFa > FMRFa.
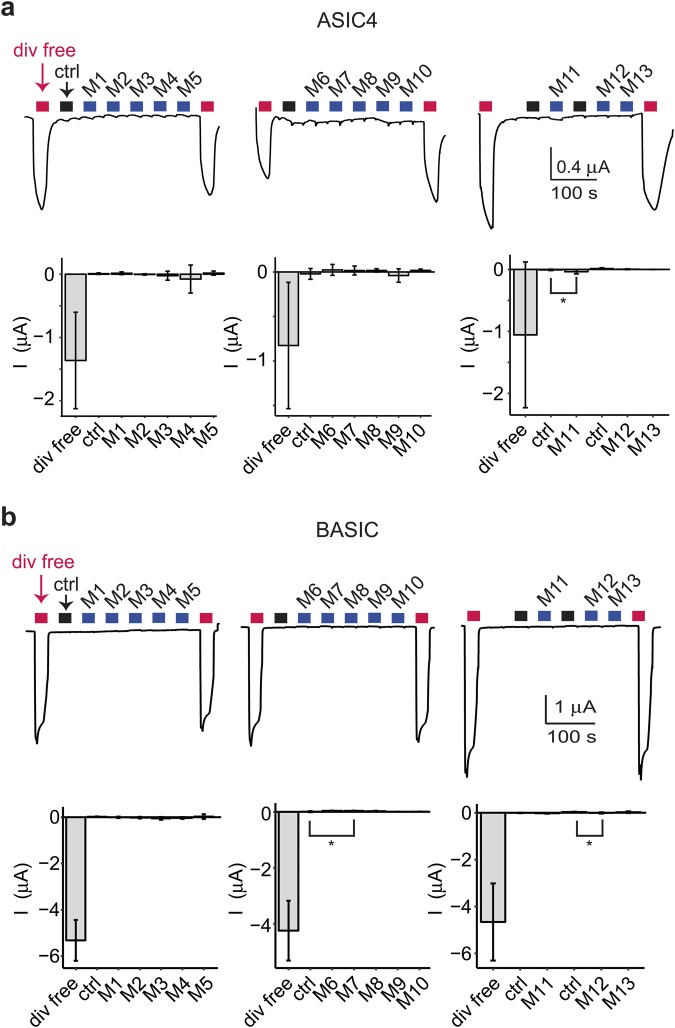
Direct effects of neuropeptides on ASIC4 and BASIC
We then screened the 109 neuropeptides on ASIC4- or BASIC-expressing oocytes to identify a direct agonist of these two channels. The peptides were applied at a pH of 7.4 in the same 13 mixes as before (Table 1). Divalent free bath solution opens ASIC4 and BASIC24,25. Therefore, we checked for channel expression with control activations with divalent free solution at the beginning and at the end of experiments. Solution containing the respective vehicle (water, NH4Cl or DMSO) was applied as a control for solution exchange artifacts. Mix 11 induced significant currents of ASIC4-expressing oocytes relative to control solution (p < 0.05; Fig. 8a). The current amplitude was tiny, however, for a high concentration (20 μM) of individual peptides (Imix11 = −0.04 ± 0.03 μA; Icontrol = −0.01 ± 0.01 μA; n = 6; p < 0.05; Fig. 8a). Therefore, we did not consider this effect size as physiologically relevant and did not investigate mix 11 on ASIC4 further. Mix 7 and mix 12 induced small but significant currents on BASIC expressing oocytes (mix 7: Imix7 = 0.03 ± 0.01 μA; Icontrol = −0.01 ± 0.02 μA, n = 6, p < 0.05; mix 12: Imix12 = -0.01 ± 0.02 μA; Icontrol = −0.03 ± 0.01 μA; n = 6; p < 0.05; Fig. 8b). The current that was induced by mix 7 however was an outward current and the current induced by mix 12 was quite small. Therefore, we decided not to further investigate the effect of mix 7 and mix 12 on BASIC. We conclude that the 109 neuropeptides do not contain a direct agonist of ASIC4 or BASIC.
Figure 8.
There is no direct agonist for ASIC4 and BASIC among the 109 neuropeptides. (a,b) Top, representative current traces of ASIC4 (a) or BASIC (b) expressing oocytes activated with divalent free solution (div free; red bars). Controls (ctrl; black bars) before each peptide mix (M; blue bars) contained the solvent in its respective concentration. Peptides were applied at pH 7.4. Bottom, bar graphs showing the mean current amplitudes; error bars show the SD. *p < 0.05 (paired Student’s t-test followed by Bonferroni correction).
Discussion
Our study was motivated by the question whether short neuropeptides act as direct agonists of ASICs, with an emphasis on the orphan receptors ASIC4 and BASIC. Since the peptides that activate established peptide-gated DEG/ENaCs contain four (FMRFamide, the ligand of FaNaC)17, seven (Hydra-RFamides, the ligands of HyNaCs)18 or nine to ten amino acids (myoinhibitory peptides, the ligands of MGIC)20, and to limit the financial costs of our screen, we restricted it to peptides that contain less than 20 amino acids. In a recent screen of 121 neuropeptides from the marine annelid Platynereis, we identified myoinhibitory peptides as agonists of MGIC20, illustrating the sensitivity of our screening system. In our new screen, we used rat ASICs and chose rat neuropeptides from NeuroPep, a comprehensive database that at its recent release was the most complete neuropeptide database21. Thus, although we cannot exclude that some small peptides were missing from our screen, it is likely that most known small neuropeptides were included. Therefore, we conclude that short neuropeptides (<20 amino acids) are unlikely direct agonists of ASICs. We cannot exclude, however, that a short peptide currently not contained in NeuroPep or a longer peptide (>20 amino acids) directly activates an ASIC.
In addition, we used our panel of peptides to screen for new modulators of ASIC1a, ASIC1b and ASIC3. To be able to screen many peptides on several ASICs, we used an application protocol that allowed us to capture at the same time modulation of activation, of steady-state desensitization and of acute desensitization. With this protocol, however, current amplitudes were somewhat variable also under control conditions, limiting the sensitivity of our screen. To keep the sensitivity of our screen as high as possible we used a significance level of 0.05 and no correction for multiple testing. Instead, we performed follow-up tests of positive mixes to reduce the false positive rate. This is illustrated by cases, in which original peptide mixes appeared to change current amplitudes but in which this change could not be confirmed by peptide submixes. Although, we cannot rule out that some peptides were active only when co-applied with each other and that this effect was lost upon un-mixing, we considered in these cases the initial change of current amplitude as a false positive result. The variability of current amplitude can at least partially be explained by a conditioning pH that desensitized approximately 50% of the channels. Due to the extreme steepness of steady-state desensitization curves of ASICs (Hill coefficient for ASIC1a ~11)6, at this pH a minor change in pH in our recording chamber would have immediately affected current amplitude. Similarly, due to the large size of oocytes and the comparatively slow solution exchange, acute desensitization of ASICs cannot be measured with a high precision and reproducibility26. Moreover, the size of effects on desensitization was small. This led us to apply a more stringent criterion to assess significance of changes in τdes (p < 0.001) than of changes in current amplitudes (p < 0.05). Finally, since we used mixes of peptides, we cannot exclude that peptides within one mix interfered with each other, potentially masking a modulatory effect. We have recently shown, however, that by screening of peptide mixes, novel peptide ligands for DEG/ENaCs can be identified20.
The limited sensitivity of our screen is also illustrated by the fact that the power of our screen to detect a relatively small change in current amplitude of 25% was high only for ASIC1b, but low for ASIC1a and ASIC3 (Table 2). The power to detect a robust change in current amplitude of 50% was high also for ASIC1a, but relatively low for ASIC3. The power to detect changes of τdes was generally high, with the exception of small changes of τdes for ASIC3 (Table 2). This limited sensitivity for ASIC3 can explain why we missed NPFF, a known modulator of ASIC3, which slows desensitization12,22,27 and which was contained in mix 3 (NPFF) (Table 1), but which has rather small effects12,22,27.
Irrespective of the limitations of our screen (uncomprehensive set of peptides, limited sensitivity, use of peptide mixes), we identified several known modulators of ASICs, dynorphin A16 and endomorphin-115, demonstrating that we were able to identify strong modulators of ASICs. Thus, although we might have missed some peptides with a small modulatory effect on ASICs, we conclude that most small neuropeptides robustly modulating ASICs are already known.
In summary, reporting mainly negative results, we believe that our screen will help to focus future research on the interaction of ASICs with known modulatory neuropeptides and advances our understanding of the interaction of ASICs with neuropeptides.
Methods
Peptides
Peptide sequences were retrieved from the online database NeuroPep on July 25, 2016. In some cases, peptide sequences were adjusted relative to the sequences supplied by NeuroPep (see Table 1): the two peptides hexarelin and cortistatin-14 were not included in our screen and sequences of ORG2766 and ACTH4–10 were corrected according to the literature. Although indicated as “rat” peptides in the database, some of the peptides have an uncertain origin. YFMRFamide, for example, is not a mammalian neuropeptide.
Neuropeptides were purchased from GenScript (New Jersey, United States) as either lyophilized powders or crystals with a purity of >95%. RPRFamide and FMRFamide were obtained from ProteoGenix (Schiltigheim, France) and kept in aqueous stock solutions. We dissolved the neuropeptides either in ultrapure degased water (mixes 1–10), in water containing 0.75–3% (w/v) NH3 (mix 11) or in dimethyl sulfoxide (DMSO) (mixes 12 and 13) to obtain a concentration of 2 to 10 mM per peptide. Up to ten peptides were then pooled to form stock solutions of the 13 mixes. The composition of each mix is indicated in Table 1. The concentration of individual peptides in the stock solution was 200 μM for mixes 1–10, 1 mM for mix 11, 5 mM for mix 12 and 3.33 mM for mix 13. Stock solutions of individual peptides or of peptide mixes were kept at −80 °C.
All other chemicals were purchased from Sigma-Aldrich (Munich, Germany) or Carl Roth GmbH (Karlsruhe, Germany).
Molecular biology and handling of oocytes
Animal care and experimental protocols for female Xenopus laevis frogs were approved by the Landesamt für Natur, Umwelt und Verbraucherschutz of North Rhine-Westphalia (LANUV NRW) and were performed in accordance with LANUV NRW guidelines. cRNA synthesis and preparation of oocytes was described previously20. Stage V and VI oocytes were injected with the following amounts of cRNA: 0.02 ng of rat ASIC1a, 0.41 ng of rat ASIC1b, 3.3 ng of rat ASIC3, 1.7–6.7 ng of rat ASIC4 or 26 ng of rat BASIC. After injection, oocytes were kept at 19 °C in Oocyte Ringer-2 (OR-2; in mM: 82 NaCl, 2.5 KCl, 1 Na2HPO4, 1 MgCl2, 1 CaCl2, 0.5 g/l PVP, 5 HEPES; pH 7.3), except for BASIC-expressing oocytes which were kept in low-Na+ OR-2 (in mM: 77.5 NMDG, 5 NaCl, 2.5 KCl, 1 Na2HPO4, 1 MgCl2, 1 CaCl2, 0.5 g/l PVP, 5 mM HEPES; pH 7.3).
Electrophysiological recordings
Electrophysiological recordings on Xenopus laevis oocytes were performed 24–72 h after injection with a TurboTec 03X two electrode voltage clamp amplifier (npi electronic, Tamm, Germany). Fast solution exchange was provided by a programmable pipetting robot (screening tool; npi electronic)28. Data were filtered at 20 Hz and sampled at 500 Hz. Data acquisition was performed using CellWorks 6.2.1 (npi electronic). The holding potential was −70 mV.
Bath solutions were freshly prepared each day by adding peptide mixes or single peptides to bath solution at a final concentration of 20 μM of each peptide. Bath solution contained the following (in mM): 140 NaCl, 1.8 CaCl2, 1 MgCl2 and 10 HEPES or MES (pH < 6.6); solutions with a pH of 7.0–7.25 contained 20 HEPES. For mix 11, bath solution contained also NH3 (final concentration 0.042%, w/v) and for mixes 12 and 13 DMSO (final concentration 0.4% DMSO in mix 12 and 0.6% DMSO in mix 13; v/v). Peptide-containing solutions and control solutions only differed in the presence or absence of neuropeptides. Peptide-containing solutions were titrated by adding NaOH and HCl immediately before the measurements. We have chosen a pH that desensitizes 50% of the channels based on values published previously by our group6,13; those pH values were verified for the specific set-up used in control experiments at the beginning of this study. Different peptide mixes were applied to half of the oocytes expressing ASIC1a, ASIC1b or ASIC3 in numerical order (e.g. from mix 1 to mix 5) and to the other half in inverse order (e.g. from mix 5 to mix 1).
Data analysis
Data was analyzed using CellWorks Reader 6.2.2 (npi electronic) or Igor Pro 5.0.3 (WaveMetrics, Portland, USA); statistical analysis was performed in Microsoft Excel or R 3.4.129 and plots were generated using the ggplot2 package30 in R. In some cases, the baseline was corrected with the software.
For ASIC1a, ASIC1b and ASIC3, p-values were determined by paired Students t-test. A t-test was performed for each peptide application; the comparison was made between peak current amplitudes after pre-application of peptide and the mean of the peak current amplitudes of the two flanking activations after pre-application of vehicle. For reasons of clarity, instead of plotting the absolute values of currents after pre-application of peptide and vehicle we plotted the ratios (I/Icontrol). Current decays were fit to a single exponential function in Igor Pro to obtain time constants of desensitization (τdes). Statistical analysis and plotting of τdes were performed in analogy to the peak currents. To detect false-positive results, we performed a second round of screening with submixes.
For ASIC4 and BASIC, p-values were also determined by paired Students t-test. In contrast to ASIC1a, ASIC1b and ASIC3, only one control activation with vehicle was performed at the beginning of each measurement. To correct for multiple testing, we applied Bonferroni correction. A t-test was performed for each peptide application; the comparison was made between current amplitudes during peptide application and current amplitudes during application of vehicle. Plots depict absolute current amplitudes.
All results are reported as mean ± standard deviation (SD). Experiments were performed with oocytes of at least two different batches of oocytes.
We estimated the statistical power of our screen for a modulatory effect on ASIC1a, ASIC1b and ASIC3 for a relative current change of 50% and of 25%. The expected current, Iexp, was thus obtained by multiplying the average control current amplitude by 0.5 or 0.25, respectively. Since the t-test compared current amplitudes after pre-application of peptide with control applications (see above), we determined the standard deviation for control currents without peptide. First, we calculated the standard deviation of control currents for each oocyte. These standard deviations were then averaged for each ASIC. Note that we assumed that the standard deviation was similar for control measurements and measurements with pre-application of peptides. Iexp was then divided by the standard deviation of the corresponding control measurements to yield Cohen’s d as a measure of the effect size: d = Iexp/SD. The ‘pwr’ package of R was used to calculate the power for specific values of d, the average number of recordings n (n = 8) and the α-level (α = 0.05) for each channel. The statistical power for relative changes of τdes was calculated in a similar way, except that the α-level was adjusted (α = 0.001). Values for the statistical power are indicated in Table 2.
Electronic supplementary material
Acknowledgements
We thank Adrienne Oslender-Bujotzek for expert technical assistance.
Author Contributions
A.S. chose peptides. A.V. performed TEVC measurements. A.S. and D.W. supervised A.V. and D.W. participated in the design of the study. A.V. and A.S. analyzed the data. A.S. and S.G. conceived the study, designed the study and coordinated the study. A.V. and S.G. drafted the manuscript and all other authors edited and approved the final manuscript.
Data Availability
All data are available from the corresponding author upon request. All relevant data are within the paper.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Axel Schmidt and Stefan Gründer jointly supervised this work.
Contributor Information
Axel Schmidt, Email: axel.schmidt1@rwth-aachen.de.
Stefan Gründer, Email: sgruender@ukaachen.de.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-36125-5.
References
- 1.Kellenberger S, Schild L. International Union of Basic and Clinical Pharmacology. XCI. structure, function, and pharmacology of acid-sensing ion channels and the epithelial Na+ channel. Pharmacol Rev. 2015;67:1–35. doi: 10.1124/pr.114.009225. [DOI] [PubMed] [Google Scholar]
- 2.Wiemuth, D. et al. BASIC–a bile acid-sensitive ion channel highly expressed in bile ducts. FASEB J.26, 4122–4130, fj.12-207043 (2012). [DOI] [PubMed]
- 3.Jasti J, Furukawa H, Gonzales EB, Gouaux E. Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature. 2007;449:316–323. doi: 10.1038/nature06163. [DOI] [PubMed] [Google Scholar]
- 4.Bartoi, T., Augustinowski, K., Polleichtner, G., Gründer, S. & Ulbrich, M. H. Acid-sensing ion channel (ASIC) 1a/2a heteromers have a flexible 2:1/1:2 stoichiometry. Proc Natl Acad Sci USA, 1324060111 (2014). [DOI] [PMC free article] [PubMed]
- 5.Waldmann R, et al. Molecular cloning of a non-inactivating proton-gated Na+ channel specific for sensory neurons. J Biol Chem. 1997;272:20975–20978. doi: 10.1074/jbc.272.34.20975. [DOI] [PubMed] [Google Scholar]
- 6.Babini E, Paukert M, Geisler HS, Gründer S. Alternative splicing and interaction with di- and polyvalent cations control the dynamic range of acid-sensing ion channel 1 (ASIC1) J Biol Chem. 2002;277:41597–41603. doi: 10.1074/jbc.M205877200. [DOI] [PubMed] [Google Scholar]
- 7.Bassilana F, et al. The acid-sensitive ionic channel subunit ASIC and the mammalian degenerin MDEG form a heteromultimeric H+-gated Na+ channel with novel properties. J Biol Chem. 1997;272:28819–28822. doi: 10.1074/jbc.272.46.28819. [DOI] [PubMed] [Google Scholar]
- 8.Lingueglia E, et al. A modulatory subunit of acid sensing ion channels in brain and dorsal root ganglion cells. J Biol Chem. 1997;272:29778–29783. doi: 10.1074/jbc.272.47.29778. [DOI] [PubMed] [Google Scholar]
- 9.Gründer S, Geissler HS, Bässler EL, Ruppersberg JP. A new member of acid-sensing ion channels from pituitary gland. Neuroreport. 2000;11:1607–1611. doi: 10.1097/00001756-200006050-00003. [DOI] [PubMed] [Google Scholar]
- 10.Sakai H, Lingueglia E, Champigny G, Mattei MG, Lazdunski M. Cloning and functional expression of a novel degenerin-like Na+ channel gene in mammals. J Physiol (Lond) 1999;519(Pt 2):323–333. doi: 10.1111/j.1469-7793.1999.0323m.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vick JS, Askwith CC. ASICs and neuropeptides. Neuropharmacology. 2015;94:36–41. doi: 10.1016/j.neuropharm.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Askwith CC, et al. Neuropeptide FF and FMRFamide potentiate acid-evoked currents from sensory neurons and proton-gated DEG/ENaC channels. Neuron. 2000;26:133–141. doi: 10.1016/S0896-6273(00)81144-7. [DOI] [PubMed] [Google Scholar]
- 13.Reimers C, et al. Identification of a cono-RFamide from the venom of Conus textile that targets ASIC3 and enhances muscle pain. Proc Natl Acad Sci USA. 2017;114:E3507–E3515. doi: 10.1073/pnas.1616232114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reiners M, et al. The Conorfamide RPRFa Stabilizes the Open Conformation of Acid-Sensing Ion Channel 3 via the Nonproton Ligand-Sensing Domain. Mol Pharmacol. 2018;94:1114–1124. doi: 10.1124/mol.118.112375. [DOI] [PubMed] [Google Scholar]
- 15.Farrag M, et al. Endomorphins potentiate acid-sensing ion channel currents and enhance the lactic acid-mediated increase in arterial blood pressure: effects amplified in hindlimb ischaemia. J Physiol. 2017;595:7167–7183. doi: 10.1113/JP275058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sherwood TW, Askwith CC. Dynorphin opioid peptides enhance acid-sensing ion channel 1a activity and acidosis-induced neuronal death. J Neurosci. 2009;29:14371–14380. doi: 10.1523/JNEUROSCI.2186-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lingueglia E, Champigny G, Lazdunski M, Barbry P. Cloning of the amiloride-sensitive FMRFamide peptide-gated sodium channel. Nature. 1995;378:730–733. doi: 10.1038/378730a0. [DOI] [PubMed] [Google Scholar]
- 18.Golubovic A, et al. A peptide-gated ion channel from the freshwater polyp Hydra. J Biol Chem. 2007;282:35098–35103. doi: 10.1074/jbc.M706849200. [DOI] [PubMed] [Google Scholar]
- 19.Assmann M, Kuhn A, Dürrnagel S, Holstein TW, Gründer S. The comprehensive analysis of DEG/ENaC subunits in Hydra reveals a large variety of peptide-gated channels, potentially involved in neuromuscular transmission. BMC Biology. 2014;12:84. doi: 10.1186/s12915-014-0084-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt A, et al. Dual signaling of Wamide myoinhibitory peptides through a peptide-gated channel and a GPCR in Platynereis. FASEB J. 2018;32:5338–5349. doi: 10.1096/fj.201800274R. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, et al. NeuroPep: a comprehensive resource of neuropeptides. Database (Oxford) 2015;2015:bav038. doi: 10.1093/database/bav038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Paukert M, Kadurin I, Pusch M, Gründer S. Strong modulation by RFamide neuropeptides of the ASIC1b/3 heteromer in competition with extracellular calcium. Neuropharmacology. 2006;50:964–974. doi: 10.1016/j.neuropharm.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 23.Raffa RB, et al. Low affinity of FMRFamide and four FaRPs (FMRFamide-related peptides), including the mammalian-derived FaRPs F-8-Famide (NPFF) and A-18-Famide, for opioid mu, delta, kappa 1, kappa 2a, or kappa 2b receptors. Peptides. 1994;15:401–404. doi: 10.1016/0196-9781(94)90195-3. [DOI] [PubMed] [Google Scholar]
- 24.Chen X, Polleichtner G, Kadurin I, Gründer S. Zebrafish Acid-sensing Ion Channel (ASIC) 4, Characterization of Homo- and Heteromeric Channels, and Identification of Regions Important for Activation by H+ J Biol Chem. 2007;282:30406–30413. doi: 10.1074/jbc.M702229200. [DOI] [PubMed] [Google Scholar]
- 25.Wiemuth, D. & Gründer, S. A single amino acid tunes Ca2+ inhibition of brain liver intestine Na+channel (BLINaC). J Biol Chem285, 30404–30410, M110.153064 (2010). [DOI] [PMC free article] [PubMed]
- 26.Gründer S, Pusch M. Biophysical properties of acid-sensing ion channels (ASICs) Neuropharmacology. 2015;94:9–18. doi: 10.1016/j.neuropharm.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 27.Deval E, et al. Effects of neuropeptide SF and related peptides on acid sensing ion channel 3 and sensory neuron excitability. Neuropharmacology. 2003;44:662–671. doi: 10.1016/S0028-3908(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 28.Baburin I, Beyl S, Hering S. Automated fast perfusion of Xenopus oocytes for drug screening. Pflugers Arch. 2006;453:117–123. doi: 10.1007/s00424-006-0125-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2017).
- 30.Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag New York 2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available from the corresponding author upon request. All relevant data are within the paper.