Abstract
The impermeability barrier provided by the outer membrane of enteric bacteria, a feature lacking in Gram-positive bacteria, plays a major role in maintaining resistance to numerous antimicrobial compounds and antibiotics. Here we demonstrate that mutational inactivation of spr, coding for a muramyl endopeptidase, significantly sensitizes Salmonella enterica serovar Typhimurium to vancomycin without any accompanying apparent growth defect or outer membrane destabilization. A similar phenotype was not achieved by deleting the genes coding for muramyl endopeptidases MepA, PbpG, NlpC, YedA, or YhdO. The spr mutant showed increased autolytic behavior in response to not only vancomycin, but also to penicillin G, an antibiotic for which the mutant displayed a wild-type MIC. A screen for suppressor mutations of the spr mutant phenotype revealed that deletion of tsp (prc), encoding a periplasmic carboxypeptidase involved in processing Spr and PBP3, restored intrinsic resistance to vancomycin and reversed the autolytic phenotype of the spr mutant. Our data suggest that Spr contributes to intrinsic antibiotic resistance in S. Typhimurium without directly affecting the outer membrane permeability barrier. Furthermore, our data suggests that compounds targeting specific cell wall endopeptidases might have the potential to expand the activity spectrum of traditional Gram-positive antibiotics.
Keywords: vancomycin, antibiotic resistance, Spr, MepS, YebA, MepM, Tsp, Prc
Introduction
Peptidoglycan (murein) constitutes a main component of the bacterial cell wall. It is composed of repeated N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) disaccharide units, cross-linked by peptide bridges. The synthesis of this mesh is the target of several classes of antibiotics, such as the β-lactams and glycopeptides. Peptidoglycan functions to maintain bacterial shape, septum formation at the point of cell division, and cell integrity upon internal turgor stress. To facilitate changes in size and shape during growth, bacteria need enzymes that can assemble and disassemble peptidoglycan. The process of re-shaping peptidoglycan involves the concerted activities of periplasmic amidases, endopeptidases, glycosylases and transpeptidases (penicillin-binding proteins, PBPs) (Sauvage et al., 2008). While the PBPs have the important function of catalyzing the formation of interpeptide bridges between overlapping GlcNAc-MurNAc polymers, the murein endopeptidases are tasked with cleaving interpeptide bridges to facilitate the incorporation of new GlcNAC-MurNAc polymers into the growing peptidoglycan mesh. The importance of correctly balancing these opposing activities is illustrated by the fact that blocking PBP activity with β-lactam antibiotics results in autolysis in Escherichia coli (Prestidge and Pardee, 1957).
The outer membrane of Gram-negative enteric bacteria, due to its relative impermeability, provides an intrinsic resistance barrier against many large compounds, including the antibiotics erythromycin (Nikaido and Vaara, 1985; Delcour, 2009), novobiocin (Anderle et al., 2008), rifampicin and vancomycin (Weeks et al., 2010; Krishnamoorthy et al., 2013). Furthermore, the increasing frequency of clinical bacterial isolates producing extended-spectrum β-lactamases is limiting the effectiveness of antibiotics that target cell wall synthesis amongst Gram-negative species (Coque et al., 2008). The search for new antibiotics to treat Gram-negative bacterial infections would be advanced by a better understanding of bacterial cell wall homeostasis at the level of peptidoglycan. Because it is a genetically amenable bacterium, E. coli has been the main focus for studies on the activities of cell wall-modulating enzymes. From these studies a consensus has emerged that, apart from PBP2 and PBP3 (Botta and Park, 1981), each of the glycolytic, endopeptic hydrolases and PBPs are individually dispensable for bacterial viability. Accordingly, inactivation of any one (or sometimes more than one) of the genes encoding these enzymes [PBP4: (Matsuhashi et al., 1977; Denome et al., 1999; Meberg et al., 2004), PBP5: (Matsuhashi et al., 1979; Nishimura et al., 1980; Spratt, 1980; Denome et al., 1999), PBP6: (Broome-Smith and Spratt, 1982; Denome et al., 1999), PBP6b: (Baquero et al., 1996), PBP7/PBP8: (Henderson et al., 1995; Denome et al., 1999), reviewed in: (van Heijenoort, 2011)] does not prevent bacterial growth under laboratory conditions. While this might imply a high degree of functional redundancy, it does not exclude the possibility that some or all of these enzymes may have unique functions under other more specific conditions.
A recent study (Singh et al., 2012) confirmed muramyl endopeptidase activity for three additional E. coli proteins; Spr, YebA and YdhO, renamed in E. coli to MepS, MepM, and MepH (Singh et al., 2015). More specifically, the study presented data implying that Spr or YebA might represent endopeptidases with less redundant functions, since it was feasible to construct an ΔsprΔyebA mutant only in an E. coli strain genetically complemented with spr (mepS) (Singh et al., 2012).
Salmonella enterica serovar Typhimurium (S. Typhimurium) is a Gram-negative enterobacterium with an increasing antibiotic resistance development in its genus (Angelo et al., 2016; Hong et al., 2016; Klemm et al., 2018). As the S. Typhimurium genome includes genes with high sequence similarity to the mepS, mepM, and mepH genes of E. coli, and given the potential importance of mepS and mepM for viability of E. coli (Singh et al., 2012), we studied the phenotypes of S. Typhimurium mutants in which these genes were deleted, either singly or in combination. We characterized Δspr, ΔyebA, and ΔydhO mutants in terms of their growth and susceptibility profiles against antimicrobials, and in addition Δspr mutant for autolytic behavior. Our findings highlight Spr as a possible new target for antibacterial treatment in order to sensitize Salmonella against Gram-positive-specific antibiotics.
Materials and Methods
Bacterial Strains
Mutants were constructed in the Salmonella enterica serovar Typhimurium SR-11 background (Sukupolvi et al., 1997), and are listed in Table 1. The S. Typhimurium strains LB5010 (Bullas and Ryu, 1983), ATCC 14028 and E. coli Top10, TG1 were used as intermediary hosts during mutant constructions or cloning. Furthermore, S. Typhimurium strains ATCC 14028s and SL1344 were also used to host a Δspr mutation. The pACYC184-derived plasmid coding for β-galactosidase was available from a previous work (Taira et al., 1991).
Table 1.
Strains used in study.
| KV82 | Salmonella enterica serovar Typhimurium strain SR-11 wild-type |
| KV141 | Salmonella enterica serovar Typhimurium strain 14028 wild-type |
| KV110 | Salmonella enterica serovar Typhimurium strain LB5010 wild-type |
| KV199 | Salmonella enterica serovar Typhimurium strain SL1344 wild-type |
| KV154 | LB5010:pSIM6 |
| KV145 | 14028:pSIM5-tet |
| KV224 | SR-11:pSIM6 |
| KV244 | SR-11 Δspr |
| KV268 | SR-11 ΔyebA |
| KV407 | SR-11 ΔydhO |
| KV267 | SR-11 ΔsprΔyebA |
| KV408 | SR-11 ΔsprΔydhO |
| KV409 | SR-11 ΔyebAΔydhO |
| KV240 | SR-11 Δspr::cat |
| KV255 | SR-11 ΔyebA::cat |
| KV403 | SR-11 ΔydhO::neo |
| KV259 | SR-11 ΔsprΔyebA::cat |
| KV404 | SR-11 ΔsprΔydhO::neo |
| KV405 | SR-11 ΔyebAΔydhO::neo |
| KV235 | LB5010 Δspr::cat |
| KV249 | 14028 ΔyebA::cat |
| KV397 | LB5010 ΔydhO::neo |
| KV386 | SR-11 Δtsp |
| KV387 | SR-11 ΔsprΔtsp |
| KV388 | SR-11 ΔsprΔyebAΔtsp |
| KV374 | SR-11 Δtsp::cat |
| KV378 | SR-11 ΔsprΔtsp::cat |
| KV382 | SR-11 ΔsprΔyebAΔtsp::cat |
| KV370 | LB5010 Δtsp::cat |
| KV83 | SR-11:pBAD30 |
| KV247 | SR-11 Δspr:pBAD30 |
| KV295 | SR-11 Δspr:pBAD30::spr |
| KV308 | SR-11 Δspr:pBAD30::sprC70S |
| KV322 | SR-11 ΔsprΔyebA:pBAD30 |
| KV305 | SR-11 ΔsprΔyebA:pBAD30::spr |
| KV289 | LB5010:pBAD30::spr |
| KV302 | LB5010:pBAD30::sprC70S |
| KV278 | Escherichia coli TG1:pBAD30spr |
| KV299 | Escherichia coli Top10:pBAD30sprC70S |
| KV418 | SR-11 ΔsprΔtsp:pBAD30 |
| KV416 | SR-11 ΔsprΔtsp:pBAD30::tsp |
| KV419 | SR-11 ΔsprΔyebAΔtsp:pBAD30 |
| KV417 | SR-11 ΔsprΔyebAΔtsp:pBAD30::tsp |
| KV413 | LB5010:pBAD30::tsp |
| KV411 | Escherichia coli Top10:pBAD30::tsp |
| KV424 | SR-11 ΔrfaC::cat |
| KV427 | SR-11 ΔrfaG::neo |
| KV428 | SR-11 ΔrfaP::cat |
| KV433 | SR-11 ΔmepA::neo |
| KV434 | SR-11 ΔpbpG::neo |
| KV435 | SR-11 ΔnlpC::neo |
| KV430 | LB5010 ΔmepA::neo |
| KV431 | LB5010 ΔpbpG::neo |
| KV432 | LB5010 ΔnlpC::neo |
| KV392 | SR-11 ΔproQ |
| KV393 | SR-11 ΔsprΔproQ |
| KV394 | SR-11 ΔsprΔyebAΔproQ |
| KV395 | SR-11 ΔhtpX |
| KV396 | SR-11 ΔsprΔhtpX |
| KV376 | SR-11 ΔproQ::cat |
| KV380 | SR-11 ΔsprΔproQ::cat |
| KV384 | SR-11 ΔsprΔyebAΔproQ::cat |
| KV377 | SR-11 ΔhtpX::cat |
| KV381 | SR-11 ΔsprΔhtpX::cat |
| KV385 | SR-11 ΔsprΔyebAΔhtpX::cat |
| KV372 | LB5010 ΔproQ::cat |
| KV373 | LB5010 ΔhtpX::cat |
| KV436 | SR-11 ΔnlpI::neo |
| KV437 | SR-11 ΔsprΔnlpI::neo |
| KV438 | SR-11 ΔsprΔyebAΔnlpI::neo |
| KV441 | SR-11:pKTH3088 |
| KV442 | SR-11 Δspr:pKTH3088 |
| KV445 | SR-11 ΔsprΔyebA:pKTH3088 |
| KV446 | SR-11 ΔsprΔtsp:pKTH3088 |
| KV448 | SR-11 ΔsprΔyebAΔtsp:pKTH3088 |
| KV449 | 14028 Δspr::cat |
| KV450 | SL1344 Δspr::cat |
| FIA1500 | SR-11 ΔsprΔyebA + Δ nt 1943335–1949114 (suppressor mutant) |
| FIA1501 | SR-11 ΔsprΔyebA + Δ nt 1943335–1949114 (suppressor mutant) |
| FIA1502 | SR-11 ΔsprΔyebA + Δ nt 1939839–1948327 (suppressor mutant) |
| FIA1503 | SR-11 ΔsprΔyebA + Δ nt 1923856–1964581 (suppressor mutant) |
| FIA1504 | SR-11 ΔsprΔyebA + Δ nt 1940175–1942805 (suppressor mutant) |
| FIA1505 | SR-11 ΔsprΔyebA + Δ nt 1939318–1964299 (suppressor mutant) |
| FIA1506 | SR-11 ΔsprΔyebA + Δ nt 1923856–1964581 (suppressor mutant) |
| FIA1507 | SR-11 ΔsprΔyebA + Δ nt 1923856–1964581 (suppressor mutant) |
| FIA1508 | SR-11 ΔsprΔyebA + Δ nt 1926281–1948787 (suppressor mutant) |
| FIA1509 | SR-11 ΔsprΔyebA + Δ nt 1923856–1964581 (suppressor mutant) |
| FIA1510 | SR-11 ΔsprΔyebA + tsp T479R (suppressor mutant) |
| FIA1511 | SR-11 ΔsprΔyebA + Δ nt 1923856–1964581 (suppressor mutant) |
| FIA1512 | SR-11 ΔsprΔyebA + Δ nt 1943335–1949114 (suppressor mutant) |
| FIA1513 | SR-11 ΔsprΔyebA + Δ nt 1943335–1949114 (suppressor mutant) |
Media and Growth Conditions
Growth media included tryptone and yeast extract (Sigma-Aldrich, 10 g/l respective 5 g/l) with 10 g/l of NaCl (LB medium), or without NaCl (TY medium). Cultures were incubated at 37°C unless otherwise stated. When needed, antibiotics were added to the growth media at the following concentrations: ampicillin 100 μg/ml; chloramphenicol 25 μg/ml; kanamycin 50 μg/ml; tetracycline 10 μg/ml. All antibiotics were purchased from Sigma-Aldrich (Sweden).
For determining growth curves, bacteria were incubated overnight in 2 ml LB at 220 rpm and 37°C. The next day, 150 μl of the culture was mixed with 850 μl of PBS and the OD600 nm measured. The bacteria were then adjusted to an OD600 nm of 0.25 (Ultrospec 1000, Pharmacia Biotech). Bacteria were further diluted 1:25 in either LB or TY broth resulting in a final OD600 nm of 0.01. 400 μl of this bacterial suspension was then loaded into wells in a Honeycomb Bioscreen plate (OY Growth Curves AB Ltd., Helsinki, Finland) in three technical replicates. Uninoculated media was used as a negative growth control. The Bioscreen C plate reader (OY Growth Curves AB Ltd.) was set to an OD of 600 nm and optical density measurement was taken every 15 min with 5 s of agitation before every measurement, up until 24 h.
PCR and Oligonucleotides
Polymerase chain reaction (PCR) was performed using an Eppendorf Mastercycler Personal. Oligonucleotides were designed using the genome of S. Typhimurium LT2 as reference (McClelland et al., 2001). For the generation of the inserts for gene deletions, the PCR was performed using Phusion High-Fidelity PCR master mix with HF buffer (New England Biolabs, United States). The cycling conditions were as following: 98°C for 1 min and 30 cycles of 98°C for 15 s, 54.5°C for 10 s, 72°C for 40 s, and 72°C for 2 min. The oligonucleotides were ordered from Sigma-Aldrich and specified in Table 2.
Table 2.
Oligonucleotides for construction of strains and diagnostic PCR.
| FSprrec | CGATATTTATCGTTAAGGACTTCAAGGGAAAACAAACAAC GTGTAGGCTGGAGCTGCTTC |
| Rsprrec | TCTCATCAGGTAAGCCAAGGGAGGTGCTGCCTGATGAAGA CATATGAATATCCTCCTTAG |
| Fspr(c) | GAA TTG TCT CAA GCT GTG CAG G |
| Rspr(c) | ATT CGG CAA AAC GGG TTC AG |
| FYebArec | TGCGAGCTGCCTGAAAGGAGATTAACGAGGAAGTGAATAC GTGTAGGCTGGAGCTGCTTC |
| RYebArec | AGC CGG CAC ACA TCG CGT ACC GGC TCT GTC AGC GCA TTT G CATATGAATATCCTCCTTAG |
| FYebA(c) | TTA GCC AAC CAG TAT GCG AGC |
| RYebA(c) | GTA GCG ACG TCT GCG TCT C |
| FYdhOrec | GTAGATTAGAATTATCAGGTTTTGTAAATCATACGCAGGC GTGTAGGCTGGAGCTGCTTC |
| RYdhOrec | AAG AAG AAG TTA TCC TGT CGT TAA ACG ACA GGA TAA AAT A CATATGAATATCCTCCTTAG |
| FYdhO(c)2 | CTG AAG CCT GTC ATT GTA ACG G |
| RYdhO(c)2 | CGA TCT CTT CCA GCG ATT TGC |
| FTsprec | ATGTCTTTGATTGTACGCGCAGAACACCTGGTGTTCTGAA GTGTAGGCTGGAGCTGCTTC |
| RTsprec | TTA AAA AAA AAC AGG CAC AAT TTT TTG TGC CTG TTT AGC GCATATGAATATCCTCCTTAG |
| FTsp(c) | TCA CCA AAG ATG GTG TCC GT |
| RTsp(c) | TAT CCT GAC GAC TTC TGC GC |
| FRfaCrec | GCAGCGGGTTCTGGAAGAGCTTCATTCGCTGTTGTCGGAA GTGTAGGCTGGAGCTGCTTC |
| RRfaCrec | TCT TTT CTC CAC AAT AGG TTT GGG ATG AGA CAG AGT CTC T CATATGAATATCCTCCTTAG |
| FRfaC(c) | AAG TGC GTA AAG GTG ATA CGG |
| RRfaC(c) | CGC TTT ATT CCA GAT CGG CTT |
| FRfaPrec | GATTTATACAGCTTACCGGAGAAGGCCGCGGATATTATTA GTGTAGGCTGGAGCTGCTTC |
| RRfaPrec | CTC ACT CAT AAA TTA CTC ACT GAG TGC ATA ATT ATT ATA A CATATGAATATCCTCCTTAG |
| FRfaP(c) | ACA CAG CCT TCC TTA CGC AA |
| RRfaP(c) | GCC AGC AGG TGT GGC AAT ATA |
| FRfaGrec | GACGGAAAAAATGCTGCCGCATGAGGCACGCACCATAGAT GTGTAGGCTGGAGCTGCTTC |
| RRfaGrec | ATC TTT ACC GCG CCA TAG TGT GGT TAA CGG CGC TTT CAG CCATATGAATATCCTCCTTAG |
| FRfaG(c) | TAC CTT TCC GTT ATT CCG GCT G |
| RRfaG(c) | GTC TCC AGC TCT CTG AAC AC |
| FMepArec | ATCGGGCACAGAATGCGGATGTAAAGACAGAGATTCCACG GTGTAGGCTGGAGCTGCTTC |
| RMepArec | AGC AGC GGG GAG ACC ATA AAC AGA TCA TAA AAA TTG TCC A CATATGAATATCCTCCTTAG |
| FMepA(c) | AGT GCC GAT CGC AGA AG |
| RMepA(c) | AAA TCC TGC CAG TAC GGC |
| FNlpCrec | CAGGTAATTTCGACGCTAAATTAATACCAAAATAAAAACA GTGTAGGCTGGAGCTGCTTC |
| RNlpCrec | ATG TTA AAA ATA GAC TAT AAA ATT TAT ATC GTC TGC GAG GCATATGAATATCCTCCTTAG |
| FNlpC(c) | CGT CGA GGG GCA TCC AAT |
| RNlpC(c) | AGT TCA ACC GGC GAT ATG TT |
| FPbpGrec | AGCTCAGGCGGTGTGCGTTACGACGCGCGTGAATCATTAT GTGTAGGCTGGAGCTGCTTC |
| RPbpGrec | TGA AGC CCG GCG GCG CGA TGC CTG CCG GGC CTG CGG CGA C CATATGAATATCCTCCTTAG |
| FPbpG(c) | TTC TGT AGC GGC AAC GCT |
| RPbpG(c) | GCG GAA ATT CTG GCA GGA A |
| FsprSacI2 | CATGAGCTCAGGAGGA CAA CAT GGT CAA ATC TCA ACC G |
| RsprHindIII | CATGAAGCTT TTA ACT GCG GCT CAG AAC TC |
| FSprC68S (Primer for making C70S) | GCAGCACTAAGAAAGGCGTCGACTCTTCC AGC TTT GTA CAG CGC ACC TTC |
| RSprC68S (Primer for making C70S) | GAA GGT GCG CTG TAC AAA GCT GGA AGA GTC GAC GCC TTT CTT AGT GCT GC |
| pBAD30Forward | TTA GCG GAT CCT ACC TGA CG |
| FTspEcoRI | CATGAATTCAGGAGGA TGT TCT GAA ACG GAG GCC A |
| RTspHindIII | CATGAAGCTT TTA CTT ATT GGC TGC CGC CT |
| FProQrec | CTGTTCATGCCTGCTGCTTGTTGGCTACGTCCGTTGTAAT GTGTAGGCTGGAGCTGCTTC |
| RProQrec | AAG CCT AAA AAA AGT GTT CAT GCC AGG CCT GGC CTC CGT TCATATGAATATCCTCCTTAG |
| FProQ(c) | GTC GCA GGA TAA TCA ACG GA |
| RProQ(c) | CGT AAT ATC TTC CAC GGC GAA G |
| FHtpXrec | CATACGATGTGGGTAATCGCATAGTGCGCTTTGTTAAATT GTGTAGGCTGGAGCTGCTTC |
| RHtpXrec | GCG TCA TTC GAC GCG CTT TTC ATA CTT GCC AGT GGG CTT ACATATGAATATCCTCCTTAG |
| FHtpX(c) | TTT CTC GTG ACT TAC CGC CT |
| RHtpX(c) | CGG TAG TGA GCG GTT TAC GTA |
| ΔnlpI-mutant | Rouf SF, 2011, J. Bac |
For routine confirmatory PCR, Phusion High-Fidelity PCR master mix with HF buffer was used. The cycling conditions were as follows: 98°C for 1 min and 30 cycles of 98°C for 15 s, annealing temperature for 10 s, 72°C for elongation, and 72°C for 2 min. Annealing temperature varied depending on the primer pairs used, and elongation time was based on the length of the expected product (30 s per kilobase). Oligonucleotide sequences are shown in Table 2.
Bacteriophage Transduction
Transducing phages (phage P22int; Schmieger, 1972) were prepared on strain LB5010 (in LB broth supplemented with D-galactose (Fluka BioChemika) to 0.2% (wt/vol)) or strain 14028s carrying the mutation of interest. The next day chloroform was added to the culture and the culture was vortexed. The culture was then centrifuged for 10 min at 18,500 g to create phase separation. The top phase was recovered and used to transfer the genetic marker. The transduction into S. Typhimurium SR-11 was done by incubating 20 μl of the P22int phage containing the genetic marker with 1ml of exponential phase culture. These were incubated at 37°C with shaking at 220 rpm for 1 h and after washing in PBS plated onto appropriate selective LB agar plates.
Construction and Isolation of Mutants
The antibiotic marker amplified from either pKD3 for chloramphenicol resistance (cat gene) or pKD4 for kanamycin resistance (neo gene) was introduced using primers with 3′-ends overlapping the borders of the gene to be deleted, and subsequently inserted into the chromosome; to replace the gene of interest, by double-stranded DNA lambda-red recombination (Datsenko and Wanner, 2000; Yu et al., 2000). As recipients we used S. Typhimurium strain LB5010 containing the pSIM6, or S. Typhimurium strain 14028 containing pSIM5-tet plasmid, each grown to an OD600 nm of approximately 0.3 at 32°C with shaking at 220 rpm in a water bath. To induce the lambda-red genes, the bacteria were transferred to a 42°C water bath shaking at 220 rpm for 15 min. After cooling on ice for 10 min, bacterial cells were made electrocompetent by washing with ice-cold deionized water four times.
Electroporation of the PCR products generated from pKD3 or pKD4 was done using a Gene Pulser (Bio-Rad, United States) by mixing 25 μl of electrocompetent cells and 0.5 μl of purified PCR product, with settings 1.8 kV, 25 F and 200 Ω. Cells were recovered in 1 ml of LB at 32°C and 220 rpm for 2 h. After recovery the culture was spun down and the pellet was spread on LB agar plates containing either chloramphenicol or kanamycin to select for recombinants. The genetic marker was subsequently transferred to wild-type S. Typhimurium SR-11 by phage P22int transduction from either the LB5010 or 14028 mutant strains.
Antibiotic markers were removed from the mutants using the plasmid pCP20. Briefly, pCP20 was transferred by P22int transduction to the recipient strain containing the antibiotic marker, with selection for colonies on LB agar plates containing ampicillin. Transductants were subcultured three times at 28°C on selective LB agar plates. The bacteria were then subcultured three more times at 37°C to select for loss of plasmid and loss of antibiotic marker. The loss of the antibiotic marker was confirmed by a diagnostic PCR.
Isolation of vancomycin-tolerant ΔsprΔyebA mutants was conducted by spreading a LB broth culture of the ΔsprΔyebA mutant on a vancomycin gradient TY agar plate. A plate of 14 cm in diameter was poured in a tilted position with 37.5 ml of TY agar containing vancomycin at 40 μg/ml. After solidification, 37.5 ml TY agar lacking antibiotic was poured on top of the solidified TY agar containing vancomycin, now in a horizontal plane. The plate was seeded with about 107 CFU of ΔsprΔyebA mutant bacteria in their logarithmic growth phase. After 16 h of incubation yielded colonies were isolated at the higher concentration end of the gradient.
Plasmid Constructions and Site-Directed Mutagenesis
For creating plasmids for genetic complementation, spr and tsp were PCR amplified using S. Typhimurium SR-11 genomic DNA as template, using oligonucleotide primers to create suitable restriction sites at each end of the amplified fragment. Enzymes used for restriction digestion were SacI or EcoRI, and HindIII (New England Biolabs) while T4 DNA ligase was used to ligate spr fragments into vector pBAD30 (Guzman et al., 1995). Following ligation, plasmids were transformed into chemically competent E. coli TG1 or Top10 cells (Invitrogen), from which the constructs were purified and electroporated into S. Typhimurium LB5010. The plasmids were then transferred into S. Typhimurium SR-11 by P22int transduction.
Whole-Genome Sequencing and Analysis
Genomic DNA was prepared from bacterial cultures using a Masterpure DNA Purification Kit (Epicenter). Libraries for sequencing were prepared using Nextera XT sample preparation and index kits (Illumina). The quality of libraries was assessed using a Tapestation 2200 (Agilent) with high-sensitivity D5000 Screentape. Sequencing of the libraries was done using a Miseq device (Illumina) using a 600-cycle V3 reagent kit. The sequences were processed and analyzed using CLC Genomics Workbench 9.0.1 (CLC bio).
Deoxycholate (DOC) Sensitivity Test
To assay detergent sensitivity, bacteria were diluted 1:100 in LB from an overnight culture, and grown in LB for 2 h at 37°C with shaking at 220 rpm. The OD600 nm of the culture was measured, and the formula [(0.484/OD600 nm) × 2.1] was used to adjust the amounts of bacteria to approximately 107 CFU/ml. The bacterial suspension was then diluted 1:100 in distilled water with sodium deoxycholate (DOC; Sigma-Aldrich, Sweden) freshly added to a final concentration of 0.5% (wt/vol). After an incubation of 30 min at room temperature, viable counts were determined from the DOC-suspension by plating dilutions on LB agar plates.
SDS-PAGE Gel Electrophoresis
Polyacrylamide-bis-acrylamide gel electrophoresis was conducted according to Laemmli (1970), using custom-made 12.5% separation (pH 8.8) and 6% stacking gel (pH 6.8). (Thermo Scientific, Sweden). For solubilization, samples were suspended in reducing protein sample buffer (0.125 M Tris-HCl pH 6.5, 3.6% SDS, 10% β-mercaptoethanol, 2% glycerol, and bromophenol blue) and heated 10 min at 97°C before application on the gel. After completed electrophoresis, gels were stained using Imperial stain.
Bacterial Membrane Fractionation
The outer membrane fraction was isolated according to Sabet and Schnaitman (1973). Bacteria from overnight LB cultures were diluted 1:100 in TY broth and grown at 37°C for 2 h to mid exponential phase. The OD600nm of the bacterial cultures were measured and then normalized. Bacteria were pelleted by centrifugation at 6,000 g for 10 min, re-suspended in PBS, cooled on ice, and disrupted by equal numbers of 10 s sonication pulses until the suspensions visibly cleared. Bacteria were removed by low–speed centrifugation (1,500 g), after which the membrane fraction was pelleted by high-speed centrifugation (18,500 g, 10 min). The waxy brownish pellet was then re-suspended in 50 mM Tris-HCl buffer containing 10 mM MgCl2 and 0.5% Triton X-100. The membrane fraction was again pelleted by centrifugation (18,500 g, 10 min), and the re-suspension and high-speed centrifugation steps were repeated. The final colorless pellet was then suspended in reducing protein sample buffer and run on a 12.5% SDS-PAGE gel as specified above.
Disk Diffusion Sensitivity Testing
Bacteria were grown overnight in LB broth at 37°C and 220 rpm. The overnight culture was diluted 1:100 in LB broth and grown for 2 h at 37°C and 220 rpm. The OD600 nm was measured of the 2 h culture and the formula [(0.484/OD600 nm) × 2.1] was used to estimate the amount of bacteria. An estimated 3 × 105 CFU/ml bacteria were then spread on large (13.7 cm diameter) TY agar plates and the antibiotic disks were placed on top of the bacteria, and the plates were incubated overnight at 37°C. The next day the diameters of the inhibitory zones were measured.
The disks, 6 mm in diameter, were made out of Whatman 3 paper, using an ordinary office paper puncher. Each disk was infused with 5 μl of an antibiotic, and then let to dry. Concentrations of the antibiotics used were; tetracycline 10 μg/ml, vancomycin 20 μg/ml, rifampicin 10 μg/ml, polymyxin B 10 μg/ml, novobiocin 10 μg/ml, and penicillin G 10 μg/ml (Sigma-Aldrich, Sweden). Every antibiotic was dissolved in water except rifampicin, which was dissolved in DMSO.
MIC Determination
A total amount of 2 × 104 CFU/ml bacteria was prepared and normalized as described above, but using TY broth. This dilution was subsequently pipetted into a 96-well plate, containing TY broth with either vancomycin or penicillin G, resulting in a final concentration of 104 CFU/ml bacteria. The highest concentration for the vancomycin MIC testing was 100 and 50 μg/ml for penicillin G. Then, the antibiotic was diluted down in steps of 1:2 from the highest concentration before an incubation over night at 37°C.
Drop-on-Lawn
Bacteria were grown overnight in LB broth at 37°C and 220 rpm. The overnight culture was diluted 1:100 in LB broth and grown for 2 h at 37°C and 220 rpm. The OD600 nm was measured of the 2 h culture and the formula [(0.484/OD600 nm) × 2.1] was used to calculate the amount of bacteria needed for 106 CFU/ml. From 106 CFU/ml a 1:2 serial dilution series was made in 1ml PBS. From each dilution a 5 μl droplet was pipetted onto TY agar plates containing none, 20 μg/ml, or 40 μg/ml vancomycin, and the TY agar plates were incubated overnight at 37°C. For genetic complementation tests TY agar plates were supplemented with 0.02% (weight/vol) L-arabinose (Sigma-Aldrich, Sweden).
β-Galactosidase (LacZ) Release Assay
To assay for the release of β-galactosidase (LacZ), pKTH3088-containing strains (KV441–KV448, Table 1) were grown overnight in LB broth at 37°C and 220 rpm. The overnight culture was diluted 1:100 in TY broth and grown for 2 h at 37°C and 220 rpm to mid exponential phase. Following incubation, the OD600 nm of the culture was measured, after which 300 μl of bacteria was added to 700 μl TY broth containing different concentrations of penicillin G or vancomycin and incubated at 37°C for 60 min. When attempting to inhibit penicillin G and vancomycin induced autolysis, 20 μg/ml tetracycline was added during this step. The bacteria were then pelleted by centrifugation and 200 μl of the supernatant was transferred into a tube containing 600 μl reaction buffer [0.001 M MgSO4 and 0.05 M β-mercaptoethanol (Sigma-Aldrich, Sweden) in 0.01 M PBS, pH 7.2], and 200 μl of 4 mg/ml ONPG (Sigma-Aldrich, Sweden) dissolved in reaction buffer. The β-galactosidase activity was stopped at given time points by adding 500 μl 0.5 M sodium carbonate. 1ml of the samples were transferred into cuvettes and the β-galactosidase activity was measured using the absorbance at 420 nm as readout. The formula to calculate the arbitrary units (AU) was implemented according to Miller (Miller, 1972).
When measuring proportional or total β-galactosidase activities in bacteria, the samples were similarly incubated overnight and diluted 1:100 the next day. After incubation for 2 h at 37°C, the OD600 nm of the culture was measured and 300 μl culture was added to 700 μl of TY broth containing 40 μg/ml penicillin G and incubated for a further 60 min at 37°C. The samples were pelleted by centrifugation and 200 μl of the supernatant was added to 600 μl of Z-buffer and 200 μl of 4 mg/ml ONPG in reaction buffer. In order to assay the amount of LacZ in the pellet, the remainder of the supernatant was discarded and the pellet resuspended in 100 μl TY broth. This suspension was added to 600 μl of reaction buffer and 200 μl of 4 mg/ml ONPG in reaction buffer supplemented with 50 μl of 0.1% SDS (Sigma-Aldrich, Sweden) and 50 μl of >99% chloroform (Sigma-Aldrich, Sweden) to allow the ONPG to penetrate into the pelleted cells. The samples were then incubated at 30°C for 20 min, sodium carbonate was added, the absorbance at 420 nm measured, and the calculations for the arbitrary units performed as above.
OD600 nm Determination Following Antibiotic Treatment
As a complement to the β-galactosidase release assay we followed alterations in OD600 nm for the same strains. The strains were grown overnight in LB broth at 37°C and 220 rpm. The overnight culture was diluted 1:100 in TY broth and grown for 2 h at 37°C and 220 rpm to mid-exponential phase. Following this incubation the OD600 nm of the culture was measured to obtain a “pre-antibiotic value.” Simultaneously, 300 μl of bacteria was added to 700 μl TY broth containing different concentrations of vancomycin or penicillin G and incubated at 37°C for 60 min. Following this second incubation the OD600 nm was measured as the “post-antibiotic value.” To quantify the effect of each antibiotic on the OD600 nm, the post-antibiotic values were divided with the pre-antibiotic values.
Viable Bacterial Counts From Broth
Strains were grown overnight in LB broth at 37°C and 220 rpm. The overnight culture was diluted 1:100 in TY broth and grown for 2 h at 37°C and 220 rpm to mid-exponential phase. Following this incubation the viable bacterial count for the input was enumerated by taking 300 μl of bacteria into 700 μl TY broth and a 1:10 serial dilution was performed in PBS. Bacteria were then spread on LB agar plates and incubated overnight at 37°C and the cfu were counted the next day, yielding the input value. In parallel 300 μl of bacteria was added to 700 μl TY broth containing either penicillin G or vancomycin and incubated at 37°C for 60 min. Strains were then serially diluted 1:10 in PBS and the bacteria were spread on LB agar plates, incubated overnight at 37°C, and cfu counted the next day, yielding the output value.
Statistical Analysis
GraphPad Prism v6.0g (GraphPad Software, Inc., United States) was used for statistical analysis.
Results
Lack of Muramyl Endopeptidases Spr, YebA, and YdhO Does Not Result in Growth Defects in S. Typhimurium
To assess any functional similarity of the S. Typhimurium homologs to the E. coli MepS, MepM and MepH proteins regarding growth phenotypes, we constructed single and all combinations of double deletion mutants of spr, yebA, and ydhO in S. Typhimurium SR-11, using allelic replacement (for details, see section “Materials and Methods”). All deletions were verified by PCR. In agreement with observations from E. coli (Singh et al., 2012) we were not successful in creating a ΔsprΔyebAΔydhO triple mutant. In agreement with observations made in E. coli (Singh et al., 2012) none of the single deletion mutants revealed any significant difference in the overall shape of their growth curves (Figures 1A,B). All single mutants had a similar logarithmic growth rate, and reached a similar optical density in stationary phase in both LB and TY medium. Even the ΔyebAΔydhO mutant grew like the wild-type parental strain. On the other hand, the ΔsprΔyebA mutant showed a somewhat decreased rate of replication in TY medium at later stages of the growth slope (Figure 1B). Taken together, these findings suggest a high degree of redundancy for the Spr, YebA, and YhdO endopeptidases in S. Typhimurium regarding growth in broth cultures.
FIGURE 1.
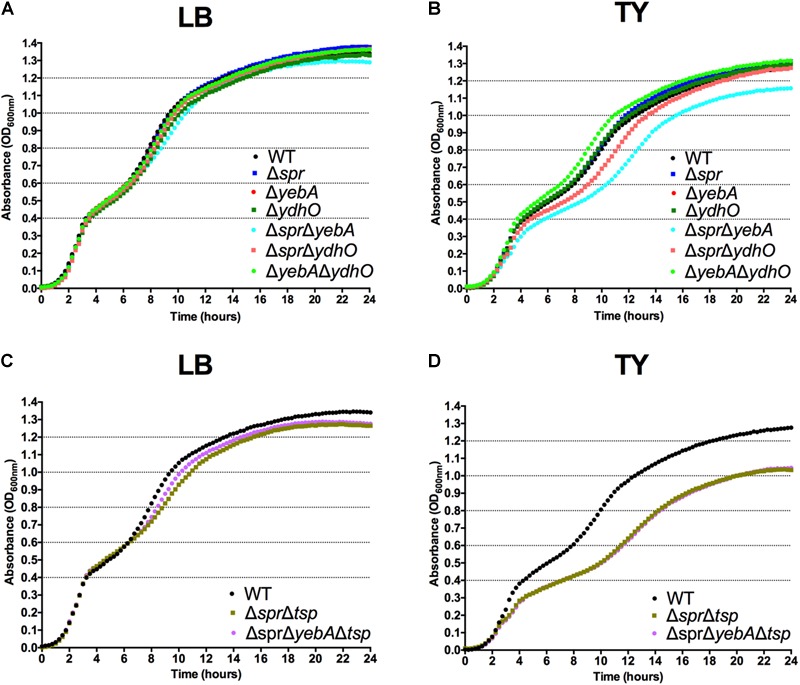
Growth characteristics of S. Typhimurium strains. Bacterial strains were diluted to an OD600 nm of 0.01 from overnight LB broth cultures into LB (A,C) or TY (B,D) broth where after the increase in bacterial density was followed for 24 h.
Deletion of Spr Results in Vancomycin Sensitivity Without an Outer Membrane Phenotype
Peptidoglycan turnover and outer membrane synthesis are connected in E. coli (Gray et al., 2015), and E. coli mutants simultaneously lacking several murein hydrolases show evidence of an outer membrane permeability barrier defect (Heidrich et al., 2002). Therefore, we assessed whether any of the murein endopeptidases Spr, YebA, or YhdO were necessary for maintaining the outer membrane permeability barrier in S. Typhimurium. To achieve this, we screened the panel of our S. Typhimurium endopeptidase mutants for possible sensitization to six different antibiotics using the disk diffusion method, as well as for detergent tolerance. The antibiotics tested were penicillin G, polymyxin B, tetracycline, rifampicin, novobiocin, and vancomycin. Wild-type S. Typhimurium is intrinsically resistant to the latter three due to the outer membrane permeability barrier (Sukupolvi et al., 1984). As comparator strains we used wild-type S. Typhimurium SR-11, and three isogenic LPS mutants expected to have an outer membrane permeability defect (Sukupolvi et al., 1984). High salt concentrations has been reported to reduce the sensitivity of E. coli to selected antibiotics (Beggs and Andrews, 1976), while low osmolarity would favor the expression of the more permeable outer membrane porin OmpF (Harder et al., 1981; Jaffe et al., 1982) and increase the probability of detecting any sensitization. Thus, sensitivity testing was conducted using low osmolarity TY medium.
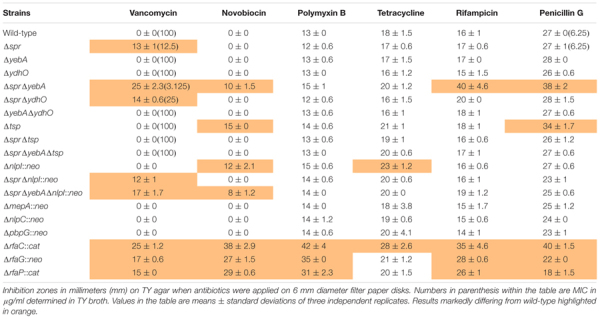
As compared to the wild-type, the three LPS mutants, ΔrfaC (waaC), ΔrfaG (waaG), and ΔrfaP (waaP) were each sensitized to polymyxin B, novobiocin, rifampicin, and vancomycin, but not to tetracycline (Table 3). The Δspr mutant was strongly sensitized to vancomycin (inhibition zone increased from 0 to 13 mm), but not to the other antibiotics tested. Subsequent MIC determinations demonstrated that the intrinsic vancomycin resistance was reduced 8-fold for the Δspr mutant and 32-fold for the ΔsprΔyebA mutant (Table 3). Also, the ΔsprΔyebA mutant revealed a moderate sensitization to novobiocin, penicillin G and rifampicin, while the ΔyebA mutant did not show any increase in sensitization to these antibiotics compared to the wild-type (Table 3).
Table 3.
Antibiotic sensitivity profiles.
 |
Any sensitization to the detergent deoxycholate (DOC) of the Δspr and ΔsprΔyebA mutants was evaluated by incubation in 0.5% DOC for 30 min. In this assay only the ΔsprΔyebA mutant showed clear evidence of sensitization (Figure 2A).
FIGURE 2.
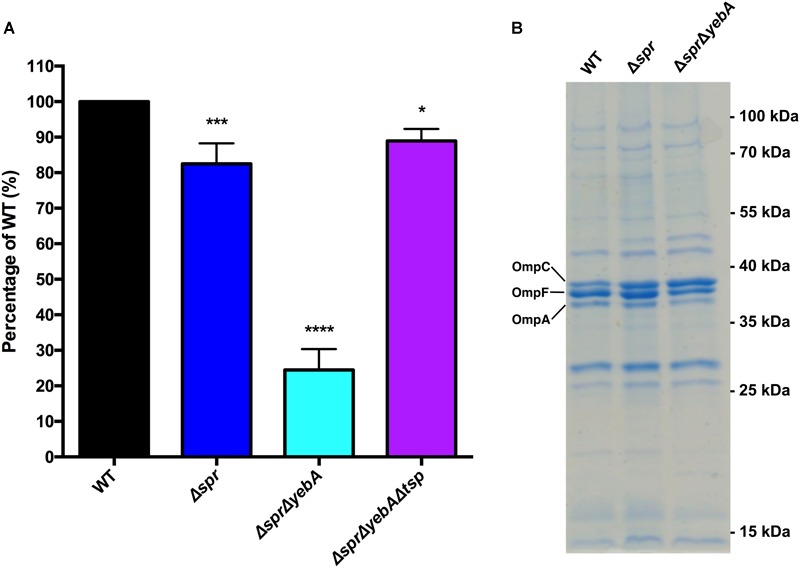
Detergent sensitivity and membrane protein profile of muramyl endopeptidase mutants. (A) For probing detergent sensitivity 105 bacteria of each strains was incubated in 0.5% DOC for 30 min, where after the bacterial viable colonies were enumerated. Results are presented as relative CFU yield in relation to the wild-type. The graph shows the mean value and the standard error of the mean of three independent replicates. Statistical test was an one-way ANOVA with Dunnett’s correction for multiple comparisons when comparing the mutants to the wild-type. ∗ = p < 0.05, ∗∗∗ = p < 0.001, and ∗∗∗∗ = p < 0.0001. (B) SDS-PAGE gel revealing the outer membrane protein profiles of the wild-type and mutant strains. The positions of pre-stained molecular weight markers are indicated at the right. The labeling of OmpC, OmpF, and OmpA is based on comparison with published data (Sukupolvi et al., 1984; Liu et al., 2016; Choi et al., 2018).
Spr Is the Only Muramyl Endopeptidase to Selectively Maintain Vancomycin Resistance
As the antibiotic sensitization profile caused by the Δspr mutation was unexpected, we next tested whether this mutant phenotype was restricted to the SR-11 line of S. Typhimurium. Hence, the Δspr mutation was introduced into the commonly used laboratory S. Typhimurium lines SL1344 and 14028. When tested for vancomycin tolerance, the MIC for the wild-type SL1344 and 14028 strains was the same as for the wild-type SR-11 line (100 μg/ml), whereas in the corresponding SL1344 and 14028 Δspr mutant strains the MIC decreased to 12.5 μg/ml, equaling that of the SR-11 Δspr mutant (Table 3).
In E. coli, overproduction of PBP7 suppresses thermosensitive growth associated with a mepS mutation (Hara et al., 1996), suggesting that PBP7 and MepS connect in parallel pathways. Hence, we deleted pbpG, coding for the PBP7 homolog in S. Typhimurium. We also created deletion mutants for the mepA and nlpC homologs in S. Typhimurium, each coding for muramyl endopeptidases. Yet, none of the three additional mutants showed sensitization to the antibiotics included in the test panel (Table 3).
In E. coli, overexpression of selected outer membrane porin proteins can result in an outer membrane permeability defect (Krishnamoorthy et al., 2016). Yet, outer membrane protein profiles of the wild-type, Δspr and ΔsprΔyebA mutants on SDS-PAGE gels did not reveal any significant differences (Figure 2B), arguing that the increased vancomycin sensitization is not caused by a major alteration in outer membrane protein composition.
Our observations combined show that deletion of spr in S. Typhimurium is associated with sensitization to vancomycin, and that this sensitization is not restricted to line SR-11. Furthermore, we note that the lack of muramyl endopeptidases YebA, YdhO, PBP7, MepA, or NlpC as such do not result in sensitization to vancomycin, nor does the spr deletion associate with a general overproduction of major porin proteins.
Vancomycin Resistance of S. Typhimurium Requires a Catalytically Active Spr
To further ensure the PCR-based confirmations of the Δspr and ΔsprΔyebA mutations, we performed whole genome sequencing on the two mutant constructs, which verified their expected genetic composition. To exclude any potential polar effects of the verified Δspr deletions as a cause of vancomycin sensitization, we applied genetic complementation. We cloned the spr gene from S. Typhimurium SR-11 under the control of the arabinose-inducible promoter in the plasmid vector pBAD30. There after we generated a point mutation in this plasmid replacing the codon for the conserved catalytic Spr cysteine residue with a codon for serine, creating a C70S alteration in the mature protein (Aramini et al., 2008; Singh et al., 2012). Introducing the cloned native spr gene into either the Δspr or ΔsprΔyebA mutant fully restored vancomycin resistance, whereas both the empty pBAD30 vector, and the plasmid coding for a catalytically inactive Spr variant, did not restore vancomycin resistance (Figure 3). We conclude that intrinsic vancomycin resistance of S. Typhimurium requires a catalytically active Spr.
FIGURE 3.
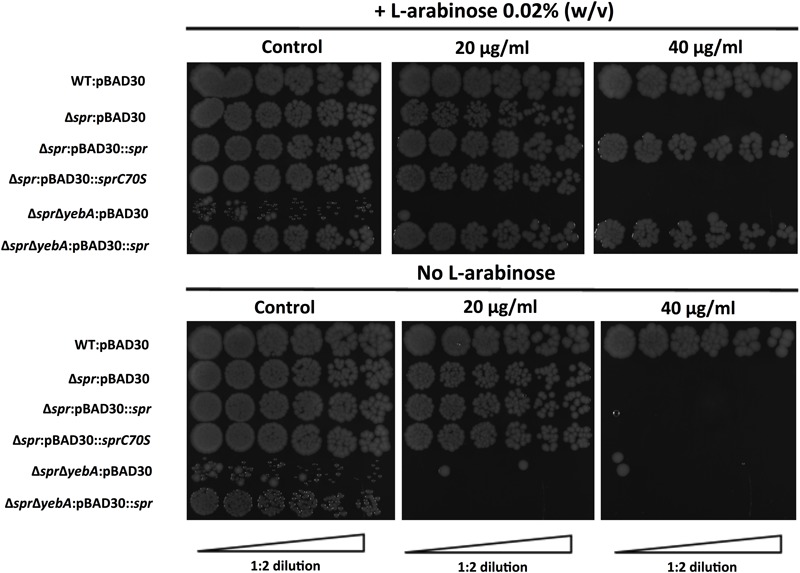
Complementation of vancomycin sensitivity in muramyl endopeptidase mutants as visualized by a drop-on-lawn assay. Wild-type, Δspr and ΔsprΔyebA mutants harboring either vector control pBAD30, pBAD30::spr or pBAD30::sprC70S were serially diluted and spread on TY agar plates, or TY agar plates supplemented with 20 μg/ml or 40 μg/ml of vancomycin, either containing L-arabinose or not. Images are representative of three replicates each.
Lack of Spr Results in S. Typhimurium Being More Prone to Autolyse
Penicillin G activates in E. coli a protein-synthesis-dependent autolysis (Prestidge and Pardee, 1957). Hence, we set out to test whether the Δspr mutation would affect any autolytic behavior of S. Typhimurium in response to cell wall synthesis inhibitors. To enable quantification of bacterial cell lysis, we adapted a β–galactosidase (LacZ) release assays (see detailed in section “Materials and Methods”). The β–galactosidase release assay is based on the pKTH3088 plasmid (Taira et al., 1991). pKTH3088 carries the E. coli lacZ gene in the medium copy pACYC184 vector, yielding a constant low yet measurable level of β–galactosidase. This β–galactosidase could be observed in whole cell lysates for all pKTH3088 containing strains, and at equal levels.
In this, we incubated a logarithmic growth phase culture of S. Typhimurium with increasing concentrations of a cell wall synthesis inhibitor for 1 h, after which the β–galactosidase activities were determined from the culture supernatants. Both the wild-type and Δspr mutant revealed an increased level of β–galactosidase release as a function of increased concentration of vancomycin, with the release being more pronounced for the Δspr mutant (Figure 4A).
FIGURE 4.
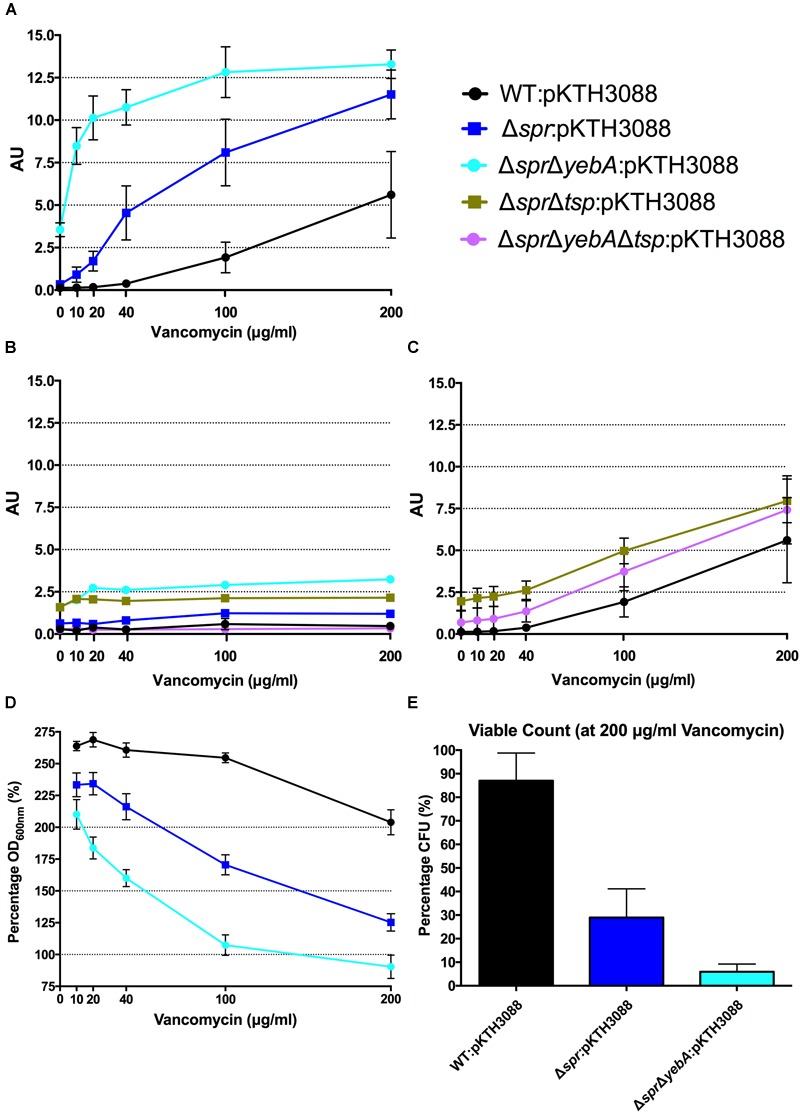
Vancomycin induced autolysis. Release of cytoplasmic β-galactosidase in the presence of increasing concentrations of vancomycin (A,C), or in combination with tetracycline (B) after 1 h of incubation at 37°C. Decrease in optical density at OD600 with increasing vancomycin concentrations (D) and viable counts at highest concentration at 200 μg/ml vancomycin after 1 h of incubation (E). Graph presents the mean values and standard error of the mean of three independent replicates.
To confirm that the vancomycin induced lysis depended on active protein synthesis, we repeated the experiments with tetracycline added into the reaction mixture, as the wild- type and the Δspr mutant exhibited identical MIC:s for tetracycline (Table 3). Adding tetracycline blocked the release of β–galactosidase from both strains (Figure 4B).
As wild-type S. Typhimurium SR-11 and the Δspr mutant had an equal MIC for penicillin G (Table 3), we next repeated the lysis assay by replacing vancomycin with penicillin G. Both the wild-type and Δspr mutant reached a similar plateau level of β–galactosidase release at a higher concentration range of penicillin G (Figure 5A). However, compared to the wild-type, the β–galactosidase release was more pronounced in the Δspr mutant at concentrations below the determined 6.25 μg/ml MIC for penicillin G (t-test: p < 0.01 each for 2 and 4 μg/ml when comparing Δspr mutant to wild-type). Yet again, as for vancomycin, the β–galactosidase release by penicillin G was blocked by addition of tetracycline (Figure 5B).
FIGURE 5.
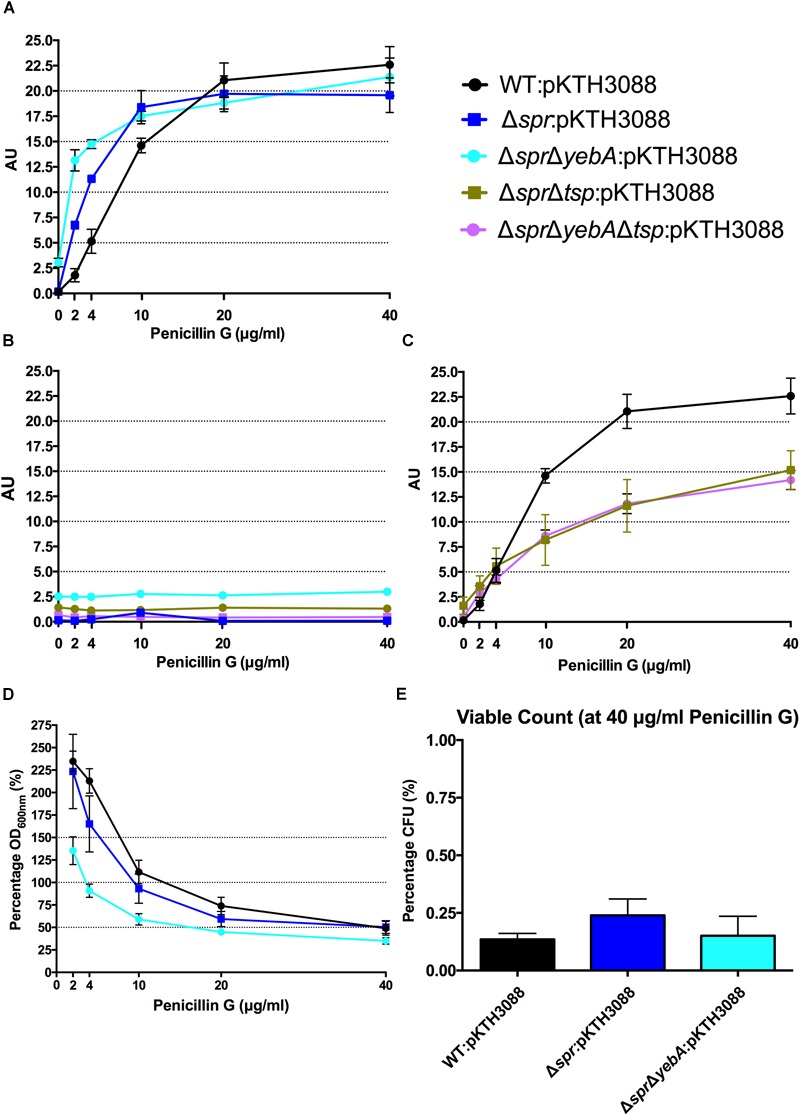
Penicillin G induced autolysis. Release of cytoplasmic β-galactosidase in the presence of increasing concentrations of penicillin G (A,C), or in combination with tetracycline (B) after 1 h of incubation at 37°C. Decrease in optical density at OD600 nm with increasing penicillin G concentrations (D) and viable counts from highest concentration at 40 μg/ml penicillin G after 1 h of incubation (E). Graph presents the mean values and standard error of the mean of three independent replicates.
When we followed the development of the optical density (OD600 nm) under the 1 h incubation with cell wall synthesis inhibitors (Figures 4D, 5D), we noted that a substantial proportion of the bacteria apparently remained unlysed. When we determined the viable count from cultures exposed to 200 μg/ml of vancomycin, representing 2-fold MIC for wild-type and 16-fold MIC for Δspr mutant bacteria, we recovered a substantial residual amount of viable bacteria from the cultures (Figure 4E). When the viable counts were measured for the penicillin G exposed cultures (containing antibiotic six times the MIC), we could barely detect any viable bacteria (Figure 5E).
The ΔsprΔyebA mutant exhibited a substantial decrease in tolerance to both vancomycin and penicillin G as compared to either the wild-type or Δspr mutant (Table 3). This increased sensitivity was associated with a significantly higher level of β–galactosidase release (relative to the wild-type or the Δspr mutant) after exposure to either antibiotic for 1 h. In combination, these data demonstrate that vancomycin induce a protein-synthesis-dependent autolysis in S. Typhimurium, and the intensity of this autolysis inversely correlated with the MIC to vancomycin. On the other hand, penicillin G evoked a more proficient autolysis in the Δspr mutant despite the wild-type and Δspr mutant had the same MIC for penicillin G.
Periplasmic Protease Tsp Suppresses Δspr-Dependant Vancomycin Sensitivity
Vancomycin-resistant mutants were selected in the ΔsprΔyebA background (for details, see section “Materials and Methods”). Twelve vancomycin-tolerant ΔsprΔyebA mutants were analyzed by whole genome sequencing. Eleven of the mutants carried overlapping deletions covering nucleotides 1,920,000–1,965,000 in the S. Typhimurium LT2 reference genome (Figure 6A). At the center of this region is tsp, encoding a periplasmic carboxypeptidase. The remaining suppressor mutant carried a point mutation within tsp itself (Figure 6A). These data suggest that inactivation of tsp is the common feature of mutations that suppress the vancomycin sensitivity phenotype of the ΔsprΔyebA mutant.
FIGURE 6.
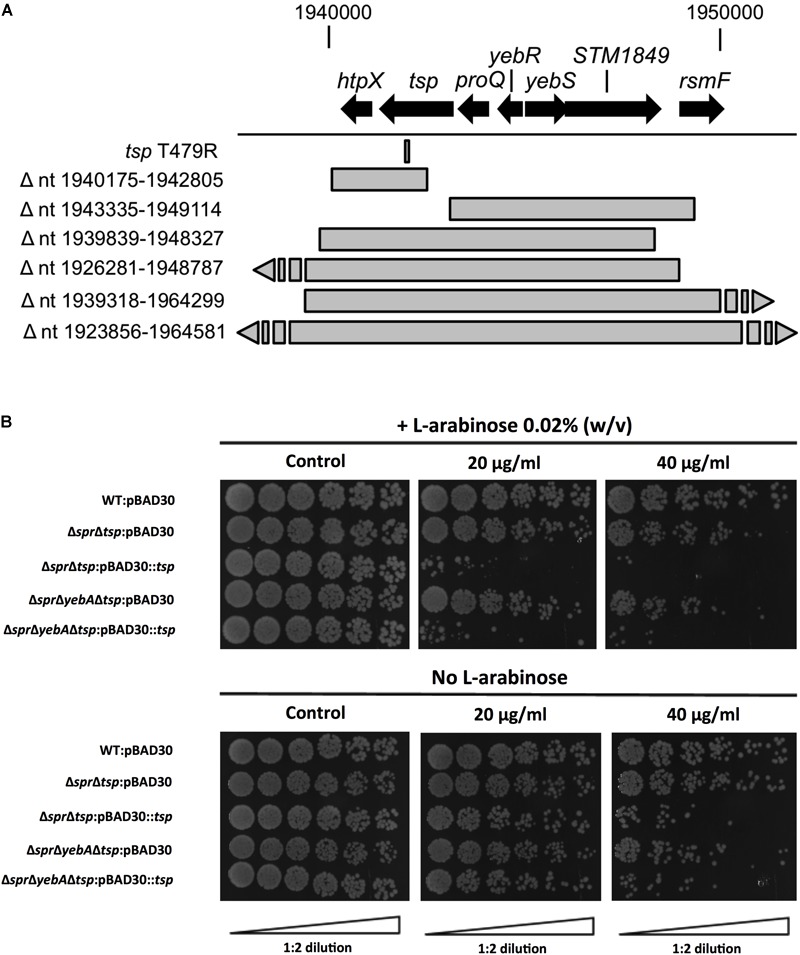
Suppression of vancomycin sensitivity in Δspr mutant backgrounds. (A) Genomic visualization of vancomycin tolerant suppressor mutations with the bars representing genomic deletions, except for tsp T479R where the bar points to the position of a point mutation. (B) Wild-type and the Δtsp suppressed mutants harboring either vector control pBAD30 or full complementation pBAD30::tsp were serially diluted and spread on TY agar plates, or TY agar plates supplemented with 20 μg/ml or 40 μg/ml of vancomycin, either containing L-arabinose or not. Images are representative of three replicates each.
To confirm the tsp mutations as suppressors, we deleted tsp, as well as the individual genes that mapped upstream and downstream of tsp (proQ and htpX) in the ΔsprΔyebA mutant. Out of these three constructs, only deletion of tsp resulted in a vancomycin resistant phenotype in the ΔsprΔyebA mutant (Table 3). In addition, when the tsp deletion was introduced into the Δspr mutant, it converted the phenotype from vancomycin sensitive to vancomycin resistant (Table 3). Conversely, when a cloned tsp gene was introduced into the vancomycin resistant ΔsprΔtsp and ΔsprΔyebAΔtsp mutants, the phenotypes were reverted to vancomycin sensitive (Figure 6B).
The Δtsp mutation also reverted the general antibiotic sensitization of the ΔsprΔyebA mutant without affecting growth (Table 3, Figures 1C,D). In addition, introduction of the tsp deletion into the Δspr and ΔsprΔyebA mutants suppressed the autolysis by reducing their release of β–galactosidase in the presence of vancomycin or penicillin G (Figures 4C, 5C). Thus, a tsp deletion acted as a general suppressor mutation for Δspr and ΔsprΔyebA mutant phenotypes. That said, the tsp deletion alone did not confer increased vancomycin resistance (Table 3).
In E. coli, Tsp co-purifies with the outer membrane lipoprotein NlpI, which assists Tsp in degrading MepS (Spr) (Singh et al., 2015). Furthermore, nlpI mutations suppress a temperature-sensitive phenotype of an E. coliΔmepS mutant (Tadokoro et al., 2004) implying a functional connection between Tsp, NlpI and MepS. Yet, when we deleted nlpI in the S. Typhimurium Δspr and ΔsprΔyebA mutants, they retained their sensitization to vancomycin (Table 3). Hence the vancomycin-sensitive phenotype of the Δspr mutant is mainly dependent on Tsp rather than on NlpI.
Discussion
In E. coli, mutants that lack any one of the muramyl endopeptidases, Spr (MepS), YebA (MepM) or YhdO (MepH), suffer no obvious growth defects (Singh et al., 2012). In agreement with this, we found that genetic deletion of the individual murein endopeptidases, Spr, YebA, or YhdO in S. Typhimurium did not affect bacterial growth in broth cultures. We also created Δspr, ΔyebA, and ΔyhdO double mutants, to test for redundancy in their contribution to S. Typhimurium growth in broth. Only for the ΔsprΔyebA mutant, did we note a minor defect in growth capacity. This phenotype was not seen with any of the single mutants, or with the ΔyebAΔyhdO double mutant, suggesting that spr might not be completely non-redundant under the growth conditions tested.
In Gram-negative enteric bacteria, the outer membrane acts as a barrier adding to intrinsic resistance to lysozyme, and to antibiotics such as novobiocin, rifampicin and vancomycin (Grundström et al., 1980; Helander et al., 1989). Lipopolysaccharide (LPS) contributes to outer membrane integrity and mutations in genes involved in LPS biosynthesis can sensitize Gram-negative bacteria simultaneous to numerous antibiotics (Grundström et al., 1980; Helander et al., 1989). Outer membrane integrity can also be disturbed by the expression of aberrant outer membrane proteins (Rhen et al., 1988) or polymyxin B nonapeptide (Vaara and Vaara, 1983; Ofek et al., 1994). Simultaneous genetic depletion of multiple murein hydrolases may also cause outer membrane destabilization (Heidrich et al., 2002) in E. coli, including vancomycin sensitization (Korsak et al., 2005).
Here we report that depletion of a single muramyl endopeptidase alone, Spr, results in vancomycin sensitization in an enteric bacterium. Significantly, the vancomycin sensitization associated with the Δspr mutation did not associate with sensitization to rifampicin or novobiocin, which would be expected in the case of classical outer membrane destabilization. Recent work on Vibrio cholerae suggested that mechanisms other than outer membrane permeability are also involved in preventing antibiotics from entering, or acting, in the periplasm (Dörr et al., 2016). Our observation that the growth of wild-type S. Typhimurium is inhibited by vancomycin, albeit at a high concentration (Table 3), suggests that also in S. Typhimurium the outer membrane barrier does not completely prevent vancomycin entry. Consequently, at high concentrations vancomycin could accumulate to a level that prevents efficient peptidoglycan cross-linking. In the Δspr mutant the capacity to ensure peptidoglycan turnover, while not yet preventing growth, could be compromised as such. In the Δspr mutant vancomycin concentrations sub-inhibitory for wild-type bacteria would further add to disturbed peptidoglycan composition and consequently lower the threshold for preventing growth.
In E. coli, blocking peptidoglycan cross-linking with penicillin G results in autolysis. Thus we argued that increased vancomycin sensitivity of the Δspr mutant could associate with an altered autolytic behavior. Hence, we set out to compare the autolytic behavior of wild-type and Δspr mutants by measuring release of the cytoplasmic enzyme β–galactosidase after vancomycin exposure. Release of β–galactosidase was more prominent for the Δspr mutant, and notably so the for the ΔsprΔyebA mutant (Figure 4A). Thus, vancomycin induced an autolysis in S. Typhimurium that inversely correlated with the vancomycin MIC. To demonstrate that the increased autolysis of the Δspr mutant was not only a reflection of a decreased MIC for vancomycin, we repeated the autolysis assay upon exposure to penicillin G, for which the wild-type and spr mutant expressed an equal MIC (Table 3). In this, the Δspr mutant revealed a more rapid onset of autolysis upon exposure to penicillin G (Figure 5A).
When recording autolysis measured as decrease in optical density, we noted that a proportion of each culture exposed to either vancomycin or penicillin G remained apparently non-lysed (Figures 4D, 5D). Even at a vancomycin concentration that was 16-fold MIC, we recovered a substantial proportion of viable Δspr bacteria at the end of the experiment (Figure 4E). Viable bacteria were also recovered from the corresponding penicillin G-exposed cultures but at 100-fold lower frequency for both wild-type and Δspr mutant bacteria (Figure 5E). This would imply that the Δspr mutant indeed is more prone to autolysis, and that the PBPs of wild-type and Δspr mutant bacteria, whether autolytic or not, are equally and irreversibly inhibited by penicillin G.
In Gram-positive bacteria vancomycin resistance is achieved through the acquisition of large genetic blocks coding for new peptidoglycan motifs, rather than through point mutations (Gardete and Tomasz, 2014; Faron et al., 2016). As the MIC for our vancomycin-sensitive ΔsprΔyebA mutant under our test conditions approached MIC values of susceptible Gram-positive pathogens such as Enterococcus faecalis and Staphylococcus aureus [Susceptible Enterococci spp. ≤4 μg/ml; susceptible1 S. aureus ≤ 2 μg/ml, “EUCAST: Clinical Breakpoints”, 2018], we set out to probe whether we could in a Gram-negative vancomycin-sensitive bacterium select for spontaneous mutations contributing to vancomycin tolerance. In this, we selected and genetically confirmed that Δspr-mediated vancomycin sensitization, including the more pronounced sensitization of the ΔsprΔyebA mutant, required a tsp-proficient genetic background (Figure 6). While corresponding Spr or Tsp proteins do not exist in enterococci, the success in isolating vancomycin-resistant mutations, including point mutations, in our ΔsprΔyebA mutant background adds to our understanding of how vancomycin resistance in a sensitized genetic background can be achieved without horizontal gene transfer. We here also demonstrate that intrinsic S. Typhimurium vancomycin resistance relies on a catalytically active Spr (Figure 3). This, and the fact that the vancomycin sensitivities of the Δspr and ΔsprΔyebA mutants are close to the clinical breakpoints of relevant clinical Gram-positive pathogens places the catalytic activity of Spr and related endopeptidases as potential target whose inhibition could potentiate treatment of enteric bacterial infections with vancomycin.
Tsp is a periplasmic endopeptidase implicated in the processing of Spr and PBP3 (Hara et al., 1989, 1991; Singh et al., 2015). In Pseudomonas aeruginosa the YebA/MepM homolog is also subjected to proteolysis (Srivastava et al., 2018). Thus, there might exist analogous protein complexes in a wide range of bacteria that regulate turnover of peptidoglycan. Depletion of any component of such a complex could distort the cell wall composition with accompanying sensitization to antibiotics. Indeed, in S. Typhimurium the tsp mutant revealed sensitization to novobiocin and penicillin, a phenotype suppressed by deleting spr (Table 3). Even so, the tsp mutant exhibited the same MIC for vancomycin as the wild-type strain (Table 3).
In summary, we have genetically defined a new pathway for intrinsic resistance to the large-molecular-weight antibiotic vancomycin that is not dependent on outer membrane permeability, in the Gram-negative pathogen S. Typhimurium. This new pathway involves the combined action of the muramyl endopeptidase Spr, together with the protease Tsp, in maintaining the peptidoglycan homeostasis essential for maintaining the cell wall integrity of the bacterium upon antibiotic challenge. These insights add to the knowledge needed to combat the increasing problem of antibiotic resistance in Gram-negative bacteria.
Author Contributions
KV, HW, DH, and MR designed the study. KV, DLH, IS, and MR performed the experiments. KV, DH, and MR wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Sandra Muschiol for making electrocompetent E. coli Top10 cells, and Edmund Loh for giving input on the manuscript. This manuscript has been uploaded as a pre-print at BioRxiv (Vesto et al., 2018).
Funding. This work was supported by Vetenskapsrådet (the Swedish Research Council) grants Dnr 4-30 16-2013 (MR), 2013-8643 (DH), 2016-04449 (DH), and 2017-03953 (DH). MR was also supported as a visiting professor in the Umeå Centre for Microbial Research (UCMR) Linnaeus Program by grant Dnr 349-2007-8673 from Vetenskapsrådet.
References
- Angelo K. M., Reynolds J., Karp B. E., Hoekstra R. M., Scheel C. M., Friedman C. (2016). Antimicrobial resistance among nontyphoidal Salmonella isolated from blood in the United States, 2003–2013. J. Infect. Dis. 214 1565–1570. 10.1093/infdis/jiw415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramini J. M., Rossi P., Huang Y. J., Zhao L., Jiang M., Maglaqui M., et al. (2008). Solution NMR structure of the NlpC/P60 domain of lipoprotein Spr from Escherichia coli: structural evidence for a novel cysteine peptidase catalytic triad. Biochemistry 47 9715–9717. 10.1021/bi8010779 [DOI] [PubMed] [Google Scholar]
- Baquero M. R., Bouzon M., Quintela J. C., Ayala J. A., Moreno F. (1996). DacD, an Escherichia coli gene encoding a novel penicillin-binding. J. Bacteriol. 178 7106–7111. 10.1128/jb.178.24.7106-7111.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs W. H., Andrews F. A. (1976). Role of ionic strength in salt antagonism of aminoglycoside action on Escherichia coli and Pseudomonas aeruginosa. J. Infect. Dis. 134 500–504. 10.1093/infdis/134.5.500 [DOI] [PubMed] [Google Scholar]
- Botta G. A., Park J. T. (1981). Evidence for involvement of penicillin-binding protein 3 in murein synthesis during septation but not during cell elongation. J. Bacteriol. 145 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broome-Smith J. K., Spratt B. G. (1982). Deletion of the penicillin-binding protein 6 gene of Escherichia coli. J. Bacteriol. 152 904–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullas L. R., Ryu J. I. (1983). Salmonella typhimurium LT2 strains which Are R- M+ for all three chromosomally located systems of DNA restriction and modification. J. Bacteriol. 156 471–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K. M., Kim M. H., Cai H., Lee Y. J., Hong Y., Ryu P. Y. (2018). Salicylic acid reduces OmpF expression, rendering Salmonella enterica Serovar Typhimurium more resistant to cephalosporin antibiotics. Chonnam Med. J. 54 17–23. 10.4068/cmj.2018.54.1.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coque T. M., Baquero F., Canton R. (2008). Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill. 13:19044. [PubMed] [Google Scholar]
- Datsenko K. A., Wanner B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcour A. H. (2009). Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta 1794 808–816. 10.1016/j.bbapap.2008.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denome S. A., Elf P. K., Henderson T. A., Nelson D. E., Young K. D. (1999). Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J. Bacteriol. 181 3981–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörr T., Delgado F., Umans B. D., Gerding M. A., Davis B. M., Waldor M. K. (2016). A transposon screen identifies genetic determinants of Vibrio cholerae resistance to high-molecular-weight antibiotics. Antimicrob. Agents Chemother. 60 4757–4763. 10.1128/AAC.00576-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EUCAST: Clinical Breakpoints (2018). EUCAST: Clinical Breakpoints 2018. Available at: http://www.eucast.org/clinical_breakpoints/ [Accessed July 6, 2018]. [Google Scholar]
- Faron M. L., Ledeboer N. A., Buchan B. W. (2016). Resistance mechanisms, epidemiology, and approaches to screening for vancomycin-resistant enterococcus in the health care setting. J. Clin. Microbiol. 54 2436–2447. 10.1128/JCM.00211-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardete S., Tomasz A. (2014). Mechanisms of vancomycin resistance in Staphylococcus aureus. J. Clin. Invest. 124 2836–2840. 10.1172/JCI68834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray A. N., Egan A. J. F., van’t Veer I. L., Verheul J., Colavin A., Koumoutsi A., et al. (2015). Coordination of peptidoglycan synthesis and outer membrane constriction during Escherichia coli cell division. eLife 4:e07118. 10.7554/eLife.07118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundström T., Normark S., Magnusson K. E. (1980). Overproduction of outer membrane protein suppresses envA-induced hyperpermeability. J. Bacteriol. 144 884–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177 4121–4130. 10.1128/jb.177.14.4121-4130.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara H., Abe N., Nakakouji M., Nishimura Y., Horiuchi K. (1996). Overproduction of penicillin-binding protein 7 suppresses thermosensitive growth defect at low osmolarity due to an spr mutation of Escherichia coli. Microb. Drug Resist. 2 63–72. 10.1089/mdr.1996.2.63 [DOI] [PubMed] [Google Scholar]
- Hara H., Nishimura Y., Kato J., Suzuki H., Nagasawa H., Suzuki A., et al. (1989). Genetic analyses of processing involving c-terminal cleavage in penicillin-binding protein 3 of Escherichia coli. J. Bacteriol. 171 5882–5889. 10.1128/jb.171.11.5882-5889.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara H., Yamamoto Y., Higashitani A., Suzuki H., Nishimura Y. (1991). Cloning, mapping, and characterization of the Escherichia coli Prc gene, which is involved in C-terminal processing of penicillin-binding protein 3. J. Bacteriol. 173 4799–4813. 10.1128/jb.173.15.4799-4813.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder K. J., Nikaido H., Matsuhashi M. (1981). Mutants of Escherichia coli that are resistant to certain beta-lactam compounds lack the ompF porin. Antimicrob. Agents Chemother. 20 549–552. 10.1128/AAC.20.4.549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich C., Ursinus A., Berger J., Schwarz H., Höltje J.-V. (2002). Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. J. Bacteriol. 184 6093–6099. 10.1128/JB.184.22.6093-6099.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helander I. M., Vaara M., Sukupolvi S., Rhen M., Saarela S., Zähringer U., et al. (1989). rfaP mutants of Salmonella typhimurium. Eur. J. Biochem. 185 541–546. 10.1111/j.1432-1033.1989.tb15147.x [DOI] [PubMed] [Google Scholar]
- Henderson T. A., Templin M., Young K. D. (1995). Identification and cloning of the gene encoding penicillin-binding protein 7 of Escherichia coli. J. Bacteriol. 177 2074–2079. 10.1128/jb.177.8.2074-2079.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Rovira A., Davies P., Ahlstrom C., Muellner P., Rendahl A., et al. (2016). Serotypes and antimicrobial resistance in Salmonella enterica recovered from clinical samples from cattle and swine in Minnesota, 2006 to 2015. PLoS One 11:e0168016. 10.1371/journal.pone.0168016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A., Chabbert Y. A., Semonin O. (1982). Role of porin proteins OmpF and OmpC in the permeation of beta-lactams. Antimicrob. Agents Chemother. 22 942–948. 10.1128/AAC.22.6.942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm E. J., Shakoor S., Page A. J., Qamar F. N., Judge K., Saeed D. K., et al. (2018). Emergence of an extensively drug-resistant Salmonella enterica Serovar Typhi clone harboring a promiscuous plasmid encoding resistance to fluoroquinolones and third-generation cephalosporins. mBio 9:e00105-18. 10.1128/mBio.00105-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsak D., Liebscher S., Vollmer W. (2005). Susceptibility to antibiotics and β-Lactamase induction in murein hydrolase mutants of Escherichia coli. Antimicrob. Agents Chemother. 49 1404–1409. 10.1128/AAC.49.4.1404-1409.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy G., Tikhonova E. B., Dhamdhere G., Zgurskaya H. I. (2013). On the role of TolC in multidrug efflux: the function and assembly of AcrAB-TolC tolerate significant depletion of intracellular TolC protein. Mol. Microbiol. 87 982–997. 10.1111/mmi.12143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy G., Wolloscheck D., Weeks J. W., Croft C., Rybenkov V. V., Zgurskaya H. I. (2016). Breaking the permeability barrier of Escherichia coli by controlled hyperporination of the outer membrane. Antimicrob. Agents Chemother. 60 7372–7381. 10.1128/AAC.01882-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- Liu Q., Liu Q., Yi J., Liang K., Hu B., Zhang X., et al. (2016). Outer membrane vesicles from flagellin-deficient Salmonella enterica Serovar Typhimurium induce cross-reactive immunity and provide cross-protection against heterologous Salmonella challenge. Sci. Rep. 6:34776. 10.1038/srep34776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuhashi M., Takagaki Y., Maruyama I. N., Tamaki S., Nishimura Y., Suzuki H., et al. (1977). Mutants of Escherichia coli lacking in highly penicillin-sensitive D-alanine carboxypeptidase activity. Proc. Natl. Acad. Sci. U.S.A. 74 2976–2979. 10.1073/pnas.74.7.2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuhashi M., Tamaki S., Curtis S. J., Strominger J. L. (1979). Mutational evidence for identity of penicillin-binding protein 5 in Escherichia coli with the major D-Alanine carboxypeptidase IA activity. J. Bacteriol. 137 644–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Sanderson K. E., Spieth J., Clifton S. W., Latreille P., Courtney L., et al. (2001). Complete genome sequence of Salmonella enterica Serovar Typhimurium LT2. Nature 413 852–856. 10.1038/35101614 [DOI] [PubMed] [Google Scholar]
- Meberg B. M., Paulson A. L., Priyadarshini R., Young K. D. (2004). Endopeptidase penicillin-binding proteins 4 and 7 play auxiliary roles in determining uniform morphology of Escherichia coli. J. Bacteriol. 186 8326–8336. 10.1128/JB.186.24.8326-8336.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Nikaido H., Vaara M. (1985). Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 49 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y., Suzuki H., Hirota Y., Park J. T. (1980). A mutant of Escherichia coli defective in penicillin-binding protein 5 and lacking D-alanine carboxypeptidase IA. J. Bacteriol. 143 531–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofek I., Cohen S., Rahmani R., Kabha K., Tamarkin D., Herzig Y., et al. (1994). Antibacterial synergism of polymyxin B nonapeptide and hydrophobic antibiotics in experimental gram-negative infections in mice. Antimicrob. Agents Chemother. 38 374–377. 10.1128/AAC.38.2.374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestidge L. S., Pardee A. B. (1957). Induction Of bacterial lysis by penicillin1. J. Bacteriol. 74 48–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhen M., O’Connor C. D., Sukupolvi S. (1988). The outer membrane permeability mutation of the virulence-associated plasmid of Salmonella typhimurium is located in a tra T-like gene. FEMS Microbiol. Lett. 52 145–153. 10.1111/j.1574-6968.1988.tb02586.x [DOI] [Google Scholar]
- Sabet S. F., Schnaitman C. A. (1973). Purification and properties of the colicin E3 receptor of Escherichia coli. J. Biol. Chem. 248 1797–1806. [PubMed] [Google Scholar]
- Sauvage E., Kerff F., Terrak M., Ayala J. A., Charlier P. (2008). The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32 234–258. 10.1111/j.1574-6976.2008.00105.x [DOI] [PubMed] [Google Scholar]
- Schmieger H. (1972). Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 119 75–88. 10.1007/BF00270447 [DOI] [PubMed] [Google Scholar]
- Singh S. K., Parveen S., SaiSree L., Reddy M. (2015). Regulated proteolysis of a cross-link–specific peptidoglycan hydrolase contributes to bacterial morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 112 10956–10961. 10.1073/pnas.1507760112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S. K., SaiSree L., Amrutha R. N., Reddy M. (2012). Three redundant murein endopeptidases catalyse an essential cleavage step in peptidoglycan synthesis of Escherichia coli K12. Mol. Microbiol. 86 1036–1051. 10.1111/mmi.12058 [DOI] [PubMed] [Google Scholar]
- Spratt B. G. (1980). Deletion of the penicillin-binding protein 5 gene of Escherichia coli. J. Bacteriol. 144 1190–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D., Seo J., Rimal B., Kim S. J., Zhen S., Darwin A. J. (2018). A proteolytic complex targets multiple cell wall hydrolases in Pseudomonas aeruginosa. mBio 9:e00972-18. 10.1128/mBio.00972-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukupolvi S., Edelstein A., Rhen M., Normark S. J., Pfeifer J. D. (1997). Development of a murine model of chronic Salmonella infection. Infect. Immun. 65 838–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukupolvi S., Vaara M., Helander I. M., Viljanen P., Mäkelä P. H. (1984). New Salmonella typhimurium mutants with altered outer membrane permeability. J. Bacteriol. 159 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro A., Hayashi H., Kishimoto T., Makino Y., Fujisaki S., Nishimura Y. (2004). Interaction of the Escherichia coli lipoprotein NlpI with periplasmic Prc (Tsp) protease. J. Biochem. 135 185–191. 10.1093/jb/mvh022 [DOI] [PubMed] [Google Scholar]
- Taira S., Riikonen P., Saarilahti H., Sukupolvi S., Rhen M. (1991). The mkaC virulence gene of the Salmonella Serovar Typhimurium 96 Kb plasmid encodes a transcriptional activator. Mol. Gen. Genet.? 228 381–384. 10.1007/BF00260630 [DOI] [PubMed] [Google Scholar]
- Vaara M., Vaara T. (1983). Sensitization of gram-negative bacteria to antibiotics and complement by a nontoxic oligopeptide. Nature 303 526–528. 10.1038/303526a0 [DOI] [PubMed] [Google Scholar]
- van Heijenoort J. (2011). Peptidoglycan hydrolases of Escherichia coli. Microbiol. Mol. Biol. Rev. 75 636–663. 10.1128/MMBR.00022-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesto K., Huseby D. L., Snygg I., Wang H., Hughes D., Rhen M. (2018). Muramyl endopeptidase spr contributes to intrinsic vancomycin resistance in Salmonella enterica Serovar Typhimurium. bioRxiv [Preprint] 10.1101/274886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks J. W., Celaya-Kolb T., Pecora S., Misra R. (2010). AcrA suppressor alterations reverse the drug hypersensitivity phenotype of a TolC mutant by inducing TolC aperture opening. Mol. Microbiol. 75 1468–1483. 10.1111/j.1365-2958.2010.07068.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Ellis H. M., Lee E. C., Jenkins N. A., Copeland N. G., Court D. L. (2000). An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 97 5978–5983. 10.1073/pnas.100127597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderle C., Stieger M., Burrell M., Reinelt S., Maxwell A., Page M., et al. (2008). Biological activities of novel gyrase inhibitors of the aminocoumarin class. Antimicrob. Agents Chemother. 52 1982–1990. 10.1128/AAC.01235-07 [DOI] [PMC free article] [PubMed] [Google Scholar]


