Abstract
Sorghum (Sorghum bicolor) is an important bioenergy crop. Its biomass mainly consists of the cellulosic and non-cellulosic polysaccharides, both which can be converted to biofuels. The biosynthesis of non-cellulosic polysaccharides involves several glycosyltransferases (GT) families including GT47. However, there was no systemic study on GT47 family in sorghum to date. Here, we identified 39 sorghum GT47 family members and showed the functional divergences of MURUS3 (MUR3) homologs. Sorghum GT47 proteins were phylogenetically clustered into four distinct subfamilies. Within each subfamily, gene structure was relatively conserved between the members. Ten gene pairs were identified from the 39 GT47 genes, of which two pairs might be originated from tandem duplication. 25.6% (10/39) of sorghum GT47 genes were homologous to Arabidopsis MUR3, a xyloglucan biosynthesis gene in primary cell walls. SbGT47_2, SbGT47_7, and SbGT47_8, three most homologous genes of MUR3, exhibited different tissue expression patterns and were selected for complementation into Arabidopsis mur3-3. Physiological and cell wall analyses showed that SbGT47_2 and SbGT47_7 may be two functional xyloglucan galactosyltransferases in sorghum. Further studies found that MUR3-like genes are widely present in the seed plants but not in the chlorophytic alga Chlamydomonas reinhardtii. Our results provide novel information for evolutionary analysis and functional dissection of sorghum GT47 family members.
Keywords: Sorghum, GT47 family, genome-wide analysis, MUR3, functional divergences
Introduction
Sorghum(Sorghum bicolor), a highly productive C4 photosynthetic grass, is the fifth most cultivated cereal crop globally due to its huge biomass yield, high nitrogen utilization efficiency and remarkable adaptability on marginal land (Taylor et al., 2010; Byrt et al., 2011). Sorghum germplasms have abundant genetic diversity and can be classified into grain, forage, energy, and sweet sorghum based on the growth characteristics and end-uses (Rooney et al., 2007). In comparison to lignocellulosic biomass crops such as switchgrass and Miscanthus, sorghum has a smaller genome (∼730 Mb) and more fermentable soluble sugars, making it an ideal model for functional analysis of C4 grasses (Paterson et al., 2009).
Sorghum biomass consists of 24–38% cellulose, 12–22% non-cellulosic polysaccharides, 17–22% lignin, and 1–22% starch (Corredor et al., 2009). Non-cellulosic polysaccharides generally interact with cellulose and lignin, contributing to the strength of plant cell walls (Scheller and Ulvskov, 2010). In dicots and non-commelinoid monocotyledons, the major non-cellulosic polysaccharides of primary walls are xyloglucans and a range of pectic polysaccharides, with lower levels of heteroxylans and heteromannans. In contrast, primary walls of grasses have much lower levels of xyloglucans and pectins, which are replaced by higher amounts of heteroxylans and, in some cases, with (1,3; 1,4)-β-glucans (Burton and Fincher, 2012). Charactirization of Arabidopsis mutants shows that a battery of glycosyltransferases (GTs) from family GT2, GT8, GT34, GT37, GT43, and GT47 are involved in the biosynthesis of non-cellulosic polysaccharides (Perrin et al., 1999; Faik et al., 2002; Madson et al., 2003; Bauer et al., 2006; Burton et al., 2006; Brown et al., 2007, 2009; Cocuron et al., 2007; Lee et al., 2007).
GT47 proteins participate in xylan and xyloglucan biosynthesis in plants. Arabidopsis IRREGULAR XYLEM10 (IRX10) and IRX10-L possess UDP-Xyl: β-(1→4)-xylosyl transferase activities in vitro and are essential for formation of xylan backbone in secondary cell walls (Brown et al., 2009; Wu et al., 2009; Jensen et al., 2014; Urbanowicz et al., 2014). FRAGILE FIBER8 (FRA8)/IRX7 is required for synthesis of the tetrasaccharide at the reducing end of glucronoxylan in secondary cell walls (Zhong et al., 2005). Other GT47 family members MURUS3 (MUR3), XYLOGLUCAN L-SIDE CHAIN GALACTOSYLTRANSFERASE2 (XLT2), XYLOGLUCAN D-SIDE-CHAIN TRANSFERASE (XDT), XYLOGLUCAN S-SIDECHAIN TRANSFERASE 1 (XST1) and XYLOGLUCAN-SPECIFIC GALACTURONOSYLTRANSFERASE 1 (XUT1) contribute to xyloglucan side chain synthesis (Madson et al., 2003; Jensen et al., 2012; Peña et al., 2012; Schultink et al., 2013; Zhu et al., 2018). Arabidopsis MUR3 adds a galactosyl unit solely to the third xylosyl residue from the non-reducing end in the XXXG xyloglucan (XyG) oligosaccharide core, producing XXLG (Reiter et al., 1997; Madson et al., 2003; Kong et al., 2015). Ectopic expression of Eucalyptus grandis MUR3 in the Arabidopsis mur3 restored the fucosylated side chains, indicating a functional conservation of XyG structure in dicots (Lopes et al., 2010). In rice, fucogalactosylated xyloglucan is retained only in specific tissues, such as the root epidermis, root tip-growing hairs and pollen tubes (Liu et al., 2015). Further, OsMUR3 overexpression in mur3.1xlt2 results in production of XXLG, XXFG and their O-acetylated forms, indicating that OsMUR3 is enzymatically equivalent to MUR3 (Liu et al., 2015).
Although the sorghum genome (v3.1.1) is recently released (McCormick et al., 2018), there are few functional studies in sorghum due to loss of high-efficient genetic transformation system. In this work, we reported the comprehensive genomic identification and phylogenetic analysis of 39 GT47 family members in sorghum, as well as their expression patterns in six different tissues. Three genes (SbGT47_2, SbGT47_7, and SbGT47_8) that are the most homologous to MUR3 were selected for functional characterization. Genetic complementation and cell wall analysis showed that they differentially rescued phenotypic defects of mur3-3. At least two MUR3-like genes (SbGT47_2 and SbGT47_7) are part of the synthetic machinery necessary to produce fucoglalactoxyloglucan in sorghum.
Materials and Methods
Identification of GT47 Proteins in Sorghum and Other Nine Species
To identify GT47 proteins in ten representative species namely green alga (Chlamydomonas reinhardtii), moss (Physomitrella patens), sorghum (S. bicolor), stiff brome (Brachypodium distachyon), rice (Oryza sativa), switchgrass (Panicum virgatum), Arabidopsis (Arabidopsis thaliana), grape (Vitis vinifera), alfalfa (Medicago truncatula) and poplar (Populus trichocarpa), Pfam (PF03016) analysis was employed to search against their corresponding genome database1. All GT47 proteins were manually verified for the presence of exostosin domain using InterProScan program2. Online software FGENESH3 was used to correct the mis-annotated GT47 genes.
Sequence Properties of Sorghum GT47 Proteins
The physicochemical parameters of sorghum GT47 proteins were calculated using DNAman software (Lynnon Biosoft, Canada). Transmembrane domain was predicted by TMHMM Server v. 2.04. The exon/intron organization of genes was generated with Gene Structure Display Server (GSDS) 5. Tandem duplications was identified based on the criteria that two partners are separated by no more than five gene loci in a range of 100 kb distance (Chai et al., 2012).
Phylogenetic Analysis
Full-length protein sequences were aligned by ClustalX (version 1.83). The phylogenetic tree was generated using the Neighbor-Joining (NJ) and Maximum Parsimony (MP) algorithms, respectively, in MEGA 4.0 (Tamura et al., 2007). Evaluation of the nodes significance was performed by bootstrap analysis with 1,000 replicates. Pairwise gap deletion mode was adopted to ensure that the divergent domains in the NJ tree.
Quantitative Real Time PCR (qRT-PCR)
Young leaf, root, upper stem (internodes 6∼8), middle stem (internodes 3∼5) and basal stem (internodes 1∼2) were sampled from 3-month-old sorghum cultivar “Keller” grown in the greenhouse (16 h light/8 h dark, 25∼28°C). Three independent plants were selected for qRT-PCR analysis as biological repeats. Total RNA was isolated with the RNeasy mini kit (Qiagen, United States) and genomic DNA was removed with RQ1 RNase-Free DNase (Promega, United States). First-strand cDNA was synthesized using oligo (dT) primer and M-MLV RT (Promega, United States). Primers were designed by Beacon Designer v7.0 (Premier Biosoft International, United States) setting primer melting temperature as 58∼60°C, primer length as 20∼24 bp and amplicon length as 90∼150 bp. All primers (Supplementary Table S2) were rechecked with online software Primer-BLAST 6 in order to determine primer specificity. qRT-PCR was conducted on a LightCycler®480 detection system (Roche, Germany) with SYBR Premix Ex Taq (TaKaRa, Japan). SbACTIN2 was used as an internal reference gene.
Subcellular Localization of SbGT47_2, SbGT47_7 and SbGT47_8
The SbGT47_2, SbGT47_7, and SbGT47_8 coding regions were individually fused with GFP at their C-terminus and expressed under control of a 35S cauliflower mosaic virus (CaMV 35S) promoter in the modified pBI221-GFP vector (Chen et al., 2003). The cytoplasmic tail and transmembrane domain of GmMan1 was fused to mCherry at its C-terminus as Golgi marker (Nelson et al., 2007). Arabidopsis transient expression assay was performed following a method described previously (Chai et al., 2015). Protoplasts from 4-week-old rosette leaves were transformed with the PEG-mediated transformation method. The fluorescence was observed using FluoView FV1000 confocal microscope (Olympus, Japan) with an excitation of 488 nm and an emission of 510 nm for GFP, with an excitation of 580 nm and an emission of 610 nm for mCherry.
Complementation of mur3-3 by SbGT47_2, SbGT47_7 or SbGT47_8 Overexpression
The SbGT47_2, SbGT47_7, and SbGT47_8 coding sequence were individually ligated into the pCAMBia1300-GFP vector (Kong et al., 2009) to generate the overexpression constructs. The resulting constructs were transformed into Agrobacterium tumefaciens strain (EHA105) by electroporation. After confirmation that the A. tumefaciens strain contained the proper gene, pCAMBia1300-gene-GFP was introduced into 5-week-old Arabidopsis mur3-3 mutant by using A. tumefaciens–mediated floral dip method (Zhang et al., 2006). T0 transgenic plants were screened on 1/2 MS plates containing 20 mg/L hygromycin, and T3 homologous lines were used for subsequent analysis.
Cell Wall Compositions
Rosette leaves of 4-week-old Arabidopsis plants and inflorescence stems of 7-week-old plants were sampled for quantification of cell wall compositions as previously described (Chai et al., 2015). To ensure the accuracy of the data, WT, mur3-3 and three independent complemented lines for each SbGT47 gene were selected. Alcohol insoluble residues (AIR) were prepared by gradient ethanol extraction of the sample powder for 30 min per gradient, and then soaked with 100% acetone for 2 h at 37°C. The starch in rosette leaves was degraded with α-amylase and amyloglucosidase (Sigma-Aldrich, United States).
To measure monosaccharide compositions, AIRs were hydrolyzed with 2 M trifluroacetic acid (TFA) at 120°C for 2 h (Balaghi et al., 2011; Yu et al., 2014). The hydrolysates were incubated with 1-phenyl-3-methyl-5-pyrazolone (PMP) for derivatization at 70°C for 30 min and extracted by chloroform for three times. The PMP derivatives were analyzed on a Waters high performance liquid chromatography (HPLC) system with a Thermo ODS-2 C18 column (4.6 × 250 mm) and a 2489 UV visible detector. To determine crystalline cellulose content, TFA-resistant materials were incubated with Updegraff reagent (acetic acid/nitric acid/water, 8:1:2 v/v) at 100°C for 30 min, and completely hydrolyzed with 72% H2SO4 (v/v) (Updegraff, 1969). Crystalline cellulose content was calculated by measurement of glucose level and a dehydration factor of 0.9 using a glucose assay kit (Cayman Chemical, United States) following our previous method (Chai et al., 2015). To detect lignin content, about 3 mg AIRs were solubilized by acetyl bromide solution, and 2 M sodium hydroxide and 0.5 M hydroxylamine hydrochloride were added to stop the reaction (Fukushima and Hatfield, 2001). Absorbance at 280 nm was recorded by an UV-visible spectrophotometer of VARIAN Cary 50 (VARIAN, United States). Percentage of acetyl bromide soluble lignin (% ABSL) was calculated by the formula (% ABSL = 0.236 × absorbance at 280 nm/weight of AIR).
MALDI-TOF MS Analysis
XyG oligosaccharides (XyGOs) were obtained by treating suspensions of the AIR mentioned above and solutions of the 4 M KOH-soluble materials in 50 mm ammonium formate, pH 5, with 2 units of xyloglucan-specific endoglucanase (XEG) as described (Pauly et al., 1999). Ethanol was added to 70% (v/v) and the soluble fraction was concentrated to dryness. The residue was dissolved in water and repeatedly freeze-dried to ensure removal of ammonium formate. The residue was then dissolved in water (1 mL) and the XyGOs enriched using graphitized carbon (Packer et al., 1998).
MALDI-TOF MS was performed in the positive ion mode using a Bruker Microflex spectrometer and workstation (Bruker, Billerica, MA, United States). Solutions (5 μL) of XyGOs (∼1 mg/mL in water) were mixed with an equal volume of 10 mM NaCl. A portion of this mixture (1 μL) was then added to 0.1 M dihydroxybenzoic acid (1 μL, 10 mg/mL in aqueous 50% acetonitrile) on the MALDI target plate and concentrated to dryness under a flow of warm air. Spectra from at least 200 laser shots were summed up to generate each mass spectrum.
Statistical Analysis
Data were statistically analyzed using the one-way analysis of variance (ANOVA). Asterisks denote significant differences between two groups of data (∗P < 0.05; ∗∗P < 0.01). Significant differences is represent by different letter in the analysis of cell wall compositions (P < 0.05).
Results
Identification and Phylogenetic Analyses of 468 GT47 Proteins From Ten Species
A total of 468 GT47 proteins were identified in an alga (38), moss (57), four members of the grass family (rice, 41; switchgrass, 75; sorghum, 39; and stiff brome, 34), and four dicots (Arabidopsis, 39; alfalfa, 48; poplar, 62; and grape, 35) (Supplementary Table S1). The 468 GT47 proteins can be divided into nine clades (GT47-A to F) in the phylogenetic analysis (Supplementary Figure S1). The numbers of GT47 proteins from sorghum and other three monocots were obviously larger than those from the dicots in GT47-A, -B, and -D2, but smaller in GT47-C1, -C2, and -C3, similar to the distribution of sorghum GT8, GT34, and GT37 family members (Rai et al., 2016). GT47-E covered almost all GT47 genes from the chlorophytic alga C. reinhardtii, but only one copy in each plant species except allotetraploid switchgrass, consistent with a recent report that the GT47 genes from C. reinhardtii are clustered into an ancient clade in phylogenetic analysis (Tan et al., 2018).
Phylogenetic Analyses of the Sorghum GT47 Proteins
Thirty-nine sorghum GT47 proteins were named as from SbGT47_1 to SbGT47_39 following the previously available nomenclature (Chai et al., 2012). These proteins contained one exostosin motif and varied from 206 to 783 amino acids (aa) in length with an average of 485 aa. It’s noteworthy that 87.2% (34/39) of GT47 proteins had a transmembrane region. The details on other parameters of nucleic acid and protein sequences were provided in Table 1 and Supplementary Table S1.
Table 1.
Sequence characteristics of 39 sorghum GT47 genes.
| Gene symbol | Gene locus | CDS(bp) | Exons | Protein |
Transmembrane | ||
|---|---|---|---|---|---|---|---|
| Length(aa) | Mol. Wt (kD) | PI | |||||
| SbGT47_1 | Sobic.001G228900.1 | 1380 | 2 | 459 | 51.10 | 8.59 | 1 |
| SbGT47_2 | Sobic.001G229000.1 | 1842 | 1 | 613 | 67.55 | 6.54 | 0 |
| SbGT47_3 | Sobic.001G229100.1 | 1473 | 1 | 490 | 54.41 | 9.14 | 0 |
| SbGT47_4 | Sobic.001G303300.1 | 1353 | 1 | 450 | 50.49 | 8.28 | 0 |
| SbGT47_5 | Sobic.001G387300.1 | 1389 | 2 | 462 | 52.03 | 7.85 | 1 |
| SbGT47_6 | Sobic.001G486900.1 | 2352 | 14 | 783 | 88.59 | 6.83 | 1 |
| SbGT47_7 | Sobic.001G506300.1 | 1851 | 1 | 616 | 69.75 | 5.81 | 1 |
| SbGT47_8 | Sobic.001G506500.1 | 1641 | 1 | 514 | 58.39 | 8.10 | 1 |
| SbGT47_9 | Sobic.001G506600.1 | 1563 | 2 | 520 | 58.52 | 8.47 | 1 |
| SbGT47_10 | Sobic.001G506700.1 | 1539 | 2 | 512 | 57.02 | 9.96 | 1 |
| SbGT47_11 | Sobic.001G506800.1 | 1590 | 2 | 529 | 58.95 | 9.78 | 0 |
| SbGT47_12 | Sobic.001G506900.1 | 1542 | 2 | 513 | 57.14 | 8.52 | 0 |
| SbGT47_13 | Sobic.001G538700.1 | 1290 | 3 | 429 | 48.55 | 9.85 | 1 |
| SbGT47_14 | Sobic.001G541600.1 | 621 | 1 | 206 | 22.70 | 6.50 | 0 |
| SbGT47_15 | Sobic.002G062100.1 | 1755 | 3 | 584 | 63.59 | 8.80 | 1 |
| SbGT47_16 | Sobic.002G342300.1 | 1371 | 3 | 500 | 55.79 | 9.70 | 1 |
| SbGT47_17 | Sobic.003G102700.1 | 1716 | 8 | 566 | 63.92 | 9.31 | 1 |
| SbGT47_18 | Sobic.003G234701.1 | 1515 | 4 | 307 | 35.59 | 10.3 | 0 |
| SbGT47_19 | Sobic.003G331000.1 | 1500 | 8 | 499 | 56.27 | 9.07 | 1 |
| SbGT47_20 | Sobic.003G360300.1 | 1281 | 8 | 426 | 48.56 | 9.91 | 1 |
| SbGT47_21 | Sobic.003G405600.1 | 1299 | 4 | 437 | 48.78 | 9.44 | 1 |
| SbGT47_22 | Sobic.003G410600.1 | 1248 | 2 | 420 | 47.28 | 6.57 | 1 |
| SbGT47_23 | Sobic.003G410700.1 | 1254 | 4 | 417 | 46.90 | 6.90 | 0 |
| SbGT47_24 | Sobic.003G410800.1 | 1263 | 4 | 420 | 47.15 | 6.90 | 1 |
| SbGT47_25 | Sobic.004G070100.1 | 1212 | 5 | 408 | 46.29 | 9.25 | 1 |
| SbGT47_26 | Sobic.004G159100.1 | 1299 | 4 | 437 | 48.93 | 8.15 | 1 |
| SbGT47_27 | Sobic.004G213500.1 | 1413 | 3 | 475 | 53.08 | 9.65 | 1 |
| SbGT47_28 | Sobic.006G059000.1 | 1293 | 4 | 435 | 48.64 | 7.15 | 1 |
| SbGT47_29 | Sobic.006G186100.1 | 1707 | 2 | 573 | 63.03 | 8.47 | 1 |
| SbGT47_30 | Sobic.006G186200.1 | 1440 | 1 | 484 | 53.04 | 8.93 | 1 |
| SbGT47_31 | Sobic.006G260900.1 | 1371 | 3 | 461 | 52.27 | 9.19 | 1 |
| SbGT47_32 | Sobic.007G139300.1 | 1572 | 2 | 528 | 59.29 | 10.3 | 1 |
| SbGT47_33 | Sobic.008G021000.1 | 1488 | 2 | 500 | 55.13 | 8.98 | 1 |
| SbGT47_34 | Sobic.008G077900.1 | 1572 | 2 | 528 | 58.96 | 6.03 | 1 |
| SbGT47_35 | Sobic.008G138100.1 | 1761 | 1 | 591 | 65.75 | 9.25 | 1 |
| SbGT47_36 | Sobic.009G162700.1 | 1269 | 2 | 427 | 47.54 | 6.83 | 1 |
| SbGT47_37 | Sobic.009G220100.1 | 1248 | 1 | 420 | 46.81 | 6.83 | 1 |
| SbGT47_38 | Sobic.009G220200.1 | 1251 | 4 | 421 | 47.51 | 6.94 | 0 |
| SbGT47_39 | Sobic.010G059400.1 | 1530 | 4 | 551 | 61.69 | 9.84 | 0 |
Molecular Wt and PI of sorghum GT47 proteins have been calculated using DNAman software. Transmembrane domain is predicted by TMHMM Server v. 2.0.
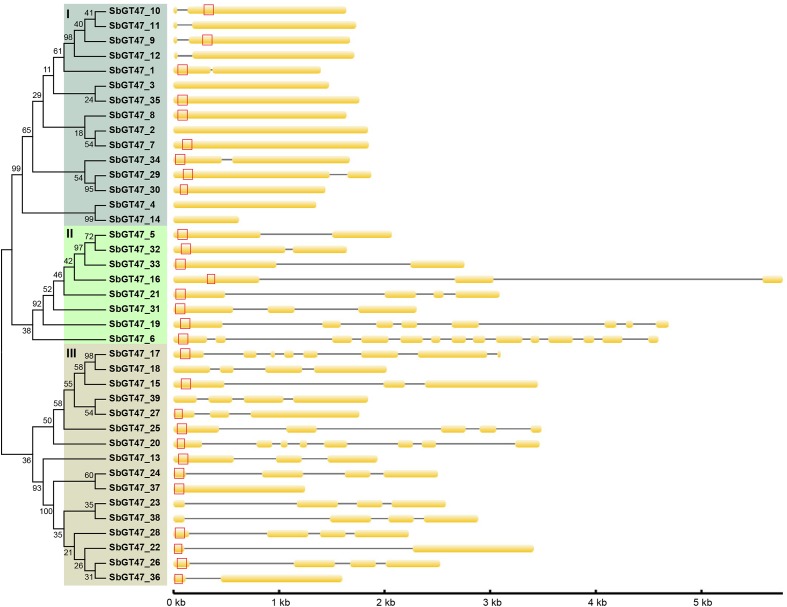
To evaluate the evolutionary relationships between sorghum GT47 members, a phylogenetic analyze was performed based on full-length protein or exostosin motif sequences. Phylogenetic trees generated by the NJ and MP algorithms were largely comparable with only minor modifications at interior branches (Figure 1 and Supplementary Figure S1). The 39 GT47 proteins were generally divided into four subfamilies (I-IV). Within each subfamily, most closely related members shared similar exon/intron structures either in terms of intron numbers or exon length, which was roughly cconsistent with the classification defined in the phylogenetic tree. For instance, the GT47 genes in subfamily I contained zero or one intron, while those in subfamily IV possessed two to three introns with the exception of SbGT47_36 and SbGT47_37. In addition, ten gene pairs were identified from the 39 GT47 genes based on the >50% bootstrap values in phylogenetic analysis (Figure 1).
FIGURE 1.
Phylogenetic relationship and gene structure of sorghum GT47 genes. Full-length proteins were aligned with Clustal X 1.83 and the phylogenetic tree was constructed using MEGA 4.0 by the method of Neighbor-Joining (NJ) with 1,000 bootstrap replicates. Orange box and black line show exon and intron, respectively. Transmembrane domain predicted by TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) is shown with red box. Gene size can be estimated by the scale (bottom).
Chromosomal Location of the 39 GT47 Genes
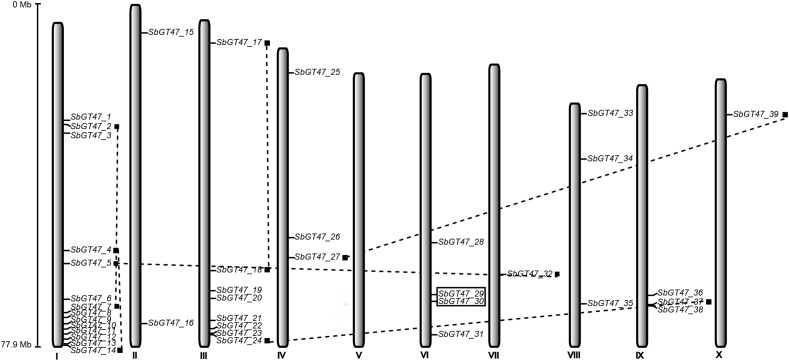
The 39 sorghum GT47 genes were located on 10 linkage groups (LG) of sorghum chromosome (Figure 2). Their distributions among the chromosomes appeared to be uneven: LG II, IV, VI, VII, VIII, IX, and X harbor no more than four GT47 genes, while relatively high densities of GT47 genes (22/39, 56.4%) were discovered in some locations on LG I and III. Ten GT47 gene pairs were widely distributed on 7 chromosomes. Of them, two counterparts of two adjacent gene pairs (SbGT47_10/SbGT47_11 and SbGT47_29/SbGT47_30) have high protein sequence similarities (>70%) calculated by the Smith-Waterman algorithm7 and were distributed within a distance less than 6.5 kb on a chromosome (Figure 2), indicating that they may originate from tandem duplication during the evolutionary process.
FIGURE 2.
Chromosomal locations of sorghum GT47 genes. Eight of ten putative gene pairs are connected by dashed lines, other two resulted from tandem duplication are encompassed in black boxes. Scale represents the length of chromosome II.
Expression Patterns of Sorghum GT47 Genes in Various Tissues
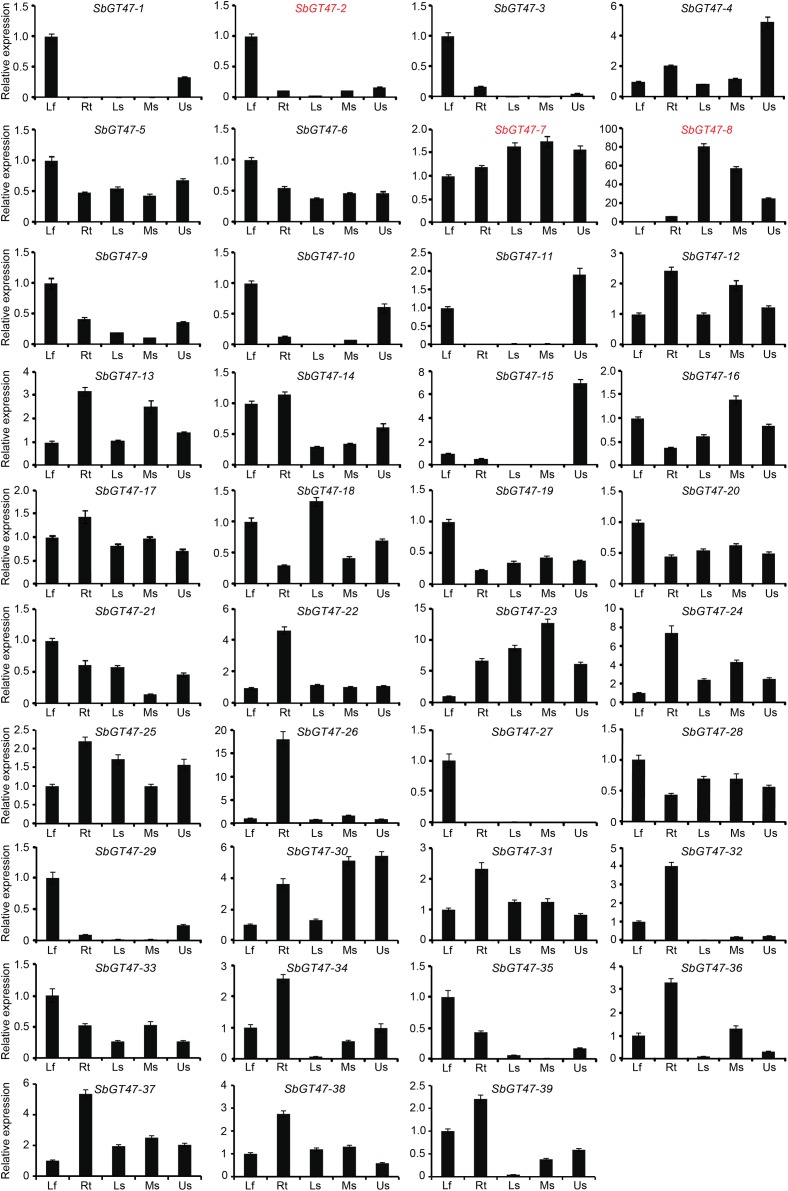
To gain insight into the tissue-specific expression patterns of the 39 sorghum GT47 genes, qRT-PCR assays were performed in young leaves, roots, basal stems, middle stems, and upper stems of sorghum plants (Figure 3). 41% (16/39) of genes displayed tissue-specific expression patterns. Of them, eight genes (SbGT47_1, SbGT47_2, SbGT47_3, SbGT47_9, SbGT47_10, SbGT47_27, SbGT47_29, and SbGT47_35) were specifically expressed in young leaves, six genes (SbGT47_22, SbGT47_24, SbGT47_26, SbGT47_32, SbGT47_34, and SbGT47_36) in roots, and two genes (SbGT47_8 and SbGT47_15) in stems. Interestingly, eight genes displayed distinct expression patterns across stem segments. SbGT47_1, SbGT47_4, SbGT47_10, SbGT47_11, SbGT47_15, and SbGT47_29 were predominantly expressed in the upper stem, whereas transcripts of SbGT47_8 and SbGT47_18 were gradually increased from the upper to basal stem.
FIGURE 3.
Tissue expression profiles of sorghum GT47 genes detected by qRT-PCR. SbGT47_2, SbGT47_7, and SbGT47_8 marked with red were selected for subsequent analysis. Lf, leaves; Rt, roots; BS, basal stems; MS, middle stems; US, upper stems. SbACTIN2 was used as an internal control. Bar represents standard deviations (SD) from three biological repeats.
Tissue expression patterns of ten pairs of genes were obviously classified into two categories (Figure 3). The first category was composed of four gene pairs (SbGT47_10/11, SbGT47_3/35, SbGT47_17/18, and SbGT47_24/18), of which two gene duplicates exhibited similar expression patterns with respect to the tissues examined, implying the functional similarity between each other. In the second category, both duplicates of six gene pairs (SbGT47_2/7, SbGT47_29/30, SbGT47_4/14, SbGT47_5/32, SbGT47_29/27, and SbGT47_23/38) shared divergent expression patterns. For example, SbGT47_2 was preferentially expressed in leaves, while SbGT47_7 was constitutively expressed in all six tissues detected. This suggests that two counterparts in the six gene pairs might have undergone substantial divergence after duplication.
Functional Analysis of Three Sorghum MUR3-Like Genes
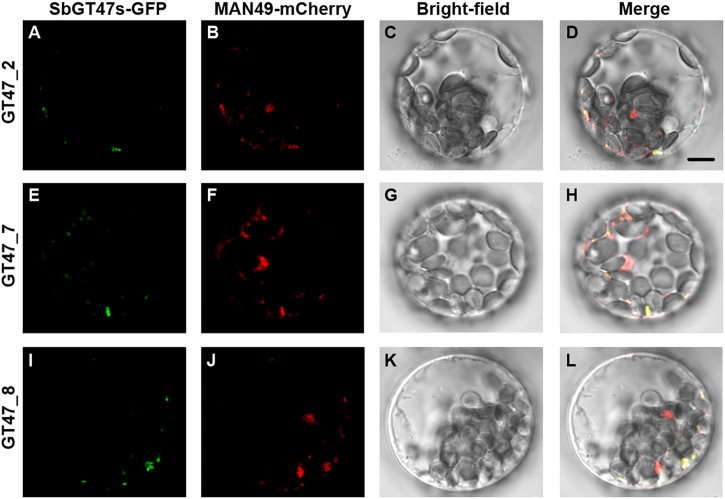
Arabidopsis MUR3 is involved in the modification side chain substitutions of xyloglucan, which has important roles in the formation of structural maintenance of primary cell walls (Madson et al., 2003; Kong et al., 2015). Here, we identified ten orthologs (SbGT47_1, _2, _3, _7, _8, _9, _10, _11, _12, and _35) of MUR3 in sorghum and found that they displayed distinct expression patterns across various tissue (Figure 3). To investigate putative divergences in their functions, SbGT47_2, SbGT47_7 and SbGT47_8, three most homologous genes of MUR3 (Supplementary Figure S1), were selected for subsequent analysis. Their subcellular localizations were first determined in Arabidopsis leave protoplasts. Similar to MUR3 (Chou et al., 2015), SbGT47_2-GFP, SbGT47_7-GFP or SbGT47_8-GFP was perfectly co-expressed with MAN49-mCherry (a Golgi marker) (Figure 4), indicating that SbGT47_2, SbGT47_7 and SbGT47_8 are located in Golgi apparatus.
FIGURE 4.
Subcellular localization of SbGT47_2, SbGT47_7 and SbGT47_8. SbGT47s-GFP and MAN49-mCherry (a Golgi marker) were transiently co-expressed in Arabidopsis leaf protoplasts. The localization of SbGT47_2 (A), SbGT47_7 (E) and SbGT47_8 (I) were detected by green fluorescence, while Golgi was marked by MAN49 with red fluorescence (B,F,J). Besides, bright-field images (C,G,K) and merged images (D,H,L) were also acquired for subcellular localization analysis. Bar = 10μm.
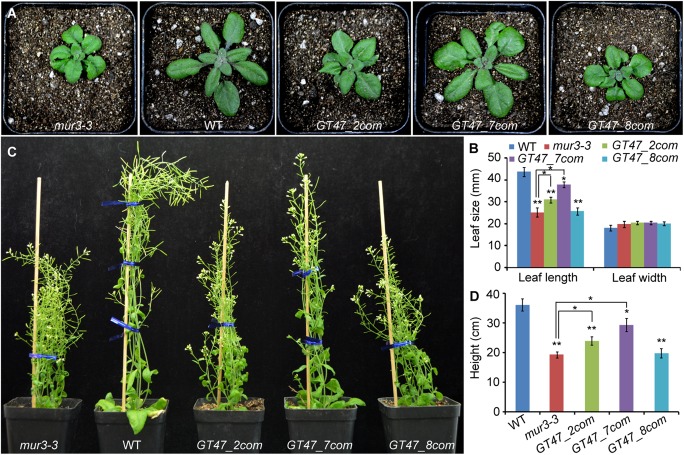
To determine the biological functions of SbGT47_2, SbGT47_7, and SbGT47_8, their overexpression constructs were individually introduced into Arabidopsis mur3-3 to generate transgenic plants called GT47_2com, GT47_7com and GT47_8com. At least 30 transgenic lines for each construct were obtained and the expression of these genes was confirmed by using RT-PCR (Supplementary Figure S2). T3 homozygous transgenic lines for each construct exhibited similar phenotypes. The cabbage-like and dwarf phenotypes of mur3-3 were largely rescued by SbGT47_7 overexpression, slightly rescued by SbGT47_2 overexpression, but not affected by SbGT47_8 overexpression, which were supported by the statistic data of leaf length and height in the transgenic plants (Figure 5). These results indicated functional divergences between sorghum SbGT47_2, SbGT47_7, and SbGT47_8. It is noteworthy that there was no observable difference for leaf width between wild-type, mur3-3 and transgenic plants (Figure 5B).
FIGURE 5.
Phenotypes of four- (A,B) and 7-week-old (C,D) WT, mur3-3 and representative transgenic plants overexpressing SbGT47_2 (GT47_2com), SbGT47_7 (GT47_7com) or SbGT47_8 (GT47_8com) in mur3-3. For each genotype, at least 20 independent plants were used for measurement of leaf size and plant height. Asterisks represent the significant difference between two groups (∗P < 0.05; ∗∗P < 0.01).
To investigate whether overexpression of SbGT47_2, SbGT47_7, or SbGT47_8 in mur3-3 alters the composition of cell walls, monosaccharide composition analysis was performed in leaves and stems. The level of fucose, a major side-chain component of xyloglucans (XyG) solubilized from primary cell wall material, was dramatically reduced in AIR of the mur3-3 leaves compared with the wild type (Table 2). Similarly, galactose content was also reduced in the mur3-3 leaves. The relative abundances of all other detected monosaccharaides (xylose, arabinose, glucose, mannose, rhamnose and glucuronic acid) were not significantly different between two genotypes. These results were consistent with a previous report (Madson et al., 2003). In leaves, ectopic expression of SbGT47_7 in mur3-3 restored fucose and galactose contents to wild type-like level, whereas only fucose content was recovered by SbGT47_2 overexpression. In contrast, the reduced fucose and galactose levels in mur3-3 were not changed by SbGT47_8 overexpression. These results strongly suggest that the three sorghum MUR3-like proteins may have different GT activities in leaves. Interestingly, mutation of MUR3 led to significant alterations of xylose, arabinose, glucose, mannose and fucose contents in stems (Table 2). The contents of the five monosaccharaides in mur3-3 were recovered by SbGT47_2 and SbGT47_7 overexpression to varying degrees, while only xylose was restored by SbGT47_8 overexpression. WT and mur3-3 showed similar galactose levels in 7-week-old inflorescence stems, possibly due to the dominance of other galactose containing polymers in secondary cell walls. Compared with the wild type, the levels of cellulose and lignin, other two major components of secondary cell walls, were markedly reduced in the mur3-3 leaves or stems. Overexpression of SbGT47_2, SbGT47_7, or SbGT47_8 in mur3-3 resulted in an increase in cellulose and lignin contents to various extents.
Table 2.
Cell wall compositions of wild-type, mur3-3 and complemented transgenic plants.
| Galacturonic | Glucuronic | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tissue | Samples | Cellulose | Xylose | Arabinose | Glucose | Mannose | Galactose | Rhamnose | acid | acid | Fucose | Lignin |
| Leaf | WT | 27.3±2.59b | 13.8±1.34a | 10.3±0.52a | 26.5±1.08a | 23.3±1.17a | 31.2±1.62a | 12.3±0.98a | 45.0±2.03a | 5.1±0.48a | 4.1±0.36a | 34.1±1.30a |
| mur3-3 | 22.4±1.36c | 14.0±3.51a | 10.1±1.54a | 29.0±2.04a | 19.7±2.31a | 25.5±1.82b | 12.5±0.85a | 43.9±4.97a | 6.0±.059a | 1.7±0.36c | 29.2±2.09b | |
| SbGT47_2com | 30.0±3.02ab | 14.1±0.68a | 10.7±0.21a | 28.6±0.27a | 20.4±2.95a | 28.4±0.87b | 12.5±0.38a | 42.6±2.91a | 5.4±0.79a | 2.7±0.05b | 32.0±2.95a | |
| SbGT47_7com | 34.1±2.89a | 11.7±1.32a | 10.1±1.07a | 27.7±1.93a | 20.5±2.10a | 29.8±1.41a | 11.2±0.74a | 43.6±3.52a | 5.8±0.37a | 3.7±0.20a | 35.1±3.78a | |
| SbGT47_8com | 27.0±0.78b | 12.3±0.75a | 11.4±0.46a | 24.8±1.74a | 23.3±2.34a | 26.8±1.89b | 12.5±0.73a | 39.7±4.11a | 5.9±0.60a | 2.2±0.21c | 34.8±3.07a | |
| Stem | WT | 484.5±28.4ab | 208.2±4.59a | 10.7±0.47d | 13.8±1.08c | 36.3±2.46a | 19.7±0.70a | 17.1±0.12a | 45.3±3.97a | 2.8±0.42a | 1.5±0.13a | 60.9±5.78a |
| mur3-3 | 333.1±25.7c | 159.7±8.62c | 20.5±1.56a | 21.2±2.65ab | 28.5±1.05b | 19.2±1.16a | 15.8±2.10a | 46.5±3.38a | 2.2±0.55a | 0.5±0.05c | 48.8±2.92c | |
| SbGT47_2com | 463.7±16.5ab | 186.2±9.28b | 18.4±0.36b | 17.1±0.77b | 29.1±1.92b | 19.1±0.23a | 16.1±0.73a | 43.8±3.17a | 2.0±0.32a | 0.7±0.10b | 51.3±3.39bc | |
| SbGT47_7com | 498.3±11.6a | 189.1±4.50b | 13.5±0.93c | 15.9±2.46bc | 30.6±2.43ab | 19.4±1.14a | 17.5±1.54a | 45.5±2.04a | 3.1±0.85a | 1.6±0.22a | 52.0±1.48bc | |
| SbGT47_8com | 435.2±19.3b | 189.6±8.12b | 20.88±0.54a | 21.7±0.98a | 29.1±2.48b | 20.3±0.14a | 17.4±1.64a | 46.0±2.51a | 2.6±0.29a | 0.5±0.10c | 55.5±2.67b |
Values are mean ± SE (n = 3) with the unit (mg/g). Means with the different letter are significantly different (One-way ANOVA, Duncan’s test, p < 0.05).
MALDI-TOF MS Analysis of XyG Structure in Three MUR3-Like Complemented Plants
To determine whether SbGT47_2, SbGT47_7 or SbGT47_8, like MUR3, affects XyG structure, XyG oligosaccharides (XyGOs) derived from leaf AIRs of 4-week-old WT, mur3-3 and complemented plants were analyzed by MALDITOF MS. Consistent with a previous report (Kong et al., 2015), all galactosylation (XXLG) and fucosylation (XXFG and XLFG) ions were lost in mur3-3 (Figure 6). In the SbGT47_2 and SbGT47_7 complemented plants, the peaks corresponding to m/z 1,247 (XXLG/XLXG), 1,393 (XXFG), 1,410 (XLLG), and 1,555 (XLFG) were restored, confirming that SbGT47_2 and SbGT47_7 possess a xyloglucan galactosyltransferase activity. In contrast, the XyG structure was not restored in SbGT47_8 complemented plants, indicating that this complemented line cannot be considered to contain a functional MUR3.
FIGURE 6.
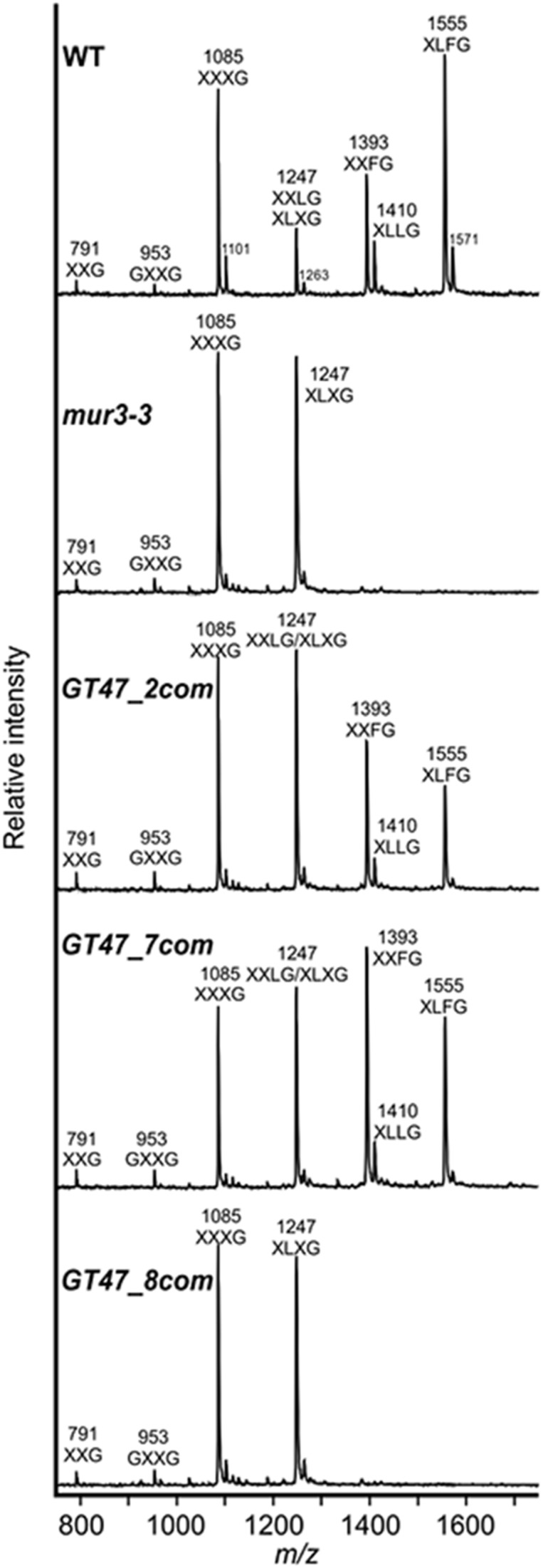
XyG MALDI-TOF mass spectra derived leaf cell wall material of 4-week-old WT, mur3-3, GT47_2com, GT47_7com, and GT47_8com. Ions representing known XyGOs are labeled using the one-letter code according to the nomenclature reported by Tuomivaara et al. (2015).
Discussion
In dicot plants, xyloglucan is one of major non-cellulosic polysaccharides in primary cell walls (Gibeaut et al., 2005). Arabidopsis MUR3 transforms the galactosyl residue from UDP-galactose to the third xylose (L side chain) of XXXG-type, forming XXLG subunits (Madson et al., 2003; Jensen et al., 2012). MUR3Gal is often substituted at O-2 with a α-L-Fucp residue (F side chain: XXFG and XLFG) by GT37 family member FUCOSYLTRANSFERASE1 (FUT1) (Perrin et al., 1999). In this study, forty-six orthologs of MUR3 were identified in the seed plants selected, one in moss (Zhu et al., 2018), but none in the chlorophytic alga C. reinhardtii (Supplementary Figure S1). Mikkelsen et al. (2014) reported that the next level of xyloglucan side-chain biosynthesis genes are present in the charophytic algae Penium margaritaceum, which are the closest living relatives of land plants (Nishiyama et al., 2018). It is possible that glalactoxyloglucan is synthesized by enzymes with the MUR3 activity within charophyte algae, similar to the origination of plant 1,4-β-xylan (Jensen et al., 2018). At least seven orthologs of MUR3 were found in each of the four monocots, and drastically more than those (1–2 orthologs) in each of the four dicots, indicating that gene duplication may play a major role in the expansion of MUR3 in monocots. In addition to MUR3, XLT2 adds galactose to the second xylosyl residue of XXXG, producing XLXG, and XUT and XDT add galacturonic acid and arabinopyranose, respectively, to the O2-position of a xylosyl residue on xyloglucan (Jensen et al., 2012; Peña et al., 2012; Zhu et al., 2018). Here, 15 orthologs of XLT2 were found in all species except algae, 6 orthologs of XUT in dicots, one ortholog of XDT in moss but no ortholog of XST in all species detected (Supplementary Figure S1). These results suggest the specificity of side chain residue in xyloglucan structure among species.
Sorghum has ten orthologs of MUR3 and eight orthologs of IRX10/IRX10L, accounting for 46% of 39 GT47 genes (Supplementary Figure S1), suggesting the functional redundancy of these genes in xyloglucan and xylan biosynthesis in sorghum. 90% (9/10) of MUR3-like genes were arranged in clusters on LG I, and displayed various tissue expression patterns (Figures 2, 3). Physiological and cell wall analysis of transgenic plants expressing SbGT47_2, SbGT47_7, or SbGT47_8, three most homologous genes of MUR3, in mur3-3 confirmed that SbGT47_2 and SbGT47_7 represent functional equivalent to MUR3 (Figures 5, 6 and Table 2). SbGT47_7 showed stronger capability complement to mur3-3 than SbGT47_2, correlating with its higher similarity to MUR3 in both nucleotide sequence and tissue expression pattern (Schmid et al., 2005). Therefore, SbGT47_7 can be considered a fully functional xyloglucan MUR3 ortholog. These results suggest that the biosynthesis of xyloglucan structure in primary cell walls of sorghum may require MUR3-like genes, at least SbGT47_2 and SbGT47_7. Also, sorghum, like rice (Liu et al., 2015), may have the ability to synthesize fucogalactoXyG in vivo.
Overexpressing tomato SlMUR3 or rice OsMUR3 in the Arabidopsis xlt2mur3.1 mutant leads to a rescue of the dwarfed growth phenotype of plants (Schultink et al., 2013; Liu et al., 2015). Here, genetic and biochemical results showed that ectopic expression of sorghum SbGT47_7 in mur3-3 fully complemented the cabbage-like phenotype and defect in XyG structure, and the levels of cellulose, xylose and lignin were also restored in the stems of complemented lines (Table 2). It is generally believed that altered abundance of secondary wall polymers in mur3 is a consequence of the stunted growth but not a direct effect of gene dysfunction (Liu et al., 2015). Thus, SbGT47_7 may indirectly affect the accumulation of cellulose, xylan and lignin in secondary cell walls of sorghum stems.
Author Contributions
HX and GC designed the experiments, interpreted the results and wrote the paper. SC, DW, and RH participated in the data mining. AD, PM, and MC helped in sorghum materials collection and qRT-PCR detection. MO’N performed the MALDI-TOF MS analysis. GZ and YK analyzed the cell wall compositions and critically revised the manuscript. All authors approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This study was supported by grants from the National Key Technology Support Program of China (2015BAD15B03-05), National Natural Science Foundation of China (31670606, 51509237, 31470291, and 31600237), Key Research and Development Program of Shandong Province (2017CXGC0309), Major Science and Technology Projects of Shandong Province (2015ZDJS03002), US Department of Energy Grant DE-FG02-12ER16324, the Taishan Scholar Program of Shandong Province (to GZ), and Youth Innovation Promotion Association of CAS (2017254).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01773/full#supplementary-material
Phylogeny of GT47 homologous proteins from ten representative species.
Identification of SbGT47_2, SbGT47_7 and SbGT47_8 complemented transgenic plants.
Protein sequences of GT47 family members from ten species (doi: 10.6084/m9.figshare.6958895).
Primers in this study.
References
- Balaghi S., Mohammadifar M. A., Zargaraan A., Gavlighi H. A., Mohammadi M. (2011). Compositional analysis and rheological characterization of gum tragacanth exudates from six species of Iranian Astragalus. Food Hydrocolloid 25 1775–1784. 10.1016/j.foodhyd.2011.04.003 [DOI] [Google Scholar]
- Bauer S., Vasu P., Persson S., Mort A. J., Somerville C. R. (2006). Development and application of a suite of polysaccharide-degrading enzymes for analyzing plant cell wall. Proc. Natl. Acad. Sci. U.S.A. 103 11417–11422. 10.1073/pnas.0604632103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. M., Goubet F., Wong V. W., Goodacre R., Stephens E., Dupree P., et al. (2007). Comparison of five xylan synthesis mutants reveals new insight into the mechanisms of xylan synthesis. Plant J. 52 1154–1168. 10.1111/j.1365-313X.2007.03307.x [DOI] [PubMed] [Google Scholar]
- Brown D. M., Zhang Z., Stephens E., Dupree P., Turner S. R. (2009). Characterization of IRX10 and IRX10-like reveals an essential role in glucuronoxylan biosynthesis in Arabidopsis. Plant J. 57 732–746. 10.1111/j.1365-313X.2008.03729.x [DOI] [PubMed] [Google Scholar]
- Burton R. A., Fincher G. B. (2012). Current challenges in cell wall biology in the cereals and grasses. Front. Plant Sci. 15:130. 10.3389/fpls.2012.00130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton R. A., Wilson S. M., Hrmova M., Harvey A. J., Shirlev N. J., Medhurst A., et al. (2006). Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-beta-D-glucans. Science 311 1940–1942. 10.1126/science.1122975 [DOI] [PubMed] [Google Scholar]
- Byrt C. S., Grof C. P. L., Furbank R. T. (2011). C4 plants as biofuel feedstocks: optimising biomass production and feedstock quality from a lignocellulosic perspective. J. Integr. Plant Biol. 53 120–135. 10.1111/j.1744-7909.2010.01023.x [DOI] [PubMed] [Google Scholar]
- Chai G., Hu R., Zhang D., Qi G., Zuo R., Cao Y., et al. (2012). Comprehensive analysis of CCCH zinc finger family in poplar (Populus trichocarpa). BMC Genomics 13:253. 10.1186/1471-2164-13-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai G., Kong Y., Zhu M., Yu L., Qi G., Tang X., et al. (2015). Arabidopsis C3H14 and C3H15 have overlapping roles in the regulation of secondary wall thickening and anther development. J. Exp. Bot. 66 2595–2609. 10.1093/jxb/erv060 [DOI] [PubMed] [Google Scholar]
- Chen P. Y., Wang C. K., Soong S. C., To K. Y. (2003). Complete sequence of the binary vector pBI121 and its application in cloning T-DNA insertion from transgenic plants. Mol. Breed. 11 287–293. 10.1023/A:1023475710642 [DOI] [Google Scholar]
- Chou Y. H., Pogorelko G., Young Z. T., Zabotina O. A. (2015). Protein-protein interactions among xyloglucan-synthesizing enzymes and formation of Golgi-localized multiprotein complexes. Plant Cell Physiol. 56 255–267. 10.1093/pcp/pcu161 [DOI] [PubMed] [Google Scholar]
- Cocuron J. C., Lerouxel O., Drakakaki G., Alonso A. P., Liepman A. H., Keegstra K., et al. (2007). A gene from the cellulose synthase-like C family encodes a beta-1,4 glucan synthase. Proc. Natl. Acad. Sci. U.S.A. 104 8550–8555. 10.1073/pnas.0703133104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corredor D. Y., Salazar J. M., Hohn K. L., Bean S., Bean B., Wang D. (2009). Evaluation and characterization of forage sorghum as feedstock for fermentable sugar production. Appl. Biochem. Biotech. 158 164–179. 10.1007/s12010-008-8340-y [DOI] [PubMed] [Google Scholar]
- Faik A., Price N. J., Raikhel N. V., Keegstra K. (2002). An Arabidopsis gene encoding an alpha-xylosyltransferase involved in xyloglucan biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 99 7797–7802. 10.1073/pnas.102644799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima R. S., Hatfield R. D. (2001). Extraction and isolation of lignin for utilization as a standard to determine lignin concentration using the acetyl bromide spectrophotometric method. J. Agric. Food Chem. 49 3133–3139. 10.1021/jf010449r [DOI] [PubMed] [Google Scholar]
- Gibeaut D. M., Pauly M., Bacic A., Fincher G. B. (2005). Changes in cell wall polysaccharides in developing barley (Hordeum vulgare) coleoptiles. Planta 221 729–738. 10.1007/s00425-005-1481-0 [DOI] [PubMed] [Google Scholar]
- Jensen J. K., Busse-Wicher M., Poulsen C. P., Fangel J. U., Smith P. J., Yang J. Y., et al. (2018). Identification of an algal xylan synthase indicates that there is functional orthology between algal and plant cell wall biosynthesis. New Phytol. 218 1049–1060. 10.1111/nph.15050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J. K., Johnson N. R., Wilkerson C. G. (2014). Arabidopsis thaliana IRX10 and two related proteins from psyllium and Physcomitrella patens are xylan xylosyltransferases. Plant J. 80 207–215. 10.1111/tpj.12641 [DOI] [PubMed] [Google Scholar]
- Jensen J. K., Schultink A., Keegstra K., Wilkerson C. G., Pauly M. (2012). RNA-seq analysis of developing nasturtium seeds (Tropaeolum majus): identification and characterization of an additional galactosyltransferase involved in xyloglucan biosynthesis. Mol. Plant 5 984–992. 10.1093/mp/sss032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y., Peña M. J., Renna L., Avci U., Pattathil S., Tuomivaara S. T., et al. (2015). Galactose-depleted xyloglucan is dysfunctional and leads to dwarfism in Arabidopsis. Plant Physiol. 167 1296–1306. 10.1104/pp.114.255943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y., Zhou G., Avci U., Gu X., Jones C., Yin Y., et al. (2009). Two poplar glycosyltransferase genes, PdGATL1.1 and PdGATL1.2, are functional orthologs to PARVUS/AtGATL1 in Arabidopsis. Mol. Plant 2 1040–1050. 10.1093/mp/ssp068 [DOI] [PubMed] [Google Scholar]
- Lee C., O’Neill M. A., Tsumuraya Y., Darvill A. G., Ye Z. H. (2007). The irregular xylem9 mutant is deficient in xylan xylosyltransferase activity. Plant Cell Physiol. 48 1624–1634. 10.1093/pcp/pcm135 [DOI] [PubMed] [Google Scholar]
- Liu L. F., Paulitz J., Pauly M. (2015). The presence of fucogalactoxyloglucan and its synthesis in rice indicates conserved functional importance in plants. Plant Physiol. 168 549–560. 10.1104/pp.15.00441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes F. J. F., Pauly M., Brommonshenkel S. H., Lau E. Y., Diola V., Passos J. L., et al. (2010). The EgMUR3 xyloglucan galactosyltransferase from Eucalyptus grandis complements the mur3 cell wall phenotype in Arabidopsis thaliana. Tree Genet. Genomes 6 745–756. 10.1007/s11295-010-0288-8 [DOI] [Google Scholar]
- Madson M., Dunand C., Li X., Verma R., Vanzin G. F., Caplan J., et al. (2003). The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15 1662–1670. 10.1105/tpc.009837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick R. F., Truong S. K., Sreedasyam A., Jenkins J., Shu S., Sims D., et al. (2018). The Sorghum bicolor reference genome: improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 93 338–354. 10.1111/tpj.13781 [DOI] [PubMed] [Google Scholar]
- Mikkelsen M. D., Harholt J., Ulvskov P., Johansen I. E., Fangel J. U., Doblin M. S., et al. (2014). Evidence for land plant cell wall biosynthetic mechanisms in charophyte green algae. Ann. Bot. 114 1217–1236. 10.1093/aob/mcu171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B. K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51 1126–1136. 10.1111/j.1365-313X.2007.03212.x [DOI] [PubMed] [Google Scholar]
- Nishiyama T., Sakayama H., de Vries J., Buschmann H., Saint-Marcoux D., Ullrich K. K., et al. (2018). The chara genome: secondary complexity and implications for plant terrestrialization. Cell 174 448–464. 10.1016/j.cell.2018.06.033 [DOI] [PubMed] [Google Scholar]
- Packer N. H., Lawson M. A., Jardine D. R., Redmond J. W. (1998). A general approach to desalting oligosaccharides released from glycoproteins. Glycoconj. J. 15 737–747. 10.1023/A:1006983125913 [DOI] [PubMed] [Google Scholar]
- Paterson A. H., Bowers J. E., Bruggmann R., Dubchak I., Grimwood J., Gundlach H., et al. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457 551–556. 10.1038/nature07723 [DOI] [PubMed] [Google Scholar]
- Pauly M., Andersen L. N., Kauppinen S., Kofod L. V., York W. S., Albersheim P., et al. (1999). A xyloglucan-specific endo-β-1,4-glucanase from Aspergillus aculeatus: expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology 9 93–100. 10.1093/glycob/9.1.93 [DOI] [PubMed] [Google Scholar]
- Peña M. J., Kong Y. Z., York W. S., O’Neill M. A. (2012). A galacturonic acid-containing xyloglucan is involved in Arabidopsis root hair tip growth. Plant Cell 24 4511–4524. 10.1105/tpc.112.103390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin R. M., DeRocher A. E., Bar-Peled M., Zeng W., Norambuena L., Orellana A., et al. (1999). Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science 284 1976–1979. 10.1126/science.284.5422.1976 [DOI] [PubMed] [Google Scholar]
- Rai K. M., Thu S. W., Balasubramanian V. K., Cobos C. J., Disasa T., Mendu V. (2016). Identification, characterization, and expression analysis of cell wall related genes in Sorghum bicolor (L.) moench, a food, fodder, and biofuel crop. Front. Plant Sci. 7:1287. 10.3389/fpls.2016.01283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter W. D., Chapple C., Somerville C. R. (1997). Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. Plant J. 12 335–345. 10.1046/j.1365-313X.1997.12020335.x [DOI] [PubMed] [Google Scholar]
- Rooney W. L., Blumenthal J., Bean B., Mullet J. E. (2007). Designing sorghum as a dedicated bioenergy feedstock. Biofuels Bioprod. Biorefining 1 147–157. 10.1002/bbb.15 [DOI] [Google Scholar]
- Scheller H. V., Ulvskov P. (2010). Hemicellulose. Annu. Rev. Plant Biol. 61 263–289. 10.1146/annurev-arplant-042809-112315 [DOI] [PubMed] [Google Scholar]
- Schmid M., Davison T. S., Henz S. R., Pape U. J., Demar M., Vingron M., et al. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37 501–506. 10.1038/ng1543 [DOI] [PubMed] [Google Scholar]
- Schultink A., Cheng K., Park Y. B., Cosgrove D. J., Pauly M. (2013). The identification of two arabinosyltransferases from tomato reveals functional equivalency of xyloglucan side chain substituents. Plant Physiol. 163 86–94. 10.1104/pp.113.221788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24 1596–1599. 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- Tan J., Miao Z., Ren C., Yuan R., Tang Y., Zhang X., et al. (2018). Evolution of intron-poor clades and expression patterns of the glycosyltransferase family 47. Planta 247 745–760. 10.1007/s00425-017-2821-6 [DOI] [PubMed] [Google Scholar]
- Taylor S. H., Hulme S. P., Rees M., Ripley B. S., Woodward F. I., Osborne C. P. (2010). Ecophysiological traits in C3 and C4 grasses: a phylogenetically controlled screening experiment. New Phytol. 185 780–791. 10.1111/j.1469-8137.2009.03102.x [DOI] [PubMed] [Google Scholar]
- Tuomivaara S. T., Yaoi K., O’Neill M. A., York W. S. (2015). Generation and structural validation of a library of diverse xyloglucan-derived oligosaccharides, including an update on xyloglucan nomenclature. Carbohydr. Res. 402 56–66. 10.1016/j.carres.2014.06.031 [DOI] [PubMed] [Google Scholar]
- Updegraff D. M. (1969). Semimicro determination of cellulose in biological materials. Anal. Biochem. 32 420–424. 10.1016/S0003-2697(69)80009-6 [DOI] [PubMed] [Google Scholar]
- Urbanowicz B. R., Pena M. J., Moniz H. A., Moremen K. W., York W. S. (2014). Two Arabidopsis proteins synthesize acetylated xylan in vitro. Plant J. 80 197–206. 10.1111/tpj.12643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Rihouey C., Seveno M., Hörnblad E., Singh S. K., Matsunaga T., et al. (2009). The Arabidopsis IRX10 and IRX10-LIKE glycosyltransferases are critical for glucuronoxylan biosynthesis during secondary cell wall formation. Plant J. 54 718–731. 10.1111/j.1365-313X.2008.03724.x [DOI] [PubMed] [Google Scholar]
- Yu L., Shi D. C., Li J. L., Kong Y. Z., Yu Y. C., Chai G. H., et al. (2014). CELLULOSE SYNTHASE-LIKE A2, a glucomannan synthase, is involved in maintaining adherent mucilage structure in Arabidopsis seed. Plant Physiol. 164 1842–1856. 10.1104/pp.114.236596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Henrigues R., Lin S. S., Niu Q. W., Chua N. H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1 641–646. 10.1038/nprot.2006.97 [DOI] [PubMed] [Google Scholar]
- Zhong R., Peña M. J., Zhou G. K., Nairn C. J., Wood-Jones A., Richardson E. A., et al. (2005). Arabidopsis fragile fiber8, which encodes a putative glucuronyltransferase, is essential for normal secondary wall synthesis. Plant Cell 17 3390–3408. 10.1105/tpc.105.035501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Dama M., Pauly M. (2018). Identification of an arabinopyranosyltransferase from Physcomitrella patens involved in the synthesis of the hemicellulose xyloglucan. Plant Direct 2:e00046 10.1002/pld3.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogeny of GT47 homologous proteins from ten representative species.
Identification of SbGT47_2, SbGT47_7 and SbGT47_8 complemented transgenic plants.
Protein sequences of GT47 family members from ten species (doi: 10.6084/m9.figshare.6958895).
Primers in this study.