Graphical abstract
Highlights
-
•
Gold rush in cryo-EM structure determination of challenging transcription complexes.
-
•
Initiation complexes of all eukaryotic RNA polymerases have been solved by cryo-EM.
-
•
Cryo-EM structures of chromatin modifiers become available at increasing pace.
Abstract
Direct electron detector technology combined with improved imaging processing procedures has dramatically increased the resolution that can be obtained by single-particle cryo-electron microscopy and cryo-electron tomography. These developments — often referred to as the `resolution revolution’ in cryo-EM — have had a profound impact on the structural biology of transcription as they allow the determination of atomic or near-atomic resolution structures of very large, flexible and often transient transcription complexes that in many cases had resisted crystal structure determination for decades. In this review, we will discuss recent advances and breakthroughs in the structural biology of transcription complexes enabled by the revolution in cryo-electron microscopy with particular focus on eukaryotic RNA polymerases and their pre-initiation complexes, but also chromatin remodelers and epigenetic regulators.
Current Opinion in Structural Biology 2018, 52:8–15
This review comes from a themed issue on Cryo electron microscopy
Edited by John Briggs and Werner Kuhlbrandt
For a complete overview see the Issue and the Editorial
Available online 14th July 2018
https://doi.org/10.1016/j.sbi.2018.07.002
0959-440X/© 2018 The Authors. Published by Elsevier Ltd. This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/4.0/).
Introduction
Gene transcription is a fundamental process of life performed by a set of highly dynamic molecular machines with RNA polymerase as a core component. Transcriptional activity is cell-type specific and undergoes tremendous changes during the cell cycle. As such, tight control over the activity of the transcription machinery is required [1]. In eukaryotes, transcriptional regulation occurs at the level of the three eukaryotic RNA polymerases, associated general and sequence-specific transcription factors, and co-activators such as Mediator. On a second level, chromatin modifying and chromatin remodeling complexes regulate the accessibility of chromatin for transcription.
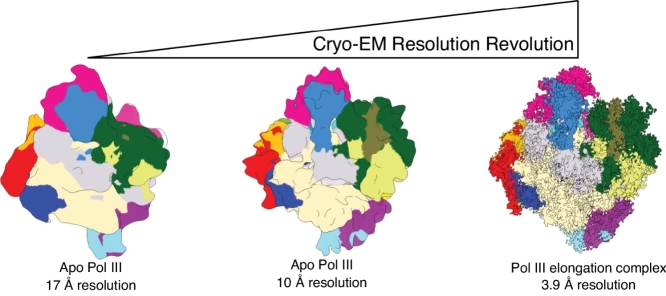
During the last decades, X-ray crystallography has been the central structural biology technique, providing a constant flow of molecular structures which gave important insight into eukaryotic transcription in the context of chromatin (see also the 100 landmark crystal structures that influenced transcription and chromatin research [2]). Despite these enormous successes, transcription complexes are nevertheless challenging targets for X-ray crystallography not only due to their size (often there are several dozens of polypeptide chains involved), but also because of their inherent flexibility, their modular structure, the transient interactions of their components and their limited accessibility [3]. Accordingly, the crystallization and X-ray structure determination of large transcription complexes is often a painstakingly long but also risky process that is not always crowned by success. During the last years, single-particle cryo-EM has increasingly gained importance as an alternative or complementary technique in the analysis of transcription complexes. Compared to X-ray crystallography, sample consumption is lower, larger complexes are more easily observed, and conformational heterogeneity can be overcome by particle classification in many cases. For many years, progress in cryo-EM was halted by the low resolution of the structures obtained due to technical limitations such as detector sensitivity. However, recent technical advances such as direct electron detectors combined with improved single-particle cryo-EM data acquisition and data processing pipelines, coined as `the resolution revolution’ [4], have caused a gold rush in the transcription field with a large number of transcription complex structures having become available only within the last few years (Figure 1). Here, we highlight the impact of the cryo-EM resolution revolution in the field of transcription and chromatin research and review recent insights on eukaryotic RNA polymerase transcription complexes and chromatin modifier complexes obtained by cryo-EM.
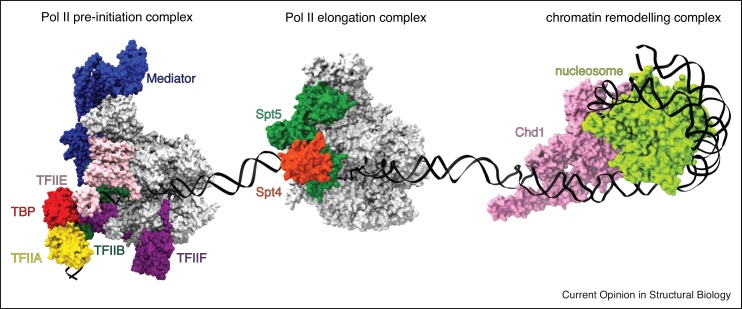
Figure 1.
Landscape of Pol II-mediated transcription. Steps of the transcription cycle from formation of pre-initiation complex (left) to elongation (middle), preceded by nucleosome removal by chromatin remodeler Chd1 (right) is depicted. In the PIC (pdb: 5fyw), Pol II is shown in grey, TBP in red, TFIIA in yellow, TFIIB in dark green, TFIIF in purple, TFIIE in pink, and Mediator in blue. In the Pol II elongation complex (pdb: 5oik), Pol II is depicted in grey, Spt5 in green and Spt4 in orange. In the nucleosome–Chd1 complex (pdb: 5o9g), the nucleosome and Chd1 are colored in light green and magenta, respectively. DNA is colored in black.
Transcription initiation of eukaryotic polymerases
In eukaryotes, three different RNA polymerases (Pol I, II, and III) catalyze DNA-dependent transcription into distinct RNA products by the same conserved mechanism also present in bacteria and archaea [5]. However, eukaryotes have evolved a large and diverse set of specific protein factors to ensure regulated promoter-dependent transcription initiation, elongation and termination. To initiate transcription, general transcription factors (GTF) recognize the promoter with the help of activators, assemble at the transcription start site, recruit their corresponding RNA polymerase to form a pre-initiation complex (PIC) and together facilitate DNA strand opening.
The crystal structure determination of the 10-subunit Pol II core enzyme and the Pol II elongation complex [6,7] were milestone achievements and served as starting point for the detailed structural analysis of Pol II transcription. At the time, the Pol II 10-subunit core structure was the largest asymmetric protein complex solved by X-ray crystallography. Moving beyond these initial crystal structures and obtaining Pol II crystal structures in different states and bound to transcription factors proved not to be an easy task. Obtaining the structure of the 12-subunit Pol II [8] and of elongating Pol II in complex with TFIIS [9] took an additional two years, while the complete complex of Pol II with TFIIB [10] was only solved six years later. Even larger assemblies of Pol II transcription complexes escaped crystallization attempts showing the limitations in resolving large and heterogeneous assemblies by X-ray crystallography due to the conformational and compositional heterogeneity of many transcription complexes.
At the time the Pol II crystal structure was solved, first cryo-EM structures of transcription complexes had already become available: TFIID together with TFIIA and TFIIB [11], TFIIH [12], and Mediator [13], an important co-activator of Pol II transcription. However, these structures generally suffered from low resolution limiting the biological conclusions that could be drawn from them. The limited resolution often also prevented accurate assignment of the cryo-EM densities and the unambiguous fitting of (partial) structures even in cases where crystal structures were available. However, the advent of direct electron detectors finally allowed overcoming this impediment.
In 2013, the Nogales lab proposed the structure of a minimal human Pol II PIC with cryo-EM map reconstructions at about 11 Å nominal resolution [14••], which was sufficient to confidently place homology models of Pol II and some GTFs. These results were followed shortly by intermediate resolution models of the yeast Pol II PICs [15,16] that, also supported by crosslinking mass-spectrometry [17], allowed the positioning of atomic models of many of the single components obtained by X-ray crystallography into the cryo-EM maps (reviewed in [18]). Finally, cryo-EM maps below 4.0 Å resolution enabled the construction of almost complete, near-atomic models of the human and yeast core Pol II PICs in different functional states [19,20].
Beyond the core Pol II PIC, several cryo-EM structures show the Pol II PIC bound to the Mediator complex [15,16,21], while structures of the multi-subunit GTFs TFIID and TFIIH were solved separately by cryo-EM [22,23]. Accordingly, we can now aim for super-assemblies and analyze them by cryo-EM as exemplified by the recent cryo-EM structure of the Pol II-TFIIH-mediator complex with a molecular mass of 2 MDa and comprising more than 46 polypeptides [24••] (Figure 1), while an even larger assembly that also comprises TFIID with a molecular mass of ∼3 MDa and >65 polypeptides at present only is a structural model that still awaits experimental validation [22]. It is obvious that the complexity and size of transcription complexes has reached a stage where X-ray crystallography is no longer the method of choice. However, in cryo-EM reconstructions the highest resolution is often only observed in the core, while more flexible peripheral subunits are observed at lower resolutions. Therefore, X-ray crystal structures often remain essential for the correct assignment as it has been also shown for the Mediator [24••,25,26].
Compared to the first high resolution 10-subunit Pol II core structure, atomic-resolution structures of 14-subunit Pol I and 17-subunit Pol III have only become available more than one decade later. This presumably reflects the additional difficulties in obtaining high-resolution crystal structures of both polymerases due to the additional subunits and the resulting increased flexibility. Shortly after the first crystal structures of apo Pol I could finally be obtained [27,28], structures of transcribing Pol I [29,30] and the promoter-binding competent Pol I [31,32] were also solved by cryo-EM, giving important further insight into its transcription mechanism. Only in the last year, three independent structures of the basal Pol I PIC comprising Pol I and core factor (CF) in an open complex were published [33•,34•,35•]. The crystal structure of CF obtained in one of the reconstructions [33•] greatly helped the complete interpretation of the cryo-EM maps. Although the structures generally agree on the overall architecture of the complex, certain flexible parts were differently positioned in the structures likely due to differences in the preparation.
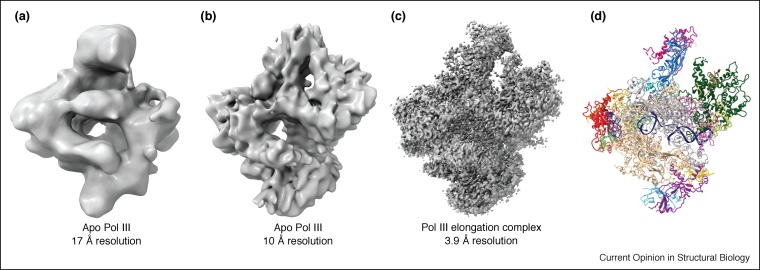
In the case of the most complex eukaryotic RNA polymerase, 17-subunit Pol III, all attempts to solve the structure by X-ray crystallography were futile, while the structure elucidation of Pol III by cryo-EM gradually proceeded over almost 10 years paralleling the resolution revolution (Figure 2). The first cryo-EM structure of Pol III [36], resolved at 17 Å, already allowed appreciating the structural differences among the three eukaryotic polymerases and the approximate positions of the TFIIE-like and TFIIF-like subcomplexes, but the low resolution limited any further insight into the mechanism. An improved structural model resolved at 10 Å allowed for a better appreciation of the architecture of the Pol III-specific sub-complexes [37] and confirmed the high inherent flexibility of the peripheral sub-complexes. Finally, a `post-cryo-EM revolution’ Pol III structure at 3.9 Å resolution allowed for atomic model building of the entire complex [38]. The same preparation awarded three distinct conformational states: one of transcribing Pol III bound to a transcription scaffold and two apo Pol III structures both at slightly lower resolution. The increased resolution finally allowed visualizing individual side chains and this information can now be used for testing mechanistic hypothesis and to compare architecture and mechanisms of Pol I, Pol II and Pol III in detail.
Figure 2.
Resolution revolution in Pol III cryo-EM structures. (a) Electron density of apo RNA polymerase III obtained from particles recorded on Kodak SO163 films using a JEOL JEM2010 microscope at a magnification of 40 000× and processed with SPIDER to 17 Å (EMDB: 1322). (b) Electron density of apo RNA polymerase III recorded on Kodak SO163 films using a CM200 microscope at a magnification of 50 000× and processed with SPIDER to 10 Å (EMDB: 1804). (c) Electron density of the RNA polymerase III elongation complex recorded using a Titan Krios microscope equipped with Falcon II direct-electron detector at a magnification of 75 000× and processed using Relion to 3.9 Å (EMDB: 3178). (d) Model of the RNA polymerase III elongation complex (pdb: 5fj8) colored as in Hoffmann et al. [38].
Only this year, the Vannini group and our group independently proposed models for a minimal Pol III PIC [39••,40••]. Both groups used yeast Pol III together with TFIIIB bound to closed or artificially opened DNA constructs, which gave rise to distinct conformational states of the initially transcribing Pol III, and the PIC in closed and spontaneously opened complexes. In this case, the power of cryo-EM to catch distinct conformations in one sample preparation allowed for proposing a mechanism for DNA melting and OC formation in Pol III.
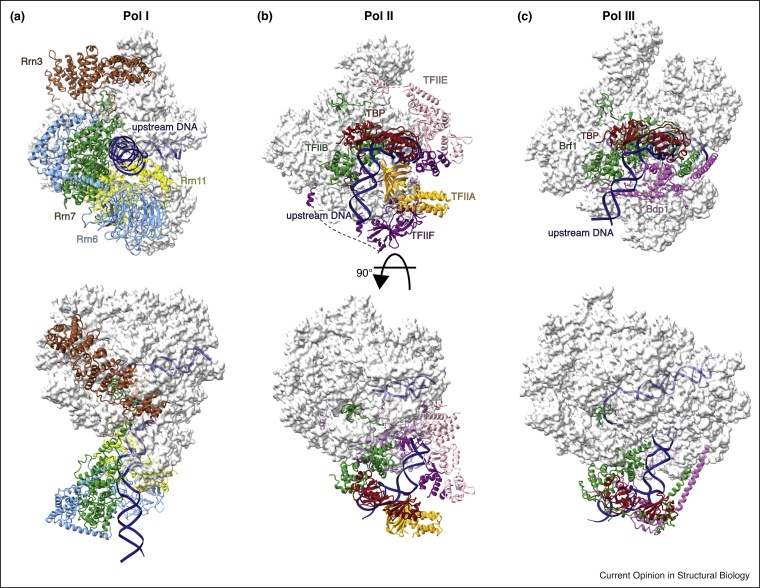
Comparing the recent PIC structures of all three eukaryotic RNA polymerases (Figure 3), a clear difference in architecture between Pol I PIC, on the one hand, and Pol II and Pol III PIC, on the other, can be appreciated. In the case of Pol II and Pol III, DNA follows a similar path and is translocated into the cleft of the polymerase upon opening, allowing the assumption that the general mechanism of DNA strand opening is conserved between Pol II and Pol III. In this process, the pre-assembled Pol III-specific subunits take over tasks that are performed by GTFs in the Pol II system thus providing also a rationale how Pol III achieves higher initiation rates than Pol II. In the case of Pol I, the highly divergent path of the upstream DNA in the open PIC suggests that Pol I translocates over the DNA by altering its position relative to the CF. This speculative mechanism rather resembles the way how bacterial polymerase achieves DNA promoter opening and might reflect the diverging complexity of regulation of transcription initiation in the three polymerases [41].
Figure 3.
High resolution structures of RNA polymerase I, II and III pre-initiation complexes (PIC). (a) Pol I PIC (pdb: 5oa1) containing Pol I, Rrn3 (sienna) and Core Factor (CF) (Rrn6: blue, Rrn7 green, Rrn11: yellow) resolved at 4.4 Å [34•]. The path of upstream DNA is different compared to the Pol II and III PICs, and TFIIB-like Rrn7 is located further upstream from the polymerase as compared to TFIIB and Brf1. (b) Pol II PIC (pdb: 5fyw) containing Pol II, TFIIA (orange), TFIIB (green), TFIIE (pink), TFIIF (purple), and TBP (red) resolved at 3.6 Å [20]. (c) Pol III PIC (pdb: 6f40) containing Pol III and TFIIIB (Brf1, green; Bdp1, magenta; TBP, red) resolved at 3.7 Å [39••].
Transcription elongation and termination
By contrast to transcription initiation, less structural information about the elongation complexes of eukaryotic RNA polymerases is available. In part, this is due to the transient nature of these complexes that involve the progressive synthesis of the RNA transcripts. The structure of transcribing yeast Pol II was solved by X-ray crystallography shortly after the Pol II apo enzyme [6,7] and the high degree of conservation among eukaryotes was demonstrated by a recent structure of the transcribing mammalian Pol II solved at 3.4 Å resolution [42]. There are also structures of elongating Pol I [29,30] and Pol III [38] solved by cryo-EM. However, in none of these structures elongation factors are present. Only last year, elongating Pol II was solved in complex with elongation factors (EFs) at high resolution [43•,44]. The structural model of Ehara et al. demonstrates how the basal EFs Elf1, Spt4/5, and TFIIS modify the polymerase surface thereby creating DNA exit and entry tunnels to keep the DNA in the active center thereby ensuring processive elongation. Nascent RNA is guided through another exit tunnel preventing interactions with single-stranded DNA that might lead to stalling. Recent attempts to obtain structural models of elongating Pol II together with Paf1C and TFIIS still suffer from low resolution [45] but there is little doubt that in the near future more structural models of elongation complexes will become available. This will likely be also the case for Pol I, which shares several EFs with Pol II [46]. Finally, Pol I, Pol II and Pol III use different mechanisms for transcription termination. Whereas Pol I and Pol II require binding of transcription termination factors, Pol III only requires a short stretch of 5 thymines on the non-template DNA strand as termination signal [47]. Considering the rapid elucidation of the other aspects of the transcription cycle in the recent years, we anticipate that cryo-EM structures will soon also contribute elucidating the transcription termination mechanisms of eukaryotic RNA polymerases.
Chromatin remodelers, transcription activators, and Polycomb group proteins
To allow for promoter recognition, PIC assembly, and eventually transcription elongation, the DNA has to be made accessible from densely packed chromatin by displacing or shifting nucleosomes and recruiting GTFs and polymerases. Both nucleosomes and their regulators are challenging targets for structural biology due to their high inherent flexibility, their posttranslational modifications and the transient interactions of the complexes they form. After the X-ray crystal structure of the nucleosome core particle was solved some 20 years ago [48], it took almost two decades to obtain cryo-EM reconstructions of the same particle at a comparable resolution [49, 50, 51]. In addition to the limited resolution that was only overcome by the advent of direct electron detectors, progress was also slowed down by the preferred orientation of nucleosomes in the ice resulting in a limited number of independent views. With the cryo-EM nucleosome structure in hand, larger complexes comprising nucleosome-bound chromatin modifiers can now be tackled.
The chromatin-remodeling helicase-like ATPases enable transcription through active chromatin [52]. These multi-modular molecular machines modify chromatin architecture by acting on the nucleosome histone octamers, sliding them along DNA, altering their structure, and catalyzing their exchange or even their eviction [53]. Early work resulted only in low resolution structures that provided limited insight into the mode of action of these enzymes [54, 55, 56]. Only very recently, the structures of two chromatin remodelers bound to nucleosomes have been solved by cryo-EM with resolutions from 4.8 Å to just below 4 Å [57•,58•]. The structural model of yeast Snf2 bound to a nucleosome showed the resting state of the ATPase bound to DNA and interacting with the N-terminal tail of the histone H4 whereas the transition state of Chd-1 bound to DNA and the H4 tail was captured by using an ATP analog. Taken together, these data led to the proposal of a functional model of a ratcheting mechanism of the helicase translocation along the DNA thereby promoting histone sliding [58•]. Moreover, the structure of another chromatin remodeler, INO80, a large (>1 MDa) and flexible complex, was solved in its apo state by cryo-EM [59]. The high variation in resolution from 4.1 Å in the core to about 12 Å in the periphery reflects the high degree of flexibility of the complex. More recently, the architecture of the yeast NuA4/TIP60 complex has been solved at 4.7 Å, giving insight into the scaffolding and regulatory mechanism of this gene regulator and DNA repair enzyme complex [60].
Whereas chromatin remodelers actively regulate transcription in active chromatin, the family of Polycomb group (PcG) proteins control transcription on the epigenetic level by placing or removing epigenetic marks such as methyl groups or ubiquitin on histones to silence chromatin [61,62]. The overall architecture of the human Polycomb repressive complex 2 (PRC2), a highly modular and flexible protein complex, was first described by a negative-stain EM structure at 21 Å resolution [63]. Only very recently, more cryo-EM structures of PRC2 bound to its co-factors AEBP2 and JARID2 [64••] and of PRC2 bound to a di-nucleosome [65••] revealed how PRC2 is activated by a methylated lysine in histone H3 (H3 K27) in one nucleosome to methylate H3 K27 of the next nucleosome, thereby allowing methylation marks to spread. The interpretation of these cryo-EM reconstructions were greatly supported by the available crystal structures of a trimeric PRC2 core complex [66, 67, 68] demonstrating (again) the complementarity between crystal and cryo-EM structures. In yeast, the 19-subunit transcription co-activator complex SAGA is recruited to the transcription start site of Pol II by activators to alter the chromatin structure by modifying nucleosomal histones. Its structure has been elucidated with one of its central subunits, Tra1, resolved at medium resolution shedding light on the modular assembly of this major epigenetic regulator [69].
Conclusions and perspectives
Cryo-EM has become an invaluable tool for the structural analysis of large transcriptional assemblies leading to a surge of publications within the last few years. Large transcription complexes such as the Pol II PIC and the PIC–core mediator complex [24••], RNA polymerase–elongation factor complexes, but also chromatin modifying complexes bound to nucleosomes whose size, conformational and compositional heterogeneity prevented crystallographic analysis can now be analyze by cryo-EM. At present most of these very large complexes are still determined at medium resolution and in many cases still rely on the availability of high-resolution X-ray structures of subunits and subcomplexes to obtain atomic structures. With the still increasing sensitivity of detectors, the introduction of the phase plate to allow for increased contrast at low defocus, improved sample preparation protocols, efforts to prevent surface denaturation of the sample using automatic sample dispenser like the Spotiton [70], and the advancement in computational analysis, we expect many high-resolution EM maps to be used for ab initio model building without prior knowledge of crystal structures.
One yet less explored application in the analysis of transcription complexes is time-resolved cryo-EM, which has enabled some recent breakthroughs in other biological applications [71]. The controlled mixing of complex components at millisecond timescales before plunge freezing can help capture more transient intermediate complexes for a more detailed understanding of the structural transitions between different conformational states.
All the structures discussed in this review were in vitro reconstitutions in many cases using recombinant subunits and complexes. Although in vitro systems offer a wealth of structural insight and lead to testable hypotheses, the vision is to observe these complexes also in their native environment, the nucleus. Recent advances in cryo-electron tomography (cryo-ET, reviewed in this issue) show promise that transcription complexes can be soon also observed in their cellular context. Cryo-ET has already allowed locating nucleosome chains [72] and lamin meshworks within the lamina [73], but hopefully soon will also picture specific transcription complexes in the nucleus such that high resolution single-particle cryo-EM structures can be positioned into the tomograms. However, preparing thin cellular samples remains challenging thus posing a serious limitation on the study of transcription complexes in their native environment. The coming years will bring more integrative studies utilizing these two methods. Nonetheless, functional studies in solution, both in vitro and in vivo, will still be required to test hypotheses derived from the structural data obtained by EM. Guided by the structural data, single-molecule approaches are also powerful to probe domain movements and complex assembly and lifetimes as well as disentangle the dynamics of the underlying processes both in vivo and in vitro.
Conflict of interest statement
The authors declare no conflicts of interest.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
We thank Herman Fung, Matthias Vorländer and Lucas Tafur for carefully reading and commenting on the manuscript. This work was funded by the ERC Advanced Grant (ERC-2013-AdG340964-POLIPIC) to JH, YS and CWM.
References
- 1.Kornberg R.D. Eukaryotic transcriptional control. Trends Biochem Sci. 1999;24:M46–M49. [PubMed] [Google Scholar]
- 2.Cramer P. A tale of chromatin and transcription in 100 structures. Cell. 2014;159:985–994. doi: 10.1016/j.cell.2014.10.047. [DOI] [PubMed] [Google Scholar]
- 3.Cramer P. Structure determination of transient transcription complexes. Biochem Soc Trans. 2016;44:1177–1182. doi: 10.1042/BST20160079. [DOI] [PubMed] [Google Scholar]
- 4.Kühlbrandt W. The resolution revolution. Science. 2014;343:1443–1444. doi: 10.1126/science.1251652. [DOI] [PubMed] [Google Scholar]
- 5.Vannini A., Cramer P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol Cell. 2012;45:439–446. doi: 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 6.Cramer P., Bushnell D.A., Kornberg R.D. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 7.Gnatt A.L., Cramer P., Fu J., Bushnell D.A., Kornberg R.D. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 8.Bushnell D.A., Kornberg R.D. Complete, 12-subunit RNA polymerase II at 4.1-Å resolution: implications for the initiation of transcription. Proc Natl Acad Sci. 2003;100:6969–6973. doi: 10.1073/pnas.1130601100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kettenberger H., Armache K.-J., Cramer P. Architecture of the RNA polymerase II–TFIIS complex and implications for mRNA cleavage. Cell. 2003;114:347–357. doi: 10.1016/s0092-8674(03)00598-1. [DOI] [PubMed] [Google Scholar]
- 10.Kostrewa D., Zeller M.E., Armache K.-J., Seizl M., Leike K., Thomm M., Cramer P. RNA polymerase II–TFIIB structure and mechanism of transcription initiation. Nature. 2009;462:323–330. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- 11.Andel F., Ladurner A.G., Inouye C., Tjian R., Nogales E. Three-dimensional structure of the human TFIID–IIA–IIB complex. Science. 1999;286:2153–2156. doi: 10.1126/science.286.5447.2153. [DOI] [PubMed] [Google Scholar]
- 12.Schultz P., Fribourg S., Poterszman A., Mallouh V., Moras D., Egly J.M. Molecular structure of human TFIIH. Cell. 2000;102:599–607. doi: 10.1016/s0092-8674(00)00082-9. [DOI] [PubMed] [Google Scholar]
- 13.Dotson M.R., Yuan C.X., Roeder R.G., Myers L.C., Gustafsson C.M., Jiang Y.W., Li Y., Kornberg R.D., Asturias F.J. Structural organization of yeast and mammalian mediator complexes. Proc Natl Acad Sci U S A. 2000;97:14307–14310. doi: 10.1073/pnas.260489497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14••.He Y., Fang J., Taatjes D.J., Nogales E. Structural visualization of key steps in human transcription initiation. Nature. 2013;495:481–486. doi: 10.1038/nature11991. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using cryo-EM the authors describe the stepwise assembly of a minimal human Pol II PIC.
- 15.Murakami K., Tsai K.-L., Kalisman N., Bushnell D.A., Asturias F.J., Kornberg R.D. Structure of an RNA polymerase II preinitiation complex. Proc Natl Acad Sci U S A. 2015;112:13543–13548. doi: 10.1073/pnas.1518255112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plaschka C., Larivière L., Wenzeck L., Seizl M., Hemann M., Tegunov D., Petrotchenko E.V., Borchers C.H., Baumeister W., Herzog F. Architecture of the RNA polymerase II–mediator core initiation complex. Nature. 2015;518:376–380. doi: 10.1038/nature14229. [DOI] [PubMed] [Google Scholar]
- 17.Mühlbacher W., Sainsbury S., Hemann M., Hantsche M., Neyer S., Herzog F., Cramer P. Conserved architecture of the core RNA polymerase II initiation complex. Nat Commun. 2014;5:4310. doi: 10.1038/ncomms5310. [DOI] [PubMed] [Google Scholar]
- 18.Sainsbury S., Bernecky C., Cramer P. Structural basis of transcription initiation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16:129–143. doi: 10.1038/nrm3952. [DOI] [PubMed] [Google Scholar]
- 19.He Y., Yan C., Fang J., Inouye C., Tjian R., Ivanov I., Nogales E. Near-atomic resolution visualization of human transcription promoter opening. Nature. 2016;533:359–365. doi: 10.1038/nature17970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plaschka C., Hantsche M., Dienemann C., Burzinski C., Plitzko J., Cramer P. Transcription initiation complex structures elucidate DNA opening. Nature. 2016;533:353–358. doi: 10.1038/nature17990. [DOI] [PubMed] [Google Scholar]
- 21.Robinson P.J., Trnka M.J., Bushnell D.A., Davis R.E., Mattei P.-J., Burlingame A.L., Kornberg R.D. Structure of a complete mediator–RNA polymerase II pre-initiation complex. Cell. 2016;166 doi: 10.1016/j.cell.2016.08.050. 1411–1422.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Louder R.K., He Y., López-Blanco J.R., Fang J., Chacón P., Nogales E. Structure of promoter–bound TFIID and model of human pre-initiation complex assembly. Nature. 2016;531:604–609. doi: 10.1038/nature17394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greber B.J., Nguyen T.H.D., Fang J., Afonine P.V., Adams P.D., Nogales E. The cryo-electron microscopy structure of human transcription factor IIH. Nature. 2017;549:414–417. doi: 10.1038/nature23903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24••.Schilbach S., Hantsche M., Tegunov D., Dienemann C., Wigge C., Urlaub H., Cramer P. Structures of transcription pre-initiation complex with TFIIH and mediator. Nature. 2017;551:204–209. doi: 10.1038/nature24282. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors describe the cryo-EM structure of Pol II PIC with TFIIH and mediator together comprising 46 polypeptides.
- 25.Nozawa K., Schneider T.R., Cramer P. Core mediator structure at 3.4 Å extends model of transcription initiation complex. Nature. 2017;545:248–251. doi: 10.1038/nature22328. [DOI] [PubMed] [Google Scholar]
- 26.Tsai K.-L., Yu X., Gopalan S., Chao T.-C., Zhang Y., Florens L., Washburn M.P., Murakami K., Conaway R.C., Conaway J.W. Mediator structure and rearrangements required for holoenzyme formation. Nature. 2017;544:196–201. doi: 10.1038/nature21393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernández-Tornero C., Moreno-Morcillo M., Rashid U.J., Taylor N.M.I., Ruiz F.M., Gruene T., Legrand P., Steuerwald U., Müller C.W. Crystal structure of the 14-subunit RNA polymerase I. Nature. 2013;502:644–649. doi: 10.1038/nature12636. [DOI] [PubMed] [Google Scholar]
- 28.Engel C., Sainsbury S., Cheung A.C., Kostrewa D., Cramer P. RNA polymerase I structure and transcription regulation. Nature. 2013;502:650–655. doi: 10.1038/nature12712. [DOI] [PubMed] [Google Scholar]
- 29.Tafur L., Sadian Y., Hoffmann N.A., Jakobi A.J., Wetzel R., Hagen W.J.H., Sachse C., Müller C.W. Molecular structures of transcribing RNA polymerase I. Mol Cell. 2016;64:1–9. doi: 10.1016/j.molcel.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neyer S., Kunz M., Geiss C., Hantsche M., Hodirnau V.-V., Seybert A., Engel C., Scheffer M.P., Cramer P., Frangakis A.S. Structure of RNA polymerase I transcribing ribosomal DNA genes. Nature. 2016;540:607–610. doi: 10.1038/nature20561. [DOI] [PubMed] [Google Scholar]
- 31.Pilsl M., Crucifix C., Papai G., Krupp F., Steinbauer R., Griesenbeck J., Milkereit P., Tschochner H., Schultz P. Structure of the initiation–competent RNA polymerase I and its implication for transcription. Nat Commun. 2016;7:12126. doi: 10.1038/ncomms12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engel C., Plitzko J., Cramer P. RNA polymerase I–Rrn3 complex at 4.8 Å resolution. Nat Commun. 2016;7:12129. doi: 10.1038/ncomms12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33•.Engel C., Gubbey T., Neyer S., Sainsbury S., Oberthuer C., Baejen C., Bernecky C., Cramer P. Structural basis of RNA polymerase I transcription initiation. Cell. 2017;169 doi: 10.1016/j.cell.2017.03.003. 120–131.e22. [DOI] [PubMed] [Google Scholar]; See annotation to Ref.[35•].
- 34•.Sadian Y., Tafur L., Kosinski J., Jakobi A.J., Wetzel R., Buczak K., Hagen W.J., Beck M., Sachse C., Müller C.W. Structural insights into transcription initiation by yeast RNA polymerase I. EMBO J. 2017;36:2698–2709. doi: 10.15252/embj.201796958. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation to Ref.[35•].
- 35•.Han Y., Yan C., Nguyen T.H.D., Jackobel A.J., Ivanov I., Knutson B.A., He Y. Structural mechanism of ATP-independent transcription initiation by RNA polymerase I. Elife. 2017;6:e27414. doi: 10.7554/eLife.27414. [DOI] [PMC free article] [PubMed] [Google Scholar]; The three papers show the highly diverging architecture of the Pol I PIC compared to the Pol II PIC.
- 36.Fernández-Tornero C., Böttcher B., Riva M., Carles C., Steuerwald U., Ruigrok R.W.W.H., Sentenac A., Müller C.W., Schoehn G. Insights into transcription initiation and termination from the electron microscopy structure of yeast RNA polymerase III. Mol Cell. 2007;25:813–823. doi: 10.1016/j.molcel.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 37.Fernández-Tornero C., Böttcher B., Rashid U.J., Steuerwald U., Flörchinger B., Devos D.P., Lindner D., Müller C.W. Conformational flexibility of RNA polymerase III during transcriptional elongation. EMBO J. 2010;29:3762–3772. doi: 10.1038/emboj.2010.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffmann N.A., Jakobi A.J., Moreno-Morcillo M., Glatt S., Kosinski J., Hagen W.J.H., Sachse C., Müller C.W. Molecular structures of unbound and transcribing RNA polymerase III. Nature. 2015;528:231–236. doi: 10.1038/nature16143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39••.Vorländer M.K., Khatter H., Wetzel R., Hagen W.J.H., Müller C.W. Molecular mechanism of promoter opening by RNA polymerase III. Nature. 2018;553:295–300. doi: 10.1038/nature25440. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation to Ref.[40••].
- 40••.Abascal-Palacios G., Ramsay E.P., Beuron F., Morris E., Vannini A. Structural basis of RNA polymerase III transcription initiation. Nature. 2018;553:301–306. doi: 10.1038/nature25441. [DOI] [PubMed] [Google Scholar]; The two papers reveal the architectural similarity of the Pol III PIC to the Pol II PIC while several functions of Pol II GTFs have been incorporated into Pol III’s additional subunits.
- 41.Viktorovskaya O.V., Schneider D.A. Functional divergence of eukaryotic RNA polymerases: unique properties of RNA polymerase I suit its cellular role. Gene. 2015;556:19–26. doi: 10.1016/j.gene.2014.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernecky C., Herzog F., Baumeister W., Plitzko J.M., Cramer P. Structure of transcribing mammalian RNA polymerase II. Nature. 2016;529:551–554. doi: 10.1038/nature16482. [DOI] [PubMed] [Google Scholar]
- 43•.Ehara H., Yokoyama T., Shigematsu H., Yokoyama S., Shirouzu M., Sekine S. Structure of the complete elongation complex of RNA polymerase II with basal factors. Science. 2017;357:921–924. doi: 10.1126/science.aan8552. [DOI] [PubMed] [Google Scholar]; This structure reveals how Pol II remains processive in elongation by altering its surface with the help of elongation factors forming entry and exit tunnels for the DNA and RNA.
- 44.Bernecky C., Plitzko J.M., Cramer P. Structure of a transcribing RNA polymerase II–DSIF complex reveals a multidentate DNA–RNA clamp. Nat Struct Mol Biol. 2017;24:809–815. doi: 10.1038/nsmb.3465. [DOI] [PubMed] [Google Scholar]
- 45.Xu Y., Bernecky C., Lee C.-T., Maier K.C., Schwalb B., Tegunov D., Plitzko J.M., Urlaub H., Cramer P. Architecture of the RNA polymerase II–Paf1C–TFIIS transcription elongation complex. Nat Commun. 2017;8:15741. doi: 10.1038/ncomms15741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y., Sikes M.L., Beyer A.L., Schneider D.A. The Paf1 complex is required for efficient transcription elongation by RNA polymerase I. Proc Natl Acad Sci U S A. 2009;106:2153–2158. doi: 10.1073/pnas.0812939106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arimbasseri A.G., Maraia R.J. Mechanism of transcription termination by RNA polymerase III utilizes a non-template strand sequence-specific signal element. Mol Cell. 2015;58:1124–1132. doi: 10.1016/j.molcel.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luger K., Mäder A.W., Richmond R.K., Sargent D.F., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 49.Chua E.Y.D., Vogirala V.K., Inian O., Wong A.S.W., Nordenskiöld L., Plitzko J.M., Danev R., Sandin S. 3.9 Å structure of the nucleosome core particle determined by phase-plate cryo-EM. Nucleic Acids Res. 2016;44:8013–8019. doi: 10.1093/nar/gkw708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song F., Chen P., Sun D., Wang M., Dong L., Liang D., Xu R.-M., Zhu P., Li G. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014;344 doi: 10.1126/science.1251413. [DOI] [PubMed] [Google Scholar]
- 51.Bilokapic S., Strauss M., Halic M. Histone octamer rearranges to adapt to DNA unwrapping. Nat Struct Mol Biol. 2018 doi: 10.1038/s41594-017-0005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Narlikar G.J., Sundaramoorthy R., Owen-Hughes T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154:490–503. doi: 10.1016/j.cell.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clapier C.R., Cairns B.R. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 54.Yamada K., Frouws T.D., Angst B., Fitzgerald D.J., Deluca C., Schimmele K., Sargent D.F., Richmond T.J. Structure and mechanism of the chromatin remodelling factor ISW1a. Nature. 2011;472 doi: 10.1038/nature09947. [DOI] [PubMed] [Google Scholar]
- 55.Nguyen V.Q., Ranjan A., Stengel F., Wei D., Aebersold R., Wu C., Leschziner A.E. Molecular architecture of the ATP-dependent chromatin–remodeling complex SWR1. Cell. 2013;154:1220–1231. doi: 10.1016/j.cell.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tosi A., Haas C., Herzog F., Gilmozzi A., Berninghausen O., Ungewickell C., Gerhold C.B., Lakomek K., Aebersold R., Beckmann R. Structure and subunit topology of the INO80 chromatin remodeler and its nucleosome complex. Cell. 2013;154:1207–1219. doi: 10.1016/j.cell.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 57•.Liu X., Li M., Xia X., Li X., Chen Z. Mechanism of chromatin remodelling revealed by the Snf2–nucleosome structure. Nature. 2017;544:440–445. doi: 10.1038/nature22036. [DOI] [PubMed] [Google Scholar]; See annotation to Ref.[58•].
- 58•.Farnung L., Vos S.M., Wigge C., Cramer P. Nucleosome–Chd1 structure and implications for chromatin remodelling. Nature. 2017;550:539–542. doi: 10.1038/nature24046. [DOI] [PMC free article] [PubMed] [Google Scholar]; By trapping two ATP-dependent helicases in two distinct states, the two papers together provide a mechanism how these enzymes act on nucleosomes.
- 59.Aramayo R.J., Willhoft O., Ayala R., Bythell-Douglas R., Wigley D.B., Zhang X. Cryo-EM structures of the human INO80 chromatin–remodeling complex. Nat Struct Mol Biol. 2018;25:37–44. doi: 10.1038/s41594-017-0003-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang X., Ahmad S., Zhang Z., Côté J., Cai G. Architecture of the Saccharomyces cerevisiae NuA4/TIP60 complex. Nat Commun. 2018;9:1147. doi: 10.1038/s41467-018-03504-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chittock E.C., Latwiel S., Miller T.C.R., Müller C.W. Molecular architecture of polycomb repressive complexes. Biochem Soc Trans. 2017;45:193–205. doi: 10.1042/BST20160173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sparmann A., van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 63.Ciferri C., Lander G.C., Maiolica A., Herzog F., Aebersold R., Nogales E. Molecular architecture of human polycomb repressive complex 2. Elife. 2012;1:e00005. doi: 10.7554/eLife.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64••.Kasinath V., Faini M., Poepsel S., Reif D., Feng X.A., Stjepanovic G., Aebersold R., Nogales E. Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science. 2018 doi: 10.1126/science.aar5700. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation to Ref.[65••].
- 65••.Poepsel S., Kasinath V., Nogales E. Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nat Struct Mol Biol. 2018;25:154–162. doi: 10.1038/s41594-018-0023-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors show PRC2 interacting with two of its cofactors and bound to a dinucleosome substrate giving insight how PRC2 spreads methylation mark across chromatin regions.
- 66.Jiao L., Liu X. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science. 2015;350 doi: 10.1126/science.aac4383. aac4383-aac4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Justin N., Zhang Y., Tarricone C., Martin S.R., Chen S., Underwood E., De Marco V., Haire L.F., Walker P.A., Reinberg D. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat Commun. 2016;7:11316. doi: 10.1038/ncomms11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brooun A., Gajiwala K.S., Deng Y.-L., Liu W., Bolaños B., Bingham P., He Y.-A., Diehl W., Grable N., Kung P.-P. Polycomb repressive complex 2 structure with inhibitor reveals a mechanism of activation and drug resistance. Nat Commun. 2016;7:11384. doi: 10.1038/ncomms11384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sharov G., Voltz K., Durand A., Kolesnikova O., Papai G., Myasnikov A.G., Dejaegere A., Shem A., Ben, Schultz P. Structure of the transcription activator target Tra1 within the chromatin modifying complex SAGA. Nat Commun. 2017;8:1556. doi: 10.1038/s41467-017-01564-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jain T., Sheehan P., Crum J., Carragher B., Potter C.S. Spotiton: a prototype for an integrated inkjet dispense and vitrification system for cryo-TEM. J Struct Biol. 2012;179:68–75. doi: 10.1016/j.jsb.2012.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frank J. Time-resolved cryo-electron microscopy: recent progress. J Struct Biol. 2017;200:303–306. doi: 10.1016/j.jsb.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mahamid J., Pfeffer S., Schaffer M., Villa E., Danev R., Kuhn Cuellar L., Forster F., Hyman A.A., Plitzko J.M., Baumeister W. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science. 2016;351:969–972. doi: 10.1126/science.aad8857. [DOI] [PubMed] [Google Scholar]
- 73.Turgay Y., Eibauer M., Goldman A.E., Shimi T., Khayat M., Ben-Harush K., Dubrovsky-Gaupp A., Sapra K.T., Goldman R.D., Medalia O. The molecular architecture of lamins in somatic cells. Nature. 2017;543:261–264. doi: 10.1038/nature21382. [DOI] [PMC free article] [PubMed] [Google Scholar]