Abstract
Human pluripotent stem cells hold great promise for improving regenerative medicine. However, a risk for tumor formation and difficulties in generating large amounts of subtype derivatives remain the major obstacles for clinical applications of stem cells. Here, we discovered that zinc finger E-box–binding homeobox 1 (ZEB1) is highly expressed upon differentiation of human embryonic stem cells (hESCs) into neuronal precursors. CRISPR/Cas9-mediated ZEB1 depletion did not impede neural fate commitment, but prevented hESC-derived neural precursors from differentiating into neurons, indicating that ZEB1 is required for neuronal differentiation. ZEB1 overexpression not only expedited neural differentiation and neuronal maturation, which ensured safer neural cell transplantation, but also facilitated the generation of excitatory cortical neurons, which were valuable for managing certain neurological disorders, such as Parkinson's disease (PD) and amyotrophic lateral sclerosis (ALS). Our study provides useful information on how human neural cells are generated, which may help in forming strategies for developing and improving replacement therapies for treating patients with neurological diseases.
Keywords: embryonic stem cell, neuron, differentiation, adhesion, transplantation, ZEB1
Introduction
Human pluripotent stem cells (PSCs),4 particularly human embryonic stem cells (ESCs) or inducible pluripotent stem cells (iPSCs), because of their ability to differentiate into all cell types, become the most reasonable sources for transplantation therapy (1–4). In recent years, progress has been made in direct neural differentiation of human PSCs, especially embryonic stem cells, into certain functional cell types (5–9), which provides an ideal resource for pathogenesis research and clinical therapies of neurodegenerative diseases, such as Parkinson's disease, amyotrophic lateral sclerosis, and other disorders in the nervous system. However, some problems hinder the application of human ESCs, such as time-consuming differentiation, low yield, heterogeneous nature of derived neuronal populations, and a risk of tumor formation upon transplantation in vivo. Therefore, further efforts are required to guarantee both efficiency of differentiation and maturation.
During the differentiation from hPSCs into neurons, hPSCs undergo substantial morphological alterations, first from adherent hESC colonies to cell aggregates, then to neural rosettes, neural stem cells, and neurons or glia, a course very similar to that occurs in epithelial-mesenchymal transition (EMT). It is highly possible that molecules related to EMT may play important roles in neural differentiation. We made a screening and found that ZEB1 was expressed along with neural differentiation.
ZEB1, also named as ZFHX1A or δEF1, together with its fellow member ZEB2 (also called ZFHX1B or SIP1), belongs to the zinc finger E-box binding homeobox family (ZFH family). ZEB1 is putatively involved in cell morphological change during EMT. Accordingly, ZEB1 has been extensively studied in various cancers (10–13). Members of the ZEB family facilitate neural fate decision through tuning TGFβ/BMP signaling or as targets of the miR-200 family (14–16) and ZEB1 has an essential role in neural stem cell survival in vitro (17). Zeb1 may be functionally redundant to Zeb2, as Zeb1 was ectopically activated in Zeb2 knockout embryos (18). However, in mouse models, Zeb1 has been shown to be strongly expressed in the progenitor cells of the ventricular zone (VZ) and the subventricular zone (SVZ) of the developing cortex and that Zeb1 is necessary for the proliferation of the VZ and SVZ progenitors (19, 20). Recently, Singh et al. (21) reported that Zeb1 is required for tangential migration of unpolarized cerebellar granule neuron progenitors, and down-regulation of Zeb1 is necessary for maturation and polarization of the cerebellar granule neurons.
However, whether ZEB1 is involved in human neural differentiation still remains unclear. Here, by using a model system of neural differentiation from hESCs (22, 23), we carried out a series of functional assays to explore the role of ZEB1 in human neural differentiation.
Results
ZEB1 was progressively expressed during neural differentiation of hESCs
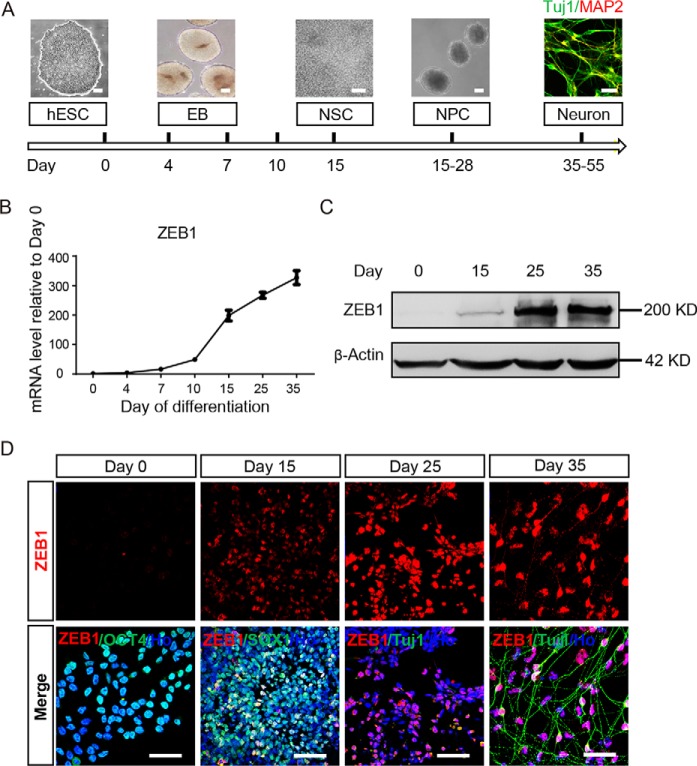
To inspect the expression of ZEB1 during neural differentiation, we differentiated hESC line WA09 (H9) as shown in Fig. 1A, and harvested cells at the indicated stages for real-time RT-PCR and Western blotting analysis. The mRNA level of ZEB1 was hardly detected in hESC (day 0). Upon neural differentiation, ZEB1 started expression and reached a peak on day 15, and remained at a high level thereafter (Fig. 1B). The expression pattern of ZEB1 was also verified by Western blotting and immunostaining together with cell type-specific markers at different stages of differentiation on samples taken from days 0, 15, 25, and 35 (Fig. 1, C and D). We also detected the expression of other fellow molecules of EMT in the same set of experiments. Except for N-CADHERIN, which was up-regulated over the progression of differentiation, no other EMT-related molecules, such as ZEB2, SNAIL, SLUG, and TWIST2 were detected (Fig. S1). These results together indicate ZEB1 may play a role during neural differentiation, which was not previously documented.
Figure 1.
ZEB1 was up-regulated in hESC-derived neural cells. A, a scheme of neural differentiation of hESCs. Scale bar: 50 μm. B, real-time PCR analysis showed the alteration of ZEB1 mRNA levels during neural differentiation of hESCs. The amount of mRNA was normalized to GAPDH levels. Bars represent mean ± S.D. of three experimental replicates. C, Western blotting showed expression of the ZEB1 protein at different stages of neural differentiation from hESCs. β-ACTIN was a loading control; n = 3. D, representative images of immunostaining results using indicated antibodies in hESC-derived neural cells. DAPI was included to show the nuclei (here and after); n = 3. Scale bar: 50 μm. hESC, human embryonic stem cell; EB, embryoid bodies; NSC, neural stem cell; E8, essential 8 medium; NIM, neural induction medium; NDM, neural differentiation medium.
ZEB1 was required for the intrinsic neuronal differentiation
To examine whether ZEB1 was required for neural and neuronal differentiation, we generated ZEB1-knockout hESCs based on a H9 hESC line that harbored a cassette of inducible Cas9 (iCas9) and a verified sgRNA targeting ZEB1 locus (sgRNA1) (Fig. S2A). We designed three sgRNAs targeting ZEB1 (Table S1), and identified the indel percentages with T7EI assays for each target in HEK293T cells (data not shown), then sgRNA1 (Table S2), which targeted the third exon (CCDS53506.1) was selected as the most efficient one. In H9ESCs, a restriction fragment length polymorphism assay revealed ∼21% indel rate (Fig. S2B). hESCs were then dissociated into single cells. By picking colonies, using restriction fragment length polymorphism analysis and sequencing (Table S3), we ultimately obtained a hESC line with a homozygous mutant of ZEB1 (Clone 9, Fig. S2, C and D). Potential off-target sites were identified at MIT, and only one possible off-target with four mismatches fall in gene coding sequences (NM_001130675) (Table S4), indicating the specificity of the selected sgRNA1 locus.
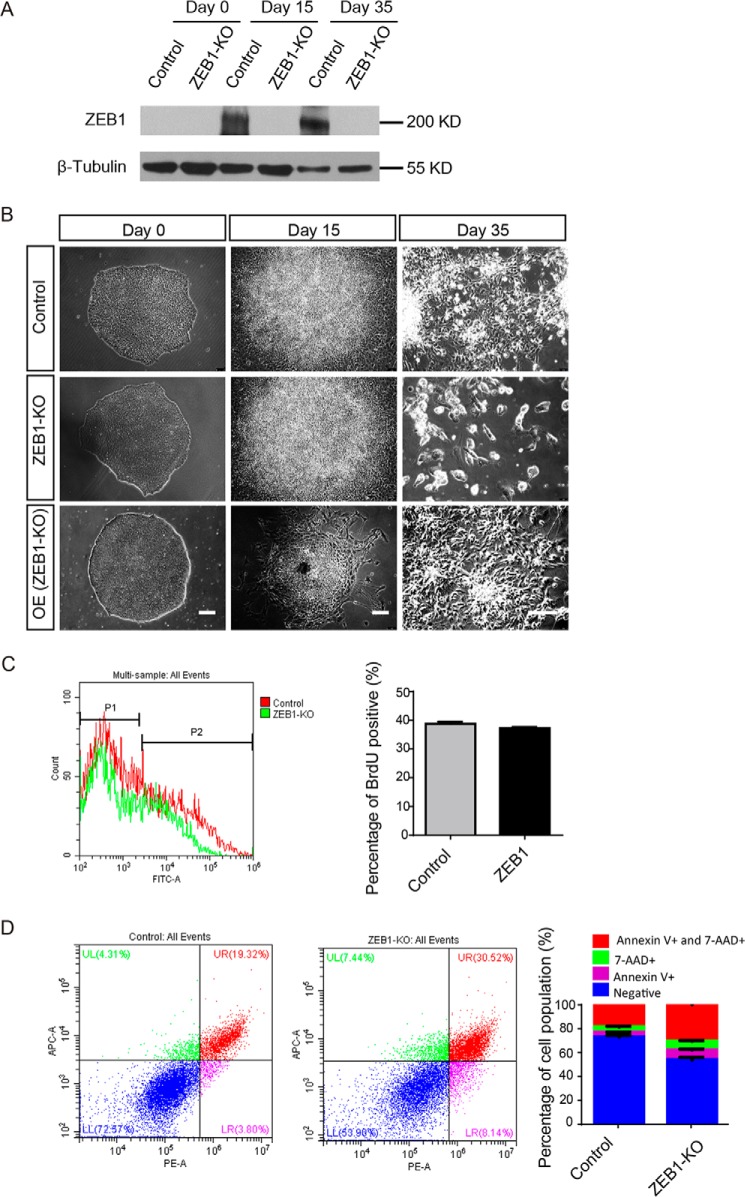
Deletion of ZEB1 was confirmed at the protein level at different stages during neural differentiation (Fig. 2A). During normal differentiation of hESCs into neurons, cells spread out and formed rossettes at the early stage, and then differentiated into neuron phenotype at the late stage (Fig. 2B, upper panel). ZEB1 knockout cells did not exhibit distinguishable differences from control cells at the hESCs stage and early phase of neural differentiation (Fig. 2B, middle panel). However, with neural differentiation proceeded, most of the ZEB1-depleted cells died at the stage when neuronal differentiation occurred (Fig. 2B, middle panel). We then overexpressed ZEB1 in ZEB1 KO hESCs cells, some cells underwent morphological changes at the early stage of differentiation, and eventually became neuron cells (Fig. 2B, lower panel). We did not observe any significant difference by evaluating cell proliferation with BrdU antibody (Fig. 2C). However, we observed more cells that were undergoing apoptosis in the ZEB1-depleted group compared with the control group using a PE Annexin V/7-AAD Kit (Fig. 2D). Re-expressing of ZEB1 in ZEB1 KO cells rescued the neural differentiation (Fig. 2B, lower panel). In addition to the rescue effects, forced expression of ZEB1 in ZEB1 KO caused discernable dispersing of the differentiated cells at the early stage of neural differentiation (Fig. 2B, right lower panel), which was consistent with the phenomenon detected in ZEB1 overexpression cell lines (as discussed below). Taken together, these data indicated that ZEB1 expression may not be critical for early neural specification, but was essential for the conversion from neural precursors to neurons.
Figure 2.
ZEB1 was necessary for neuronal differentiation. A, Western blotting showed ZEB1 protein levels at days 0, 15, and 35 of neural differentiation from hESCs with or without ZEB1 knockout. β-TUBULIN was the loading control. B, phase-contrast images of the indicated cells during different days of neural differentiation; n = 3. Scale bar: 50 μm. C, left, flow cytometry analysis with BrdU antibody using cells after 4 days attachment of spheres for neuronal differentiation (day 30). Right, statistical analysis of the left results; n = 3. D, left, cell apoptosis and death analysis with PE Annexin V/7-AAD Kit. Right, statistical analysis to show the percentage of viable cells (Negative) or cells in early apoptosis (Annexin V+, p < 0.001), in late apoptosis or already dead (7-AAD+ and Annexin V+ and 7-AAD+, p < 0.001); n = 3. ZEB1-KO, ZEB1 knockout. OE (ZEB1-KO), overexpress ZEB1 in ZEB1-knockout cells.
Forced expression of ZEB1 promoted the neural differentiation of hESCs
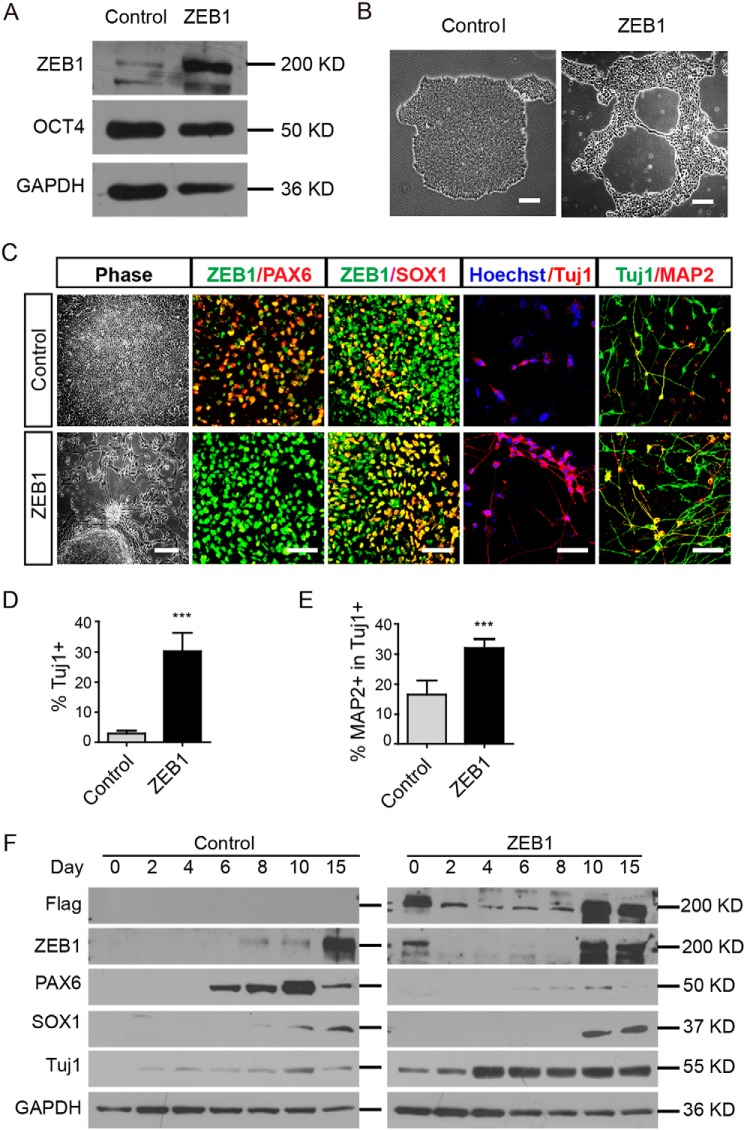
We next overexpressed ZEB1 (with 3× FLAG fusion to the N terminus) in hESCs with a lentivirus tet-on 3G inducible expression system (Fig. S3A). The insertion of a mNeonGreen-P2A-blasticidin (BSD) fragment facilitated picking of single colony hESCs (Fig. S3B). Selected colonies of H9ESC lines were verified using GFP or antibodies against ZEB1 or FLAG upon doxycycline induction (Fig. S3, C and D and Fig. 3A). Forced expression of GFP itself in hESCs neither affected the morphology of the hESC colonies nor changed the expression of pluripotent marker such as OCT4 (Fig. S3C). However, when overexpressing ZEB1 in hESCs, although the expression patterns of OCT4, NANOG, and SOX2 were not changed, these colonies exhibited unique fragmentation upon long-term culture (Fig. 3B, Figs. S3D and S4). For some hESC colonies, continual overexpression of ZEB1 elicited neurite-like out-growth, which was immunostained for the neuron-specific antibody TUJ1 (Fig. S3E).
Figure 3.
Sustained overexpression of ZEB1 in hESC-induced neural differentiation. A, Western blotting using the indicated antibodies in selected clone #2 of hESCs. GAPDH was the loading control. B, representative phase-contrast images showing different morphologies of H9ESC colonies with (ZEB1) or without (Control) doxycycline in the medium; n = 3. Scale bar: 100 μm. C, images showing the phenotypic alterations of cells at various stages of neural differentiation after overexpression of ZEB1. Typical rosettes in control and extended neurite-like structures from colonies with ZEB1 overexpression were observed. Alterations of PAX6 and SOX1 on day 15, TUJ1 and MAP2 on day 25 were shown as well. Scale bar, 100 μm. D and E, quantification of TUJ1+ cells and MAP2-positive cells. ***, p < 0.001; n = 3. F, Western blotting showed the dynamic expression of ZEB1, PAX6, SOX1, and TUJ1 during early neural differentiation in the control and ZEB1 overexpressing cells. GAPDH was a loading control; n = 3.
Extra amount of ZEB1 accelerated the neural differentiation of hESCs
To further investigate the effect of ZEB1 in neural differentiation, we induced the expression of ZEB1 once the hESCs were lifted for differentiation. At day 15, most of the WT cell aggregates formed neural rosettes within colonies (Fig. 3C, left panel). However, in the colonies of hESCs with ZEB1 overexpression, only a small number of rosettes were identified. Quite a few cells migrated from the colonies and extended the neurite-like structures surrounding the cells (Fig. 3C, left panel).
Decreasing neural rosettes may reflect a result of inhibited neural differentiation or alteration of the progress of differentiation. We then stained the cells with neuron-specific and dendrites antibodies TUJ1 and MAP2 to address this issue. In the control group, only sporadic TUJ1+ cells were detected (∼2%) on day 15 (Fig. 3, C and D, p < 0.001), and the vast majority of TUJ1+ neurons did not appear until day 25. In the ZEB1-overexpressed group, however, TUJ1+ cells appeared as early as day 15 (up to 30%) (Fig. 3D, p < 0.001), and the number of TUJ1+ cells continually increased when neural differentiation proceeded. One week after the cells were attached for neuronal differentiation, only 16% of TUJ1+ cells express more mature neuronal marker MAP2 in the control group, whereas approximately 32% among the TUJ1+ cells began to express MAP2 in the ZEB1-overexpressed group (Fig. 3, C and E, p < 0.001). Thus, we deemed that forced expression of ZEB1 promoted neural differentiation by altering the phase of neural differentiation.
The formation of neural rosettes accompanying the precocious TUJ1+ and MAP2+ cells indicated that neural differentiation was accelerated. Furthermore, we examined the expression of early neural markers such as PAX6, SOX1, as well as neuronal markers TUJ1 and MAP2 by immunofluorescence staining or Western blotting analysis. In the control group, PAX6 was expressed as the earliest transcription factor of neural differentiation from days 6 to 8 on, which was followed by SOX1 expression from days 8 to 10 on (Fig. 3, C and F). The early presence of extra ZEB1 repressed PAX6 but increased the expression of SOX1 on days 10–15, endowing cells becoming TUJ1+ neuronal-like cells directly within 15 days of differentiation (Fig. 3, C and F). Bypassing the early stage of neural differentiation was also accompanied by down-regulation of various genes, such as ZEB1 itself and PAX6, which likely ensured later smooth transition to an intrinsic neuronal differentiation program (Fig. 3F).
ZEB1 promoted neuronal maturation
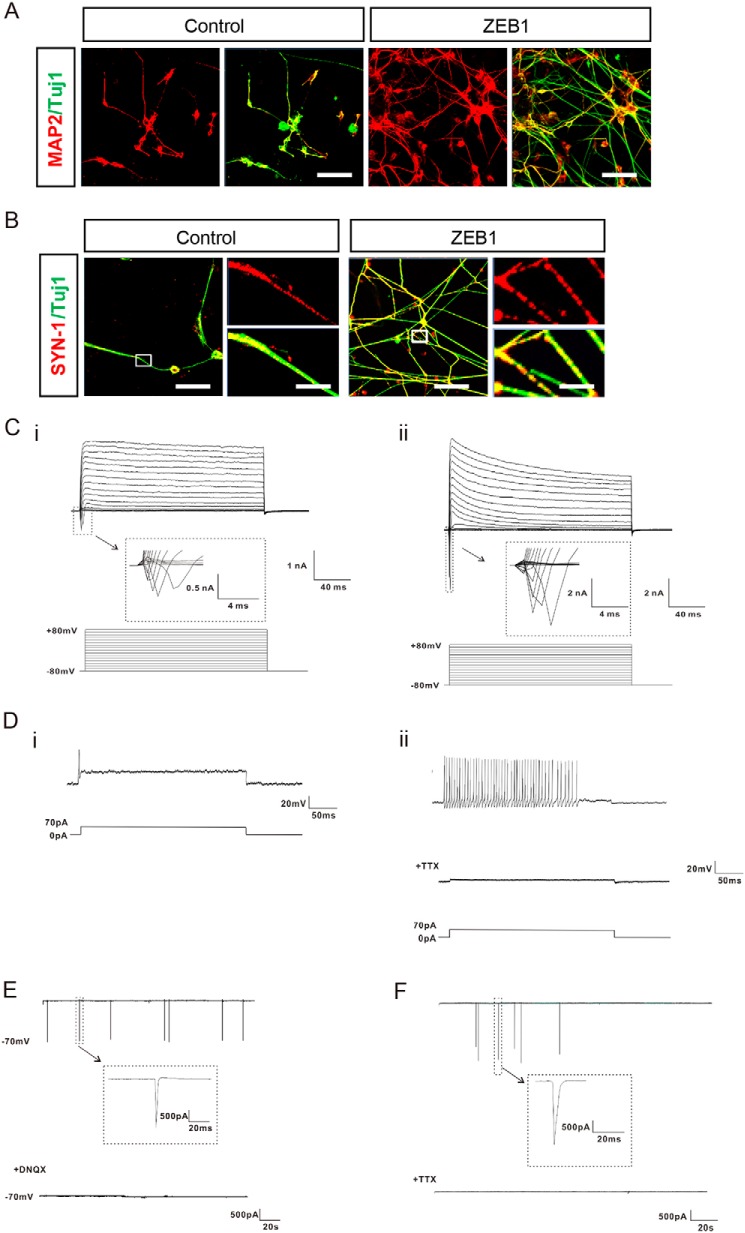
Deletion of ZEB1 impeded neuronal differentiation, whereas forced ZEB1 expression expedited this process, which prompted us to examine whether ZEB1 could also promote neuronal maturation. We thus continued the neuronal differentiation experiment by placing the hESCs-derived neural progenitor cells on laminin substrate from day 26 and cultured in neural differentiation medium supplemented with trophic factors such as BDNF, GDNF, IGF1, and cAMP. Four weeks later, the majority of cells in both control and ZEB1-overexpressed groups became elaborately branched and expressed the neuronal marker MAP2 in the cell body and in the neurites (Fig. 4A). Considering that the synaptic connection between neurons was an integral component of functional maturity, we then examined the localization of synaptic proteins such as SYNAPSIN-1 in differentiated neurons by immunofluorescent staining. Although SYNAPSIN-1 was diffusely distributed in the cytoplasm of neurons in the control group, it aggregated as punctuated throughout the more complex dendritic arborizations of neurons when ZEB1 was induced during the neuronal differentiation (Fig. 4B).
Figure 4.
ZEB1 promoted neuronal maturation. A, immunostaining for MAP2 along with TUJ1 showed the neuronal differentiation of hESCs with or without overexpression of ZEB1 at 8 weeks of neural differentiation; n = 3. B, representative immunofluorescent images showing the localization of SYNAPSIN-1 (SYN-1) in the cells. The boxed areas at the left were enlarged as shown in the right panel, respectively; n = 3. Scale bar: 50 m (each left panel) and 5 μm (each enlarged right panel). C–E, whole cell patch recordings indicated that neurons derived from hESCs after 8 weeks of differentiation were electrophysiological active; n = 3. C, representative traces of whole cell currents were elicited by stepwise depolarizations from −80 to 80 mV from a holding potential of −70 mV in neurons of (i) control and (ii) ZEB1 group, respectively. The inset shows sodium currents. D, representative traces of action potentials (APs) from a 70 pA current injection were shown for neurons of control (i) and the ZEB1 group (ii), respectively. (i), APs in control neurons exhibited single or typically diminished amplitude in peak value. (ii), repetitive trains of APs on the similar spikes were detected in ZEB1-overexpressed neurons from neural differentiation. The fired APs could be blocked completely by TTX (1 μm). E, representative voltage-clamp recordings of spontaneous postsynaptic currents (PSCs) from neurons of the ZEB1 group after 8 weeks of differentiation. The GluR currents could be blocked with DNQX (20 μm). Cells were held at −70 mV. F, after 10 weeks of neural differentiation without forced expression of ZEB1, spontaneous PSCs held at −70 mV were detected in the neurons from the control group, and the synaptic activity could be blocked by TTX (1 μm).
To further examine whether overexpression of ZEB1 increased the functionality of hESCs-derived neurons, we employed whole cell voltage-clamp to record electrophysiological activities at week 8 of neural differentiation. Cells from both control and ZEB1-overexpressed groups displayed full-blown Na+/K+ currents, and these inward currents could be completely blocked by tetrotodoxin (TTX) (1 μm) (Fig. 4C, i and ii). However, neurons from the control group failed to fire multiple action potentials (APs) in response to a 70 pA current pulse, or displayed single AP or APs with typically faded amplitude in peak value (Fig. 4D, i), whereas strains of sequential APs on the similar spikes were detected in neurons from the ZEB1-overexpressed group (Fig. 4D, ii). Neurons in the ZEB1-overexpressed group also showed spontaneous inward synaptic currents at 8 weeks of differentiation, which were almost completely eliminated by the non-N-methyl-d-aspartate antagonist DNQX (20 μm) (Fig. 4E). Neurons from the control group did not exhibit spontaneous synaptic currents until 10 weeks of differentiation (Fig. 4F). These data further validated that ZEB1 promoted the functional maturation and synaptogenesis of the hESCs-derived neurons.
ZEB1 facilitated the differentiation of excitatory neurons
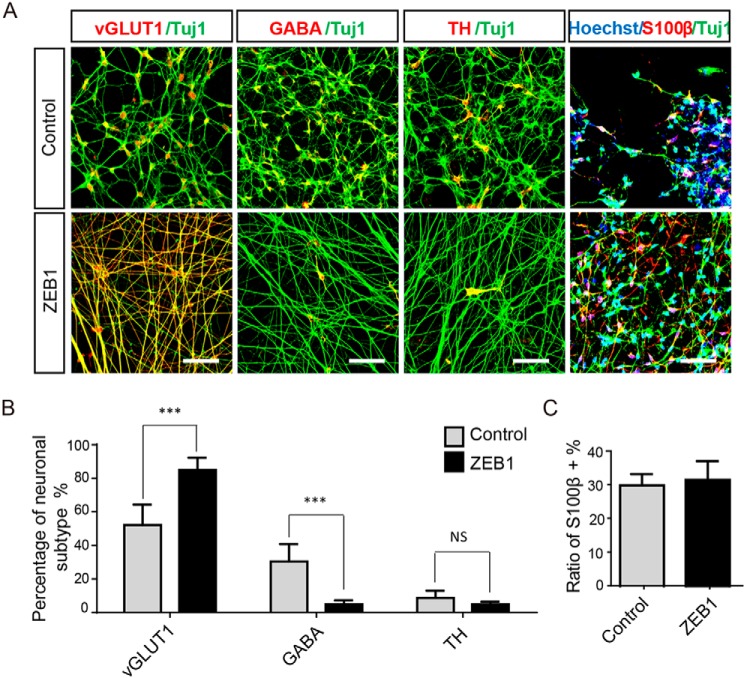
To identify which specific types of neurons or glia were differentiated when ZEB1 was overexpressed, we labeled the cells with various markers at week 8 of neural differentiation. In the control group, hESCs were randomly differentiated into excitatory, GABAergic, and dopaminergic neurons at a proportion of 52.2 (±9.0%), 30.5 (±5.5%), and 8.6% (±3.7%), according to immunostaining for vGLUT1, GABA, or TH, respectively (Fig. 5, A and B). When ZEB1 was overexpressed during neural differentiation, the majority of the cells were distinctly positive for vGLUT1 (84.9 ± 10.2%) (Fig. 5, A and B, p < 0.001). Only a small proportion of cells were stained with GABA (4.8 ± 7.9%) and TH (4.8 ± 6.8%) (Fig. 5, A and B). In both groups, we detected a similar proportion of astrocytes (∼30%) (Fig. 5A) but no oligodendrocytes (data not shown), as revealed by immunostaining for S100β/TUJ1 and myelin basic protein (data not shown), indicating that ZEB1 preferably affected the neuronal subtype specification but not glia fate choice. These results collectively demonstrated that forced expression of ZEB1 specifically promoted hESCs to differentiate into excitatory neuron subtypes.
Figure 5.
Forced expression of ZEB1 facilitated differentiation of excitatory neurons. A, representative immunofluorescent images of specific subtypes of neurons in control and the ZEB1-overexpression group at 8 weeks of neural differentiation from hESCs using the indicated antibodies; n = 3. Scale bar: 50 μm. B, quantification and statistical analysis of different types of neurons as indicated in A respect to the number of TUJ1+ cells; n = 3. Scale bar: 50 μm. C, statistical analysis of the S100β-positive cells respected to total cells. For statistical analysis, cells were counted under ×20 objective by selecting 10 fields randomly for each group from each experiment. Bars represent mean ± S.D. of three experimental replicates. ***, p < 0.001; **, p < 0.01. NS, not significant.
ZEB1 reduced the overgrowth of hESCs-derived progenitors upon transplantation into mouse brains
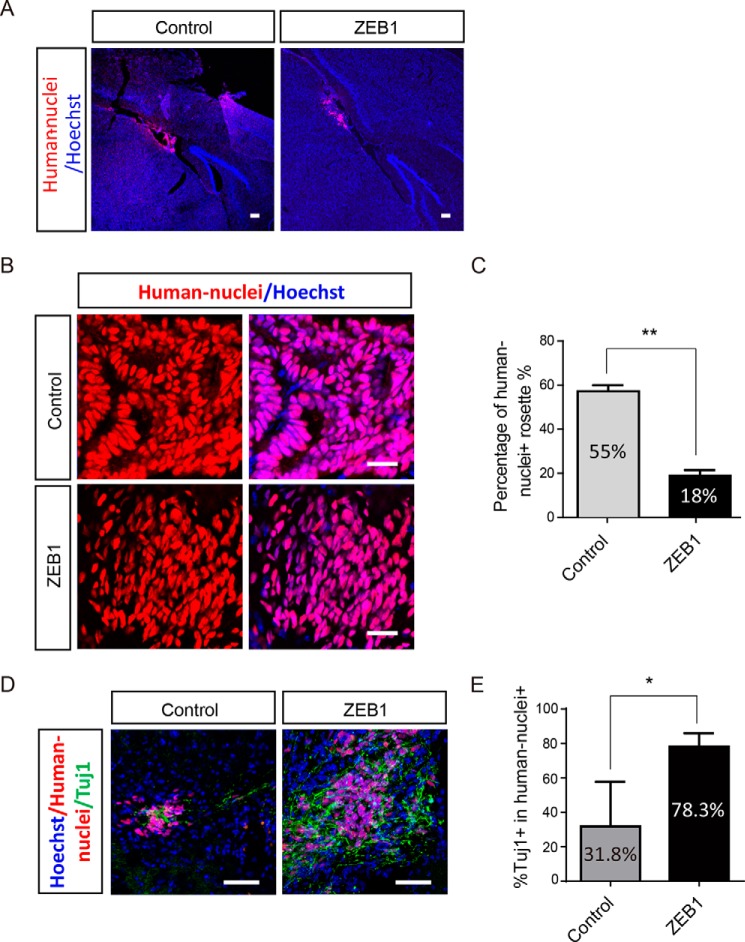
Because ZEB1 accelerated neural differentiation and promoted functional maturation of hESCs-derived neurons, it thus became a plausible lever for safety control of hESCs in regenerative medicine. We then examined whether overexpression of ZEB1 in hESCs-derived neural progenitor cells could reduce the chance of tumor formation upon transplanting into brains of neonatal pups of NOD/SCID mice. Eight weeks after transplantation of hESC-derived neural stem cells (day 15 of differentiation), mice were perfused for brain sections and immunostaining. Monoclonal antibodies that specifically recognized human nuclei were used to identify the transplanted human cells. Graft human cells were found in the cerebral cortex or nearby hippocampus and there was no difference for their distribution in the mouse brain between two groups (Fig. 6A). In the control group, transplanted cells formed substantial rosette-like overgrowth in the resident brain regions (presence in 55.2% of the brain slices) (Fig. 6, B and C). However, in the ZEB1-overexpressed group, this percentage was only 18.3% (Fig. 6C, p < 0.01), and most of the graft cells dispersed across the brain (Fig. 6, B and C). Consistently, much more of grafts differentiated into neurons, with more TUJ1+ cells in ZEB1 group relative to control group (Fig. 6, D and E, p < 0.05). These data indicate that ZEB1 overexpression promoted the differentiation of hESC-derived neural stem cells and potentially improved safety when used for therapy. In accordance with the in vitro results (Fig. 3, C and F), lack of rosettes in the ZEB1 group in vivo also reflected the loss of a PAX6+ fate in the ZEB1-overexpressed cells.
Figure 6.
Overexpression of ZEB1 decreased the risks of tumor formation of transplanted cells derived from hESCs. A, representative images showing the distribution of transplanted cells in vivo based on a human nuclei antibody immunostaining along with Hoechst staining (nuclei) 10 weeks after transplantation of hESCs-derived cells. Scale bar: 2 mm (left panel), 50 μm (middle and right panels). Arrows indicate the region of graft cells distribution with human nuclei staining. B, immunostaining images showing the difference of grafted cells from control and ZEB1-overexpressed groups in vivo. Scale bar: 50 μm. C, statistical analysis of the percentage of human nuclei-positive rosettes-like structure in the control and ZEB1 overexpression groups as shown in B. D, representative images of grafted cells (human nuclei-positive) expressing the neuronal marker βШ-tubulin (TUJ1) after transplantation. Arrows indicate the typical co-labeled neurons; n = 3. Scale bar: 50 μm. E, statistical analysis for the percentage of brain slices with TUJ1+ cells in the two groups as representatively shown in D. For the statistical analysis, 100–200 brain slices from 9 mice were analyzed, and the brain slices without human cells were discarded directly. **, p < 0.01; *, p < 0.05. Bars represent mean ± S.D. of three experimental replicates.
ZEB1 knockout prevented neural precursors differentiate into neurons
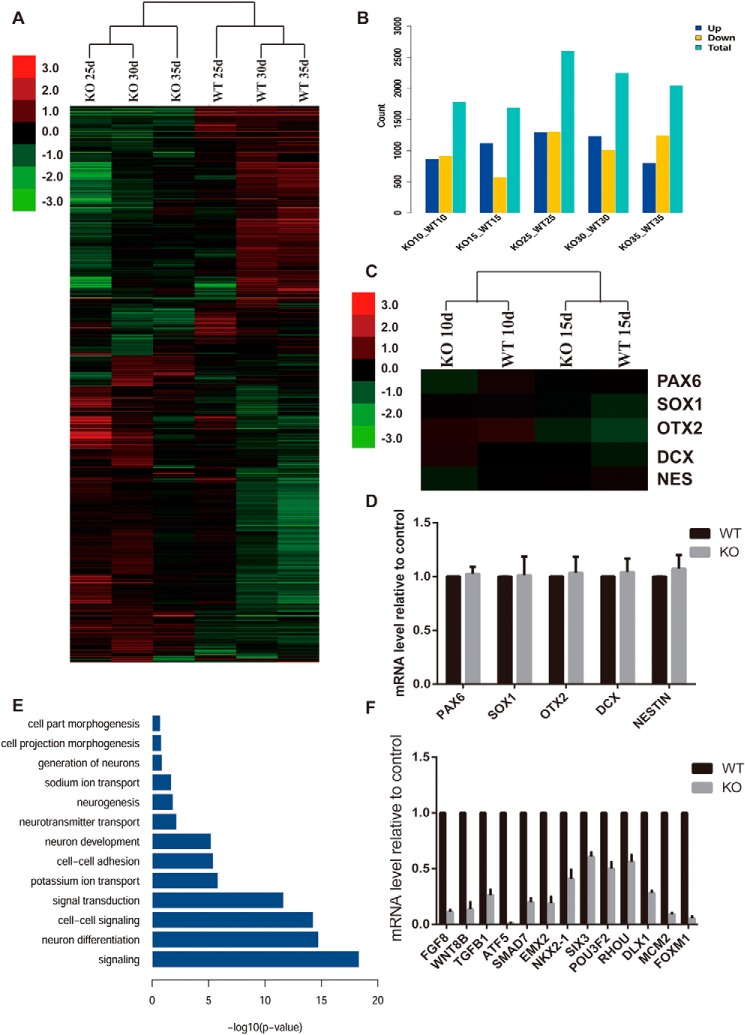
To elucidate the mechanism of ZEB1 function during the differentiation from hESCs into neurons, we collected RNA samples of different stages and examined genome-wide changes during the process by performing a RNA sequence analysis. Both clustering and principle component analysis indicated that the genome expression pattern occurred with significant changes since day 25 differentiation, as the 25-day samples' genome expression pattern were more close to 30 and 35 days, far from 15- and 10-day samples (Fig. S5, A and B). When we turned to the different expression genes (DEGs) at different stages, we found more DEGs starting at day 25 (Fig. 7B). Meanwhile, the genome expression pattern of the day 25 WT could be easily clustered with days 30 and 35 WT, and the ZEB1 KO cell lines clustered together (Fig. 7A). To further validate the ZEB1 functioning stage, we picked some of the neuronal related genes and found there was no significant difference in expression levels between WT and KO cell lines at the early differentiation stage (Fig. 7C), which were then confirmed by real-time PCR (Fig. 7D). As 25 day seemed to be a coteau, we subjected DEGs at 25 days into gene ontology (GO) analysis, and found most of the genes accumulated to neuronal related items (Fig. 7E), most of these genes were down-regulated after ZEB1 KO, such as FGF8, NKX2.1, SIX3, and DLX1 (Fig. 7F). Besides, we also observed some DEGs subjected to cell adhesion items in GO analysis, and found most of them were down-regulated in the ZEB1 KO cell line (Fig. S5C).
Figure 7.
Whole genome RNA sequence analysis of gene expression profiles. A, hierarchical clustering analysis of the whole genome profiles during the differentiation. RNA were harvested at the indicated time points. B, histogram showing the DEGs between WT and ZEB1 KO cell lines at the indicated time points. C, hierarchical clustering analysis of specific neuronal related genes at early stage between the 2 cell lines. D, quantitative real-time PCR analysis confirm the genes expression referred in C. E, GO analysis of the significantly differentially expressed genes at 25 days of the differentiation. F, quantitative real-time PCR analysis confirm the genes expression picked from DEGs related to neuronal development.
Discussion
In human neural differentiation, ZEB transcription factors, particularly ZEB2, as direct targets of miR-200 family members play essential roles in fate specification of the neuroectoderm (14). In this study, we found that when ZEB1 was depleted, although these cells could commence an early neural differentiation, they failed to differentiate further into neurons and died within a few days. Based on our data and previous literature, we speculated that during neural differentiation of hESCs, two members of ZEB family, ZEB1 and ZEB2, synergistically regulated the progression of cell fate conversion, with ZEB2 functioning primarily at the early stage of neural specification, whereas ZEB1 exclusively affecting the neuronal conversion during late neural differentiation.
Although not necessary for early neural specification, forced expression of ZEB1 during the early stage did expedite the progress of neuronal conversion, as evidenced by positive staining of a series of markers of neural differentiation and electrophysiological recordings. Overexpression of ZEB1 in hESCs significantly inhibited the expression of PAX6 but increased the expression of TUJ1, a conventional neuron marker, thus prompting a comprehensive revision of cell fate determination differed from the original opinion, which believed PAX6 pioneered a cohort of neural determinants (24). The expression of SOX1, another neural fate determinant that was usually expressed following PAX6 during neural differentiation of hESCs (24, 25), however, did not shift to earlier expression, suggesting the temporary alteration of cell fate did not completely perplex the intrinsic program of neural differentiation. Interestingly, ZEB1 was not detected at the protein level in the following days once it was overexpressed in hESCs at the initiation of neural differentiation. This might be due to an instigation of a quality control system within cells in response to the forced expression of ZEB1 and the sudden shuffle of cell fate. In both groups, ZEB1 was expressed at high levels from days 10 to 15 on, indicating its endogenous expression and intrinsic function. These data collectively supported a notion that cells undergoing cell fate rearrangement were able to gear to the intrinsic neural differentiation program when endogenous ZEB1 start to take over.
ZEB1 may play an important role in regulating the differentiation of ESCs into neurons, particularly in the transition from NPCs to neurons, as ZEB1 KO made no significant difference in the expression of neuronal related genes at the early stage, but after 25 days of differentiation, we detected the remarkable decrease for genes that associated with neuronal differentiation (FGF8, NKX2.1, SIX3, and DLX1). ZEB1 KO also resulted in down-regulation of cell adhesion–related genes, indicating a low adherence ability of ZEB1 KO cells, which may be another reason that blocks NPC cells differentiating into neurons in ZEB1 KO cell line.
We noticed that ZEB1 overexpression promoted neuronal maturation for over 2 weeks in advance, as compared with regular neural differentiation. As it is known that astrocytes promote neuronal maturation both in vivo and in vitro (26, 27), we doubt whether ZEB1 accelerates neuronal maturation through an indirect regulation of astrocytes. Whether ZEB1 was overexpressed or not, a similar proportion of S100β+ cells were identified at 8 weeks of neural differentiation. Thus, the presence of the astrocytic lineage in the culture unlikely caused a significant difference of the spontaneous synaptic currents of the hESCs-derived neurons, and the extra ZEB1 expression during neural differentiation autonomously accelerated neuronal maturation and promoted the occurrence of synaptic currents.
ZEB1 may also play a role in neuronal fate determination, as forced expression of ZEB1 almost exclusively induced hESCs into vGLUT1 + glutamatergic excitatory neurons in which ZEB1 may directly interact with key transcription factors involving excitatory fate. Nevertheless, it is also highly possible that the acceleration of neuronal differentiation itself accounts for the augmented generation of excitatory neurons. As noted above, forced expression of ZEB1 granted the neural stem cells for directed neuronal conversion and bypassed certain stages of neural differentiation. This may let neural stem cells miss competence to certain morphogens, such as Sonic Hedgehog (SHH), which is critical for ventral patterning and GABAergic neuron differentiation (22, 28–30). Thus, ZEB1 appears to be an essential player that ensures appropriate cellular constitution of brains for proper function.
Transplantation of ESC-derived cells even at the late stage of differentiation still bear risks of overgrowth in brains (31). Expedited neuronal differentiation is valuable for efficient differentiation into functional cells as well as for transplantation safety. In a set of proof of concept for neural cell transplantation, we did observe aggregates of cell overgrowth in brains that received transplantation of regular hESC-derived neural progenitors, which is in accordance with the previous report (31). Nevertheless, mice transplanted with hESCs-derived ZEB1 overexpression neural progenies contained much fewer aggregates, suggesting increased safety from overgrowth with ZEB1 overexpression. Thus, ZEB1 and the relevant signaling pathway could be informative to optimize the procedures of differentiating hESCs into neural cells for therapy.
Overall, we identified an important regulator of neural differentiation that coupled the neural differentiation progression and fate choice. However, the molecular mechanism under it still remains elusive and needs further research.
Materials and methods
Ethics statement, animals
All mouse studies were approved by the Ethics Committee of the Institute of Zoology (research license number AEI-06-07-2014), Chines Academy of Sciences. NOD/SCID mice were purchased from Vital River Laboratory Animal Technology Corp. (Beijing, China) and then raised in a sterile room.
Culture and differentiation of human ESCs
The H9ESC line (WiCell Institute, Madison, WI, passages 29–50) was cultured with feeder-free media. Neural differentiation from hESCs was performed as described previously (9). See the Supplemental Experimental Procedures for additional details.
Generation of the ZEB1-knockout hESC line using iCRISPR
To generate loss-of-function mutations of ZEB1 in human ESCs, we first constructed the CRISPR/Cas9-inducible (iCas9) hESC line by inserting a CRISPR/Cas9-inducible expression cassette in the AAVS1 genomic locus with TALEN technology (32, 33). For gRNA transfection and transduction in hESCs, we followed the published protocols (34, 35). In brief, iCas9 hESCs were treated with doxycycline (2 mg/ml) for 1 or 2 days before and during transfection. Cells were dissociated into single cells using Accutase and transfected the sgRNA using Lipofectamine RNAiMAX (Life Technologies) following the manufacturer's instructions. The details were described in the supporting “Experimental procedures”.
Construction of the ZEB1 overexpression hES cell line
To overexpress ZEB1 in H9ESCs and ZEB1-depleted H9ESCs, the tetracycline inducible tet-on lentiviral expression vectors were used. For the plasmid construction please see the supporting “Experimental procedures”
Preparation of lentiviruses and transduction were conducted as previously described (24). hESCs were treated with ROCK inhibitor (1 μm, Calbiochem, 688001) for 1 day before transduction. Cells were dissociated with 0.5 mm EDTA on the following day (UltraPureTM, 15575-020, Gibco). Cell pellets were incubated with concentrated virus at 37 °C for 1 h, then replated onto Matrigel substrate overnight with E8 medium. Two days later, neomycin (50 μg/ml, Sigma, G8168) and blasticidin (2 μg/ml, Sigma, 15205) were added to the culture medium to select drug-resistant clones. To obtain stable transduced monoclonal lines, hESCs were dissociated into single cells with Accutase and the mNeonGreen-positive cells were sorted by FACS. hESCs were treated with the ROCK inhibitor (1 μm) for all the processes. After 1–10 days of cultivation, mNeonGreen-positive colonies were identified and picked.
Real-time quantitative RT-PCR
Total RNA was isolated manually with TRIzol reagent (Invitrogen, 15596-026). cDNA was synthesized using 1 μg of RNA using a ReverAid First Strand cDNA Synthesis Kit according to the manufacturer's instruction (Fermentas, K1662). Quantitative RT-PCR was conducted with SYBR Premix ExTaq (Takara, RR430A) and the primers were listed in Table S5.
Western blot analysis
Cell pellets were lysed in RIPA lysis buffer (Thermo, 89900) supplemented with protease inhibitor (Roche Applied Science), then quantified using a BCA protein assay kit (Thermo, 23225). After electrophoresis, proteins were transferred to nitrocellulose membranes. The following primary antibodies were used: ZEB1 (Millipore, ABN285), ZEB2 (Santa Cruz, sc48789), N-CADHERIN (Abcam, ab18203), SNAIL (Cell Signaling Technology, number 3879), SLUG (Cell Signaling Technology, number 9585), TWIST2 (Abcam, ab50887), FLAG (Sigma, F3165), OCT4 (Abcam, ab19857), NANOG (R&D Systems, AF1997), SOX2 (Millipore, AB5603), PAX6 (Developmental Studies Hybridoma Bank), SOX1 (Millipore, MAB3369), NESTIN (Millipore, MAB5326), βIII-tubulin (TUJ1, Covance, PRB-435P), GAPDH (Abcam, ab9485), β-actin (Proteintech, 60008-1-Ig),and β-tubulin (Cell Signaling Technology, number 2148).
Immunofluorescence staining
Primary antibodies used for immunostaining were ZEB1 (Millipore, ABN285), OCT4 (Abcam, ab19857), NANOG (R&D Systems, AF1997), SOX2 (R&D Systems, AF2018), PAX6 (Developmental Studies Hybridoma Bank), SOX1 (R&D Systems, AF3369), NESTIN (Millipore, MAB5326), βШ-tubulin (TUJ1, Covance, PRB-435P; Sigma, T8660), MAP2 (Abcam, ab11267), S100β (Millipore, 04-1054), SYNAPSIN1 (Synaptic System, 106011), vGLUT1 (Synaptic System, 135302), GAD65&67 (Millipore, AB1511), GABA (Sigma, A0310), TH (Millipore, MAB318), and human nuclei (Millipore, MAB1281). Stained cells were mounted and observed on a LSM 780 META microscope (Zeiss, Berlin, Germany).
Proliferation assay
BrdU labeling of proliferating cells was performed according to previously published protocols (36). Differentiated cells were treated with BrdU for 24 h and analyzed using a Cell Proliferation Assay kit (Calbiochem, K306) according to the manufacturer's instructions. Results were analyzed with a CytoFLEX Flow cytometry (Beckman Coulter, Suzhou, China).
Apoptosis assay
Apoptosis analysis was performed using the Annexin V-PE/7-ADD Apoptosis Detection Kit (BD Pharmingen 559763) according to the manufacturer's instructions.
Electrophysiology
Whole cell patch clamp recordings of hESCs-derived neurons that were differentiated for 8–10 weeks were performed as described previously (26). The transmitter receptor blockers, TTX (1 μm), 4-aminopyridine (1 mm), bicuculline (20 μm), and DNQX (20 μm) were used in the bath solution for the detection of action potentials and spontaneous excitatory postsynaptic currents. Data were analyzed using pClamp version 9.2 software (Molecular Devices).
Transplantation of hESCs-derived cells in vivo
Differentiated cells (1 × 105) from control or ZEB1 groups were transplanted into the lateral ventricles of the brain of P1–2 pups (ice anesthetized), respectively. For the ZEB1 group, 2 mg/ml of doxycycline was administered orally in the drinking water to induce the expression of ZEB1. The mice were perfused (anesthetized with 40 mg/kg of chloral hydrate) with 0.9% saline followed by 4% paraformaldehyde 10 weeks post-transplantation. Consecutive coronal brain sections (30 μm) were sliced using a Leica SM 2000R Sliding Microtome and fixed in 4% paraformaldehyde overnight followed by dehydration in 0.1 m PBS containing 30% sucrose for 2 days at 4 °C. Immunofluorescence staining for the brain sections were performed as protocols used for cultured cells except using 1% Triton-100 to permeabilize.
Statistical analysis
Statistical analyses were executed with two-tailed Student's t tests. Data are presented as the mean ± S.D., and values were considered statistically significant at p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***), as described in the figure legends. Diagrams were created by GraphPad Prism 5 software.
RNA-seq analysis
For RNA-seq, cells at the indicated time points during differentiation were harvested, and total RNA was extracted with TRIzol reagent (1559618, Ambion, USA). After testing the quality of RNA by an Agilent 2100, 0.2 μg of each sample was sent to Annoroad Genomics for RNA-seq analysis. Raw data and processed data were uploaded to the NCBI Gene Expression Omnibus database under accession number GSE118990. RPKMs at the current sequencing depth were evaluated by using the jackets of the total comparison reads in RSeQC, and a relative error rate was utilized to measure the accuracy of the evaluated RPKM. Unsupervised heat map clustering analysis was performed by using Cluster 3.0. The clustering results were visualized and exported by TreeView 1.1.6r4. A principle component analysis image was generated by R(3.3.3)/Bioconductor (3.4) with “edgeR” and “limma” packages. Kyoto encyclopedia of genes and genomes (KEGG) and GO enrichment analyses were executed by R/Bioconductor with the “clusterProfiler” package.
Author contributions
Y. J., H. W., and B. H. conceptualization; Y. J., L. Y., H. W., and B. H. resources; Y. J., L. Y., L. X., X. L., W. Z., D. D., M. D., H. W., and B. H. formal analysis; Y. J., H. W., and B. H. supervision; Y. J., L. Y., X. L., H. W., and B. H. funding acquisition; Y. J., L. Y., L. X., X. L., W. Z., D. D., M. D., D. Z., H. W., and B. H. investigation; Y. J., L. Y., X. L., W. Z., and M. D. methodology; Y. J. and L. Y. writing-original draft; Y. J., L. Y., X. L., H. W., and B. H. project administration; Y. J. and L. Y. writing-review and editing; L. Y., X. L., M. D., D. Z., H. W., and B. H. validation; L. X., W. Z., and M. D. data curation; X. L., D. D., D. Z., H. W., and B. H. software; D. D., D. Z., H. W., and B. H. visualization.
Supplementary Material
Acknowledgments
We thank Xiangxiang Jiang for bioinformatics analysis and Annoroad Gene Technology Co. Ltd. (Beijing) for technical assistance with the RNA sequence experiment.
This work was supported by Natural Science Foundation of China Grants 31501102, 81225004, and 31171430. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5, Tables S1–S5, and “Experimental procedures”.
The data were deposited to the NCBI Gene Expression Omnibus database under accession number GSE118990.
- PSC
- pluripotent stem cell
- ESC
- embryonic stem cell
- EMT
- epithelial-mesenchymal transition
- VZ
- ventricular zone
- SVZ
- subventricular zone
- TTX
- tetrotodoxin
- AP
- action potential
- DNQX
- 6,7-dinitroquinoxaline-2,3-dione
- DEG
- different expression gene
- GO
- gene ontology
- KO
- knockout
- sgRNA
- single guide RNA
- NPC
- neural precursors
- NOD/SCID
- non-obese diabetic/severe combined immunodeficiency
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- TH
- tyrosine hydroxylase
- RPKMs
- reads per kilobase per million mapped reads.
References
- 1. Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S., and Jones J. M. (1998) Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 10.1126/science.282.5391.1145 [DOI] [PubMed] [Google Scholar]
- 2. Takahashi K., and Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 3. Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., and Yamanaka S. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- 4. Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., Slukvin I. I., and Thomson J. A. (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 10.1126/science.1151526 [DOI] [PubMed] [Google Scholar]
- 5. Chambers S. M., Fasano C. A., Papapetrou E. P., Tomishima M., Sadelain M., and Studer L. (2009) Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275–280 10.1038/nbt.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li X. J., Du Z. W., Zarnowska E. D., Pankratz M., Hansen L. O., Pearce R. A., and Zhang S. C. (2005) Specification of motoneurons from human embryonic stem cells. Nat. Biotechnol. 23, 215–221 10.1038/nbt1063 [DOI] [PubMed] [Google Scholar]
- 7. Liu Y., Liu H., Sauvey C., Yao L., Zarnowska E. D., and Zhang S. C. (2013) Directed differentiation of forebrain GABA interneurons from human pluripotent stem cells. Nat. Protoc. 8, 1670–1679 10.1038/nprot.2013.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Watanabe K., Kamiya D., Nishiyama A., Katayama T., Nozaki S., Kawasaki H., Watanabe Y., Mizuseki K., and Sasai Y. (2005) Directed differentiation of telencephalic precursors from embryonic stem cells. Nat. Neurosci. 8, 288–296 10.1038/nn1402 [DOI] [PubMed] [Google Scholar]
- 9. Zhang S. C., Wernig M., Duncan I. D., Brüstle O., and Thomson J. A. (2001) In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat. Biotechnol. 19, 1129–1133 10.1038/nbt1201-1129 [DOI] [PubMed] [Google Scholar]
- 10. Chaffer C. L., Marjanovic N. D., Lee T., Bell G., Kleer C. G., Reinhardt F., D'Alessio A. C., Young R. A., and Weinberg R. A. (2013) Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell 154, 61–74 10.1016/j.cell.2013.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Spaderna S., Schmalhofer O., Hlubek F., Berx G., Eger A., Merkel S., Jung A., Kirchner T., and Brabletz T. (2006) A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology 131, 830–840 10.1053/j.gastro.2006.06.016 [DOI] [PubMed] [Google Scholar]
- 12. Spaderna S., Schmalhofer O., Wahlbuhl M., Dimmler A., Bauer K., Sultan A., Hlubek F., Jung A., Strand D., Eger A., Kirchner T., Behrens J., and Brabletz T. (2008) The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res. 68, 537–544 10.1158/0008-5472.CAN-07-5682 [DOI] [PubMed] [Google Scholar]
- 13. Wellner U., Schubert J., Burk U. C., Schmalhofer O., Zhu F., Sonntag A., Waldvogel B., Vannier C., Darling D., zur Hausen A., Brunton V. G., Morton J., Sansom O., Schüler J., Stemmler M. P., et al. (2009) The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 11, 1487–1495 10.1038/ncb1998 [DOI] [PubMed] [Google Scholar]
- 14. Du Z. W., Ma L. X., Phillips C., and Zhang S. C. (2013) miR-200 and miR-96 families repress neural induction from human embryonic stem cells. Development 140, 2611–2618 10.1242/dev.092809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nitta K. R., Tanegashima K., Takahashi S., and Asashima M. (2004) XSIP1 is essential for early neural gene expression and neural differentiation by suppression of BMP signaling. Dev. Biol. 275, 258–267 10.1016/j.ydbio.2004.08.010 [DOI] [PubMed] [Google Scholar]
- 16. Postigo A. A., Depp J. L., Taylor J. J., and Kroll K. L. (2003) Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. EMBO J. 22, 2453–2462 10.1093/emboj/cdg226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sabourin J. C., Ackema K. B., Ohayon D., Guichet P. O., Perrin F. E., Garces A., Ripoll C., Charité J., Simonneau L., Kettenmann H., Zine A., Privat A., Valmier J., Pattyn A., and Hugnot J. P. (2009) A mesenchymal-like ZEB1(+) niche harbors dorsal radial glial fibrillary acidic protein-positive stem cells in the spinal cord. Stem Cells 27, 2722–2733 10.1002/stem.226 [DOI] [PubMed] [Google Scholar]
- 18. Miyoshi T., Maruhashi M., Van De Putte T., Kondoh H., Huylebroeck D., and Higashi Y. (2006) Complementary expression pattern of Zfhx1 genes Sip1 and δEF1 in the mouse embryo and their genetic interaction revealed by compound mutants. Dev. Dyn. 235, 1941–1952 10.1002/dvdy.20799 [DOI] [PubMed] [Google Scholar]
- 19. Liu Y., El-Naggar S., Darling D. S., Higashi Y., and Dean D. C. (2008) Zeb1 links epithelial-mesenchymal transition and cellular senescence. Development 135, 579–588 10.1242/dev.007047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yen G., Croci A., Dowling A., Zhang S., Zoeller R. T., and Darling D. S. (2001) Developmental and functional evidence of a role for Zfhep in neural cell development. Brain Res. Mol. Brain Res. 96, 59–67 10.1016/S0169-328X(01)00267-4 [DOI] [PubMed] [Google Scholar]
- 21. Singh S., Howell D., Trivedi N., Kessler K., Ong T., Rosmaninho P., Raposo A. A., Robinson G., Roussel M. F., Castro D. S., and Solecki D. J. (2016) Zeb1 controls neuron differentiation and germinal zone exit by a mesenchymal-epithelial-like transition. eLife 5, e12717 10.7554/eLife.12717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiang Y., Zhang M. J., and Hu B. Y. (2012) Specification of functional neurons and glia from human pluripotent stem cells. Protein Cell 3, 818–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang S. C. (2006) Neural subtype specification from embryonic stem cells. Brain Pathol. 16, 132–142 10.1111/j.1750-3639.2006.00008.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang X., Huang C. T., Chen J., Pankratz M. T., Xi J., Li J., Yang Y., Lavaute T. M., Li X. J., Ayala M., Bondarenko G. I., Du Z. W., Jin Y., Golos T. G., and Zhang S. C. (2010) Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell 7, 90–100 10.1016/j.stem.2010.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pankratz M. T., Li X. J., Lavaute T. M., Lyons E. A., Chen X., and Zhang S. C. (2007) Directed neural differentiation of human embryonic stem cells via an obligated primitive anterior stage. Stem Cells 25, 1511–1520 10.1634/stemcells.2006-0707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Johnson M. A., Weick J. P., Pearce R. A., and Zhang S. C. (2007) Functional neural development from human embryonic stem cells: accelerated synaptic activity via astrocyte coculture. J. Neurosci. 27, 3069–3077 10.1523/JNEUROSCI.4562-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ullian E. M., Harris B. T., Wu A., Chan J. R., and Barres B. A. (2004) Schwann cells and astrocytes induce synapse formation by spinal motor neurons in culture. Mol. Cell. Neurosci. 25, 241–251 10.1016/j.mcn.2003.10.011 [DOI] [PubMed] [Google Scholar]
- 28. Liu H., and Zhang S. C. (2011) Specification of neuronal and glial subtypes from human pluripotent stem cells. Cell. Mol. Life Sci. 68, 3995–4008 10.1007/s00018-011-0770-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ma L., Hu B., Liu Y., Vermilyea S. C., Liu H., Gao L., Sun Y., Zhang X., and Zhang S. C. (2012) Human embryonic stem cell-derived GABA neurons correct locomotion deficits in quinolinic acid-lesioned mice. Cell Stem Cell 10, 455–464 10.1016/j.stem.2012.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wichterle H., Turnbull D. H., Nery S., Fishell G., and Alvarez-Buylla A. (2001) In utero fate mapping reveals distinct migratory pathways and fates of neurons born in the mammalian basal forebrain. Development 128, 3759–3771 [DOI] [PubMed] [Google Scholar]
- 31. Aubry L., Bugi A., Lefort N., Rousseau F., Peschanski M., and Perrier A. L. (2008) Striatal progenitors derived from human ES cells mature into DARPP32 neurons in vitro and in quinolinic acid-lesioned rats. Proc. Natl. Acad. Sci. U.S.A. 105, 16707–16712 10.1073/pnas.0808488105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., and Zhang F. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hockemeyer D., Wang H., Kiani S., Lai C. S., Gao Q., Cassady J. P., Cost G. J., Zhang L., Santiago Y., Miller J. C., Zeitler B., Cherone J. M., Meng X., Hinkley S. J., Rebar E. J., et al. (2011) Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 29, 731–734 10.1038/nbt.1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. González F., Zhu Z., Shi Z. D., Lelli K., Verma N., Li Q. V., and Huangfu D. (2014) An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell 15, 215–226 10.1016/j.stem.2014.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhu Z., González F., and Huangfu D. (2014) The iCRISPR platform for rapid genome editing in human pluripotent stem cells. Methods Enzymol. 546, 215–250 10.1016/B978-0-12-801185-0.00011-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Crews L., Adame A., Patrick C., Delaney A., Pham E., Rockenstein E., Hansen L., and Masliah E. (2010) Increased BMP6 levels in the brains of Alzheimer's disease patients and APP transgenic mice are accompanied by impaired neurogenesis. J. Neurosci. 30, 12252–12262 10.1523/JNEUROSCI.1305-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.