Abstract
The activity of X box–binding protein 1 (XBP1), a master transcriptional regulator of endoplasmic reticulum (ER) homeostasis and the unfolded protein response (UPR), is controlled by a two-step noncanonical splicing reaction in the cytoplasm. The first step of nuclease cleavage by inositol-requiring enzyme 1 (IRE1), a protein kinase/endoribonuclease, is conserved in all eukaryotic cells. The second step of RNA ligation differs biochemically among species. In yeast, tRNA ligase 1 (Trl1) and tRNA 2′-phosphotransferase 1 (Tpt1) act through a 5′-PO4/3′-OH pathway. In metazoans, RNA 2′,3′-cyclic phosphate and 5′-OH ligase (RtcB) ligate XBP1 exons via a 3′-PO4/5′-OH reaction. Although RtcB has been identified as the primary RNA ligase, evidence suggests that yeast-like ligase components may also operate in mammals. In this study, using mouse and human cell lines along with in vitro splicing assays, we investigated whether these components contribute to XBP1 splicing during ER stress. We found that the mammalian 2′-phosphotransferase Trpt1 does not contribute to XBP1 splicing even in the absence of RtcB. Instead, we found that 2′,3′-cyclic nucleotide phosphodiesterase (CNP) suppresses RtcB-mediated XBP1 splicing by hydrolyzing 2′,3′-cyclic phosphate into 2′-phosphate on the cleaved exon termini. By contrast, RNA 3′-terminal cyclase (RtcA), which converts 2′-phosphate back to 2′,3′-cyclic phosphate, facilitated XBP1 splicing by increasing the number of compatible RNA termini for RtcB. Taken together, our results provide evidence that CNP and RtcA fine-tune XBP1 output during ER stress.
Keywords: X-box binding protein 1 (XBP1), signaling, RNA splicing, unfolded protein response (UPR), stress response, gene regulation, RNA processing, CNP, RtcA, RtcB, Trpt1
Introduction
The endoplasmic reticulum (ER)2 serves many diverse functions that include the synthesis, translocation, folding, and modification of secretory and membrane proteins (1). The protein-rich environment makes this organelle highly susceptible to physiological or pathological perturbations of proteostasis, which can trigger ER stress. An adaptive signaling network known as the unfolded protein response (UPR) has evolved to regulate ER function in response to such perturbations (2). The mammalian UPR consists of three parallel signaling mechanisms: (i) noncanonical splicing through IRE1α, (ii) translation control by pancreatic endoplasmic reticulum kinase (PERK), and (iii) proteolysis and translocation of activating transcription factor 6 (ATF6) (2, 3). Collectively, signaling initiated by these three ER-resident sensors leads to inhibition of protein loads, degradation of unfolded proteins, and an increase of ER chaperones to restore ER function (4).
The IRE1 branch of the UPR is conserved from yeast to mammals (2). The primary signaling output for this branch is a basic leucine zipper transcription factor, XBP1 in metazoans and Hac1 in yeast (5, 6). During ER stress, XBP1/Hac1 mRNA undergoes two-step unconventional splicing to generate a mature transcript that encodes full-length active XBP1/Hac1 protein. In the first step, yeast IRE1 cleaves a 252-nucleotide intron from Hac1 precursor mRNA (7), whereas mammalian IRE1α cleaves a 26-nucleotide intron from the XBP1u mRNA (8, 9). Precise removal of the introns extends the reading frame to encode a longer and active form of the transcription factors. In the second splicing step, yeast and mammalian cells utilize different RNA ligases to rejoin cleaved exons. Upon IRE1 cleavage, HAC1/XBP1 splice sites terminate with 2′,3′-cyclic phosphate and 5′-OH, respectively (10, 11). In yeast, a multifunctional enzyme, Trl1, works through a 5′-PO4/3′-OH (5′–3′) ligation mechanism (12). The reaction in yeast involves four steps to complete the ligation: (i) hydrolysis of 2′,3′-cyclic phosphate bond, (ii) phosphorylation of the 5′-hydroxyl group, (iii) ligation of the two ends, and (iv) removal of 2′-phosphate from the junction (13–16). Trl1 catalyzes the first three steps of the 5′–3′ pathway using different enzyme activities that reside in three independent functional domains (cyclic phosphodiesterase, polynucleotide kinase, and RNA ligase) (14). To complete the reaction, the 2′-phosphotransferase Tpt1 removes 2′-phosphate of the noncanonical 2′-phosphomonoester, 3′,5′-phosphodiester linkage (16, 17). By contrast, mammalian cells utilize RtcB in a single-step 3′-PO4/5′-OH (3′–5′) ligation reaction to reunite IRE1-cleaved XBP1 exons (18–21). Despite a mechanistically different ligation step among species, noncanonical mRNA splicing during the UPR results in functionally active proteins, XBP1s or HAC1. Both transcription factors act to elicit a robust UPR transcriptional program that increases protein folding capacity and reestablishes homeostasis in the ER (5, 6).
Several labs, including ours, have recently identified RtcB as a UPR RNA ligase responsible for XBP1 splicing during ER stress in metazoans (18–20). Although it is evident that RtcB contributes to the majority of RNA ligase activity in mammalian cells, existing evidence suggested that a yeast-like 5′–3′ RNA ligase may also operate in mammalian cells. For example, a previous biochemical study showed that HeLa cell extracts possessed a ligase activity that utilized γ-phosphate of ATP to form the phosphodiester bond at the tRNA exon-exon junction, leaving a 2′-phosphate (22). However, the identity of such RNA ligase remains unknown. It has also been shown that mammalian genomes encode some proteins that share parts of the enzymatic activities required for the yeast 5′–3′ RNA ligation. These proteins include 2′,3′-cyclic nucleotide phosphodiesterase (CNP), Clp1 (RNA 5′-kinase), and Trpt1 (2′-phosphotransferase). Moreover, CNP was shown to complement the loss of Trl1's cyclic phosphodiesterase activity in yeast (23). Similarly, human Clp1 and Trpt1 were able to rescue the corresponding mutations in yeast (24, 25). Thus, these proteins may participate in noncanonical RNA splicing as a parallel pathway in mammals. Although a role for these proteins in the UPR has not yet been demonstrated, genetic studies clearly show that mutations in CNP (26) and Clp1 (27, 28) are implicated in neurodegeneration. CNP-deficient mice were shown to develop axonal swelling and neurodegeneration throughout the brain that led to hydrocephaly and premature death (26). Mutations abolishing Clp1 kinase activity also resulted in neurodegeneration in mice (29), humans (27, 28), and zebrafish (27). Given the known enzymatic activities of CNP and Clp1, molecular deficits associated with 5′–3′ RNA ligation (e.g. XBP1 splicing) may contribute to degenerative phenotypes seen in the nervous system.
In contrast, Trpt1−/− mice showed an overall normal phenotype (30). Although the 2′-phosphotransferase activities were not detectable in Trpt1-null cells, no XBP1 splicing defect was found (31). These results suggested that a 5′–3′ RNA ligation pathway may not contribute significantly to the outcome of XBP1 splicing. One caveat for this interpretation is that RtcB-mediated 3′–5′ and putative 5′–3′ RNA ligation pathways may be redundant in this regard. In this scenario, the major RNA ligase RtcB could potentially mask the contribution of 5′–3′ RNA ligation or Trpt1. Consistent with this possibility, we observed residual XBP1 splicing activity in RtcB conditional knockout cells (18). Moreover, the residual activity was nearly abolished by genetic rescue with a ligase-dead RtcB, suggestive of a compensatory unknown RNA ligase (18).
In this study, we set to further investigate the function of putative 5′–3′ RNA ligation in mammalian cells. First, we performed Trl1 genetic rescue, showing that 5′–3′ RNA ligase was sufficient to rescue RtcB deficiency in mammalian cells and that Trpt1 was required for an operational 5′–3′ RNA ligation pathway. However, by generating cells deficient for both RtcB and Trpt1, we found that Trpt1 did not have a contribution to the endogenous XBP1 splicing. Next, we examined the role of CNP, another component of the 5′–3′ pathway. We demonstrated that CNP acts to negatively regulate RtcB-dependent XBP1 splicing instead of working in parallel. Furthermore, we showed that RNA 3′-terminal cyclase (RtcA) counteracts CNP to enhance XBP1 splicing. Collectively, our results elucidate a new mechanism that controls the level of XBP1 splicing during ER stress.
Results
Yeast 5′–3′ RNA ligase Trl1 is able to rescue RtcB deficiency
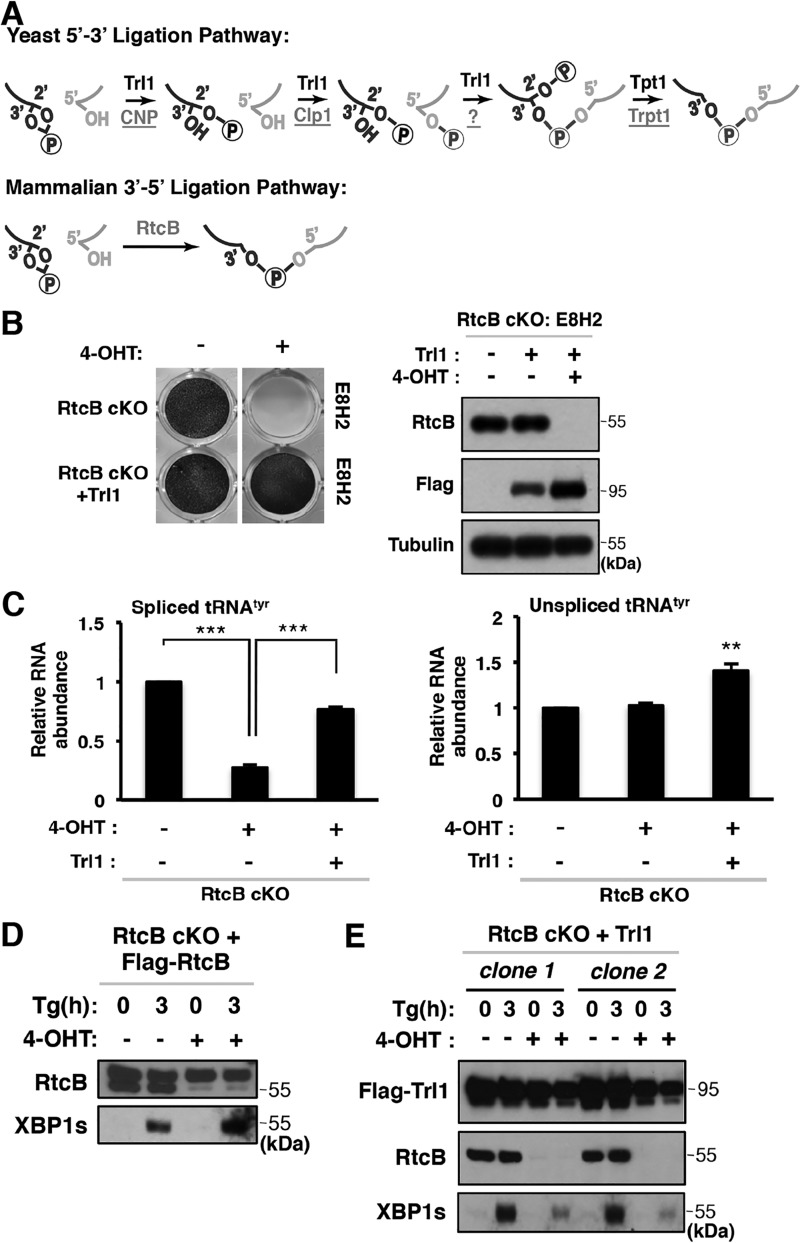
Two distinct biochemical pathways have been described to mediate noncanonical RNA splicing downstream of endonucleolytic cleavage in yeast and mammalian cells (Fig. 1A) (32, 33). Although it is evident that RtcB acts as a major RNA ligase in mammals, earlier biochemical evidence suggested that a yeast-like RNA ligation pathway may operate in mammalian cells (22, 27). Consistent with this idea, putative mammalian candidate proteins (CNP, Clp1, and Trpt1) that can act sequentially in the 5′–3′ pathway have been identified (Fig. 1A, top). As a first step to test this hypothesis, we carried out a genetic rescue experiment using the known multifunctional yeast 5′–3′ RNA ligase Trl1. We had previously generated an RtcB conditional knockout (RtcB cKO) mouse ES cell line (E8H2) (18). Upon 4-OHT–induced RtcB deletion, E8H2 cells failed to survive due to RNA ligation defects in tRNA splicing. As shown in Fig. 1B, overexpression of Trl1 in RtcB cKO cells was able to rescue the viability of RtcB-deficient cells upon 4-OHT treatment. In line with this cell viability result, the defect of tyrosine tRNA splicing in RtcB-depleted cells (4-OHT–treated E8H2) was also rescued by yeast Trl1 (Fig. 1C). As shown previously (18), FLAG-RtcB cDNA expression completely rescued XBP1 splicing in E8H2 cells (Fig. 1D). In comparison, Trl1 expression in E8H2 cells partially rescued XBP1s protein expression upon ER stress when RtcB was depleted (Fig. 1E). Taken together, our data show that yeast 5′–3′ RNA ligase can function in tRNA and XBP1 splicing in mammalian cells.
Figure 1.
Genetic rescue of RtcB-mediated RNA ligation by yeast Trl1. A, schematic representation of yeast (5′–3′ ligation) and mammalian (3′–5′ ligation) RNA ligation pathways and mammalian proteins involved in a putative 5′–3′ pathway. Yeast proteins are denoted in black, and mammalian proteins are underlined. B, images of methylene blue staining show that RtcB cKO ES cells survived in the absence of RtcB when yeast Trl1 was expressed. Western blotting confirmed that RtcB was depleted in E8H2 RtcB cKO ES cells after 4-OHT treatment and the rescued cells expressed Trl1. C, RT-qPCR analysis showed that the level of spliced tRNATyr dropped significantly in RtcB-depleted cells, whereas Trl1-expressing RtcB cKO cells exhibited a significantly higher level of tRNATyr. Expression levels were normalized to GAPDH mRNA levels and to the untreated control E8H2 cells. The error bars represent S.E. The results were subjected to two-tailed t tests. **, p < 0.01; ***, p < 0.001. D, FLAG-RtcB serves as a positive control for a complete rescue of XBP1s expression in E8H2. In RtcB Western blotting, the top bands are FLAG-RtcB, and the bottom bands are endogenous RtcB. ER stress was induced with thapsigargin (Tg) for 3 h. E, Western blots show that yeast Trl1 protein partially rescued the expression of XBP1s protein.
Mammalian Trpt1 is required for Trl1-mediated rescue of RtcB cKO cells
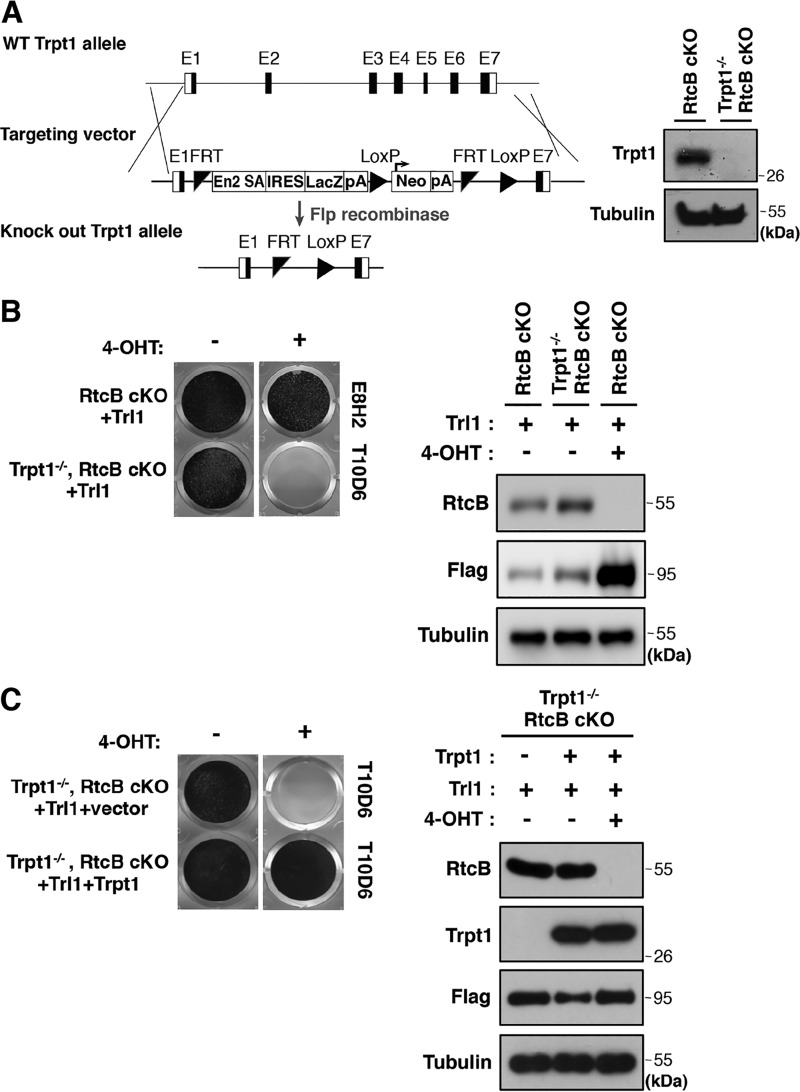
Yeast Trl1 contains three of four enzymatic activities that are required to fulfill a functional 5′–3′ RNA ligation pathway (14). A 2′-phosphotransferase (Trpt1 in mammals) is essential to remove 2′-phosphate at Trl1-ligated splice junctions (34). To test whether Trl1 genetic rescue in RtcB cKO cells requires Trpt1, we generated a Trpt1 and RtcB double knockout ES cell line (T10D6) using a gene-targeting strategy (Fig. 2A, left). Western blot analysis confirmed that no Trpt1 was expressed in T10D6 cells (Fig. 2A, right). A previous study showed that Trpt1-null mouse embryonic fibroblasts have no detectable 2′-phosphotransferase activity (31). In contrast with Trpt1+/+ E8H2 cells, Trl1 failed to rescue the cell viability in Trpt1−/− RtcB cKO T10D6 cells when both proteins were absent (Fig. 2B). As a control, we carried out genetic rescue experiments using both Trl1- and Trpt1-expressing PiggyBac vectors in T10D6 cells (Fig. 2C). When both Trl1 and Trpt1 were reintroduced, RtcB deficiency was successfully rescued. Thus, our results demonstrate that a fully functional 5′–3′ yeast-like RNA ligation pathway is required to compensate for RtcB function in mammalian cells.
Figure 2.
Mammalian 2′-phosphotransferase Trpt1 is required for Trl1-mediated rescue of RtcB cKO ES cells. A, representation of the mouse Trpt1 genomic locus, the targeting construct, and the targeted locus. After FLP recombinase–mediated recombination, the NeoR cassette is removed. In the Trpt1 knockout alleles, exons 2–6 are deleted. The Western blot shows the loss of Trpt1 expression in the generated Trpt1−/−, RtcB cKO ES cell line (T10D6). B, methylene blue staining shows the survival of RtcB cKO cells but not Trpt1, RtcB double KO cells with Trl1 expression in the presence of 4-OHT for 7 days. Western blots show that RtcB was depleted in Trl1 rescued cells and Trl1 was up-regulated after prolonged 4-OHT selection. C, methylene blue staining shows the genetic rescue of Trpt1, RtcB double KO T10D6 cells with Trl1 and Trpt1 after 4-OHT treatment. Western blotting verified the expression of Trpt1, RtcB, and Trl1 proteins.
Trpt1 does not significantly contribute to endogenous XBP1 splicing
We previously showed that RtcB-depleted cells maintained a low level of XBP1 splicing and that expression of ligase-dead RtcB in RtcB cKO cells caused a nearly complete loss of XBP1 splicing (18). One interpretation is that an additional RNA ligase might compensate for RtcB function. Given a previous report that describes the activity of a yet-to-be-identified 5′–3′ RNA ligase in HeLa cell extract (22), we set out to test whether putative components of a mammalian 5′–3′ RNA ligation pathway contribute to endogenous XBP1 splicing.
It has been previously shown that mice deficient for Trpt1 have no obvious defect in the UPR-induced XBP1 splicing (31). As described in Fig. 2, Trpt1 is essential for a 5′–3′ RNA ligation pathway to operate in mammalian cells. Considering the recent finding that RtcB plays a predominant role in mammalian RNA ligation reactions (18–20), we reasoned that a role for the 5′–3′ RNA pathway could potentially be masked by RtcB in the previous Trpt1 knockout mouse study (31). Therefore, we took advantage of our Trpt1 and RtcB double KO cells to reexamine this possibility. Using RT-PCR analysis, we compared levels of spliced XBP1 mRNA in Trpt1+/+ and Trpt1−/− RtcB cKO cells after tunicamycin induction. We did not observe a significant difference in XBP1 splicing between Trpt1+/+ (E8H2) and Trpt1−/− (T10D6) before or after RtcB was depleted (Fig. 3, A and B). Because it has been suggested that the splice junction 2′-phosphate interferes with translation (31, 35), we monitored XBP1s protein levels using Western blot analysis (Fig. 3C). Upon induction of ER stress by tunicamycin or DTT, XBP1s protein levels were not further down-regulated in the absence of Trpt1 (Fig. 3C). These results indicated that the remaining XBP1s was not due to a mammalian 5′–3′ RNA ligation pathway but might be due to residual RtcB activity after 4-OHT treatment or an unknown 3′–5′ RNA ligase. We also compared tRNATyr splicing in Trpt1+/+ and Trpt1−/− RtcB cKO cells (Fig. 3D). Consistent with XBP1 splicing, we did not observe a significant difference in spliced tRNATyr levels between Trpt1+/+ and Trpt1−/− RtcB cKO cells. Thus, Trpt1 does not have any significant contribution to endogenous noncanonical RNA splicing in mammalian cells.
Figure 3.
Trpt1 does not affect XBP1 or tRNATyr splicing even in the absence of RtcB in ES cells. A, RT-PCR analysis comparing spliced and unspliced XBP1 mRNA levels in RtcB cKO and Trpt1−/−, RtcB cKO cells. RtcB was depleted following a 3-day 4-OHT treatment. To induce ER stress, the cells were treated with tunicamycin (TUNI) for 4 h. The ladder is denoted with L. B, RT-qPCR analysis to quantify XBP1 splicing. Both spliced and total XBP1 levels were normalized to GAPDH. C, XBP1s protein levels were analyzed using chemiluminescence (top) and quantitative (bottom) Western blotting. The cells were treated with tunicamycin for 6 h or with DTT for 4 h followed by a 2-h recovery. Relative intensities of XBP1s are shown below the blot. D, RT-qPCR analysis of spliced and unspliced tRNATyr levels shows that spliced tRNATyr levels did not differ significantly between E8H2 and T10D6 cells. Error bars represent S.E. CTL, control.
CNP inhibits RtcB-mediated XBP1 splicing in vitro and in vivo
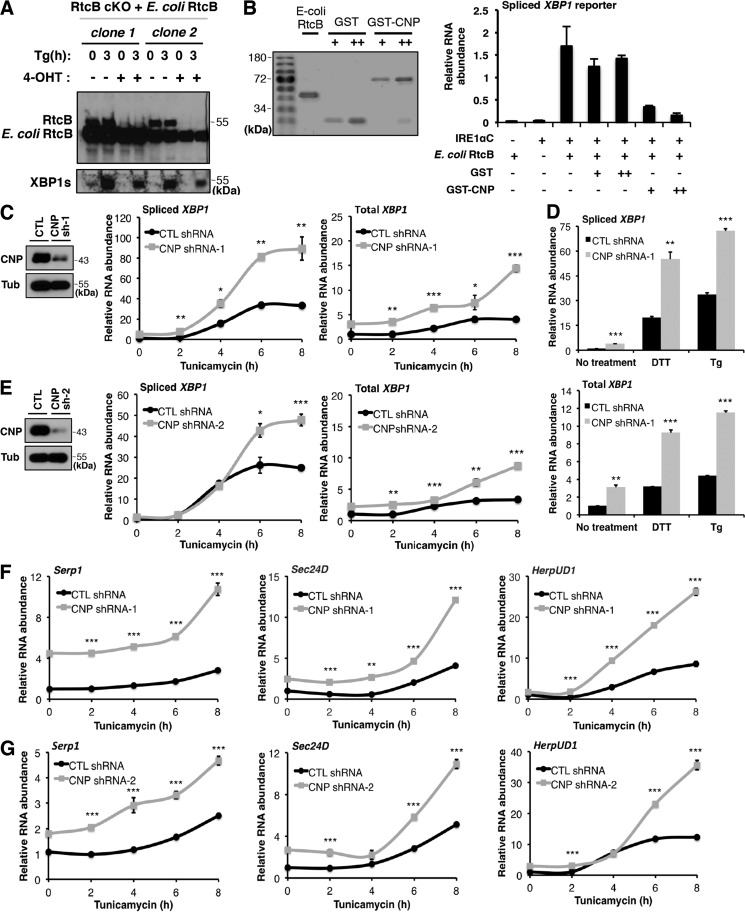
Our results thus far show that Trpt1, an essential enzyme of the 5′–3′ RNA ligase pathway, is not involved in XBP1 splicing. This finding raises the question of how other putative components such as CNP and Clp1 are involved in the regulation of noncanonical RNA splicing. CNP is the only mammalian enzyme known to hydrolyze the 2′,3′-cyclic phosphate bond, giving rise to RNA termini with 2′-PO4 and 3′-OH, in the first step of 5′–3′ RNA ligation (36). RtcB requires RNA substrates with either 2′,3′-cyclic phosphate or 3′-PO4 and 2′-OH at the 3′-end and those with 5′-OH at the 5′-end to complete a ligation reaction (21, 37, 38). Thus, if CNP hydrolysis occurs at cleaved RNA termini, those RNA termini cannot be ligated by RtcB due to different substrate requirements for 3′–5′ and 5′–3′ RNA ligases. Consequently, CNP is expected to inhibit RtcB-mediated XBP1 splicing by converting cleaved exons to an incompatible form rather than positively contributing to XBP1 splicing. To test this hypothesis, we first investigated the effect of CNP on XBP1 splicing in vitro. To efficiently reconstitute in vitro XBP1 splicing reactions, we chose to purify recombinant His-tagged Escherichia coli RtcB protein because E. coli RtcB is soluble and highly active, and pure RtcB is easily generated (Fig. 4, A and B). Importantly, we showed that E. coli RtcB is fully functional by genetic rescue experiments in RtcB cKO ES cells (Fig. 4A). Using recombinant RtcB and IRE1α cytoplasmic domain (IRE1αC; from a commercial source), we reconstituted the XBP1 splicing reaction in vitro (Fig. 4B). By adding increasing amounts of purified GST-CNP or GST proteins, RT-qPCR analysis showed that GST-CNP but not GST alone inhibited RtcB-dependent XBP1 splicing (Fig. 4B). Thus, CNP can suppress RtcB-mediated ligation in vitro.
Figure 4.
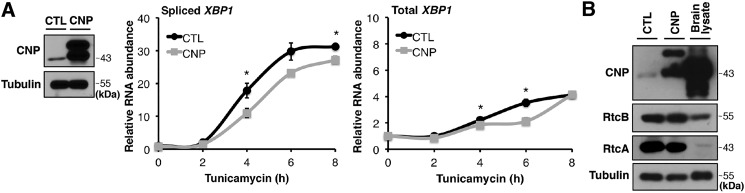
CNP inhibits RtcB-mediated XBP1 splicing in vitro and in vivo. A, E. coli RtcB rescued mammalian XBP1 splicing when endogenous RtcB was depleted. B, recombinant CNP inhibited XBP1 splicing in vitro. Coomassie Blue staining shows affinity-purified E. coli RtcB, GST, and GST-CNP proteins. The purified proteins were diluted to have similar protein concentrations and added to the in vitro splicing reaction of an XBP1u intron–containing in vitro transcribed RNA as described in Lu et al. (18). The RNA products were purified and analyzed by RT-qPCR. The results were normalized to the total reporter RNA levels in the reactions. C, knockdown of CNP by shRNA-1 (sh-1) in HEK293T cells. CNP down-regulation was verified by Western blotting. Upon ER stress induction by tunicamycin for the indicated times, spliced and total XBP1 mRNA levels were quantified by RT-qPCR. D, as in C, ER stress was induced in CNP knockdown cells by DTT for 2 h and thapsigargin (Tg) for 3 h. Spliced (top panel) and total (bottom panel) XBP1 mRNA levels were quantified. E, to control for off-target effects of RNAi, the experiment in C was repeated with a different shRNA (CNP shRNA-2 (sh-2)). F and G, up-regulation of XBP1 target genes in CNP knockdown cells. Expression of previously identified XBP1 targets (Serp1, Sec24D, and HerpUD1) were analyzed by RT-qPCR. GAPDH was used for normalization. The results are shown as mean ± S.E. (error bars) and were subjected to two-tailed t tests. *, p < 0.05; **, p < 0.01; ***, p < 0.001. CTL, control.
To test whether CNP plays an inhibitory role in endogenous XBP1 splicing during ER stress, we first knocked down endogenous CNP protein in HEK293T cells using PiggyBac transgenics (39) to stably express shRNA against CNP. When comparing the control with CNP shRNA–expressing cells, quantitative analysis of XBP1 splicing showed that during a time course of ER stress induction by tunicamycin, down-regulation of CNP significantly increased XBP1 splicing (Fig. 4C). Similar results were seen with other UPR inducers, including DTT and thapsigargin (Fig. 4D). To rule out off-target effects of shRNA, we verified the effect of CNP knockdown on XBP1 splicing with a different shRNA (Fig. 4E). In addition to the up-regulation of spliced XBP1 levels, we also observed a consistent increase of total XBP1 mRNA levels upon CNP knockdown. This observation is consistent with the existence of a known positive feedback loop of XBP1 on its own transcription (6, 40). Changes of total XBP1 mRNA levels were previously observed in cells defective in XBP1 splicing (18). Furthermore, we tested whether the observed increase of XBP1s had an effect on other known XBP1 target genes (6, 41). Using RT-qPCR, we showed that Serp1, Sec24D, and HerpUD1 mRNA levels increased upon CNP depletion (Fig. 4, F and G).
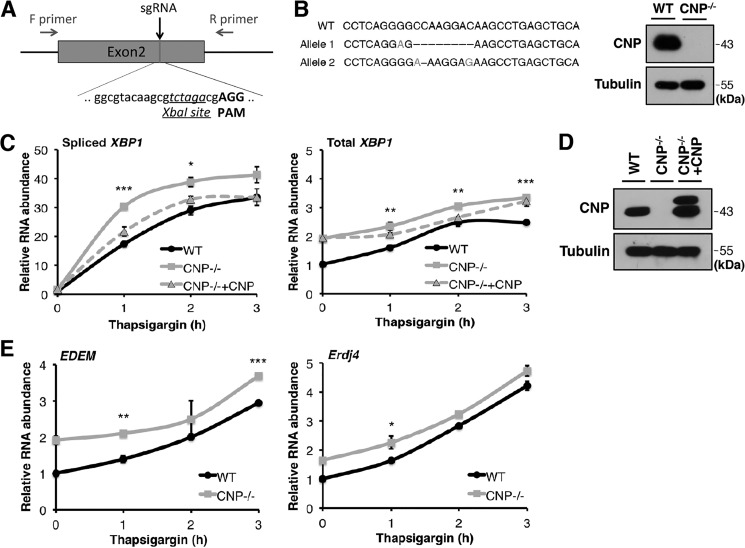
To further validate the role of CNP in XBP1 splicing in mammalian cells, we designed an sgRNA targeting exon 2 of the mouse CNP gene (Fig. 5A). By coexpression of CNP sgRNA and Cas9 in mouse ES cells, we generated CNP KO cells (Fig. 5B). The sgRNA we designed specifically targets mouse but not human CNP cDNA, thus allowing us to carry out a rescue experiment with a human CNP cDNA expression vector. Using CNP KO and hCNP-rescued KO cells, we compared the induction of XBP1 splicing during ER stress. After thapsigargin treatment for 1, 2, and 3 h, we observed more robust XBP1 splicing in CNP−/− cells and XBP1 splicing levels approximating WT levels in hCNP-introduced CNP−/− cells (Fig. 5, C and D). Similarly, CNP knockout in mouse ES cells lead to up-regulated expression of EDEM and Erdj4, both known to be XBP1 target genes (Fig. 5E). In summary, with two different experimental approaches and in two different cell types, our data demonstrate that CNP negatively regulates XBP1 splicing during the UPR.
Figure 5.
Up-regulation of XBP1s in CNP-deficient mouse ES cells. A, design of an sgRNA targeting an XbaI site in exon 2 of mouse CNP gene. Resulting clones were genotyped by PCR amplification of exon 2 and scanned by XbaI digestion. F, forward; R, reverse. B, a CNP−/− ES cell clone was identified with frameshift mutations in both alleles. Western blotting showed the loss of CNP protein. C, RT-qPCR analysis of spliced and total XBP1 mRNA levels in WT, CNP−/−, and genetically rescued CNP−/− ES cells. ER stress was induced with thapsigargin for 1, 2, and 3 h. D, CNP protein levels in the samples analyzed in C. Transfected hCNP was tagged with 3xFLAG. We observed that the translation was initiated from the start codons of both FLAG and CNP; thereby two bands were observed in the Western blot. E, expression of XBP1 target genes EDEM and Erdj4 were analyzed by RT-qPCR. Error bars represent S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Overexpression of CNP attenuates XBP1 splicing in HEK293T cells
In light of our CNP loss-of-function results, we next tested whether CNP overexpression would lead to down-regulation of XBP1 splicing during the UPR in 293T cells. We were able to express exogenous FLAG-tagged CNP at a significantly higher level than the endogenous protein in 293T cells. Consistent with our hypothesis, we showed that increased CNP levels attenuated XBP1 splicing (Fig. 6A). However, we noted that the effect of CNP overexpression on XBP1 splicing in cells was modest compared with that seen in the in vitro assay (Fig. 4B).
Figure 6.
The effect of CNP overexpression on XBP1 splicing. A, HEK293T cells were transfected with control cDNA (FLAG-Sox2) or FLAG-CNP–expressing PB transposon. Upon ER stress, spliced and total XBP1 mRNA levels were analyzed by RT-qPCR. Error bars represent S.E. *, p < 0.05. Overexpression of CNP was confirmed by Western blot analysis. B, Western blot analysis of CNP, RtcB, and RtcA proteins in parental and CNP-overexpressing HEK293T cells and brain lysate. CTL, control.
Several reasons may account for the modest effect of CNP overexpression. First, in our experimental system, we did not achieve a comparably high level of CNP expression as reported in the brain tissue (Fig. 6B and Refs. 36 and 42). The extremely abundant CNP protein in the nervous system likely has a strong inhibitory effect on XBP1 splicing. Alternatively, RtcA has been described to counteract CNP by converting RNA termini of 2′-PO4 and 3′-OH back to a form with 2′,3′-cyclic phosphate in vitro (43–45). Noticeably, RtcB and RtcA have lower levels of expression in the brain compared with 293T cells (Fig. 6B). The presence of RtcA in 293T cells may have contributed to a less pronounced effect of CNP overexpression.
Regulation of XBP1 splicing by RtcA
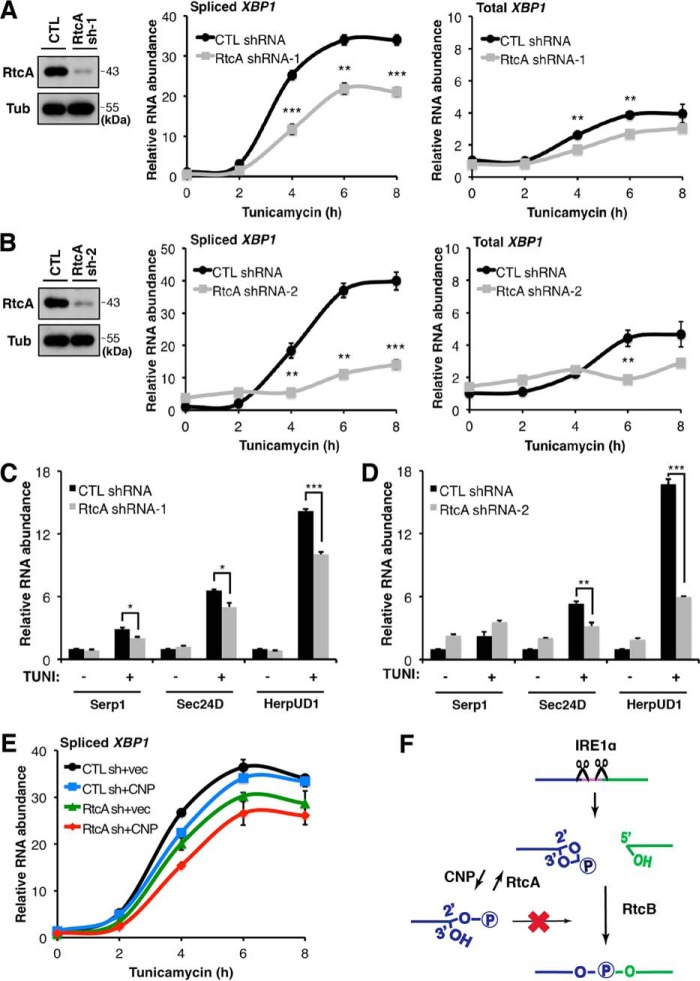
To investigate the possibility of RtcA counteracting CNP in the regulation of XBP1 splicing, we knocked down endogenous RtcA protein in 293T cells and tested levels of XBP1 splicing during ER stress. As shown in Fig. 4, ER stress was induced by tunicamycin, and XBP1 splicing was monitored in a time course. Our results showed that XBP1 splicing significantly decreases in RtcA–down-regulated cells compared with the control (Fig. 7, A and B). Moreover, mRNA levels of previously characterized XBP1 target genes, Serp1, Sec24D, and HerpUD1, were also down-regulated upon RtcA knockdown (Fig. 7, C and D).
Figure 7.
Regulation of XBP1 splicing by RtcA. A and B, RtcA knockdown attenuated XBP1 splicing. 293T cells were transfected with control (CTL) and two different RtcA shRNAs (sh). RtcA knockdown was verified by Western blotting. Spliced and total XBP1 mRNA levels were analyzed by RT-qPCR. C and D, expression levels of XBP1 target genes were down-regulated in RtcA knockdown cells following tunicamycin treatment for 8 h. RT-qPCR analysis was performed as described in Fig. 4, F and G. Error bars represent S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001. E, RT-qPCR analysis of XBP1 splicing in 293T cells transfected with a combination of shRNA and cDNA-expressing vectors. Down-regulation of RtcA and CNP overexpression in combination exhibited a more dramatic effect on XBP1 splicing. F, a working model depicting how IRE1α–RtcB-mediated XBP1 splicing is regulated by CNP and RtcA during ER stress. Tub, tubulin; vec, vector.
If RtcA and CNP counteracted each other to regulate the substrate availability for RtcB ligation, then a combination of CNP overexpression and RtcA down-regulation should enhance inhibitory effects on XBP1 splicing. To test this hypothesis, we coexpressed CNP cDNA and RtcA shRNA in 293T cells. Compared with the controls, the combination of RtcA shRNA and CNP cDNA expression achieved a maximal inhibitory effect on XBP1 splicing (Fig. 7E).
In summary, the data presented here support a working model in which mammalian components that possess individual enzymatic activities corresponding to a single step of the yeast 5′–3′ RNA ligation pathway exhibit regulatory functions to control the substrate availability for the RtcB-mediated ligation reaction downstream of IRE1α cleavage during the UPR. In this regulation, CNP and RtcA counteract each other to fine-tune the appropriate level of compatible RNA substrates for RtcB and control the signaling output of spliced XBP1 mRNA (Fig. 7F).
Discussion
The UPR contributes to the pathophysiological process of a variety of human diseases ranging from diabetes to neurodegeneration to cancer (46–50). Genetic manipulations of this pathway have revealed a complex picture in which modulation of a UPR signaling output can have context-specific and even opposite effects on the disease progression in animal models. For example, XBP1 knockout in pancreatic β-cells led to reduced insulin secretion and loss of β-cells (51). However, overexpression of XBP1 surprisingly induced β-cell apoptosis and down-regulated insulin expression (52). In contrast, XBP1 knockout in the nervous system promoted neuroprotection in ALS (53) and Huntington's disease mouse models (54), whereas XBP1s overexpression by adeno-associated virus improved neuron and oligodendrocyte survival in Parkinson's disease (55) and spinal cord injury models (56). Importantly, changes in XBP1 gene dosage in XBP1+/− animals appeared to be sufficient to affect the degeneration of the retina (57) and spiral ganglion cells (58). Furthermore, a polymorphism in human XBP1 promoter that impairs its transcriptional feedback has been associated with risk of Alzheimer's disease (59) and bipolar disorder (60). In addition, XBP1 has been shown to be transcriptionally up-regulated by brain-derived neurotrophic factor and brain activity (50). Taken together, fine-tuning of spliced XBP1 levels is critical for specific cell types to cope with the cellular stress associated with disease pathogenesis. During ER stress, XBP1 splicing is a hallmark and the primary signaling output of the conserved IRE1 UPR branch (5, 6). This noncanonical mRNA splicing consists of two essential steps: the cleavage of precursor XBP1 mRNA by IRE1α and the ligation of cleaved XBP1 exons. In principle, regulations in either step can alter the level of XBP1 splicing. In the past, much attention has been focused on the regulation of IRE1 activity during ER stress in part because RtcB was only recently identified as the mammalian UPR RNA ligase (18–20). In this study, by focusing on mammalian proteins involved in a parallel yeast-like 5′–3′ RNA ligation reaction, we demonstrate that CNP and RtcA regulate the output of XBP1 splicing by enzymatic conversion of compatible substrates for RNA ligation. More importantly, the fine-tuning of XBP1 splicing by CNP and RtcA significantly alters the expression levels of direct targets of XBP1. Thus, this regulation can play a critical role in the progression of disease states (e.g. neurodegeneration) associated with ER stress.
Prior to this study, the role of a purported 5′–3′ RNA ligation pathway in the UPR remained elusive (61). Using cells deficient for both RtcB and Trpt1, we show that a 5′–3′ RNA ligation pathway, if it exists, does not act as a parallel/compensatory mechanism to generate a significant amount of spliced XBP1 even in the absence of the primary ligase RtcB. Instead, the components in the pathway have a regulatory function to modulate levels of RtcB-mediated ligation. We show that the 2′,3′-cyclic phosphodiesterase component of the 5′ RNA ligation pathway, CNP, inhibits XBP1 splicing. CNP is expressed as one of the most abundant and long-lived proteins in the nervous system (36, 42). CNP knockout mice suffer from axonal loss and neurodegeneration (26). Given its inhibitory role in XBP1 splicing shown in this study, we hypothesize that upon ER stress, CNP desensitizes XBP1 splicing in the nervous system. The IRE1–RtcB–XBP1 axis of the UPR is likely hyperactivated in CNP knockout mice. Hyperactivation of XBP1 splicing provides a plausible explanation for the neurodegenerative phenotype observed with CNP deficiency. However, no analysis of the UPR has been reported for the CNP knockout mice. It is therefore important to quantitatively analyze both acute and chronic ER stress responses, including XBP1 splicing, to further test this hypothesis.
In addition, our study shows that RtcA with an opposite enzyme activity facilitates XBP1 splicing during the UPR. Thus, the balancing act between CNP and RtcA is crucial for maintenance of appropriate levels of XBP1s; hence, skewing this balance in one direction influences the UPR output and cell fate decision. Consistent with our hypothesis, genetic alterations of RtcA appear to have an opposite neuronal phenotype compared with CNP knockout (62). Loss of RtcA function increased axon regeneration in Drosophila, whereas its overexpression decreased the regeneration capacity after injury. Similarly, RtcB was also shown to inhibit axon regeneration in the GABA motor neurons of Caenorhabditis elegans (63).
Clp1, another putative component in the 5′–3′ pathway, has a kinase activity that is capable of phosphorylating the 5′-OH in cleaved exons. Interestingly, mice expressing a kinase-dead Clp1 exhibited a progressive loss of lower motor neurons (29). Homozygous mutations in Clp1 have also been identified in human patients with severe motor sensory defects, cerebral neurodegeneration, and microcephaly (27, 28). Thus, a deficiency in 5′–3′ RNA ligation components appears to manifest itself in the nervous system. To define the precise role of Clp1 in UPR regulation, future biochemical and genetic experiments are necessary.
Experimental procedures
Plasmid construction
Vectors for sgRNA or shRNA were generated with oligos from Integrated DNA Technologies (Table S1). Phosphorylated DNA oligos were annealed and ligated into a BbsI site in the PiggyBac (PB) vector (39) with mouse U6 promoter to express shRNA or sgRNA, respectively. For cDNA-expressing PB vectors, cDNAs amplified with Q5 polymerase from New England Biolabs were cloned downstream of CAG promoter between EcoRI and XhoI sites in a PB vector. For bacterial protein expression plasmids, cDNAs were cloned into pGEX-KG (for GST tag) or pET28b.
Cell culture and ER stress induction
HEK293T cells were cultured in Dulbecco's modified Eagle's medium (4.5g/liter d-glucose; Gibco) with 10% fetal bovine serum (Benchmark FBS, Gemini) and 1× penicillin-streptomycin-glutamine (Gibco). Mouse AB2.2 ES cells (mESCs) were cultured with knockout Dulbecco's modified Eagle's medium with 15% fetal bovine serum, 1× penicillin-streptomycin-glutamine, 100 μm β-mercaptoethanol, and mouse leukemia inhibitory factor. mESCs were routinely cultured on feeder cells. For phenotype analysis, mESCs were cultured on a gelatinized surface without feeder cells. For the cell survival experiments, RtcB deletion was induced by 4-OHT treatment at a final concentration of 1 μm in E8H2 and T10D6 mESCs as described previously (18). Viable cells were stained on the plates with 2% methylene blue in 70% ethanol and destained with distilled water.
For stable shRNA transfection, HEK293T cells were cotransfected with shRNA-expressing PB vectors and a PBase-expressing plasmid using Lipofectamine 2000 (Life Technologies). Transfected cells were selected with 2 μg/ml puromycin for shRNA expression. For stable cDNA-expressing cells, 293T cells were selected with 10 μg/ml blasticidin. After 2-day selection and 1-day drug-free recovery, the stably transfected HEK293T cells were subjected to UPR induction with 0.2 μg/ml tunicamycin, 1 mm DTT, or 0.4 μg/ml thapsigargin for the indicated times. For UPR induction in mESCs, 2 μg/ml tunicamycin, 0.15 μg/ml thapsigargin, or 2 mm DTT was used.
Gene targeting in mESCs
To generate Trpt1 knockout mESCs, we obtained Trpt1 gene–targeting vector (DPGS00168_A_D11) from the Knock Out Mouse Project (KOMP) through BACPAC Resources, Children's Hospital Oakland Research Institute (64). The E8H2 RtcB cKO cell line was electroporated with the linearized targeting vector, and after neomycin selection, this cassette was removed with transient FLP expression in the first-round targeted clones. The mESCs homozygous for both targeted alleles were obtained by the repeated gene targeting process described above.
To generate CNP knockout mESCs, we coelectroporated a CNP sgRNA–expressing PB vector with a Cas9 expression vector from Addgene. Individual ES clones were screened for loss of the XbaI site in the genome, and the positive clones were verified by DNA sequencing and Western blot analysis.
RNA extraction and quantitative PCR analysis
Total RNA was extracted using TRIzol according to the manufacturer's protocol (Life Technologies). Following DNase I (Roche Applied Science) treatment, cDNA was synthesized with oligo(dT) or tRNATyr-specific primer 3918 using a Thermo Fisher reverse transcription kit following the company's protocol. RT-qPCRs were performed with Power SYBR Green PCR Mix (Life Technologies) to quantify different RNAs with primers listed in Table S1.
Western blotting
Antibodies used for Western blotting in this study include rabbit anti-RtcB (19809-1-AP, Proteintech), mouse anti-β-tubulin (DSHB, E7), mouse anti-CNP (sc-166019, Santa Cruz Biotechnology), rabbit anti-XBP1s (619502, BioLegend), rabbit anti-Trpt1 (31) (Ron Lab, University of Cambridge), mouse anti-FLAG (F7425, Sigma), rabbit anti-RtcA (15996-1-AP, Proteintech), and horseradish peroxidase–conjugated secondary antibodies (Santa Cruz Biotechnology and Bio-Rad). XBP1s-immunoreactive bands were quantified with the Odyssey IR Imaging System (LI-COR Biosciences).
Recombinant protein purification
BL21 (DE3) E. coli cells that expressed His-E. coli RtcB, GST-CNP, and GST were grown at 37 °C in Luria-Bertani medium to mid-log phase. After 3-h induction with 0.2 mm isopropyl 1-thio-β-d-galactopyranoside at 37 °C, GST fusion protein–expressing cells were collected by centrifugation and lysed by sonication in 20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, 1 mm PMSF. The lysate was incubated with glutathione-Sepharose beads and then washed with a high-salt buffer (450 mm NaCl, 20 mm Na2HPO4, pH 7.3, 0.5 mm EDTA, 1% Triton, 1 mm PMSF, 1 mm DTT) four times and subsequently with a low-salt buffer (150 mm NaCl, 20 mm Na2HPO4, pH 7.3, 0.5 mm EDTA, 0.01% Triton, 1 mm PMSF, 1 mm DTT). The GST proteins were eluted with 10 mm GSH in PBS. Bacteria expressing His-E. coli RtcB were collected by centrifugation and lysed by sonication in 20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm PMSF, 20 mm imidazole. The lysate was applied to nickel beads and then washed with a high-salt buffer (450 mm NaCl, 20 mm Na2HPO4, pH 7.3, 1% Triton, 1 mm PMSF, 20 mm imidazole) four times and subsequently with a low-salt buffer (150 mm NaCl, 20 mm Na2HPO4, pH 7.3, 0.01% Triton, 1 mm PMSF, 20 mm imidazole). His-E. coli RtcB protein was then eluted with 200 mm imidazole in PBS. All purified proteins were dialyzed in 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 10% glycerol, 0.1 mm MgCl2, 0.1 mm MnCl2, 0.1 mm DTT.
In vitro XBP1 splicing reaction
The in vitro XBP1u splicing reaction was carried out as described previously (18). 10 ng of in vitro transcribed V5-XBP1uin-Rn transcript (MAXIscript T7 kit by Life Technologies), 1 μg of GST-IRE1αC (Sino Biological), purified E. coli RtcB, GST-CNP, and GST proteins were incubated in kinase buffer (2 mm ATP, 2 mm GTP, 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm MnCl2, 1 mm MgCl2, 5 mm β-mercaptoethanol) at 37 °C for 2 h. RNA was purified from the reactions with and RNA Clean & Concentrator kit (Zymo Research) for RT-qPCR analysis.
Statistical analysis
The data in this study are shown as means with S.E. Comparisons between groups were made by Student's t test (two-tailed, unpaired). p values less than 0.05 were considered significant.
Author contributions
I. U. and X. W. conceptualization; I. U. and Y. L. resources; I. U., Y. L., and X. W. formal analysis; I. U. validation; I. U., Y. L., and X. W. investigation; I. U. visualization; I. U. and Y. L. methodology; I. U. and X. W. writing-original draft; I. U. and X. W. writing-review and editing; X. W. funding acquisition; X. W. project administration.
Supplementary Material
Acknowledgments
We thank Robert Holmgren, Lilien Voong, and Christina Liu for comments on the manuscript and Ron Lab from the University of Cambridge for sharing Trpt1 antibody.
This work was supported by NIGMS, National Institutes of Health Grant 5R01GM120307 (to X. W.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1.
- ER
- endoplasmic reticulum
- XBP1
- X box–binding protein 1
- UPR
- unfolded protein response
- IRE1
- inositol-requiring enzyme 1
- Trl1
- tRNA ligase 1
- Tpt1
- tRNA 2′-phosphotransferase 1
- CNP
- 2′,3′-cyclic nucleotide phosphodiesterase
- XBP1s
- spliced form of X-box binding protein 1
- KO
- knockout
- cKO
- conditional knockout
- RtcA
- RNA 3′-terminal cyclase
- ES
- embryonic stem
- 4-OHT
- 4-hydroxytamoxifen
- GST
- glutathione S-transferase
- qPCR
- quantitative PCR
- shRNA
- short hairpin RNA
- sgRNA
- single guide RNA
- hCNP
- human CNP
- EDEM
- ER degradation–enhancing α-mannosidase-like protein
- PB
- PiggyBac
- mESC
- mouse AB2.2 ES cell
- PMSF
- phenylmethylsulfonyl fluoride.
References
- 1. Braakman I., and Bulleid N. J. (2011) Protein folding and modification in the mammalian endoplasmic reticulum. Annu. Rev. Biochem. 80, 71–99 10.1146/annurev-biochem-062209-093836 [DOI] [PubMed] [Google Scholar]
- 2. Walter P., and Ron D. (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]
- 3. Ron D., and Walter P. (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8, 519–529 10.1038/nrm2199 [DOI] [PubMed] [Google Scholar]
- 4. Hetz C., and Papa F. R. (2018) The unfolded protein response and cell fate control. Mol. Cell 69, 169–181 10.1016/j.molcel.2017.06.017 [DOI] [PubMed] [Google Scholar]
- 5. Acosta-Alvear D., Zhou Y., Blais A., Tsikitis M., Lents N. H., Arias C., Lennon C. J., Kluger Y., and Dynlacht B. D. (2007) XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol. Cell 27, 53–66 10.1016/j.molcel.2007.06.011 [DOI] [PubMed] [Google Scholar]
- 6. Lee A. H., Iwakoshi N. N., and Glimcher L. H. (2003) XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23, 7448–7459 10.1128/MCB.23.21.7448-7459.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cox J. S., and Walter P. (1996) A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87, 391–404 10.1016/S0092-8674(00)81360-4 [DOI] [PubMed] [Google Scholar]
- 8. Calfon M., Zeng H., Urano F., Till J. H., Hubbard S. R., Harding H. P., Clark S. G., and Ron D. (2002) IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96 10.1038/415092a [DOI] [PubMed] [Google Scholar]
- 9. Yoshida H., Matsui T., Yamamoto A., Okada T., and Mori K. (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891 10.1016/S0092-8674(01)00611-0 [DOI] [PubMed] [Google Scholar]
- 10. Gonzalez T. N., Sidrauski C., Dörfler S., and Walter P. (1999) Mechanism of non-spliceosomal mRNA splicing in the unfolded protein response pathway. EMBO J. 18, 3119–3132 10.1093/emboj/18.11.3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peebles C. L., Gegenheimer P., and Abelson J. (1983) Precise excision of intervening sequences from precursor tRNAs by a membrane-associated yeast endonuclease. Cell 32, 525–536 10.1016/0092-8674(83)90472-5 [DOI] [PubMed] [Google Scholar]
- 12. Sidrauski C., Cox J. S., and Walter P. (1996) tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell 87, 405–413 10.1016/S0092-8674(00)81361-6 [DOI] [PubMed] [Google Scholar]
- 13. Abelson J., Trotta C. R., and Li H. (1998) tRNA splicing. J. Biol. Chem. 273, 12685–12688 10.1074/jbc.273.21.12685 [DOI] [PubMed] [Google Scholar]
- 14. Sawaya R., Schwer B., and Shuman S. (2003) Genetic and biochemical analysis of the functional domains of yeast tRNA ligase. J. Biol. Chem. 278, 43928–43938 10.1074/jbc.M307839200 [DOI] [PubMed] [Google Scholar]
- 15. Greer C. L., Peebles C. L., Gegenheimer P., and Abelson J. (1983) Mechanism of action of a yeast RNA ligase in tRNA splicing. Cell 32, 537–546 10.1016/0092-8674(83)90473-7 [DOI] [PubMed] [Google Scholar]
- 16. McCraith S. M., and Phizicky E. M. (1990) A highly specific phosphatase from Saccharomyces cerevisiae implicated in tRNA splicing. Mol. Cell. Biol. 10, 1049–1055 10.1128/MCB.10.3.1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Spinelli S. L., Consaul S. A., and Phizicky E. M. (1997) A conditional lethal yeast phosphotransferase (tpt1) mutant accumulates tRNAs with a 2′-phosphate and an undermodified base at the splice junction. RNA 3, 1388–1400 [PMC free article] [PubMed] [Google Scholar]
- 18. Lu Y., Liang F. X., and Wang X. (2014) A synthetic biology approach identifies the mammalian UPR RNA ligase RtcB. Mol. Cell 55, 758–770 10.1016/j.molcel.2014.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kosmaczewski S. G., Edwards T. J., Han S. M., Eckwahl M. J., Meyer B. I., Peach S., Hesselberth J. R., Wolin S. L., and Hammarlund M. (2014) The RtcB RNA ligase is an essential component of the metazoan unfolded protein response. EMBO Rep. 15, 1278–1285 10.15252/embr.201439531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jurkin J., Henkel T., Nielsen A. F., Minnich M., Popow J., Kaufmann T., Heindl K., Hoffmann T., Busslinger M., and Martinez J. (2014) The mammalian tRNA ligase complex mediates splicing of XBP1 mRNA and controls antibody secretion in plasma cells. EMBO J. 33, 2922–2936 10.15252/embj.201490332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Englert M., Sheppard K., Aslanian A., Yates J. R. 3rd, and Söll D. (2011) Archaeal 3′-phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proc. Natl. Acad. Sci. U.S.A. 108, 1290–1295 10.1073/pnas.1018307108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zillmann M., Gorovsky M. A., and Phizicky E. M. (1991) Conserved mechanism of tRNA splicing in eukaryotes. Mol. Cell. Biol. 11, 5410–5416 10.1128/MCB.11.11.5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schwer B., Aronova A., Ramirez A., Braun P., and Shuman S. (2008) Mammalian 2′,3′ cyclic nucleotide phosphodiesterase (CNP) can function as a tRNA splicing enzyme in vivo. RNA 14, 204–210 10.1261/rna.858108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ramirez A., Shuman S., and Schwer B. (2008) Human RNA 5′-kinase (hClp1) can function as a tRNA splicing enzyme in vivo. RNA 14, 1737–1745 10.1261/rna.1142908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hu Q. D., Lu H., Huo K., Ying K., Li J., Xie Y., Mao Y., and Li Y. Y. (2003) A human homolog of the yeast gene encoding tRNA 2′-phosphotransferase: cloning, characterization and complementation analysis. Cell. Mol. Life Sci. 60, 1725–1732 10.1007/s00018-003-3107-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lappe-Siefke C., Goebbels S., Gravel M., Nicksch E., Lee J., Braun P. E., Griffiths I. R., and Nave K. A. (2003) Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat. Genet. 33, 366–374 10.1038/ng1095 [DOI] [PubMed] [Google Scholar]
- 27. Schaffer A. E., Eggens V. R., Caglayan A. O., Reuter M. S., Scott E., Coufal N. G., Silhavy J. L., Xue Y., Kayserili H., Yasuno K., Rosti R. O., Abdellateef M., Caglar C., Kasher P. R., Cazemier J. L., et al. (2014) CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. Cell 157, 651–663 10.1016/j.cell.2014.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Karaca E., Weitzer S., Pehlivan D., Shiraishi H., Gogakos T., Hanada T., Jhangiani S. N., Wiszniewski W., Withers M., Campbell I. M., Erdin S., Isikay S., Franco L. M., Gonzaga-Jauregui C., Gambin T., et al. (2014) Human CLP1 mutations alter tRNA biogenesis, affecting both peripheral and central nervous system function. Cell 157, 636–650 10.1016/j.cell.2014.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hanada T., Weitzer S., Mair B., Bernreuther C., Wainger B. J., Ichida J., Hanada R., Orthofer M., Cronin S. J., Komnenovic V., Minis A., Sato F., Mimata H., Yoshimura A., Tamir I., et al. (2013) CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature 495, 474–480 10.1038/nature11923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Harding H. P., Calfon M., Urano F., Novoa I., and Ron D. (2002) Transcriptional and translational control in the mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 18, 575–599 10.1146/annurev.cellbio.18.011402.160624 [DOI] [PubMed] [Google Scholar]
- 31. Harding H. P., Lackey J. G., Hsu H. C., Zhang Y., Deng J., Xu R. M., Damha M. J., and Ron D. (2008) An intact unfolded protein response in Trpt1 knockout mice reveals phylogenic divergence in pathways for RNA ligation. RNA 14, 225–232 10.1261/rna.859908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Popow J., Schleiffer A., and Martinez J. (2012) Diversity and roles of (t)RNA ligases. Cell. Mol. Life Sci. 69, 2657–2670 10.1007/s00018-012-0944-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Phizicky E. M., and Hopper A. K. (2010) tRNA biology charges to the front. Genes Dev. 24, 1832–1860 10.1101/gad.1956510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Culver G. M., McCraith S. M., Consaul S. A., Stanford D. R., and Phizicky E. M. (1997) A 2′-phosphotransferase implicated in tRNA splicing is essential in Saccharomyces cerevisiae. J. Biol. Chem. 272, 13203–13210 10.1074/jbc.272.20.13203 [DOI] [PubMed] [Google Scholar]
- 35. Ogle J. M., Brodersen D. E., Clemons W. M. Jr, Tarry M. J., Carter A. P., and Ramakrishnan V. (2001) Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897–902 10.1126/science.1060612 [DOI] [PubMed] [Google Scholar]
- 36. Drummond G. I., Iyer N. T., and Keith J. (1962) Hydrolysis of ribonucleoside 2′,3′-cyclic phosphates by a diesterase from brain. J. Biol. Chem. 237, 3535–3539 [Google Scholar]
- 37. Tanaka N., and Shuman S. (2011) RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J. Biol. Chem. 286, 7727–7731 10.1074/jbc.C111.219022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Popow J., Englert M., Weitzer S., Schleiffer A., Mierzwa B., Mechtler K., Trowitzsch S., Will C. L., Lührmann R., Soll D., and Martinez J. (2011) HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science 331, 760–764 10.1126/science.1197847 [DOI] [PubMed] [Google Scholar]
- 39. Lu Y., Lin C., and Wang X. (2009) PiggyBac transgenic strategies in the developing chicken spinal cord. Nucleic Acids Res. 37, e141 10.1093/nar/gkp686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yoshida H., Oku M., Suzuki M., and Mori K. (2006) pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J. Cell Biol. 172, 565–575 10.1083/jcb.200508145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shoulders M. D., Ryno L. M., Genereux J. C., Moresco J. J., Tu P. G., Wu C., Yates J. R. 3rd, Su A. I., Kelly J. W., and Wiseman R. L. (2013) Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell Rep. 3, 1279–1292 10.1016/j.celrep.2013.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vogel U. S., and Thompson R. J. (1988) Molecular structure, localization, and possible functions of the myelin-associated enzyme 2′,3′-cyclic nucleotide 3′-phosphodiesterase. J. Neurochem. 50, 1667–1677 10.1111/j.1471-4159.1988.tb02461.x [DOI] [PubMed] [Google Scholar]
- 43. Das U., and Shuman S. (2013) 2′-Phosphate cyclase activity of RtcA: a potential rationale for the operon organization of RtcA with an RNA repair ligase RtcB in Escherichia coli and other bacterial taxa. RNA 19, 1355–1362 10.1261/rna.039917.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Filipowicz W., Konarska M., Gross H. J., and Shatkin A. J. (1983) RNA 3′-terminal phosphate cyclase activity and RNA ligation in HeLa cell extract. Nucleic Acids Res. 11, 1405–1418 10.1093/nar/11.5.1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Filipowicz W. (2016) RNA 3′-terminal phosphate cyclases and cyclase-like proteins. Postepy Biochem. 62, 327–334 [PubMed] [Google Scholar]
- 46. Wang M., and Kaufman R. J. (2016) Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 529, 326–335 10.1038/nature17041 [DOI] [PubMed] [Google Scholar]
- 47. Oakes S. A., and Papa F. R. (2015) The role of endoplasmic reticulum stress in human pathology. Annu. Rev. Pathol. 10, 173–194 10.1146/annurev-pathol-012513-104649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sha H., He Y., Yang L., and Qi L. (2011) Stressed out about obesity: IRE1α-XBP1 in metabolic disorders. Trends Endocrinol. Metab. 22, 374–381 10.1016/j.tem.2011.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cubillos-Ruiz J. R., Bettigole S. E., and Glimcher L. H. (2017) Tumorigenic and immunosuppressive effects of endoplasmic reticulum stress in cancer. Cell 168, 692–706 10.1016/j.cell.2016.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hetz C., and Mollereau B. (2014) Disturbance of endoplasmic reticulum proteostasis in neurodegenerative diseases. Nat. Rev. Neurosci. 15, 233–249 10.1038/nrn3689 [DOI] [PubMed] [Google Scholar]
- 51. Lee A. H., Heidtman K., Hotamisligil G. S., and Glimcher L. H. (2011) Dual and opposing roles of the unfolded protein response regulated by IRE1α and XBP1 in proinsulin processing and insulin secretion. Proc. Natl. Acad. Sci. U.S.A. 108, 8885–8890 10.1073/pnas.1105564108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Allagnat F., Christulia F., Ortis F., Pirot P., Lortz S., Lenzen S., Eizirik D. L., and Cardozo A. K. (2010) Sustained production of spliced X-box binding protein 1 (XBP1) induces pancreatic β cell dysfunction and apoptosis. Diabetologia 53, 1120–1130 10.1007/s00125-010-1699-7 [DOI] [PubMed] [Google Scholar]
- 53. Hetz C., Thielen P., Matus S., Nassif M., Court F., Kiffin R., Martinez G., Cuervo A. M., Brown R. H., and Glimcher L. H. (2009) XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes Dev. 23, 2294–2306 10.1101/gad.1830709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vidal R. L., Figueroa A., Court F. A., Thielen P., Molina C., Wirth C., Caballero B., Kiffin R., Segura-Aguilar J., Cuervo A. M., Glimcher L. H., and Hetz C. (2012) Targeting the UPR transcription factor XBP1 protects against Huntington's disease through the regulation of FoxO1 and autophagy. Hum. Mol. Genet. 21, 2245–2262 10.1093/hmg/dds040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sado M., Yamasaki Y., Iwanaga T., Onaka Y., Ibuki T., Nishihara S., Mizuguchi H., Momota H., Kishibuchi R., Hashimoto T., Wada D., Kitagawa H., and Watanabe T. K. (2009) Protective effect against Parkinson's disease-related insults through the activation of XBP1. Brain Res. 1257, 16–24 10.1016/j.brainres.2008.11.104 [DOI] [PubMed] [Google Scholar]
- 56. Valenzuela V., Collyer E., Armentano D., Parsons G. B., Court F. A., and Hetz C. (2012) Activation of the unfolded protein response enhances motor recovery after spinal cord injury. Cell Death Dis. 3, e272 10.1038/cddis.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ryoo H. D., Domingos P. M., Kang M. J., and Steller H. (2007) Unfolded protein response in a Drosophila model for retinal degeneration. EMBO J. 26, 242–252 10.1038/sj.emboj.7601477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Oishi N., Duscha S., Boukari H., Meyer M., Xie J., Wei G., Schrepfer T., Roschitzki B., Boettger E. C., and Schacht J. (2015) XBP1 mitigates aminoglycoside-induced endoplasmic reticulum stress and neuronal cell death. Cell Death Dis. 6, e1763 10.1038/cddis.2015.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu S. Y., Wang W., Cai Z. Y., Yao L. F., Chen Z. W., Wang C. Y., Zhao B., and Li K. S. (2013) Polymorphism −116C/G of human X-box-binding protein 1 promoter is associated with risk of Alzheimer's disease. CNS Neurosci. Ther. 19, 229–234 10.1111/cns.12064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kakiuchi C., Iwamoto K., Ishiwata M., Bundo M., Kasahara T., Kusumi I., Tsujita T., Okazaki Y., Nanko S., Kunugi H., Sasaki T., and Kato T. (2003) Impaired feedback regulation of XBP1 as a genetic risk factor for bipolar disorder. Nat. Genet. 35, 171–175 10.1038/ng1235 [DOI] [PubMed] [Google Scholar]
- 61. Weitzer S., Hanada T., Penninger J. M., and Martinez J. (2015) CLP1 as a novel player in linking tRNA splicing to neurodegenerative disorders. Wiley Interdiscip. Rev. RNA 6, 47–63 10.1002/wrna.1255 [DOI] [PubMed] [Google Scholar]
- 62. Song Y., Sretavan D., Salegio E. A., Berg J., Huang X., Cheng T., Xiong X., Meltzer S., Han C., Nguyen T. T., Bresnahan J. C., Beattie M. S., Jan L. Y., and Jan Y. N. (2015) Regulation of axon regeneration by the RNA repair and splicing pathway. Nat. Neurosci. 18, 817–825 10.1038/nn.4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kosmaczewski S. G., Han S. M., Han B., Irving Meyer B., Baig H. S., Athar W., Lin-Moore A. T., Koelle M. R., and Hammarlund M. (2015) RNA ligation in neurons by RtcB inhibits axon regeneration. Proc. Natl. Acad. Sci. U.S.A. 112, 8451–8456 10.1073/pnas.1502948112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Skarnes W. C., Rosen B., West A. P., Koutsourakis M., Bushell W., Iyer V., Mujica A. O., Thomas M., Harrow J., Cox T., Jackson D., Severin J., Biggs P., Fu J., Nefedov M., et al. (2011) A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342 10.1038/nature10163 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.