Abstract
MicroRNAs (miRNAs) are noncoding RNAs that regulate gene expression at the post-transcriptional level and are involved in the regulation of the formation, maintenance, and function of skeletal muscle. Using miRNA sequencing and bioinformatics analysis, we previously found that the miRNA miR-664-5p is significantly differentially expressed in longissimus dorsi muscles of Rongchang pigs. However, the molecular mechanism by which miR-664-5p regulates myogenesis remains unclear. In this study, using flow cytometry, 5-ethynyl-2′-deoxyuridine staining, and cell count and immunofluorescent assays, we found that cell-transfected miR-664-5p mimics greatly promoted proliferation of C2C12 mouse myoblasts by increasing the proportion of cells in the S- and G2-phases and up-regulating the expression of cell cycle genes. Moreover, miR-664-5p inhibited myoblast differentiation by down-regulating myogenic gene expression. In contrast, miR-664-5p inhibitor repressed myoblast proliferation and promoted myoblast differentiation. Mechanistically, using dual-luciferase reporter gene experiments, we demonstrated that miR-664-5p directly targets the 3′-UTR of serum response factor (SRF) and Wnt1 mRNAs. We also observed that miR-664-5p inhibits both mRNA and protein levels of SRF and Wnt1 during myoblast proliferation and myogenic differentiation, respectively. Furthermore, the activating effect of miR-664-5p on myoblast proliferation was attenuated by SRF overexpression, and miR-664-5p repressed myogenic differentiation by diminishing the accumulation of nuclear β-catenin. Of note, miR-664-5p's inhibitory effect on myogenic differentiation was abrogated by treatment with Wnt1 protein, the key activator of the Wnt/β-catenin signaling pathway. Collectively, our findings suggest that miR-664-5p controls SRF and canonical Wnt/β-catenin signaling pathways in myogenesis.
Keywords: microRNA (miRNA), muscle, myogenesis, Wnt pathway, cell proliferation, cell differentiation, post-transcriptional regulation
Introduction
Skeletal muscle is a vital organ, occupying 30–40% of the body, and proper muscle formation and function are required for a healthy life. Myogenesis is a complex process (1), which includes the Myf5+/Pax7+ myogenic progenitor derived from somatic mesoderm developing into myoblasts (2), and determined myoblasts must withdraw from the cell cycle before differentiation into multinucleate myotubes. The whole myogenic lineage progression is regulated by myogenic regulation factors (MRFs),3 including myogenic factor 5 (Myf5), myoblast determination protein (MyoD), muscle-specific regulatory factor 4 (MRF4 and also known as MYF6), and myogenin (MyoG) (3, 4). Besides MRFs, it is necessary to study the underlying mechanism of regulation on myogenesis by novel genes, including microRNAs (miRNAs).
miRNA, transcribed within long primary transcripts, is cleaved twice to form functional 21-nucleotide RNAs and plays a crucial role in physiological processes (5). It silences genes by base pairing to the mRNA of the target genes, thus contributing to mRNA degradation and the inhibition of translation at the post-transcription level (6). In most cases, miRNA binds to the 3′ UTR of gene mRNA, but it was also demonstrated that miRNA binds to the 5′ UTR (7, 8) or the coding sequence (9–11). Increasing evidence has shown that miRNA plays a key role in skeletal muscle formation (12, 13). Skeletal muscle-specific knockouts of a key enzyme in the maturation of pre-miRNA resulted in a decrease in muscle mass and disordered muscle formation (14). In addition, some skeletal muscle-specific miRNAs, such as miR-1, miR-133 (15), and miR-206 (16), have been demonstrated to act as vital modulators in myogenesis. However, the effects and regulatory mechanisms of novel miRNAs on skeletal muscle formation still need to be further explored.
Serum response factor (SRF), a member of the MADs (MCM1, Agamous, Deficiens) box superfamilies, controls the transcription of muscle-specific genes. The serum response element, located in the promoter of the genes, was usually bound by SRF protein (17, 18). It has been demonstrated that SRF could regulate the cell cycle and cell apoptosis by controlling the expression of c-fos (19, 20), which is a proto-oncogene and plays an important role in cellular processes (21). Moreover, increasing evidence indicated that MRFs combined with other transcription factors, including myocyte enhancer factor-2 (MEF2) and SRF, whose expressions were not restricted in control myogenesis (22). Furthermore, SRF is abundantly expressed in smooth muscle and skeletal muscle, and Li et al. (23) observed that the skeletal muscle-specific depletion of SRF by Cre recombinase led to perinatal lethality and skeletal muscle hypoplasia. The above findings indicated that SRF plays a vital role in skeletal muscle. In addition, although miR-133, a skeletal muscle-specific miRNA, targeted SRF to enhance C2C12 cells proliferation (15), the information on other new miRNAs regulating myogenesis through SRF is still limited.
The wingless-type mouse mammary tumor virus integration site family member 1 (Wnt1), a member of the Wnt protein family, is a secreted glycoprotein and involved in the canonical Wnt/β-catenin signaling pathway (24). Previous studies indicated that the Wnt signaling pathway had an essential role in skeletal muscle development during the embryonic period by regulating the expression of MRFs (25, 26) and inducing the myogenic differentiation of stem cells in adult muscle (27). β-Catenin is a critical component in canonical Wnt signaling, and it could be degraded by a complex composed of axin, adenomatous polyposis coli, and GSK-3β (28). However, when Wnt proteins bind to the Frizzled receptor to activate the signaling pathway, the degradation complex is broken up, which leads to the accumulation of β-catenin in the cytoplasm (29). β-Catenin translocated into nucleus from the cytoplasm to influence gene transcription as a coactivator (30), but there were few studies about miRNA regulating skeletal muscle development through Wnt signaling.
In this study, based on miRNA sequencing analysis of 35- and 287-day porcine longissimus dorsi muscle (31), we discovered that miR-664-5p showed a significant expressional difference, and the effects and mechanism of miR-664-5p on myoblast proliferation and differentiation were explored. Our findings revealed that miR-664-5p promotes proliferation and inhibits differentiation in myoblasts by respectively targeting SRF and Wnt1, suggesting that miR-664-5p is a novel key regulatory factor in skeletal muscle formation.
Results
Profile of miR-664-5p during myogenesis
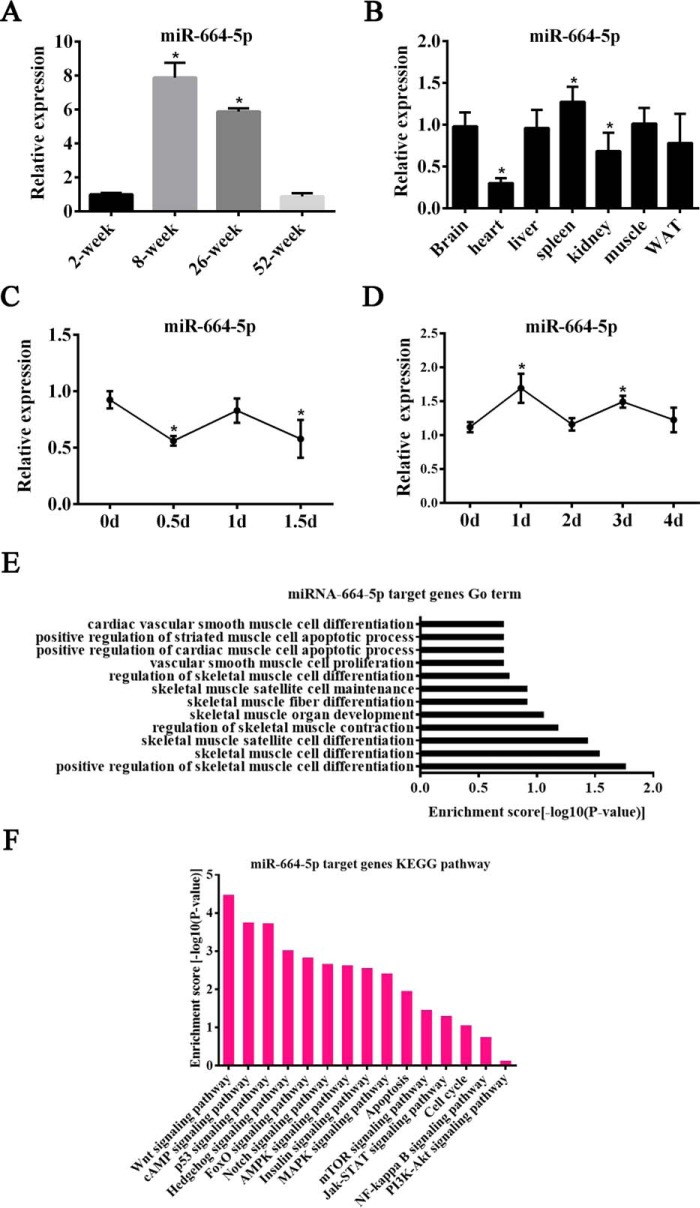
Our previous study demonstrated that miR-664-5p showed a differential expression in different growth stages (Fig. S1A), and thus we detected miR-664-5p expression in the skeletal muscle of 2-, 8-, 26-, and 52-week-old mice. The results showed that the miR-664-5p expression was the highest in 8-week-old mice (Fig. 1A). Although sequence alignment of mature miR-664-5p among multiple species, including mice and pig, showed that miR-664-5p was poorly conserved in the seed sequence (Fig. S1B), the similar expression profiles of miR-664-5p in myogenesis between mice and pigs implied that miR-664-5p played a crucial role in skeletal muscle development. In addition, the levels of miR-664-5p were detected in muscle, fat, kidney, spleen, liver, heart, and brain tissues derived from 8-week-old mice, and it was found that miR-664-5p was moderately expressed in skeletal muscle (Fig. 1B). Moreover, the expression trends of miR-664-5p were first decreased, then increased, and finally decreased in proliferating cells (Fig. 1C), but they first increased, then decreased, and finally increased in differentiating cells (Fig. 1D). We further performed GO term analysis for miR-664-5p target genes and found that miR-664-5p may be involved in skeletal muscle development, especially muscle cell proliferation and differentiation (Fig. 1E). Moreover, KEGG pathway analysis revealed that miR-664-5p may regulate biological processes through the Wnt signaling pathway (Fig. 1F). Hence, we propose that miR-664-5p is a potential regulator in myogenesis.
Figure 1.
Levels of miR-664-5p in different tissues from mice and during C2C12 cell myogenesis. A, relative expression of miR-664-5p in skeletal muscle from 2-, 8-, 26-, and 52-week-old mice. *, p < 0.01, versus 2-week-old mice. B, tissue expression profile of miR-664-5p in mice. *, p < 0.05, versus brain. C, RT-qPCR was performed to detect the expression of miR-664-5p in proliferating myoblasts. *, p < 0.05, versus 0 days. D, RT-qPCR analysis of miR-664-5p expression after inducing myoblast differentiation. *, p < 0.05, versus 0 days. E, GO term analysis of the miR-664-5p target genes. F, KEGG pathway analysis of the miR-664-5p target genes. Results are representative of the mean ± S.E. of three independent experiments.
miR-664-5p promotes myoblast proliferation
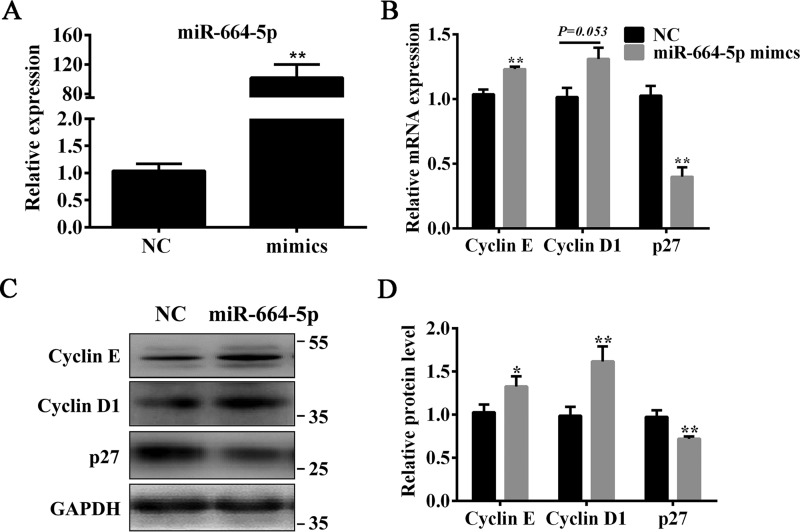
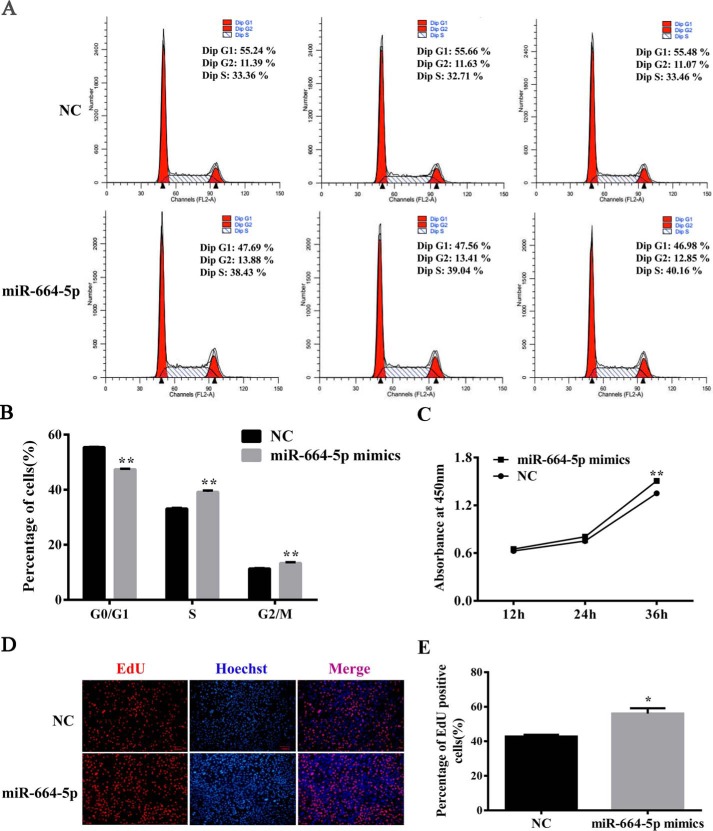
To understand the role of miR-664-5p in myoblast proliferation, C2C12 cells were transfected with mimics or the negative control (NC). The results showed that miR-664-5p mimics markedly increased the levels of miR-664-5p (Fig. 2A), the mRNA (Fig. 2B), and protein (Fig. 2, C and D) of cell cycle–related genes, including cyclin E and cyclin D1, but significantly decreased the mRNA (Fig. 2B) and protein (Fig. 2, C and D) levels of p27. Using cell cycle analysis by a flow cytometer, miR-664-5p increased the number of cells in the S-phase, indicating that miR-664-5p promoted myoblast DNA replication (Fig. 3, A and B). In addition, by a cell count assay (CCK-8), miR-664-5p increased the cell number after transfection for 1 day (Fig. 3C). Moreover, the EdU staining assay indicated that miR-664-5p increased S-phage cells (Fig. 3, D and E). To further confirm the function of miR-664-5p on myoblast proliferation, we transfected myoblasts with the miR-664-5p inhibitor. The results showed that the miR-664-5p inhibitor decreased the levels of cell cycle marker genes (cyclin E and cyclin D1) and increased the levels of cell cycle marker inhibiting gene p27 (Fig. S2, B–D). The EdU staining assay revealed that EdU-labeled cells were less in miR-664-5p inhibitor-treated cells than NC-treated cells (Fig. S2, E and F). Moreover, the total number of cells decreased according to the CCK8 assay (Fig. S2G). Collectively, the results showed that miR-664-5p promoted myoblast proliferation.
Figure 2.
miR-664-5p up-regulates the expression levels of cell cycle genes. A, overexpression efficiency of miR-664-5p mimics after transfection for 24 h. B, RT-qPCR was used to detect the cell cycle genes cyclin E, cyclin D1, and p27. C, Western blot analysis of cyclin E, cyclin D1, and p27 after transfection with miR-664-5p mimics. D, protein quantitative analysis of cyclin E, cyclin D1, and p27. Values are expressed as the mean ± S.E. (n = 3). *, p < 0.05; **, p < 0.01, versus NC.
Figure 3.
miR-664-5p promotes the cell cycle process and increases the number of total cells as well as S-phase cells. A, cell cycle analysis of myoblast by flow cytometry. B, statistical results of flow cytometry. C, CCK-8 analysis after treatment with miR-664-5p mimics during myoblast proliferation. D, EdU staining assay. Myoblasts in the S-phase were stained with EdU in red, and cell nuclei were dyed with Hoechst in blue. E, quantification ratio of EdU-positive cells/total cells. Values are expressed as the mean ± S.E. (n = 3). *, p < 0.05; **, p < 0.01, versus NC.
miR-664-5p targets SRF in proliferating cells
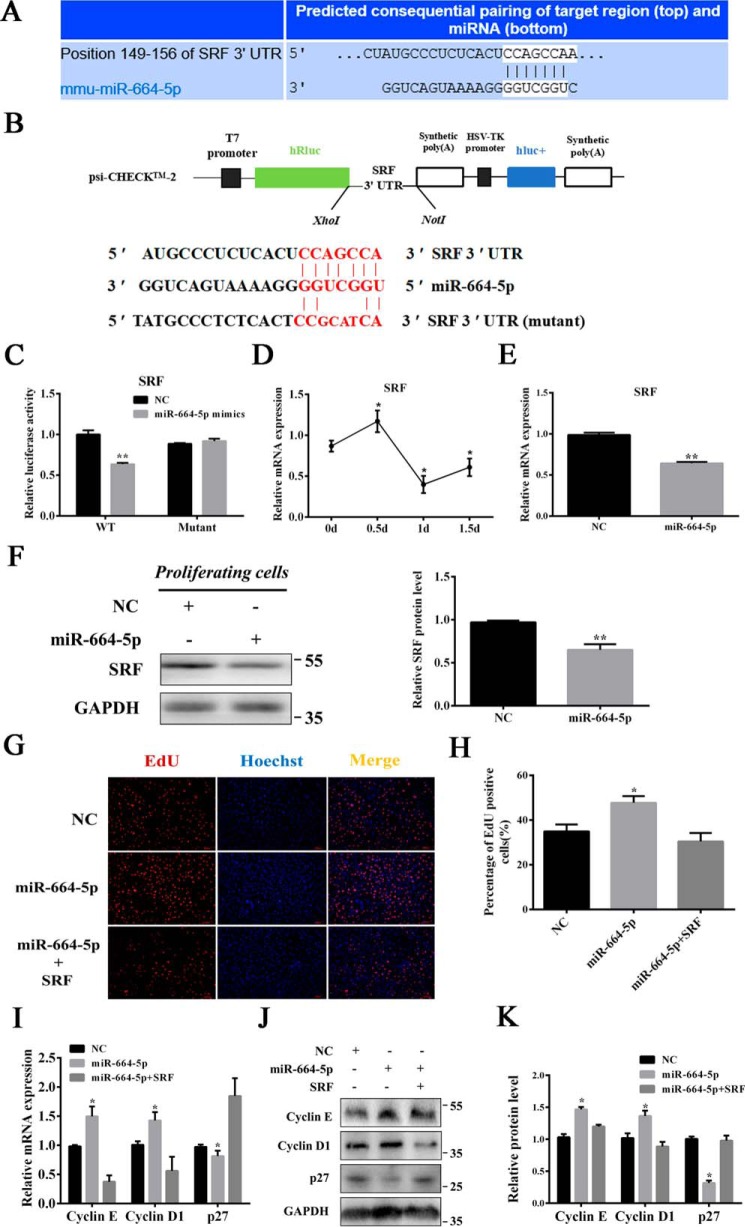
To illuminate the regulatory mechanism of miR-664-5p on myoblast proliferation, TargetScan was used to predict the target genes and found that SRF may be targeted by miR-664-5p (Fig. 4A). We constructed WT and mutant dual-luciferase reporter psiCHECK-2.0–SRF 3′ UTR (Fig. 4B). The dual-luciferase reporter assay revealed that miR-664-5p could bind to the 3′ UTR of SRF mRNA, which led to a decrease in the relative luciferase activity in response to miR-664-5p mimics (Fig. 4C). We also found that the level of SRF showed a reverse tendency with miR-664-5p in proliferating myoblasts (Fig. 4D). Moreover, for myoblasts transfected with miR-664-5p mimics at 2 days, the levels of SRF mRNA and protein were markedly down-regulated, indicating that miR-664-5p inhibited SRF expression (Fig. 4, E and F). Furthermore, overexpression of SRF significantly inhibited myoblast proliferation (Fig. S3). We also performed rescue experiments. The results indicated that the overexpression of SRF attenuated miR-664-5p–induced effects on myoblast proliferation by decreasing EdU-positive cells (Fig. 4, G and H) and down-regulated the levels of cell cycle marker genes, including cyclin E and cyclin D1, but it significantly increased the levels of p27 (Fig. 4, I–K).
Figure 4.
miR-664-5p targets SRF during myoblast proliferation. A, SRF was predicted to be a target of miR-664-5p by TargetScan software. B, WT and mutant psiCHECK-2.0–SRF vectors. C, relative luciferase activity of SRF responding to miR-664-5p mimics. D, expression pattern of SRF during myoblast proliferation. *, p < 0.05, versus 0 days. E, relative mRNA expression after transfection with miR-664-5p mimics. F, change of SRF protein expression after transfection for 24 h. G, EdU staining assay. Myoblasts in the S-phase were stained with EdU in red, and cell nuclei were dyed with Hoechst in blue. H, quantification ratio of EdU-positive cells/total cells. I, RT-qPCR was used to detect the cell cycle genes cyclin E, cyclin D1 and p27. J, Western blot analysis of cyclin E, cyclin D1, and p27. K, protein quantitative analysis of cyclin E, cyclin D1 and p27. Values are expressed as the mean ± S.E. (n = 3). *, p < 0.05; **, p < 0.01, versus NC.
miR-664-5p inhibits myogenic differentiation
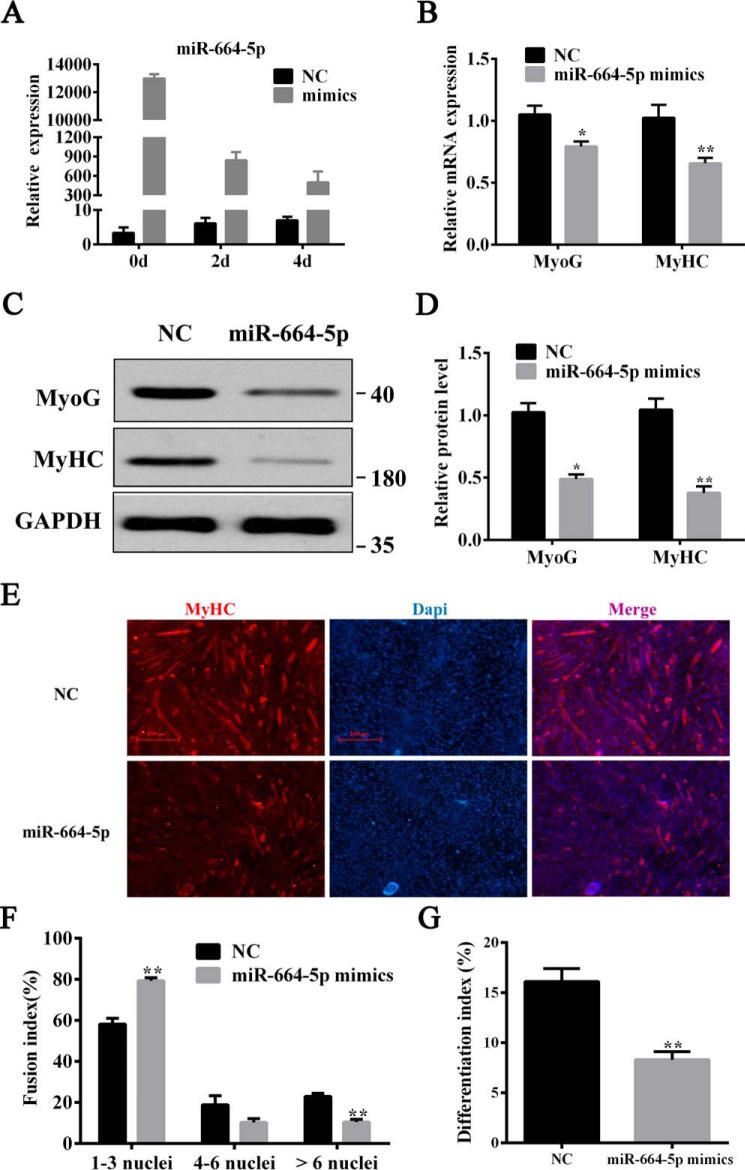
After transfection with mimics, miR-664-5p was markedly up-regulated at 0, 2, and 4 days after myogenic induction (Fig. 5A). By qPCR and Western blot analysis, we discovered that miR-664-5p mimics significantly suppressed either the mRNA (Fig. 5B) or the protein level of myogenic markers (Fig. 5, C and D), including MyoG and MyHC at the 4th day of differentiation. Additionally, miR-664-5p obviously decreased the number and size of myotubes in miR-664-5p–treated cells as determined using an immunofluorescent assay (Fig. 5E). By calculating the number of cell nuclei in myotubes, we discovered that miR-664-5p mimic–treated myoblasts formed less myotubes with less than six nuclei and had a lower differentiation index than the negative control (Fig. 5, F and G). To further explore the function of miR-664-5p on myogenic differentiation, myoblasts were transfected with NC or miR-664-5p inhibitor. The results showed that miR-664-5p inhibitor dramatically promoted the mRNA expression of myogenic genes, including MyoG and MyHC, which was consistent with the protein level (Fig. S4, A–D). Furthermore, the immunofluorescent assay revealed that myoblasts treated with miR-664-5p inhibitor had more multinucleated myotubes and a greater differentiation index relative to cells that were transfected with the negative control (Fig. S4, E–G).
Figure 5.
miR-664-5p inhibits myogenic differentiation. A, overexpression efficiency of miR-664-5p mimics at 0, 2, and 4 days. B, relative mRNA expression of MyoG and MyHC after induction for 4 days. C, protein expression of MyoG and MyHC after induction for 4 days. D, quantitative results of protein expression in C. E, immunofluorescence staining of MyHC after induction for 4 days. F, statistical results of the myoblast fusion index after induction for 4 days. G, differentiation index of myoblast after induction for 4 days. Values are expressed as the means ± S.E. (n = 3). *, p < 0.05; **, p < 0.01, versus NC.
miR-664-5p inhibits myogenic differentiation through targeting Wnt1 and modulating the canonical Wnt/β-catenin signaling pathway
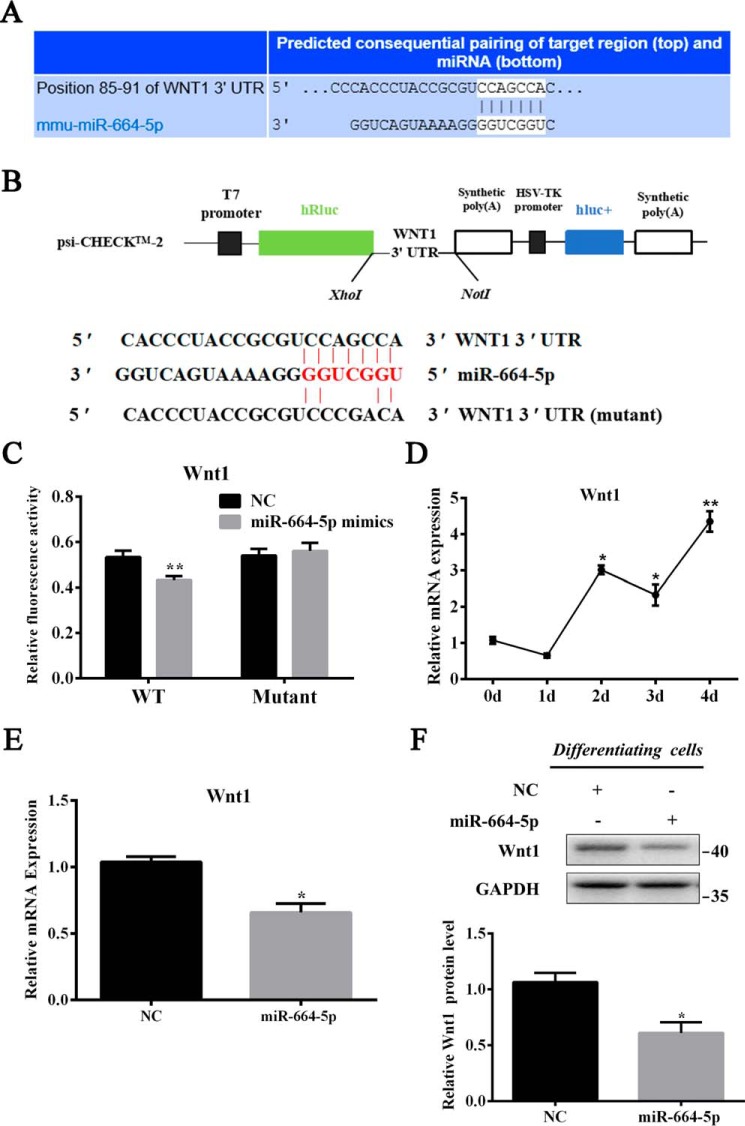
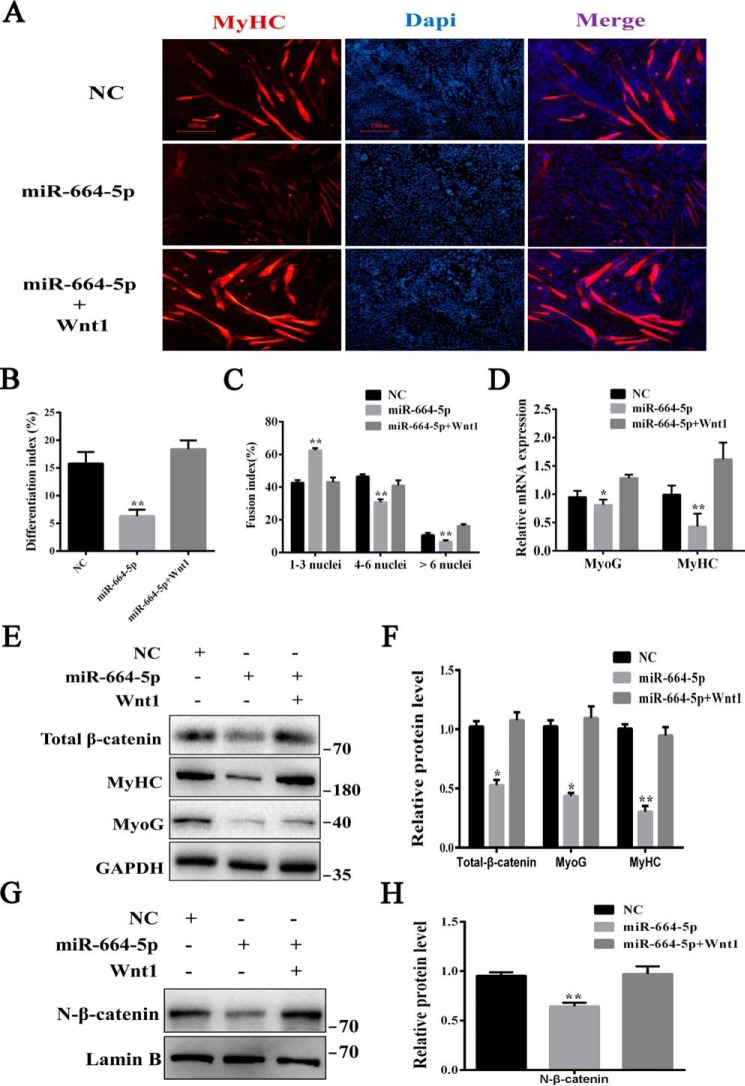
To understand the regulatory mechanism of myogenesis by miR-664-5p, we predicted the target genes of miR-664-5p by TargetScanMouse 6.2 again. From thousands of predicted targets, Wnt1, due to its essential role in skeletal muscle development, attracted our attention (Fig. 6A). We amplified the 3′ UTR from myoblast cDNA using the added restriction site of XhoI and NotI and connected it with the psiCHECK-2–vector (Fig. 6B). The dual-luciferase reporter assay revealed that miR-664-5p mimics remarkably inhibited the luciferase activity of the wild psiCHECK-2–Wnt1 3′ UTR reporter, although the consequence of the mutant reporter vector did not change (Fig. 6C). Moreover, to further confirm the relevance between miR-664-5p and Wnt1, we detected the expression of Wnt1 during myogenic differentiation (Fig. 6D) and changes in Wnt1 expression after transfection with mimics (Fig. 6, E and F). Consistently, miR-664-5p mimics decreased significantly not only in the mRNA level (Fig. 6E) but also in the protein level (Fig. 6F). In addition, we performed rescue experiments on myoblast differentiation. The results showed that the Wnt1 treatment restored miR-664-5p–induced inhibitory effects on myogenic differentiation by increasing the MyHC-positive cells, differentiation index, and multinucleated myotube fusion index, and it up-regulated the expression of MyoG and MyHC (Fig. 7, A–D). All of these results demonstrated that Wnt1 was a direct target gene of miR-664-5p in differentiating cells.
Figure 6.
miR-664-5p targets Wnt1 during myogenic differentiation. A, Wnt1 is a candidate target gene of miR-664-5p. B, construction of the dual-luciferase reporter psiCHECK-2.0–Wnt1 3′ UTR. C, dual-luciferase reporter assay was performed by cotransfecting miR-664-5p mimics and WT or mutant vectors. The relative luciferase activity was presented as Renilla luciferase/firefly luciferase. D, relative expression profile of miR-664-5p during myogenic differentiation. *, p < 0.05; **, p < 0.01 versus 0 days. E, relative mRNA expression of Wnt1 after treatment with miR-664-5p mimics. F, Western blot analysis of Wnt1 protein expression after transfection with miR-664-5p mimics and the statistical results of Wnt1 protein expression at 4 days after myogenic induction. Values are expressed as the mean ± S.E. (n = 3). *, p < 0.05; **, p < 0.01, versus NC.
Figure 7.
miR-664-5p inhibits myogenic differentiation through the Wnt/β-catenin signaling pathway. To induce the Wnt/β-catenin signaling pathway, myoblasts transfected with miR-664-5p mimics were grown in the presence of 50 ng/ml Wnt1 protein. A, immunofluorescence staining of MyHC after induction for 4 days. B, differentiation index of myoblasts after induction for 4 days. C, statistical results of the myoblast fusion index after induction for 4 days. D, relative mRNA expression of MyoG and MyHC after induction for 4 days. E, total protein was extracted, and Western blot analysis was performed to detect the protein expression of MyoG, MyHC, and β-catenin. F, quantification of the protein level in E. G, nuclear protein was extracted, and N-β-catenin relative to lamin B was detected by Western blotting. H, quantification of the protein level in G. Values are expressed as the mean ± S.E. (n = 3). *, p < 0.05; **, p < 0.01, versus NC.
As Wnt1 is an activator of the canonical Wnt/β-catenin signaling pathway, we detected the levels of total (Fig. 7, E and F) and nuclear β-catenin (Fig. 7, G and H). The results showed that miR-664-5p down-regulated the levels of both the total and nuclear β-catenin, implying that miR-664-5p inhibited myogenic differentiation through the Wnt/β-catenin signaling pathway. Moreover, after treatment with 50 ng/ml Wnt1 or 10 mm LiCl, which was also used to activate the signaling pathway (Fig. S5, A–D), the expressions of MyoG and MyHC were significantly recovered, indicating that the inhibitory effect caused by miR-664-5p was weakened, and the differentiation was rescued. Taken together, the results suggested that miR-664-5p inhibited myogenic differentiation by blocking the Wnt/β-catenin signaling pathway.
SRF and Wnt1 are respectively targeted by miR-664-5p during proliferation and differentiation in myoblasts
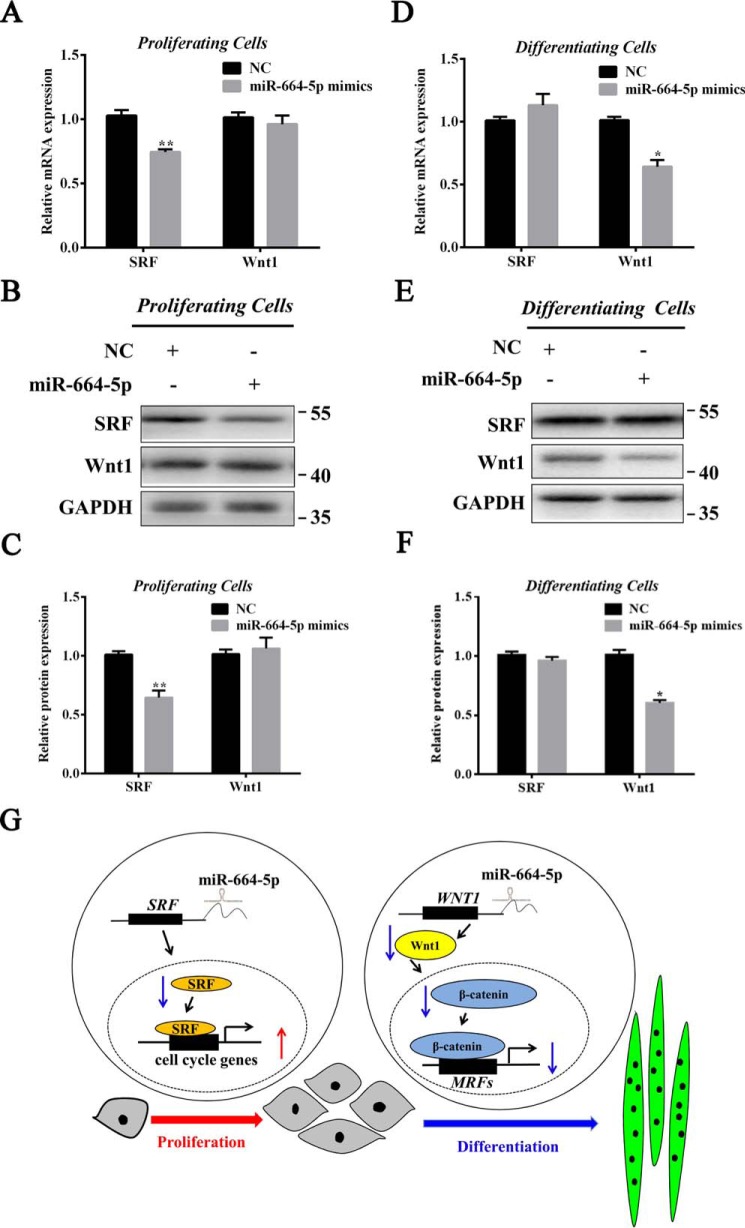
To further investigate the effect of miR-664-5p on Wnt1 in proliferation and SRF in differentiation, the levels of the two genes were detected during myogenesis. The results indicated that the level of SRF was significantly down-regulated by miR-664-5p mimics during myoblast proliferation (Fig. 8, A–C), whereas the level of Wnt1 was significantly down-regulated during differentiation (Fig. 8, D–F). We therefore suggested the mechanism by which miR-664-5p inhibited skeletal muscle formation through targeting SRF and the Wntβ–catenin signaling pathway (Fig. 8G).
Figure 8.
SRF and Wnt1 are respectively targeted by miR-664-5p during myoblast proliferation and differentiation. A–C, mRNA and protein expressions of Wnt1 and SRF in proliferating cells after transfection for 1 day. D–F, mRNA and protein expressions of Wnt1 and SRF in differentiating cells after induction for 4 days. G, proposed mechanism by which miR-664-5p inhibits myogenesis through the targeting of SRF and the Wntβ-catenin signaling pathway. Values are expressed as the mean ± S.E. (n = 3). *, p < 0.05; **, p < 0.01, versus NC.
Discussion
Our present study first found that miR-664-5p plays a vital role in regulating myoblast proliferation and differentiation. In particular, the miR-664-5p mimics promoted myoblast proliferation by targeting SRF, although they inhibited myoblast differentiation through targeting Wnt1. Overall, our findings provided evidence to suggest that miR-664-5p may be a novel negative regulator for the control of skeletal muscle formation.
As a family of endogenous noncoding RNA, miRNA plays a vital role in myogenesis. Based on our miRNA sequencing assay on the longissimus dorsi muscle of 35- and 287-day-old pigs (31), miR-664-5p was singled out because of its significant differential expression. We further found that miR-664-5p was a poorly conserved miRNA among mice, humans, and pigs, but the similar expression profiles of miR-664-5p on myogenesis between mice and pigs imply a similar effect of miR-664-5p on myogenesis between mice and pigs and suggest that miR-664-5p might be a novel valuable regulator in myogenesis.
Skeletal muscle formation is determined by the proliferation and differentiation of myocytes. Importantly, microRNA is implicated in skeletal muscle formation. To investigate the role of miR-664-5p in myoblast proliferation, miR-664-5p mimics and inhibitor were evaluated. We found that miR-664-5p contributed to myoblast proliferation by increasing the number of S-phase cells and up-regulating the levels of cyclin D1 and cyclin E genes both at the mRNA and protein levels. A previous study revealed that up-regulation of miR-664-5p could inhibit cervical cancer cell growth (32), and miR-664 promoted osteosarcoma cells proliferation via the down-regulation of FOXO4 (33), indicating that miR-664 was involved in regulation of cell proliferation. Moreover, microRNA has been identified as having an important regulator in myoblast proliferation. miR-1 is a muscle-specific miRNA and has been identified as a significant role in skeletal muscle cell proliferation by targeting HDAC4 (15); miR-432 was reported to inhibit the proliferation of myoblasts by targeting E2F3 and P55PIK (31). Meanwhile, in this study, miR-664-5p mimics inhibited myoblast differentiation by down-regulating the levels of MyHC and MyoG. Moreover, immunofluorescence confirmed that miR-664-5p mimics inhibited but miR-664-5p inhibitor promoted the formation of myotubes. Taken together, we thought that miR-664-5p displayed a regulatory role in myogenesis by promoting cellular proliferation and inhibiting differentiation. Interestingly, miRNAs were demonstrated to have adverse effects on cell proliferation and differentiation. For example, miR-1 and miR-206 targeted PAX7 to facilitate myogenic differentiation by restricting their proliferative potential (34); miR-29 reduced the proliferation and promoted differentiation by the down-regulation of Akt3 (35); and miR-133 promoted differentiation and repressed proliferation by forming a feedback circuit with the ERK1/2 pathway (36). In this study, miR-664-5p was demonstrated to promote myoblast proliferation and to inhibit myoblast differentiation.
Generally, miRNA functions by regulating the expression of target genes (37). Therefore, we predicted that miR-664-5p might target the SRF, which is a highly expressed regulator in myogenesis. As an important transcription factor of muscle genes, SRF could recruit a variety of partner proteins, including members of the myocardin family of transcriptional coactivators (38–41). Moreover, SRF was mediated by miRNAs. miR-133, a muscle-specific microRNA, inhibited myogenic differentiation by targeting SRF and metastasis-associated lung adenocarcinoma transcript 1 (Malat1), a well-known long noncoding RNA, which competes with miR-133 to regulate the serum response factor in myogenesis (42). Although SRF plays an essential role for in skeletal muscle development (23), the literature was not implicated in the relationship between SRF and muscle cell proliferation. Interestingly, miR-133 enhances myoblast proliferation by repressing SRF (15), and the TGF-β–miR-122–fibronectin 1/serum response factor signaling cascade has been identified in hepatic fibrogenesis (44); SRF was also an upstream transcription factor of miR-29b and is involved in NSCLC invasion and metastasis (45). In this study, we verified the binding of miR-664-5p and the 3′ UTR of SRF by the dual-luciferase reporter assay. Furthermore, we demonstrated that miR-664-5p mimics resulted in the marked decrease of SRF mRNA and protein levels during proliferation. Meanwhile, our rescue experiment further confirmed that SRF was a vital target gene of miR-664-5p.
In this study, Wnt1 is another target gene identified by the dual-luciferase reporter assay. Previous studies have revealed that Wnt1 was implicated in oncogenesis by promoting cell proliferation (46, 47). Interestingly, our results showed that a notable decrease of the levels in both mRNA and protein in differentiating cells, but not in proliferating cells, showed that the miR-664-5p targets Wnt1 during the differentiation phase. Wnt1 is a member of the Wnt/β-catenin signaling pathway that plays a key role in adult tissue development (48) and stem cell maintenance (49). Most importantly, microRNA and the Wnt/β-catenin signaling pathway were also implicated in myoblast differentiation. miR-145a-5p promoted myoblast differentiation by enhancing Wnt/β-catenin signaling pathway gene expression (50); miR-143-3p inhibited myoblast differentiation by down-regulating the expression of genes involved in the endogenous Wnt/β-catenin signaling pathway (51). A recent study has shown that Wnt3a-mediated Wnt/β-catenin signaling plays a critical role in C2C12 cell differentiation (52). In this study, miR-664-5p mimics inhibited the levels of nuclear β-catenin, implying that miR-664-5p inactivated Wnt/β-catenin signaling by the degradation of β-catenin. When treated with 50 ng/ml Wnt1 protein or 10 mm LiCl, the inhibitory effect of miR-664-5p almost disappeared by increasing the MyHC positive cells, differentiation index, and fusion index. However, compared with Wnt1, we need to point out that the LiCl used for activating Wnt/β-catenin is not specified. Based on our findings, miR-664-5p inhibited myoblast differentiation by targeting Wnt1 through repressing the Wnt/β-catenin signaling.
Additionally, it is very interesting why miR-664-5p promotes proliferation and inhibits differentiation in myoblasts by targeting SRF and Wnt1, respectively. We thought that the reasons may be related to the secondary structure and epigenetic modification of target gene mRNAs during different cell physiological stages. However, the effects of the secondary structure and epigenetic modification of target gene mRNAs on miR-664-5p need to be further explored during myogenesis.
In summary, our results revealed that miR-664-5p, a novel regulator factor, promoted myoblast proliferation by targeting SRF and inhibited differentiation by blocking the Wnt1/β-catenin signaling pathway, respectively. These findings contribute to a better understanding of myogenesis controlled by microRNA and provide new potential insights into regulatory approaches in skeletal muscle formation.
Experimental procedures
Animals and animal care
C57/BL male mice were purchased from the Animal Center at Xi'an Jiaotong University. All animals used in this study were fed according to the protocols approved by the Animal Care and Use Committee of China. Skeletal muscle tissues were isolated from 2-, 8-, 26-, and 52-week-old mice.
Cell culture and transfection
Human embryonic kidney cell line (HEK293T) and C2C12 myoblasts were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C and 5% CO2 to proliferate. Cells myogenic differentiation was initiated by switching the growth medium (GM) to differentiation medium (DM), which was composed of high-glucose DMEM and 2% horse serum, when the cells reached confluence. Both GM and DM were changed every other day. For myoblast proliferation, myoblasts were seeded in 12- or 6-well plates, and miRNA mimics or the NC (Ribobio, China) were transfected into cells at 40% density using X-tremeGENE siRNA Transfection Reagent (Roche Applied Science) and Opti-MEM (Gibco) culture medium according to the manufacturer's protocol. The concentrations of miRNA mimics and NC were 50 nm. Cells were harvested 24 h after transfection. For myogenic differentiation, cells were transfected when the density of myoblasts reached 70%. When cells grew to confluence after transfection, myogenic differentiation was initiated by switching to DM. The transfection protocol of the miR-664-5p inhibitor (miR21417153538, Ribobio, Guangzhou, China) was the same as mimics, but the concentration of miR-664-5p inhibitor was 100 nm according to the manufacturer's instruction. Furthermore, the pcDNA3.1 vector that induced the overexpression of SRF was used to recover the effects of miR-664-5p on proliferation. The pcDNA3.1 plasmids were transfected into cells at 40% density using X-tremeGENE HP DNA Transfection Reagent (Roche Applied Science) according to the manufacturer's instructions. The 50 ng/ml Wnt1 protein (Abcam) and 10 mm LiCl (Sigma) were used to activate the Wnt/β-catenin signaling pathway and to rescue the effects of miR-664-5p on differentiation.
RNA isolation and real-time qPCR
The total RNA of cells and different tissues was extracted with RNAiso Plus (TaKaRa) according to the manufacturer's instruction. cDNA synthesis was performed with reverse transcription kits (Takara, Otsu, Japan) according to the standard processes. RT-qPCR was performed using the iQTM5 system (Bio-Rad) with a 10-μl reaction volume. The expressions of all genes were normalized to GAPDH, and the primer sequences used for qPCR are listed in Table S1. For miRNA expression analysis, we conducted specific reverse transcription with RT primers of miR-664-5p and small RNA U6 (Takara, Otsu, Japan), which usually was a reference gene for miRNA, to synthesis cDNA. For real-time qPCR, every reaction was performed in triplicate using SYBR Green kits on a iQTM5 system (Bio-Rad). miRNA primers were purchased from Ribobio (Guangzhou, China).
Western blotting
The total protein was extracted from myoblasts using RIPA (Applygen Technologies Inc., China) and protease inhibitor mix (Cwbio, China) after washing the cells with PBS three times. All extraction processes were according to the standard protocols. The protein concentration was measured using the BCA protein assay kit (Cwbio, China). Twenty micrograms of protein was electrophoresed on a 10% SDS-polyacrylamide gel followed by shifting protein to a polyvinylidene fluoride membrane (Cell Signaling Technology (CST), Boston), and then the membrane was blocked with 5% BSA at 4 °C for 2 h, incubated with antibodies (1:1000) against cyclin E (Santa Cruz Biotechnology), cyclin D1 (Santa Cruz Biotechnology), p27 (Santa Cruz Biotechnology), MyHC (Abcam), MyoG (Abcam), Wnt1 (CST), SRF (Santa Cruz Biotechnology), β-catenin (ProteinTech, China), lamin B (CST), and GAPDH (BOSTER, China).
Target prediction and luciferase activity assay
The target genes of miR-664-5p were predicted with TargetScan 6.2. For the dual-reporter assay, we amplified its target genes, the SRF, and Wnt1 3′ UTR WT and mutant type from myoblast cDNAs containing miR-664-5p–binding sites and cloned them into the psiCHECK-2–reporter vector (Promega). The primers (Table S2) were synthesized with restriction sites of XhoI or NotI, respectively. The 3′ UTR mutants of Wnt1 and SRF were produced using overlap PCR (53). HEK293T was seeded in a 48-well plate and cotransfected with miRNA mimics or the negative control with psiCHECK-2–SRF (or Wnt1)-reporter vector or mutant vector. After 48 h of transfection, the relative luciferase activity of Renilla compared with firefly was measured.
Flow cytometry, EdU, and CCK-8 assays
Flow cytometry, EdU incorporation, and cell count kit 8 (CCK-8) assays were performed as described previously (31). Myoblasts were transfected with NC and mimics when the cell densities were 40%. Then 24 h after transfection, the myoblasts were used to perform proliferation-related assays.
Bioinformatics analysis
The prediction of miRNA target genes was performed by using some online software, including miRbase (http://www.mirbase.org/)4 (54), TargetScan (http://www.targetscan.org/vert_71/)4 (55), miRDB (http://www.mirdb.org/miRDB/)4 (43). The GO term analysis and the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis were performed using Gene Ontology Consortium (http://geneontology.org/)4 (56, 57) and DAVID Bioinformatics Resources (https://david.ncifcrf.gov/)4 (58, 59), respectively.
Immunofluorescent assay
After inducing differentiation for 4 days, cells were used to detect the formation of myotubes by staining MyHC. The assay was performed according to the standard procedure. Myoblasts were first fixed with 4% paraformaldehyde and blocked with 5% BSA for 30 min. The cells were then incubated with MyHC primary antibody (1:200) overnight at 4 °C followed by washing with PBS three times. Next, the samples were incubated with Alexa Fluor 488–conjugated anti-mouse IgG (1:200) for 1 h at room temperature and DAPI for 10 min. Images were obtained from a Leica fluorescent microscope. The fusion index was counted by comparing the MyHC-positive cells to total nuclei.
Statistical analysis
All charts were created using GraphPad Prism 6.0 and the data represent the mean ± S.E. The significance of differences between the groups was assessed using the Student's t test or one-way analysis (*, p < 0.05; **, p < 0.01).
Author contributions
R. C., Y. S., and T. Y. data curation; R. C., N. Q., M. M., Q. Z., and Y. S. investigation; R. C., M. M., Q. Z., and W. D. methodology; R. C. writing-original draft; N. Q. and X. C. formal analysis; N. Q. validation; Y. W., G. T., and X. C. software; W. D. and W. P. supervision; G. Y. and W. P. resources; W. P. funding acquisition; W. P. writing-review and editing.
Supplementary Material
This work was supported by National Natural Science Foundation Grants 31572366 and 31872979, National Basic Research Programs of China Grant 2015CB943102, and the National Key Research and Development Program of China Grant 2017YFD0502002. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5 and Tables S1 and S2.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- MRF
- myogenic regulation factor
- SRF
- serum response factor
- miRNA
- microRNA
- MyoG
- myogenin
- EdU
- 5-ethynyl-2′-deoxyuridine
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- NC
- negative control
- GO
- Gene Ontology
- DMEM
- Dulbecco's modified Eagle's medium
- DM
- differentiation medium
- GM
- growth medium
- qPCR
- quantitative PCR.
References
- 1. Yun K., and Wold B. (1996) Skeletal muscle determination and differentiation: story of a core regulatory network and its context. Curr. Opin. Cell Biol. 8, 877–889 10.1016/S0955-0674(96)80091-3 [DOI] [PubMed] [Google Scholar]
- 2. Buckingham M., Bajard L., Chang T., Daubas P., Hadchouel J., Meilhac S., Montarras D., Rocancourt D., and Relaix F. (2003) The formation of skeletal muscle: from somite to limb. J. Anat. 202, 59–68 10.1046/j.1469-7580.2003.00139.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Buckingham M. (2001) Skeletal muscle formation in vertebrates. Curr. Opin. Genet. Dev. 11, 440–448 10.1016/S0959-437X(00)00215-X [DOI] [PubMed] [Google Scholar]
- 4. Buckingham M. (2006) Myogenic progenitor cells and skeletal myogenesis in vertebrates. Curr. Opin. Genet. Dev. 16, 525–532 10.1016/j.gde.2006.08.008 [DOI] [PubMed] [Google Scholar]
- 5. Bartel D. P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 6. Ambros V. (2004) The functions of animal microRNAs. Nature 431, 350–355 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- 7. Lee I., Ajay S. S., Yook J. I., Kim H. S., Hong S. H., Kim N. H., Dhanasekaran S. M., Chinnaiyan A. M., and Athey B. D. (2009) New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res. 19, 1175–1183 10.1101/gr.089367.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou H., and Rigoutsos I. (2014) miR-103a-3p targets the 5′ UTR of GPRC5A in pancreatic cells. RNA 20, 1431–1439 10.1261/rna.045757.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen H., Jiang S., Wang L., Wang L., Wang H., Qiu L., and Song L. (2016) Cgi-miR-92d indirectly regulates TNF expression by targeting CDS region of lipopolysaccharide-induced TNF-α factor 3 (CgLITAF3) in oyster Crassostrea gigas. Fish Shellfish Immun. 55, 577–584 10.1016/j.fsi.2016.06.036 [DOI] [PubMed] [Google Scholar]
- 10. Polioudakis D., Abell N. S., and Iyer V. R. (2015) miR-191 regulates primary human fibroblast proliferation and directly targets multiple oncogenes. PLoS ONE 10, e126535 10.1371/journal.pone.0126535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shi L., Zhou B., Li P., Schinckel A. P., Liang T., Wang H., Li H., Fu L., Chu Q., and Huang R. (2015) MicroRNA-128 targets myostatin at coding domain sequence to regulate myoblasts in skeletal muscle development. Cell. Signal. 27, 1895–1904 10.1016/j.cellsig.2015.05.001 [DOI] [PubMed] [Google Scholar]
- 12. Sokol N. S. (2012) The role of microRNAs in muscle development. Curr. Top. Dev. Biol. 99, 59–78 10.1016/B978-0-12-387038-4.00003-3 [DOI] [PubMed] [Google Scholar]
- 13. Goljanek-Whysall K., Sweetman D., and Münsterberg A. E. (2012) microRNAs in skeletal muscle differentiation and disease. Clin. Sci. 123, 611–625 10.1042/CS20110634 [DOI] [PubMed] [Google Scholar]
- 14. O'Rourke J. R., Georges S. A., Seay H. R., Tapscott S. J., McManus M. T., Goldhamer D. J., Swanson M. S., and Harfe B. D. (2007) Essential role for Dicer during skeletal muscle development. Dev. Biol. 311, 359–368 10.1016/j.ydbio.2007.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen J. F., Mandel E. M., Thomson J. M., Wu Q., Callis T. E., Hammond S. M., Conlon F. L., and Wang D. Z. (2006) The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38, 228–233 10.1038/ng1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anderson C., Catoe H., and Werner R. (2006) MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Res. 34, 5863–5871 10.1093/nar/gkl743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dalton S., Marais R., Wynne J., and Treisman R. (1993) Isolation and characterization of SRF accessory proteins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 340, 325–332 10.1098/rstb.1993.0074 [DOI] [PubMed] [Google Scholar]
- 18. Price M. A., Hill C., and Treisman R. (1996) Integration of growth factor signals at the c-fos serum response element. Philos Trans R Soc Lond B Biol Sci. 351, 551–559 10.1098/rstb.1996.0054 [DOI] [PubMed] [Google Scholar]
- 19. Treisman R. (1995) Journey to the surface of the cell: fos regulation and the SRE. EMBO J. 14, 4905–4913 10.1002/j.1460-2075.1995.tb00173.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Norman C., Runswick M., Pollock R., and Treisman R. (1988) Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell 55, 989–1003 10.1016/0092-8674(88)90244-9 [DOI] [PubMed] [Google Scholar]
- 21. van Delft J. H., Mathijs K., Staal Y. C., van Herwijnen M. H., Brauers K. J., Boorsma A., and Kleinjans J. C. (2010) Time series analysis of benzo[a]pyrene-induced transcriptome changes suggests that a network of transcription factors regulates the effects on functional gene sets. Toxicol. Sci. 117, 381–392 10.1093/toxsci/kfq214 [DOI] [PubMed] [Google Scholar]
- 22. L'Honore A., Lamb N. J., Vandromme M., Turowski P., Carnac G., and Fernandez A. (2003) MyoD distal regulatory region contains an SRF binding CArG element required for MyoD expression in skeletal myoblasts and during muscle regeneration. Mol. Biol. Cell 14, 2151–2162 10.1091/mbc.e02-07-0451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li S., Czubryt M. P., McAnally J., Bassel-Duby R., Richardson J. A., Wiebel F. F., Nordheim A., and Olson E. N. (2005) Requirement for serum response factor for skeletal muscle growth and maturation revealed by tissue-specific gene deletion in mice. Proc. Natl. Acad. Sci. U.S.A. 102, 1082–1087 10.1073/pnas.0409103102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sethi J. K., and Vidal-Puig A. (2010) Wnt signalling and the control of cellular metabolism. Biochem. J. 427, 1–17 10.1042/BJ20091866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Münsterberg A. E., Kitajewski J., Bumcrot D. A., McMahon A. P., and Lassar A. B. (1995) Combinatorial signaling by Sonic hedgehog and Wnt family members induces myogenic bHLH gene expression in the somite. Genes Dev. 9, 2911–2922 10.1101/gad.9.23.2911 [DOI] [PubMed] [Google Scholar]
- 26. Tajbakhsh S., Borello U., Vivarelli E., Kelly R., Papkoff J., Duprez D., Buckingham M., and Cossu G. (1998) Differential activation of Myf5 and MyoD by different Wnts in explants of mouse paraxial mesoderm and the later activation of myogenesis in the absence of Myf5. Development 125, 4155–4162 [DOI] [PubMed] [Google Scholar]
- 27. Sun Z., Wang C., Shi C., Sun F., Xu X., Qian W., Nie S., and Han X. (2014) Activated Wnt signaling induces myofibroblast differentiation of mesenchymal stem cells, contributing to pulmonary fibrosis. Int. J. Mol. Med. 33, 1097–1109 10.3892/ijmm.2014.1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Katoh M., and Katoh M. (2007) WNT signaling pathway and stem cell signaling network. Clin. Cancer Res. 13, 4042–4045 10.1158/1078-0432.CCR-06-2316 [DOI] [PubMed] [Google Scholar]
- 29. Grumolato L., Liu G., Mong P., Mudbhary R., Biswas R., Arroyave R., Vijayakumar S., Economides A. N., and Aaronson S. A. (2010) Canonical and noncanonical Wnts use a common mechanism to activate completely unrelated coreceptors. Gene. Dev. 24, 2517–2530 10.1101/gad.1957710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Abu-Elmagd M., Robson L., Sweetman D., Hadley J., Francis-West P., and Münsterberg A. (2010) Wnt/Lef1 signaling acts via Pitx2 to regulate somite myogenesis. Dev. Biol. 337, 211–219 10.1016/j.ydbio.2009.10.023 [DOI] [PubMed] [Google Scholar]
- 31. Ma M., Wang X., Chen X., Cai R., Chen F., Dong W., Yang G., and Pang W. (2017) MicroRNA-432 targeting E2F3 and P55PIK inhibits myogenesis through PI3K/AKT/mTOR signaling pathway. RNA Biol. 14, 347–360 10.1080/15476286.2017.1279786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang Y., Liu H., Wang X., and Chen L. (2015) Up-regulation of microRNA-664 inhibits cell growth and increases cisplatin sensitivity in cervical cancer. Int. J. Clin. Exp. Med. 8, 18123–18129 [PMC free article] [PubMed] [Google Scholar]
- 33. Chen B., Bao Y., Chen X., Yi J., Liu S., Fang Z., Zheng S., and Chen J. (2015) Mir-664 promotes osteosarcoma cells proliferation via downregulating of FOXO4. Biomed. Pharmacother. 75, 1–7 10.1016/j.biopha.2015.08.012 [DOI] [PubMed] [Google Scholar]
- 34. Chen J. F., Tao Y., Li J., Deng Z., Yan Z., Xiao X., and Wang D. Z. (2010) microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J. Cell Biol. 190, 867–879 10.1083/jcb.200911036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wei W., He H. B., Zhang W. Y., Zhang H. X., Bai J. B., Liu H. Z., Cao J. H., Chang K. C., Li X. Y., and Zhao S. H. (2013) miR-29 targets Akt3 to reduce proliferation and facilitate differentiation of myoblasts in skeletal muscle development. Cell Death Dis. 4, e668 10.1038/cddis.2013.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Feng Y., Niu L. L., Wei W., Zhang W. Y., Li X. Y., Cao J. H., and Zhao S. H. (2013) A feedback circuit between miR-133 and the ERK1/2 pathway involving an exquisite mechanism for regulating myoblast proliferation and differentiation. Cell Death Dis. 4, e934 10.1038/cddis.2013.462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bartel D. P. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hauschka S. D. (2001) Myocardin. a novel potentiator of SRF-mediated transcription in cardiac muscle. Mol. Cell 8, 1–2 10.1016/S1097-2765(01)00297-0 [DOI] [PubMed] [Google Scholar]
- 39. Wang Z., Wang D. Z., Hockemeyer D., McAnally J., Nordheim A., and Olson E. N. (2004) Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature 428, 185–189 10.1038/nature02382 [DOI] [PubMed] [Google Scholar]
- 40. Wang D. Z., Li S., Hockemeyer D., Sutherland L., Wang Z., Schratt G., Richardson J. A., Nordheim A., and Olson E. N. (2002) Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc. Natl. Acad. Sci. U.S.A. 99, 14855–14860 10.1073/pnas.222561499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang Z., Wang D. Z., Pipes G. C., and Olson E. N. (2003) Myocardin is a master regulator of smooth muscle gene expression. Proc. Natl. Acad. Sci. U.S.A. 100, 7129–7134 10.1073/pnas.1232341100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Han X., Yang F., Cao H., and Liang Z. (2015) Malat1 regulates serum response factor through miR-133 as a competing endogenous RNA in myogenesis. FASEB J. 29, 3054–3064 10.1096/fj.14-259952 [DOI] [PubMed] [Google Scholar]
- 43. Wong N., and Wang X. (2015) miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 43, D146–D152 10.1093/nar/gku1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zeng C., Wang Y. L., Xie C., Sang Y., Li T. J., Zhang M., Wang R., Zhang Q., Zheng L., and Zhuang S. M. (2015) Identification of a novel TGF-β-miR-122-fibronectin 1/serum response factor signaling cascade and its implication in hepatic fibrogenesis. Oncotarget 6, 12224–12233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang H. Y., Tu Y. S., Long J., Zhang H. Q., Qi C. L., Xie X. B., Li S. H., and Zhang Y. J. (2015) SRF-miR-29b-MMP2 axis inhibits NSCLC invasion and metastasis. Int. J. Oncol. 47, 641–649 10.3892/ijo.2015.3034 [DOI] [PubMed] [Google Scholar]
- 46. Wang R., Zheng J., Zhang D. S., Yang Y. H., and Zhao Z. F. (2015) Wnt1-induced MAFK expression promotes osteosarcoma cell proliferation. Genet. Mol. Res. 14, 7315–7325 10.4238/2015.July.3.7 [DOI] [PubMed] [Google Scholar]
- 47. Song P., Zheng J. X., Liu J. Z., Xu J., Wu L. Y., Liu C., Zhu Q., and Wang Y. (2014) Effect of the Wnt1/β-catenin signalling pathway on human embryonic pulmonary fibroblasts. Mol. Med. Rep. 10, 1030–1036 10.3892/mmr.2014.2261 [DOI] [PubMed] [Google Scholar]
- 48. van Amerongen R., and Nusse R. (2009) Towards an integrated view of Wnt signaling in development. Development 136, 3205–3214 10.1242/dev.033910 [DOI] [PubMed] [Google Scholar]
- 49. Nusse R. (2008) Wnt signaling and stem cell control. Cell Res. 18, 523–527 10.1038/cr.2008.47 [DOI] [PubMed] [Google Scholar]
- 50. Du J., Li Q., Shen L., Lei H., Luo J., Liu Y., Zhang P., Pu Q., Zhang Y., Shuai S., Li X., Zhang S., and Zhu L. (2016) miR-145a-5p promotes myoblast differentiation. Biomed. Res. Int. 2016, 5276271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Du J., Zhang Y., Shen L., Luo J., Lei H., Zhang P., Pu Q., Liu Y., Shuai S., Li Q., Li X., Zhang S., and Zhu L. (2016) Effect of miR-143–3p on C2C12 myoblast differentiation. Biosci. Biotechnol. Biochem. 80, 706–711 [DOI] [PubMed] [Google Scholar]
- 52. Abraham S. T. (2016) A role for the Wnt3a/β-catenin signaling pathway in the myogenic program of C2C12 cells. In Vitro Cell Dev. Biol. Anim. 52, 935–941 10.1007/s11626-016-0058-5 [DOI] [PubMed] [Google Scholar]
- 53. Higuchi R., Krummel B., and Saiki R. K. (1988) A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16, 7351–7367 10.1093/nar/16.15.7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kozomara A., and Griffiths-Jones S. (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Agarwal V., Bell G. W., Nam J. W., and Bartel D. P. (2015) Predicting effective microRNA target sites in mammalian mRNAs. eLife 4, 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. The Gene Ontology Consortium (2017) Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 45, D331–D338 10.1093/nar/gkw1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Huang D. W, Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 59. Huang D. W., Sherman B. T., and Lempicki R. A. (2009) Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.