Abstract
ClpB, a bacterial homologue of heat shock protein 104 (Hsp104), can disentangle aggregated proteins with the help of the DnaK, a bacterial Hsp70, and its co-factors. As a member of the expanded superfamily of ATPases associated with diverse cellular activities (AAA+), ClpB forms a hexameric ring structure, with each protomer containing two AAA+ modules, AAA1 and AAA2. A long coiled-coil middle domain (MD) is present in the C-terminal region of the AAA1 and surrounds the main body of the ring. The MD is subdivided into two oppositely directed short coiled-coils, called motif-1 and motif-2. The MD represses the ATPase activity of ClpB, and this repression is reversed by the binding of DnaK to motif-2. To better understand how the MD regulates ClpB activity, here we investigated the roles of motif-1 in ClpB from Thermus thermophilus (TClpB). Using systematic alanine substitution of the conserved charged residues, we identified functionally important residues in motif-1, and using a photoreactive cross-linker and LC-MS/MS analysis, we further explored potential interacting residues. Moreover, we constructed TClpB mutants in which functionally important residues in motif-1 and in other candidate regions were substituted by oppositely charged residues. These analyses revealed that the intra-subunit pair Glu-401–Arg-532 and the inter-subunit pair Asp-404–Arg-180 are functionally important, electrostatically interacting pairs. Considering these structural findings, we conclude that the Glu-401–Arg-532 interaction shifts the equilibrium of the MD conformation to stabilize the activated form and that the Arg-180–Asp-404 interaction contributes to intersubunit signal transduction, essential for ClpB chaperone activities.
Keywords: protein aggregation, ATPases associated with diverse cellular activities (AAA), chaperone, protein conformation, protein cross-linking, protein dynamic, structure-function, mass spectrometry (MS), disaggregation, Hsp104
Introduction
ClpB/Hsp104 is a protein disaggregase required for thermotolerance of cells (1–3). To disentangle aggregated proteins, ClpB/Hsp104 cooperates with DnaK/Hsp70 chaperone and its co-factors, DnaJ/Hsp40, and the nucleotide exchange factor, GrpE (4–7).
As a member of an expanded superfamily of ATPases associated with diverse cellular activities (AAA+), ClpB/Hsp104 contains two AAA+ modules, AAA1 and AAA2, in a polypeptide and forms a hexameric ring-like structure, in which each AAA+ module constitutes each layer of a two layered ring (8). At the top of the ring, highly mobile globular N-terminal domains (ND)3 contribute to bind protein aggregates (9, 10), and long coiled-coil middle domains (MD) protruding from the C-terminal small subdomain of the AAA1 modules surround the main body of the ring (Fig. 1a) (8). The MD is subdivided into two oppositely-directed short coiled-coils, motif-1 and motif-2.
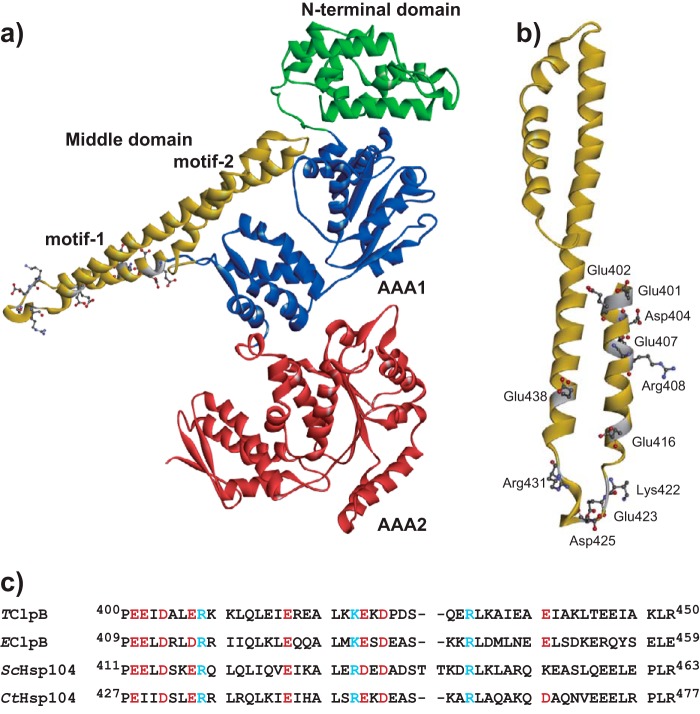
Figure 1.
Positions of the mutated residues of TClpB. a, structure of TClpB (PDB code 1QVR) is shown. The N-terminal domain, AAA1, middle domain, and AAA2 are colored in green, blue, yellow, and red, respectively. b, structure of the TClpB middle domain. The amino acid residues, Glu-401, Glu-402, Asp-404, Glu-407, Arg-408, Glu-416, Lys-422, Glu-423, Asp-425, Arg-431, and Glu-438, which were replaced by Ala, are shown as sticks. c, amino acid sequences of the middle domain motif-1 of ClpB/Hsp104 from T. thermophilus (TClpB), E. coli (EClpB), S. cerevisiae (ScHsp104), and Chaetomium thermophilum (CtHsp104) are aligned. The positively and negatively charged conserved residues, which were replaced by Ala, are shown in blue and red, respectively.
Both the AAA1 and AAA2 modules bind and hydrolyze ATP in a complex cooperative manner that contains intra- and inter-ring cooperation (11–13). The structural changes of the ring, induced by the cooperative ATPase cycle, promote disentangling of aggregated substrates by threading them into the central pore of the ring. Recent cryo-electron microscopy (cryo-EM) and high-speed atomic force microscopy analyses demonstrated that the structural inter-conversion between the hexameric ring, spiral, and twisted half-spiral structures occurred during the ATPase cycle and contributed to the disaggregation reaction (14–17).
Ordinarily, the MD represses the ATPase activity of ClpB/Hsp104 by an unknown mechanism (18, 19). In addition, the binding affinity of ClpB/Hsp104 for aggregation is not high enough to start threading (20). To perform disaggregation, the recruitment of ClpB/Hsp104 to the aggregates and the cancellation of repression are required (21). DnaK/Hsp70 consists of an N-terminal nucleotide-binding domain (NBD) and a substrate-binding domain (SBD) (22, 23). The NBD controls the conformation of the SBD and its binding affinity for substrate proteins in an ATPase cycle-dependent manner (24, 25). In the high-affinity conformation, the NBD can bind to the edge of MD motif-2 of ClpB/Hsp104 (21, 26). This interaction causes recruitment of ClpB/Hsp104 to aggregates and relieves repression by the MD.
Mutations at the edge of motif-1 and motif-2, E432A and Y503D of Escherichia coli ClpB (EClpB), are known to stabilize the repressed and activated conformations, and these mutants are termed repressed and hyperactive mutants, respectively (18, 27). Single particle reconstitution of cryo-EM images of these mutants revealed that the conformations of the MD in the repressed and hyperactive mutants are horizontal and tilted to the main body, respectively (28). In the horizontal conformation, both edges of the MD interact with the MD edges of neighboring subunits in a head-to-tail manner. The interaction between the DnaK NBD and the motif-2 of ClpB MD is thought to disrupt the inter-MD interactions and induce the MD into the tilted conformation.
Although the role of MD motif-2 in binding DnaK NBD is clear, the roles of motif-1 should also be investigated to understand the mechanisms involved in controlling MD conformation, and in the regulation of ClpB activity by the MD. To clarify these issues, here we identified functionally important residues in motif-1 and their targets for interaction. We also identified the nucleotide dependence of the interactions.
Results
Effects of alanine substitutions of conserved charged residues in the ClpB MD motif-1
To identify important residues in the MD motif-1, we substituted the conserved charged residues, Glu-401, Glu-402, Asp-404, Glu-407, Arg-408, Glu-416, Lys-422, Glu-423, Asp-425, Arg-431, and Glu-438, in motif-1 with Ala and tested the effects on ClpB activities (Fig. 1, b and c).
At 55 °C, WT TClpB hydrolyzed ATP at a rate of 48 min−1, and the rate was stimulated by the presence of κ-casein up to 83 min−1 (Fig. 2a). All of the tested Ala mutants showed similar or at most 2-fold higher ATPase activity to that of the WT, in both the absence and presence of κ-casein (Fig. 2a).
Figure 2.
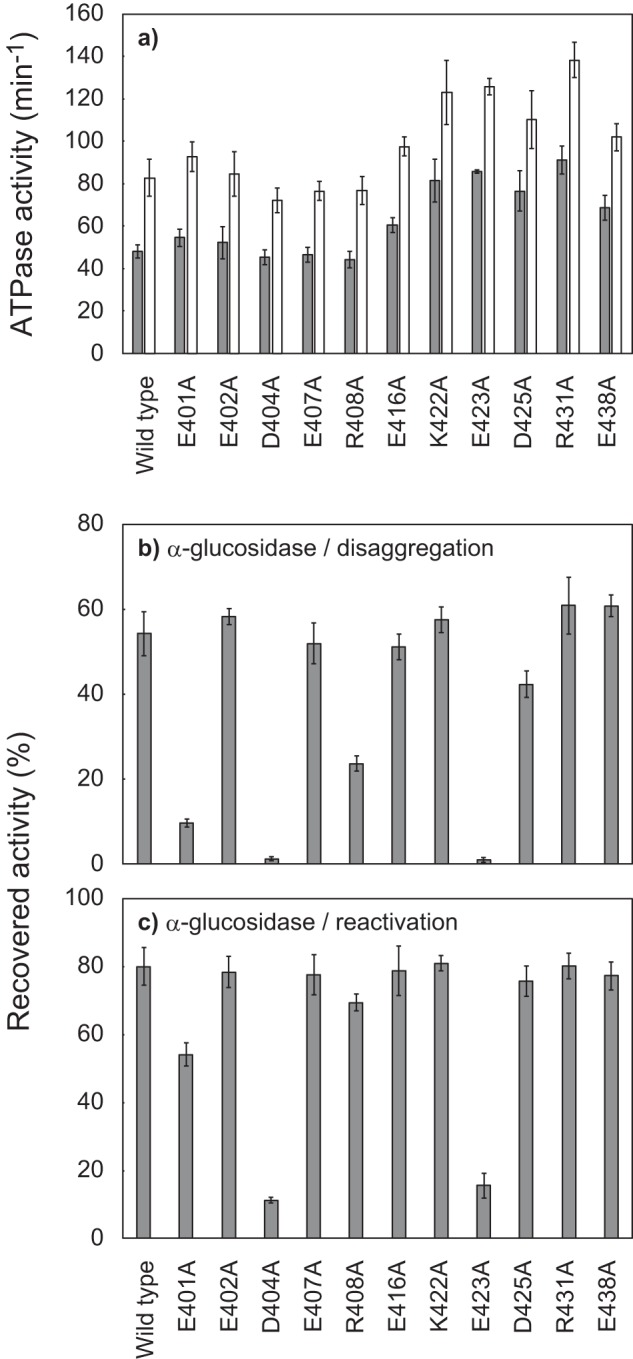
Effects of alanine substitutions of conserved charged residues in TClpB middle domain motif-1. a, ATPase activities of TClpB mutants, in the absence (gray bar) or the presence (open bar) of 0.1 mg/ml κ-casein, are shown as turnover number per monomer. The measurements were performed in the presence of 3 mm ATP at 55 °C using an ATP-regeneration system. b, disaggregation activities of TClpB mutants. α-Glucosidase (final concentration, 0.2 μm monomer) was heat-aggregated by incubation at 73 °C for 10 min in the presence of 3 mm ATP. TDnaK (0.6 μm), TDnaJ (0.2 μm), TGrpE (0.1 μm dimer), and TClpB mutants (0.05 μm hexamers) were added to the reaction mixture, and the mixture was subsequently incubated at 55 °C for 90 min. After incubation, the recovered enzymatic activities were measured and are shown as a percentage of the activities before heat aggregation. c, chaperone activities of TClpB mutants for the reactivation of soluble, denatured proteins were measured by using α-glucosidase as a substrate. The experimental procedures were essentially the same as for b, except that the TDnaK, TDnaJ, and TGrpE were added prior to the heat treatment, and the substrate was denatured but not aggregated. a–c, error bars represent standard deviations of three or more independent experiments.
The disaggregation activities were measured by using α-glucosidase from Bacillus stearothermophilus, which was completely aggregated by heat treatment at 73 °C for 10 min (29). The WT TClpB could reactivate 54% of the aggregated α-glucosidase by incubation with TDnaK, TDnaJ, TGrpE (TKJE), and ATP at 55 °C for 90 min (Fig. 2b). Although the E402A, E407A, E416A, K422A, R431A, and E438A mutants showed almost the same disaggregation activity as that of WT, the reactivation yields of E401A, R408A, and D425A mutants decreased to 10, 24, and 42%, respectively, and the D404A and E423A mutants completely lost disaggregation activity.
When the α-glucosidase was heat-treated in the presence of TKJE and ATP, the substrate protein lost enzyme activity (denatured); however, it remained soluble (29). The soluble, denatured protein can be reactivated by incubation at 55 °C in the presence of TKJE, ATP, and TClpB supplemented after the 73 °C heat treatment. In this case, WT TClpB could reactivate 80% of soluble, denatured α-glucosidase (Fig. 2c). As in the case of disaggregation, D404A and E423A mutations severely inhibited the reactivation activity. However, R408A and D425A mutants could reactivate the soluble, denatured α-glucosidase at almost the same efficiency as that of the WT, and the reactivation yield by the E401A mutant also increased, reaching 54%.
Here, we identified several important (Glu-401, Arg-408, and Asp-425) and essential (Asp-404 and Glu-423) residues for the chaperone activity of ClpB. Among these residues, the importance of Glu-423 (corresponding to Glu-432 of EClpB) has been reported, and its roles have been proposed previously (18, 28). Thus, hereafter, we focused on Asp-404 and Glu-401. The former is a newly identified essential residue, and the latter is an amino acid that is more important than Arg-408 and Asp-425.
Glu-401 and Asp-404 interact with neighboring subunits
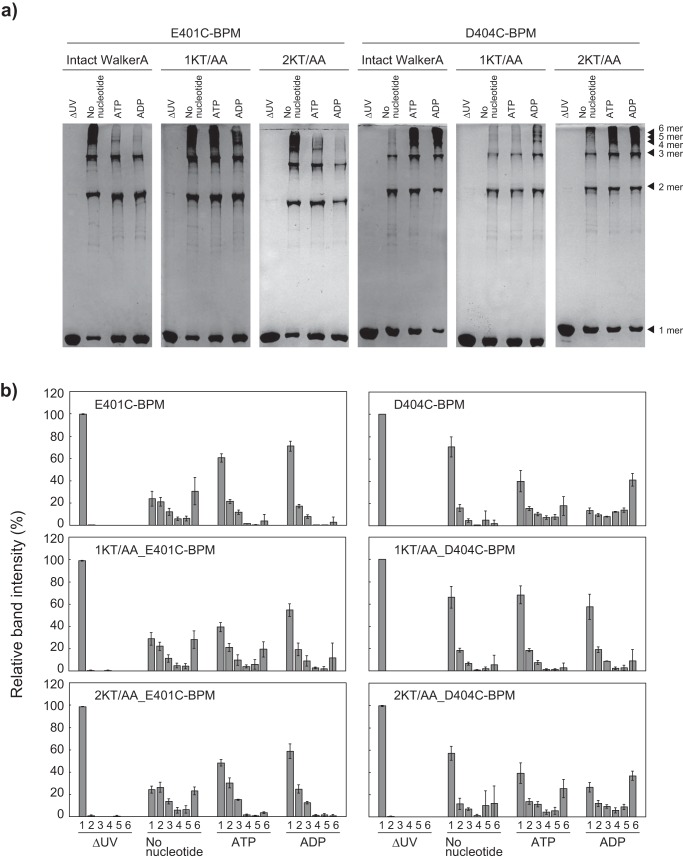
To identify the interaction targets of Glu-401 and Asp-404, we used the bidirectional photoreactive cross-linker, benzophenone-4-maleimide (BPM). The BPM-labeled TClpB mutants, E401C-BPM and D404C-BPM, were UV-irradiated in the absence or presence of nucleotides and analyzed by SDS-PAGE (Fig. 3, a and b). By the UV irradiation, the intensity of the band representing noncross-linked BPM-labeled TClpB decreased, and several ladder bands with higher molecular weights appeared (Fig. 3a). As the reaction mixture did not contain proteins other than BPM-labeled TClpB, inter-subunit cross-linking would occur, and each ladder band corresponded to a multimer of BPM-labeled TClpB. We quantified the intensities of bands corresponding to 1–5- and over 6-mer (Fig. 3b). It should be noted that the over 6-mer bands consisted of a few closely positioned bands, which would correspond to linear 6-mer, closed-ring 6-mer, or 7-mer that would appear transiently (17). When the E401C-BPM was UV-irradiated in the absence of nucleotide, the intensity of the noncross-linked 1-mer band decreased to 24%, and 31% of E401C-BPM appeared as over 6-mer band. However, intensities of the over 6-mer bands decreased to less than 4% when the UV irradiation was performed in the presence of ATP or ADP (Fig. 3, a and b). Also, in the case of D404C-BPM, similar ladder bands appeared after UV irradiation (Fig. 3, a and b). However, the nucleotide dependence was opposite that seen in the case of E401C-BPM. These results indicated that Glu-401 and Asp-404 interacted with neighboring subunits, in different ways, and these interactions were influenced by the nucleotide states.
Figure 3.
Essential charged residues in the middle domain motif-1 involved in nucleotide-dependent inter-subunit interactions. To form covalent bonds between the essential charged residues in motif-1 and the closely positioned C–H bond, BPM-labeled TClpB mutants were UV-irradiated at 365 nm for 90 min in the absence or presence of 3 mm nucleotides. The cross-linked samples were analyzed by SDS-PAGE (5% polyacrylamide). a, gel images; b, intensities of bands corresponding to 1–5- and over 6-mers of E401C-BPM and D404C-BPM, with or without Walker-A mutations are shown. b, error bars represent standard deviations of three or more independent experiments.
To identify the responsible nucleotide-binding domain, similar experiments using E401C-BPM and D404C-BPM bearing mutations in the Walker-A consensus sequences of AAA1 or AAA2 were also performed (Fig. 3, a and b). Conserved Lys–Thr pairs in the Walker-A sequence (GXXGXGKT) were replaced by Ala–Ala, and the mutated AAA module could not bind nucleotides (30). In both cases of E401C-BPM and D404C-BPM, Walker-A mutations involving AAA1 were more effective than those in AAA2 in canceling the nucleotide dependence of cross-linking efficiency (Fig. 3, a and b).
Mass spectrometric analysis of cross-linked TClpB-BPM
To identify the site of cross-linkage, we performed mass spectrometric analysis of cross-linked TClpBs by using LC-MS/MS (Table 1). To simplify the interpretation of MS data, ND-truncated TClpB (TClpB_ΔN) was used in this analysis. The TClpB_ΔN_E401C-BPM, UV-irradiated in the absence of nucleotides, was cleaved by trypsin and analyzed by LC-MS/MS. We detected some peptides cross-linked by BPM. Peptides comprising the sequence from Gly-218 to Lys-225 (218GDVPEGLK225) of TClpB were most frequently detected as the cross-linkage targets of E401C-BPM. The peptides 519LEVTEEDIAEIVSRWTGIPVSK540 and 381AIDLIDEAAARLR393 were also detected. When the UV irradiation was performed in the presence of ATP, a similar set of peptides was detected. In this condition, the peptides 519LEVTEEDIAEIVSR532 (a part of 519LEVTEEDIAEIVSRWTGIPVSK540) and 392LRMALESAPCEIDALERKK410 were also detected. In the cases of TClpB_ΔN_D404C-BPM, UV-irradiated in the absence or presence of ATP, peptides containing the sequence 218GDVPEGLK225, 381AIDLIDEAAARLR393, 409KKLQLEIER417, and 701NTVIILTSNLGSPLILEGLQK721 were detected as the cross-linkage targets. A peptide 835VQVDVGPAGLVFAVPAR851 was detected only when the UV irradiation was performed in the presence of ATP. It should be noted that the peptide, 701NTVIILTSNLGSPLILEGLQK721 might be mis-annotated, because the peptide was also detected in the analysis without UV irradiation.
Table 1.
Cross-linked peptide pairs detected by LC-MS/MS
| TClpB | Nucleotide | Peptide including Cys labeled with BPM | Peptide including photo cross-linked residue | Theoretical peptide mass | No. of detected spectra |
|---|---|---|---|---|---|
| Da | |||||
| E401C | No nucleotide | 394MALESAPCEIDALER408 | 218GDVPEGLK225 | 2756.27924 | 12 |
| 394MALESAPCEIDALER408 | 215IVKGDVPEGLK225 | 3096.52666 | 10 | ||
| 394MALESAPCEIDALERK409 | 215IVKGDVPEGLK225 | 3224.62162 | 7 | ||
| 392LRMALESAPCEIDALERK409 | 218GDVPEGLK225 | 3153.55935 | 5 | ||
| 394MALESAPCEIDALER408 | 519LEVTEEDIAEIVSRWTGIPVSK540 | 4413.15145 | 8 | ||
| 394MALESAPCEIDALER408 | 381AIDLIDEAAARLR393 | 3368.64995 | 5 | ||
| ATP | 394MALESAPCEIDALER408 | 218GDVPEGLK225 | 2756.27924 | 7 | |
| 394MALESAPCEIDALER408 | 215IVKGDVPEGLK225 | 3096.52666 | 11 | ||
| 394MALESAPCEIDALERK409 | 218GDVPEGLK225 | 2884.37419 | 2 | ||
| 394MALESAPCEIDALER408 | 519LEVTEEDIAEIVSR532 | 3544.67080 | 2 | ||
| 394MALESAPCEIDALER408 | 519LEVTEEDIAEIVSRWTGIPVSK540 | 4413.15145 | 12 | ||
| 394MALESAPCEIDALERK409 | 519LEVTEEDIAEIVSRWTGIPVSK540 | 4541.24640 | 2 | ||
| 394MALESAPCEIDALER408 | 381AIDLIDEAAARLR393 | 3368.64995 | 4 | ||
| 394MALESAPCEIDALER408 | 392LRMALESAPCEIDALERKK410 | 4114.99543 | 2 | ||
| D404C | No nucleotide | 394MALESAPEEICALER408 | 218GDVPEGLK225 | 2770.29489 | 2 |
| 394MALESAPEEICALER408 | 215IVKGDVPEGLK225 | 3110.54231 | 5 | ||
| 394MALESAPEEICALERKK410 | 215IVKGDVPEGLK225 | 3366.73222 | 2 | ||
| 394MALESAPEEICALER408 | 381AIDLIDEAAARLR393 | 3382.66560 | 11 | ||
| 394MALESAPEEICALER408 | 409KKLQLEIER417 | 3112.56919 | 2 | ||
| 394MALESAPEEICALER408 | 701NTVIILTSNLGSPLILEGLQK721a | 4179.16016 | 4 | ||
| ATP | 394MALESAPEEICALER408 | 215IVKGDVPEGLK225 | 3110.54231 | 6 | |
| 394MALESAPEEICALERK409 | 218GDVPEGLK225 | 2898.38984 | 2 | ||
| 394MALESAPEEICALER408 | 381AIDLIDEAAARLR393 | 3382.66560 | 7 | ||
| 394MALESAPEEICALER408 | 409KKLQLEIER417 | 3112.56919 | 2 | ||
| 394MALESAPEEICALER408 | 835VQVDVGPAGLVFAVPAR851 | 3650.82315 | 2 | ||
| 394MALESAPEEICALER408 | 701NTVIILTSNLGSPLILEGLQK721a | 4179.16016 | 4 | ||
a MALESAPEEICALER-NTVIILTSNLGSPLILEGLQK cross-linked signals were also detected in no UV-irradiation samples.
Candidates for residues interacting with Glu-401 and Asp-404
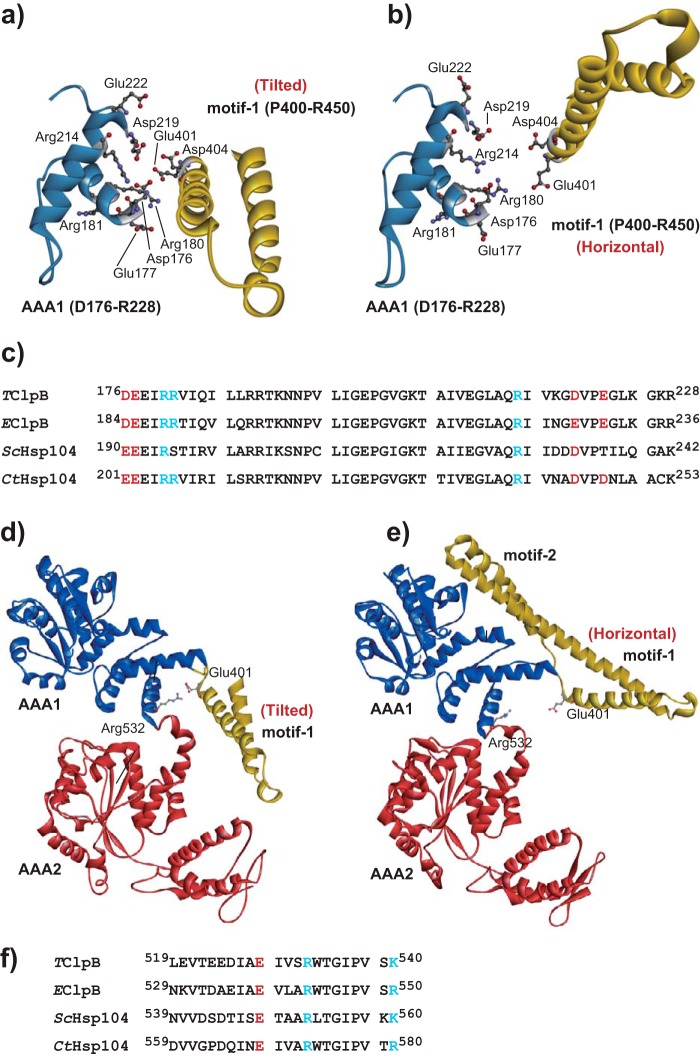
To confirm the plausibility of the mass spectrometric analysis, we constructed hexameric structural models of TClpB by homology modeling using recently solved cryo-EM structures of EClpB hexamers as templates (16). Fig. 4 shows interfaces between an MD motif-1 and the AAA1 of the neighboring subunit in the TClpB models. The MDs in Fig. 4, a and b, correspond to the tilted (chain D of the PDB code 5og1) and the horizontal (chain C of the PDB code 5ofo) conformation, respectively. Although inter-subunit interactions involving Glu-401 and Asp-404 were not identified in these models, the B3-helix (Asp-176–Leu-187) and the loop between the B4-helix and b3-strand (Gly-218–Lys-227) of the AAA1 large sub-domain of the neighboring subunit were found to be proximal to Glu-401 and Asp-404. This was consistent with the result that the 218GDVPEGLK225 peptide was found as the primary cross-linking target of both E401C-BPM and D404C-BPM. On the surface of the helix and the loop, some conserved charged residues, Asp-176, Glu-177, Arg-180, Arg-181, Arg-214, Asp-219, and Glu-222, were found (Fig. 4, a–c). In addition, only in the tilted conformation Glu-401 was located near Arg-532 in the 519LEVTEEDIAEIVSRWTGIPVSK540 peptide that was detected as a cross-linking target for E401C-BPM (Fig. 4, d and e). Furthermore, Arg-532 was highly conserved among various ClpB/Hsp104s (Fig. 4f). Considering the hexameric model, structural regions corresponding to the other detected peptides, 381AIDLIDEAAARLR393, 392LRMALESAPCEIDALERKK410, 409KKLQLEIER417, 835VQVDVGPAGLVFAVPAR851, did not seem to interact with either Glu-401 or Asp-404. Thus, we considered these conserved charged eight residues as candidates for target residues that functionally interact with Glu-401 and/or Asp-404.
Figure 4.
Structural models of TClpB. The structural models of the TClpB hexamer were constructed by homology modeling using cryo-EM structures of EClpB with or without casein (PDB codes 5og1 and 5ofo) as templates. a and b, interfaces between AAA1 (light blue) and middle domain motif-1 (yellow) of neighboring subunits. AAA1 and motif-1 correspond to chains C and D (the middle domain is in the tilted conformation) of 5og1 (a), and chains B and C (the middle domain is in the horizontal conformation) of 5ofo (b). Amino acid residues, Glu-401, Asp-404, Asp-176, Glu-177, Arg-180, Arg-181, Arg-214, Asp-219, and Glu-222, which were replaced by Ala, are shown as sticks. c, amino acid sequences of a part of AAA1 of ClpB/Hsp104 from T. thermophilus (TClpB), E. coli (EClpB), S. cerevisiae (ScHsp104), and C. thermophilum (CtHsp104) were aligned. The positively and negatively charged conserved residues, which were replaced by Ala, are shown in blue and red, respectively. d and e, TClpB structural model constructed based on chain D of 5og1 (d) and chain C of 5ofo (e). Amino acid residues Glu-401 and Arg-532 are shown as sticks. f, amino acid sequences of a part of AAA1 of TClpB, EClpB, ScHsp104, and CtHsp104 were aligned. Positively and negatively charged conserved residues are shown in blue and red, respectively.
Effects of alanine substitutions of candidate residues
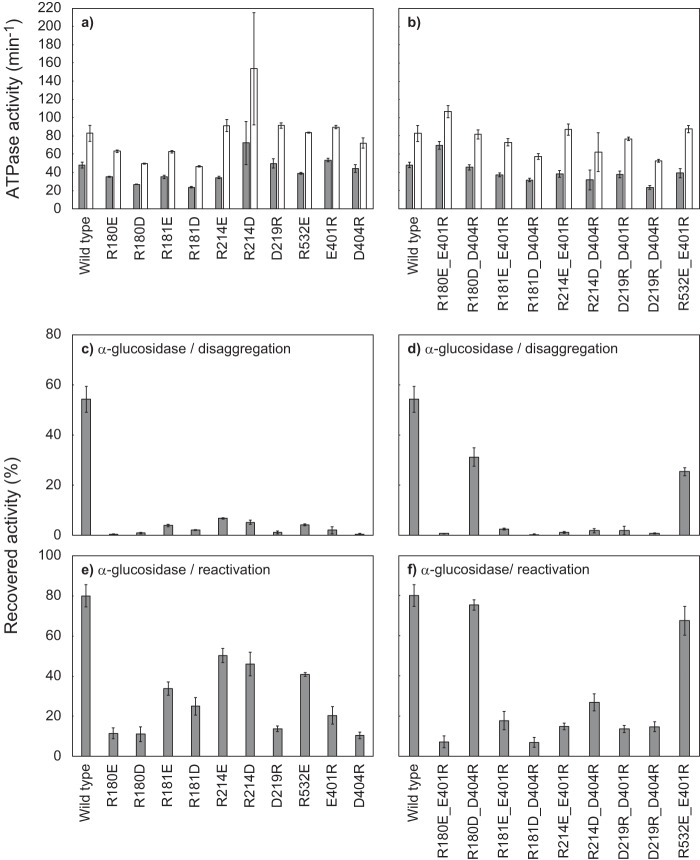
We substituted these candidate residues with Ala and tested the effects on ClpB activities. D176A and E177A mutants showed 2–4 times higher ATPase activity than the WT, whereas the activities of the other mutants were similar to or slightly lower than the WT (Fig. 5a). D176A, E177A, and E222A mutants could reactivate aggregated and soluble, denatured α-glucosidase with efficiencies comparable with that of WT (Fig. 5, b and c). However, the other mutants hardly reactivated the aggregated α-glucosidase. The reactivation yields of soluble, denatured α-glucosidase by R180A, R181A, R214A, and D219A mutants were also relatively low, ∼20–30%, whereas that of R532A was considerably high at 61%, similar to the case of E401A (Fig. 5, b and c).
Figure 5.
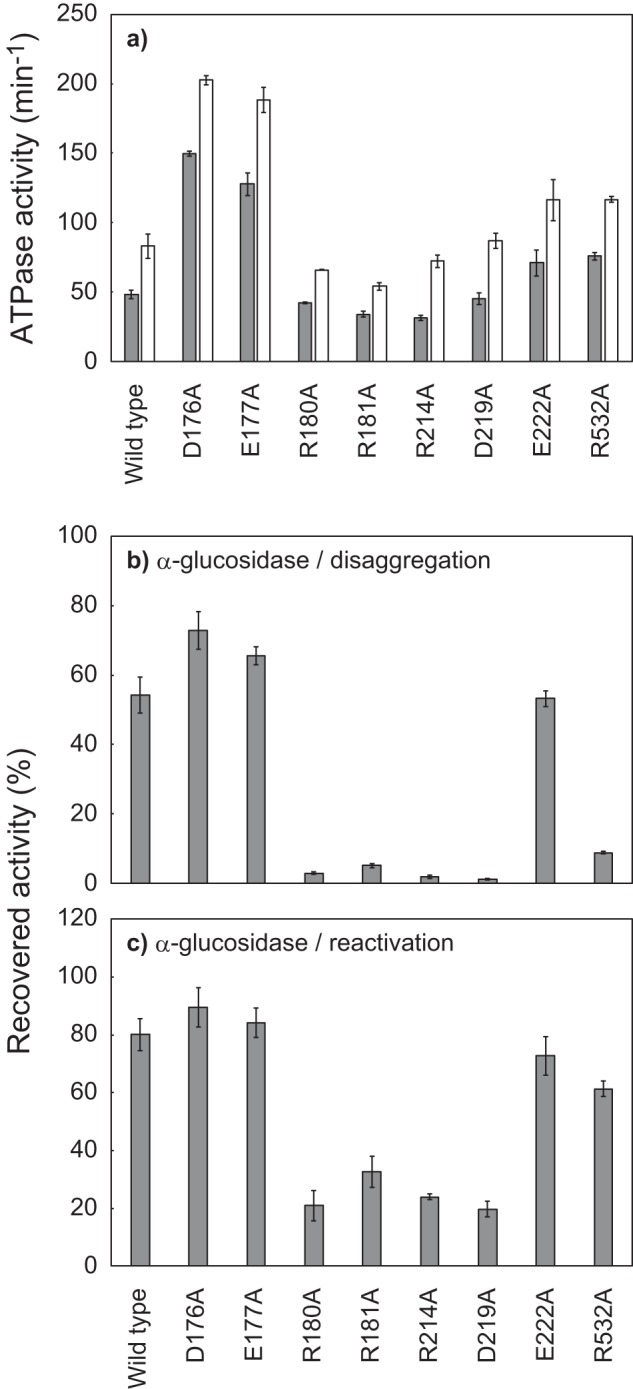
Effects of alanine substitutions of the charged residues in the region of AAA1 that potentially interact with Glu-401 and Asp-404. a, ATPase activities of TClpB mutants in the absence (gray bar) or presence (open bar) of 0.1 mg/ml κ-casein are shown. b, disaggregation activities of TClpB mutants were measured using α-glucosidase as a substrate. c, chaperone activities of TClpB mutants for the reactivation of soluble, denatured proteins were measured by using α-glucosidase as a substrate. Experimental procedures for a–c were the same as for Fig. 2, a–c. Error bars represent standard deviations of three or more independent experiments.
Effects of charge-inversion mutations
Because the effects of the R180A, R181A, R214A, D219A, and R532A mutations were similar to those of the E401A and D404A mutations, these residues might interact with Glu-401 and/or Asp-404. To test this possibility, we prepared TClpB mutants in which these charged residues were substituted by amino acids having the opposite charge, and the mutations were combined. All of the single and combined charge-inverted mutants tested here showed comparable ATPase activities to that of the WT (48–150%) and 1.5–2.7-fold ATPase stimulation by κ-casein (Fig. 6, a and b). Although all the single charge-inverted mutants hardly reactivated the heat-aggregated α-glucosidase (the reactivation yields were less than 7%), the R180D_D404R and R532E_E401R double mutants could reactivate it to 31 and 25%, respectively (Fig. 6, c and d). Such recovery of disaggregation activity was not observed with any other combinations of charge-inverted mutations. Concerning the reactivation of soluble, denatured α-glucosidase, some single and combined charge-inverted mutants resulted in some extent of reactivation: R181E (34%), R181D (25%), R214E (50%), R214D (46%), R532E (41%), E401R (20%), and R214D_D404R (27%) (Fig. 6, e and f). However, compared with these mutants, R180D_D404R and R532E_E401R showed an outstanding reactivation yield of 75 and 68%, respectively, and these approached the WT level, 80%.
Figure 6.
Effects of charge-inversion of potentially interacting pairs of amino acid residues. a and b, ATPase activities of TClpB mutants having charge-inversion mutations, in the absence (gray bar) or presence (open bar) of 0.1 mg/ml κ-casein, are shown. c and d, disaggregation activities of TClpB mutants were measured using α-glucosidase as a substrate. e and f, chaperone activities of TClpB mutants for the reactivation of soluble, denatured proteins were measured using α-glucosidase as a substrate. a, c, and e, activities of TClpB mutants having a single charge-inversion mutation. b, d, and f, activities of TClpB mutants having combined charge-inversion mutations. a–f, experimental procedures were the same as Fig. 2, a–c, and error bars represent standard deviations of three or more independent experiments.
Discussion
Here, we tested the effects of alanine substitution of conserved charged residues in the MD motif-1 of TClpB. Some Ala mutants, especially E401A, D404A, and E423A, lost their disaggregation activities, whereas their ATPase activities were maintained. Among them, Glu-423 (Glu-432 of EClpB) was previously identified as a functionally-important residue and thought to be responsible for destabilizing the repressed state of ClpB (18, 28). In the recent cryo-EM structure of Hsp104 from Saccharomyces cerevisiae (ScHsp104), Glu-401 (Glu-412 of ScHsp104) was found to interact with Arg-180 (Arg-194 of ScHsp104), and the substitution of this residue to Lys caused a loss of chaperone activity (15). The functional importance of Asp-404 has not been reported.
The cross-linked products of E401C-BPM and D404C-BPM behaved as multimers of TClpB, indicating that these residues interact with adjacent subunits. The multimeric cross-linking efficiencies of E401C-BPM and D404C-BPM were higher in the absence and presence of nucleotides, respectively. In addition, the nucleotide dependences observed in both of the mutants were weakened when nucleotide binding to AAA1 was inhibited. Moreover, LC-MS/MS analysis revealed that both E401C-BPM and D404C-BPM were cross-linked to the loop from Gly-218 to Lys-225 in AAA1. Some residues in this loop were located 5–10 Å from Glu-401 and Asp-404 in the TClpB hexameric models constructed based on the cryo-EM structures of EClpB (16). These results suggested that Glu-401 and Asp-404 are located near the loop when AAA1 is in the vacant or nucleotide-bound state, respectively. Nucleotide binding to AAA1 would cause sliding and/or rotational motions of the MD that could change the relative positions of these two residues at the surface of the neighboring AAA1.
In addition to the loop, the B3-helix from Asp-176 to Leu-187 was also found to be located ∼5–10 Å away from Glu-401 and Asp-404. We tested the effects of alanine substitution of conserved charged residues in these loop and helix, and we found that Arg-180, Arg-181, Arg-214, and Asp-219 were essential residues for the disaggregation activity of TClpB. Similar to E401A and D404A, the Ala mutants of these four residues lost their disaggregation activities without losing ATPase activity. Glu-401, Asp-404, and these four residues might share a common role in disaggregation activity. The Ala substitutions of Asp-176 and Glu-177 caused the stimulation of ATPase activity and improvement of disaggregation activity, indicating that these two residues contributed to other repressive roles. Although in the cryo-EM structures of ScHsp104 and EClpB, interaction between Asp-176 (Glu-190 and Asp-184 of ScHsp104 and EClpB, respectively) and Arg-408 (Arg-419 and Arg-417 of ScHsp104 and EClpB, respectively) was observed (14, 16), the effects of alanine substitution of these residues were opposite.
By charge exchange experiments, we tested whether functional direct interactions between the charged residues of motif-1 and neighboring AAA1 exist. The R180D_D404R double mutant showed comparable disaggregation activity compared with the WT, whereas the individual single mutants and the double mutants with other combinations lost almost all disaggregation activity. This result directly indicated that inter-subunit electrostatic interaction between Arg-180 and Asp-404 was indispensable for disaggregation activity. In the cryo-EM structure of ScHsp104, certain electrostatic interactions between charged residues in AAA1 and the MD motif-1 of neighboring subunits were shown as follows: Glu-190 and Arg-419, Arg-194 and Glu-412, Arg-353 and Glu-427, Arg-366 and Asp-434 (14, 15). However, the combination of Arg-180 and Glu-404 of TClpB, corresponding to Arg-194 and Asp-415 of ScHsp104, was not found in the list. This discrepancy might be due to differences between species.
Although any additional charge-changing mutations on the loop and the helix could not restore the disaggregation activity of E401R, the E401R_R532E double mutant showed substantial disaggregation activity. Arg-532 is located at the C-terminal region of AAA1 and was close to Glu-401 of the same subunit when the MD was in the tilted conformation. In addition, the 519LEVTEEDIAEIVSRWTGIPVSK540 peptide, including Arg-532, was found as a cross-linking target for E401C-BPM but not for D404C-BPM. These results indicated that Glu-401 and Arg-532 in the same subunit constitute a functionally-important electrostatic interacting pair. It should be noted that the efficiency of inter-subunit cross-linking of E401C-BPM was relatively low in the presence of nucleotides. The cross-link between E401C-BPM and Arg-532 in the same subunit should not cause inter-subunit cross-linkage, but rather it should compete with it. Thus, the Glu-401–Arg-532 interaction would be more likely to occur in the presence of nucleotides. Indeed, the peptides containing Arg-532 were detected as cross-linking targets of E401C-BPM more frequently in the presence of ATP than in the absence of nucleotide. This interaction might support the MD in forming a tilted conformation when the AAA1 bound ATP. The other important residue on the edge of motif-1, Glu-423 (Glu-432 of EClpB), was thought to destabilize the repressive horizontal conformation of the MD by preventing stable interaction between motif-1 and motif-2 of the adjacent subunits (28). The Glu-401–Arg-532 pair and Glu-423 would function complementarily.
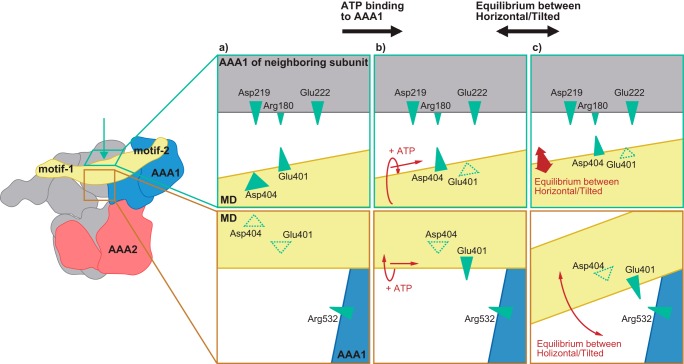
Fig. 7 shows a possible model that was constructed based on this study. In this model, MD conformation is controlled as follows. 1) ATP binding to the AAA1 causes sliding and/or rotational motion of the MD. 2) The structural change enables Glu-401 to interact with Arg-532. This interaction shifts the equilibrium of the MD conformation between the horizontal and the tilted to the tilted. Because of this shift, when ClpB was bound to DnaK, ClpB would maintain the activated form, in which the MD is tilted, sufficiently to perform disaggregation. 3) In the activated form, the Arg-180–Asp-404 electrostatic interaction contributes to inter-subunit signal transduction that is essential for the chaperone activities of ClpB. In contrast to the mutations involving Arg-180 and Asp-404, the effects of single mutations of Glu-401 and Arg-532 on the reactivation of soluble, denatured proteins were not so severe. The equilibrium shift of MD conformations caused by these residues might contribute, in particular, to continuous generation of the mechanical power required to disentangle large insoluble aggregations.
Figure 7.
Schematic model of ATP-dependent changes in interactions between the middle domain motif-1 and AAA1 of neighboring subunits. a, Glu-401, but not Asp-404, is located near the loop, including Asp-219 and Glu-222. b, ATP binding to AAA1 causes sliding and/or rotational motion of the middle domain in which the Asp-404 moves close to the loop instead of Glu-401. c, structural change enables Glu-401 to interact with Arg-532. This interaction shifts the equilibrium of the middle domain conformation between horizontal and tilted to tilted conformation. The shift would support DnaK-induced middle domain tilting (i.e. ClpB activation). In the activated form, the Arg-180–Asp-404 electrostatic interaction might contribute to inter-subunit signal transduction that is essential for the chaperone activities of ClpB.
Experimental procedures
Plasmids
The recombinant plasmid pMCB1 containing the ClpB gene from Thermus thermophilus was used as a mutagenesis template (5). Site-directed mutagenesis was performed by the overlap–extension PCR method using Ex Taq DNA polymerase (31, 32). Mutations were confirmed by DNA sequence analysis.
Proteins
Rabbit pyruvate kinase and chicken heart lactate dehydrogenase were purchased from Oriental Yeast (Tokyo, Japan). α-Glucosidase from B. stearothermophilus was purchased from Merck (Darmstadt, Germany). TClpB and its mutants were expressed in E. coli BL21 (DE3) and purified as described previously (33), except that mutants with a cysteine residue were purified in the presence of 1 mm DTT. Purified TClpB mutants bearing a cysteine residue were stored in a buffer containing 1 mm tris(2-carboxyethyl)phosphine hydrochloride (TCEP), at −80 °C. TDnaK, TDnaJ, and TGrpE were expressed in E. coli BL21 (DE3) carrying pMDK6 (TDnaK), pMDJ10 (TDnaJ), and pMGE3 (TGrpE), respectively, and purified as described previously (29, 34–36). Concentrations of substrate proteins were expressed as monomers, and those of chaperones were expressed as the monomers for TDnaK and TDnaJ, dimer for TGrpE, and hexamers for TClpB, and its mutants.
Measurement of ATPase activity
The ATPase activities of the WT and mutant TClpB were measured at 55 °C using an ATP-regenerating system containing MOPS-NaOH (pH 7.5), 150 mm KCl, 5 mm MgCl2, 2 mm NADH, 50 μg/ml pyruvate kinase, 50 μg/ml lactate dehydrogenase, 2.5 mm phosphoenolpyruvate, and 3 mm ATP. The reaction was initiated by the addition of 0.05 μm WT or mutant TClpB hexamers. Changes in absorbance at 340 nm were monitored using a V-650 spectrophotometer (Jasco, Tokyo, Japan).
Reactivation of aggregated α-glucosidase
α-Glucosidase (0.2 μm monomer) in a mixture containing 50 mm MOPS-NaOH (pH 7.5), 150 mm KCl, 5 mm MgCl2, 3 mm ATP, and 5 mm TCEP were heat-aggregated by incubation at 73 °C for 10 min. Subsequently, chaperones (final concentration of 0.6 μm TDnaK, 0.2 μm TDnaJ, 0.1 μm TGrpE, and 0.05 μm WT or mutant TClpB hexamers) were added to the reaction mixture. After incubation at 55 °C for 90 min, the enzymatic activities were measured as described previously (10). Disaggregation activities were expressed as percentages of the recovered enzymatic activities compared with that before heat treatment.
Reactivation of soluble, denatured proteins
To measure activities for reactivation of soluble, denatured proteins, essentially the same procedure as the disaggregation activity measurements was performed. TDnaK, TDnaJ, and TGrpE were added to the reaction mixtures, prior to the heat treatment. By heat treatment, the substrate proteins were denatured but not aggregated.
Labeling of TClpB mutant with BPM
TClpB mutants (1–10 μm hexamer) were mixed with more than 60 molar eq of benzophenone-4-maleimide (BPM) (Merck) in 50 mm potassium phosphate buffer (pH 7.5) and incubated at 55 °C for 2 h. This buffer was replaced by the reaction buffer (50 mm MOPS-NaOH (pH 7.5), 20 mm KCl, 5 mm MgCl2) by using a disposable gel-filtration column, NAP-10 (GE Healthcare, Little Chalfont, UK).
Photocross-linking of TClpB mutants
The BPM-labeled TClpB mutants (0.86 μm) dissolved in buffer (50 mm MOPS-NaOH (pH 7.5), 20 mm KCl, 5 mm MgCl2) were irradiated with a 365 nm UV lamp (8 watts) for 90 min at room temperature in the absence or presence of 3 mm nucleotides using an LM-20E trans-illuminator (Analytik Jena, Jena, Germany). After photocross-linking, samples were concentrated by TCA precipitation and then analyzed by SDS-PAGE on a 5.5% polyacrylamide gel and stained with Coomassie Brilliant Blue. The gel images were acquired by using ImageQuant LAS 4010 system (GE Healthcare). The intensities of bands corresponding to 1–5-and over 6-mers of TClpB were quantified by using ImageQuant TL version 8.1 (GE Healthcare).
LC-MS/MS analysis
The cross-linked samples were diluted ≥5-fold with PTS solution (12 mm sodium deoxycholate, 12 mm sodium N-lauroyl sarcosinate, and 100 mm Tris-HCl (pH 9.0)) (37), reduced with 10 mm DTT for 30 min, and alkylated with 55 mm iodoacetamide for 30 min at room temperature. The solution was diluted 5-fold with 50 mm NH4HCO3, and proteins were digested by 0.25 μg of Lys-C (FUJIFILM Wako Pure Chemical, Osaka, Japan) for 3 h at room temperature. The proteins were further digested by 0.5 μg of Trypsin Gold (Promega, Fitchburg, WI) at 37 °C overnight. After digestion, the solution was acidified with 0.5% TFA (final concentration) and added to an equal volume of ethyl acetate. After vigorous mixing and centrifugation at 15,700 × g for 2 min, the aqueous phase was collected and dried using a centrifugal evaporator. The peptides were dissolved in 2% acetonitrile and 0.1% TFA solution and desalted with C18 Stage Tip (Nikkyo Technos, Tokyo, Japan).
Nano-LC-MS/MS analysis was performed on an Easy-nLC 1000 nanoLC system and a Q-Exactive mass spectrometer (ThermoFisher Scientific, Waltham, MA). The separation column used for the nano-LC was a 12.5 cm × 75 μm capillary column packed with 3-μm C18-silica particles (Nikkyo Technos). The separation was conducted using a 10–50% linear acetonitrile gradient at 40 min in the presence of 0.1% formic acid. The LC-MS/MS data were acquired in data-dependent acquisition mode controlled by Xcalibur 4.0 (ThermoFisher Scientific). The settings of data-dependent acquisition were as follows: the resolution was 70,000 for a full MS scan and 17,500 for MS2 scan; the AGC target was 3.0E6 for a full MS scan and 5.0E5 for MS2 scan; the maximum IT was 60 ms for both a full MS scan and MS2 scan; the scan range was 310–1,500 m/z for a full MS scan and 200–2,000 m/z for MS2 scan; and the top 10 signals were selected for MS2 scan per one full MS scan. For the detection of cross-linked peptides, the raw data were analyzed by pLink 2.0 software (38). The pLink 2.0 settings were as follows: linker mass was 295.085 Da (C17H11NO3·H2O), maximum missed cleavages was 3, peptide length was 5–60, mass tolerance was ± 5 ppm, separate false discovery rate was ≤1%.
Construction of structural models of TClpB hexamer
The structural models of TClpB hexamers were constructed by homology modeling by using Discovery Studio 4.5 (Dassault Systemes, Velizy-Villacoublay, France). The recently solved cryo-EM structures of EClpB hexamers (PDB codes 5ofo and 5og1) were used as templates (16).
Author contributions
S. S., K. W., T. N., E. U., and Y.-h. W. data curation; S. S., K. W., T. N., E. U., and Y.-h. W. formal analysis; S. S., K. W., T. N., E. U., and Y.-h. W. validation; S. S., K. W., K. H., T. N., and E. U. investigation; S. S., K. W., T. N., and Y.-h. W. visualization; S. S., K. W., T. N., and Y.-h. W. writing-original draft; T. N., H. T., and Y.-h. W. conceptualization; T. N. and Y.-h. W. methodology; T. N., H. T., and Y.-h. W. writing-review and editing; Y.-h. W. resources; Y.-h. W. supervision; Y.-h. W. funding acquisition; Y.-h. W. project administration.
This work was supported by JSPS KAKENHI Grants 24241048 and 26440085. The authors declare that they have no conflicts of interest with the contents of this article.
- ND
- N-terminal domain
- MD
- middle domain
- BPM
- benzophenone-4-maleimide
- NBD
- nucleotide-binding domain
- SBD
- substrate-binding domain
- PDB
- Protein Data Bank
- TCEP
- tris(2-carboxyethyl)phosphine.
References
- 1. Sanchez Y., and Lindquist S. L. (1990) HSP104 required for induced thermotolerance. Science 248, 1112–1115 10.1126/science.2188365 [DOI] [PubMed] [Google Scholar]
- 2. Thomas J. G., and Baneyx F. (1998) Roles of the Escherichia coli small heat shock proteins IbpA and IbpB in thermal stress management: comparison with ClpA, ClpB, and HtpG in vivo. J. Bacteriol. 180, 5165–5172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weibezahn J., Tessarz P., Schlieker C., Zahn R., Maglica Z., Lee S., Zentgraf H., Weber-Ban E. U., Dougan D. A., Tsai F. T., Mogk A., and Bukau B. (2004) Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 119, 653–665 10.1016/j.cell.2004.11.027 [DOI] [PubMed] [Google Scholar]
- 4. Glover J. R., and Lindquist S. (1998) Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94, 73–82 10.1016/S0092-8674(00)81223-4 [DOI] [PubMed] [Google Scholar]
- 5. Motohashi K., Watanabe Y., Yohda M., and Yoshida M. (1999) Heat-inactivated proteins are rescued by the DnaK.J-GrpE set and ClpB chaperones. Proc. Natl. Acad. Sci. U.S.A. 96, 7184–7189 10.1073/pnas.96.13.7184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goloubinoff P., Mogk A., Zvi A. P., Tomoyasu T., and Bukau B. (1999) Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc. Natl. Acad. Sci. U.S.A. 96, 13732–13737 10.1073/pnas.96.24.13732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zolkiewski M. (1999) ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. A novel multi-chaperone system from Escherichia coli. J. Biol. Chem. 274, 28083–28086 10.1074/jbc.274.40.28083 [DOI] [PubMed] [Google Scholar]
- 8. Lee S., Sowa M. E., Watanabe Y. H., Sigler P. B., Chiu W., Yoshida M., and Tsai F. T. (2003) The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell 115, 229–240 10.1016/S0092-8674(03)00807-9 [DOI] [PubMed] [Google Scholar]
- 9. Barnett M. E., Nagy M., Kedzierska S., and Zolkiewski M. (2005) The amino-terminal domain of ClpB supports binding to strongly aggregated proteins. J. Biol. Chem. 280, 34940–34945 10.1074/jbc.M505653200 [DOI] [PubMed] [Google Scholar]
- 10. Mizuno S., Nakazaki Y., Yoshida M., and Watanabe Y. H. (2012) Orientation of the amino-terminal domain of ClpB affects the disaggregation of the protein. FEBS J 279, 1474–1484 10.1111/j.1742-4658.2012.08540.x [DOI] [PubMed] [Google Scholar]
- 11. Fernández-Higuero J. Á., Acebrón S. P., Taneva S. G., Del Castillo U., Moro F., and Muga A. (2011) Allosteric communication between the nucleotide binding domains of caseinolytic peptidase B. J. Biol. Chem. 286, 25547–25555 10.1074/jbc.M111.231365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Franzmann T. M., Czekalla A., and Walter S. G. (2011) Regulatory circuits of the AAA+ disaggregase Hsp104. J. Biol. Chem. 286, 17992–18001 10.1074/jbc.M110.216176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yamasaki T., Oohata Y., Nakamura T., and Watanabe Y. H. (2015) Analysis of the cooperative ATPase cycle of the AAA+ chaperone ClpB from Thermus thermophilus by using ordered heterohexamers with an alternating subunit arrangement. J. Biol. Chem. 290, 9789–9800 10.1074/jbc.M114.617696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yokom A. L., Gates S. N., Jackrel M. E., Mack K. L., Su M., Shorter J., and Southworth D. R. (2016) Spiral architecture of the Hsp104 disaggregase reveals the basis for polypeptide translocation. Nat. Struct. Mol. Biol. 23, 830–837 10.1038/nsmb.3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gates S. N., Yokom A. L., Lin J., Jackrel M. E., Rizo A. N., Kendsersky N. M., Buell C. E., Sweeny E. A., Mack K. L., Chuang E., Torrente M. P., Su M., Shorter J., and Southworth D. R. (2017) Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science 357, 273–279 10.1126/science.aan1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deville C., Carroni M., Franke K. B., Topf M., Bukau B., Mogk A., and Saibil H. R. (2017) Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase. Sci. Adv. 3, e1701726 10.1126/sciadv.1701726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Uchihashi T., Watanabe Y. H., Nakazaki Y., Yamasaki T., Watanabe H., Maruno T., Ishii K., Uchiyama S., Song C., Murata K., Iino R., and Ando T. (2018) Dynamic structural states of ClpB involved in its disaggregation function. Nat. Commun. 9, 2147 10.1038/s41467-018-04587-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oguchi Y., Kummer E., Seyffer F., Berynskyy M., Anstett B., Zahn R., Wade R. C., Mogk A., and Bukau B. (2012) A tightly regulated molecular toggle controls AAA+ disaggregase. Nat. Struct. Mol. Biol. 19, 1338–1346 10.1038/nsmb.2441 [DOI] [PubMed] [Google Scholar]
- 19. Lipińska N., Zíe¸tkiewicz S., Sobczak A., Jurczyk A., Potocki W., Morawiec E., Wawrzycka A., Gumowski K., Ślusarz M., Rodziewicz-Motowid;̸o S., Chruściel E., and Liberek K. (2013) Disruption of ionic interactions between the nucleotide binding domain 1 (NBD1) and middle (M) domain in Hsp100 disaggregase unleashes toxic hyperactivity and partial independence from Hsp70. J. Biol. Chem. 288, 2857–2869 10.1074/jbc.M112.387589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Acebrón S. P., Martín I., del Castillo U., Moro F., and Muga A. (2009) DnaK-mediated association of ClpB to protein aggregates. A bichaperone network at the aggregate surface. FEBS Lett. 583, 2991–2996 10.1016/j.febslet.2009.08.020 [DOI] [PubMed] [Google Scholar]
- 21. Hayashi S., Nakazaki Y., Kagii K., Imamura H., and Watanabe Y. H. (2017) Fusion protein analysis reveals the precise regulation between Hsp70 and Hsp100 during protein disaggregation. Sci. Rep. 7, 8648 10.1038/s41598-017-08917-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Flaherty K. M., DeLuca-Flaherty C., and McKay D. B. (1990) Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature 346, 623–628 10.1038/346623a0 [DOI] [PubMed] [Google Scholar]
- 23. Zhu X., Zhao X., Burkholder W. F., Gragerov A., Ogata C. M., Gottesman M. E., and Hendrickson W. A. (1996) Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272, 1606–1614 10.1126/science.272.5268.1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kityk R., Kopp J., Sinning I., and Mayer M. P. (2012) Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol. Cell 48, 863–874 10.1016/j.molcel.2012.09.023 [DOI] [PubMed] [Google Scholar]
- 25. Zhuravleva A., Clerico E. M., and Gierasch L. M. (2012) An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell 151, 1296–1307 10.1016/j.cell.2012.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rosenzweig R., Moradi S., Zarrine-Afsar A., Glover J. R., and Kay L. E. (2013) Unraveling the mechanism of protein disaggregation through a ClpB–DnaK interaction. Science 339, 1080–1083 10.1126/science.1233066 [DOI] [PubMed] [Google Scholar]
- 27. Haslberger T., Weibezahn J., Zahn R., Lee S., Tsai F. T., Bukau B., and Mogk A. (2007) M domains couple the ClpB threading motor with the DnaK chaperone activity. Mol. Cell 25, 247–260 10.1016/j.molcel.2006.11.008 [DOI] [PubMed] [Google Scholar]
- 28. Carroni M., Kummer E., Oguchi Y., Wendler P., Clare D. K., Sinning I., Kopp J., Mogk A., Bukau B., and Saibil H. R. (2014) Head-to-tail interactions of the coiled-coil domains regulate ClpB activity and cooperation with Hsp70 in protein disaggregation. Elife 3, e02481 10.7554/eLife.02481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Watanabe Y. H., and Yoshida M. (2004) Trigonal DnaK–DnaJ complex versus free DnaK and DnaJ: heat stress converts the former to the latter, and only the latter can do disaggregation in cooperation with ClpB. J. Biol. Chem. 279, 15723–15727 10.1074/jbc.M308782200 [DOI] [PubMed] [Google Scholar]
- 30. Watanabe Y. H., Motohashi K., and Yoshida M. (2002) Roles of the two ATP binding sites of ClpB from Thermus thermophilus. J. Biol. Chem. 277, 5804–5809 10.1074/jbc.M109349200 [DOI] [PubMed] [Google Scholar]
- 31. Higuchi R., Krummel B., and Saiki R. K. (1988) A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16, 7351–7367 10.1093/nar/16.15.7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., and Pease L. R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 10.1016/0378-1119(89)90358-2 [DOI] [PubMed] [Google Scholar]
- 33. Watanabe Y. H., Takano M., and Yoshida M. (2005) ATP binding to nucleotide binding domain (NBD)1 of the ClpB chaperone induces motion of the long coiled-coil, stabilizes the hexamer, and activates NBD2. J. Biol. Chem. 280, 24562–24567 10.1074/jbc.M414623200 [DOI] [PubMed] [Google Scholar]
- 34. Motohashi K., Yohda M., Endo I., and Yoshida M. (1996) A novel factor required for the assembly of the DnaK and DnaJ chaperones of Thermus thermophilus. J. Biol. Chem. 271, 17343–17348 10.1074/jbc.271.29.17343 [DOI] [PubMed] [Google Scholar]
- 35. Motohashi K., Yohda M., Odaka M., and Yoshida M. (1997) K+ is an indispensable cofactor for GrpE stimulation of ATPase activity of DnaK x DnaJ complex from Thermus thermophilus. FEBS Lett. 412, 633–636 10.1016/S0014-5793(97)00847-8 [DOI] [PubMed] [Google Scholar]
- 36. Mizutani T., Nemoto S., Yoshida M., and Watanabe Y. H. (2009) Temperature-dependent regulation of Thermus thermophilus DnaK/DnaJ chaperones by DafA protein. Genes Cells 14, 1405–1413 10.1111/j.1365-2443.2009.01357.x [DOI] [PubMed] [Google Scholar]
- 37. Masuda T., Saito N., Tomita M., and Ishihama Y. (2009) Unbiased quantitation of Escherichia coli membrane proteome using phase transfer surfactants. Mol. Cell. Proteomics 8, 2770–2777 10.1074/mcp.M900240-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang B., Wu Y. J., Zhu M., Fan S. B., Lin J., Zhang K., Li S., Chi H., Li Y. X., Chen H. F., Luo S. K., Ding Y. H., Wang L. H., Hao Z., Xiu L. Y., Chen S., Ye K., He S. M., and Dong M. Q. (2012) Identification of cross-linked peptides from complex samples. Nat. Methods 9, 904–906 10.1038/nmeth.2099 [DOI] [PubMed] [Google Scholar]