Abstract
RidA is a conserved and broadly distributed protein that has enamine deaminase activity. In a variety of organisms tested thus far, lack of RidA results in the accumulation of the reactive metabolite 2-aminoacrylate (2AA), an obligate intermediate in the catalytic mechanism of several pyridoxal 5′-phosphate (PLP)-dependent enzymes. This study reports the characterization of variants of the biosynthetic serine/threonine dehydratase (EC 4.3.1.19; IlvA), which is a significant generator of 2AA in the bacteria Salmonella enterica, Escherichia coli, and Pseudomonas aeruginosa and the yeast Saccharomyces cerevisiae. Two previously identified mutations, ilvA3210 and ilvA3211, suppressed the phenotypic growth consequences of 2AA accumulation in S. enterica. Characterization of the respective protein variants suggested that they affect 2AA metabolism in vivo by two different catalytic mechanisms, both leading to an overall reduction in serine dehydratase activity. To emphasize the physiological relevance of the in vitro enzyme characterization, we sought to explain in vivo phenotypes using these data. A simple mathematical model describing the impact these catalytic deficiencies had on 2AA production was generally supported by our data. However, caveats arose when kinetic parameters, determined in vitro, were used to predict formation of the isoleucine precursor 2-ketobutyrate and model in vivo (growth) behaviors. Altogether, our data support the need for a holistic approach, including in vivo and in vitro analyses, to generate data used in understanding and modeling metabolism.
Keywords: pyridoxal phosphate, reactive nitrogen species (RNS), bacterial metabolism, Salmonella enterica, stress, 2-aminoacrylate, mathematical modeling, metabolic network, RidA, serine/threonine dehydratase
Introduction
Microbial metabolic networks are robust systems able to absorb perturbations caused by internal and external stress (1–4). Metabolic robustness provides an evolutionary advantage to an organism exposed to sudden and frequent environmental shifts, but the integration of pathways can complicate the ability to predict metabolic potential or model flux distribution based solely upon genomic cataloging (5–7). The extensive integration of metabolic pathways within the network can lead to unanticipated solutions to metabolic perturbations that are often uncovered by genetic analyses (8–10). This point is exemplified by the observable differences between Salmonella enterica and Escherichia coli with respect to 2-aminoacrylate (2AA)2 stress and thiamine biosynthesis, despite the conservation of all the component enzymes in each (8, 11). The observation that organisms containing the same components may alter the configuration of those parts indicates the need for genetic and biochemical approaches to supplement genomic cataloging in efforts to model cellular metabolism (5, 12).
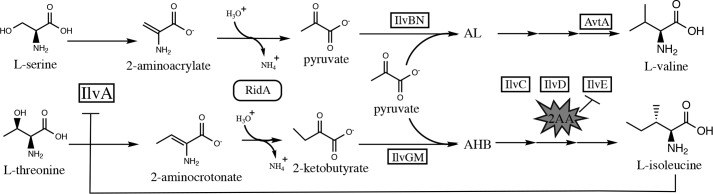
A number of pyridoxal 5′-phosphate (PLP)-dependent β-eliminase enzymes produce and release the reactive molecule 2AA. 2AA then inactivates target PLP enzymes through covalent modification of active-site bound PLP (13–16). RidA subfamily members, belonging to the broader Rid (YjgF/YER057c/UK114) protein family, are enamine/imine deaminases that reduce 2AA accumulation (13, 14, 17, 18). Strains of S. enterica lacking RidA do not grow on minimal pyruvate medium or minimal glucose medium containing serine. These growth defects are due to 2AA generated by the biosynthetic serine/threonine dehydratase (IlvA; EC 4.3.1.19) from serine. S. enterica ridA mutants show a mild growth defect in minimal glucose medium, attributable to 2AA generated from endogenously generated serine. Inclusion of serine increases endogenous serine pools and results in greater 2AA production. A ridA mutant fails to grow because of the increased 2AA-dependent damage to multiple enzymes (19–22). This global stress is best prevented by mechanisms that reduce 2AA production (e.g. allosteric inhibition of IlvA by isoleucine) (14, 23–27). Lack of growth on minimal pyruvate medium has been attributed to 2AA-dependent damage of a single enzyme, the branched-chain amino acid aminotransferase (IlvE; EC 2.6.1.42) (21). WT S. enterica, grown on pyruvate, experiences a flux bias away from isoleucine biosynthesis and toward valine, although sufficient isoleucine is still produced for growth (28). However, ridA mutants undergo 2AA-dependent damage of IlvE, the penultimate enzyme in the isoleucine synthesis pathway, and experience an isoleucine limitation (14, 21). An overview of the role of RidA in the context of branched-chain amino acid biosynthesis is depicted in Fig. 1.
Figure 1.
2-Aminoacrylate in the context of branched-chain amino acid metabolism. The pathways for branched-chain amino acid biosynthesis and 2-aminoacrylate hydrolysis are shown schematically. Enzymes are indicated next to the step they catalyze. IlvA catalyzes the first committed step in isoleucine biosynthesis using threonine as a substrate. IlvA can also dehydrate serine to form 2-aminoacrylate. RidA activity increases the rate of hydrolysis for both enamine products of IlvA. Elimination of RidA causes accumulation of 2-aminoacrylate, which can damage IlvE. (2AC would also accumulate but is not known to have deleterious effects.) Importantly, due to AvtA, absence of active IlvE prevents isoleucine but not valine biosynthesis. IlvA is sensitive to feedback inhibition by isoleucine, as indicated in the schematic. Intermediates in isoleucine and valine biosynthesis compete for enzymes IlvC, IlvD, and IlvE. Abbreviations: AL, 2-acetolactate; AHB, 2-aceto-3-hydroxybutyrate.
Suppressor mutations that restored growth to a ridA mutant on minimal pyruvate mapped to ilvA and encoded variant IlvA proteins (26). The suppressor alleles of ilvA, ilvA3210 and ilvA3211, restored WT levels of IlvE activity and allowed isoleucine biosynthesis sufficient for growth. Isolation and initial analysis of these suppressors was prior to the elucidation of RidA function and the role of 2AA in causing the phenotypes of a ridA mutant. Notably, the suppressor mutations were unable to restore growth of a ridA mutant on medium containing serine, indicating the IlvA variants still produce 2AA when serine levels are elevated.
Based on the current understanding of the RidA paradigm, two scenarios could explain how the IlvA variants restored growth of a ridA mutant on pyruvate medium but not in the presence of serine. First, the IlvA variants could have decreased affinity for the 2AA precursor, l-serine. Second, the catalytic turnover of the variants could be reduced, maintaining sufficient 2-aminocrotonate production for isoleucine biosynthesis but minimizing 2AA formation. In either scenario, 2AA production by the variants would be reduced but not eliminated. The goal of this work was to biochemically characterize the IlvA variants and better understand the synthesis and impact of 2AA formation by IlvA in the metabolic context of branched-chain amino acid synthesis. We further addressed the feasibility of using kinetic and metabolite concentration data to model the factors controlling IlvA-dependent 2AA metabolism and its impact in vivo.
Results
ilvA3210 and ilvA3211 alleles suppress one 2AA-dependent growth defect
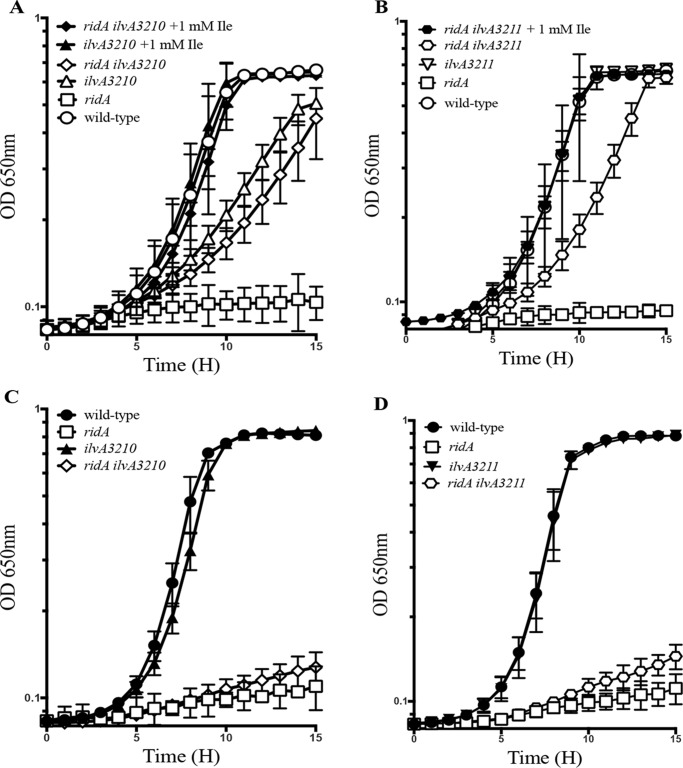
Growth of strains with WT ilvA, ilvA3210, and ilvA3211 was quantified, and the data are shown in Fig. 2. The ridA mutant failed to grow with pyruvate as a sole carbon source (Fig. 2, A and B) or when minimal glucose medium included serine (Fig. 2, C and D). As reported previously, the ilvA3210 and ilvA3211 alleles significantly improved growth of the ridA mutant on pyruvate but not in minimal glucose with serine (21). On pyruvate medium, (i) growth of the ridA ilvA3210 and ridA ilvA3211 mutants was not fully restored to WT levels, and (ii) the ilvA3210 allele caused a growth defect in an otherwise WT strain. Both of these phenotypes were corrected by the addition of isoleucine (Fig. 2). In glucose medium containing 5 mm serine, the ridA ilvA3210 and ridA ilvA3211 strains grew only slightly better than a ridA mutant (Fig. 2, C and D). In total, these data were consistent with the following hypotheses: (i) IlvAA142T and IlvAG191S have reduced dehydratase activity; (ii) the reduced serine dehydratase activity of the IlvA variants decreases 2AA available to damage cellular targets; and (iii) ilvA3210 decreases flux enough to cause a starvation for isoleucine on pyruvate, even in the absence of a ridA mutation.
Figure 2.
Mutations in ilvA restore growth to ridA mutants under some conditions. Strains were grown at 37 °C in minimal medium containing 50 mm pyruvate as the sole carbon source (A and B) or with 11 mm glucose as the sole carbon source and supplemented with 5 mm l-serine (C and D). Growth is shown for WT (DM9404), ridA (DM3480), ilvA3210 (DM15899), ridA ilvA3210 (DM15898), ilvA3211 (DM15936), and ridA ilvA3211 (DM15937) as indicted in the legends. Data are the mean, and error bars represent the 95% confidence interval of three biological replicates.
ilvA alleles restore branched-chain amino acid aminotransferase activity
Threonine dehydratase (IlvA) and branched-chain amino acid aminotransferase (IlvE) activity was assayed in the crude extracts of strains containing four different ilvA alleles (Table 1, white shading) and stains carrying the same ilvA alleles in addition to a ridA mutation (Table 1, gray shading). Analysis of the strains with a WT ridA provides insight into the impact of the IlvA variants on the biosynthetic pathway. Each of the ilvA alleles caused a decrease in total threonine dehydratase activity, with ilvA3210 (IlvAA142T, DM15898) and ilvA3211 (IlvAG191S, DM15936) causing an ∼10-fold decrease. Despite the decrease in threonine dehydratase activity, the strains had WT growth level on minimal glucose medium, emphasizing the excess capacity the cells have to synthesize isoleucine. IlvA and IlvE are encoded in an operon (ilvGMEDA), and the ratio of their activity suggests the status of isoleucine synthesis and/or starvation in the cell (29). IlvE activity was significantly increased in the ilvA3210 strain, suggesting that the decreased activity of IlvAA142T caused a limitation of isoleucine, which resulted in induction of the operon. Consistent with this model, previous work showed by Western blotting that more IlvE protein accumulates in the ilvA3210 strain than in the WT strain (21). These data correlate with the growth defect of DM15898 (ilvA3210) on pyruvate, where flux through the isoleucine synthesis pathway is less than on glucose medium. Although threonine dehydratase activity measured in crude lysates was slightly higher for IlvAA142T than IlvAG191S, assuming a similar induction across the ilv operon, the data suggest the specific activity of IlvAA142T in vivo is no greater than that of IlvAG191S. In total, the results show that the capacity of the biosynthetic pathway exceeds what is needed to satisfy growth and supports a hypothesis that allosteric regulation is a major driver controlling metabolic efficiency.
Table 1.
IlvA activity and 2AA accumulation differs among ilvA mutantsa
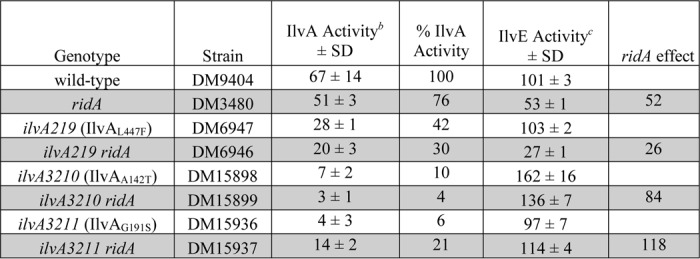
a Indicated strains were grown to stationary phase in minimal glucose medium containing 0.67 mm glycine.
b Threonine deaminase (IlvA) acitivty was measured from crude lysate extracts. Data represent the mean and S.D. from three biological replicates by measuring 2KB formation (nmol 2KB min−1 mg−1). % IlvA activity represents the ratio of IlvA activity for each strain relative to WT (DM9404).
c Branched-chain amino acid aminotransferase (IlvE) activity was measured from crude lysate extracts. Data represent the mean and S.D. from three biological replicates by measuring 2-keto-3-methylvalerate (KMV) formation (nmol KMV min−1 mg−1). ridA effect represents the ratio of IlvE activity in strains without RidA compared with their RidA+ parent and is a measure of relative 2AA accumulation.
The impact of eliminating RidA in the strains described above is shown by the gray shading in Table 1. IlvE is irreversibly damaged by 2AA and can be used as a proxy for the amount of this metabolite that accumulates in vivo (14). A ridA mutant (DM3480) accumulates 2AA, which results in the ∼50% reduction in IlvE activity, compared with WT (DM9404) (Table 1). The S. enterica IlvAL447F variant, encoded by the ilvA219 allele, is insensitive to inhibition by isoleucine (30) and, despite having decreased threonine dehydratase activity, increases 2AA production in vivo (14, 27). This results in a severe decrease in IlvE activity (∼75%) in the ridA ilvA219 mutant (DM6946). In contrast, although the ridA mutant strains expressing the IlvAA142T or IlvAG191S variants had lower IlvA activity, IlvE activity was similar, or slightly higher, than the parental strains (Table 1). This latter result suggested that unlike the IlvAL447F variant, the two suppressor variants of IlvA generated less 2AA in vivo.
The data presented in Table 1 and Fig. 2 suggested a simple correlation between IlvE activity and growth on pyruvate. IlvE activity of ∼100 nmol of KMV min−1 mg−1 (measured on glucose medium) supports adequate isoleucine synthesis for growth in minimal pyruvate medium. By this scenario, ridA derivatives of WT or ilvA219 strains have lower IlvE activity, which constricts isoleucine synthesis and prevents growth. The restored activity of IlvE in the ridA ilvA3210 and ridA ilvA3211 mutants overcomes the growth defect. Notably, this simple model does not explain why ilvA3210, but not ilvA3211, resulted in an isoleucine requirement in minimal pyruvate media.
Characterization of purified IlvA variants
Because IlvA uses serine and threonine as competing substrates, it was possible that differential substrate specificities influenced the in vivo effects described above. Kinetic parameters were determined from purified preparations of the four IlvA protein variants, using either serine or threonine substrate (Fig. 3). The threonine saturation curves for all four proteins were fit using the equation by Michaelis-Menten. The saturation curves using serine substrate showed mild positive cooperativity, with Hill coefficients (h) greater than 1. Because the fit of the curves was close using the equation from Michaelis-Menten, the slight cooperative effects were not considered in determining the kinetic values of the proteins. This tactic simplified the comparisons between the results with serine and threonine substrates.
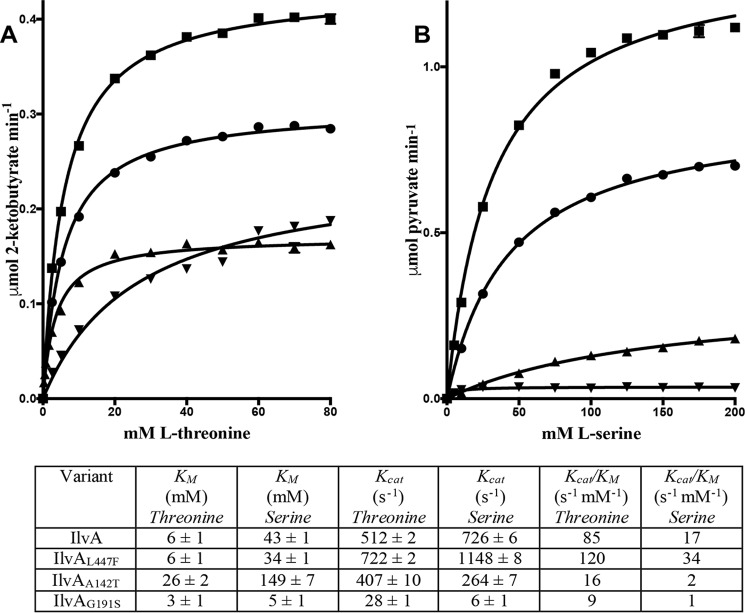
Figure 3.
Kinetic characterization of IlvA variants. A and B, saturation curves for IlvA (circles), IlvAL447F (squares), IlvAA142T (upward-facing triangles), and IlvAG191S (downward-facing triangles) measuring the initial rate of 2-ketobutyrate formation versus l-threonine concentration (A) or the initial rate of pyruvate formation versus l-serine concentration (B). Error bars represent the standard deviation of four replicates. The kinetic parameters used to plot the hyperbolic line describing the data (Km and kcat) are provided in the table containing the values and their respective standard deviations. The specificity constant (kcat/Km) was determined for each enzyme with each of the two substrates.
Considering the WT protein, the values obtained for the Michaelis-Menten constant (Km) and catalyst rate constant (kcat) with serine or threonine were consistent with values previously observed for S. enterica IlvA (31). The IlvAL447F variant had a lower Km value for serine and a higher kcat value with both threonine and serine substrates. Compared with WT enzyme, the IlvAA142T variant had significantly higher Km and lower kcat values for both substrates, whereas the IlvAG191S variant had significantly lower Km and kcat values for both substrates. The specificity constant (kcat/Km) of an enzyme can be used to assess preference for different substrates. The kcat/Km values showed that all four IlvA proteins were more effective at using threonine than serine (Fig. 3). However, the ratio of specificity constants (threonine kcat/Km versus serine kcat/Km) increases from 5 for IlvA to 8 and 9 for IlvAA142T and IlvAG191S, respectively. This suggests that IlvAA142T and IlvAG191S have a slightly stronger preference for threonine over serine than WT enzyme.
The relative in vitro stability of each enzyme variant was determined by measuring enzyme activity over 8 h in the presence of 200 mm serine. IlvAA142T was the least stable and had an 80% reduction in activity after 8 h (40 versus 184 nmol of pyruvate min−1). IlvAG191S was moderately stable with a 40% reduction in activity after 8 h (18 versus 30 nmol of pyruvate min−1). In contrast, WT IlvA (716 versus 777 nmol of pyruvate min−1) and IlvAL447F (1,043 versus 1,162 nmol pyruvate min−1) showed only a 10% loss in activity over the same time.
Modeling the competing synthesis of 2AA and 2AC in vivo
Use of threonine and serine by IlvA generates two significant metabolites, 2-aminocrotonate (2AC) and 2-aminoacrylate, respectively. 2AC, which is not known to be detrimental if it accumulates, is deaminated to 2-ketobutyrate, a precursor of isoleucine. In contrast, 2AA is detrimental if it accumulates before its conversion to pyruvate, as described above. Thus, the two products of IlvA have opposing impacts on the growth phenotypes, and therefore differences in how the IlvA variants impacted production of both 2AC and 2AA in vivo was significant. The kinetic information of each of the four IlvA enzymes (Fig. 3) was used to explore the relationship between formation of 2AA and 2AC in cells with different ilvA alleles. Reported concentrations of endogenous metabolites in E. coli were used to estimate the concentration of l-serine (67 μm) and l-threonine (180 μm) (32). The concentration of IlvA (20 μm) was based on the concentration of other PLP-dependent enzymes in E. coli (33). For simplicity, known cooperative and allosteric behaviors of IlvA were excluded from the calculations (34, 35). Equation 1 predicts product formation for a specified substrate (serine or threonine), accounting for competitive inhibition by the alternative substrate (36). VProd is the rate of product production (μm/s); kcat (Sub) is the turnover number for the substrate (s−1); [Et] is IlvA active-site concentration (μm); [Sub] is substrate concentration (μm); [CompSub] is competing substrate concentration (μm); Km, (Sub) is the Michaelis-Menten constant for the substrate (μm); and Km (CompSub) is the Michaelis-Menten constant for the competing substrate (μm).
| (Eq. 1) |
Using Equation 1, the amount of 2AC and 2AA generated during steady-state growth was estimated for each of the IlvA variants (Table 2). Consistent with the conclusion from the data in Fig. 3, Equation 1 predicts each IlvA enzyme generates more 2AC than 2AA. Furthermore, the variants that suppressed the ridA mutant phenotype on pyruvate produced dramatically less 2AA and 2AC than the WT protein. The growth of strains with these variants on minimal medium indicated that these variants produced sufficient 2AC to satisfy the isoleucine requirement of the strain. These data suggest that despite being simplified, Equation 1 does an adequate job of accounting for the effect of the IlvA variants in vivo and suggests the mechanism they use to suppress the ridA phenotype on pyruvate. The ilvA alleles that restored growth to a ridA mutant failed to restore growth when exogenous serine was added, indicating detrimental levels of 2AA still accumulated under these conditions. Equation 1 was used to calculate the impact of 5 mm serine on the products of IlvA. Assuming endogenous serine matched the external concentration, the data showed a significant increase in 2AA production in all strains (Table 2). The strains carrying ilvA3210 and ilvA3211 were calculated to have 2AA levels well above what accumulated in a WT strain without serine addition (85 and 29 μm/s for ilvA3210 and ilvA3211, respectively). Interestingly, the amount of 2AC was essentially unchanged in this condition.
Table 2.
Estimated endogenous product synthesis rates for IlvA variantsa
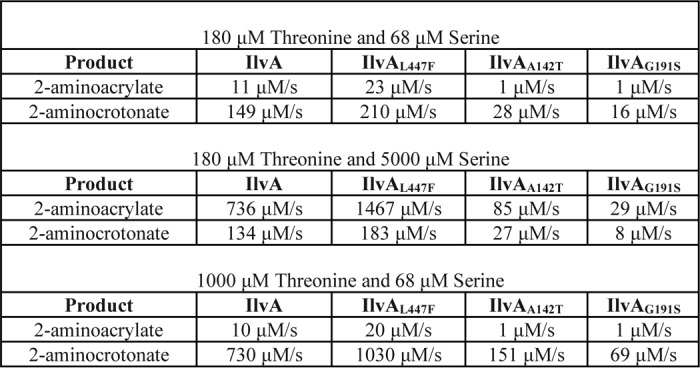
a Rates of 2-aminoacrylate and 2-aminocrotonate synthesis were estimated using the competing substrate concentrations shown and Equation 1. During calculation, IlvA active-site concentrations for all variants were provided as 10 μm (33). Appropriate Km and kcat values for each variant enzyme were taken from Fig 3.
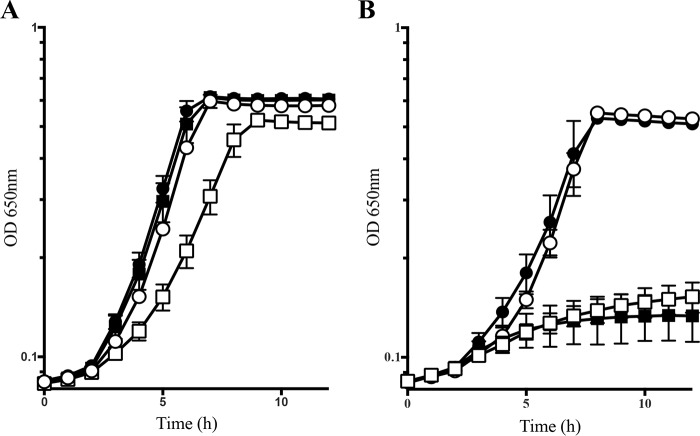
A hallmark of the RidA paradigm is that the effects are prevented by the addition of isoleucine, which inhibits IlvA and prevents formation of the toxic 2AA. Addition of threonine was similarly shown to reduce the formation of 2AA, a result that was previously suggested to be due to competitive inhibition of serine catalysis by IlvA. However, for threonine to cause a 10-fold decrease in 2AA production by competitive inhibition, 55 mm l-threonine would be required (Equation 1). In contrast, addition of 1 mm l-threonine eliminated the growth defect of a ridA mutant on minimal glucose medium (Fig. 4A) (26). Modeling of this condition with Equation 1 showed that 1 mm threonine did not affect the level of 2AA, but rather it significantly increased the production of 2AC. Based on this analysis, we hypothesized that the exogenous threonine increased isoleucine biosynthesis, which allosterically inhibited IlvA, decreasing 2AA production (Fig. 1). A ridA mutant expressing the isoleucine feedback-resistant variant has a growth defect when grown in minimal glucose medium due to the increased 2AA formed. In this strain, l-threonine did not restore growth on minimal medium, a result that is consistent with threonine acting to increase isoleucine synthesis and resulting in the allosteric inhibition of IlvA to allow growth (Fig. 4).
Figure 4.
ilvA219 prevents effect of threonine on a ridA mutant growth. A, growth is shown for S. enterica WT (DM9404; circles) and ridA (DM3480; squares) strains. B, growth is shown for S. enterica ilvA219 (DM6947; circles) and ridA ilvA219 (DM6946; squares) strains. Growth analysis was performed at 37 °C in minimal medium containing 11 mm glucose as the sole carbon source alone (open symbols) or supplemented with 1 mm l-threonine (open symbols). Data are the mean, and error bars represent the 95% confidence interval of three biological replicates.
Discussion
Strains lacking ridA in S. enterica have several metabolic defects that result from the accumulation of 2AA in the absence of RidA deaminase activity. This study sought to use the biochemical characteristics of IlvA variants to better understand the mechanism by which they reversed one defect, growth with pyruvate as a carbon source. Analyzing the role of IlvA in ridA phenotypes is complicated by, among other things, the fact that the enzyme generates both 2AA (from serine) and 2AC (from threonine). The former is potentially damaging, and the latter is a metabolite necessary for synthesis of isoleucine. Therefore, variants that minimize 2AA formation cannot decrease 2AC so much that isoleucine synthesis is compromised. Kinetic analysis of four IlvA proteins, two of which suppressed a ridA growth defect, provided data to address the impact of 2AA and 2AC on growth. Based on these data and a simplified equation to characterize product formation in vivo, several conclusions were made.
IlvA variants able to suppress ridA phenotypes decrease 2AA production
Strains with IlvAA142T or IlvAG191S had significantly less threonine deaminase activity than WT in crude extracts. Equation 1 used data from Fig. 3 to estimate that 2AA production from either IlvAA142T or IlvAG191S was reduced 10-fold from WT IlvA (Table 2). IlvE activity in ridA ilvA3210 and ridA ilvA3211 strains was not statistically significantly different from the isogenic parents (ilvA3210 and ilvA3211, respectively), allowing the conclusion that a 10-fold reduction in 2AA eliminated detectable damage to the target IlvE. Exogenous serine (5 mm) in the growth media could still lead to significant 2AA production (Table 2), which then eliminates growth (Fig. 2). The fact that the IlvA variants with reduced 2AA production also had decreased 2AC production highlights the difficulty in altering enzymes to impact a single substrate.
IlvAA142T and IlvAG191S were compromised catalytically and had increased preference for threonine substrate
An expanded two-state model accounting for cooperative interactions within E. coli IlvA has been described previously (34, 35). Because the saturation curves using threonine or serine substrate could be fit using the hyperbolic equation by Michaelis-Menten, the majority of enzyme active sites (in the absence of isoleucine) exist in the active R-state (35). This simplified determination and comparison of Km and kcat values for each enzyme. Increased Km and lower kcat values suggested that IlvAA142T bound each substrate less well than the WT enzyme and generated both products more slowly. Alternatively, IlvAG191S bound both substrates more readily (lower Km) but was compromised in dehydratase activity (lower kcat). Thus, both enzyme variants were defective in generation of 2KB and pyruvate, but by different mechanisms. These findings highlight two features of enzymes that could impact 2AA production and stress in different organisms. Furthermore, the ratio of kcat/Km values for threonine versus serine indicated that IlvAA142T and IlvAG191S had a greater preference for threonine over serine than WT enzyme. PLP enzymes, such as IlvA, are well known for their substrate and reaction promiscuity (37–39). This work shows how changes that result in greater specificity toward a single substrate can be undesirable because they can also hamper the maximal rate of catalysis.
Exogenous threonine increases flux toward isoleucine, indirectly controlling 2AA production
A previous report found that mutations increasing threonine biosynthesis could suppress the growth defect of a ridA mutant in the presence of serine (26). Restoration of IlvE activity suggested that threonine competed with serine for the active site of IlvA and thus reduced the amount of 2AA produced (26). Implementation of Equation 1 suggested this was an unlikely explanation for the mechanism of threonine, because the amount of threonine required to reduce 2AA generation 10-fold would be unrealistically high (55 mm). The data supported a model in which isoleucine synthesis was increased when threonine was provided (i.e. increased AC production and no change in 2AA). Analysis of the impact of threonine on the growth of an ilvA219 ridA mutant supported this model. The ilvA219 allele encodes a variant of IlvA (IlvAL447F) insensitive to inhibition by isoleucine. If threonine addition reduced 2AA production via competitive inhibition, the ilvA219 ridA mutant should still be rescued by threonine supplementation. The finding that the ilvA219 ridA strain was not rescued by threonine supported the model implicating isoleucine biosynthesis (and allosteric inhibition of the 2AA generator, IlvA) as the relevant consequence of threonine addition.
Kinetic models can complement in vivo analyses
Assays of IlvA activity in crude extracts were the starting point for understanding how the IlvA variants impacted ridA phenotypes. Unexpectedly, there were several instances where the growth data, crude extract assays, and activity measurements using pure protein seemed inconsistent. First, purified IlvAL447F was not significantly different in catalytic capacity from the WT protein, yet in crude extract, it produced half the activity. Second, data from crude extracts confirmed the growth observation that strains expressing the IlvAA142T variant were starving for isoleucine, based on the apparent induction of the ilv operon (i.e. increased IlvE activity). However, the total activity of IlvAA142T in crude extracts was no lower than that of IlvAG191S. This point was of interest because strains with IlvAG191S have sufficient isoleucine synthesis for growth. The disparity in isoleucine requirement between ilvA3210 and ilvA3211 strains was somewhat reconciled by in vitro assays with purified protein, which showed IlvAA142T had lower affinity for threonine and was less stable over time. This could suggest that IlvAA142T was unable to produce sufficient 2AC in vivo, where threonine concentration is expected to be 2 orders of magnitude lower than that used in crude assays (20 mm). An inherent difficulty still exists in using in vitro determinations to precisely calculate metabolic flux and model in vivo behaviors. This difficulty encompasses technical limitations, where features like oligomerization, protein stability, small molecule binding affinity, and enzymatic activity are not effectively recapitulated in vitro (40). Discrepancies between in vitro assays and in vivo behavior are also impacted by transcriptional and post-transcriptional systems of regulation. These differences serve as a reminder that changes in flux caused by enzyme variants can result in unexpected regulatory shifts, as the cells respond to the perturbation. Altogether, the limited effectiveness in using in vitro data to correctly predict growth behavior differences between WT, ilvA3211, and ilvA3210 strains underscores technical limitations and physiological complexity. Despite its simplicity, Equation 1 proved useful in predicting the physiological consequence of threonine addition on 2AA metabolism and thus refined a previous model. In total, the data and discussion herein emphasize the need for a holistic approach for understanding and modeling metabolism, as we seek to define subtle and integrated responses of the cell to metabolic stress and cellular perturbation.
Experimental procedures
Bacterial strains, media, and chemicals
Strains used in this work are provided in Table 3 and are all derivatives of Salmonella enterica serovar Typhimurium LT2. Minimal medium was no-carbon E supplemented with 1 mm MgSO4 (41), trace minerals (42), and 11 mm d-glucose or 50 mm pyruvate as the sole carbon source. Difco nutrient broth (NB) (8 g/liter) containing NaCl (5 g/liter) was used as rich medium. Difco BiTek agar (15 g/liter) was added for solid medium. Superbroth (32 g of tryptone, 20 g of yeast extract, 5 g of sodium chloride, and 5 ml of 1 n sodium hydroxide/liter) was used for purifying proteins. Antibiotics were used at the following concentrations for rich (or minimal) medium: kanamycin, 50 (12.5) μg/ml; chloramphenicol, 20 (5) μg/ml; and ampicillin, 150 (7.5) μg/ml. All chemicals were obtained from Sigma.
Table 3.
Bacterial strains, plasmids, and primers
Genetic techniques
Strains were constructed via transductional crosses using the high-frequency general transducing mutant of bacteriophage P22 (HT105/1, int-201) (43). Methods for performing transductions, isolation of cells from phage contamination, and identifying phage-free recombinants were previously described (44, 45). Briefly, recipient cells (∼108 CFU) and transducing phage (∼109 PFU) were preincubated for 1 h at 37 °C prior to plating on selective media. Transductants were purified by isolation using nonselective green indicator plates, and putative phage-free clones were identified as light green colonies. These clones were checked for phage sensitivity by cross-streaking with phage P22. Transductional crosses in E. coli were performed using bacteriophage P1vir, as described previously (8). E. coli BL21-AI ΔilvA823 was constructed by transduction of the ilvA723::kan locus from the Keio collection into the BL21-AI strain background (46). The kanamycin cassette was then resolved using flippase (FLP) recombinase, as previously described by Datsenko and Wanner (47), to create E. coli BL21-AI ΔilvA823.
Molecular techniques
The ilvA locus was amplified from LT2 strains DM7608 (ilvA3211) and DM7610 (ilvA3210) by PCR with Q5 high-fidelity DNA polymerase (New England Biolabs) using primers IlvA_BspQI_LT2_F and IlvA_BspQI_LT2_R (Table 3). The PCR product was purified, digested with BspQI, and ligated into the modified pET28b, pSAPKO-CH, as described by Galloway et al. (48). The resulting plasmids were named pDM1461 and pDM1543, which encoded IlvAA142T and IlvAG191S fused to a C-terminal penta-histidine tag, respectively. ilvA219 was amplified from LT2 strain DM6947 by PCR with Q5 high-fidelity DNA polymerase (New England Biolabs) using primers IlvA_NdeI_LT2_F and IlvA_XhoI_LT2_R (Table 1). The PCR product was purified, digested with NdeI and XhoI (New England Biolabs), and ligated into predigested pET20b (Novagen), digested with the same enzymes. The resulting plasmid was named pDM1420 and encoded IlvAL447F fused to a C-terminal hexa-histidine tag. Constructs were transformed into E. coli DH5α, and transformants were screened for vectors containing the correct insert. Inserts were confirmed through sequence analysis, performed by Eton Biosciences.
Protein purification
The three plasmids generated above and pDM1578, which encoded S. enterica IlvA fused to a C-terminal hexa-histidine tag and was generated previously (13), were each inserted into an E. coli BL21-AI strain (Invitrogen) lacking the native ilvA locus. The resulting strains were inoculated into 2 ml of LB containing either ampicillin (pET20b-ilvA and pET20b-ilvA219) or kanamycin (pSapKOCH-ilvA3210 and pSapKOCH-ilvA3211) and allowed to reach full density. These were subcultured into 50 ml of superbroth and grown for 3 h before inoculating 1.5 liters of superbroth, containing the appropriate drug. Cultures were grown at 30 °C shaking (180 rpm) to an A650 of 0.6, and expression was induced by the addition of 0.2% arabinose (pET20b-ilvA and pET20b-ilvA219) or 0.2% arabinose and 0.2 mm isopropyl 1-thio-β-d-galactopyranoside (pSapKOCH-ilvA3210 and pSapKOCH-ilvA3211) and allowed to grow an additional 16 h. Cells were harvested at 4 °C by centrifugation (7,000 × g), and pellets were frozen at −80 °C until purification. During purification, cells were resuspended in 2.5 ml of buffer (100 mm KPO4, pH 8, 100 mm NaCl, 20 mm imidazole, and 10% glycerol) for every 1 g of cell pellet. Resuspension was incubated on ice with 1 mg/ml lysozyme and 0.125 mg/ml DNase 30 min prior to cell disruption using a One-Shot Cell Disruptor at 18,000 p.s.i. (Constant Systems). After disruption, phenylmethylsulfonyl fluoride was added to 1 mm. Lysates were clarified by centrifugation (48,000 × g) and filtered through a 0.45-μm filter. Filtered lysates were bound to a 5-ml HisTrapTM HP column and purified using the manufacturer's protocol (GE Healthcare). The column was washed using resuspension buffer (100 mm KPO4, pH 8, 100 mm NaCl, 20 mm imidazole, and 10% glycerol) and then eluted with elution buffer (100 mm KPO4, pH 8, 100 mm NaCl, 500 mm imidazole, and 10% glycerol), in a linear gradient over the course of 10 column volumes. The fractions were assessed for purity, pooled, and concentrated using a 30,000 molecular weight cutoff Ultra-15 centrifugal filter (Amicon). Samples were desalted using a PD-10 desalting column (GE Healthcare) and eluted in storage buffer (100 mm KPO4, pH 8.0, 100 mm NaCl, 10 μm PLP, 10% glycerol). Protein concentration was determined by the BCA assay (Pierce). Protein was frozen in liquid nitrogen and stored at −80 °C.
Quantification of growth
A 2-μl aliquot of cells from an overnight NB culture was used to inoculate 198 μl of growth medium. 96-Well plates were incubated at 37 °C in a microplate reader (model EL808; Bio-Tek Instruments) with low shaking, and growth was monitored as the change in optical density at 650 nm (OD650) over time. Unless stated, growth experiments were performed in biological triplicate. Results were plotted using GraphPad Prism 7.0d, with curves (log10-format) representing averages and standard error of the means for the replicates.
Dehydratase (IlvA) assays
Crude assay
Full-density NB cultures were inoculated (1:100) into 100 ml minimal medium containing 0.67 mm glycine and grown at 37 °C shaking to stationary phase (12 h) before being pelleted (10,000 × g, 10 min) and stored as pellets at −80 °C. Cell pellets were resuspended in 2 ml of buffer (50 mm KPO4, pH 8.0, 0.4 mm DTT). Lysates were prepared using one pass through a One Shot cell disrupter (Constant Systems) at 18,000 psi, and clarified by centrifugation (20 min @ 16,000 × g at 4 °C). Threonine dehydratase assays were carried out as described previously (27, 49) and reported as micromoles 2-ketobutyrate formed per mg of protein per min. Protein concentrations were estimated by the method from Bradford (50). The experiment contained three biological replicates for each strain tested and the average with standard deviation is reported.
Purified enzyme assay
Pyruvate and 2-ketobutyrate formation was assayed by tracking the absorbance at 230 nm (13, 51) at room temperature (25 °C) for 2 mins. Assays were carried out in quadruplicate, using 100 mm potassium phosphate buffer, pH 8.0. To obtain values within the range of detection for the instrument, reactions used 100 nm of IlvA, IlvAL447F, and IlvAA142T for threonine assays and 200 nm of IlvA, IlvAL447F, and IlvAA142T for serine assays; 1 μm IlvAG191S was used for both assays. The indicated concentrations of serine and threonine were used to start each 200 μl reaction, which were assayed continuously in a quartz 96-well plate at room temperature by a SpectraMax Plus (Molecular Devices) supporting SoftMax Pro 7.0 software.
Protein stability
Pyruvate formation with purified protein was assayed as described above. Assays were carried out every 2 h, for a total of 8 h with 200 mm l-serine used to start each reaction. Each measurement was performed in duplicate, using 200 nm IlvA, IlvAL447F, and IlvAA142T and 1 μm IlvAG191S. Enzyme preparations were kept on ice between measurements.
Branched-chain amino acid aminotransferase (IlvE) assays
Full-density NB cultures were inoculated (1:100) into 5 ml of minimal medium containing 0.67 mm glycine, grown at 37 °C shaking to stationary phase (12 h) before being pelleted (10,000 × g, 2 min), and stored as pellets at −20 °C. IlvE activity was assayed according to previously described methods (27). Cells were permeabilized in buffer (50 mm KPO4, pH 8, 50 μm PLP, 10 mm 2-ketoglutarate) containing 10% PopCulture Reagent (Novagen). The reaction was started with 20 mm l-isoleucine. The product, KMV, was derivatized by 2,4-dinitrophenylhydrazine (DNPH), producing a chromophore with absorbance of 540 nm. KMV was quantified using a standard curve from known quantities of KMV derivatized with DNPH and normalized to total protein content (50).
Kinetic data analysis
Initial velocity calculations were estimated over the course of 2-min reactions tracking ΔmOD230 min−1 for various concentrations of l-threonine or l-serine. Standard curves for absorbance at 230 nm for known amounts of 2-ketobutyrate and pyruvate were used to transform ΔmOD230 min−1 to μmol of 2-KB min−1 and μmol of pyruvate min−1. A nonlinear regression fit (Equation 2) to a graph of the substrate concentration versus initial velocity was used to fit the data.
| (Eq. 2) |
In the absence of cooperative effects (h = 1), this reduced to the equation by Michaelis-Menten (Equation 3), from which each IlvA variant's Michaelis-Menten constant (Km) and maximal velocity (Vmax) values for each substrate could be determined. Calculations were performed using GraphPad Prism 7.0d.
| (Eq. 3) |
The turnover number (kcat) was obtained using the active-site concentration (Et) (Equation 4).
| (Eq. 4) |
Because IlvA is a dimer of dimers, containing two active sites per enzyme, Et is equal to half of the enzyme concentration used in the assay (52, 53).
Author contributions
A. J. B. and D. M. D. conceptualization; A. J. B. formal analysis; A. J. B. methodology; A. J. B. writing-original draft; A. J. B. and D. M. D. writing-review and editing; D. M. D. resources; D. M. D. supervision; D. M. D. funding acquisition; D. M. D. project administration.
Acknowledgment
We thank Kelsey Hodge-Hanson for constructing plasmid pDM1461.
This work was supported by National Institutes of Health Competitive Grants Program Award GM095837 (to D. M. D.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- 2AA
- 2-aminoacrylate
- PLP
- pyridoxal 5′-phosphate
- KMV
- 2-keto-3-methylvalerate
- DNPH
- 2,4-dinitrophenylhydrazine
- 2-KB
- 2-ketobutyrate
- 2AC
- 2-aminoacrylate
- NB
- nutrient broth.
References
- 1. Koenigsknecht M. J., and Downs D. M. (2010) Thiamine biosynthesis can be used to dissect metabolic integration. Trends Microbiol. 18, 240–247 10.1016/j.tim.2010.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Koenigsknecht M. J., Lambrecht J. A., Fenlon L. A., and Downs D. M. (2012) Perturbations in histidine biosynthesis uncover robustness in the metabolic network of Salmonella enterica. PLoS ONE 7, e48207 10.1371/journal.pone.0048207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Albert R., Jeong H., and Barabasi A. L. (2000) Error and attack tolerance of complex networks. Nature 406, 378–382 10.1038/35019019 [DOI] [PubMed] [Google Scholar]
- 4. Bazurto J. V., and Downs D. M. (2011) Plasticity in the purine-thiamine metabolic network of Salmonella. Genetics 187, 623–631 10.1534/genetics.110.124362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bazurto J. V., and Downs D. M. (2016) Metabolic network structure and function in bacteria goes beyond conserved enzyme components. Microb. Cell 3, 260–262 10.15698/mic2016.06.509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cardinale S., and Arkin A. P. (2012) Contextualizing context for synthetic biology–identifying causes of failure of synthetic biological systems. Biotechnol. J. 7, 856–866 10.1002/biot.201200085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kittleson J. T., Wu G. C., and Anderson J. C. (2012) Successes and failures in modular genetic engineering. Curr. Opin. Chem. Biol. 16, 329–336 10.1016/j.cbpa.2012.06.009 [DOI] [PubMed] [Google Scholar]
- 8. Bazurto J. V., Farley K. R., and Downs D. M. (2016) An unexpected route to an essential cofactor: Escherichia coli relies on threonine for thiamine biosynthesis. MBio 7, e01840–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mehta V., Athar M., Jha P. C., Panchal M., Modi K., and Jain V. K. (2016) Efficiently functionalized oxacalix[4]arenes: synthesis, characterization and exploration of their biological profile as novel HDAC inhibitors. Bioorg. Med. Chem. Lett. 26, 1005–1010 10.1016/j.bmcl.2015.12.044 [DOI] [PubMed] [Google Scholar]
- 10. Kim J., Kershner J. P., Novikov Y., Shoemaker R. K., and Copley S. D. (2010) Three serendipitous pathways in E. coli can bypass a block in pyridoxal-5′-phosphate synthesis. Mol. Syst. Biol. 6, 436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Borchert A. J., and Downs D. M. (2017) The response to 2-aminoacrylate differs in Escherichia coli and Salmonella enterica, despite shared metabolic components. J. Bacteriol. pii: e00140-17 10.1128/JB.00140-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Downs D. M., Bazurto J. V., Gupta A., Fonseca L. L., and Voit E. O. (2018) The three-legged stool of understanding metabolism: integrating metabolomics with biochemical genetics and computational modeling. AIMS Microbiol. 4, 289–303 10.3934/microbiol.2018.2.289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lambrecht J. A., Flynn J. M., and Downs D. M. (2012) Conserved YjgF protein family deaminates reactive enamine/imine intermediates of pyridoxal 5′-phosphate (PLP)-dependent enzyme reactions. J. Biol. Chem. 287, 3454–3461 10.1074/jbc.M111.304477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lambrecht J. A., Schmitz G. E., and Downs D. M. (2013) RidA proteins prevent metabolic damage inflicted by PLP-dependent dehydratases in all domains of life. MBio 4, e00033–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Flynn J. M., Christopherson M. R., and Downs D. M. (2013) Decreased coenzyme A levels in ridA mutant strains of Salmonella enterica result from inactivated serine hydroxymethyltransferase. Mol. Microbiol. 89, 751–759 10.1111/mmi.12313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Flynn J. M., and Downs D. M. (2013) In the absence of RidA, endogenous 2-aminoacrylate inactivates alanine racemases by modifying the pyridoxal 5′-phosphate cofactor. J. Bacteriol. 195, 3603–3609 10.1128/JB.00463-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hodge-Hanson K. M., and Downs D. M. (2017) Members of the Rid protein family have broad imine deaminase activity and can accelerate the Pseudomonas aeruginosa d-arginine dehydrogenase (DauA) reaction in vitro. PLoS ONE 12, e0185544 10.1371/journal.pone.0185544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Downs D. M., and Ernst D. C. (2015) From microbiology to cancer biology: the Rid protein family prevents cellular damage caused by endogenously generated reactive nitrogen species. Mol. Microbiol. 96, 211–219 10.1111/mmi.12945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ernst D. C., Anderson M. E., and Downs D. M. (2016) l- 2,3-diaminopropionate generates diverse metabolic stresses in Salmonella enterica. Mol. Microbiol. 101, 210–223 10.1111/mmi.13384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lambrecht J. A., Browne B. A., and Downs D. M. (2010) Members of the YjgF/YER057c/UK114 family of proteins inhibit phosphoribosylamine synthesis in vitro. J. Biol. Chem. 285, 34401–34407 10.1074/jbc.M110.160515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Christopherson M. R., Schmitz G. E., and Downs D. M. (2008) YjgF is required for isoleucine biosynthesis when Salmonella enterica is grown on pyruvate medium. J. Bacteriol. 190, 3057–3062 10.1128/JB.01700-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Enos-Berlage J. L., Langendorf M. J., and Downs D. M. (1998) Complex metabolic phenotypes caused by a mutation in yjgF, encoding a member of the highly conserved YER057c/YjgF family of proteins. J. Bacteriol. 180, 6519–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ernst D. C., and Downs D. M. (2016) 2-Aminoacrylate stress induces a context-dependent glycine requirement in ridA strains of Salmonella enterica. J. Bacteriol. 198, 536–543 10.1128/JB.00804-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ernst D. C., and Downs D. M. (2018) Mmf1p couples amino acid metabolism to mitochondrial DNA maintenance in Saccharomyces cerevisiae. MBio 9, e00084–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hodge-Hanson K. M., Zoino A., and Downs D. M. (2018) Expression of PLP-independent racemases can reduce 2-aminoacrylate stress in Salmonella enterica. J. Bacteriol. 200, e00751–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Christopherson M. R., Lambrecht J. A., Downs D., and Downs D. M. (2012) Suppressor analyses identify threonine as a modulator of ridA mutant phenotypes in Salmonella enterica. PLoS ONE 7, e43082 10.1371/journal.pone.0043082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schmitz G., and Downs D. M. (2004) Reduced transaminase B (IlvE) activity caused by the lack of yjgF is dependent on the status of threonine deaminase (IlvA) in Salmonella enterica serovar Typhimurium. J. Bacteriol. 186, 803–810 10.1128/JB.186.3.803-810.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barak Z., Chipman D. M., and Gollop N. (1987) Physiological implications of the specificity of acetohydroxy acid synthase isozymes of enteric bacteria. J. Bacteriol. 169, 3750–3756 10.1128/jb.169.8.3750-3756.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen J. W., Bennett D. C., and Umbarger H. E. (1991) Specificity of attenuation control in the ilvGMEDA operon of Escherichia coli K-12. J. Bacteriol. 173, 2328–2340 10.1128/jb.173.7.2328-2340.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. LaRossa R. A., and Van Dyk T. K. (1987) Metabolic mayhem caused by 2-ketoacid imbalances. Bioessays 7, 125–130 10.1002/bies.950070308 [DOI] [PubMed] [Google Scholar]
- 31. Hofler J. G., and Burns R. O. (1978) Threonine deaminase from Salmonella typhimurium. Effect of regulatory ligands on the binding of substrates and substrate analogues to the active sites and the differentiation of the activator and inhibitor sites from the active sites. J. Biol. Chem. 253, 1245–1251 [PubMed] [Google Scholar]
- 32. Bennett B. D., Kimball E. H., Gao M., Osterhout R., Van Dien S. J., and Rabinowitz J. D. (2009) Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599 10.1038/nchembio.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Albe K. R., Butler M. H., and Wright B. E. (1990) Cellular concentrations of enzymes and their substrates. J. Theor. Biol. 143, 163–195 10.1016/S0022-5193(05)80266-8 [DOI] [PubMed] [Google Scholar]
- 34. Eisenstein E. (1995) Allosteric regulation of biosynthetic threonine deaminase from Escherichia coli: effects of isoleucine and valine on active-site ligand binding and catalysis. Arch. Biochem. Biophys. 316, 311–318 10.1006/abbi.1995.1042 [DOI] [PubMed] [Google Scholar]
- 35. Gallagher D. T., Chinchilla D., Lau H., and Eisenstein E. (2004) Local and global control mechanisms in allosteric threonine deaminase. Methods Enzymol. 380, 85–106 10.1016/S0076-6879(04)80004-1 [DOI] [PubMed] [Google Scholar]
- 36. Segel I. H., and Segel L. D. (1992) An alternative substrate is not the same as a dead end inhibitor. Biochem. Educ. 20, 155–157 10.1016/0307-4412(92)90060-Y [DOI] [Google Scholar]
- 37. Lal P. B., Schneider B. L., Vu K., and Reitzer L. (2014) The redundant aminotransferases in lysine and arginine synthesis and the extent of aminotransferase redundancy in Escherichia coli. Mol. Microbiol. 94, 843–856 10.1111/mmi.12801 [DOI] [PubMed] [Google Scholar]
- 38. Toney M. D. (2005) Reaction specificity in pyridoxal phosphate enzymes. Arch. Biochem. Biophys. 433, 279–287 10.1016/j.abb.2004.09.037 [DOI] [PubMed] [Google Scholar]
- 39. Toney M. D. (2011) Controlling reaction specificity in pyridoxal phosphate enzymes. Biochim. Biophys. Acta 1814, 1407–1418 10.1016/j.bbapap.2011.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kuznetsova I. M., Turoverov K. K., and Uversky V. N. (2014) What macromolecular crowding can do to a protein. Int. J. Mol. Sci. 15, 23090–23140 10.3390/ijms151223090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vogel H. J., and Bonner D. M. (1956) Acetylornithase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218, 97–106 [PubMed] [Google Scholar]
- 42. Balch W. E., and Wolfe R. S. (1976) New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl. Environ. Microbiol. 32, 781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schmieger H. (1972) Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 119, 75–88 10.1007/BF00270447 [DOI] [PubMed] [Google Scholar]
- 44. Chan R. K., Botstein D., Watanabe T., and Ogata Y. (1972) Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II Properties of a high transducing lysate. Virology 50, 883–898 10.1016/0042-6822(72)90442-4 [DOI] [PubMed] [Google Scholar]
- 45. Downs D. M., and Petersen L. (1994) apbA, a new genetic locus involved in thiamine biosynthesis in Salmonella typhimurium. J. Bacteriol. 176, 4858–4864 10.1128/jb.176.16.4858-4864.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., and Mori H. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006 2, 2006.0008 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Datsenko K. A., and Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Galloway N. R., Toutkoushian H., Nune M., Bose N., and Momany C. (2013) Rapid cloning for protein crystallography using type IIS restriction enzymes. Cryst. Growth Des. 13, 2833–2839 10.1021/cg400171z [DOI] [Google Scholar]
- 49. Burns R. O. (1971) l-Threonine deaminase-biosynthetic (Salmonella typhimurium). Methods Enzymol. 17, 555–560 10.1016/0076-6879(71)17097-8 [DOI] [Google Scholar]
- 50. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 51. Davis L. (1965) A spectrophotometric method for the assay of threonine dehydratase. Anal. Biochem. 12, 36–40 10.1016/0003-2697(65)90139-9 [DOI] [PubMed] [Google Scholar]
- 52. Burns R. O., and Zarlengo M. H. (1968) Threonine deaminase from Salmonella typhimurium. I. Purification and properties. J. Biol. Chem. 243, 178–185 [PubMed] [Google Scholar]
- 53. Zarlengo M. H., Robinson G. W., and Burns R. O. (1968) Threonine deaminase from Salmonella typhimurium. II. The subunit structure. J. Biol. Chem. 243, 186–191 [PubMed] [Google Scholar]