Abstract
Mitochondrial oxidants (or reactive oxygen species) participate in a myriad of physiological and pathological processes. They are, however, quite hard to measure due to their chemical nature and specific subcellular location. Here, we review techniques to measure mitochondrial oxidants in biological systems as well as the results of their activity, highlighting conditions to be considered, controls and recommended practices. We will delineate experimental setups that use combined strategies to convincingly demonstrate the biological effects of mitochondrial oxidants, using the imperfect methodology available today.
Keywords: Mitochondria, Oxidants, Reactive oxygen species, Probes, Methods
1. Introduction
Mitochondria produce oxidants (or reactive oxygen species) such as the superoxide radical anion (O2•-) and hydrogen peroxide (H2O2) as a result of the immense variety of redox reactions that are located in this organelle [2], [20], [34], [51]. Consequently, mitochondria also developed efficient antioxidant systems and regulatory pathways that modulate oxidant production [20], [51]. The balance between production and removal determines oxidant release from the organelle, which is modified by the substrate in use, physiological energy metabolism regulation and a variety of pathological conditions [20], [48], [9]. Interestingly, a large body of data shows that oxidants not only lead to varying degrees of oxidative damage in mitochondria and in other cellular components (when antioxidant systems are overwhelmed), but also act as signaling molecules [21], [45].
Given the varied and pivotal roles of mitochondrial oxidants, measuring these species, mitochondrial redox state and the consequences of oxidants in this organelle is an important element in many current scientific studies. Unfortunately, although the literature has described many tools to measure mitochondrial oxidants, none is foolproof nor can be used in the absence of controls. Furthermore, most mitochondrial redox state measurement techniques are artifact-prone due to the reactive nature of the oxidants they measure and conditions specific to mitochondrial biology such as the presence of a variable inner membrane potential, changes in mitochondrial morphology and mass. The aim of this review is to highlight experimental conditions, controls and recommended practices when measuring mitochondrial redox state. We will start by quickly reviewing characteristics of mitochondrial oxidant production and removal. We will then address some important particularities of animal and cell culture models that pertain to studies of mitochondrial oxidants. Next, we will focus on techniques available, discussing their capabilities and limitations, as well as necessary controls. The review will end by discussing ideal strategy designs to study biological effects of mitochondrial oxidants using the imperfect techniques available currently.
2. Mitochondrial oxidants and antioxidants – a very brief overview
Characteristics of mitochondrial oxidant production and antioxidant mechanisms have been extensively reviewed elsewhere [2], [20], [34], [51], [9] and are not the aim of this review, which focusses instead on mitochondrial oxidant measurement techniques. It suffices to say here that many mitochondrial components, including electron transport chain complexes (such as complexes I, II and III) and redox enzymes (such as glycerol 3 phosphate, pyruvate, alfa-ketoglutarate, dihydroorotate, very long chain acyl-CoA dehydrogenases and the electron transfer flavoprotein) can generate oxidants such as O2•- and H2O2 within mitochondria. The rate of oxidant production is modulated by oxygen consumption velocity in the organelle: higher mitochondrial oxygen consumption rates are generally, but not universally, associated with lower oxidant production [48], since intermediates generating these oxidants turn over more quickly, reverse electron transport is unfavorable, and local oxygen tensions are lower [51].
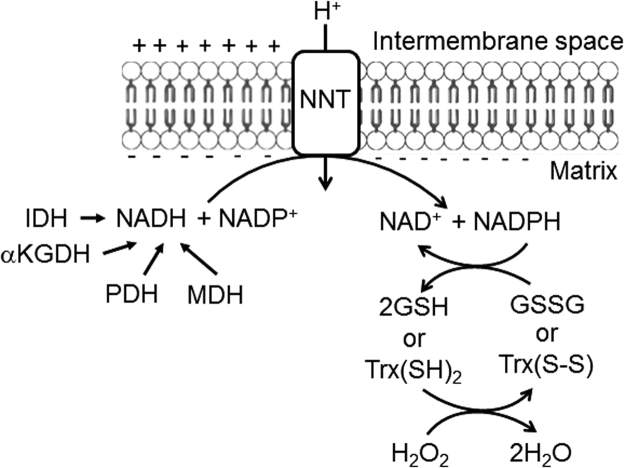
Superoxide radical anions generated in both the matrix and the intermembrane space are rapidly converted to H2O2 due to the presence of superoxide dismutases in both compartments [38]. Because mitochondria contain both labile iron and metal-containing enzymes, the highly reactive hydroxyl radical, generated from H2O2 via the Fenton reaction, has been shown to be produced in the organelle [53], although its short half-life hampers its detection in complex biological models. Since H2O2 has the ability to generate damaging hydroxyl radicals, these organelles have evolved efficient systems to remove peroxides [2], [20], [34], [51], including catalases and thiol peroxidases such as glutathione and thioredoxin peroxidases. Reduced glutathione and thioredoxin are necessary for the function of thiol peroxidases, and are maintained reduced by electrons donated from mitochondrial NADPH. Mitochondrial NADPH is mostly produced by the mitochondrial nicotinamide nucleotide transhydrogenase (NNT, Fig. 1), an inner membrane protein described in more detail below. Curiously, this enzyme is deficient in one of the most used laboratory animal rodent models.
Fig. 1.
Mitochondrial nicotinamide nucleotide transhydrogenase (NNT). NNT uses the inner membrane proton gradient as a driving force to transfer electrons to NADP+ from NADH, produced by many intramitochondrial enzymes including pyruvate, isocitrate, malate and α-ketoglutarate dehydrogenases (PDH, IDH, MDH and αKGDH, respectively). This NNT activity generates mitochondrial matrix NADPH coupled to the entry of a proton into the matrix, and is deficient in the widely-used C57BL/6 J mouse. Intramitochondrial NADPH reduces oxidized glutathione and thoredoxin [GSSG and Trx(S-S) to GSH and Trx(SH)2], which allows for the removal of H2O2 through the activities of glutathione and thioredoxin peroxidases. As a result, C57BL/6 J mouse mitochondria are unable to adequately remove oxidants and prone to oxidative damage.
3. Animal models
Animal models are often used to study the effects of disease, diet, aging or other interventions on mitochondrial oxidants in different tissues. Laboratory rodents constitute the bulk of these animal models and are also often used to obtain isolated mitochondria or tissue samples for in vitro redox state studies.
Some particularities regarding mitochondrial redox state should be considered when using laboratory rodents. The first pertains to the animal strain used. In 2005, while investigating reasons for impaired insulin secretion in the widely-used C57BL/6J mouse, Toye et al. [50] discovered this strain harbored a spontaneous missense mutation in exon 1 and a multi-exon deletion (exons 7–11) in the nuclear gene that encodes for NNT. NNT is an inner mitochondrial membrane protein which uses the proton gradient as a driving force to transfer electrons from NADH to NADP+ (Fig. 1). In the absence of this protein, NADP+ reduction in mitochondria is hampered, resulting in low NADPH availability for antioxidant systems, increased oxidized glutathione levels, poor ability to remove added oxidants and higher H2O2 release rates [29], [36], [42]. Thus, the lack of NNT in C57BL/6J mice has a strong impact on mitochondrial redox state.
Importantly, the C57BL/6J mouse is the most widely used inbred laboratory mouse strain today (https://www.jax.org/strain/000664, consulted September 2nd, 2018), and is also the background used to generate many genetically modified mouse strains, which means the redox effects in most of these studies should be analyzed in light of the hampered mitochondrial antioxidant system these animals have. Furthermore, mispairings and mislabeling of C57BL/6 mouse strains are not uncommon, and can result in widely different biological effects [5].
Given the wide use of this animal model, we don´t believe it is feasible to avoid it altogether, but recommend testing C57BL/6 mice for the presence of the Nnt mutation, as well as considering biological implications of the lack of NNT when using this widely-adopted spontaneous mutant. This characteristic of C57BL/6 mice also brings to light the peculiar nature of inbred laboratory animal strains and their tendency to develop specific genetic drifts over time, emphasizing the importance of using congenic animals to determine the effects of singe gene modifications whenever possible.
Another aspect to keep in mind regarding animal studies and mitochondrial redox state is the diet used by these animals. Because of the central role of mitochondria in energy metabolism, mitochondrial oxidants are often studied following nutritional interventions such as caloric restriction or overnutrition. We find that many studies in which calories are restricted do not use supplemented diets to account for micronutrient intake decreases [10]. Non-supplemented diets can result in micronutrient malnutrition, including low levels of vitamins and minerals important to maintain redox balance such as copper, iron, manganese, selenium, riboflavin and thiamin. As a result, many studies that see changes in mitochondrial physiology with caloric restriction may in fact describe effects of lower micronutrient, and not caloric, intake [11].
Overnutrition studies such as studies involving high fat diets may also promote malnutrition due to decreases in total food intake promoted by fat-induced satiety. High fat dietary interventions in the literature are also confounded by the fact that the low fat diets they are compared to are quite variable, and often not a known-component diet which differs only in fat and energy contents, thus promoting changes in micronutrient availability [24]. Overall, dietary interventions should be formulated in animal redox studies so as not to involve unwanted changes in micronutrient content that lead to differences in electron transport chain function or impair the synthesis of antioxidant system components. Diets should also be checked to ensure no alterations in added antioxidant preservatives, such as butylated hydroxytoluene (BHT), occur within the experimental protocol.
4. Cell culture models
Cultured cells are often used in studies involving mitochondrial oxidant production, both for in situ experiments and as sources of isolated organelles or homogenates. A particularity of mammalian cell cultures that these cells are grown at oxygen concentrations similar to the solubility of oxygen in media (~200 μM), while the concentration of oxygen in mammalian tissues depends on proximity to blood vessels and diffusion across cell layers, and is thus usually much lower (~1–30 μM). Careful experiments in isolated mitochondria by Hoffman and Brookes [22] have determined that mitochondrial H2O2 release rates are highly sensitive to changes in oxygen concentrations within the expected physiological in vivo concentration range, while completely saturated at the concentrations in which cells are cultured. Experiments under more physiological oxygen concentration growth conditions are possible, although they require specialized incubators and are thus probably unrealistic for most studies. However, researchers should keep this important experimental difference in mind when designing protocols, and particularly when comparing in vivo animal tissue effects to those in cultured cells.
Another consideration in cell cultures pertains to the adaptations these cultures undergo during many generations of replication under laboratory conditions, which usually involve growth in large relative quantities of media with high glucose content. These high glucose levels allow the cells to generate ATP mainly through glucose fermentation to lactate, and many cell lines grown in this manner have significant loss of mitochondrial function, which can predictably affect mitochondrial redox state and signaling. This low mitochondrial activity in cell lines can be reversed by cultivating cells in non-fermentable substrates or galactose, which reduces glycolytic efficiency, thus stimulating respiration [47]. Again, this is not a necessary experimental condition for every cell culture mitochondrial oxidant study, but a characteristic that should be kept in mind when using cell lines.
5. Measuring mitochondrial oxidants – extramitochondrial probes
A common manner to measure mitochondrial oxidants is to follow the release of these oxidants into the extramitochondrial environment using probes that are oxidized and change absorbance or fluorescence. This form of measurement follows an increase in signal over time, theoretically representing an increase in oxidants detected, and is much more widely used than measuring single point markers of oxidative damage in the system. Because the oxidant must be released from mitochondria for this form of measurement, it must be relatively stable, membrane diffusible and long-lived [9]. Therefore this form of measurement is usually restricted to detection of mitochondrial H2O2.
When using isolated mitochondrial preparations or fractions of mitochondria, H2O2 peroxide release can be measured in a relatively straightforward manner using horseradish peroxidase and probes such as Amplex Red, which is oxidized to fluorescent resorufin [56]. Amplex Red has been widely adopted because it is stable and highly responsive. It produces fluorescence increases that correlate linearly with H2O2 additions within biological ranges of the oxidant which makes quantification of H2O2 release possible. However, it does not exclusively detect H2O2, even when used with horseradish peroxidase [14], as the system is also responsive to peroxynitrite. A control that can be employed to exclude the participation of peroxynitrite or other interferants in the signal is to add excess catalase to remove H2O2, and thus estimate the response that is H2O2-dependent and -independent.
Another methodological consideration when using Amplex red is that resorufin, its product, can undergo light-mediated photochemical oxidation when in the presence of reductants such as NADH [55]. This possibility should be considered when using NADH as an added substrate (and therefore not separated from the probe by the mitochondrial membrane), when mitochondrial membrane integrity is compromised (releasing NADH) and when conducting continuous fluorescence measurements (which expose the samples to large amounts of light).
While adequate for studies in isolated mitochondrial samples, methods that included added enzymes such as horseradish peroxidase are limited to measurements of oxidants released from membrane-bound biological systems, and therefore cannot detect cytosolic oxidants inside intact cells, which is a measurement conducted very frequently. The probe most widely used in intact cells is dichlorodihydrofluorescein diacetate (DCFH-DA), a membrane-permeable molecule which is metabolized by live cells to DCFH carboxylate anion, which can be oxidized to its easily detectable fluorescent product dichlorofluorescein (DCF). Although it is easy to obtain a fluorescent signal from this probe, it has many shortcomings (extensively reviewed by [25]), including: (i) DCF formation is not specific to any particular oxidant species. (ii) The probe itself can generate O2•-, thus building up fluorescence over time even in the absence of more oxidants [4]. (iii) The probe´s fluorescence does not increase in direct proportion to the amount of oxidants. For example, double DCF fluorescence does not indicate double the amount of the oxidant measured, making quantifications quite difficult. (iv) DCF is not directly oxidized by H2O2 released from mitochondria, requiring secondary reactions catalyzed by metal ions or heme proteins. (v) Of particular relevance to studies following mitochondrial oxidant generation is the fact that DCF formation is pH-sensitive, doubling fluorescence increases within physiological ranges around pH 7 [54]. Changes in mitochondrial energy metabolism are often accompanied by modifications in the local production of CO2 and lactate, among other effects that alter intracellular pH, which may modify DCF detection independently of the presence of oxidants. Overall, DCF is a questionable method to detect mitochondrial H2O2 release, and should not be used insolation for such measurements.
An alternative system to measure mitochondrially-generated H2O2 inside cells are membrane-permeable boronate esters attached to a fluorophore, which interact directly with intracellular H2O2 and other peroxides to form fluorescent products [25], [32], [57]. These compounds have the disadvantage of reacting with H2O2 with low second-order rate constants, that compete with endogenous detoxifying systems. However, even the small quantity of oxidants that escape from antioxidant systems and are detectable by boronate esters are already a strong indication of oxidant production. Mitochondrially-targeted boronate esters have been developed for detection within these organelles [15], [17], [57].
Dihydroethidium and dihydrorhodamine 123 are also often-used probes to measure oxidants in the cytosol. Dihydroethidium specifically generates 2-hydroxyethidium when oxidized by O2•-, which could allow it to be an oxidant-specific probe. However, other fluorescent dihydroethidium products, particularly ethidium, are also formed in vivo, so total fluorescence is not an indicator of O2•-. These different fluorescent products can be separated by HPLC for specific measurements [19]. Interestingly, dihydroethidium and dihydrorhodamine 123 oxidation products are accumulated in mitochondria when oxidized, since they have positive charges in this form [43], [7], and therefore their use will also require the considerations we will discuss under the next topic for mitochondrially-accumulated probes.
Another class of probes to detect oxidants in cells are genetically encoded fluorescent proteins which are sensitive to oxidant levels, generally by means of structural modifications involving disulfides (reviewed in detail by [31]). Examples of these proteins are redox-sensitive green fluorescent proteins (roGFPs), and HyPer, which have the advantage of being ratiometric probes, thus eliminating concerns regarding expression levels and unequal probe distribution. These proteins can be targeted to specific intracellular compartments, and molecularly engineered for different types of detection. Expression of fluorescent proteins in small intracellular spaces (such as the mitochondrial intermembrane space) can hamper the detection of fluorescence, but oxidized and reduced forms of these proteins can also be separated in gels and quantified, overcoming this caveat [23]. Recently, peroxiredoxins in combination with GFP and YFPs have also been proposed to be good scaffolds for the design of intracellular peroxide probes [52]. While encoded protein probes are very interesting and bring exciting new possibilities, some care must be expended when considering studies with them: (i) All fluorescent proteins can undergo photobleaching and produce oxidants under illumination [39]. (ii) As mentioned previously, accumulation of these probes in specific environments can affect whole cell fluorescence. (iii) Some of these proteins are pH-sensitive [41], and changes in pH have been erroneously interpreted as changes in O2•- levels in the past [44]. These caveats, as well as possible lack of selectivity to specific oxidants, should be considered when using encoded protein sensors to measure mitochondrially-generated oxidants.
6. Measuring mitochondrial oxidants – mitochondrial probes
Another class of probes used to measure changes in oxidant levels are cationic probes which accumulate in mitochondria by means of their inner membrane potential [16], [57], including dihidrorhodamine, Mitotracker Red CMX Ros and MitoSOX. A first point to consider when using cationic probes is that their accumulation in mitochondria is dependent on both the plasma membrane potential and the mitochondrial inner membrane potential [6]. Mitochondrial matrix concentrations of the probe at 37 °C can thus be estimated using the Nernst equation:
were [Probe+]matrix is the probe concentration in the mitochondrial matrix, [Probe+]extracellular is the concentration of the probe added to the extracellular medium, ΔΨm is the mitochondrial inner membrane potential and ΔΨp is the plasma membrane inner membrane potential.
From this equation, we can see that the accumulation of cationic probes is exponentially and equally dependent on mitochondrial and plasma membrane inner membrane potentials (Fig. 2). This means that changes in any of these potentials will alter the accumulation this class of redox-sensitive probes, independently of possible modifications in oxidant production. It also follows that the detection of oxidants will be affected if the probe is present at different quantities.
Fig. 2.
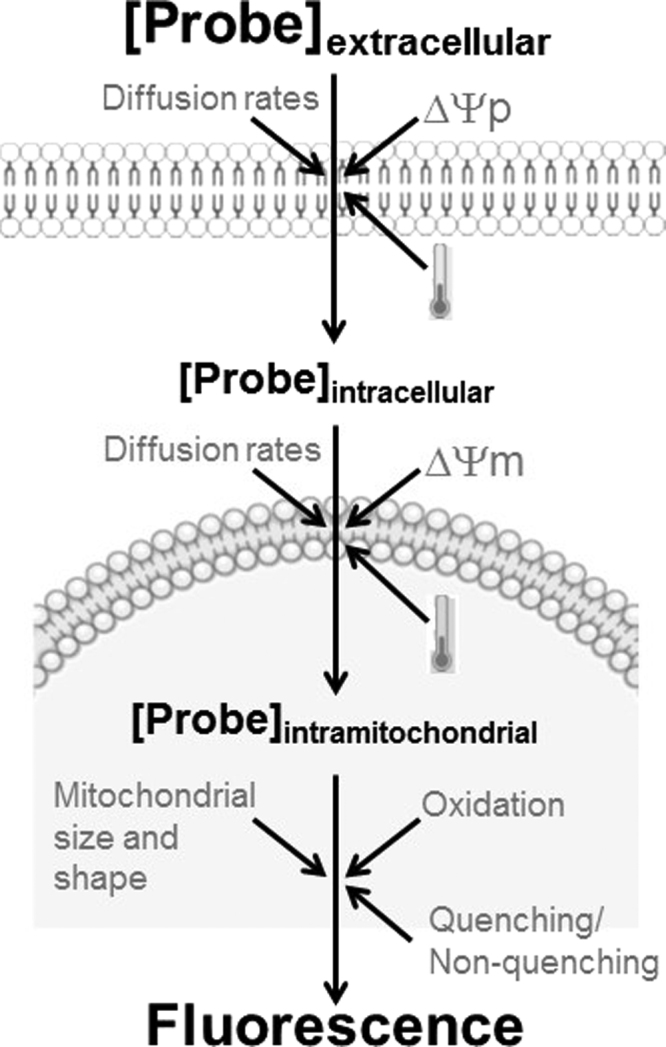
Oxidant-independent conditions that affect the fluorescence of mitochondrially-targeted probes. The accumulation of mitochondrially-targeted probes is dependent on both the plasma membrane (Δψp) and mitochondrial inner membrane (Δψm) potentials, as well as temperature, diffusion rates of the probe through these membranes and incubation time. The fluorescence of the probes within mitochondria depends on probe oxidation, quenching as a result of high intramitochondrial concentrations, and mitochondrial size and shape. Therefore, multiple parameters affect the loading and fluorescence of mitochondrial oxidant probes and should be considered when conducting measurements with these indicators.
Adding to this complication is the fact that fluorescence of these probes is not necessarily proportional to the total quantity of these probes in a cell. When concentrations of these probes are high in a given cellular microenvironment (such as mitochondria), the fluorescence of the oxidized form is decreased relative to that expected for the probe concentration [18], [37]. This loss of fluorescence of the probes when accumulated in mitochondria is known as fluorescence quenching and is possibly related to a spectral shift in absorption and emission spectra and stacking of probe molecules in the mitochondrial microenvironment, reducing their ability to absorb and emit light. Overall, this means the fluorescence of molecules accumulated in mitochondria is often lower than that of the same quantity of the molecule in the cytosol, and that total cell fluorescence can decrease with more mitochondrial uptake of the probe. On the other hand, when low concentrations of probes are used, quenching does not happen, and higher mitochondrial accumulation results in higher cellular fluorescence [6]. It is thus important to know when quenching and non-quenching conditions occur in cells. Particularly for oxidant-sensitive mitochondrial probes, the quenching mode should be avoided since the aim is to measure the fluorescent oxidized version of the probe. For this, concentrations used should as low as possible and titrated for each cell type and condition. Using low probe concentrations also prevents mitochondrial toxicity, such as respiratory chain inhibition, promoted by the probe´s accumulation in mitochondria [57].
Another confounding factor involving cationic mitochondrially-accumulated probes is that fluorescence responses are also affected by mitochondrial size and shape [26]. Mitochondria have been recently found to be highly dynamic, changing size and shape continuously, in a manner altered by many redox-sensitive factors and nutrient availability (reviewed by [27]). As a result, organelle morphology and dynamics should also be considered when using mitochondrially-accumulated oxidant probes (Fig. 2). A suggested control that helps to normalize both for changes in membrane potentials and size and shape is to use an oxidant-insensitive mitochondrially-accumulated probe as a normalizing factor when analyzing whole cell fluorescence (cytometry or microscopy) or mitochondrial fluorescence (confocal localized microscopy). Overall, we suggest this class of probes has too many artifacts associated with them to be used in isolation as evidence of mitochondrial oxidant production (summarized in Fig. 2). While they can provide a useful suggestion of changes in mitochondrial oxidants, other techniques and experimental approaches should be used in association.
Murphy´s group [12], [8] has developed an elegant bypass for the concerns listed above: a mitochondrially-accumulated probe, MitoB, that reacts with H2O2 to form a stable phenol, MitoP. The two forms can be extracted from the tissue or cells and quantified by liquid chromatography-tandem mass spectrometry, providing a snapshot of in vivo mitochondrial H2O2 [28]. While this method is technically far superior to the others discussed, it does not allow for continuous time-scans, which are often desired by researchers.
7. Is measuring mitochondrial oxidants necessary?
While many studies follow fluorescent markers in order to estimate mitochondrial oxidant production or release over time, we saw above that there are several technical limitations that make real-time oxidant measurements using fluorescent indicators untrustworthy, especially when dealing with whole cell, tissue, or animal measurements. Indeed, we would like to urge researchers to consider the actual necessity of this form of measurement in their experimental setup.
First, consider instead the use of a probe isolated from the cells or tissue and quantified at specific time-points such as the western blot or mass spectrometry experimental setups described above. These provide far more dependable data, since they isolate the indicator from the biological setup that generated the oxidants, avoiding artifacts generated by the large complexity inherent to biological systems.
Second, consider if you need a direct measurement of mitochondrial oxidants altogether. You do not always need to measure an oxidant to understand biological phenomena that involve this chemical species. Molecules modified by oxidants are good indictors of their presence, including oxidatively-modified lipids, proteins and nucleic acids in mitochondria, which can undergo specific modifications with specific oxidants. In fact, aconitase, a citric acid cycle enzyme, is sensitive to inactivation by superoxide radicals as well as some reactive nitrogen species [49]. Other small molecule markers in mitochondria are good indicators of redox state and redox balance, such as oxidized versus reduced glutathione levels [13], [30] or NADP+/NADPH [3], [42]. All of these measurements are quantitative and provide solid information regarding the redox state of mitochondria.
In addition to measuring molecules that are modified by the action of oxidants, and effective manner to link mitochondrial oxidants to a biological process is by using mitochondrially-targeted antioxidants to remove these oxidants (reviewed by [57]). Two groups of mitochondrial antioxidants have been extensively tested in this sense: MitoQ, which contains ubiquinone as the antioxidant (reviewed by [33], [35]) and SkQ molecules, which contain plastoquinone (reviewed by [46]). Loss of a biological effect when using these antioxidants is a strong indication that mitochondrial oxidants are involved, although controls should include the mitochondrial target (usually triphenylphosphonium cations) in the absence of the antioxidant and the antioxidant in the absence of the mitochondrial target [1].
Conversely, specifically generating O2•− within mitochondria using mitochondrially-targeted paraquat [40] can identify mitochondrial oxidant-triggered effects. Mitochondrial oxidant production can also be modulated by stimulating or inhibiting the electron transport chain at specific points [48]. Beware that inhibition of electron transport chain activity does not usually inhibit mitochondrial oxidant production. In fact, respiratory inhibition usually enhances oxidant generation by causing accumulation of reduced intermediates capable of promoting monoelectronic oxygen reduction to produce O2•- [51].
Overall, measuring oxidant biomarkers and following the effects of pro- or anti-oxidant molecules can be a solid and less artifact-prone manner to uncover biological effects of mitochondrial oxidants. These strategies should be considered when developing experimental designs.
8. Strategic design in studies involving mitochondrial oxidants
As we saw, most techniques to measure mitochondrial oxidants have significant caveats and are prone to many artifacts, most notably when studies involve cultured cells or tissues. As a result, care must be taken when designing experimental strategies in mitochondrial redox biology.
While fluorescent probes are often used, we stress the importance of the following precautions: (i) Keep in mind that these probes are very rarely specifically oxidized by a single reactive oxygen species. (ii) Do not equate fluorescence increases to a proportional increase in oxidant content – the response curves of these probes are not usually linear. Instead, indicate the data as relative fluorescence. (iii) Use mitochondrially-loaded probes and whole cell fluorescence measurements (cytometry, suspension fluorimetry or whole cell fluorescence microscopy) in the presence of a loading normalizer such as a mitochondrial fluorophore which does not require oxidation to be detected. (iv) Consider that changes in fluorescence may be caused by changes in pH, inner membrane potentials, and/or mitochondrial morphology, independently of changes in oxidant levels. (v) Finally, and most importantly, do not use fluorescent probes to measure oxidants as the only experimental evidence for the presence of these species in the process you are studying.
Indeed, the ideal experimental strategy to study mitochondrial oxidants includes a combination of techniques and methodological approaches, which together bring strength and reliability to the findings:
-
•
Measure oxidants using extracellular probes (for diffusible oxidants) or, ideally, probes that involve quantification after removal from the biological system.
-
•
In addition to (or maybe even instead of) oxidant measurements in the system, use measurements of oxidized products, including biological macromolecules and/or small molecule redox markers such as glutathione.
-
•
Change mitochondrial oxidant levels using mitochondrially-targeted oxidants, antioxidants and/or electron transport modulators and measure the biological outcome of these modifications.
Through the combination of these different approaches, more trustworthy mitochondrial redox results can be obtained in the future, bringing substantial new mechanistic insights into the roles of mitochondrial oxidants.
Acknowledgements
Prof. Kowaltowski is supported by the Centro de Pesquisa, Inovação e Difusão de Processos Redox em Biomedicina (Redoxoma) Grant 13/07937-8, from the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Conselho Nacional de Pesquisa (CNPq), and Coordenação de Aperfeiçoamento do Pessoal do Ensino Superior (CAPES) Finance Code 001 and PROEX 1888/2016. The author wishes to thank Prof. Ohara Augusto, Dr. Phablo Abreu and Pâmela Kakimoto for critical reading of the manuscript.
References
- 1.Adlam V.J., Harrison J.C., Porteous C.M., James A.M., Smith R.A., Murphy M.P., Sammut I.A. Targeting an antioxidant to mitochondria decreases cardiac ischemia-reperfusion injury. FASEB J. 2005;19:1088–1095. doi: 10.1096/fj.05-3718com. [DOI] [PubMed] [Google Scholar]
- 2.Balaban R.S., Nemoto S., Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Blacker T.S., Duchen M.R. Investigating mitochondrial redox state using NADH and NADPH autofluorescence. Free Radic. Biol. Med. 2016;100:53–65. doi: 10.1016/j.freeradbiomed.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonini M.G., Rota C., Tomasi A., Mason R.P. The oxidation of 2′,7′-dichlorofluorescin to reactive oxygen species: a self-fulfilling prophesy? Free Radic. Biol. Med. 2006;40:968–975. doi: 10.1016/j.freeradbiomed.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 5.Bourdi M., Davies J.S., Pohl L.R. Mispairing C57BL/6 substrains of genetically engineered mice and wild-type controls can lead to confounding results as it did in studies of JNK2 in acetaminophen and concanavalin A liver injury. Chem. Res. Toxicol. 2011;24:794–796. doi: 10.1021/tx200143x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brand M.D., Nicholls D.G. Assessing mitochondrial dysfunction in cells. Biochem. J. 2011;435:297–312. doi: 10.1042/BJ20110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Budd S.L., Castilho R.F., Nicholls D.G. Mitochondrial membrane potential and hydroethidine-monitored superoxide generation in cultured cerebellar granule cells. FEBS Lett. 1997;415:21–24. doi: 10.1016/s0014-5793(97)01088-0. [DOI] [PubMed] [Google Scholar]
- 8.Cairns A.G., McQuaker S.J., Murphy M.P., Hartley R.C. Targeting mitochondria with small molecules: the preparation of MitoB and MitoP as exomarkers of mitochondrial hydrogen peroxide. Methods Mol. Biol. 2015;1265:25–50. doi: 10.1007/978-1-4939-2288-8_3. [DOI] [PubMed] [Google Scholar]
- 9.Cardoso A.R., Chausse B., da Cunha F.M., Luévano-Martínez L.A., Marazzi T.B., Pessoa P.S., Queliconi B.B., Kowaltowski A.J. Mitochondrial compartmentalization of redox processes. Free Radic. Biol. Med. 2012;52:2201–2208. doi: 10.1016/j.freeradbiomed.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 10.Cerqueira F.M., Kowaltowski A.J. Commonly adopted caloric restriction protocols often involve malnutrition. Ageing Res. Rev. 2010;9:424–430. doi: 10.1016/j.arr.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Cerqueira F.M., Kowaltowski A.J. Mitochondrial metabolism in aging: effect of dietary interventions. Ageing Res. Rev. 2013;12:22–28. doi: 10.1016/j.arr.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 12.Cochemé H.M., Quin C., McQuaker S.J., Cabreiro F., Logan A., Prime T.A., Abakumova I., Patel J.V., Fearnley I.M., James A.M., Porteous C.M., Smith R.A., Saeed S., Carré J.E., Singer M., Gems D., Hartley R.C., Partridge L., Murphy M.P. Measurement of H2O2 within living Drosophila during aging using a ratiometric mass spectrometry probe targeted to the mitochondrial matrix. Cell Metab. 2011;13:340–350. doi: 10.1016/j.cmet.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de la Asuncion J.G., Millan A., Pla R., Bruseghini L., Esteras A., Pallardo F.V., Sastre J., Viña J. Mitochondrial glutathione oxidation correlates with age-associated oxidative damage to mitochondrial DNA. FASEB J. 1996;10:333–338. doi: 10.1096/fasebj.10.2.8641567. [DOI] [PubMed] [Google Scholar]
- 14.Dębski D., Smulik R., Zielonka J., Michałowski B., Jakubowska M., Dębowska K., Adamus J., Marcinek A., Kalyanaraman B., Sikora A. Mechanism of oxidative conversion of Amplex® Red to resorufin: pulse radiolysis and enzymatic studies. Free Radic. Biol. Med. 2016;95:323–332. doi: 10.1016/j.freeradbiomed.2016.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dickinson B.C., Chang C.J. A targetable fluorescent probe for imaging hydrogen peroxide in the mitochondria of living cells. J. Am. Chem. Soc. 2008. 2008;130:9638–9639. doi: 10.1021/ja802355u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickinson B.C., Srikun D., Chang C.J. Mitochondrial-targeted fluorescent probes for reactive oxygen species. Curr. Opin. Chem. Biol. 2010;14:50–56. doi: 10.1016/j.cbpa.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dickinson B.C., Lin V.S., Chang C.J. Preparation and use of MitoPY1 for imaging hydrogen peroxide in mitochondria of live cells. Nat. Protoc. 2013;8:1249–1259. doi: 10.1038/nprot.2013.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emaus R.K., Grunwald R., Lemasters J.J. Rhodamine 123 as a probe of transmembrane potential in isolated rat-liver mitochondria: spectral and metabolic properties. Biochim. Biophys. Acta. 1986;850:436–448. doi: 10.1016/0005-2728(86)90112-x. [DOI] [PubMed] [Google Scholar]
- 19.Fernandes D.C., Gonçalves R.C., Laurindo F.R. Measurement of superoxide production and NADPH oxidase activity by HPLC analysis of dihydroethidium oxidation. Methods Mol. Biol. 2017;1527:233–249. doi: 10.1007/978-1-4939-6625-7_19. [DOI] [PubMed] [Google Scholar]
- 20.Figueira T.R., Barros M.H., Camargo A.A., Castilho R.F., Ferreira J.C., Kowaltowski A.J., Sluse F.E., Souza-Pinto N.C., Vercesi A.E. Mitochondria as a source of reactive oxygen and nitrogen species: from molecular mechanisms to human health. Antioxid. Redox Signal. 2013;18:2029–2074. doi: 10.1089/ars.2012.4729. [DOI] [PubMed] [Google Scholar]
- 21.Fleury C., Mignotte B., Vayssière J.L. Mitochondrial reactive oxygen species in cell death signaling. Biochimie. 2002;84:131–141. doi: 10.1016/s0300-9084(02)01369-x. [DOI] [PubMed] [Google Scholar]
- 22.Hoffman D.L., Brookes P.S. Oxygen sensitivity of mitochondrial reactive oxygen species generation depends on metabolic conditions. J. Biol. Chem. 2009;284:16236–16245. doi: 10.1074/jbc.M809512200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu J., Dong L., Outten C.E. The redox environment in the mitochondrial intermembrane space is maintained separately from the cytosol and matrix. J. Biol. Chem. 2008;283:29126–29134. doi: 10.1074/jbc.M803028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kakimoto P.A., Kowaltowski A.J. Effects of high fat diets on rodent liver bioenergetics and oxidative imbalance. Redox Biol. 2016;8:216–225. doi: 10.1016/j.redox.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalyanaraman B., Darley-Usmar V., Davies K.J., Dennery P.A., Forman H.J., Grisham M.B., Mann G.E., Moore K., Roberts L.J., 2nd, Ischiropoulos H. Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Radic. Biol. Med. 2012;52:1–6. doi: 10.1016/j.freeradbiomed.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kowaltowski A.J., Cosso R.G., Campos C.B., Fiskum G. Effect of Bcl-2 overexpression on mitochondrial structure and function. J. Biol. Chem. 2002;277:42802–42807. doi: 10.1074/jbc.M207765200. [DOI] [PubMed] [Google Scholar]
- 27.Liesa M., Shirihai O.S. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 2013;17:491–506. doi: 10.1016/j.cmet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Logan A., Shabalina I.G., Prime T.A., Rogatti S., Kalinovich A.V., Hartley R.C., Budd R.C., Cannon B., Murphy M.P. In vivo levels of mitochondrial hydrogen peroxide increase with age in mtDNA mutator mice. Aging Cell. 2014;13:765–768. doi: 10.1111/acel.12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopert P., Patel M. Nicotinamide nucleotide transhydrogenase (Nnt) links the substrate requirement in brain mitochondria for hydrogen peroxide removal to the thioredoxin/peroxiredoxin (Trx/Prx) system. J. Biol. Chem. 2014;289:15611–15620. doi: 10.1074/jbc.M113.533653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marí M., Morales A., Colell A., García-Ruiz C., Fernández-Checa J.C. Mitochondrial glutathione, a key survival antioxidant. Antioxid. Redox Signal. 2009;11:2685–2700. doi: 10.1089/ars.2009.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer A.J., Dick T.P. Fluorescent protein-based redox probes. Antioxid. Redox Signal. 2010;13:621–650. doi: 10.1089/ars.2009.2948. [DOI] [PubMed] [Google Scholar]
- 32.Miller E.W., Albers A.E., Pralle A., Isacoff E.Y., Chang C.J. Boronate-based fluorescent probes for imaging cellular hydrogen peroxide. J. Am. Chem. Soc. 2005;127:16652–16659. doi: 10.1021/ja054474f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy M.P., Smith R.A. Targeting antioxidants to mitochondria by conjugation to lipophilic cations. Ann. Rev. Pharmacol. Toxicol. 2007;47:629–656. doi: 10.1146/annurev.pharmtox.47.120505.105110. [DOI] [PubMed] [Google Scholar]
- 34.Murphy M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy M.P. Understanding and preventing mitochondrial oxidative damage. Biochem. Soc. Trans. 2016;44:1219–1226. doi: 10.1042/BST20160108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Navarro C.D.C., Figueira T.R., Francisco A., Dal'Bó G.A., Ronchi J.A., Rovani J.C., Escanhoela C.A.F., Oliveira H.C.F., Castilho R.F., Vercesi A.E. Redox imbalance due to the loss of mitochondrial NAD(P)-transhydrogenase markedly aggravates high fat diet-induced fatty liver disease in mice. Free Radic. Biol. Med. 2017;113:190–202. doi: 10.1016/j.freeradbiomed.2017.09.026. [DOI] [PubMed] [Google Scholar]
- 37.Nicholls D.G., Ward M.W. Mitochondrial membrane potential and neuronal glutamate excitotoxicity: mortality and millivolts. Trends Neurosci. 2000;23:166–174. doi: 10.1016/s0166-2236(99)01534-9. [DOI] [PubMed] [Google Scholar]
- 38.Okado-Matsumoto A., Fridovich I. Subcellular distribution of superoxide dismutases (SOD) in rat liver: cu,zn-sod in mitochondria. J. Biol. Chem. 2001;276:38388–38393. doi: 10.1074/jbc.M105395200. [DOI] [PubMed] [Google Scholar]
- 39.Remington S.J. Fluorescent proteins: maturation, photochemistry and photophysics. Curr. Opin. Struct. Biol. 2006;16:714–721. doi: 10.1016/j.sbi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 40.Robb E.L., Gawel J.M., Aksentijević D., Cochemé H.M., Stewart T.S., Shchepinova M.M., Qiang H., Prime T.A., Bright T.P., James A.M., Shattock M.J., Senn H.M., Hartley R.C., Murphy M.P. Selective superoxide generation within mitochondria by the targeted redox cycler MitoParaquat. Free Radic. Biol. Med. 2015;89:883–894. doi: 10.1016/j.freeradbiomed.2015.08.021. [DOI] [PubMed] [Google Scholar]
- 41.Roma L.P., Duprez J., Takahashi H.K., Gilon P., Wiederkehr A., Jonas J.C. Dynamic measurements of mitochondrial hydrogen peroxide concentration and glutathione redox state in rat pancreatic β-cells using ratiometric fluorescent proteins: confounding effects of pH with HyPer but not roGFP1. Biochem. J. 2012;441:971–978. doi: 10.1042/BJ20111770. [DOI] [PubMed] [Google Scholar]
- 42.Ronchi J.A., Figueira T.R., Ravagnani F.G., Oliveira H.C., Vercesi A.E., Castilho R.F. A spontaneous mutation in the nicotinamide nucleotide transhydrogenase gene of C57BL/6J mice results in mitochondrial redox abnormalities. Free Radic. Biol. Med. 2013;63:446–456. doi: 10.1016/j.freeradbiomed.2013.05.049. [DOI] [PubMed] [Google Scholar]
- 43.Ronot X., Benel L., Adolphe M., Mounolou J.C. Mitochondrial analysis in living cells: the use of rhodamine 123 and flow cytometry. Biol. Cell. 1986;57:1–7. doi: 10.1111/j.1768-322x.1986.tb00458.x. [DOI] [PubMed] [Google Scholar]
- 44.Schwarzländer M., Wagner S., Ermakova Y.G., Belousov V.V., Radi R., Beckman J.S., Buettner G.R., Demaurex N., Duchen M.R., Forman H.J., Fricker M.D., Gems D., Halestrap A.P., Halliwell B., Jakob U., Johnston I.G., Jones N.S., Logan D.C., Morgan B., Müller F.L., Nicholls D.G., Remington S.J., Schumacker P.T., Winterbourn C.C., Sweetlove L.J., Meyer A.J., Dick T.P., Murphy M.P. The 'mitoflash' probe cpYFP does not respond to superoxide. Nature. 2014;514:E12–E14. doi: 10.1038/nature13858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sena L.A., Chandel N.S. Physiological roles of mitochondrial reactive oxygen species. Mol. Cell. 2012;48:158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skulachev V.P. Cationic antioxidants as a powerful tool against mitochondrial oxidative stress. Biochem. Biophys. Res. Commun. 2013;441:275–279. doi: 10.1016/j.bbrc.2013.10.063. [DOI] [PubMed] [Google Scholar]
- 47.Swerdlow R.H., E L, Aires D., Lu J. Glycolysis-respiration relationships in a neuroblastoma cell line. Biochim. Biophys. Acta. 2013;1830:2891–2898. doi: 10.1016/j.bbagen.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tahara E.B., Navarete F.D., Kowaltowski A.J. Tissue-, substrate-, and site-specific characteristics of mitochondrial reactive oxygen species generation. Free Radic. Biol. Med. 2009;46:1283–1297. doi: 10.1016/j.freeradbiomed.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 49.Tórtora V., Quijano C., Freeman B., Radi R., Castro L. Mitochondrial aconitase reaction with nitric oxide, S-nitrosoglutathione, and peroxynitrite: mechanisms and relative contributions to aconitase inactivation. Free Radic. Biol. Med. 2007;42:1075–1088. doi: 10.1016/j.freeradbiomed.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 50.Toye A.A., Lippiat J.D., Proks P., Shimomura K., Bentley L., Hugill A., Mijat V., Goldsworthy M., Moir L., Haynes A., Quarterman J., Freeman H.C., Ashcroft F.M., Cox R.D. A genetic and physiological study of impaired glucose homeostasis control in C57BL/6J mice. Diabetologia. 2005;48:675–686. doi: 10.1007/s00125-005-1680-z. [DOI] [PubMed] [Google Scholar]
- 51.Turrens J.F. Mitochondrial formation of reactive oxygen species. J. Physiol. 2003;552:335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Laer K., Dick T.P. Utilizing natural and engineered peroxiredoxins as intracellular peroxide reporters. Mol. Cells. 2016;39:46–52. doi: 10.14348/molcells.2016.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vasquez-Vivar J., Kalyanaraman B., Kennedy M.C. Mitochondrial aconitase is a source of hydroxyl radical. An electron spin resonance investigation. Reson. Investig. J. Biol. Chem. 2000;275:14064–14069. doi: 10.1074/jbc.275.19.14064. [DOI] [PubMed] [Google Scholar]
- 54.Wrona M., Wardman P. Properties of the radical intermediate obtained on oxidation of 2′,7′-dichlorodihydrofluorescein, a probe for oxidative stress. Free Radic. Biol. Med. 2006;41:657–667. doi: 10.1016/j.freeradbiomed.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 55.Zhao B., Ranguelova K., Jiang J., Mason R.P. Studies on the photosensitized reduction of resorufin and implications for the detection of oxidative stress with Amplex Red. Free Radic. Biol. Med. 2011;51:153–159. doi: 10.1016/j.freeradbiomed.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou M., Diwu Z., Panchuk-Voloshina N., Haugland R.P. A stable nonfluorescent derivative of resorufin for the fluorometric determination of trace hydrogen peroxide: applications in detecting the activity of phagocyte NADPH oxidase and other oxidases. Anal. Biochem. 1997;253:162–168. doi: 10.1006/abio.1997.2391. [DOI] [PubMed] [Google Scholar]
- 57.Zielonka J., Joseph J., Sikora A., Hardy M., Ouari O., Vasquez-Vivar J., Cheng G., Lopez M., Kalyanaraman B. Mitochondria-targeted triphenylphosphonium-based compounds: syntheses, mechanisms of action, and therapeutic and diagnostic applications. Chem. Rev. 2017;117:10043–10120. doi: 10.1021/acs.chemrev.7b00042. [DOI] [PMC free article] [PubMed] [Google Scholar]