HIV diagnostics have played a central role in the remarkable progress in identifying, staging, initiating, and monitoring infected individuals on life-saving antiretroviral therapy. They are also useful in surveillance and outbreak responses, allowing for assessment of disease burden and identification of vulnerable populations and transmission “hot spots,” thus enabling planning, appropriate interventions, and allocation of appropriate funding.
KEYWORDS: CD4, enzyme immunoassay, HIV incidence, HIV rapid tests, dried blood spots, drug resistance, early infant diagnosis, point-of-care testing, quality assurance, viral load
SUMMARY
HIV diagnostics have played a central role in the remarkable progress in identifying, staging, initiating, and monitoring infected individuals on life-saving antiretroviral therapy. They are also useful in surveillance and outbreak responses, allowing for assessment of disease burden and identification of vulnerable populations and transmission “hot spots,” thus enabling planning, appropriate interventions, and allocation of appropriate funding. HIV diagnostics are critical in achieving epidemic control and require a hybrid of conventional laboratory-based diagnostic tests and new technologies, including point-of-care (POC) testing, to expand coverage, increase access, and positively impact patient management. In this review, we provide (i) a historical perspective on the evolution of HIV diagnostics (serologic and molecular) and their interplay with WHO normative guidelines, (ii) a description of the role of conventional and POC testing within the tiered laboratory diagnostic network, (iii) information on the evaluations and selection of appropriate diagnostics, (iv) a description of the quality management systems needed to ensure reliability of testing, and (v) strategies to increase access while reducing the time to return results to patients. Maintaining the central role of HIV diagnostics in programs requires periodic monitoring and optimization with quality assurance in order to inform adjustments or alignment to achieve epidemic control.
INTRODUCTION
The Joint United Nations Program on HIV/AIDS (UNAIDS) estimated that there were 36.7 million HIV-infected persons and 1.8 million new infections globally by the end of 2016 (1). The sub-Saharan Africa (SSA) region alone accounted for approximately 69.5% (25.5 million) of the global HIV infections, with only 54.1% (13.8 million) having access to antiretroviral (ARV) therapy (ART) (2, 3) to suppress viral replication, prevent opportunistic infections, and prolong the lives of people living with HIV/AIDS (PLHIV). As part of the strategy to achieve an “AIDS-free generation” (4, 5), the 2016 World Health Organization (WHO) HIV treatment guidelines recommend that ART be initiated in all individuals living with HIV regardless of the clinical stage or CD4 cell count in order to preserve patients’ immune systems, control HIV replication, and reduce further transmission (6). HIV prevention and the use of ART have reduced new HIV infections by 14%, from 2.1 million in 2013 to 1.8 million in 2016 (3, 7). However, at this rate, the decline will still fall short of the United Nations’ set target of <500,000 new infections globally by 2020 (8). To accelerate the attainment of this goal, UNAIDS set ambitious 90-90-90 targets to be achieved by 2020, with the first 90 defined as 90% of HIV-infected persons knowing their status, the second 90 defined as 90% of patients with a diagnosis of HIV infection receiving ART, and the third 90 defined as 90% of ART-treated patients having viral suppression (9, 10). There are challenges in reaching these targets for developing countries, which include government policies, financial provision, data-driven programming, operational coordination among health care agencies and implementing partners, and laboratory infrastructure (11, 12). However, recent U.S. President’s Emergency Plan for AIDS Relief (PEPFAR)-sponsored population-based HIV impact assessments have demonstrated that these targets are achievable (13). The first and third 90 targets specifically require the delivery of high-quality diagnostic testing to identify infections, monitor the effectiveness of ART, and provide timely assessment of HIV/AIDS control and the emergence of drug resistance (DR) at both the individual and population levels.
Currently, serologic testing algorithms using mostly rapid tests is used in resource-limited settings (RLS) to diagnose HIV infections. Serology-based incidence assays are widely used for estimating the rate of new infections in cross-sectional surveys, combined with viral load (VL) measurement in an algorithm. The new rapid incidence assay has generated significant excitement because it may provide an opportunity to identify hot spots in real time when used in program settings. Appropriate deployment of simple point-of-care (POC) CD4 technologies to provide same-day results to patients has had a significant impact on enrolling and retaining patients in ART programs (14, 15). Similarly, ART programs have implemented VL determination technologies in centralized and decentralized facilities to assess the effectiveness of ART. Furthermore, dried blood spot (DBS) and whole-blood-based POC methodologies have performed well in the area of early infant diagnosis (EID). Because of the rapid scale-up of ART (16), transmitted drug resistance (TDR) is on the rise in RLS. As such, testing for DR to determine the occurrence of mutations in order to provide patients with optimal treatment without an unnecessary switch to second-line and perhaps third-line ART regimens with higher costs is critical.
Accurate diagnosis of HIV infection is key and represents the entry point of infected patients into the treatment cascade. Furthermore, it has taken on renewed importance in the era of “test and treat” with ART (17). WHO guidelines for HIV testing services (HTS) underscored the importance of quality management systems. With the push to expand HIV testing to achieve the UNAIDS 90-90-90 targets, it is both programmatically and ethically imperative, and a priority for ministries of health (MOH) and national AIDS control programs, to implement robust quality management systems to support the implementation and delivery of accurate testing results to everyone tested. In this report, we review various aspects of HIV diagnostics, including advances in testing technologies to improve diagnosis and expand access, with a focus on RLS. Furthermore, we examine strategies to employ new methods and to critically assess/evaluate and support quality management systems for accurate and reliable diagnosis.
STAGES OF HIV INFECTION
There are three main stages following HIV infection in an untreated individual, characterized by clinical symptoms and biological markers that also offer the opportunity for use in diagnosis and monitoring using laboratory testing. The first stage is the acute phase, characterized by rapid multiplication and spread of the virus in the body, which may take about 2 to 4 weeks following infection (18). During this stage, there is a burst of viral replication, with shedding and peaking of p24 antigen (Ag) in blood. During the acute stage, some people experience flu-like symptoms, such as headache, fever, and rashes, for several weeks (19). The second stage is the chronic or asymptomatic stage, during which the virus continues to multiply but at low levels, and the infected individual may not experience any clinical symptoms. The host immune system also starts producing antibodies (Ab), which coincides with a decline in the VL to a steady state. Also, there is a decline in p24 antigen levels as the VL drops, due to p24 antigen bound by antibodies to form an antibody-p24 antigen complex, thereby reducing the level of free p24 antigen in blood. The period from infection to the appearance of Ab (seroconversion) is known as the “window period.” If a patient remains untreated, as viral replication continues, CD4 cells, which serve as host target cells for viral replication, are gradually destroyed, leading to a decline of CD4 cell numbers (18). The third stage, the AIDS phase, with continual viral replication and depletion of CD4 cells, leads to a weakened host immune system. The final stage is characterized by opportunistic infections and other clinical symptoms (20).
The biological markers HIV RNA, p24 antigen, HIV antibodies, and CD4 cells, appearing at various stages following infection, have been exploited for laboratory diagnostics for HIV for various applications, including (i) the determination of an individual’s serostatus (antibodies), (ii) distinguishing of recent from long-term infection (antibodies, p24 antigen, and VL), (iii) EID using RNA and DNA, (iv) staging and monitoring of disease progression (CD4), (v) identification and monitoring of treatment effectiveness or failure (VL), and (vi) identification of DR mutations (DRMs) for a specific ART regimen failure (RNA and DNA).
LABORATORY DIAGNOSTIC NETWORK
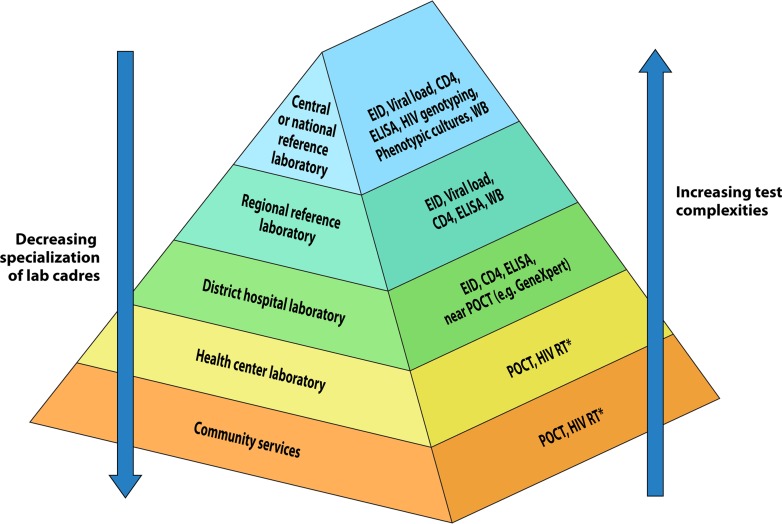
The laboratory diagnostic network is key in each country’s health care delivery services in order to improve the health of its population. The laboratory diagnostic network is typically organized in a tiered structure that aligns with the different layers of health care delivery (e.g., referral hospitals, regional hospitals, district hospitals, and health centers). The tiered laboratory network is a hierarchical structure, with the national reference or central laboratory at the top of the structure, followed by regional/provincial, district, and health center laboratories. This tiered structure matches the level of test complexities that can be carried out at each level, with complex and advanced tests usually being reserved for national and regional laboratories (Fig. 1). Similarly, training and the levels of specialized laboratory cadres align with this structure. At the community level, lay counselors and community health care workers primarily perform POC testing. POC testing is an extension of the tiered laboratory diagnostic network to the community where facility-based laboratories are typically absent. Coordination and oversight from the national reference laboratory are crucial to ensure standardized training of the different cadres within the tiered laboratory network. Furthermore, evaluations or verification of new diagnostics, including POC testing, is usually performed at the national reference laboratory prior to their placement within the laboratory diagnostic network. Also, the national reference laboratory should provide oversight and monitoring of quality assurance (QA) testing within the diagnostic network.
FIG 1.
The tiered laboratory diagnostic network showing the different laboratory tiers, community services, and tests performed at each tier. EID, early infant diagnosis; POCT, point-of-care testing (instrument based). *, HIV RT is the HIV rapid test and refers to strip-like devices.
ADVANCES IN SEROLOGIC DIAGNOSIS
AIDS was first recognized in the early 1980s. Initially, simple serologic tests for HIV antibodies using culture-derived viral antigen preparations were developed to diagnose HIV infections and to safeguard blood and blood product supplies (21). Over the next 3 decades, a wide spectrum of serologic assays was developed for simple/rapid testing (22, 23), high-throughput screening (24, 25), supplementary confirmation (26), epidemiological surveillance (27–29), and incidence determination (30–33). Assays with different testing formats, antigen designs, and signal detection amplification chemistries were formulated and evaluated in the field to assess their testing accuracy, viral strain coverage, and field applicability to ensure that infected patients could be quickly and accurately identified and linked to health care services, with minimal loss to follow-up. The progress in the development, application, and benefit of these serologic methodologies has been extraordinary, as evidenced by the identification of HIV-contaminated blood from blood supplies in the developed world (34), reduced testing times for rapid tests and supplemental confirmatory tests, and their direct impact (e.g., early ART initiation and reduced loss to follow-up) on improving health care delivery in RLSs.
Generations of Enzyme Immunoassays
Since the mid-1980s, there have been five generations of enzyme immunoassays (EIAs) using different antigen preparations and detection chemistries to provide accurate screening for blood banks and centralized laboratories with a high specimen volume (Fig. 2). The first-generation assays used antigens derived from whole viral lysates from HIV-positive cultures for the detection of IgG antibodies. Due to the presence of impurities in the crude antigen lysate preparations, these assays had relative low specificity and high false positivity. Later confirmatory tests, such as immunofluorescence assays or Western blotting (WB), with high specificity were introduced to eliminate false positivity (35–37). In the second-generation assays, synthetic peptides or recombinant proteins derived from the immunodominant regions (IDR) of HIV-1 proteins and gp36 of HIV-2 were used to increase sensitivity and reduce false positivity. The third-generation assays, such as the Genetic Systems HIV-1/HIV-2 Plus O EIA, used a sandwich format and a variety of antigens to capture HIV-1 and -2 antibodies in serum. These antigens included recombinant p24, gp160 derived from HIV-1 group M, a recombinant peptide from HIV-2 gp36 IDR, and a synthetic peptide from HIV-1 group O (38). In addition to IgG, the third-generation assays also detected early HIV-1 IgM antibodies and further reduced the window period. The fourth-generation EIAs, such as the Abbott Architect HIV Ag/Ab Combo assay, used fully automated chemiluminescent microparticle technology to simultaneously detect HIV-1 p24 antigen and antibodies to HIV-1 (groups M, N, and O) and HIV-2. The detection of p24 antigen by the Abbott Architect HIV Ag/Ab Combo assay shortens the window period and increases the chances of early detection of HIV infection. The detection instrument provides random-access capability so that specimens can be tested on arrival without delays and generates results in 30 min with a throughput of >150 tests per h. These assays are best suited for facilities handling high volumes of blood bank screening tests (39–41). However, in these tests, the p24 antigen and HIV-1/2 antibodies detected are not individually distinguished. Fifth-generation EIAs, such as the Bio-Rad BioPlex 2200 HIV Ag-Ab assay, used multiple sets of magnetic beads coated with p24 monoclonal antibodies and epitopes specific for HIV-1 (groups M, N, and O) and HIV-2 (42). Acutely infected persons with p24 antigen in the window period can be identified in a single test and can be referred specifically for early intervention to further prevent HIV transmission. Individuals infected with HIV-1 or HIV-2 can also be identified for quick confirmation and linked to HIV-1- or HIV-2-specific ART.
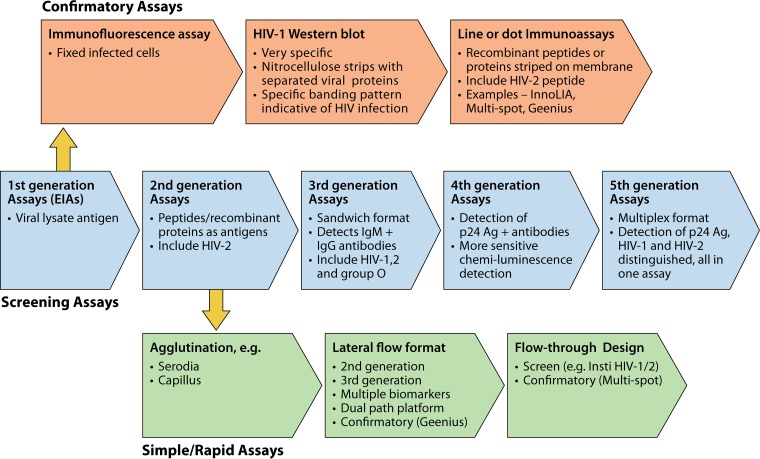
FIG 2.
Historical evolution of serologic assays for HIV diagnosis. Shown are five generations of screening assays using an EIA format for high-throughput processing. Supplemental assays for confirmation of infection used immunofluorescence, WB, and, more recently, simple line or dot immunoassays. Rapid assays for POC testing were initially agglutination tests and later of a lateral flow format and flowthrough design.
Supplemental Confirmatory Tests
Despite progress with the operational characteristics and high sensitivity of EIA-based screening technologies, challenges with false positivity persisted (39–41). As such, for diagnosis of HIV infection, supplemental testing of an initially EIA-reactive specimen was required for confirmation of HIV infection prior to initiating patients on ART (Fig. 2). Specimens that were reactive by an EIA were retested using a more specific supplemental test. A common supplemental test used was WB. A WB assay was performed using a gradient-purified HIV lysate electrophoresed and impregnated onto nitrocellulose strips (Fig. 3A). For a specimen run on a WB to be considered positive, it had to contain antibodies reactive to p24 antigen and one or more antibodies against the envelope glycoproteins (gp160/gp120 and/or gp41). The WB banding patterns varied depending on the extents of antigen glycosylation and antibody maturation.
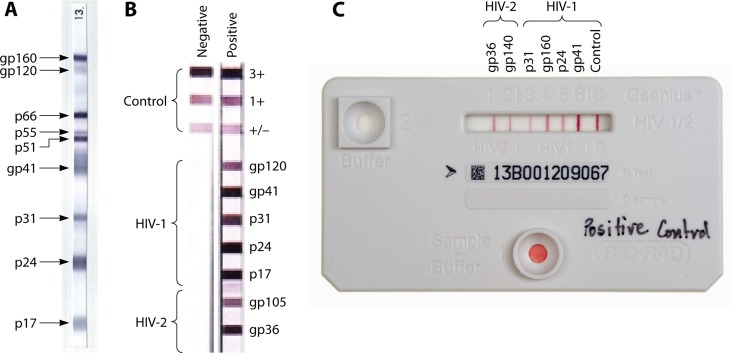
FIG 3.
Illustration of three supplemental confirmatory assays, Western blotting (WB) (A), the Inno-LIA line blot (B), and the Geenius cartridge (C). WB uses gradient-purified viral lysates as separated antigens impregnated onto nitrocellulose strips. The lateral flow Inno-LIA uses combinations of five and two synthetic or recombinant antigens from HIV-1 and -2, respectively, and the Geenius assay with the dual-path platform (DPP) uses four and two synthetic or recombinant antigens for HIV-1 and -2, respectively. Three and one internal control bands are included in the Inno-LIA and Geenius assay formats, respectively. The banding patterns of a negative specimen and an HIV-1 and -2 dually positive specimen for Inno-LIA is shown in panel B, and the banding pattern of a dually positive specimen for Geenius is shown in panel C.
In general, the WB banding pattern is complex, and its interpretation requires well-trained, experienced, and competent laboratory staff. To avoid misinterpretation, new line blots that used purified HIV-1 and HIV-2 recombinant proteins and peptides were developed (Fig. 3B). The Inno-LIA HIV-1/2 strip (43) contains three positive-control lines (strong, moderate, and weak), five HIV-1 antigen lines (gp120, gp41, p31, p24, and p17), and two HIV-2 antigen lines (gp105 and gp36). Optimal clarity of banding is often produced after overnight incubation. The test cartridge of the Bio-Rad Multispot test contains only four spots, a procedural control, HIV-1 recombinant gp41 protein, HIV-1 gp41 peptides, and HIV-2 gp36 peptides, to simplify interpretation of results. In 2013, the U.S. Centers for Disease Control and Prevention (CDC) proposed the use of the Multispot test on samples that were reactive with third- or fourth-generation EIAs in order to confirm and differentiate HIV-1 and HIV-2 infections (44). Bio-Rad recently developed the Geenius HIV-1/2 supplemental assay to replace the Multispot assay (43, 45, 46). The new Geenius assay uses a closed lateral flow cartridge with a dual-path platform (DPP) to detect antibodies to recombinant or synthetic peptides for HIV-1 (p41, gp160, p24, and p31) and HIV-2 (gp36 and gp140) (Fig. 3C). It has a simple 3-step procedure with a total testing time of 30 min. Interpretation can also be made with the aid of an automated Geenius reader.
HIV RAPID TESTING
Development of Rapid Tests
The first HIV rapid test assays to be developed were the MicroGenesys agglutination test and the Abbott Murex single-use diagnostic system. A primary concern of these early HIV rapid tests was the considerably high rates of false-positive results (47) and false-negative results (48). The subsequent use of different formats, such as lateral flow devices (Determine HIV-1/2, Unigold, StatPak, and OraQuick HIV-1/2), flowthrough cartridges (Insti HIV-1/2), and modified agglutination assays (Serodia and Capillus HIV-1/2), greatly simplified and improved the testing procedure, with increased sensitivity and specificity. Rapid testing is usually completed in about 20 to 30 min, thus making them ideal for testing and counseling in primary health care sites and mobile clinics. The Insti HIV-1/HIV-2 test requires only 1 min, with an accuracy comparable to those of other rapid tests (49). Nonlaboratory staff can perform most rapid assays with standard training (with a minimum set of competencies as defined by the national program) using finger-pricked blood, plasma, or serum (22, 50–54). Assays such as the OraQuick Advance rapid HIV-1/2 assay use oral fluid, an alternative specimen type to finger-pricked whole blood, to provide the HIV serostatus of an individual.
In 2017, more than 85 million people were tested in PEPFAR-supported countries alone using mostly HIV rapid tests, and the majority of tests were performed by lay counselors or nurses (55). Although rapid tests, in general, are slightly less sensitive than EIAs because of the shorter duration of antigen/antibody incubation, their specificity is higher than that of EIAs (56). In RLS, HIV testing algorithms include a combination of two or three rapid tests, usually in a serial algorithm, depending on HIV prevalence; this has produced suitable sensitivities and specificities and improved the accuracy of testing results in POC settings (57). Countries decide on a testing algorithm, serial or parallel, to apply to their programs and use the right combination of HIV rapid tests to improve testing accuracy. To reduce costs, a serial algorithm is often adopted, with specimens initially reactive by the first test being subjected to a second test (a different rapid test) with higher specificity. The quick turnaround time (TAT) of less than an hour allows the provision of counseling and testing during the same visit. Together with other HIV POC testing for VL, EID, and CD4, health care providers at the district or community level can provide multiple testing and treatment services to patients during same-day visits (49, 58, 59). Achieving this requires comprehensive implementation of national policies, resources, training, education, health care infrastructure, and strategic partnerships with stakeholders.
Decentralization of HIV Rapid Testing To Increase Access
HIV rapid tests became available in the early 1990s to fulfill the needs to promptly determine the serostatus of persons prior to surgical operations, organ transplantation, and maternal labor/delivery. Simple rapid tests offered the advantage that nonlaboratory staff in primary health care centers could also provide diagnostic services to patients residing in remote areas in RLS. The decentralization of HIV diagnostic services with POC testing has been transformative, as this has significantly increased access to services; however, it also posed huge implementation challenges with the sheer scale involved. There are constraints related to decentralization, including the lack of national guidelines, training, waste disposal, human resources, cold chains, inventory management, and QA monitoring (Table 1). With a high burden of HIV in many RLS, it is critical that the population have access to HIV diagnostic testing and other related services. Recent data demonstrate an increased risk of acquiring HIV for women during pregnancy and postpartum periods; therefore, it is critical to establish a retesting policy and ensure that resources are in place to allow the identification of recently acquired HIV infections in women during pregnancy and breastfeeding periods (60). Decentralization of services, including the use of POC testing for CD4, VL, and EID, further increases the opportunity to achieve the UNAIDS 90-90-90 goals.
TABLE 1.
Comparison of strengths and challenges for testing conducted in centralized laboratories and POC sitesa
| Testing consideration | Centralized laboratories |
POC sites |
||
|---|---|---|---|---|
| Strength(s) | Challenge(s) | Strength(s) | Challenge(s) | |
| Patient care | High throughput | Limited rural access | Increased patient access | Low throughput |
| Long TAT | Short TAT | |||
| High rate of loss to follow-up | Early ART initiation | |||
| Implementation | Access to LIS | Sophisticated instrumentation | Fewer infrastructure needs | Difficulty in instrument service |
| Specialized training | Remote QC potential with connectivity | Limited access to LIS | ||
| Less hands-on time | Simple test performance | More hands-on time per test | ||
| Sample referral strengthens integrated disease system | Associated cost to maintain specimen referral network | No specimen referral required | ||
| Procurement and supply chain | Procurement system easy to establish for relatively small numbers of laboratories | More system strengthening needed | Reagents do not require cold chain | Strategic planning for scale-up |
| Instrument maintenance contract usually inadequate and not adhered to | Complicated reagent delivery to vast number of POC sites | |||
| QA | Standard enrollment into EQA program or ability for interlaboratory comparison | Associated cost with maintaining QA program | Simple testing; may have built-in internal control | Tester training |
| Acquisition of QC materials | ||||
| Dedicated QA staff | ||||
| False sense of simplicity equated to not requiring QA checks | ||||
| Waste management | Easy to manage centrally for facility-based management, e.g., with availability of incinerator | Bulky and more waste | Less bulky; smaller quantities of waste | Usually lacks facilities for processing of waste prior to disposal |
| Human resources | Small number of staff needed | Special training and QA on sophisticated instrument operation | Simple testing could be performed by nonlaboratory staff | Multitasking of staff on testing and clinical care |
| Need for proper training | ||||
ART, antiretroviral treatment; LIS, laboratory information system; QC, quality control; QA, quality assurance; TAT, turnaround time; POC, point of care; EQA, external quality assessment.
HIV Self-Testing
Achieving the UNAIDS target of 90% of people living with HIV knowing their status will require innovative strategies outside traditional facility- and community-based testing approaches in order to reach all populations. HIV self-testing is an innovative approach to reach groups at high risk of HIV infection or youth populations that do not have access to facility-based services. It will allow individuals who want to know their HIV status to collect their own specimen (oral fluid or whole blood), perform an HIV test, and interpret the results privately or in the presence of someone whom they trust (61). HIV self-testing empowers patients and will allow access to populations that cannot be reached using existing services. Many at-risk persons refrain from testing due to fear of discrimination or stigmatization (62); thus, home-based testing provides them an avenue to access testing in privacy. In Malawi, acceptability was high in antenatal clinics where self-testing was offered to pregnant women and their male partners (63). The WHO has recommended HIV self-testing as an additional testing approach, and 40 countries have adopted and included it in their national policies (64). The WHO has prequalified three HIV rapid tests for HIV self-testing, the Autotest VIH and Insti HIV-1/HIV-2 antibody tests, which use whole blood as the specimen type, and the Oraquick rapid HIV-1/2 antibody test, which uses oral fluid (65). Similarly, the FDA has approved the Autotest VIH and Insti HIV-1/HIV-2 antibody tests for whole blood and the Oraquick Advance rapid HIV-1/2 antibody test for oral fluid. The level of access to HIV self-testing kits can vary with settings and countries. The kits can be provided to clients in health facilities, or some users can procure the kits over the counter in pharmacies or grocery stores to perform finger-pricking at home using a lancet provided in the kit. The test kits include inserts with instructions on how to conduct the test and interpret the result. Individuals who test reactive following HIV self-testing should seek further testing from a trained person using an approved algorithm for confirmation of a positive status and then be linked to treatment and care services. Individuals who have nonreactive results but have known recent HIV exposure or are at a high risk of infection should repeat self-testing 6 weeks later to exclude the possibility of being in the window period (61).
Rapid Test Prequalification and Postmarket Surveillance
Prequalification of in vitro diagnostics, such as HIV rapid test kits, is an important quality step to ascertain claims by the manufacturer on the performance of a particular test kit and whether or not it meets set standards. It is usually performed by an independent and credible institution. For example, WHO prequalification of a rapid test kit entails the quality assessment of the test using standardized procedures to ensure that it is in compliance with WHO prequalification requirements. The WHO prequalification procedure for a particular rapid test kit includes (i) presubmission of a form by the manufacturer of the rapid test kit to apply for WHO prequalification, (ii) review of the dossier by the WHO to understand the product, (iii) inspection of the test kit manufacturing site to assess compliance with quality standards, and (iv) laboratory evaluation of the test kit to assess operational and performance characteristics (66). The WHO uses its Collaborating Centers of Excellence or independent laboratories, including the Centers for Disease Control and Prevention, to perform laboratory evaluations. Following the completion of the above-mentioned procedures, a determination is made regarding whether or not the test kit meets WHO prequalification requirements. Test kits conforming to WHO prequalification requirements are included in the list of prequalified products (67). The MOH together with partners such as the PEPFAR and the Global Fund can access the WHO-prequalified list of rapid test kits for use in their HIV programs in RLS. HIV programs procuring these test kits perform in-country evaluations to determine the suitable combination of rapid tests for their national testing algorithms for diagnosis of HIV infection. These different steps ensure that test kits are safe and reliable and perform optimally for use in programs.
Once the test kits have been procured, postmarket surveillance (PMS) should be implemented to monitor the performance of the test kits after they have been shipped by the manufacturer. PMS should be done once test kits arrive in the country, prior to the shipment of test kits to different sites for programmatic use. This can be achieved by using quality control materials and performance indicators to monitor each new lot received in the country and whether it is performing as expected. Following the distribution of the test kits to different program sites, PMS should be routinely conducted and monitored as test kits are used in the field to ensure that they are performing as intended (68). The importance of PMS was demonstrated when the WHO removed the Standard Diagnostics Bioline HIV-1/2 3.0 rapid HIV test kit from its list of prequalified rapid test kits due to multiple batches that yielded excessive invalid results (69). PMS is an important quality assurance monitoring process that should be routinely performed. PMS also challenges manufacturers to maintain high standards in ensuring a quality product and thereby minimizes disruption of services due to recalls of poorly performing products.
INCIDENCE TESTING
HIV prevention and treatment programs underwent a significant global expansion in the past decade. Based on the UNAIDS 90-90-90 targets, two-thirds of individuals living with HIV knew their status at the end of 2016. Of the two-thirds who knew their status, 77% were on ART with 82% viral suppression (70). Despite this progress, the decline in the number of new infections has been slow. The number of new infections in 2010 (1.9 million) fell by only 11% to 1.8 million in 2016, as modeled by UNAIDS (71). In order to achieve the UNAIDS goal of eliminating HIV as a public health threat by 2030, individuals who do not know their status need to be diagnosed and put on ART, and populations with high rates of new infections need to be identified and initiated on ART to break the cycle of transmission.
HIV incidence testing requires recent infections to be distinguished from long-term infections to facilitate measurement of the incidence rate. Furthermore, this approach is a useful tool to allow country programs to monitor the impact of the scale-up of combination prevention packages (HIV testing and counseling, ART, and voluntary medical male circumcision) to prevent new infections. Identifying recent infections in a given high-risk population, including those in defined geographic areas, provides reliable data for better planning, resource allocation, and targeted interventions (72, 73). Longitudinal follow-up of at-risk HIV-negative individuals, with retesting to detect seroconversions, is considered the gold standard to estimate incidence. However, this approach is very expensive and difficult to implement in most countries. Alternative methods include modeling, back-calculations from AIDS case reporting, age-based prevalence determinations, and prevalence determinations with multiple rounds of longitudinal surveys to estimate HIV incidence (74–76). In last few years, several laboratory-based approaches have been developed to distinguish recent from long-term HIV infections in cross-sectional surveys based on the evolution of maturating antibodies following infection. In 1998, a serologic laboratory-based strategy, using cross-sectionally collected specimens and sensitive/less-sensitive (LS) EIAs, was used to measure antibody titers to estimate incidence (30). Subsequent methodology optimization, based on pre- and postseroconversion specimens, resulted in a series of laboratory-based assays to detect recent HIV-1 infections (Fig. 4).
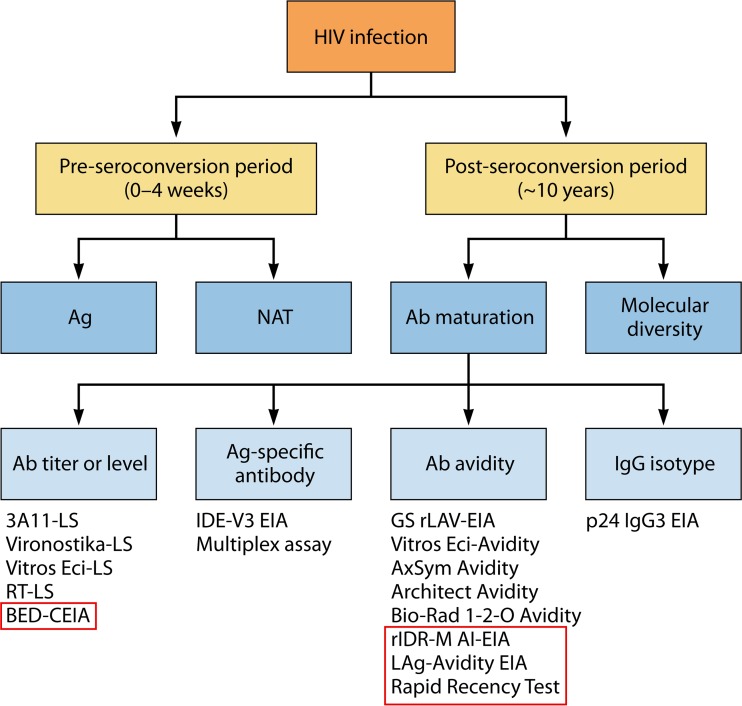
FIG 4.
Methods used for incidence measurement cover pre- or postseroconversion periods. Methods used for preseroconversion specimens include detection of p24 antigen (Ag) and RNA by nucleic acid amplification tests. Methods used for postseroconversion specimens are based on antibody (Ab) maturation and viral genetic diversity. Commercial or in-house assays for the four antibody maturation groups are listed. Assays highlighted in red boxes are methods that use peptides or recombinant antigens derived from multiple subtypes to broaden subtype coverage. Abbreviations: LS, less sensitive; EIA, enzyme immunoassay; BED-CEIA: BED-Capture EIA; IDE, immunodominant epitope; rIDR-M, recombinant immunodominant region of HIV-1 gp41 group M; LAg, limiting antigen; NAT, nucleic acid testing.
Limited Usefulness of Assays during the Preseroconversion Period
The preseroconversion phase is usually 2 to 4 weeks following exposure and is characterized by the presence of viral RNA and/or p24 antigen. Methods for detecting these two viral components (viral RNA or p24 antigen) for incidence determination have been reported (77, 78). However, the duration of p24 antigen/viral RNA prior to detection of antibodies is short during the preseroconversion period. Therefore, the use of this method would require a large number of specimens to attain the needed confidence in incidence measurement. For instance, even in Swaziland (now known as the Kingdom of eSwatini), with an HIV-1 prevalence as high as 32% and an incidence of 2.5 per 100 persons per year, only 1 person in every 1,000 seronegative persons was found to have HIV-1 RNA (79). If the HIV-1 incidence in a given study population is low, the probability of finding individuals during this acute phase with viral RNA or p24 would be even lower. The large sample size requirement and operational cost limit the utility of this approach.
Detection of Recent Infections during the Postseroconversion Period
Detection of recent infections using postseroconversion specimens can be achieved by examining antibody maturation and viral genetic diversity (Fig. 4). Antibody maturation can be further characterized based on the development of antibody titers/levels, antibody avidity, and IgG isotypes. Early in incidence assay development, the antibody titer was the primary target based on the premise that recently infected persons would have lower antibody titers than persons with long-term infections. Initially, the Abbott HIV-1 subtype B-based 3A11 assay was modified to be “LS” by using a 20,000-fold-diluted serum specimen with a reduced antigen/antibody reaction time (30). An optical density (OD) value of the diluted specimen together with a predetermined cutoff obtained from a calibrator specimen were used to distinguish recent from long-term infections. This assay was found to work well with specimens obtained from regions with predominantly subtype B viruses but poorly with specimens from countries with predominantly non-subtype B viruses. The assay overestimated the incidence in specific populations due to antigenic differences between subtypes, resulting in significant differences in mean durations of recent infection (“recency window period”) and misclassification rates (31, 33). However, several additional commercial assays were modified as less-sensitive assays to detect recent infections, including Vironostika-LS, Vitros-ECi-LS, and multiple rapid tests, such as Serodia, Unigold, Determine, OraQuick, and SeroStrip (80–84). None of them were widely adopted due to limitations of commercial assays using antigens derived primarily from HIV-1 subtype B, which led to ongoing research into and development of several new approaches. The first one included the development of the BED-Capture EIA, a de novo assay specifically developed to detect and distinguish recent from long-term infections using increasing levels of HIV-specific gp41 antibodies as a proportion of total IgG (31). This assay included the design of a multisubtype branched gp41 peptide (termed “BED”) to address the antigenic variability of subtypes based on data from the Los Alamos sequence database. An additional unique feature was the capture format of the assay, which captured both HIV-specific IgG (HIV-IgG) and non-HIV-IgG, followed by the detection of gp41-specific antibodies using a biotin-labeled BED peptide. The capture format allowed measurement of increasing proportions of HIV-IgG during antibody maturation. This approach changed the dynamic range of the assay, compared to diagnostic assays, making it less sensitive during the early phase of seroconversion, when the ratio of HIV-IgG to total IgG is low. Since the ratio of HIV-IgG to total IgG in the blood determined the signal, the outcome was not affected by dilution of the specimen, a major advantage over LS assays, which depended heavily on the precision of a 1:20,000 dilution, which was difficult to achieve. Following optimization and calibration of the assay (32), it was commercialized as a kit and was widely used in the United States and several other countries, including South Africa, China, Thailand, Ethiopia, India, Indonesia, and Kenya, for surveillance of recent infections and estimating HIV-1 incidence (85–95). However, ongoing studies also indicated that the BED assay results were confounded by individuals with low CD4 levels (those with AIDS), elite controllers, those with high levels of total IgG (as found in Africa), and those on ART (96–99), resulting in elevated HIV-1 incidence rates. UNAIDS recommended that the BED assay not be used for incidence surveillance (100). Due to the absence of any other good laboratory-based methods, solutions were proposed, including the use of the BED assay in case-based surveillance, which allowed the removal of misclassified cases or adjustments based on misclassification rates in a given population (96, 101–110). However, misclassification rates, which contribute to overestimations of incidence, are not constant but vary with time, place, and population (99). Parallel developments of other approaches include a two-well assay based on the differential detection of antibodies to gp41 IDR and gp120 V3 loop antigens (111, 112). Recognizing the diversity of HIV-1 antigens and the corresponding immune responses, the assay incorporated 2 sequences of IDR peptides and 5 sequences of V3 peptides. The investigators used the in-house assay to detect recently infected persons in France (112–114). However, the assay is not commercially available due to the complexity of the assay, limiting its wider application or evaluation. In 2004, Wilson et al. described the development of an EIA to detect IgG3 antibodies to p24 as a marker to detect recent HIV-1 infection (115). However, the presence of p24-IgG3 was not consistent among recently infected persons, limiting the use or commercial development of the assay.
Antibody Avidity
Antibody avidity (binding strength of antibodies) has been recognized as a reliable marker for maturation of antibodies and has been used to identify and distinguish recent infections (with low Ab avidity) from long-term infections (with high avidity) for many viral infections, including cytomegalovirus (CMV), rubella, hepatitis B/C, and HIV infections (72, 116–122). Most early efforts were focused on modifying commercial HIV assays to measure avidity indexes using different chaotropes or dissociation reagents. Although these assays provided useful information, limitations included the use of antigens that are derived mainly from HIV-1 subtype B, the need for an automated platform such as Abbott Axsym or Architect, the requirement for duplicate wells to calculate the avidity index, and increased variability coming from two wells (122).
To address antigenic diversity, a chimeric recombinant gp41 protein (rIDR-M) that included multiple gp41 sequences in a single protein was developed (123). A novel, single-well avidity assay was developed using wells coated with a limiting amount of antigen. Limiting the amount of antigen forced bivalent antibodies to bind monovalently, thus facilitating the separation of recent infections (weak antibodies) from long-term infections (strong antibodies), all in one well (Fig. 5A). The limiting-antigen (LAg) avidity EIA worked as well as or better than the two-well assays that measured antibody avidity indexes but benefited from a calibrator specimen to minimize variability and determine the cutoff for recency classification (33, 124). Again, the assay was optimized, characterized, and subsequently transferred to commercial partners for development of an LAg-Avidity EIA kit.
FIG 5.
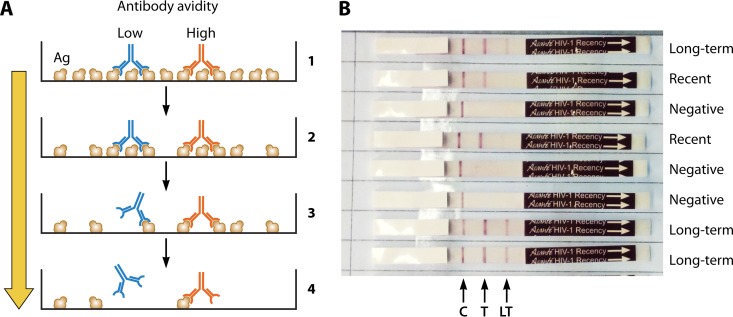
Schematic representations of the assay principle for a limiting-antigen (LAg) assay to differentiate low- and high-avidity antibodies (A) and a rapid incidence-prevalence assay (B). (A) At high antigen concentrations, bivalent antibodies of both low avidity (blue) and high avidity (red) bind to microwells coated with antigen, whereas at limiting antigen concentrations, only the high-avidity antibodies bind due to monovalent binding and are retained after washing. (B) Two empirically optimized antigen concentrations are incorporated onto the nitrocellulose strip for diagnosis of HIV infection and detection of recent infection. C, control line; T, test line at a high antigen concentration; LT, low antigen concentration to differentiate recent from long-term infections. The binding of antibody is determined with lateral flow technology. The control band indicated by the arrow is an internal IgG procedural control. Seronegative samples are reactive only for this control band. Persons with recent infections will show C and T lines but not the LT line, and those with long-term infections (>1 year) are reactive to all three lines, including the LT line.
The LAg-Avidity EIA was found to have clean antibody kinetics, separating recent from long-term infection with less persistence of low avidity beyond 1 year (33). It also had the lowest misclassification rate compared to other incidence assays. The assay was validated in Swaziland, where the LAg-based incidence in a cross-sectional survey was compared to the observed incidence in a cohort (125). That study as well as work from independent groups, such as the Consortium for Performance and Evaluation of Incidence Assays (CEPHIA), demonstrated that almost all antibody-based assays will misclassify a certain proportion of persons as having been recently infected who otherwise are infected for a long time but are either on ART or identified as elite controllers (126). Both these groups are characterized by a low VL, which contributes to low antibody avidity, resulting in misclassification in some cases. Therefore, a recent infection testing algorithm has been proposed, which includes LAg EIA testing followed by VL testing of those classified as having been recently infected. Final classification of recency is based on recent LAg (normalized OD [ODnormalized] of <1.5) combined with a VL of ≥1,000 copies/ml to identify true cases of recent infection. Other investigators have used the presence of antiretrovirals (ARVs) in the blood, in addition to VL, to reclassify recent LAg cases as part of the algorithm. Application of these approaches has shown that estimated HIV-1 incidence rates are plausible and provide meaningful data for risk factor analysis of the acquisition of new infections in multiple studies. Currently, the LAg-Avidity EIA, in an algorithm with VL testing (and, in some surveys, with ARV), is the preferred method for detection of recent infections and estimation of incidence. The assay is widely used in many countries for population-based HIV impact assessment (PHIA) surveys with promising results (127, 128).
Rapid Incidence Assay
A rapid incidence test that can diagnose HIV infection and distinguish recent from long-term infections in a single test was recently developed (129), using the same concept of limiting antigen. To overcome limitations of commercial rapid tests, used primarily in a less-sensitive format, rIDR-M was used in limiting concentrations, in addition to a diagnostic line, in a rapid, lateral flow strip format (Fig. 5B) (129). The assay is now commercially available as the Asanté HIV-1 rapid recency assay (Sedia Biosciences Corporation). The assay was validated in laboratory settings, with sensitivity and specificity of >99% and high correlation with LAg-Avidity EIA results (130). As a POC test, the rapid recency assay has the potential to significantly change the outcome in the fight against HIV/AIDS and play a key role in prevention and treatment programs. For instance, the use of this test would allow diagnoses and identification of recent infections of individuals using HIV testing services, in antenatal clinics, and in surveillance. The use of such a tool in case-based incidence surveillance, with a focus on persons with newly diagnosed infections, can help identify geographic hot spots and high-incidence populations in real time for targeted prevention and interruption of ongoing transmission. Additionally, it would allow quick initiation of contact tracing of partners of the index case for testing and initiation of treatment. Several pilot projects are under way in Malawi, Vietnam, Central America, and other areas to assess the utility of the rapid recency assay. The rapid incidence assay is being recommended for wider use in many countries to detect new infections among adolescent girls and young women and in routine programs to detect hot spots of new infections.
Role of Genetic Diversity Approaches in Incidence
The premise of the genetic diversity approach is that only a small subset of HIV variant founders is established in the new host after transmission, followed by the expansion of the genetic profile with increased accumulation of mutations in the HIV genomes with viral replication. Persons with recent infections are thought to possess less HIV genetic diversity than persons with long-term infections. Genetic diversity can be estimated by direct sequencing, which is labor-intensive and cost-prohibitive. Simplified assays, such as evaluation of ambiguous nucleotide calls (131, 132), the Hamming distance assay (133), and the high-resolution melting diversity assay (134), have been recently shown in many small studies to be promising (135). However, there are complications due to infections with multiple variants in newly infected persons (e.g., blood transfusion-related infections or infections in intravenous drug users) and among those with advanced AIDS. Moreover, none of the methods has been widely applied or commercialized.
HIV TESTING USING DBS SPECIMENS
DBS specimens are an alternative specimen type to whole blood and plasma for diagnosis and monitoring of patients infected with HIV. DBS specimens are easy to collect, process, and package and can be transported at ambient temperature (136, 137). DBS specimens have been evaluated and showed results comparable to those with plasma and are widely used in programs for HIV antibody testing (138), HIV incidence determination (139), HIV DR monitoring (140–143), surveillance (144), VL testing (145), EID (146–148), and proficiency testing (PT) (149). Different types of commercially available filter paper, including Ahlstrom 226, Munktell TFN, and Whatman 903, have been assessed and shown to be suitable for the collection of DBS specimens for EID, thus providing options for use in RLS (150). However, only Whatman 903 filter paper is optimized and recommended for testing for recent infection with BED or the LAg-Avidity EIA. The use of DBS specimens would increase access for populations where laboratory services are not available. For example, within the tiered laboratory network, where complex molecular tests such as VL tests, DR tests, and EID are found mostly in laboratories at the national, provincial, and district levels, the use of DBS specimens will ensure that the specimens can be collected by trained health care workers in remote areas and transported to a specialized laboratory for testing.
ENSURING QUALITY OF TESTING
Current Status of Quality Assurance Programs in RLS
The rapid expansion of HIV testing in the past decade has not been matched by quality assurance. The lack of adequate quality assurance practices could negatively impact the accuracy of tests results, especially those conducted in remote areas (151). In line with WHO recommendations on task shifting to support universal access to HIV/AIDS prevention, treatment, and care services, many RLS countries have revised their national policies to use community counselors to perform HIV rapid testing (152–154). Countries have the primary responsibilities to develop policies to implement and monitor QA of HIV testing. An unfortunate consequence of low-quality HIV rapid testing is wrong diagnosis, with an enormous adverse impact on individuals, families, communities, health care workers, and programs. The resulting unnecessary anxiety and treatment undermine the credibility of entire HIV programs (155–157).
To prevent enrolling misdiagnosed persons into lifelong ART, the WHO recommends the retesting of all clients determined to be HIV positive with a second specimen prior to ART initiation (57). Factors often attributed to misdiagnosis include staff inexperience due to high turnover rates, the use of a suboptimal algorithm, clerical errors, inadequate training and competency assessments, and a lack of supervisory oversight (158–160). Improper or inadequate documentation of testing records could also lead to erroneous result reporting and is often found in lower-level health care facilities. To document testing properly, the use of a standardized paper-based HIV testing logbook nationwide is a useful approach (68). The logbook captures much relevant quality information, including selection of preprinted simple test results (i.e., without the need for handwriting for reactive or nonreactive test results), retesting decisions, test kit names, lot numbers, and expiration dates. The logbook also provides a summary tabulation of the collective results entered for a page to allow testers to better self-monitor the concordance or discordance between results of two rapid tests and to identify and capture abnormalities. Since the log is paper-based, the information captured from different testing sites cannot be aggregated in a timely manner for regional or national program managers to analyze for any trends and corrective actions, if needed. Thus, when applicable, the country should progress to the use of electronic filing and reporting methods to better record and report testing results.
Periodic site assessments and refresher training using standardized tools can narrow the quality gaps at primary sites. Due to limited resources and workforce constraints, reference laboratories at the regional and national levels should focus on newly established testing sites and those with known low-performance records. Participation in a PT program, in particular, has been found to greatly enhance problem-solving skills of testers. Some countries emphasize the importance of the use of PT materials to train testers under the supervision of experienced staff (161, 162). National reference laboratories can produce and distribute PT materials. PT programs using a cost-effective alternative approach, such as the dried tube specimen (DTS) approach, have been properly documented (151). DTS are an alternative to serum or plasma samples for use in PT panels to monitor the quality of testing. DTS are prepared from well-characterized specimens by mixing small volumes of the characterized plasma or serum sample with a color dye and allowing the mixtures to air dry. The resultant pellet is visible and is then reconstituted with phosphate-buffered saline (PBS)–Tween 20 and used for testing according to the instructions (151). DTS are temperature stable, and results obtained are comparable to those with plasma or serum samples. DTS are also used as PT materials to monitor the quality of other HIV biomarkers, such as VL (163).
Enabling Environment for Quality-Assured HIV Rapid Testing
Although HIV testing services (HTS) QA guidelines for rapid testing existed for many RLS countries (5, 6), they were deficient in policies for implementation (159, 164–166). In 2015, the WHO released a handbook, Improving the Quality of HIV-Related Point-of-Care Testing: Ensuring Reliability and Accuracy of Test Results (167), to address QA program weaknesses (8, 168). This handbook emphasizes the importance of continuous quality improvement for POC testing, the need to strengthen quality practices by quality officers and testers, the provision of new quality strategies, and the use of a comprehensive package with established new tools. The handbook describes and emphasizes the processes of the quality assurance cycle (QAC): planning, implementing, and sustaining (167). These new recommendations are being translated into national policies and guidelines in many RLS countries (161, 169). For instance, South Africa has developed an HTS policy and guidelines with emphasis on the importance of completing the QAC to prevent testing errors along the “diagnostic continuum” (169). Kenya highlighted the importance of competency demonstration and certification of testers by the National AIDS/STD Control Program and defined the specific tasks of stakeholders at the facility and country levels to address problems of personnel shortages (161).
The U.S. CDC and the WHO developed a systematic and multimodular training curriculum (170, 171). Most RLS countries have adopted this curriculum with revisions to suit their respective implementation needs. These country-specific revisions include training processes, durations of training (1 to 10 days), teaching approaches (lectures versus practical sessions), and delivery formats (158, 159, 166, 172). Due to the large number of persons to be trained, most RLS countries opted for a top-down approach through a training-of-trainers (TOT) model that allows the rapid expansion of standardized training to staff in the region, district, community, and outreach program. This training includes pre- and posttraining quizzes to assess the performance skills and knowledge gained. For example, in a health facility in eastern Kenya, a training assessment showed that of the 91% of nurses who received HTS training, 79% reported self-perceived competency and 70% demonstrated competency in performing rapid testing (173). While the study sample size was small, this report identified the existence of a gap between training and testing performance and the need to enhance training methods, including hands-on sessions, and to ensure that core competencies are adequately assessed.
Many countries established guidelines recommending the provision of periodic refresher courses to maintain tester competency. Refresher training can involve a high work burden and requires extra resources, and thus, it is important that the country’s policies are supported with adequate financial and human resources. Because Internet- and mobile-based technologies have become widely accessible in RLS, distance-learning programs can greatly improve training by reducing the time that trainees need to be away from their health facility while receiving off-site training. Examples of these cost-effective programs include the Project Extension for Community Healthcare Outcomes (ECHO) (174) and Supercourse (175) public health training programs currently in use in several African countries. Project ECHO, an example of a virtual community of practice, brings together subject matter experts, health care practitioners, and peer groups virtually to share knowledge and practical experience, including problem-solving skills, in real time to impact programs (176).
Promoting Community Partnerships for the Uptake of Quality Measures
The severe shortage of government health workers in RLS countries has highlighted the need for the participation of community volunteers. Many of the health care tasks that do not require specialized training could be delegated to community volunteers to increase access to HIV services in underserved or hard-to-reach communities (177). In 2012, a systematic review described a wide range of nonprofessional cadres of health workers to assist HIV treatment programs (178). These volunteers included care coordinators, peer health workers, field officers, health extension workers, HIV/AIDS lay counselors, adherence supporters, and home-based care workers. The WHO has recommended the involvement of community volunteers or quality corps (Q-Corps) volunteers to support the rollout of quality activities and to ensure that the QAC is completed (167, 179). Several PEPFAR-supported countries have implemented this initiative, with improved HIV testing in remote areas. Q-Corps are further stratified into two tiers, level I and level II, based on different roles, with Q-Corps level II being higher than level I and having more responsibilities (Table 2). The Q-Corps volunteers may be new health science graduates or part-time laboratory volunteers who are trained on specific QA tasks under the guidance of the MOH, implementation partners, and other managerial agencies at various health care levels (Table 2). The Q-Corps can be involved in the distribution and collection of PT results, supportive site supervision, training and competency assessment of site staff, and distribution and review of a standardized logbook or electronic system.
TABLE 2.
Responsibilities and working relationship between Q-Corps volunteers and the supporting MOH or other managerial authoritiesa
| Organization or program | Working relationships and/or responsibility(ies) |
||
|---|---|---|---|
| Program activities/management | Q-Corps level II | Q-Corps level I | |
| Responsible organizations | MOH, NRL, and regional and district agencies | Local implementation partners, district laboratory staff, QA officers, and coordinators | Local implementation partners |
| PT program | PT network establishment | Panel distribution to testing sites | |
| PT panel preparation | Assistance with data report and corrective action | ||
| PT outcome assessment | |||
| HTS program QA | National testing logbook customization, outcome review, and corrective action plans | Logbook utilization and review for data integrity | Logbook distribution and entry assistance |
| Monthly data assembly collection | |||
| Training programs | Planning, development, and conduct of national training curriculum and programs | Assistance with training material distribution | Participation and assistance with simple training courses |
| Supervisory site visits and corrective action provision | Assistance with internal audits | ||
| Assistance with competency assessment | |||
Q-Corps level II is higher, with additional responsibilities for data review, than level I. Abbreviations: MOH, ministry of health; NRL, national reference laboratory; Q-Corps, quality corps; HTS, HIV testing services; PT, proficiency testing; QA, quality assurance.
In Cameroon, Q-Corps were successfully employed to help distribute DTS-based rapid test PT panels to remote testing sites and contributed to reducing the PT result turnaround time from 30 days to 5 days (59, 180). This was a remarkable accomplishment, especially in areas with poor transportation and road infrastructures. With the proper training and guidance, Q-Corps can be used to help advance laboratory testing quality.
In countries where there are established partnerships with nongovernmental organizations (NGOs) for health care service delivery, HIV testing quality-related needs may not be delegated to nonprofessional community volunteers. It is critical for the managers of the MOH and the national reference laboratory to develop a supportive policy environment with appropriate regulations for NGOs to support the scale-up of QA programs. Q-Corps volunteer programs should be guided and built through sound work regulations and quality improvement frameworks. It is also beneficial for governments to establish collaborative partnership with NGOs, community or professional associations whose members can be trained to oversee the program and provide supportive supervision and input (181). Moreover, while partnering with communities and other NGOs, appropriate coordination should be built at either the regional, district, or community level with delegated responsibilities.
National Program for Competency Assessment and Tester Certification
A review of POC testing outside the traditional laboratory environment clearly demonstrated gaps that should be addressed by implementing stepwise quality programs to improve tester and site competency. Implementation of a national certification program ensures adherence to quality standards across POC sites, lends credence to the sites, and inspires confidence in patients and clinical staff. Currently, only a few RLS countries have competency-based certification programs. For instance, a 2007 act to provide for the registration and regulation of the health laboratory practitioners of Tanzania specifies requirements for certification, recertification, and decertification of testers (182). However, this act was not fully implemented due to the lack of resources. National guidelines for HIV counseling and testing in Namibia also outlined certification and recertification requirements (162). It was intended for nonlaboratorians and staff conducting HIV testing in all facilities. Zimbabwe’s national policy mandating HIV testing by laboratory technicians has recently been revised to extend to nonlaboratorians performing rapid HIV testing (183). When establishing a national certification program, it is critical for the government to allocate resources, infrastructure, and personnel for implementation and to identify independent organizations, such as laboratory professional associations, academic partners, or a public health institute, with demonstrated high standards to act as the certifying body. The certifying bodies can assess the competency of testers, maintain the certification data, and provide reports to ensure impartiality and transparency.
Optimizing the Process for National Certification of Rapid Testing Sites
Until recently, there have been few or no programs to systematically monitor and assess compliance to the quality of HIV rapid testing sites. Although in most countries, periodic supportive supervisory visits have been conducted using program-specific assessment tools, they are often not standardized, and they do not comprehensively cover quality processes. Furthermore, the site supervision teams often do not include trained technical staff. In order to identify gaps for targeted intervention and resource allocations needed by national programmers, the CDC, the WHO, and several implementing partners developed a stepwise process for improving rapid testing (SPI-RT) checklist (167). The checklist contains 70 audit inquiries with preassigned scores and covers seven QA areas, including personnel training and certification, the physical facility, safety, the pretesting phase, the testing phase, the posttesting phase, and external QA (EQA) assessment. The checklist is categorized into five levels to represent the extent of readiness of the sites for national site certification, level 0 (<40%), level 1 (40% to 59%), level 2 (60% to 79%), level 3 (80% to 89%), and level 4 (90% to 100%). Level 0 indicates that the site is in poor readiness and requires immediate remediation in all seven quality system essentials, while level 4 indicates that the site is eligible for national site certification. Sites are assessed using the checklist and scored as one of the different levels.
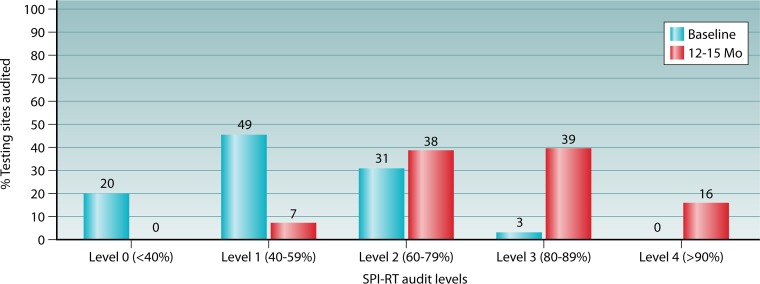
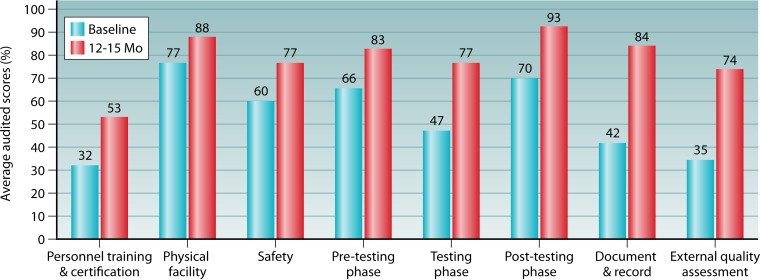
In 2014, nine countries in Africa and the Caribbean (Cameroon, Ethiopia, Uganda, Kenya, Malawi, Tanzania, Zambia, South Africa, and Jamaica) initiated training of MOH staff and Q-Corps volunteers and used an earlier version of the SPI-RT checklist that covered the seven areas described above, including a section on documents and records, to audit 1,740 testing sites. The quarterly audited sites included several testing modalities, such as voluntary counseling and testing (VCT), provider-initiated testing and counseling (PITC), prevention of mother-to-child transmission (PMTCT), and treatment centers and laboratories. The median SPI-RT score at the baseline audits of these sites was found to be 55.6% (level 1). Of these sites, 968 (55.6%) sites in six countries (Cameroon, Uganda, Malawi, Tanzania, Zambia, and Jamaica) had corresponding scores at baseline and 12 to 15 months later to allow for pairwise comparison over time. It was found that at the baseline audits, 20% of the sites were at level 0, 46% were at level 1, 31% were at level 2, 3% were at level 3, and none of the sites were at level 4 (Fig. 6). After 12 to 15 months of implementation of corrective actions with provision of supervisions, 38%, 39%, and 16% of the sites had moved up to levels 2, 3, and 4, respectively, and none of the sites remained at level 0. At the baseline audits, high scores were obtained for the physical facility (77%), safety (60%), the pretesting phase (66%), and the posttesting phase (70%) (Fig. 7). After 12 to 15 months of remediation, there was a >20% increase in the audit score for the testing phase (77%), the posttesting phase (93%), documents and records (84%), and the EQA program (74%). The areas with moderate improvement were the physical facility (88%), safety (77%), and the pretesting phase (83%). Although noticeable improvement was observed for EQA, there were inconsistent PT panel distributions by the national PT provider and deficiencies in the documentation of PT participation, which were previously found to be prevalent in many RLS countries (157–159, 166). The inconsistent PT panel distribution was corrected in Cameroon by using the innovative Q-Corps (180).
FIG 6.
Performance levels from the baseline audit to the 12- to 15-month audit among 968 testing sites in five African countries (Cameroon, Uganda, Malawi, Tanzania, and Zambia) and one Caribbean country (Jamaica). Shown are SPI-RT scores at level 0 (<40%), level 1 (40 to 59%), level 2 (60 to 79%), level 3 (80 to 89%), and level 4 (>90%). The numbers on the top of each bar represent the percentages of the total sites.
FIG 7.
Average audit scores (percent) of 968 HIV testing sites in five African countries (Cameroon, Uganda, Malawi, Tanzania, and Zambia) and one Caribbean country (Jamaica) using the SPI-RT checklist on eight quality components.
Our experience in RLS, including expansion of testing at thousands of testing sites, demonstrates that QA measures should be appropriate for the environment where testing is being conducted. Laboratory accreditation, offered by the College of American Pathologists (CAP) and the ISO, may not be feasible in RLS where HIV diagnosis is offered. However, development of novel approaches, including a strong training curriculum and ongoing monitoring, can ensure that HIV testing is accurate. Data from multiple countries in PEPFAR-supported programs indicate that the positive predictive value of HIV diagnosis can be 99% or higher.
MOLECULAR METHODOLOGIES FOR DETECTION AND QUANTIFICATION OF VIRAL LOAD
Viral Load and ART
The efficacy of ART for individuals infected with HIV can be monitored by using parameters such as increases in CD4 cell counts, VL suppression, and improvements in clinical symptoms. The 2013 WHO consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection strongly recommended VL testing for use in monitoring of patients on ART and for confirmation of treatment failure (184). VL monitoring is the preferred approach because it is an early and accurate marker of ART success compared to immunological and clinical monitoring markers and can facilitate detection of DR mutations early (185–187). VL monitoring of a patient’s response to treatment is also a good indicator for assessing adherence to ART and allows the effective use of differentiated service delivery models to manage virally suppressed and nonsuppressed patients (188). In a multicountry observational study, patients with adherence counseling showed viral resuppression when the VL was used to identify patients requiring adherence support, while delayed implementation of routine VL monitoring was associated with the accumulation of DR (189). In the United States, VL is assessed at multiple time points, prior to ART initiation, within 2 to 4 weeks post-ART, and every 3 to 4 months thereafter for the first 2 years, which can be extended to every 6 months after 2 years if viral suppression is consistently observed (190). In contrast, in 2013, in SSA, only 20% of ART-treated patients had access to routine VL testing, and clinicians often received results too late to assist in better patient management (9). The WHO has recommended that VL testing be carried out 6 and 12 months after ART initiation and once every 12 months thereafter, with >1,000 copies/ml being the threshold for treatment failure in RLS (6). However, some countries have implemented routine VL monitoring with different thresholds for virologic failure. For example, Botswana treatment guidelines recommend a threshold of >400 copies/ml as virologic failure (191). A systematic review of adult virologic outcomes in response to an ART program in SSA showed 78%, 76%, and 67% viral suppression after 6, 12, and 24 months, respectively, suggesting that the first-line regimens were effective (187). Also, in a recent PEPFAR PHIA, adult viral suppression rates of 85%, 78%, and 71% were observed in Zimbabwe, Malawi, and Zambia, respectively (192).
Providing timely VL results has been challenging in RLS, partly because VL assays are complex and performed at higher-tiered laboratories in urban areas, in addition to some patients residing in hard-to-access areas. Currently, whole blood or plasma specimens, when collected from distant sites, are shipped within a few hours through an established specimen referral transportation network to centralized laboratories, where they are tested in a cost-efficient manner (193, 194). The scale-up of HIV VL testing is key for routine monitoring of patients on ART, especially as programs transition from CD4-based monitoring to VL monitoring requiring advanced molecular laboratories. One key factor that can limit access to VL testing is the high cost for testing. VL testing costs range from US$30 to US$125 per test (195, 196), and more than half of the cost can be attributed to the cost of regents and consumables (197). To increase access to VL testing and improve patient management, the Roche Global Access Program significantly reduced the price of VL testing reagents for low- and middle-income countries in 2014 (198). The Global Fund also negotiated significant cost reductions with major manufacturers for instruments and VL testing and EID reagents (199). One of the challenges with some of the agreements is that manufacturers sometimes require the ordering of guaranteed volumes of test reagents. Although welcomed as progress, the guaranteed-volume approach tends to exclude countries with lower prevalences or those countries making significant strides to curb their epidemic. It is almost counterintuitive and could be perceived as punitive. There should be harmonization of these prices for universal access that would lead to sustained VL testing scale-up, and the cost reduction would render the monitoring of ART programs more cost-effective (200).
Commercial Assays Suitable for Centralized Laboratories
There has been great progress in the development of assays to quantify HIV loads in blood since the beginning of the pandemic. During the early phase of the pandemic, quantitative cultures of peripheral blood mononuclear cells or plasma were used to measure VL (201, 202). This methodology was labor-intensive, lacked reproducibility, and often yielded negative cultures for patients with CD4 counts of <200 cells/µl (201). The limitations of culture techniques led to the development of simple and more-accurate PCR-based and other molecular methodologies to accurately measure VL.
Since the first PCR-based commercial VL assay in 1996, several commercial assays have been manufactured. These assays are FDA approved, “Conformite Europeene” (CE) marked, and/or WHO prequalified. In earlier stages, the users often experienced amplification contamination-associated false positivity and large VL variations. However, the methods had been greatly improved by the utilization of contamination control strategies, internal calibration controls, process automation, and real-time detection technology approaches (203–205). With a common set of standards of known VL employed by all manufacturers, VL determination standardization became feasible. Manufacturers also examined assay sensitivity and specificity using collections of clinical specimens. The quantification of HIV-1 VL in plasma was thus standardized and expressed as the number of HIV-1 RNA copies present per milliliter of plasma or as log10 units per milliliter of plasma. Independent collaborative evaluations by end users in different geographic locations with different subtypes and recombinant circulating forms further validated the assay performance and the comparability of results using different assays.
A list of 12 major commercial VL assays with their respective performance characteristics, including detection ranges, gene targets, testing times, random-access modality, plasma volumes, and DBS application, is shown in Table 3. With the exception of the Cavidi assay, the other 11 assays are nucleic acid tests (NATs), since detection is based on the amplification of nucleic acid or signal amplification with the initiation target derived from viral RNA. The detected VL range is smaller with the Cavidi ExaVir load assay (200 to 6 × 105 copies/ml) and the Biocentric Generic assay (300 to 5 × 106 copies/ml), while the other 10 NAT-based assays have a wider range of approximately <50 to 107 copies/ml. The low detection limits according to all manufacturers enable the threshold of >1,000 copies/ml recommended by the WHO for ART monitoring in RLS.
TABLE 3.
Characteristics of viral load platforms commonly found in centralized laboratoriesa
| Purpose | Manufacturer and trade name | VL range (copies/ml plasma) | Viral genome(s) detected | Testing time | Random access | Plasma vol (µl) | DBS utility |
|---|---|---|---|---|---|---|---|
| Real-time RT-PCR | Abbott RealTime | 150–107 | int | 7 h | No | 200 | Yes |
| 75–107 | 500 | ||||||
| 40–107 | 600 | ||||||
| 40–107 | 1,000 | ||||||
| Beckman Coulter DxN Veris | 235–1.06 × 107 | pol | 1.5 h | Yes | 175 | NA | |
| 35–1.06 × 107 | 1,000 | ||||||
| Biocentric Generic HIV Charge Virale | 390–5 × 106 | LTR | 7 h | No | 250 | Yes | |
| Cepheid GeneXpert | 40–107 | LTR | 1.5 h | Yes | 1,000 | Yes | |
| Qiagen Artus RG | 45-4.5 × 107 | LTR | 5 h | No | 200 | NA | |
| Roche Cobas CAP/CTM, v2.0 | 20–107 | LTR, gag | 7 h | No | 1,000 | Yes | |
| Siemens Versant kPCR 1.5 | 35–11 × 107 | Pol | 6 h | No | 500 | NA | |
| T7 RNA polymerase | bioMérieux NucliSENS Easy Q, v2.0 | 25–108 | gag | 2 h | No | 100 | Yes |
| 20–2 × 107 | 500 | ||||||
| 10–107 | 1,000 | ||||||
| Hologic Aptima Quant Dx | 13–107 | LTR, pol | 3 h | Yes | 500 | NA | |
| Signal amplification | Siemens Versant bDNA, v3.0 | 75–500,000 | pol | 7 h | No | 1,000 | NA |
| Enzyme based | Cavidi ExaVir, v3, or Ziva | 200–6 × 105 | 2 days | No | 1,000 | No |
Abbreviations: VL, viral load; RT-PCR, reverse transcriptase PCR; bDNA, branched DNA; DBS, dried blood spot; pol, polymerase region of the HIV genome; int, integrase region; gag, group-specific antigen region; LTR, long terminal repeat region.
The NATs can be divided into three groups according to their amplification principles and signal detections. The first group contains seven assays using the reverse transcriptase PCR (RT-PCR) principle to copy viral RNA and amplify the resultant cDNA. The assays using RT-PCR include Abbott RealTime HIV-1 PCR, the Beckman Coulter DXN Veris HIV-1 assay, the Biocentric Generic HIV-1 viral load assay, the Cepheid GeneXpert HIV-1 viral load assay, the Roche Cobas CAP/CTM HIV-1 viral load assay, and Siemens Versant HIV-1 RNA 1.5 (kinetic PCR [kPCR]). The second group, with the bioMérieux NucliSens EasyQ HIV-1 v2.0 assay and the Hologic Aptima HIV-1 Quant assay, uses the T7 RNA polymerase for amplification. The third group, with the Siemens Versant HIV-1 RNA 3.0 (bDNA [branched DNA]) assay, uses signal amplification methodology.
In the RT-PCR-based assays, plasma RNA is first reverse transcribed. The resultant cDNA is amplified by PCR using HIV-specific primer pairs derived from several conserved long terminal repeat (LTR), gag, int, and pol regions of the viral genomes to broaden subtype coverage. The generation of nascent DNA in each cycle of amplification is quantified in real time using sequence-specific fluorescent probes. The two most widely used assays in RLS are the Abbott RealTime HIV-1 PCR and Roche Cobas TaqMan assays. Abbott, Roche, bioMérieux, and Siemens provide proprietary automated plasma RNA processors and amplification/detection devices. Barcoded specimens are tracked and recorded by the instrument to minimize hands-on time and transcriptional errors. Typically, the footprints of these automated RNA processors are large. Specimens are processed in a batchwise manner to yield VL results in 6 to 8 h. Improvements have been made to the Beckman Coulter DxN Veris (206) and Cepheid GeneXpert (207) systems using cartridge-based plasma RNA processing and PCR to achieve the one-step “sample-in and result-out” process in 1.5 h. Individual plasma specimens can be loaded into the cartridge and run in a random-access manner without a time delay. This kind of instrument may be useful in district laboratories near primary health care locations with technical staff trained to prepare plasma. Further modification of the cartridge to include the plasma preparation step will make these instruments directly applicable to POC testing. The Biocentric Generic assay is run in an “open” manner. The manufacturer provides PCR reagents and quantitation controls but not the automated RNA extractor and PCR/detection device. The end users must procure their own real-time amplification/detection device. Although this open system allows end users the flexibility to conduct in-house assays to study other biological agents of interest, it requires the end users to manually calculate VL results and may be open for result variability.
The second group includes the bioMérieux NucliSens and Hologic Aptima assays. The NucliSens and Aptima assays are commonly known as a nucleic acid sequence-based amplification (NASBA) assay and a transcription-mediated amplification (TMA) assay, respectively. The RNA is first reverse transcribed using the reverse transcriptases derived from avian myeloblastosis virus and murine leukemia virus for the NucliSens and Aptima assays, respectively. The primers used for reverse transcription contain a sequence complementary to HIV-1 and a sequence complementary to the T7 RNA polymerase transcription promoter. Thus, the ensuing cDNA molecules serve as the templates to generate hundreds of nascent RNA molecules in each run of the amplification process catalyzed by T7 RNA polymerase at a constant temperature of 41°C. Quantifiable signals are produced within 1 h. The importance of the isothermic temperature used in this assay cannot be overemphasized. At 41°C, double-stranded DNA molecules do not serve as a template for T7 RNA polymerase. Thus, when DBS specimens are used for VL determination with NASBA and TMA assays, only the viral RNA, not the proviral DNA, is quantified.
The Siemens Versant HIV-1 RNA 3.0 bDNA assay is the sole NAT listed in Table 3 that does not use nucleic acid amplification. Instead, it uses a signal amplification process to yield a VL range of between 75 and 500,000 copies/ml. The viral RNA is first immobilized by capture oligonucleotide probes on the microwell surface. The size and structure of the captured RNA grow after three steps of cross-hybridization with oligonucleotide probes to form a large immobilized branched complex containing multiple alkaline phosphatase (AP) moieties. The three sets of probes are (i) target probes complementary to multiple regions of the pol gene, (ii) amplifier probes, and (iii) AP label probes. AP catalyzes the added enzyme substrate to generate chemiluminescence to derive the VL using a Bayer system 340 analyzer.
Finally, the recent Cavidi ExaVir load assay distinctively measures the enzymatic activity of RT from plasma HIV particles. The viral RT in the plasma is isolated using a gel separation step, and its enzymatic activity is measured using an inexpensive enzyme-linked immunosorbent assay (ELISA) reader. The measured enzymatic activity is then translated into viral RNA copies using a special conversion factor provided by the manufacturer. The VL detection dynamic range is between 200 and 6 × 105 copies/ml, a range smaller than that of NAT-based assays but adequate for ART monitoring. In a longitudinal follow-up study of ART patients with RNA levels of >400 copies/ml in Kenya, ExaVir v.2 was found to exhibit 100% sensitivity compared to the reference Roche v 1.5 assay (208). Recently, the sensitivity of a new ExaVir v.3 system was 96 to 100% at 2,000 copies/ml (209–211). The reagent cost is low compared to those of most VL test kits, and the ELISA equipment used in this assay is commonly used for various types of serologic testing. However, this assay requires cold-chain transportation to maintain the integrity of the viral enzyme prior to testing, and it takes 2 days to complete the test. Nevertheless, this assay is not affected by HIV sequence diversity and is capable of detecting both HIV-1 and HIV-2. Furthermore, the new fully automated version may be suitable for district or regional hospital laboratories with medium volumes of patients on ART.
VL Tests and Genetic Diversity
Genetic diversity may pose a challenge for the quantification of VL. The majority of the VL assays are based on PCR amplification and hybridization using primers/probes derived from the conserved regions of the HIV genome. However, the presence of one or two nucleotide substitutions at the 3′ end of a primer may cause VL underestimation by more than 100-fold (212). Earlier versions of VL tests were developed based mostly on subtype B viruses. These tests often failed with non-B subtypes (213–216). Most of the current assays have been improved by using primers/probes targeting multiple conserved regions of HIV-1 group M subtypes (subtypes A to H) and circulating recombinant forms (217, 218).
Since genetic diversity can significantly affect VL determinations, it is important that the communities of assay developers, clinicians, and laboratorians stay vigilant to quickly recognize abnormal VL determinations. Laboratories should compare different evaluation results of VL test kits and identify the ones that correctly quantify and match with strains circulating in the country. It is also recommended that one assay be used throughout the course of VL monitoring of the patient (219).
Decentralization of VL Testing
Decentralization of VL testing would be key in enabling the monitoring of all patients on ART, including those in rural areas and small treatment centers, in order to evaluate the success of the UNAIDS third 90 goal. The VL assay is complex, and current conventional platforms can be used at central reference, regional, district, or hospital laboratories within the tiered laboratory network (Fig. 1). The current conventional platform has a large footprint and a high throughput, allowing testing of several specimens a day. Decentralized laboratories for VL testing would need to be supported by strong systems, including a sample referral network, an EQA program, human resources, a quality management system, a laboratory information system (LIS), and a supply chain. For example, Ethiopia has a decentralized laboratory network for VL determination and EID together with an effective specimen referral network to ensure that patient specimens are efficiently transported to viral testing laboratories (220–222).
POC testing can be used to increase access to VL testing in rural areas and testing sites near the residence of patients. POC testing should be part of an existing laboratory diagnostic network. The current POC VL instrument has a small footprint and low throughput (223). Careful consideration should be taken with the introduction of POC VL testing within an existing laboratory network. These considerations include (i) site-level patient testing volumes, (ii) accessibility to remote sites, (iii) human resources and training, (iv) EQA and connectivity coverage, (v) the supply chain and equipment maintenance, and (vi) integration with other diseases, such as tuberculosis, and operational diagnostic function, such as EID or VL testing. Near-POC VL devices would still require reasonably renovated facilities with at least some minimum requirements (running water, reagent storage capacity, electricity, and waste disposal availability) in lower-tiered laboratories or in the community for their placement. In some settings, they may still require an efficient specimen referral system to transport specimens. As POC tests are introduced into existing laboratory networks, they should be accompanied by effective quality measures to monitor the quality of testing (179).
One of the challenges to guard against with the decentralization of VL testing is the temptation of “flooding” countries with conventional or POC VL instruments that may arise from multiple partners, with each partner procuring for the program. The lack of coordination among partners can result in a costly and less efficient system and can be burdensome to the entire laboratory network (224). Country and site targets should be used in conjunction with instrument capacity to inform what type and quantity of conventional or POC instruments should be procured or leased as well as their placement. The WHO reported the underutilization of conventional VL platforms, with only a 36.5% capacity of the existing platforms being used in the reported countries (224). Similarly, other studies have reported underutilization of POC or near-POC instruments (222, 225). Decentralized VL testing, if properly planned, offers several advantages, including (i) cost-efficient, flexible, and close monitoring of patients; (ii) faster turnaround times for patient results; and (iii) early detection of treatment failures for better patient management.
POC VL Testing Platforms
If properly implemented, POC VL testing locations can improve access to VL testing and improve patient care management. The POC VL system retains some unique features, which include (i) easy operation by nonlaboratory staff, (ii) short result turnaround time, (iii) built-in plasma isolation, (iv) reagent inclusivity and a self-contained cartridge, (v) accurate detection of VL of ≤1,000 copies/ml, (vi) low cost, (vii) a small footprint, (viii) connectivity or the ability to be electronically linkable, and (ix) low maintenance. The pipeline of POC VL platforms is promising (223), and five assays with advanced development and their performance characteristics are summarized in Table 4. Furthermore, the WHO has prequalified the GeneXpert platform for VL testing, and it is already being implemented in some field programs (226). The features of some POC VL platforms are summarized below.
TABLE 4.
Characteristics of major viral load assays intended for use at POC or near POC sitesa
| Manufacturer and trade name | Targeted virus(es)b | HIV-1 VL range (copies/ml)c | TAT | Plasma vol (µl) | Cold chain needed | Reference(s) |
|---|---|---|---|---|---|---|
| Alere q HIV-1/2 VL plasma | HIV-1, -2 | NA | 1 h | 50 | No | 233 |
| SAMBA Semi-Q | HIV-1 | 1,000 (cutoff) | 1.5 h | 200 | No | 227, 228 |
| Cavidi Ziva | HIV-1, -2 | 200–6 × 105 | 2 days | 200–1,000 | Yes | 210, 211 |
| Cepheid GeneXpert | HIV-1 | 40–107 | 1.5 h | 1,000 | No | 234 |
| Roche Liat | HIV-1, -2 | 57–1.5 × 106 | 0.5 h | 150 | Yes | 231, 232 |
Abbreviations: TAT, turnaround time from blood draw to VL report; NA, information not available.
Although HIV-1 and -2 are detected in three assays, they do not yield discrete VL for HIV-1 and -2.
The HIV-2 detection range of the Roche Liat and Alere q platforms have not been specified by the manufacturers.
Simple amplification-based assay.
The SAMBA (simple amplification-based assay) Semi Q platform manufactured by Diagnostic for the Real World (DRW) is a semiquantitative assay using the proprietary isothermic method to amply HIV-1 nucleic acid from an input volume of plasma and to capture amplified HIV targets onto the solid phase of a dipstick. Detector probes labeled with multiple hapten moieties and a colored antihapten detection conjugate are used to amplify the signal and yield a visible blue band within 90 min. It provides a qualitative one- or two-line result when the VL of the tested specimen is below or over 1,000 copies/ml, respectively, to determine ART success or failure (227, 228). It has been shown to detect all major HIV-1 group M subtypes (subtypes A to K), circulating recombinant forms, and groups N and O (228). Compared to the Roche Cobas AmpliPrep/Cobas TaqMan HIV-1 test, v2.0, with 488 clinical samples from London, Malawi, and Uganda, the accuracies of the SAMBA were 99%, 96.9%, and 95%, respectively (227). Further analysis of the Malawi and Uganda results showed that a SAMBA cutoff of 1,000 copies/ml was able to distinguish between suppressed and nonsuppressed patients. Also, a comparison of SAMBA results from 150 specimens from Ukraine with the those of the Abbott RealTime assay yielded 98% concordance (229). Importantly, the specimens were tested on-site in laboratories in district hospitals and health care facilities in Malawi and health care units in Uganda, with results being available the same day (111). Other features of the instrument include the possession of Bluetooth connectivity, and thus, the results can be electronically transmitted. To limit the interference of cellular nucleic acids, the manufacturer has developed a miniaturize leukodepletion column that can deplete more than 99.9% of white blood cells (WBCs) from 100 µl venous whole blood (230). The new device with the built-in WBC-depleting component significantly improved the assay for VL testing.
Roche Cobas Liat platform.
The Liat HIV quantitative plasma assay uses a pencil-sized and self-contained Liat tube with chaotropic lysis buffer and magnetic beads to extract RNA from 150 µl plasma. The tube contains reagents for RNA extraction and real-time RT-PCR quantitation in multiple segments and is processed by an actuator motor to control each step of the VL determination process. This assay also uses an internal armored HIV RNA to help provide an accurate VL calculation. The dynamic detection range is 57 to 1.5 × 106 copies (231), with a turnaround time of 30 to 35 min, to detect HIV-1 group M (subtypes A to H) and group O and HIV-2. The evaluation results of the Liat HIV quantitative plasma assay compared to the reference Roche CAP CTM v2.0 and Abbott RealTime HIV-1 assays showed good performance. Compared with the Roche platform, the clinical sensitivity of the Liat assay at a cutoff of 1,000 copies/ml was 100%, and the specificity was 88.2% (232). One advantage of the Liat assay over SAMBA Semi Q is its quantitative output. This device has full connectivity with Health Level-7 (HL7) communication protocols and has wireless and Ethernet capabilities. Currently, the storage and shipping of Liat tubes require cold-chain transportation, and the shelf life is only 6 months.
Alere q platform.
The Alere q assay uses microarray-based technology to yield results in 1 h. It utilizes a cartridge containing chaotropic salts and heat and mechanical disruption to extract RNA from 0.5 to 1 ml of plasma. HIV RNA is captured by HIV-1- and HIV-2-specific biotin-containing capture oligonucleotides and streptavidin-coated Sepharose particles. This technology uses real-time fluorescence and amplification to determine VL. In addition to target spots in the microarray to calculate VL, there are also control spots for positive hybridization and negative hybridization and internal controls to identify potential errors. No cold-chain storage is needed. The throughput of the analyzer is low because it runs only one specimen at a time. In addition to the plasma cartridge, there is also a whole-blood cartridge with a direct 25-µl capillary blood intake (233).
GeneXpert platform.
The Cepheid GeneXpert platform is available in a 1-, 2-, 16-, 48-, or 80-module configuration, thus providing facilities options to purchase the module best suited for their patient volume. The hands-on time for the HIV-1 VL assay is about 1 min, and the result can be obtained in 92 min. The device can process specimens in a random-access manner, an advantage at field sites, to yield rapid results without the need for batching of specimens prior to testing.
In Malawi, comparison of GeneXpert HIV-1 VL assay results with Abbott RealTime HIV-1 quantitative assay results showed high concordance (234). Similarly, this test also showed concordance with Roche Cobas TaqMan and Abbott RealTime assay results in India, the United States, and Europe (226, 235). Furthermore, the TAT from specimen collection to receipt of results was less than a day, compared to the 7 to 10 days that were required for the Roche Cobas TaqMan assay. During testing, the instrument encountered some problems, such as cartridge leakage; thus, there should be careful monitoring of cartridges to promptly identify defective or broken ones that can leak plasma samples (226). The Xpert HIV-1 VL assay with GeneXpert has been WHO prequalified (236).
DBS for VL Expansion
Until recently, most VL testing was often conducted in urban-based centralized laboratories because of the dedicated space, the relative ease of management, and the presence of better-trained technical staff. Whole-blood specimens from rural ART health care clinics are collected and transported at ambient temperature to laboratories within 6 h after blood draw (237). Plasma can be separated from whole blood and transported under a cold chain to centralized laboratories for testing. There is a need to scale up VL testing in order to provide access to all eligible patients on ART. One alternative sample type is DBS specimens to increase access to VL testing, especially in remote settings, because of its ease of collection under field conditions, storage, and transportation to centralized laboratories (238, 239).
Several studies have shown that DBS specimens are a convenient specimen type for VL testing. Cell-free virion-encapsulated HIV RNA is stable in a dried state for several weeks (137, 168, 240). There have been improvements in assay optimization to measure VL using DBS specimens. In a review of 13 publications on DBS VL testing, Smit and colleagues found that 52 to 100% of DBS samples yielded VL values within 0.5 log units of those of paired plasma samples (241). The sensitivity of DBS specimens for the detection of VL of >5,000 copies/ml was close to 100% compared to plasma VL testing. However, the sensitivity decreased to 78 to 100% when the VL threshold was 1,000 copies/ml. WHO recommendations then used of a threshold of 5,000 copies/ml to indicate treatment failure (242). The major reason for VL variation between DBS and plasma VL specimens is primarily the presence of proviral DNA and cell-associated RNA in DBS specimens (243). When obtaining VL from DBS specimens, most researchers used guanidinium-based agents from Roche to isolate nucleic acids from DBS filter paper discs. This process extracted not only RNA but also DNA from DBS specimens and resulted in an overestimation of VL. In a study with specimens from Equatorial Guinea,10.5% false-positive VL (>1,000 copies) were noted (244). The implications may be that when the VL is low, the amount of proviral DNA would become significant, and persons with VL lower than the threshold (1,000 copies/ml) may have elevated VL and be considered HIV nonsuppressed.
It is noteworthy that although the bioMérieux EasyMag extraction method also yields both RNA and DNA, its isothermal amplification is RNA dependent, and the copurified proviral DNA does not affect the VL results. This is well demonstrated by the higher specificities of identifying treatment failure by the NASBA than by the Roche method at a cutoff of 800 copies/ml (245). The Abbott DBS sample-processing approach (246) preferentially isolates RNA and obviates the proviral DNA problem (246). In a Kenyan study, plasma specimens were first processed with the Qiagen RNA-specific chromatographic isolation method, and although the Roche RT-PCR assay was used, the VL results obtained in this manner yielded 100% sensitivity and specificity for VL of <1,000 copies/ml (145). These reports underscored the importance of choosing an appropriate RNA-specific isolation or an RNA-specific amplification method to eliminate misclassification problems contributed by cellular proviral DNA.
A “free-virus elution” method was recently developed by Roche for DBS specimens (247). It uses a simple PBS solution to differentially elute plasma from DBS specimens. Using DBS specimens from 196 patients with and without ART, it was shown that the new method correctly identified 64 patients previously misclassified by using the guanidinium-based method (247). Other studies have shown a high correlation between matching DBS and plasma specimen VL results (248–251). Furthermore, DBS specimens are suitable for infants for whom a venous blood draw may be challenging, including the required volume, as opposed to a finger or heel prick. DBS VL results can be reliably used to monitor patients on ART and would increase access to VL testing, as it overcomes logistic challenges (collection, storage, and cold-chain transportation) posed by plasma samples.
HIV-2 VL Assays
HIV-1 and HIV-2 are the etiological agents of AIDS, and both viruses have similar modes of transmission, including sexual and, rarely, mother-to-child transmission. However, individuals infected with HIV-2 have slower progression to disease and have lower plasma VL (252, 253). Epidemiologically, HIV-2 is prevalent and mostly confined to West Africa (7). HIV-2 is phylogenetically classified into eight different subtypes (subtypes A to H), with subtypes A and B predominating (254). Unlike HIV-1 with several WHO-prequalified commercial VL assays, there are no WHO-prequalified HIV-2 commercial VL assays. As a result, in-house HIV-2 VL assays have been developed and used to monitor patients on treatment (253, 255).
The Cavidi assay to measure viral reverse transcriptase activity can measure HIV-1 and HIV-2. All the other commercial assays listed in Table 3 were manufactured specifically for HIV-1. HIV-2 commercial kits have been lagging, presumably because of the small number of HIV-2-infected persons. The new Liat (231) and Alere q (256) assays, ideal for POC locations, could also detect HIV-2. A couple of assays were recently developed using in-house HIV-2-specific primers with a low detection limit of 10 copies/ml (257).
VIROLOGIC METHODOLOGIES FOR EARLY INFANT DIAGNOSIS
EID Guidelines, Current Practices, and Bottlenecks
Globally, more than 2.1 million children under the age of 15 years were living with HIV and accounted for about 6% of all people living with HIV in 2016 (71). In the same year, there were 160,000 new HIV infections in children, decreases of 33% and 72% from the numbers in 2013 and 2002, respectively. Despite these achievements, eliminating pediatric HIV infections remains a major public health challenge. Progression to AIDS is aggressive in infants who acquire HIV infection in utero or around the time of delivery and are not on ART. Studies have shown that if these infants are left untreated, about 35% and 52% of the infants die before the ages of 1 and 2 years, respectively (258). With PMTCT interventions, including early ART intervention in infected pregnant women and prophylactic ARV treatment in exposed infants, vertical transmission of HIV was drastically reduced from 35% to less than 2% in developed countries (259, 260). Early diagnosis of infection in HIV-exposed infants (HEI) provides a unique opportunity to strengthen these infants’ follow-up and linkage to ART as well as the provision of prophylaxis, such as co-trimoxazole, for opportunistic infections and ART. Early identification of infants using serologic methods cannot establish the presence of infection because of passively acquired maternal antibodies in HEI. Virologic assays targeting viral nucleic acids are the methods of choice to minimize misdiagnosis (261, 262). Over several years, the colorimetric Roche Amplicor HIV-1 DNA qualitative assay was utilized as the gold standard in RLS countries (263). However, many quantitative and qualitative assays with high sensitivity and specificity using either plasma or DBS specimens have become available (Tables 2 and 3). However, pediatric testing and treatment continue to remain a challenge. In 2016, the rate of access to EID was only 43% for HEI tested by the second month of age, and of the 2.1 million infants living with HIV globally, only 43% received ART (264). Even when EID tests are conducted, a significant proportion of the results are not returned to the clinic or returned late, resulting in delayed diagnosis and ART initiation. Point-of-care testing and birth testing provide opportunities to improve EID services through rapid identification of infected infants and immediate initiation of treatment (57).
The WHO recommends the testing of all exposed infants at the 4- to 6-week postnatal immunization visit and requires the detection assay to have a minimum sensitivity of at least 95% and a specificity of 98% or higher (265). Importantly, ART should be initiated for children who test positive as soon as possible to minimize HIV-associated mortality. When the first EID test is positive, the infant should be initiated on ART while a second specimen is collected for confirmation of infection. Several factors can influence the identification of infected infants, including the VL in mothers, the timing of vertical transmission, ongoing exposure due to breastfeeding, the timing of the test (e.g., birth testing), maternal ART, and prophylactic intervention regimens (57). The 2016 WHO guidelines also recommended the use of birth testing (0 to 2 days) to identify infants who had in utero transmission in addition to testing at 4 to 6 weeks following birth. However, national programs should optimize testing at 4 to 6 weeks as a priority to identify perinatally infected infants with the greatest risk of early mortality before expanding birth testing. While a study from South Africa demonstrated the potential of birth testing in increasing the yield of HIV-infected infants being initiated on treatment, infants who tested negative were less likely to return for a follow-up EID (266). A negative result at birth testing for infected infants could be due to late transmission during labor or breastfeeding postnatally. The level of linkage of HIV-infected infants identified at birth was lower than that with routine EID testing and underscored the need to immediately initiate HIV-infected infants on ART. Since infants who test negative at birth are less likely to return for a follow-up EID, together with operational challenges following the introduction of birth testing, it is cost-effective to focus on EID at 4 to 6 weeks, which also coincides with established immunization visits and where staff have been trained to perform EID (267). WHO guidelines recommend the provision of PMTCT option B+ of lifelong ART to pregnant and breastfeeding HIV-infected women and that all exposed infants receive a course of prophylactic ART as soon as possible after birth to suppress viral replication. The WHO reaffirmed the use of EID at 4 to 6 weeks after birth, a second EID test when breastfeeding ends, and an HIV serologic test at 18 months to provide the final diagnosis (57).
POC for EID
DBS-based EID in RLS countries are conducted mostly in centralized facilities because of the relative ease in overall support for laboratory operations and quality management (66). Most of the conventional platforms for VL testing serve a dual role and are used for both VL and EID testing. Despite concerted efforts from various donors, agencies, and implementing partners, most of the EID programs in RLS countries continue to display challenges, including (i) the low testing coverage of eligible infants, (ii) the old age of tested infants, and (iii) the prolonged TAT (239, 268). These challenges can prevent early initiation of ART and lead to high infant morbidity and mortality rates (269–276). With the availability of POC technologies that can readily use capillary whole blood, the infection status could be determined in 1 to 2 h, thus enabling the provision of prompt care and ART initiation to infected infants during a same-day visit. All POC assays have undergone or are in the process of field evaluations to determine the ease of operation and assay accuracy compared to established reference assays, such as Roche CAP/CTM or Abbott RealTime assays (Table 5). Two POC EID tests, Alere q HIV-1/2 Detect and Cepheid GeneXpert, have been WHO prequalified and are being used in field settings (277).
TABLE 5.
Performance characteristics of major EID POC methodologiesa
| Manufacturer and trade nameb | Target | Amplification method and/or detection | Means of specimen processing | Specimen vol and type | LOD (copies/ml) | Operational time (h) | Field study site | No. of infants (median age) | Sensitivity, specificity (%) | Operator | Reference(s) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Alere q HIV-1/2 Detect | RNA | RT-PCR, fluorescence | Cartridge | 25 µl blood | 1 | Mozambique | 827 (5.6 wk) | 98.5, 99.9 | Nurse | 278 | |
| RNA | RT-PCR, fluorescence | Cartridge | 25 µl plasma | 2,491 | 1 | South Africa | 1,098 (6.7 wk) | 96.8, 100e | Laboratory staff | 279 | |
| Cepheid GeneXpert qualitative | TNA | RT-PCR, fluorescence | Cartridge | 100 µl whole blood | 278 | 1.5 | South Africa | 300 (0–18 mo) | 98.69, 100 | Laboratory staff | 234, 281 |
| 1 DBS | 688 | Malawi | 200 (1–6 mod ) | 100c , 100c | |||||||
| NWGHF Lynx p24 Ag | p24 | p24 antigen | NA | 80 µl plasma | NA | 1 | South Africa | 389 | 95.8, >99.4 | NA | 372 |
| Roche Liat | RNA | RT-PCR, fluorescence | Liat tube | 150 µl plasma | 80 | 1.5 | South Africa | NA | NA | NA | 232 |
| SAMBA Qual whole blood (SAMBA IIf ) | TNA | Dipstick, random access | Cartridges | 100 µl blood | 433 | 1.5 | NA | NA | NA | NA | 227, 230 |
Abbreviations: RT-PCR, reverse transcriptase PCR; TNA, total nucleic acid; NA, not applicable because no field study was reported at the time of preparation of the manuscript; NWGHF, Northwestern Global Health Foundation; LOD, limit of detection.
In alphabetical order.
Samples from 4 of the 200 infants yield invalid results and were not included in the sensitivity/specificity calculations. An Abbott RealTime assay using 2 DBS specimens was used as the reference.
No median age was provided.
After some testing errors were corrected upon retesting.
Can be run with the less-automated SAMBA I platform with a similar LOD (Table 3).
Alere q HIV-1/2 Detect.
The Alere q HIV-1/2 Detect assay is conducted in a small portable analyzer and uses a cartridge to process 25 µl capillary blood to produce results within 1 h. In a study conducted at 4 primary health clinics and a central hospital in Mozambique, consisting of 65 infected infants with known infection and 762 uninfected exposed infants with a median age of 1.4 months, the sensitivity and specificity were found to be 98.5% and 99.9%, respectively (278). That study also demonstrated the feasibility of the test being performed by clinic nurses after short pretesting training of 2 h. In another study in South Africa, involving 1,065 HIV-exposed infants with a median age of 47 days, the sensitivity and specificity of the Alere q HIV-1/2 Detect assay were 96.9% and 100%, respectively (279, 280). That study also found that birth testing in a subset of infants younger than 1 week of age yielded slightly a lower sensitivity of 93.3%, compared to the sensitivity of 96.9% for the routine standard of care, with high error rates (due to sample collection or operator error) that were subsequently resolved with additional testing. Testing was carried out by laboratory staff, and this highlights the critical need to routinely monitor these assays, even simplified assays. The current Alere q analyzer processes one specimen at a time; thus, an upgrade to multiple-specimen processing and random-access capacity would greatly improve its POC utility.
GeneXpert for HIV-1 qualitative detection.
The Cepheid GeneXpert HIV-1 Qual assay, similar to its quantitative VL version, has been evaluated in field settings in Malawi using DBS specimens, and results were concordant with those of the Abbott RealTime qualitative assay (234). This instrument can also use capillary whole blood, and other performance characteristics of the assay are shown in Table 5. The GeneXpert systems are manufactured in different configurations, including GeneXpert I, II, IV, XV1, Omni, Infinity-48s, and Infinity-80 (223). Although not yet commercially available, GeneXpert Omni is considered the true POC platform, with its flexibility to be moved around and used beside the patient, while the other GeneXpert systems are near-POC systems, requiring laboratory or facility settings for operability. GeneXpert has been WHO prequalified for use in diagnosing infants using whole blood or DBS specimens (281).
Lynx p24 antigen assay.
The Lynx p24 antigen assay is manufactured by Northwestern Global Health Foundation and is a strip-based assay with a battery-driven processor that yields results in less than an hour. It consists of a plasma separator to isolate plasma from 80 µl capillary blood. The detection of p24 requires a heat shock step to dissociate it from immune complexes. Detection is done by a colorimetric process using monoclonal anti-p24 antibodies. Similar to the SAMBA or HIV rapid test kits, it incorporates an “internal control” line and a “test” line. For a valid test result, the control line must appear, followed by interpretation of the test line: if the test line appears, then the test is positive, while the absence of a test line indicates that the test is negative. If the control line does not appear, the test is invalid. The performance of the Lynx p24 antigen assay was evaluated in Mozambique using heel-pricked whole blood and showed a sensitivity of 71.9% and a specificity of 99.6% compared to the gold-standard Roche Cobas AmpliPrep/Cobas TaqMan (CAP/CTM 96) HIV-1 qualitative test or the Roche Amplicor HIV-1 DNA test, v1.5, using corresponding DBS samples collected from the infants (282). Similar results were also observed when the Lynx p24 antigen assay was used in Zambia, with a sensitivity of 70% and a specificity of 100% compared to the Roche Cobas TaqMan assay (283). The Lynx p24 antigen assay is promising because of its high specificity; it does not require transportation or processing of specimens prior to testing, and the test can be performed in front of parents/guardians by nonlaboratory staff in facilities.
SAMBA HIV-1 qualitative assay.
The SAMBA HIV-1 Qual whole-blood test manufactured by DRW processes 100 µl of capillary blood with full automation in a SAMBA II device to produce results in less than 2 h. The working principle of using a dipstick display is similar to that of its semiquantitative VL counterparts (Table 5). It can detect HIV-1 groups M, N, and O with a limit of detection of 433 copies/ml and had 100% concordance of results with laboratory gold-standard results (284). It can process samples in a random-access manner and thus is highly advantageous for throughput and POC use.
SURVEILLANCE OF HIV DRUG RESISTANCE
There has been a rapid expansion of ART programs, with 20.9 million HIV-infected individuals having access to ART as of June 2017 (3), and with the expansion are fears or the inevitability of the emergence of HIV DR. If treatment programs are not optimally implemented or managed, there will be emergence and transmission of drug-resistant HIV strains with significant adverse public health impacts. Widespread resistance can compromise treatment programs, as it limits treatment options, with implications for first- and second-line therapy (285). In the realm of maternal and child health, it is critical for clinicians to be aware of HIV drug resistance in the HIV-positive women whom they serve to allow them to select an ART regimen that will prevent transmission of drug-resistant strains of the virus to the child. HIV DR can be categorized into two groups: (i) acquired resistance, whereby an infected individual on ART develops DRMs and, (ii) transmitted DR, when a drug-naive individual is infected with a virus having HIV DRMs. In surveys conducted between 2014 and 2016, rates of acquired DRMs of adults failing nonnucleoside reverse transcriptase inhibitor therapy (VL of >1,000 copies/ml) after 12 to 24 months were 59.7%, 47.3%, and 76.0% in Cameroon, Zambia, and Guatemala, respectively (286). Similarly, a review of studies with children failing first-line treatment showed high levels of DRMs of 90.4%, 67.6%, 91%, and 96.5% in Tanzania, Zimbabwe, South Africa, and Rwanda, respectively (286).
Recent PHIA surveys have shown high viral suppression rates of 91.0%, 89.0%, and 86.0% in Malawi, Zambia, and Zimbabwe, respectively, and suggest that first-line regimens are effective (13). It is important to monitor and prevent the emergence of DR mutations so that patients can continue to benefit from these lifesaving medications. The WHO recommended that countries in RLS implement HIV DR early-warning indicator surveys and classify resistance to each drug class as <5%, 5 to 15%, or >15% to gather information and inform national treatment programs with ART expansion (287, 288).
HIV DR testing can be conducted phenotypically or genotypically. Phenotypic assays are performed in vitro and measure susceptibility to ARVs in cell culture as a function of the concentration of the ARV required to inhibit the replication of HIV-1 by 50% (50% inhibitory concentration [IC50]). Due to the complexity of phenotypic assay protocols and the duration required to complete them, they are typically recommended for persons with known or suspected complex DR mutation patterns, particularly resistance to protease inhibitors (190), or for DR research (289). This assay requires 3 ml of plasma with a minimal VL of 500 copies/ml and 2 to 4 weeks for reporting, and it is not expensive (289).
Genotypic testing involves sequencing of the protease, reverse transcriptase, or integrase regions of the HIV genome and identification of DR mutations of clinical significance to ARVs. Genotype testing is recommended as the preferred method of resistance testing to guide therapy in patients with suboptimal virologic responses while on first- or second-line regimens in the United States (190). While it remains an integral part of standard HIV care and management in resource-rich countries, it is unaffordable in most resource-constrained countries due to its high cost and the need for advanced laboratory infrastructure and analytical skills. To monitor and support surveillance of DR in low- and middle-income countries, the WHO recommends population-based approaches to determine the transmitted DR threshold in recently infected populations, DR in ART-initiating populations, and acquired DR in ART-receiving populations (237).
Genotyping Methodologies
There are three basic types of genotyping methods: (i) Sanger-based sequencing, (ii) deep sequencing, and (iii) allele-specific discrimination assays (Table 6).
TABLE 6.
Major HIV-1 drug resistance detection methodologiesa
| Principle | Methodology | DRM variant detection rate (%) | Salient features | Challenges | Reference(s) |
|---|---|---|---|---|---|
| Phenotyping | PhenoSense | 20 | Patient-derived PR and RT (and INT in an additional assay) gene segments are cloned into a vector, and the IC50 of ART drugs for new recombinant virus is measured in vitro; 3 ml of plasma with a viral load of >500 copies/ml is needed | Long TAT (2–4 wk) and high cost | |
| Genotyping | |||||
| Sanger-based sequencing | Trugene/long-read Towers sequencer | 20 | FDA approved, well standardized for HIV-1 subtype B, closed sequencing system to cover PR codons 4–99 and RT codons 38–248, proven DBS utility | Not optimized for non-B subtypes, labor-intensive, high cost ($170/sample), integrase gene not included | 373, 374 |
| ViroSeq/ABI 3100 | 20 | FDA approved, well standardized for HIV-1 subtype B, semiopen sequencing system to cover Prot codons 1–99 and RT codons 1–335, proven DBS utility | Same as above, high cost ($210/sample) | 375 | |
| In-house, ABI 3100b | 20 | Optimized for subtype B, non-B subtypes, and CRFs; lower reagent cost ($40/sample) than commercial Sanger-based methods; proven DBS utility; ABI 3100 open-system sequencer | High overall operational complexity, expensive instrument maintenance | 290, 293, 294 | |
| Deep sequencing | Illumina MiSeq | 2 | FDA-approved sequencing platform, good for most subtypes and detects INT DRMs, uses barcoded primers to pool multiple samples for high-throughput (96 samples/run) sequencing, quick turnaround and low cost ($10/sample), can be adopted in national or multinational centralized laboratories | Relatively new, needs more validation with samples from developing countries and DBS | 296, 297 |
| Roche/454 GS Junior pyrosequencing | 5 | 3 amplicons to cover PR and RT genes, pools 48 samples with unique barcodes per sequencing run, rapid analysis (1 h) with a reduced cost of $20 per sample, sequencing errors near known homopolymer sites are reduced | Sequencing errors after homopolymer sites; Roche/454 may be phased out soon, and thus, the method may become obsolete | 295, 298, 299 | |
| Ion PGM 200 sequencing | 1 | 3 PCR segments, 1,657 bp (from gag p2 to 5′ RT), 2,002 bp (from 3′ RT to INT), and a 480-bp env gene (C2V3); covers DRMs from both subtype B and non-B subtypes and includes INT DRMs; also yields HIV-1 CCR5 coreceptor tropism genotypes | Newly established and requires independent validation and applications | 300 | |
| Multiplex allele-specific assay | In-house, Luminexb | 1.56–12.5 | Uses allele-specific primer extension to estimate % DRMs using suspension array, high throughput, low cost ($40/sample), quick TAT, DBS applicable, high applicability for personnel in RLS without the need for establishing a sequencing facility and skills | Subtype sequence information prerequisite prior to optimizing the assay; current assays are optimized for 20 DRM loci in subtypes B and C | 302, 303 |
Abbreviations: DRM, drug resistance mutation; Prot, protease; RT, reverse transcriptase; INT, integrase; env, envelope gene; CRFs, circulating recombinant forms; DBS, dried blood spot; C2V3, constant 2 and variable 3 domains of the env gene.
This is an in-house assay from the CDC.
Sanger sequencing assay.
Sanger sequencing targets the protease, reverse transcriptase, or integrase region of the HIV-1 genome for sequencing using the traditional Sanger sequencing principle, which is the selective incorporation of chain-terminating dideoxynucleotides during in vitro replication by DNA polymerase. The most commonly used commercial assays are the FDA-approved Trugene and ViroSeq assays. The reagent kits are costly and have been mostly used in the United States with plasma specimens or peripheral mononuclear cells. To reduce cost, in-house and DBS-applicable assays were developed using degenerate amplification and sequencing primers (290, 291). For instance, the in-house assay developed by the CDC has been used for population-based surveys using plasma specimens from Nigeria (292) and DBS specimens from Vietnam (290), Malawi (290), Nigeria (290), Kenya (142), and Uganda (293). The reagent cost of the assay is 20 to 25% lower than that of the commercial test kit method. Complete genotyping kits are available to WHO-designated and CDC-supported PEPFAR genotyping laboratories (294). It is worth noting that while the success rate of DR genotyping can be >90% using plasma with VL of >1,000 copies/ml (141, 295), the success rate with the use of finger-prick-derived DBS specimens is much lower (293). General recommendations for the preparation of DBS specimens and transportation for population DR surveillance are as follows: (i) venous blood should be used to prepare the DBS specimen; (ii) a finger-prick-based DBS specimen can be used if the VL is ≥50,000 copies/ml; (iii) the DBS specimen should be kept frozen prior to its shipment to a centralized DR laboratory in-country or overseas; (iv) postal or courier transportation can be done at ambient temperature, but the specimen should be delivered within 6 days (293); and (v) a DBS specimen received by the centralized laboratory should be kept frozen prior to genotyping.
Deep sequencing.
The three different types of deep sequencing instruments currently in use include Illumina MiSeq (296, 297), the Roche 454/GS Junior pyrosequencer (295, 298, 299), and the Ion PGM 200 sequencer (300). The operational principles of these methods are similar and can be completed to yield DRM reports in 1 to 2 days. DR-associated genes are amplified using patient-specific barcoded primers, and thus, amplified products from 48 to 96 patients can be pooled in a single deep sequencing run to drastically reduce the operational cost. Because of the clonal nature of amplified viral sequences, the sensitivity for detecting minor variants can be extended down from 20% by Sanger methods to 1% to 5%. One study using the Roche 454 GS Junior sequencer documented a 35% increased finding of DRMs compared to Sanger-based assays (298). A study involving 11 laboratories in Austria, Belgium, France, Germany, Italy, South Africa, Spain, and Switzerland confirmed the robustness and reproducibility of 454-asociated pyrosequencing (299). The same 454-asociated pyrosequencing platform was also successfully used in a study conducted by the Public Health Agency of Canada using 48 DBS samples selected from a transmitted DR surveillance study in Mexico (301).
All the above-mentioned genotyping assays require the performance of conventional sequencing on individual specimens or new high-throughput next-generation sequencing on pooled specimens. A common requirement for the application of these assays is the establishment of a complex high-maintenance molecular laboratory with DNA sequencers together with highly skilled laboratory personnel, thus making these assays most suitable for centralized national or regional molecular laboratories where samples from the lower health care facilities can be pooled and DRM can be determined with high-quality management.
Allele-specific discrimination assay.
Allele-specific discrimination assays do not require the use of sequencers and have been evaluated in RLS. For example, the multiplex allele-specific (MAS) array assay has been optimized for 20 DRM loci for subtype B (302). A study with a subtype C-specific MAS assay optimized for DBS specimens obtained from Honduras showed a high level of agreement (99.3%) of detected major DRMs compared with data generated by Sanger-based sequencing, and the low-abundance variants were verified by Roche 454 GS-based deep sequencing (303). The results from MAS assays are easy to interpret, and if optimized for known circulating viruses in a specific country or region, the assay can yield rapid DRM surveillance data to inform ART policy in the country or region.
Ensuring and Monitoring Quality of Molecular Testing in RLS
The implementation of a comprehensive quality management system that depends on continuous quality improvement is critical for ensuring the quality of HIV molecular testing. The scale-up of VL testing and EID has led to the decentralization of molecular laboratories in order to meet country needs (220). These laboratories are encouraged to participate in strengthening laboratory management toward accreditation to improve laboratories and then be enrolled into the WHO/Stepwise Laboratory Quality Improvement Process towards Accreditation for formal recognition using the 5-star grading system (304–306). Furthermore, some laboratories are aiming for international ISO15189 accreditation and ensuring compliance to standards in the laboratory (307, 308). Specimen integrity tracking can be accomplished through the use of data loggers during the specimen transport and storage phases. The data logger tracks and records the temperature of specimens or reagents during transportation and flags out-of-range temperatures to prevent erroneous results.
Internal and External Quality Control Materials
Strict QA standards, including quality control samples, are required to ascertain in vitro diagnostic claims (309). Quality control materials, including internal and external controls, are useful for laboratories to monitor their routine operations (310). For commercial VL quality control, noninfectious armored HIV-1 RNA is usually processed with patient specimens to monitor and calibrate equipment and identify problems with reagent and amplification contamination (311–313). The internal quality control and other kit controls are usually lot specific and are not suitable for the determination of lot-to-lot consistency or relative performance among different testing platforms. The instrument software captures and flags erroneous testing processes or specimens. False-negative results due to amplification inhibition and false-positive results due to low-level contamination in isolated specimens can be readily identified (314), followed by retesting of the affected specimens.
External quality control can provide the laboratory with a better understanding of testing variations and help identify systematic and random errors. Because of the high procurement costs, the use of external quality control to monitor variation across different kit lots, instruments, and laboratories is limited in RLS. External plasma RNA controls can be obtained from the Virology Quality Assessment (VQA) program, Accrometrix, and Seracare. However, they require cold-chain transportation and are expensive (315, 316).
Monitoring laboratory performance using Levey-Jennings charts could identify some systematic and random abnormal testing events. Interlaboratory comparison of the same method can be a valuable tool to compare and monitor the collective performance of laboratories within a laboratory network in multiple countries or regions. Assay manufacturers typically have a limited number of lots released concurrently; laboratories using the same external control can compare performances. Countries with multiple and different platforms can monitor performance and identify laboratories performing out of range in real time rather than waiting for an EQA panel (317, 318). Lot monitoring is critical to ensure that reagents are performing as intended. The importance of lot monitoring was recently underscored when certain VL reagent lots were found to be defective (319, 320). National laboratory programs should put into place a mechanism for systematic lot verification and work with manufacturers to manage situations where there are lot failures. Manufacturers should also perform lot verifications prior to shipment to minimize the impact of a recall if faulty lots could have been identified before shipment.
VL and EID PT Programs in RLS Countries
PT is a useful EQA method used to monitor the quality of VL testing, EID, or DR testing in a laboratory. Laboratories participating in PT programs have been shown to consistently perform better than those not enrolled in a PT program (163). HIV-1 DNA and RNA VL PT started in the 1990s to ensure the quality of results from clinical trials and are typically provided two to six times per year. For instance, the VQA program, through the support of the U.S. National Institutes of Health, provides qualitative DNA, VL, and DR panels, which are available four, six, and two times annually, respectively. The VL panel from the College of American Pathologists is available two times annually (321).
Most of these materials are based on subtype B viruses and not easily accessible to many RLS laboratories due to the high cost and the need to maintain a cold chain during shipment. To alleviate some of these challenges, the CDC initiated a free program for DBS-based EID PT and DTS-based VL PT provisions to RLS countries in 2006 and 2010, respectively. EID PT panels are prepared using filter papers spotted with seronegative whole blood spiked with known concentrations of viral DNA copies. Similar to DTS PT for HIV serology (151), the VL PT panel tubes contain dried materials with known VL and food dye for visualization (322). The DBS and DTS panels can be shipped through regular mail at ambient temperature. In 2012, 141 laboratories in 41 countries were enrolled in the EID PT scheme, and 114 laboratories in 44 countries were enrolled in the DTS VL PT scheme (322). The success of the DBS EID PT program was replicated by the National Health Laboratory Service (NHLS) in South Africa in 2013 and the Cheikh Anta Diop University (CADU) Afriqualab PT program in Dakar, Senegal, in 2014. Currently, both the NHLS and CADU serve 50 laboratories in 20 African countries. Afriqualab provides DTS VL PT panels to 24 regional laboratories. In addition, the WHO encourages laboratories to seek WHO accreditation for HIV DR testing to ensure compliance with quality standards, including receiving PT panels (323).
VL/EID Dashboard for Program Improvement and Accountability
Most often, implemented QA programs are limited to EQA programs such as PT programs, retesting of a subset of samples previously tested and supportive supervision, and the use of QC materials by testing sites (164–166, 324–326). Monitoring and evaluation of key indicators allow the identification of deficiencies or gaps to be remedied in the laboratory or program as well as assessment of laboratory or program improvement over time, thus providing accountability in the laboratory program. There is an increasing use of VL/EID dashboards, LIS, instrument connectivity, and cloud-based remote monitoring and tracking of key laboratory or program indicators. In Mozambique, mobile phone connectivity has been used to monitor the quality of POC platforms (327).
The utilization of a VL database in Uganda allows monitoring of the number of patients with ART failure (VL of >1,000 copies/ml) as well as the specimen rejection rate by the receiving laboratories in real time (328). Web-based tools, such as the HIV Infant Tracking System (HITSystem), have been used by multiple EID hospitals in Kenya (329). HITSystem requires a full-time coordinator and one or two staff in each of the participating laboratories for daily data entry. This system tracks a set of key indicators of the EID cascade, including infant age at DBS collection, time of laboratory receipt of samples, return of PCR results, mother notification, ART initiation, and infant retesting at 9 and 18 months. The system thus could provide data analysis for each and all sites for comparative analysis on a real-time basis to identify site-specific and overall program deficiencies.
Countries have developed dashboards at the national level for overall monitoring and improvement of programs. The dashboard enables viewing data at the regional and site-specific levels. For example, Kenya, Uganda, and Malawi have developed VL and EID dashboards that allow the programs to monitor several parameters for deficiencies and improvements (328, 330, 331), and similar systems are also being used in China and Cambodia.
CD4 COUNTING METHODOLOGIES
Role of CD4 and Testing Guidelines in Staging and Monitoring Patients on ART
In 1981, the CDC described the presence of Pneumocystis pneumonia in three homosexual men in Los Angeles, CA (332), and Gottlieb et al. described Pneumocystis pneumonia and mucosal candidiasis in four homosexual men with a “virtual elimination” of the Leu-3+ (CD4) helper/inducer subset (333). As reports of severe immune deficiency increased, CD4 cell counts became a critical part of the care of HIV-infected patients and were used as a surrogate marker for staging of patients and as an indicator for disease progression.
Prior to 2015, CD4 T cell count was the primary laboratory-based test used to guide the initiation of patients on ART and to monitor treatment response. WHO global ART guidelines for adults and adolescents in 2002 recommended that ART be initiated in adults with clinical stage IV disease or with a CD4 cell count of <200 cells/mm3 (334). While these guidelines acknowledged that CD4 testing was not widely available in RLS and advocated for the development of new technologies that could be used in such settings, they included immunological failure, defined as a fall in CD4 counts of over 30% from the peak value or a return to or a fall below the pretherapy baseline level, as one of the criteria for switching from first- to second-line ART. In 2003, the WHO revised the guidelines and recommended ART initiation in adults and adolescents with clinical stage III disease and consideration of using CD4 cell counts of <350 cells/mm3 to guide decision-making (335). These guidelines also recommended that district- and regional-level hospitals should have the capacity to perform CD4 testing and that patients on ART should be monitored by CD4 testing every 6 to 12 months, if available.
By 2006, the guidelines recommended ART initiation in all patients with WHO stage IV disease and all patients with CD4 counts of ≤200 cells/mm3, regardless of clinical staging, and consideration of ART for patients with CD4 counts of between 200 and 350 cells/mm3 before the count dropped to below 200 cells/mm3. In these guidelines, the WHO advocated for wider availability of affordable POC CD4 testing to increase access to CD4 results. CD4 testing was recommended at baseline, ART initiation, and suspected treatment failure and every 6 months prior to and after ART initiation (336). Between 2006 and 2010, several studies demonstrated that early initiation of ART led to reductions in AIDS-related morbidity and mortality (337–342). Following this, the WHO updated the ART guidelines to recommend that, in addition to patients with clinical stage III and IV disease, irrespective of the CD4 count, asymptomatic patients with CD4 counts of ≤350 cells/mm3 should be initiated on ART (343). In 2010, the WHO stated that all patients should have access to CD4 testing and recommended the use of VL testing to confirm suspected treatment failures. CD4 testing continued to be recommended at baseline, at ART initiation, during treatment, at treatment failure, and every 6 months after ART initiation. In 2013, the WHO raised the CD4 cutoff and recommended that ART be initiated in patients with CD4 counts of ≤500 cells/mm3, prioritizing those with CD4 counts of ≤350 cells/mm3 (184). VL testing was recommended as the preferred monitoring approach to diagnose and confirm treatment failure, with CD4 testing being recommended in areas with an insufficient VL testing capacity. The frequency of CD4 testing for HIV-infected patients remained at baseline, at ART initiation, at treatment failure, every 6 to 12 months pretherapy, and every 6 months posttherapy (184).
The evolution of the WHO guidelines on CD4 cutoffs for treatment initiation clearly illustrated the fundamental role that CD4 testing has played in HIV patient care over the years. With strong evidence from the Strategic Timing of Antiretroviral Therapy, TEMPRANO, and HPTN 052 trials demonstrating substantial clinical benefits of early ART initiation, particularly when CD4 cell counts are above 500 cells/mm3 (5, 344, 345), the WHO released new guidelines in 2016 recommending the initiation of ART for all PLHIV irrespective of CD4 counts (57). While the guidelines no longer require a CD4 test prior to ART initiation, they recommend that individuals with CD4 counts of ≤350 cells/mm3 be given priority for ART initiation, especially where resources may not be available for initiation of treatment of all infected patients. In the 2016 guidelines, HIV VL testing remained the preferred method for ART monitoring, and CD4 testing is recommended at baseline, every 6 months until patients are stable on ART, and in areas where VL testing is not available. Together, these recommendations represent a shift in the utilization of CD4 cell counts in HIV care and treatment, ultimately leading to a reduction in the volume of CD4 testing in RLS. Despite this shift in routine clinical monitoring, CD4 testing remains an important clinical test for assessing the risk of disease progression, informing decisions on prophylaxis, and prioritizing assessment for opportunistic infections.
Flow Cytometry and Pipeline of Commercial CD4 Instruments and Assays
Researchers and clinicians have developed CD4 T cell flow cytometry as a clinical laboratory test for identifying and monitoring HIV-infected patients. Flow cytometry allows the quantification and analysis of single cells using light scatter to determine cellular size and internal complexity and using laser excitation of fluorochromes bound to cellular components to identify cells of interest. The first CD4 assays utilized a dual-platform approach in which a flow cytometer was used to determine the percentage of CD4 T cells and a hematology analyzer was used to provide the total lymphocyte count. Both values were then used to calculate the absolute CD4 T cell count (346). Volumetric flow cytometry delivers a precise volume of the sample through the flow cell, and bead-based counting (i.e., adding a known concentration of fluorescent beads to each sample) enabled determinations of CD4 percentages and CD4 absolute counts to be performed on one instrument, thus reducing the processing complexity and variability of results (347, 348).
The need for robust CD4 testing platforms and assays that could be utilized in settings with limited infrastructure and human and financial resources was emphasized early in the HIV epidemic. The Becton, Dickinson (BD) FACSCount system, released in 1994, was the first dedicated instrument for clinical CD4 cell enumeration (349). It is a closed system using solely BD reagents. The BD FACSCount instrument is a color flow cytometer that is used with FACSCount reagent kits to measure CD4, CD3, and CD8 cell counts. Additional reagents, such as the BD FACSCount CD4 reagent, subsequently became available to measure both CD4 percentages and absolute CD4 cell counts. The FACSCount system was designed for ease of operation, as it featured automatic gating and premeasured reagent tubes. Due to the simplicity of the instrument and the moderate cost, the FACSCount system remains the most widely utilized benchtop CD4 analyzer in RLS (350).
One of the limitations of FACSCount was its relatively low throughput, with only 30 to 80 specimens that could be analyzed per day (350). This led to the production and standardization of high-throughput flow cytometers with dedicated CD4 reagents and software packages, such as the Beckman Coulter Epics XL instrument with the TetraONE system and reagents and the BD FACSCalibur instrument with the BD Tritest and BD Trucount tubes. These instruments are open systems, and reagents are not restricted to those provided by the manufacturer. They are also multicolor cytometers that have the capacity to accommodate CD4 testing needs in high-prevalence settings. Both cytometers have automatic loading options and off-board staining platforms available to automate processing and increase the throughput for high-volume laboratories. While these instruments remain widely used and often serve as the reference instruments for evaluations of new CD4 technologies, their suitability for use in only centralized referral laboratories limited patient access in smaller communities and rural areas.
A wave of new CD4 technology entered the market, including the Millipore-Guava Auto CD4%/CD4 system, the Apogee Auto40 flow cytometer, and the Partec CyFlow counter (223). These instruments are volumetric benchtop cytometers that are capable of measuring both CD4 percentages and absolute counts without the use of reference beads. The Apogee Auto40 cytometer was designed to be used in military environments and is therefore a practical cytometer for RLS. Similarly, the Partec CyFlow counter was designed with “align-free” technology and lyophilized reagents, reducing the level of service required and the need for cold-chain transportation and storage.
The Beckman Coulter Aquios CL instrument with Tetra-1 was released in 2015 and is marketed as the first “load-and-go” cytometer. This is a large benchtop cytometer that is fully automated and reduces hands-on time for laboratorians. All reagents are barcoded and tracked on the instrument, along with specialized quality control materials, which are analyzed onboard and flagged if out of range or expired. The Beckman Coulter Aquios CL instrument represents a new age of clinical flow cytometry that automates specimen processing and tracks the validity of quality control and the shelf-life of reagents.
Expansion of CD4 Services Using POC Assays
While complex automated instruments save time in large reference laboratories with highly trained staff and good infrastructure, they are not well suited for remote geographical regions and laboratories with nontechnical staff and minimal infrastructure (Table 1). Weak specimen transport and supply chain systems, limited infrastructure at lower-tiered laboratories, and shortages of trained staff have been challenges for flow cytometry-based CD4 assays in high-burden settings. Alternative strategies to overcome these challenges include the manufacture of more community-accessible, affordable, and simple-to-perform POC CD4 instruments that would afford quality HIV care in RLS (351–353).
The first POC CD4 instrument to reach the market was the PointCare Now system in 2007. This device was designed for RLS, as it used a light-emitting diode instead of a laser. It has heat-stable reagents, daily quality controls, and a short testing time of 8 min per sample. However, independent field evaluation of the PointCare Now system showed an unacceptable high mean relative bias of +153 cells/µl for absolute CD4 counts (354). Subsequently, many POC CD4 instruments and assays with significant improvement were developed to support HIV treatment programs in Africa (14, 355, 356). The Alere Pima CD4 system has been the most widely used and distributed POC CD4 device (14, 356–363). It is small and easily transportable and uses an all-in-one cassette and static image analysis to provide absolute CD4 counts. It is compatible with the Alere connectivity solution, which sends results across a mobile network from the analyzer to a remote server to be viewed and analyzed via the Alere data point website. The centralized data point allows comprehensive and fast program management of supply stock, error analysis, and QC monitoring for multiple Pima analyzers located at different sites. Several evaluations have shown acceptable concordance between the results of the Pima POC analyzer and reference cytometers (357, 359, 360, 362, 363). The Pima analyzer has exerted a profound impact and changed the CD4 testing landscape in Africa and other continents.
The Partec CyFlow CD4 MiniPOC and BD FACSPresto systems are POC devices that provide both absolute CD4 counts and percentages, but they are not as widely distributed as the Pima analyzer. The Partec CyFlow CD4 MiniPOC system utilizes the same volumetric cytometry principles as the Partec CyFlow counter as well as lyophilized reagents and controls, allowing room-temperature storage. A laboratory and field evaluation in Senegal demonstrated good agreement between the MiniPOC system and the reference FACSCount cytometer (364).
The BD FACSPresto system is an image-based analyzer with an all-in-one cassette that, unlike the Alere Pima system, offers an off-board incubation time of 18 min and a shorter read time of 4 min on the instrument and is able to accommodate up to 10 specimens at one time. Laboratory and field evaluations conducted in Kenya (216) and South Africa (217) showed acceptable agreement between the results of the BD FACSPresto and reference CD4 instruments. However, a positive bias of 40 to 67 cells/µl for the FACSPresto system was reported in both studies (365, 366).
Another POC CD4 technology is the EMD Millipore Muse Auto CD4/CD4% system (367, 368). This technology replaces the discontinued Guava Auto CD4%/CD4 system. It utilizes volumetric cytometry and microcapillary technology similar to those of the Guava system. The instrument is preloaded with autogating software, and its assay reagents permit the determination of both CD4 counts and percentages.
POC CD4 testing has many benefits over centralized testing, including increased access to testing, reduced TAT, and minimal infrastructure requirements (Table 1). The introduction of POC devices such as Alere Pima, Partec CyFlow CD4 MiniPOC, and BD FACSPresto provides national programs the opportunity to decentralize and expand access to CD4 testing. The use of POC devices has been shown to significantly reduce the TAT of CD4 results and subsequently the time to initiation of ART, doubled linkage to care, and increased patient retention in health care (14, 15, 355, 356, 369). There are several promising POC systems for CD4 in the pipeline (223). Following standard evaluations of platforms that enter the market with WHO prequalification, they will offer options for use in CD4 testing.
Monitoring Quality of CD4 Testing
Despite these advantages of POC technology, significant challenges surrounding QA, implementation, service maintenance, and supply chain management persist. Ensuring uninterrupted provision of accurate and reliable CD4 testing will require a substantial and sustained effort to identify needs and gaps within the care cascade, proper implementation of POC and laboratory-based testing to fill these gaps, and continuous monitoring and oversight to identify and resolve quality issues. CD4 testing facilities should be enrolled into EQA schemes, for example, UK NEQAS (United Kingdom National External Quality Assessment Services), to monitor the quality of CD4 testing. Also, the implementation of connectivity would enable monitoring and improving QA, service, and maintenance at multiple sites. Also, the use of the WHO Stepwise Process for Improving Point-of-Care Testing (SPI-POCT) to monitor POC testing sites for CD4 would identify gaps and allow monitoring of improvement of sites to maintain quality testing (167).
Role of CD4 Counts in the Era of “Test and Start” and VL Testing
Over several years, CD4 counts have played a pivotal role in making clinical decisions in the management of HIV/AIDS. First, CD4 counts were used to determine the eligibility of patients for initiation of ART. As our understanding of better management of AIDS patients with ART improved, ART initiation moved from the use of CD4 counts of less than 200 cells/µl to counts of 350 cells/µl and, later, 500 cells/µl. The increasing threshold of CD4 counts was meant to avoid patients presenting with already advanced progression to disease before ART initiation. Second, CD4 counts were used to monitor and classify disease progression and survival outcomes. Third, CD4 counts were used to monitor the effectiveness of ART as well as inform a switch to second-line treatment after ART failure. Fourth, CD4 counts have been used to assess immune reconstitution and advise on the initiation or discontinuation of prophylaxis for opportunistic infections.
In the advent of “test and start” and VL testing, there is uncertainty as to the role of CD4 counts. The 2016 WHO guidelines recommended diagnosis and initiation of a positive individual into ART regardless of CD4 counts. Furthermore, the guidelines recommended the use of VL testing for monitoring the effectiveness of ART. As the scale-up of VL testing has increased, there has been a decline in CD4 testing in some countries (370). Nonetheless, CD4 counts still have a role to play in the management of patients. As recommended by WHO 2016 guidelines, CD4 counts should still be used for monitoring ART patients in situations where there is no capacity for VL testing. For some RLS, full transition to a test-and-start regimen remains challenging, and in these scenarios, CD4 counts can be used to prioritize patients. Also, CD4 cell counts are useful for monitoring patients with advanced disease (<200 cells/µl) and informing clinical decisions to prevent mortality. The number of patients presenting with advanced disease is still high and is between 34 and 41% in some SSA countries (371).
Despite the huge investment in laboratory diagnostic strengthening, including POC testing, over the past 2 decades to support CD4 testing, coupled with the era of test-and-start and VL testing, determination of CD4 counts still has its place in the care and management of patients, especially those with advanced disease. Thus, national programs will need to consider this as they make strategic decisions to provide better patient management. The rapidly evolving role of CD4 counts, especially in the last decade, can be challenging for any robust program and more so for programs in RLS, and stakeholder support would be critical.
CONCLUSION
HIV diagnostics have evolved significantly over the past 3 decades from first-generation to fifth-generation assays, with each generation aimed at overcoming a deficiency of the previous one in order to provide improved diagnosis offered by these tests. Similarly, the advent of HIV molecular testing shortened the window period and also advanced from initial designs based primarily on HIV subtype B to encompass non-B subtypes in order to overcome barriers with HIV genetic diversity. In addition to tackling HIV genetic diversity, we have adjusted to program needs to improve access to testing, misdiagnosis, and their impact on patients. National programs routinely evaluate test kits to establish suitable testing algorithms using combinations of tests for accurate diagnosis, for example, selecting the optimal combination of HIV rapid test kits in a serial or parallel testing algorithm for improved diagnosis. New robust and simple assays are available to distinguish recent HIV-1 infection from long-term infection, permitting estimation of incidence, impact assessment, and real-time surveillance. Program challenges with decentralization have led to adaptations of approaches, including task shifting, in order to optimize the use of various diagnostic tools, such as the use of both professional laboratory and nonlaboratory personnel (nurses, lay counselors, and community workers) to provide and expand diagnosis. Ensuring the quality of diagnostic assays (at the facility and community levels) has required a combination of both traditional and innovative strategies, including positive and negative quality control materials, proficiency testing or an EQA program, site and tester certification, connectivity, and the use of Q-Corps and community volunteer involvement in monitoring quality results in nonlaboratory settings. The selection of appropriate diagnostics in an evolving field with frequently changing guidelines has been challenging but necessary in order to improve the quality of patient care with a better understanding of the epidemic. Such has been the impact of the rollout of HIV POC testing on expanding and increasing access to testing. Diagnostics have played a critical role in the fight against HIV/AIDS, and achieving epidemic control will require responsiveness and adaptation of HIV diagnostics to overcome program challenges in identifying HIV status and monitoring HIV-infected patients on ART.
ACKNOWLEDGMENTS
This work has been supported by the President’s Emergency Plan for AIDS Relief (PEPFAR) through the Centers for Disease Control and Prevention.
All authors, except B.S.P., report no conflicts. As an inventor of the BED-Capture EIA, LAg-Avidity EIA, and rapid incidence test, B.S.P. receives a portion of royalties received by the CDC as per policy of the U.S. Government.
The findings and conclusions of this report are those of the authors and do not necessarily represent the official position of the funding agencies.
Biographies

Bharat S. Parekh, Ph.D., obtained his B.Sc. and M.Sc. from the University of Mumbai, India, before moving to the United States, where he earned his doctorate in Biochemistry/Virology from Louisiana State University, Baton Rouge, working on lentiviruses. He did his postdoctoral research at Scripps Research Institute in La Jolla, CA, working on hemorrhagic fever viruses. Prior to joining the CDC, he was a Senior Scientist/Project Leader at Bio-Rad Laboratories in California, where he was engaged in research and development of HIV and human T lymphotrophic virus (HTLV) diagnostic assays, including confirmatory Western blot assays. As a Team Lead of the HIV Serology/Incidence Team, Dr. Parekh currently provides critical laboratory support to the CDC’s global HIV work as part of the PEPFAR in 40+ countries. HIV-1 incidence assays and quality assurance tools developed in his laboratory are used worldwide to estimate HIV-1 incidence to measure the impact of HIV programs and ensure accuracy of HIV-related diagnostic testing.

Chin-Yih Ou, Ph.D., received his M.S. in microbiology at National Taiwan University (1976) and Ph.D. at the Oak Ridge National Laboratory, TN (1982). In the late 1980s, as the chief of the molecular laboratory in the Global AIDS Program, U.S. CDC, he collaborated with the scientists in Cetus to pioneer the novel application of PCR to directly determine HIV infection in adults and infants. In addition, with the combination of the strengths of the CDC’s epidemiology and molecular programs, he helped establish the new HIV study frontier of molecular epidemiology. He served as the laboratory manager for the Clinton Health Access Initiatives and the CDC’s Global AIDS Program in China in 2008 to 2009 and 2009 to 2014, respectively, prior to his retirement in 2014.

Peter N. Fonjungo, Ph.D., is the Associate Chief for the Africa Region at the International Laboratory Branch, Division of Global HIV and TB, Centers for Disease Control and Prevention (CDC). He provides technical assistance in support of strengthening public health systems and quality service delivery for President’s Emergency Plan for AIDS Relief (PEPFAR) programs in more than 40 countries. He received a master of science in Molecular Biology from the University of Aberdeen and earned his Ph.D. in Molecular Immunology at the University of Edinburgh, United Kingdom. He completed a postdoctoral program at the CDC with a focus on HIV genetic diversity and implications for HIV diagnosis as well as HIV drug resistance. He has served as Laboratory Director at three CDC field stations (Cameroon, Botswana, and Ethiopia) and served in clinical trials teams, including a combination prevention intervention trial for HIV infection and the safety and efficacy of preexposure prophylaxis for the prevention of HIV infection.

Mireille B. Kalou, M.D., M.P.H., earned her medical doctor degree from the University of Abidjan, Côte d’Ivoire, and a master’s degree in Public Health from the University of California, Berkeley. She first joined the U.S. Centers for Disease Control and Prevention (CDC) in 1998, where she served as a research fellow in Côte d’Ivoire and subsequently as the laboratory director in Cameroon. In 2007, she joined the CDC Division of Global HIV/TB, where she led efforts to improve the quality and accuracy of HIV testing and to strengthen laboratory systems in PEPFAR-supported countries. She designed several training programs and evaluated over 50 HIV laboratory tests and testing algorithms for HIV diagnostics. She provided technical assistance to ministries of health and other implementing partners to improve laboratory services and systems. Since 2013, she has served as project coordinator for the PEPFAR-funded continuous quality improvement initiative for HIV rapid testing.

Erin Rottinghaus obtained a bachelor of science in Biology from Truman State University in Kirksville, MO, and a Ph.D. in Integrated Biomedical Science from The Ohio State University, where she studied the response of the aging immune system to Mycobacterium tuberculosis. She has worked at the Centers for Disease Control and Prevention in the Division of Global HIV/AIDS and TB (International Laboratory Branch) in various capacities since 2009 and is currently leading the Laboratory Data Monitoring Team. Dr. Rottinghaus has always had an interest in infectious diseases and has found her passion working in public health, where innovation and research are utilized to make evidence-based decisions to improve lives.

Adrian Puren, M.B.B.Ch., Ph.D., was trained and held a lectureship at the University of the Witwatersrand, Johannesburg, South Africa, before taking on various positions at the National Institute for Communicable Disease, Johannesburg. He was appointed as Deputy Director and Head of Virology in 1999 and Head of the Centre for HIV and STIs in 2017. As head of virology, he focused on developing and implementing a range of viral diagnostics in support of surveillance and diagnostics. He serves as the quality assurance technical manager for the NICD and in this capacity has provided support to the National Department of Health’s implementation and quality assurance of HIV rapid testing. He has supported various initiatives to improve quality of testing, including the PEPFAR-supported African Centre for International Laboratory Training (ACILT) and Southern African Development Community (SADC) implementation of minimum standards for laboratories. He heads the regional and national endpoint diagnostics laboratory for HVTN-supported vaccine trials.

Heather Alexander, Ph.D., earned a doctorate in Microbiology and Molecular Genetics from Emory University in Atlanta, GA. She joined the U.S. Centers for Disease Control and Prevention in 2005, where she served as a Microbiologist in the Division of Tuberculosis Elimination, International Research and Programs Branch, for five years. From 2010 to 2017, Dr. Alexander was the TB/HIV Unit Lead and then Clinical Monitoring Team Lead in the International Laboratory Branch of the Division of Global HIV and TB. Since October 2017, Dr. Alexander has served as the Chief of the International Laboratory Branch in the Division of Global HIV and TB.

Mackenzie Hurlston Cox is a Health Scientist within the HIV Viral Load and Early Infant Diagnosis Team in the International Laboratory Branch (ILB) at the Division of Global HIV and TB, Center for Global Health, U.S. Centers for Disease Control and Prevention (CDC), Atlanta, GA. She received her bachelor of arts in Middle Eastern Studies from Emory University and her master of science in Public Health from the Rollins School of Public Health, Emory University. From 2008 to 2012, she worked as a research specialist and lead research specialist at the Emory Vaccine Center in Dr. Eric Hunter’s laboratory. She has served in in her current role since 2012; during this time, she has focused on evaluation of new molecular technologies for HIV load and early infant diagnosis as well as development of quality assurance materials and training resources suited for use in resource-limited settings.

John N. Nkengasong is Director of the Africa Centers for Disease Control and Prevention. He served as the deputy principal director (acting) of the Center for Global Health, U.S. Centers for Disease Control and Prevention (CDC). Between 2005 and 2017, he was Chief of the International Laboratory Branch at the U.S. CDC. He has received numerous awards for his work, including, but not limited to, the U.S. Secretary of Health and Human Services Award for excellence in Public Health Protection Research, the Sheppard Award, and the William Watson Medal of Excellence, the highest recognition awarded by the U.S. CDC. He is also recipient of the knight of honour medal by the government of Cote d’Ivoire and on 19 June 2017 was knighted as the officer of Lion by the president of Senegal, H. E. Macky Sall, for his significant contributions to public health. He has authored over 200 peer-reviewed articles in international journals and published several book chapters.
REFERENCES
- 1.UNAIDS. 2017. Fact sheet—World AIDS Day 2017. UNAIDS, Geneva, Switzerland: www.unaids.org/sites/default/files/media_asset/UNAIDS_FactSheet_en.pdf. Accessed 2 April 2018. [Google Scholar]
- 2.UNAIDS. 2015. Treatment 2015. UNAIDS, Geneva, Switzerland: www.unaids.org/sites/default/files/media_asset/JC2484_treatment-2015_en_1.pdf. Accessed 7 March 2018. [Google Scholar]
- 3.UNAIDS. 2017. Fact sheet July 2017. UNAIDS, Geneva, Switzerland: www.unaids.org/sites/default/files/media_asset/UNAIDS_FactSheet_en.pdf. Accessed 11 November 2017. [Google Scholar]
- 4.Cohen MS, McCauley M, Gamble TR. 2012. HIV treatment as prevention and HPTN 052. Curr Opin HIV AIDS 7:99–105. doi: 10.1097/COH.0b013e32834f5cf2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grinsztejn B, Hosseinipour MC, Ribaudo HJ, Swindells S, Eron J, Chen YQ, Wang L, Ou SS, Anderson M, McCauley M, Gamble T, Kumarasamy N, Hakim JG, Kumwenda J, Pilotto JH, Godbole SV, Chariyalertsak S, de Melo MG, Mayer KH, Eshleman SH, Piwowar-Manning E, Makhema J, Mills LA, Panchia R, Sanne I, Gallant J, Hoffman I, Taha TE, Nielsen-Saines K, Celentano D, Essex M, Havlir D, Cohen MS, HPTN 052-ACTG Study Team. 2014. Effects of early versus delayed initiation of antiretroviral treatment on clinical outcomes of HIV-1 infection: results from the phase 3 HPTN 052 randomised controlled trial. Lancet Infect Dis 14:281–290. doi: 10.1016/S1473-3099(13)70692-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO. 2015. Guideline on when to start antiretroviral therapy and on pre-exposure prophylaxis for HIV. WHO, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/186275/1/9789241509565_eng.pdf. Accessed 31 January 2016. [PubMed] [Google Scholar]
- 7.UNAIDS. 2017. Fact sheet—latest statistics on the status of the AIDS epidemic: global HIV statistics. UNAIDS, Geneva, Switzerland: www.unaids.org/en/resources/fact-sheet. Accessed 3 August 2017. [Google Scholar]
- 8.UNAIDS. 2016. 2016 United Nations political declaration on ending AIDS sets world on the fast-track to end the epidemic by 2030. UNAIDS, Geneva, Switzerland: www.unaids.org/sites/default/files/20160608_PS_HLM_Political_Declaration_final.pdf. Accessed 4 January 2018. [Google Scholar]
- 9.UNAIDS. 2014. Ending the AIDS epidemic by 2030. UNAIDS, Geneva, Switzerland: www.unaids.org/sites/default/files/media_asset/JC2686_WAD2014report_en.pdf. Accessed 31 January 2016. [Google Scholar]
- 10.UNAIDS. 2014. 90-90-90: an ambitious treatment target to help end the AIDS epidemic. UNAIDS, Geneva, Switzerland: http://www.unaids.org/sites/default/files/media_asset/90-90-90_en.pdf. Accessed 22 September 2017. [Google Scholar]
- 11.Belec L, Bonn JP. 2011. Challenges in implementing HIV laboratory monitoring in resource-constrained settings: how to do more with less. Future Microbiol 6:1251–1260. doi: 10.2217/fmb.11.121. [DOI] [PubMed] [Google Scholar]
- 12.Aleku GA, Adoga MP, Agwale SM. 2014. HIV point-of-care diagnostics: meeting the special needs of sub-Saharan Africa. J Infect Dev Ctries 8:1231–1243. doi: 10.3855/jidc.4664. [DOI] [PubMed] [Google Scholar]
- 13.PEPFAR. 2017. PEPFAR latest global results. PEPFAR, Washington, DC: https://www.pepfar.gov/documents/organization/264882.pdf. Accessed 3 August 2017. [Google Scholar]
- 14.Jani IV, Sitoe NE, Alfai ER, Chongo PL, Quevedo JI, Rocha BM, Lehe JD, Peter TF. 2011. Effect of point-of-care CD4 cell count tests on retention of patients and rates of antiretroviral therapy initiation in primary health clinics: an observational cohort study. Lancet 378:1572–1579. doi: 10.1016/S0140-6736(11)61052-0. [DOI] [PubMed] [Google Scholar]
- 15.Desai MA, Okal DO, Rose CE, Ndivo R, Oyaro B, Otieno FO, Williams T, Chen RT, Zeh C, Samandari T. 2017. Effect of point-of-care CD4 cell count results on linkage to care and antiretroviral initiation during a home-based HIV testing campaign: a non-blinded, cluster-randomised trial. Lancet HIV 4:e393–e401. doi: 10.1016/S2352-3018(17)30091-7. [DOI] [PubMed] [Google Scholar]
- 16.Onywera H, Maman D, Inzaule S, Auma E, Were K, Fredrick H, Owiti P, Opollo V, Etard JF, Mukui I, Kim AA, Zeh C. 2017. Surveillance of HIV-1 pol transmitted drug resistance in acutely and recently infected antiretroviral drug-naive persons in rural western Kenya. PLoS One 12:e0171124. doi: 10.1371/journal.pone.0171124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cambiano V, Rodger AJ, Phillips AN. 2011. ‘Test-and-treat’: the end of the HIV epidemic? Curr Opin Infect Dis 24:19–26. doi: 10.1097/QCO.0b013e3283422c8c. [DOI] [PubMed] [Google Scholar]
- 18.CDC. About HIV/AIDS. CDC, Atlanta, GA: www.cdc.gov/actagainstaids/basics/whatishiv.html. Accessed 20 September 2017. [Google Scholar]
- 19.Schacker T, Collier AC, Hughes J, Shea T, Corey L. 1996. Clinical and epidemiologic features of primary HIV infection. Ann Intern Med 125:257–264. [DOI] [PubMed] [Google Scholar]
- 20.CDC. 1992. 1993 revised classification system for HIV infection and expanded surveillance case definition for AIDS among adolescents and adults. MMWR Recommend Rep 41(RR-17):961–962. [PubMed] [Google Scholar]
- 21.Jaffe HW, Sarngadharan MG, DeVico AL, Bruch L, Getchell JP, Kalyanaraman VS, Haverkos HW, Stoneburner RL, Gallo RC, Curran JW. 1985. Infection with HTLV-III/LAV and transfusion-associated acquired immunodeficiency syndrome. Serologic evidence of an association. JAMA 254:770–773. [PubMed] [Google Scholar]
- 22.Franco-Paredes C, Tellez I, del Rio C. 2006. Rapid HIV testing: a review of the literature and implications for the clinician. Curr HIV/AIDS Rep 3:169–175. [DOI] [PubMed] [Google Scholar]
- 23.Pai NP, Tulsky JP, Cohan D, Colford JM Jr, Reingold AL. 2007. Rapid point-of-care HIV testing in pregnant women: a systematic review and meta-analysis. Trop Med Int Health 12:162–173. doi: 10.1111/j.1365-3156.2006.01812.x. [DOI] [PubMed] [Google Scholar]
- 24.M’Boup S, Kanki PJ, Mhalu FS. 1991. Laboratory diagnosis of HIV-1 and HIV-2 in Africa. AIDS 5(Suppl 1):S93–S101. [PubMed] [Google Scholar]
- 25.Constantine NT, Kabat W, Zhao RY. 2005. Update on the laboratory diagnosis and monitoring of HIV infection. Cell Res 15:870–876. doi: 10.1038/sj.cr.7290361. [DOI] [PubMed] [Google Scholar]
- 26.Auwanit W, Ayuthaya PI, Balachandra K, Jayavasu C, Phanthumachinda B, Ikuta K, Yamanishi K, Kanai K. 1990. Immunofluorescence, enzyme-linked immunosorbent assay, particle agglutination and Western blot for the detection of antibody to human immunodeficiency virus type 1. Southeast Asian J Trop Med Public Health 21:53–59. [PubMed] [Google Scholar]
- 27.Melbye M, Biggar RJ, Chermann JC, Montagnier L, Stenbjerg S, Ebbesen P. 1984. High prevalence of lymphadenopathy virus (LAV) in European haemophiliacs. Lancet ii:40–41. [DOI] [PubMed] [Google Scholar]
- 28.Melbye M, Biggar RJ, Ebbesen P, Sarngadharan MG, Weiss SH, Gallo RC, Blattner WA. 1984. Seroepidemiology of HTLV-III antibody in Danish homosexual men: prevalence, transmission, and disease outcome. Br Med J (Clin Res Ed) 289:573–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melbye M, Froebel KS, Madhok R, Biggar RJ, Sarin PS, Stenbjerg S, Lowe GD, Forbes CD, Goedert JJ, Gallo RC. 1984. HTLV-III seropositivity in European haemophiliacs exposed to factor VIII concentrate imported from the USA. Lancet ii:1444–1446. [DOI] [PubMed] [Google Scholar]
- 30.Janssen RS, Satten GA, Stramer SL, Rawal BD, O’Brien TR, Weiblen BJ, Hecht FM, Jack N, Cleghorn FR, Kahn JO, Chesney MA, Busch MP. 1998. New testing strategy to detect early HIV-1 infection for use in incidence estimates and for clinical and prevention purposes. JAMA 280:42–48. [DOI] [PubMed] [Google Scholar]
- 31.Parekh BS, Kennedy MS, Dobbs T, Pau CP, Byers R, Green T, Hu DJ, Vanichseni S, Young NL, Choopanya K, Mastro TD, McDougal JS. 2002. Quantitative detection of increasing HIV type 1 antibodies after seroconversion: a simple assay for detecting recent HIV infection and estimating incidence. AIDS Res Hum Retroviruses 18:295–307. doi: 10.1089/088922202753472874. [DOI] [PubMed] [Google Scholar]
- 32.Dobbs T, Kennedy S, Pau CP, McDougal JS, Parekh BS. 2004. Performance characteristics of the immunoglobulin G-capture BED-enzyme immunoassay, an assay to detect recent human immunodeficiency virus type 1 seroconversion. J Clin Microbiol 42:2623–2628. doi: 10.1128/JCM.42.6.2623-2628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duong YT, Qiu M, De AK, Jackson K, Dobbs T, Kim AA, Nkengasong JN, Parekh BS. 2012. Detection of recent HIV-1 infection using a new limiting-antigen avidity assay: potential for HIV-1 incidence estimates and avidity maturation studies. PLoS One 7:e33328. doi: 10.1371/journal.pone.0033328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glynn SA, Kleinman SH, Schreiber GB, Busch MP, Wright DJ, Smith JW, Nass CC, Williams AE. 2000. Trends in incidence and prevalence of major transfusion-transmissible viral infections in US blood donors, 1991 to 1996. Retrovirus Epidemiology Donor Study (REDS). JAMA 284:229–235. [DOI] [PubMed] [Google Scholar]
- 35.Lyons SF, McGillivray GM, Coppin AP, Schoub BD. 1985. Limitations of enzyme immunoassay tests for detection of HTLV-III/LAV antibodies. S Afr Med J 68:575–576. [PubMed] [Google Scholar]
- 36.Constantine NT, Fox E, Abbatte EA, Woody JN. 1989. Diagnostic usefulness of five screening assays for HIV in an East African city where prevalence of infection is low. AIDS 3:313–317. [DOI] [PubMed] [Google Scholar]
- 37.Zeh C, Oyaro B, Vandenhoudt H, Amornkul P, Kasembeli A, Bondo P, Mwaengo D, Thomas TK, Hart C, Laserson KF, Ondoa P, Nkengasong JN. 2011. Performance of six commercial enzyme immunoassays and two alternative HIV-testing algorithms for the diagnosis of HIV-1 infection in Kisumu, Western Kenya. J Virol Methods 176:24–31. doi: 10.1016/j.jviromet.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 38.Louie B, Pandori MW, Wong E, Klausner JD, Liska S. 2006. Use of an acute seroconversion panel to evaluate a third-generation enzyme-linked immunoassay for detection of human immunodeficiency virus-specific antibodies relative to multiple other assays. J Clin Microbiol 44:1856–1858. doi: 10.1128/JCM.44.5.1856-1858.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nandi S, Maity S, Bhunia SC, Saha MK. 2014. Comparative assessment of commercial ELISA kits for detection of HIV in India. BMC Res Notes 7:436. doi: 10.1186/1756-0500-7-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ravanshad M, Mahboudi F, Sabahi F, Bayanolhagh S. 2006. Indeterminate human immunodeficiency virus Western blot results in Iranian patients with discordant screening assay results. Saudi Med J 27:1130–1133. [PubMed] [Google Scholar]
- 41.Esteban JI, Shih JW, Tai CC, Bodner AJ, Kay JW, Alter HJ. 1985. Importance of Western blot analysis in predicting infectivity of anti-HTLV-III/LAV positive blood. Lancet ii:1083–1086. [DOI] [PubMed] [Google Scholar]
- 42.Salmona M, Delarue S, Delaugerre C, Simon F, Maylin S. 2014. Clinical evaluation of BioPlex 2200 HIV Ag-Ab, an automated screening method providing discrete detection of HIV-1 p24 antigen, HIV-1 antibody, and HIV-2 antibody. J Clin Microbiol 52:103–107. doi: 10.1128/JCM.02460-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mor O, Mileguir F, Michaeli M, Levy I, Mendelson E. 2014. Evaluation of the Bio-Rad Geenius HIV 1/2 assay as an alternative to the INNO-LIA HIV 1/2 assay for confirmation of HIV infection. J Clin Microbiol 52:2677–2679. doi: 10.1128/JCM.01184-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.CDC. 1989. Interpretation and use of the Western blot assay for serodiagnosis of human immunodeficiency virus type 1 infections. MMWR Morb Mortal Wkly Rep 38:1–7. [PubMed] [Google Scholar]
- 45.Malloch L, Kadivar K, Putz J, Levett PN, Tang J, Hatchette TF, Kadkhoda K, Ng D, Ho J, Kim J. 2013. Comparative evaluation of the Bio-Rad Geenius HIV-1/2 confirmatory assay and the Bio-Rad Multispot HIV-1/2 rapid test as an alternative differentiation assay for CLSI M53 algorithm-I. J Clin Virol 58(Suppl 1):e85–e91. doi: 10.1016/j.jcv.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 46.Keating SM, Kassanjee R, Lebedeva M, Facente SN, MacArthur JC, Grebe E, Murphy G, Welte A, Martin JN, Little S, Price MA, Kallas EG, Busch MP, Pilcher CD. 2016. Performance of the Bio-Rad Geenius HIV1/2 supplemental assay in detecting “recent” HIV infection and calculating population incidence. J Acquir Immune Defic Syndr 73:581–588. doi: 10.1097/QAI.0000000000001146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gray RH, Makumbi F, Serwadda D, Lutalo T, Nalugoda F, Opendi P, Kigozi G, Reynolds SJ, Sewankambo NK, Wawer MJ. 2007. Limitations of rapid HIV-1 tests during screening for trials in Uganda: diagnostic test accuracy study. BMJ 335:188. doi: 10.1136/bmj.39210.582801.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aghokeng AF, Mpoudi-Ngole E, Dimodi H, Atem-Tambe A, Tongo M, Butel C, Delaporte E, Peeters M. 2009. Inaccurate diagnosis of HIV-1 group M and O is a key challenge for ongoing universal access to antiretroviral treatment and HIV prevention in Cameroon. PLoS One 4:e7702. doi: 10.1371/journal.pone.0007702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Juarez SI, Nunez AE, Aranda MM, Mojica D, Kim AA, Parekh B. 2016. Field evaluation of four rapid tests for diagnosis of HIV infection in Panama. J Clin Microbiol 54:1127–1129. doi: 10.1128/JCM.02654-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Machado AA, Martinez R, Haikal AA, Rodrigues da Silva MC. 2001. Advantages of the rapid HIV-1 test in occupational accidents with potentially contaminated material among health workers. Rev Inst Med Trop Sao Paulo 43:199–201. doi: 10.1590/S0036-46652001000400004. [DOI] [PubMed] [Google Scholar]
- 51.Nogueira SA, Lambert JS, Albuquerque AL, Rodrigues R, Reis S, Bornia R, Dias M, Barbosa R, Sztanjbock D, Santos AL, Blattner W, Constantine NT. 2001. Assessment of a rapid HIV test strategy during labor: a pilot study from Rio de Janeiro, Brazil. J Hum Virol 4:278–282. [PubMed] [Google Scholar]
- 52.Granade TC, Parekh BS, Phillips SK, McDougal JS. 2004. Performance of the OraQuick and Hema-Strip rapid HIV antibody detection assays by non-laboratorians. J Clin Virol 30:229–232. doi: 10.1016/j.jcv.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 53.Wouters K, Fransen K, Beelaert G, Kenyon C, Platteau T, Van Ghyseghem C, Collier I, Buyze J, Florence E. 2014. Use of rapid HIV testing in a low threshold centre in Antwerp, Belgium, 2007-2012. Int J STD AIDS 25:936–942. doi: 10.1177/0956462414526705. [DOI] [PubMed] [Google Scholar]
- 54.Sema Baltazar C, Raposo C, Jani IV, Shodell D, Correia D, Goncalves da Silva C, Kalou M, Patel H, Parekh B. 2014. Evaluation of performance and acceptability of two rapid oral fluid tests for HIV detection in Mozambique. J Clin Microbiol 52:3544–3548. doi: 10.1128/JCM.01098-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.PEPFAR. 2017. PEPFAR latest global results. Nov 2017 PEPFAR, Washington, DC: www.pepfar.gov/documents/organization/276321.pdf. Accessed 4 January 2018. [Google Scholar]
- 56.Koblavi-Deme S, Maurice C, Yavo D, Sibailly TS, N’Guessan K, Kamelan-Tano Y, Wiktor SZ, Roels TH, Chorba T, Nkengasong JN. 2001. Sensitivity and specificity of human immunodeficiency virus rapid serologic assays and testing algorithms in an antenatal clinic in Abidjan, Ivory Coast. J Clin Microbiol 39:1808–1812. doi: 10.1128/JCM.39.5.1808-1812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.WHO. 2016. Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection. Recommendations for a public health approach, 2nd ed WHO, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/208825/1/9789241549684_eng.pdf?ua=1. Accessed 22 September 2016. [Google Scholar]
- 58.Alemnji GA, Guevara G, Parris K, Kalou M, Behel SK, Parekh B, Nkengasong JN, Albalak R. 16 June 2016. Improving the quality of and access to HIV rapid testing in the Caribbean region: program implementation, outcomes, and recommendations. AIDS Res Hum Retroviruses doi: 10.1089/AID.2015.0353. [DOI] [PubMed] [Google Scholar]
- 59.Fonjungo PN, Boeras DI, Zeh C, Alexander H, Parekh BS, Nkengasong JN. 2016. Access and quality of HIV-related point-of-care diagnostic testing in global health programs. Clin Infect Dis 62:369–374. doi: 10.1093/cid/civ866. [DOI] [PubMed] [Google Scholar]
- 60.Thomson KA, Hughes J, Baeten JM, John-Stewart G, Celum C, Cohen CR, Ngure K, Kiarie J, Mugo N, Heffron R. 5 March 2018. Increased risk of female HIV-1 acquisition throughout pregnancy and postpartum: a prospective per-coital act analysis among women with HIV-1 infected partners. J Infect Dis doi: 10.1093/infdis/jiy113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.WHO. 2016. HIV testing services: WHO recommends HIV self-testing. WHO, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/251549/1/WHO-HIV-2016.21-eng.pdf?ua=1. Accessed 22 September 2017. [Google Scholar]
- 62.Chanda MM, Ortblad KF, Mwale M, Chongo S, Kanchele C, Kamungoma N, Fullem A, Dunn C, Barresi LG, Harling G, Barnighausen T, Oldenburg CE. 2017. HIV self-testing among female sex workers in Zambia: a cluster randomized controlled trial. PLoS Med 14:e1002442. doi: 10.1097/QAD.0000000000001740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choko AT, Kumwenda MK, Johnson CC, Sakala DW, Chikalipo MC, Fielding K, Chikovore J, Desmond N, Corbett EL. 2017. Acceptability of woman-delivered HIV self-testing to the male partner, and additional interventions: a qualitative study of antenatal care participants in Malawi. J Int AIDS Soc 20:21610. doi: 10.7448/IAS.20.1.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.WHO. 2017. Number of countries adopting HIV self-testing policies rises sharply. WHO, Geneva, Switzerland: www.who.int/hiv/mediacentre/news/hiv-self-testing-increases/en/. [Google Scholar]
- 65.UNITAID. 2017. Market and technology landscape: HIV rapid diagnostic tests for self-testing. WHO, Geneva, Switzerland: www.who.int/hiv/pub/vct/unitaid-landscape-hivst-2017.pdf?ua=1. Accessed 22 September 2017. [Google Scholar]
- 66.WHO. 2017. Overview of the WHO prequalification of in vitro diagnostics assessment: WHO prequalification of in vitro diagnostics. WHO, Geneva, Switzerland: www.who.int/diagnostics_laboratory/evaluations/170728_pqdx_007_overview_document_v7.pdf?ua=1. Accessed 22 September 2017. [Google Scholar]
- 67.WHO. 2017. WHO list of prequalified in vitro diagnostic products. WHO, Geneva, Switzerland: www.who.int/diagnostics_laboratory/evaluations/170816_prequalified_product_list.pdf?ua=1. Accessed 6 October 2017. [Google Scholar]
- 68.Parekh BS, Kalou MB, Alemnji G, Ou CY, Gershy-Damet GM, Nkengasong JN. 2010. Scaling up HIV rapid testing in developing countries: comprehensive approach for implementing quality assurance. Am J Clin Pathol 134:573–584. doi: 10.1309/AJCPTDIMFR00IKYX. [DOI] [PubMed] [Google Scholar]
- 69.WHO. 2011. Field safety notice no. 1. WHO, Geneva, Switzerland: www.who.int/diagnostics_laboratory/procurement/111201_productalert_for_product0027_mx012.pdf. Accessed 22 September 2017. [Google Scholar]
- 70.UNAIDS. 2017. Ending AIDS: progress towards the 90-90-90 targets. UNAIDS, Geneva, Switzerland: www.unaids.org/sites/default/files/media_asset/Global_AIDS_update_2017_en.pdf. Accessed 22 September 2017. [Google Scholar]
- 71.UNAIDS. 2017. UNAIDS data 2017. UNAIDS, Geneva, Switzerland: www.unaids.org/sites/default/files/media_asset/2017_data-book_en.pdf. Accessed 25 August 2017. [Google Scholar]
- 72.Parekh BS, McDougal JS. 2005. Application of laboratory methods for estimation of HIV-1 incidence. Indian J Med Res 121:510–518. [PubMed] [Google Scholar]
- 73.McDougal JS, Pilcher CD, Parekh BS, Gershy-Damet G, Branson BM, Marsh K, Wiktor SZ. 2005. Surveillance for HIV-1 incidence using tests for recent infection in resource-constrained countries. AIDS 19(Suppl 2):S25–S30. doi: 10.1097/01.aids.0000172874.90133.7a. [DOI] [PubMed] [Google Scholar]
- 74.Calzavara L, Burchell AN, Major C, Remis RS, Corey P, Myers T, Millson P, Wallace E, Polaris Study Team. 2002. Increases in HIV incidence among men who have sex with men undergoing repeat diagnostic HIV testing in Ontario, Canada. AIDS 16:1655–1661. doi: 10.1097/00002030-200208160-00011. [DOI] [PubMed] [Google Scholar]
- 75.Gregson S, Donnelly CA, Parker CG, Anderson RM. 1996. Demographic approaches to the estimation of incidence of HIV-1 infection among adults from age-specific prevalence data in stable endemic conditions. AIDS 10:1689–1697. doi: 10.1097/00002030-199612000-00014. [DOI] [PubMed] [Google Scholar]
- 76.Saidel T, Sokal D, Rice J, Buzingo T, Hassig S. 1996. Validation of a method to estimate age-specific human immunodeficiency virus (HIV) incidence rates in developing countries using population-based seroprevalence data. Am J Epidemiol 144:214–223. doi: 10.1093/oxfordjournals.aje.a008916. [DOI] [PubMed] [Google Scholar]
- 77.Brookmeyer R, Quinn T, Shepherd M, Mehendale S, Rodrigues J, Bollinger R. 1995. The AIDS epidemic in India: a new method for estimating current human immunodeficiency virus (HIV) incidence rates. Am J Epidemiol 142:709–713. [DOI] [PubMed] [Google Scholar]
- 78.Pilcher CD, Fiscus SA, Nguyen TQ, Foust E, Wolf L, Williams D, Ashby R, O’Dowd JO, McPherson JT, Stalzer B, Hightow L, Miller WC, Eron JJ Jr, Cohen MS, Leone PA. 2005. Detection of acute infections during HIV testing in North Carolina. N Engl J Med 352:1873–1883. doi: 10.1056/NEJMoa042291. [DOI] [PubMed] [Google Scholar]
- 79.Justman J, Reed JB, Bicego G, Donnell D, Li K, Bock N, Koler A, Philip NM, Mlambo CK, Parekh BS, Duong YT, Ellenberger DL, El-Sadr WM, Nkambule R. 2017. Swaziland HIV Incidence Measurement Survey (SHIMS): a prospective national cohort study. Lancet HIV 4:e83–e92. doi: 10.1016/S2352-3018(16)30190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rawal BD, Degula A, Lebedeva L, Janssen RS, Hecht FM, Sheppard HW, Busch MP. 2003. Development of a new less-sensitive enzyme immunoassay for detection of early HIV-1 infection. J Acquir Immune Defic Syndr 33:349–355. doi: 10.1097/00126334-200307010-00009. [DOI] [PubMed] [Google Scholar]
- 81.Constantine NT, Sill AM, Jack N, Kreisel K, Edwards J, Cafarella T, Smith H, Bartholomew C, Cleghorn FR, Blattner WA. 2003. Improved classification of recent HIV-1 infection by employing a two-stage sensitive/less-sensitive test strategy. J Acquir Immune Defic Syndr 32:94–103. doi: 10.1097/00126334-200301010-00014. [DOI] [PubMed] [Google Scholar]
- 82.Soroka SD, Granade TC, Candal D, Parekh BS. 2005. Modification of rapid human immunodeficiency virus (HIV) antibody assay protocols for detecting recent HIV seroconversion. Clin Diagn Lab Immunol 12:918–921. doi: 10.1128/CDLI.12.8.918-921.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li H, Ketema F, Sill AM, Kreisel KM, Cleghorn FR, Constantine NT. 2007. A simple and inexpensive particle agglutination test to distinguish recent from established HIV-1 infection. Int J Infect Dis 11:459–465. doi: 10.1016/j.ijid.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 84.Sill AM, Kreisel K, Deeds BG, Wilson CM, Constantine NT, Peralta L, HIV/AIDS Interventions Adolescent Trials Network. 2007. Calibration and validation of an oral fluid-based sensitive/less-sensitive assay to distinguish recent from established HIV-1 infections. J Clin Lab Anal 21:40–45. doi: 10.1002/jcla.20144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nesheim S, Parekh B, Sullivan K, Bulterys M, Dobbs T, Lindsay M, Cashat-Cruz M, Byers B, Lee F. 2005. Temporal trends in HIV type 1 incidence among inner-city childbearing women in Atlanta: use of the IgG-capture BED-enzyme immunoassay. AIDS Res Hum Retroviruses 21:537–544. doi: 10.1089/aid.2005.21.537. [DOI] [PubMed] [Google Scholar]
- 86.Jiang Y, Wang M, Ni M, Duan S, Wang Y, Feng J, Xiao Y, Dong Y, Wang D, Han M, Xiang L, Ma L, Zhou Q. 2007. HIV-1 incidence estimates using IgG-capture BED-enzyme immunoassay from surveillance sites of injection drug users in three cities of China. AIDS 21(Suppl 8):S47–S51. doi: 10.1097/01.aids.0000304696.62508.8a. [DOI] [PubMed] [Google Scholar]
- 87.Hu DJ, Vanichseni S, Mock PA, Young NL, Dobbs T, Byers RH Jr, Choopanya K, van Griensven F, Kitayaporn D, McDougal JS, Tappero JW, Mastro TD, Parekh BS. 2003. HIV type 1 incidence estimates by detection of recent infection from a cross-sectional sampling of injection drug users in Bangkok: use of the IgG capture BED enzyme immunoassay. AIDS Res Hum Retroviruses 19:727–730. doi: 10.1089/088922203769232511. [DOI] [PubMed] [Google Scholar]
- 88.Barnighausen T, Tanser F, Gqwede Z, Mbizana C, Herbst K, Newell ML. 2008. High HIV incidence in a community with high HIV prevalence in rural South Africa: findings from a prospective population-based study. AIDS 22:139–144. doi: 10.1097/QAD.0b013e3282f2ef43. [DOI] [PubMed] [Google Scholar]
- 89.Kana K, Tabprasit S, Chuenchitra T, Sirisopana N, Rangsin R. 2009. HIV-1 incidence estimates among young Thai men using IgG-capture BED-enzyme immunoassay (BED-CEIA) during 2005-2006. J Med Assoc Thai 92(Suppl 1):S112–S116. [PubMed] [Google Scholar]
- 90.Lakshmi V, Sudha T, Dandona R, Teja VD, Kumar GA, Dandona L. 2009. Application of human immunodeficiency virus type 1 BED enzyme immunoassay on dried blood spots in India. J Med Microbiol 58:312–317. doi: 10.1099/jmm.0.005249-0. [DOI] [PubMed] [Google Scholar]
- 91.Kim AA, McDougal JS, Hargrove J, Rehle T, Pillay-Van Wyk V, Puren A, Ekra A, Borget-Alloue MY, Adje-Toure C, Abdullahi AS, Odawo L, Marum L, Parekh BS. 2010. Evaluating the BED capture enzyme immunoassay to estimate HIV incidence among adults in three countries in sub-Saharan Africa. AIDS Res Hum Retroviruses 26:1051–1061. doi: 10.1089/aid.2009.0218. [DOI] [PubMed] [Google Scholar]
- 92.McNicholl JM, McDougal JS, Wasinrapee P, Branson BM, Martin M, Tappero JW, Mock PA, Green TA, Hu DJ, Parekh B, Thai-US BED Assay Validation Working Group. 2011. Assessment of BED HIV-1 incidence assay in seroconverter cohorts: effect of individuals with long-term infection and importance of stable incidence. PLoS One 6:e14748. doi: 10.1371/journal.pone.0014748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Morineau G, Magnani R, Nurhayati A, Bollen L, Mustikawati DE. 2011. Is the bed capture enzyme immunoassay useful for surveillance in concentrated epidemics? The case of female sex workers in Indonesia. Southeast Asian J Trop Med Public Health 42:634–642. [PubMed] [Google Scholar]
- 94.Hall HI, Song R, Rhodes P, Prejean J, An Q, Lee LM, Karon J, Brookmeyer R, Kaplan EH, McKenna MT, Janssen RS, HIV Incidence Surveillance Group. 2008. Estimation of HIV incidence in the United States. JAMA 300:520–529. doi: 10.1001/jama.300.5.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wolday D, Meles H, Hailu E, Messele T, Mengistu Y, Fekadu M, Parekh BS, Wuhib T. 2007. Temporal trends in the incidence of HIV infection in antenatal clinic attendees in Addis Ababa, Ethiopia, 1995-2003. J Intern Med 261:132–137. doi: 10.1111/j.1365-2796.2006.01740.x. [DOI] [PubMed] [Google Scholar]
- 96.McDougal JS, Parekh BS, Peterson ML, Branson BM, Dobbs T, Ackers M, Gurwith M. 2006. Comparison of HIV type 1 incidence observed during longitudinal follow-up with incidence estimated by cross-sectional analysis using the BED capture enzyme immunoassay. AIDS Res Hum Retroviruses 22:945–952. doi: 10.1089/aid.2006.22.945. [DOI] [PubMed] [Google Scholar]
- 97.Westreich D, Pettifor A, Karita E, Price M, Fiamma A, Fiscus S, Cohen M. 2007. Overestimation of the South African HIV incidence using the BED IgG assay? S Afr Med J 97:476, 478. (Reply, 97:478, 480.) [PubMed] [Google Scholar]
- 98.Hayashida T, Gatanaga H, Tanuma J, Oka S. 2008. Effects of low HIV type 1 load and antiretroviral treatment on IgG-capture BED-enzyme immunoassay. AIDS Res Hum Retroviruses 24:495–498. doi: 10.1089/aid.2007.0150. [DOI] [PubMed] [Google Scholar]
- 99.Hallett TB, Ghys P, Barnighausen T, Yan P, Garnett GP. 2009. Errors in ‘BED’-derived estimates of HIV incidence will vary by place, time and age. PLoS One 4:e5720. doi: 10.1371/journal.pone.0005720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Anonymous. 2006. UNAIDS Reference Group on estimates, modelling and projections—statement on the use of the BED assay for the estimation of HIV-1 incidence for surveillance or epidemic monitoring. Wkly Epidemiol Rec 81:40. [PubMed] [Google Scholar]
- 101.Hargrove JW, Humphrey JH, Mutasa K, Parekh BS, McDougal JS, Ntozini R, Chidawanyika H, Moulton LH, Ward B, Nathoo K, Iliff PJ, Kopp E. 2008. Improved HIV-1 incidence estimates using the BED capture enzyme immunoassay. AIDS 22:511–518. doi: 10.1097/QAD.0b013e3282f2a960. [DOI] [PubMed] [Google Scholar]
- 102.Hargrove JW. 2009. BED estimates of HIV incidence must be adjusted. AIDS 23:2061–2062. (Reply, 23:2066–2068.) doi: 10.1097/QAD.0b013e32832f3d8b. [DOI] [PubMed] [Google Scholar]
- 103.Hargrove J, Humphrey J, ZVITAMBO Study Group. 2010. Short communication: simplified estimation of the long-term specificity of the BED assay to improve estimates of HIV incidence. AIDS Res Hum Retroviruses 26:977–979. doi: 10.1089/aid.2010.0009. [DOI] [PubMed] [Google Scholar]
- 104.McDougal JS. 2009. BED estimates of HIV incidence must be adjusted. AIDS 23:2064–2065. (Reply, 23:2066–2068.) doi: 10.1097/QAD.0b013e32832eff6e. [DOI] [PubMed] [Google Scholar]
- 105.Marinda ET, Hargrove J, Preiser W, Slabbert H, van Zyl G, Levin J, Moulton LH, Welte A, Humphrey J. 2010. Significantly diminished long-term specificity of the BED capture enzyme immunoassay among patients with HIV-1 with very low CD4 counts and those on antiretroviral therapy. J Acquir Immune Defic Syndr 53:496–499. doi: 10.1097/QAI.0b013e3181b61938. [DOI] [PubMed] [Google Scholar]
- 106.Braunstein SL, Nash D, Kim AA, Ford K, Mwambarangwe L, Ingabire CM, Vyankandondera J, van de Wijgert JH. 2011. Dual testing algorithm of BED-CEIA and AxSYM avidity index assays performs best in identifying recent HIV infection in a sample of Rwandan sex workers. PLoS One 6:e18402. doi: 10.1371/journal.pone.0018402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hargrove J, van Schalkwyk C, Eastwood H. 2012. BED estimates of HIV incidence: resolving the differences, making things simpler. PLoS One 7:e29736. doi: 10.1371/journal.pone.0029736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Laeyendecker O, Brookmeyer R, Oliver AE, Mullis CE, Eaton KP, Mueller AC, Jacobson LP, Margolick JB, Brown J, Rinaldo CR, Quinn TC, Eshleman SH. Multicenter Aids Cohort Study. 2012. Factors associated with incorrect identification of recent HIV infection using the BED capture immunoassay. AIDS Res Hum Retroviruses 28:816–822. doi: 10.1089/aid.2011.0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Longosz AF, Serwadda D, Nalugoda F, Kigozi G, Franco V, Gray RH, Quinn TC, Eshleman SH, Laeyendecker O. 2014. Impact of HIV subtype on performance of the limiting antigen-avidity enzyme immunoassay, the bio-rad avidity assay, and the BED capture immunoassay in Rakai, Uganda. AIDS Res Hum Retroviruses 30:339–344. doi: 10.1089/aid.2013.0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mastro TD, Kim AA, Hallett T, Rehle T, Welte A, Laeyendecker O, Oluoch T, Garcia-Calleja JM. 2010. Estimating HIV incidence in populations using tests for recent infection: issues, challenges and the way forward. J HIV AIDS Surveill Epidemiol 2:1–14. [PMC free article] [PubMed] [Google Scholar]
- 111.Barin F, Meyer L, Lancar R, Deveau C, Gharib M, Laporte A, Desenclos JC, Costagliola D. 2005. Development and validation of an immunoassay for identification of recent human immunodeficiency virus type 1 infections and its use on dried serum spots. J Clin Microbiol 43:4441–4447. doi: 10.1128/JCM.43.9.4441-4447.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pillonel J, Barin F, Laperche S, Bernillon P, Le Vu S, Brunet S, Thierry D, Desenclos JC, Transfusion-Transmissible Agents Working Group of the French Society of Blood Transfusion. 2008. Human immunodeficiency virus type 1 incidence among blood donors in France, 1992 through 2006: use of an immunoassay to identify recent infections. Transfusion 48:1567–1575. doi: 10.1111/j.1537-2995.2008.01739.x. [DOI] [PubMed] [Google Scholar]
- 113.Semaille C, Barin F, Cazein F, Pillonel J, Lot F, Brand D, Plantier JC, Bernillon P, Le Vu S, Pinget R, Desenclos JC. 2007. Monitoring the dynamics of the HIV epidemic using assays for recent infection and serotyping among new HIV diagnoses: experience after 2 years in France. J Infect Dis 196:377–383. doi: 10.1086/519387. [DOI] [PubMed] [Google Scholar]
- 114.Le Vu S, Le Strat Y, Barin F, Pillonel J, Cazein F, Bousquet V, Brunet S, Thierry D, Semaille C, Meyer L, Desenclos JC. 2010. Population-based HIV-1 incidence in France, 2003-08: a modelling analysis. Lancet Infect Dis 10:682–687. doi: 10.1016/S1473-3099(10)70167-5. [DOI] [PubMed] [Google Scholar]
- 115.Wilson KM, Johnson EI, Croom HA, Richards KM, Doughty L, Cunningham PH, Kemp BE, Branson BM, Dax EM. 2004. Incidence immunoassay for distinguishing recent from established HIV-1 infection in therapy-naive populations. AIDS 18:2253–2259. doi: 10.1097/00002030-200411190-00005. [DOI] [PubMed] [Google Scholar]
- 116.Kanengisser-Pines B, Hazan Y, Pines G, Appelman Z. 2009. High cytomegalovirus IgG avidity is a reliable indicator of past infection in patients with positive IgM detected during the first trimester of pregnancy. J Perinat Med 37:15–18. doi: 10.1515/JPM.2009.012. [DOI] [PubMed] [Google Scholar]
- 117.Lagrou K, Bodeus M, Van Ranst M, Goubau P. 2009. Evaluation of the new architect cytomegalovirus immunoglobulin M (IgM), IgG, and IgG avidity assays. J Clin Microbiol 47:1695–1699. doi: 10.1128/JCM.02172-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mubareka S, Richards H, Gray M, Tipples GA. 2007. Evaluation of commercial rubella immunoglobulin G avidity assays. J Clin Microbiol 45:231–233. doi: 10.1128/JCM.01243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vauloup-Fellous C, Ursulet-Diser J, Grangeot-Keros L. 2007. Development of a rapid and convenient method for determination of rubella virus-specific immunoglobulin G avidity. Clin Vaccine Immunol 14:1416–1419. doi: 10.1128/CVI.00312-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rodella A, Galli C, Terlenghi L, Perandin F, Bonfanti C, Manca N. 2006. Quantitative analysis of HBsAg, IgM anti-HBc and anti-HBc avidity in acute and chronic hepatitis B. J Clin Virol 37:206–212. doi: 10.1016/j.jcv.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 121.Klimashevskaya S, Obriadina A, Ulanova T, Bochkova G, Burkov A, Araujo A, Stramer SL, Tobler LH, Busch MP, Fields HA. 2007. Distinguishing acute from chronic and resolved hepatitis C virus (HCV) infections by measurement of anti-HCV immunoglobulin G avidity index. J Clin Microbiol 45:3400–3403. doi: 10.1128/JCM.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Suligoi B, Galli C, Massi M, Di Sora F, Sciandra M, Pezzotti P, Recchia O, Montella F, Sinicco A, Rezza G. 2002. Precision and accuracy of a procedure for detecting recent human immunodeficiency virus infections by calculating the antibody avidity index by an automated immunoassay-based method. J Clin Microbiol 40:4015–4020. doi: 10.1128/JCM.40.11.4015-4020.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wei X, Liu X, Dobbs T, Kuehl D, Nkengasong JN, Hu DJ, Parekh BS. 2010. Development of two avidity-based assays to detect recent HIV type 1 seroconversion using a multisubtype gp41 recombinant protein. AIDS Res Hum Retroviruses 26:61–71. doi: 10.1089/aid.2009.0133. [DOI] [PubMed] [Google Scholar]
- 124.Duong YT, Kassanjee R, Welte A, Morgan M, De A, Dobbs T, Rottinghaus E, Nkengasong J, Curlin ME, Kittinunvorakoon C, Raengsakulrach B, Martin M, Choopanya K, Vanichseni S, Jiang Y, Qiu M, Yu H, Hao Y, Shah N, Le LV, Kim AA, Nguyen TA, Ampofo W, Parekh BS. 2015. Recalibration of the limiting antigen avidity EIA to determine mean duration of recent infection in divergent HIV-1 subtypes. PLoS One 10:e0114947. doi: 10.1371/journal.pone.0114947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Parekh BDY, Mavenger Y, Dobbs T, Chang J, Rasberry C, Emel LM, Sibandze D, Makhanya M, Sexton C, Justman J, Donnell D, Reed J, Bock N, Moore J, Nkengasong J, Ellenberger D, Birx D, Bicego G. 2012. Performance of new LAg-avidity EIA to measure HIV-1 incidence in a cross-sectional population: Swaziland HIV Incidence Measurement Survey (SHIMS), abstr LPBE 27. Abstr 2012 IAS Conf, Washington, DC. [Google Scholar]
- 126.Kassanjee R, Pilcher CD, Busch MP, Murphy G, Facente SN, Keating SM, McKinney E, Marson K, Price MA, Martin JN, Little SJ, Hecht FM, Kallas EG, Welte A, Consortium for the Evaluation and Performance of HIV Incidence Assays. 2016. Viral load criteria and threshold optimization to improve HIV incidence assay characteristics. AIDS 30:2361–2371. doi: 10.1097/QAD.0000000000001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.ICAP. 5 March 2018, accession date Population-based HIV impact assessments. ICAP, New York, NY: http://icap.columbia.edu/global-initiatives/the-phia-project. [Google Scholar]
- 128.CDC. 5 March 2018, accession date Five African countries approaching control of their HIV epidemics. CDC, Atlanta, GA: www.cdc.gov/globalhivtb/who-we-are/features/phia_lesotho_uganda.html. [Google Scholar]
- 129.Granade TC, Nguyen S, Kuehl DS, Parekh BS. 2013. Development of a novel rapid HIV test for simultaneous detection of recent or long-term HIV type 1 infection using a single testing device. AIDS Res Hum Retroviruses 29:61–67. doi: 10.1089/aid.2012.0121. [DOI] [PubMed] [Google Scholar]
- 130.Parekh BS, Shanmugam V, Dobbs T, Kim A, Nkengasong J. 2012. Performance evaluation of Asante Rapid Recency assay for HIV diagnosis and detection of recent infection: potential for surveillance and prevention, abstr TUPEC0849 Abstr 9th IAS Conf HIV Sci, Paris, France. [Google Scholar]
- 131.Kouyos RD, von Wyl V, Yerly S, Boni J, Rieder P, Joos B, Taffe P, Shah C, Burgisser P, Klimkait T, Weber R, Hirschel B, Cavassini M, Rauch A, Battegay M, Vernazza PL, Bernasconi E, Ledergerber B, Bonhoeffer S, Gunthard HF, Swiss HIV Cohort Study. 2011. Ambiguous nucleotide calls from population-based sequencing of HIV-1 are a marker for viral diversity and the age of infection. Clin Infect Dis 52:532–539. doi: 10.1093/cid/ciq164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Andersson E, Shao W, Bontell I, Cham F, Cuong DD, Wondwossen A, Morris L, Hunt G, Sonnerborg A, Bertagnolio S, Maldarelli F, Jordan MR. 2013. Evaluation of sequence ambiguities of the HIV-1 pol gene as a method to identify recent HIV-1 infection in transmitted drug resistance surveys. Infect Genet Evol 18:125–131. doi: 10.1016/j.meegid.2013.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Park SY, Love TM, Nelson J, Thurston SW, Perelson AS, Lee HY. 2011. Designing a genome-based HIV incidence assay with high sensitivity and specificity. AIDS 25:F13–F19. doi: 10.1097/QAD.0b013e328349f089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cousins MM, Swan D, Magaret CA, Hoover DR, Eshleman SH. 2012. Analysis of HIV using a high resolution melting (HRM) diversity assay: automation of HRM data analysis enhances the utility of the assay for analysis of HIV incidence. PLoS One 7:e51359. doi: 10.1371/journal.pone.0051359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cousins MM, Konikoff J, Sabin D, Khaki L, Longosz AF, Laeyendecker O, Celum C, Buchbinder SP, Seage GR III, Kirk GD, Moore RD, Mehta SH, Margolick JB, Brown J, Mayer KH, Kobin BA, Wheeler D, Justman JE, Hodder SL, Quinn TC, Brookmeyer R, Eshleman SH. 2014. A comparison of two measures of HIV diversity in multi-assay algorithms for HIV incidence estimation. PLoS One 9:e101043. doi: 10.1371/journal.pone.0101043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gruner N, Stambouli O, Ross RS. 13 March 2015. Dried blood spots—preparing and processing for use in immunoassays and in molecular techniques. J Vis Exp doi: 10.3791/52619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Monleau M, Butel C, Delaporte E, Boillot F, Peeters M. 2010. Effect of storage conditions of dried plasma and blood spots on HIV-1 RNA quantification and PCR amplification for drug resistance genotyping. J Antimicrob Chemother 65:1562–1566. doi: 10.1093/jac/dkq205. [DOI] [PubMed] [Google Scholar]
- 138.Gwinn M, Pappaioanou M, George JR, Hannon WH, Wasser SC, Redus MA, Hoff R, Grady GF, Willoughby A, Novello AC. 1991. Prevalence of HIV infection in childbearing women in the United States. Surveillance using newborn blood samples. JAMA 265:1704–1708. [PubMed] [Google Scholar]
- 139.Schlusser KE, Pilcher C, Kallas EG, Santos BR, Deeks SG, Facente S, Keating SM, Busch MP, Murphy G, Welte A, Quinn T, Eshleman SH, Laeyendecker O. 2017. Comparison of cross-sectional HIV incidence assay results from dried blood spots and plasma. PLoS One 12:e0172283. doi: 10.1371/journal.pone.0172283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.WHO. 2010. WHO manual for HIV drug resistance testing using dried blood spot specimens. WHO, Geneva, Switzerland: www.who.int/hiv/pub/drugresistance/dried_blood_spots/en/. Accessed 20 September 2017. [Google Scholar]
- 141.Rottinghaus EK, Ugbena R, Diallo K, Bassey O, Azeez A, Devos J, Zhang G, Aberle-Grasse J, Nkengasong J, Yang C. 2012. Dried blood spot specimens are a suitable alternative sample type for HIV-1 viral load measurement and drug resistance genotyping in patients receiving first-line antiretroviral therapy. Clin Infect Dis 54:1187–1195. doi: 10.1093/cid/cis015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Inzaule S, Yang C, Kasembeli A, Nafisa L, Okonji J, Oyaro B, Lando R, Mills LA, Laserson K, Thomas T, Nkengasong J, Zeh C. 2013. Field evaluation of a broadly sensitive HIV-1 in-house genotyping assay for use with both plasma and dried blood spot specimens in a resource-limited country. J Clin Microbiol 51:529–539. doi: 10.1128/JCM.02347-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bertagnolio S, Parkin NT, Jordan M, Brooks J, Garcia-Lerma JG. 2010. Dried blood spots for HIV-1 drug resistance and viral load testing: a review of current knowledge and WHO efforts for global HIV drug resistance surveillance. AIDS Rev 12:195–208. [PubMed] [Google Scholar]
- 144.UNAIDS, WHO. 2009. Guidelines for using HIV testing technologies in surveillance. WHO, Geneva, Switzerland: http://www.who.int/hiv/pub/surveillance/hiv_testing_technologies_surveillance.pdf. Accessed 20 September 2017. [Google Scholar]
- 145.Ouma KN, Basavaraju SV, Okonji JA, Williamson J, Thomas TK, Mills LA, Nkengasong JN, Zeh C. 2013. Evaluation of quantification of HIV-1 RNA viral load in plasma and dried blood spots by use of the semiautomated Cobas Amplicor assay and the fully automated Cobas Ampliprep/TaqMan assay, version 2.0, in Kisumu, Kenya. J Clin Microbiol 51:1208–1218. doi: 10.1128/JCM.03048-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sherman GG, Stevens G, Jones SA, Horsfield P, Stevens WS. 2005. Dried blood spots improve access to HIV diagnosis and care for infants in low-resource settings. J Acquir Immune Defic Syndr 38:615–617. doi: 10.1097/01.qai.0000143604.71857.5d. [DOI] [PubMed] [Google Scholar]
- 147.Creek TL, Sherman GG, Nkengasong J, Lu L, Finkbeiner T, Fowler MG, Rivadeneira E, Shaffer N. 2007. Infant human immunodeficiency virus diagnosis in resource-limited settings: issues, technologies, and country experiences. Am J Obstet Gynecol 197:S64–S71. doi: 10.1016/j.ajog.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 148.Stevens W, Erasmus L, Moloi M, Taleng T, Sarang S. 2008. Performance of a novel human immunodeficiency virus (HIV) type 1 total nucleic acid-based real-time PCR assay using whole blood and dried blood spots for diagnosis of HIV in infants. J Clin Microbiol 46:3941–3945. doi: 10.1128/JCM.00754-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Garcia A, Subbarao S, Zhang G, Parsons L, Nkengasong J, Ou CY, Ellenberger D. 2014. Impact of proficiency testing program for laboratories conducting early diagnosis of HIV-1 infection in infants in low- to middle-income countries. J Clin Microbiol 52:773–780. doi: 10.1128/JCM.03097-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Masciotra S, Khamadi S, Bile E, Puren A, Fonjungo P, Nguyen S, Girma M, Downing R, Ramos A, Subbarao S, Ellenberger D. 2012. Evaluation of blood collection filter papers for HIV-1 DNA PCR. J Clin Virol 55:101–106. doi: 10.1016/j.jcv.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 151.Parekh BS, Anyanwu J, Patel H, Downer M, Kalou M, Gichimu C, Keipkerich BS, Clement N, Omondi M, Mayer O, Ou CY, Nkengasong JN. 2010. Dried tube specimens: a simple and cost-effective method for preparation of HIV proficiency testing panels and quality control materials for use in resource-limited settings. J Virol Methods 163:295–300. doi: 10.1016/j.jviromet.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 152.Van Rie A, Clouse K, Hanrahan C, Selibas K, Sanne I, Williams S, Kim P, Bassett J. 2014. High uptake of systematic HIV counseling and testing and TB symptom screening at a primary care clinic in South Africa. PLoS One 9:e105428. doi: 10.1371/journal.pone.0105428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kayigamba FR, Van Santen D, Bakker MI, Lammers J, Mugisha V, Bagiruwigize E, De Naeyer L, Asiimwe A, Schim Van Der Loeff MF. 2016. Does provider-initiated HIV testing and counselling lead to higher HIV testing rate and HIV case finding in Rwandan clinics? BMC Infect Dis 16:26. doi: 10.1186/s12879-016-1355-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kennedy CE, Fonner VA, Sweat MD, Okero FA, Baggaley R, O’Reilly KR. 2013. Provider-initiated HIV testing and counseling in low- and middle-income countries: a systematic review. AIDS Behav 17:1571–1590. doi: 10.1007/s10461-012-0241-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.WHO. 2015. Consolidated guidelines on HIV testing services. Annex 14. A report on the misdiagnosis of HIV status. WHO, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/180231/1/WHO_HIV_2015.33_eng.pdf?ua=1. [Google Scholar]
- 156.Mulinda N, Johnstone K, Newton K. 2011. Case report: HIV test misdiagnosis. Malawi Med J 23:122–123. [PMC free article] [PubMed] [Google Scholar]
- 157.Shanks L, Klarkowski D, O’Brien DP. 2013. False positive HIV diagnoses in resource limited settings: operational lessons learned for HIV programmes. PLoS One 8:e59906. doi: 10.1371/journal.pone.0059906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mwangala S, Moland KM, Nkamba HC, Musonda KG, Monze M, Musukwa KK, Fylkesnes K. 2015. Task-shifting and quality of HIV testing services: experiences from a national reference hospital in Zambia. PLoS One 10:e0143075. doi: 10.1371/journal.pone.0143075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mwangala S, Musonda KG, Monze M, Musukwa KK, Fylkesnes K. 2016. Accuracy in HIV rapid testing among laboratory and non-laboratory personnel in Zambia: observations from the National HIV Proficiency Testing System. PLoS One 11:e0146700. doi: 10.1371/journal.pone.0146700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Johnson CC, Dalal S, Baggaley R, Taegtmeyer M. 2017. A public health approach to addressing and preventing misdiagnosis in the scale-up of HIV rapid testing programmes. J Int AIDS Soc 20:22190. doi: 10.7448/IAS.20.7.22190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ministry of Health of Kenya. 2015. The Kenya HIV testing services guidelines 2015. Ministry of Health of Kenya, Nairobi, Kenya: https://aidsfree.usaid.gov/sites/default/files/hts_policy_kenya_2015.pdf. [Google Scholar]
- 162.Ministry of Health and Social Services, Republic of Namibia. 2011. National guidelines for HIV counselling and testing in Namibia, p 82 Ministry of Health and Social Services, Republic of Namibia, Windhoek, Namibia. [Google Scholar]
- 163.Nguyen S, Ramos A, Chang J, Li B, Shanmugam V, Boeras D, Nkengasong JN, Yang C, Ellenberger D. 2015. Monitoring the quality of HIV-1 viral load testing through a proficiency testing program using dried tube specimens in resource-limited settings. J Clin Microbiol 53:1129–1136. doi: 10.1128/JCM.02780-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Beber AM, Sabido M, Vieira JM, Bazzo ML, Benzaken AS. 2015. External quality assessment in the voluntary counseling and testing centers in the Brazilian Amazon using dried tube specimens: results of an effectiveness evaluation. Rev Soc Bras Med Trop 48(Suppl 1):87–97. doi: 10.1590/0037-8682-0106-2014. [DOI] [PubMed] [Google Scholar]
- 165.Louis FJ, Anselme R, Ndongmo C, Buteau J, Boncy J, Dahourou G, Vertefeuille J, Marston B, Balajee SA. 2013. Evaluation of an external quality assessment program for HIV testing in Haiti, 2006-2011. Am J Clin Pathol 140:867–871. doi: 10.1309/AJCPYWX49IZSQKFS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mashauri FM, Siza JE, Temu MM, Mngara JT, Kishamawe C, Changalucha JM. 2007. Assessment of quality assurance in HIV testing in health facilities in Lake Victoria zone, Tanzania. Tanzan Health Res Bull 9:110–114. [DOI] [PubMed] [Google Scholar]
- 167.WHO. 2015. Improving the quality of HIV-related point of care testing: ensuring the reliability and accuracy of test results. WHO, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/199799/1/9789241508179_eng.pdf. [Google Scholar]
- 168.Kane CT, Ndiaye HD, Diallo S, Ndiaye I, Wade AS, Diaw PA, Gaye-Diallo A, Mboup S. 2008. Quantitation of HIV-1 RNA in dried blood spots by the real-time NucliSENS EasyQ HIV-1 assay in Senegal. J Virol Methods 148:291–295. doi: 10.1016/j.jviromet.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 169.Health Department of the Republic of South Africa. 2015. National HIV counselling and testing policy guidelines, May 2015. Health Department of the Republic of South Africa, Pretoria, South Africa: https://www.health-e.org.za/wp-content/uploads/2015/07/HCT-Guidelines-2015.pdf. [Google Scholar]
- 170.Yao K, Wafula W, Bile EC, Cheignsong R, Howard S, Demby A, Nkengasong J. 2010. Ensuring the quality of HIV rapid testing in resource-poor countries using a systematic approach to training. Am J Clin Pathol 134:568–572. doi: 10.1309/AJCPOPXR8MNTZ5PY. [DOI] [PubMed] [Google Scholar]
- 171.WHO. 2005. HIV rapid test training package. WHO, Geneva, Switzerland. [Google Scholar]
- 172.Kanal K, Chou TL, Sovann L, Morikawa Y, Mukoyama Y, Kakimoto K. 2005. Evaluation of the proficiency of trained non-laboratory health staffs and laboratory technicians using a rapid and simple HIV antibody test. AIDS Res Ther 2:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Smith J, Odera DN, Chege D, Muigai EN, Patnaik P, Michaels-Strasser S, Howard AA, Yu-Shears J, Dohrn J. 2016. Identifying the gaps: an assessment of nurses’ training, competency, and practice in HIV care and treatment in Kenya. J Assoc Nurses AIDS Care 27:322–330. doi: 10.1016/j.jana.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 174.Zigmond J. 2013. Teaching by telementoring. Project ECHO advancing physicians’ skill sets. Mod Healthc 43:28–35. [PubMed] [Google Scholar]
- 175.Freese KE, Documet P, Lawrence JJ, Linkov F, Laporte RE, Stall RD. 2014. Public health education using Supercourse: a computer-based learning resource for health-care professionals in the southern province of Zambia. Public Health Rep 129:100–106. doi: 10.1177/003335491412900116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Struminger B, Arora S, Zalud-Cerrato S, Lowrance D, Ellerbrock T. 2017. Building virtual communities of practice for health. Lancet 390:632–634. doi: 10.1016/S0140-6736(17)31666-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Celletti F, Wright A, Palen J, Frehywot S, Markus A, Greenberg A, de Aguiar RA, Campos F, Buch E, Samb B. 2010. Can the deployment of community health workers for the delivery of HIV services represent an effective and sustainable response to health workforce shortages? Results of a multicountry study. AIDS 24(Suppl 1):S45–S57. doi: 10.1097/01.aids.0000366082.68321.d6. [DOI] [PubMed] [Google Scholar]
- 178.Wouters E, Van Damme W, van Rensburg D, Masquillier C, Meulemans H. 2012. Impact of community-based support services on antiretroviral treatment programme delivery and outcomes in resource-limited countries: a synthetic review. BMC Health Serv Res 12:194. doi: 10.1186/1472-6963-12-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Fonjungo PN, Osmanov S, Kuritsky J, Ndihokubwayo JB, Bachanas P, Peeling RW, Timperi R, Fine G, Stevens W, Habiyambere V, Nkengasong JN. 2016. Ensuring quality: a key consideration in scaling-up HIV-related point-of-care testing programs. AIDS 30:1317–1323. doi: 10.1097/QAD.0000000000001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Njukeng P. 2014. A progress report: implementation of HIV rapid test quality improvement initiative in Cameroon, p 14. Global Health Systems Solutions, Limbe, Cameroon. [Google Scholar]
- 181.Theobald S, Hawkins K, Kok M, Rashid S, Datiko DG, Taegtmeyer M. 2016. Close-to-community providers of health care: increasing evidence of how to bridge community and health systems. Hum Resour Health 14:32. doi: 10.1186/s12960-016-0132-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Parliament of the United Republic of Tanzania. 2007. The Health Laboratory Practitioners Act, 2007, p 27. United Republic of Tanzania. http://ihi.eprints.org/729/1/ihi.eprints.pdf_%2833%29.pdf. Accessed 5 November 2018.
- 183.Ziyambi Z, Osewe P, Taruberekera N. 2002. Evaluation of the performance of non-laboratory staff in the use of simple rapid HIV antibody assays at New Start voluntary counselling and testing (VCT) centres, abstr MoPeB3110 Abstr 14th Int AIDS Conf, Barcelona, Spain. [Google Scholar]
- 184.WHO. 2013. Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection: recommendations for a public health approach. WHO, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/85321/1/9789241505727_eng.pdf?ua=1. Accessed 22 September 2017. [PubMed] [Google Scholar]
- 185.Bartlett JA, Shao JF. 2009. Successes, challenges, and limitations of current antiretroviral therapy in low-income and middle-income countries. Lancet Infect Dis 9:637–649. doi: 10.1016/S1473-3099(09)70227-0. [DOI] [PubMed] [Google Scholar]
- 186.Novitsky V, Essex M. 2012. Using HIV viral load to guide treatment-for-prevention interventions. Curr Opin HIV AIDS 7:117–124. doi: 10.1097/COH.0b013e32834fe8ff. [DOI] [PubMed] [Google Scholar]
- 187.Barth RE, van der Loeff MF, Schuurman R, Hoepelman AI, Wensing AM. 2010. Virological follow-up of adult patients in antiretroviral treatment programmes in sub-Saharan Africa: a systematic review. Lancet Infect Dis 10:155–166. doi: 10.1016/S1473-3099(09)70328-7. [DOI] [PubMed] [Google Scholar]
- 188.Working Group on Modelling of Antiretroviral Therapy Monitoring Strategies in Sub-Saharan Africa, Phillips A, Shroufi A, Vojnov L, Cohn J, Roberts T, Ellman T, Bonner K, Rousseau C, Garnett G, Cambiano V, Nakagawa F, Ford D, Bansi-Matharu L, Miners A, Lundgren JD, Eaton JW, Parkes-Ratanshi R, Katz Z, Maman D, Ford N, Vitoria M, Doherty M, Dowdy D, Nichols B, Murtagh M, Wareham M, Palamountain KM, Chakanyuka Musanhu C, Stevens W, Katzenstein D, Ciaranello A, Barnabas R, Braithwaite RS, Bendavid E, Nathoo KJ, van de Vijver D, Wilson DP, Holmes C, Bershteyn A, Walker S, Raizes E, Jani I, Nelson LJ, Peeling R, Terris-Prestholt F, Murungu J, Mutasa-Apollo T, Hallett TB, Revill P. 2015. Sustainable HIV treatment in Africa through viral-load-informed differentiated care. Nature 528:S68–S76. doi: 10.1038/nature16046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Bonner K, Mezochow A, Roberts T, Ford N, Cohn J. 2013. Viral load monitoring as a tool to reinforce adherence: a systematic review. J Acquir Immune Defic Syndr 64:74–78. doi: 10.1097/QAI.0b013e31829f05ac. [DOI] [PubMed] [Google Scholar]
- 190.US DHHS. 2015. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. US DHHS, Washington, DC: http://aidsinfo.nih.gov/contentfiles/lvguidelines/AdultandadolescentGL.pdf. [Google Scholar]
- 191.Gaolathe T, Wirth KE, Holme MP, Makhema J, Moyo S, Chakalisa U, Yankinda EK, Lei Q, Mmalane M, Novitsky V, Okui L, van Widenfelt E, Powis KM, Khan N, Bennett K, Bussmann H, Dryden-Peterson S, Lebelonyane R, El-Halabi S, Mills LA, Marukutira T, Wang R, Tchetgen EJ, DeGruttola V, Essex M, Lockman S, Botswana Combination Prevention Project Study Team. 2016. Botswana’s progress toward achieving the 2020 UNAIDS 90-90-90 antiretroviral therapy and virological suppression goals: a population-based survey. Lancet HIV 3:e221–e230. doi: 10.1016/S2352-3018(16)00037-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.PEPFAR. 2016. The epidemic is becoming controlled in several key African countries. PEPFAR, Washington, DC: www.pepfar.gov/documents/organization/264882.pdf. Accessed 6 October 2017. [Google Scholar]
- 193.CLSI. 2004. Protocols for determination of limits of detection and limits of quantitation, approved guideline. CLSI document EP17; CLSI, Wayne, PA. [Google Scholar]
- 194.Bonner K, Siemieniuk RA, Boozary A, Roberts T, Fajardo E, Cohn J. 2014. Expanding access to HIV viral load testing: a systematic review of RNA stability in EDTA tubes and PPT beyond current time and temperature thresholds. PLoS One 9:e113813. doi: 10.1371/journal.pone.0113813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ondoa P, Shamu T, Bronze M, Wellington M, Boender TS, Manting C, Steegen K, Luethy R, Rinke de Wit T. 2014. Performance and logistical challenges of alternative HIV-1 virological monitoring options in a clinical setting of Harare, Zimbabwe. Biomed Res Int 2014:102598. doi: 10.1155/2014/102598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Médecins Sans Frontières. 2013. How low can we go? Pricing for HIV viral load testing in low- and middle-income countries. Médecins Sans Frontières, Geneva, Switzerland: www.msfaccess.org/content/issue-brief-how-low-can-we-go. Accessed 31 January 2016. [Google Scholar]
- 197.Lynch S. 2014. Viral load testing—are cost and complexity out-dated argument?, abstr 5082 Abstr 2014 Treat Prev Workshop, Vancouver, Canada. [Google Scholar]
- 198.UNAIDS. 2014. Press release: Landmark HIV diagnostic access program will save $150m and help achieve new global goals on HIV. UNAIDS, Geneva, Switzerland: http://www.unaids.org/en/resources/presscentre/pressreleaseandstatementarchive/2014/september/20140925prviralload. Accessed 31 January 2016. [Google Scholar]
- 199.Gund G. 2017. HIV viral load and early infant diagnosis selection and procurement information tool. The Global Fund, Geneva, Switzerland: www.theglobalfund.org/media/5765/psm_viralloadearlyinfantdiagnosis_content_en.pdf. Accessed 6 October 2017. [Google Scholar]
- 200.Keebler D, Revill P, Braithwaite S, Phillips A, Blaser N, Borquez A, Cambiano V, Ciaranello A, Estill J, Gray R, Hill A, Keiser O, Kessler J, Menzies NA, Nucifora KA, Vizcaya LS, Walker S, Welte A, Easterbrook P, Doherty M, Hirnschall G, Hallett TB. 2014. Cost-effectiveness of different strategies to monitor adults on antiretroviral treatment: a combined analysis of three mathematical models. Lancet Glob Health 2:e35–e43. doi: 10.1016/S2214-109X(13)70048-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Coombs RW, Collier AC, Allain JP, Nikora B, Leuther M, Gjerset GF, Corey L. 1989. Plasma viremia in human immunodeficiency virus infection. N Engl J Med 321:1626–1631. doi: 10.1056/NEJM198912143212402. [DOI] [PubMed] [Google Scholar]
- 202.Ho DD, Moudgil T, Alam M. 1989. Quantitation of human immunodeficiency virus type 1 in the blood of infected persons. N Engl J Med 321:1621–1625. doi: 10.1056/NEJM198912143212401. [DOI] [PubMed] [Google Scholar]
- 203.Higuchi R, Dollinger G, Walsh PS, Griffith R. 1992. Simultaneous amplification and detection of specific DNA sequences. Nat Biotechnol 10:413–417. doi: 10.1038/nbt0492-413. [DOI] [PubMed] [Google Scholar]
- 204.Bacich DJ, Sobek KM, Cummings JL, Atwood AA, O’Keefe DS. 2011. False negative results from using common PCR reagents. BMC Res Notes 4:457. doi: 10.1186/1756-0500-4-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Champlot S, Berthelot C, Pruvost M, Bennett EA, Grange T, Geigl EM. 2010. An efficient multistrategy DNA decontamination procedure of PCR reagents for hypersensitive PCR applications. PLoS One 5:e13042. doi: 10.1371/journal.pone.0013042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Barnabas RV, Carabin H, Garnett GP. 2002. The potential role of suppressive therapy for sex partners in the prevention of neonatal herpes: a health economic analysis. Sex Transm Infect 78:425–429. doi: 10.1136/sti.78.6.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Cepheid. Xpert HIV-1 viral load. Cepheid, Sunnyvale, CA: http://www.cepheid.com/en/cepheid-solutions/clinical-ivd-tests/virology/xpert-hiv-1-viral-load. Accessed 5 November 2018. [Google Scholar]
- 208.Sivapalasingam S, Wangechi B, Marshed F, Laverty M, Essajee S, Holzman RS, Valentine F. 2009. Monitoring virologic responses to antiretroviral therapy in HIV-infected adults in Kenya: evaluation of a low-cost viral load assay. PLoS One 4:e6828. doi: 10.1371/journal.pone.0006828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Elbeik T, Alvord WG, Trichavaroj R, de Souza M, Dewar R, Brown A, Chernoff D, Michael NL, Nassos P, Hadley K, Ng VL. 2002. Comparative analysis of HIV-1 viral load assays on subtype quantification: Bayer Versant HIV-1 RNA 3.0 versus Roche Amplicor HIV-1 Monitor version 1.5. J Acquir Immune Defic Syndr 29:330–339. doi: 10.1097/00126334-200204010-00002. [DOI] [PubMed] [Google Scholar]
- 210.Greengrass VL, Plate MM, Steele PM, Denholm JT, Cherry CL, Morris LM, Hearps A, Crowe SM. 2009. Evaluation of the Cavidi ExaVir Load assay (version 3) for plasma human immunodeficiency virus type 1 load monitoring. J Clin Microbiol 47:3011–3013. doi: 10.1128/JCM.00805-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Gare J, Kelly-Hanku A, Ryan CE, David M, Kaima P, Imara U, Lote N, Crowe SM, Hearps AC. 2015. Factors influencing antiretroviral adherence and virological outcomes in people living with HIV in the Highlands of Papua New Guinea. PLoS One 10:e0134918. doi: 10.1371/journal.pone.0134918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Korn K, Weissbrich B, Henke-Gendo C, Heim A, Jauer CM, Taylor N, Eberle J. 2009. Single-point mutations causing more than 100-fold underestimation of human immunodeficiency virus type 1 (HIV-1) load with the Cobas TaqMan HIV-1 real-time PCR assay. J Clin Microbiol 47:1238–1240. doi: 10.1128/JCM.02204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Rouet F, Chaix M-L, Nerrienet E, Ngo-Giang-Huong N, Plantier J-C, Burgard M, Peeters M, Damond F, Ekouevi DK, Msellati P, Ferradini L, Rukobo S, Maréchal V, Schvachsa N, Wakrim L, Rafalimanana C, Rakotoambinina B, Viard J-P, Seigneurin J-M, Rouzioux C. 2007. Impact of HIV-1 genetic diversity on plasma HIV-1 RNA quantification: usefulness of the Agence Nationale de Recherches sur le SIDA second-generation long terminal repeat-based real-time reverse transcriptase polymerase chain reaction test. J Acquir Immune Defic Syndr 45:380–388. doi: 10.1097/QAI.0b013e3180640cf5. [DOI] [PubMed] [Google Scholar]
- 214.Triques K, Coste J, Perret JL, Segarra C, Mpoudi E, Reynes J, Delaporte E, Butcher A, Dreyer K, Herman S, Spadoro J, Peeters M. 1999. Efficiencies of four versions of the Amplicor HIV-1 Monitor test for quantification of different subtypes of human immunodeficiency virus type 1. J Clin Microbiol 37:110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Mouinga-Ondémé A, Mabika-Mabika A, Alalade P, Mongo AD, Sica J, Liégeois F, Rouet F. 2014. Significant impact of non-B HIV-1 variants genetic diversity in Gabon on plasma HIV-1 RNA quantitation. J Med Virol 86:52–57. doi: 10.1002/jmv.23770. [DOI] [PubMed] [Google Scholar]
- 216.Luft LM, Gill MJ, Church DL. 2011. HIV-1 viral diversity and its implications for viral load testing: review of current platforms. Int J Infect Dis 15:e661–e670. doi: 10.1016/j.ijid.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 217.Aghokeng AF, Monleau M, Eymard-Duvernay S, Dagnra A, Kania D, Ngo-Giang-Huong N, Toni TD, Toure-Kane C, Truong LX, Delaporte E, Chaix ML, Peeters M, Ayouba A, ANRS 12186 Study Group. 2014. Extraordinary heterogeneity of virological outcomes in patients receiving highly antiretroviral therapy and monitored with the World Health Organization public health approach in sub-Saharan Africa and southeast Asia. Clin Infect Dis 58:99–109. doi: 10.1093/cid/cit627. [DOI] [PubMed] [Google Scholar]
- 218.Tang N, Huang S, Salituro J, Mak WB, Cloherty G, Johanson J, Li YH, Schneider G, Robinson J, Hackett J Jr, Swanson P, Abravaya K. 2007. A RealTime HIV-1 viral load assay for automated quantitation of HIV-1 RNA in genetically diverse group M subtypes A-H, group O and group N samples. J Virol Methods 146:236–245. doi: 10.1016/j.jviromet.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 219.van Rensburg EJ, Tait K, Watt A, Schall R. 2011. Comparative evaluation of the Roche Cobas AmpliPrep/Cobas TaqMan HIV-1 version 2 test using the TaqMan 48 analyzer and the Abbott RealTime HIV-1 assay. J Clin Microbiol 49:377–379. doi: 10.1128/JCM.01285-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Fonjungo PN, Girma M, Melaku Z, Mekonen T, Tanuri A, Hailegiorgis B, Tegbaru B, Mengistu Y, Ashenafi A, Mamo W, Abreha T, Tibesso G, Ramos A, Ayana G, Freeman R, Nkengasong JN, Zewdu S, Kebede Y, Abebe A, Kenyon TA, Messele T. 2013. Field expansion of DNA polymerase chain reaction for early infant diagnosis of HIV-1: the Ethiopian experience. Afr J Lab Med 2:31. doi: 10.4102/ajlm.v2i1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Kebede Y, Fonjungo PN, Tibesso G, Shrivastava R, Nkengasong JN, Kenyon T, Kebede A, Gadde R, Ayana G. 2016. Improved specimen-referral system and increased access to quality laboratory services in Ethiopia: the role of the public-private partnership. J Infect Dis 213(Suppl 2):S59–S64. doi: 10.1093/infdis/jiv576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Kebede A, Kebede Y, Desale A, Mulugeta A, Yaregal Z, Gebreegziabxier A, Tedla Y, Zeh C, Ayana G. 2016. Quality assurance for point-of-care testing: Ethiopia’s experience. Afr J Lab Med 5:452. doi: 10.4102/ajlm.v5i2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.UNITAID. 2015. HIV/AIDS diagnostics technology landscape, 5th ed UNITAID, Geneva, Switzerland: www.unitaid.org/assets/UNITAID_HIV_Nov_2015_Dx_Landscape-1.pdf. Accessed 20 November 2017. [Google Scholar]
- 224.Habiyambere V, Ford N, Low-Beer D, Nkengasong J, Sands A, Pérez González M, Fernandes P, Milgotina E. 2016. Availability and use of HIV monitoring and early infant diagnosis technologies in WHO member states in 2011-2013: analysis of annual surveys at the facility level. PLoS Med 13:e1002088. doi: 10.1371/journal.pmed.1002088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Omrani AS, Al-Otaibi MF, Al-Ateah SM, Al-Onazi FM, Baig K, El-Khizzi NA, Albarrak AM. 2014. GeneXpert MTB/RIF testing in the management of patients with active tuberculosis; a real life experience from Saudi Arabia. Infect Chemother 46:30–34. doi: 10.3947/ic.2014.46.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Nash M, Ramapuram J, Kaiya R, Huddart S, Pai M, Baliga S. 2017. Use of the GeneXpert tuberculosis system for HIV viral load testing in India. Lancet Glob Health 5:e754–e755. doi: 10.1016/S2214-109X(17)30247-4. [DOI] [PubMed] [Google Scholar]
- 227.Ritchie AV, Ushiro-Lumb I, Edemaga D, Joshi HA, De Ruiter A, Szumilin E, Jendrulek I, McGuire M, Goel N, Sharma PI, Allain JP, Lee HH. 2014. SAMBA HIV semiquantitative test, a new point-of-care viral-load-monitoring assay for resource-limited settings. J Clin Microbiol 52:3377–3383. doi: 10.1128/JCM.00593-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Lee HH, Dineva MA, Chua YL, Ritchie AV, Ushiro-Lumb I, Wisniewski CA. 2010. Simple amplification-based assay: a nucleic acid-based point-of-care platform for HIV-1 testing. J Infect Dis 201(Suppl 1):S65–S72. doi: 10.1086/650385. [DOI] [PubMed] [Google Scholar]
- 229.Goel N, Ritchie AV, Mtapuri-Zinyowera S, Zeh C, Stepchenkova T, Lehga J, De Ruiter A, Farleigh LE, Edemaga D, So R, Sembongi H, Wisniewski C, Nadala L, Schito M, Lee H. 2017. Performance of the SAMBA I and II HIV-1 Semi-Q tests for viral load monitoring at the point-of-care. J Virol Methods 244:39–45. doi: 10.1016/j.jviromet.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 230.Titchmarsh L, Zeh C, Verpoort T, Allain JP, Lee H. 2015. Leukodepletion as a point-of-care method for monitoring HIV-1 viral load in whole blood. J Clin Microbiol 53:1080–1086. doi: 10.1128/JCM.02853-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Tanriverdi S, Chen L, Chen S. 2010. A rapid and automated sample-to-result HIV load test for near-patient application. J Infect Dis 201(Suppl 1):S52–S58. doi: 10.1086/650387. [DOI] [PubMed] [Google Scholar]
- 232.Scott L, Gous N, Carmona S, Stevens W. 2015. Laboratory evaluation of the Liat HIV Quant (IQuum) whole-blood and plasma HIV-1 viral load assays for point-of-care testing in South Africa. J Clin Microbiol 53:1616–1621. doi: 10.1128/JCM.03325-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Steinmetzer K, Seidel T, Stallmach A, Ermantraut E. 2010. HIV load testing with small samples of whole blood. J Clin Microbiol 48:2786–2792. doi: 10.1128/JCM.02276-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Ceffa S, Luhanga R, Andreotti M, Brambilla D, Erba F, Jere H, Mancinelli S, Giuliano M, Palombi L, Marazzi MC. 2016. Comparison of the Cepheid GeneXpert and Abbott M2000 HIV-1 real time molecular assays for monitoring HIV-1 viral load and detecting HIV-1 infection. J Virol Methods 229:35–39. doi: 10.1016/j.jviromet.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 235.Jordan JA, Plantier JC, Templeton K, Wu AH. 2016. Multi-site clinical evaluation of the Xpert HIV-1 viral load assay. J Clin Virol 80:27–32. doi: 10.1016/j.jcv.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 236.WHO. 2017. WHO prequalification of in vitro diagnostics: Xpert HIV-1 viral load with GeneXpert Dx, GeneXpert Infinity-48, GeneXpert Infinity-48s and GeneXpert Infinity-80. WHO, Geneva, Switzerland: www.who.int/diagnostics_laboratory/evaluations/pq-list/hiv-vrl/170720_final_pq_report_pqdx_0192_0193_0194_0195_070-00.pdf?ua=1. Accessed 14 May 2018. [Google Scholar]
- 237.WHO. 2014. Interim technical update: Technical and operational considerations for implementing HIV viral load testing. WHO, Geneva, Switzerland: http://apps.who.int/iris/bitstream/handle/10665/128121/9789241507578_eng.pdf;jsessionid=6C832044202A0CA8C151CB804C43E2E3?sequence=1. Accessed 31 January 2016. [Google Scholar]
- 238.Koethe JR, Westfall AO, Luhanga DK, Clark GM, Goldman JD, Mulenga PL, Cantrell RA, Chi BH, Zulu I, Saag MS, Stringer JS. 2010. A cluster randomized trial of routine HIV-1 viral load monitoring in Zambia: study design, implementation, and baseline cohort characteristics. PLoS One 5:e9680. doi: 10.1371/annotation/dbc3ee9b-dab4-494b-87d1-7c32b8d684b5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Kiyaga C, Sendagire H, Joseph E, McConnell I, Grosz J, Narayan V, Esiru G, Elyanu P, Akol Z, Kirungi W, Musinguzi J, Opio A. 2013. Uganda’s new national laboratory sample transport system: a successful model for improving access to diagnostic services for early infant HIV diagnosis and other programs. PLoS One 8:e78609. doi: 10.1371/journal.pone.0078609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Vidya M, Saravanan S, Rifkin S, Solomon SS, Waldrop G, Mayer KH, Solomon S, Balakrishnan P. 2012. Dried blood spots versus plasma for the quantitation of HIV-1 RNA using a real-time PCR, m2000rt assay. J Virol Methods 181:177–181. doi: 10.1016/j.jviromet.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 241.Smit PW, Sollis KA, Fiscus S, Ford N, Vitoria M, Essajee S, Barnett D, Cheng B, Crowe SM, Denny T, Landay A, Stevens W, Habiyambere V, Perriens JH, Peeling RW. 2014. Systematic review of the use of dried blood spots for monitoring HIV viral load and for early infant diagnosis. PLoS One 9:e86461. doi: 10.1371/journal.pone.0086461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.WHO. 2010. Towards universal access: scaling up priority HIV/AIDS interventions in the health sector. WHO, Geneva, Switzerland: http://whqlibdoc.who.int/publications/2010/9789241500395_eng.pdf. [Google Scholar]
- 243.Mbida AD, Sosso S, Flori P, Saoudin H, Lawrence P, Monny-Lobe M, Oyono Y, Ndzi E, Cappelli G, Lucht F, Pozzetto B, Oukem-Boyer OO, Bourlet T. 2009. Measure of viral load by using the Abbott real-time HIV-1 assay on dried blood and plasma spot specimens collected in 2 rural dispensaries in Cameroon. J Acquir Immune Defic Syndr 52:9–16. doi: 10.1097/QAI.0b013e3181aeccbc. [DOI] [PubMed] [Google Scholar]
- 244.Alvarez P, Martín L, Prieto L, Obiang J, Vargas A, Avedillo P, Rojo P, Fernández McPhee C, Benito A, Ramos JT, Holguín Á. 2015. HIV-1 variability and viral load technique could lead to false positive HIV-1 detection and to erroneous viral quantification in infected specimens. J Infect 71:368–376. doi: 10.1016/j.jinf.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 245.Mercier-Delarue S, Vray M, Plantier JC, Maillard T, Adjout Z, de Olivera F, Schnepf N, Maylin S, Simon F, Delaugerre C. 2014. Higher specificity of nucleic acid sequence-based amplification isothermal technology than of real-time PCR for quantification of HIV-1 RNA on dried blood spots. J Clin Microbiol 52:52–56. doi: 10.1128/JCM.01848-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Marconi A, Balestrieri M, Comastri G, Pulvirenti FR, Gennari W, Tagliazucchi S, Pecorari M, Borghi V, Marri D, Zazzi M. 2009. Evaluation of the Abbott real-time HIV-1 quantitative assay with dried blood spot specimens. Clin Microbiol Infect 15:93–97. doi: 10.1111/j.1469-0691.2008.02116.x. [DOI] [PubMed] [Google Scholar]
- 247.Wu X, Crask M, Ramirez H, Landas T, Do TD, Honisch C, Will S, Baum PD. 2015. A simple method to elute cell-free HIV from dried blood spots improves their usefulness for monitoring therapy. J Clin Virol 65:38–40. doi: 10.1016/j.jcv.2015.01.022. [DOI] [PubMed] [Google Scholar]
- 248.Zeh C, Ndiege K, Inzaule S, Achieng R, Williamson J, Chih-Wei Chang J, Ellenberger D, Nkengasong J. 2017. Evaluation of the performance of Abbott m2000 and Roche COBAS Ampliprep/COBAS Taqman assays for HIV-1 viral load determination using dried blood spots and dried plasma spots in Kenya. PLoS One 12:e0179316. doi: 10.1371/journal.pone.0179316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Fajardo E, Metcalf CA, Chaillet P, Aleixo L, Pannus P, Panunzi I, Trivino L, Ellman T, Likaka A, Mwenda R. 2014. Prospective evaluation of diagnostic accuracy of dried blood spots from finger prick samples for determination of HIV-1 load with the NucliSENS Easy-Q HIV-1 version 2.0 assay in Malawi. J Clin Microbiol 52:1343–1351. doi: 10.1128/JCM.03519-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Napierala Mavedzenge S, Davey C, Chirenje T, Mushati P, Mtetwa S, Dirawo J, Mudenge B, Phillips A, Cowan FM. 2015. Finger prick dried blood spots for HIV viral load measurement in field conditions in Zimbabwe. PLoS One 10:e0126878. doi: 10.1371/journal.pone.0126878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Schmitz ME, Agolory S, Junghae M, Broyles LN, Kimeu M, Ombayo J, Umuro M, Mukui I, Alwenya K, Baraza M, Ndiege K, Mwalili S, Rivadeneira E, Ngʼangʼa L, Yang C, Zeh C, for the VL-DBS Study Group. 2017. Field evaluation of dried blood spots for HIV-1 viral load monitoring in adults and children receiving antiretroviral treatment in Kenya: implications for scale-up in resource-limited settings. J Acquir Immune Defic Syndr 74:399–406. doi: 10.1097/QAI.0000000000001275. [DOI] [PubMed] [Google Scholar]
- 252.Marlink R, Kanki P, Thior I, Travers K, Eisen G, Siby T, Traore I, Hsieh CC, Dia MC, Gueye EH. 1994. Reduced rate of disease development after HIV-2 infection as compared to HIV-1. Science 265:1587–1590. [DOI] [PubMed] [Google Scholar]
- 253.Popper SJ, Sarr AD, Gueye-Ndiaye A, Mboup S, Essex ME, Kanki PJ. 2000. Low plasma human immunodeficiency virus type 2 viral load is independent of proviral load: low virus production in vivo. J Virol 74:1554–1557. doi: 10.1128/JVI.74.3.1554-1557.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Pieniazek D, Rayfield M, Hu DJ, Nkengasong JN, Soriano V, Heneine W, Zeh C, Agwale SM, Wambebe C, Odama L, Wiktor SZ. 2004. HIV-2 protease sequences of subtypes A and B harbor multiple mutations associated with protease inhibitor resistance in HIV-1. AIDS 18:495–502. doi: 10.1097/00002030-200402200-00016. [DOI] [PubMed] [Google Scholar]
- 255.Adje-Toure CA, Cheingsong R, Garcia-Lerma JG, Eholie S, Borget MY, Bouchez JM, Otten RA, Maurice C, Sassan-Morokro M, Ekpini RE, Nolan M, Chorba T, Heneine W, Nkengasong JN. 2003. Antiretroviral therapy in HIV-2-infected patients: changes in plasma viral load, CD4+ cell counts, and drug resistance profiles of patients treated in Abidjan, Cote d’Ivoire. AIDS 17(Suppl 3):S49–S54. doi: 10.1097/00002030-200317003-00007. [DOI] [PubMed] [Google Scholar]
- 256.Chang MWK, Raugi DN, Smith RA, Ba S, Seydi M, Steinmetzer K, Coombs RW, Gottlieb GS. 2015. The POC Alere q HIV-1/2 Detect test for detection and quantification of HIV-2, abstr 614. Abstr 2015 Conf Retrovir Oppor Infect. www.croiconference.org/sessions/poc-alere-q-hiv-12-detect-test-detection-and-quantification-hiv-2. Accessed 4 January 2018. [Google Scholar]
- 257.Chang M, Gottlieb GS, Dragavon JA, Cherne SL, Kenney DL, Hawes SE, Smith RA, Kiviat NB, Sow PS, Coombs RW. 2012. Validation for clinical use of a novel HIV-2 plasma RNA viral load assay using the Abbott m2000 platform. J Clin Virol 55:128–133. doi: 10.1016/j.jcv.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Newell ML, Coovadia H, Cortina-Borja M, Rollins N, Gaillard P, Dabis F, Ghent International AIDS Society Working Group on HIV Infection in Women and Children. 2004. Mortality of infected and uninfected infants born to HIV-infected mothers in Africa: a pooled analysis. Lancet 364:1236–1243. doi: 10.1016/S0140-6736(04)17140-7. [DOI] [PubMed] [Google Scholar]
- 259.Siberry GK. 2014. Preventing and managing HIV infection in infants, children, and adolescents in the United States. Pediatr Rev 35:268–286. doi: 10.1542/pir.35-7-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Mnyani CN, Simango A, Murphy J, Chersich M, McIntyre JA. 2014. Patient factors to target for elimination of mother-to-child transmission of HIV. Global Health 10:36. doi: 10.1186/1744-8603-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Laure F, Courgnaud V, Rouzioux C, Blanche S, Veber F, Burgard M, Jacomet C, Griscelli C, Brechot C. 1988. Detection of HIV1 DNA in infants and children by means of the polymerase chain reaction. Lancet ii:538–541. [DOI] [PubMed] [Google Scholar]
- 262.Rogers MF, Ou C-Y, Rayfield M, Thomas PA, Schoenbaum EE, Abrams E, Krasinski K, Selwyn PA, Moore J, Kaul A, Grimm KT, Bamji M, Schochetman G, New York City Collaborative Study of Maternal HIV Transmission and Montefiore Medical Center HIV Perinatal Transmission Study Group. 1989. Use of the polymerase chain reaction for early detection of the proviral sequences of human immunodeficiency virus in infants born to seropositive mothers. N Engl J Med 320:1649–1654. doi: 10.1056/NEJM198906223202503. [DOI] [PubMed] [Google Scholar]
- 263.Stevens W, Sherman G, Downing R, Parsons LM, Ou CY, Crowley S, Gershy-Damet GM, Fransen K, Bulterys M, Lu L, Homsy J, Finkbeiner T, Nkengasong JN. 2008. Role of the laboratory in ensuring global access to ARV treatment for HIV-infected children: consensus statement on the performance of laboratory assays for early infant diagnosis. Open AIDS J 2:17–25. doi: 10.2174/1874613600802010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.UNICEF. 2017. Children and AIDS: statistical update. UNICEF, New York, NY: http://data.unicef.org/wp-content/uploads/2017/11/HIVAIDS-Statistical-Update-2017.pdf. Accessed 5 May 2018. [Google Scholar]
- 265.WHO. 2010. Diagnosis of HIV infection in infants and children. WHO, Geneva, Switzerland: http://www.who.int/hiv/pub/paediatric/diagnosis/en/. Accessed 20 September 2017. [Google Scholar]
- 266.Dunning L, Kroon M, Fourie L, Ciaranello A, Myer L. 2017. Impact of birth HIV-PCR testing on the uptake of follow-up early infant diagnosis services in Cape Town, South Africa. Pediatr Infect Dis J 36:1159–1164. doi: 10.1097/INF.0000000000001677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Lilian RR, Johnson LF, Moolla H, Sherman GG. 2014. A mathematical model evaluating the timing of early diagnostic testing in HIV-exposed infants in South Africa. J Acquir Immune Defic Syndr 67:341–348. doi: 10.1097/QAI.0000000000000307. [DOI] [PubMed] [Google Scholar]
- 268.Sutcliffe CG, van Dijk JH, Hamangaba F, Mayani F, Moss WJ. 2014. Turnaround time for early infant HIV diagnosis in rural Zambia: a chart review. PLoS One 9:e87028. doi: 10.1371/journal.pone.0087028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Aliyu MH, Blevins M, Parrish DD, Megazzini KM, Gebi UI, Muhammad MY, Ahmed ML, Hassan A, Shepherd BE, Vermund SH, Wester CW. 2014. Risk factors for delayed initiation of combination antiretroviral therapy in rural north central Nigeria. J Acquir Immune Defic Syndr 65:e41–e49. doi: 10.1097/QAI.0b013e31829ceaec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Motswere-Chirwa C, Voetsch A, Lu L, Letsholathebe V, Lekone P, Machakaire E, Legwaila K, Matambo S, Maruping M, Kolobe T, Petlo C, Lebelonyane R, Glenshaw M, Dale H, Davis M, El Halabi S, Pelletier A, Centers for Disease Control and Prevention. 2014. Follow-up of infants diagnosed with HIV—early infant diagnosis program, Francistown, Botswana, 2005-2012. MMWR Morb Mortal Wkly Rep 63:158–160. [PMC free article] [PubMed] [Google Scholar]
- 271.Chatterjee A, Tripathi S, Gass R, Hamunime N, Panha S, Kiyaga C, Wade A, Barnhart M, Luo C, Ekpini R. 2011. Implementing services for early infant diagnosis (EID) of HIV: a comparative descriptive analysis of national programs in four countries. BMC Public Health 11:553. doi: 10.1186/1471-2458-11-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Chiduo MG, Mmbando BP, Theilgaard ZP, Bygbjerg IC, Gerstoft J, Lemnge M, Katzenstein TL. 2013. Early infant diagnosis of HIV in three regions in Tanzania; successes and challenges. BMC Public Health 13:910. doi: 10.1186/1471-2458-13-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Mugambi ML, Deo S, Kekitiinwa A, Kiyaga C, Singer ME. 2013. Do diagnosis delays impact receipt of test results? Evidence from the HIV early infant diagnosis program in Uganda. PLoS One 8:e78891. doi: 10.1371/journal.pone.0078891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Woldesenbet SA, Jackson D, Goga AE, Crowley S, Doherty T, Mogashoa MM, Dinh TH, Sherman GG. 2015. Missed opportunities for early infant HIV diagnosis: results of a national study in South Africa. J Acquir Immune Defic Syndr 68:e26–e32. doi: 10.1097/QAI.0000000000000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Anoje C, Aiyenigba B, Suzuki C, Badru T, Akpoigbe K, Odo M, Odafe S, Adedokun O, Torpey K, Chabikuli ON. 2012. Reducing mother-to-child transmission of HIV: findings from an early infant diagnosis program in south-south region of Nigeria. BMC Public Health 12:184. doi: 10.1186/1471-2458-12-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Hanna LE, Siromany VA, Annamalai M, Karunaianantham R, Swaminathan S. 2015. Challenges in the early diagnosis of HIV infection in infants: experience from Tamil Nadu, India. Indian Pediatr 52:307–309. doi: 10.1007/s13312-015-0629-7. [DOI] [PubMed] [Google Scholar]
- 277.WHO. 2016. WHO prequalification of in vitro diagnostics. Product: Alere q HIV-1/2 Detect, WHO reference number PQDx 0226-032-00. WHO, Geneva, Switzerland: www.who.int/diagnostics_laboratory/evaluations/pq-list/hiv-vrl/160613PQPublicReport_0226-032-00AlereHIVDetect_v2.pdf. Accessed 2 November 2017. [Google Scholar]
- 278.Jani IV, Meggi B, Mabunda N, Vubil A, Sitoe NE, Tobaiwa O, Quevedo JI, Lehe JD, Loquiha O, Vojnov L, Peter TF. 2014. Accurate early infant HIV diagnosis in primary health clinics using a point-of-care nucleic acid test. J Acquir Immune Defic Syndr 67:e1–e4. doi: 10.1097/QAI.0000000000000250. [DOI] [PubMed] [Google Scholar]
- 279.Hsiao N-Y, Kroon M, Dunning L, Myer L. 2015. Evaluation of the Alere q for point-of-care early infant HIV diagnosis in South Africa, abstr 34. Abstr 2015 Conf Retrovir Oppor Infect, Seattle, WA. [Google Scholar]
- 280.Hsiao NY, Dunning L, Kroon M, Myer L. 2016. Laboratory evaluation of the Alere q point-of-care system for early infant HIV diagnosis. PLoS One 11:e0152672. doi: 10.1371/journal.pone.0152672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.WHO. 2016. WHO prequalification of in vitro diagnostics. Product: Xpert HIV-1 Qual assay, WHO reference number PQDx 0259-070-00. WHO, Geneva, Switzerland: www.who.int/diagnostics_laboratory/evaluations/pq-list/hiv-vrl/160613PQPublicReport_0259-0700-00_XpertQualHIV_v2.pdf. Accessed 22 November 2017. [Google Scholar]
- 282.Meggi B, Bollinger T, Mabunda N, Vubil A, Tobaiwa O, Quevedo JI, Loquiha O, Vojnov L, Peter TF, Jani IV. 2017. Point-of-care p24 infant testing for HIV may increase patient identification despite low sensitivity. PLoS One 12:e0169497. doi: 10.1371/journal.pone.0169497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Chilikwazi M, Sutcliffe C, Thuma P, Sinywimaanzi K, Matakala H, Munachoonga P, Moss W. 2017. Feasibility of using the Lynx point-of-care test for early infant HIV diagnosis in rural Zambia. BMJ Glob Health 2(Suppl 2):A48.2. doi: 10.1136/bmjgh-2016-000260.128. [DOI] [Google Scholar]
- 284.Ritchie AV, Goel N, Sembongi H, Lehga J, Farleigh LE, Edemaga D, Wisniewski CA, Lee HH. 2016. Performance evaluation of the point-of-care SAMBA I and II HIV-1 Qual whole blood tests. J Virol Methods 237:143–149. doi: 10.1016/j.jviromet.2016.08.017. [DOI] [PubMed] [Google Scholar]
- 285.Bertagnolio S, Perno CF, Vella S, Pillay D. 2013. The impact of HIV drug resistance on the selection of first- and second-line ART in resource-limited settings. J Infect Dis 207(Suppl 2):S45–S48. doi: 10.1093/infdis/jit121. [DOI] [PubMed] [Google Scholar]
- 286.WHO. 2017. HIV drug resistance report 2017. WHO, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/255896/1/9789241512831-eng.pdf?ua=1. Accessed 19 October 2017. [Google Scholar]
- 287.Bennett DE, Myatt M, Bertagnolio S, Sutherland D, Gilks CF. 2008. Recommendations for surveillance of transmitted HIV drug resistance in countries scaling up antiretroviral treatment. Antivir Ther 13(Suppl 2):25–36. [PubMed] [Google Scholar]
- 288.Bennett DE, Jordan MR, Bertagnolio S, Hong SY, Ravasi G, McMahon JH, Saadani A, Kelley KF. 2012. HIV drug resistance early warning indicators in cohorts of individuals starting antiretroviral therapy between 2004 and 2009: World Health Organization global report from 50 countries. Clin Infect Dis 54(Suppl 4):S280–S289. doi: 10.1093/cid/cis207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Zhang J, Rhee SY, Taylor J, Shafer RW. 2005. Comparison of the precision and sensitivity of the Antivirogram and PhenoSense HIV drug susceptibility assays. J Acquir Immune Defic Syndr 38:439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Zhou Z, Wagar N, DeVos JR, Rottinghaus E, Diallo K, Nguyen DB, Bassey O, Ugbena R, Wadonda-Kabondo N, McConnell MS, Zulu I, Chilima B, Nkengasong J, Yang C. 2011. Optimization of a low cost and broadly sensitive genotyping assay for HIV-1 drug resistance surveillance and monitoring in resource-limited settings. PLoS One 6:e28184. doi: 10.1371/journal.pone.0028184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Manasa J, Danaviah S, Pillay S, Padayachee P, Mthiyane H, Mkhize C, Lessells RJ, Seebregts C, de Wit TF, Viljoen J, Katzenstein D, De Oliveira T. 30 March 2014. An affordable HIV-1 drug resistance monitoring method for resource limited settings. J Vis Exp doi: 10.3791/51242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Diallo K, Zheng DP, Rottinghaus EK, Bassey O, Yang C. 2015. Viral genetic diversity and polymorphisms in a cohort of HIV-1-infected patients eligible for initiation of antiretroviral therapy in Abuja, Nigeria. AIDS Res Hum Retroviruses 31:564–575. doi: 10.1089/aid.2014.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Parry CM, Parkin N, Diallo K, Mwebaza S, Batamwita R, DeVos J, Bbosa N, Lyagoba F, Magambo B, Jordan MR, Downing R, Zhang G, Kaleebu P, Yang C, Bertagnolio S. 2014. Field study of dried blood spot specimens for HIV-1 drug resistance genotyping. J Clin Microbiol 52:2868–2875. doi: 10.1128/JCM.00544-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.ATCC. 2014. Surveillance of HIV drug resistance. ATCC, Manassas, VA: https://www.atcc.org/~/media/PDFs/Micro%20Flyers/Whitepapers/HIV_Whitepaper.ashx. Accessed 5 November 2018. [Google Scholar]
- 295.Dudley DM, Chin EN, Bimber BN, Sanabani SS, Tarosso LF, Costa PR, Sauer MM, Kallas EG, O’Connor DH. 2012. Low-cost ultra-wide genotyping using Roche/454 pyrosequencing for surveillance of HIV drug resistance. PLoS One 7:e36494. doi: 10.1371/journal.pone.0036494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Dudley DM, Bailey AL, Mehta SH, Hughes AL, Kirk GD, Westergaard RP, O’Connor DH. 2014. Cross-clade simultaneous HIV drug resistance genotyping for reverse transcriptase, protease, and integrase inhibitor mutations by Illumina MiSeq. Retrovirology 11:122. doi: 10.1186/s12977-014-0122-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Lapointe HR, Dong W, Lee GQ, Bangsberg DR, Martin JN, Mocello AR, Boum Y, Karakas A, Kirkby D, Poon AF, Harrigan PR, Brumme CJ. 2015. HIV drug resistance testing by high-multiplex “wide” sequencing on the MiSeq instrument. Antimicrob Agents Chemother 59:6824–6833. doi: 10.1128/AAC.01490-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Vandenhende MA, Bellecave P, Recordon-Pinson P, Reigadas S, Bidet Y, Bruyand M, Bonnet F, Lazaro E, Neau D, Fleury H, Dabis F, Morlat P, Masquelier B. 2014. Prevalence and evolution of low frequency HIV drug resistance mutations detected by ultra deep sequencing in patients experiencing first line antiretroviral therapy failure. PLoS One 9:e86771. doi: 10.1371/journal.pone.0086771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Simen BB, Braverman MS, Abbate I, Aerssens J, Bidet Y, Bouchez O, Gabriel C, Izopet J, Kessler HH, Stelzl E, Di Giallonardo F, Schlapbach R, Radonic A, Paredes R, Recordon-Pinson P, Sakwa J, St John EP, Schmitz-Agheguian GG, Metzner KJ, Däumer MP, 454 HIV Alphastudy Group. 2014. An international multicenter study on HIV-1 drug resistance testing by 454 ultra-deep pyrosequencing. J Virol Methods 204:31–37. doi: 10.1016/j.jviromet.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 300.Gibson RM, Meyer AM, Winner D, Archer J, Feyertag F, Ruiz-Mateos E, Leal M, Robertson DL, Schmotzer CL, Quiñones-Mateu ME. 2014. Sensitive deep-sequencing-based HIV-1 genotyping assay to simultaneously determine susceptibility to protease, reverse transcriptase, integrase, and maturation inhibitors, as well as HIV-1 coreceptor tropism. Antimicrob Agents Chemother 58:2167–2185. doi: 10.1128/AAC.02710-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Ji H, Li Y, Graham M, Liang BB, Pilon R, Tyson S, Peters G, Tyler S, Merks H, Bertagnolio S, Soto-Ramirez L, Sandstrom P, Brooks J. 2011. Next-generation sequencing of dried blood spot specimens: a novel approach to HIV drug-resistance surveillance. Antivir Ther 16:871–878. doi: 10.3851/IMP1839. [DOI] [PubMed] [Google Scholar]
- 302.Zhang G, Cai F, Zhou Z, DeVos J, Wagar N, Diallo K, Zulu I, Wadonda-Kabondo N, Stringer JS, Weidle PJ, Ndongmo CB, Sikazwe I, Sarr A, Kagoli M, Nkengasong J, Gao F, Yang C. 2013. Simultaneous detection of major drug resistance mutations in the protease and reverse transcriptase genes for HIV-1 subtype C by use of a multiplex allele-specific assay. J Clin Microbiol 51:3666–3674. doi: 10.1128/JCM.01669-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Zhang G, Cai F, de Rivera IL, Zhou Z, Zhang J, Nkengasong J, Gao F, Yang C. 2016. Simultaneous detection of major drug resistance mutations of HIV-1 subtype B viruses from dried blood spot specimens by multiplex allele-specific assay. J Clin Microbiol 54:220–222. doi: 10.1128/JCM.02833-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Yao K, Maruta T, Luman ET, Nkengasong JN. 16 September 2014. The SLMTA programme: transforming the laboratory landscape in developing countries. Afr J Lab Med doi: 10.4102/ajlm.v3i1.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Ndihokubwayo JB, Maruta T, Ndlovu N, Moyo S, Yahaya AA, Coulibaly SO, Kasolo F, Turgeon D, Abrol AP. 2016. Implementation of the World Health Organization Regional Office for Africa stepwise laboratory quality improvement process towards accreditation. Afr J Lab Med 5:280. doi: 10.4102/ajlm.v5i1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Yao K, Luman ET, SLMTA Collaborating Authors. 3 November 2014. Evidence from 617 laboratories in 47 countries for SLMTA-driven improvement in quality management systems. Afr J Lab Med . doi: 10.4102/ajlm.v3i2.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Zeh CE, Inzaule SC, Magero VO, Thomas TK, Laserson KF, Hart CE, Nkengasong JN, KEMRI/CDC HIV Research Laboratory. 2010. Field experience in implementing ISO 15189 in Kisumu, Kenya. Am J Clin Pathol 134:410–418. doi: 10.1309/AJCPZIRKDUS5LK2D. [DOI] [PubMed] [Google Scholar]
- 308.Gachuki T, Sewe R, Mwangi J, Turgeon D, Garcia M, Luman ET, Umuro M. 2014. Attaining ISO 15189 accreditation through SLMTA: a journey by Kenya’s National HIV Reference Laboratory. Afr J Lab Med 3:216. doi: 10.4102/ajlm.v3i2.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.NCCLS. 1991. Internal quality control testing: principles and definitions, vol 11, no. 6. NCCLS document C24-A. NCCLS, Villanova, PA. [Google Scholar]
- 310.CDC. 2009. Guide to implementation of services for early diagnosis of HIV in infants in resource-limited settings: book 3. Guide for laboratorians. CDC, Atlanta, GA. [Google Scholar]
- 311.Roche Molecular Systems. 2013. Cobas AmpliPrep/Cobas TaqMan HIV-1 test, version 2.0, package insert. Roche Molecular Systems, Branchburg, NJ. [Google Scholar]
- 312.Hologic. 2014. Package insert, IUO, Aptima HIV-1 Quant DX assay. Gen-Probe Incorporated, San Diego, CA. [Google Scholar]
- 313.Abbott Molecular Inc. 2012. Abbott RealTime HIV-1, package insert. Abbott Molecular Inc, Des Plaines, IL. [Google Scholar]
- 314.Rosenstraus M, Wang Z, Chang S-Y, DeBonville D, Spadoro JP. 1998. An internal control for routine diagnostic PCR: design, properties, and effect on clinical performance. J Clin Microbiol 36:191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Seracare. 2014. Accurun RNA controls. Seracare, Milford, MA. [Google Scholar]
- 316.ThermoFisher Scientific. AcroMetrix transplant & infectious disease testing quality controls. ThermoFisher Scientific, Waltham, MA: https://www.thermofisher.com/us/en/home/clinical/diagnostic-testing/quality-controls-assurance-program/molecular-quality-controls/infectious-disease-testing-controls.html. Accessed 5 Novemeber 2018. [Google Scholar]
- 317.ThermoFisher Scientific. 2015. Lablink XL quality assurance program, 10017437-2MTL-MAS catalog 2015-03. Thermo Fisher Scientific, Waltham, MA. [Google Scholar]
- 318.Qconnect. 2015. 2015 catalogue. Qconnect, Fitzroy, Melbourne, Australia: http://online.fliphtml5.com/mkgo/mcyp/#p=1. Accessed 5 November 2018. [Google Scholar]
- 319.Abbott. 2017. Abbott field safety notice: Abbott RealTime HIV-1 assay. Abbott, Chicago, IL: www.who.int/diagnostics_laboratory/procurement/170808_abbott_fa_am_aug2017_224c.pdf. Accessed 4 January 2018. [Google Scholar]
- 320.Abbott. 2017. Updated urgent field safety notice: Abbott RealTime HIV-1 assay. Abbott, Chicago, IL: www.who.int/diagnostics_laboratory/procurement/170808_abbott_fa_am_aug2017_224c.pdf. Accessed 4 January 2018. [Google Scholar]
- 321.College of American Pathologists. 2017. 2017 surveys and anatomic pathology education programs. College of American Pathologists, Northfield, IL: http://www.cgikk.com/CAP-2017-Media/2017-surveys-catalog.pdf. Accessed 4 January 2018. [Google Scholar]
- 322.Ramos A, Nguyen S, Garcia A, Subbarao S, Nkengasong JN, Ellenberger D. 2013. Generation of dried tube specimen for HIV-1 viral load proficiency test panels: a cost-effective alternative for external quality assessment programs. J Virol Methods 188:1–5. doi: 10.1016/j.jviromet.2012.11.036. [DOI] [PubMed] [Google Scholar]
- 323.WHO. 2014. WHO drug resistance accredited laboratories. WHO, Geneva, Switzerland: www.who.int/hiv/topics/drugresistance/AccreditedLabsFeb2014.pdf?ua=1. Accessed 22 November 2017. [Google Scholar]
- 324.Smit PW, Mabey D, van der Vlis T, Korporaal H, Mngara J, Changalucha J, Todd J, Peeling RW. 2013. The implementation of an external quality assurance method for point-of-care tests for HIV and syphilis in Tanzania. BMC Infect Dis 13:530. doi: 10.1186/1471-2334-13-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Chalermchan W, Pitak S, Sungkawasee S. 2007. Evaluation of Thailand national external quality assessment on HIV testing. Int J Health Care Qual Assur 20:130–140. doi: 10.1108/09526860710731825. [DOI] [PubMed] [Google Scholar]
- 326.Kyaw LL, Nozaki I, Wada K, Oo KY, Tin HH, Yoshihara N. 2015. Ensuring accurate testing for human immunodeficiency virus in Myanmar. Bull World Health Organ 93:42–46. doi: 10.2471/BLT.14.138909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Chongo P, Sitoe N, Viegas S, Pinto I, Macave A, Sitoe F, Vubil A, Mabunda N, Meggi B, Gudo ES, Jani IV. 2016. Quality assurance for point-of-care testing in Mozambique’s National Health Service. Afr J Lab Med 5:445. doi: 10.4102/ajlm.v5i2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Ministry of Health of Uganda. 4 January 2018, accession date Uganda viral load dashboard. Ministry of Health of Uganda, Kampala, Uganda: https://vldash.cphluganda.org/. [Google Scholar]
- 329.Finocchario-Kessler S, Odera I, Okoth V, Bawcom C, Gautney B, Khamadi S, Clark K, Goggin K. 2015. Lessons learned from implementing the HIV infant tracking system (HITSystem): a Web-based intervention to improve early infant diagnosis in Kenya. Healthc (Amst) 3:190–195. doi: 10.1016/j.hjdsi.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Ministry of Health of Kenya. 4 January 2018, accession date Kenya viral load dashboard. Ministry of Health of Kenya, Nairobi, Kenya: https://eid.nascop.org/. [Google Scholar]
- 331.Ministry of Health of Malawi. 4 January 2018, accession date Viral load national dashboard. Ministry of Health of Malawi, Lilongwe, Malawi: www.eidmalawi.org/viraloverall.php. [Google Scholar]
- 332.CDC. 1981. Pneumocystis pneumonia—Los Angeles. MMWR Morb Mortal Wkly Rep 30:250–252. [PubMed] [Google Scholar]
- 333.Gottlieb MS, Schroff R, Schanker HM, Weisman JD, Fan PT, Wolf RA, Saxon A. 1981. Pneumocystis carinii pneumonia and mucosal candidiasis in previously healthy homosexual men: evidence of a new acquired cellular immunodeficiency. N Engl J Med 305:1425–1431. doi: 10.1056/NEJM198112103052401. [DOI] [PubMed] [Google Scholar]
- 334.WHO. 2002. Scaling up antiretroviral therapy in resource-limited settings: guidelines for a public health approach. WHO, Geneva, Switzerland. [PubMed] [Google Scholar]
- 335.WHO. 2004. Scaling up antiretroviral therapy in resource limited settings: treatment guidelines for a public health approach, 2003 revision. WHO, Geneva, Switzerland. [Google Scholar]
- 336.WHO. 2006. Antiretroviral therapy for HIV infection in adults and adolescents: recommendations for a public health approach. WHO, Geneva, Switzerland. [PubMed] [Google Scholar]
- 337.Badri M, Bekker LG, Orrell C, Pitt J, Cilliers F, Wood R. 2004. Initiating highly active antiretroviral therapy in sub-Saharan Africa: an assessment of the revised World Health Organization scaling-up guidelines. AIDS 18:1159–1168. doi: 10.1097/00002030-200405210-00009. [DOI] [PubMed] [Google Scholar]
- 338.When To Start Consortium, Sterne JA, May M, Costagliola D, de Wolf F, Phillips AN, Harris R, Funk MJ, Geskus RB, Gill J, Dabis F, Miro JM, Justice AC, Ledergerber B, Fatkenheuer G, Hogg RS, Monforte AD, Saag M, Smith C, Staszewski S, Egger M, Cole SR. 2009. Timing of initiation of antiretroviral therapy in AIDS-free HIV-1-infected patients: a collaborative analysis of 18 HIV cohort studies. Lancet 373:1352–1363. doi: 10.1016/S0140-6736(09)60612-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Strategies for Management of Antiretroviral Therapy Study Group, Emery S, Neuhaus JA, Phillips AN, Babiker A, Cohen CJ, Gatell JM, Girard PM, Grund B, Law M, Losso MH, Palfreeman A, Wood R. 2008. Major clinical outcomes in antiretroviral therapy (ART)-naive participants and in those not receiving ART at baseline in the SMART study. J Infect Dis 197:1133–1144. doi: 10.1086/586713. [DOI] [PubMed] [Google Scholar]
- 340.Moh R, Danel C, Messou E, Ouassa T, Gabillard D, Anzian A, Abo Y, Salamon R, Bissagnene E, Seyler C, Eholie S, Anglaret X. 2007. Incidence and determinants of mortality and morbidity following early antiretroviral therapy initiation in HIV-infected adults in West Africa. AIDS 21:2483–2491. doi: 10.1097/QAD.0b013e3282f09876. [DOI] [PubMed] [Google Scholar]
- 341.Severe P, Juste MA, Ambroise A, Eliacin L, Marchand C, Apollon S, Edwards A, Bang H, Nicotera J, Godfrey C, Gulick RM, Johnson WD Jr, Pape JW, Fitzgerald DW. 2010. Early versus standard antiretroviral therapy for HIV-infected adults in Haiti. N Engl J Med 363:257–265. doi: 10.1056/NEJMoa0910370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Wong KH, Chan KC, Cheng KL, Chan WK, Kam KM, Lee SS. 2007. Establishing CD4 thresholds for highly active antiretroviral therapy initiation in a cohort of HIV-infected adult Chinese in Hong Kong. AIDS Patient Care STDS 21:106–115. doi: 10.1089/apc.2006.0037. [DOI] [PubMed] [Google Scholar]
- 343.WHO. 2010. Antiretroviral therapy for HIV infection in adults and adolescents: recommendations for a public health approach. WHO, Geneva, Switzerland. [PubMed] [Google Scholar]
- 344.INSIGHT START Study Group, Lundgren JD, Babiker AG, Gordin F, Emery S, Grund B, Sharma S, Avihingsanon A, Cooper DA, Fatkenheuer G, Llibre JM, Molina JM, Munderi P, Schechter M, Wood R, Klingman KL, Collins S, Lane HC, Phillips AN, Neaton JD. 2015. Initiation of antiretroviral therapy in early asymptomatic HIV infection. N Engl J Med 373:795–807. doi: 10.1056/NEJMoa1506816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.TEMPRANO ANRS 12136 Study Group, Danel C, Moh R, Gabillard D, Badje A, Le Carrou J, Ouassa T, Ouattara E, Anzian A, Ntakpe JB, Minga A, Kouame GM, Bouhoussou F, Emieme A, Kouame A, Inwoley A, Toni TD, Ahiboh H, Kabran M, Rabe C, Sidibe B, Nzunetu G, Konan R, Gnokoro J, Gouesse P, Messou E, Dohoun L, Kamagate S, Yao A, Amon S, Kouame AB, Koua A, Kouame E, Ndri Y, Ba GO, Daligou M, Ackoundze S, Hawerlander D, Ani A, Dembele F, Kone F, Guehi C, Kanga C, Koule S, Seri J, Oyebi M, Mbakop N, Makaila O, Babatunde C, Babatounde N, et al. 2015. A trial of early antiretrovirals and isoniazid preventive therapy in Africa. N Engl J Med 373:808–822. doi: 10.1056/NEJMoa1507198. [DOI] [PubMed] [Google Scholar]
- 346.Nicholson JK. 1994. Immunophenotyping specimens from HIV-infected persons: laboratory guidelines from the Centers for Disease Control and Prevention. Cytometry 18:55–59. doi: 10.1002/cyto.990180111. [DOI] [PubMed] [Google Scholar]
- 347.O’Gorman MR, Gelman R. 1997. Inter- and intrainstitutional evaluation of automated volumetric capillary cytometry for the quantitation of CD4- and CD8-positive T lymphocytes in the peripheral blood of persons infected with human immunodeficiency virus. Site Investigators and the NIAID New CD4 Technologies Focus Group. Clin Diagn Lab Immunol 4:173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Reimann KA, O’Gorman MR, Spritzler J, Wilkening CL, Sabath DE, Helm K, Campbell DE, NIAID DAIDS New Technologies Evaluation Group. 2000. Multisite comparison of CD4 and CD8 T-lymphocyte counting by single- versus multiple-platform methodologies: evaluation of Beckman Coulter flow-count fluorospheres and the tetraONE system. Clin Diagn Lab Immunol 7:344–351. doi: 10.1128/CDLI.7.3.344-351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Strauss K, Hannet I, Engels S, Shiba A, Ward DM, Ullery S, Jinguji MG, Valinsky J, Barnett D, Orfao A, Kestens L. 1996. Performance evaluation of the FACSCount system: a dedicated system for clinical cellular analysis. Cytometry 26:52–59. doi:. [DOI] [PubMed] [Google Scholar]
- 350.UNITAID. 2014. HIV/AIDS diagnostics technology landscape, 4th ed UNITAID, Geneva, Switzerland: https://unitaid.eu/assets/UNITAID-HIV_Diagnostic_Landscape-4th_edition.pdf. Accessed 4 January 2018. [Google Scholar]
- 351.WHO. 2011. WHO prequalification of diagnostics programme: Pima CD4 test. Number: PQDx 0099-032-00. WHO, Geneva, Switzerland: www.who.int/diagnostics_laboratory/evaluations/111208_0099_032_00_public_report_v2.pdf. Accessed 4 January 2018. [Google Scholar]
- 352.Wynberg E, Cooke G, Shroufi A, Reid SD, Ford N. 2014. Impact of point-of-care CD4 testing on linkage to HIV care: a systematic review. J Int AIDS Soc 17:18809. doi: 10.7448/IAS.17.1.18809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Zachariah R, Reid SD, Chaillet P, Massaquoi M, Schouten EJ, Harries AD. 2011. Viewpoint: why do we need a point-of-care CD4 test for low-income countries? Trop Med Int Health 16:37–41. doi: 10.1111/j.1365-3156.2010.02669.x. [DOI] [PubMed] [Google Scholar]
- 354.Bergeron M, Daneau G, Ding T, Sitoe NE, Westerman LE, Stokx J, Jani IV, Coetzee LM, Scott L, De Weggheleire A, Boel L, Stevens WS, Glencross DK, Peter TF. 2012. Performance of the PointCare NOW system for CD4 counting in HIV patients based on five independent evaluations. PLoS One 7:e41166. doi: 10.1371/journal.pone.0041166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Faal M, Naidoo N, Glencross DK, Venter WD, Osih R. 2011. Providing immediate CD4 count results at HIV testing improves ART initiation. J Acquir Immune Defic Syndr 58:e54–e59. doi: 10.1097/QAI.0b013e3182303921. [DOI] [PubMed] [Google Scholar]
- 356.Mtapuri-Zinyowera S, Chiyaka ET, Mushayi W, Musuka G, Naluyinda-Kitabire F, Mushavi A, Chikwasha V. 2013. PIMA point of care CD4+ cell count machines in remote MNCH settings: lessons learned from seven districts in Zimbabwe. Infect Dis (Auckl) 6:51–60. doi: 10.4137/IDRT.S12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Diaw PA, Daneau G, Coly AA, Ndiaye BP, Wade D, Camara M, Mboup S, Kestens L, Dieye TN. 2011. Multisite evaluation of a point-of-care instrument for CD4(+) T-cell enumeration using venous and finger-prick blood: the PIMA CD4. J Acquir Immune Defic Syndr 58:e103–e111. doi: 10.1097/QAI.0b013e318235b378. [DOI] [PubMed] [Google Scholar]
- 358.Galiwango RM, Lubyayi L, Musoke R, Kalibbala S, Buwembo M, Kasule J, Serwadda D, Gray RH, Reynolds SJ, Chang LW. 2014. Field evaluation of PIMA point-of-care CD4 testing in Rakai, Uganda. PLoS One 9:e88928. doi: 10.1371/journal.pone.0088928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Glencross DK, Coetzee LM, Faal M, Masango M, Stevens WS, Venter WF, Osih R. 2012. Performance evaluation of the Pima point-of-care CD4 analyser using capillary blood sampling in field tests in South Africa. J Int AIDS Soc 15:3. doi: 10.1186/1758-2652-15-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Jani IV, Sitoe NE, Chongo PL, Alfai ER, Quevedo JI, Tobaiwa O, Lehe JD, Peter TF. 2011. Accurate CD4 T-cell enumeration and antiretroviral drug toxicity monitoring in primary healthcare clinics using point-of-care testing. AIDS 25:807–812. doi: 10.1097/QAD.0b013e328344f424. [DOI] [PubMed] [Google Scholar]
- 361.Mwau M, Adungo F, Kadima S, Njagi E, Kirwaye C, Abubakr NS, Okubi LA, Waihenya M, Lusike J, Hungu J. 2013. Evaluation of PIMA point of care technology for CD4 T cell enumeration in Kenya. PLoS One 8:e67612. doi: 10.1371/journal.pone.0067612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Sukapirom K, Onlamoon N, Thepthai C, Polsrila K, Tassaneetrithep B, Pattanapanyasat K. 2011. Performance evaluation of the Alere PIMA CD4 test for monitoring HIV-infected individuals in resource-constrained settings. J Acquir Immune Defic Syndr 58:141–147. doi: 10.1097/QAI.0b013e31822866a2. [DOI] [PubMed] [Google Scholar]
- 363.Wade D, Daneau G, Aboud S, Vercauteren GH, Urassa WS, Kestens L. 2014. WHO multicenter evaluation of FACSCount CD4 and Pima CD4 T-cell count systems: instrument performance and misclassification of HIV-infected patients. J Acquir Immune Defic Syndr 66:e98–e107. doi: 10.1097/QAI.0000000000000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Wade D, Diaw PA, Daneau G, Camara M, Dieye TN, Mboup S, Kestens L. 2013. CD4 T-cell enumeration in a field setting: evaluation of CyFlow counter using the CD4 easy count kit-dry and Pima CD4 systems. PLoS One 8:e75484. doi: 10.1371/journal.pone.0075484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Bwana P, Vojnov L, Adhiambo M, Akinyi C, Mwende J, Prescott M, Mwau M. 2015. The BD FACSPresto point of care CD4 test accurately enumerates CD4+ T cell counts. PLoS One 10:e0145586. doi: 10.1371/journal.pone.0145586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Coetzee LM, Moodley K, Glencross DK. 2016. Performance evaluation of the Becton Dickinson FACSPresto near-patient CD4 instrument in a laboratory and typical field clinic setting in South Africa. PLoS One 11:e0156266. doi: 10.1371/journal.pone.0156266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Mulry J. 2016. Unique features of the Muse Auto CD4/CD4% system. MOJ Immunol 4:00134 http://medcraveonline.com/MOJI/MOJI-04-00134.php. Accessed 4 January 2018. [Google Scholar]
- 368.Mossoro-Kpinde CD, Kouabosso A, Mboumba Bouassa R-S, Longo JDD, Kokanzo E, Féissona R, Grésenguet G, Bélec L. 2016. Performance evaluation of the touchscreen-based Muse Auto CD4/CD4% single-platform system for CD4 T cell numeration in absolute number and in percentage using blood samples from children and adult patients living in the Central African Republic. J Transl Med 14:326. doi: 10.1186/s12967-016-1082-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Vojnov L, Markby J, Boeke C, Harris L, Ford N, Peter T. 2016. POC CD4 testing improves linkage to HIV care and timeliness of ART initiation in a public health approach: a systematic review and meta-analysis. PLoS One 11:e0155256. doi: 10.1371/journal.pone.0155256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Lecher S, Williams J, Fonjungo PN, Kim AA, Ellenberger D, Zhang G, Toure CA, Agolory S, Appiah-Pippim G, Beard S, Borget MY, Carmona S, Chipungu G, Diallo K, Downer M, Edgil D, Haberman H, Hurlston M, Jadzak S, Kiyaga C, MacLeod W, Makumb B, Muttai H, Mwangi C, Mwangi JW, Mwasekaga M, Naluguza M, Ng’Ang’A LW, Nguyen S, Sawadogo S, Sleeman K, Stevens W, Kuritsky J, Hader S, Nkengasong J. 2016. Progress with scale-up of HIV viral load monitoring—seven sub-Saharan African countries, January 2015-June 2016. MMWR Morb Mortal Wkly Rep 65:1332–1335. doi: 10.15585/mmwr.mm6547a2. [DOI] [PubMed] [Google Scholar]
- 371.Auld AF, Shiraishi RW, Oboho I, Ross C, Bateganya M, Pelletier V, Dee J, Francois K, Duval N, Antoine M, Delcher C, Desforges G, Griswold M, Domercant JW, Joseph N, Deyde V, Desir Y, Van Onacker JD, Robin E, Chun H, Zulu I, Pathmanathan I, Dokubo EK, Lloyd S, Pati R, Kaplan J, Raizes E, Spira T, Mitruka K, Couto A, Gudo ES, Mbofana F, Briggs M, Alfredo C, Xavier C, Vergara A, Hamunime N, Agolory S, Mutandi G, Shoopala NN, Sawadogo S, Baughman AL, Bashorun A, Dalhatu I, Swaminathan M, Onotu D, Odafe S, Abiri OO, Debem HH, Tomlinson H, et al. 2017. Trends in prevalence of advanced HIV disease at antiretroviral therapy enrollment—10 countries, 2004-2015. MMWR Morb Mortal Wkly Rep 66:558–563. doi: 10.15585/mmwr.mm6621a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Parpia ZA, Elghanian R, Nabatiyan A, Hardie DR, Kelso DM. 2010. p24 antigen rapid test for diagnosis of acute pediatric HIV infection. J Acquir Immune Defic Syndr 55:413–419. doi: 10.1097/QAI.0b013e3181f1afbc. [DOI] [PubMed] [Google Scholar]
- 373.Grant RM, Kuritzkes DR, Johnson VA, Mellors JW, Sullivan JL, Swanstrom R, D’Aquila RT, Van Gorder M, Holodniy M, Lloyd RM Jr, Reid C, Morgan GF, Winslow DL. 2003. Accuracy of the TRUGENE HIV-1 genotyping kit. J Clin Microbiol 41:1586–1593. doi: 10.1128/JCM.41.4.1586-1593.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Hallack R, Doherty LE, Wethers JA, Parker MM. 2008. Evaluation of dried blood spot specimens for HIV-1 drug-resistance testing using the Trugene HIV-1 genotyping assay. J Clin Virol 41:283–287. doi: 10.1016/j.jcv.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 375.Eshleman SH, Crutcher G, Petrauskene O, Kunstman K, Cunningham SP, Trevino C, Davis C, Kennedy J, Fairman J, Foley B, Kop J. 2005. Sensitivity and specificity of the ViroSeq human immunodeficiency virus type 1 (HIV-1) genotyping system for detection of HIV-1 drug resistance mutations by use of an ABI PRISM 3100 genetic analyzer. J Clin Microbiol 43:813–817. doi: 10.1128/JCM.43.2.813-817.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]