Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has been successfully applied in recent years for first-line identification of pathogens in clinical microbiology because it is simple to use, rapid, and accurate and has economic benefits in hospital management. The range of clinical applications of MALDI-TOF MS for bacterial isolates is increasing constantly, from species identification to the two most promising applications in the near future: detection of antimicrobial resistance and strain typing for epidemiological studies.
KEYWORDS: antimicrobial resistance, MALDI-TOF
SUMMARY
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has been successfully applied in recent years for first-line identification of pathogens in clinical microbiology because it is simple to use, rapid, and accurate and has economic benefits in hospital management. The range of clinical applications of MALDI-TOF MS for bacterial isolates is increasing constantly, from species identification to the two most promising applications in the near future: detection of antimicrobial resistance and strain typing for epidemiological studies. The aim of this review is to outline the contribution of previous MALDI-TOF MS studies in relation to detection of antimicrobial resistance and to discuss potential future challenges in this field. Three main approaches are ready (or almost ready) for clinical use, including the detection of antibiotic modifications due to the enzymatic activity of bacteria, the detection of antimicrobial resistance by analysis of the peak patterns of bacteria or mass peak profiles, and the detection of resistance by semiquantification of bacterial growth in the presence of a given antibiotic. This review provides an expert guide for MALDI-TOF MS users to new approaches in the field of antimicrobial resistance detection, especially possible applications as a routine diagnostic tool in microbiology laboratories.
INTRODUCTION
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has recently been introduced in clinical microbiology laboratories for identifying microorganisms (1–4). Although the initial investment in a mass spectrometer is relatively high, the cost of identifying a single isolate is lower than with previously used biochemical or molecular techniques (5, 6). In addition, the use of MALDI-TOF MS enables accurate identification at least 24 h earlier than phenotypic methods, even those that are automated (7–9). Among the advantages, the simplified workflow is one of the key points for the rapid acceptance of MALDI-TOF MS in laboratories and for constant growth in the range of applications.
Bacterial resistance to antibiotics has increased in recent years. In 2013, the Centers for Disease Control and Prevention (CDC) published a report outlining the top 18 drug-resistant threats to the United States: carbapenem-resistant Enterobacteriaceae are considered an urgent threat, and multidrug-resistant Acinetobacter baumannii and Pseudomonas aeruginosa, fluconazole-resistant Candida, extended-spectrum-β-lactamase (ESBL)-producing Enterobacteriaceae, vancomycin-resistant enterococci, and methicillin-resistant Staphylococcus aureus are considered serious threats (https://www.cdc.gov/drugresistance/biggest_threats.html). Tests for the rapid diagnosis of infections produced by these microorganisms are therefore urgently needed. The time spent until microbiological results are provided is inversely proportional to the medical value (10), so the sooner information on antimicrobial resistance is obtained, the better the antimicrobial therapy that will be prescribed. The time to result is especially important to prevent unnecessary treatments and avoid the transmission of these bacteria of particular concern in hospital settings (11–13).
Five main approaches have been introduced as rapid means of detecting antimicrobial resistance (14–17) (Table 1). (i) Some of the techniques used are based on molecular genetics analysis. The sensitivity and specificity can be higher than those of previously used techniques: they deliver results in less than 1 h or within a few hours and have the advantage of being able to be applied directly to clinical samples. The disadvantages are the cost (usually higher); the lack of universal primers for detecting groups of resistance mechanisms, such as β-lactamases; the lack of proof of the biochemical activity of an enzyme, as detection of part of a gene sequence does not provide information regarding the phenotype associated with the enzyme; and the difficulty in performing the technique (by untrained or nonexperienced users) (18–20). One of these commercially available techniques is the GeneXpert system (Cepheid, France), which can detect carbapenemases like IMP, VIM, NDM, KPC, and OXA-48 with the Xpert Carba-R kit (21) and can also detect methicillin-resistant Staphylococcus aureus (MRSA) by the Xpert MRSA test (22). The Check-Direct CPE kit with the BD Max instrument detects KPC, VIM, NDM, and OXA-48 carbapenemases (23). (ii) Other methods for the rapid detection of antimicrobial resistance include biochemical methods such as the Carba NP test. These methods are based on the color change of the buffer in the presence of a pH indicator. For β-lactam antibiotics, the pH indicator changes its color when the hydrolysis of the antibiotic by bacterial resistance enzymes takes place. They are easy to perform, inexpensive, and universally applicable to different enzymes, and the results are delivered within 1 to 3 h. The disadvantages are the need for subjective interpretation of results, the low sensitivity relative to molecular methods, and the need to isolate the microorganism. It has shown good sensitivity with most carbapenemases but not with OXA-48-like enzymes, mainly because of the weak carbapenemase activity, or with OXA-23- and OXA-24-type enzymes in Acinetobacter baumannii (24). The Carba NP test has been commercialized through the Rapidec Carba NP system (bioMérieux, France) (25, 26), facilitating the procedure and hands-on time. However, false-positive and -negative results have been detected for OXA-48-like variants, in a more significant number than with the in-house Carba NP test (27). To date, the different variations of the Carba NP test has been used to detect not only β-lactamase resistance but also aminoglycoside and polymyxin resistance, all mediated by enzymes (28, 29). For the detection of aminoglycoside and polymyxin resistance, the principle of the assay is the detection of bacterial growth due to the fermentation process that acidifies the medium and makes the pH indicator change its color. (iii) Rapid immunoassays based on monoclonal antibodies have recently been developed for detecting carbapenemases (30). This technology has shown high sensitivity and specificity for detecting carbapenemase producers directly from bacterial colonies within 15 min. It has also shown promising results with biological samples such as positive blood cultures and urine samples (detection limit of 106 CFU/ml), as recently evaluated (31, 32). The main disadvantage of this method is that it does not yield quantitative results (33). This immunochromatographic assay could also be applied in the future to detect different types of resistance enzymes other than β-lactamases. (iv) The BYG Carba test is an innovative concept for the detection of carbapenemase-producing Enterobacteriaceae through an electrochemical assay that measures variations in conductivity in a polymer electrode due to the redox potential generated by imipenem hydrolysis by a carbapenemase enzyme. The measurement and analysis are reported in real time. The BYG Carba test has proven to be highly sensitive for discriminating between carbapenemase producers and nonproducers. It has the advantage of delivering results in 30 min without incubation at 37°C, and the sensitivity and specificity are higher than those of biochemical assays, although this requires further clinical validation (34–36). For the moment, the electrodes, the reader, and the software are made in-house and are not commercially available. (v) Finally, mass spectrometry analysis can be used to detect antimicrobial resistance mediated by all possible resistance mechanisms. Two main technologies are used: liquid chromatography-tandem mass spectrometry (LC-MS/MS) and MALDI-TOF MS. LC-MS/MS has been developed mainly to detect β-lactamases (37–40). The different procedures consist of either detecting the enzyme directly from bacteria or detecting the hydrolysis of the antibiotic by the β-lactamase enzyme. If detecting the enzyme, a first step of trypsin digestion and formic acid extraction should be performed. The main advantage compared to MALDI-TOF MS is the high sensitivity that can be achieved, and the main disadvantage of using this methodology is that microbiology laboratories do not usually have this technology. On the contrary, MALDI-TOF MS has several advantages: the use of solid samples; the low cost per test, despite the initial high investment in the equipment, as the savings for microbial identification are around 50% annually (5); the possibility of identifying microorganisms and detecting antimicrobial resistance with the same equipment; the simplicity of the procedure; and the automated interpretation of the spectra generated (41). The major disadvantages are the need for a high bacterial load, which precludes the use of the technique with clinical samples, except those with high burdens, such as positive blood cultures and urine samples (42–44), and the lack of commercially available kits with European Conformity (CE) regulatory clearance for in vitro diagnostics (IVD) or Food and Drug Administration (FDA) clearance for IVD and developed software for the majority of techniques described in this document. In cases where no certified commercial kit is available, we recommend performing an internal validation of the procedure and using reagents with the mark of the European Pharmacopoeia (EP) reference standard or U.S. Pharmacopeia (USP) convention so that the maximum level of quality is guaranteed.
TABLE 1.
Comparison of the different technologies for rapid detection of antimicrobial resistance
| Technology | LOD (CFU/ml)a | Sensitivity (%)b | Specificity (%)b | TATc | Price/sample ($) | References |
|---|---|---|---|---|---|---|
| Molecular genetics | 102–2 × 104 | 96–99 | 96–99 | 1–3 h | 5–50d | 21–23 |
| Biochemical methods | 6 × 108 | 90–99 | 96–100 | 30 min–2 h | 1–5e | 24–29 |
| Immunoassays | 104–106 | 96–100 | 97–100 | 20 min | 7–15f | 30–33 |
| Electrochemical assays | 106–109 | 95–96 | 100 | 5–30 min | 1 (plus $100 for the homemade reader)g |
34–36 |
| MALDI-TOF MS | 105–106 | 98–100 | 92–100 | 30 min–3 h | 1–10 | 44, 53–55, 60–66, 132 |
LOD, limit of detection, expressed in CFU per milliliter, of the different technologies.
Ranges of sensitivity and specificity of the different technologies.
TAT, turnaround time of the different technologies.
The price is lower if performing in-house methods and maximum if using commercial kits.
The price is lower if performing in-house methods and maximum if using the Rapidec Carba NP system (bioMérieux).
The price increases if resistance detection is focused on more than one cassette. The prices for OXA-48 K-SeT and KPC K-SeT are around $7/sample, the price for O.K.N. K-SeT is around $14/sample (detection of OXA-48, KPC, and NDM in the same test), and the price for O.K.N.V. K-SeT is around $15/sample (detection of OXA-48, KPC, NDM, and VIM in the same test).
The price is around $1/sample, the electrodes are made in-house and cost around $2/electrode, and the reader is also produced in-house, at about $100.
The MALDI-TOF MS method is a rapid and simple procedure that combines the universal advantages of phenotypic assays with the rapidity and accuracy of molecular assays. Since the first studies reported in 2011 (45–47), further developments in the method, along with advances in the equipment and software, have led to the implementation of this new technology in clinical laboratories for the detection of antimicrobial resistance. In this review, we consider the different possible applications of MALDI-TOF MS in this respect.
Three main approaches are used to detect antimicrobial resistance by MALDI-TOF MS: the detection of antimicrobial resistance by measuring antibiotic modifications due to the enzymatic activity of bacteria, analysis of the peak patterns of bacteria, and semiquantification of bacterial growth in the presence of a given antibiotic. Each of these approaches has advantages and disadvantages that we consider further here. However, in our opinion, each has the potential to be automated and applied in a clinical setting. The first two applications are currently in use with the new MALDI Biotyper IVD (Bruker Daltonik, GmbH, Germany).
DETECTION OF ANTIMICROBIAL RESISTANCE MECHANISMS BY MEASURING THE ENZYMATIC ACTIVITY OF BACTERIA
One of the main resistance mechanisms is modification of the antibiotic structure by bacterial enzymes, which renders the antibiotic inactive (47–51) (Table 2). If the modification involves a change in the molecular mass of the native molecule, the reaction can be monitored by mass spectrometry. The first step in this process is the accurate determination of the structure of the antibiotic and the reaction metabolite(s). The antibiotic structure is studied by observing the mass peaks obtained in the MALDI-TOF MS spectrum and assigning a possible ionic form to each mass peak. Antibiotics and their degradation products are usually analyzed in the mass range between 100 and 1,000 Da (47). One of the mass peaks most likely to be present is that of the molecular ion peak that represents the exact mass of the antibiotic. In the ionization process, the matrix usually adds a proton to the antibiotic, but it can also provide sodium and potassium ions, thus increasing the mass of the antibiotic by 1, 23, and 39 Da, respectively (47). Other reactions that can take place during ionization of antibiotics include decarboxylation, decarbonylation, and loss of other groups of molecules. These reactions usually take place during hydrolysis of β-lactams, as the newly hydrolyzed molecule is rendered unstable by the opening of the β-lactam ring and tends to break into different fragments, yielding compounds of different molecular weights. The mass peak patterns are unique to each antibiotic and can be used to detect antibiotic resistance.
TABLE 2.
Structural determination of β-lactam antibiotics (ampicillin, piperacillin, cefotaxime, ceftazidime, ceftriaxone, cefpodoxime, cefepime, ertapenem, meropenem, and imipenem) and their corresponding hydrolyzed forms, defining the susceptibility and resistance patternsa by MALDI-TOF MSb
| Antibiotic | MW (g/mol)c |
Susceptibility pattern (Da) |
Resistance pattern (Da) |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [M + H]+ |
[M + Na]+ |
[M + 2Na]+ |
[M + 3Na]+ |
[M + K]+ |
[M + Na + K]+ |
[M + Cd + H]+ |
[M − X + H]+ |
[M – Xe − CO + H]+ |
[M – X – CO − CO2 + H]+ |
[Mhydrol + H]+ |
[Mhydrol + Na]+ |
[Mhydrol + 2Na]+ |
[Mhydrol + Na + K]+ |
[Mhydrol − CO2 + H]+ |
[Mhydrol − CO2 + Na]+ |
[Mhydrol − CO2 + K]+ |
[Mhydrol − X + H]+ |
[Mhydrol − X − CO2 + H]+ |
|||
| Ampicillin | 349.4 | 350.4 | 372.4 | 394.4 | 368.4 | 390.4 | 412.4 | 324.4 | |||||||||||||
| Piperacillin | 517.5 | 518.5 | 540.5 | 562.5 | 536.5 | 558.5 | 580.5 | ||||||||||||||
| Cefotaxime | 455.5 | 456.5 | 478.5 | 396.5 | 414.5 | 370.5 | |||||||||||||||
| Ceftazidime | 546.6 | 547.6 | 468.6 | 486.6 | 442.6 | ||||||||||||||||
| Ceftriaxone | 554.6 | 555.6 | 577.6 | 616.6 | 396.6 | 368.6 | 324.6 | 414.6 | 370.6 | ||||||||||||
| Cefpodoxime | 427.5 | 428.5 | 450.5 | 473.5 | 466.5 | 414.5 | 370.5 | ||||||||||||||
| Cefepime | 480.6 | 481.6 | 296.6 | 414.6 | 370.6 | ||||||||||||||||
| Ertapenem | 475.5 | 476.5 | 498.5 | 520.5 | 542.5 | 514.5 | 536.5 | 494.5 | 516.5 | 538.5 | 554.5 | 450.5 | 472.5 | 488.5 | |||||||
| Meropenem | 383.4 | 384.5 | 406.5 | 428.5 | |||||||||||||||||
| Imipenem | 299.4 | 300.4 | 489.4 | ||||||||||||||||||
The terms susceptibility and resistance by MALDI-TOF MS refer to the characteristic mass peak pattern of the antibiotic without modifications and to its corresponding hydrolyzed form. Mhydrol, hydrolysis.
MW, molecular weight.
Imipenem is complexed with the matrix (α-cyano-4-hydroxy-cinnamic acid), resulting in a peak that is the summation of their molecular masses.
X, acetyl for cefotaxime, pyridine for ceftazidime, triazin-ythiol for ceftriaxone, methoxy for cefpodoxime, and pyrrolidine for cefepime.
Detection of β-Lactamases
Direct detection of β-lactamase activity by MALDI-TOF MS by measuring the mass changes in the antibiotic is one of the fields in which the greatest advances have been achieved in recent years (52). A very similar method is used in all reported studies. A fresh bacterial culture is suspended in a buffer containing a β-lactam and incubated at 37°C. At the end of the reaction, the mixture is centrifuged; the supernatant is measured in a suitable matrix, usually α-cyano-4-hydroxy-cinnamic acid (HCCA); and the spectrum obtained is then analyzed for fragmentation of the compound.
Regarding the detection of cephalosporin resistance, Oviaño et al. validated a procedure for the direct detection and classification of ESBL and AmpC producers in positive blood cultures (53). The antibiotics cefotaxime and ceftazidime were used to detect resistance, and clavulanic acid was added for accurate differentiation between class A (ESBL) and class C β-lactamases. An incubation time of 90 min was established. A series of different enzymes were analyzed for optimization of the assay, those encoded by blaTEM-1, blaSHV-12, blaTEM-29, blaCTX-M-32, blaCTX-M-14, blaCTX-M-15, blaFOX-4, blaCMY-2, blaDHA-1, blaDHA-6, and blaDHA-7. For clinical validation, 140 positive blood cultures were studied for detection of β-lactamases. The use of cefotaxime resulted in a shorter turnaround time and higher sensitivity, although ceftazidime is useful for detecting AmpC producers. This was the first study to report the detection of β-lactam hydrolysis activity by MALDI-TOF MS directly from positive blood cultures.
Jung et al. validated a procedure for detecting β-lactam resistance in 2.5 h in Enterobacteriaceae in positive blood cultures (54). The antibiotics cefotaxime and ampicillin were used, and data were analyzed using an in-house-developed program. The reproducibility of the measurements was calculated, and comparison between paired blood cultures and plated isolates was made. The results were expressed as logarithm of the quantification of resistance (logRQ) values using the formula logRQ = log(sum of hydrolyzed peak intensities)/(sum of nonhydrolyzed peak intensities) and displayed in a box plot diagram. Eighty-five positive blood cultures were used for validating the detection of cefotaxime resistance. The lowest logRQ values were obtained for chromosomal AmpC, SHV, and TEM producers, and the highest values were obtained for AmpC hyperproducers and ESBL isolates. Higher logRQ values were also observed for plated microorganisms than for blood cultures; nevertheless, the assay provides accurate classification of susceptible and resistant isolates from blood cultures. The sensitivity and specificity of this method were 100% and 91.5%, respectively. The findings of this study are of great interest, as they represent the first step toward automation of the hydrolysis procedure, which will overcome the need for visual inspection of the spectra of the antibiotics and subjective interpretation of the results.
Oviaño et al. evaluated the sensitivities, specificities, and turnaround times for different antibiotics (cefotaxime, ceftazidime, ceftriaxone, cefpodoxime, and cefepime) and also the performance of the automated MALDI Biotyper (MBT) Selective Testing of β-Lactamase Activity (STAR-BL) module for Compass software (Bruker Daltonik) relative to qualitative interpretation of spectra for detecting β-lactamase resistance (ESBL and AmpC) by MALDI-TOF MS (Bruker Daltonik) (55). Regarding the antibiotics evaluated, ceftriaxone yielded 70% more positive results in relation to detecting resistance at the same incubation time than cefotaxime, 80% more than ceftazidime, and 20% more than cefpodoxime, with 100% specificity. Cefepime revealed 100% sensitivity but only 27% specificity. For the same incubation time, the automated software yielded on average 41% more positive results in relation to detecting resistance than qualitative interpretation of spectra. β-Lactamase resistance was detected after 30 min of incubation with ceftriaxone as the antibiotic marker in 100 genotypically characterized clinical isolates, with 100% sensitivity and specificity. This study reported the first structural determination of ceftriaxone by MALDI-TOF MS (Table 2).
The main advantage of measuring the activity of enzymes by MALDI-TOF MS is the potential power to detect all types of carbapenemases, even those that are considered rare (e.g., IMI, GES, FRI, and DIM, etc.) and are not included in commercially available molecular tests. Besides, with current clinical breakpoints defined by the EUCAST or the Clinical and Laboratory Standards Institute (CLSI), nearly 20% of carbapenemase-producing isolates can be missed, especially OXA-48-like enzyme producers (56). One of the major shortcomings of carbapenemase detection in previous papers was the lower sensitivity for detecting OXA-48 producers than for detecting other families of carbapenemases. This is due to the low catalytic efficiency of this family of enzymes in hydrolyzing carbapenems (57). Based on the structure of OXA-48 β-lactamases, carboxylation of the active site of lysine is important for enzyme activity (58). The addition of NH4HCO3 increased the kcat/Km values of the OXA-58 enzyme for imipenem by more than 5-fold (59). Applying this principle, Papagiannitsis et al. improved the detection of OXA-48-like enzyme producers from 76% to 98% by adding NH4HCO3 to the reaction buffer and using meropenem as an antibiotic marker of resistance, with an incubation time of 2.5 h (60). The assay was validated using 124 Enterobacteriaceae and 37 Pseudomonas aeruginosa isolates. A novelty introduced by the group led by Hrabák (60) is the use of 10 mg/ml 2,5-dihydroxybenzoic acid (DHB) as the matrix. This change in the procedure allows better identification of the hydrolysis products of meropenem. Automatic interpretation of the MALDI-TOF MS assay results by the MBT STAR-BL module (Bruker Daltonik) was generally consistent with the results obtained by manual analysis. The sensitivity of detection of blaOXA-48 by MALDI-TOF MS has since reached the same level as for other carbapenemase enzymes.
Another problematic aspect was the turnaround time for β-lactamase detection. In developing the method, Lasserre et al. used a 20-min incubation time with imipenem to detect carbapenemase producers and applied a cutoff MS ratio (mass peaks of the metabolite/imipenem plus the metabolite of ≥0.82) to classify 223 strains, 77 carbapenemase producers and 146 nonproducers, as isolates harboring carbapenemase enzymes, yielding 100% sensitivity and specificity (61). The peak of the imipenem metabolite was previously described as being at 254 Da by Kempf et al. (62) using an Ultraflex I instrument (Bruker Daltonik). However, several other authors seem hesitant to ensure the appearance of such a metabolite, as it cannot be visualized under standard MALDI-TOF MS conditions (47, 63–65). Lasserre et al. estimated the cost per test to be less than $0.10. To date, this is the fastest assay performed by MALDI-TOF MS and with imipenem as the standard antibiotic. Both groups used imipenem that contains cilastatin (Tienam; MSD, France) for MALDI-TOF MS measurements. Although the stability issue seems to be solved with the addition of cilastatin, we recommend using imipenem alone, as the complexation of imipenem with cilastatin and not with the matrix can make proper visualization of the mass peaks in the spectrum difficult.
In an attempt to improve the procedure, Monteferrante et al. developed a protocol involving cellular lysis and enzyme extraction from a defined number of bacterial cells, followed by the addition of an antibiotic (imipenem or ertapenem), in a series of 260 isolates (208 carbapenemase producers and 52 non-carbapenemase producers), yielding 100% sensitivity and specificity (64). Standardization was achieved by prior adjustment of the cells to a turbidity of a 3.0 McFarland standard. Comparison of the uses of imipenem and ertapenem revealed higher sensitivity and specificity and a higher hydrolysis rate for imipenem. However, imipenem is less stable in solution, and MS signals are about 5-fold lower than those for ertapenem. This group used the Ultraflex III instrument (Bruker Daltonik), which is a MALDI-TOF/TOF MS technology, so results may vary with respect to those obtained with the Microflex technology (MALDI-TOF MS) usually used in routine microbiology laboratories, especially regarding the detection of the imipenem metabolite. The longer flight tube used with the Ultraflex equipment provides a higher resolution. In this case, the authors described the appearance of the hydrolyzed decarboxylated form at 274 Da and the sodium adduct at 296 Da. The use of a standard number of cells and subsequent extraction was previously developed by Hrabák et al. (46). The main consequences are that it is a less technician-dependent technique but much more laborious and slower, with a possible hands-on time of 120 min relative to the 30 min required for the direct method using whole cells.
With the aim of establishing rapid and accurate procedures for detecting antimicrobial-resistant bacteria directly from positive blood cultures, Oviaño et al. developed a universal method for detecting carbapenemase producers among 119 Gram-negative bacilli, including Enterobacteriaceae, Pseudomonas spp., and Acinetobacter spp., in 30 min, adding imipenem to a buffer containing NH4HCO3, ZnCl2, and sodium dodecyl sulfate (SDS) (66). NH4HCO3 proved useful for detecting OXA-48 producers, as previously demonstrated by Papagiannitsis et al. with meropenem (60). Zn2+ was essential for some metallo-β-lactamase-producing Pseudomonas species, and SDS was essential for satisfactory reactivity for Acinetobacter spp., as previously demonstrated by Hrabák et al. (65). The method of extraction from the blood culture using the Sepsityper kit (Bruker Daltonik) was also modified from that recommended by the manufacturer for identification purposes, reducing the amount of lysis and thus improving the yield. The overall sensitivity and specificity were 98% and 100%, respectively. Analysis was performed using the STAR-BL module for MALDI-TOF Biotyper Compass software (Bruker Daltonik), which automatically provides a result for sensitivity or resistance, calculated as a logRQ value or a ratio of hydrolysis of the antibiotic. The logRQ value is the logarithm of the intensity of the area under the curve (AUC) of the internal standard and the AUC of imipenem. Reserpine was used as the internal standard, as it is a stable molecule in the mass range of study (608 Da). This compound is lyophilized into the MALDI matrix and then dissolved and finally added to the sample, in the same way as it is done for a conventional matrix. This method is ready to use in clinical practice. Until this work, OXA-type carbapenemases in A. baumannii from plate cultures required longer incubation times for detection and frequently remained undetected in blood cultures. In addition, this method unifies carbapenemase detection for all types of Gram-negative bacilli, and it does not require an initial identification step or any change in the procedure to detect resistance.
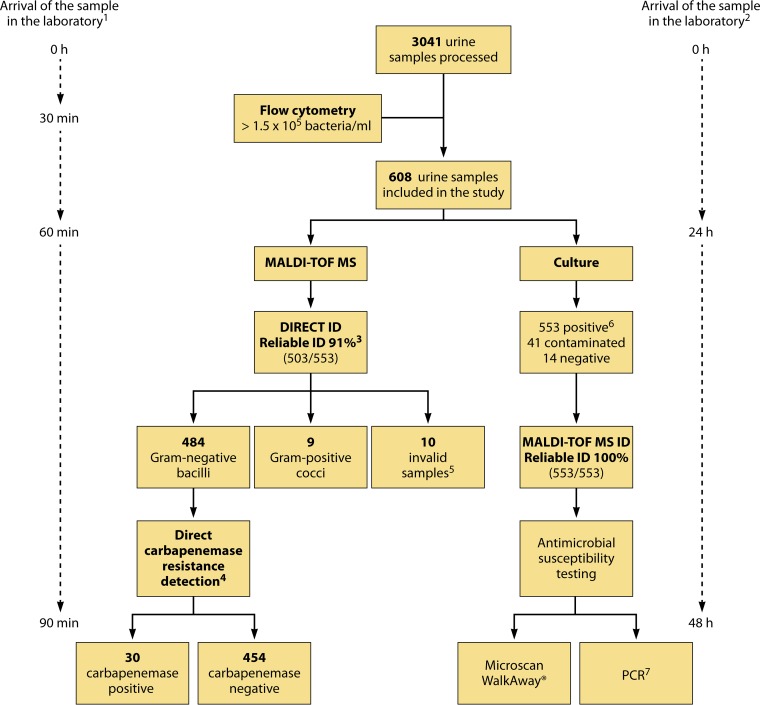
Oviaño et al. developed an interpretative automated MALDI-TOF MS-based method for the direct detection of carbapenemase-producing Enterobacteriaceae in clinical urine samples within 90 min of sample reception (44). A total of 3,041 urine samples were processed by flow cytometry, and a cutoff value of ≥1.5 × 105 bacteria/ml was used to select samples for the study. A novel protocol was developed for extracting bacteria from urine samples by using the Sepsityper kit (Bruker Daltonik). Carbapenem resistance was detected using imipenem, and the results were automatically interpreted with MALDI-TOF Biotyper Compass software (Bruker Daltonik). The assay yielded direct, reliable identification of 91% (503/553) of the samples and showed 100% sensitivity (30/30) and specificity (454/454) for detecting carbapenemase activity. The main advantage of the method is that the results are obtained 24 to 48 h earlier than with conventional methods (Fig. 1). However, it also has some disadvantages, such as the large volume (10 ml) of urine required, the need for bacterial counts higher than 1.5 × 105 bacteria/ml, and the exclusive identification of monomicrobial infections. This study demonstrated for the first time the potential detection of carbapenemase-producing Gram-negative bacilli directly from urine samples by using a standardized procedure and automated software.
FIG 1.
Workflow diagram showing the detection of carbapenemase activity directly from clinical urine samples. (Republished from reference 44 with permission of the British Society for Antimicrobial Chemotherapy.) 1, expected time for direct identification of bacteria and detection of carbapenemase susceptibility in urine samples by MALDI-TOF MS. 2, expected time for identification of bacteria and detection of carbapenemase susceptibility in urine samples by routine microbiological processing. 3, according to the manufacturer, a score of <1.7 indicates unreliable identification, a score of between 1.7 and 2.0 indicates genus identification, and a score of ≥2.0 indicates species identification. 4, carbapenemase activity was detected directly in urine samples and was measured by MALDI-TOF MS and automated spectrum interpretation. LogRQ values below 0.2 represent negative strains, values above 0.4 indicate carbapenemase activity, and values between 0.2 and 0.4 represent an ambiguous hydrolysis pattern. 5, invalid samples were those in which the amount of pellet obtained was not sufficient to carry out direct identification of bacteria and the carbapenem resistance assay. 6, urine samples were considered positive with ≥104 CFU/ml of one microorganism, whereas samples with two or more microorganisms were considered contaminated. 7, Gram-negative bacilli in positive urine samples were characterized by PCR and further sequencing of carbapenemase genes.
All these previous studies were performed using MALDI-TOF MS (Bruker Daltonik). Carvalhaes et al. performed a study evaluating the detection of carbapenemase activity using ertapenem and Vitek MS (bioMérieux) for detection of antimicrobial resistance (67). Sensitivity was evaluated according to the incubation time, and it was found that it was dependent on the type of carbapenemase, as classes A and B were the fastest, achieving 62% sensitivity in 15 min and 87% in 60 min. Vitek MS proved useful for the detection of antimicrobial resistance; however, there is a need for more studies validating the procedure. No studies regarding automated interpretation of spectra have been performed using Vitek MS.
The findings of these studies enable us to conclude that the method shows great potential for use in all microbiology laboratories as a routine method for detecting β-lactamase activity.
This field of work is progressing very rapidly, and since the last review, written by Hrabák et al. (52) in 2013, some notable methodological advances have been made. (i) Incubation times have been reduced from around 3 h to 30 min by using ceftriaxone for β-lactamase detection (ESBL and AmpC) and by using imipenem for carbapenemase detection (55, 61, 66). (ii) A larger number of antibiotics have been tested, and ceftriaxone, cefpodoxime, and cefepime can now be analyzed (55). (iii) Visual interpretation has been replaced by automatic acquisition and interpretation of spectra, thereby eliminating subjective measurement of mass peaks and making previous expertise in mass spectrometry unnecessary, thus facilitating the implementation of MALDI-TOF MS in routine in clinical microbiology laboratories (66). New antibiotics not previously evaluated by MS must first be properly analyzed by structural determination, and once the mass peaks of the antibiotic and the hydrolysis products are established, automated interpretation can be performed so that the logRQ value of the antibiotic is provided. To establish a threshold for positivity, hydrolysis peaks must have an AUC of at least 40%, and positive and negative controls must be used in every assay (66). Negative controls are necessary so that spontaneous hydrolysis is not interpreted as a positive reaction (especially for unstable compounds like imipenem). Besides, positive and negative controls can be used for normalization of the hydrolysis values so that intra-assay variability is minimized. This is especially important for assays in which the amount of cells added has not been previously standardized and positivity is related to the value of the positive control and not to the absolute value of the sample. (iv) Finally, detection of carbapenemase activity directly from clinical samples without enrichment of the bacteria in specific cultivation medium is already being performed with urine samples. Although large amounts of bacteria are used in the analysis (44), similar to those required for identification (42), this test could potentially be used with other types of clinical samples in the future (68).
Detection of the AAC(6′)-Ib-cr Enzyme
MALDI-TOF MS has proven useful for detecting β-lactamase activity, either narrow-spectrum, ESBL, AmpC-like, or carbapenemase-type enzymes, as we note above. Using the same concept of detecting antimicrobial resistance mechanisms by measuring the enzymatic activity of bacteria, any type of resistance that produces a change in the mass of the antibiotic can be detected. Detection of the AAC(6′)-Ib-cr enzyme highlights the ability of MALDI-TOF MS to detect resistance determinants other than β-lactamases, by monitoring the acetylation reaction in the substrate, in this case the fluoroquinolones ciprofloxacin and norfloxacin (69).
The AAC(6′)-Ib-cr enzyme is the most prevalent plasmid-mediated quinolone resistance mechanism in Enterobacteriaceae (70, 71). This resistance mechanism provides by itself low MICs that are not included in the clinical category of resistance; however, it facilitates the selection of higher-level resistance during treatment and results in limited therapeutic options for patients with bacteria containing the AAC(6′)-Ib-cr enzyme (51). In addition, detection is important from an epidemiological point of view and as a surrogate marker for specific bacterial clones. At present, this antimicrobial resistance mechanism is detected mainly by a PCR-based assay.
Pardo et al. developed a method of detecting the presence of the AAC(6′)-Ib-cr enzyme by measuring acetyltransferase activity in a series of 113 strains of Enterobacteriaceae, 64 of which harbored the AAC(6')-Ib-cr enzyme (72). MALDI-TOF MS measurements were performed after 4 h of incubation with norfloxacin. Results were analyzed by considering the ratio of the sum of areas under the curve of the different forms of acetylated norfloxacin and of those of native norfloxacin by using Vitek MS research-use-only (RUO) software (bioMérieux).
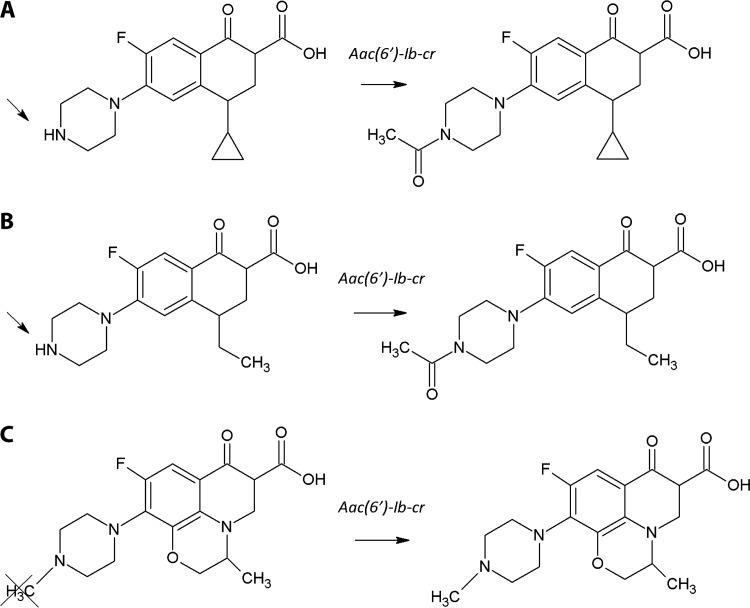
Oviaño et al. studied the presence of the AAC(6′)-Ib-cr enzyme in a collection of 81 isogenic Escherichia coli control strains [10 carrying the AAC(6′)-Ib-cr enzyme] during exposure to ciprofloxacin, norfloxacin, and levofloxacin and a further 36 clinical isolates [25 carrying the AAC(6′)-Ib-cr enzyme in addition to different combinations of quinolone resistance mechanisms] (73). Acetylation yields increases of 43 Da in the masses of ciprofloxacin and norfloxacin, but not of levofloxacin, which can be observed by visual inspection of the mass peaks in the spectra (Fig. 2). On the basis of the characteristic peak patterns of the acetylated and nonacetylated forms of ciprofloxacin and norfloxacin, AAC(6′)-Ib-cr-producing isolates and nonproducing isolates were clearly differentiated after an incubation time of 30 min. The presence of other determinants of quinolone resistance had no impact on the acetylation reaction or on the results obtained by MALDI-TOF MS. Total disappearance of the nonacetylated forms did not occur in any of the isolates tested, and both acetylated and nonacetylated forms coexisted in the same spectrum, in contrast to what usually happens when β-lactam antibiotics are observed in the presence of a β-lactamase (47). Those authors confirmed that the AAC(6′)-Ib-cr acetyltransferase enzyme confers only a low level of resistance, as previously reported, and described the structural determination of compounds other than β-lactam antibiotics with great success.
FIG 2.
Acetylation reaction in fluoroquinolones. (Republished from reference 73 with permission of the British Society for Antimicrobial Chemotherapy.) Shown are effects of the aac(6′)-Ib-cr gene on the chemical structures of ciprofloxacin (A), norfloxacin (B), and levofloxacin (C). Acetylation occurs at the amino nitrogen on the piperazinyl substituent of fluoroquinolones, a structural feature shared by ciprofloxacin and norfloxacin but not levofloxacin. Acetylation yields an increase of 43 Da in the mass of the antibiotic, which can be measured by MS.
In order to improve this procedure and for the purpose of automation, Oviaño et al. established an algorithm that enables the quantification of acetylation in relation to enzyme activity applied on a series of 122 clinical isolates, 15 of which harbored the AAC(6′)-Ib-cr enzyme (74). The MALDI Biotyper PeakShift prototype (Bruker Daltonik) was used for automation of the interpretation of spectra and semiquantification of the ratio of acetylation by logarithm intensity quantification (logIQ), the same principle that was used for measuring the hydrolysis of β-lactam antibiotics by β-lactamases. This method was 100% sensitive and specific for the detection of the AAC(6′)-Ib-cr enzyme. The automated interpretation of spectra led us to conclude that the use of norfloxacin as a resistance marker in MS will enhance detection as more-complete acetylation of the isolates is achieved, as the MICs are also higher for norfloxacin than for ciprofloxacin.
The findings of these previous studies indicate the potential use of MALDI-TOF MS to detect other mechanisms of resistance that are not β-lactam related, based on structural modification of antibiotics.
Detection of Aminoglycoside-Modifying Enzymes
Aminoglycoside resistance based on enzymatic modification of the antibiotic is one of the most common mechanisms of resistance in this group of antibiotics and is highly prevalent in both Gram-positive and Gram-negative microorganisms (75–77). Aminoglycoside-modifying enzymes display acetyltransferase, phosphotransferase, and nucleotidyltransferase activities. In addition, the substrate is not always of the same functional group, as acylation can affect hydroxyl and/or and amino groups, giving rise to more-complex reactions (78, 79). Zimmermann developed a rapid (<3-h) method of detecting N-acetyltransferase-mediated aminoglycoside resistance in Gram-negative bacilli by using a liquid chromatography-electrospray ionization-time of flight (LC-ESI-TOF) method (80). Isolates were characterized as AAC-6′ carriers. However, the mass shifts on the acetylated aminoglycoside molecules were not detected by MALDI-TOF MS. We had the same experience (data not shown). Further studies are required to finally rule out the possibility of detection of aminoglycoside resistance by MALDI-TOF MS. We acknowledge that some molecules will not be detected by MALDI-TOF MS, as negatively charged molecules can be detected only in a negative-ion mode of acquisition that is not available with conventional instruments like Microflex (Bruker Daltonik) or Vitek MS (bioMérieux). Additional research will confirm whether we will finally be able to detect these resistance mechanisms by MALDI-TOF MS, or we will have to rule out the use of this technology for detecting aminoglycoside-modifying enzymes.
DETECTION OF ANTIMICROBIAL RESISTANCE BY ANALYSIS OF THE MASS PEAK PROFILES OF THE BACTERIA
In the application of MALDI-TOF MS for the detection of antimicrobial resistance by analysis of the mass peak profiles of bacteria, susceptible and resistant microorganisms of the same species are differentiated on the basis of their spectra. Some mass peaks are associated with a resistance pattern of an isolate due to the expression of a specific protein involved in the directed or associated resistance phenotype and may be useful for differentiating between susceptible and resistant isolates. Although this technique has been successfully applied in some studies that are explained further below (81–83), it must be validated in clinical settings to ensure the reproducibility of the results. The main advantage of this methodology is its simplicity, as the sample preparation procedure is the same as or quite similar to that used for identification. The mass peaks (biomarkers) identified can also be automatically analyzed in real-time workflows in laboratories, at no additional expense.
Detection of MRSA
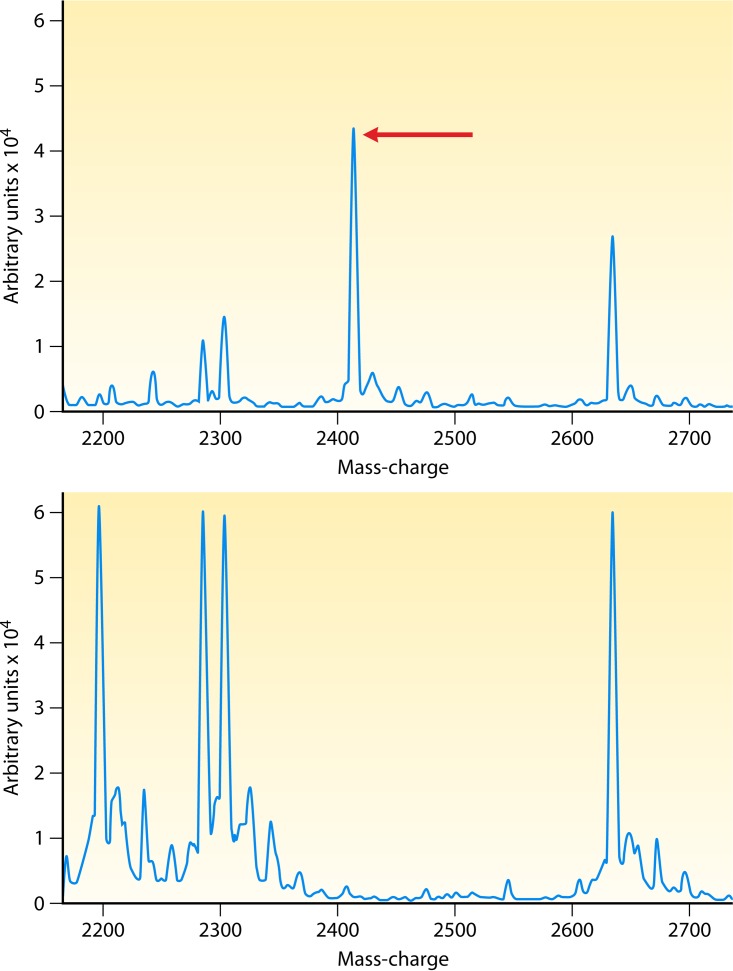
Many studies that have investigated how best to discriminate between methicillin-susceptible S. aureus (MSSA) and MRSA have obtained different sensitivity values as well as variable findings in regard to the mass peaks used to differentiate these two groups (84–87). Other studies have stressed the lack of correlation between any peak profiles and MRSA or MSSA (88, 89). The possible clinical application of this methodology was in dispute until Josten et al. detected a small peptide called phenol-soluble modulin (PSM-mec) that is encoded on the type II, III, and VIII staphylococcal cassette chromosome mec element (SCCmec) cassettes present in the genomes of health care-associated MRSA strains by MALDI-TOF MS (81). This peptide is excreted by agr-positive strains, and the presence of the peptide can be detected in the MALDI-TOF MS spectra of whole cells as a peak at 2,415 Da (Fig. 3). PSM-mec is detected by MALDI-TOF MS with high sensitivity and specificity values of 0.95 and 1, respectively. Rhoads et al. reported that the sensitivities of the 2,415-Da peak for mecA carriage in S. aureus and Staphylococcus epidermidis were low (37% and 6%, respectively), whereas the specificities were high (≥98%) (90).
FIG 3.
MALDI-TOF spectra showing the 2,415-Da mass peak representing PSM-mec of methicillin-resistant S. aureus. (Republished from reference 90 with permission of the publisher.) Shown are MALDI-TOF mass spectra for 2 MRSA strains. One strain is a USA 100 strain (top), carrying the mecA gene, and the other strain is a USA 300 strain (bottom), which lacks the mecA gene. The x axis depicts mass/charge ratios, and the y axis is arbitrary units.
This method could be used to establish a sound approach to distinguishing between MRSA and MSSA with higher specificity. However, sensitivity is in this case dependent on the prevalence of the cassettes in each geographical area, thus limiting the application to those areas with a high prevalence of cassettes II, III, and VIII. Besides, negative results should be confirmed by molecular methods. In areas with a low prevalence of the above-mentioned cassettes, phenotypic or molecular methods should not be replaced by MALDI-TOF MS given the worse accuracy of this method (91).
Detection of β-Lactamases
In a study reporting a method of real-time analysis of outbreaks of infections caused by carbapenemase-producing Enterobacteriaceae, Lau et al. observed a mass peak at 11,109 Da corresponding to a specific protein (p019) carried on the pKpQIL plasmid after a simple formic acid extraction step (82). This mass peak was used as a biomarker to identify KPC-producing Klebsiella pneumoniae, with excellent results. The insertion containing p019 has been found only in plasmids carrying blaKPC to date (92). In a later study, Youn et al. applied the above-described method of identifying KPC-producing Enterobacteriaceae along with different extraction methods (plate extraction and full formic acid extraction) and different acquisition methods (manual and automated), demonstrating 96% sensitivity and 99% specificity under the best conditions, for plate extraction and interpretative automation analysis, respectively (93). Gaibani et al. demonstrated that the 11,109-Da peak was detected in 88.2% of the KPC producers, confirming previous results (94).
Papagiannitsis et al. described the identification of CMY-2 β-lactamases by detection of the mass peak at 39.850 Da by MALDI-TOF MS (95). The preparation of the sample involved a labor-intensive method with extraction of the periplasmic proteins performed by a modified sucrose method that lasts around 22 h. This group also performed some preliminary analyses for detection of ACC and DHA enzymes. However, these methods could hardly be used by routine microbiology laboratories.
In any case, caution should be applied when using this type of approach, as protein expression may depend on specific clones or growth terms, and the data cannot be extrapolated to all β-lactamases.
Detection of cfiA-Positive Bacteroides fragilis
Carbapenemase production encoded by the cfiA gene in Bacteroides fragilis is increasingly recognized in clinical isolates (96–98). The detection and control of B. fragilis and resistance of the species to carbapenems are of great concern in relation to prescribing the most appropriate antibiotic therapy, as this is one of the first-choice treatments.
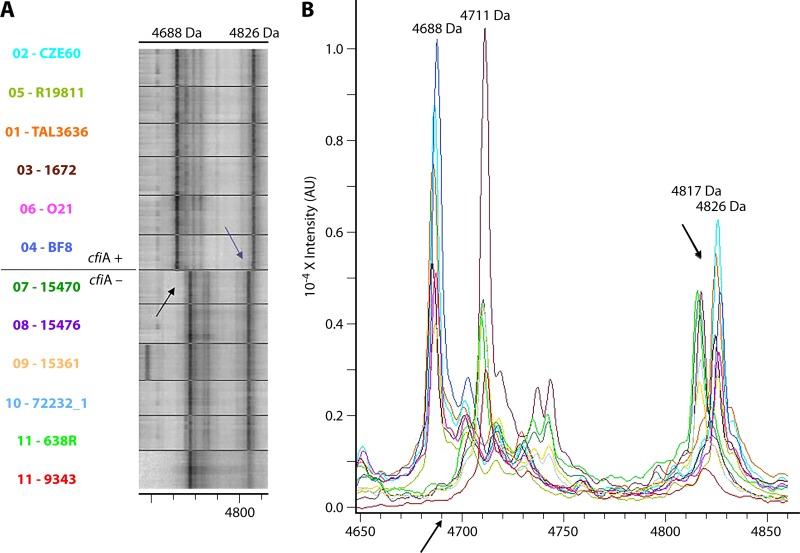
Wybo et al. differentiated cfiA-positive Bacteroides fragilis in 248 clinical samples by grouping the samples into 2 clusters (one with the cfiA gene, encoding carbapenemase production, and the other without the gene) in a matrix calculated from the mass peak profiles (83). The relatedness of the spectra was determined by using the composite correlation index (CCI) tool of the MALDI Biotyper (Bruker Daltonik). The CCI is a mathematical algorithm used to compare and distinguish MALDI mass spectra, where CCI values of around 1 represent a close relationship between spectra, matching an intense red color in the matrix, whereas a value close to zero represents a distant relationship, with a blue color in the matrix. Nagy et al. analyzed 28 isolates (9 cfiA producers and 19 nonproducers) to validate a similar method based on the presence or absence of a series of mass peaks in the range of 4,000 to 5,500 Da observed in MALDI-TOF mass spectra, with 100% accuracy (99). The ability to compare spectra by using the MALDI-TOF MS graphical matrix was successfully applied by these authors to detect antimicrobial resistance (Fig. 4).
FIG 4.
Representative spectra of cfiA-positive and cfiA-negative Bacteroides fragilis strains. (Republished from reference 99 with permission of the publisher.) Shown is an example of the clear differences in the MS peaks in the interval from 4,650 to 4,850 Da between cfiA-positive and -negative strains. (A) Visualization by a gel/stack view. (B) Visualization by mass spectra. Peaks are shifted from 4,711 and 4,817 Da (cfiA negative) to 4,688 and 4,826 Da (cfiA positive), respectively. AU, arbitrary units.
Porin Detection
The interaction between β-lactamase (ESBL, AmpC, or carbapenemase) production and porin loss in bacteria is important in carbapenem resistance (100, 101). Klebsiella pneumoniae produces two main porins (OmpK35 and OmpK36) (102). While most clinical isolates of K. pneumoniae express both the OmpK35 and OmpK36 porins, most ESBL-producing K. pneumoniae clinical isolates produce only OmpK36 (103). Loss of porins is one of the factors contributing to antimicrobial resistance and may favor the selection of additional mechanisms of resistance. OmpK35 and OmpK36 are especially related to resistance to β-lactams and quinolones (104).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) is the most common method used to analyze porins. However, SDS-PAGE is laborious, time-consuming, and not available in most clinical laboratories. MALDI-TOF MS has arisen as a new technique to detect the presence and absence of porins and to help in the detection of antimicrobial resistance. Disappearance of the mass peak related to the porin would imply resistance to the antibiotics that penetrate the porin. On the contrary, the presence of the mass peak related to the porin would not strictly mean susceptibility, as quantification of the expression level cannot be performed yet, and we cannot ensure that the porin has a natural behavior or has reduced expression.
Cai et al. described a new method for the rapid detection of porins in K. pneumoniae. MALDI-TOF MS was performed with a Microflex LT mass spectrometer (Bruker Daltonik) operating in positive-linear-ion mode between 15,000 Da and 80,000 Da. MALDI-TOF MS analysis showed that mass peaks at 38,000 Da and 19,000 Da could be used as indicators of the OmpK36 porin (105). In a similar study, Hu et al. identified mass peaks at 35,000, 37,000, and 38,000 Da, corresponding to OmpA, OmpC, and OmpF, respectively, in the mass spectrum of E. coli ATCC 25922 (106). The OmpC and OmpF porins were easily differentiated by MALDI-TOF MS but were indistinguishable by SDS-PAGE. In K. pneumoniae isolates, mass peaks were observed at 36,000 and 38,600 Da, corresponding to the OmpA and OmpK36 porins. Porin OmpK35 was not detected by the SDS-PAGE method, but it was apparently detected by MALDI-TOF MS. Further studies must be conducted to check the reproducibility of the technique and to expand this novel approach to other microorganisms. However, these studies demonstrate the usefulness of MALDI-TOF MS in saving time and reducing the analytical effort involved in microbiology laboratories, opening the possibility for a future application of these techniques in clinical practice.
Detection of Polymyxin Resistance
As carbapenemase-producing Enterobacteriaceae have become more prevalent, the use of polymyxin has gradually increased, and the resistance of these bacteria has subsequently also increased (107, 108). Acquired resistance to polymyxins in Enterobacteriaceae is due mainly to modifications in the polymyxin target, the lipopolysaccharide. These modifications may be brought about by chromosome-encoded mechanisms or by the acquisition of the plasmid-borne phosphoethanolamine transferase gene mcr-1. Dortet et al. developed a novel MALDI-TOF MS method for detecting polymyxin resistance in Enterobacteriaceae within 15 min (109). This process is similar to that used for identifying bacteria but with a modified atypical matrix, which enables observation of the peaks corresponding to intact lipid A and lipid A modified by either chromosome-encoded or plasmid-mediated mechanisms. Independently of the resistance mechanism involved, a mass peak at 1,919 Da corresponding to the addition of a phosphoethanolamine group to the 1,796-Da nonmodified lipid A has been detected for all polymyxin-resistant isolates, including E. coli and K. pneumoniae. In the case of resistance due to the mcr-1 gene, an additional mass peak was observed at 1,821 Da. Although no information has been given regarding the nature of this mass peak, it appears to be specific to the MCR-1-producing isolates, independently of the species considered. As far as we are aware, no other studies on polymyxin resistance have been carried out by direct detection of lipid A by MALDI-TOF MS, and further research is needed to confirm the validity of this novel approach and broaden the number of Enterobacteriaceae and nonfermenter microorganisms, such as Acinetobacter baumannii, with which the method can be used. This application is potentially of great interest because of the rapidity and simplicity of the procedure relative to previous methods used to identify lipid A that needed a laborious extraction procedure (110). The atypical matrix must be fully characterized so that it does not complicate the identification process and to enable further automation of the process.
DETECTION OF ANTIMICROBIAL RESISTANCE BY MEASUREMENT OF THE EFFECTS OF THE ANTIBIOTIC ON MICROORGANISM GROWTH
For the application of MALDI-TOF MS for the detection of antimicrobial resistance by measurement of the effects of the antibiotic on microorganism growth, susceptible and resistant microorganisms of the same species are differentiated on the basis of the relative growth of the isolate in the presence of a particular antibiotic, as in other nonproteomic approaches (111, 112).
Quantitative Resistance Profile
First, Sparbier et al. measured the protein synthesis of bacteria growing in an isotope-labeled amino acid medium (113). The incorporation of heavy amino acids increases the molecular weight of the newly synthesized proteins, causing peak shifts in the mass spectral profiles that can be automatically detected by a software algorithm. This assay is called MBT-Resist (MALDI Biotyper resistance test), with stable isotope-labeled amino acids. Bacteria are incubated with the antibiotic in an isotope-labeled medium, lysed, and spiked with an internal standard for quantification of bacterial growth. In this case, lysine was the amino acid that was isotope labeled. Ten MRSA and 10 MSSA isolates were analyzed for the detection of methicillin resistance by calculating the ratio between the sum of areas of the “heavy” peaks and that of the areas of the “normal” peaks. The assay provides results in 3 h, being able to differentiate between MRSA and MSSA.
Next, Lange et al. developed the MBT-ASTRA (MALDI Biotyper antibiotic susceptibility test rapid assay), which facilitates measurement of the quantity of peptides and small proteins within a spectrum (114). These quantities are correlated with the number of microorganisms and, therefore, with the growth of a microorganism. If the microorganism is unable to grow in the presence of a certain antibiotic, it is susceptible to that antibiotic, and the spectrum obtained will be almost flat, with no mass peaks, while resistant isolates will grow, and the corresponding mass peaks will be visible in the spectrum.
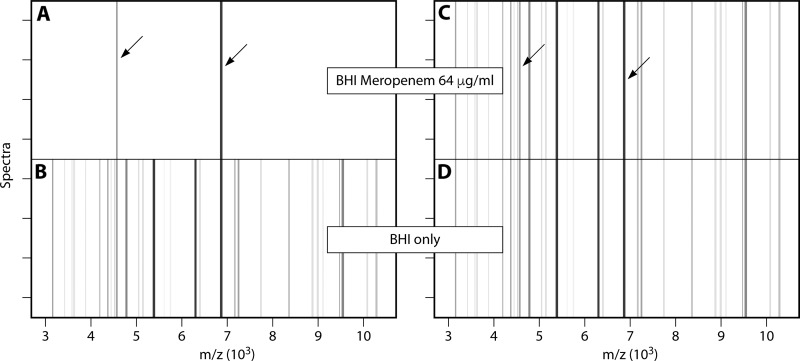
The bacterial growth ratio is calculated by measuring the whole mass peak profile of a microorganism with and without the antibiotic analyzed. The use of an internal standard enables quantification as the ratio of the AUCs of a microorganism incubated with and without the antibiotic: relative growth = AUCBHI + meropenem/AUCBHI. In this case, meropenem was used for the evaluation, and brain heart infusion (BHI) broth was used to culture the microorganisms. The results were analyzed by box plots and pseudogels. The use of meropenem at a concentration of 8 μg/ml to detect carbapenem resistance in 108 K. pneumoniae isolates yielded a sensitivity of 97.3% and a specificity of 93.5%. As illustrated in Fig. 5, for susceptible bacteria, no visible mass peaks were observed (Fig. 5A and B) (the internal standard is shown for reference purposes), whereas for resistant bacteria, the whole mass spectrum was observed (Fig. 5C and D).
FIG 5.
Pseudogel view of the mass spectra of susceptible and resistant K. pneumoniae strains after growth in the presence of meropenem. (Republished from reference 114 with permission.) Shown are pseudogel views of the mass range between 3 and 10 kDa of susceptible (A and B) and resistant (C and D) K. pneumoniae strains after incubation in the absence (bottom panels) or presence (top panels) of meropenem (64 µg/ml) for 1 h. For each incubation, four spectra acquired from two different spots are shown. Internal-standard peaks are marked by arrows.
Jung et al. also evaluated this method for detecting resistance against cefotaxime, piperacillin-tazobactam, gentamicin, and ciprofloxacin (115), by analyzing a series of 30 spiked blood cultures and 99 patient-derived blood cultures. Bacterial cells extracted from positive blood culture bottles were used to prepare an inoculum of 5 × 106 CFU/ml. The isolates were evaluated using a concentration of antibiotic that was 1 dilution higher than the EUCAST susceptibility breakpoint for Enterobacteriaceae. This is referred to as the MBT-ASTRA breakpoint concentration. The assay yielded almost 100% sensitivity and specificity for all antibiotics evaluated, with poorer results for piperacillin-tazobactam. Identification of bacteria and analysis of susceptibility are possible within 4 h. In another study, Maxson et al. applied this method for the first time to study the susceptibilities of 35 S. aureus isolates to several antibiotics, such as ciprofloxacin, oxacillin, cefepime, and vancomycin (116). The incubation time was established as 2 h, and the resistance breakpoints for classifying the isolates were based on current guidelines from the CLSI (117). Interpretative automation was performed with MBT-ASTRA software (Bruker Daltonik), and results were displayed in box-and-whisker plots. In this case, the assay showed an overall accuracy of 95%.
Idelevich et al. developed an interesting method based on the MBT-ASTRA but with microdroplet incubation carried out directly on the MALDI-TOF MS target (118). The isolates (24 K. pneumoniae and 24 P. aeruginosa isolates) were incubated in a wet chamber, and the broth was supplemented or not supplemented with the antibiotic (meropenem) evaluated. Using 6-µl microdroplets and incubation times of 4 h for K. pneumoniae and 5 h for P. aeruginosa, the rate of efficiency was maximum, with almost 100% sensitivity. This study is of the utmost importance because it has simplified the procedure and reduced the time required for processing. However, the overall time needed to deliver the results is still too long for the method to be considered a rapid technique for clinical application.
The main advantage of the MBT-ASTRA is the universality of the method for detecting resistance to different antibiotics and different resistance mechanisms. The main disadvantage is the incubation time (2 to 4 h), which is much longer than those of the other MALDI-TOF MS-based approaches used to detect resistance. The incubation time cannot be reduced, as the microorganisms must grow before the susceptibility or resistance of the microorganism can be evaluated. These advantages and disadvantages are also shared by the MBT-Resist method. The difference in the analysis is that the MBT-Resist assay compares single peak shifts derived from the incorporation of heavy labeled amino acids during bacterial protein synthesis with the usual mass peak profile of the bacteria analyzed. This analysis requires a previous species-specific test that is not necessary for the MBT-ASTRA, as it analyzes the whole mass profile of the bacteria for evaluation of susceptibility or resistance.
Quantitative Resistance Profiles Applied to Mycobacteria and Yeast
Ceyssens et al. used the MBT-ASTRA to detect susceptibility to rifampin, isoniazid, linezolid, and ethambutol in Mycobacterium tuberculosis and to detect susceptibility to clarithromycin and rifabutin in nontuberculous mycobacteria (119). Although molecular techniques can rapidly detect resistance genes in M. tuberculosis, they have not yet replaced the need for culture-based susceptibility assays to provide information about the full spectrum of available antituberculous drugs. Molecular techniques have not been widely developed for the detection of resistance in nontuberculous mycobacteria (120, 121), and broth microdilution is recommended as the gold-standard method (122). However, Cesyssens et al. correctly identified 156/156 M. tuberculosis and 65/66 nontuberculous mycobacterium drug resistance profiles by using the MBT-ASTRA. Although this novel application of the MBT-ASTRA yielded excellent results for M. tuberculosis, routine use of the technique in clinical settings is unlikely. First, MALDI-TOF MS requires a larger biomass of sample than molecular techniques, which leads to longer turnaround times, especially for slow-growing mycobacteria. Second, turnaround times were not significantly different for M. tuberculosis testing, although the MBT-ASTRA method delivered results a week earlier than routine susceptibility methods for nontuberculous mycobacteria. On average, with the MALDI-TOF MS method, an incubation time of 6 days was needed for both M. tuberculosis and nontuberculous mycobacteria. Thus, the method still needs prolonged incubation and a sustained processing time, and there is a greater risk of exposure. Moreover, the technique has not yet been standardized.
Regarding the study of fungi, Marinach et al. used MALDI-TOF MS to detect fluconazole resistance in Candida albicans based on monitoring modifications in the mass peak profile of the yeast in the presence or absence of different concentrations of the antifungal agent (123). The yeast cells were incubated with the antifungal agent for 15 h. Protein extraction was performed prior to acquisition of the spectra. The spectra were acquired for 12-fold dilutions starting at 128 µg/ml. The minimal profile change concentration (MPCC), corresponding to the lowest fluconazole concentration at which a mass spectrum profile change can be detected, was established. When relating the value of the MPCC obtained by MALDI-TOF MS with the susceptibility category assigned by using CLSI criteria, only 1 out of 16 isolates was incorrectly classified. Although excellent as a proof of concept, routine application of this method in clinical practice is unlikely due to the long processing time (as the samples must be incubated overnight). De Carolis et al. applied the protocol to study the susceptibilities of Candida and Aspergillus to caspofungin (124). These researchers used the CCI matrix tool of the MALDI Biotyper to analyze the relationships between the spectra generated at different concentrations of the antifungal. The MPCC was obtained from the matrix, and an agreement of 94.1% was reached in establishing the clinical categories of susceptibility in the Candida isolates tested. Finally, Vella et al. used a simplified version of the above-described approach, with an incubation time of 3 h and only three breakpoint concentrations of caspofungin: 0 µg/ml (null), 0.03 µg/ml (intermediate), and 32 µg/ml (maximal). The overall agreement was 98% for comparison of the MALDI-TOF MS methodology with CLSI-established categories of susceptibility (125). The latter approach shows very promising results for further clinical application.
AUTOMATION IN ROUTINE DIAGNOSIS
Although technological advances have been introduced in microbiology laboratories, there is still a real or perceived idea that microbiological assays cannot be automated. In comparison with clinical biochemistry, which has traditionally been highly automated, clinical microbiological analysis still requires expert staff and prolonged processing times (126, 127). Microbiological analysis is often considered too complex to automate because of the various types of samples involved, and automation is considered too expensive for microbiology laboratories. Although complex molecular tests are frequently used in reference laboratories or in virology laboratories, bacteriology is increasingly benefiting from the introduction of these techniques in routine laboratories (128, 129). As a result of the growing demand for microbiological analysis of samples, increased workloads, the shortage of trained personnel, the need for quality standards, and reductions in financial resources, automation has become a real possibility in recent years in clinical microbiology laboratories (130). The emergence of MALDI-TOF MS technology is a major factor enabling automation in microbiology laboratories (131). MALDI-TOF MS is easy to perform and highly automated because it is technically very simple. The ability to perform more tests, with minimal staffing, with less expertise, with high-quality standards, and with guaranteed traceability throughout the process, can be achieved by the use of MALDI-TOF MS.
Ready-To-Use Techniques in Clinical Practice
Throughout this review, we have exposed the different available software packages for analysis of spectra for the detection of antimicrobial resistance. However, not all these software packages have been approved for clinical use. Here, we review the different available products for the various principles that have been described for MALDI-TOF MS and their regulatory status (Table 3).
TABLE 3.
Summary of the different MALDI-TOF MS principles for detection of antimicrobial resistance and available analysis software
| MALDI-TOF MS principle | Application | Software | Target(s) | Commercially available kit | Regulatory statusb | Advantage(s) | Disadvantage(s) |
|---|---|---|---|---|---|---|---|
| Detection of enzymatic activity | Detection of β-lactamases (ESBL and AmpC, etc.) | MBT-STAR BL module for Compass softwarea | Ratio between the sum of areas of the hydrolyzed antibiotic and the intact form for ampicillin, piperacillin, ceftazidime, cefotaxime, ertapenem, and meropenem | No | RUO | Easy and fast technique (∼30 min); detects resistance directly from the clinical sample; does not require further interpretation of spectra | No identification of the β-lactamase type; does not provide information regarding the MIC |
| Detection of carbapenemases | Ratio between the area under the peak of the internal standard and intact imipenem | MBT STAR-Carba kit | CE-IVD | ||||
| Detection of the AAC(6′)-Ib-cr enzyme | MBT PeakShift prototypec | Ratio between the sum of areas of the acetylated antibiotic and the intact form for ciprofloxacin and norfloxacin | No | RUO | Easy and fast technique (∼30 min); more simple than the reference method (molecular technique) | Does not provide information regarding the MIC; developed skills for interpretation of spectra needed | |
| Measurement of growth in the presence of an antibiotic | Detection of resistance by measuring protein synthesis in the presence of isotope-labeled amino acids | MBT-Resistc | Ratio between the sum of areas of the isotope-labeled peaks and the “normal” peaks | No | RUO | Provides information on susceptibility and resistance to different antibiotics | Requirement for isotope-labeled medium; labor-intensive methodology; time-consuming methodology (∼3 h to deliver results) |
| Detection of resistance by semiquantification of bacterial growth at the breakpoint concn | MBT-ASTRAc | Ratio between the sum of areas of the peak profile of a microorganism incubated with and without antibiotic | No | RUO | Provides information on susceptibility and resistance to different antibiotics; easy handling using the microdroplet assay | Time-consuming methodology (∼3 h to deliver results) | |
| Measurement of resistance by analysis of the peak pattern of bacteria | Detection of KPC-producing K. pneumoniae | MBT Subtyping module for Compass softwarea | Identification of the 11.109-Da mass peak; identification of the 2.415-Da mass peak; identification of a series of mass peaks in the range of 4,000–5,500 Da | Not necessaryd | RUO | No more steps are needed other than the minimum needed for identification; information provided after high-confidence identification; does not require further interpretation of spectra | Limited to a few resistance mechanisms; specificity is high, but sensitivity is related to prevalences in different geographic areas |
| Detection of MRSA | |||||||
| Detection of cfiA-positive B. fragilis | |||||||
Compass software (Bruker Daltonik GmbH, Germany) is an intuitive and integrated software for mass spectrometry analysis that allows modular application in packages. The MALDI Biotyper (MBT) Selective Testing of β-Lactamase Activity (STAR-BL) and the MBT Subtyping modules are two modules of the software.
RUO, research use only; CE-IVD, European Conformity for in vitro diagnostics.
MBT PeakShift prototype (Bruker Daltonik), MBT-Resist (Bruker Daltonik), and the MBT-ASTRA (Bruker Daltonik) are RUO software not incorporated into Compass software.
No commercially available kit is necessary for analysis of mass peak profiles, as the procedure is the same as the one used for identification, and no extra reagents are employed.
Compass software (Bruker Daltonik) is an intuitive and integrated software for mass spectrometry analysis that allows a modular application in packages. It is included in the Microflex MALDI-TOF MS Biotyper IVD test. The MBT STAR-BL and the MBT Subtyping modules are two modules of the software. Vitek MS (bioMérieux) does not include any platform for detection of antimicrobial resistance in an automatable manner. Comparison of a profile spectrum acquired from an antibiotic with profile signatures stored in a reference library by visual inspection enables rapid determination of antibiotic susceptibility but requires certain expertise in interpreting mass spectra. Automated methods will allow and facilitate the implementation of the technique in clinical laboratories and support its application by users who are not trained in mass spectrometry techniques. The only commercially available kit for use with software for automated interpretation of spectra is the MBT STAR-Carba IVD kit (Bruker Daltonik) in the MBT STAR-BL software module (Bruker Daltonik). Samples are analyzed using imipenem to detect carbapenemase enzymes. According to the manufacturer, the kit contains enough reagents to test 20 to 58 samples depending on the assay batch size. The kit contains all the reagents required to perform the assay, except for a positive and a negative control, which must be provided by the user and used in all assays. Previous calibration of the equipment must be carried out in the mass range of spectrum acquisition, i.e., in a low-mass range (100 to 1,000 Da). The MBT STAR antibiotic calibration standard (MBT STAR-ACS), which contains a mixture of four small peptides that provide characteristic and well-defined MALDI-TOF mass spectrum signals, can be used for this purpose. The Carba assay can be used with microorganisms from plate cultures or from positive blood cultures. If using blood cultures, previous extraction with the Sepsityper kit (Bruker Daltonik) is necessary, and the procedure used is a slightly modified version of that used for identification. For a standard workflow, incubation of the blood samples with the antibiotic for 30 min is sufficient. However, for testing Acinetobacter spp., an incubation time of 60 min is recommended. The software automatically interprets the spectra by normalizing the intensities of the sample mass peaks relative to signal intensities obtained from positive- and negative-control strains. A report is generated with a color-coded plot that enables evaluation along with the logRQ value. Quantification reduces the risk of misclassifying isolates due to a lack of experience in the visual interpretation of mass spectra. Use of the software also reduces intra- and interassay (observer-related) variability, thus yielding standardized results. Moreover, the software enables the identification of the same target and assay of the same colonies being tested to detect carbapenemase activity. Dortet et al. evaluated the commercialized MBT STAR-Carba IVD kit with two in-house MALDI-TOF MS methods and Rapidec Carba NP for detecting carbapenemase activity in 175 isolates, including 120 carbapenemase producers (132). Overall, all systems have very similar performances, with sensitivities ranging from 95% to 100% and specificities ranging from 98.2% to 100%. The advantages of the STAR-Carba IVD kit are the turnaround time, the best for all systems evaluated (<1 h); the automated interpretation of results, which can be confusing for Rapidec Carba NP, especially for OXA-48-like enzyme producers with poor expression (133); and the possibility of coupling the software to the laboratory system so that results are directly connected and technical errors are minimized.
Regarding the detection of antimicrobial resistance by analysis of the mass peak profiles of bacteria, the commercially available MALDI Biotyper Subtyping software module (Bruker Daltonik) enables the detection of proxies for specific resistance mechanisms in an automated workflow (134). The prerequisite for automation of the process for identifying resistance markers is high confidence (score of >2.0) in identification of the bacterium by the MALDI Biotyper. The process is quite simple, as no additional sample preparation is required after the samples have been directly transferred to MALDI target plates. Once the bacterium has been identified, the software automatically performs typing, comparing the spectrum obtained against a reference library and reporting the results. This module enables the identification of KPC-producing K. pneumoniae by searching for the p019 protein-related peak (11.109 Da) in the sample spectrum, which, if present, indicates that the sample is a presumptive blaKPC-positive sample. This module can also be used to detect methicillin-resistant S. aureus by detecting the PSM-mec peak (2.415 Da). Finally, the module can also be used to detect cfiA-positive B. fragilis by the detection of specific peaks associated with the protein expressed.
Automation has been achieved in the field of resistance detection by measuring the effects of the antibiotic on microorganism growth; however, these software packages are categorized as RUO and have not yet been included as modules for Compass software (Bruker Daltonik). The same happens for the detection of the AAC(Ib)-cr enzyme, which is available as prototype software and has not yet been released for the public in a commercialized software package.
The MALDI-TOF MS process of deposition of microorganisms on the MALDI target has already been developed by some companies. Chudejova et al. evaluated four different sample preparation methods: manual deposition, in-target extraction with formic acid, wet deposition (in-target extraction with the order altered, where formic acid is applied first and the colony is applied last), and automatic deposition using MALDI Colonyst (Biovendor Instruments, Brno, Czech Republic) (135). For clinical validation, the lowest scores were obtained by manual deposition of the target, and higher identification scores were obtained using automated processing by MALDI Colonyst for all tested microbial groups (Gram-positive bacteria, Gram-negative bacteria, anaerobes, and yeasts). Bruker Daltonik developed MALDI Biotyper Galaxy for automated deposition of colonies on the MALDI-TOF target. The system is coupled with on-board barcode reading so that the targets are matched to their corresponding projects, and traceability is guaranteed for every sample. The system also reduces contamination and frees personnel from routine work. There has not yet been a publication evaluating this system. Broyer et al. evaluated a benchtop instrument (bioMérieux, France) for automated processing of positive blood cultures (136). The yield was 83% correct identifications compared to manual methods. This prototype instrument is based on an “all-in-one” extraction strip. It allows less than 5 min of personnel time and provides identification results in less than 30 min. Moreover, it can be performed on demand and in small batches of positive blood cultures, allowing 24/7 service.
In the future, as different procedures achieve clinical validation, more applications could be integrated into the software and will be available for the user. MALDI-TOF MS should occupy an outstanding place in the microbiology laboratory, coupled with automated systems for seeding, incubation, and digitized reading of plate cultures, thus yielding an integrated operating model with different innovative combinations and capacities.
FUTURE CHALLENGES
As demonstrated above, MALDI-TOF MS has gained value as a multifaceted instrument for clinical microbiology laboratories. Although tremendous progress in MALDI-TOF MS has been made in the past decade, particularly regarding the identification of microorganisms, antimicrobial resistance detection by MALDI-TOF MS has been proposed as a possible application. However, several major challenges must be overcome to enable further progress in clinical proteomics diagnostics in microbiology laboratories.
The following bottlenecks have been recognized: (i) limited MS sensitivity (>105 CFU/ml) for direct detection in clinical samples (43–45), (ii) the limited number of resistance mechanisms that can be identified by MALDI-TOF MS if only assays involving enzymatic activity of bacteria are chosen and a delayed response time if the effects of antibiotics on bacterial growth are measured, (iii) the need for public libraries of MALDI-TOF MS spectra for analysis of bacterial mass peak profiles, (iv) the lack of established and standardized routines for MALDI-TOF MS-based technology in detecting antimicrobial resistance, (v) the limited range of automated applications and commercially available kits, and (vi) the lack of studies on the cost-effectiveness of incorporating resistance detection by MALDI-TOF MS.
The balance between costs and savings is one of the main advantages of the new technologies relative to traditional methods (137, 138). The most important clinical benefits of MALDI-TOF MS-based identification observed in the literature are the earlier administration of correct antimicrobial therapy, reduced morbidity and mortality, reduction in lengths of hospital stays, and reduced overall costs per hospitalized patient. However, no clinical benefits have yet been reported regarding the implementation of MALDI-TOF MS for resistance testing. This should be one of the main cornerstones of further research. In our opinion, the application of MALDI-TOF MS for detecting antimicrobial resistance will supplement the results obtained for the identification of microorganisms, providing more-accurate results to clinicians.
One of the first studies to evaluate the clinical impact of MALDI-TOF MS on patient outcomes was conducted by Huang et al. (139). These researchers proposed integrating MALDI-TOF MS into the workflow of an antimicrobial stewardship program. This study included 501 patients with bacteremia or candidemia for evaluation and 245 patients in the intervention group. For patients included in the MALDI-TOF MS group, the time to organism identification and the time to effective antibiotic therapy were reduced, optimal antibiotic therapy was provided, the mortality rate was lower, and the length of intensive care unit stay and the risk of recurrent bacteremia were reduced.
Clerc et al. conducted a prospective observational study regarding the clinical impact of MALDI-TOF MS for identifying positive blood cultures grown in samples from patients with bacteremia caused by Gram-negative bacilli compared with the information obtained by use of Gram staining (140). In 35% of the episodes (n = 202), the information generated by MALDI-TOF MS had a clinical impact by modifying empirical therapy. The most remarkable consequence was the correct broadening of the antibiotic spectrum in 43.7% of cases. In those cases in which MALDI-TOF MS did not have a documented impact, modification of therapy would have been possible for 24% of cases. Factors involved in not taking MALDI-TOF MS results into account included younger age of the patient, admission of the patient in an intensive care unit, male sex, and a lack of confidence shown by medical staff in the use of an unfamiliar technology. Verroken et al. evaluated the impact on antimicrobial prescription of a workflow in which the identification was provided by MALDI-TOF MS (141). Partial susceptibility results were performed regarding the previously obtained identification results and were provided on the same day as the identification of blood cultures positive for Enterobacteriaceae, Pseudomonas aeruginosa, and S. aureus. The mean time to identification was reduced by around 65%, the mean time to complete susceptibility results was reduced by 27%, and the mean time to optimal antimicrobial treatment was decreased to 16 h. Beganovic et al. evaluated the impact of MALDI-TOF MS in combination with antimicrobial stewardship intervention on patient outcomes (142). The primary outcome was the time to optimal therapy, which was significantly reduced by the use of MALDI-TOF MS. The mean time to microbiological clearance, the mean length of hospital stay, the mean length of stay in a critical care unit, the number of patients with recurrence of the same bacteremia, and the mean length of time of antimicrobial therapy were also reduced. Osthoff et al. evaluated the impact of MALDI-TOF MS on the management of bacteremic patients, by applying a single-center, open-label, controlled clinical trial with 425 patients during 1 year (143). Surprisingly, rapid identification by MALDI-TOF MS directly from positive blood cultures in this study did not affect the duration of intravenous antimicrobial therapy or the length of hospital stay; however, identification of contaminated blood cultures led to a shorter duration of therapy (mean time, 4.8 versus 7.5 days).
Further cost-effective multicenter studies are required to evaluate the impact of resistance testing by MALDI-TOF MS using the above-reported methodologies and others yet to be developed. It will be very interesting to observe the clinical impact of these determinations and the situations in which the use of MALDI-TOF MS will be preferred for detecting antimicrobial resistance or when other similar tests, such as immunochromatographic techniques, biochemical tests, and/or molecular tests, should be used. Although further knowledge is needed to optimize the routine application of antimicrobial resistance detection by MALDI-TOF MS, the simplicity, rapidity, and accuracy of this technology will enable MALDI-TOF MS to be included in the workflow of all antimicrobial stewardship programs.
Another potential approach that should be followed up is the detection of high-risk clones associated with multidrug-resistant bacteria. In this approach, the relatedness between clusters of bacteria can be measured faster and more cost-effectively than by molecular methods, and more information regarding antimicrobial resistance can be obtained directly or indirectly, i.e., by detecting biomarkers associated with resistance to antimicrobial agents (144–146). We should also take into account that whole-genome sequencing is evolving very rapidly, especially with nanopore technology (147, 148). The main advantage that MALDI-TOF MS offers is that information will be obtained at no additional cost besides those already incurred for identification purposes. Specific guidelines need to be standardized for the different species in order to interpret spectra for epidemiological purposes (149–152). In the future, we will see if MALDI-TOF MS has the ability to displace molecular techniques in infection control measures.
Automation in microbiology laboratories will enable the application of telebacteriology with expert systems and instrument interfaces and, thus, the creation of new routines (153, 154), for example, the use of robotic incubation and assignment via screen technology for specific follow-up for growing colonies in plate cultures. This technique could even be applied to positive blood cultures, with automated reading at any time of day and with subsequent downstream applications such as MALDI-TOF MS identification and antibiotic resistance detection. Samples could be examined in real time, and those requiring a rapid response in relation to antimicrobial therapy could be identified.
The effectiveness of MALDI-TOF MS makes this technology absolutely necessary in current microbiology laboratories, not only for the identification of microorganisms, as established above, but also for the detection of antimicrobial resistance and other applications. While advances are being made in automated technology, we must gain the maximum clinical benefit in its application. Even more importantly, the optimal workflow must be determined, particularly in relation to those clinical samples for which the greatest benefit will be obtained, the most suitable isolates, and the most suitable time of the day for performing the assays. Other questions that must be addressed are whether it is necessary for procedures to be carried out 24/7 in microbiology laboratories, whether it will be possible to stop routine identification to perform resistance assays, and what type of information is most urgent. One of the main issues that must be resolved is how to prioritize the different techniques that can be performed with MALDI-TOF MS by developing comparative studies of cost-benefit relative to other phenotypic, molecular, or biochemical assays. Although automation will speed up procedures and improve quality standards, the future of most microbiology laboratories will depend on the acquisition of at least two pieces of equipment to facilitate the workflow and to enable handling the growth in demand for diagnostic techniques. However, justifying the acquisition of a second instrument is not easy for nontertiary hospitals with limited budgets. In our experience, the use of MALDI-TOF MS for resistance detection has important value in having a positive impact on health care assistance, so it should justify to a certain extent the slowdown of microorganism identification results. In this regard, we are aware that MALDI-TOF MS cannot replace conventional antimicrobial susceptibility testing, and it must be done in addition to routine testing. This is mainly because the sample processing time is longer for applying MALDI-TOF MS techniques, because the whole process is not automated so far, and because the antibiotic MIC is not known for most of the above-described methods. Besides, it has the inconvenience of requiring technologists’ time. Because of that, we think that the most appropriate position for antimicrobial resistance detection by MALDI-TOF MS is as a rapid technique to solve or confirm individual cases of antimicrobial resistance, such as carbapenemase detection, in a similar way as biochemical methods, like the Carba NP method, and molecular techniques are applied. Thus, the number of tests per day would not delay the results of identification in a significant way. However, as we have cautioned above, each laboratory should evaluate its own situation and evaluate the diagnostic demand for improving health care assistance.
Manufacturers must be able to rapidly adapt to evolution in the laboratory, by improving systems and adapting novel protocols. The time-consuming hands-on portion of the process should need further automation so that we can reduce the preanalytical steps. We think that this is possible by simplifying centrifugation steps and the automated deposition of the analyte on the MALDI-TOF MS target. It is very important that users keep their equipment supervised by expert engineers so that maintenance is performed according to the technical specificities and results are optimized to the maximum level. If manufacturer recommendations are not followed strictly, users could observe poor spectral quality, with variations in signal intensities, signal-to-noise ratios, resolution, and so on, that can finally result in false-positive or -negative results.
In our opinion, the second most interesting application of MALDI-TOF MS in microbiology laboratories is for the detection of high-risk clones harboring specific antimicrobial resistance mechanisms, as it actually simplifies currently used typing methods, and what is more important, no extra hands-on time is needed beyond identification. However, this technique should still be applied with caution. Antimicrobial resistance is a complex process that is the result of, but not limited to, chromosomic mutations, acquisition of resistance determinants, different levels of expression, efflux pumps, and loss of porins, etc. MALDI-TOF MS will not replace any of the preexisting techniques for detection of antimicrobial resistance but will add accurate and valuable information at a very low cost in a limited time.
ACKNOWLEDGMENTS
This work was supported by the Fondo de Investigación Sanitaria (grant PI15/00860 to G.B.), integrated in the Plan Nacional de I+D and funded by the Instituto de Salud Carlos III (ISCIII). The results of this work have been funded by the Spanish Network of Research in Infectious Diseases (REIPI) (grant RD16/0016/0006), integrated in the National Plan for Scientific Research, Development, and Technological Innovation 2013–2016 and funded by the ISCIII-General Subdirection of Assessment and Promotion of the Research-European Regional Development Fund (FEDER) A Way of Making Europe.
We also thank the 6 anonymous reviewers of the manuscript for their suggestions and comments.
Biographies

Marina Oviaño, Ph.D., graduated in Chemistry in the Faculty of Chemistry in Oviedo, Spain. She completed her residence in clinical microbiology at the University Hospital A Coruña and her doctoral studies in the Health Sciences University of A Coruña, Spain. Dr. Oviaño served as a researcher at the National Health System in Spain (2016 to 2017), and at present, she works as a clinical microbiologist at the Microbiology Department of the University Hospital A Coruña. She has been working on research in the field of bacterial resistance to antibiotics, focused on the use of MALDI-TOF MS as a rapid technology for diagnostic use.

Germán Bou received his Ph.D. from the Molecular Biology Center (CSIC)-Autonoma University, Madrid, Spain. He also completed a residence in clinical microbiology at Ramon y Cajal Hospital. Afterwards, he was granted a postdoctoral position with the Fulbright Scholarship Program to work in the Department of Laboratory Medicine and Pathology at Mayo Clinic, Rochester, MN. Dr. Bou then served as an investigator at the National Health System in Spain (2001 to 2005) and as a consultant in clinical microbiology (2005 to 2010), and at present, he is the Head of the Microbiology Department of the University Hospital A Coruña. Since 2009, he has been associate professor of medical microbiology at Santiago of Compostela University. Dr. Bou’s research focuses on the understanding of the molecular basis for antimicrobial resistance in human pathogens, developing rapid tests for detecting resistant organisms, and designing and developing bacterial vaccines. So far, he has published more than 200 international peer-reviewed manuscripts on these topics as well as obtained 7 related patents. Recently, he was appointed with the honorary title of ESCMID fellow for professional excellence and service to society.
REFERENCES
- 1.Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, Raoult D. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 2.Singhal N, Kumar M, Kanaujia PK, Virdi JS. 2015. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol 6:791. doi: 10.3389/fmicb.2015.00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou M, Yang Q, Kudinha T, Sun L, Zhang R, Liu C, Yu S, Xiao M, Kong F, Zhao Y, Xu YC. 2017. An improved in-house MALDI-TOF MS protocol for direct cost-effective identification of pathogens from blood cultures. Front Microbiol 8:1824. doi: 10.3389/fmicb.2017.01824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodríguez-Sánchez B, Sánchez-Carrillo C, Ruiz A, Marín M, Cercenado E, Rodríguez-Créixems M, Bouza E. 2014. Direct identification of pathogens from positive blood cultures using matrix-assisted laser desorption-ionization time-of-flight mass spectrometry. Clin Microbiol Infect 20:O421–O427. doi: 10.1111/1469-0691.12455. [DOI] [PubMed] [Google Scholar]
- 5.Tran A, Alby K, Kerr A, Jones M, Gilligan PH. 2015. Cost savings realized by implementation of routine microbiological identification by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 53:2473–2479. doi: 10.1128/JCM.00833-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dhiman N, Hall L, Wohlfiel SL, Buckwalter SP, Wengenack NL. 2011. Performance and cost analysis of matrix-assisted laser desorption ionization–time of flight mass spectrometry for routine identification of yeast. J Clin Microbiol 49:1614–1616. doi: 10.1128/JCM.02381-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lévesque S, Dufresne PJ, Soualhine H, Domingo MC, Bekal S, Lefebvre B, Tremblay C. 2015. A side by side comparison of Bruker biotyper and VITEK MS: utility of MALDI-TOF MS technology for microorganism identification in a public health reference laboratory. PLoS One 10:e0144878. doi: 10.1371/journal.pone.0144878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee TF, Lee H, Chen CM, Du SH, Cheng YC, Hsu CC, Chung MY, Teng SH, Teng LJ, Hsueh PR. 2013. Comparison of the accuracy of matrix-assisted laser desorption ionization–time of flight mass spectrometry with that of other commercial identification systems for identifying Staphylococcus saprophyticus in urine. J Clin Microbiol 51:1563–1566. doi: 10.1128/JCM.00261-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oviaño M, Rodríguez-Sánchez B, Gómara M, Alcalá L, Zvezdanova E, Ruíz A, Velasco D, Gude MJ, Bouza E, Bou G. 2017. Direct identification of clinical pathogens from liquid culture media by MALDI-TOF MS analysis. Clin Microbiol Infect 24:624–629. doi: 10.1016/j.cmi.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 10.Seah VXF, Ong RYL, Lim ASY, Chong CY, Tan NWH, Thoon KC. 2017. Impact of a carbapenem antimicrobial stewardship program on patient outcomes. Antimicrob Agents Chemother 61:e00736-17. doi: 10.1128/AAC.00736-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morency-Potvin P, Schwartz DN, Weinstein RA. 2017. Antimicrobial stewardship: how the microbiology laboratory can right the ship. Clin Microbiol Rev 30:381–407. doi: 10.1128/CMR.00066-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goff DA, Jankowski C, Tenover FC. 2012. Using rapid diagnostic tests to optimize antimicrobial selection in antimicrobial stewardship programs. Pharmacotherapy 32:677–687. doi: 10.1002/j.1875-9114.2012.01137.x. [DOI] [PubMed] [Google Scholar]
- 13.Maurer FP, Christner M, Hentschke M, Rohde H. 2017. Advances in rapid identification and susceptibility testing of bacteria in the clinical microbiology laboratory: implications for patient care and antimicrobial stewardship programs. Infect Dis Rep 9:6839. doi: 10.4081/idr.2017.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howden BP, Davies JK, Johnson PD, Stinear TP, Grayson ML. 2010. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin Microbiol Rev 23:99–139. doi: 10.1128/CMR.00042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hrabák J, Chudáčková E, Papagiannitsis CC. 2014. Detection of carbapenemases in Enterobacteriaceae: a challenge for diagnostic microbiological laboratories. Clin Microbiol Infect 20:839–853. doi: 10.1111/1469-0691.12678. [DOI] [PubMed] [Google Scholar]
- 16.Dortet L, Cuzon G, Plésiat P, Naas T. 2016. Prospective evaluation of an algorithm for the phenotypic screening of carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother 71:135–140. doi: 10.1093/jac/dkv308. [DOI] [PubMed] [Google Scholar]
- 17.Nordmann P, Poirel L. 2014. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect 20:821–830. doi: 10.1111/1469-0691.12719. [DOI] [PubMed] [Google Scholar]
- 18.Mohareb AM, Dugas AF, Hsieh YH. 2016. Changing epidemiology and management of infectious diseases in US EDs. Am J Emerg Med 34:1059–1065. doi: 10.1016/j.ajem.2016.03.024. [DOI] [PubMed] [Google Scholar]
- 19.Ellington MJ, Findlay J, Hopkins KL, Meunier D, Alvarez-Buylla A, Horner C, McEwan A, Guiver M, McCrae LX, Woodford N, Hawkey P. 2016. Multicentre evaluation of a real-time PCR assay to detect genes encoding clinically relevant carbapenemases in cultured bacteria. Int J Antimicrob Agents 47:151–154. doi: 10.1016/j.ijantimicag.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 20.Lee TD, Adie K, McNabb A, Purych D, Mannan K, Azana R, Ng C, Tang P, Hoang LM. 2015. Rapid detection of KPC, NDM, and OXA-48-like carbapenemases by real-time PCR from rectal swab surveillance samples. J Clin Microbiol 53:2731–2733. doi: 10.1128/JCM.01237-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burillo A, Marín M, Cercenado E, Ruiz-Carrascoso G, Pérez-Granda MJ, Oteo J, Bouza E. 2016. Evaluation of the Xpert Carba-R (Cepheid) assay using contrived bronchial specimens from patients with suspicion of ventilator-associated pneumonia for the detection of prevalent carbapenemases. PLoS One 11:e0168473. doi: 10.1371/journal.pone.0168473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yarbrough ML, Warren DK, Allen K, Burkholder D, Daum R, Donskey C, Knaack D, LaMarca A, May L, Miller LG, Parenti DM, Peterson L, Tan TY, Widen R, Hernandez DR, Wolk DM, Burnham CA. 2017. Multicenter evaluation of the Xpert MRSA NxG assay for detection of methicillin-resistant Staphylococcus aureus in nasal swabs. J Clin Microbiol 56:e01381-17. doi: 10.1128/JCM.01381-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antonelli A, Arena F, Giani T, Colavecchio OL, Valeva SV, Paule S, Boleij P, Rossolini GM. 2016. Performance of the BD MAX instrument with Check-Direct CPE real-time PCR for the detection of carbapenemase genes from rectal swabs, in a setting with endemic dissemination of carbapenemase-producing Enterobacteriaceae. Diagn Microbiol Infect Dis 86:30–34. doi: 10.1016/j.diagmicrobio.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 24.Dortet L, Agathine A, Naas T, Cuzon G, Poirel L, Nordmann P. 2015. Evaluation of the RAPIDEC CARBA NP, the Rapid CARB Screen and the Carba NP test for biochemical detection of carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother 70:3014–3022. doi: 10.1093/jac/dkv213. [DOI] [PubMed] [Google Scholar]
- 25.Nordmann P, Jayol A, Dobias J, Poirel L. 2017. Rapid aminoglycoside NP test for rapid detection of multiple aminoglycoside resistance in Enterobacteriaceae. J Clin Microbiol 55:1074–1079. doi: 10.1128/JCM.02107-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jayol A, Dubois V, Poirel L, Nordmann P. 2016. Rapid detection of polymyxin-resistant Enterobacteriaceae from blood cultures. J Clin Microbiol 54:2273–2277. doi: 10.1128/JCM.00918-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chong PM, McCorrister SJ, Unger MS, Boyd DA, Mulvey MR, Westmacott GR. 2015. MALDI-TOF MS detection of carbapenemase activity in clinical isolates of Enterobacteriaceae spp., Pseudomonas aeruginosa, and Acinetobacter baumannii compared against the Carba-NP assay. J Microbiol Methods 111:21–23. doi: 10.1016/j.mimet.2015.01.024. [DOI] [PubMed] [Google Scholar]
- 28.Kabir MH, Meunier D, Hopkins KL, Giske CG, Woodford N. 2016. A two-centre evaluation of RAPIDEC CARBA NP for carbapenemase detection in Enterobacteriaceae, Pseudomonas aeruginosa and Acinetobacter spp. J Antimicrob Chemother 71:1213–1216. doi: 10.1093/jac/dkv468. [DOI] [PubMed] [Google Scholar]
- 29.Österblad M, Lindholm L, Jalava J. 2016. Evaluation of two commercial carbapenemase gene assays, the Rapidec Carba NP test and the in-house Rapid Carba NP test, on bacterial cultures. J Antimicrob Chemother 71:2057–2059. doi: 10.1093/jac/dkw077. [DOI] [PubMed] [Google Scholar]
- 30.Erdem F, Abulaila A, Aktas Z, Oncul O. 2017. Comparison of the novel OXA-48 and KPC K-SeT assay, and Blue-Carba test for the detection of carbapenemase-producing Enterobacteriaceae using PCR as a reference method. Clin Lab 63:515–522. doi: 10.7754/Clin.Lab.2016.160911. [DOI] [PubMed] [Google Scholar]
- 31.Pasteran F, Denorme L, Ote I, Gomez S, De Belder D, Glupczynski Y, Bogaerts P, Ghiglione B, Power P, Mertens P, Corso A. 2016. Rapid identification of OXA-48 and OXA-163 subfamilies in carbapenem-resistant Gram-negative bacilli with a novel immunochromatographic lateral flow assay. J Clin Microbiol 54:2832–2836. doi: 10.1128/JCM.01175-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wareham DH, Shah R, Betts JW, Phee LM, Momin MH. 2016. Evaluation of an immunochromatographic lateral flow assay (OXA-48 K-SeT) for rapid detection of OXA-48 like carbapenemases in Enterobacteriaceae. J Clin Microbiol 54:471–473. doi: 10.1128/JCM.02900-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glupczynski Y, Jousset A, Evrard S, Bonnin RA, Huang TD, Dortet L, Bogaerts P, Naas T. 2017. Prospective evaluation of the OKN K-SeT assay, a new multiplex immunochromatographic test for the rapid detection of OXA-48-like, KPC and NDM carbapenemases. J Antimicrob Chemother 72:1955–1960. doi: 10.1093/jac/dkx089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bogaerts P, Yunus S, Massart M, Huang TD, Glupczynski Y. 2016. Evaluation of the BYG Carba test, a new electrochemical assay for rapid laboratory detection of carbapenemase-producing Enterobacteriaceae. J Clin Microbiol 54:349–358. doi: 10.1128/JCM.02404-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bogaerts P, Oueslati S, Meunier D, Nonhoff C, Yunus S, Massart M, Denis O, Woodford N, Hopkins KL, Naas T, Dortet L, Huang TD, Glupczynski Y. 2017. Multicentre evaluation of the BYG Carba v2.0 test, a simplified electrochemical assay for the rapid laboratory detection of carbapenemase-producing Enterobacteriaceae. Sci Rep 7:9937. doi: 10.1038/s41598-017-09820-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noël A, Huang TD, Berhin C, Hoebeke M, Bouchahrouf W, Yunus S, Bogaerts P, Glupczynski Y. 2017. Comparative evaluation of four phenotypic tests for detection of carbapenemase-producing Gram-negative bacteria. J Clin Microbiol 55:510–518. doi: 10.1128/JCM.01853-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen JL, Xu YH, She TT. 2015. LC-MS/MS: a rapid and simple new method for the determination of carbapenem β-lactamases. Genet Mol Res 14:14457–14468. doi: 10.4238/2015.November.18.8. [DOI] [PubMed] [Google Scholar]
- 38.Trip H, Mende K, Majchrzykiewicz-Koehorst JA, Sedee NJ, Hulst AG, Jansen HJ, Murray CK, Paauw A. 2015. Simultaneous identification of multiple β-lactamases in Acinetobacter baumannii in relation to carbapenem and ceftazidime resistance, using liquid chromatography-tandem mass spectrometry. J Clin Microbiol 53:1927–1930. doi: 10.1128/JCM.00620-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carricajo A, Verhoeven PO, Guezzou S, Fonsale N, Aubert G. 2014. Detection of carbapenemase-producing bacteria by using an ultraperformance liquid chromatography-tandem mass spectrometry method. Antimicrob Agents Chemother 58:1231–1234. doi: 10.1128/AAC.01540-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carvalhaes CG, Cayô R, Assis DM, Martins ER, Juliano L, Juliano MA, Gales AC. 2013. Detection of SPM-1-producing Pseudomonas aeruginosa and class D β-lactamase-producing Acinetobacter baumannii isolates by use of liquid chromatography-mass spectrometry and matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 51:287–290. doi: 10.1128/JCM.02365-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Opota O, Croxatto A, Prod’hom G, Greub G. 2015. Blood culture-based diagnosis of bacteremia: state of the art. Clin Microbiol Infect 21:313–322. doi: 10.1016/j.cmi.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 42.Yan Y, He Y, Maier T, Quinn C, Shi G, Li H, Stratton CW, Kostrzewa M, Tang YW. 2011. Improved identification of yeast species directly from positive blood culture media by combining Sepsityper specimen processing and Microflex analysis with the matrix-assisted laser desorption ionization Biotyper system. J Clin Microbiol 49:2528–2532. doi: 10.1128/JCM.00339-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferreira L, Sánchez-Juanes F, González-Avila M, Cembrero-Fuciños D, Herrero-Hernández A, González-Buitrago JM, Muñoz-Bellido JL. 2010. Direct identification of urinary tract pathogens from urine samples by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 48:2110–2115. doi: 10.1128/JCM.02215-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oviaño M, Ramírez CL, Barbeyto LP, Bou G. 2017. Rapid direct detection of carbapenemase-producing Enterobacteriaceae in clinical urine samples by MALDI-TOF MS analysis. J Antimicrob Chemother 72:1350–1354. doi: 10.1093/jac/dkw579. [DOI] [PubMed] [Google Scholar]
- 45.Burckhardt I, Zimmermann S. 2011. Using matrix-assisted laser desorption ionization–time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J Clin Microbiol 49:3321–3324. doi: 10.1128/JCM.00287-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hrabák J, Walková R, Studentová V, Chudácková E, Bergerová T. 2011. Carbapenemase activity detection by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 49:3222–3227. doi: 10.1128/JCM.00984-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sparbier K, Schubert S, Weller U, Boogen C, Kostrzewa M. 2012. Matrix-assisted laser desorption ionization–time of flight mass spectrometry-based functional assay for rapid detection of resistance against β-lactam antibiotics. J Clin Microbiol 50:927–937. doi: 10.1128/JCM.05737-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Page MI. 1999. The reactivity of beta-lactams, the mechanism of catalysis and the inhibition of beta-lactamases. Curr Pharm Des 5:895–913. [PubMed] [Google Scholar]
- 49.Naas T, Nordmann P. 1999. OXA-type beta-lactamases. Curr Pharm Des 5:865–879. [PubMed] [Google Scholar]
- 50.Shaw KJ, Rather PN, Hare RS, Miller GH. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev 57:138–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jacoby GA, Strahilevitz J, Hooper DC. 2014. Plasmid-mediated quinolone resistance. Microbiol Spectr 2:PLAS-0006-2013. doi: 10.1128/microbiolspec.PLAS-0006-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hrabák J, Chudácková E, Walková R. 2013. Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry for detection of antibiotic resistance mechanisms: from research to routine diagnosis. Clin Microbiol Rev 26:103–114. doi: 10.1128/CMR.00058-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oviaño M, Fernández B, Fernández A, Barba MJ, Mouriño C, Bou G. 2014. Rapid detection of Enterobacteriaceae producing extended spectrum beta-lactamases directly from positive blood cultures by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Clin Microbiol Infect 20:1146–1157. doi: 10.1111/1469-0691.12729. [DOI] [PubMed] [Google Scholar]
- 54.Jung JS, Popp C, Sparbier K, Lange C, Kostrzewa M, Schubert S. 2014. Evaluation of matrix-assisted laser desorption ionization–time of flight mass spectrometry for rapid detection of β-lactam resistance in Enterobacteriaceae derived from blood cultures. J Clin Microbiol 52:924–930. doi: 10.1128/JCM.02691-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oviaño M, Gómara M, Barba MJ, Revillo MJ, Barbeyto LP, Bou G. 2017. Towards the early detection of β-lactamase-producing Enterobacteriaceae by MALDI-TOF MS analysis. J Antimicrob Chemother 72:2259–2262. doi: 10.1093/jac/dkx127. [DOI] [PubMed] [Google Scholar]
- 56.Huang TD, Poirel L, Bogaerts P, Berhin C, Nordmann P, Glupczynski Y. 2014. Temocillin and piperacillin/tazobactam resistance by disc diffusion as antimicrobial surrogate markers for the detection of carbapenemase-producing Enterobacteriaceae in geographical areas with a high prevalence of OXA-48 producers. J Antimicrob Chemother 69:445–450. doi: 10.1093/jac/dkt367. [DOI] [PubMed] [Google Scholar]
- 57.Poirel L, Potron A, Nordmann P. 2012. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother 67:1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 58.Leonard DA, Hujer AM, Smith BA, Schneider KD, Bethel CR, Hujer KM, Bonomo RA. 2008. The role of OXA-1 β-lactamase Asp (66) in the stabilization of the active-site carbamate group and in substrate turnover. Biochem J 410:455–462. doi: 10.1042/BJ20070573. [DOI] [PubMed] [Google Scholar]
- 59.Verma V, Testero SA, Amini K, Wei W, Liu J, Balachandran N, Monoharan T, Stynes S, Kotra LP, Golemi-Kotra D. 2011. Hydrolytic mechanism of OXA-58 enzyme, a carbapenem hydrolyzing class D β-lactamase from Acinetobacter baumannii. J Biol Chem 286:37292–37303. doi: 10.1074/jbc.M111.280115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Papagiannitsis CC, Študentová V, Izdebski R, Oikonomou O, Pfeifer Y, Petinaki E, Hrabák J. 2015. Matrix-assisted laser desorption ionization–time of flight mass spectrometry meropenem hydrolysis assay with NH4HCO3, a reliable tool for direct detection of carbapenemase activity. J Clin Microbiol 53:1731–1735. doi: 10.1128/JCM.03094-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lasserre C, De Saint Martin L, Cuzon G, Bogaerts P, Lamar E, Glupczynski Y, Naas T, Tandé D. 2015. Efficient detection of carbapenemase activity in Enterobacteriaceae by matrix-assisted laser desorption ionization–time of flight mass spectrometry in less than 30 minutes. J Clin Microbiol 53:2163–2171. doi: 10.1128/JCM.03467-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kempf M, Bakour S, Flaudrops C, Berrazeg M, Brunel JM, Drissi M, Mesli E, Touati A, Rolain JM. 2012. Rapid detection of carbapenem resistance in Acinetobacter baumannii using matrix-assisted laser desorption ionization-time of flight mass spectrometry. PLoS One 7:e31676. doi: 10.1371/journal.pone.0031676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rotova V, Papagiannitsis CC, Skalova A, Chudejova K, Hrabak J. 2017. Comparison of imipenem and meropenem antibiotics for the MALDI-TOF MS detection of carbapenemase activity. J Microbiol Methods 137:30–33. doi: 10.1016/j.mimet.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 64.Monteferrante CG, Sultan S, Ten Kate MT, Dekker LJ, Sparbier K, Peer M, Kostzrewa M, Luider TM, Goessens WH, Burgers PC. 2016. Evaluation of different pretreatment protocols to detect accurately clinical carbapenemase-producing Enterobacteriaceae by MALDI-TOF. J Antimicrob Chemother 71:2856–2867. doi: 10.1093/jac/dkw208. [DOI] [PubMed] [Google Scholar]
- 65.Hrabák J, Studentová V, Walková R, Zemlicková H, Jakubu V, Chudácková E, Gniadkowski M, Pfeifer Y, Perry JD, Wilkinson K, Bergerová T. 2012. Detection of NDM-1, VIM-1, KPC, OXA-48, and OXA-162 carbapenemases by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 50:2441–2443. doi: 10.1128/JCM.01002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oviaño M, Sparbier K, Barba MJ, Kostrzewa M, Bou G. 2016. Universal protocol for the rapid automated detection of carbapenem-resistant gram-negative bacilli directly from blood cultures by matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF/MS). Int J Antimicrob Agents 48:655–660. doi: 10.1016/j.ijantimicag.2016.08.024. [DOI] [PubMed] [Google Scholar]
- 67.Carvalhaes CG, da Silva AC, Streling AP, Cayô R, Rockstroh AC, Machado AM, Gales AC. 2015. Detection of carbapenemase activity using VITEK MS: interplay of carbapenemase type and period of incubation. J Med Microbiol 64:946–947. doi: 10.1099/jmm.0.000102. [DOI] [PubMed] [Google Scholar]
- 68.Bishop B, Geffen Y, Plaut A, Kassis O, Bitterman R, Paul M, Neuberger A. 2017. The use of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for rapid bacterial identification in patients with smear-positive bacterial meningitis. Clin Microbiol Infect 24:171–174. doi: 10.1016/j.cmi.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 69.Robicsek A, Strahilevitz J, Jacoby GA, Macielag M, Abbanat D, Park CH, Bush K, Hooper DC. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat Med 12:83–88. doi: 10.1038/nm1347. [DOI] [PubMed] [Google Scholar]
- 70.Warburg G, Korem M, Robicsek A, Engelstein D, Moses AE, Block C, Strahilevitz J. 2009. Changes in AAC(6′)-Ib-cr prevalence and fluoroquinolone resistance in nosocomial isolates of Escherichia coli collected from 1991 through 2005. Antimicrob Agents Chemother 53:1268–1270. doi: 10.1128/AAC.01300-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Briales A, Rodríguez-Martínez JM, Velasco C, de Alba PD, Rodríguez-Baño J, Martínez-Martínez L, Pascual A. 2012. Prevalence of plasmid mediated quinolone resistance determinants qnr and AAC(6′)-Ib-cr in Escherichia coli and Klebsiella pneumoniae producing extended-spectrum β-lactamases in Spain. Int J Antimicrob Agents 39:431–434. doi: 10.1016/j.ijantimicag.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 72.Pardo CA, Tan RN, Hennequin C, Beyrouthy R, Bonnet R, Robin F. 2016. Rapid detection of AAC(6′)-Ib-cr production using a MALDI-TOF MS strategy. Eur J Clin Microbiol Infect Dis 35:2047–2051. doi: 10.1007/s10096-016-2762-1. [DOI] [PubMed] [Google Scholar]
- 73.Oviaño M, Rodríguez-Martínez JM, Pascual Á, Bou G. 2017. Rapid detection of the plasmid-mediated quinolone resistance determinant AAC(6′)-Ib-cr in Enterobacteriaceae by MALDI-TOF MS analysis. J Antimicrob Chemother 72:1074–1080. doi: 10.1093/jac/dkw552. [DOI] [PubMed] [Google Scholar]
- 74.Oviaño M, Gómara M, Barba MJ, Sparbier K, Pascual Á, Bou G. 2017. Quantitative and automated MALDI-TOF MS-based detection of the plasmid-mediated quinolone resistance determinant AAC(6′)-Ib-cr in Enterobacteriaceae. J Antimicrob Chemother 72:2952–2954. doi: 10.1093/jac/dkx218. [DOI] [PubMed] [Google Scholar]
- 75.Fernández-Martínez M, Miró E, Ortega A, Bou G, González-López JJ, Oliver A, Pascual A, Cercenado E, Oteo J, Martínez-Martínez L, Navarro F, Spanish Network for the Research in Infectious Diseases. 2015. Molecular identification of aminoglycoside-modifying enzymes in clinical isolates of Escherichia coli resistant to amoxicillin/clavulanic acid isolated in Spain. Int J Antimicrob Agents 46:157–163. doi: 10.1016/j.ijantimicag.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 76.Haldorsen BC, Simonsen GS, Sundsfjord A, Samuelsen O, Norwegian Study Group on Aminoglycoside Resistance. 2014. Increased prevalence of aminoglycoside resistance in clinical isolates of Escherichia coli and Klebsiella spp. in Norway is associated with the acquisition of AAC(3)-II and AAC(6')-Ib. Diagn Microbiol Infect Dis 78:66–69. doi: 10.1016/j.diagmicrobio.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 77.Yao H, Liu D, Wang Y, Zhang Q, Shen Z. 2017. High prevalence and predominance of the aph(2″)-If gene conferring aminoglycoside resistance in Campylobacter. Antimicrob Agents Chemother 61:e00112-17. doi: 10.1128/AAC.00112-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Davies J, Wright GD. 1997. Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol 5:234–240. doi: 10.1016/S0966-842X(97)01033-0. [DOI] [PubMed] [Google Scholar]
- 79.Ramirez MS, Tolmasky ME. 2010. Aminoglycoside modifying enzymes. Drug Resist Updat 13:151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zimmermann S. 2013. Detecting aminoglycoside resistance by mass spectrometry, poster P-1549. Abstr 23rd Eur Congr Clin Microbiol Infect Dis, Berlin, Germany. [Google Scholar]
- 81.Josten M, Dischinger J, Szekat C, Reif M, Al-Sabti N, Sahl HG, Parcina M, Bekeredjian-Ding I, Bierbaum G. 2014. Identification of agr-positive methicillin-resistant Staphylococcus aureus harbouring the class A mec complex by MALDI-TOF mass spectrometry. Int J Med Microbiol 304:1018–1023. doi: 10.1016/j.ijmm.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 82.Lau AF, Wang H, Weingarten RA, Drake SK, Suffredini AF, Garfield MK, Chen Y, Gucek M, Youn JH, Stock F, Tso H, DeLeo J, Cimino JJ, Frank KM, Dekker JP. 2014. A rapid matrix-assisted laser desorption ionization–time of flight mass spectrometry-based method for single-plasmid tracking in an outbreak of carbapenem-resistant Enterobacteriaceae. J Clin Microbiol 52:2804–2812. doi: 10.1128/JCM.00694-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wybo I, De Bel A, Soetens O, Echahidi F, Vandoorslaer K, Van Cauwenbergh M, Piérard D. 2011. Differentiation of cfiA-negative and cfiA-positive Bacteroides fragilis isolates by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 49:1961–1964. doi: 10.1128/JCM.02321-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Edwards-Jones V, Claydon MA, Evason DJ, Walker J, Fox AJ, Gordon DB. 2000. Rapid discrimination between methicillin-sensitive and methicillin-resistant Staphylococcus aureus by intact cell mass spectrometry. J Med Microbiol 49:295–300. doi: 10.1099/0022-1317-49-3-295. [DOI] [PubMed] [Google Scholar]
- 85.Du Z, Yang R, Guo Z, Song Y, Wang J. 2002. Identification of Staphylococcus aureus and determination of its methicillin resistance by matrix assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Chem 74:5487–5491. doi: 10.1021/ac020109k. [DOI] [PubMed] [Google Scholar]
- 86.Bernardo K, Pakulat N, Macht M, Krut O, Seifert H, Fleer S, Hünger F, Krönke M. 2002. Identification and discrimination of Staphylococcus aureus strains using matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Proteomics 2:747–753. doi:. [DOI] [PubMed] [Google Scholar]
- 87.Wang YR, Chen Q, Cui SH, Li FQ. 2013. Characterization of Staphylococcus aureus isolated from clinical specimens by matrix assisted laser desorption/ionization time-of-flight mass spectrometry. Biomed Environ Sci 26:430–436. doi: 10.3967/0895-3988.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 88.Szabados F, Kaase M, Anders A, Gatermann SG. 2012. Identical MALDI TOF MS-derived peak profiles in a pair of isogenic SCCmec-harboring and SCCmec-lacking strains of Staphylococcus aureus. J Infect 65:400–405. doi: 10.1016/j.jinf.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 89.Lasch P, Fleige C, Stämmler M, Layer F, Nübel U, Witte W, Werner G. 2014. Insufficient discriminatory power of MALDI-TOF mass spectrometry for typing of Enterococcus faecium and Staphylococcus aureus isolates. J Microbiol Methods 100:58–69. doi: 10.1016/j.mimet.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 90.Rhoads DD, Wang H, Karichu J, Richter SS. 2016. The presence of a single MALDI-TOF mass spectral peak predicts methicillin resistance in staphylococci. Diagn Microbiol Infect Dis 86:257–261. doi: 10.1016/j.diagmicrobio.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 91.Lee AS, de Lencastre H, Garau J, Kluytmans J, Malhotra-Kumar S, Peschel A, Harbarth S. 2018. Methicillin-resistant Staphylococcus aureus. Nat Rev Dis Primers 4:18033. doi: 10.1038/nrdp.2018.33. [DOI] [PubMed] [Google Scholar]
- 92.Partridge SR. 2014. Tn4401 carrying blaKPC is inserted within another insertion in pKpQIL and related plasmids. J Clin Microbiol 52:4448–4449. doi: 10.1128/JCM.02426-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Youn JH, Drake SK, Weingarten RA, Frank KM, Dekker JP, Lau AF. 2016. Clinical performance of a matrix-assisted laser desorption ionization–time of flight mass spectrometry method for detection of certain blaKPC-containing plasmids. J Clin Microbiol 54:35–42. doi: 10.1128/JCM.01643-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gaibani P, Galea A, Fagioni M, Ambretti S, Sambri V, Landini MP. 2016. Evaluation of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of KPC-producing Klebsiella pneumoniae. J Clin Microbiol 54:2609–2613. doi: 10.1128/JCM.01242-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Papagiannitsis CC, Kotsakis SD, Tuma Z, Gniadkowski M, Miriagou V, Hrabak J. 2014. Identification of CMY-2-type cephalosporinases in clinical isolates of Enterobacteriaceae by MALDI-TOF MS. Antimicrob Agents Chemother 58:2952–2957. doi: 10.1128/AAC.02418-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sóki J, Edwards R, Hedberg M, Fang H, Nagy E, Nord CE, ESCMID Study Group on Antimicrobial Resistance in Anaerobic Bacteria. 2006. Examination of cfiA-mediated carbapenem resistance in Bacteroides fragilis strains from a European antibiotic susceptibility survey. Int J Antimicrob Agents 28:497–502. doi: 10.1016/j.ijantimicag.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 97.Ferløv-Schwensen SA, Sydenham TV, Hansen KCM, Hoegh SV, Justesen US. 2017. Prevalence of antimicrobial resistance and the cfiA resistance gene in Danish Bacteroides fragilis group isolates since 1973. Int J Antimicrob Agents 50:552–556. doi: 10.1016/j.ijantimicag.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 98.Schwensen SA, Acar Z, Sydenham TV, Johansson ÅC, Justesen US. 2017. Phenotypic detection of the cfiA metallo-β-lactamase in Bacteroides fragilis with the meropenem-EDTA double-ended Etest and the ROSCO KPC/MBL Confirm kit. J Antimicrob Chemother 72:437–440. doi: 10.1093/jac/dkw436. [DOI] [PubMed] [Google Scholar]
- 99.Nagy E, Becker S, Sóki J, Urbán E, Kostrzewa M. 2011. Differentiation of division I (cfiA-negative) and division II (cfiA-positive) Bacteroides fragilis strains by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Med Microbiol 60:1584–1590. doi: 10.1099/jmm.0.031336-0. [DOI] [PubMed] [Google Scholar]
- 100.Zhang Y, Jiang X, Wang Y, Li G, Tian Y, Liu H, Ai F, Ma Y, Wang B, Ruan F, Rajakumar K. 2014. Contribution of β-lactamases and porin proteins OmpK35 and OmpK36 to carbapenem resistance in clinical isolates of KPC-2-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 58:1214–1217. doi: 10.1128/AAC.02045-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rumbo C, Gato E, López M, Ruiz de Alegría C, Fernández-Cuenca F, Martínez-Martínez L, Vila J, Pachón J, Cisneros JM, Rodríguez-Baño J, Pascual A, Bou G, Tomás M, Spanish Group of Nosocomial Infections and Mechanisms of Action and Resistance to Antimicrobials, Spanish Society of Clinical Microbiology and Infectious Diseases, Spanish Network for Research in Infectious Diseases. 2013. Contribution of efflux pumps, porins, and β-lactamases to multidrug resistance in clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 57:5247–5257. doi: 10.1128/AAC.00730-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Doménech-Sánchez A, Martínez-Martínez L, Hernández-Allés S, del Carmen Conejo M, Pascual A, Tomás JM, Albertí S, Benedí VJ. 2003. Role of Klebsiella pneumoniae OmpK35 porin in antimicrobial resistance. Antimicrob Agents Chemother 47:3332–3335. doi: 10.1128/AAC.47.10.3332-3335.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hamzaoui Z, Ocampo-Sosa A, Martinez MF, Landolsi S, Ferjani S, Maamar E, Saidani M, Slim A, Martinez-Martinez L, Boubaker IB. 2 April 2018. Role of association of OmpK35 and OmpK36 alteration and bla(ESBL) and/or bla(AmpC) in conferring carbapenem resistance among non-producing carbapenemase Klebsiella pneumoniae. Int J Antimicrob Agents doi: 10.1016/j.ijantimicag.2018.03.020. [DOI] [PubMed] [Google Scholar]
- 104.Hamzaoui Z, Ocampo-Sosa A, Maamar E, Fernandez Martinez M, Ferjani S, Hammami S, Harbaoui S, Genel N, Arlet G, Saidani M, Slim A, Boutiba-Ben Boubaker I, Martinez-Martinez L. 2018. An outbreak of NDM-1-producing Klebsiella pneumoniae, associated with OmpK35 and OmpK36 porin loss in Tunisia. Microb Drug Resist 24:1137–1147. doi: 10.1089/mdr.2017.0165. [DOI] [PubMed] [Google Scholar]
- 105.Cai JC, Hu YY, Zhang R, Zhou HW, Chen GX. 2012. Detection of OmpK36 porin loss in Klebsiella spp. by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 50:2179–2182. doi: 10.1128/JCM.00503-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hu YY, Cai JC, Zhou HW, Zhang R, Chen GX. 2015. Rapid detection of porins by matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Front Microbiol 6:784. doi: 10.3389/fmicb.2015.00784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 108.Bartolleti F, Seco BMS, Capuzzo Dos Santos C, Felipe CB, Lemo MEB, Alves TDS, Passadore LF, Mimica MJ, Sampaio SCF, Zavascki AP, Sampaio JLM. 2016. Polymyxin B resistance in carbapenem-resistant Klebsiella pneumoniae, São Paulo, Brazil. Emerg Infect Dis 22:1849–1851. doi: 10.3201/eid2210.160695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dortet L, Filloux A, Larrouy-Maumus G. 2017. Rapid detection and discrimination of plasmid- and chromosome-mediated resistance to polymyxins in Enterobacteriaceae using MALDI-TOF, oral session OS112. Abstr 27th Eur Congr Clin Microbiol Infect Dis, Vienna, Austria. [Google Scholar]
- 110.Beceiro A, Llobet E, Aranda J, Bengoechea JA, Doumith M, Hornsey M, Dhanji H, Chart H, Bou G, Livermore DM, Woodford N. 2011. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob Agents Chemother 55:3370–3379. doi: 10.1128/AAC.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Santiso R, Tamayo M, Fernández JL, del Carmen Fernández M, Molina F, Villanueva R, Gosálvez J, Bou G. 2009. Rapid and simple determination of ciprofloxacin resistance in clinical strains of Escherichia coli. J Clin Microbiol 47:2593–2595. doi: 10.1128/JCM.00367-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li H, Balan P, Vertes A. 2016. Molecular imaging of growth, metabolism, and antibiotic inhibition in bacterial colonies by laser ablation electrospray ionization mass spectrometry. Angew Chem Int Ed Engl 55:15035–15039. doi: 10.1002/anie.201607751. [DOI] [PubMed] [Google Scholar]
- 113.Sparbier K, Lange C, Jung J, Wieser A, Schubert S, Kostrzewa M. 2013. MALDI Biotyper-based rapid resistance detection by stable-isotope labeling. J Clin Microbiol 51:3741–3748. doi: 10.1128/JCM.01536-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lange C, Schubert S, Jung J, Kostrzewa M, Sparbier K. 2014. Quantitative matrix-assisted laser desorption ionization–time of flight mass spectrometry for rapid resistance detection. J Clin Microbiol 52:4155–4162. doi: 10.1128/JCM.01872-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jung JS, Hamacher C, Gross B, Sparbier K, Lange C, Kostrzewa M, Schubert S. 2016. Evaluation of a semiquantitative matrix-assisted laser desorption ionization–time of flight mass spectrometry method for rapid antimicrobial susceptibility testing of positive blood cultures. J Clin Microbiol 54:2820–2824. doi: 10.1128/JCM.01131-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Maxson T, Taylor-Howell CL, Minogue TD. 2017. Semi-quantitative MALDI-TOF for antimicrobial susceptibility testing in Staphylococcus aureus. PLoS One 12:e0183899. doi: 10.1371/journal.pone.0183899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Clinical and Laboratory Standards Institute. 2016. Performance standards for antimicrobial susceptibility testing, 26th ed. Document M100-S26D. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 118.Idelevich EA, Sparbier K, Kostrzewa M, Becker K. 2018. Rapid detection of antibiotic resistance by MALDI-TOF mass spectrometry using a novel direct-on-target microdroplet growth assay. Clin Microbiol Infect 24:738–743. doi: 10.1016/j.cmi.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 119.Ceyssens PJ, Soetaert K, Timke M, Van den Bossche A, Sparbier K, De Cremer K, Kostrzewa M, Hendrickx M, Mathys V. 2017. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for combined species identification and drug sensitivity testing in mycobacteria. J Clin Microbiol 55:624–634. doi: 10.1128/JCM.02089-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shallom SJ, Moura NS, Olivier KN, Sampaio EP, Holland SM, Zelazny AM. 2015. New real-time PCR assays for detection of inducible and acquired clarithromycin resistance in the Mycobacterium abscessus group. J Clin Microbiol 53:3430–3437. doi: 10.1128/JCM.01714-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Blakemore R, Story E, Helb D, Kop J, Banada P, Owens MR, Chakravorty S, Jones M, Alland D. 2010. Evaluation of the analytical performance of the Xpert MTB/RIF assay. J Clin Microbiol 48:2495–2501. doi: 10.1128/JCM.00128-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ Jr, Winthrop K, ATS Mycobacterial Diseases Subcommittee, American Thoracic Society, Infectious Disease Society of America. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 123.Marinach C, Alanio A, Palous M, Kwasek S, Fekkar A, Brossas JY, Brun S, Snounou G, Hennequin C, Sanglard D, Datry A, Golmard JL, Mazier D. 2009. MALDI-TOF MS-based drug susceptibility testing of pathogens: the example of Candida albicans and fluconazole. Proteomics 9:4627–4631. doi: 10.1002/pmic.200900152. [DOI] [PubMed] [Google Scholar]
- 124.De Carolis E, Vella A, Florio AR, Posteraro P, Perlin DS, Sanguinetti M, Posteraro B. 2012. Use of matrix-assisted laser desorption ionization–time of flight mass spectrometry for caspofungin susceptibility testing of Candida and Aspergillus species. J Clin Microbiol 50:2479–2483. doi: 10.1128/JCM.00224-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vella A, De Carolis E, Vaccaro L, Posteraro P, Perlin DS, Kostrzewa M, Posteraro B, Sanguinetti M. 2013. Rapid antifungal susceptibility testing by matrix-assisted laser desorption ionization–time of flight mass spectrometry analysis. J Clin Microbiol 51:2964–2969. doi: 10.1128/JCM.00903-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Murphy MJ. 2013. Automation in UK clinical biochemistry. Ann Clin Biochem 50:285–286. doi: 10.1177/0004563213486632. [DOI] [PubMed] [Google Scholar]
- 127.Armbruster DA, Overcash DR, Reyes J. 2014. Clinical chemistry laboratory automation in the 21st century—amat Victoria curam (Victory loves careful preparation). Clin Biochem Rev 35:143–153. [PMC free article] [PubMed] [Google Scholar]
- 128.Ledeboer NA, Dallas SD. 2014. The automated clinical microbiology laboratory: fact or fantasy? J Clin Microbiol 52:3140–3146. doi: 10.1128/JCM.00686-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bourbeau PP, Ledeboer NA. 2013. Automation in clinical microbiology. J Clin Microbiol 51:1658–1665. doi: 10.1128/JCM.00301-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dauwalder O, Landrieve L, Laurent F, de Montclos M, Vandenesch F, Lina G. 2016. Does bacteriology laboratory automation reduce time to results and increase quality management? Clin Microbiol Infect 22:236–243. doi: 10.1016/j.cmi.2015.10.037. [DOI] [PubMed] [Google Scholar]
- 131.Buchan BW, Ledeboer NA. 2014. Emerging technologies for the clinical microbiology laboratory. Clin Microbiol Rev 27:783–822. doi: 10.1128/CMR.00003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dortet L, Tandé D, de Briel D, Bernabeu S, Lasserre C, Gregorowicz G, Jousset AB, Naas T. 2018. MALDI-TOF for the rapid detection of carbapenemase-producing Enterobacteriaceae: comparison of the commercialized MBT STAR-Carba IVD kit with two in-house MALDI-TOF techniques and the RAPIDEC CARBA NP. J Antimicrob Chemother 73:2352–2359. doi: 10.1093/jac/dky209. [DOI] [PubMed] [Google Scholar]
- 133.Hoyos-Mallecot Y, Naas T, Bonnin RA, Patino R, Glaser P, Fortineau N, Dortet L. 2017. OXA-244-producing Escherichia coli isolates, a challenge for clinical microbiology laboratories. Antimicrob Agents Chemother 61:e00818-17. doi: 10.1128/AAC.00818-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kostrzewa M. 2018. Application of the MALDI Biotyper to clinical microbiology: progress and potential. Expert Rev Proteomics 15:193–202. doi: 10.1080/14789450.2018.1438193. [DOI] [PubMed] [Google Scholar]
- 135.Chudejova K, Bohac M, Skalova A, Rotova V, Papagiannitsis CC, Hanzlickova J, Bergerova T, Hrabák J. 2017. Validation of a novel automatic deposition of bacteria and yeasts on MALDI target for MALDI-TOF MS-based identification using MALDI Colonyst robot. PLoS One 12:e0190038. doi: 10.1371/journal.pone.0190038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Broyer P, Perrot N, Rostaing H, Blaze J, Pinston F, Gervasi G, Charles MH, Dachaud F, Dachaud J, Moulin F, Cordier S, Dauwalder O, Meugnier H, Vandenesch F. 2018. An automated sample preparation instrument to accelerate positive blood cultures microbial identification by MALDI-TOF mass spectrometry (VitekMS). Front Microbiol 9:911. doi: 10.3389/fmicb.2018.00911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sanders GD, Neumann PJ, Basu A, Brock DW, Feeny D, Krahn M, Kuntz KM, Meltzer DO, Owens DK, Prosser LA, Salomon JA, Sculpher MJ, Trikalinos TA, Russell LB, Siegel JE, Ganiats TG. 2016. Recommendations for conduct, methodological practices, and reporting of cost-effectiveness analyses second panel on cost-effectiveness in health and medicine. JAMA 316:1093–1103. doi: 10.1001/jama.2016.12195. [DOI] [PubMed] [Google Scholar]
- 138.Higgins AM, Harris AH. 2012. Health economic methods: cost-minimization, cost-effectiveness, cost-utility, and cost-benefit evaluations. Crit Care Clin 28:11–24. doi: 10.1016/j.ccc.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 139.Huang AM, Newton D, Kunapuli A, Gandhi TN, Washer LL, Isip J, Collins CD, Nagel JL. 2013. Impact of rapid organism identification via matrix-assisted laser desorption/ionization time-of-flight combined with antimicrobial stewardship team intervention in adult patients with bacteremia and candidemia. Clin Infect Dis 57:1237–1245. doi: 10.1093/cid/cit498. [DOI] [PubMed] [Google Scholar]
- 140.Clerc O, Prod’hom G, Vogne C, Bizzini A, Calandra T, Greub G. 2013. Impact of matrix-assisted laser desorption ionization time-of-flight mass spectrometry on the clinical management of patients with Gram-negative bacteremia: a prospective observational study. Clin Infect Dis 56:1101–1107. doi: 10.1093/cid/cis1204. [DOI] [PubMed] [Google Scholar]
- 141.Verroken A, Defourny L, Le Polain de Waroux O, Belkhir L, Laterre PF, Delmée M, Glupczynski Y. 2016. Clinical impact of MALDI-TOF MS identification and rapid susceptibility testing on adequate antimicrobial treatment in sepsis with positive blood cultures. PLoS One 11:e0156299. doi: 10.1371/journal.pone.0156299. (Erratum, 11:e0160537. doi:.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Beganovic M, Costello M, Wieczorkiewicz SM. 2017. Effect of matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) alone versus MALDI-TOF MS combined with real-time antimicrobial stewardship interventions on time to optimal antimicrobial therapy in patients with positive blood cultures. J Clin Microbiol 55:1437–1445. doi: 10.1128/JCM.02245-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Osthoff M, Gürtler N, Bassetti S, Balestra G, Marsch S, Pargger H, Weisser M, Egli A. 2017. Impact of MALDI-TOF-MS-based identification directly from positive blood cultures on patient management: a controlled clinical trial. Clin Microbiol Infect 23:78–85. doi: 10.1016/j.cmi.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 144.Khennouchi NC, Loucif L, Boutefnouchet N, Allag H, Rolain JM. 2015. MALDI-TOF MS as a tool to detect a nosocomial outbreak of extended-spectrum-β-lactamase- and ArmA methyltransferase-producing Enterobacter cloacae clinical isolates in Algeria. Antimicrob Agents Chemother 59:6477–6483. doi: 10.1128/AAC.00615-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Egli A, Tschudin-Sutter S, Oberle M, Goldenberger D, Frei R, Widmer AF. 2015. A matrix-assisted laser desorption/ionization time of flight mass-spectrometry (MALDI-TOF MS) based typing of extended-spectrum β-lactamase producing E. coli—a novel tool for real-time outbreak investigation. PLoS One 10:e0120624. doi: 10.1371/journal.pone.0120624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cabrolier N, Sauget M, Bertrand X, Hocquet D. 2015. Matrix-assisted laser desorption ionization–time of flight mass spectrometry identifies Pseudomonas aeruginosa high-risk clones. J Clin Microbiol 53:1395–1398. doi: 10.1128/JCM.00210-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.You Y, Kou Y, Niu L, Jia Q, Liu Y, Davies MR, Walker MJ, Zhu J, Zhang J. 2018. Complete genome sequence of a Streptococcus pyogenes serotype M12 scarlet fever outbreak isolate from China, compiled using Oxford Nanopore and Illumina sequencing. Genome Announc 6:e00389-18. doi: 10.1128/genomeA.00389-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zahra R, Javeed S, Malala B, Babenko D, Toleman MA. Analysis of Escherichia coli STs and resistance mechanisms in sewage from Islamabad, Pakistan indicates a difference in E. coli carriage types between South Asia and Europe. J Antimicrob Chemother, in press. [DOI] [PubMed] [Google Scholar]
- 149.Albrethsen J. 2007. Reproducibility in protein profiling by MALDI-TOF mass spectrometry. Clin Chem 53:852–858. doi: 10.1373/clinchem.2006.082644. [DOI] [PubMed] [Google Scholar]
- 150.Williams TL, Andrzejewski D, Lay JO, Musser SM. 2003. Experimental factors affecting the quality and reproducibility of MALDI TOF mass spectra obtained from whole bacteria cells. J Am Soc Mass Spectrom 14:342–351. doi: 10.1016/S1044-0305(03)00065-5. [DOI] [PubMed] [Google Scholar]
- 151.Spinali S, van Belkum A, Goering RV, Girard V, Welker M, Van Nuenen M, Pincus DH, Arsac M, Durand G. 2015. Microbial typing by matrix-assisted laser desorption ionization–time of flight mass spectrometry: do we need guidance for data interpretation? J Clin Microbiol 53:760–765. doi: 10.1128/JCM.01635-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sauget M, Valot B, Bertrand X, Hocquet D. 2017. Can MALDI-TOF mass spectrometry reasonably type bacteria? Trends Microbiol 25:447–455. doi: 10.1016/j.tim.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 153.Rhoads DD, Sintchenko V, Rauch CA, Pantanowitz L. 2014. Clinical microbiology informatics. Clin Microbiol Rev 27:1025–1047. doi: 10.1128/CMR.00049-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Croxatto A, Prod’hom G, Faverjon F, Rochais Y, Greub G. 2016. Laboratory automation in clinical bacteriology: what system to choose? Clin Microbiol Infect 22:217–235. doi: 10.1016/j.cmi.2015.09.030. [DOI] [PubMed] [Google Scholar]