Abstract
Background
Walking catfish (Clarias batrachus) is a freshwater fish capable of air-breathing and locomotion on land. It usually inhabits various low-oxygen habitats, burrows inside the mudflat, and sometimes “walks” to search for suitable environments during summer. It has evolved accessory air-breathing organs for respiring air and corresponding mechanisms to survive in such challenging environments. Thereby, it serves as a great model for understanding adaptations to terrestrial life.
Results
Comparative genomics with channel catfish (Ictalurus punctatus) revealed specific adaptations of C. batrachus in DNA repair, enzyme activator activity, and small GTPase regulator activity. Comparative analysis with 11 non-air-breathing fish species suggested adaptive evolution in gene expression and nitrogenous waste metabolic processes. Further, myoglobin, olfactory receptor related to class A G protein-coupled receptor 1, and sulfotransferase 6b1 genes were found to be expanded in the air-breathing walking catfish genome, with 15, 15, and 12 copies, respectively, compared to non-air-breathing fishes that possess only 1–2 copies of these genes. Additionally, we sequenced and compared the transcriptomes of the gill and the air-breathing organ to characterize the mechanism of aerial respiration involved in elastic fiber formation, oxygen binding and transport, angiogenesis, ion homeostasis and acid-base balance. The hemoglobin genes were expressed dramatically higher in the air-breathing organ than in the gill of walking catfish.
Conclusions
This study provides an important genomic resource for understanding the adaptive mechanisms of walking catfish to terrestrial environments. It is possible that the coupling of enhanced abilities for oxygen storage and oxygen transport through genomic expansion of myoglobin genes and transcriptomic up-regulation of hemoglobin and angiogenesis-related genes are important components of the molecular basis for adaptation of this aquatic species to terrestrial life.
Electronic supplementary material
The online version of this article (10.1186/s12864-018-5355-9) contains supplementary material, which is available to authorized users.
Keywords: Fish, Genome, Adaptation, Evolution, Duplication, Air-breathing organ
Background
The walking catfish (Clarias batrachus) is a freshwater teleost species with air-breathing capability native to Southeast Asia, where it is widely used as an aquaculture species due to its high economic value as food [1]. Unfortunately, this species also is categorized as endangered because of over-exploitation and habitat alterations in its native India and Bangladesh [2–4]. On the other hand, it is an invasive species in the United States, currently found in over ten states on the eastern and western coasts (http://maps.iucnredlist.org/map.html?id=166613), but with established populations likely only in Florida [5]. C. batrachus was imported into Florida from Thailand in the early 1960s [6]. It has been thought to be damaging to native fish populations; however, there is little evidence to support this except that they do invade aquaculture facilities and can cause severe damage to cultured fish populations (Florida Museum, University of Florida 2017; https://www.floridamuseum.ufl.edu/fish/discover/species-profiles/clarias-batrachus/). Their air breathing capability allows them to disperse quickly across terrestrial environments, a feature that most native fish do not have.
A combination of traits - such as high fecundity, adaptation to adverse ecological conditions, and in particular the ability to “walk” between isolated water bodies - make this fish an especially successful invasive species. It is able to inhabit various low-oxygen habitats such as swamps and wetlands, and burrows inside the mudflat during summer periods [3, 7, 8]. When the original habitat dries up or after a heavy rainfall, the walking catfish can make snake-like movements to move from one body of water to another by pulling its body across land with the pectoral fins [3, 8, 9]. The accessory air-breathing organ is another key innovation to survival during its terrestrial walk to the next aquatic environment. This structure is derived from the gill, and the air-breathing organ in particular consists of suprabranchial chambers, gill fans and arborescent organs [10, 11]. Clarias can breathe air as well as using gills for respiration in water [8]. Its “walking” abilities allow Clarias to cope with respiration challenges without a lung in the terrestrial environment, as well as adaptation to extreme environmental challenges such as high ammonia as well as hypoxic and desiccation stresses [12]. This makes C. batrachus a perfect model for studying the evolution of adaptations such as terrestrial dispersal, aerial respiration and high tolerance to hypoxia and ammonia.
Recent genome projects have demonstrated that comparative genomic analysis combined with transcriptomic analysis allow the elucidation of the genomic basis for adaptation to terrestrial life in mangrove rivulus (Kryptolebias marmoratus) and mudskippers (Bolelphthalmus pectinirostris, Scartelaos histophorus, Periophthalmodon schlosseri and Periophthalmus magnuspinnatus) [13, 14]. Mangrove rivulus mainly utilizes its skin and mudskippers mainly utilize their buccal cavity to breathe air [15, 16], while C. batrachus utilizes an accessory air-breathing organ. It is of great interest to determine the genomic basis of adaptations of aquatic species to the terrestrial environment based on the genome sequence of C. batrachus and the characterization of some of its genomic features that are potentially linked to terrestrial adaptations.
Results
Genome assembly and annotation
The statistics for the draft genome sequence assembly are shown in Table 1. The final assembly contained 10,041 scaffolds, with a scaffold N50 of 361.2 kb. The assembly covered a total of 821 Mb, similar to the genome size of 854 Mb estimated from ALLPATHS-LG, but slightly smaller than the estimated 900 Mb based on the Feulgen densitometry method [17] and the 1.17 Gb based on the bulk fluorometric assay method [18].
Table 1.
Summary statistics for walking catfish (Clarias batrachus) genome sequencing, assembly and annotation
| Genome sequencing | ||||
| Library | # of reads | Read length | Trimmed data | Genome coverage |
| Paired-end 180 bp | 426 M | 100 bp | 39.2 Gb | 46X |
| Mate-pair 3 kb | 483 M | 100 bp | 32.1 Gb | 38X |
| Genome assembly | ||||
| Total | N50 | Longest | Assembled size | |
| Contigs | 87,962 | 19.0 kb | 194 kb | 747.7 Mb |
| Scaffolds | 10,041 | 361.2 kb | 2843 kb | 821.8 Mb |
| Genome annotation | ||||
| Number of genes | Repetitive elements content | |||
| 22,914 | 30.3% | |||
The completeness of the genome assembly was assessed by mapping the 248 core eukaryotic genes (CEGs) from CEGMA v2.5 [19] to the genome sequence. The draft genome sequence covered 95.2% of the CEGs (Additional file 1: Table S1). When the 3023 genes from vertebrate BUSCO orthologues [20] were mapped to the genome assembly, the draft genome sequence included 83.9% of these genes (Additional file 1: Table S1). As our objective was to identify additional gene copies or novel genes in the walking catfish not found in non-air-breathing fishes, which may account for its adaptations for partial living on land, this level of completeness is reasonable, although a small percentage of missing genes may reduce the capacity of identifying more such genes. The assembly was assessed also to be accurate. The 5 longest scaffolds (1.3 Mb–2.2 Mb) assembled using a second software, ABySS, had 99.4% alignments with the genome sequence assembled using ALLPATHS-LG (Additional file 1: Table S2).
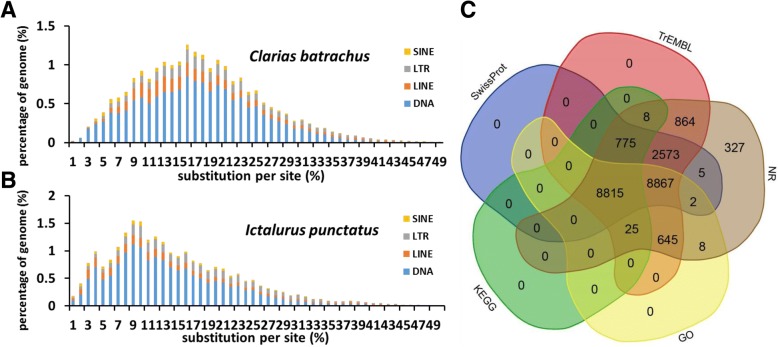
The C. batrachus genome had a GC content of 39.2%, similar to those of other fish species [21, 22]. Repetitive elements comprised 30.3% of the genome (Table 1, Additional file 1: Table S3). Although the contents of repetitive elements in the C. batrachus genome were similar to those in the channel catfish genome [23], it appeared that the number of substitutions per site for C. batrachus repetitive elements exhibited a peak at about 16% (Fig. 1a), higher than that of channel catfish repetitive elements (~ 10%, Fig. 1b), indicating that its repetitive elements had a longer evolutionary history and/or have been more active through evolution than those of the channel catfish genome [24].
Fig. 1.
Annotation of the walking catfish genome. The distribution of repetitive elements and their contents are shown for Clarias batrachus (a) and Ictalurus punctatus (b). The average number of substitutions per site for each fragment was estimated using the Jukes-Cantor formula. SINE, short interspersed elements; LTR, long terminal repeats; LINE, long interspersed elements. c Venn diagram showing the number of homologues of the predicted genes from the C. batrachus genome in various databases: NR (non-redundant protein database), TrEMBL, and SwissProt
A total of 22,914 genes were annotated from the C. batrachus genome sequence, of which 19,834 genes (86.6%) were supported by RNA-Seq evidence from the gill and the air-breathing organ. Among the identified protein-coding genes, the majority (22,587, 98.6%) were supported by matches from at least two publicly available databases including the non-redundant protein database, SwissProt and TrEMBL subsets of the UniProt database [25], KEGG and GO terms (Fig. 1c).
Comparative genomic analysis
To identify genes that are specific to the C. batrachus genome, we first compared the genes between the walking catfish and channel catfish (Fig. 2a, detailed methods described in the “Methods”). They both belong to Order Siluriformes, and therefore shared the highest number of orthogroups compared to other fish species in this study (Additional file 1: Table S4), but the walking catfish possesses the air-breathing organ while the channel catfish does not. A total of 1854 genes were present in the walking catfish, but absent from channel catfish (Additional file 1: Table S5). These genes were enriched for “DNA repair”, “enzyme activator activity” and “small GTPase regulator activity” (Additional file 1: Table S6), which may be associated with its adaptation to the terrestrial life, such as responding to increased DNA damage and accelerated metabolic processes. Small GTPases are well-known for maintaining cell adhesion, cell migration, gene transcription and cytogenesis [26, 27], and one of their critical modulators, namely “guanyl-nucleotide exchange factor activity”, was also found to be significantly enriched (Additional file 1: Table S6). Furthermore, small GTPases were also reported to be under selection in the alkaline-tolerant population compared with the flowing freshwater population of Amur ide Leuciscus waleckii, reflecting their roles in regulating ion transport and acid-base balance under extreme environmental conditions [28].
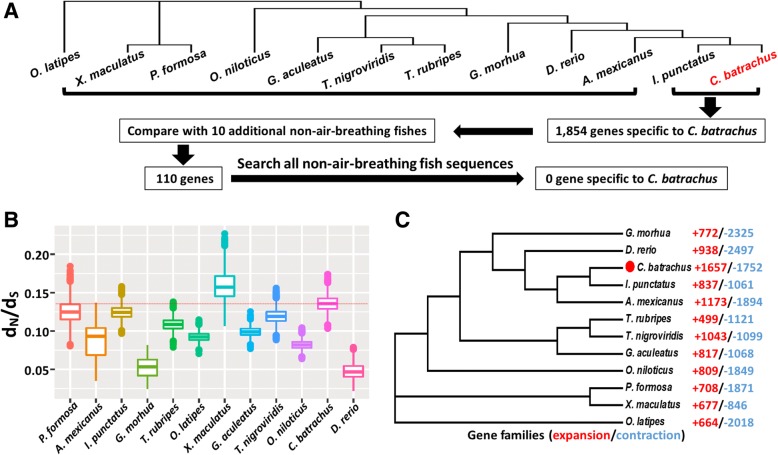
Fig. 2.
Comparisons of genomic features of Clarias batrachus with those non-air-breathing fish species. a Schematic presentation of comparative genomic analyses of C. batrachus against non-air-breathing teleost fishes. In the catfish lineage, the protein sequences of the C. batrachus and I. punctatus genomes were comparatively subtracted, resulting in the identification of 1854 genes specific to the C. batrachus genome; similarly, 10 additional non-air-breathing fishes were added to compare with C. batrachus genome, resulting in the identification of 110 genes that were only present in the C. batrachus genome. The names of these 110 C. batrachus specific genes were searched with all existing sequences from non-air-breathing fish species in the NCBI database, resulting in no genes specific to the C. batrachus genome. b Comparison of the values of dN/dS ratio among various fish species against the ancestor estimated from 150 randomly picked single-copy genes with 10,000 bootstrap replicates. The red line represents the average dN/dS value in C. batrachus, noting that it is the second most rapidly evolving genome. c The number of gene families exhibiting expansion (red) / contraction (blue). C. batrachus is marked with red solid circle, showing that it has the largest number of expanded gene families. D. rerio, Danio rerio; G. aculeatus, Gasterosteus aculeatus; T. nigroviridis, Tetraodon nigroviridis; T. rubripes, Takifugu rubripes; O. latipes, Oryzias latipes; G. morhua, Gadus morhua; A. mexicanus, Astyanax mexicanus; O. niloticus, Oreochromis niloticus; X. maculatus, Xiphophorus maculatus; P. formosa, Poecilia formosa; I. punctatus, Ictalurus punctatus
To further narrow down the list of genes potentially present in the walking catfish but absent in non-air-breathing fishes, the status of the 1854 genes were determined in 10 additional non-air-breathing fish species (Fig. 2a). Only 110 genes were then found to be present in the walking catfish, but absent in the 10 non-air-breathing fishes. When this list of genes was further investigated by comparison with sequences from all non-air-breathing fish species in the NCBI databases, no genes were found to be specific to the walking catfish (Fig. 2a). Although it is possible that the genome sequence assembly is incomplete and that unique and specific genes of the walking catfish could have been missed, it is unlikely that the walking catfish harbors a large number of specific genes for air-breathing.
Next, we also determined the ratio of non-synonymous substitutions to synonymous substitutions using 705 single-copy orthologous genes in the C. batrachus and the other 11 non-air-breathing fishes. As shown in Fig. 2b, C. batrachus apparently evolved rapidly with the second highest dN/dS ratio, next only to X. maculatus, suggesting that parts of its genome are under strong selection. Of the 705 single-copy genes, 132 were positively selected (Additional file 1: Table S7). These genes were mainly enriched in “mitochondrial intermembrane space”, “nucleoplasm part”, “RNA polymerase II transcription factor complex” and “nuclear DNA-directed RNA polymerase complex” (Additional file 1: Table S8), indicating the accelerated evolution of genes involved in regulation of gene expression in C. batrachus. The overrepresented pathway “gene expression” included a list of genes related to transcription factors (med6, med14, gtf2e2, mnat1 and nfyc), RNA binding protein (paip1), mRNA splicing factors (cstf2, sf3b2, rbm8a and cpsf5), chromatin binding (noc2l) and translation initiation factor (eif3m). Additionally, the GO term “cellular nitrogen compound metabolic process” was also found to be enriched for genes under positive selection (Additional file 1: Table S8).
Gene expansion
In addition to analysis of positive selection on single-copy genes, levels of gene family dynamics, including expansion due to gene duplication and contraction due to gene loss, were studied. The C. batrachus genome shows signs of expansion in 1657 families and contraction in 1752 families (Fig. 2c). Among the 12 studied fish genomes, it had the largest number of expanded gene families, suggesting that its adaptation to terrestrial lifestyle may have been mediated partly by gene family expansion. Among the 1657 expanded gene families, three families were significantly expanded (P-value = 0) in C. batrachus: myoglobin (mb), olfactory receptor related to class A G protein-coupled receptor 1 (ora1) and sulfotransferase 6b1 (sult6b1).
The myoglobin gene exists in almost all vertebrate species with one-to-two copies in the genome, except for the seven copies in the West African lungfish (Protopterus annectens, obviously air-breathing) genome [29–35]. We found a huge expansion of myoglobin, fifteen copies of the gene, in the C. batrachus genome (Fig. 3a, Additional file 1: Table S9). Multiple sequence alignments showed some diversities among them (Additional file 1: Figure S1). These 15 copies of the myoglobin gene were located on six scaffolds. We do not have information of their chromosomal locations, but the sequence analysis indicated that tandem duplications exist in three of the six scaffolds. Based on the flanking genes and syntenic analysis, the fifteen myoglobin genes of C. batrachus may be located on different chromosomes. The ora1 gene also was found to be significantly expanded in the C. batrachus genome with 15 copies, while there is only a single copy in most teleost species (Fig. 3b, Additional file 1: Figure S2, Additional file 1: Table S9). The 15 copies of ora1 genes in C. batrachus were found on the same scaffold, suggesting tandem duplications, and these fifteen genes displayed high sequence similarities (Additional file 1: Figure S3). The sult6b1 gene also was found to be highly expanded in C. batrachus with twelve copies, as compared to 1–2 copies in non-air-breathing teleost fishes (Fig. 3c, Additional file 1: Table S9).
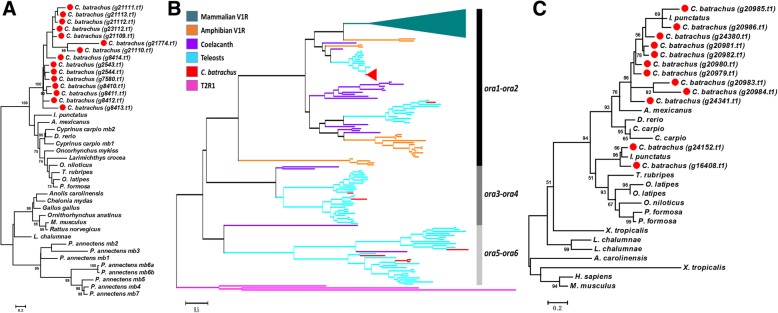
Fig. 3.
Maximum likelihood phylogenetic trees of expanded gene families in the Clarias batrachus genome. a Phylogenetic tree of myoglobin (mb) in vertebrates showing gene expansion of mb in the C. batrachus genome. The red solid circles represent the genes in the C. batrachus genome. Bootstrap support values (1000 replications) are indicated on the nodes. b Phylogenetic tree of the olfactory receptor related to class A G protein-coupled receptor (ora) gene family in vertebrates showing the expansion of ora1 in the C. batrachus genome with taste receptor of type 2 member 1 (T2R1) as an outgroup. The three clades (ora1-ora2, ora3-ora4 and ora5-ora6) formed from six members are indicated on the right of the figure. The dark green solid triangle represents the expansion of vomeronasal type 1 receptors (V1Rs) in mammals. The red solid triangle represents the 15 copies in the C. batrachus genome. The term “teleosts” here is used to indicate the non-air-breathing fish species discussed in this study. The detailed phylogenetic tree with species names and sequence names is displayed in the Additional file 1: Figure S2. c Phylogenetic tree of sulfotransferase 6b1 (sult6b1) in vertebrates showing gene expansion of sult6b1 in the C. batrachus genome. The red solid circles represent the genes in the C. batrachus genome. Bootstrap support values (1000 replications) are indicated on the nodes. D. rerio, Danio rerio; G. aculeatus, Gasterosteus aculeatus; T. nigroviridis, Tetraodon nigroviridis; T. rubripes, Takifugu rubripes; O. latipes, Oryzias latipes; G. morhua, Gadus morhua; A. mexicanus, Astyanax mexicanus; O. niloticus, Oreochromis niloticus; X. maculatus, Xiphophorus maculatus; P. formosa, Poecilia formosa; I. punctatus, Ictalurus punctatus; C. carpio, Cyprinus carpio; P. annectens, Protopterus annectens; M. musculus, Mus musculus; H. sapiens, Homo sapiens; X. tropicalis, Xenopus tropicalis; L. chalumnae, Latimeria chalumnae; A. carolinensis, Anolis carolinensis
Expression of significantly expanded gene families
The expression of the various copies of the mb and sult6b1 genes in C. batrachus was analyzed using RNA-Seq datasets (Table 2). All 15 myoglobin genes were expressed, but in different tissues and at different levels. The brain had an overall high level of expression, consistent with the results in common carp and West African lungfish [32, 35]. The expression levels in the air-breathing organ are of particular interest: two copies of the myoglobin gene, g21774.t1 and g8414.t1, were expressed at high levels, at least 10 times higher than other copies. In all the other tissues, only one copy, g8414.t1, was expressed at very high levels. Interestingly, synteny analyses indicated that g8414.t1 is orthologous to the single-copy myoglobin gene in other fish species (Additional file 1: Figure S4). Its high expression in all tissues suggested that its function may be conserved in evolution. We suggest that these gene copies may be evolved as inducible genes to cope with hypoxic environments such as the terrestrial condition, and thus are related to the adaptation to the transition from water to land. For the sult6b1 gene, all 12 copies were expressed, but with some tissue-specificity. For instance, g20983.t1 and g24341.t1 were expressed only in the gill, g24152.t1 and g16408.t1 were expressed only in the gill and the air-breathing organ, and g24380.t1 was expressed only in the gill and liver. All 12 copies were expressed in the gill, but the most highly expressed copies were g20980.t1, g20985.t1 and g20986.t1. Notably, all these highly expressed copies were tandem duplicates on the same scaffold (Table 2). The ora1 gene is known to be tissue-specific and expressed only in the olfactory epithelium in other fish species [36]. In regards to the five tissues examined with RNA-Seq, no transcripts of ora1 were detected, consistent with the tissue-specific expression of this gene only in olfactory epithelium in other fish species [36].
Table 2.
Expression of myoglobin and sulfotransferase 6b1 genes (FPKM) in the air-breathing organ, gill, brain, head kidney and liver of Clarias batrachus as determined by analysis of RNA-Seq datasets. RNA-Seq datasets from air-breathing organ and gill were generated from this project; those from brain (SRR2057993), head kidney (SRR2057995), and liver (SRR2052655) were downloaded from NCBI. Each continuous bold and italic gene cluster indicated tandemly duplicated genes. FPKM, Fragments Per Kilobase per Million mapped fragments
| Gene name | Gene ID | Air-breathing organ | Gill | Brain | Head kidney | Liver |
|---|---|---|---|---|---|---|
| Myoglobin | g21109.t1 | 0 | 0 | 0 | 2.91 | 2.84 |
| g21110.t1 | 0 | 0 | 8.48 | 0.85 | 4.14 | |
| g21111.t1 | 0.25 | 0.18 | 3.14 | 1.57 | 1.53 | |
| g21112.t1 | 0.59 | 0.05 | 1.45 | 1.55 | 0 | |
| g21113.t1 | 0.09 | 0 | 4.48 | 9.88 | 2.92 | |
| g21774.t1 | 4.52 | 5.07 | 0 | 1.02 | 0 | |
| g23112.t1 | 0.46 | 0.76 | 0 | 0.58 | 0 | |
| g2543.t1 | 0 | 0 | 26.84 | 25.61 | 5.11 | |
| g2544.t1 | 0 | 0 | 15.71 | 12.23 | 3.31 | |
| g7580.t1 | 0 | 0 | 0 | 2.32 | 0 | |
| g8410.t1 | 0 | 0 | 17.4 | 23.41 | 3.31 | |
| g8411.t1 | 0 | 0 | 0 | 0.59 | 0 | |
| g8412.t1 | 0 | 0.56 | 16.23 | 13.52 | 3.27 | |
| g8413.t1 | 0 | 0.16 | 0 | 0 | 0 | |
| g8414.t1 | 6.18 | 139.88 | 838.66 | 643.35 | 145 | |
| Sulfotransferase 6b1 | g16408.t1 | 0.2 | 1.16 | 0 | 0 | 0 |
| g20979.t1 | 2.78 | 1.22 | 3.96 | 1.94 | 1.95 | |
| g20980.t1 | 23.9 | 28.5 | 13.36 | 18.93 | 2.09 | |
| g20981.t1 | 3.42 | 3.79 | 6.16 | 3.34 | 1.28 | |
| g20982.t1 | 2.91 | 2.42 | 1.57 | 1.62 | 1.28 | |
| g20983.t1 | 0 | 0.58 | 0 | 0 | 0 | |
| g20984.t1 | 0.54 | 0.53 | 0.57 | 1.43 | 1.75 | |
| g20985.t1 | 9.37 | 16.57 | 5.6 | 4.94 | 0.26 | |
| g20986.t1 | 24.61 | 14.81 | 89.49 | 44.81 | 0 | |
| g24152.t1 | 0.13 | 0.53 | 0 | 0 | 0 | |
| g24341.t1 | 0 | 0.06 | 0 | 0 | 0 | |
| g24380.t1 | 0 | 0.31 | 0 | 0 | 15.45 |
Comparative transcriptomic analysis between gill and the air-breathing organ
To understand the transcriptomic differences between the gill and the air-breathing organ, we sequenced the transcriptomes of the gill and the air-breathing organ (Additional file 1: Table S10), analyzed the expression levels of all transcripts (Additional file 1: Table S11), and determined the differentially expressed genes (DEGs, Additional file 1: Table S12). A total of 813 DEGs were identified between the gill and the air-breathing organ, of which 335 and 478 genes were up-regulated and down-regulated in the air-breathing organ, respectively. These results were validated by determining the expression levels of a fraction of these genes using qRT-PCR (Additional file 1: Figure S5). All the DEGs were subjected to GO and pathway enrichment analyses, and the significantly over-represented GO terms and pathways in the DEGs are listed in Additional file 1: Table S13 and Additional file 1: Table S14, respectively. As summarized in Table 3, a total of 51 genes belonging to five functional groups were highly and differentially expressed between the gill and the air-breathing organ.
Table 3.
A list of genes differentially expressed between the gill and the air-breathing organ in Clarias batrachus
| Category name | Gene name | Gene ID | FDR |
|---|---|---|---|
| Acid-base balance | Solute carrier family 4 member 1 | g12078.t1 | 0 |
| Carbonic anhydrase | g8816.t1 | 0 | |
| Carbonic anhydrase 2 | g3019.t1 | 1.60E-07 | |
| Carbonic anhydrase 4 | g7074.t1 | 7.21E-07 | |
| Carbonic anhydrase 9 | g411.t1 | 5.37E-05 | |
| Carbonic anhydrase 6 | g456.t1 | 7.39E-04 | |
| Ion homeostasis | Sodium/potassium-transporting ATPase subunit alpha-1 | g15272.t1 | 0 |
| Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | g16367.t1 | 0 | |
| Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | g5688.t1 | 0 | |
| Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | g9634.t1 | 0 | |
| Sodium potassium-transporting ATPase subunit beta-233 | g9311.t1 | 0 | |
| Adenosylhomocysteinase 3 | g1970.t1 | 2.72E-10 | |
| Adenosylhomocysteinase 3 | g1011.t1 | 1.19E-08 | |
| Calsequestrin-1 | g3495.t1 | 5.23E-04 | |
| Calsequestrin-2 | g9400.t1 | 3.67E-02 | |
| Pendrin | g2970.t1 | 0 | |
| Sodium/hydrogen exchanger 3 | g3908.t1 | 0 | |
| Elastic fiber formation | Transforming growth factor beta-2 | g2430.t1 | 8.22E-03 |
| Fibulin-1 | g4468.t1 | 9.66E-08 | |
| Transforming growth factor beta-3 | g14668.t1 | 2.68E-02 | |
| Fibulin-2 | g20129.t1 | 5.73E-03 | |
| Bone morphogenetic protein 4 | g189.t1 | 8.79E-06 | |
| Fibronectin | g6260.t1 | 0 | |
| Fibronectin | g12205.t1 | 9.01E-03 | |
| Latent-transforming growth factor beta-binding protein 3 | g14988.t1 | 8.22E-03 | |
| Oxygen binding and transport | Hemoglobin subunit alpha | g20835.t1 | 0 |
| Hemoglobin subunit beta | g20836.t1 | 3.55E-11 | |
| Hemoglobin subunit alpha | g20837.t1 | 0 | |
| Hemoglobin subunit beta | g20838.t1 | 0 | |
| Hemoglobin subunit beta | g21168.t1 | 0 | |
| Hemoglobin subunit alpha | g21169.t1 | 0 | |
| Hemoglobin subunit beta | g21170.t1 | 0 | |
| Hemoglobin subunit alpha | g21171.t1 | 0 | |
| Angiogenesis | Sphingosine 1-phosphate receptor 1 | g5232.t1 | 7.89E-03 |
| Fibronectin | g6260.t1 | 8.79E-06 | |
| Semaphorin-3E | g5331.t1 | 6.86E-05 | |
| C-X-C chemokine receptor type 4 | g23603.t1 | 0 | |
| Neuropilin-1a | g3757.t1 | 5.00E-02 | |
| Cadherin-5 | g21848.t1 | 7.36E-03 | |
| BMP-binding endothelial regulator protein | g7031.t1 | 2.71E-02 | |
| Bone morphogenetic protein 4 | g189.t1 | 5.73E-03 | |
| Bone morphogenetic protein 5 | g11279.t1 | 2.76E-03 | |
| Bone morphogenetic protein 8A | g14806.t1 | 9.26E-03 | |
| Bone morphogenetic protein receptor type-1A | g14843.t1 | 9.45E-05 | |
| Transforming growth factor beta-2 | g2430.t1 | 8.22E-03 | |
| Transforming growth factor beta-3 | g14668.t1 | 2.68E-02 | |
| Rho-related GTP-binding protein RhoB | g5763.t1 | 1.23E-09 | |
| Thrombospondin-1 | g5480.t1 | 3.33E-02 | |
| Mothers against decapentaplegic homolog 6 | g4737.t1 | 6.93E-13 | |
| Mothers against decapentaplegic homolog 6 | g4739.t1 | 3.88E-11 | |
| Mothers against decapentaplegic homolog 3 | g3356.t1 | 4.28E-03 |
Of the five groups of DEGs, two groups were highly expressed in the gill: six genes for acid-base balance, and 11 genes for ion homeostasis (Table 3), reflecting the critical role that the gill plays in acid-base and ion regulation. Three groups of genes were highly expressed in the air-breathing organ: eight “elastic fiber formation” genes, eight hemoglobin genes, and 18 genes involved in angiogenesis (Table 3). The eight hemoglobin genes (four alpha subunit genes and four beta subunit genes) were found to be dramatically up-regulated in the air-breathing organ, as compared to in the gill which is primarily an aquatic respiratory organ (Additional file 1: Table S12), demonstrating that the air-breathing organ is highly committed to the respiratory processes for oxygen transport.
Discussion
In this study, we sequenced and assembled the genome sequence from walking catfish, which provided a comprehensive understanding of this species at the genomic and evolutionary levels. Comparative analysis with 11 non-air-breathing fish species suggested its adaptive evolution in terms of gene expression and nitrogenous waste metabolic processes. It has been well documented that different organisms can achieve diverse and specific responses to multiple environmental stresses by regulating gene expression to maintain homeostasis [37–39]. In addition, ammonia is the main nitrogenous waste in fishes, which is highly toxic and needs to be excreted promptly or converted to other less-toxic chemicals. C. batrachus usually inhabits water bodies with high levels of ammonia and sometimes dwells inside mudflats or “walks” on the land, during which excretion of ammonia directly into the aqueous environment through the gill is impossible [7]. To adapt to the hyper-ammonia stress, C. batrachus is highly tolerant to external ammonia and can convert ammonia into non-essential amino acids and less-toxic urea through the ornithine-urea cycle (OUC) [7, 40–42]. Interestingly, the gene argininosuccinate synthase (ass) encoding one of the key enzymes in the OUC was found to be under positive selection in comparison with non-air-breathing fish species, implying the necessity of this adaptive strategy for air-breathing walking catfish for survival in hyper-ammonia environments. Further, the wide use of urea as the main nitrogenous waste product in amphibians, some turtles and mammals has been hypothesized to be a key evolutionary process for transition from water to land [43, 44], suggesting the importance of urea excretion among the adaptations of walking catfish to the terrestrial life.
Although no specific genes that are present only in the air-breathing walking catfish were found, three important genes (mb, ora1 and sult6b1) were found to be significantly expanded in the genome - with 15, 15, and 12 copies, respectively - compared to non-air-breathing fishes that possess only 1–2 copies of these genes. We believe that the noted gene expansions are real, not caused by inaccuracies of genome assembly. In spite of being a draft genome sequence, the genome assembly is of high quality. The same assembly results, especially in the regions containing the noted gene expansions, were achieved by using different de novo genome assemblers (ABySS and ALLPATHS-LG), suggesting the accuracy of the assembly, and therefore the accuracy for assessment of tandem duplications. The genome sequencing was conducted using DNA template from a single individual, and thus two allelic variations of sequences are expected. Multiple sequence alignments (Additional file 1: Figure S1, Additional file 1: Figure S3) and phylogenetic trees (Fig. 3, Additional file 1: Figure S2) indicated that the amino acid sequences of the duplicated genes are divergent. Additionally, duplicates of mb and sult6b1 are located on different scaffolds with different flanking genes, all of which also have transcriptomic evidence (Table 2).
Gene expansion may be a “handy” approach for genome evolution to rapidly adapt to environments, especially stressful conditions. Tandem gene duplication usually results from unequal crossing over, but the fixation of duplicated genes is a time-consuming process, which is determined by the functions of the gene duplicates [45, 46]. If the functions are beneficial and essential, strong purifying selection would prevent duplicated genes from pseudogenization or neofunctionalization during a long-term evolutionary process, resulting in very similar sequences among those duplicates [45]. As predicted by Susumo Ohno (1970) [47], our study provides support for the view that tandem duplications provide important evolutionary mechanisms for adaptation and diversification [48]. It appeared that the “sand-bagging” style of gene expansion could be a possible mechanism for evolution of aquatic genomes to cope with stressful environments, especially those that pose life-or-death consequences. In a recent study, Xu et al. [28] reported drastic expansion of egg-coat proteins and natriuretic peptide receptors in Amur ide Leuciscus waleckii that lives under extremely alkaline conditions.
The expansion of myoglobin genes in C. batrachus may be consistent with its frequent exposure to low-oxygen habitats and occasional terrestrial migration. Myoglobin, as an oxygen binding protein predominantly in skeletal and cardiac muscles, is able to bind and store oxygen and facilitate the delivery of oxygen to peripheral tissues [49, 50]. Millikan [51] reported that myoglobin maintains balance in periods of fluctuating oxygen supply and demand through rapid oxygenation and deoxygenation. Additionally, myoglobin maintains a steady level of oxygenation to the mitochondria during muscle contraction [49, 52]. Although the copy number for myoglobin is not expanded in mammals, many studies indicate that it is highly regulated, with higher levels of expression in the skeletal muscle of hypoxia-tolerant animals such as deep-diving and high-elevation mammals compared to surface and lowland relatives [53, 54]. In fish species, comparative studies have been conducted between mb-high sea raven (Hemitripterus americanus) and mb-low ocean pout (Macrozoarces americanus) [55] and between mb-present icefsh (Chionodraco rastrospinosus) and mb-absent icefish (Chaenocephalus aceratus) [56]. Both studies indicated that myoglobin plays a critical role in maintaining oxygen consumption in the heart and enhances cardiac performance. In addition to those functions of oxygen storage and transport, myoglobin also was found to be involved in protecting mitochondrial respiration from nitric-oxide (NO) inhibition [57] and in scavenging of reactive oxygen species (ROS) [58]. Especially during hypoxia and subsequent re-oxygenation periods, the production of ROS increased significantly [59, 60]. One prominent example was found in common carp (Cyprinus carpio), in which the additional myoglobin isoform mb2 played a protective role against ROS in the brain [32, 61]. Similarly in West African lungfish, notable myoglobin expression in the brain was observed, and the cell-level experiments also suggested a key role of myoglobin in protecting the tissues from ROS [35].
Olfaction is an important sense for fish to recognize odorants due to the great ability of water to carry chemical compounds, and their reduced visual ability in turbid environments. Some fishes, including catfishes, have chemoreceptors on their barbels and anterior surfaces of the body. Unlike mammals possessing a main olfactory epithelium (MOE) and a vomeronasal organ (VNO) to express different types of chemoreceptors, fish only have MOE [62–65]. The separation of MOE and VNO in terrestrial vertebrates may have resulted evolutionarily from the segregation of distinct classes of neurons that were differentially positioned in the MOE of aquatic vertebrates [65]. Furthermore, the ora genes in fish species are homologs of the vomeronasal receptor 1 (v1r) in mammals [66]. Surprisingly, the ora genes are very conserved in fish species, with very rare gene duplication events [36], while mammalian genomes harbor hundreds of v1r genes [67]. In most cases, fish species possess six ora genes with ora1-ora2, ora3-ora4 and ora5-ora6 forming three phylogenetic clades (Fig. 3b, Additional file 1: Figure S2), suggesting a close evolutionary relationship within each gene pair [36]. In the C. batrachus genome, we identified all six ora genes, but ora1 was expanded with fifteen tandem copies. Interestingly, the expansions of v1r in mammals also were clustered as tandem duplications [68] and fell within the ora1-ora2 gene-pair clade of teleost species (Fig. 3b, Additional file 1: Figure S2), suggesting that the gaining of ora3-ora6 genes in aquatic species might be due to the aquatic lifestyle [36]. Also, coelacanth (Latimeria chalumnae), an ancient lobe-finned fish that is thought to be evolutionarily close to tetrapods, not only possesses all the ora genes, but also experienced an expansion in the ora1-ora2 gene-pair clade [69, 70] (Fig. 3b, Additional file 1: Figure S2), which is similar to C. batrachus. These related observations may suggest that the expansion of ora1 genes in C. batrachus might be associated with adaptation for transition from water to land, allowing recognition of airborne chemicals to help better detect threats and locate water sources.
Sulfotransferase 6b1 encodes a key enzyme for the process of detoxifying and eliminating xenobiotics. Aquatic habitats are increasingly polluted world-wide, and such contaminants adversely affect the health of aquatic animals [71]. C. batrachus, as an air-breathing fish, not only suffers from the same toxins in the water as other aquatic animals, but also endures higher concentrations of toxic chemicals in drying water bodies as well as from the land. To counteract the toxic effects of these xenobiotics, complex enzyme-based mechanisms are needed to detoxify and eliminate these chemical compounds. Sulfotransferases function by conjugation of a sulfate group on the target xenobiotics to increase their hydrophilicity for excretion [72]. C. batrachus demonstrated an overall higher tolerance to three widely distributed xenobiotics than two other air-breathing fish species, the Asian stinging catfish (Heteropneustes fossilis) and spotted snakehead (Channa punctatus) [73]. Rainbow trout (Oncorhynchus mykiss) fry and Japanese flounder (Paralichthys olivaceus) showed significantly increased expression of sult6b1 after exposure to diesel and the water-accommodated fraction of crude oil, respectively, indicating that sult6b1 does function in eliminating toxic chemicals in fish species [74, 75]. Taken together, the expansion of sult6b1 may play crucial roles in protecting C. batrachus from the deleterious effects of different xenobiotics from the aquatic and terrestrial environments.
It has been suggested that air breathing has evolved as an adaptation for fish to cope with hypoxic conditions, and consequently it provided an essential first step to terrestrial habitation in the evolution of vertebrates [76–80]. Consequently, we sequenced and compared the transcriptomes of the gill and the air-breathing organ to investigate the mechanism of aerial respiration. It showed that acid-base balance and ion homeostasis related genes were up-regulated in the gill, while elastic fiber formation, oxygen binding and transport, and angiogenesis genes were up-regulated in the air-breathing organ. Acid-base regulation in vertebrates is coupled to carbon dioxide (CO2) excretion through the reversible hydration/dehydration reactions of CO2 and the acid-base equivalents H+ and HCO3− by carbonic anhydrase (CA) (Table 3). It is always linked to ion regulation because acid-base compensation depends on the transfer of H+ and HCO3− in exchange for Na+ and Cl− across the gill, respectively [81–83]. Both acid-base balance and ion homeostasis contribute greatly to maintaining the well-balanced conditions for efficient aerial respiration by the air-breathing organ.
Elastic fibers are important structural components of the arborescent organ [84, 85], while hemoglobin genes and genes involved in angiogenesis apparently provide a functional basis for C. batrachus to cope with low oxygen in the terrestrial environment. The hemoglobin genes were expressed dramatically higher in the air-breathing organ than in the gill of walking catfish (Additional file 1: Table S12), suggesting their important roles in sufficient oxygen supply during air-breathing activities. Additionally, 18 genes involved in angiogenesis were found differentially expressed in the air-breathing organ (Table 3). It is also well documented that angiogenesis plays a critical role in respiratory function for accessory air-breathing organs of fishes [76, 86, 87]. Also, the air-breathing organ of C. batrachus is highly vascularized on the surface, and the capillaries extensively bulge out onto the surface to facilitate gas exchange between blood and atmospheric air [10, 88]. Consequently, heightened angiogenesis may be one additional adaptation for the air-breathing organ to maintain high efficiency of air exchange. Overall, it appears that the strategy during adaptive evolution of C. batrachus to the transition from aquatic to terrestrial environment may be through the coupling of high expression of hemoglobin and angiogenesis genes for oxygen transport with expansion of myoglobin genes for oxygen uptake and storage in the peripheral tissues.
Conclusions
The walking catfish is an aquatic species but can move about on land without a lung. As such, it is a remarkable model to investigate the transition from the aquatic to the terrestrial environment, and the adaptation to terrestrial life. Through whole-genome sequencing analysis, we did not find any specific genes that were present in this air-breathing fish, but absent in non-air-breathing fishes. However, highly suggestive gene family expansions (mostly in tandem) were found within the C. batrachus genome. Of particular interest is the expansion of the oxygen-storage protein myoglobin gene, with 15 copies, while non-air-breathing fishes have only one to two copies of this gene. West African lungfish was found to harbor seven copies of the myoglobin gene, and this expansion of myoglobin genes was believed to be crucially important for its adaptation to survive hypoxic periods [35]. Therefore, it is likely that the expansion of myoglobin genes may be a possible mechanism for the water-to-land transition. Additionally, the olfactory receptor related to class A G protein-coupled receptor 1 and the sulfotransferase 6b1 genes were found to be highly expanded, with the former being related to the olfactory sense and the latter to provide resistance to xenobiotics.
The coupling of enhanced oxygen transport, and oxygen uptake and storage may be important for the water-to-land transition. Hemoglobin genes were found to be expressed at much higher levels in the air-breathing organ of C. batrachus than in its gill. While the hemoglobin genes are also highly duplicated, the number of paralogous copies in the air-breathing C. batrachus is not larger than that in the non-air-breathing fishes. Instead, regulation appeared to be at the transcriptional level, where hemoglobin RNAs were transcribed many times more highly in the air-breathing organ, ensuring the greater capacity for oxygen transport. In addition, many genes involved in angiogenesis were found to be expressed at much higher levels in the air-breathing organ than in the gill of C. batrachus, providing the structural basis for expanded blood vessel systems for gas exchange. Taken together, the evolution for the water-to-land transition seemed to involve mostly expanded oxygen storage genes through gene duplications and transcriptional up-regulation of oxygen transport genes.
Methods
C. batrachus samples and genome sequencing
One wild walking catfish (Clarias batrachus) was collected from Florida, USA in June of 2014, and the fish was euthanized with MS-222 (200 mg/l) before blood sampling. Genomic DNA was extracted from blood cells using the DNeasy Blood and Tissue kit (Qiagen, CA). One short-insert (180 bp) paired-end library and one long-insert (3 kb) library were constructed. Each library was subjected to one lane of 2 × 100 bp read-length run on an Illumina HiSeq 2500 sequencer at HudsonAlpha (Huntsville, AL, USA).
Genome assembly and assessment
After raw reads were evaluated in FastQC v0.11.4 [89], low-quality bases and adapter sequences were trimmed from the raw sequences using cutadapt v1.8.1 [90], and then reads with length shorter than 30 bases after trimming were removed. The genome sequence was assembled by ABySS v1.5.2 [91] with k-mers ranging from 40 to 70 in size and ALLPATHS-LG [92]. Finally, k-mer size of 61 yielded the best assembly results using ABySS. To increase scaffold length, we selected assembled sequences with longer contig N50 from ALLPATHS-LG for scaffolding by SSPACE v3.0 [93]. Finally, the paired-end reads were utilized to fill the gaps in the scaffolds with Gapfiller v1.10 [94]. Genome size was estimated in the ALLPATHS-LG using trimmed paired-end reads.
To assess the quality of the assembly results, CEGMA v2.5 (Core Eukaryotic Genes Mapping Approach) [19] was employed to evaluate the completeness of the C. batrachus draft genome sequence. In other words, 248 highly conserved core eukaryotic genes (CEGs) from six genomes of model systems (Homo sapiens, Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis thaliana, Saccharomyces cerevisiae and Schizosaccharomyces pombe) [19] were mapped to the genome assembly to display the percentage of the CEGs present in the C. batrachus genome. Another assessment procedure, BUSCO v1.22 (Benchmarking Universal Single-Copy Orthologs) [20], was used to evaluate the completeness of genome assembly by 3023 genes selected from orthologous groups with single-copy orthologs in > 90% of available vertebrate genomes. Then, the five longest scaffolds of assembly resulting from another assembler (ABySS) were mapped against the genome sequence using NUCmer in MUMmer v3.23 [95] to evaluate the aligned identity.
Genome annotation
A de novo repeat library was constructed using RepeatModeler v1.0.8 (http://www.repeatmasker.org/RepeatModeler.html), which contains two de novo repeats-finding programs, RECON [96] and RepeatScout [97]. Next, RepeatMasker v4.0.6 (http://www.repeatmasker.org/) was used to predict and categorize repeat sequences in the C. batrachus genome with the repeat library constructed from RepeatModeler. The Jukes-Cantor model [98] was used to estimate the average number of substitutions per site for each fragment based on the divergence levels from the results of RepeatMasker. For the subsequent genome annotation, the genome sequence was masked with “N” in the repeat regions except for low-complexity DNA or simple repeats.
AUGUSTUS v3.2.1 [99] was used for the ab initio predictions of genes on the repeat-masked genome. Gene model parameter sets for AUGUSTUS were trained from genes in zebrafish (Danio rerio). The predicted genes with length less than 30 amino acids were removed. The remaining predicted amino acid sequences were aligned to entries in the NCBI non-redundant (nr) protein database and SwissProt and TrEMBL subsets of the UniProt database [25] by BLASTP with an E-value cut-off of 1 × 10− 5 to identify homologous genes. Functional categories of GO terms were determined by Blast2GO version 4.0.7 [100], and the KEGG Automatic Annotation Server (KAAS) (http://www.genome.jp/tools/kaas/) BBH (bi-directional best hit) method [101] was used to perform a biological pathway analysis.
Comparative genomic analysis
The protein sequences of channel catfish (Ictalurus punctatus; NCBI version IpCoco_1.2) [23] were downloaded for comparison with those of C. batrachus to determine C. batrachus-specific genes in the catfish lineage. The methodology was based on the one used in the channel catfish genome paper [23]. First, the proteins from both catfishes were sent to OrthoFinder v1.0.2 [102] for an all-to-all BLASTP comparison with an E -value threshold of 1 × 10− 5 and subsequent clustering into orthogroups based on the MCL algorithm. Next, a further round of BLASTP searches was performed using the genes not included in the orthogroups to query against the genes in the orthogroups within the same species with an E -value threshold of 1 × 10− 10. In the end, reciprocal BLASTP searches between them with an E -value threshold of 1 × 10− 5 were performed using genes with no hits from last step as queries. The remaining genes in C. batrachus were considered as species-specific genes and kept for a further GO-term overrepresentation test using PANTHER version 11 [103] with the best homologous gene ID from zebrafish.
Protein sequences of an additional 10 teleost fish species including zebrafish (Danio rerio; Ensembl version GRCz10), three-spined stickleback (Gasterosteus aculeatus; Ensembl version BROAD S1), green spotted pufferfish (Tetraodon nigroviridis; Ensembl version TETRAODON8.0), Japanese pufferfish (Takifugu rubripes; Ensembl version FUGU4.0), medaka (Oryzias latipes; Ensembl version HdrR), Atlantic cod (Gadus morhua; Ensembl version fadMor1), Mexican cave fish (Astyanax mexicanus; Ensembl version AstMex102), Nile tilapia (Oreochromis niloticus; Ensembl version Orenil1.0), southern platyfish (Xiphophorus maculatus; Ensembl version Xipmac4.4.2) and amazon molly (Poecilia formosa; Ensembl version Poecilia_formosa-5.1.2) were downloaded for inferring orthologues. The longest protein sequence was selected for each gene among the eleven sequenced fish species (channel catfish included). After combination with C. batrachus protein sequences, all the sequences were sent to OrthoFinder v1.0.2 [102] to identify orthologues and orthogroups among these species. Genes that are present in the C. batrachus genome but absent from the non-air-breathing fishes were obtained. Next, these genes specific to C. batrachus were searched with all existing sequences from non-air-breathing fishes in the NCBI database to find the genes that are present only in the C. batrachus genome.
Single-copy genes were extracted from all the species to construct a phylogenetic tree. Multiple sequence alignments were performed using MUSCLE v3.8.31 [104] for protein alignments and PAL2NAL [105] for codon alignments. We used Gblock v0.91b [106] to eliminate poorly aligned positions and divergent regions of the alignments. Final alignments with length shorter than 50 amino acids for protein alignments and 150 bp for codon alignments were removed. AMAS [107] was performed to combine all the refined alignments into a concatenated alignment. PartitionFinder v2.0.0 was used to determine the best substitution model for each gene with the parameter of -rcluster-percent = 20.0 [108]. Then we used the rapid bootstrap algorithm with a thorough ML search (−f a) and 100 bootstrap replicates in RAxML v8.2.9 [109] to construct a maximum likelihood tree for those single-copy genes.
To determine positively-selected genes in C. batrachus, the single-copy genes were collected for analyzing the dN/dS ratio. The values of dN, dS and dN/dS ratio were estimated using the codeml program in the PAML package version 4.9 [110]. Sequence alignments with dS value greater than 2 were removed to avoid distortion of the dN/dS ratio by saturation of synonymous substitutions [111]. The values of dN/dS between each species branch and the ancestral branch from 150 randomly picked genes were estimated with 10,000 bootstrap replicates to evaluate the magnitude of natural selection acting on each species. Then, a branch-site model [112] was used to designate C. batrachus as a “foreground” branch and the rest of the species as “background”. A likelihood ratio test (LRT) was computed to compare a model that allows sites to be under positive selection (ω > 1) on the foreground branch with the null model that allows sites to be under negative selection (ω < 1) and to evolve neutrally (ω = 1) with a posterior probability greater than 0.95 based on Bayes Empirical Bayes (BEB) results [113]. After an FDR multiple-testing correction, the positively selected genes (FDR < 0.05) were selected for further GO-term enrichment analysis by Blast2GO version 4.0.7 [100] with the whole reference gene set as the background for statistical analysis. After annotating C. batrachus genes with the best homologous zebrafish genes by BLASTP and Ensembl BioMart [114], the Reactome pathway database v60 [115] was used for further pathway enrichment analysis.
Gene family analysis
Orthologous genes were sent to the CAFÉ v3.0 [116] program to assess gene family expansion and contraction (−r 1000 -s). A family-wide P-value of less than 0.01 and a branch-specific P-value of less than 0.001 was utilized to identify gene family expansion in the C. batrachus genome. The expanded families in the C. batrachus genome were searched against the NCBI database to exclude false-positive expansions due to the limited number of species in previous analyses, during which the numbers of genes in tetraploid species were divided by two for direct comparisons.
For those significantly expanded genes, phylogenetic trees were constructed to display the gene expansions. The accession numbers of all the protein sequences used in the phylogenetic analyses are listed in Additional file 1: Table S15. Multiple sequence alignments were performed using ClustalW [117] in MEGA6 [118], and ProtTest v3.4 [119] was utilized to select the best model for constructing phylogenetic trees (Additional file 1: Table S15). Phylogenetic analysis was conducted using MEGA6 with the maximum likelihood method. Bootstrapping with 1000 replications was conducted to evaluate the phylogenetic tree. RNA-Seq datasets from gill and the air-breathing organ in this study and also from brain (SRR2057993), head kidney (SRR2057995) and liver (SRR2052655) in the public database were mapped to the genome sequence to estimate the Fragments Per Kilobase per Million mapped fragments (FPKM) metric for those expanded genes respectively by TopHat 2.0.10 and Cufflinks 2.1.1 [120–122].
Comparative transcriptomic analysis between the gill and the air-breathing organ
Wild C. batrachus individuals (70–136 g) were collected from Miami, Florida, USA in October of 2015. Tissue samples were collected after euthanasia using MS-222. The tissue samples from gill and air-breathing organ were kept in the RNAlater solution (Ambion) to prevent RNA degradation. Total RNAs were extracted from tissues of five individuals using the RNeasy Plus Universal Mini kit (Qiagen, CA) according to manufacturer’s instructions, and then the RNAs from five samples were mixed in equal amounts for RNA-Seq at HudsonAlpha (Huntsville, AL, USA). Standard Poly-A libraries were prepared, and 125 bp paired-end reads were generated using Illumina HiSeq 2500 sequencing platform. Raw reads were filtered with the parameters of base quality ≥20 and trimmed length ≥ 36 bp by Trimmomatic v0.32 [123]. All the trimmed reads from both tissues were mapped to 22,914 coding sequences predicted from genome assembly by the CLC Genomics Workbench software package [124]. The parameters for mapping were set as 90% or greater sequence identity with a maximum of two mismatches. The number of total mapped reads on each contig and reads per kilobase per million mapped reads (RPKM) were collected. After normalization of RPKM values, fold-changes were estimated to exhibit differentially expressed patterns of gene expression between the air-breathing organ and gill transcriptomes with a P-value < 0.05 using proportions-based Kal’s test [125] in the CLC Genomics Workbench software package. Transcripts with fold-change values greater than 2 were regarded as differentially expressed genes for subsequent analysis. Blast2GO version 4.0.7 [100] was used with default settings for the over-representation analysis of GO terms among the differentially expressed genes in the air-breathing organ and gill, and Reactome pathway database v60 [115] was used for further pathway enrichment analysis to indicate the functional differences between air-breathing organ and gill.
Real-time PCR validation of differentially expressed genes
To confirm the accuracy of the RNA-Seq analysis, quantitative real-time PCR (qRT-PCR) analysis was conducted. Total RNAs were extracted from the gill and air-breathing organ using the RNeasy Plus Universal Mini kit (Qiagen, CA) following the manufacturer’s instructions. After quantification with a Nanodrop spectrophotometer (Thermo Scientific), cDNA was synthesized with a final concentration of 50 ng/μL using the iScript cDNA Synthesis Kit (Quanta BioSciences) based on the manufacturer’s protocol. The primers used in qRT-PCR are listed in Additional file 1: Table S16. Amplification was performed on a CFX96 real-time PCR Detection System (Bio-Rad, CA). The thermal cycling profile consisted of an initial denaturation at 95 °C for 30 s, 40 cycles of denaturation at 94 °C for 5 s and an appropriate annealing/extension temperature at 60 °C for 10 s, and 72 °C for 5 s, followed by dissociation curve analysis to validate the specificity of amplified products. The 28S ribosomal RNA (rRNA) [126] (accession number JK488212) was used as the reference gene. Relative fold-changes for each gene were calculated in the Relative Expression Software Tool (REST) version 2009 [127] based on the values of cycle threshold (Ct) from real-time PCR.
Additional file
Figure S1. Multiple sequence alignment of myoglobin genes in the genomes of Clarias batrachus, Danio rerio, Ictalurus punctatus and Astyanax mexicanus. Figure S2. Detailed phylogenetic tree of olfactory receptor related to class A G protein-coupled receptor (ora) with species names and sequence names for Figure 3b. Figure S3. Multiple sequence alignment of olfactory receptor related to class A G protein-coupled receptor 1 (ora1) in the genomes of Clarias batrachus, Danio rerio, Ictalurus punctatus and Astyanax mexicanus. Figure S4. Syntenic analysis of myoglobin gene (mb) using genomic information of Danio rerio, Ictalurus punctatus and Clarias batrachus. Figure S5. Comparison of relative fold changes between air-breathing organ and gill in Clarias batrachus after normalization to 28S rRNA using RNA-Seq datasets and qRT-PCR. Table S1. Completeness of genome assembly assessed by CEGMA and BUSCO. Table S2. Mapping of five longest ABySS scaffolds to genome assembly. Table S3. Repetitive elements in the Clarias batrachus genome. Table S4. Orthogroups shared between species by OrthoFinder. Table S5. Specific genes in the Clarias batrachus genome compared with that of channel catfish (Ictalurus punctatus). Table S6. GO terms significantly enriched in the specific genes in the Clarias batrachus genome compared with that of channel catfish (Ictalurus punctatus). Table S7. Positively selected genes in the Clarias batrachus genome compared with those of 11 non-air-breathing teleost fish. Table S8. GO terms significantly enriched in the positively selected genes in the Clarias batrachus genome. Table S9. Expanded genes in the Clarias batrachus genome. Table S10. Summary of transcriptome sequencing data. Table S11. Expression values (RPKM) of all the genes in the transcriptomes of the gill and the air-breathing organ. Table S12. Differentially expressed genes in the transcriptome of the air-breathing organ compared with that of the gill. Table S13. GO terms significantly enriched in the differentially expressed genes comparing the transcriptome of the air-breathing organ with that of the gill. Table S14. Pathways significantly enriched in the differentially expressed genes comparing the transcriptome of the air-breathing organs with that of the gill. Table S15. Accession numbers of protein sequences and models used in the construction of phylogenetic trees of expanded genes. Table S16. Primer sequences used for qRT-PCR. (ZIP 5238 kb)
Acknowledgements
We want to thank Kelly Gestring from Florida Fish and Wildlife Conservation Commission for his help with the sampling, and Reviewer 3 for the thoughtful and constructive suggestions on the improvement of this manuscript. All the computational work was performed by using Auburn University high performance computer CASIC and Hopper systems. Ning Li is supported by the scholarship from the China Scholarship Council (CSC).
Funding
This project was partially supported by Agriculture and Food Research Initiative (AFRI) Competitive Grants 2009–35205-05101, 2012–67015-19410, and 2017–67015-26295 from the USDA National Institute of Food and Agriculture (NIFA) Animal Genomics, Genetics, and Breeding Program.
Availability of data and materials
The C. batrachus genome project has been deposited at GenBank under the BioProject PRJNA477835. The genome assembly is under accession QMIH00000000. Original sequencing reads are available from the Sequence Read Archive (SRA) under the accessions SRR7440017-SRR7440020.
Abbreviations
- BUSCO
Benchmarking Universal Single-Copy Orthologs
- CEG
Core Eukaryotic Gene
- DEG
Differentially Expressed Gene
- FPKM
Fragments Per Kilobase per Million mapped fragments
- GO
Gene Ontology
- LINE
Long Interspersed Elements
- LRT
Likelihood Ratio Test
- LTR
Long Terminal Repeats
- MOE
Main Olfactory Epithelium
- NR
Non-Redundant
- OUC
Ornithine Urea Cycle
- qRT-PCR
Quantitative Real-Time PCR
- ROS
Reactive Oxygen Species
- RPKM
Reads Per Kilobase per Million mapped reads
- rRNA
Ribosomal RNA
- SINE
Short Interspersed Elements
- VNO
Vomeronasal Organ
Authors’ contributions
NL prepared the samples for sequencing, conducted most of the experiments, data analysis, and prepared the manuscript. LB, ZY, and SL coordinated genome sequencing, genome assembly, annotation and comparative genomic analysis. TZ, XX, QZ, YaL, and YY coordinated transcriptome sequencing and comparative transcriptomic analysis. YuL and KW coordinated phylogenetic analysis and evolutionary analysis. YJ, SG and QF participated in the whole process of data analysis and generation of figures and Tables. RD, QL, AM and DG provided suggestions to improve the whole project and assisted in manuscript revision. ZL supervised the entire study and provided assistance in manuscript preparation. All authors have read and approved the manuscript.
Ethics approval and consent to participate
All procedures involving the handling and treatment of fish were approved by the Institutional Animal Care and Use Committee (IACUC) at Auburn University prior to the initiation of the experiments. All animal procedures were carried out according to the Guide for the Care and Use of Laboratory Animals and the Animal Welfare Act in the United States.
Consent for publication
Not applicable.
Competing interests
ZL is an associate editor of the journal.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ning Li, Email: nzl0016@auburn.edu.
Lisui Bao, Email: lzb0021@auburn.edu.
Tao Zhou, Email: tzz0016@auburn.edu.
Zihao Yuan, Email: zzy0012@auburn.edu.
Shikai Liu, Email: szl0021@auburn.edu.
Rex Dunham, Email: dunhara@auburn.edu.
Yuanning Li, Email: yzl0084@auburn.edu.
Kun Wang, Email: wk8910@gmail.com.
Xiaoyan Xu, Email: xzx0015@auburn.edu.
Yulin Jin, Email: yzj0025@auburn.edu.
Qifan Zeng, Email: qzz0011@auburn.edu.
Sen Gao, Email: szg0041@auburn.edu.
Qiang Fu, Email: qzf0005@auburn.edu.
Yang Liu, Email: yzl0173@auburn.edu.
Yujia Yang, Email: yzy0042@auburn.edu.
Qi Li, Email: qili66@ouc.edu.cn.
Axel Meyer, Email: axel.meyer@uni-konstanz.de.
Dongya Gao, Email: gaodong@auburn.edu.
Zhanjiang Liu, Email: johnliu@syr.edu.
References
- 1.Srivastava S, Kushwaha B, Prakash J, Kumar R, Nagpure N, Agarwal S, Pandey M, Das P, Joshi C, Jena J. Development and characterization of genic SSR markers from low depth genome sequence of Clarias batrachus (Magur) J Genet. 2016;95(3):603–609. doi: 10.1007/s12041-016-0672-8. [DOI] [PubMed] [Google Scholar]
- 2.Dahanukar N, Raut R, Bhat A. Distribution, endemism and threat status of freshwater fishes in the Western Ghats of India. J Biogeogr. 2004;31(1):123–136. doi: 10.1046/j.0305-0270.2003.01016.x. [DOI] [Google Scholar]
- 3.Islam MN, Islam MS, Alam MS. Genetic structure of different populations of walking catfish (Clarias batrachus L.) in Bangladesh. Biochem Genet. 2007;45(9–10):647–662. doi: 10.1007/s10528-007-9102-1. [DOI] [PubMed] [Google Scholar]
- 4.Khedkar GD, Reddy ACS, Mann P, Ravinder K, Muzumdar K. Clarias batrachus (Linn. 1758) population is lacking genetic diversity in India. Mol Biol Rep. 2010;37(3):1355–1362. doi: 10.1007/s11033-009-9517-3. [DOI] [PubMed] [Google Scholar]
- 5.Allen DJ. Clarias batrachus. The IUCN Red List of Threatened Species 2011: e.T166613A6247551. (http://dx.doi.org/10.2305/IUCN.UK.2011-1.RLTS.T166613A6247551.en). Accessed 17 Dec 2016.
- 6.Courtenay W Jr, Hensley D, Taylor J, McCann J. Distribution of exotic fishes in North America. Wiley; 1986. p. 675–698.
- 7.Saha N, Ratha B. Functional ureogenesis and adaptation to ammonia metabolism in Indian freshwater air-breathing catfishes. Fish Physiol Biochem. 2007;33(4):283–295. doi: 10.1007/s10695-007-9172-3. [DOI] [Google Scholar]
- 8.Das B. The bionomics of certain air-breathing fishes of India, together with an account of the development of their air-breathing organs. Phil Trans R Soc Lond B. 1928;216:183–219. doi: 10.1098/rstb.1928.0003. [DOI] [Google Scholar]
- 9.Courtenay WR, Sahlman HF, Miley WW, Herrema DJ. Exotic fishes in fresh and brackish waters of Florida. Biol Conserv. 1974;6(4):292–302. doi: 10.1016/0006-3207(74)90008-1. [DOI] [Google Scholar]
- 10.Munshi J. The accessory respiratory organs of Clarias batrachus (Linn.) J Morphol. 1961;109(2):115–139. doi: 10.1002/jmor.1051090202. [DOI] [PubMed] [Google Scholar]
- 11.Lewis S. The morphology of the accessory air-breathing organs of the catfish, Clarias batrachus: a SEM study. J Fish Biol. 1979;14(2):187–191. doi: 10.1111/j.1095-8649.1979.tb03509.x. [DOI] [Google Scholar]
- 12.Saha N, Ratha B. Ureogenesis in Indian air-breathing teleosts: adaptation to environmental constraints. Comp Biochem Physiol A Mol Integr Physiol. 1998;120(2):195–208. doi: 10.1016/S1095-6433(98)00026-9. [DOI] [Google Scholar]
- 13.Kelley JL, Yee M-C, Brown AP, Richardson RR, Tatarenkov A, Lee CC, Harkins TT, Bustamante CD, Earley RL. The genome of the self-fertilizing mangrove rivulus fish, Kryptolebias marmoratus: a model for studying phenotypic plasticity and adaptations to extreme environments. Genome Biol Evol. 2016;8(7):2145–2154. doi: 10.1093/gbe/evw145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.You X, Bian C, Zan Q, Xu X, Liu X, Chen J, Wang J, Qiu Y, Li W, Zhang X, et al. Mudskipper genomes provide insights into the terrestrial adaptation of amphibious fishes. Nat Commun. 2014;5:5594. doi: 10.1038/ncomms6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Randall D, Ip Y, Chew S, Wilson J. Air breathing and ammonia excretion in the giant mudskipper, Periophthalmodon schlosseri. Physiol Biochem Zool. 2004;77(5):783–788. doi: 10.1086/423745. [DOI] [PubMed] [Google Scholar]
- 16.Wright PA. Environmental physiology of the mangrove rivulus, Kryptolebias marmoratus, a cutaneously breathing fish that survives for weeks out of water. Integr Comp Biol. 2012;52(6):792–800. doi: 10.1093/icb/ics091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jianxun C, Xiuhai R, Qixing Y. Nuclear DNA content variation in fishes. Cytologia. 1991;56(3):425–429. doi: 10.1508/cytologia.56.425. [DOI] [Google Scholar]
- 18.Hinegardner R, Rosen DE. Cellular DNA content and the evolution of teleostean fishes. Am Nat. 1972:621–44.
- 19.Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23(9):1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 20.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 21.Vinogradov AE. Genome size and GC-percent in vertebrates as determined by flow cytometry: the triangular relationship. Cytometry. 1998;31(2):100–109. doi: 10.1002/(SICI)1097-0320(19980201)31:2<100::AID-CYTO5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 22.Tarallo A, Angelini C, Sanges R, Yagi M, Agnisola C, D’Onofrio G. On the genome base composition of teleosts: the effect of environment and lifestyle. BMC Genomics. 2016;17(1):173. doi: 10.1186/s12864-016-2537-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z, Liu S, Yao J, Bao L, Zhang J, Li Y, Jiang C, Sun L, Wang R, Zhang Y, et al. The channel catfish genome sequence provides insights into the evolution of scale formation in teleosts. Nat Commun. 2016;7:11757. doi: 10.1038/ncomms11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li WH, Graur D. Fundamentals of molecular evolution. 1st ed. Sinauer Associates; 1991.
- 25.Boeckmann B, Bairoch A, Apweiler R, Blatter M-C, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, Phan I. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31(1):365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toksoz D, Merdek K. The rho small GTPase: functions in health and disease. Histol Histopathol. 2001;17(3):915–927. doi: 10.14670/HH-17.915. [DOI] [PubMed] [Google Scholar]
- 27.Narumiya S. The small GTPase rho: cellular functions and signal transduction. J Biochem. 1996;120(2):215–228. doi: 10.1093/oxfordjournals.jbchem.a021401. [DOI] [PubMed] [Google Scholar]
- 28.Xu J, Li J-T, Jiang Y, Peng W, Yao Z, Chen B, Jiang L, Feng J, Ji P, Liu G. Genomic basis of adaptive evolution: the survival of Amur ide (Leuciscus waleckii) in an extremely alkaline environment. Mol Biol Evol. 2016;34(1):145–59. [DOI] [PMC free article] [PubMed]
- 29.Fuchs C, Burmester T, Hankeln T. The amphibian globin gene repertoire as revealed by the Xenopus genome. Cytogenet Genome Res. 2006;112(3–4):296–306. doi: 10.1159/000089884. [DOI] [PubMed] [Google Scholar]
- 30.Sidell BD, O'Brien KM. When bad things happen to good fish: the loss of hemoglobin and myoglobin expression in Antarctic icefishes. J Exp Biol. 2006;209(10):1791–1802. doi: 10.1242/jeb.02091. [DOI] [PubMed] [Google Scholar]
- 31.Hoffmann FG, Opazo JC, Storz JF. Differential loss and retention of cytoglobin, myoglobin, and globin-E during the radiation of vertebrates. Genome Biol Evol. 2011;3:588–600. doi: 10.1093/gbe/evr055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fraser J, de Mello LV, Ward D, Rees HH, Williams DR, Fang Y, Brass A, Gracey AY, Cossins AR. Hypoxia-inducible myoglobin expression in nonmuscle tissues. Proc Natl Acad Sci U S A. 2006;103(8):2977–2981. doi: 10.1073/pnas.0508270103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roesner A, Mitz SA, Hankeln T, Burmester T. Globins and hypoxia adaptation in the goldfish, Carassius auratus. FEBS J. 2008;275(14):3633–3643. doi: 10.1111/j.1742-4658.2008.06508.x. [DOI] [PubMed] [Google Scholar]
- 34.Schwarze K, Campbell KL, Hankeln T, Storz JF, Hoffmann FG, Burmester T. The globin gene repertoire of lampreys: convergent evolution of hemoglobin and myoglobin in jawed and jawless vertebrates. Mol Biol Evol. 2014;31(10):2708–2721. doi: 10.1093/molbev/msu216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koch J, Lüdemann J, Spies R, Last M, Amemiya CT, Burmester T. Unusual diversity of myoglobin genes in the lungfish. Mol Biol Evol. 2016;33(12):3033–3041. doi: 10.1093/molbev/msw159. [DOI] [PubMed] [Google Scholar]
- 36.Saraiva LR, Korsching SI. A novel olfactory receptor gene family in teleost fish. Genome Res. 2007;17(10):1448–1457. doi: 10.1101/gr.6553207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murray JI, Whitfield ML, Trinklein ND, Myers RM, Brown PO, Botstein D. Diverse and specific gene expression responses to stresses in cultured human cells. Mol Biol Cell. 2004;15(5):2361–2374. doi: 10.1091/mbc.e03-11-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Nadal E, Ammerer G, Posas F. Controlling gene expression in response to stress. Nature Rev Genet. 2011;12(12):833–845. doi: 10.1038/nrg3055. [DOI] [PubMed] [Google Scholar]
- 39.Atkinson B. Changes in eukaryotic gene expression in response to environmental stress. Academic Press; 2012.
- 40.Saha N, Dutta S, Haussinger D. Changes in free amino acid synthesis in the perfused liver of an air-breathing walking catfish, Clarias batrachus infused with ammonium chloride: a strategy to adapt under hyperammonia stress. J Exp Zool. 2000;286(1):13–23. doi: 10.1002/(SICI)1097-010X(20000101)286:1<13::AID-JEZ2>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 41.Saha N, Das L. Stimulation of ureogenesis in the perfused liver of an Indian air-breathing catfish, Clarias batrachus, infused with different concentrations of ammonium chloride. Fish Physiol Biochem. 1999;21(4):303–311. doi: 10.1023/A:1007856432423. [DOI] [Google Scholar]
- 42.Saha N, Dutta S, Bhattacharjee A. Role of amino acid metabolism in an air-breathing catfish, Clarias batrachus in response to exposure to a high concentration of exogenous ammonia. Comp Biochem Physiol B Biochem Mol Biol. 2002;133(2):235–250. doi: 10.1016/S1096-4959(02)00145-8. [DOI] [PubMed] [Google Scholar]
- 43.Wright PA. Nitrogen excretion: three end products, many physiological roles. J Exp Biol. 1995;198(2):273–281. doi: 10.1242/jeb.198.2.273. [DOI] [PubMed] [Google Scholar]
- 44.Amemiya CT, Alfoldi J, Lee AP, Fan S, Philippe H, Maccallum I, Braasch I, Manousaki T, Schneider I, Rohner N, et al. The African coelacanth genome provides insights into tetrapod evolution. Nature. 2013;496(7445):311–316. doi: 10.1038/nature12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J. Evolution by gene duplication: an update. Trends Ecol Evol. 2003;18(6):292–298. doi: 10.1016/S0169-5347(03)00033-8. [DOI] [Google Scholar]
- 46.Kimura M. The neutral theory of molecular evolution. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- 47.Ohno S. Evolution by gene duplication. New York: Springer-Verlag; 1970. [Google Scholar]
- 48.Meyer A, Schartl M. Gene and genome duplications in vertebrates: the one-to-four (−to-eight in fish) rule and the evolution of novel gene functions. Curr Opin Cell Biol. 1999;11(6):699–704. doi: 10.1016/S0955-0674(99)00039-3. [DOI] [PubMed] [Google Scholar]
- 49.Millikan G. Muscle hemoglobin. Physiol Rev. 1939;19(4):503–523. doi: 10.1152/physrev.1939.19.4.503. [DOI] [Google Scholar]
- 50.Wittenberg JB. Myoglobin-facilitated oxygen diffusion: role of myoglobin in oxygen entry into muscle. Physiol Rev. 1970;50(4):559–636. doi: 10.1152/physrev.1970.50.4.559. [DOI] [PubMed] [Google Scholar]
- 51.Millikan G. Experiments on muscle haemoglobin in vivo; the instantaneous measurement of muscle metabolism. Proc R Soc Lond [Biol]. 1937:218–41.
- 52.Wittenberg B, Wittenberg J, Caldwell P. Role of myoglobin in the oxygen supply to red skeletal muscle. J Biol Chem. 1975;250(23):9038–9043. [PubMed] [Google Scholar]
- 53.Reynafarje B. Myoglobin content and enzymatic activity of muscle and altitude adaptation. J Appl Physiol. 1962;17(2):301–305. doi: 10.1152/jappl.1962.17.2.301. [DOI] [PubMed] [Google Scholar]
- 54.Helbo S, Fago A. Functional properties of myoglobins from five whale species with different diving capacities. J Exp Biol. 2012;215(19):3403–3410. doi: 10.1242/jeb.073726. [DOI] [PubMed] [Google Scholar]
- 55.Driedzic WR, Stewart JM, Scott DL. The protective effect of myoglobin during hypoxic perfusion of isolated fish hearts. J Mol Cell Cardiol. 1982;14(11):673–677. doi: 10.1016/0022-2828(82)90164-X. [DOI] [PubMed] [Google Scholar]
- 56.Acierno R, Agnisola C, Tota B, Sidell B. Myoglobin enhances cardiac performance in antarctic icefish species that express the protein. Am J Physiol Regul Integr Comp Physiol. 1997;273(1):R100–R106. doi: 10.1152/ajpregu.1997.273.1.R100. [DOI] [PubMed] [Google Scholar]
- 57.Brunori M. Nitric oxide moves myoglobin Centre stage. Trends Biochem Sci. 2001;26(4):209–210. doi: 10.1016/S0968-0004(01)01824-2. [DOI] [PubMed] [Google Scholar]
- 58.Flögel U, Gödecke A, Klotz L-O, Schrader J. Role of myoglobin in the antioxidant defense of the heart. FASEB J. 2004;18(10):1156–1158. doi: 10.1096/fj.03-1382fje. [DOI] [PubMed] [Google Scholar]
- 59.Li C, Jackson RM. Reactive species mechanisms of cellular hypoxia-reoxygenation injury. Am J Physiol Cell Physiol. 2002;282(2):C227–C241. doi: 10.1152/ajpcell.00112.2001. [DOI] [PubMed] [Google Scholar]
- 60.Bickler PE, Buck LT. Hypoxia tolerance in reptiles, amphibians, and fishes: life with variable oxygen availability. Annu Rev Physiol. 2007;69:145–170. doi: 10.1146/annurev.physiol.69.031905.162529. [DOI] [PubMed] [Google Scholar]
- 61.Helbo S, Dewilde S, Williams DR, Berghmans H, Berenbrink M, Cossins AR, Fago A. Functional differentiation of myoglobin isoforms in hypoxia-tolerant carp indicates tissue-specific protective roles. Am J Physiol Regul Integr Comp Physiol. 2012;302(6):R693–R701. doi: 10.1152/ajpregu.00501.2011. [DOI] [PubMed] [Google Scholar]
- 62.Bargmann CI. Olfactory receptors, vomeronasal receptors, and the organization of olfactory information. Cell. 1997;90(4):585–587. doi: 10.1016/S0092-8674(00)80518-8. [DOI] [PubMed] [Google Scholar]
- 63.Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat Rev Neurosci. 2004;5(4):263–278. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- 64.Yoshihara Y. Molecular genetic dissection of the zebrafish olfactory system. In: Meyerhof W, Korsching SI, editors. Chemosensory Systems in Mammals, fishes, and insects. Springer. 2008. pp. 1–19. [Google Scholar]
- 65.Cao Y, Oh BC, Stryer L. Cloning and localization of two multigene receptor families in goldfish olfactory epithelium. Proc Natl Acad Sci U S A. 1998;95(20):11987–11992. doi: 10.1073/pnas.95.20.11987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pfister P, Rodriguez I. Olfactory expression of a single and highly variable V1r pheromone receptor-like gene in fish species. Proc Natl Acad Sci U S A. 2005;102(15):5489–5494. doi: 10.1073/pnas.0402581102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Young JM, Massa HF, Hsu L, Trask BJ. Extreme variability among mammalian V1R gene families. Genome Res. 2010;20(1):10–18. doi: 10.1101/gr.098913.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kurzweil VC, Getman M, Green ED, Lane RP. Dynamic evolution of V1R putative pheromone receptors between Mus musculus and Mus spretus. BMC Genomics. 2009;10(1):74. doi: 10.1186/1471-2164-10-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Syed AS, Korsching SI. Positive Darwinian selection in the singularly large taste receptor gene family of an ‘ancient’fish, Latimeria chalumnae. BMC Genomics. 2014;15(1):650. doi: 10.1186/1471-2164-15-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nikaido M, Noguchi H, Nishihara H, Toyoda A, Suzuki Y, Kajitani R, Suzuki H, Okuno M, Aibara M, Ngatunga BP. Coelacanth genomes reveal signatures for evolutionary transition from water to land. Genome Res. 2013;23(10):1740–1748. doi: 10.1101/gr.158105.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Van der Oost R, Beyer J, Vermeulen NP. Fish bioaccumulation and biomarkers in environmental risk assessment: a review. Environ Toxicol Pharmacol. 2003;13(2):57–149. doi: 10.1016/S1382-6689(02)00126-6. [DOI] [PubMed] [Google Scholar]
- 72.Mulder GJ. Conjugation reactions in drug metabolism: an integrated approach. Boca Raton: CRC Press; 2003. [Google Scholar]
- 73.Farah MA, Ateeq B, Ali MN, Sabir R, Ahmad W. Studies on lethal concentrations and toxicity stress of some xenobiotics on aquatic organisms. Chemosphere. 2004;55(2):257–265. doi: 10.1016/j.chemosphere.2003.10.063. [DOI] [PubMed] [Google Scholar]
- 74.Mos L, Cooper GA, Serben K, Cameron M, Koop BF. Effects of diesel on survival, growth, and gene expression in rainbow trout (Oncorhynchus mykiss) fry. Environ Sci Technol. 2008;42(7):2656–2662. doi: 10.1021/es702215c. [DOI] [PubMed] [Google Scholar]
- 75.Zhu L, Qu K, Xia B, Sun X, Chen B. Transcriptomic response to water accommodated fraction of crude oil exposure in the gill of Japanese flounder, Paralichthys olivaceus. Marine Poll Bull. 2016;106(1):283–291. doi: 10.1016/j.marpolbul.2015.12.022. [DOI] [PubMed] [Google Scholar]
- 76.Martin K. Theme and variations: amphibious air-breathing intertidal fishes. J Fish Biol. 2014;84(3):577–602. doi: 10.1111/jfb.12270. [DOI] [PubMed] [Google Scholar]
- 77.Janis CM, Farmer C. Proposed habitats of early tetrapods: gills, kidneys, and the water-land transition. Zool J Linnean Soc. 1999;126(1):117–126. doi: 10.1111/j.1096-3642.1999.tb00609.x. [DOI] [Google Scholar]
- 78.Randall DJ. The evolution of air breathing in vertebrates. Cambridge: Cambridge University Press; 1981. [Google Scholar]
- 79.Romer AS. Major steps in vertebrate evolution. Science. 1967;158(3809):1629–1637. doi: 10.1126/science.158.3809.1629. [DOI] [PubMed] [Google Scholar]
- 80.Inger RF. Ecological aspects of the origins of the tetrapods. Evolution. 1957;11(3):373–376. doi: 10.1111/j.1558-5646.1957.tb02907.x. [DOI] [Google Scholar]
- 81.Henry RP, Heming TA. Carbonic anhydrase and respiratory gas exchange. Fish Physiol. 1998;17:75–111. doi: 10.1016/S1546-5098(08)60259-9. [DOI] [Google Scholar]
- 82.Randall D, Val A. The role of carbonic anhydrase in aquatic gas exchange. In: Heisler N, editor. Mechanisms of Systemic Regulation Springer. 1995. pp. 25–39. [Google Scholar]
- 83.Gilmour K, Perry S. Carbonic anhydrase and acid–base regulation in fish. J Exp Biol. 2009;212(11):1647–1661. doi: 10.1242/jeb.029181. [DOI] [PubMed] [Google Scholar]
- 84.Ahmed A, Mohamed K, Ahmed S-A, Masoud F. Anatomical, light and scanning electron microscopic studies on the air breathing dendretic organ of the sharp tooth catfish (Clarias gariepinus) J Vet Anat. 2008;1(1):29–37. [Google Scholar]
- 85.Ikpegbu E, Nlebedum U, Nnadozie O, Agbakwuru O. Histological observations on the dendretic organ of the farmed adult African catfish (Clarias gariepinus) from eastern Nigeria. J Agric Sci. 2013;58(2):139–146. [Google Scholar]
- 86.Luo W, Cao X, Xu X, Huang S, Liu C, Tomljanovic T. Developmental transcriptome analysis and identification of genes involved in formation of intestinal air-breathing function of dojo loach, Misgurnus anguillicaudatus. Sci Rep. 2016;6:31845. doi: 10.1038/srep31845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jiang Y, Feng S, Xu J, Zhang S, Li S, Sun X, Xu P. Comparative transcriptome analysis between aquatic and aerial breathing organs of Channa argus to reveal the genetic basis underlying bimodal respiration. Mar Genomics. 2016;29:89–96. doi: 10.1016/j.margen.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 88.Chandra S, Banerjee TK. Histopathological analysis of the respiratory organs of the air-breathing catfish Clarias batrachus (Linn.) exposed to the air. Acta Zool Taiwanica. 2003;14(1):45–64. [Google Scholar]
- 89.Andrews S. FastQC: A quality control tool for high throughput sequence data. 2010. (Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc).
- 90.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 91.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19(6):1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gnerre S, MacCallum I, Przybylski D, Ribeiro FJ, Burton JN, Walker BJ, Sharpe T, Hall G, Shea TP, Sykes S, et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A. 2011;108(4):1513–1518. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27(4):578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 94.Boetzer M, Pirovano W. Toward almost closed genomes with GapFiller. Genome Biol. 2012;13(6):R56. doi: 10.1186/gb-2012-13-6-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Delcher AL, Salzberg SL, Phillippy AM. Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinformatics. 2003; chapter 10, unit 10.3. [DOI] [PubMed]
- 96.Bao Z, Eddy SR. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 2002;12(8):1269–1276. doi: 10.1101/gr.88502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21(suppl 1):i351–i358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- 98.Jukes TH, Cantor CR. Evolution of protein molecules. Mammalian Protein Metabolism. 1969;3(21):132. [Google Scholar]
- 99.Stanke M, Steinkamp R, Waack S, Morgenstern B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 2004;32(Suppl 2):W309–W312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 101.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35(Suppl 2):W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Emms D, Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16(1):157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD. PANTHER version 11: expanded annotation data from gene ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2016;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34(Suppl 2):W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56(4):564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 107.Borowiec ML. AMAS: a fast tool for alignment manipulation and computing of summary statistics. PeerJ. 2016;4:e1660. doi: 10.7717/peerj.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 2016;34(3):772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- 109.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 111.Gojobori T. Codon substitution in evolution and the “saturation” of synonymous changes. Genetics. 1983;105(4):1011–1027. doi: 10.1093/genetics/105.4.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22(12):2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 113.Yang Z, Wong WS, Nielsen R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22(4):1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 114.Kinsella RJ, Kähäri A, Haider S, Zamora J, Proctor G, Spudich G, Almeida-King J, Staines D, Derwent P, Kerhornou A. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database. 2011;2011:bar030. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fabregat A, Sidiropoulos K, Garapati P, Gillespie M, Hausmann K, Haw R, Jassal B, Jupe S, Korninger F, McKay S. The reactome pathway knowledgebase. Nucleic Acids Res. 2016;44(D1):D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Han MV, Thomas GW, Lugo-Martinez J, Hahn MW. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol Biol Evol. 2013;30(8):1987–1997. doi: 10.1093/molbev/mst100. [DOI] [PubMed] [Google Scholar]
- 117.Thompson JD, Gibson T, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2003;(1):2–3. [DOI] [PubMed]
- 118.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27(8):1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.CLC Genomics Workbench (https://www.qiagenbioinformatics.com/).
- 125.Kal AJ, van Zonneveld AJ, Benes V, van den Berg M, Koerkamp MG, Albermann K, Strack N, Ruijter JM, Richter A, Dujon B. Dynamics of gene expression revealed by comparison of serial analysis of gene expression transcript profiles from yeast grown on two different carbon sources. Mol Biol Cell. 1999;10(6):1859–1872. doi: 10.1091/mbc.10.6.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mohindra V, Tripathi RK, Singh A, Singh RK, Lal KK. Identification of candidate reference genes for quantitative expression analysis by real-time PCR for hypoxic stress in Indian catfish, Clarias batrachus (Linnaeus, 1758) Int Aquat Res. 2014;6(2):61. doi: 10.1007/s40071-014-0061-y. [DOI] [Google Scholar]
- 127.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30(9):e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Multiple sequence alignment of myoglobin genes in the genomes of Clarias batrachus, Danio rerio, Ictalurus punctatus and Astyanax mexicanus. Figure S2. Detailed phylogenetic tree of olfactory receptor related to class A G protein-coupled receptor (ora) with species names and sequence names for Figure 3b. Figure S3. Multiple sequence alignment of olfactory receptor related to class A G protein-coupled receptor 1 (ora1) in the genomes of Clarias batrachus, Danio rerio, Ictalurus punctatus and Astyanax mexicanus. Figure S4. Syntenic analysis of myoglobin gene (mb) using genomic information of Danio rerio, Ictalurus punctatus and Clarias batrachus. Figure S5. Comparison of relative fold changes between air-breathing organ and gill in Clarias batrachus after normalization to 28S rRNA using RNA-Seq datasets and qRT-PCR. Table S1. Completeness of genome assembly assessed by CEGMA and BUSCO. Table S2. Mapping of five longest ABySS scaffolds to genome assembly. Table S3. Repetitive elements in the Clarias batrachus genome. Table S4. Orthogroups shared between species by OrthoFinder. Table S5. Specific genes in the Clarias batrachus genome compared with that of channel catfish (Ictalurus punctatus). Table S6. GO terms significantly enriched in the specific genes in the Clarias batrachus genome compared with that of channel catfish (Ictalurus punctatus). Table S7. Positively selected genes in the Clarias batrachus genome compared with those of 11 non-air-breathing teleost fish. Table S8. GO terms significantly enriched in the positively selected genes in the Clarias batrachus genome. Table S9. Expanded genes in the Clarias batrachus genome. Table S10. Summary of transcriptome sequencing data. Table S11. Expression values (RPKM) of all the genes in the transcriptomes of the gill and the air-breathing organ. Table S12. Differentially expressed genes in the transcriptome of the air-breathing organ compared with that of the gill. Table S13. GO terms significantly enriched in the differentially expressed genes comparing the transcriptome of the air-breathing organ with that of the gill. Table S14. Pathways significantly enriched in the differentially expressed genes comparing the transcriptome of the air-breathing organs with that of the gill. Table S15. Accession numbers of protein sequences and models used in the construction of phylogenetic trees of expanded genes. Table S16. Primer sequences used for qRT-PCR. (ZIP 5238 kb)
Data Availability Statement
The C. batrachus genome project has been deposited at GenBank under the BioProject PRJNA477835. The genome assembly is under accession QMIH00000000. Original sequencing reads are available from the Sequence Read Archive (SRA) under the accessions SRR7440017-SRR7440020.