Abstract
Promoter single nucleotide polymorphisms (SNPs) of the ABCB1 gene, encoding the placental efflux transporter P-glycoprotein, can alter its expression and affect fetal exposure to therapeutics and environmental xenobiotics. SNPs are not arrayed as independent variants but as combinations forming defined haplotypes. Recently, we defined the haplotypes encompassing ABCB1 promoter SNPs and found that ABCB1 haplotypes differentially affect its promoter activity. The mechanism(s) by which ABCB1 haplotypes alter its promoter activity are not known. We hypothesize that the haplotype-dependent differences in ABCB1 promoter activity are due to haplotype-specific alterations in transcription factor (TF) binding. To test our hypothesis, we used a TF binding profile array and determined whether differences in TF binding exist across different ABCB1 haplotypes. TFs showing significant haplotype binding differences were mechanistically evaluated using small interfering RNA (siRNA) in cultured human placental cells. Our data indicate significant haplotype-dependent differences in TF binding. Our siRNA studies showed that the regulatory effects of TFs on promoter activity are also haplotype dependent. Our data provide a mechanistic explanation for the differential effects of ABCB1 haplotypes on its promoter activity and underscore the importance of evaluating genetic variants in the context of haplotypes rather than individual SNPs when investigating their effects on gene/protein expression and disease risk.
Keywords: ABCB1 expression, P-glycoprotein, SNPs, haplotypes, transcription factors
Introduction
P-glycoprotein (P-gp), encoded by the ABCB1 gene, is a promiscuous efflux transporter that interacts with numerous structurally diverse substrates (Kim, 2002; Schinkel and Jonker, 2003; Ceckova-Novotna et al., 2006). P-gp was first discovered in cancer cells, associated with the phenomenon of multiple drug resistance (Juliano and Ling, 1976). However, it is now known that P-gp is highly expressed in normal tissues including liver, lower gastrointestinal tract, and kidneys; endothelial cells that make up the blood–brain barrier; and in other tissue–blood barriers such as the testes and placenta (Kim, 2002).
In the placenta, P-gp is found on the apical membrane of the syncytiotrophoblasts (Young et al., 2003). By utilizing ATP hydrolysis, P-gp actively extrudes its substrates from the trophoblasts back to the maternal circulation, thus limiting their entry into the fetal circulation (Nakamura et al., 1997). As such, variability in placental P-gp expression/activity could significantly influence maternal and fetal exposure to many prescribed medications and environmental agents that are P-gp substrates.
There is a large interindividual variability in placental P-gp levels/activity (Hemauer et al., 2010), which could be a result of variability in ABCB1 expression levels. The ABCB1 promoter contains many single nucleotide polymorphisms (SNPs) that form specific haplotypes (defined SNP combinations) that differentially affect ABCB1 promoter activity (Takane et al., 2004; Speidel et al., 2018). However, the exact mechanism(s) by which these haplotypes exert their effect on ABCB1 promoter activity has not been determined.
The ABCB1 promoter is rich with regulatory regions, and many cis-elements have been identified, including a heat shock element and a partial estrogen response element (Chin et al., 1990; Shi et al., 2014). In addition, other binding sites for important transcription factors (TFs) that regulate ABCB1 promoter, including the transcriptional regulatory proteins Sp1, AP-1, and p53, have been identified (reviewed in Labialle et al., 2002). Previous work has shown that Sp1 binds to two different regions of the ABCB1 promoter and, depending on which of the two sites it occupies, it plays either a transcriptional-activating or -repressing role (Cornwell and Smith, 1993). In a recent study, we performed an in silico bioinformatics analysis of the ABCB1 promoter region, which further confirmed the presence of these binding sites and identified additional putative TF binding sites in the promoter (Speidel et al., 2018). These sites include glucocorticoid receptors, vitamin D receptors, aryl hydrocarbon receptors, and others (Speidel et al., 2018).
To date, a comprehensive evaluation of the effects of TF binding on the ABCB1 promoter activity has not been conducted. In addition, the effect of ABCB1 promoter haplotypes on TF binding and their regulatory effect on ABCB1 promoter activity have not been considered. Several ABCB1 haplotypes include SNPs within or in proximity to known or predicted TF binding sites. For example, the G-1157aA SNP (rs28381797) found in several ABCB1 haplotypes (Speidel et al., 2018) is in a binding domain of the Sp1 TF. Similarly, the G-1459aA SNP (rs12720464) found in other ABCB1 haplotypes (Speidel et al., 2018) is in a shared binding domain for GATA1 and GATA2 TFs.
Given that ABCB1 haplotypes have been shown to differentially alter its promoter activity (Takane et al., 2004; Speidel et al., 2018), it is plausible that these haplotypes provoke structural changes capable of modifying the recruitment and/or binding of different transcriptional regulators, resulting in altered ABCB1 expression.
We, therefore, hypothesized that haplotype-dependent differences observed in ABCB1 promoter activity are due to haplotype-specific alterations in TF binding. We tested our hypothesis using four ABCB1 promoter haplotypes with significantly different promoter activities that we identified in a previous study from our laboratory (Speidel et al., 2018). To test our hypothesis, we used a TF binding profile array and determined whether differences in TF binding across the haplotypes exist.
TFs that showed significant differences in binding to different haplotypes were identified and selected for in-depth in vitro mechanistic studies. We also included other TFs which we identified from our bioinformatics analysis (Speidel et al., 2018) and from review of the literature. Their role in regulating ABCB1 promoter activity was evaluated using small interfering RNA (siRNA) in cultured human placental cells. Our data indicate that TF binding, as well as their regulatory effect on promoter activity, is haplotype dependent.
Materials and Methods
Cell culture
The human trophoblastic 3A placental cell line (CRL-1584) was purchased from American Type Culture Collection (Manassas, VA). Nuclear extracts containing TFs were isolated from cultured cells and used to determine the effect of different haplotypes on TF binding. The 3A cells were also used in additional experiments involving siRNA studies as the host for ABCB1 promoter haplotype luciferase reporter construct transfection. These studies were conducted to further characterize the effect of individual TFs on ABCB1 promoter activity. Cells were maintained in 75 cm2 flasks with Minimal Essential Medium with Earle's salts and l-glutamine (Gibco, Grand Island, NY) supplemented with 10% FBS in 5% CO2 at 37°C. Cells were passaged at 85% confluency (2–3 days) and subcultured at a 3:1 ratio. A solution of 0.25% Trypsin (w/v)—0.53 mM EDTA solution was used to detach the cells. The cells were detached for subculture or for transfer to six well plates for transfection. Nuclear extracts were collected from 3A cells following the manufacturer's protocol using the nuclear extraction kit (Signosis, Santa Clara, CA).
ABCB1 promoter haplotype luciferase reporter construct generation
We generated luciferase reporters using the NanoLuc™ Luciferase vector system (Promega, Madison, WI) to determine the effects of ABCB1 promoter haplotypes on promoter activity as we had done previously (Speidel et al., 2018). For the current study, four haplotypes were evaluated (Table 1), namely, the ancestral haplotype 1 representing the reference promoter activity (100%), haplotypes 4 and 29 with significantly higher basal promoter activity than haplotype 1 (390% and 350%, respectively), and haplotype 30 with significantly lower basal promoter activity than haplotype 1 (6%). In brief, constructs representing the four ABCB1 promoter haplotypes were generated by inserting the appropriate promoter sequences into the NanoLuc pNL1.1 vector (Promega) after double-digestion with the restriction enzymes KpnI-HF and NheI-HF (New England Biolabs, Ipswich, MA). The reporter constructs were then used to transform competent Escherichia coli DH5α cells (New England Biolabs) and plated on 100 μg/mL ampicillin Luria-Bertani (LB) agar plates. Individual colonies were selected and grown in LB medium containing 100 μg/mL ampicillin for 18–24 h at 37°C.
Table 1.
ABCB1 Haplotypes Evaluated for TF Binding and Promoter Activity
| Haplotypea | ABCB1 promoter haplotype structureb |
|---|---|
| 1 | −1572aA/−1517aT/−1459aG/−1157aG/−1017aT/−684aA/−274aG/−41aA/−240G/−129T/−43A/133C |
| 4 | −1572aA/−1517aT/−1459aG/−1157aG/−1017aT/−684aA/−274aG/−41aA/−240A/−129T/−43A/133C |
| 29 | −1572aA/−1517aT/−1459aA/−1157aG/−1017aT/−684aA/−274aG/−41aA/−240G/−129C/−43A/133C |
| 30 | −1572aA/−1517aT/−1459aG/−1157aA/−1017aC/−684aA/−274aG/−41aA/−240G/−129T/−43A/133C |
The (−) without the “a” are within or after exon 1 before the transcription start site. Bold denotes variant(s) present in the haplotype (see Speidel et al., 2018 for additional details). Variants present in the different haplotypes are G-240A (rs35265821); G-1459aA (rs12720464); T-129C (rs3213619); G-1157aA (rs28381797); and T-1017aC (rs28746504).
Haplotype 1 (ancestral haplotype) is used as a reference for TF binding and promoter activity comparisons. Haplotypes 4 and 29 have significantly higher basal promoter activity, and haplotype 30 has significantly lower basal promoter activity than haplotype 1 (Speidel et al., 2018).
Small “a” nomenclature represents nucleotide before the nontranscribed exon 1.
TF, transcription factor.
Plasmids were isolated using the endotoxin-free ZR Plasmid Miniprep™-Classic kit (Zymo Research Corp., Irvine, CA) and quantified at 260 nm using a DS-11 spectrophotometer (DeNovix, Inc., Wilmington, DE). The isolated plasmids were sequenced to verify the presence of the proper promoter haplotype in the reporter plasmid and to ensure no additional mutations were introduced during the preparation. Isolated plasmids were stored at −20°C to maintain plasmid integrity until transfection.
TF binding assay
TF binding to the ABCB1 promoter haplotypes was determined with the Promoter Binding TF Profiling Plate Array I (Signosis) using the nuclear extracts isolated from placental 3A cells. This array provides an assay for rapid determination of binding of 48 TFs as detailed in the product manual. In brief, an ABCB1 promoter DNA fragment, corresponding to a specific haplotype, competes with biotin-labeled DNA oligos for TFs present in the nuclear extract from placental 3A cells. If a TF binding site is present in the ABCB1 promoter DNA fragment tested, a decrease in the formation of biotin-labeled probe–TF complex for that TF occurs. Using streptavidin conjugated with horseradish peroxidase and a chemiluminescent substrate, a luminescence signal was detected. The intensity of luminescence correlates with the degree of TF binding to the corresponding ABCB1 promoter, where a strong luminescence signal indicates low binding between the TF and the tested ABCB1 promoter, whereas a weak luminescence signal indicates strong binding between the TF and the tested ABCB1 promoter.
TF knockdown with siRNA
To further characterize the relationship between TFs and different ABCB1 haplotypes on promoter activity, the effect of selected TFs on ABCB1 promoter activity was investigated using siRNAs (Table 2). The siRNAs were cotransfected into 3A cells with the luciferase reporter constructs representing the haplotypes tested. Transfections were performed between passages 6 and 8 with cells at low confluency (≤40%). For each transfection, cells grown in a 24-well plate were treated with a mixture of 600 ng promoter haplotype plasmid DNA, 60 ng firefly luciferase control plasmid pGL4.53 PGK (Promega), 2.5 pmol siRNA, and 2 μL Lipofectamine 3000 transfection reagent (Invitrogen, Carlsbad, CA). After transfection, cells were allowed to recover for 40 h before harvest.
Table 2.
List of siRNAs and Their Corresponding Targets Evaluated in This Study
| siRNA ID | Gene symbola | TFb | Gene name |
|---|---|---|---|
| s19772 | CEBPZ | C/EBP | CCAAT/Enhancer Binding Protein, Zeta |
| s3489 | CREB1 | CREB | cAMP Responsive Element Binding Protein 1 |
| s5594 | GATA1 | GATA1 | GATA Binding Protein 1 |
| s10064 | PAX-5 | Pax-5 | Paired Box Gene 5 |
| s279 | STAT1 | STAT1 | Signal Transducer and Activator of Transcription 1 |
| s13531 | STAT4 | STAT4 | Signal Transducer and Activator of Transcription 4 |
| s13318 | SP1 | Sp1 | Specificity Protein 1 |
| s14778 | VDR | VDR | Vitamin D Receptor |
| s4825 | ESR1 | ERα | Estrogen Receptor 1, Alpha |
| s9528 | NFYA | NFY | Nuclear Transcription Factor Y, Alpha |
| s5596 | GATA2 | GATA2 | GATA Binding Protein 2 |
| s9501 | NFIX | NF-1 | Nuclear Factor I/X (CCAAT-Binding Transcription Factor) |
| s743 | STAT3 | STAT3 | Signal Transducer and Activator of Transcription 3 |
| s14006 | TFAP2B | AP2 | Transcription Factor AP-2 Beta |
| s21070 | NFAT5 | NFAT | Nuclear Factor of Activated T cells 5 |
| s16909 | NR1I2 | PXR | Pregnane X Receptor |
| s3494 | ATF2 | ATF2 | Activating Transcription Factor 2 |
| s605 | TP53 | p53 | Tumor Protein p53 |
| s12479 | SATB1 | SATB1 | Special AT-Rich Sequence Binding Protein 1 |
| s13826 | TBP | TFIID | TATA-Box Binding Protein |
Genes targeted by the siRNA listed according to Thermo Fisher Scientific.
TFs targeted by siRNA (PROMO; Signosis, Santa Clara, CA).
siRNA, small interfering RNA; TF, transcription factor.
Nano-Glo Dual-Luciferase Assay to determine the effect of siRNAs on the activity of ABCB1 promoter haplotypes
The Nano-Glo® Dual-Luciferase® Reporter Assay (Promega) was performed according to the manufacturer's instructions. In brief, 3A cells were harvested using 400 μL Passive Lysis Buffer 40–48 h after transfection. Lysates were then either used immediately or stored at −80°C for later analysis. Luciferase activity was measured according to the manufacturer's recommendations, and luminescence was measured in triplicate using a Tecan GenIOS Pro plate reader (Tecan, Durham, NC). Luminescence was measured as relative light units (RLU) by normalization against the cotransfected firefly luciferase. Each experiment was repeated at least three times.
Statistical analysis
The nonparametric Kruskal–Wallis one-way analysis of variance was used to compare luminescence values corresponding to the effects of different siRNA knockdowns on ABCB1 haplotype promoter activity. Post hoc analysis using Dunnett's method was used to compare each siRNA TF knockdown against control within the same haplotype. p-values <0.05 were considered significant.
Results
Determination of ABCB1 promoter haplotype TF binding profiles
Nuclear extracts were isolated from placental 3A cells that express P-gp. ABCB1 promoter DNA fragments representing the four haplotypes tested were then individually used in the Promoter Binding TF Profiling Plate Array. The ABCB1 promoter haplotype 1 was used as a reference to identify the binding profile of 48 TFs present in the array to the ABCB1 promoter region. The TF binding activity, measured by chemiluminescent signals, is inversely correlated with the binding activities of TFs evaluated. A strong signal indicates weak TF–DNA binding and a weak signal indicates a strong DNA–TF complex formation. The differences observed in TF binding between the haplotypes are shown in Table 3 and visually in the heatmap (Fig. 1). In Table 3, the haplotype effect on TF binding activity is expressed as a ratio of RLU of the tested haplotype over the RLU of haplotype 1. A ratio <1.0 indicates an increased binding activity for a TF with the evaluated haplotype compared with that observed with haplotype 1, whereas a ratio >1.0 indicates a decreased haplotype binding activity for a TF than that observed with haplotype 1. For haplotype 1, the signal ranged from 417 RLU for glucocorticoid/progesterone (GR/PR), indicating a strong TF binding (or multiple binding sites for this TF) to 5125 RLU for TATA-box binding protein (TFIID), demonstrating weak binding (or lack of a binding site) in the ABCB1 haplotype 1 promoter. In Figure 1, strong binding is represented by red, and weak binding is denoted by green. It should be noted that a strong binding activity reflects a high affinity of the TF to one or more binding sites on the promoter.
Table 3.
Haplotype Effect on TF Binding Activity Expressed as a Ratio of RLU of the Tested Haplotype Over the RLU of Haplotype 1
| Haplotype construct | ||||
|---|---|---|---|---|
| TFs | Haplotype 1a | Haplotype 4b | Haplotype 29b | Haplotype 30b |
| TFIID | 5125 | 0.22 | 0.63 | 0.55 |
| TR | 4703 | 0.16 | 1.04 | 0.51 |
| NF-1 | 4686 | 0.29 | 1.62 | 0.87 |
| ER | 4499 | 0.13 | 0.63 | 0.52 |
| Stat3 | 4254 | 0.36 | 0.49 | 0.60 |
| TCF/LEF | 4061 | 0.31 | 0.36 | 0.40 |
| Brn-3 | 3974 | 0.14 | 0.62 | 0.48 |
| C/EBP | 3851 | 0.08 | 0.20 | 0.33 |
| Myb | 3438 | 0.18 | 0.58 | 0.40 |
| YY1 | 3345 | 0.32 | 1.38 | 0.53 |
| NF-E2 | 3233 | 0.25 | 1.02 | 0.35 |
| Pax-5 | 3216 | 0.23 | 2.10 | 0.48 |
| Oct4 | 3169 | 0.26 | 0.62 | 0.83 |
| FAST-1 | 2940 | 0.20 | 0.41 | 0.45 |
| GAS/ISRE | 2856 | 0.66 | 0.83 | 0.35 |
| GATA | 2855 | 0.63 | 1.21 | 0.39 |
| SATB1 | 2849 | 0.45 | 0.26 | 0.65 |
| PXR | 2733 | 0.33 | 1.76 | 0.67 |
| Sp1 | 2473 | 0.22 | 0.31 | 1.02 |
| CAR | 2408 | 0.22 | 1.14 | 0.42 |
| Pit | 2394 | 0.61 | 1.04 | 0.68 |
| Ets | 2366 | 0.18 | 0.81 | 0.63 |
| MEF2 | 2202 | 0.16 | 0.44 | 0.69 |
| Stat1 | 2083 | 0.72 | 1.05 | 0.20 |
| E2F-1 | 2076 | 0.38 | 0.72 | 0.42 |
| p53 | 2028 | 0.24 | 1.11 | 0.54 |
| CDP | 1950 | 1.00 | 1.11 | 2.20 |
| Stat6 | 1898 | 0.45 | 0.60 | 1.07 |
| CREB | 1653 | 0.29 | 1.32 | 2.89 |
| IRF | 1646 | 0.51 | 0.56 | 0.65 |
| SMAD | 1552 | 0.34 | 0.69 | 1.14 |
| CBF | 1429 | 0.50 | 0.64 | 0.55 |
| NFAT | 1418 | 0.31 | 0.75 | 1.04 |
| ATF2 | 1380 | 0.51 | 0.66 | 0.72 |
| HNF4 | 1361 | 0.39 | 0.80 | 0.94 |
| SRF | 1335 | 0.24 | 2.99 | 1.17 |
| EGR | 1276 | 0.69 | 0.74 | 0.92 |
| HIF | 1272 | 0.56 | 0.66 | 0.57 |
| Pbx1 | 1218 | 0.84 | 1.70 | 0.71 |
| NFkB | 1217 | 0.45 | 0.61 | 0.88 |
| Stat5 | 1178 | 0.93 | 1.18 | 1.25 |
| PPAR | 1133 | 0.51 | 0.77 | 0.74 |
| Myc-Max | 1091 | 0.57 | 1.69 | 1.01 |
| AP1 | 1058 | 1.96 | 2.05 | 1.66 |
| Stat4 | 826 | 1.32 | 1.30 | 1.97 |
| AR | 762 | 1.04 | 2.08 | 1.51 |
| AP2 | 471 | 0.97 | 1.45 | 4.24 |
| GR/PR | 417 | 1.13 | 1.12 | 3.04 |
TF binding activity to haplotype 1 measured with chemiluminescence and expressed as RLU. RLU values are inversely correlated with the binding activities of TFs evaluated.
The change in binding activity is expressed as the ratio of RLU of the tested haplotype over RLU of haplotype 1. A ratio <1.0 indicates stronger TF promoter binding than to haplotype 1, whereas a ratio >1.0 indicates lower promoter binding for the TF than to haplotype 1.
RLU, relative light units; TF, transcription factor.
FIG. 1.
Heatmap representing the TF binding profiles for four ABCB1 promoter haplotypes evaluated. The heatmap was generated using Heatmapper (Babicki et al., 2016) from the RLU values representing the binding activity for 48 TFs to the ABCB1 promoter haplotypes evaluated. Each color-tile represents the average RLU value from two independent assays for TF binding to an ABCB1 promoter fragment. RLU, relative light units; TF, transcription factor. Color images available online at www.liebertpub.com/dna
Overall, our data indicate clear haplotype-dependent differences in TF binding. For example, the C/EBP had a 12.5-fold lower signal when binding with haplotype 4 than with haplotype 1, indicating much stronger binding activity with haplotype 4 than with haplotype 1. Alternatively, the signal from AP2 binding activity is increased more than fourfold for haplotype 30 relative to haplotype 1, indicating decreased binding activity with haplotype 30 compared with haplotype 1. Other TFs had low binding affinity to haplotype 1, but strong binding to one or more of the other haplotypes evaluated as indicated by the low ratio values in Table 3 and as depicted in the heatmap (Fig. 1). Examples include thyroid hormone receptor (TR), estrogen receptor (ER), signal transducer and activator of transcription 3 (Stat3), runt-related transcription factor 2 (TCF/LEF), and TFIID. With the TATA box-binding TFIID, we observed a fivefold stronger binding affinity with haplotype 4 than the minimal affinity observed with haplotype 1. Some other TFs appeared to exhibit a strong binding activity to the ABCB1 promoter regardless of the haplotype tested (e.g., peroxisome proliferator activated receptor). An interesting observation was the variability in binding affinity of the GR/PR receptor depending on the haplotype evaluated. Although there are numerous GR/PR binding domains on the ABCB1 promoter, as indicated by its strong binding observed with the ancestral haplotype 1 and haplotypes 4 and 29, its binding affinity with haplotype 30 was significantly decreased by threefold compared with that of the ancestral haplotype.
Effect of TF binding on ABCB1 promoter activity
To investigate the potential regulatory effects of specific TFs on ABCB1 expression, we used Ambion® Silencer® Select siRNAs to target 20 individual TFs (Table 2). These TFs were chosen for in-depth evaluation based on the results obtained from the TF binding array data and based on our in silico bioinformatics analysis performed previously on the ABCB1 promoter (Speidel et al., 2018) and from a literature search that identified additional TFs known to bind to the ABCB1 promoter (Labialle et al., 2002; Saeki et al., 2011; Gromnicova et al., 2012; Rigalli et al., 2015). The siRNAs that target specific TFs were cotransfected into 3A cells with the various haplotype reporter constructs and a luciferase transfection control plasmid. The effect of individual siRNAs on the haplotype-dependent promoter activity was then evaluated by comparing the activity with the corresponding basal promoter activity (without siRNA treatment).
The siRNAs studied provoked significant haplotype-dependent changes in ABCB1 promoter activity (Table 4 and Fig. 2). In Table 4, the activity of the promoter with each siRNA tested is compared with that of the nontreated control. A value >1.0 indicates higher promoter activity than the control, whereas a value <1.0 indicates a promoter activity lower than the control. A value equal to 1 indicates no effect for the tested siRNA on promoter activity. Although several of the siRNAs tested induced significant changes in promoter activity, the effect was not consistent across the haplotypes. For example, siRNA knockdown of Pax-5 produced a significant increase in promoter activity for haplotype 1. However, knockdown of Pax-5 induced a significant decrease in promoter activity for haplotype 29. For haplotypes 4 and 30, Pax-5 knockdown produced only a slight nonsignificant decrease in promoter activity. With haplotype 29, which has a high basal promoter activity, the knockdown of CREB, GATA1, Pax-5, Sp1, NFYA, and ATF2 with siRNA significantly decreased the promoter activity (Table 4). Overall, our data indicate that siRNA knockdown of certain TFs resulted in upregulating ABCB1 promoter activity, whereas knockdown of others led to a downregulation of promoter activity. Importantly, the effect of an individual siRNA was not always consistent across the haplotypes tested, but rather haplotype dependent.
Table 4.
Effects of siRNA-Mediated TF Knockdown for 20 Selected TFs
| Haplotype 1 | Haplotype 4 | Haplotype 29 | Haplotype 30 | |
|---|---|---|---|---|
| C/EBP | 0.80 ± 0.17 | 0.67 ± 0.06 | 0.65 ± 0.09 | 2.40 ± 0.61 |
| CREB | 0.95 ± 0.10 | 0.53 ± 0.04 | 0.44 ± 0.02 | 1.44 ± 0.38 |
| GATA1 | 1.28 ± 0.25 | 1.44 ± 0.39 | 0.43 ± 0.04 | 0.99 ± 0.12 |
| PAX-5 | 1.72 ± 0.08 | 0.86 ± 0.07 | 0.47 ± 0.06 | 0.88 ± 0.10 |
| STAT1 | 1.20 ± 0.25 | 0.94 ± 0.12 | 0.64 ± 0.09 | 1.08 ± 0.13 |
| STAT4 | 1.35 ± 0.11 | 1.01 ± 0.17 | 0.49 ± 0.04 | 0.99 ± 0.14 |
| SP1 | 1.04 ± 0.27 | 0.52 ± 0.07 | 0.49 ± 0.08 | 0.76 ± 0.12 |
| VDR | 1.41 ± 0.26 | 1.02 ± 0.11 | 0.72 ± 0.04 | 1.43 ± 0.18 |
| ER1 | 1.44 ± 0.09 | 0.89 ± 0.04 | 0.89 ± 0.05 | 1.10 ± 0.18 |
| NFYA | 1.00 ± 0.08 | 0.68 ± 0.08 | 0.37 ± 0.04 | 1.25 ± 0.18 |
| AP2 | 2.33 ± 0.32 | 1.59 ± 0.24 | 1.15 ± 0.12 | 0.96 ± 0.10 |
| ATF2 | 1.27 ± 0.15 | 1.02 ± 0.11 | 0.44 ± 0.03 | 1.01 ± 0.12 |
| GATA2 | 1.11 ± 0.15 | 1.14 ± 0.12 | 0.68 ± 0.08 | 1.80 ± 0.30 |
| NF-1 | 1.15 ± 0.06 | 1.13 ± 0.19 | 0.54 ± 0.05 | 1.16 ± 0.11 |
| NFAT5 | 2.00 ± 0.17 | 1.06 ± 0.09 | 0.51 ± 0.06 | 1.54 ± 0.25 |
| P53 | 0.73 ± 0.16 | 0.68 ± 0.09 | 0.50 ± 0.04 | 0.82 ± 0.12 |
| SATB1 | 1.41 ± 0.11 | 0.60 ± 0.08 | 0.92 ± 0.15 | 1.20 ± 0.11 |
| STAT3 | 1.35 ± 0.17 | 1.06 ± 0.12 | 0.56 ± 0.03 | 1.35 ± 0.19 |
| TBP | 1.85 ± 0.16 | 0.92 ± 0.06 | 1.46 ± 0.16 | 1.46 ± 0.20 |
| PXR | 1.48 ± 0.07 | 1.07 ± 0.08 | 0.71 ± 0.05 | 1.22 ± 0.16 |
| Control | 1.00 ± 0.09 | 1.00 ± 0.14 | 1.00 ± 0.10 | 1.00 ± 0.13 |
Bold values indicate significant difference (p < 0.05) for tested siRNA vs. control.
The change in promoter activity is expressed as the ratio of RLU for the siRNA over the RLU of the control within the same haplotype. A ratio <1.0 indicates a decrease in promoter activity with the TF knockdown, whereas a ratio >1.0 indicates an increase in promoter activity after TF knockdown.
RLU, relative light units; siRNA, small interfering RNA; TF, transcription factor.
FIG. 2.
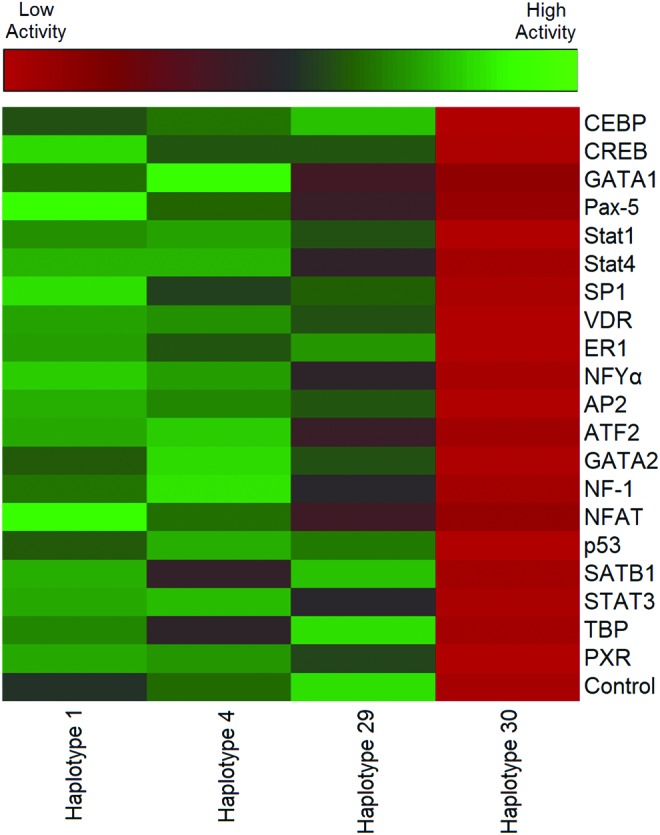
Heatmap representing ABCB1 promoter activities before and after treatment with siRNA. The heatmap represents ABCB1 promoter activity after siRNA knockdown of 20 different transcription factors. The heatmap was generated from the RLU values before (control) and after treatment with different siRNAs. Each tile represents the average RLU value from three independent assays for the promoter activity after treatment with a specific siRNA. RLU, relative light units; siRNA, small interfering RNA. Color images available online at www.liebertpub.com/dna
Discussion
The efflux transporter protein P-gp, located on the apical membrane of the placental trophoblasts, plays a major role in the transfer of xenobiotics across the human placenta. Alteration in P-gp expression can, therefore, have serious consequences for the fetus if the mother is exposed to xenobiotics that are P-gp substrates. Although variability in P-gp could be due to a number of factors, genetic variability in the promoter of the ABCB1 gene could alter its expression and consequently P-gp levels (Takane et al., 2004; Speidel et al., 2018). Recently, we comprehensively defined the haplotypes encompassing the common promoter SNPs of the ABCB1 gene and demonstrated that the activity of the ABCB1 promoter is haplotype dependent (Speidel et al., 2018). To define the underlying mechanisms, in this study we investigated whether the variability in promoter activity was due to alteration in TF binding.
To determine the putative TFs involved in regulating ABCB1 promoter activity, we used the Signosis Promoter Binding TF Profiling Array I plate, which provides the ability to evaluate the binding activity of 48 common TFs known to affect the expression of many genes. Our data indicated that certain TFs that demonstrated a strong binding activity to the ancestral haplotype 1 exhibited lower binding affinity with the other haplotypes. Other TFs had a low binding affinity to haplotype 1, but a strong binding affinity to one or more of the other haplotypes evaluated. These data indicate a haplotype-dependent difference in TF binding to the ABCB1 promoter. Our data also show that other TFs appeared to exhibit a strong binding activity to the ABCB1 promoter regardless of the haplotype tested, suggesting that these TFs may be essential for regulating ABCB1 promoter activity, a possibility that needs to be confirmed by future studies.
A noteworthy observation from our data is that, although there is no TATA box in the ABCB1 promoter (van Groenigen et al., 1993), we observed a fivefold stronger binding affinity for the TATA box binding TFIID with haplotype 4 than the minimal affinity observed with haplotype 1. Although the exact mechanisms for such variabilities remain to be elucidated, it is possible that structural changes due to SNPs forming specific haplotypes resulted in changes in ABCB1 promoter 3D structure and, consequently, differentially affected TF binding, including TFIID. It is known that the architecture of a promoter, dictated by its sequence, determines TF binding to that promoter (Reményi et al., 2004). TFIID is a protein complex composed of TATA Box Binding Protein (TBP) and several subunits called TATA binding protein-associated factors (TAFs), which add promoter selectivity, especially if there is no TATA box sequence for TBP to bind to (Louder et al., 2016). It is, therefore, possible that structural changes associated with haplotype 4 facilitated the binding of TFIID and/or its associated TAFs. Consistent with this possibility, our bioinformatics analysis revealed the presence of several putative TFIID binding sites on this promoter (Speidel et al., 2018).
The potential regulatory function of different TFs on ABCB1 promoter activity across different haplotypes was evaluated using siRNAs that preferentially block individual TFs (Table 4). Our data indicate that although some siRNA-mediated TF knockdown demonstrated universal alterations in promoter activity across the different haplotypes evaluated, the knockdown effect of others was not always consistent across the haplotypes. For example, siRNA-mediated knockdown of p53 resulted in a decrease in ABCB1 promoter activity with all haplotypes tested (Table 4 and Fig. 2). However, AP2 siRNA knockdown led to a significant increase in promoter activity of haplotype 1 but no change in activity of any of the other three haplotypes. Knockdown of Pax-5 exhibited a differential effect depending on the haplotype where it led to a significant increase in activity of haplotype 1, a significant decrease in activity of haplotype 29, and nonsignificant change with haplotypes 4 and 30, suggesting that Pax-5 can act as either an activator or a repressor depending on the haplotype.
Targeting TBP with siRNA induced a significant increase in promoter activity with haplotype 1 but had no effect with the other haplotypes tested. These findings are consistent with recent data from our laboratory that indicated that different haplotypes alter the binding of TFs to the MGMT promoter and, subsequently, affect its promoter activity and expression level (Xu et al., 2016). The siRNA studies with haplotype 30, which has very low basal promoter activity, revealed that none of the tested siRNAs had any effect on its activity (Fig. 2). A possible explanation may be that TFs other than those evaluated are involved in the regulation of haplotype 30 promoter activity. Another explanation could be that other non-cis-acting elements are driving the phenotype of this ABCB1 promoter haplotype.
A noteworthy observation was that the TF binding activity does not always correlate with the regulatory function as determined by siRNA. For example, we found that the knockdown of ER1 had no significant effect on promoter activity across the different haplotypes (Table 4 and Fig. 2). Although these observations are difficult to interpret, it is well documented that eukaryotic gene expression regulation is combinatorial in nature involving multiple proteins and different signaling pathways (Pique-Regi et al., 2011; Vazquez-Santillan et al., 2015). Therefore, it is possible that changes in promoter sequence and associated 3D structure due to haplotype effects allow other TFs to compensate for the effect of blocked TF and still drive the expression.
In conclusion, our data indicate that ABCB1 promoter haplotypes can affect promoter activity by altering TF binding. Our data also show that the regulatory effects of TFs are haplotype dependent. These results provide a possible mechanistic explanation for the observed differential effects of ABCB1 haplotypes on its promoter activity. Our results also underscore the importance of evaluating genetic variants in the context of haplotypes rather than individual SNPs when investigating their effects on genes and proteins expression and disease risk since SNPs are not arrayed as independent variants in the genome but as combinations forming defined haplotypes. The information generated from our studies has significant translational implications, particularly for pregnant women undergoing treatment with P-gp substrate medications.
Acknowledgments
This work was supported by grants from the National Institutes of Health (T-32-ES007254 to J.T.S.) and the John Sealy Memorial Endowment Fund for Biomedical Research (to S.Z.A.-R.). Additional partial support was provided by the NIEHS Center in Environmental Toxicology at UTMB funded through P30 ES006676, The Molecular Genomics Core at UTMB, the Institute for Translational Sciences UTMB, supported in part by a Clinical and Translational Science Award 8UL1TR000071 from the National Center for Research Resources, now the National Center for Advancing Translational Sciences, and the National Institutes of Health grants R01 DA 030998-01 and U54 HD047891.
Disclosure Statement
No competing financial interests exist.
References
- Babicki S., Arndt D., Marcu A., Liang Y., Grant J.R., Maciejewski A., et al. (2016). Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res 44, W147–W153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceckova-Novotna M., Pavek P., and Staud F. (2006). P-glycoprotein in the placenta: expression, localization, regulation and function. Reprod Toxicol 22, 400–410 [DOI] [PubMed] [Google Scholar]
- Chin K.V., Tanaka S., Darlington G., Pastan I., and Gottesman M.M. (1990). Heat shock and arsenite increase expression of the multidrug resistance (MDR1) gene in human renal carcinoma cells. J Biol Chem 265, 221–226 [PubMed] [Google Scholar]
- Cornwell M.M., and Smith D.E. (1993). SP1 activates the MDR1 promoter through one of two distinct G-rich regions that modulate promoter activity. J Biol Chem 268, 19505–19511 [PubMed] [Google Scholar]
- Gromnicova R., Romero I., and Male D. (2012). Transcriptional control of the multi-drug transporter ABCB1 by transcription factor Sp3 in different human tissues. PLoS One 7,e48189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemauer S.J., Nanovskaya T.N., Abdel-Rahman S.Z., Patrikeeva S.L., Hankins G.D.V., and Ahmed M.S. (2010). Modulation of human placental P-glycoprotein expression and activity by MDR1 gene polymorphisms. Biochem Pharmacol 79, 921–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano R., and Ling V. (1976). A surface glycoprotein modulating drug permeability in Chinese hamster ovary cell mutants. Biochim Biophys Acta 455, 152–162 [DOI] [PubMed] [Google Scholar]
- Kim R.B. (2002). Drugs as P-glycoprotein substrates, inhibitors, and inducers. Drug Metab Rev 34, 47–54 [DOI] [PubMed] [Google Scholar]
- Labialle S., Gayet L., Marthinet E., Rigal D., and Baggetto L.G. (2002). Transcriptional regulators of the human multidrug resistance 1 gene: recent views. Biochem Pharmacol 64, 943–948 [DOI] [PubMed] [Google Scholar]
- Louder R.K., He Y., López-Blanco J.R., Fang J., Chacón P., and Nogales E. (2016). Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature 531, 604–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Ikeda S., Furukawa T., Sumizawa T., Tani A., Akiyama S., et al. (1997). Function of P-glycoprotein expressed in placenta and mole. Biochem Biophys Res Commun 235, 849–853 [DOI] [PubMed] [Google Scholar]
- Pique-Regi R., Degner J., and Pai A. (2011). Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res 3, 447–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reményi A., Schöler H.R., and Wilmanns M. (2004). Combinatorial control of gene expression. Nat Struct Mol Biol 11, 812–815 [DOI] [PubMed] [Google Scholar]
- Rigalli J.P., Ciriaci N., Arias A., Ceballos M.P., Villanueva S.S.M., Luquita M.G., et al. (2015). Regulation of multidrug resistance proteins by genistein in a hepatocarcinoma cell line: impact on sorafenib cytotoxicity. PLoS One 10,e0119502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki M., Kurose K., Hasegawa R., and Tohkin M. (2011). Functional analysis of genetic variations in the 5′-flanking region of the human MDR1 gene. Mol Genet Metab 102, 91–98 [DOI] [PubMed] [Google Scholar]
- Schinkel A.H., and Jonker J.W. (2003). Mammalian drug efflux transporters of the ATP binding cassette (ABC) family: an overview. Adv Drug Deliv Rev 55, 3–29 [DOI] [PubMed] [Google Scholar]
- Shi J.F., Yang N., Ding H.J., Zhang J.X., Hu M.L., Leng Y., et al. (2014). ERα directly activated the MDR1 transcription to increase paclitaxel-resistance of ERα-positive breast cancer cells in vitro and in vivo. Int J Biochem Cell Biol 53, 35–45 [DOI] [PubMed] [Google Scholar]
- Speidel J.T., Xu M., and Abdel-Rahman S.Z. (2018). Differential effect of ABCB1 haplotypes on promoter activity. Pharmacogenet Genomics 28, 69–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takane H., Kobayashi D., and Hirota T. (2004). Haplotype-oriented genetic analysis and functional assessment of promoter variants in the MDR1 (ABCB1) gene. J Pharmacol Exp Ther 311, 1179–1187 [DOI] [PubMed] [Google Scholar]
- van Groenigen M., Valentijn L.J., and Baas F. (1993). Identification of a functional initiator sequence in the human MDR1 promoter. Biochim Biophys Acta 1172, 138–146 [DOI] [PubMed] [Google Scholar]
- Vazquez-Santillan K., Melendez-Zajgla J., Jimenez-Hernandez L., Martínez-Ruiz G., and Maldonado V. (2015). NF-κB signaling in cancer stem cells: a promising therapeutic target? Cell Oncol 38, 327–339 [DOI] [PubMed] [Google Scholar]
- Xu M., Cross C.E., Speidel J.T., and Abdel-Rahman S.Z. (2016). MGMT DNA repair gene promoter/enhancer haplotypes alter transcription factor binding and gene expression. Cell Oncol 39, 435–447 [DOI] [PubMed] [Google Scholar]
- Young A.M., Allen C.E., and Audus K.L. (2003). Efflux transporters of the human placenta. Adv Drug Deliv Rev 55, 125–132 [DOI] [PubMed] [Google Scholar]



