Abstract
Triple-negative breast cancer (TNBC) has few therapeutic options, and alternative approaches are urgently needed. Stimulator of IFN genes (STING) is becoming an exciting target for therapeutic adjuvants. However, STING resides inside the cell, and the intracellular delivery of CDNs, such as cGAMP, is required for the optimal activation of STING. We show that liposomal nanoparticle-delivered cGAMP (cGAMP-NP) activates STING more effectively than soluble cGAMP. These particles induce innate and adaptive host immune responses to preexisting tumors in both orthotopic and genetically engineered models of basal-like TNBC. cGAMP-NPs also reduce melanoma tumor load, with limited responsivity to anti–PD-L1. Within the tumor microenvironment, cGAMP-NPs direct both mouse and human macrophages (M), reprograming from protumorigenic M2-like phenotype toward M1-like phenotype; enhance MHC and costimulatory molecule expression; reduce M2 biomarkers; increase IFN-γ–producing T cells; augment tumor apoptosis; and increase CD4+ and CD8+ T cell infiltration. Activated T cells are required for tumor suppression, as their depletion reduces antitumor activity. Importantly, cGAMP-NPs prevent the formation of secondary tumors, and a single dose is sufficient to inhibit TNBC. These data suggest that a minimal system comprised of cGAMP-NP alone is sufficient to modulate the tumor microenvironment to effectively control PD-L1–insensitive TNBC.
Keywords: Immunology, Therapeutics
Keywords: Breast cancer, Cancer immunotherapy, Innate immunity
Nanoparticle-incorporated cGAMP is sufficient to modulate the tumor microenvironment and promote both innate and adaptive antitumor immunity in preclinical mouse models.
Introduction
Checkpoint inhibitors (e.g., ipilimumab, pembrolizumab, and nivolumab) are approved mAbs for cancer treatment (1–3). Although checkpoint blockade has been groundbreaking in expanding treatment options for multiple solid tumors, less than one-third of patients have a vigorous response (1, 2, 4). Retrospective clinical studies have reported that both the presence of tumor-infiltrating lymphocytes (TILs) and expression of programmed death ligand 1 (PD-L1) are correlated with responsiveness to immunotherapy in multiple cancer types, including melanoma (4) and breast cancer (5).
Among triple-negative breast cancers (TNBCs), basal-like TNBC is a heterogeneous disease that exhibits a high rate of recurrence, poor prognosis, and few effective treatment options. Basal-like TNBC represents a further challenge, as it lacks human epidermal growth factor receptor 2 (HER2), estrogen receptor, and progesterone receptor, which are effective therapeutic targets among hormone-responsive or HER2+ breast cancers. As a result, basal-like TNBC has few options for targeted therapy and is accompanied by poor prognosis (6). An ongoing nonrandomized phase Ib trial showed that only 18.5% of TNBC patients were responsive to checkpoint blockade immunotherapy (7), suggesting alternative approaches are urgently needed for this disease.
Immunotherapy can be augmented by targeting and reprogramming macrophages in tumors (8). Macrophages display a spectrum of activation statuses in cancer, with two extreme ends. Classically activated M1-like macrophages (M1 cells) are known to modulate host defense against pathogens and trigger antitumor immunity. In contrast, alternatively activated M2-like macrophages (M2 cells) can mediate a protumorigenic response by promoting angiogenesis and suppressing cytotoxic immune response (9). Tumor-associated macrophages (TAMs) have been reported to exhibit an “M2-like” functional profile (10), and the reprogramming of M2-like cells or TAMs toward M1-like cells by innate immune activation can be beneficial for tumor control (11).
The innate immune pathway involving stimulator of IFN genes (STING) has emerged as a critical pathway that may enhance antitumor immunity through the induction of type I IFN (IFN-I) (12). Cyclic dinucleotides (CDNs), including cyclic [G(3′,5′)pA(3′,5′)p] (cGAMP), are agonists for the STING receptor, and they drive IFN-I proinflammatory responses, which can enhance both innate and adaptive immunity (13). Intratumoral (IT) injection of STING agonists has shown efficacy in mouse models (14), and phase I clinical trials (ClinicalTrials.gov, NCT02675439 and NCT03172936) have been initiated. cGAMP also induces proinflammatory cytokines and chemokines, which are expected to cause macrophage maturation and CD8+ T cell cross-priming for adaptive tumor killing (15). However, IT injection is not always feasible in clinical practice due to nonuniform drug diffusion and difficult-to-control drug distribution. Additionally, STING resides within the cytosol of the cell (16), necessitating intracellular delivery of CDNs to ensure optimal biological activity (17). Thus, it is important to overcome these barriers for the practical and affordable implementation of CDN-based adjuvants in a clinical setting.
This study uses liposomal nanoparticles (NPs) to deliver cGAMP intracellularly to amplify innate immune activation and antitumor efficacy. Liposomes were the first nanomedicines to be approved by the FDA (18). This study demonstrates the amplified efficacy of liposome formulation relative to soluble cGAMP as an antitumor therapeutic that obviates the need of IT injection. We evaluated the efficacy in 3 preclinical models that are insensitive to PD-L1 checkpoint blockade, including an orthotopically transplanted model [C3(1)Tag model] for basal-like TNBC that is insensitive to most therapies, an aggressive B16F10 subcutaneous melanoma that is moderately responsive to checkpoint inhibitors, and a spontaneous genetic engineered mouse (GEM) model of basal-like TNBC [C3(1)Tag GEM] that is also resistant to most therapies. This last model offers several advantages for cancer drug development in a clinical setting, such as spontaneous primary and/or secondary tumors, heterogeneous immunosuppressive repertoires more predictive of human responses, and an array of tumor-stromal interactions during tumor progression (19, 20). We used these models to study the impact of liposomal NP-delivered cGAMP (cGAMP-NP) on macrophage reprograming and T cell activation and showed that antitumor immunity was achieved in a STING-dependent fashion that does not require prior identification of the tumor antigens. This work advances the understanding of immune activation via liposomal delivery of cGAMP in the treatment of PD-L1–insensitive tumors.
Results
Liposomal cGAMP-NPs are strong IFN-I inducers.
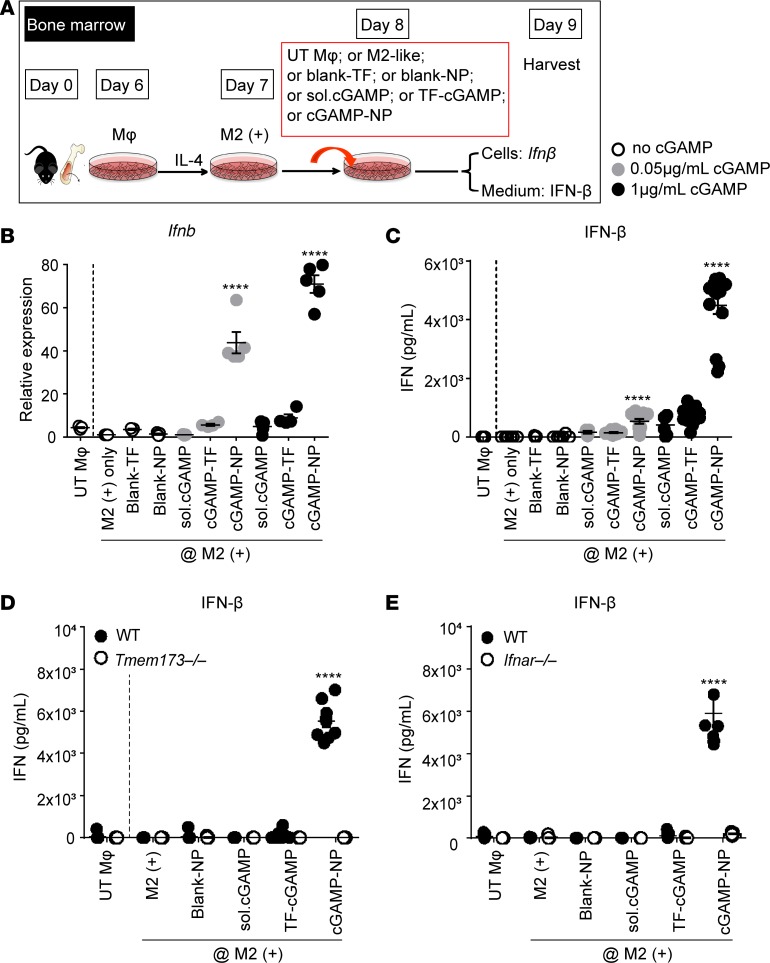
Liposomal cGAMP-NPs were produced with a particle size of 85 ± 27 nm and a positive zeta potential (14.8 ± 4.6 mV). After phase transition of the lipids, cGAMP was passively encapsulated into NPs with high encapsulation efficiency (43.11% ± 5.42%) and drug loading (2.15% ± 0.27% by weight) (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.120638DS1). NPs were spherical and uniformly distributed (under transmission electron microscopy [TEM]) (Supplemental Figure 1B). Upon cGAMP activation of STING in myeloid and antigen-presenting cells (APCs), such as macrophages, TBK1/IRF3 signaling-dependent IFN-I is released (21). To determine the activation of IFN-I production by cGAMP-NP, we first assayed for IFN production by in vitro–polarized M2 cells (Figure 1, A–C). We tested M2-like cells because these are frequently found at tumor sites (22). Among these cells, cGAMP-NP induced significantly higher IFN-β mRNA (Figure 1B) and protein (Figure 1C) levels when compared with transfection agent (TF), empty NPs, soluble cGAMP, or TF-delivered cGAMP. We confirmed that IFN-I production induced by cGAMP-NP is dependent on the STING, since bone marrow–derived macrophages (BMDMs) from Sting–/– mice (Sting is also known as Tmem173) failed to induce IFN-β (Figure 1D). In addition, deficiency in the IFN receptor (IFNAR) is known to disrupt the IFN-I–positive feedback loop (23) and Ifnar–/– BMDMs also failed to respond to cGAMP-NP (Figure 1E).
Figure 1. Liposomal cGAMP-NPs drive type I IFN production in a STING-dependent manner.
(A) BMDMs from C57BL/6J mice were cultured in IL-4 to induce an M2+ phenotype or left untreated (UT Mφ), followed by treatment with cGAMP delivered as soluble (sol.) cGAMP, mixed with transfection reagent (TF), or encapsulated in NP or blank-NP. (B) cGAMP-NP induced dose-dependent Ifnb transcripts and (C) IFN-β protein. (D) cGAMP-NP–induced IFN-β in M2+ cells was deficient in the absence of STING (in Tmem173–/– mice) or (E) IFN receptor (in Ifnar–/– mice). Experiments in B–E were repeated 3 times (B and D, n = 5/group; C and E, n = 15/group). Statistical significance was determined by 1-way ANOVA with a Tukey’s post hoc test (vs. M2+ only). ****P < 0.0001.
Tumor suppression by cGAMP-NP in transplanted models of TNBC and melanoma.
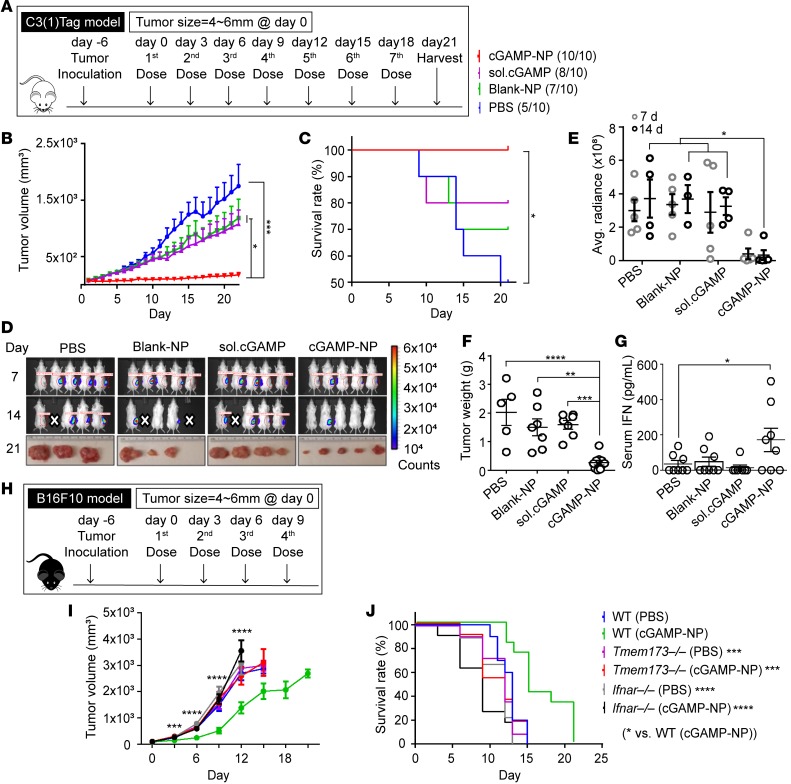
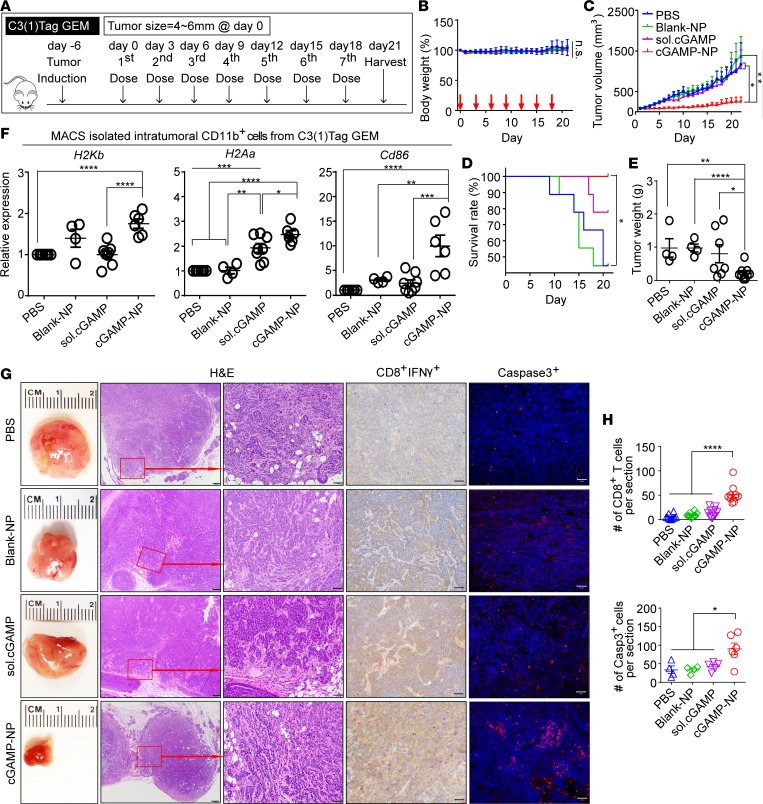
We next explored the antitumor therapy in engrafted C3(1)Tag orthotopic TNBC and B16F10 melanoma. A mammary cell line was derived from the C3(1)/SV40 Tag FVB/NJ–transgenic mice [hereafter referred to as C3(1)Tag mice] and was used to inoculate FVB/NJ female mice (24, 25). When these tumors were 4–6 mm in 1 dimension, mice were given the first of 7 i.v. injections of cGAMP-NP (Figure 2A). To monitor systemic inflammatory response, we collected sera from treated tumor-bearing mice at 6 hours and 24 hours and detected the cytokine levels (IL-6, TNF, and IFN-β). Levels of proinflammatory cytokines were upregulated 6 hours after treatment but dropped back to baseline levels, which were indistinguishable from those of control groups (Supplemental Figure 2, A–C). Surviving mice showed no difference in body weight loss, except that there was only one time point (endpoint) where the weight was statistical greater in PBS group, likely caused by the growing tumor mass in PBS-treated control mice (Supplemental Figure 2D). Compared with PBS, blank-NP, and soluble cGAMP, cGAMP-NP treatment significantly reduced tumor growth, as measured by a digital caliper (Figure 2B), and increased survival (Figure 2C). As an additional approach, bioluminescence via an in vivo imaging system (IVIS) was used to measure tumor size (Figure 2D, top and middle), and tumors were excised at the end of the experiment at day 21 (Figure 2D, bottom). There was no mouse death reported within hours after treatment. If tumor size reached the criteria for euthanasia, the animal in question would be removed from the study; this generally happened to mice in control groups without cGAMP-NP treatment (labeled as white cross in Figure 2D). cGAMP-NP–treated tumor-bearing mice yielded the lowest average radiance of tumor mass at days 14 and 21, as determined by IVIS (Figure 2E), and the lowest tumor weight at day 21 when tumors were harvested (Figure 2F). These data confirm the effectiveness of cGAMP-NP therapy for tumor suppression. The cGAMP-NP injection activated the known target of STING, as serum IFN-β was increased 6 hours after the first cGAMP-NP injection (Figure 2G).
Figure 2. Liposomal cGAMP-NPs suppress established tumor growth in a STING/IFNAR-dependent manner.
(A–G) Luciferase-expressing C3(1)Tag cells were used to generate orthotopic basal-like TNBC tumors. When tumors were 4–6 mm in 1 dimension, mice were treated with 7 doses of cGAMP-NP (i.v.) administration. (B) Tumor volume, (C) survival rate, (D, top and middle) bioluminescence imaging of tumor growth was monitored, and (E) the radiance efficiency was quantified. (D) Gross morphology (bottom) and (F) tumor weight were monitored on day 21. (G) Sera were collected from C3(1)Tag-bearing mice after the first dose of cGAMP-NP treatment and assayed for IFN-β by ELISA. (H–J) B16F10 cells were used to generate melanomas in C57BL/6J WT, Tmem173–/–, and Ifnar–/– mice, followed by 4 doses of cGAMP-NP (i.v.) administration. (I) Tumor volume and (J) survival rate were monitored. Data in B, C, E–G, I, and J were repeated and pooled from 2 experiments (n = 10 mice/group). Images in D (n = 5 mice/group) are representative of 2 independent experiments. Dead mice (indicated with white cross) were removed from the study when the tumors reached the criteria for euthanasia. Statistical significance was determined by 1-way ANOVA with a Tukey’s post hoc test (B, E, F, G, and I) or log-rank test (C and J). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
We next evaluated the effect of cGAMP-NP on the transplanted B16F10 melanoma model in C57BL/6 mice. cGAMP-NPs were given 6 days after the melanoma was introduced into the animals. We evaluated cGAMP-NP treatment in WT, Tmem173–/–, and Ifnar–/– mice maintained on a C57BL/6 background (Figure 2H). cGAMP-NP did not cause weight loss in B16F10 melanoma mice (Supplemental Figure 2E), but it did reduce tumor volume and increase survival (Figure 2, I and J). Consistent with a STING/IFNAR-dependent antitumor effect, cGAMP-NP failed to significantly slow tumor growth or increase mean survival in Tmem173–/– and Ifnar–/– mice. This indicates that the antitumor effects of cGAMP-NP is mediated by STING activation.
Myeloid cells uptake cGAMP-NPs in culture and in mice.
To examine the uptake of NP by cells, we conducted in vitro NP cellular uptake studies by incubating isolated mouse BMDMs, bone marrow–derived DCs (BMDCs), and tumor cells with fluorescent-labeled NPs (acyl 12:0 NBD-PC; green), wherein cells were stained with LysoTracker (red) for live-cell microscopy imaging and flow cytometry (Supplemental Figure 3, A and B, and Supplemental Video 1). For BMDMs, fluorescent NPs colocalized with LysoTracker+ subcellular organelles, indicating vesicular internalization and intracellular transport. BMDMs were labeled with the highest intensity of NPs during a 5-hour incubation, followed by BMDCs, with little uptake by C3(1)Tag cells (Supplemental Figure 3A). We also confirmed that NPs were located and accumulated in the cytosol of BMDMs by confocal microscopy images (Supplemental Figure 3C and Supplemental Video 2).
We next assessed in vivo NPs biodistribution and trafficking in the C3(1)Tag model (Supplemental Figure 4). Among all collected tissues, high radiance was observed in the spleen, liver, tumor, and lung 6 hours after administration, but this level diminished with time in spleen, liver, and lung over 72 hours. NP accumulation in the tumor significantly increased up to 48 hours and then started to decrease at 72 hours (Supplemental Figure 4A and quantitated in Supplemental Figure 4B). During the first 72 hours, the tumor tissues reproducibly contained a steady level of macrophages, neutrophils, and CD4+ T cells, while CD8+ T cells and DCs fluctuated over time but were nevertheless present (Supplemental Figure 4, C and D). NPs were found associated with immune cells (Supplemental Figure 4E).
cGAMP-NP treatment polarizes M2 macrophages toward M1 macrophages.
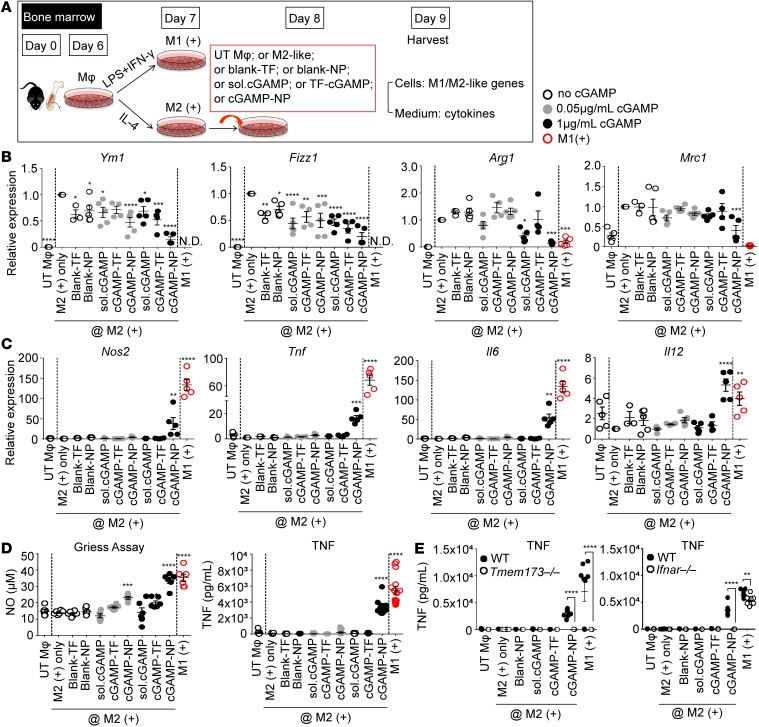
The above results indicate that cGAMP-NP is taken up by mouse macrophages, and the literature shows that macrophage polarization to M2-like cells can be an important factor that reduces effective antitumor immunity (26–29). Therefore, we assessed the impact of cGAMP-NP on mouse M2 cells, which were polarized by the addition of IL-4 (Figure 3A). Mouse M1-like cells generated by the addition of LPS and IFN-γ were included as controls. Blank TF or blank-NPs did not alter M-2-like biomarkers while soluble cGAMP and TF-delivered cGAMP caused an occasional reduction of M2 biomarkers. By contrast, cGAMP-NP consistently downregulated M2 biomarkers, Ym1, Fizz1, and Arg1 transcripts (Figure 3B), while increasing M1 biomarkers, Nos1, Tnf, Il6, and Il12b transcripts (Figure 3C), TNF protein, and nitric oxide (30) (Figure 3D). A higher concentration of cGAMP-NP also reduced the M2 marker Mrc1 (Figure 3B). The induction of TNF expression by cGAMP-NP was lost in Tmem173–/– and Ifnar–/– BMDMs when compared with those from WT mice (P < 0.0001; Figure 3E), indicating STING/IFNAR dependency.
Figure 3. Liposomal cGAMP-NPs polarize murine M2-like to M1-like macrophages in vitro.
(A) C57BL/6J BMDMs were cultured in LPS plus IFN-γ for 24 hours to induce an M1-like phenotype and IL-4 to induce an M2-like phenotype. UT Mφ, untreated control. On day 7, M2 cells (shown as groups between two dash lines in B–D) were treated with 0.05 and 1 μg/ml cGAMP delivered as soluble (sol.) cGAMP, mixed with TF or encapsulated in NP, TF alone, or blank-NP (n = 5/group). (B) On day 8, cells were harvested and assayed for the expression of M2 (Ym1, Fizz1, Arg1, and Mrc1) or (C) M1 (Tnf, Il6, Il12b, and Nos2) biomarkers. (D) Supernatants were collected for nitric oxide production (n = 6/group) and TNF detection by ELISA (n = 15/group). (E) Secreted TNF response to cGAMP-NP and controls was measured in WT, Tmem173–/–, and Ifnar–/– cells (n = 6/group). (B–E) Pooled data from 3 experiments. Statistical significance was determined by 1-way ANOVA with a Tukey’s post hoc test (B–D; vs. M2+ only) or 2-tailed Student’s t test (E) to compare WT (black) and Tmem173–/– or Ifnar–/– cells (white) with treatment. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
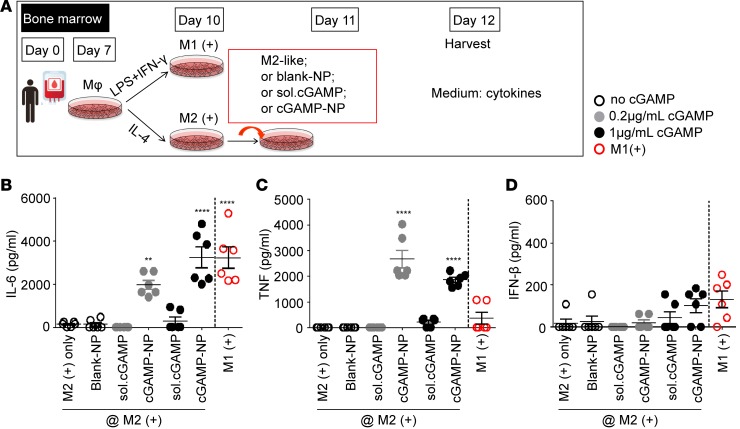
In addition to mouse macrophages, we have investigated the impact of cGAMP-NP on human macrophages. Similarly, human PBMC-derived macrophages were first polarized to either M2 cells by supplementing with IL-4 or to M1 cells by supplementing with LPS and IFN-γ (Figure 4A). cGAMP-NP significantly increased IL-6 and TNF proteins, and slightly induced IFN-β production (no significant difference), while blank-NPs or soluble cGAMP did not alter these M1 biomarkers (Figure 4, B–D).
Figure 4. Liposomal cGAMP-NPs polarize human M2-like to M1-like macrophages in vitro.
(A) PBMCs were cultured to induce human macrophage polarization. On day 7, M1 cells were stimulated with LPS plus IFN-γ while M2 cells were stimulated with IL-4 for activation. On day 11, M2 cells (shown as groups on the left side of the dash line in B–D) were treated with 0.2 and 1 μg/ml cGAMP delivered as soluble (sol.) cGAMP or encapsulated in NP or blank-NP alone. (B–D) On day 12, supernatants were collected for IL-6, TNF, and IFN-β detection by ELISA (n = 6/group). Data were pooled data from 2 experiments. Statistical significance was determined by 1-way ANOVA with a Tukey’s post hoc test (B–D; vs. M2+ only). **P < 0.01; ****P < 0.0001.
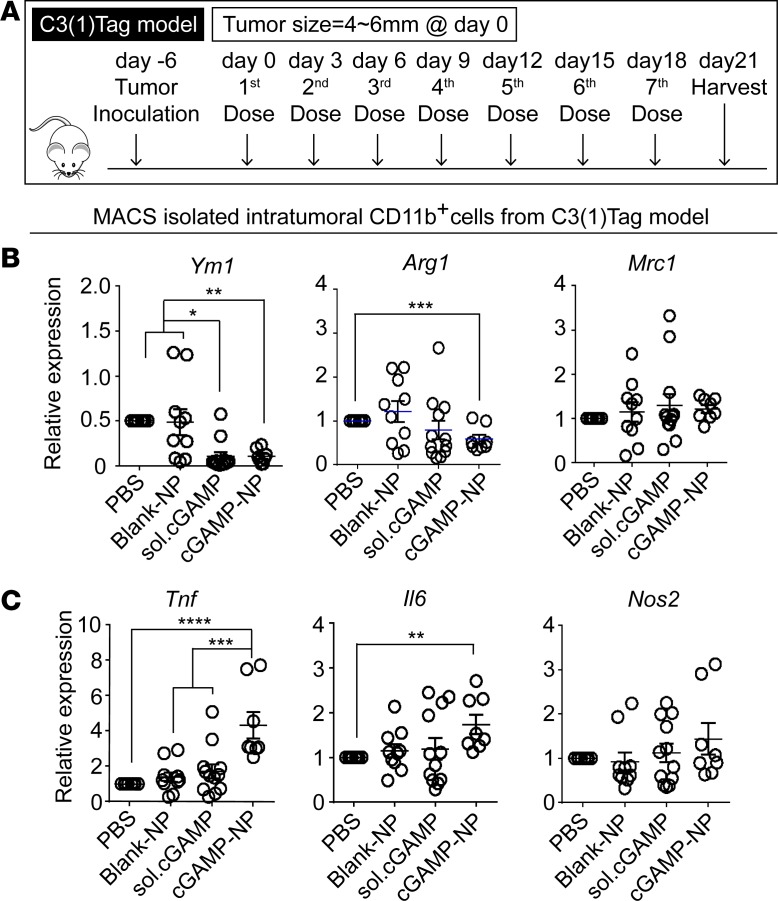
To address the in vivo effect of cGAMP-NP on the spectrum of in vivo macrophage polarization, we detected M1/M2 gene expression in isolated CD11b+ macrophages by magnetic-activated microbead sorting (MACS) of cells from transplanted C3(1)Tag mammary tumors undergoing the last cGAMP-NP treatment (Figure 5A). Importantly, cGAMP-NP significantly decreased M2-associated genes, Arg1 and Ym1 (Figure 5B), and increased M1-associated, genes Il6 and Tnf, compared with control groups (Figure 5C). It had no effect on Mrc1 and Nos2.
Figure 5. Liposomal cGAMP-NPs polarize M2-like to M1-like macrophages in tumor tissue.
(A) Treatment regimen in the C3(1)Tag orthotopic model. (B) On day 21, CD11b+ cells were isolated from C3(1)Tag tumors by MACS sorting to detect M2 (Ym1, Arg1, and Mrc1) or (C) M1 (Tnf, Il6, and Nos2) biomarkers. (B and C) Data pooled from 2 experiments (n = 10 mice/group). Statistical significance was determined by 1-way ANOVA with a Tukey’s post hoc test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
cGAMP-NP treatment enhances macrophage surface proteins that are known to induce antitumor cytotoxic CD8+ T cells.
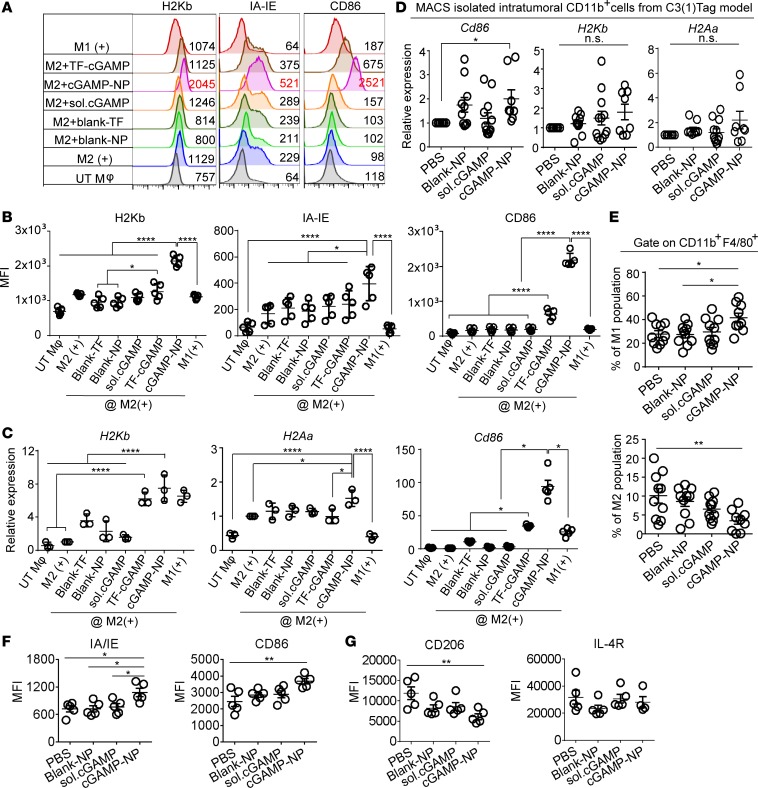
We next examined the impact of cGAMP-NP on surface molecules that are important for the activation of CD8 cytotoxic cells, including major histocompatibility complex class I (MHCI) and class II (MHCII) and costimulatory molecule (CD86) expression. These IFN-I–inducible proteins/genes were measured by flow cytometry (Figure 6, A and B) or RT-PCR (Figure 6C) in M2-polarized macrophages. All 3 molecules were significantly enhanced in response to cGAMP-NP treatment compared with untreated control, whereas less or no change was detected with blank TF, blank-NPs, soluble cGAMP, or TF reagent–delivered cGAMP. To assess if these changes occurred in vivo, we examined H2Kb, H2Aa, and Cd86 gene expression in CD11b+ macrophages sorted from C3(1)Tag tumors. Among all treatment groups, cGAMP-NP induced the highest expression levels of all 3 markers, with a significant increase of Cd86 expression compared with PBS control (Figure 6D). Our flow cytometry analysis showed that cGAMP-NP induced a significantly higher percentage of M1 population (CD45+CD11b+F4/80+CD206-IA/IE+) accompanied by reduced M2 (CD45+CD11b+F4/80+CD206+IA/IE-) population when compared with control treatments (Figure 6E). These C3(1)Tag TAMs also expressed higher levels of M1 biomarkers (IA/IE [MHCII] and CD86) while expressing lower intensity of a M2 biomarker (CD206) (Figure 6, F and G).
Figure 6. Liposomal cGAMP-NPs enhance MHC and costimulatory molecules in M2-like macrophages.
(A–C) BMDMs were induced to M1+ or M2+ in vitro, followed by treatment with soluble cGAMP, cGAMP delivered by transfection agent, or by liposomal NP. (A) The expression of surface MHC and costimulatory molecules (H2Kb, IA-IE, and CD86) in cGAMP-NP–stimulated M2+ cells detected by flow cytometry. Dashed lines in A indicate expression level on M2+ cells; numbers to the right indicate representative MFI. (B) Composite MFI from n = 5/group. (C) H2bKb, H2Aa (IA-IE), and Cd86 mRNA expression (n = 3/group) in cGAMP-NP–stimulated M2+ cells detected by real-time PCR. (D) The expression of H2bKb, H2Aa, and Cd86 in intratumoral CD11b+ cells isolated from orthotopic C3(1)Tag tumors by MACS sorting. (E–G) C3(1)Tag tumors were processed for flow cytometry analysis to detect the percentage of M1 cells (CD45+CD11b+F4/80+CD206–IA/IE+) and M2 cells (CD45+CD11b+F4/80+CD206+IA/IE–) in E and the expression of M1 (IA/IE and CD86) and M2 (CD206 and IL-4R) biomarkers in F and G. Data in A are representative of 3 experiments. Data in B and C were pooled from 3 experiments (n = 3–5/group). Data in D–G were pooled from 2 experiments (D, n = 8 mice/group; E, n = 10 mice/group; F and G, n = 5 mice/group). (B–G) Statistical significance was determined by 1-way ANOVA with a Tukey’s post hoc test. *P < 0.05; **P < 0.01; ****P < 0.0001.
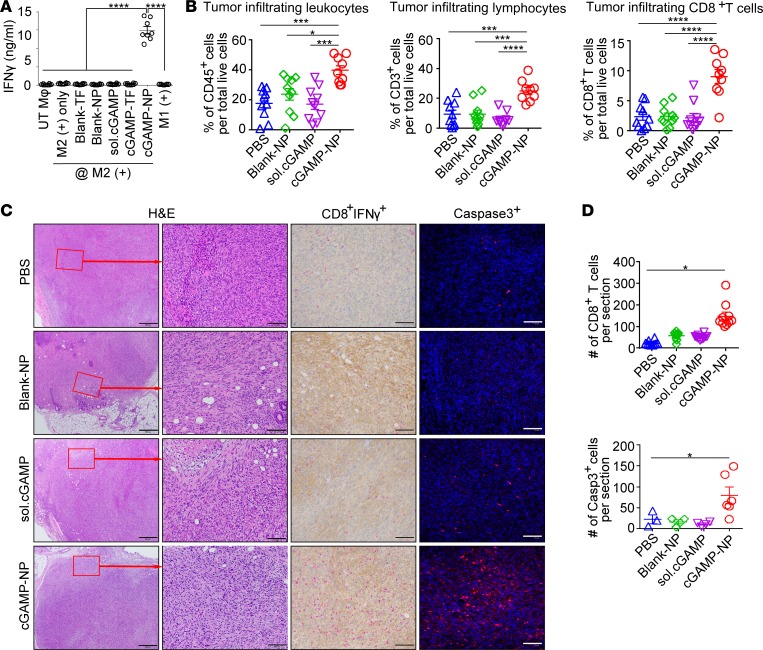
To evaluate the in vitro stimulation of T cell by cGAMP-NP–treated macrophages, we detected IFN-γ production from isolated splenic CD3+ T cells by coculturing with B16F10 tumor antigen–pulsed macrophages as direct cell-to-cell contact activation. By using this method, cGAMP-NP treatment induced robust secretion of IFN-γ, as detected by the ELISA assay, while a minimal amount of IFN-γ was induced by control treatments (blank-TF, blank-NP, TF, or soluble form of cGAMP) (Figure 7A). The amount of in vivo tumor-infiltrating leukocytes, lymphocytes, and CD8+ T cells in C3(1)Tag tumors were determined by flow cytometry (Figure 7B), and all 3 populations were significantly increased by cGAMP-NP treatment compared with other control groups. Furthermore, the presence of cytotoxic T cells and cell death in histological samples from C3(1)Tag tumors were also analyzed. A blinded assessment by a trained pathologist of H&E-stained sections showed that these tumors were stage III invasive breast cancer that had spread to the peripheral mammary tissue (Figure 7C). Immunohistochemical analysis of IFN-γ–producing CD8+ T cells were infrequent in all control groups. However, cGAMP-NP treatment induced a dramatic increase of infiltrating CD8+ cells and IFN-γ expression, along with a significant increase in caspase-3+ cells (Figure 7C, quantitation shown in Figure 7D), suggesting enhanced cytotoxic T cells and in situ cell apoptosis.
Figure 7. Liposomal cGAMP-NPs increase CD8+ cytotoxic T cells and apoptosis.
(A) In vitro stimulation of T cells was evaluated by directly coculturing splenic isolated CD3+ T cells with cGAMP-NP–treated B16F10 tumor antigen–pulsed macrophages (using blank-NP, TF, or soluble form as control treatments). IFN-γ secretion was determined by ELISA assay after 3 days of coculture. (B–D) C3(1)Tag orthotopic tumors were harvested on day 21. (B) The percentages of tumor-infiltrating leukocytes (CD45+ cells), CD3+ lymphocytes, and CD8+ T cells were detected by flow cytometry. (C) Representative H&E staining (top, scale bar: 500 μm; bottom: scale bar: 100 μm), IHC staining (red, CD8; brown, IFN-γ; scale bar: 100 μm), and IF staining (caspase-3+, red; nucleus, blue; scale bar: 100 μm) in tumors harvested on day 21. (D) Quantitative analysis of CD8+ T cells and caspase-3+ cells as shown in C. Data in A, B, and D were pooled from 2 experiments (A, n = 8/group; B, n = 10 mice/group; D, n = 5–10/group). Images in C are representative of 2 experiments (n = 10 mice/group). Statistical significance was determined by 1-way ANOVA (A and B) or 2-tailed Student’s t test (D). *P < 0.05; ***P < 0.001; ****P < 0.0001.
cGAMP-NP is an effective cancer immunotherapy in tumors that show limited or no response to PD-L1 intervention.
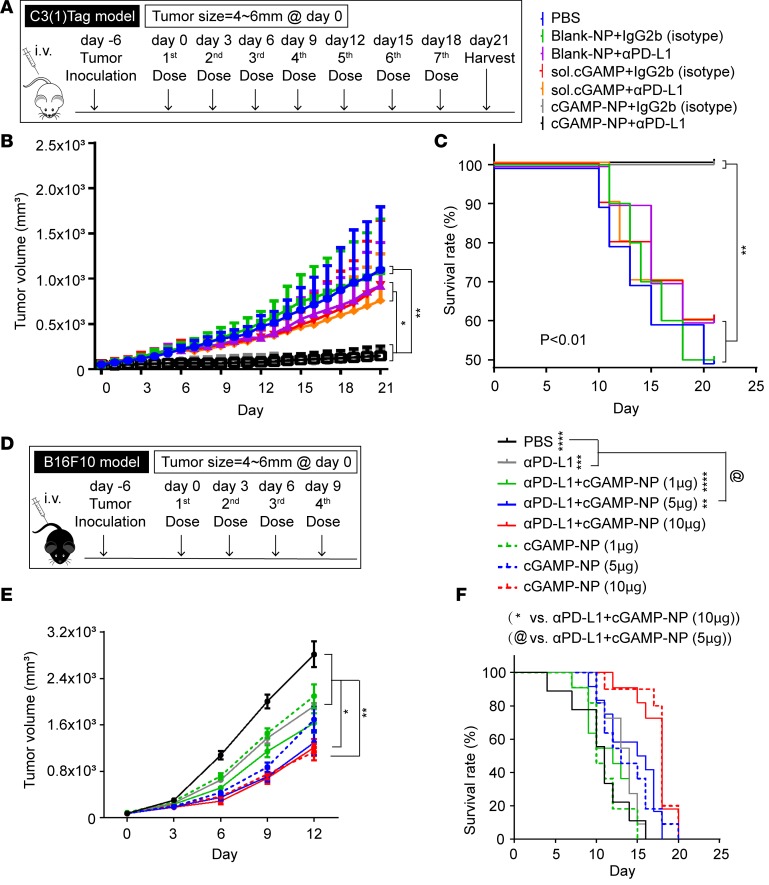
Programmed death receptor-1 (PD-1) is a key immune-checkpoint receptor on activated T cells (31). The binding of PD-1 to its ligand PD-L1 stimulates IL-10 secretion (32) but dampens IL-2 production and T cell proliferation in a tumor microenvironment (33, 34), and targeting of the PD-1/PD-L1 inhibitory pathway is used for immunotherapy (35, 36). However, some cancers, such as TNBC, are unresponsive to PD-1–targeted therapies. Indeed, anti–PD-1, anti–PD-L1, and a combination of both antibodies were ineffective in reducing C3(1)Tag orthotopic tumors (Supplemental Figure 5, A–C). Since checkpoint blockade has achieved partial response rate in melanoma (37–39), we tested the effectiveness of anti–PD-L1 in B16F10 model and found a 20%–25% response rate in the mice treated with 50 or 100 μg/mouse dosages (Supplemental Figure 5, D–G). We next tested the efficacy of cGAMP-NP with or without anti–PD-L1 treatment in C3(1)Tag orthotopic TNBC (Figure 8A). cGAMP-NPs administered with an isotype control IgG significantly suppressed tumor growth and increased survival rate without additive improvement when combined with anti–PD-L1 antibody, suggesting that cGAMP NPs alone was effective in controlling PD-L1–insensitive tumor burden (Figure 8, B and C). A similar test with the B16F10 melanoma (Supplemental Figure 5, H–J) also showed limited response to anti–PD-L1 while cGAMP-NP effectively controlled melanoma growth and prolonged mice survival when delivered alone. The addition of anti–PD-L1 to cGAMP-NP showed improvement over cGAMP-NP, but the difference did not reach statistical significance. To titrate the amount of cGAMP-NP to enhance the tumor suppression by anti–PD-L1, we used a combination of cGAMP-NP at 1 μg/dose (low), 5 μg/dose (medium), or 10 μg/dose (high), with a fixed amount of anti–PD-L1 (100 μg per mouse) into B16F10 tumor-bearing mice (Figure 8D). The addition of medium to high dose of cGAMP-NP showed significant improvement over anti–PD-L1 alone (Figure 8, E and F). The combination of cGAMP-NP and anti-PD-L1 did not significantly enhance the anti-tumor response over cGAMP-NP alone. However, there is a trend of lowered tumor volume when aPDL1 is used with a combination of 1ug or 5 ug of cGAMP-NP.
Figure 8. Liposomal cGAMP induces tumor rejection in tumors with limited or no sensitivity to PD-L1 immunotherapy.
(A–C) C3(1)Tag cells were inoculated in FVB/NJ mice (n = 10 mice/group). When the tumor reached 4–6 mm in 1 dimension (designated as day 0), mice received PBS, blank-NP plus IgG2b, blank-NP plus anti–PD-L1, soluble cGAMP plus IgG2b, soluble cGAMP plus anti–PD-L1, cGAMP-NP plus IgG2b, or cGAMP-NP plus anti–PD-L1. cGAMP-NPs were administered (i.v.) 7 times, using blank-NP and soluble cGAMP as controls. Anti–PD-L1 was administered (i.p.) 10 minutes after each cGAMP injection, using IgG2b as an isotype control. (B) Tumor volume and (C) survival rate were monitored. (D–F) B16F10 cells were inoculated in C57BL/6J mice (n = 10 mice/group). Six days later (designated as day 0), mice received PBS, α-PD-L1 (100μg/mouse), α-PD-L1+cGAMP-NP (1μg/mouse), α-PD-L1+cGAMP-NP (5μg/mouse), α-PD-L1+cGAMP-NP (10μg/mouse), cGAMP-NP (1μg/mouse), cGAMP-NP (5μg/mouse), or cGAMP-NP (10μg/mouse). Anti–PD-L1 antibodies were administered i.p., and cGAMP-NPs were administered i.v. Tumor volume (E) and survival rate (F) were monitored. Data in B, C, E, and F were pooled from 2 independent experiments. Statistical significance was determined by 1-way ANOVA with a Tukey’s post hoc test (B and E) or log-rank test (C and F). *P < 0.05; **P < 0.01.
cGAMP-NP is an effective cancer immunotherapy in a GEM model.
GEM models of cancers develop spontaneous tumors in immunocompetent animals and are considered superior models for testing therapeutics than orthotopic-transplanted or xenotransplanted models of tumor (19, 20). We examined the efficacy of cGAMP-NPs in a C3(1)Tag GEM model in which both Rb and p53 were inactivated by SV40 T(t) antigen expression. C3(1)Tag GEM best represents human basal-like TNBC that showed chemoresistant or limited-responsiveness to chemotherapeutic treatments (40, 41). These GEM mice showed ductal carcinoma-like lesions by 8 weeks of age, which became invasive carcinomas at 18 weeks of age (24). cGAMP-NP treatment was initiated when the primary tumors were 4–6 mm in 1 dimension (Figure 9A), and the treatment significantly inhibited tumor growth, leading to 100% survival at day 20 when compared with significant death among control mice receiving either blank-NP or soluble cGAMP (Figure 9, B–E). Similar to the orthotopic model, cGAMP-NPs increased H2Kb, H2Aa, and Cd86 among TAMs (Figure 9F). Furthermore, increased IFN-γ+ secretion, CD8+ T cells, and caspase-3+ cells were noted throughout the tumors (Figure 9, G and H), suggesting an enhanced tumor killing response.
Figure 9. Liposomal cGAMP-NPs inhibit tumor growth in an established genetically engineered model of basal-like TNBC.
(A) C3(1)Tag genetically engineered model (GEM) mice with spontaneous single basal-like TNBC were recruited when tumor size (≈4–6 mm) was met in any dimension (n = 8–10 mice/group). cGAMP-NPs were administered (i.v.) 7 times, using PBS, blank-NP, or sol. cGAMP as controls. (B) Total body weight, (C) tumor volume, and (D) survival rate were monitored for 21 days, and (E) tumor weight was measured upon sacrifice on day 21. (F) CD11b+ TAM cells were isolated from tumors by MACS sorting and analyzed for immune activation markers H2bKb, H2Aa, and Cd86. (G) Representative images of gross tumor morphology (first column from left to right): H&E staining (second column, scale bar: 200 μm; third column, scale bar: 50 μm), IHC staining (red, CD8; brown, IFN-γ; scale bar: 50 μm) (fourth row), and IF staining (red, caspase-3; blue, nucleus; scale bar: 50 μm) (fifth row) of harvested tumors. (H) Quantitative analysis of CD8+ T cells and caspase-3+ cells as shown in G. (B–F) Data were pooled from 9 mice/group. Images in G are representative of 2 experiments. Data in H were pooled from 2 experiments (n = 5–10/group). Statistical significance was determined by 1-way ANOVA with a Tukey’s post hoc test (B, C, E, and F), log-rank test (D) or 2-tailed Student’s t test (H). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
cGAMP-NP causes M1-like polarization and requires T cells for its efficacy.
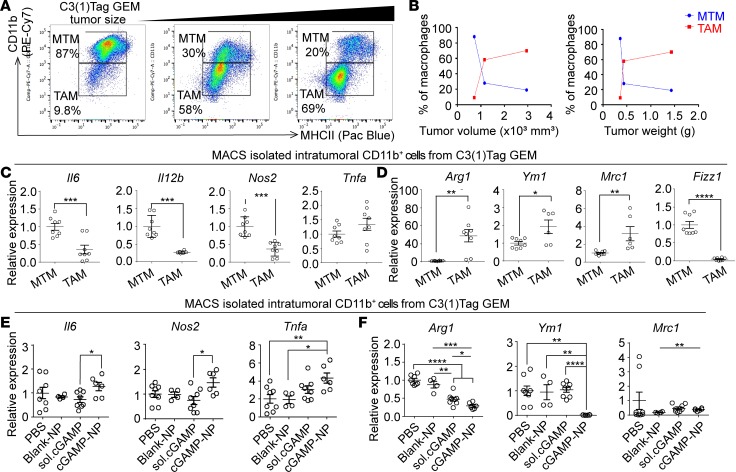
Previous reports defined MHCII+CD11blo as TAMs and MHCII+CD11bhi as mammary tissue macrophages (MTMs) on F4/80+ gated population in breast tumors (8, 42, 43). Consistent with these findings, we demonstrated a shift of MHCII+CD11bhi cells toward MHCII+CD11blo population in C3(1)Tag GEM tumors with increasing tumor size (Figure 10A). A quantitation of TAMs expansion confirmed an association with increased tumor volume (Figure 10B). TAMs expressed high levels of Arg1, Ym1, and Mrc1 (M2-like genes) but low levels of Il6, Il12b, and Nos2 (M1-like genes) when compared with those in MTMs, suggesting that TAMs exhibit a M2-like polarized phenotype in tumor-bearing animals (Figure 10, C and D). Similar to the findings in orthotopic model, cGAMP-NP treatment significantly upregulated M1 biomarkers (Il6, Nos2, and Tnfα) and downregulated M2-like biomarkers (Arg2 and Ym1) in CD11b+ macrophages sorted from C3(1)Tag GEM tumors (Figure 10, E and F).
Figure 10. cGAMP-NPs enhance M1-like markers and reduce M2-like markers in basal-like TNBC GEM.
(A and B) Flow cytometric analysis of macrophage populations (gated on CD45+ leukocytes) from pooled C3(1)Tag GEM tumors (n = 4 mice/group). (C and D) The expression of M1 (C) and M2 (D) biomarkers in MACS-sorted CD11b+ cells from mammary fat tissue and C3(1)Tag GEM tumors (n = 8–10 mice/group). (E and F) The expression of M1 (E) and M2 (F) biomarkers in MACS-sorted CD11b+ TAMs isolated from animals treated with cGAMP-NP and controls (n = 8–10 mice/group). Data in A are representative of 2 independent experiments. Data in B and C–F were pooled from 2 individual experiments. Statistical significance was determined by 2-tailed Student’s t test (C and D) or 1-way ANOVA with a Tukey’s post hoc test (E and F). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
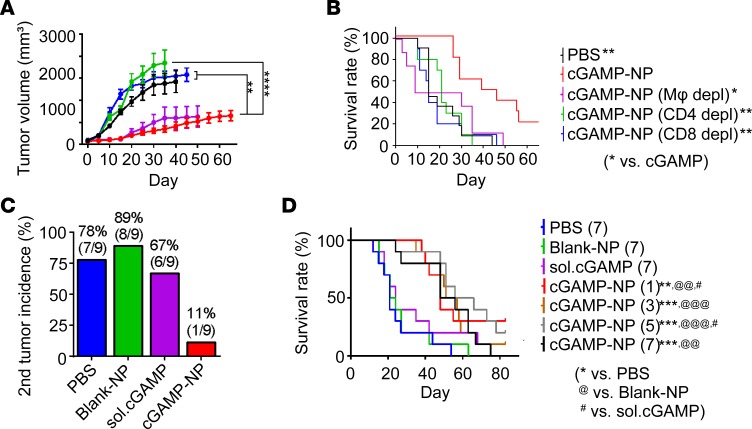
To understand the underlying in vivo cellular mechanisms for cGAMP-NP–induced antitumor effects, we studied the roles of effector CD4+ and CD8+ T cells and macrophages in cGAMP-NP–mediated immunotherapy by chemical and antibody depletion in the orthotopic C3(1)Tag model, which were monitored for 60 days. The depletion specificity was confirmed by injecting blank vector (Encapsome) that was used to deliver clodronate or isotype antibody (IgG2b), neither of which affected the tumor burden (Supplemental Figure 6). Depletion of either CD4+ or CD8+ T cells resulted in enhanced tumor growth, implying that both T cell populations were responsible for tumor suppression (Figure 11, A and B). Depletion of macrophages by clodronate did not eliminate antitumor activity of cGAMP-NP but did cause reduced survival. We observed morbidity after injection in macrophage depleted mice, such as dehydration, limited movement and grooming. A possible reason for the latter is likely a systemic depletion of macrophages in major immune organs, including liver, spleen, and bone marrow, by the i.v. administration of clodronate-containing liposomes resulting in toxicity (44).
Figure 11. cGAMP-NPs increase antitumor T cells, prevent secondary tumor, and are effective in 1 dose.
(A and B) Long-term study of C3(1)Tag orthotopic tumors undergoing cGAMP-NP (i.v.) therapy in mice depleted of macrophages (Clodrosome; i.v.) or CD4 (GK1.5; i.p.) or CD8 (clone 2.43; i.p.) T cells. (C) The percentage of C3(1)Tag GEM mice (n = 8–10 mice/group) that spontaneously developed secondary tumors upon treatment with cGAMP-NP (i.v.) or controls. (D) Survival rate of mice with C3(1)Tag orthotopic tumors that received PBS (7 doses), blank-NP (7 doses), soluble cGAMP (7 doses), and cGAMP-NP (1, 3, 5, or 7 doses). (A, B, and D) Pooled data from 2 individual experiments (n = 10 mice/group). Statistical significance was determined by 1-way ANOVA with a Tukey’s post hoc test (A) or log-rank test (B and D). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
These data show the involvement of adaptive immune cells that are typically associated with immune memory. To examine if antitumor memory was elicited, we examined for secondary tumors, which were shown to form in C3(1)Tag GEM mice (45). Mice treated with cGAMP-NPs showed a dramatic and marked drop of secondary tumor initiation that was 6~8 times lower than that in untreated control mice or mice treated with either blank-NP or soluble cGAMP (Figure 11C). Finally, we reduced the number of doses (1–7 doses) of cGAMP-NPs and found that 1 dose of cGAMP-NPs significantly increased survival for at least 80 days after treatment (Figure 11D). This shows the robustness of the cGAMP-NP as an immunotherapy.
Discussion
Various immunotherapies have been advanced for clinical cancer vaccines and therapies, the most prominent are immune checkpoint blockade for solid tumors (4) and chimeric antigen receptor (CAR) T cell therapy for leukemia (46). However, only a subpopulation of patients achieved successful responses to the former (47), while the potential toxicity and restricted application of CAR T cell therapy in blood cancers have been noted (48, 49). Thus, alternative antitumor therapies harnessing both innate and adaptive immune cells to target difficult-to-treat cancer types are urgently needed. The STING pathway was first discovered as an essential regulator to recognize viral DNA for effective host defense (50, 51) and has emerged as a potentially important pathway for cancer development and therapy (52). However, STING is located inside the cell, while current commercially available STING agonists are formulated in a soluble form, which may not gain ready access to the intracellular STING. Indeed, we showed less effectiveness of soluble cGAMP compared with cGAMP-NP in tumors, likely due to limited tumor penetration and poor cell membrane permeability of free cGAMP. In agreement with this, one study used 60 times higher doses of i.v.-delivered soluble cGAMP than the amount used in this report to achieve antitumor response (53). However, overdosage of immunotherapies may generate systemic cytokine-associated toxicity, which can lead to inflammation and even lethality. Others tried IT (14) or intramuscular injection (17) of soluble STING agonists to maximize the therapeutic effect. However, the former is not feasible for primary and metastatic tumors of internal organs, while the latter is not the typical route for the delivery of anticancer therapy. By contrast, cGAMP-NPs were delivered by a conventional i.v. route and showed significant improvement over soluble cGAMP in the suppression of established PD-1– and PD-L1–insensitive cancer models.
Our study demonstrates that cGAMP-NP has multiple effects on the immune system. It drives proinflammatory cytokines (e.g., IL-6 and TNF) and nitric oxygen species that promote M1-like polarization (30). It also induces IFN-I, which promotes MHC and costimulatory molecules expression, accompanied by increased CD8+ T cell (21). Moreover, IFN-I is known to effect tumor cells by inhibiting proliferation (54), inducing apoptosis (55), and modulating migration and cell surface antigen expression (56). We confirmed that cGAMP-NP efficacies are STING dependent, as the treatment is ineffective in mice deficient in the STING pathway.
Our study indicates that cGAMP-NP is superior to soluble cGAMP in converting M2-like cells to M1-like cells and in eliciting antitumor CD4 and CD8 T cells. Thus, it heightens both innate and adaptive limbs of the antitumor immune response. Recent studies suggest that TAMs function as reliable biomarkers to predict patient responses to certain treatment strategy (21). At the primary tumor site, TAMs promote angiogenesis (57), drive tumor cell invasion and metastasis (58), exhibit immunosuppressive properties by releasing antiinflammatory cytokines, and directly suppress cytotoxic CD8+ T cell responses to tumor cells by the expression of inhibitor ligands PD-L1 and B7-H4 (59, 60). TAMs also enable tumor cell migration and seeding to induce the metastatic cascade and contribute to inflammation-induced tumor initiation. Our study shows that, in untreated animals, TAMs are increased in C3(1)Tag tumors and this increase is directly associated with tumor growth. cGAMP-NP is more effective than soluble cGAMP in the reprogramming of TAMs from M2 cells toward an M1-like phenotype and in enhancing the presence of tumor-associated cytotoxic T cells that are important in the containment of tumors.
Our study shows that NPs are consistently associated with immune cells, particularly macrophages, neutrophils, and CD4+ T cells, when trafficking to tumor sites. It has been reported that cGAS and STING expression levels are extremely low in neutrophils (61), which leads us to primarily focus on macrophages and T cells in this work. However, it is worthwhile to explore the role of neutrophils in response to STING-independent cytosolic nucleic acid sensing for future therapeutic application, since neutrophils also constitute a major population of leukocytes in human.
While immunotherapy holds much promise, a major challenge is that many cancers remain resistant to existing cancer immunotherapy. Here, we show that the efficacy of cGAMP-NP alone is significantly superior to that of soluble cGAMP or checkpoint blockade in both melanoma and TNBC models. A recent paper shows similar findings in the B16F10 melanoma model (62). However, our work shows that cGAMP-NP is therapeutic in basal-like TNBC, even in the setting of a genetically engineered basal-like TNBC mouse model that has little response to most treatment. Furthermore, cGAMP-NP elicits T cell response and antitumor memory that prevent the development of secondary tumors. Compared with the administration of multiple doses required in other reports, a single dose of cGAMP-NP treatment is sufficient to suppress tumor growth in a long-term study. These results suggest that the liposomal cGAMP-NP described here may achieve an effective antitumor immune response in difficult-to-treat cancers that have limited or no response to PD-L1–targeted treatments. Thus, cGAMP-NP may represent a promising new immunotherapy for basal-like TNBC and melanoma.
Methods
Synthesis and characterization of cGAMP Soy-PC-DOTAP liposome.
All materials used for liposome fabrication were purchased from Avanti Lipids Polar Inc., unless otherwise indicated. Vaccine grade 3′3′-cGAMP was purchased from Invivogen. The fabrication of liposome NPs was modified according to previous reports (63, 64). Briefly, a 100:1 (weight ratio) mixture of hydrogenated (soy)L-α-phosphatidylcholine (Soy-PC) and 1,2-dioleoyl-3-trimethyl-ammonium-propane (DOTAP) was dissolved in chloroform and dried by a rotary evaporator (Buchi R-200) to form lipids film. The dry film was then reconstituted in a Krebs-Henseleit buffer for 45 minutes in a 60°C water bath (Buchi B-490), either without cGAMP, as blank liposome control, or with cGAMP. Liposomes were freeze thawed 3 times, followed by consecutively extruding them through polycarbonate membrane filters 6 times with an Avanti Mini-Extruder/Heating Block. Membrane filters (100 nm) were used for different batches of NPs. Free cGAMP was removed by passing through a disposable PD-10 column (GE Healthcare). The eluted NPs solution was frozen at –80°C before lyophilization. Particle size, zeta potential, and polydispersion index were characterized by a Malvern ZetaSizer Nano series. TEM images of Soy-PC-DOTAP (negatively stained with 2% uranyl acetate) were acquired using a JEOL 100CX II TEM (JEOL, Japan). cGAMP loading was determined by performance liquid chromatography (HPLC). Briefly, cGAMP liposomes were first suspended in HPLC-grade methane, and water at a 2:3 (v/v) was then added. This mixture was vigorously vortexed and ultrasonicated for 2 minutes. An aliquot of the water phase was injected into an isocratic 60:40 (v/v) water/methanol mobile phase, operating at 0.6 ml/min. After passing through an Aquasil C18, 150 × 4.6 mm column (Thermo Fisher Scientific), the absorbance of the eluent was measured at 256 nm. A standard curve of cGAMP dissolved in water was subjected to the same conditions, and cGAMP loading in the liposomes (μg cGAMP/mg liposomes) was determined. Blank liposomes subjected to the same process were used as a background correction. The encapsulation efficiency (%) and drug loading (%) of the cGAMP Soy-PC-DOTAP liposomes were calculated based on the following equations: encapsulation efficiency (%) = mass of cGAMP in liposomes/initial amount of feeding cGAMP × 100% and drug loading (%) = mass of cGAMP in liposomes/total mass of loaded liposomes × 100%.
Cell preparation.
The C3(1)/Tag cell line was provided by Mouse Phase I Unit (Lineberger Comprehensive Cancer Center [LCCC], University of North Carolina at Chapel Hill). The B16F10 cell line was provided by Shelton Earp (University of North Carolina at Chapel Hill). C3(1)/Tag-luc cells were generated by stably transducing cells with retrovirus expressing pMMP-Luc-Neo construct. All cell lines were maintained in DMEM (Gibco) with 10% FBS and 1% penicillin/streptomycin at 37°C and 5% CO2.
Murine BMDMs were isolated from 8-week-old C57BL/6 WT, Tmem173–/–, and Ifnar–/–mice. Briefly, all femurs and tibia were harvested and cut at the epiphysis level, and bone marrow was flushed out by injecting 10 ml DMEM medium (Gibco) for 3 times. The cell suspension was filtered by 70-μm nylon BD Falcon cell strainers, centrifuged, and then lysed by ACK buffer (Gibco). For BMDM culture, macrophage growth medium (MGM; DMEM + 10%FBS + 30%L929 medium) was added to resuspend the cell pellets. Cells were plated at 5 × 105 cells/10 ml in petri dishes. On day 4, 10 ml MGM was added. On day 6, cells were induced into macrophages and harvest for polarization study. Macrophages at 5 × 105 cells/ml were cultured in LPS (500 ng/ml) plus IFN-γ (25 ng/ml) to induce an M1 phenotype (M1+) or IL-4 (25 ng/ml) to induce an M2 phenotype (M2+) or left as untreated macrophage control. For in vitro murine macrophage skewing study, on day 7, M2 cells were treated with 3′3′ cGAMP delivered as soluble drug or transfected by Lipofectamine 3000 (Thermo Fisher Scientific) or encapsulated in liposome NPs or equivalent amount of blank liposomes. On day 8, cells were harvested for gene expression analysis (M1/M2 biomarkers and costimulatory molecules) and flow cytometry; supernatants were collected for cytokine detection (ELISA/Luminex assay) and nitric oxide analysis using Griess Reagent System (Promega). Cell viability was determined by MTT assay.
Murine splenic T cells were harvested from spleens and purified by using the EasySep mouse CD3 T Cell Isolation Kit (STEMCELL Technology) according to the manufacturer’s instructions. T cells were cultured in RPMI 1640 plus 10% FBS for further experiments.
Human peripheral blood mononuclear cells (PBMCs) were isolated from buffy coats (Gulf Coast Regional Blood Center, Houston, Texas, USA). On day 0, PBMCs were first isolated by density gradient centrifugation, washed in HBSS (Gibco), and then lysed in LymphoPrep (STEMCELL Technology). After washing with HBSS, PBMCs were directly seeded into plates and cultured in RPMI 1640 plus 10% human AB serum (Gemcell) at 37°C and 5% CO2. After 2-hour incubation, nonadherent lymphocytes were removed, and adherent monocytes were left to further induce macrophage polarization using Macrophage Generation Medium DXF (PromoCell). On day 6, 50% volume of fresh medium was added into wells. On day 7, cells were either induced to activated hM1 cells in supplement of IFN-γ (50 ng/ml) plus LPS (10 ng/ml) or induced to activated hM2 cells in supplement of IL-4 (20 ng/ml). On day 9, nonadherent cells collected from supernatant in each well were collected and suspended in fresh medium and then added back to adherent cells. On day 10, both hM1 and hM2 cells were ready for use.
In vitro cellular uptake assay.
1% (w/w) fluorescent lipid analog 1-acyl-2-{12-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]dodecanoyl}-sn-glycero-3-phosphocholine (acyl 12:0 NBD-PC) (Avanti Lipids Polar Inc.) was incorporated into lipid bilayer prior to study the cellular internalization detected by live-cell imaging, confocal microscopy, and FACS analysis. The cellular uptake assay was performed as follows. Murine BMDMs, BMDCs, and C3(1)Tag tumor cells were incubated with 100 μg/ml NBD-PC–labeled liposomes. Cells were stained with LysoTracker Red DND-99 (Life Technologies) for 2 hours prior to live-cell imaging according to the manufacturer’s instructions, and images were captured every 5 minutes for 24 hours by the Olympus IX70 Live Cell System equipped with a ×20/0.04 Ph1 LCPlanFL objective. Videos were compiled by Volocity version 6.3 (PerkinElmer). For live-cell confocal microscopy, NBD-PC liposome–treated cells were collected at 0, 2, and 5 hours and stained with CellMask Deep Red plasma membrane stain (Thermo Fisher Scientific) for 10 minutes according to the manufacturer’s instructions. Samples were examined by an Olympus FV1000-MPE microscope (Olympus). For FACS assay, NBD-PC liposome–treated cells were harvested at 0, 2, and 5 hours and fixed with ice-cold 2% PFA before being run through the cytometer.
Tumor antigen pulsed–macrophage coculture with T cells.
Tumor cells at 90% confluence were heated in 42°C water bath for 1 hour, followed by recovery for 2 hours at 37°C. Cells were then digested by trypsin and washed twice with PBS. After being enumerated and resuspended in PBS at 1 × 107 cells/ml, tumor cells were frozen in liquid nitrogen for 10 minutes and then thawed at 4°C for 3 cycles. The frozen-thawed resultants were centrifuged at 12,000 g for 15 minutes, and the supernatant was preserved as tumor cell lysate at –80°C. To induce tumor cell lysate-pulsed macrophages, 1.2 × 106 polarized M1/M2 macrophages were incubated with the tumor cell lysates (macrophages/tumor cells = 5:1) along with cGAMP-NP treatment for 24 hours (using blank-NP, TF, or soluble cGAMP as controls). Then, splenic CD3+ T cells were isolated and added to coculture with pulsed macrophages (macrophages/T cells = 1:2). After 3 days coculture, supernatants were collected for cytokine detection by ELISA.
Tumor models.
C57BL/6, Tmem173–/–, Ifnar–/–, FVB/NJ, and C3(1)Tag mice were purchased from The Jackson Laboratory. All mice were maintained in specific pathogen–free facilities at University of North Carolina at Chapel Hill.
For orthotopic implantation of the C3(1)Tag model, C3(1)Tag-luc cells (5 × 105 cells/mouse) were trypsinized and resuspended in a 100-μl PBS/Matrigel (BD Biosciences) mixture (1:1 volume) and injected into the lower right inguinal mammary fat pat of 8- to 10-week-old FVB/NJ female mice.
For subcutaneous implantation of the B16F10 model, B16F10 cells (2 × 105 cells/mouse) were trypsinized and resuspended in a 100-μl PBS/Matrigel (BD Biosciences) mixture (1:1 volume) and injected into the right flank of 8- to 10-week-old C57BL/6 mice.
For the C3(1)Tag GEM, the 5′ flanking regulatory region of the gene encoding the C3(1) component of the rat prostate steroid-binding protein was used to target the expression of SV40 large T antigen (Tag) to the epithelium of the mammary glands, resulting in models of mammary carcinomas that histologically resemble the human diseases. All female C3(1)Tag GEM mice were established in FVB/NJ background and were bred in-house and used for experiments when the tumor size (≈4 × 6 mm) was met in any dimension (21).
Tumor-bearing mice (size ≈ 4 × 6 mm) were randomized into treatment groups immediately before treatment. Liposome cGAMP-NPs (10 μg loaded cGAMP/dose) were delivered i.v. in 100 μl PBS every 3 days, starting on day 0. An equivalent amount of PBS, soluble PAMPs, and blank-NPs was delivered as control. Tumor size was measured daily by a digital caliper. Tumor volume was calculated according to the equation (length × width2) × 0.5. Mouse survival rate was recorded. Mice showing the following symptoms were humanely euthanized: (a) weight loss greater than 20%; (b) ulcerated tumor; (c) tumor size reaching 2 cm in any dimension with a single tumor or 1.3 cm in any dimension with multiple tumors in mice with grafted tumors; and (d) tumor size reaching 1.3 cm in any dimension with a single tumor or 1 cm in any dimension with multiple tumors in mice with spontaneous tumors. Unless otherwise specified, at day 21, mice were euthanized for tumor processing and measuring of tumor weight. Serum samples were collected for cytokine and chemokine detection. Tumor tissues were collected for cell isolation and histological studies. To monitor toxicity, we closely checked for mouse behavior, body weight changes, and survival to assess the systemic cytokine-associated toxicity over time course of treatment. There was no mouse death observed within hours after treatment. Mice were sacrificed if the tumor size reached the criteria for euthanasia, which generally occurred in mice in control groups without cGAMP-NP treatment.
Macrophage and T cell depletion in TNBC.
To deplete the macrophages, mice were i.p. injected with 200 μl in 5 mg/ml m-Clodrosome (Encapsula NanoSciences) twice per week for 2 weeks prior to orthotopic injection and maintained thereafter until the experimental endpoint. Control mice received empty liposomes (Encapsome) following the same protocol. To deplete CD4+ T cells and CD8+ T cells, mice were i.p. injected with control rat IgG2b antibody, anti-CD4, or anti-CD8a (0.5 mg/mouse; Supplemental Table 1). Injections were administered every day for 3 days prior to orthotopic injection and twice a week thereafter until the experimental endpoint.
IVIS study.
Fluorescence imaging was performed using the IVIS Kinetic Optical System (PerkinElmer) to evaluate the in vivo tissue biodistribution of liposome NPs. DiD-labeled Soy-PC-DOTAP liposomes, representing 1% (w/w) of DiD, were i.v. injected into tumor-bearing mice at 0.6 mg/mouse (cGAMP equivalent). Mice (n = 3) were sacrificed at 6, 24, 48, and 72 hours after administration. At each time point, major organs, including tumor, brain, heart, lung, liver, spleen, kidney, thymus, and pancreas, were harvested. Their ex vivo images were obtained with the following parameters: exposure time = 0.5 s; binning = 4 (medium); f/stop = 2; excitation filter = 640 nm; emission filter = 680 nm. Acquired images were analyzed with Living Imaging software (Caliper Life Sciences) to quantify the distribution of DiD-labeled liposomes in each type of organ by average radiant efficiency ([photons/s/cm2/steradian]/[μW/cm2]). Tumor tissues were further processed for FACS staining (a) to characterize the immune cell components (macrophage, DCs, neutrophils, and CD4+ and CD8+ T cells) and (b) to evaluate the cellular trafficking of DiD-labeled liposomes in each type of immune cells.
Bioluminescence imaging was performed using the IVIS Lumina Optical System (PerkinElmer) to monitor the tumor growth in orthotopic C3(1)/Tag mice after treatment. Mice were placed in the dorsal positions to obtain whole-body optical images at day 7 and 14. Acquired images were analyzed by Living Image software (Caliper Life Sciences) to quantify luciferase activity of orthotopic C3(1)Tag tumors by average radiance (photons/s/cm2/ steradian).
Preparation of single-cell suspensions from tumor tissues.
The resected mouse tumors were digested with 200 U/ml Collagenase IV (Gibco) and 100 μg/ml DNase I (Calbiochem) and mechanically dissociated with surgical scissors in dissociation buffer (RPMI + 10%FBS) for 1 hour in a 37°C shaking incubator (150 rpm). After dissociation, samples were transferred to ice to stop the reaction. The tumor suspension was strained and washed with FACS buffer and centrifuged at 4°C (1,532 g). Red blood cells were removed with ACK lysing buffer. The samples were washed, resuspended in MACS buffer on ice throughout the rest of the sorting process. Briefly, cells were enriched and sorted by CD11b+ MACS beads (Miltenyi Biotec) according to the manufacturer’s instructions. An aliquot of sorted cells was collected for Cytospin. Both the MACS-sorted and unsorted cells were further counted and used for FACS staining.
FACS.
Single-cell suspensions from either tissue or cell culture were kept on ice, stained with Live/Dead dye (Tonbo Biosciences), and blocked with anti-CD16/32 (Fc block, eBioscience). The following antibodies were used to stain cell surface molecules: CD45, CD11b, F4/80, H2Kb, IA/IE, CD86, CD11c, CD206, IL-4R, CD3, CD4, and CD8 (Supplemental Table 1). M1 cells (CD45+CD11b+F4/80+CD206–IA/IE+) and M2 cells (CD45+CD11b+F4/80+CD206+IA/IE–) were defined. All data were collected using a Cyan ADP (Beckman Coulter) or LSRII (BD Biosciences) flow cytometer and analyzed using FlowJo software (Tree Star).
RNA extraction and RT-PCR analysis.
Total RNA was harvested using TRIzol (Invitrogen) according to the manufacturer’s protocol. The RNA concentration was measured by using a NanodropND-1000 spectrophotometer (Thermo Fisher Scientific). cDNA was synthesized by oligo(dT20)-primed reverse transcription using M-MLV Reverse Transcriptase (Invitrogen) according to the manufacturer’s instructions. Real-time PCR (RT-PCR) for murine costimulatory molecules (H2Kb, H2-Aa, and Cd86), M1 markers (Tnfa, Il6, Il12b, Nos2, and Ifnb), M2 markers (Ym1, Fizz1, Arg1, and Mrc1), Pd-l1 and Actb (used for normalization) was performed using TaqMan primer/probe sets and master mix (Applied Biosystems).
Cytokine detection.
Supernatants either from cell culture or dissociated tumor tissues were centrifuged at 9,576 g for 10 minutes, and then the clear liquid phase was carefully collected to remove cell debris. Serum was also collected to determine in vivo systematic cytokine changes. Samples were transferred to a 96-well plate on ice, and ELISA kits were used to measure IL-6, IL-12, TNF-α, and IFN-γ (all from BD Biosciences) and IFN-β (BioLegend). The absorbance was measured by a microplate reader (BioTek). Supernatants or serum samples were also analyzed by the 32-Plex Luminex Bead Panel (Invitrogen) in the Immunology Unit of the Regional Biocontainment Laboratory at Duke University.
Histology.
The cellular morphology of TAMs and peritoneal macrophages was analyzed by Cytospin followed by H&E staining. Briefly, 100-ml single-cell suspensions were transferred to Cytospin funnels and centrifuged in a Cytocentrifuge (Thermo Fisher Scientific) for 5 minutes (1,064 g). The slides were then stained with a Diff-Quik kit (Baxter Merz & Dade) according to the manufacturer’s instructions.
Tumor tissues were fixed in 10% paraformaldehyde (PFA), dehydrated, subsequently paraffin embedded, and sectioned within the LCCC Animal Histopathology Core Facility at the University of North Carolina at Chapel Hill. H&E staining, IHC, and immunofluorescent (IF) were performed. CD8, IFN-γ and caspase-3 antibodies used for IHC and IF staining (Supplemental Table 1). All histology images were captured by Olympus BX43F (Olympus) or Olympus BX61 (Olympus). Quantification for caspase-3 staining was done by using an open-source software project Fiji (ImageJ, NIH).
Statistics.
All statistical analyses were performed with GraphPad Prism 6.0. For cellular study, tumor study, and image quantifications, statistical significance was assayed by 2-tailed Student’s t test or 1-way ANOVA with a Tukey’s post hoc test. Kaplan-Meier survival curves were calculated, and significance was determined by the log-rank test. All data points were included in the analyses and presented as mean ± SEM. A P value less than 0.05 was considered significant.
Study approval.
All animal care and studies were approved by the Institutional Animal Care and Use Committee at the University of North Carolina at Chapel Hill (15-113).
Author contributions
NC and JPYT designed the experiments, analyzed data, and wrote the manuscript, with critical input from KMA, EMB, and RDJ. RWS assisted with animal experiments. RDJ assisted with human cell cultures. CND assisted in confocal imaging and live-cell imaging. SAM assisted with the histological analysis. KJP assisted with NP design. BMJ aided with flow cytometry. QL and LM assisted with NPs characterization. HY, DBD, and LH provided critical discussion. KPM aided with human immune cell culture. All contributing authors have agreed to the submission of this manuscript for publication.
Supplementary Material
Acknowledgments
We acknowledge the technical support from the University of North Carolina LCCC Mouse Phase I Unit, Animal Studies Core, Microscopy Services Laboratory, Flow Cytometry Core, and Small Animal Imaging Core (BRIC-SAI). The authors would like to thank Dawud Hilliard for IHC, Michael Collier and Naihan Chen for HPLC, and Jose Roques for providing C3(1)/Tag colonies. This work was supported by NIH grants U54CA198999 and U19AI109784 and LCCC grant 2017-T2-12 to JPYT and North Carolina Biotechnology Center grant 2018-BIG-6504 to JPYT, KMA, and RDJ. NC was supported by the National Cancer Institute F99/K00 fellowship F99CA223019, RDJ was supported by NIH grant T32-AI007151, RWS was supported by NIH grant 5T32AI007273, CND was supported by National Institute of Allergy and Infectious Diseases grant T32-AI07273, BMJ was supported by NIH grant T32-HL007106, and HY was supported by NIH grant P30-CA016086-40.
Version 1. 11/15/2018
Electronic publication
Footnotes
LM’s present address is: Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2018, American Society for Clinical Investigation.
Reference information: JCI Insight. 2018;3(22):e120638. https://doi.org/10.1172/jci.insight.120638.
Contributor Information
Ning Cheng, Email: Ning_Cheng@unc.edu.
Rebekah Watkins-Schulz, Email: rebekahw@email.unc.edu.
Hong Yuan, Email: yuanh@med.unc.edu.
Qi Liu, Email: qiliu67@unc.edu.
Lei Miao, Email: mysky_05@mit.edu.
Leaf Huang, Email: leafh@unc.edu.
References
- 1.Robert C, et al. Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med. 2015;372(26):2521–2532. doi: 10.1056/NEJMoa1503093. [DOI] [PubMed] [Google Scholar]
- 2.Larkin J, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med. 2015;373(1):23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khalil DN, Smith EL, Brentjens RJ, Wolchok JD. The future of cancer treatment: immunomodulation, CARs and combination immunotherapy. Nat Rev Clin Oncol. 2016;13(6):394. doi: 10.1038/nrclinonc.2016.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herbst RS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emens LA, et al. Inhibition of PD-L1 by MPDL3280A leads to clinical activity in patients with metastatic triple-negative breast cancer. Cancer Res. 2015;75:abstr PD1–6. [Google Scholar]
- 6.Toft DJ, Cryns VL. Minireview: Basal-like breast cancer: from molecular profiles to targeted therapies. Mol Endocrinol. 2011;25(2):199–211. doi: 10.1210/me.2010-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nanda R, et al. Pembrolizumab in patients with advanced triple-negative breast cancer: phase Ib KEYNOTE-012 study. J Clin Oncol. 2016;34(21):2460–2467. doi: 10.1200/JCO.2015.64.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franklin RA, et al. The cellular and molecular origin of tumor-associated macrophages. Science. 2014;344(6186):921–925. doi: 10.1126/science.1252510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Palma M, Lewis CE. Macrophage regulation of tumor responses to anticancer therapies. Cancer Cell. 2013;23(3):277–286. doi: 10.1016/j.ccr.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 10.Roland CL, et al. Inhibition of vascular endothelial growth factor reduces angiogenesis and modulates immune cell infiltration of orthotopic breast cancer xenografts. Mol Cancer Ther. 2009;8(7):1761–1771. doi: 10.1158/1535-7163.MCT-09-0280. [DOI] [PubMed] [Google Scholar]
- 11.Genard G, Lucas S, Michiels C. Reprogramming of tumor-associated macrophages with anticancer therapies: radiotherapy versus chemo- and immunotherapies. Front Immunol. 2017;8:828. doi: 10.3389/fimmu.2017.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corrales L, Gajewski TF. Molecular pathways: targeting the stimulator of interferon genes (STING) in the immunotherapy of cancer. Clin Cancer Res. 2015;21(21):4774–4779. doi: 10.1158/1078-0432.CCR-15-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo M, et al. A STING-activating nanovaccine for cancer immunotherapy. Nat Nanotechnol. 2017;12(7):648–654. doi: 10.1038/nnano.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corrales L, et al. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 2015;11(7):1018–1030. doi: 10.1016/j.celrep.2015.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.González-Navajas JM, Lee J, David M, Raz E. Immunomodulatory functions of type I interferons. Nat Rev Immunol. 2012;12(2):125–135. doi: 10.1038/nri3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, et al. cGAS is essential for the antitumor effect of immune checkpoint blockade. Proc Natl Acad Sci USA. 2017;114(7):1637–1642. doi: 10.1073/pnas.1621363114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi J, Kantoff PW, Wooster R, Farokhzad OC. Cancer nanomedicine: progress, challenges and opportunities. Nat Rev Cancer. 2017;17(1):20–37. doi: 10.1038/nrc.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Day CP, Merlino G, Van Dyke T. Preclinical mouse cancer models: a maze of opportunities and challenges. Cell. 2015;163(1):39–53. doi: 10.1016/j.cell.2015.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharpless NE, Depinho RA. The mighty mouse: genetically engineered mouse models in cancer drug development. Nat Rev Drug Discov. 2006;5(9):741–754. doi: 10.1038/nrd2110. [DOI] [PubMed] [Google Scholar]
- 21.Coffelt SB, de Visser KE. Immune-mediated mechanisms influencing the efficacy of anticancer therapies. Trends Immunol. 2015;36(4):198–216. doi: 10.1016/j.it.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11(10):889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 23.Ma F, et al. Positive feedback regulation of type I IFN production by the IFN-inducible DNA sensor cGAS. J Immunol. 2015;194(4):1545–1554. doi: 10.4049/jimmunol.1402066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green JE, et al. The C3(1)/SV40 T-antigen transgenic mouse model of mammary cancer: ductal epithelial cell targeting with multistage progression to carcinoma. Oncogene. 2000;19(8):1020–1027. doi: 10.1038/sj.onc.1203280. [DOI] [PubMed] [Google Scholar]
- 25.Zhou B, et al. Erythropoietin promotes breast tumorigenesis through tumor-initiating cell self-renewal. J Clin Invest. 2014;124(2):553–563. doi: 10.1172/JCI69804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41(1):49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruffell B, Affara NI, Coussens LM. Differential macrophage programming in the tumor microenvironment. Trends Immunol. 2012;33(3):119–126. doi: 10.1016/j.it.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allavena P, Mantovani A. Immunology in the clinic review series; focus on cancer: tumour-associated macrophages: undisputed stars of the inflammatory tumour microenvironment. Clin Exp Immunol. 2012;167(2):195–205. doi: 10.1111/j.1365-2249.2011.04515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Georgoudaki AM, et al. Reprogramming tumor-associated macrophages by antibody targeting inhibits cancer progression and metastasis. Cell Rep. 2016;15(9):2000–2011. doi: 10.1016/j.celrep.2016.04.084. [DOI] [PubMed] [Google Scholar]
- 30.Wang N, Liang H, Zen K. Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Front Immunol. 2014;5:614. doi: 10.3389/fimmu.2014.00614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Topalian SL, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong H, Zhu G, Tamada K, Chen L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat Med. 1999;5(12):1365–1369. doi: 10.1038/70932. [DOI] [PubMed] [Google Scholar]
- 33.Freeman GJ, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192(7):1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Im SJ, et al. Defining CD8 + T cells that provide the proliferative burst after PD-1 therapy. Nature. 2016;537(7620):417–421. doi: 10.1038/nature19330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen L, Han X. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J Clin Invest. 2015;125(9):3384–3391. doi: 10.1172/JCI80011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Algazi AP, et al. Clinical outcomes in metastatic uveal melanoma treated with PD-1 and PD-L1 antibodies. Cancer. 2016;122(21):3344–3353. doi: 10.1002/cncr.30258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luke JJ, Flaherty KT, Ribas A, Long GV. Targeted agents and immunotherapies: optimizing outcomes in melanoma. Nat Rev Clin Oncol. 2017;14(8):463–482. doi: 10.1038/nrclinonc.2017.43. [DOI] [PubMed] [Google Scholar]
- 39.Gong J, Chehrazi-Raffle A, Reddi S, Salgia R. Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: a comprehensive review of registration trials and future considerations. J Immunother Cancer. 2018;6(1):8. doi: 10.1186/s40425-018-0316-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song G, et al. Effects of tumor microenvironment heterogeneity on nanoparticle disposition and efficacy in breast cancer tumor models. Clin Cancer Res. 2014;20(23):6083–6095. doi: 10.1158/1078-0432.CCR-14-0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bennett C, Tomlinson C, Hollingshead M, Green J. Accelerating combination drug therapies for triple negative breast cancer through comparative pre-clinical testing of human xenografts and a mouse breast cancer model. Cancer Res. 2009;69:2166. doi: 10.1158/0008-5472.SABCS-09-2166. [DOI] [Google Scholar]
- 42.Liu Y, Cao X. The origin and function of tumor-associated macrophages. Cell Mol Immunol. 2015;12(1):1–4. doi: 10.1038/cmi.2014.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tymoszuk P, et al. In situ proliferation contributes to accumulation of tumor-associated macrophages in spontaneous mammary tumors. Eur J Immunol. 2014;44(8):2247–2262. doi: 10.1002/eji.201344304. [DOI] [PubMed] [Google Scholar]
- 44.van Rooijen N, Hendrikx E. Liposomes for specific depletion of macrophages from organs and tissues. Methods Mol Biol. 2010;605:189–203. doi: 10.1007/978-1-60327-360-2_13. [DOI] [PubMed] [Google Scholar]
- 45.Maroulakou IG, Anver M, Garrett L, Green JE. Prostate and mammary adenocarcinoma in transgenic mice carrying a rat C3(1) simian virus 40 large tumor antigen fusion gene. Proc Natl Acad Sci USA. 1994;91(23):11236–11240. doi: 10.1073/pnas.91.23.11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med. 2011;365(8):725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541(7637):321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 48.Neelapu SS, et al. Chimeric antigen receptor T-cell therapy - assessment and management of toxicities. Nat Rev Clin Oncol. 2018;15(1):47–62. doi: 10.1038/nrclinonc.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Champiat S, et al. Management of immune checkpoint blockade dysimmune toxicities: a collaborative position paper. Ann Oncol. 2016;27(4):559–574. doi: 10.1093/annonc/mdv623. [DOI] [PubMed] [Google Scholar]
- 50.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455(7213):674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhong B, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29(4):538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 52.Woo SR, et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41(5):830–842. doi: 10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li T, et al. Antitumor activity of cGAMP via stimulation of cGAS-cGAMP-STING-IRF3 mediated innate immune response. Sci Rep. 2016;6:19049. doi: 10.1038/srep19049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Balkwill F, Watling D, Taylor-Papadimitriou J. Inhibition by lymphoblastoid interferon of growth of cells derived from the human breast. Int J Cancer. 1978;22(3):258–265. doi: 10.1002/ijc.2910220307. [DOI] [PubMed] [Google Scholar]
- 55.Choi EA, et al. Stat1-dependent induction of tumor necrosis factor-related apoptosis-inducing ligand and the cell-surface death signaling pathway by interferon beta in human cancer cells. Cancer Res. 2003;63(17):5299–5307. [PubMed] [Google Scholar]
- 56.Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu Rev Immunol. 2004;22:329–360. doi: 10.1146/annurev.immunol.22.012703.104803. [DOI] [PubMed] [Google Scholar]
- 57.Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124(2):263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 58.Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4(1):71–78. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 59.Ostuni R, Kratochvill F, Murray PJ, Natoli G. Macrophages and cancer: from mechanisms to therapeutic implications. Trends Immunol. 2015;36(4):229–239. doi: 10.1016/j.it.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 60.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141(1):39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xia P, et al. Sox2 functions as a sequence-specific DNA sensor in neutrophils to initiate innate immunity against microbial infection. Nat Immunol. 2015;16(4):366–375. doi: 10.1038/ni.3117. [DOI] [PubMed] [Google Scholar]
- 62.Wilson DR, Sen R, Sunshine JC, Pardoll DM, Green JJ, Kim YJ. Biodegradable STING agonist nanoparticles for enhanced cancer immunotherapy. Nanomedicine. 2018;14(2):237–246. doi: 10.1016/j.nano.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tep K, et al. Formulation and evaluation of ATP-containing liposomes including lactosylated ASGPr ligand. J Liposome Res. 2009;19(4):287–300. doi: 10.3109/08982100902838682. [DOI] [PubMed] [Google Scholar]
- 64.Levchenko TS, Hartner WC, Verma DD, Bernstein EA, Torchilin VP. ATP-loaded liposomes for targeted treatment in models of myocardial ischemia. Methods Mol Biol. 2010;605:361–375. doi: 10.1007/978-1-60327-360-2_25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.