Abstract
Background and Aims
Mesenteric lymph nodes are sites in which translocated bacteria incite and progress immunological responses. For this reason, understanding the microbiome of mesenteric lymph nodes in inflammatory bowel disease is important. The bacterial profile of Crohn’s disease mesenteric lymph nodes has been analysed using culture-independent methods in only one previous study. This study aimed to investigate the mesenteric lymph node microbiota from both Crohn’s disease and ulcerative colitis patients.
Methods
Mesenteric lymph nodes were collected from Crohn’s disease and ulcerative colitis patients undergoing resection. Total DNA was extracted from mesenteric lymph nodes and assessed for the presence of bacterial DNA [16S]. All work was completed in a sterile environment using aseptic techniques. Samples positive for 16S DNA underwent next-generation sequencing, and the identity of bacterial phyla and species were determined.
Results
Crohn’s disease mesenteric lymph nodes had a distinctly different microbial profile to that observed in ulcerative colitis. The relative abundance of Firmicutes was greater in nodes from ulcerative colitis patients, whereas Proteobacteria were more abundant in Crohn’s disease. Although species diversity was reduced in the mesenteric lymph nodes of patients with Crohn’s disease, these lymph nodes contained greater numbers of less dominant phyla, mainly Fusobacteria.
Conclusion
This study confirms that there are distinct differences between the Crohn’s disease and ulcerative colitis mesenteric lymph node microbiomes. Such microbial differences could aid in the diagnosis of Crohn’s disease or ulcerative colitis, particularly in cases of indeterminate colitis at time of resection, or help explain their mechanisms of development and progression.
Keywords: Inflammatory bowel disease, microbiome, mesenteric lymph nodes
1. Introduction
Inflammatory bowel diseases [IBDs], which include Crohn’s disease [CD] and ulcerative colitis [UC], are thought to result from an overly aggressive immune response in genetically susceptible individuals to an environmental factor, such as gut commensals.1,2 The highest prevalence rates worldwide of CD and UC are found in Europe, with 322 and 505 cases per 100000 persons, respectively.3
The human gut microbiota has been investigated to a great extent, in both health and disease, particularly for gastrointestinal diseases, such as IBD. The earliest of these studies investigated bacterial content in faecal matter using conventional culture methods.4–11 Following this, the microbial profile of mucosal tissue was explored because it was believed to be a more accurate representation of the gut microbiota. Indeed, it has since been proven, by recent comprehensive studies, that significant differences exist between the bacterial groups found in the mucosa and those in faecal matter.12,13 Culture methods were used once again, to ascertain the mucosal tissue microbial profile.14–17 These studies provided a preliminary insight into the human gut microbiota; however, it has been estimated that up to 70–80% of intestinal bacteria are unculturable.18,19 For this reason, the development of culture-independent methods has significantly advanced our knowledge of gastrointestinal flora.20–22 These techniques have been used to great effect in establishing a gut microbiota profile for health and for IBD.
In healthy individuals, the phyla Firmicutes [a phylum including Enteroccocaceae, Clostridiaceae, and Ruminococcaceae] and Bacteroidetes [a phylum including Bacteroidaceae, Porphyromonadaceae, Prevotellaceae, Rikenellaceae, and Flavobacteriaceae] dominate, with lower abundance of other phyla, mainly Proteobacteria [a phylum including Enterobacteriaceae, and Burkholderiaceae] and Actinobacteria [a phylum including Mycobacteriaceae, Micrococcaceae, and Streptomycineae].23–25 The IBD gut microbiota displays dysbiosis when compared with that of healthy individuals, and there is a marked reduction in bacterial diversity, related particularly to the Firmicutes and Bacteroidetes phyla.26,27 This coincides with an increase in Gammaproteobacteria [which includes Enterobacteriaceae and Pseudomonadaceae].26 The CD microbiome displays greater dysbiosis than that found in UC - specifically, a greater reduction in microbial diversity, with a more altered and less stable microbiome composition.28 Inflamed mucosal tissue from CD patients contains higher levels of Bacteroidetes and Fusobacteria [includes Fusobacterium nucleatum], whereas Proteobacteria and Firmicutes [in particular Clostridiaceae, Enterobacteriaceae, and Ruminococcaceae] are more frequently observed in inflamed UC mucosa.29,30 Despite these changes, the microbiome of UC patients has been described as one that is more similar to healthy individuals.31,32
Although mesenteric lymph nodes [MLNs] have been associated with initiation of immunological responses to bacterial translocation,33–35 the importance of the MLN microbiome in IBD remains unclear.
To our knowledge, only one study has assessed the microbial content of MLNs in CD using culture-independent methods, reporting a bacterial profile similar to that of the CD gut microbiome, and no study has described the microbiome of UC patient MLNs.36 This is despite various culture-dependent studies demonstrating bacterial translocation to MLNs and mesenteric fat in IBD.37–39 Consistently, the most common organism detected in the MLNs in IBD was Escherichia coli37,38,40; however, the full diversity of the MLN microbiota has not yet been elucidated. Addressing this gap in knowledge may prove valuable in better understanding these distinct diseases. Our group has recently highlighted the potential role of the mesentery in IBD and has demonstrated that surgical recurrence rates for ileocolic CD patients are significantly reduced when the mesentery, and MLNs, are removed during resection.41,42 As the similarity of the MLN and mucosal microbiome in CD has been comprehensively reported previously, and selective bacterial translocation from the gut to the nodes effectively refuted,36 our interest concerned the variation in bacteria in MLNs from IBD patients. Therefore, we aimed to investigate and compare the microbial profile of MLNs from CD and UC patients diagnosed pathologically, using culture-independent methods for the first time.
2. Methods
2.1. Ethical approval
Ethical approval was obtained from the Research Ethics Committee of University Hospital Limerick [UHL].
2.2. Inclusion and exclusion criteria
All adult patients [≥18 years] undergoing resection for histopathologically diagnosed CD or UC in UHL from October 2015 to September 2016 were recruited. Patients were excluded if they had been diagnosed with colorectal cancer previously or were suspected of having colorectal cancer at the time of operation, so as not to interfere with pathologic analysis.
2.3. Mesenteric lymph node harvest
Where possible, IBD patients in UHL undergo resections that include the mesentery,41 allowing for greater MLN yields. Diseased intestine, along with associated mesentery, was resected and placed immediately in a sterile surgical tray. The resected specimen was kept in the sterile surgical area until the lymph nodes could be harvested. Lymph nodes were harvested from resected specimens in a sterile environment, which was outside the sterile surgical area but under the same atmospheric pressure, using aseptic techniques and sterile surgical instruments. The lymph nodes were identified and cut from the mesentery by a trained colorectal surgeon. Nodes were placed, without handling, into sterile microtubes. Although desirable, it was not possible to utilize nodes as morphological controls [e.g. nodes halved and sent for both 16S testing and pathological analysis]. Allprotect Tissue Reagent [Qiagen, UK] was added to each specimen tube until the tissue was fully immersed. A separate control sample of Allprotect Tissue Reagent was also obtained. Samples were kept at room temperature for a maximum of 2 h before transfer to long-term storage at –80°C.
2.4. DNA extraction
Lymph nodes were thawed on ice for 30 min before processing. All work was performed in sterile conditions in a Class II biosafety cabinet, using aseptic techniques and DNase-free instruments and consumables. Excess Allprotect Tissue Reagent was removed, and nodes were washed in 1 mL sterile phosphate-buffered saline (PBS; pH 7.4 [137 mM NaCl; 2.68 mM KCl; 9.94 mM Na2HPO4; 1.76 mM KH2PO4]). MLNs were weighed, and samples of ~25 mg were used for DNA extraction. Total DNA was extracted from samples using a QIAamp cador pathogen Mini Kit [Qiagen, UK], using the manufacturer’s instructions. In brief, tissue lysis buffer [180 µL; Buffer ATL] was added to the node tissue and the sample was homogenized for 1 min using a motorized pestle. Proteinase K enzyme [20 µL] was added to this to commence enzymatic tissue digestion. Samples were incubated overnight at 56°C with constant shaking. Following this, digested tissue [200 µL] mixed with sterile PBS [200 µL] was transferred to Pathogen Lysis Tubes S [Qiagen, UK], pre-prepared with tissue lysis buffer [Buffer ATL] and anti-foaming reagent [Reagent DX] [100 µL total] for mechanical disruption of hard-to-lyse bacteria by glass beads. This was completed by vigorous shaking for 10 min. Buffer VXL [100 µL], which ensures the inactivation of nucleases when in the presence of proteinase [100 µL], was added to the pre-treated samples [enzymatically digested and mechanically disrupted], mixed well, and left to incubate at room temperature for 15 min. Binding reagent [Buffer ACB; 350 µL] was added to samples, mixed well, and transferred to spin columns. DNA was then isolated following a series of centrifugations using QIAamp spin columns and buffer solutions. DNA was eluted in 100 µL elution buffer [Buffer AVE] [2 × 10-min elutions], and quantity and quality were assessed using the 260/280 function on the Spectrostar Nano plate reader LVis plate function [BMG Labtech]. Extracted DNA was stored at –20°C for subsequent polymerase chain reaction [PCR].
2.5. Polymerase chain reaction amplification of 16S DNA
The presence or absence of bacteria in MLNs was confirmed by PCR amplification of 16S [bacterial] DNA. All work was carried out on a dedicated clean PCR bench, in sterile conditions and using sterile and DNase-free instruments and consumables. Suitable positive [extracted DNA from a combined mixture of bacterial species] and negative [no template DNA and extracted DNA from PBS false extraction] controls were included. The PCR reaction included HotStarTaq Master Mix [25 µL], 500 ng template DNA, 0.5 µM 16S rRNA Forward primer, and 0.5 µM 16S rRNA Reverse primer. The total volume of the PCR reaction was 50 µL; the remaining volume was made up with PCR grade water [HotStarTaq Master Mix Kit, Invitrogen]. The forward and reverse primers were ready-made primers obtained from Integrated DNA Technologies [IDT] and are designed to incorporate a large portion of the 16S gene: For [5-AGA GTT TGA TCC TGG CTC AG-3] and Rev [5-ACG GCT ACC TTG TTA CGA CTT-3]. Reactions were performed using a Kyratec Thermal Cycler [Kyratec, Australia]: heat activation step at 95°C for 15 min; 30 cycles of denaturation at 94°C for 30 s, annealing at 57.49°C for 30 s and extension at 72°C for 60 s; and the final extension step at 72°C for 10 min. Amplicons were extracted using GenElute PCR Clean-Up Kit [Sigma Aldrich] as per the manufacturer’s instructions. Amplified DNA was eluted in 50 µL elution buffer [from kit], and presence, quantity, and quality were assessed using the 260/280 function on the Spectrostar Nano plate reader. DNA quality was also assessed by visualization on a 1% agarose gel. Lymph nodes were deemed as “16S negative” when: [i] no quantity of DNA was detected on the Spectrostar Nano plate reader, [ii] the 260/280 ratio and graph profiles were not similar to that expected of DNA content, and [iii] a large quantity of the amplified product was ran on a 1% agarose gel, and no DNA band was visualized. Amplified DNA was stored at –20°C for future species identification through sequencing.
2.6. Next-generation sequencing
Library preparation and sequencing were completed in the Teagasc Sequencing Centre [Cork, Ireland] as follows: DNA was quantified using the Qubit High Sensitivity Kit [Life Technologies], standardized, and then used as a template for PCR. PCR primers and conditions are essentially as outlined in the Illumina 16S Metagenomic Sequencing Library preparation guide [Illumina] with the following exceptions: For the initial 16S PCR, the process was performed using Kapa Robust [Roche] in 50 µl reaction volumes, and 30 cycles were used in the PCR. The V3–V4 variable regions were amplified by the primers. These regions are the suggested target for the Illumina 16S Metagenomic Sequencing system as previously found to be the optimum primer pair for sequencing-based species diversity studies.43,44 Products were then cleaned with an appropriate volume of Ampure beads and eluted in 30 µl per sample. This was then used as the template for the index PCRs as outlined in the protocol [Illumina]. Library quantification, normalization, and pooling were as outlined in the protocol. After pooling, the sample was re-quantified by qPCR using the Kapa Library Quantification Kit for Illumina [Roche] and run on an Agilent high-sensitivity chip [Agilent]. Library denaturation and MiSeq sample loading were then performed according to the manufacturer’s instructions [Illumina]. PhiX was spiked into the final pool at 20% [v/v], and sequencing was performed using a 500-cycle V2 chemistry kit [Illumina]. Previous studies using the Illumina MiSeq system and primer pairs targeting the V3–V4 variable regions have identified bacteria to species level.45,46 The V3–V4 variable regions are commonly used to investigate the gut microbiome and to detect intestinal bacterial species.45–48
2.7. Bioinformatics
Illumina reads were filtered on the basis of quality [removal of low-quality nucleotides at the 3′ end, in addition to bases where quality was below 20 in a trimming window] and length [removal of sequences with <200 bp] with PRINSEQ,49 and the paired-end reads with a minimum overlap of 20 bp were joined using Fastq-join.50 A pipeline [a series of data-processing events] was generated, using clean reads only [i.e. filtered reads described above], to cluster sequences [from next-generation sequencing data] with 97% identity to obtain Operative Taxonomic Units [OTUs] using a closed-reference USEARCH v7.0 algorithm [Edgar RC. 2010] and remove chimeric OTUs against the Genome OnLine Database [GOLD]. An OTU represents a group of similar 16S rRNA sequences that theoretically reflect shared species identity. However, while a 97% threshold is sufficient for diversity studies, similarity thresholds of >98% are required to identify species definitively. The taxonomic assignment of these OTUs was obtained against the Ribosomal Database Project [RDP; http://rdp.cme.msu.edu/].51 Beta-diversity was determined using QIIME,52 and additional analysis were performed with the R package, phyloseq.53
2.8. Statistical analysis
Data are presented as mean ± 95% confidence interval [CI]. To determine phyla, family, and species OTUs, data were statistically analysed by PERMANOVA for beta-diversity. All other statistical analyses were completed in SPSS v22 [SPSS Inc., Chicago, USA]. An independent two-tailed t-test was used to compare parametric variables, and a Mann–Whitney U test was utilized for non-parametric comparisons. A Kruskal-Wallis test was used to determine the most abundant bacterial genus in MLNs from patients with CD or UC. Species richness estimates were generated using the Chao 1 estimator and the Abundance-based Coverage Estimator [ACE], and community and species diversity were estimated using the Shannon Diversity index. A 5% level of significance was used for all statistical tests.
3. Results
3.1. Patient and MLN characteristics
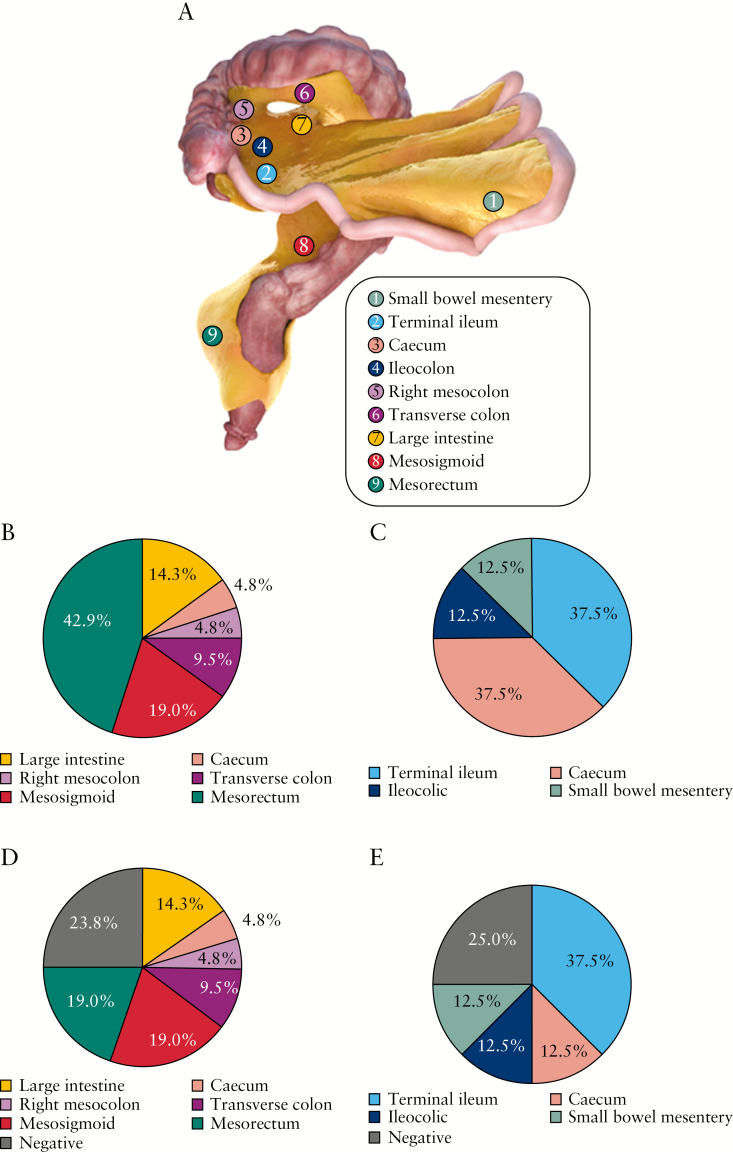
Thirteen pathologically diagnosed IBD patients [CD: n = 5, 38%; UC: n = 8, 62%] were recruited [Supplementary Table 1]. These underwent 15 surgical procedures for IBD. All patients had intravenous antibiotics administered with induction of anaesthesia, ~15 min before the start of the surgical procedure, as per standard of care. MLNs were taken from locations along the small bowel mesentery and mesocolon [Figure 1A–C]. No biases were observed regarding the number of lymph nodes or lymph node diameter irrespective of sampling location. There was no evidence of abscesses in the MLNs collected. Furthermore, histopathologic analysis of resection specimens did not identify the presence of mesenteric abscesses. In total, 25% and 24% of MLNs collected from CD and UC patients, respectively, did not contain bacterial DNA [Figure 1D, E]. Demographics of patients with bacterial DNA–positive nodes are provided in Table 1.
Figure 1.
Mesenteric lymph node [MLN] mapping. [A] Digital image of the small and large bowel with associated continuous mesentery. Source locations of MLNs are mapped on the mesentery. [B] Proportions of MLNs taken from each location of ulcerative colitis [UC] mesentery. The majority of lymph nodes were taken from the mesorectum [42.9%]. [C] Proportions of MLNs taken from each location of Crohn’s disease [CD] mesentery. The majority of lymph nodes were taken from the ileocolic region [87.5%]. [D] Proportions of 16S PCR–positive MLNs taken from each location of UC mesentery. [E] Proportions of 16S PCR–positive MLNs taken from each location of CD mesentery.
Table 1.
Demographics of patients with PCR-positive lymph nodes. All patients received antibiotics at induction of anaesthesia.
| Patient ID | Disease | Gender | Age at op | Age at dx | Disease duration | Operation | Disease behaviour | Disease location | Smoking status | Antibiotics | Anti-TNF | Immunosuppressants | Steroids | 5-ASA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | UC | F | 29 | 28 | 12 | TC | N/A | E3 | Ex | N | Y | N | Y | Y |
| 2 | UC | F | 26 | 19 | 87 | TC | N/A | E3 | N | N | Y | Y | N | Y |
| 3 | CD | F | 33 | 27 | 70 | IC | B1 | L1 | N | N | N | N | Y | Y |
| 5 | UC | M | 33 | 15 | 214 | TC | N/A | E3 | N | N | Y | Y | N | N |
| 6 | UC | F | 23 | 21 | 23 | TC | N/A | E2 | Ex | Y | Y | N | Y | Y |
| 6 | UC | F | 24 | 21 | 28 | CP | N/A | E2 | Ex | N | N | N | N | N |
| 7 | UC | F | 36 | 36 | 6 | TC | N/A | E2 | Ex | N | Y | N | Y | N |
| 7 | UC | F | 36 | 36 | 10 | CP | N/A | E2 | Ex | N | Y | Y | N | N |
| 8 | CD | M | 39 | 38 | 10 | IC | B2 | L1 | Y | N | N | N | Y | Y |
| 11 | CD | M | 38 | 37 | 2 | IC | B3 | L1/L4 | Y | N | N | N | Y | N |
| 13 | CD | F | 76 | 49 | 331 | IC | B2 | L3 | Ex | Y | N | Y | Y | N |
| 15 | CD | F | 33 | 17 | 202 | IC | B3 | L1 | Y | N | Y | N | Y | Y |
Key: Disease: UC = ulcerative colitis, CD = Crohn’s disease, N = non-IBD. Gender: M = male, F = female. Dx = diagnosis. Operation: TC = total colectomy, IC = ileocolic resection, CP = completion proctectomy. Montreal classification: Disease behaviour: B1 = inflammatory, B2 = stricturing, B3 = penetrating. Disease location: Crohn’s disease – L1 = terminal ileum ± caecum, L2 = colonic, L3 = ileocolic, L4 = upper gastrointestinal tract. Ulcerative colitis – E1 = proctitis, E2 = left-sided colitis, E3 = pan-colitis. Smoking status: Ex = ex-smoker, N = non-smoker, Y = smoker. Antibiotics: N = no, Y = on antibiotics <5 days prior to surgery. Medications: N = no, Y = yes.
3.2. The microbiome of MLNs from CD patients was distinctly different to that of MLNs from UC patients
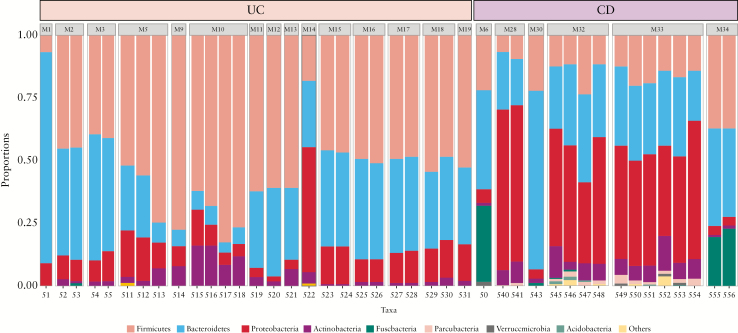
Bacteria from 14 different phyla were detected, irrespective of disease cohort [Figure 2]. The majority of OTUs belonged to Firmicutes and Bacteroidetes, followed by Proteobacteria and Actinobacteria. Other phyla, generally less studied and only defined in the last two decades [e.g. Parcubacteria, Verrucomicrobia], were also detected. MLNs were obtained from various locations within the mesentery from Patients 5, 6, and 7, such as the mesorectum [M8] and mesosigmoid [M9 and M10] from Patient 5, the mesorectum [M11 and M12] and right mesocolon [M13] from Patient 6, and the mesentery associated with the caecum [M16], transverse mesocolon [M17 and M18], mesosigmoid [M15 and M19], and mesorectum [M14] from Patient 7 [Figure 2]. The microbial profiles of MLNs taken from the same patient were similar, irrespective of sampling location. Furthermore, the microbiomes of MLNs taken from areas of mesentery that were determined to be disease-free [i.e. those with no features of macroscopic inflammation] at time of harvest were similar to those nodes taken from areas of diseased mesentery [i.e. those which displayed features of inflammation and disease] [Patient 5, UC, disease-free nodes: M8 and diseased nodes: M9 and M10; Patient 13, CD, disease-free nodes: M32 and diseased nodes: M33; Figure 2]. There was no readily apparent association between disease behaviour and bacterial profiles.
Figure 2.
Relative abundance of predominant bacterial phyla in mesenteric lymph nodes [MLNs] from inflammatory bowel disease [IBD] patients. There was a distinct difference in the profile of phyla from MLNs of Crohn’s disease [CD] and ulcerative colitis [UC] patients. S = sample number, M = merge of samples of same node.
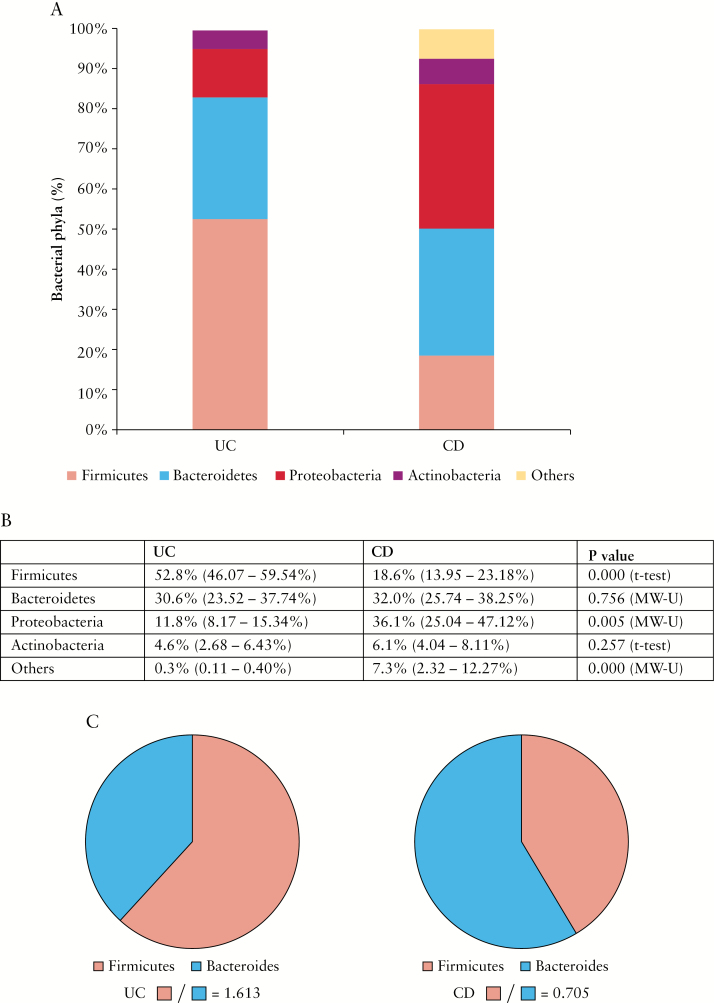
The relative abundance of Firmicutes was greater in UC MLNs than in those of CD patients [52.8% ± 6.73% vs 18.6% ± 4.65%; p < 0.0001, independent t-test] [Figure 3]. In contrast, MLNs from CD patients contained greater abundance of Proteobacteria [36.1% ± 11.06% vs 11.8% ± 3.63%; p = 0.005, Mann–Whitney U test] and greater numbers of less dominant phyla, mainly Fusobacteria [7.3% ± 4.98% vs 0.3% ± 0.15%; p < 0.0001, Mann–Whitney U test] [Figure 3A, B]. Similar levels of Bacteroidetes and Actinobacteria were detected in the two cohorts [Figure 3A, B]. The ratio of Firmicutes to Bacteroidetes was increased in UC MLNs [1.613], but decreased in CD MLNs [0.705] [Figure 3C].
Figure 3.
Abundance of phyla in pooled mesenteric lymph nodes [MLNs] of Crohn’s disease [CD] and ulcerative colitis [UC] patients. [A] Quantities [abundance %] of major bacterial phyla Firmicutes, Bacteroides, Proteobacteria, and Actinobacteria in pooled MLNs of UC and CD patients. [B] Statistical analysis of phyla abundance with 95% confidence interval [CI]. MLNs of UC patients had higher proportions of Firmicutes [Mean ± 95% CI: 52.8% ± 6.73% vs 18.6% ± 4.65%; p < 0.0001, independent t-test], whereas MLNs of CD patients had higher proportions of Proteobacteria [36.1% ± 11.06% vs 11.8% ± 3.63%; p = 0.005, Mann–Whitney U test] and other type bacteria, comprising mainly Fusobacteria [7.3% ± 4.98% vs 0.3% ± 0.15%; p < 0.0001, Mann–Whitney U test]. Independent t-tests were used to compare normally distributed [parametric] data, and Mann–Whitney U tests were used to compare non-parametric data. [C] The ratio of Firmicutes to Bacteroidetes in MLNs from UC [1.613] and CD [0.705] patients.
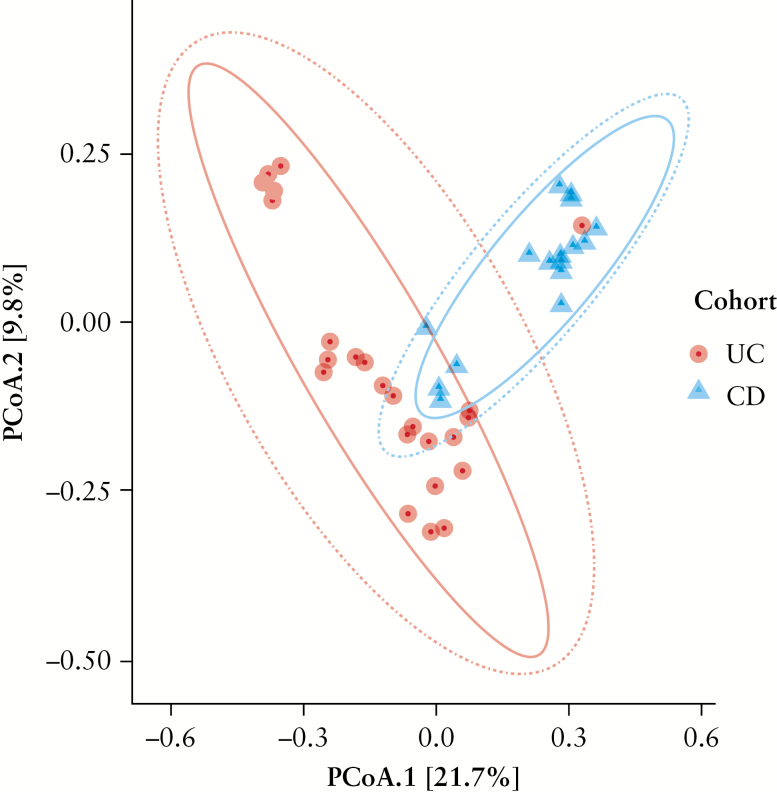
Interestingly, although Firmicutes was the most abundant phylum in UC MLNs, the most abundant bacterial genus was Bacteroides [phylum Bacteroidetes], followed by Faecalibacterium [phylum Firmicutes] [p < 0.0001, Kruskal–Wallis test]. In CD MLNs, the most abundant bacterial genus was Flavobacterium, followed by Bacteroides, both members of phylum Bacteroidetes [p < 0.0001, Kruskal–Wallis test]. Although Faecalibacterium [i.e. Faecalibacterium prausnitzii], an important component of the healthy gut microbiome, was abundant in UC, it was present to a lesser extent in CD [10.9% ± 4.26% vs 1.2% ± 1.15%; p < 0.0001, Mann–Whitney U test]. Definitively, a Principal Coordinates Analysis [PCoA] of unweighted Unifrac distance demonstrated that the bacterial compositions of UC and CD MLNs were distinctly different [Figure 4].
Figure 4.
Principal coordinate analysis [PCoA] of Unifrac difference. The chart is based on unweighted unifrac distances and displays variation between cohort samples.
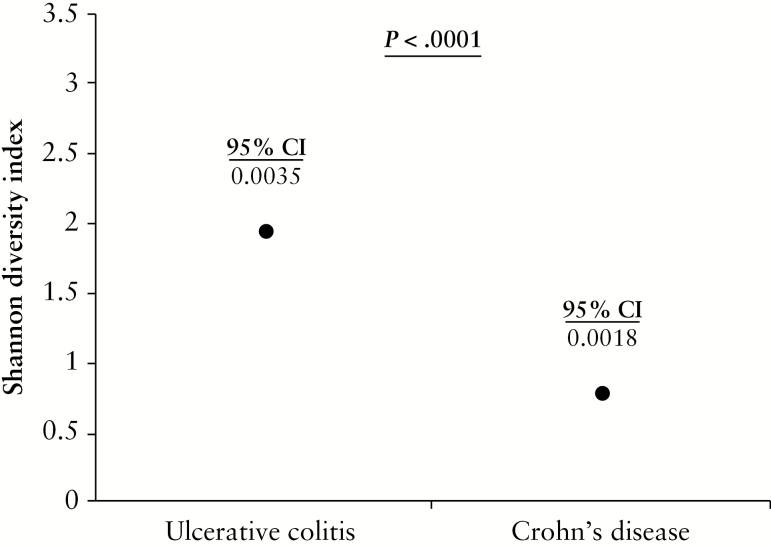
3.3. Diversity and species richness was reduced in Crohn’s disease MLNs
Species diversity was estimated using the Shannon Diversity index. MLNs from UC patients displayed significantly greater microbial diversity than those of CD patients [1.95 ± 0.0035 vs 0.79 ± 0.0018; p < 0.0001, Mann–Whitney U test] [Figure 5]. We determined species richness using the Chao 1 and ACE estimators [Supplementary Table 2]. The Chao 1 estimator provides investigators with a lower bound estimate for species richness.54 This indicated greater species richness in CD MLNs than in those of UC [237 vs 183] [Supplementary Table 2]. ACE is a more comprehensive estimate of species richness, derived from the number of rare and common species present in a sample, while also considering how many more undiscovered species there may be.54 This showed greater species richness in UC than in CD MLNs [175–178 vs 148–156] [Supplementary Table 2], similar to actual raw species counts observed.
Figure 5.
Microbial diversity of mesenteric lymph nodes [MLNs] from Crohn’s disease [CD] and ulcerative colitis [UC] patients. Shannon Diversity Indices for MLNs of UC and CD patients defined using data from 16S sequencing data at species level. Data are presented as mean ± 95% confidence interval [CI]. MLNs from UC patients demonstrated significantly greater microbial diversity than those of CD patients [1.95 ± 0.0035 vs 0.79 ± 0.0018; p < 0.0001, Mann–Whitney U test]. As data were found to be non-parametric [not-normally distributed], Mann–Whitney U tests were used to compare data.
4. Discussion
This is the first study to investigate and report distinct differences between the microbial profiles of MLNs from UC and CD patients. Aided somewhat by the advantageous availability of resected material from UC patients, this report also represents the first time that the microbiome of MLNs from UC patients has been elucidated. In comparison with the reported normal microbiome,23,24 the UC MLN microbial profile appears imbalanced in favour of elevated Firmicutes relative to Bacteroidetes. Crohn’s disease MLN dysbiosis is reflected by overabundance of Proteobacteria [e.g. pathogens such as Escherichia, Shigella, Helicobacter, and Salmonella], a decreased Firmicutes:Bacteroidetes ratio, and a reduction in species diversity and richness. The MLN bacterial profiles remained unchanged irrespective of extent of disease or sampling location. Notably, MLNs from UC patients who underwent completion proctectomy at least 6 months following their total colectomy were free of bacterial DNA [Patients 10, 12, and 14, Supplementary Table 1]. During this time, patients had an end ileostomy in situ. This suggests that, in these patients, diversion of the faecal stream allowed clearance of bacterial DNA from MLNs by the host immune system.
It has been reported previously that the UC and CD faecal and mucosal microbiota are distinct from one another.28–30 Our results now confirm that this variation extends to the MLN microbiome. This variation differentiates clearly between the diseases, representing new diagnostic potential, and may also offer targets for innovative therapeutic developments. Approximately 10–15% of IBD cases are recorded as indeterminate colitis, meaning a definitive diagnosis of CD or UC cannot be made from the resection specimen, or from biopsies at colonoscopy.55,56 We believe that the distinctive UC and CD MLN microbiota could enable definitive diagnoses at resection, potentially revolutionizing prophylactic treatment decisions, patient aftercare and survival.
In healthy individuals, the gut microbiota is comprised mainly of Firmicutes and Bacteroidetes [~50% and ~40%, respectively]23,24; thus the ratio of Firmicutes to Bacteroidetes is often used as an indicator of gut microbiota balance. In this study, the relative abundance of Firmicutes was greater than Bacteroidetes in MLNs from UC patients. Inversely, Bacteroidetes had a greater relative abundance than Firmicutes in CD MLNs. The ratio of Firmicutes to Bacteroidetes in the MLN microbiome from both CD and UC patients has not been characterized previously. However, it has been studied to some extent in IBD patient–derived faeces.57,58 Most of those studies reported an overall decrease in the ratio relative to that of healthy controls.57,58 Conversely, there have also been reports of increases in the ratio.59 This disparity could be explained by geographical, dietary, or treatment factors. In addition, such studies have often reported their results based on UC and CD patients combined, which could confound the results because there was presumption that both diseases share similar microbiomes. One previous study has described a decrease in Firmicutes relative to Bacteroidetes in UC and CD mucosal tissue when studied separately.60 This differs from the profile that we have observed in UC MLNs. This further differentiates UC and CD and emphasizes the need, when analyzing the microbiome of MLNs, to segregate IBD-derived samples.
Mechanistically, the gut microbiota reportedly restricts translocation of pathogenic bacteria to the MLN.61 This may be compromised in CD but not UC, because the MLN microbiota of CD patients display an overabundance of Proteobacteria [known to contain numerous pathogenic species]. Likewise, the clearance of harmful bacteria from MLNs could also be reduced.61 The MLNs are sites in which immunological responses can commence and proliferate.33–35 It may be that CD MLNs are host to bacterial types that trigger more aggressive responses than those of UC MLNs, such as members of the Proteobacteria phylum. Conversely, Faecalibacterium, which mediates anti-inflammatory effects,62 is present in CD MLNs in low proportions compared with the proportions in UC MLNs. Is it possible that the pathologic features of CD, which differ greatly to that of UC with regard to the increased level of mesenteric involvement, are a consequence of the CD MLN microbiota? It is reasonable to argue that future work could usefully investigate the role of various members of Proteobacteria or Faecalibacterium, or indeed the ratios of one to the other, in the MLNs and in immunological responses in both CD and UC. In conclusion, the distinctive UC and CD MLN microbiota provide us with a novel diagnostic tool for defining indeterminate colitis, an opportunity to understand the mechanisms mediating each disease, and the prospect of improving patient outcomes.
Funding
This work was supported by the Graduate Entry Medical School [University of Limerick] Strategic Research Fund.
Conflict of Interest
The authors declare no competing personal or financial interests and have nothing to disclose.
Author Contributions
MGK collected and processed mesenteric lymph nodes, performed experiments and was involved in study design, clinical data collection, data analysis, and drafting of the manuscript. JCC was involved in study design, sample collection and drafting of the manuscript. PAK aided in laboratory analysis and drafting of the manuscript. PDC completed sample library preparation and next-generation sequencing. RCR completed bioinformatics analysis. KMcD was involved in study design and drafting of the manuscript. CPD initiated the study concept and design, and was involved in data analysis and drafting of the manuscript.
Supplementary Material
Acknowledgments
We thank Dara Walsh for assistance with digitally sculpted images, Dr Paul Tibbitts for aid in collecton of samples, and Dr Fiona Crispie and Laura Finnegan [Teagasc] for help in completing the next-generation sequencing.
Glossary
Abbreviations
- MLNs
mesenteric lymph nodes
- IBD
inflammatory bowel disease
- CD
Crohn’s disease
- UC
ulcerative colitis
- DNA
deoxyribonucleic acid
- PCR
polymerase chain reaction
- UHL
University Hospital Limerick
- RNA
ribonucleic acid
- PBS
phosphate-buffered saline
- IDT
Integrated DNA Technologies
- OTU
operational taxonomic unit
- GOLD
Genome OnLine Database
- RDP
Ribosomal Database Project
- CI
confidence interval
- PCoA
Principal Coordinates Analysis.
References
- 1. Sartor RB. Mechanisms of disease: Pathogenesis of Crohn’s disease and ulcerative colitis. Nat Clin Pract Gastroenterol Hepatol 2006;3:390–407. [DOI] [PubMed] [Google Scholar]
- 2. Baumgart DC, Sandborn WJ. Crohn’s disease. Lancet 2012;380:1590–605. [DOI] [PubMed] [Google Scholar]
- 3. Molodecky NA, Soon IS, Rabi DM, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 2012;142:46–54.e42. [DOI] [PubMed] [Google Scholar]
- 4. Cregan J, Hayward NJ. The bacterial content of the healthy human small intestine. Br Med J 1953;1:1356–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tabaqchali S, Booth CC. Relationship of the intestinal bacterial flora to absorption. Br Med Bull 1967;23:285–90. [DOI] [PubMed] [Google Scholar]
- 6. Finegolld SM. Intestinal bacteria. The role they play in normal physiology, pathologic physiology, and infection. Calif Med 1969;110:455–9. [PMC free article] [PubMed] [Google Scholar]
- 7. Hamilton JD, Dyer NH, Dawson AM, et al. Assessment and significance of bacterial overgrowth in the small bowel. QJM: Int J Med 1970;39:265–86. [PubMed] [Google Scholar]
- 8. Gorbach SL, Neale G, Levitan R, Hepner GW. Alterations in human intestinal microflora during experimental diarrhoea. Gut 1970;11:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aries V, Crowther JS, Drasar BS, Hill MJ, Williams RE. Bacteria and the aetiology of cancer of the large bowel. Gut 1969;10:334–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gorbach SL, Nahas L, Plaut AG, et al. Studies of intestinal microflora. Gastroenterology 1968;54:575–87. [PubMed] [Google Scholar]
- 11. Keighley MR, Arabi Y, Dimock F, et al. Influence of inflammatory bowel disease on intestinal microflora. Gut 1978;19:1099–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zoetendal EG, von Wright A, Vilpponen-Salmela T, et al. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol 2002;68:3401–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Marteau P, Pochart P, Doré J, et al. Comparative study of bacterial groups within the human cecal and fecal microbiota. Appl Environ Microbiol 2001;67:4939–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hartley CL, Neumann CS, Richmond MH. Adhesion of commensal bacteria to the large intestine wall in humans. Infect Immun 1979;23:128–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nelson DP, Mata LJ. Bacterial flora associated with the human gastrointestinal mucosa. Gastroenterology 1970;58:56–61. [PubMed] [Google Scholar]
- 16. Peach S, Lock MR, Katz D, Todd IP, Tabaqchali S. Mucosal-associated bacterial flora of the intestine in patients with Crohn’s disease and in a control group. Gut 1978;19:1034–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Plaut AG, Gorbach SL, Nahas L, Weinstein L, Spanknebel G, Levitan R. Studies of intestinal microflora. 3. The microbial flora of human small intestinal mucosa and fluids. Gastroenterology 1967;53:868–73. [PubMed] [Google Scholar]
- 18. Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science 2005;308:1635–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host–bacterial mutualism in the human intestine. Science 2005;307:1915–20. [DOI] [PubMed] [Google Scholar]
- 20. Ji B, Nielsen J. From next-generation sequencing to systematic modeling of the gut microbiome. Front Genet 2015;6:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dunne C. Adaptation of bacteria to the intestinal niche: Probiotics and gut disorder. Inflamm Bowel Dis 2001;7:136–45. [DOI] [PubMed] [Google Scholar]
- 22. Schirmer M, Franzosa EA, Lloyd-Price J, et al. Dynamics of metatranscription in the inflammatory bowel disease gut microbiome. Nat Microbiol 2018;3:337–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hong PY, Croix JA, Greenberg E, Gaskins HR, Mackie RI. Pyrosequencing-based analysis of the mucosal microbiota in healthy individuals reveals ubiquitous bacterial groups and micro-heterogeneity. PLoS ONE 2011;6: e25042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Faith JJ, Guruge JL, Charbonneau M, et al. The long-term stability of the human gut microbiota. Science 2013;341:1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Frank DN, St. Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 2007;104:13780–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel diseases: Current status and the future ahead. Gastroenterology 2014;146:1489–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pascal V, Pozuelo M, Borruel N, et al. A microbial signature for Crohn’s disease. Gut 2017;66:813–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Forbes JD, Van Domselaar G, Bernstein CN. Microbiome survey of the inflamed and noninflamed gut at different compartments within the gastrointestinal tract of inflammatory bowel disease patients. Inflamm Bowel Dis 2016;22:817–25. [DOI] [PubMed] [Google Scholar]
- 30. Lavelle A, Lennon G, O’Sullivan O, et al. Spatial variation of the colonic microbiota in patients with ulcerative colitis and control volunteers. Gut 2015;64:1553–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morgan XC, Tickle TL, Sokol H, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 2012;13:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Halfvarson J, Brislawn CJ, Lamendella R, et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol 2017;2:17004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mowat AM.Anatomical basis of tolerance and immunity to intestinal antigens. Nat Rev Immunol 2003;3:331–41. [DOI] [PubMed] [Google Scholar]
- 34. Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 2004;303:1662–5. [DOI] [PubMed] [Google Scholar]
- 35. Wei B, Velazquez P, Turovskaya O, et al. Mesenteric B cells centrally inhibit CD4+ T cell colitis through interaction with regulatory T cell subsets. Proc Nat Acad Sci U S A 2005;102:2010–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. O’Brien CL, Pavli P, Gordon DM, Allison GE. Detection of bacterial DNA in lymph nodes of Crohn’s disease patients using high throughput sequencing. Gut 2014;63:1596–606. [DOI] [PubMed] [Google Scholar]
- 37. Laffineur G, Lescut D, Vincent P, et al. Bacterial translocation in Crohn’s disease. Gastroenterol Clin Biol 1992;16:777–81 [PubMed] [Google Scholar]
- 38. Sedman PC, Macfie J, Sagar P, et al. The prevalence of gut translocation in humans. Gastroenterology 1994;107:643–9. [DOI] [PubMed] [Google Scholar]
- 39. Peyrin-Biroulet L, Gonzalez F, Dubuquoy L, et al. Mesenteric fat as a source of C reactive protein and as a target for bacterial translocation in Crohn’s disease. Gut 2012;61:78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. O’Boyle CJ, MacFie J, Mitchell CJ, et al. Microbiology of bacterial translocation in humans. Gut 1998;42:29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Coffey JC, Kiernan MG, Sahebally SM, et al. Inclusion of the mesentery in ileocolic resection for Crohn’s disease is associated with reduced surgical recurrence. J Crohns Colitis 2018, published online Jan 4 DOI: 10.1096/ecco-jcc/jjx187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Coffey JC, O’Leary DP, Kiernan MG, Faul P. The mesentery in Crohn’s disease: Friend or foe?Curr Op Gastroenterol 2016;32:267–73. [DOI] [PubMed] [Google Scholar]
- 43. Klindworth A, Pruesse E, Schweer T, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing–based diversity studies. Nucleic Acids Res 2013;41:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. PCR Amplicon, PCR Clean-up, and Index PCR. In: 16S Metagenomic Sequencing Library Preparation; 2013. https://web.uri.edu/gsc/files/16s-metagenomic-library-prep-guide-15044223-b.pdf Accessed March 8, 2018. [Google Scholar]
- 45. Shin J, Lee S, Go M-J, et al. Analysis of the mouse gut microbiome using full-length 16S rRNA amplicon sequencing. Sci Rep 2016;6:29681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cronin O, Barton W, Skuse P, et al. A prospective metagenomic and metabolomic analysis of the impact of exercise and/or whey protein supplementation on the gut microbiome of sedentary adults. mSystems 2018;3:pii: e00044–18. doi: 10.1128/mSystems.00044-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mondot S, Lepage P, Seksik P, et al. Structural robustness of the gut mucosal microbiota is associated with Crohn’s disease remission after surgery. Gut 2016;65:954–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dong LN, Wang JP, Liu P, et al. Faecal and mucosal microbiota in patients with functional gastrointestinal disorders: Correlation with toll-like receptor 2/toll-like receptor 4 expression. World J Gastroenterol 2017;23:6665–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011;27:863–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Aronesty E. Ea-utils: Command-line Tools for Processing Biological Sequencing Data; 2011. Available online at: https://github.com/Expression Analysis/ea-utils Accessed September 4, 2018. [Google Scholar]
- 51. Cole JR, Wang Q, Fish JA, et al. Ribosomal database project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res 2014;42:D633–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McMurdie PJ, Holmes S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chao A, Chiu CH. Species richness: Estimation and comparison. Wiley StatsRef: Statistics Reference Online 2016:1–26. http://chao.stat.nthu.edu.tw/wordpress/paper/119.pdf Accessed September 4, 2018.
- 55. Price AB. Overlap in the spectrum of non-specific inflammatory bowel disease–‘colitis indeterminate’. J Clin Pathol 1978;31:567–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Guindi M, Riddell RH. Indeterminate colitis. J Clin Pathol 2004;57:1233–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sokol H, Seksik P, Furet JP, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis 2009;15:1183–9. [DOI] [PubMed] [Google Scholar]
- 58. Bamola VD, Ghosh A, Kapardar RK, et al. Gut microbial diversity in health and disease: Experience of healthy Indian subjects, and colon carcinoma and inflammatory bowel disease patients. Microb Ecol Health Dis 2017;28:1322447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Santoru ML, Piras C, Murgia A, et al. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci Rep 2017;7:9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kabeerdoss J, Jayakanthan P, Pugazhendhi S, Ramakrishna BS. Alterations of mucosal microbiota in the colon of patients with inflammatory bowel disease revealed by real time polymerase chain reaction amplification of 16S ribosomal ribonucleic acid. Indian J Med Res 2015;142:23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Diehl GE, Longman RS, Zhang JX, et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX3CR1hi cells. Nature 2013;494:116–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Quévrain E, Maubert MA, Michon C, et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut 2016;65:415–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.