Abstract
Genetic factors are an important cause of idiopathic sensorineural hearing impairment (SNHI). From the epidemiological perspective, mutations of three deafness genes: GJB2, SLC26A4, and MT-RNR1, are much more prevalent than those of other genes worldwide. However, mutation spectra of common deafness genes differ remarkably across different populations. Here, we performed comprehensive genetic examination and haplotype analyses in 188 unrelated Mongolian families with idiopathic SNHI, and compared their mutation spectra and haplotypes to those of other European and Asian cohorts. We confirmed genetic diagnoses in 18 (9.6%) of the 188 families, including 13 with bi-allelic GJB2 mutations, three with bi-allelic SLC26A4 mutations, and two with homoplasmic MT-RNR1 m.1555A>G mutation. Moreover, mono-allelic mutations were identified in 17 families (9.0%), including 14 with mono-allelic GJB2 mutations and three with mono-allelic SLC26A4 mutations. Interestingly, three GJB2 mutations prevalent in other populations, including c.35delG in Caucasians, c.235delC in East Asians, and c.-23+1G>A in Southwest and South Asians, were simultaneously detected in Mongolian patients. Haplotype analyses further confirmed founder effects for each of the three mutations, indicating that each mutation derived from its ancestral origin independently. By demonstrating the unique spectra of deafness-associated mutations, our findings may have important clinical and scientific implications for refining the molecular diagnostics of SNHI in Mongolian patients, and for elucidating the genetic relationships among Eurasian populations.
Introduction
Hearing impairment is the most common inherited sensory defect. It is estimated that permanent sensorineural hearing impairment (SNHI) occurs in approximately 1.9 per 1000 live births [1], and with late-onset SNHI included, the disorder may affect 2% of school-age children [2, 3]. More than 50% of SNHI cases in children are attributed to genetic causes, and are therefore classified as hereditary hearing impairment (HHI) [4]. To date, more than 100 genes have been identified as causally related to HHI (http://hereditaryhearingloss.org).
Among the plethora of HHI genes, mutations in three: GJB2 (MIM *121011), SLC26A4 (MIM *605646), and the mitochondrial 12S rRNA gene (MT-RNR1; MIM *561000), are particularly prevalent in deaf patients across different populations [4]. Predominant mutations in these genes differ significantly across populations. For instance, c.35delG, c.167delT, and c.235delC are the most common GJB2 mutations in Caucasians [5–8], Ashkenazi Jews [9], and East Asians [10–12], respectively; whereas the GJB2 c.-23+1G>A mutation was identified uniquely in Southwest [13, 14] and South Asians [15, 16]. Similarly, predominant SLC26A4 mutations differ among populations, including p.T416P and c.1001G>A in Caucasians [17, 18], p.H723R in Japanese [19] and Koreans [20], and c.919-2A>G in Han Taiwanese [21] and Han Chinese [22]. These findings underscore the indispensability of collecting regional data when genetic examination for SNHI is performed in a specific population.
The genetics of SNHI in the Mongolian population have been documented in several previous studies [23–26]. However, most of these studies were conducted in cohorts recruited from the Inner Mongolia region of China; only limited numbers of Mongolian patients were included in these studies, and the admixture of other ethnic populations could not be excluded because of inter-population marriage [23–25]. Hearing-impaired patients from Mongolia have been the subject of only one previous study [26]. However, this study focused on GJB2 mutations, and the contribution of other deafness genes to SNHI in these patients was not addressed [26].
The scientific value of investigating genetic diseases in Mongolian patients also lies in the geographic location of Mongolia. As an intersection between the European, Middle Eastern, and East Asian civilizations, dissecting the genetic underpinnings of Mongolians may offer insights into the genetic diversity and genetic relationships among the Eurasian populations [27–30]. In this study, we performed comprehensive mutation screening of three common HHI genes in a large cohort of Mongolian patients, and then conducted haplotype analyses to decipher the origins of SNHI-related mutations with reference to other European and Asian populations.
Methods
Subjects
From November 2016 to January 2018, a total of 188 unrelated Mongolian families with idiopathic bilateral SNHI were recruited from the EMJJ Otolaryngology Hospital and the Department of Otolaryngology, National Center for Maternal and Child Health, Ulaanbaatar, Mongolia. Patients were excluded if they (1) were aged more than 40 years, (2) had conductive or mixed-type hearing impairment, (3) had previous noise or ototoxic medical exposure, (4) had a history of perinatal insults, such as prematurity or kernicterus, or (5) had no complete records of their medical history available.
For the proband of each family, comprehensive family history, personal medical history, physical examination, audiological results, and imaging results were ascertained. The audiological results were evaluated with pure tone audiograms or auditory brainstem response, depending on age or neurological status [31]. For imaging studies, non-contrast temporal bone high-resolution computed tomography, with contiguous axial and coronal sections of 1-mm thickness, was obtained to investigate the structure of the inner ear [32–34].
Genetic examination
Dried blood spot specimens were collected from the patients and their family members, and genomic DNA was extracted using a MagCore HF16 Automatic DNA/RNA Purification system (RBC Bioscience Corp., Taiwan) with a MagCore Genomic DNA Tissue Kit (RBC Bioscience Corp., Taiwan) according to the manufacturer's instructions [35, 36]. We standardized a genetic examination protocol for mutation screening of three common deafness genes: GJB2, SLC26A4, and MT-RNR1 [37, 38] in all patients. Sanger sequencing was performed on both exons of GJB2. Real-time PCR was performed on two mutation hotspots of SLC26A4 (c.919-2A>G and p.H723R), and on the m.1555A>G mutation of MT-RNR1. Patients with enlarged vestibular aqueduct (EVA), a common inner ear malformation caused by recessive SLC26A4 mutations [39, 40], were further subjected to a next-generation sequencing (NGS)-based diagnostic panel targeting all the exons of SLC26A4. The NGS data were filtered and analyzed as previously described [41]. The Deafness Variation Database (http://deafnessvariationdatabase.org/) and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) were used to identify known causative variants. All subjects and/or their parents provided informed consent before genetic testing, and all procedures were approved by the Research Ethics Committees of National Taiwan University Hospital, National Center for Maternal and Child Health of Mongolia, and the EMJJ Otolaryngology Hospital of Mongolia.
Haplotype analyses
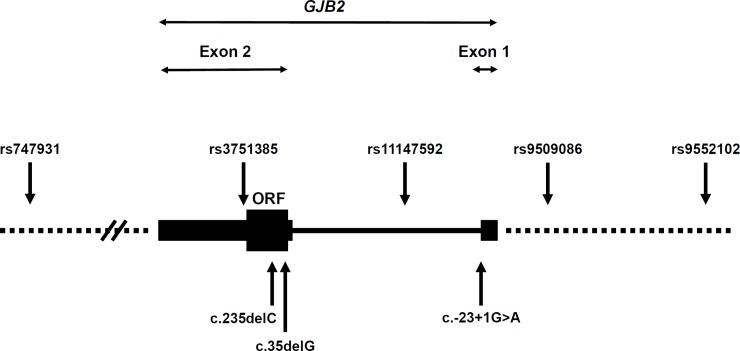
Five single-nucleotide polymorphisms (SNPs) within or in the vicinity of GJB2, namely rs747931, rs3751385, rs11147592, rs9509086, and rs9552102, were selected and genotyped by Sanger sequencing (Fig 1). Haplotypes were constructed with these five SNP markers in the mutant alleles from patients with GJB2: c.-23+1G>A, c.35delG, or c.235delC mutations, and compared to those in the wild-type alleles from 47 Mongolian controls with normal hearing. To investigate genetic relationships with other populations, we further selected 14 c.235delC homozygotes, comprising 11 Han Taiwanese and three Han Chinese from our cohort [36, 37], and determined their haplotypes. Meanwhile, population-specific and mutation-specific haplotype structures were also generated from the data of the 1000 Genomes Project using the LDlink web-based tool [42].
Fig 1. Positions of the single-nucleotide polymorphisms (SNPs) we genotyped, relative to the GJB2 gene.
The GJB2 gene consists of two exons (coding exon, thick black box; untranslated regions, thin black boxes; intron, thin line). The relative positions of the five SNPs (rs747931, rs3751385, rs11147592, rs9509086, and rs9552102) and the three GJB2 mutations (c.235delC, c.35delG, and c.-23+1G>A) are shown by arrows.
Statistical analyses
Differences between groups were tested with Fisher’s exact test (SPSS 22.0 software, IBM SPSS, Armonk, NY). Corresponding p values < 0.05 were interpreted as being statistically significant.
Results
In the 188 unrelated Mongolian families with SNHI, nine different mutant GJB2 alleles and three mutant SLC26A4 alleles were identified (Table 1). The allele frequency of GJB2 mutations (10.6%; 40/376) was higher than that of SLC26A4 mutations (2.4%; 9/376) and that of the mitochondrial m.1555A>G mutation (1.1%; 2/188). More prevalent GJB2 mutations included c.-23+1G>A (allele frequency = 3.2%; 12/376), c.235delC (2.1%; 8/376), and c.35delG (1.6%; 6/376); whereas the most prevalent SLC26A4 mutation was c.919-2A>G (2.1%). The SLC26A4 c.2168A>G mutation, common in the East Asian populations [19–21], was not detected in Mongolian patients in this study. Except for GJB2 c.235delC, none of these variants was identified in the 47 normal-hearing controls. Because of the limited number of the normal-hearing controls, there was no difference in the allele frequencies of these variants between the patient and control groups (Fisher’s exact test, all p > 0.05). However, as all these variants have been previously shown to be disease-causing in other populations, they were interpreted as causative pathogenic mutations in this study.
Table 1. Mutant alleles detected in the 188 deaf families and 47 normal-hearing controls.
| Nucleotide change | Amino acid change | Allele no. in patients (%)# | Allele no. in controls (%)† |
|---|---|---|---|
| GJB2 | |||
| c.-23+1G>A | NA | 12 (3.2) | 0 (0) |
| c.235delC | p.Leu79Cysfs*3 | 8 (2.1) | 1 (1.1) |
| c.35delG | p.Gly12Valfs*2 | 6 (1.6) | 0 (0) |
| c.109G>A | p.Val37Ile | 4 (1.0) | 0 (0) |
| c.299_300delAT | p.His100Argfs*14 | 4 (1.0) | 0 (0) |
| c.35dupG | p.Val13Cysfs*35 | 2 (0.5) | 0 (0) |
| c.560_605dup | p.Cys202* | 2 (0.5) | 0 (0) |
| c.269T>C | p.Leu90Pro | 1 (0.3) | 0 (0) |
| c.508_511dupAACG | p.Ala171Glufs*40 | 1 (0.3) | 0 (0) |
| Total | 40 (10.6)K | 1 (1.1)K | |
| SLC26A4 | |||
| c.919-2A>G | NA | 7 (2.1) | 0 (0) |
| c.281C>T | p.Thr94Ile | 1 (0.3) | 0 (0) |
| c.2027T>A | p.Leu676Gln | 1 (0.3) | 0 (0) |
| Total | 9 (2.4) | 0 (0) | |
| MT-RNR1 | |||
| m.1555A>G | NA | 2 (1.1) | 0 (0) |
# 376 GJB2, 376 SLC26A4, and 188 MT-RNR1 alleles.
† 94 GJB2, 94 SLC26A4, and 47 MT-RNR1 alleles.
K p < 0.01 by Fisher’s exact test.
NA, Not available.
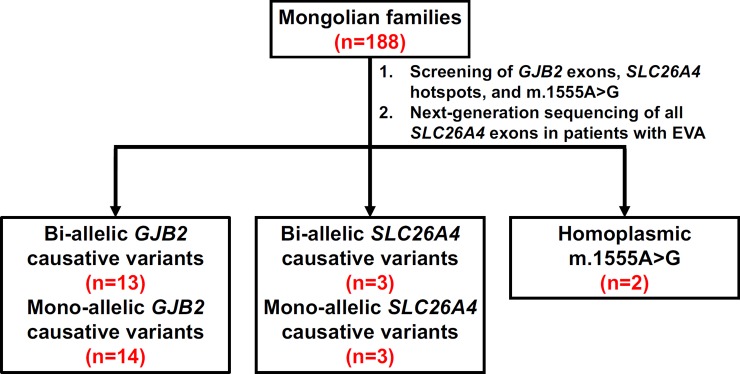
Definite genetic diagnosis was achieved in 18 of the 188 families (9.6%), including 13 with bi-allelic GJB2 mutations, three with bi-allelic SLC26A4 mutations, and two with homoplasmic m.1555A>G mutations (Table 2 & Fig 2). In addition, mono-allelic GJB2 and SLC26A4 mutations were detected in the probands of 14 and three families, respectively. All the probands in the three families with mono-allelic SLC26A4 mutations showed EVA on imaging studies. According to previous reports [43–46], a second occult mutant SLC26A4 allele, which could not be detected using current sequencing techniques, might exist in these three families, and presumably SNHI in the affected members could be attributed to SLC26A4 mutations.
Table 2. Genetic results of the 188 Mongolian families.
| Genes | Variants | Numbers | Percentage (%) |
|---|---|---|---|
| GJB2 | Bi-allelic | ||
| c.-23+1G>A/c.235delC | 3 | 1.6 | |
| c.-23+1G>A/c.35delG | 2 | 1.0 | |
| c.-23+1G>A/c.299_300delAT | 2 | 1.0 | |
| c.35delG/c.35dupG | 1 | 0.5 | |
| c.-23+1G>A/c.269T>C | 1 | 0.5 | |
| c.-23+1G>A/c.559_604dup | 1 | 0.5 | |
| c.235delC/c.235delC | 1 | 0.5 | |
| c.235delC/c.299_300delAT | 1 | 0.5 | |
| c.235delC/c.559_604dup | 1 | 0.5 | |
| Mono-allelic | |||
| c.109G>A/WT | 4 | 2.1 | |
| c.-23+1G>A/WT | 3 | 1.6 | |
| c.35delG/WT | 3 | 1.6 | |
| c.35dupG/WT | 1 | 0.5 | |
| c.235delC/WT | 1 | 1.0 | |
| c.299_300delAT/WT | 1 | 0.5 | |
| c.508_511dupAACG/WT | 1 | 0.5 | |
| SLC26A4 | Bi-allelic | ||
| c.919-2A>G/c.919-2A>G | 1 | 0.5 | |
| c.919-2A>G/c.281C>T | 1 | 0.5 | |
| c.919-2A>G/c.2027T>A | 1 | 0.5 | |
| Mono-allelic | |||
| c.919-2A>G/WT | 3 | 1.6 | |
| MT-RNR1 | m.1555A>G | 2 | 1.1 |
WT, wild-type.
Fig 2. Summary of genetic results in the 188 Mongolian families with sensorineural hearing impairment.
Bi-allelic and mono-allelic GJB2 mutations were identified in 13 and 14 families, respectively. Bi-allelic and mono-allelic SLC26A4 mutations were identified in three and three families, respectively. Homoplasmic m.1555A>G mutation was detected in two families. EVA, enlarged vestibular aqueduct.
To investigate whether prevalent mutations in the Mongolian patients derived from common origins, we performed haplotype analyses by genotyping five SNPs, in the patients and in 47 normal-hearing Mongolian controls (Table 3). Our results revealed that all six chromosomes with the GJB2 c.35delG mutation segregated exclusively with the A-A-C-G-T haplotype (Fisher’s exact test, p = 0.002; compared to the 94 control chromosomes), and all eight chromosomes with the GJB2 c.235delC mutation segregated exclusively with the A-G-T-T-A haplotype (Fisher’s exact test, p < 0.001; compared to the 94 control chromosomes). On the other hand, the GJB2 c.-23+1G>A mutation was associated with a major haplotype G-G-C-T-A (9/12 chromosomes) and a minor haplotype A-G-C-T-A (3/12 chromosomes), which also differed in haplotype distribution as compared to the control chromosomes (Fisher’s exact test, p < 0.001). These results indicated founder effects for all the three prevalent GJB2 mutations in the Mongolian patients.
Table 3. Haplotype analyses of GJB2 alleles in the Mongolian patients.
| Haplotype | c.-23+1G>A | c.35delG | c.235delC | wild-type |
|---|---|---|---|---|
| A-A-C-G-T | 0 | 6* | 0 | 31 |
| A-G-C-T-A | 3 | 0 | 0 | 27 |
| A-G-T-T-A | 0 | 0 | 8* | 19 |
| G-A-C-G-T | 0 | 0 | 0 | 7 |
| G-G-C-T-A | 9* | 0 | 0 | 6 |
| A-A-C-G-A | 0 | 0 | 0 | 3 |
| A-A-C-T-A | 0 | 0 | 0 | 1 |
| Total | 12 | 6 | 8 | 94 |
Differences between mutant and wild-type alleles for each haplotype were tested with Fisher’s exact test.
*p < 0.05
Population-specific haplotype structures, generated from the data of the 1000 Genomes Project, revealed that the Mongolian population shares a closer genetic background with East Asians than with the Europeans and South Asians (S1 Table). The three predominant GJB2 haplotypes, namely A-A-C-G-T, A-G-C-T-A, and A-G-T-T-A, were the same in the Mongolian and East Asian populations; whereas a common GJB2 haplotype in the European and South Asian populations, G-G-C-T-A, was relatively rare in the Mongolians and East Asians.
However, when focused on mutation-specific haplotype structures, the GJB2 c.235delC mutation in Mongolians originated from a common ancestor with other East Asians, whereas the GJB2 c.35delG originated from a common ancestor with the Europeans (Table 4). Of the 14 c.235delC homozygotes selected from our cohort, 21 of the 22 Taiwanese chromosomes and five of the six Chinese chromosomes with GJB2 c.235delC shared the same A-G-T-T-A haplotype with the 8 Mongolian chromosomes with c.235delC. An additional eight chromosomes with the c.235delC mutation were identified from East Asians in the 1000 Genomes Project database, and all were determined by LDlink to segregate the A-G-T-T-A haplotype as well. Both lines of evidence indicated a common founder for GJB2 c.235delC in Mongolians and that in East Asians. Similarly, in the 1000 Genomes Project database, nine and two chromosomes with the c.35delG mutation were identified from the Europeans and Admixed Americans, respectively; and 10 of the 11 chromosomes shared the same A-A-C-G-T haplotype with the 6 Mongolian chromosomes with c.35delG, indicating a common founder for GJB2 c.35delG in the Mongolians and Europeans. Unfortunately, the haplotype structures with the GJB2 c.-23+1G>A mutation could not be determined from the 1000 Genomes Project database to investigate the common ancestry of this mutation in Mongolians and South Asians.
Table 4. Mutation-specific haplotype structures of GJB2 c.235delC and c. 35delG in different populations.
| Haplotype | Mongolians | Taiwanese | Chinese | East Asians |
| c. 235delC | ||||
| A-G-T-T-A | 8 | 21 | 5 | 8* |
| G-G-T-T-A | 0 | 1 | 1 | 0* |
| Haplotype | Mongolians | Europeans | Admixed Americans | |
| c. 35delG | ||||
| A-A-C-G-T | 6 | 8* | 2* | |
| G-A-C-G-T | 0 | 1* | 0* | |
* The haplotype structures were determined from the data of the 1000 Genomes Project using the LDlink tool.
Discussion
Our results unraveled a unique genetic profile in Mongolian patients with SNHI as compared to other European and Asian populations. Notably, three GJB2 mutations that are prevalent in other populations, including c.35delG in Caucasians [5–7], c.235delC in East Asians [10–12], and c.-23+1G>A in Southwest and South Asians [13–16], were simultaneously detected in Mongolian patients. To our knowledge, this is the first study in the literature to identify these three common GJB2 mutations with significant allele frequencies in a single ethnic group.
Haplotype analyses further confirmed founder effects for each of the three mutations, indicating that each mutation derived from its individual ancestral origin independently. It was reported that the c.35delG mutation stemmed from the Volgo-Ural region of Central Asia approximately 11,800 years ago [47] and then spread throughout Europe along the two Neolithic population transportation routes [48]. The c.235delC mutation arose near the Baikal Lake or the Altai-Sayan region approximately 11,500 years ago, and then spread into East Asia [49–51]. The c.-23+1G>A mutation was estimated to occur approximately 800 years ago, and spread into Mongolia, Siberia, or South Asia with the Turkic migration in the 13th–14th centuries [52]. The concurrence of the c.35delG, c.235delC, and c.-23+1G>A mutations in the Mongolian patients might reflect the geographic location of Mongolia as a crossroads in this migration. In addition, the territorial expansion of the Mongol Empire in the 13th century might also have enhanced gene flow between Mongolians and other populations who lived in Europe, Central Asia, East Asia, and the Indian subcontinent [30].
Prior to the current study, several studies have investigated the genetics of SNHI in Mongolian patients (Table 5). Most of these studies were performed on patients recruited from the Inner Mongolia region of China or northwest China, and included patients of non-Mongolian ethnicity. Dai et al. sequenced coding exons of SLC26A4 in 135 patients from Inner Mongolia, and found that 12.6% (17/135) patients carried bi-allelic SLC26A4 mutations. However, only 31 of the 135 patients were Mongolians, and the authors did not classify their genetic results by ethnicity [23]. Yang et al. screened the coding regions of GJB2, SLC26A4, and MT-RNR1 in 189 deaf patients from northwest China, of whom only 19 were Mongolians. The authors did not identify any GJB2 mutations, and reported the SLC26A4 c.919-2A>G mutation as the most common deafness mutation in the Mongolian patients [24]. Liu et al. screened nine common mutations of GJB2, SLC26A4, MT-RNR1, and GJB3 in 738 deaf children recruited from the Inner Mongolia region of China, including 216 Mongolians. The authors also reported a higher prevalence of SLC26A4 mutations than that of GJB2 mutations [25]. In contrast to these previous reports, our study showed that GJB2 mutations are more prevalent than SLC26A4 mutations in Mongolian patients with SNHI. This difference between our study and previous reports might result from the more thorough sequencing strategy we adopted, since the GJB2 c.-23+1G>A mutation, which is prevalent in Mongolians but rare in other East Asian populations, was not targeted in previous reports. It is also notable that the genetic structure of the Mongolian people who live in China could have been influenced by that of other races through inter-population marriage.
Table 5. Summary of previous studies and our study on the genetic results of Mongolian patients.
| Reference | Patients | Target regions | Main results |
|---|---|---|---|
| Dai et al. [23] | 135 patients from the Inner Mongolia region of China, including 94 Han Chinese, 31 Mongolians, 7 Manchurians, and three Hui | The coding exons of SLC26A4 | 12.6% (17/135) patients carried bi-allelic SLC26A4 mutations. The most common mutation was c.919-2A>G. Mutations in the Mongolian patients were not specified. |
| Tekin et al. [26] | 534 Mongolian patients from Mongolia | The coding exon (exon 2) of GJB2 and the c.23+1G>A mutation in intron 1 | 24 (4.5%) and 29 (5.4%) patients carried bi- and mono-allelic GJB2 mutations, respectively. The most common mutations were GJB2 c.23+1G>A (3.5%) and c.235delC (1.5%). |
| Yang et al. [24] | 189 patients from the northwest of China, including 121 Tibetans, 49 Tu, and 19 Mongolians | The coding regions of GJB2, SLC26A4, and MT-RNR1 | The most common mutation in the Mongolian patients was SLC26A4 c.919-2A>G. No GJB2 mutations were detected in the Mongolian patients, one of whom was found to carry the MT-RNR1 m.1555A>G mutation. |
| Liu et al. [25] | 738 patients from the Inner Mongolia region of China, including 486 Han Chinese, 216 Mongolians, 24 Manchurians, 6 Hui, and 6 Daur | Nine common mutations in four deafness genes, including GJB2, SLC26A4, GJB3, and MT-RNR1 | Among the 216 Mongolian patients, 36 had GJB2 mutations and 42 had SLC26A4 mutations. |
| This study | 188 unrelated Mongolian patients from Mongolia | All exons of GJB2 and SLC26A4, and the coding region of MT-RNR1 | Definite genetic diagnosis was achieved in 18 (9.6%) of the 188 patients, including 13 with bi-allelic GJB2 mutations, 3 with bi-allelic SLC26A4 mutations, and two with homoplasmic MT-RNR1 m.1555A>G mutation. |
In the present study, definite genetic diagnoses could be achieved in 18 (9.6%) of the 188 families, comprising 13 (6.9%) with bi-allelic GJB2 mutations, three (1.6%) with bi-allelic SLC26A4 mutations, and two (1.1%) with homoplasmic m.1555A>G mutations, by screening the three common deafness genes. We additionally identified mono-allelic GJB2 or SLC26A4 mutations in 17 families (9.0%); however, given the recessive inheritance pattern of mutations in these two genes, the genetic results in these families were regarded as unconfirmed. Accordingly, the rate of confirmed results in our study is lower than that documented in other studies on European [53, 54] and Asian [37, 55–57] populations. This low rate is consistent with the results of the study by Tekin et al., who reported a low rate of 4.5% with bi-allelic GJB2 mutations in Mongolian patients from a deaf school in Ulaanbaatar [26]. Liu et al. detected GJB2 mutations in 36 (16.6%) of their 216 Mongolian patients from the Inner Mongolia region of China [25]. However, these comprised patients with both bi-allelic and mono-allelic mutations, and the actual percentage of patients with confirmed GJB2 mutations might be considerably lower than 16.6% [25].
The relatively low mutation rates of deafness genes in Mongolians have been ascribed to lower assortative mating rate and decreased genetic fitness of the deaf in Mongolia as compared to other populations [26]. It has been suggested that the introduction of sign language, establishment of residential schools for the deaf, and appearance of intense assortative mating among the deaf might have relaxed the genetic selection against deafness and contributed to high frequency of GJB2 deafness in many western populations [58]. Indeed, low frequency of < 10% of deafness-associated GJB2 mutations has been reported in several populations with lower socioeconomic status, such as patients in Sudan [59], Kenya [59], Indonesia [60], and Cameroon [61].
Notably, two hearing-impaired patients in this study were identified to have the mitochondrial m.1555A>G mutation. Their medical history revealed that both patients were exposed to aminoglycosides. According to a recent report, the Asia-Pacific region is the largest market for the aminoglycoside industry, probably owing to a high incidence rate of tuberculosis (http://www.grandviewresearch.com/industry-analysis/aminoglycoside-market). To prevent aminoglycoside-induced hearing loss, it might be reasonable to perform mutational screening of the MT-RNR1 gene before the initiation of antibiotic therapy, especially in regions where aminoglycosides are frequently used [62, 63].
By comprehensively sequencing both GJB2 exons, we identified bi-allelic and mono-allelic recessive GJB2 mutations in 13 (6.9%) and 14 (7.4%) of the 188 Mongolian families, respectively. This finding is also consistent with Tekin et al., who reported mono-allelic GJB2 mutations in 5.4% of their Mongolian patients [26]. Patients with mono-allelic recessive GJB2 mutation might have occult mutations in the non-coding regions of GJB2, such as untranslated exon 1, intron 1, promoter, enhancer, or other regulatory elements, leading to compound heterozygosity, which has been observed previously for GJB2 mutations [64, 65]. Alternatively, mutations in other gap junction genes might modulate the pathogenicity of GJB2 mutations and contribute to hearing impairment via digenic or polygenic inheritance [66–68]. The third possibility is that, instead of GJB2 mutations, hearing impairment is in fact caused by mutations in other deafness genes, and these “mono-allelic” patients are incidental carriers of certain GJB2 variants that are prevalent in the population [69].
The major strength of this study lies in the demonstration of the mutation spectra of all the three common HHI genes in a single large cohort of pure Mongolian ethnicity. However, some limitations of this study merit further discussion. Genetic mutations, with confirmed and unconfirmed results counted together, were only identified in 35 (18.6%) of the 188 families; for the other families, the etiology remained unclear. It is conceivable that in a certain portion of these families SNHI may be caused by mutations in other deafness genes, especially in families with multiple affected members. In addition, SNHI of acquired causes, such as perinatal insults or congenital cytomegalovirus (cCMV) infection, may be more prevalent in developing countries like Mongolia than in industrialized countries [70]. Recently, NGS technology, which enables the sequencing of a large number of genes simultaneously, has proven to be a powerful tool for addressing the genetically heterogeneous disorder HHI [38, 71, 72]. We are currently using an NGS-based diagnostic panel to analyze the genetic etiology in the undiagnosed families of our Mongolian cohort [38, 41], and have now identified causative MYO15A mutations in a multiplex family. The employment of NGS-based genetic examination and cCMV screening may help improve the evaluation, diagnosis, and management of Mongolian patients with SNHI.
In conclusion, by performing comprehensive genetic examination and haplotype analyses in a large Mongolian cohort with SNHI, this study throws light on the genetic epidemiology of HHI in Mongolians. It also provides insights into the development of the unique mutation spectra observed in Mongolians. These findings may have clinical implications for the refinement of molecular diagnostics in Mongolian patients, as well as scientific implications for the delineation of genetic relationships among the Eurasian populations.
Supporting information
(DOCX)
Acknowledgments
This study was supported by the Taiwanese-Mongolian Joint Research Project (Grant No.: MOST 105-2923-B-002-006 and MOST 107-2314-B-002-137-MY3, the Ministry of Science and Technology of Taiwan). We would like to thank National Center for High-performance Computing (NCHC) of National Applied Research Laboratories (NARLabs) of Taiwan for providing computational resources and storage resources. We also wish to thank all subjects and their family members for participating in the present study.
Data Availability
All relevant data are within the manuscript. A supplementary table (S1 Table) is included in the manuscript. There are no additional Supporting Information files.
Funding Statement
This study was supported by the Taiwanese-Mongolian Joint Research Project (Grant No.: MOST 105-2923-B-002-006 and MOST 107-2314-B-002-137-MY3, the Ministry of Science and Technology of Taiwan, https://www.most.gov.tw/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Morton CC, Nance WE. Newborn hearing screening—a silent revolution. N Engl J Med. 2006;354(20):2151–64. 10.1056/NEJMra050700 [DOI] [PubMed] [Google Scholar]
- 2.Wake M, Tobin S, Cone-Wesson B, Dahl HH, Gillam L, McCormick L, et al. Slight/mild sensorineural hearing loss in children. Pediatrics. 2006;118(5):1842–51. 10.1542/peds.2005-3168 [DOI] [PubMed] [Google Scholar]
- 3.Feder KP, Michaud D, McNamee J, Fitzpatrick E, Ramage-Morin P, Beauregard Y. Prevalence of Hearing Loss Among a Representative Sample of Canadian Children and Adolescents, 3 to 19 Years of Age. Ear Hear. 2017;38(1):7–20. 10.1097/AUD.0000000000000345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hilgert N, Smith RJ, Van Camp G. Forty-six genes causing nonsyndromic hearing impairment: which ones should be analyzed in DNA diagnostics? Mutation research. 2009;681(2–3):189–96. 10.1016/j.mrrev.2008.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gurtler N, Kim Y, Mhatre A, Muller R, Probst R, Lalwani AK. GJB2 mutations in the Swiss hearing impaired. Ear Hear. 2003;24(5):440–7. 10.1097/01.AUD.0000090440.84513.B3 [DOI] [PubMed] [Google Scholar]
- 6.Denoyelle F, Weil D, Maw MA, Wilcox SA, Lench NJ, Allen-Powell DR, et al. Prelingual deafness: high prevalence of a 30delG mutation in the connexin 26 gene. Hum Mol Genet. 1997;6(12):2173–7. [DOI] [PubMed] [Google Scholar]
- 7.Frei K, Szuhai K, Lucas T, Weipoltshammer K, Schofer C, Ramsebner R, et al. Connexin 26 mutations in cases of sensorineural deafness in eastern Austria. Eur J Hum Genet. 2002;10(7):427–32. 10.1038/sj.ejhg.5200826 [DOI] [PubMed] [Google Scholar]
- 8.Stinckens C, Kremer H, van Wijk E, Hoefsloot LH, Huygen PL, Standaert L, et al. Longitudinal phenotypic analysis in patients with connexin 26 (GJB2) (DFNB1) and connexin 30 (GJB6) mutations. Ann Otol Rhinol Laryngol. 2004;113(7):587–93. 10.1177/000348940411300714 [DOI] [PubMed] [Google Scholar]
- 9.Morell RJ, Kim HJ, Hood LJ, Goforth L, Friderici K, Fisher R, et al. Mutations in the connexin 26 gene (GJB2) among Ashkenazi Jews with nonsyndromic recessive deafness. N Engl J Med. 1998;339(21):1500–5. 10.1056/NEJM199811193392103 [DOI] [PubMed] [Google Scholar]
- 10.Abe S, Usami S, Shinkawa H, Kelley PM, Kimberling WJ. Prevalent connexin 26 gene (GJB2) mutations in Japanese. J Med Genet. 2000;37(1):41–3. 10.1136/jmg.37.1.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwa HL, Ko TM, Hsu CJ, Huang CH, Chiang YL, Oong JL, et al. Mutation spectrum of the connexin 26 (GJB2) gene in Taiwanese patients with prelingual deafness. Genet Med. 2003;5(3):161–5. 10.1097/01.GIM.0000066796.11916.94 [DOI] [PubMed] [Google Scholar]
- 12.Park HJ, Hahn SH, Chun YM, Park K, Kim HN. Connexin26 mutations associated with nonsyndromic hearing loss. Laryngoscope. 2000;110(9):1535–8. 10.1097/00005537-200009000-00023 [DOI] [PubMed] [Google Scholar]
- 13.Sirmaci A, Akcayoz-Duman D, Tekin M. The c.IVS1+1G>A mutation in the GJB2 gene is prevalent and large deletions involving the GJB6 gene are not present in the Turkish population. J Genet. 2006;85(3):213–6. [DOI] [PubMed] [Google Scholar]
- 14.Mahdieh N, Nishimura C, Ali-Madadi K, Riazalhosseini Y, Yazdan H, Arzhangi S, et al. The frequency of GJB2 mutations and the Delta (GJB6-D13S1830) deletion as a cause of autosomal recessive non-syndromic deafness in the Kurdish population. Clin Genet. 2004;65(6):506–8. 10.1111/j.1399-0004.2004.00262.x [DOI] [PubMed] [Google Scholar]
- 15.Bajaj Y, Sirimanna T, Albert DM, Qadir P, Jenkins L, Bitner-Glindzicz M. Spectrum of GJB2 mutations causing deafness in the British Bangladeshi population. Clin Otolaryngol. 2008;33(4):313–8. 10.1111/j.1749-4486.2008.01754.x [DOI] [PubMed] [Google Scholar]
- 16.Padma G, Ramchander PV, Nandur UV, Padma T. GJB2 and GJB6 gene mutations found in Indian probands with congenital hearing impairment. J Genet. 2009;88(3):267–72. [DOI] [PubMed] [Google Scholar]
- 17.Campbell C, Cucci RA, Prasad S, Green GE, Edeal JB, Galer CE, et al. Pendred syndrome, DFNB4, and PDS/SLC26A4 identification of eight novel mutations and possible genotype-phenotype correlations. Hum Mutat. 2001;17(5):403–11. 10.1002/humu.1116 [DOI] [PubMed] [Google Scholar]
- 18.Coyle B, Reardon W, Herbrick JA, Tsui LC, Gausden E, Lee J, et al. Molecular analysis of the PDS gene in Pendred syndrome. Hum Mol Genet. 1998;7(7):1105–12. [DOI] [PubMed] [Google Scholar]
- 19.Tsukamoto K, Suzuki H, Harada D, Namba A, Abe S, Usami S. Distribution and frequencies of PDS (SLC26A4) mutations in Pendred syndrome and nonsyndromic hearing loss associated with enlarged vestibular aqueduct: a unique spectrum of mutations in Japanese. Eur J Hum Genet. 2003;11(12):916–22. 10.1038/sj.ejhg.5201073 [DOI] [PubMed] [Google Scholar]
- 20.Park HJ, Lee SJ, Jin HS, Lee JO, Go SH, Jang HS, et al. Genetic basis of hearing loss associated with enlarged vestibular aqueducts in Koreans. Clin Genet. 2005;67(2):160–5. 10.1111/j.1399-0004.2004.00386.x [DOI] [PubMed] [Google Scholar]
- 21.Wu CC, Yeh TH, Chen PJ, Hsu CJ. Prevalent SLC26A4 mutations in patients with enlarged vestibular aqueduct and/or Mondini dysplasia: a unique spectrum of mutations in Taiwan, including a frequent founder mutation. Laryngoscope. 2005;115(6):1060–4. 10.1097/01.MLG.0000163339.61909.D0 [DOI] [PubMed] [Google Scholar]
- 22.Dai P, Li Q, Huang D, Yuan Y, Kang D, Miller DT, et al. SLC26A4 c.919-2A>G varies among Chinese ethnic groups as a cause of hearing loss. Genet Med. 2008;10(8):586–92. [DOI] [PubMed] [Google Scholar]
- 23.Dai P, Yuan Y, Huang D, Zhu X, Yu F, Kang D, et al. Molecular etiology of hearing impairment in Inner Mongolia: mutations in SLC26A4 gene and relevant phenotype analysis. J Transl Med. 2008;6:74 10.1186/1479-5876-6-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang XL, Bai-Cheng X, Chen XJ, Pan-Pan B, Jian-Li M, Xiao-Wen L, et al. Common molecular etiology of patients with nonsyndromic hearing loss in Tibetan, Tu nationality, and Mongolian patients in the northwest of China. Acta Otolaryngol. 2013;133(9):930–4. 10.3109/00016489.2013.795288 [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Ao L, Ding H, Zhang D. Genetic frequencies related to severe or profound sensorineural hearing loss in Inner Mongolia Autonomous Region. Genet Mol Biol. 2016;39(4):567–72. 10.1590/1678-4685-GMB-2015-0218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tekin M, Xia XJ, Erdenetungalag R, Cengiz FB, White TW, Radnaabazar J, et al. GJB2 mutations in Mongolia: complex alleles, low frequency, and reduced fitness of the deaf. Ann Hum Genet. 2010;74(2):155–64. 10.1111/j.1469-1809.2010.00564.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zerjal T, Xue Y, Bertorelle G, Wells RS, Bao W, Zhu S, et al. The genetic legacy of the Mongols. Am J Hum Genet. 2003;72(3):717–21. 10.1086/367774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merriwether DA, Hall WW, Vahlne A, Ferrell RE. mtDNA variation indicates Mongolia may have been the source for the founding population for the New World. Am J Hum Genet. 1996;59(1):204–12. [PMC free article] [PubMed] [Google Scholar]
- 29.Kolman CJ, Sambuughin N, Bermingham E. Mitochondrial DNA analysis of Mongolian populations and implications for the origin of New World founders. Genetics. 1996;142(4):1321–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bai H, Guo X, Zhang D, Narisu N, Bu J, Jirimutu J, et al. The genome of a Mongolian individual reveals the genetic imprints of Mongolians on modern human populations. Genome Biol Evol. 2014;6(12):3122–36. 10.1093/gbe/evu242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin PH, Hsu CJ, Lin YH, Lin YH, Lee HY, Wu CC, et al. Etiologic and Audiologic Characteristics of Patients With Pediatric-Onset Unilateral and Asymmetric Sensorineural Hearing Loss. JAMA Otolaryngol Head Neck Surg. 2017;143(9):912–9. 10.1001/jamaoto.2017.0945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sennaroglu L, Saatci I. A new classification for cochleovestibular malformations. Laryngoscope. 2002;112(12):2230–41. 10.1097/00005537-200212000-00019 [DOI] [PubMed] [Google Scholar]
- 33.McClay JE, Tandy R, Grundfast K, Choi S, Vezina G, Zalzal G, et al. Major and minor temporal bone abnormalities in children with and without congenital sensorineural hearing loss. Arch Otolaryngol Head Neck Surg. 2002;128(6):664–71. [DOI] [PubMed] [Google Scholar]
- 34.Wu CC, Chen YS, Chen PJ, Hsu CJ. Common clinical features of children with enlarged vestibular aqueduct and mondini dysplasia. Laryngoscope. 2005;115(1):132–7. 10.1097/01.mlg.0000150691.85387.3f [DOI] [PubMed] [Google Scholar]
- 35.Lu CY, Tsao PN, Ke YY, Lin YH, Lin YH, Hung CC, et al. Concurrent hearing, genetic, and cytomegalovirus screening in newborns, Taiwan. J Pediatr. 2018;199:144–150. 10.1016/j.jpeds.2018.02.064 [DOI] [PubMed] [Google Scholar]
- 36.Wu CC, Tsai CH, Hung CC, Lin YH, Lin YH, Huang FL, et al. Newborn genetic screening for hearing impairment: a population-based longitudinal study. Genet Med. 2017;19(1):6–12. 10.1038/gim.2016.66 [DOI] [PubMed] [Google Scholar]
- 37.Wu CC, Chen PJ, Chiu YH, Lu YC, Wu MC, Hsu CJ. Prospective mutation screening of three common deafness genes in a large Taiwanese Cohort with idiopathic bilateral sensorineural hearing impairment reveals a difference in the results between families from hospitals and those from rehabilitation facilities. Audiol Neurootol. 2008;13(3):172–81. 10.1159/000112425 [DOI] [PubMed] [Google Scholar]
- 38.Wu CC, Lin YH, Lu YC, Chen PJ, Yang WS, Hsu CJ, et al. Application of massively parallel sequencing to genetic diagnosis in multiplex families with idiopathic sensorineural hearing impairment. PloS one. 2013;8(2):e57369 10.1371/journal.pone.0057369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Everett LA, Glaser B, Beck JC, Idol JR, Buchs A, Heyman M, et al. Pendred syndrome is caused by mutations in a putative sulphate transporter gene (PDS). Nat Genet. 1997;17(4):411–22. 10.1038/ng1297-411 [DOI] [PubMed] [Google Scholar]
- 40.Li XC, Everett LA, Lalwani AK, Desmukh D, Friedman TB, Green ED, et al. A mutation in PDS causes non-syndromic recessive deafness. Nat Genet. 1998;18(3):215–7. 10.1038/ng0398-215 [DOI] [PubMed] [Google Scholar]
- 41.Lin YH, Lin YH, Lu YC, Liu TC, Chen CY, Hsu CJ, et al. A novel missense variant in the nuclear localization signal of POU4F3 causes autosomal dominant non-syndromic hearing loss. Sci Rep. 2017;7(1):7551 10.1038/s41598-017-08236-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31(21):3555–7. 10.1093/bioinformatics/btv402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Azaiez H, Yang T, Prasad S, Sorensen JL, Nishimura CJ, Kimberling WJ, et al. Genotype-phenotype correlations for SLC26A4-related deafness. Human Genet. 2007;122(5):451–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi BY, Madeo AC, King KA, Zalewski CK, Pryor SP, Muskett JA, et al. Segregation of enlarged vestibular aqueducts in families with non-diagnostic SLC26A4 genotypes. J Med Genet. 2009;46(12):856–61. 10.1136/jmg.2009.067892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu CC, Lu YC, Chen PJ, Yeh PL, Su YN, Hwu WL, et al. Phenotypic analyses and mutation screening of the SLC26A4 and FOXI1 genes in 101 Taiwanese families with bilateral nonsyndromic enlarged vestibular aqueduct (DFNB4) or Pendred syndrome. Audiol Neurootol. 2010;15(1):57–66. 10.1159/000231567 [DOI] [PubMed] [Google Scholar]
- 46.Lin YH, Wu CC, Lin YH, Lu YC, Chen CS, Liu TC, et al. Targeted Next-Generation Sequencing Facilitates Genetic Diagnosis and Provides Novel Pathogenetic Insights into Deafness with Enlarged Vestibular Aqueduct. J Mol Diagn. 2018;[Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 47.Dzhemileva LU, Posukh OL, Barashkov NA, Fedorova SA, Teryutin FM, Akhmetova VL, et al. Haplotype Diversity and Reconstruction of Ancestral Haplotype Associated with the c.35delG Mutation in the GJB2 (Cx26) Gene among the Volgo-Ural Populations of Russia. Acta Naturae. 2011;3(3):52–63. [PMC free article] [PubMed] [Google Scholar]
- 48.Van Laer L, Coucke P, Mueller RF, Caethoven G, Flothmann K, Prasad SD, et al. A common founder for the 35delG GJB2 gene mutation in connexin 26 hearing impairment. J Med Genet. 2001;38(8):515–8. 10.1136/jmg.38.8.515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dzhemileva LU, Barashkov NA, Posukh OL, Khusainova RI, Akhmetova VL, Kutuev IA, et al. Carrier frequency of GJB2 gene mutations c.35delG, c.235delC and c.167delT among the populations of Eurasia. J Hum Genet. 2010;55(11):749–54. 10.1038/jhg.2010.101 [DOI] [PubMed] [Google Scholar]
- 50.Yan D, Park HJ, Ouyang XM, Pandya A, Doi K, Erdenetungalag R, et al. Evidence of a founder effect for the 235delC mutation of GJB2 (connexin 26) in east Asians. Hum Genet. 2003;114(1):44–50. 10.1007/s00439-003-1018-1 [DOI] [PubMed] [Google Scholar]
- 51.Ohtsuka A, Yuge I, Kimura S, Namba A, Abe S, Van Laer L, et al. GJB2 deafness gene shows a specific spectrum of mutations in Japan, including a frequent founder mutation. Hum Genet. 2003;112(4):329–33. 10.1007/s00439-002-0889-x [DOI] [PubMed] [Google Scholar]
- 52.Barashkov NA, Dzhemileva LU, Fedorova SA, Teryutin FM, Posukh OL, Fedotova EE, et al. Autosomal recessive deafness 1A (DFNB1A) in Yakut population isolate in Eastern Siberia: extensive accumulation of the splice site mutation IVS1+1G>A in GJB2 gene as a result of founder effect. J Hum Genet. 2011;56(9):631–9. 10.1038/jhg.2011.72 [DOI] [PubMed] [Google Scholar]
- 53.Hutchin T, Coy NN, Conlon H, Telford E, Bromelow K, Blaydon D, et al. Assessment of the genetic causes of recessive childhood non-syndromic deafness in the UK—implications for genetic testing. Clin Genet. 2005;68(6):506–12. 10.1111/j.1399-0004.2005.00539.x [DOI] [PubMed] [Google Scholar]
- 54.Yan D, Xiang G, Chai X, Qing J, Shang H, Zou B, et al. Screening of deafness-causing DNA variants that are common in patients of European ancestry using a microarray-based approach. PloS one. 2017;12(3):e0169219 10.1371/journal.pone.0169219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo YF, Liu XW, Guan J, Han MK, Wang DY, Zhao YL, et al. GJB2, SLC26A4 and mitochondrial DNA A1555G mutations in prelingual deafness in Northern Chinese subjects. Acta Otolaryngol. 2008;128(3):297–303. 10.1080/00016480701767382 [DOI] [PubMed] [Google Scholar]
- 56.Dai P, Yu F, Han B, Liu X, Wang G, Li Q, et al. GJB2 mutation spectrum in 2,063 Chinese patients with nonsyndromic hearing impairment. J Transl Med. 2009;7:26 10.1186/1479-5876-7-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Usami S, Nishio SY, Nagano M, Abe S, Yamaguchi T, Deafness Gene Study C. Simultaneous screening of multiple mutations by invader assay improves molecular diagnosis of hereditary hearing loss: a multicenter study. PloS one. 2012;7(2):e31276 10.1371/journal.pone.0031276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nance WE, Liu XZ, Pandya A. Relation between choice of partner and high frequency of connexin-26 deafness. Lancet. 2000;356(9228):500–1. 10.1016/S0140-6736(00)02565-4 [DOI] [PubMed] [Google Scholar]
- 59.Gasmelseed NM, Schmidt M, Magzoub MM, Macharia M, Elmustafa OM, Ototo B, et al. Low frequency of deafness-associated GJB2 variants in Kenya and Sudan and novel GJB2 variants. Hum Mutat. 2004;23(2):206–7. [DOI] [PubMed] [Google Scholar]
- 60.Snoeckx RL, Djelantik B, Van Laer L, Van de Heyning P, Van Camp G. GJB2 (connexin 26) mutations are not a major cause of hearing loss in the Indonesian population. Am J Med Genet A. 2005;135(2):126–9. 10.1002/ajmg.a.30726 [DOI] [PubMed] [Google Scholar]
- 61.Trotta L, Iacona E, Primignani P, Castorina P, Radaelli C, Del Bo L, et al. GJB2 and MTRNR1 contributions in children with hearing impairment from Northern Cameroon. Int J Audiol. 2011;50(2):133–8. 10.3109/14992027.2010.537377 [DOI] [PubMed] [Google Scholar]
- 62.Gurtler N, Schmuziger N, Kim Y, Mhatre AN, Jungi M, Lalwani AK. Audiologic testing and molecular analysis of 12S rRNA in patients receiving aminoglycosides. Laryngoscope. 2005;115(4):640–4. 10.1097/01.mlg.0000161355.28073.f5 [DOI] [PubMed] [Google Scholar]
- 63.O'Sullivan ME, Perez A, Lin R, Sajjadi A, Ricci AJ, Cheng AG. Towards the Prevention of Aminoglycoside-Related Hearing Loss. Front Cell Neurosci. 2017;11:325 10.3389/fncel.2017.00325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matos TD, Caria H, Simoes-Teixeira H, Aasen T, Nickel R, Jagger DJ, et al. A novel hearing-loss-related mutation occurring in the GJB2 basal promoter. J Med Genet. 2007;44(11):721–5. 10.1136/jmg.2007.050682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Azaiez H, Chamberlin GP, Fischer SM, Welp CL, Prasad SD, Taggart RT, et al. GJB2: the spectrum of deafness-causing allele variants and their phenotype. Hum Mutat. 2004;24(4):305–11. 10.1002/humu.20084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lerer I, Sagi M, Ben-Neriah Z, Wang T, Levi H, Abeliovich D. A deletion mutation in GJB6 cooperating with a GJB2 mutation in trans in non-syndromic deafness: A novel founder mutation in Ashkenazi Jews. Hum Mutat. 2001;18(5):460. [DOI] [PubMed] [Google Scholar]
- 67.Pallares-Ruiz N, Blanchet P, Mondain M, Claustres M, Roux AF. A large deletion including most of GJB6 in recessive non syndromic deafness: a digenic effect? Eur J Hum Genet. 2002;10(1):72–6. 10.1038/sj.ejhg.5200762 [DOI] [PubMed] [Google Scholar]
- 68.del Castillo I, Villamar M, Moreno-Pelayo MA, del Castillo FJ, Alvarez A, Telleria D, et al. A deletion involving the connexin 30 gene in nonsyndromic hearing impairment. N Engl J Med. 2002;346(4):243–9. 10.1056/NEJMoa012052 [DOI] [PubMed] [Google Scholar]
- 69.Kim SY, Kim AR, Kim NK, Lee C, Kim MY, Jeon EH, et al. Unraveling of Enigmatic Hearing-Impaired GJB2 Single Heterozygotes by Massive Parallel Sequencing: DFNB1 or Not? Medicine (Baltimore). 2016;95(14):e3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goderis J, De Leenheer E, Smets K, Van Hoecke H, Keymeulen A, Dhooge I. Hearing loss and congenital CMV infection: a systematic review. Pediatrics. 2014;134(5):972–82. 10.1542/peds.2014-1173 [DOI] [PubMed] [Google Scholar]
- 71.Shearer AE, DeLuca AP, Hildebrand MS, Taylor KR, Gurrola J 2nd, Scherer S, et al. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci U S A. 2010;107(49):21104–9. 10.1073/pnas.1012989107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brownstein Z, Friedman LM, Shahin H, Oron-Karni V, Kol N, Abu Rayyan A, et al. Targeted genomic capture and massively parallel sequencing to identify genes for hereditary hearing loss in Middle Eastern families. Genome biol. 2011;12(9):R89 10.1186/gb-2011-12-9-r89 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the manuscript. A supplementary table (S1 Table) is included in the manuscript. There are no additional Supporting Information files.