Abstract
Colorectal cancer (CRC) is ranked the third most common cancer in human worldwide. However, the exact mechanisms of CRC are not well established. Furthermore, there may be differences between mechanisms of CRC in the Asian and in the Western populations. In the present study, we utilized a liquid chromatography-mass spectrometry (LC-MS) metabolomic approach supported by the 16S rRNA next-generation sequencing to investigate the functional and taxonomical differences between paired tumor and unaffected (normal) surgical biopsy tissues from 17 Malaysian patients. Metabolomic differences associated with steroid biosynthesis, terpenoid biosynthesis and bile metabolism could be attributed to microbiome differences between normal and tumor sites. The relative abundances of Anaerotruncus, Intestinimonas and Oscillibacter displayed significant relationships with both steroid biosynthesis and terpenoid and triterpenoid biosynthesis pathways. Metabolites involved in serotonergic synapse/ tryptophan metabolism (Serotonin and 5-Hydroxy-3-indoleacetic acid [5-HIAA]) were only detected in normal tissue samples. On the other hand, S-Adenosyl-L-homocysteine (SAH), a metabolite involves in methionine metabolism and methylation, was frequently increased in tumor relative to normal tissues. In conclusion, this study suggests that local microbiome dysbiosis may contribute to functional changes at the cancer sites. Results from the current study also contributed to the list of metabolites that are found to differ between normal and tumor sites in CRC and supported our quest for understanding the mechanisms of carcinogenesis.
Introduction
Colorectal cancer (CRC) is the third most commonly occurring cancer in men and the second most commonly occurring cancer in women worldwide; almost 55 percent of CRC cases occur in more developed regions [1]. The estimated 2012 age-standardized incidence rates (per 100 000 population) in Southeast Asia region are 8.9 and 6.3 cases in men and women, respectively [1]. The estimated 2012 age-standardized mortality rates (per 100 000 population) in the region are 6.3 and 4.4 cases in men and women, respectively [1]. More than 90% of colorectal carcinomas are adenocarcinomas originating from epithelial cells of the colorectal mucosa; most colorectal adenocarcinomas (~70%) are diagnosed as moderately differentiated while well and poorly differentiated carcinomas account for only 10% and 20%, respectively [2].
The right and left colons, which are developed distinctively from the embryological mid-gut and hindgut, and are joined at the proximal 2/3 and distal 1/3 of the transverse colon. Hence, anatomically, the blood supply, innervation, lymphatic drainage, and lumen environment are different between right (ascending, proximal through the hepatic flexure) and left (descending, distal to the hepatic flexure) colons [3]. Primary CRCs are more frequent on the left colon but a tendency for right-shift of the primary CRC site has been noted in recent years [4]. Patients with right-sided CRC were older, mostly female, more frequently presenting with advanced tumor stages with larger tumor sizes, more often poorly differentiated tumors, and different molecular biological tumor patterns [5]. Drewes et al. confirmed in the Malaysian cohort (designated as MAL2) that invasive biofilms, Bacteroides fragilis and several oral pathogens are enriched in CRC tissues [6].
A deeper understanding on colonic metabolism is needed to identify cancer-related biomarkers to elucidate the cancerous cell progression in CRCs. In addition to genomics and proteomics, which are eminent tools for cancer studies, metabolomics is emerging as a tool to discover biomarkers and unravel pathological processes [7]. The metabolic fingerprints of specific cellular processes and/or low-weighted molecule profiles are prioritized in metabolomics. Valuable scientific insight including in toxicity studies has occurred through metabolomic data generated through nuclear magnetic resonance (NMR), gas chromatography-mass spectrometry (GC-MS), capillary electrophoresis-mass spectrometry (CE-MS), and more recently, liquid chromatography-mass spectrometry (LC-MS) [8,9]. LC-MS is suited for the analysis of chemically diverse low molecular weight compounds produced during human metabolic processes [9]. For this current study, LC-Quadrupole Time-of-Flight (Q-TOF) MS was chosen to explore potential metabolomic biomarkers distinguishing healthy from cancerous tissues in the human colon. This technology has been proven to significantly improve mass accuracy and resolution, besides displaying high sensitivity, good isotopic fidelity, reproducibility of retention time, and optimization of data acquisition.
In this study, 17 paired surgical biopsy tissues that are subset of the MAL2 cohort [6] were included. Biofilm quantification and 16S rRNA sequencing have been performed and 16S rRNA data were analyzed using Resphera Insight, a clinical-grade proprietary analysis protocol [6]. Thus, in the current study, metabolomic analysis of paired normal–tumor tissues from patients with colorectal cancer to identify differences metabolomic profiles between cancerous and adjacent non-cancerous tissues obtained from the colon. In addition, 16S rRNA data were reanalyzed to discover possible association between metabolomic differences and microbiome aberrations. A hallmark of cancer is metabolic reprogramming. However, the underlying mechanism of metabolic reprogramming in cancer is complex and not well understood. Since many lifestyle-related factors have been linked to CRC and limited studies of this nature have been conducted in the Southeast Asian population, this study was expected to present a different perspective on metabolomics and the microbiome of CRC in this population.
Materials and Methods
Ethical statement
The University of Malaya Medical Centre (UMMC, Kuala Lumpur, Malaysia) Medical Ethics Committee (Ref. No. 1066.38) and the Johns Hopkins Institutional Review Board approved this study. All biopsy tissues were collected after obtaining informed and written consents from the patients.
Sample collection
Samples were collected consecutively from 2013 to 2014. The details of colon tissues collection method though a standard mechanical bowel preparation have been previously described [6,10]. Exclusion criteria include those individuals who have received pre-operative radiation, chemotherapy treatment or had a past history of CRC or inflammatory bowel disease. Standard pre-operative intravenous antibiotics (cefotetan, clindamycin/ gentamicin or cefoperazone/ metronidazole) were administered in all cases and none of the patients received any pre-operative oral antibiotics. Tissue from the proximal colon through the hepatic flexure is considered as right (ascending) colon, whereas distal to the hepatic flexure as left (descending) colon. During surgery, excess colon tumor (adenomas and cancers) and paired tumor-free (herein referred to as “normal”) tissues were collected for analysis. Detailed demography and characteristics of the colonic tissue of the study patients are as summarized in Table 1. Statistical significance of age, racial and gender distributions between patients presenting with left and right colorectal tumors were assessed using one-way ANOVA (IBM SPSS Statistics version 22.0) with p-value < 0.05 considered significant.
Table 1. Demographic information and characteristics of colonic tissues.
| Patient ID | Gender | Race | Age (During surgery) | Type | Location | Anatomic Stage (TNM) | Biofilm (Normal) | Biofilm (CRC) |
|---|---|---|---|---|---|---|---|---|
| S035 | Male | Indian | 74 | Moderately Differentiated Adenocarcinoma | Left | IIIB | No | No |
| S036 | Female | Chinese | 49 | Moderately Differentiated Adenocarcinoma | Left | IVB | No | No |
| S037 | Female | Chinese | 57 | Moderately Differentiated Adenocarcinoma | Left | IIIA | Yes | Yes |
| S038 | Male | Malay | 49 | Moderately Differentiated Adenocarcinoma | Left | IIIB | Yes | Yes |
| S039 | Female | Chinese | 75 | Poorly Differentiated Adenocarcinoma | Right | IIA | No | Yes |
| S040 | Female | Chinese | 84 | Moderately Differentiated Adenocarcinoma | Left | I | Yes | Yes |
| S043 | Female | Malay | 52 | Moderately Differentiated Adenocarcinoma | Left | IIA | No | No |
| S044 | Male | Indian | 41 | Well Differentiated Adenocarcinoma | Right | IVB | No | Yes |
| S045 | Female | Chinese | 50 | Adenocarcinoma | Right | I | Yes | Yes |
| S046 | Female | Chinese | 52 | Adenocarcinoma (Mucinous) | Left | IIA | Yes | Yes |
| S048 | Female | Malay | 74 | Moderately Differentiated Adenocarcinoma | Left | IVA | No | No |
| S051 | Female | Indian | 63 | Moderately Differentiated Adenocarcinoma | Right | IIA | Yes | Yes |
| S053 | Male | Chinese | 54 | Moderately Differentiated Adenocarcinoma | Left | I | No | No |
| S056 | Male | Indian | 76 | Moderately Differentiated Adenocarcinoma | Right | IV | Yes | Yes |
| S057 | Male | Chinese | 73 | Moderately Differentiated Adenocarcinoma | Left | IIIB | No | No |
| S058 | Female | Chinese | 67 | Moderately Differentiated Adenocarcinoma | Left | IIA | No | No |
| S060 | Male | Chinese | 72 | Moderately Differentiated Adenocarcinoma | Right | IIA | Yes | Yes |
Samples in this Table are from the MAL2 cohort first reported in Drewes et al [5]. Raw sequences from the MAL2 cohort have been deposited in the NCBI SRA repository (BioProject accession no. PRJNA325650). Primary sequencing data are available upon request.
Metabolite extraction
The tissue, disposable pestle and 1.5 ml-centrifuge tube in liquid nitrogen were chilled in liquid nitrogen. The tissue was pulverized in the presence of liquid nitrogen to fine powder. Metabolites were extracted from tissue samples by the Bligh and Dyer extraction method [11]. Briefly, 100 μl of chloroform (HPLC grade; Friendemann Schmidt Chemical, Australia) and 200 μl of methanol (HPLC grade; Friendemann Schmidt Chemical, Australia) were added to the fine powder and resuspended by vigorous vortexing. The mixture was stored at room temperature for 30 min. Subsequently, 100 μl of chloroform and 100 μl of water were added and mixed. The tube was centrifuged at 12,000 xg for 10 min. The biphasic solutions were separated into two separate tubes without disturbing the protein precipitate at the interface. The samples were vacuum concentrated to dryness in a Refrigerated CentriVap concentrator (Labconco, USA) at 4°C. The samples were reconstituted with 20 μl of mobile phase (95% water:5% ACN), vortexed and centrifuged at 12,000 xg for 10 minutes at 4°C.
Untargeted metabolomics by LC/MS
The samples were analyzed on an Agilent 1260 Infinity-6540 UHD Accurate-Mass Quadrupole-Time-of-Flight (Q-TOF) LC/MS system coupled with Dual Agilent Jet Stream Electrospray Ionization source (Agilent Technologies, USA). The injection volume was 3 μl of sample and separation was using a Zorbax Eclipse plus C-18 Rapid Resolution High Throughput (RRHT) 2.1x 100mm 1.8 μm column (Agilent Technologies, USA). The separation was performed at a flow rate of 0.45 mL/min with linear gradient program. Mobile phase A composed of 0.1% formic acid in Milli-Q water and mobile phase B composed of 0.1% formic acid in acetonitrile (HPLC grade; Friendemann Schmidt Chemical, Australia) The gradient program was set as follows: t = 0 min, 5% B; t = 2 min, 5% B; t = 15 min, 98% B; t = 18min, 98%; t = 20 min, 5% B and the final stop time, t = 25 min, 5% B. For positive ionization mode, two reference masses of (i) 121.0509 m/z and (ii) 922.0098 m/z were measured continuously while for negative ionization mode, the reference masses were (i) 112.9855 and (ii) 1033.9881. Reference mass correction was enabled. The gas temperature was maintained at 300°C, drying gas flow was set at the rate of 8 L/min, sheath gas temperature and sheath gas flow at 350°C and 11 L/min respectively. The capillary voltage was 3500 V. The nebulizer pressure was set at 35 psi. The MassHunter Workstation software B.05.01 (Agilent Technologies, USA) was applied for instrument control and data acquisition. The data was analyzed using the Mass Profiler Professional software version 12.6.1 (Agilent Technologies, USA and Strand Life Sciences, USA). We compared the relative abundances between paired tumor and adjacent normal tissue samples using the non-parametric two-sided Wilcoxon signed rank test. Differences were considered significant with p-value < 0.05. The procedure has been deposited at protocols.io: http://dx.doi.org/10.17504/protocols.io.u4ceysw.
DNA extraction
Genomic DNA was isolated using the MasterPure DNA Purification Kit (Epicentre/Illumina). The primers, S-D-Bact-0341-b-S-17 forward (5′-NNNNCCTACGGGNGGCWGCAG-3′) and S-D-Bact-0785-a-A-21 reverse (5′-GACTACHVGGGTATCTAATCC-3′), including Illumina-compatible adapters, were used to amplify the V3-V4 region of the 16S rRNA gene [6].
Analysis of 16S rRNA amplicon sequence data
The 16S sequencing data were quality-trimmed using Sickle (github.com/najoshi/sickle) using the following parameters: -q 20 –l 200. Merging of overlapping paired-end sequences was performed using MeFit with default parameters [12]. Filtering of chimeric sequences, de novo clustering of 16S rRNA sequences into Operational Taxonomic Units (OTUs) at 97% similarity threshold and removal of singleton OTUs were conducted using Micca (version 1.7.0) [13]. Taxonomic assignment of the representative OTUs was performed using the Bayesian LCA-based taxonomic classification method with a 1e-100 cut-off e-value and 100 bootstrap replications, against NCBI 16S microbial database [14,15]. Taxonomic assignment at each level was accepted only with a minimum confidence score of 80. Multiple sequence alignment of the OTU representative sequences was performed using PASTA [16]. A phylogenetic tree was constructed using FastTree (version 2.1.8) under the GTR+CAT model [17].
The rarefaction depth value was set at 72290 and subsequent computation of alpha and beta diversities was performed using QIIME (version 1.9.1) [18]. Briefly, alpha diversity was evaluated based on the following metrics: observed species and Shannon diversity index. Non-parametric two-sample t-test was used to compare the alpha diversity metrics between the normal and tumor samples (i.e. using Monte Carlo permutations to calculate the p-value). Principle coordinates analysis (PCoA) using unweighted UniFrac distance metric was performed to visualize separation of samples. Non-parametric statistical analysis of the distance metric was performed using ANOSIM with 1000 permutations. Graphs were generated using both phyloseq R package and PhyloToAST software [19,20].
Functional profiling based on KEGG pathways was conducted using Piphillin [21]. To generate the microbial community correlation networks, the Kendall’s tau correlation coefficients between rarefied abundances of different bacterial genera were calculated using the SparCC software [22]. The statistical significance of each pairwise comparison was examined by bootstrapping with 500 iterations. Only negative and positive correlations of values ≤ -0.7 and ≥ 0.7, respectively, and with pseudo p-values of less than 0.01, were considered. The networks were visualized using Cytoscape software 3.6.1 [23].
Pearson’s correlation analysis of bacterial genera against KEGG pathways, and bacterial genera against metabolites, was performed on rarefied abundance data using microbiomeSeq R package (https://github.com/umerijaz/microbiomeSeq.git). Correlations with p-values < 0.01 were considered significant.
Genus-level OTUs, KEGG functional pathways and metabolomics compounds with a minimum relative abundance of 0.001% and a detection frequency of at least 25% in all samples, were compared between matched normal and tumor samples by using two-sided Wilcoxon signed rank test. Data with p-values < 0.01 were considered significant.
Results
Among the 17 patients, 11 had left-sided CRC and 6 had right-sided CRC (Table 1). The mean age of patients with left-sided CRC at time of surgery was 60.9 (95% CI: 54.2–67.6) years old while the mean age of patients with right-sided CRC was 62.8 (95% CI: 47.6–78.1) years old. The difference between ages of patients at time of surgery was not statistically significant (one-way ANOVA, p-value ≥ 0.05). In the group of left-sided CRC patients, 4 were male and 7 were female. The right-sided group comprised of 3 male and 3 female patients. Racial distribution within the left-sided CRC group was 7 Chinese (64%), 3 Malay (27%) and 1 Indian (9%). On the other hand, right-sided CRC group comprised of 3 Chinese (50%) and 3 Indian (50%). The difference between racial distributions of the two groups was also not statistically significant (one-way ANOVA, p-value ≥ 0.05). It is noted that 4 out of 11 left-sided tumors were biofilm positive (36.4%), whereas the six right-sided tumors were biofilm positive (100%). However, 6/8 subjects with biofilm at the site of tumor also had biofilm at adjacent unaffected site (75%).
Metabolomics
In total, 708 compounds present in more than one sample were annotated (S1 Table). Among these, only 158 compounds had minimum relative abundance of 0.001% and detection frequency of at least 25% in all samples. Table 2 shows 36 compounds found in Kyoto Encyclopedia of Genes and Genomes (KEGG)/ LIPID MAPS Proteome (LMP)/ NIH Human Microbiome Project (HMP) databases and were detected in normal tissues only. In addition, 14 compounds were significantly more frequently found to be higher in normal mucosa than paired tumor tissues (Table 3). Diketospirilloxanthin, which is involve in carotenoid biosynthesis, was detected only in normal tissues (2-tailed Fisher’s exact test, p-value = 0.007). 5-hydroxyindoleacetic acid (5-HIAA) and serotonin, which are involved in tryptophan metabolism and serotonergic synapses, were also found only in normal tissues (Fisher’s exact test, p-value = 0.044). Serotonin, together with glycocholic acid and cortolone-3-glucuronide, found in normal tissues only (Fisher’s exact test, p-value = 0.044), are involved in bile secretion. Furthermore, the level of spermidine, which also plays a role in bile secretion, was significantly more frequently increased in normal mucosa compared to paired tumor tissues (Wilcoxon signed rank test, p-value = 0.021) (Table 3). Other metabolites that were present in normal tissues only include PE-Cer(d14:1(4E)/23:0) (Fisher’s exact test, p-value = 0.018), 26-O-beta-D-glucopyranosyl-3beta,26-dihydroxy-25(R)-furosta-5,20(22)-dien-3-O-alpha-L-rhamnopyranosyl(1–2)-beta-D-glucopyranoside (p-value = 0.018), ganglioside GA1 (d18:1/9Z-18:1) (p-value = 0.044) and Pro Arg Ile (p-value = 0.044). Phenanthrene-4,5-dicarboylate and m-coumaric acid have roles in aromatic compound degradation. Notably, many of these compounds are listed in the KEGG database to be implicated in diverse metabolic functions.
Table 2. Metabolites detected only in normal adjacent mucosa.
N, normal adjacent mucosal tissue; L, left-sided colon; R, right-sided colon.
| Compound | Frequency (n = 17) | KEGG ID | LMP ID | Description |
|---|---|---|---|---|
| Dolichyl beta-D-glucosyl phosphate | 2 | C01246 | LMPR03080014 | Metabolic pathways; N-Glycan biosynthesis |
| Diketospirilloxanthin/ 2,2'-Diketospirilloxanthin | 7 | C15885 | LMPR01070154 | Metabolic pathways; Carotenoid biosynthesis |
| Glycocholic acid | 1 | C01921 | LMST05030001 | Metabolic pathways, Bile secretion; Cholesterol metabolism, Secondary bile acid biosynthesis, Primary bile acid biosynthesis |
| Cortolone-3-glucuronide | 2 | C03033 | - | Metabolic pathways, Bile secretion; Pentose and glucuronate interconversions |
| Coproporphyrin | 1 | C03263 | - | Metabolic pathways, Biosynthesis of secondary metabolites, Porphyrin and chlorophyll metabolism |
| Pyropheophorbide a | 1 | C18064 | - | Biosynthesis of secondary metabolites, Porphyrin and chlorophyll metabolism |
| 5-HIAA / 5-Hydroxy-3-indoleacetic acid | 2 | C05635 | - | Metabolic pathways, Tryptophan metabolism, Serotonergic synapse |
| Serotonin | 4 | C00780 | - | Alkaloids; Metabolic pathways, Bile secretion, Tryptophan metabolism, Serotonergic synapse; Taste transduction, Synaptic vesicle cycle, Neuroactive ligand-receptor interaction, Gap junction, cAMP signaling pathway, Inflammatory mediator regulation of TRP channels |
| Tetrahydrofolic acid | 1 | C00101 | - | Metabolic pathways, Microbial metabolism in diverse environments; Aminoacyl-tRNA biosynthesis, One carbon pool by folate, Antifolate resistance, Methane metabolism, Carbon fixation pathways in prokaryotes, Folate biosynthesis, Carbon metabolism, Glycine, serine and threonine metabolism |
| Phenanthrene-4,5-dicarboxylate | 2 | C18252 | - | Microbial metabolism in diverse environments; Polycyclic aromatic hydrocarbon degradation |
| trans,trans-Farnesol | 4 | C01126 | LMPR0103010001 | Terpenoids/Sesquiterpenoids (C15); Biosynthesis of secondary metabolites; Insect hormone biosynthesis, Sesquiterpenoid and triterpenoid biosynthesis, Terpenoid backbone biosynthesis, Biosynthesis of antibiotics |
| Adrenosterone | 1 | C05285 | - | Steroid hormone biosynthesis |
| 2-Methoxyestrone 3-sulfate | 3 | C08358 | LMST05020006 | Steroid hormone biosynthesis |
| 2-Methoxyestrone 3-glucuronide | 1 | C11132 | LMST05010010 | Steroid hormone biosynthesis |
| Veratridine | 1 | C06544 | - | Alkaloids |
| Cardiopetalidine | 1 | C08667 | - | Alkaloids |
| makisterone B | 1 | C08829 | LMST01031017 | Terpenoids/Steroids |
| Convallamaroside | 1 | C08892 | - | Terpenoids/Steroids |
| Quassin | 1 | C08778 | LMPR0106110002 | Terpenoids/Triterpenoids (C30) |
| Cucurbitacin A | 1 | C08793 | LMST01010103 | Terpenoids/Triterpenoids (C30) |
| Cucurbitacin B | 1 | C08794 | LMST01010104 | Terpenoids/Triterpenoids (C30) |
| Cucurbitacin L | 1 | C08802 | LMST01010112 | Terpenoids/Triterpenoids (C30) |
| 26-O-beta-D-glucopyranosyl-3beta,26-dihydroxy-25(R)-furosta-5,20(22)-dien-3-O-alpha-L-rhamnopyranosyl(1–2)-beta-D-glucopyranoside | 6 | - | LMST01070008 | - |
| PE-Cer(d14:1(4E)/23:0) | 6 | - | LMSP03020010 | - |
| CDP-DG(12:0/12:0) | 3 | C13891 | LMGP13010001 | - |
| Pro Arg Ile | 5 | - | - | - |
| PIP2(16:0/20:3(5Z,8Z,11Z)) | 3 | C00626 | - | - |
| Deoxy-5-methylcytidylate | 4 | C03495 | - | - |
| Ganglioside GA1 (d18:1/9Z-18:1) | 5 | C06136 | - | - |
| Losartan | 2 | C07072 | - | - |
| Probucol | 2 | C07373 | - | - |
| Oxyphencyclimine | 2 | C07851 | - | - |
| Lauryl hydrogen sulfate | 3 | C11166 | - | - |
| Telithromycin | 3 | C12009 | - | - |
| Leucomycin A3 | 2 | C12662 | - | - |
| 9alpha-Fluoro-11beta,16alpha,17alpha,21-tetrahydroxypregn-4-ene-3,20-dione | 2 | C14638 | - | - |
Table 3. Metabolites found to be significantly different in paired normal and tumor tissues by Wilcoxon signed rank test.
| Compound | Frequency (increased in normal mucosa) (n = 17) | Frequency (increased in tumor) (n = 17) | KEGG ID | LMP ID | z-score | p-value | Description |
|---|---|---|---|---|---|---|---|
| m-Coumaric acid | 14 | 3 | C12621 | - | -3.107 | 0.002 | Microbial metabolism in diverse environments, Degradation of aromatic compounds, Phenylalanine metabolism |
| N(alpha)-t-Butoxycarbonyl-L-leucine | 12 | 2 | C04301 | - | -3.053 | 0.002 | - |
| PA(18:4(6Z,9Z,12Z,15Z)/20:4(5Z,8Z,11Z,14Z)) | 2 | 11 | - | LMGP10010446 | -2.760 | 0.006 | - |
| Formylmethionyl-leucyl-phenylalanine methyl ester | 10 | 1 | C11221 | - | -2.756 | 0.006 | - |
| Arg Arg Met | 2 | 11 | - | - | -2.691 | 0.007 | - |
| Acetyl-maltose | 13 | 4 | C02130 | - | -2.380 | 0.017 | - |
| Spermidine | 11 | 6 | C00315 | - | -2.312 | 0.021 | Metabolic pathways, ABC transporters, Bile secretion, Arginine and proline metabolism, beta-Alanine metabolism, Glutathione metabolism, Phenylpropanoid biosynthesis |
| Trp Pro Cys | 11 | 5 | - | - | -2.291 | 0.022 | - |
| Indoleacrylic acid | 13 | 4 | - | - | -2.249 | 0.025 | - |
| SAH / S-Adenosyl-L-homocysteine | 2 | 12 | C00021 | - | -2.222 | 0.026 | Metabolic pathways, Biosynthesis of amino acids, Cysteine and methionine metabolism |
| Ranitidine | 8 | 3 | C07233 | - | -2.197 | 0.028 | - |
| Arg Glu Leu | 8 | 3 | - | - | -2.172 | 0.030 | - |
| 3-Amino-3-(4-hydroxyphenyl)propanoate | 6 | 1 | C04368 | - | -2.059 | 0.040 | Tyrosine metabolism |
| Pirbuterol | 7 | 1 | C07807 | - | -2.059 | 0.040 | - |
Table 4 shows 16 compounds that were detected in cancerous tissues only. Among these, 7 compounds were involved in biosynthesis of antibiotics (Macrolides/ Type II polyketide) (Fisher’s exact test, p-value = 0.044). In addition, Amphotericin B, a Type I polyketide antifungal agent, was also found in tumor but not normal tissues. Cinnamyl benzoate was also detected only in tumor tissues (Fisher’s exact test, p-value = 0.044). Similarly, only 3 out of 14 compounds found to be differentially present in paired normal-tumor tissues were significantly more frequently elevated in tumor tissues (Table 3). Among these was S-adenosyl-L-homocysteine (SAH) that is involved in cysteine and methionine biosynthesis.
Table 4. Metabolites detected only in colorectal tumor.
T, colorectal tumor; L, left-sided colon; R, right-sided colon.
| Compound | Frequency (n = 17) | KEGG ID | LMP ID | Description |
|---|---|---|---|---|
| FAD / Flavin adenine dinucleotide | 1 | C00016 | - | Metabolic pathways, Biosynthesis of secondary metabolites; Vitamin digestion and absorption, Riboflavin metabolism |
| PE(18:3(6Z,9Z,12Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) | 1 | C00350 | - | Metabolic pathways, Biosynthesis of secondary metabolites; Glycosylphosphatidylinositol (GPI)-anchor biosynthesis, Autophagy, Glycerophospholipid metabolism, Pathogenic Escherichia coli infection, Retrograde endocannabinoid signaling, Kaposi’s sarcoma-associated herpesvirus infection |
| Presqualene diphosphate | 1 | C03428 | LMPR0106010003 | Biosynthesis of antibiotics, Metabolic pathways, Biosynthesis of secondary metabolites; Sesquiterpenoid and triterpenoid biosynthesis, Carotenoid biosynthesis, Steroid biosynthesis |
| 6,8a-Seco-6,8a-deoxy-5-oxoavermectin ''2b'' aglycone | 1 | C11953 | - | Biosynthesis of 12-, 14- and 16-membered macrolides |
| Avermectin A1b monosaccharide | 1 | C11966 | - | Biosynthesis of antibiotics, Biosynthesis of 12-, 14- and 16-membered macrolides |
| 5-O-beta-D-Mycaminosyltylonolide | 1 | C12002 | - | Biosynthesis of antibiotics, Biosynthesis of 12-, 14- and 16-membered macrolides |
| Tetracenomycin B3 | 1 | C12369 | - | Biosynthesis of antibiotics, Biosynthesis of type II polyketide products |
| Stealthin C | 2 | C12392 | - | Biosynthesis of antibiotics, Biosynthesis of type II polyketide products |
| antioside | 2 | C08848 | LMST01120016 | Terpenoids/Steroids |
| Sarsaparilloside | 2 | C08911 | - | Terpenoids/Steroids |
| Glaucarubin | 2 | C08760 | - | Terpenoids/Triterpenoids (C30) |
| Amphotericin B | 3 | C06573 | LMPK06000002 | Type I polyketide structures |
| Cinnamyl benzoate | 5 | - | - | - |
| Verapamil | 2 | C07188 | - | - |
| Enalkiren | 2 | C07466 | - | - |
| Xamoterol | 4 | C11775 | - | - |
Pre- and post-processing 16S data and microbiota composition at phylum level
A total of 5,372,109 reads were generated for 17 pairs of normal-tumor samples. Following quality trimming and merging of overlapping paired-end reads, 5,120,010 sequences were retained, ranging from 82,879 to 245,890 per sample with an average read length of 457.6 ± 3.3 bp (S2 Table). Of 1144 OTUs acquired by de novo clustering of the merged overlapping paired-end sequences, 662 were taxonomically assigned down to genus level with an 80% confidence threshold.
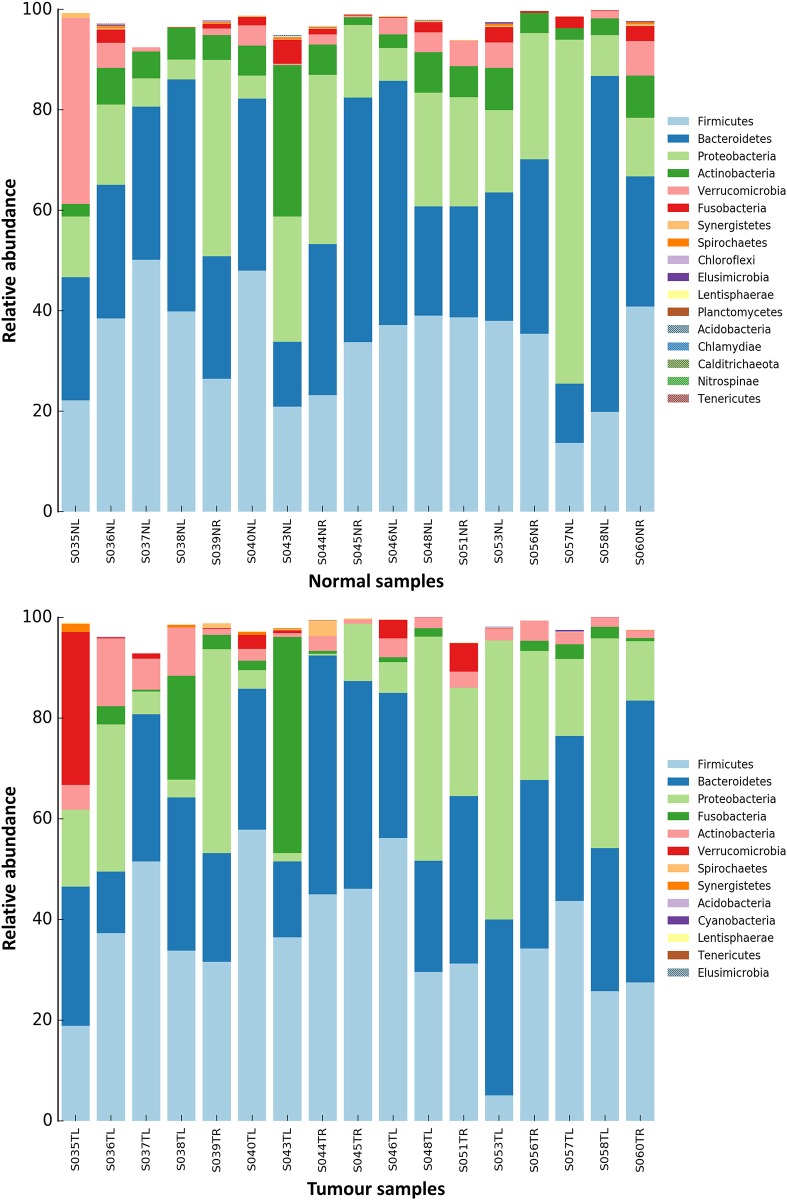
In both tumor and normal tissues, Firmicutes, Bacteroidetes and Proteobacteria constituted the three most predominant phyla, at 36%, 30.7% and 19.5% of relative abundances, respectively, in the former samples, whilst the latter showed 33.3%, 31.5% and 19.7% of abundances (S3 Table). In the tumor samples, several bacterial phyla were shown to be nearly or completely absent including Calditrichaeota, Chlamydiae, Chloroflexi, Elusimicrobia and Planctomycetes (Fig 1).
Fig 1. Bacterial phyla composition in normal and tumor samples.
Alpha and beta diversity
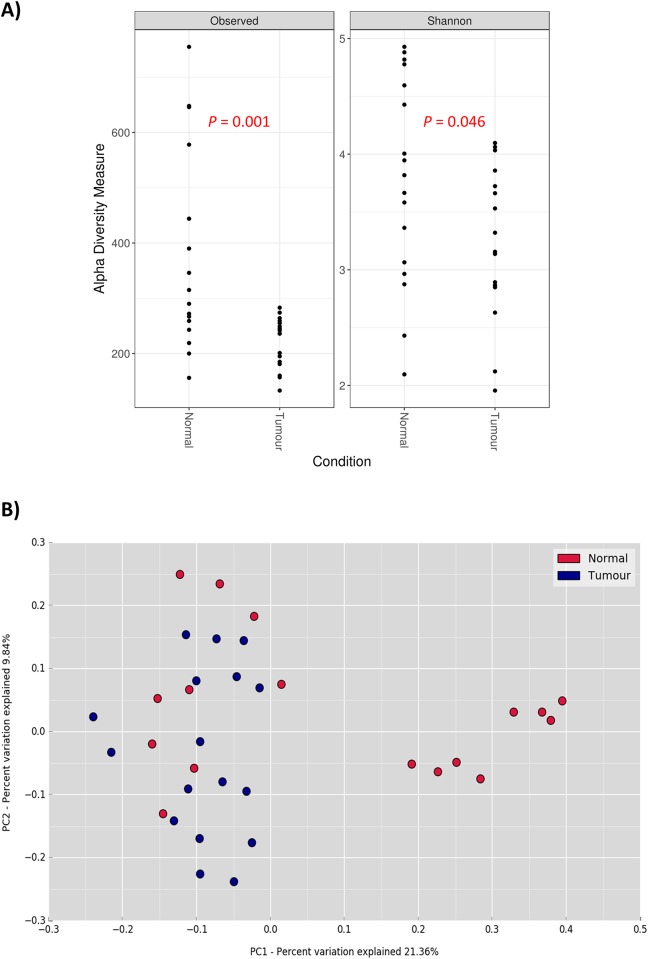
To estimate and compare the alpha diversity of the colorectal microbial community derived from both tumor samples and normal tissue counterparts, we employed observed species and Shannon diversity indexes. The tumor samples had significantly reduced species richness and microbial diversity than the matched normal samples (observed species, p-value = 0.001; Shannon diversity, p-value = 0.046) (Fig 2A). To assess the overall difference of bacterial community between tumor and normal samples, PCoA based on unweighted UniFrac distance was performed. Two significantly distinct clusters were revealed (p-value = 0.003, R = 0.18) (Fig 2B). Eight normal samples were confined in one cluster and only two of these samples had been reported previously to have biofilm. Another cluster, interestingly, contained all tumor samples and the remaining normal samples [6]. The unweighted UniFrac distance PCoA indicates that histological condition of the samples has a more significant impact on the clustering than biofilm status. These normal samples that shared similar bacterial composition with the tumor samples could be a possible early indication of carcinogenesis, which merits further investigation.
Fig 2.
(A) Comparison of alpha diversity between normal and tumor samples based on species richness and Shannon diversity indexes. (B) PCoA plot of unweighted UniFrac distance of normal and tumor samples. Statistical testing using ANOSIM method revealed significant separation between normal and tumor samples (p-value = 0.003, R = 0.18).
Taxonomic differences between tumor and paired normal tissue samples
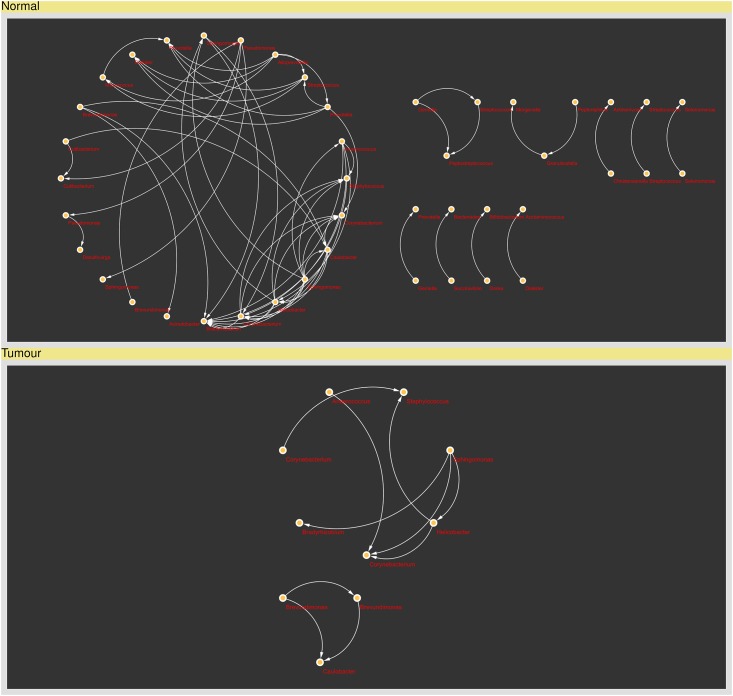
To identify bacterial genera that were significantly different between tumor and normal samples, genus-level OTUs that were present in at least 25% of total samples and with a minimum relative abundance of 0.001% were evaluated using Wilcoxon signed rank test. Of 358 OTUs tested, 24 were significantly enriched in normal tissue samples in comparison to their respective tumor counterparts (Table 5). These 24 OTUs represented 21 bacterial genera, in which Alistipes (median, 1.894 versus 0.024), Oscillibacter (median, 1.234 versus 0.009), Bacteroides (median, 0.847 versus 0.097), Pseudoflavonifractor (median, 0.012 versus 0) and Succinivibrio (median, 0.23 versus 0) being identified as the top five most differentially abundant genera. In addition, Christensenella, Dialister and Pseudomonas were assigned to two different OTUs, respectively. To examine if bacterial genera significantly affected by tissue status would deliver an impact on microbial interactions, we performed a co-occurrence network analysis based on Kendall’s tau correlation coefficient. As depicted in Fig 3, the density of interactions between different bacterial genera was profoundly less in the tumor samples as compared with the microbial interactions observed in the normal tissue counterparts. This is consistent with the abundances of several bacterial genera that were significantly decreased in the tumor samples such as Succinivibrio, Bacteroides, Christensenella, Pseudomonas, Bifidobacterium, Dialister, Prevotella and Actinomyces.
Table 5. List of genus-level OTUs that significantly differed between tumor samples and paired normal tissues by Wilcoxon signed rank testing.
| OTU | Genus | Mean normal (%) | Mean tumor (%) | Median normal (%) | Median tumor (%) | p-value |
|---|---|---|---|---|---|---|
| DENOVO80 | Actinomyces | 0.341 | 0.066 | 0 | 0 | 0.009 |
| DENOVO7 | Alistipes | 3.141 | 0.396 | 1.894 | 0.024 | 0.001 |
| DENOVO105 | Anaerotruncus | 0.272 | 0.1 | 0.004 | 0 | 0.007 |
| DENOVO9 | Bacteroides | 2.733 | 0.343 | 0.847 | 0.097 | 0.002 |
| DENOVO468 | Bifidobacterium | 0.014 | 0 | 0.001 | 0 | 0.005 |
| DENOVO171 | Christensenella | 0.056 | 0.028 | 0.015 | 0 | 0.004 |
| DENOVO74 | Christensenella | 0.344 | 0.156 | 0.151 | 0 | 0.006 |
| DENOVO23 | Collinsella | 1.282 | 0.644 | 1.108 | 0.307 | 0.007 |
| DENOVO73 | Desulfovibrio | 0.414 | 0.204 | 0.256 | 0 | 0.003 |
| DENOVO244 | Dialister | 0.029 | 0 | 0.007 | 0 | 0.005 |
| DENOVO48 | Dialister | 0.743 | 0.35 | 0.037 | 0 | 0.008 |
| DENOVO200 | Intestinimonas | 0.061 | 0.03 | 0.026 | 0 | 0.006 |
| DENOVO25 | Megasphaera | 1.401 | 0.432 | 0.216 | 0 | 0.003 |
| DENOVO153 | Mitsuokella | 0.094 | 0.026 | 0.001 | 0 | 0.008 |
| DENOVO85 | Morganella | 0.315 | 0.033 | 0.116 | 0 | 0.008 |
| DENOVO352 | Negativibacillus | 0.041 | 0.029 | 0.021 | 0 | 0.008 |
| DENOVO14 | Oscillibacter | 2.085 | 1.027 | 1.234 | 0.09 | 0.001 |
| DENOVO732 | Parabacteroides | 0.012 | 0.001 | 0.003 | 0 | 0.004 |
| DENOVO5 | Prevotella | 5.036 | 2.734 | 1.285 | 0.066 | 0.009 |
| DENOVO611 | Pseudoflavonifractor | 0.018 | 0.007 | 0.012 | 0 | 0.002 |
| DENOVO404 | Pseudomonas | 0.082 | 4.07 × 10−4 | 0.022 | 0 | 0.004 |
| DENOVO20 | Pseudomonas | 1.352 | 0.025 | 0.112 | 0.003 | 0.006 |
| DENOVO113 | Rhodococcus | 0.132 | 0.001 | 0.004 | 0 | 0.006 |
| DENOVO31 | Succinivibrio | 1.447 | 0.699 | 0.23 | 0 | 0.002 |
Fig 3. Co-occurrence network analysis of genus-level OTUs.
Nodes corresponds to bacterial genera while edges represent positive correlations of at least 0.7 and with p-values less than 0.05. No negative correlations could be identified in this network analysis.
Comparative functional differences of colorectal bacterial communities in tumor and paired normal tissue samples
We next employed Piphillin for the functional prediction of colorectal microbial communities between tumor and normal tissue samples, revealing 286 KEGG pathways. Eight KEGG pathways exhibited significant differences between both groups by Wilcoxon signed rank testing (p-value < 0.01, Table 6). The microbial communities of tumor samples had significant enrichments in both fatty acid biosynthesis and glycerolipid metabolism, while the normal microbial subsets were significantly enriched for pathways associated with citrate cycle, steroid biosynthesis, C5-branched dibasic acid metabolism, pantothenate and CoA biosynthesis, and sesquiterpenoid and triterpenoid biosynthesis.
Table 6. List of KEGG pathways that significantly differed between tumor samples and paired normal tissues by Wilcoxon signed rank testing.
| KO identifier | Pathway description | Mean normal (%) | Mean tumor (%) | Median normal (%) | Median tumor (%) | p-value |
|---|---|---|---|---|---|---|
| ko00020 | Citrate cycle (TCA cycle) | 0.627 | 0.571 | 0.619 | 0.592 | 0.009 |
| ko00061 | Fatty acid biosynthesis | 0.447 | 0.477 | 0.456 | 0.485 | 0.002 |
| ko00100 | Steroid biosynthesis | 0.004 | 0.002 | 0.002 | 9.77 × 10−5 | 0.002 |
| ko00561 | Glycerolipid metabolism | 0.257 | 0.279 | 0.258 | 0.273 | 0.004 |
| ko00660 | C5-Branched dibasic acid metabolism | 0.267 | 0.254 | 0.27 | 0.255 | 0.009 |
| ko00770 | Pantothenate and CoA biosynthesis | 0.539 | 0.519 | 0.536 | 0.504 | 0.005 |
| ko00909 | Sesquiterpenoid and triterpenoid biosynthesis | 0.009 | 0.005 | 0.005 | 3.02 × 10−4 | 0.001 |
| Ko04976 | Bile secretion | <0.001 | <0.001 | <0.001 | <0.001 | 0.008 |
| ko05131 | Shigellosis | 0.001 | 4.79 × 10−4 | 0.001 | 4.73 × 10−5 | 0.002 |
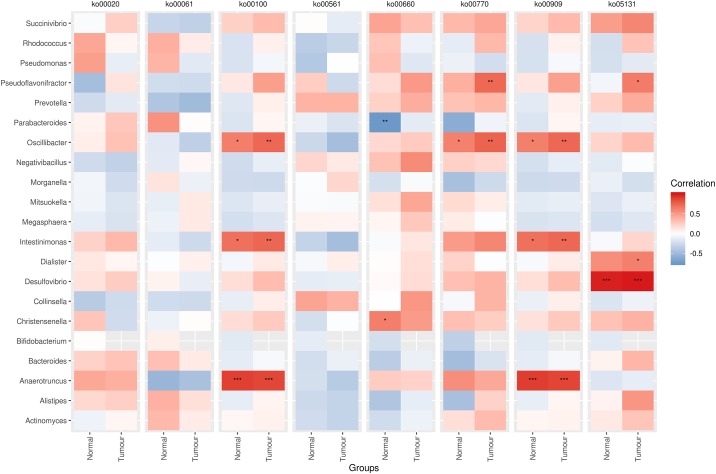
Pearson’s correlation analysis of bacterial genera and KEGG pathways that significantly differed between the tumor and normal tissue samples revealed several significant associations. The abundances of Anaerotruncus, Intestinimonas and Oscillibacter exhibited significant relationships with both steroid biosynthesis and sesquiterpenoid and triterpenoid biosynthesis pathways in both tumor and normal tissue samples with Anaerotruncus showing the most significant associations (p-values < 0.0001) (Fig 4; S4 Table). Oscillibacter was also significantly associated with pantothenate and CoA biosynthesis (p-value < 0.01, normal group; p-value < 0.001, tumor group). The C5-branched dibasic acid metabolism pathway in the normal group was positively associated with Christensenella and inversely correlated to Parabacteroides, with p-values < 0.01 and < 0.001, respectively. The shigellosis pathway that significantly differed between both tumor and normal samples was strongly correlated to the abundance of Desulfovibrio bacteria in each group, respectively (p-values < 0.0001).
Fig 4. Pearson’s correlation between bacterial genera and KEGG pathways with significant differences.
ko00020: Citrate cycle (TCA cycle); ko00061: Fatty acid biosynthesis; ko00100: Steroid biosynthesis; ko00561: Glycerolipid metabolism; ko00660: C5-Branched dibasic acid metabolism; ko00770: Pantothenate and CoA biosynthesis; ko00909: Sesquiterpenoid and triterpenoid biosynthesis; ko05131: Shigellosis.
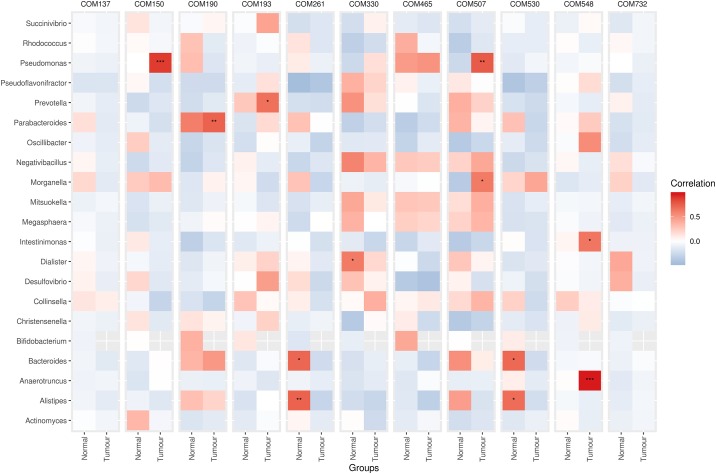
Pearson’s correlation analysis of metabolites and bacterial genera that significantly differed between the tumor and normal tissue samples also revealed several significant associations. In the tumor group, PE(P-16:0/0:0) exhibited significant relationships with Anaerotruncus (p-value < 0.0001) and Intestinimonas (p-value < 0.01) (Fig 5; S5 Table). Pseudomona correlated significantly with 6-methoxyquinoline (p-value < 0.0001) and N(alpha)-t-butoxycarbonyl-L-leucine (p-value < 0.001), which also correlated significantly with Morganella (p-value < 0.01). Parabacteroides and Prevotella correlated significantly with Antillatoxin B (p-value < 0.001) and Arg Arg Met (p-value < 0.01) respectively. On the other hand, in the normal samples, Alistipes and Bacteroides showed significantly association with both creatine (p-value < 0.001; 0.01) and PA(18:4(6Z,9Z,12Z,15Z)/20:4(5Z,8Z,11Z,14Z)) (p-value < 0.01). Dialister was also found to be significantly associated with formylmethionyl-leucyl-phenylalanine methyl ester (p-value < 0.01).
Fig 5. Pearson’s correlation between bacterial genera and metabolites with significant differences.
COM137, 5α-Androstan-3β-ol-17-one sulfate; COM150: 6-Methoxyquinoline; COM190: Antillatoxin B; COM193: Arg Arg Met; COM261: Creatine; COM330: Formylmethionyl-leucyl-phenylalanine methyl ester; COM465: m-Coumaric acid; COM507: N(alpha)-t-Butoxycarbonyl-L-leucine; COM530: PA(18:4(6Z,9Z,12Z,15Z)/20:4(5Z,8Z,11Z,14Z)); COM548: PE(P-16:0/0:0); COM732: Val Arg Phe.
Discussion
Metabolomics enables a large-scale, qualitative, and quantitative study of metabolites in a system biological approach. Unlike mRNAs and proteins, biosynthesis of metabolites is complex and requires advanced instrumentations such as MS, NMR spectroscopy, and laser-stimulated fluorescence (LSF) spectroscopy. Notably, each of these instruments has their unique strengths and limitations. Although NMR is highly selective and non-destructive and is the gold standard in metabolite structural elucidation, it has relatively lower sensitivity compared to other technologies [24]. In contrast, LSF is one of the most sensitive techniques, but it is not chemically selectivity and this limits its usefulness in structural identification of metabolites in complex biological systems [24]. On the other hand, MS, which provides good balance of sensitivity and selectivity, is frequently used in metabolomic analyses of complex biological samples [24]. Coupling chromatography to MS provides a great resolution for metabolomics identification and quantification. Currently, GC, LC, and capillary electrophoresis (CE) have been incorporated into MS-based metabolomics. GC-MS, which is suitable for the analysis of volatile, thermally stable, and energetically stable compounds, is extensively used for routine primary metabolite studies of common but important metabolite classes such as amino acids, organic acids and free fatty acids. CE-MS is inherently low in sensitivity, poor in reproducibility, and may be affected by electrochemical reactions of metabolites. Recently, Büscher et al. compared the performances of GC-MS, LC-MS, and CE-MS in application to quantitative metabolomics, and demonstrated that CE-MS was the least effective platform for analyzing complex biological samples [25]. Thus, LC-MS was chosen in this study for discovering unknown metabolites by untargeted metabolomics based on the wider range of compounds it can analyze.
Tian et al. analyzed the metabolomic signatures of CRC tissues and their adjacent non-involved tissues from Chinese patients using high-resolution magic-angle spinning (HRMAS) 1H NMR spectroscopy in combination with GC-FID/MS [26]. In that study, tissue metabolic phenotypes (in energy metabolism, membrane biosynthesis and degradations, osmotic regulation, and proteins and nucleotides metabolism) was able to discriminated CRC tissues from adjacent non-involved tissues [26]. More recently, Satoh et al., using CE-MS metabolome for profiling paired tumor and normal tissue from Japanese patients with CRC, found that S-adenosylmethionine (SAM) was the most up-regulated metabolite in tumor tissue [27]. The LC-MS-based metabolomics approach of this study provides additional information that complements our current understanding of the metabolomic differences between CRC tissues and adjacent non-involved tissues. In this study, it was shown that diverse metabolic pathways (such as N-glycan biosynthesis, carotenoid biosynthesis, cholesterol metabolism, bile acid metabolism, pentose and glucuronate interconversions, biosynthesis of secondary metabolites, amino acid metabolism and steroid hormone biosynthesis) differs between tumor and normal tissues. Consistent with previous studies [26,27], molecular evidence from this study suggests that cancer cells may alter their metabolism for the production of macromolecular precursors in CRC. The finding of SAM by Satoh et al. and subsequently SAH in our study to be frequently elevated in tumor tissues highlighted the importance of cysteine and methionine metabolism in carcinogenesis. Methionine, an essential amino acid in protein synthesis, is the precursor to SAM required by a variety of methyltransferases for the methylation of DNA, RNA, proteins, and lipids [28]. When SAM releases activated methyl group in methylation reactions, it is transformed into SAH that is further hydrolyzed to homocysteine [28]. Sibani et al. has demonstrated that there was positive correlations between SAM, SAH, and DNA hypomethylation with cellular transformation under folate-adequate conditions in pre-neoplastic small intestine of multiple intestinal neoplasia (Min) mice [29], thus illustrating the importance of SAM and SAH in DNA methylation and colorectal carcinogenesis. Furthermore, several cancer cells utilize SAM for hyperactive polyamine synthesis [28]. In turn, polyamine putrescine reacts with a decarboxylated form of SAM to form spermidine and spermine [28]. Therefore, this may explain the elevation of SAH and depletion of spermidine at tumor sites compared surrounding non-affected mucosa (Table 3).
Serotonin, 5-hydroxytryptamine (5-HT) is mainly synthesized at the gastrointestinal (GI) tract and it is closely associated with GI function and physiology as extensively reviewed by Manocha and Khan [30]. In intestinal enterochromaffin (EC) cells, conversion of dietary tryptophan is the first step in the biosynthesis of serotonin, has been implicated in various GI diseases and functional disorders [30]. Alteration in serotonin signaling is associated with celiac disease, CRC, and diverticular disease [30]. The absence of serotonin at tumor sites compared to corresponding adjacent non-involved sites may suggest increased catabolism of serotonin by cancerous cells. Serotonin is essential for the growth of s.c. colon cancer allografts in vivo by acting as a regulator of angiogenesis which reduces the expression of matrix metalloproteinase 12 (MMP-12)—an endogenous inhibitor of angiogenesis—in tumor-infiltrating macrophages [31]. The intricate interactions of the gut microbiota, food consumed, and intestinal cells together will impact the serotonin production, secretion, and degradation, and, hence, may be accountable for the impaired function of serotonin in GI diseases [30]. Thus, modulation of tryptophan metabolism, such as the production of serotonin can be used as a potential therapeutic strategy for CRC in the future.
2,2'-Diketospirilloxanthin is a naturally occurring carotenoid. Carotenoids are organic pigments produced by many plants and algae, as well as by various bacteria and fungi [32]. Natural carotenoids, because of their antioxidant properties, have been suggested to have anticarcinogenic activity [32]. On the other hand, a prospective Multiethnic Cohort Study based on quantitative food frequency questionnaires did not find any significant association between intake of individual and total carotenoids and CRC risk [33]. The detection of 2,2'-diketospirilloxanthin at cancerous sites but not adjacent non-involved sites suggests that the carotenoid may have local protective effects on epithelial tissues.
The detection of metabolites involved in antibiotics biosynthesis in CRC tissues suggests a role of microbiota structure and composition in colorectal carcinogenesis. Supported by data from meta-omics analyses and mechanistic studies in vitro and in vivo, bacteria, such as Fusobacterium nucleatum, enterotoxigenic Bacteroides fragilis, and colibactin-producing Escherichia coli, may be potentiators for CRC development [34]. In addition, it has been demonstrated that functional predictions from 16S rRNA gene sequences and metabolomics support that colonic mucosal biofilm contributes to antibiotic biosynthesis leading to alteration of the cancer metabolome to regulate cellular proliferation and colon cancer growth potentially affecting cancer development and progression [6,10,35]. The study of microbiome differences between tumor and surrounding non-affected tissues in this study has highlighted both taxonomical and predicted functional differences between normal and cancerous tissues. Predicted functions implied from the 16S rRNA gene sequences are consistent with findings from metabolomics analysis showing depletion of bacteria genera involve in steroid biosynthesis, terpenoid biosynthesis, and bile secretion in tumor relative to paired normal tissues. In addition, significant correlation was found between Anaerotruncus, Intestinimonas and Oscillibacter and steroid and terpenoid biosyntheses. However, the abundance of bacteria genera associated with bile secretion pathway was low (<0.001%). Nevertheless, the human host is known to produce large, conjugated and hydrophilic bile acids. Members of the intestinal microbiome may utilize bile acids and their conjugates resulting in smaller, unconjugated and hydrophobic bile acids. These unconjugated bile acids induce oncogenesis in colonic epithelial cells by altering muscarinic 3 receptor (M3R) and Wnt/ β-catenin signaling and thus act potential promoters of colon cancer [36]. Interestingly, many naturally occurring triterpenoids have been shown to exhibit cytotoxicity against tumor cells, as well as demonstrated to have anticancer efficacy in vivo [37].
In conclusion, this study expanded our insight into localized metabolic and microbiome differences between tumor and normal colonic tissues in CRC patients. Besides providing deeper understanding of the pathogenic process of colorectal carcinogenesis, these functional metabolites have potential implications in both the drug discovery process and in precision medicine. Future large-scale meta-analysis could be carried out by comparing the current and other datasets collected from different parts of the world to explore the association between the geographical factors with the metabolic differences in the CRC patients.
Supporting information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank Drs. Elizabeth Wick (University of California, San Francisco, Department of Surgery) and Cynthia L. Sears (Johns Hopkins University School of Medicine) for their initiation of this project. We thank Dr. Sears for helpful comments in the review of the manuscript.
Data Availability
This data is available at the NIH Common Fund’s Metabolomics Data Repository and Coordinating Center (supported by NIH grant, U01-DK097430) website, the Metabolomics Workbench, http://www.metabolomicsworkbench.org, where it has been assigned Project ID PR000726. The data can be accessed directly via it’s Project DOI: 10.21228/M8TX0N. The raw 16S sequences are available in the NCBI SRA repository, where it has been assigned BioProject accession no. PRJNA325650.
Funding Statement
This work was funded by the University of Malaya Research Grant RP016B-13HTM to WFW, University of Malaya-Ministry of Education High Impact Research Grant UM.C/625/1/HIR/MoE/CHAN/13/4 (Account No. H-50001-A000029) to JV, and Swami Institute for International Medical Education Award of Johns Hopkins University School of Medicine. EGC was supported by ShenZhen’s Sanming Project (Grant No: SZSM201510050 to EGC). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, et al. GLOBOCAN 2012 v1.1, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet]. Lyon, France: International Agency for Research on Cancer; 2014. http://globocan.iarc.fr, accessed on 16/01/2018. [Google Scholar]
- 2.Fleming M, Ravula S, Tatishchev SF, Wang HL. Colorectal carcinoma: Pathologic aspects. J Gastrointest Oncol. 2012. September;3(3):153–73. 10.3978/j.issn.2078-6891.2012.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen H, Yang J, Huang Q, Jiang MJ, Tan YN, Fu JF, et al. Different treatment strategies and molecular features between right-sided and left-sided colon cancers. World J Gastroenterol. 2015. June 7;21(21):6470–8. 10.3748/wjg.v21.i21.6470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saltzstein SL, Behling CA. Age and time as factors in the left-to-right shift of the subsite of colorectal adenocarcinoma: a study of 213,383 cases from the California Cancer Registry. J Clin Gastroenterol. 2007. February;41(2):173–7. 10.1097/01.mcg.0000225550.26751.6a [DOI] [PubMed] [Google Scholar]
- 5.Warschkow R, Sulz MC, Marti L, Tarantino I, Schmied BM, Cerny T, et al. Better survival in right-sided versus left-sided stage I—III colon cancer patients. BMC Cancer. 2016. July 28;16:554 10.1186/s12885-016-2412-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drewes JL, White JR, Dejea CM, Fathi P, Iyadorai T, Vadivelu J, et al. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes. 2017. November 29;3:34 10.1038/s41522-017-0040-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishiumi S, Kobayashi T, Ikeda A, Yoshie T, Kibi M, Izumi Y, et al. A novel serum metabolomics-based diagnostic approach for colorectal cancer. PLoS One. 2012;7(7):e40459 10.1371/journal.pone.0040459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guertin KA, Moore SC, Sampson JN, Huang WY, Xiao Q, Stolzenberg-Solomon RZ, et al. Metabolomics in nutritional epidemiology: identifying metabolites associated with diet and quantifying their potential to uncover diet-disease relations in populations. Am J Clin Nutr. 2014. July;100(1):208–17. 10.3945/ajcn.113.078758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukui Y, Itoh K. A Plasma Metabolomic Investigation of Colorectal Cancer Patients by Liquid Chromatography-Mass Spectrometry. The Open Analytical Chemistry Journal. 2010; 4:1–9. 10.2174/1874065001004010001 [DOI] [Google Scholar]
- 10.Dejea CM, Wick EC, Hechenbleikner EM, White JR, Mark Welch JL, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci U S A. 2014. December 23;111(51):18321–6. 10.1073/pnas.1406199111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959. August;37(8):911–7. 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- 12.Parikh HI, Koparde VN, Bradley SP, Buck GA, Sheth NU. MeFiT: merging and filtering tool for illumina paired-end reads for 16S rRNA amplicon sequencing. BMC Bioinformatics. 2016. December 1;17(1):491 10.1186/s12859-016-1358-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albanese D, Fontana P, De Filippo C, Cavalieri D, Donati C. MICCA: a complete and accurate software for taxonomic profiling of metagenomic data. Sci Rep. 2015. May 19;5:9743 10.1038/srep09743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao X, Lin H, Revanna K, Dong Q. A Bayesian taxonomic classification method for 16S rRNA gene sequences with improved species-level accuracy. BMC Bioinformatics. 2017. May 10;18(1):247 10.1186/s12859-017-1670-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018. January 4;46(D1):D8–D13. 10.1093/nar/gkx1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mirarab S, Nguyen N, Guo S, Wang LS, Kim J, Warnow T. PASTA: Ultra-Large Multiple Sequence Alignment for Nucleotide and Amino-Acid Sequences. J Comput Biol. 2015. May;22(5):377–86. 10.1089/cmb.2014.0156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010. March 10;5(3):e9490 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010. May;7(5):335–6. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013. April 22;8(4):e61217 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dabdoub SM, Fellows ML, Paropkari AD, Mason MR, Huja SS, Tsigarida AA, Kumar PS. PhyloToAST: Bioinformatics tools for species-level analysis and visualization of complex microbial datasets. Sci Rep. 2016. June 30;6:29123 10.1038/srep29123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwai S, Weinmaier T, Schmidt BL, Albertson DG, Poloso NJ, Dabbagh K, DeSantis TZ. Piphillin: Improved Prediction of Metagenomic Content by Direct Inference from Human Microbiomes. PLoS One. 2016. November 7;11(11):e0166104 10.1371/journal.pone.0166104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedman J, Alm EJ. Inferring correlation networks from genomic survey data. PLoS Comput Biol. 2012;8(9):e1002687 10.1371/journal.pcbi.1002687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003. November;13(11):2498–504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lei Z, Huhman DV, Sumner LW. Mass spectrometry strategies in metabolomics. J Biol Chem. 2011. July 22;286(29):25435–42. 10.1074/jbc.R111.238691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Büscher JM, Czernik D, Ewald JC, Sauer U, Zamboni N. Cross-platform comparison of methods for quantitative metabolomics of primary metabolism. Anal Chem. 2009. March 15;81(6):2135–43. 10.1021/ac8022857 [DOI] [PubMed] [Google Scholar]
- 26.Tian Y, Xu T, Huang J, Zhang L, Xu S, Xiong B, et al. Tissue Metabonomic Phenotyping for Diagnosis and Prognosis of Human Colorectal Cancer. Sci Rep. 2016. February 15;6:20790 10.1038/srep20790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Satoh K, Yachida S, Sugimoto M, Oshima M, Nakagawa T, Akamoto S, et al. Global metabolic reprogramming of colorectal cancer occurs at adenoma stage and is induced by MYC. Proc Natl Acad Sci U S A. 2017. September 12;114(37):E7697–E7706. 10.1073/pnas.1710366114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borrego SL, Fahrmann J, Datta R, Stringari C, Grapov D, Zeller M, et al. Metabolic changes associated with methionine stress sensitivity in MDA-MB-468 breast cancer cells. Cancer Metab. 2016. May 2;4:9 10.1186/s40170-016-0148-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sibani S, Melnyk S, Pogribny IP, Wang W, Hiou-Tim F, Deng L, Trasler J, James SJ, Rozen R. Studies of methionine cycle intermediates (SAM, SAH), DNA methylation and the impact of folate deficiency on tumor numbers in Min mice. Carcinogenesis. 2002. January;23(1):61–5. [DOI] [PubMed] [Google Scholar]
- 30.Manocha M, Khan WI. Serotonin and GI Disorders: An Update on Clinical and Experimental Studies. Clin Transl Gastroenterol. 2012. April 26;3:e13 10.1038/ctg.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nocito A, Dahm F, Jochum W, Jang JH, Georgiev P, Bader M, et al. Serotonin regulates macrophage-mediated angiogenesis in a mouse model of coloncancer allografts. Cancer Res. 2008. July 1;68(13):5152–8. 10.1158/0008-5472.CAN-08-0202 [DOI] [PubMed] [Google Scholar]
- 32.Nishino H, Murakosh M, Ii T, Takemura M, Kuchide M, Kanazawa M, et al. Carotenoids in cancer chemoprevention. Cancer Metastasis Rev. 2002;21(3–4):257–64. [DOI] [PubMed] [Google Scholar]
- 33.Park SY, Nomura AM, Murphy SP, Wilkens LR, Henderson BE, Kolonel LN. Carotenoid intake and colorectal cancer risk: the multiethnic cohort study. J Epidemiol. 2009;19(2):63–71. 10.2188/jea.JE20080078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brennan CA, Garrett WS. Gut Microbiota, Inflammation, and Colorectal Cancer. Annu Rev Microbiol. 2016. September 8;70:395–411. 10.1146/annurev-micro-102215-095513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson CH, Dejea CM, Edler D, Hoang LT, Santidrian AF, Felding BH, Ivanisevic J, et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 2015. June 2;21(6):891–7. 10.1016/j.cmet.2015.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farhana L, Nangia-Makker P, Arbit E, Shango K, Sarkar S, Mahmud H, Hadden T, Yu Y, Majumdar AP. Bile acid: a potential inducer of colon cancer stem cells. Stem Cell Res Ther. 2016. December 1;7(1):181 10.1186/s13287-016-0439-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bishayee A, Ahmed S, Brankov N, Perloff M. Triterpenoids as potential agents for the chemoprevention and therapy of breast cancer. Front Biosci (Landmark Ed). 2011. January 1;16:980–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
This data is available at the NIH Common Fund’s Metabolomics Data Repository and Coordinating Center (supported by NIH grant, U01-DK097430) website, the Metabolomics Workbench, http://www.metabolomicsworkbench.org, where it has been assigned Project ID PR000726. The data can be accessed directly via it’s Project DOI: 10.21228/M8TX0N. The raw 16S sequences are available in the NCBI SRA repository, where it has been assigned BioProject accession no. PRJNA325650.