Abstract
Premise of the Study
Gypsophila paniculata (baby's breath; Caryophyllaceae) is a herbaceous perennial that has invaded much of northern and western United States and Canada, outcompeting and crowding out native and endemic species. Microsatellite primers were developed to analyze the genetic structure of invasive populations.
Methods and Results
We identified 16 polymorphic nuclear microsatellite loci for G. paniculata out of 73 loci that successfully amplified from a primer library created using Illumina sequencing technology. Microsatellite primers were developed to amplify di‐, tri‐, and tetranucleotide repeats and tested in three invasive populations in Michigan.
Conclusions
These markers will be useful in characterizing the genetic structure of invasive populations throughout North America to aid targeted management efforts, and in native Eurasian populations to better understand invasion history. Five of these developed primers also amplified in G. elegans.
Keywords: Caryophyllaceae, genetic diversity, Gypsophila paniculata, invasive species, microsatellites
The herbaceous perennial forb Gypsophila paniculata L. (Caryophyllaceae) was introduced to North America in the late 1800s (Darwent and Coupland, 1966). Invasive populations have since been documented throughout the northern and western United States and Canada, specifically in agriculture fields, rangeland, roadsides, and sandy coastlines along the Great Lakes (Darwent, 1975; Emery and Doran, 2013). Despite its wide range, little information exists on how populations throughout North America are related or spreading. Due to its aggressiveness, negative impacts on native biota (Emery and Doran, 2013), and a lack of data regarding its spread, it is important to develop molecular markers that can characterize the genetic structure of invasive populations of G. paniculata. These markers were developed to investigate invasions within the Lake Michigan coastal dune system where an 1800‐acre infestation has occurred, so that land managers can use population genetic approaches to inform their management efforts. These markers and optimized protocols can also be used to characterize populations of G. paniculata throughout its invasive and native ranges to further assess its invasion history and spread.
Calistri et al. (2014) examined the genetic relationship of five Gypsophila Boiss. spp. (including G. paniculata) within their native range and 13 commercial hybrid strains using a combination of amplified fragment length polymorphisms (AFLPs), inter‐simple sequence repeats (ISSRs), target region amplification polymorphisms (TRAP), and universal chloroplast simple sequence repeats (cpSSRs). However, the majority of these markers are dominant and thus do not fully distinguish between homozygotes and heterozygotes, a characteristic that would allow for fine‐scale population genetic analyses (Freeland et al., 2011). Thus, the development of microsatellite markers for G. paniculata is necessary to adequately characterize invasive populations throughout North America.
METHODS AND RESULTS
Microsatellite library development, assembly, and identification
Adventitious buds growing from the caudex of five G. paniculata plants were collected from Sleeping Bear Dunes National Lakeshore (hereafter Sleeping Bear Dunes or SBDNL) in 2015 to develop the microsatellite library. Tissue was stored in indicator silica until DNA extraction. Genomic DNA was extracted using DNeasy Plant Mini Kits (QIAGEN, Hilden, Germany), with modifications including extra wash steps with AW2 buffer (QIAGEN). Extracted DNA was run through OneStep PCR Inhibitor Removal Columns (Zymo Research, Irvine, California, USA) twice to remove any inhibitory compounds (e.g., phenolics, humic acid) that could interfere with downstream processes, and was checked using a NanoDrop 2000 (Thermo Fisher Scientific, Waltham, Massachusetts, USA).
For microsatellite library development, each sample was diluted to 50 ng/μL and submitted to the Cornell Life Science Core Laboratory Center Sequencing and Genotyping Facility where microsatellite libraries were created and sequenced using a 2 × 250 paired‐end format on an Illumina MiSeq (Illumina, San Diego, California, USA; Appendix S1). Raw sequence files for the microsatellite library have been deposited to NCBI's Sequence Read Archive (BioProject no. PRJNA431197). A total of 58,907 contigs containing microsatellite loci were obtained (Appendix S2), and details on filtering, assembly, and microsatellite primer identification are expanded upon in the Supporting Information (Appendix S1). MSATCOMMANDER (version 1.0.3; Faircloth, 2008) identified 3892 potentially unique primers that yielded products of 150–450 bp, had a GC content between 30–70%, and had a melting temperature (T m) between 58–62°C, with an optimum of 60°C (Appendix S3).
Primer optimization
Prior to PCR optimization, contigs containing potential primers were aligned using Clustal Omega (Sievers et al., 2011) to ensure they were targeting unique microsatellite regions. We tested 107 primer pairs that consisted of either tetrameric, trimeric, or dimeric motifs, and yielded products between 150–300 bp. Of these, 73 successfully amplified (Appendix S4), and 16 were polymorphic and easily scorable (Appendix S5). DNA from leaf tissue collected in 2016 from three populations (Zetterberg Preserve, SBDNL, Petoskey State Park) along eastern Lake Michigan was used for primer optimization (Fig. 1). We collected tissue samples from a minimum of 30 individual plants from each population, and herbarium voucher specimens were collected for each location (Appendix 1). Tissue storage and DNA extraction methods are the same as previously stated.
Figure 1.
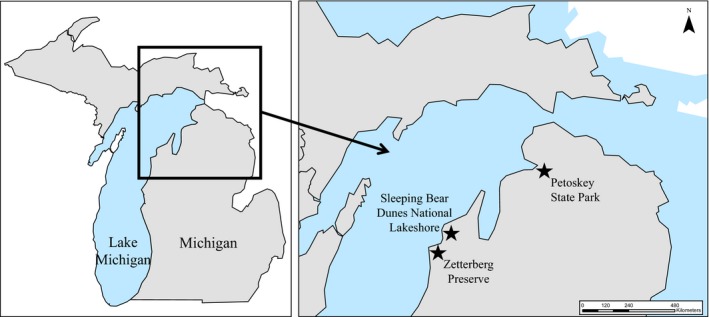
Map of Gypsophila paniculata sampling locations in the northwest corner of Michigan's Lower Peninsula.
PCR reactions consisted of 1× Taq + KCl buffer (Thermo Fisher Scientific), 2.0–2.5 mM MgCl2 (Table 1) (Thermo Fisher Scientific), 300 μM dNTP (New England BioLabs, Ipswich, Massachusetts, USA), 0.08 mg/mL bovine serum albumin (BSA; Thermo Fisher Scientific), 0.4 μM forward primer fluorescently labeled with either FAM, VIC, NED, or PET (Applied Biosystems, Foster City, California, USA), 0.4 μM reverse primer (Integrated DNA Technologies, Coralville, Iowa, USA), 0.25 units of recombinant Taq DNA polymerase 5 U/μL (Thermo Fisher Scientific), and a minimum of 50 ng of DNA template, all in a 10.0‐μL reaction volume. The thermal cycle profile consisted of 94°C for 5 min; 35 cycles of 94°C for 1 min, 62°C annealing temperature for 1 min, 72°C for 1 min; and a final elongation step of 72°C for 10 min. Successful amplification was determined by visualizing the amplicons on a 2% agarose gel stained with ethidium bromide. Fragment analysis of the amplicons was performed on an ABI 3130xl Genetic Analyzer (Applied Biosystems).
Table 1.
Characteristics of 16 polymorphic nuclear microsatellite loci developed for Gypsophila paniculata
| Locusa | Primer sequences (5′–3′) | Repeat motif | Allele size range (bp) | mM MgCl2 | Fluorescent label | GenBank accession no. |
|---|---|---|---|---|---|---|
| BB_3335 | F: TCCACCAAACTCTTAAACTGCC | (AGG)5 | 215–244 | 2.5 | NED | MH704701 |
| R: CACAGACACAAAGGATCCAACC | ||||||
| BB_3913 | F: GGCTGTCGGGTAATAAACACAG | (ACAG)5 | 159–171 | 2.0 | PET | MH704702 |
| R: TCCCAACTCAAGTCATAGCCTAG | ||||||
| BB_5567 | F: GGCTAGGGAAAGTAGGAAGACC | (ATT)5 | 198–222 | 2.0 | VIC | MH704703 |
| R: CGTGTCCTGTTTCTCCATGATC | ||||||
| BB_4443 | F: TAGGGTGGGTGCTTGTACTAAC | (AAG)16 | 171–211 | 2.0 | NED | MH704704 |
| R: AAAGTGGTGCTGCAGAAGAATC | ||||||
| BB_21680 | F: ACTACACACAGACTCGATCCTC | (AAAG)5 | 199–218 | 2.0 | PET | MH704705 |
| R: CTTTGATTGTTTGGTGTAAGTTGC | ||||||
| BB_3968 | F: CATGGAGGACAATGAGAAGACG | (AGG)6 | 207–219 | 2.0 | FAM | MH704706 |
| R: ACGGTGGTAATGAAGTTTGGTG | ||||||
| BB_1355 | F: GCTGATCTTTGTCGTCAGGAAG | (AAAC)5 | 220–224 | 2.0 | NED | MH704711 |
| R: ACTCTAGGTGTTAGGAAGGCAC | ||||||
| BB_5151 | F: TCCACCTTATAACTCACCACCC | (ACC)5 | 205–210 | 2.0 | PET | MH704712 |
| R: TGAGGAAGGATAACAGCTCTCG | ||||||
| BB_14751 | F: CCTCAAACCCTAACAATGCTCC | (AAG)12 | 195–248 | 2.5 | FAM | MH704713 |
| R: TCAGCCGATCCTCTAACACG | ||||||
| BB_4258 | F: TCACAAGAGGCCCAATTTCTTC | (AAT)5 | 178–195 | 2.5 | VIC | MH704714 |
| R: ACTTGAACCCGAACCTATACCC | ||||||
| BB_6627 | F: CAAACTCAACCAACCAGACACC | (AAAC)5 | 151–155 | 2.0 | FAM | MH704715 |
| R: CACCTCAGCAACAACAGAGTG | ||||||
| BB_5021 | F: ATTGTCGGTGGTCATTGGTTTC | (AC)8 | 162–207 | 2.0 | VIC | MH704707 |
| R: CTTAGTCCGCAGTGTAAACAAAG | ||||||
| BB_7213 | F: TTGCATTCCCACCATTTCATCC | (AC)7 | 161–248 | 2.5 | PET | MH704708 |
| R: AGCCAACCTCGTATTA21ATTGCC | ||||||
| BB_2888 | F: CTTCATTCATGTACAAGAGCGC | (AC)16 | 219–232 | 2.5 | FAM | MH704709 |
| R: AGAACTGGCTATGGATCGAAATG | ||||||
| BB_8681 | F: ATCTCCAGTTTCCGTGATTTGC | (ACC)8 | 204–222 | 2.5 | NED | MH704710 |
| R: TACGTCACAAGAGCTTTCAACC | ||||||
| BB_31555 | F: TGTATAACTGAGATAACCCAGACG | (AC)7 | 150–156 | 2.0 | VIC | MH704716 |
| R: TTGTTACCTTGTTCCGGCAAAG |
Optimal annealing temperature was 62°C for all loci except for BB_2888, where it was 63°C.
Microsatellite marker data analysis
Alleles were scored using GeneMapper version 5 (Applied Bio‐systems), and MICRO‐CHECKER version 2.2.3 (van Oosterhout et al., 2004, 2006) was used to identify null alleles and potential scoring errors from stuttering or large allele dropout. There was no significant evidence of null alleles (P > 0.05) in the Zetterberg Preserve and SBDNL populations. However, null alleles were suggested for loci BB_3968, BB_5021, and BB_8681 in the Petoskey State Park population (Table 2). Homozygote excess for the Petoskey State Park population is not surprising, given this population's reduced number of alleles at each locus and small comparative population size. We characterized genetic diversity by examining the number of alleles and expected and observed heterozygosity for each locus averaged over each population (Table 2) using the package STRATAG in the R statistical program (Archer et al., 2016). The number of observed alleles ranged from one to 10. Some loci were monomorphic for one population but polymorphic when analysis included all populations (e.g., BB_4258).
Table 2.
Genetic properties of the 16 developed polymorphic nuclear microsatellite loci for three populations of Gypsophila paniculata.a
| Locus | Zetterberg Preserve (n = 30) | Sleeping Bear Dunes (n = 33) | Petoskey State Park (n = 30) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | H o b | H e | F IS | r | A | H o b | H e | F IS | r | A | H o b | H e | F IS | r | |
| BB_3335 | 8 | 0.70 | 0.82 | 0.09 | 0.07 | 7 | 0.80 | 0.80 | −0.01 | 0.00 | 3 | 0.50 | 0.56 | 0.02 | 0.05 |
| BB_3913 | 4 | 0.74 | 0.68 | −0.04 | −0.06 | 4 | 0.59 | 0.61 | 0.05 | 0.00 | 2 | 0.13 | 0.13 | −0.06 | −0.06 |
| BB_5567 | 4 | 0.63 | 0.60 | −0.04 | −0.04 | 4 | 0.63 | 0.63 | 0.01 | −0.01 | 3 | 0.53 | 0.62 | 0.14 | 0.05 |
| BB_4443 | 10 | 0.60 | 0.64 | 0.03 | 0.03 | 8 | 0.76 | 0.77 | −0.02 | 0.00 | 5 | 0.80 | 0.71 | −0.06 | −0.09 |
| BB_21680 | 4 | 0.53 | 0.56 | 0.01 | 0.02 | 3 | 0.52 | 0.56 | 0.02 | 0.05 | 4 | 0.67* | 0.58 | 0.23 | −0.10 |
| BB_3968 | 3 | 0.50 | 0.42 | −0.14 | −0.11 | 4 | 0.35* | 0.49 | 0.15 | 0.13 | 2 | 0.04 | 0.04 | — | 0.28 |
| BB_1355 | 2 | 0.17 | 0.26 | 0.37 | 0.13 | 2 | 0.14 | 0.13 | −0.07 | −0.08 | 2 | 0.37 | 0.51 | 0.29 | 0.13 |
| BB_5151 | 2 | 0.20 | 0.18 | −0.10 | −0.11 | 2 | 0.49 | 0.51 | 0.11 | 0.00 | 2 | 0.03 | 0.03 | — | −0.02 |
| BB_14751 | 6 | 0.79 | 0.71 | −0.06 | −0.07 | 6 | 0.76 | 0.76 | −0.02 | −0.01 | 3 | 0.20 | 0.29 | 0.21 | 0.12 |
| BB_4258 | 3 | 0.40 | 0.33 | −0.11 | −0.22 | 1 | 0.00 | 0.00 | — | 0.00 | 1 | — | — | — | 0.00 |
| BB_6627 | 2 | 0.30 | 0.26 | −0.16 | −0.16 | 2 | 0.46 | 0.49 | −0.03 | 0.04 | 1 | — | — | — | 0.00 |
| BB_5021 | 5 | 0.77 | 0.73 | 0.00 | −0.05 | 5 | 0.54 | 0.60 | 0.15 | 0.02 | 1 | — | — | — | 0.30 |
| BB_7213 | 5 | 0.63 | 0.61 | −0.05 | −0.02 | 3 | 0.59 | 0.58 | −0.13 | 0.04 | 2 | 0.37 | 0.38 | 0.04 | 0.01 |
| BB_2888 | 6 | 0.90 | 0.80 | −0.11 | −0.07 | 6 | 0.71 | 0.82 | 0.20 | 0.06 | 2 | 0.57 | 0.50 | −0.13 | −0.08 |
| BB_8681 | 4 | 0.47 | 0.51 | 0.23 | 0.01 | 4 | 0.38 | 0.36 | −0.07 | −0.05 | 3 | 0.41 | 0.53 | 0.28 | 0.17 |
| BB_31555 | 4 | 0.60 | 0.67 | 0.07 | 0.05 | 4 | 0.46 | 0.59 | 0.11 | 0.12 | 2 | 0.23 | 0.26 | 0.10 | 0.04 |
— = data not available because only one allele present at locus or because A > 1 is due to rare allele; A = number of alleles; F IS = fixation index; H e = expected heterozygosity; H o = observed heterozygosity; n = number of individuals sampled; r = null allele frequency.
aVoucher and locality information for the populations are provided in Appendix 1.
bSignificant deviation from Hardy–Weinberg equilibrium (*P < 0.05).
The Zetterberg Preserve population displayed slightly higher heterozygosity values (0.17–0.90) than Sleeping Bear Dunes (0.14–0.80), but the Petoskey State Park population (0.03–0.80) had much lower heterozygosity values in comparison. A probability test for Hardy–Weinberg equilibrium (HWE), calculation of the fixation index (F IS), and linkage disequilibrium were performed in GENEPOP 4.2 (Raymond and Rousset, 1995; Rousset, 2008). The default parameters for Markov chain Monte Carlo (MCMC) iterations were used to calculate HWE. All loci were in HWE except locus BB_3968 for SBDNL and locus BB_21680 for Petoskey State Park (Table 2). The F IS estimates were calculated using the probability model following Robertson and Hill (1984). Statistical tests for genetic linkage disequilibrium were performed using the log likelihood ratio statistic (G‐test) and MCMC algorithm by Raymond and Rousset (1995). Two pairs of loci were significantly out of linkage disequilibrium (P < 0.05) for both the Zetterberg Preserve and Sleeping Bear Dunes populations (BB_5021 and BB_2888, BB_3913 and BB_1355). Out of 16 loci, five successfully amplified in the related G. elegans M. Bieb. (BB_4443, BB_4258, BB_7213, BB_5151, BB_1355) (Table 3).
Table 3.
Results of cross‐amplification of microsatellite loci isolated from Gypsophila paniculata and tested in 12 G. elegans individuals.a
| Locus | Allele size range (bp) | GYEL 01 | GYEL02 | GYEL03 | GYEL04 | GYEL07 | GYEL08 | GYEL09 | GYEL12 | GYEL13 | GYEL15 | GYEL18 | GYEL19 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BB_3335 | |||||||||||||
| BB_3913 | |||||||||||||
| BB_5567 | |||||||||||||
| BB_4443 | 152 | X | X | X | X | X | X | X | X | X | X | X | X |
| BB_21680 | |||||||||||||
| BB_3968 | |||||||||||||
| BB_1355 | 209–222 | X | X | X | X | X | X | X | X | X | X | X | X |
| BB_5151 | 202–205 | X | X | X | X | X | X | X | X | X | X | X | X |
| BB_14751 | |||||||||||||
| BB_4258 | 160–169 | X | X | X | X | X | X | X | X | X | X | X | X |
| BB_6627 | |||||||||||||
| BB_5021 | |||||||||||||
| BB_7213 | 161–171 | X | X | X | X | X | X | X | X | X | X | X | X |
| BB_2888 | |||||||||||||
| BB_8681 | |||||||||||||
| BB_31555 |
X = successful amplification in each G. elegans individual (n = 12).
Gypsophila elegans tissue was sourced from individuals grown from seed purchased from Everwilde Farms (#FGYPELE‐05, lots 36863 and 39619) in a greenhouse at the AWRI‐GVSU facilities.
CONCLUSIONS
The 16 microsatellite primers developed for G. paniculata provide a tool for estimating genetic diversity and structure of invasive populations, which will aid in understanding its invasion history, identifying source populations, and examining dispersal patterns. Although we developed these markers to specifically study the Lake Michigan dune system populations, G. paniculata is invasive throughout North America. With these markers, we can begin to understand the invasion of G. paniculata in North America in order to improve management efforts and prevent the further spread of this species.
DATA ACCESSIBILITY
A summary of the microsatellite library development and sequence analysis protocols provided to us by the Department of Ecology and Evolutionary Biology at Cornell University are in Appendix S1. FASTA sequences for the 16 microsatellite primers developed here are in Appendix S5. The FASTA file listing all identified contigs containing microsatellite regions is in Appendix S2. Potential primer pairs for the identified microsatellite‐containing contigs are in Appendix S3. The 107 G. paniculata–specific primer pairs tested during primer optimization are in Appendix S4. Raw sequence files for the microsatellite library have been deposited to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (BioProject no. PRJNA431197), and microsatellite sequences have been deposited to GenBank (Table 1). Voucher specimens for each population have been deposited into the Grand Valley State University Herbarium (GVSC), Grand Valley State University Department of Biology, Allendale, Michigan, USA (Appendix 1).
Supporting information
APPENDIX S1. Microsatellite library development and sequencing methods for Gypsophila paniculata.
APPENDIX S2. A total of 58,907 contigs potentially containing microsatellite regions from Gypsophila paniculata.
APPENDIX S3. Information about 7904 unique microsatellite primer pairs identified by MSATCOMMANDER version 1.0.3 for Gypsophila paniculata.
APPENDIX S4. One hundred and seven Gypsophila paniculata–specific microsatellite primer pairs tested during primer optimization.
APPENDIX S5. Sequences for the 16 Gypsophila paniculata microsatellite loci.
ACKNOWLEDGMENTS
The authors thank the Grand Valley State University Center for Scholarly and Creative Excellence Catalyst Grant (C.G.P.) and the Environmental Protection Agency–Great Lakes Restoration Initiative Grant (C.G.P., grant no. 00E01934) for financial support. The authors also thank E. K. Rice for help in field sampling and M. Kienitz for help in the laboratory.
APPENDIX 1. Voucher and location information for Gypsophila paniculata populations used in this study.
| Voucher specimen accession no.a | Collection locality | Geographic coordinates | N |
|---|---|---|---|
| VPSMI551, VPSMI554, VPSMI569–VPSMI571 | Petoskey State Park, Emmet County, MI, USA | 45.40288418°N, 84.91271857°W | 30 |
| VSBMI001–VSBMI016 | Sleeping Bear Dunes National Lakeshore, Leelanau County, MI, USA | 44.87372°N, 86.06170°W | 33 |
| VZPMI001–VZPMI005 | Zetterberg Preserve at Point Betsie, Benzie County, MI, USA | 44.68231°N, 86.25322°W | 30 |
N = number of individuals sampled.
Vouchers deposited at Grand Valley State University Herbarium (GVSC), Grand Valley State University Department of Biology, Allendale, Michigan, USA.
Leimbach‐Maus, H. B. , Parks S. R., and Partridge C. G.. 2018. Microsatellite primer development for the invasive perennial herb Gypsophila paniculata (Caryophyllaceae). Applications in Plant Sciences 6(12): e1203.
LITERATURE CITED
- Archer, F. I. , Adams P. E., and Schneiders B. B.. 2016. STRATAG: An R package for manipulating, summarizing and analysing population genetic data. Molecular Ecology Resources 17: 5–11. [DOI] [PubMed] [Google Scholar]
- Calistri, E. , Buiatti M., and Bogani P.. 2014. Characterization of Gypsophila species and commercial hybrids with nuclear whole‐genome and cytoplasmic molecular markers. Plant Biosystems 150: 1–11. [Google Scholar]
- Darwent, A. L. 1975. The biology of Canadian weeds. 14. Gypsophila paniculata L. Canadian Journal of Plant Science 55: 1049–1058. [Google Scholar]
- Darwent, A. L. , and Coupland R. T.. 1966. Life history of Gypsophila paniculata . Weeds 14: 313–318. [Google Scholar]
- Emery, S. M. , and Doran P. J.. 2013. Presence and management of the invasive plant Gypsophila paniculata (baby's breath) on sand dunes alters arthropod abundance and community structure. Biological Conservation 161: 174–181. [Google Scholar]
- Faircloth, B. C. 2008. MSATCOMMANDER: Detection of microsatellite repeat arrays and automated, locus‐specific primer design. Molecular Ecology Resources 8: 92–94. [DOI] [PubMed] [Google Scholar]
- Freeland, J. R. , Petersen S. D., and Kirk H.. 2011. Molecular ecology, 2nd ed Wiley‐Blackwell, Hoboken, New Jersey, USA. [Google Scholar]
- van Oosterhout, C. , Hutchinson W. F., Wills D. P. M., and Shipley P.. 2004. MICRO‐CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Resources 4: 535–538. [Google Scholar]
- van Oosterhout, C. , Weetman D., and Hutchinson W. F.. 2006. Estimation and adjustment of microsatellite null alleles in nonequilibrium populations. Molecular Ecology Resources 6: 255–256. [Google Scholar]
- Raymond, M. , and Rousset F.. 1995. GENEPOP (version 4.2): Population genetics software for exact tests and ecumenicism. Journal of Heredity 86: 248–249. [Google Scholar]
- Robertson, A. , and Hill W. G.. 1984. Deviations from Hardy‐Weinberg proportions: Sampling variances and use in estimation of inbreeding coefficients. Genetics 107: 703–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset, F. 2008. GENEPOP'007: A complete reimplementation of the GENEPOP software for Windows and Linux. Molecular Ecology Resources 8: 103–106. [DOI] [PubMed] [Google Scholar]
- Sievers, F. , Wilm A., Dineen D. G., Gibson T. J., Karplus K., Li W., Lopez R., et al. 2011. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
APPENDIX S1. Microsatellite library development and sequencing methods for Gypsophila paniculata.
APPENDIX S2. A total of 58,907 contigs potentially containing microsatellite regions from Gypsophila paniculata.
APPENDIX S3. Information about 7904 unique microsatellite primer pairs identified by MSATCOMMANDER version 1.0.3 for Gypsophila paniculata.
APPENDIX S4. One hundred and seven Gypsophila paniculata–specific microsatellite primer pairs tested during primer optimization.
APPENDIX S5. Sequences for the 16 Gypsophila paniculata microsatellite loci.
Data Availability Statement
A summary of the microsatellite library development and sequence analysis protocols provided to us by the Department of Ecology and Evolutionary Biology at Cornell University are in Appendix S1. FASTA sequences for the 16 microsatellite primers developed here are in Appendix S5. The FASTA file listing all identified contigs containing microsatellite regions is in Appendix S2. Potential primer pairs for the identified microsatellite‐containing contigs are in Appendix S3. The 107 G. paniculata–specific primer pairs tested during primer optimization are in Appendix S4. Raw sequence files for the microsatellite library have been deposited to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (BioProject no. PRJNA431197), and microsatellite sequences have been deposited to GenBank (Table 1). Voucher specimens for each population have been deposited into the Grand Valley State University Herbarium (GVSC), Grand Valley State University Department of Biology, Allendale, Michigan, USA (Appendix 1).


