Abstract
Contamination of water by meat production is an important and extensive environmental problem and even threat to human health. Biodegradation is a major mechanism which removes the pollutants from the environment. Therefore, the present study aimed to isolate and characterize a COD degrading bacteria which can effectively degrade slaughter wastewater. Six COD degrading bacteria were isolated from slaughtering waste water and sludge in Hunan a meat product Co., Ltd. And the COD degradation rate of each strain was determined by potassium permanganate method. Through observing morphologically and analyzing sequence to 16S rDNA, the highest COD degradation strain was Bacillus velezensis by preliminarily identified and classified, reaching 11.80%. The suitable conditions of the growth of Bacillus velezensis strain were 37 °C, pH 7.0, the peptone concentration 1.5%, and the yeast extract concentration 0.8%.
Keywords: Degrading bacteria, Bacillus velezensis, Identification, Characterization, Slaughter wastewater
1. Introduction
Slaughterhouse waste water can cause serious pollution of the surrounding water environment if it is discharged directly and without disposal. The health of humans, livestock and poultry, and other aquatic organisms can be seriously endangered because of the large number of virulence factors and microorganisms in the waste water that are harmful to human health. If slaughter wastewater is drained into the surrounding water, the dissolved oxygen in the water will be rapidly depleted, causing the fish and aquatic organisms to die of lack of oxygen, because the outward slaughtering wastewater containing harmful substances are mostly easy biodegradable organic matter. At the same time, pathogenic microorganisms will also be produced in large quantities with the waste water discharged into the water, which seriously endangers the health of human beings. Therefore, it is necessary to deal with the wastewater from slaughtering meat processing, remove the pollution caused by waste water and completely eliminate the harm of water environment, so as to protect the ecological environment and human health (He and Xing, 2006, Chávez et al., 2005, Del et al., 2016, Tong, 2014).
Among the various methods proposed for the removal of these compounds, those based on microbial activities are of greater priority (Samaei et al., 2013). Microbial treatment method is a low cost, no environmental pollution and thorough purification method compared with chemical treatment and physical treatment method in the treatment of slaughterhouse wastewater, will be a main method to solve the slaughterhouse wastewater pollution in the future (Yue et al., 2015, Sun, 2008, Jia et al., 2015, Kundu et al., 2014). The most important is that to have excellent ability of degrading bacteria in microbial treatment method in the process of implementation, using microorganism to degrade organic pollutants in slaughter wastewater is the better method (Fan et al., 2003, Semblante et al., 2015, Wu et al., 2013, Song et al., 2016, Bustillo-Lecompte and Mehrvar, 2015).
Up to now, a large number of bacterial strains capable of degrading COD have been isolated (Del et al., 2016, Tong, 2014). Most reports have shown that COD-degrading bacteria isolated from different contaminated sites are belonging to various bacterial species, such as ammonia bacteria, nitrifying bacteria and denitrifying bacteria (Piubeli et al., 2012, Shehzadi et al., 2014). Overall, slaughter wastewater contains a large amount of nitrogen-containing organic matter such as blood, animal internal organs and hair, and is easy to breed a large number of harmful organisms. Rapid and efficient degradation of slaughter wastewater is of great significance for environmental protection.
In this study, six high COD degrading bacteria were isolated from slaughter wastewater and identified preliminarily and investigated their ability to remove COD from water. Then, one of them with highest ability to remove COD from water was chosen, which of their physiological and biochemical characteristics were preliminarily studied in this paper.
2. Materials and methods
2.1. Isolation source
The slaughter wastewater and sludge contaminated with COD strain were the source for bacterial isolation. Samples were collected into 1000 ml sterilized glass bottles from eight sites in a Hunan meat product Co., Ltd on June 2016, and immediately carried to the laboratory and stored in a refrigerator prior at 4 °C to use.
2.2. Enrichment culture of strain
The slaughterhouse wastewater and sludge mixed samples were taken for 10 ml and filtered with gauze respectively. The samples were diluted and diluted to 10−1, 10−2, 10−3, 10−4, 10−5, 10−6, 10−7 respectively. In the sterile operating table, samples with different dilution concentrations were evenly coated in LB solid medium for 200 μL, and cultured in 32 °C incubator for 5–7 d (Su et al., 2017, Lu and Wang, 2016).
2.3. Isolation and purification of strains
The colonies were randomly picked enrichment lines separation, then were isolated single colonies obtained further separated to obtain single colonies of pure purification line, then pick a single colony is connected to the LB liquid medium, then add 4 °C refrigerator spare (Su et al., 2017, Lu and Wang, 2016, Duan et al., 2016).
2.4. Determination of degradation ability of strain COD
Ammonium sulfate was added to LB culture medium, and the concentration of ammonium sulfate in culture medium was 0.3 g/100 ml under aseptic condition and was used to replace slaughterhouse wastewater, because it was closer to the content of reducing substance in real slaughter wastewater. The 10 ml medium with purified strains were isolated from the slaughter waste water were inoculated in 90 ml LB medium, shaker culture at 32 °C, at regular intervals sampling and determination of COD in the medium by potassium permanganate method to calculate the degradation rate and draw the degradation curve (Zhang et al., 2016).
2.5. Molecular biological identification of high COD degrading bacteria
The bacteria with the highest degradation rate of COD were observed by colony morpHology and Gram staining. Preliminary identification of the strains was conducted according to Bergey's Manual of Systema Bacteriology (Su et al., 2017, Feng et al., 2012). Genetic identification of the isolated bacteria was done using 16S rDNA. Bacterial DNA was extracted using Ezup Column Bacteria Genomic DNA Purification Kit (Sangon Biotech, China), and bacterial 16S rDNA was amplified with universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGCTACCTTGTTACGACTT-3′), which had been used by other researchers for similar purposes (Molina et al., 2009). PCR was performed using 1 μL of pooled DNA as a template using primers 27F and 1492R and 25 μL Prime mastermix and 20 μL sterile water in a 100 μL reaction. The PCR program began with 4 min at 94 °C followed by 32 cycles of 30 s at 94 °C, 30 s at 60 °C, and 2 min at 72 °C, with a final elongation of 10 min at 72 °C. Phylogenetic analysis of clone sequences was conducted in MEGA version 5 (Tamura et al., 2011).
2.6. Physiological and biochemical characteristics of COD degrading bacteria
According to the test results of isolated strains of COD degrading ability, evaluation index for OD strains in different culture time. The growth temperature of the highest degradation rate of COD strain, the growth of pH, peptone and yeast extract concentration characteristics were studied to determine the optimum growth conditions (Zhang et al., 2017, Li et al., 2015, Tan et al., 2016).
2.6.1. Initial temperature test
The effect of different temperature on bacterial growth was determined by incubation of bacterial cells with 100 ml base culture in 250 ml conical flask with different initial temperature values (32 °C, 35 °C, 37 °C and 40 °C), 120 rpm and 7.0 pH. The growth was monitored by determinations of OD600 from 2 to 24 h as blank with basic medium.
2.6.2. Initial pH test
The effect of different temperature on bacterial growth was determined by incubation of bacterial cells with 100 ml base culture in 250 ml conical flask with different initial pH values (5.0, 6.0, 7.0 and 8.0) at 37 °C, 120 rpm. The growth was monitored by determinations of OD600 from 2 to 24 h as blank with basic medium.
2.6.3. Initial peptone concentration test
Peptone is the main nitrogen source and carbon source for microbial growth. The effect of different temperature on bacterial growth was determined by incubation of bacterial cells with 100 ml base culture in 250 ml conical flask with different initial peptone concentration (1.0%, 1.2%, 1.5%, 1.8% and 2.0%) at 37 °C, 120 rpm and 7.0 pH. The growth was monitored by determinations of OD600 from 2 to 24 h as blank with basic medium.
2.6.4. Initial yeast extracts concentration test
Yeast extract is also a major source of nitrogen and carbon for the growth of microorganisms. The effect of different temperature on bacterial growth was determined by incubation of bacterial cells with 100 ml base culture in 250 ml conical flask with different initial yeast extracts concentration (0.5%, 0.8%, 1.0% and 1.2%) at 37 °C, 120 rpm and 7.0 pH. The growth was monitored by determinations of OD600 from 2 to 24 h as blank with basic medium.
3. Results and analysis
3.1. Isolation and purification of strains
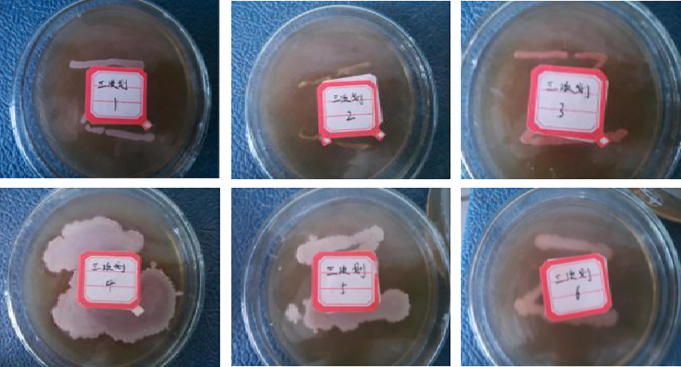
Six pure strains were isolated and purified from the enrichment culture plate, and the line culture was shown in Fig. 1.
Fig. 1.
Purified the isolated strain.
3.2. Test results of degradation rate of strain COD
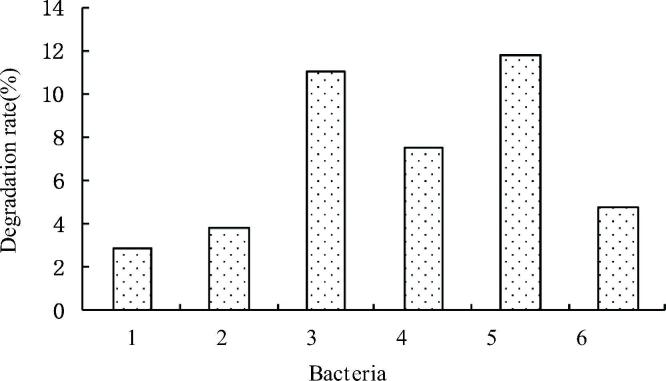
The COD degrading bacteria purification rate of six pure strains were detected by Potassium Permanganate method, and the results were shown in Fig. 2. Six strains of COD bacteria have different degradation ability, of which No. 1 degradation ability is relatively weak, the degradation ability of 5 strains of 11.79% is the strongest. Therefore, the strains of number 5 were preliminarily identified, and their physiological characteristics were further studied.
Fig. 2.
Degradation rate of the six strains.
3.3. Molecular identification
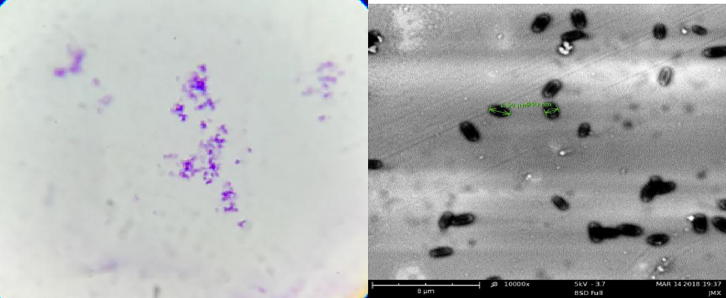
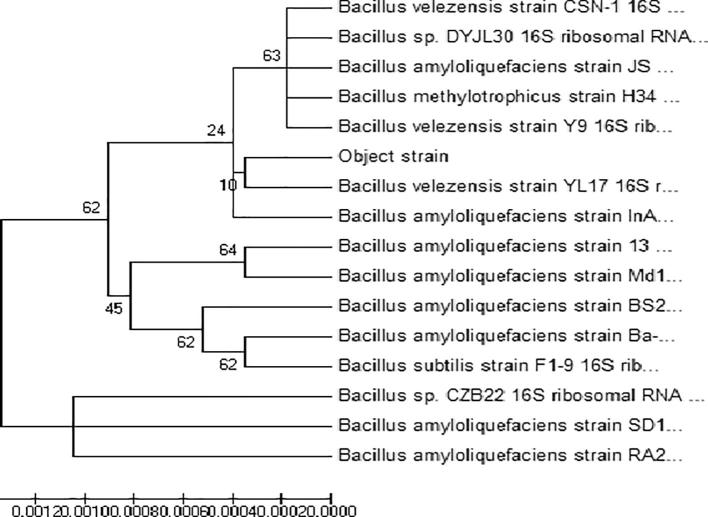
The 16S rDNA gene was amplified via PCR using a universal bacterial primer set, which was obtained from the isolates. On the basis of the observation of the morphology, Gram staining, the determination of biochemistry as well as the sequent analyse of 16S rDNA, the No. 5 bacteria was identified as Bacillus velezensis strain (Cai et al., 2017). The Gram staining and phylogenetic tree of No. 5 bacteria showed in Fig. 3 and Fig. 4.
Fig. 3.
Gram staining of NO.5 strain.
Fig. 4.
Neighbor-joining pHylogenetic tree constructed from 16S rDNA gene sequence.
3.4. Biological characteristics of high COD degrading bacteria
3.4.1. Optimum growth temperature
Temperature, as one of the important factors, affects the growth and reproduction of microorganisms, determine the high COD degradation bacteria optimum growth temperature can provide the basis for better using the strain. The growth curve of Bacillus velezensis strain is shown in Fig. 5 at different culture temperatures, the biomass has obvious lag period at 5 h, entered the logarithmic growth phase, and then into the slowly stable after 8 h. It was found that when the temperature was 32 °C, 35 °C, 37 °C and 40 °C, the growth curve of Bacillus velezensis strain were similar. However the biomass was the highest at 37 °C, 37 °C was chosen as the optimum fermentation temperature.
Fig. 5.
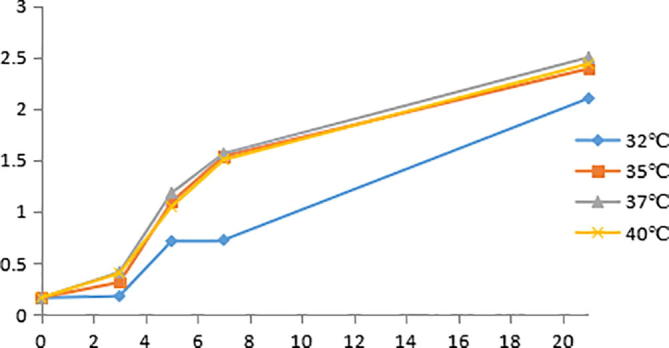
Growth curves of Bacillus velezensis strain at different temperatures.
3.4.2. Strain optimum growth pH
As can be seen from Fig. 6, the growth of Bacillus velezensis strain was inhibited at pH 5 and 8 after 6 h, while was increased at pH 6 and 7. And it was very slow due to the inhibition of the environment after 8 h at pH 6 and 7. It was found that the growth curve of Bacillus velezensis strain were similar at pH 6 and pH 7 However the biomass was the highest at pH 7, pH 7 was chosen as the optimum fermentation pH.
Fig. 6.
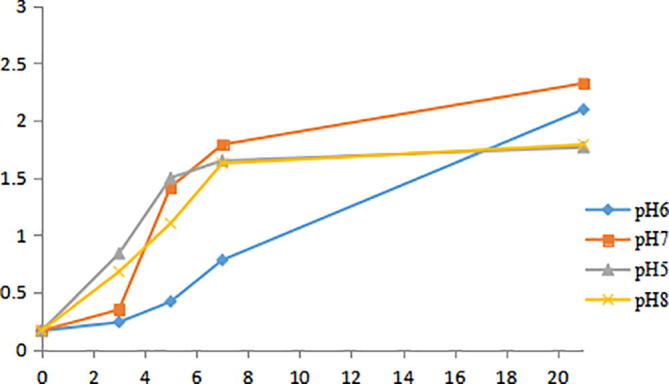
Growth curves of Bacillus velezensis strain at different pH.
3.4.3. Effect of peptone concentration on the growth of the strain
Peptone is the main nitrogen source and carbon source for microbial growth. The effect of peptone with different concentration in culture medium on the growth of Bacillus velezensis strain was studied in this paper. The results are shown in Fig. 7, it can be seen that the growth curve of strain had no significant effect in a certain concentration range of peptone, but the biomass of Bacillus velezensis strain is relatively high at the 1.5% peptone. Therefore, 1.5% peptone was chosen as the optimum fermentation peptone concentration.
Fig. 7.
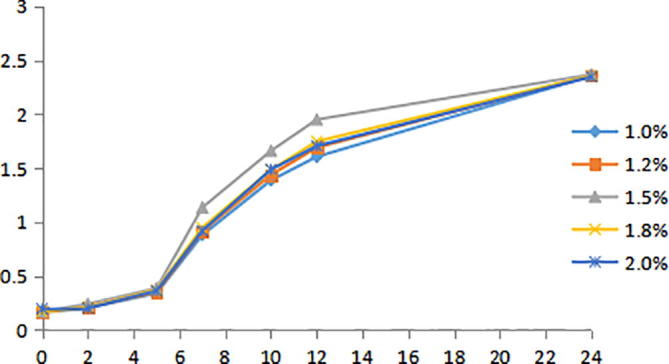
Growth curves of NO.3 strain at different concentration of peptone.
3.4.4. Effect of yeast extracts concentration on the growth of the strain
Yeast extract is also a major source of nitrogen and carbon for the growth of microorganisms. The effect of yeast extract at different concentrations on the growth of Bacillus velezensis strain was shown in Fig. 8. The growth curve of Bacillus velezensis strain was affected by the concentration of yeast extract is not obvious, but the biomass of Bacillus velezensis strain was higher than in other group in 0.8% concentration, so the culture concentration of yeast extract in the medium can choose 0.8% was chosen as the optimum fermentation yeast extracts concentration.
Fig. 8.
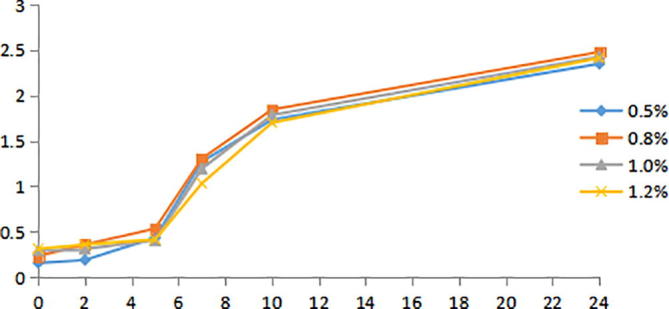
Growth curves of Bacillus velezensis strain at different concentration of yeast extract.
4. Discussions
Slaughter wastewater contains a large amount of nitrogen-containing organic matter such as blood, animal internal organs and hair, and is easy to breed a large number of harmful organisms. COD is an important factor to measure the total amount of organic matter in water. Rapid and efficient degradation of slaughter wastewater is of great significance for environmental protection. A large number of bacterial strains capable of degrading COD have been isolated (Del et al., 2016, Tong, 2014, Piubeli et al., 2012, Shehzadi et al., 2014). Most reports have shown that COD-degrading bacteria isolated from different contaminated sites are belonging to various bacterial species, such as ammonia bacteria, nitrifying bacteria and denitrifying bacteria.
It is reported that Bacillus has the function of purifying water. In this study, the Bacillus velezensis was identified for the colony morphology and the molecular characteristics by the enrichment and separation from slaughter wastewater in a Hunan meat product Co., Ltd. which was effectively degraded COD. Moreover, Bacillus velezensis was first found by Spanish scholars in Magaraga in southern Spain, identifying by constructing phylogenetic tree of 16S rDNA gene sequence analysis (Meng and Hao, 2017). Some scientific researchers were shown that Bacillus velezensis can produce a variety of digestive enzymes such as amylase, protease, gelatinase, glucanase, and cellulase, and degradate strong organic matter decomposition. Moreover, Bacillus velezensis can inhibit the growth and reproduction of harmful microorganisms, and can decompose organic substances, organic sulfides, and organic nitrogen (Wu et al., 2013, Meng and Hao, 2017, Rodgersvieira et al., 2015, Samaei et al., 2013).
The growth of the Bacillus velezensis was studied in terms of the effects of temperature, pH, peptone and yeast extract concentration characteristics, and as a result, Different conditions had great effects on the COD removal rate of Bacillus velezensis. The effects of temperature, pH, and the growth of pH, peptone and yeast extract concentration were studied to determine the optimum growth conditions (Zhang et al., 2017, Li et al., 2015, Tan et al., 2016, Meng and Hao, 2017, Rodgersvieira et al., 2015). Which were selected to do single factor test for further study of the optimum conditions for biomass, which was an ability to degrade COD because of production-associated strain.
COD degradation is limited by various conditions in slaughter wastewater and sludge (Su et al., 2017, Molina et al., 2009, Tamura et al., 2011), which propose that they may have been evolved through adaptation and selective pressure of natural ecosystems, which has resulted in selection of more potentiated organisms in biodegradation. The combination of COD degrading bacteria can degrade a variety of organic species mixture greatly improve the removal rate of COD, due to the presence of strains capable of decomposition and degradation of surfactants, hydrocarbons, phenols, fatty acids, ketones and other easily decomposed organic matter (Samaei et al., 2013, Taheri et al., 2017, Al-Enazi et al., 2018, Gao et al., 2017, Khan et al., 2018, Xie et al., 2018, Hajare and More, 2017). Moreover, Microbial immobilization technology was occure, this technology was beneficial to increase the number of microorganisms in the reactor, facilitate the solid-liquid separation after the reaction, facilitate nitrogen removal, remove high-concentration organic matter or difficult to biodegrade substances, improve system processing capacity and adaptability, and so on. As a result, they have received increasing attention in research on immobilized microorganism for wastewater treatment (Bai et al., 2010, Li et al., 2014, Ali et al., 2015). A high efficiency and stability biological carrier-immobilized Bacillus velezensis strain system was constructed to remove the chemical oxygen demand (COD) based on simultaneous adsorption and biodegradation (SAB) for the treatment of slaughter wastewater in the further research, the development and utilization of the genetic resources of Bacillus velezensis should be further studied.
5. Conclusions
In conclusion, this work provides relevant information about the efficiency of 6 COD strains by separating from slaughtering waste water. The results showed that the highest COD degradation rate was No. 5 strain, reaching 11.79%. And molecular Biological identification, colony morphology and Gram staining showed as Bacillus velezensis strain. The suitable conditions of the growth of Bacillus velezensis strain were 37 °C, pH 7.0, the peptone concentration 1.5%, and the yeast extract concentration 0.8%.
Acknowledgments
This work was supported by the National Key Technology R&D Program (2015BAD05B02), the National Natural Science Foundation of China (31501538, 51774128 and 21605046), the Natural Science Foundation of Hunan Province of China (2015JJ2049, 2015JJ3062, 2016JJ3053, 2017JJ4032, 2018JJ4061, 2018JJ2090 and 2018JJ4009), the key program of Hunan Provincial Department of science and technology (2016NK2096), Huxiang Youth Talent Support Program (2015RS4051),the Scientific Research Fund of Hunan Provincial Education Department (16C0470, YB2016B034, 17A055 and 17C0487), the China of Postdoctoral Science Foundation (2016T90769, 2016M592456, 2015M580707), Zhu zhou Key Science & Technology Program of Hunan Province (2017 and 2018), the China of Undergraduate Innovative Experiment Program (201511535003), Hunan Province Undergraduate Innovative Experiment Program (2015) and Green Packaging and Security Special Research Fund of China Packaging Federation (2017ZBLY14).
Footnotes
Peer review under responsibility of King Saud University.
References
- Al-Enazi N.M., Awaad A.S., Zain M.E., Alqasoumi S.I. Antimicrobial, antioxidant and anticancer activities of Laurencia catarinensis, Laurencia majuscula and Padina pavonica extracts. Saudi Pharm. J. 2018;26(1):44–52. doi: 10.1016/j.jsps.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M., Oshiki M., Rathnayake L., Ishii S., Satoh H., Okabe S. Rapid and successful start-up of anammox process by immobilizing the minimal quantity of biomass in PVA-SA gel beads. Water Res. 2015;79:147. doi: 10.1016/j.watres.2015.04.024. [DOI] [PubMed] [Google Scholar]
- Bai X., Ye Z.F., Li Y.F., Zhou L.C., Yang L.Q. Preparation of crosslinked macroporous PVA foam carrier for immobilization of microorganisms. Process Biochem. 2010;45(1):60–66. [Google Scholar]
- Bustillo-Lecompte C.F., Mehrvar M. Slaughterhouse wastewater characteristics, treatment, and management in the meat processing industry: a review on trends and advances. J. Environ. Manage. 2015;161:287–302. doi: 10.1016/j.jenvman.2015.07.008. [DOI] [PubMed] [Google Scholar]
- Cai X.C., Xi H., Liang L., Liu C.H., Xue Y.R., Yu X.Y. Rifampicin-resistance mutations in the rpoB Gene in Bacillus velezensis CC09 have pleiotropic effects. Front. Microbiol. 2017;8:178. doi: 10.3389/fmicb.2017.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez P.C., Castillo L.R., Dendooven L., Escamilla-Silva E.M. Poultry slaughter wastewater treatment with an up-flow anaerobic sludge blanket (UASB) reactor. Bioresource Technol. 2005;96(15):1730–1736. doi: 10.1016/j.biortech.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Del N.V., Damianovic M.H.Z., Moura R.B., Pozzi E., Pries E.C., Foresti E. Poultry slaughterhouse wastewater treatment plant for high quality effluent. Water Sci. Technol. 2016;73(2):309–316. doi: 10.2166/wst.2015.494. [DOI] [PubMed] [Google Scholar]
- Duan J., Huo X., Du W.J., Liang J.D., Wang D.Q., Yang S.C. Biodegradation of kraft lignin by a newly isolated anaerobic bacterial strain, Acetoanaerobium sp.WJDL-Y2. Lett. Appl. Microbiol. 2016;62(1):55–62. doi: 10.1111/lam.12508. [DOI] [PubMed] [Google Scholar]
- Fan, W.Y., Cheng, Y.J, Sun J.F., Sun, L., Tong, Y., 2003. Study on processes for treating slaughterhouse wastewater. J. Shenyang Instit. Chem. Technol 03, 230–232.
- Feng, G.E., Guo, K., Zhou, G.C., Zhang, H.J., Liu, J.N., Dai, Y.J., 2012. Isolation and Identification of Bacteria in the Activated Sludge from Four Sewage Treatment Plants in Nanjing City and Its Antibiotic Resistance Analysis. Environmental Science. 26(9), 906–13.
- Gao W., Baig A.Q., Ali H., Sajjad W., Farahani M.R. Margin based ontology sparse vector learning algorithm and applied in biology science. Saudi J. Bio. Sci. 2017;24(1):132–138. doi: 10.1016/j.sjbs.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajare A.A., More H.N. Design of the lyophilization process of a doxorubicin formulation based on thermal properties. Indian J. Pharm. Sci. 2017;79(6):907–913. [Google Scholar]
- He Y.W., Xing H.G. The urban water pollution status quo and countermeasures in China. Water Conserv. Sci. Technol. Econ. 2006;12(1):44–45. [Google Scholar]
- Jia Y.P., Jia X.Q., Zong Q., Heng H.K., Yu Y.Q. Research progress in the application of biological technology to slaughterhouse wastewater treatment. Indus. Water Treat. 2015;35(3):10–12. [Google Scholar]
- Khan A., Jan G., Khan A., Jan F.G., Danish M. Evaluation of antioxidant and antimicrobial activities of Bergenia ciliata Sternb (Rhizome) crude extract and fractions. Pak. J. Pharm. Sci. 2018;31(1):31–35. [PubMed] [Google Scholar]
- Kundu P., Debsarkar A., Mukherjee S., Kumar S. Artificial neural network modelling in biological removal of organic carbon and nitrogen for the treatment of slaughterhouse wastewater in a batch reactor. Environ. Technol. 2014;35(9–12):1296–1306. doi: 10.1080/09593330.2013.866698. [DOI] [PubMed] [Google Scholar]
- Li D., Yan H.G., Yuan L. Screening, identification and study of the fermentation properties of yeast strains for exclusive use of blueberry wine. Sichuan Food Ferment. 2015;51(5):75–79. [Google Scholar]
- Li T., Ren Y., Wei C. Study on preparation and properties of PVA-SA-PHB-AC composite carrier for microorganism immobilization. J. Appl. Polym. Sci. 2014;131(3):1082–1090. [Google Scholar]
- Lu W.W., Wang L.P. Isolation and identification of yeasts for purification of soy oligosaccharides and preliminary application of its fermentation characteristics. Food Ferment. Ind. 2016;42(3):168–171. [Google Scholar]
- Meng Q., Hao J.J. Optimizing the application of bacillus velezensis, BAC03 in controlling the disease caused by Streptomyces scabies. Biocontrol. 2017;62(4):535–544. [Google Scholar]
- Molina M.C., Gonzalez V., Bautista L.F., Sanz R., Simarro R., Sánchez Irene, Sanz J.L. Isolation and genetic identification of PAH degrading bacteria from a microbial consortium. Biodegradation. 2009;20:789–800. doi: 10.1007/s10532-009-9267-x. [DOI] [PubMed] [Google Scholar]
- Piubeli F., Grossman M.J., Fantinatti-Garboggini F., Durrant L.R. Enhanced reduction of cod and aromatics in petroleum-produced water using indigenous microorganisms and nutrient addition. Int. Biodeter. Biodegr. 2012;68(2):78–84. [Google Scholar]
- Rodgersvieira E.A., Zhang Z., Adrion A.C., Gold A., Aitken M.D. Identification of anthraquinone-degrading bacteria in soil contaminated with polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 2015;81(11):3775–3781. doi: 10.1128/AEM.00033-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaei M., Mortazavi S.B., Bakhshi B., Jafari A.J. Isolation, genetic identification, and degradation characteristics of n-Hexadecane degrading bacteria from tropical areas in Iran. Fresen. Environ. Bull. 2013;22(4):1304–1312. [Google Scholar]
- Semblante G.U., Hai F.I., Xia H., Ball A.S., Price W.E., Nghiem L.D. Trace organic contaminants in biosolids: impact of conventional wastewater and sludge processing technologies and emerging alternatives. J. Hazard. Mater. 2015;300:1–17. doi: 10.1016/j.jhazmat.2015.06.037. [DOI] [PubMed] [Google Scholar]
- Shehzadi M., Afzal M., Khan M.U., Islam E., Mobin A., Anwar S. Enhanced degradation of textile effluent in constructed wetland system using Typha domingensis and textile effluent-degrading endophytic bacteria. Water Res. 2014;58(58C):152–159. doi: 10.1016/j.watres.2014.03.064. [DOI] [PubMed] [Google Scholar]
- Song Y.Q., Zhang L., Li N.H. Screening the flocculent bacteria and optimizing the fermentation conditions in treating slaughter-releasing sewage. J. Saf. Environ. 2016;16(3):211–215. [Google Scholar]
- Su F.X., Zhang J., Zhang X.J. Screening and fermentation characteristics of indigenous yeast strains from spontaneously fermented pepino fruit (Solanum muricatum Ait.) Food Sci. 2017;38(4):100–106. [Google Scholar]
- Sun C.M. AIr-flotation-hydrolysis acidification-two-stage biological contact oxidation process in treating the slaughterhouse wastewater. Environ. Sci. Surv. 2008;5:019. [Google Scholar]
- Taheri M., Irandoust K., Noorian F., Bagherpour F. The effect of aerobic exercise program on cholesterol, blood lipids and cigarette withdrawal behavior of smokers. Acta Medica Mediterr. 2017;33(4):597–600. [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.D., Wang W.W., Zhu M.J., Liao Y.Z., Yao Y.F. Screening and characterization of aroma yeast with thermo-tolerant and salt-tolerance. Food Ferment. Ind. 2016;42(3):92–96. [Google Scholar]
- Tong X.W. Research progress of slaughtering wastewater treatment technology. Environ. Sustain. Dev. 2014;39(5):204–207. [Google Scholar]
- Wu N.J., Zhou X.L., Shang F. Experimental study on the treatment of simulated printing and dyeing wastewater by anaerobic hydrolysis-biological contact oxidation process. J. Shanghai Univ. Electric Power. 2013;29(4):370–373. [Google Scholar]
- Xie Y., Ge S., Jiang S., Liu Z., Chen L., Wang L., Chen J., Qin L., Peng W. Study on biomolecules in extractives of Camellia oleifera fruit shell by GC-MS. Saudi J. Bio. Sci. 2018;25(2):234–236. doi: 10.1016/j.sjbs.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue W.L., Xiong M., Li F., Wang G. The isolation and characterization of the novel chlorothalonil-degrading strain Paracoccus sp. XF-3 and the cloning of the chd gene. J. Biosci. Bioeng. 2015;120(5):544–548. doi: 10.1016/j.jbiosc.2015.03.013. [DOI] [PubMed] [Google Scholar]
- Zhang L.Y., Zhang X.Y., Zhang W. Screening and identification of chitinase producing strain and its inhibition action to dominant moulds in corn stalk. Chinese J. Animal Nutr. 2017;29(3):970–978. [Google Scholar]
- Zhang W.Y., Liu P.C., Zheng F.J. Separation, screening and application of efficient degradation microorganisms for improving COD removing rates from organic chemical wastewater. Ind. Water Treatment. 2016;36(8):52–54. [Google Scholar]