Abstract
Background
GPI-anchoring is a prevalent Glycosylphosphatidylinositol modification process of posttranslational protein and is necessary for cell wall integrity in eukaryotes. To date, the function of GPI anchored-related protein remains unknown in phytopathogenic fungi.
Results
We here characterized the functions of MoPer1, a homolog of Saccharomyces cerevisiae ScPer1, from the rice blast fungus Magnaporthe oryzae. Transcriptional analysis demonstrated that MoPER1 was significantly upregulated during conidiation and infection. We found that the ∆Moper1 mutant was defective in conidiation and appressoria formation, and MoPer1 was involved in osmotic stress response and maintaining the cell wall integrity. Pathogenicity assays indicated that deletion of MoPEP1 significant reduction in virulence. Microscopic examination of the lesions revealed that the invasive hyphae of ∆Moper1 mutants were mostly restricted to the primary infected leaf sheath cells.
Conclusions
Our results indicated that MoPer1 is necessary for growth, conidiogenesis, and pathogenicity of the fungus. Our study facilitated to deep elucidate the pathogenic molecular mechanism of M. oryzae, and also provided a very helpful reference value for developing effective fungicide pointed at as the gene for target.
Electronic supplementary material
The online version of this article (10.1186/s12284-018-0255-9) contains supplementary material, which is available to authorized users.
Keywords: MoPer1, Growth, Conidiogenesis, Pathogenicity
Introduction
The fungal cell wall play important roles in maintaining cell integrity during polarized growth (Klis et al., 2002). The main components of the fungal cell wall are glycoproteins and polysaccharides. Part of the cell wall protein must be anchored by glycosylphosphatidylinositol (GPI) after translated, and then bound to the cell wall to perform its normal biological function (Bernard and Latge, 2001; Bowman and Free, 2006; Free, 2013). GPI anchors, which generally contain four core components: ethanolamine phosphate, mannose, glucosamine and phosphatidylinositol, which are synthesized in the endoplasmic reticulum (ER). After being linked to the target protein, the lipid moieties will be further reconstructed by a series of modifications, mainly including three steps, inositol deacylation is performed first, then the acyl chain at the sn-2 site of the diacylglycerol is cleaved to form a lyso-GPI, and finally introduce a saturated 26-carbonyl chain at the sn-2 site (Orlean and Menon, 2007; Fujita and Jigami, 2008; Fujita and Kinoshita, 2012). Genes involved in this process have been characterized in mammals and yeast.
In Saccharomyces cerevisiae, after GPI anchored to the target protein, the acyl group of its inositol residue is removed by the inositol deacylase Bst1. Deletion of the BST1 gene delays the formation of GPI-anchored proteins (Tanaka et al., 2004; Fujita et al., 2006b). In the second step, the acyl chain at the sn-2 site is cleaved by the Per1 (processing in the ER) protein which performs the function of GPI phospholipase A2 and then lyso-GPI is formed (Fujita et al., 2006a). Biological function of the Per1 is similar to Bst1, which is also necessary for the maturation of GPI-anchored proteins and loss of these two genes caused defects to the integrity of cell wall. In addition, Per1 also plays a very important role in the transport of GPI-anchored proteins (Fujita et al., 2006a). Following the formation of lyso-GPI, the 26-carbon fatty acid is linked to the sn-2 site under the action of the Gup1 protein (Sipos et al., 1997). Lipid moieties are modified and the mature GPI-anchored protein adheres to the cell wall to perform a series of biological functions including cell signaling transduction, cell-cell information exchange, cell adhesion and host defense response (Kinoshita et al., 1995).
To date, except Saccharomyces cerevisiae, the function of GPI-anchored proteins in the lipid remodeling process in fungi is only reported in the human pathogenic filamentous fungus Aspergillus fumigatus. Deletion of AfPERA, a homlogues gene of Saccharomyces cerevisiae PER1, growth was slower, the produce of conidia was reduced, resistance to triazole fungicides was enhanced and toxicity to mice was lost in the Aspergillus fumigatus (Chung et al., 2014). Further study found that the cell wall components of ΔAfperA mutant changed, and the content of beta-glucan and chitin increased, leading to significant influence on cell wall integrity (Chung et al., 2014). Although it is recognized that lipid remodeling of GPI-anchored proteins plays an important role in the cell wall integrity of Saccharomyces cerevisiae and Aspergillus fumigatus, the biological functions have not been reported in plant pathogenic fungi.
Rice blast, caused by Magnaporthe oryzae, is a destructive disease during rice cultivation that severely threatens the production of rice crops worldwide (Talbot, 2003; Dean et al., 2012). Here, we define MoPer1, a ScPer1 homologue of M. oryzae, and for the first time elucidate its function in phytopathogenic fungi. Our results indicated that MoPer1 play important roles in growth, conidiogenesis, invasive hyphae growth and pathogenicity in M. oryzae.
Results
Identification and expression of MoPER1
Examination of the M. oryzae genome database at the Broad Institute (http://fungidb.org/fungidb/) revealed that MGG_04527 shares 33% identity and 44% similarity to the S. cerevisiae Per1; therefore, we named the protein MoPer1. To determine whether MoPer1 can complement ScPer1 function, we expressed MoPER1 in a ∆Scper1 mutant through the yeast expression vector pYES2. Transformants carrying the MoPER1 gene exhibited better growth on medium containing 20 μg/ml calcofluor white (CFW) compared to the ∆Scper1 mutant, and was similar to wild type BY4741 strain (Additional file 1: Figure S1), suggesting that MoPer1 is a functional paralog of ScPer1.
Before testing the functions of MoPER1, we evaluated its transcription profile. The expression of MoPER1 was higher in conidia and early infection stages than mycelium, with the highest level being detected in the conidia stage (> 2.4-fold; Fig. 1). These observations suggested that MoPER1 is over-expressed during early stage of infection.
Fig. 1.
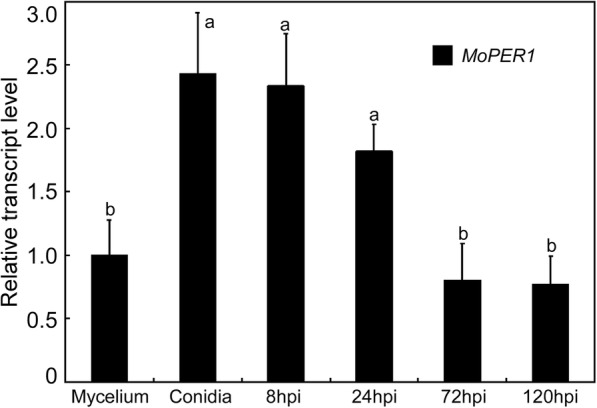
transcription profiles of MoPER1 at different stages of fungal development. The phase-specific expression of MoPER1 was quantified by quantitative real-time polymerase chain reaction (qRT-PCR), with the synthesis of cDNA from each sample including infectious growth, vegetative growth and conidia. Hpi: hour post inoculation
MoPER1 deletion affects hyphal growth
A MoPER1 deletion mutant was generated by replacing the MoPER1 coding region with the hygromycin resistance cassette (HPH) (Additional file 2: Figure S2A). Putative mutant (∆Moper1) was screened and confirmed by Southern blot analysis (Additional file 2: Figure S2B). Two gene deletion mutants, ∆Moper1#25 and ∆Moper1#29, were selected for further analysis. Furthermore, a complementation strain (∆Moper1/MoPER1) contained the ORF encoded by MoPER1 (Additional file 2: Figure S2C) was also generated. The resulting transformant were normal in growth, conidiation and infection (Table 1 and Figs. 5, 7) and considered as complemented strain.
Table 1.
Comparison of mycological characteristics among strains
| Strain | Mycelial growtha (cm) | Conidiationb (× 104/cm2) | Abnormal conidial ratec (%) | Appressorial formationd(%) |
|---|---|---|---|---|
| CM PDA OM SDC | ||||
| Guy11 | 4.20 ± 0.10A 4.50 ± 0.10A 3.83 ± 0.06A 3.77 ± 0.06A | 3.61 ± 0.23A | 2.67 ± 1.15A | 92.22 ± 3.85A |
| ΔMoper1#25 | 3.70 ± 0.10B 4.17 ± 0.06B 2.57 ± 0.21B 3.30 ± 0.10B | 1.26 ± 0.21B | 30.25 ± 5.15B | 8.89 ± 1.92B |
| ΔMoper1#29 | 3.70 ± 0.06B 4.23 ± 0.06B 2.50 ± 0.10B 3.33 ± 0.06B | 1.16 ± 0.56B | 30.89 ± 3.29B | 10.67 ± 3.33B |
| ΔMoper1/MoPER1 | 4.16 ± 0.07A 4.43 ± 0.06A 3.80 ± 0.10A 3.70 ± 0.09A | 3.75 ± 0.38A | 3.48 ± 1.67A | 93.33 ± 2.36A |
aDiameter of hyphal radii at day 7 after incubation on CM, PDA, OM and SDC agar plates at room temperature
bNumber of conidia harvested from a 9 cm SDC plate at day 10 after incubation at room temperature
cPrecentage of abnormal conidial harvested from a 9 cm SDC plate at day 10 after incubation at room temperature
dPrecentage of appressorium formation on artificial surface at 24 h post-incubation at room temperature
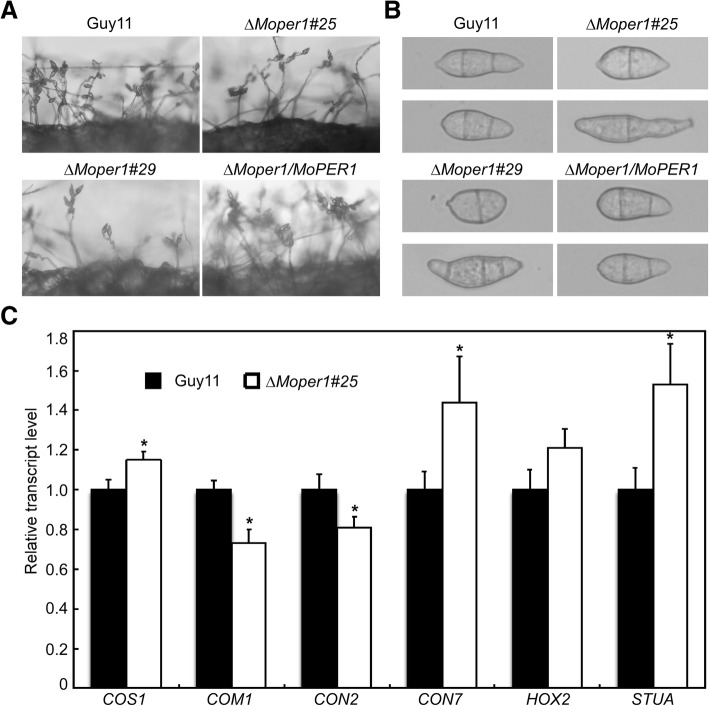
Fig. 5.
MoPer1 is required for normal conidia formation. a Conidia formation was observed under a light microscope 24 h at room temperature after induction of conidiation under cover slips. b Morphological observations of conidia. c Expression analysis of conidiation-related genes by qRT-PCR in the ∆Moper1 mutant. Asterisks represent significant differences (p < 0.05)
Fig. 7.
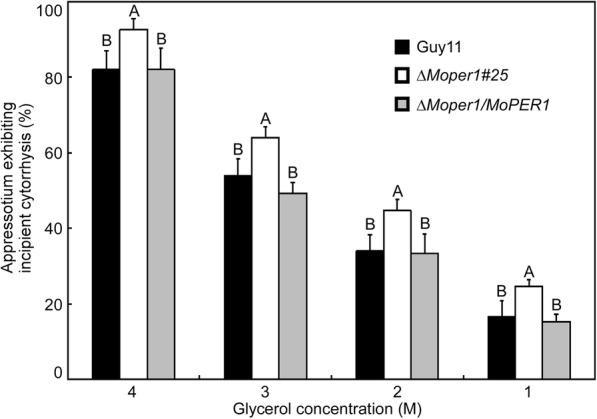
Collapsed appressoria were observed in mutant strain. For each glycerol concentration, at least 100 appressoria were observed and the numbers of collapsed appressoria were counted
We evaluated the growth of ∆Moper1 mutant on CM, PDA, OM, and SDC media. The ∆Moper1 mutants showed obviously smaller colony diameter than the wild-type strain (Guy11) and the complemented strain ∆Moper1/MoPER1 on all media types (Table 1). These results indicated that MoPer1 plays a role in hyphal growth.
MoPer1 is important in stress responses
To investigate whether ∆Moper1 exhibited any defects under different conditions of stress, ∆Moper1 mutant strains were exposed to 0.7 M NaCl and 0.6 M KCl. Surprisingly, the ∆Moper1 mutants showed weaker growth inhibition than Guy11 and the complemented strain in NaCl- containing CM (Fig. 2a) and the inhibition of the ∆Moper1 mutant was 8% less than Guy11 (Fig. 2b). However, no distinct difference was observed for the ∆Moper1 mutant when compared with Guy11 in KCl- containing CM (Fig. 2a, b). These findings suggested that MoPer1 contribute to the osmotic stress response of the fungus.
Fig. 2.
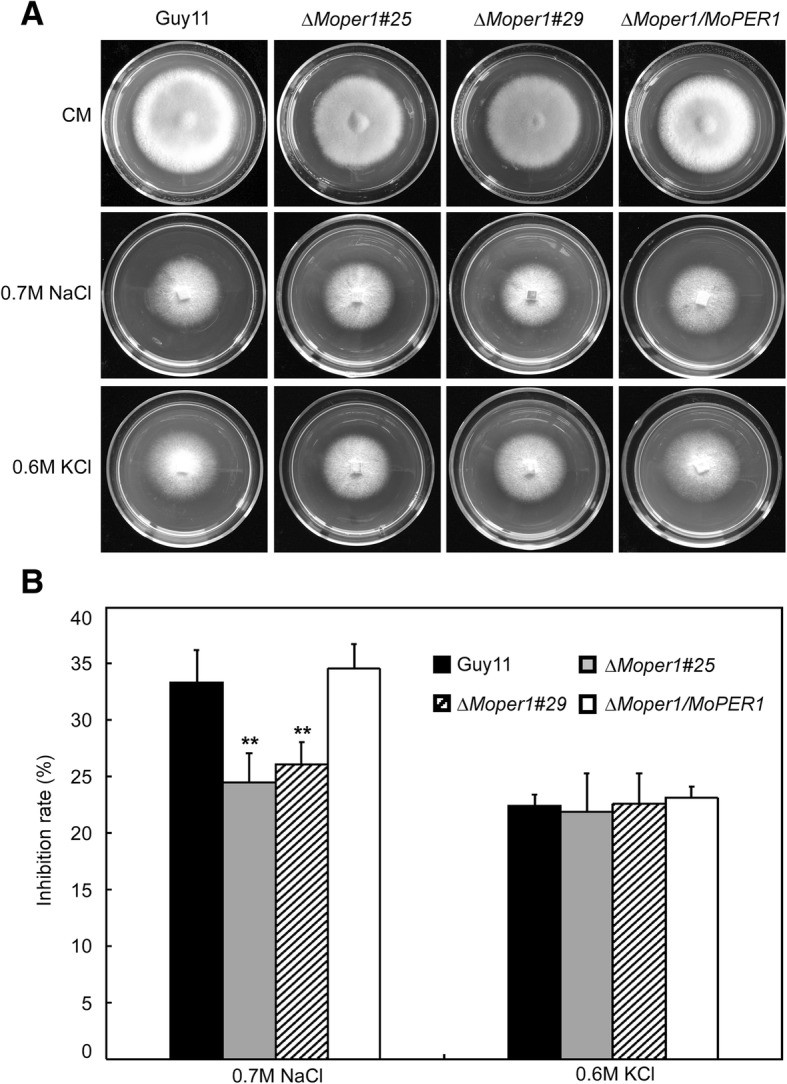
∆Moper1 mutants iron stress assessment. a The ∆Moper1 mutants are less sensitive to iron stress than Guy11. Colonies of the wild-type Guy11, the ∆Moper1 mutants and the complemented strains were grown on CM plates with 0.7 M NaCl or 0.6 M KCl and cultured at 28 °C for 7 days. b The growth inhibition rate is estimated relative to the growth rate of each untreated control [Inhibition rate = (the diameter of untreated strain – the diameter of treated strain) / (the diameter of untreated strain × 100%)]. Three repeats were performed and similar results obtained. Error bars represent the standard deviations and lowercase respresent significant differences (p < 0.01)
MoPer1 is required for cell wall integrity
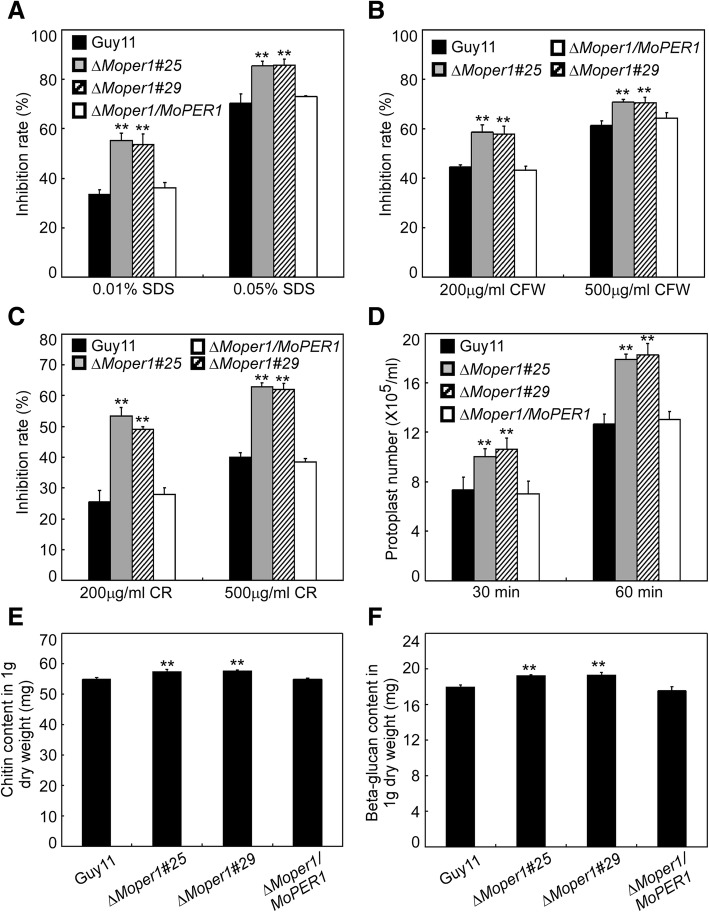
To examine the role of MoPer1 in cell wall integrity, mycelial growth was measured on CM containing sodium dodecyl sulfate (SDS), CFW and congo red (CR), all of which are cell wall-perturbing agents. The sensitivity of the ∆Moper1 mutant strains were significant higher to these agents than the wild-type strain Guy11 (Fig. 3a, b and c). Then, We examined the effects of lytic enzymes (10 mg/mL) on the ∆Moper1 mutant. More protoplasts were found in the ∆Moper1 mutant than in the controls after incubation for 30 and 60 min (Fig. 3d). We further quantified the accumulation of chitin and β-glucan in the cell wall, the results showed that the content of chitin and β-glucan in the ∆Moper1 mutant was higher than the wild-type strain Guy11 (Fig. 3e and f). These results indicated that MoPer1 is involved in maintaining the cell wall integrity.
Fig. 3.
The MoPER1 deletion mutant had defects in cell wall integrity. a-c The wild type strain Guy11 and the ∆Moper1 strain were inoculated on CM medium with or without SDS (0.01% and 0.05% w/v), CFW (200 and 500 μg/ml) and CR (200 and 500 μg/ml), and cultured at 28 °C for 7 days. The growth inhibition rate is estimated relative to the growth rate of each untreated control [Inhibition rate = (the diameter of untreated strain – the diameter of treated strain)/(the diameter of untreated strain × 100%)]. Three repeats were performed and similar results obtained. d Protoplast released under the treatment of cell-wall-degrading enzymes, The released protoplast was quantified at 30 min intervals. e and f Quantification of the chitin and β-glucan content of the mutant by ELISA
MoPer1 mediates resistance to the triazole fungicides
We tested susceptibility of the ∆Moper1 mutant to the triazole antifungal drugs difenoconazole (DCZ). We found that the sensitivity of the ∆Moper1 mutant to DCZ at low concentrations was not significantly different from that of the wild-type strain Guy11 (Fig. 4a and b); however, the mutant was more sensitive at high concentrations (Fig. 4a), inhibition of the ∆Moper1 mutant was 12.9–18.1% and 5.6–7.3% higher than Guy11 in 5 and 10 μg/ml DCZ-containing CM, respectively (Fig. 4b).
Fig. 4.
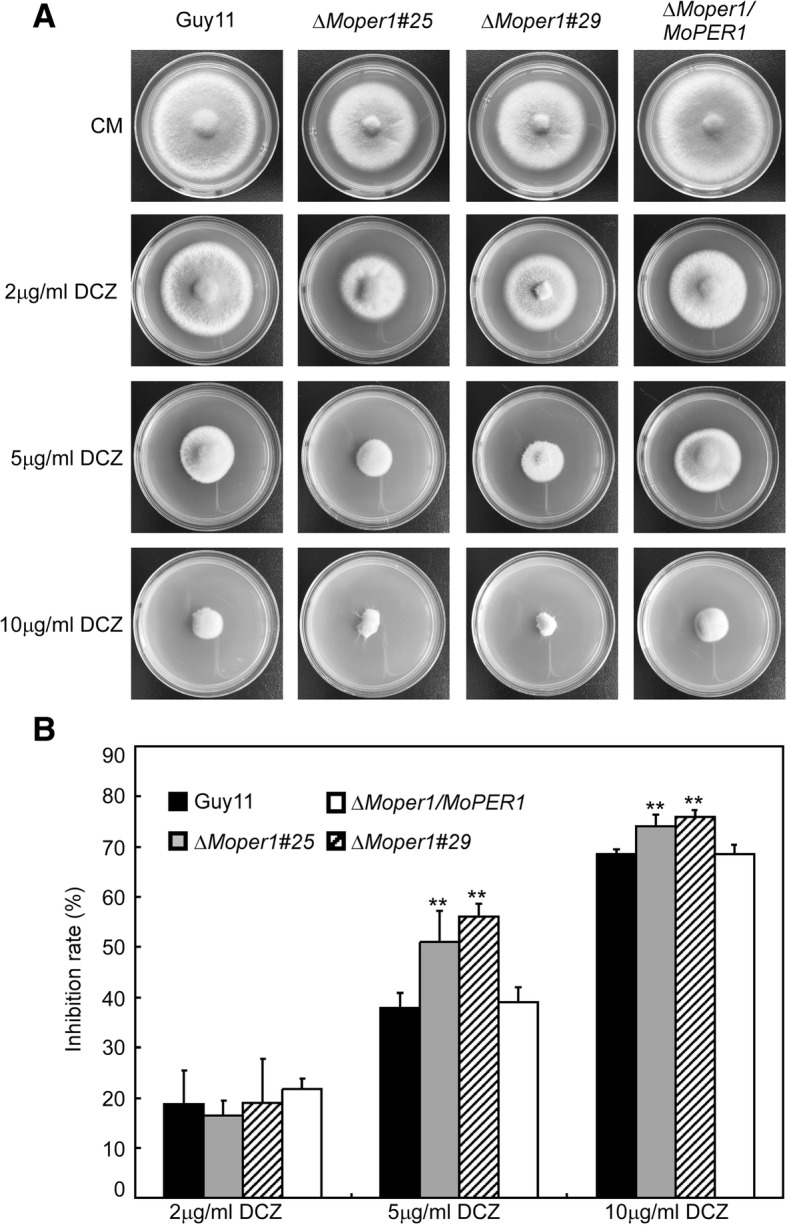
∆Moper1 mutants increased susceptibility to the antifungal triazole drug, difenoconazole (DCZ). a The wild type strain Guy11 and the ∆Moper1 strain were inoculated on CM medium with or without DCZ (2, 5 and 10 μg/ml), and cultured at 28 °C for 7 days. b The growth inhibition rate is estimated relative to the growth rate of each untreated control [Inhibition rate = (the diameter of untreated strain – the diameter of treated strain)/(the diameter of untreated strain × 100%)]. Three repeats were performed and similar results obtained
MoPER1 is involved in conidiogenesis and appressorium formation
Since conidia play an important role during M. oryzae infection, we measured the conidia production of the ∆Moper1 mutants. We found that the conidia production was significantly reduced on SDC medium (Fig. 5a). Conidium number was approximately one third that of the wild-type and complemented strain ∆Moper1/MoPER1 (Table 1). We also found that some of the conidia produced by the ∆Moper1 mutant were abnormal in morphological character (Fig. 5b) and the proportion reached 30% (Table 1). We further examined the expression of six conidiation-related genes. The expression level of MoCOM1 and MoCON2 (Yang et al., 2010; Shi and Leung, 1995) were significantly lower in the ∆Moper1 mutant than in the Guy11 strain (Fig. 5c), In contrast, the transcript levels of MoCOS1, MoCON7 and MoSTUA (Zhuangzhi Zhou et al., 2009; Nishimura et al., 2009; Shi and Leung, 1995) were significantly increased in the ∆Moper1 mutant (Fig. 5c), indicating that MoPer1 is involved in the regulation of the expression of conidiation-related genes. Next, we examined appressorium formation in the ∆Moper1 mutant. The ∆Moper1 mutant formed normal germ tubes. Microscopic examination revealed that the rate of appressorium formation in ∆Moper1 was significantly reduced than Guy11, which was only about 10% while the wild type was more than 90% (Table 1). These results suggested that MoPer1 plays critical role in conidiogenesis and appressorium formation.
MoPer1 is required for full virulence
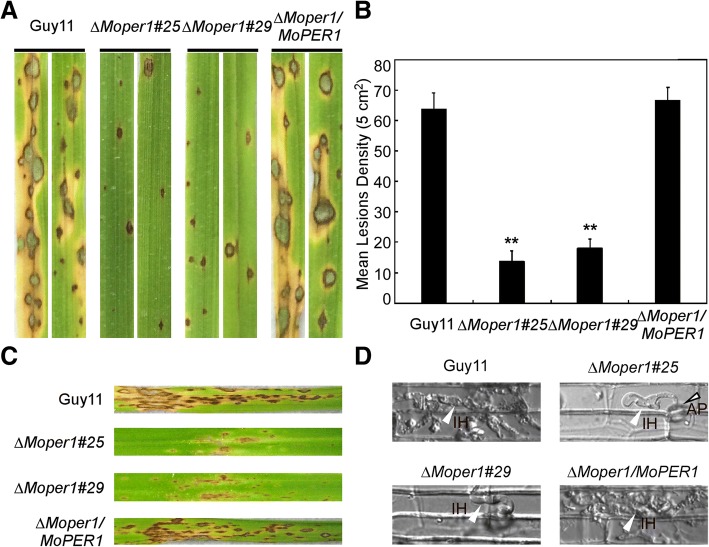
To determine whether MoPer1 is involved in pathogenicity, conidial suspensions of the ∆Moper1 mutant, wild-type, and complemented strain were sprayed onto 2-week-old and injected onto 4-week-old rice seedlings (cv. CO-39). When observed 7 (spraying assay) or 5 (injection assay) days post-infection, the ∆Moper1 mutant produced tiny and restricted lesions on rice leaves compared to Guy11, which caused spreading lesions (Fig. 6a and c). Statistical analysis indicated that the lesion numbers of ∆Moper1#25 and ∆Moper1#29 mutants were reduced by 78.5% and 71.7% (Fig. 6b), respectively. Taken together, these results indicated that MoPer1 is involved in pathogenicity.
Fig. 6.
pathogenicity assay of the mutant. a Leaf spraying assay. Four milliliters of conidia suspension (5 × 104 spores/ml) of each strain were sprayed on two-week old rice seedlings. Diseased leaves were photographed at 7 day after inoculation. b Lesion density was performed by counting lesion numbers of unit area. c Leaf injecting assay. Conidia suspension (5 × 104 spores/ml) of each strain were injected on two-week old rice seedlings. Diseased leaves were photographed at 5 day after inoculation. d Excised rice sheath from four-week old rice seedlings was inoculated with conidial suspension (5 × 104 spores/ml). Infectious growth was observed 48 h after inoculation
MoPER1 deletion impairs appressorium turgor pressure
To penetrate the rice leaf cuticle during infection, a high appressorium internal turgor pressure is required (Talbot and Foster, 2001). To elucidate the mechanism underlying virulence in the ∆Moper1 mutant, we examined the appressorium turgor in the ∆Moper1 mutant and wild-type and compared cell collapse rate in 1, 2, 3 and 4 M glycerol at 24 hpi. We found that the appressoria of the ∆Moper1 mutant showed an increased collapse rate in 1, 2, 3, and 4 M glycerol compared with those of wild type (Fig. 7), indicating that defects of appressorium turgor pressure in the ∆Moper1 mutant might lead to failed penetration.
∆Moper1 mutant is defective in infectious hyphal growth on plants
To further explore why the ∆Moper1 mutant showed reduced virulence on host plants, we used excised leaf sheath assay to examine infectious hyphae within the host cells. At 48 h post-inoculation (hpi), the ∆Moper1 mutant were mostly blocked in the primary infected leaf sheath cells (Fig. 6d), in contrast to the free spread of invasive hyphae of wild-type Guy11 and complemented strains (Fig. 6d). Further, infectious hyphae growth on barley was also evaluated by using an ‘invasive hypha type’ assay (Wang et al., 2013) at 48 hpi using spore suspensions; four types (type 1, no penetration; type 2, with a penetration peg; type 3, with a single invasive hypha; and type 4, with extensive hyphal growth) of invasive hyphae were observed in barley tissues (Fig. 8a). In the wild-type and complemented strains, more than 60% of the cells showed type 4 growth; few strains showed type 1 and type 2 invasive hyphal growth. In contrast, less than 10% of the cells showed type 4 and more than 70% showed types 1 and 2 invasive hyphal growth in the ∆Moper1 mutant (Fig. 8b). These results indicated that MoPer1 is require for invasive hyphae growth.
Fig. 8.
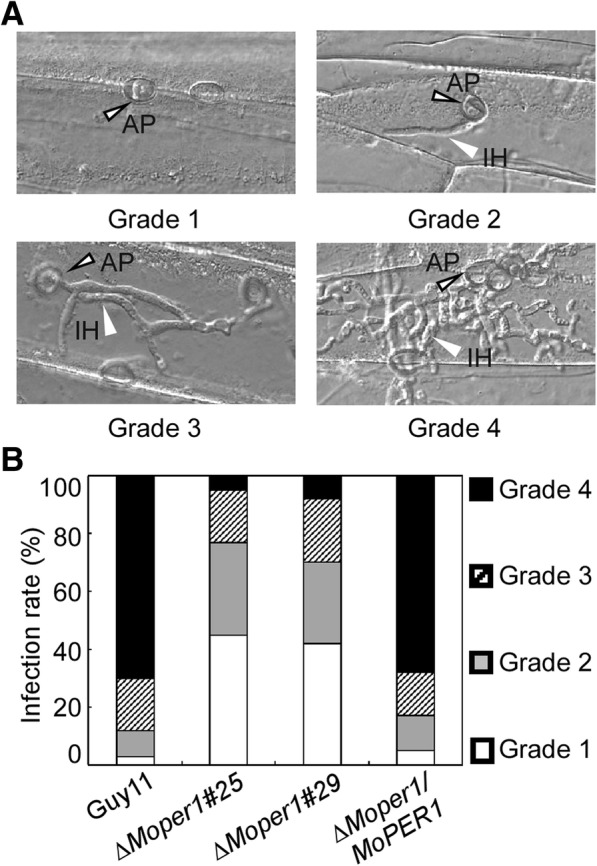
The infectious hyphae growth on barley leaves. a Excised barley leaves from 7-day-old barley seedings were inoculated with conidial suspension (5 × 104 spores/ml). Infectious growth was observed at 24 h post-inoculation (hpi). b Statistical analysis for each type of infectious hyphal shape, for each tested strain; 100 infecting hyphae were counted per replicate and the experiment was repeated three times
Discussion
In the present study, we characterized a homolog of S. cerevisiae Per1, MoPer1, in M. oryzea. We found that MoPer1 was required for growth, conidiation and plant infection of M. oryzea.
Similar to A. fumigatus (Chung et al., 2014), deletion of MoPER1 lead to a reduced growth rate on different media in M. oryzeae. We found that the ∆Moper1 mutant exhibited a high sensitivity to different cell wall stressor (SDS, CFW, CR and lytic enzymes) and triazole fungicides, and the content of chitin and β-glucan of the ∆Moper1 mutant was higher than the wild-type Guy11. These phenotypes were similar to the ∆Afper1 mutant (Chung et al., 2014). Moreover, while expressed MoPER1 gene of M. oryzea in ∆Scper1 mutant and ScPER1 gene of S. cerevisiae in ∆AfperA mutant were able to full and partial restore defects of cell wall integrity (Chung et al. 2014), respectively. These results indicated that Per1 protein share conserved functions in maintaining the cell wall integrity.
Like most fungal pathogens, conidiogenesis and appressorium formation play important role during the infection stage. The ∆Moper1 mutant produce less conidia ith part abnormality, exhibiting distinct defects in conidiation. We also found that two = conidiation`` 445.
related genes MoCOM1 and MoCON2 were significantly reduced expression in the ∆Moper1 mutant, and previous studies have shown that the loss of MoCOM1 or MoCON2 in M. oryzae reduces sporulation and produces misshapen conidia (Yang et al., 2010; Shi and Leung, 1995), which indicated that MoPer1 was involved in sporulation and conidium morphology by regulating the expression of MoCOM1 and MoCON2. Further, consistent with our results, ∆AfperA mutant also decreased the conidia production in A. fumigatus (Chung et al., 2014). These results indicated that MoPer1 and its homologs are responsible for the regulation of asexual development.
Disruption of MoPER1 leads to a defect in infection-related morphogenesis in M. oryzae. The ∆Moper1 mutant showed reduced pathogenicity on rice. Considering that the infection of rice blast fungus needs mature appressorium, we first tested the appressorium formation of the ∆Moper1 mutant and found that the appressorium forming ability of the ∆Moper1 mutant at the hydrophobic interface was significantly decreased. Per1 is involved in the maturation and normal functioning of cell wall proteins (Fujita et al., 2006a), whose homolog may be signal molecules sensing host surface signal to form appressorium in M. oryzae. Therefore, we speculated that the reasons of decreasing formation of appressoria was the mutant reduced the sensory ability of the conidia to the host. Moreover, we examined the appressorium turgor pressure. We found that the collapse rate of appressoria of the ∆Moper1 mutant was more than that of Guy11, suggesting that appressorium turgor in the ∆Moper1 mutant was decreased. It is reasonable to speculate that the imbalanced turgor due to absence of cell wall integrity of the mutant. Further, invasive growth of the ∆Moper1 mutant was inhibited in rice sheath, and the expansion ratio was markedly lower than that for wild-type Guy11 in barley epidermis. We concluded that these defects caused the decreased pathogenicity of the ∆Moper1 mutant.
Conclusion
Collectively, we identified an important pathogenic factor MoPer1, our results indicate that MoPer1 plays an important role in growth, cell wall integrity, conidiation, host infection and pathogenicity in M. oryzae.
Materials and methods
Strains and culture conditions
M. oryzae Guy11 strain was used as the wild type in this study. All strains were cultured on complete medium (CM) agar plates for 3–15 days at 28 °C (Talbot et al., 1993). Fungal mycelia were harvested from liquid CM and used for genomic DNA and RNA extractions. Protoplasts were prepared and transformed as described (Sweigard et al., 1992). Transformants were selected on TB3 medium (3 g of yeast extract, 3 g of casamino acids, 200 g of sucrose, 7.5 g of agar in 1 L of distilled water) with 300 μg/ml hygromycin B (Roche) or 200 μg/ml zeocin (Invitrogen). For conidiation, strain blocks were maintained on rice decoction and corn agar media at 28 °C for 7 days in the dark followed by 3 days of continuous illumination under fluorescent light (H. F. Zhang et al., 2009).
Yeast ∆Scper1 mutant complementation
The full length of MoPER1 cDNA were amplified using primers FL21/FL22 (Additional file 3: Table S1). The PCR product was disgested with HindIII/XhoI and cloned into the pYES2 vector (Invitrogen, Shanghai, China), and transformed into the ∆Scper1 mutant. Colonies were selected on SD medium without uracil. As a control, the ∆Scper1 mutant were transformed with the empty pYES2. Yeast cells were incubated on liquid YPD medium (2% glucose, 2% peptone, and 1% yeast extract), and aliquots (5 μl) of 10-fold serial dilution were grown in SD (galactose) and SD-CFW (galactose+ 20 μg/ml CFW) plates at 30 °C for 4 days and photographed.
MoPER1 gene disruption and ∆Moper1 mutant complementation
The ligation PCR approach (Zhao et al., 2004) was used to generate the MoPER1 gene replacement constructs. Approximately 1-kb upstream and downstream flanking sequences of MoPER1 gene were amplified by PCR with primer pairs FL11/FL12 and FL13/FL14 (Additional file 3: Table S1), respectively. The resulting PCR products of primer pairs FL11/FL12 and primer pairs FL13/FL14 were digested with XhoI/EcoRV and XbaI/SacI, respectively, then purified and orderly ligated to vector pCX62. The MoPER1 gene replacement constructs were transformed into protoplasts of Guy11. Putative ∆Moper1 mutants were identified by PCR and further confirmed by Southern blot analyses. For complementation assays, the full length except stop codon of MoPER1 gene including native promoter was amplified and cloned into the bleomycin-resistant vector pYF11 by the yeast in vivo recombination approach (Bruno et al., 2004; X. Zhou et al., 2011) and transformed into the ∆Moper1 mutant.
Vegetative growth, stress response, protoplast release assay and content of chitin and β-glucan determination
Vegetative growth of ∆Mo per1 and Guy11 was measured on CM medium (50 mL 20 x nitrate salts, 1 mL trace elements, 10 g glucose, 2 g peptone, 1 g yeast extract, 1 g casamino acids, 1 mL vitamin solution, 15 g agar in 1 L distilled water), PDA medium (200 g boiled patato, 20 g glucose and 15 g agar in 1 L distilled water), OM medium (30 g oat meal and 10 g agar in 1 L distilled water) and SDC medium (100 g straw, 40 g corn powder, 15 g agar in 1 L distilled water) for 7 days. Mycelia plugs of equal size, from 5-day-old CM plates were transferred into liquid CM. The mycelia were cultured with shaking (150 rpm) at 28 °C for 2 days. All growth assays were repeated three times, with three replicates each time.
Mycelia plugs (5 mm × 5 mm) were placed onto the freshly prepared CM agar plates with NaCl (0.7 M), KCl (0.6 M), SDS (0.01% and 0.05%), CFW (200 and 500 μg/ml), CR (200 and 500 μg/ml) and difenoconazole (2, 5 and 10 μg/ml) and cultured in the dark at 28 °C to determine their effects on fungal growth. The size of the colonies were measured and photographed after 7 days of incubation. The inhibition rate was determined by the percent decrease in the colony diameter (Haifeng Zhang et al., 2014). The experiment was repeated three times with three replicates each time.
For protoplast release assays, mycelia were cultured in liquid CM for 48 h and harvested by filtration, then was resuspended by 0.7 M NaCl with lysing enzyme (7.5 mg/ml, Sigma-Aldrich, Louis, USA) and placed in a shaker (70 rpm) at 28 °C. Lysis activity was stopped after 30 and 60 min of incubation, and protoplast were counted under a light microscope using a hemocytometer.
The fungus chitin ELISA kit (Chundu Biotechnology, Wuhan, China) was used to examined the content of chitin of M. oryzae. Mycelial smples ground into powder after freeze-dry, weighed equal quality, then washed with PBS and centrifugated (3000 rpm, 20 min) to obtain supernatant. Microtiter plate was coat by purified fungus chitin antibody to make solid-phase antibody. The supernatant was added to the microwell, combined with horseradish peroxidase (HPR) labeled, and become antibody-antigen-enzyme-antibody complex. Then washing completed, add TMB substrate solution, TMB substrate become blue color at HRP enzyme-catalyzed, reaction was terminated by the addition of a sulphuric acid solution and the color change was measured spectrophotometrically at a wavelength of 450 nm. The concentration of chitin in the samples was then determined by comparing the O.D. of the samples to the standard curve. The content of β-glucan was detected by the fungus chitin ELISA kit (Chundu Biotechnology, Wuhan, China) referred to the above method.
Nucleic acid manipulation and southern blotting
The standard Southern blot protocol was utilized (Sambrook and Russell, 2001). The target gene probe and HPH probe were amplified with primer pairs FL17/FL18 (for MoPER1) (Additional file 3: Table S1) and FL1111/FL1112 (for HPH), respectively. Probe labeling, hybridization and detection were preformed with the DIG High Prime DNA Labeling and Detection Starter Kit (Roche Applied Science, Penzberg, Germany). Total RNA was isolated from frozen fungal mycelia using the RNA extraction kit (Invitrogen, USA). To measure the relative abundance of gene transcripts, RNAs were extracted from mycelia grown in CM liquid medium for 2 days at 28 °C in a 150 rpm orbital shaker. To measure the relative abundance of MoPER1 transcripts during diverse fungal development stages, the total RNA samples were extracted from mycelia grow in CM liquid medium, conidia and plants inoculated with the conidia of Guy11 (1 × 108 spores/ml) for 8, 24, 72 and 120 h, respectively, by the method described above. The crude RNA was pretreated with DNase I (TaKaRa) and was then reverse transcriptase (Invitrogen, Carlsbad, CA, U.S.A). qRT-PCR was performed on the ABI 7500 real-time PCR system (Applied Biosystems, Foster City, CA, U.S.A) according to the manufacturer’s instructions. A 20 μl reaction volume contained 2 μl of reverse transcription product, 10 μl of SYBR premix Ex Taq (2 x), 0.4 μl of ROX reference dye (50 x, SYBR PrimeScript RT-PCR kit; TaKaRa), and 0.4 μl of each primer (10 μl). The stable-expression ACTIN gene (MGG_03982) amplified by primers FL4738 and FL4739 (Additional file 3: Table S1) was used as internal control. Normalization and comparison of mean Ct value were performed as described (Livak and Schmittgen, 2001). The experiment was repeated three times with three replicates each time.
Conidiation and appressorium formation
For conidiation, 10-day-old conidia were collected with 5 ml of distilled water, filtered through three layers of lens paper and counted with a haemacytometer under a microscope. Conidial germination and appressorium formation were measured on a hydrophobic surface. Conidial suspensions of 30 μl (5 × 104 spores/ml) were dropped onto a hydrophobic surface and placed in a moistened box at 28 °C (Zhang et al., 2011). Appressorium formation rate was counted at 24 h post-inoculation (hpi) under the microscope, more than 200 appressoria were counted for each strain. Photographs were taken at 24 hpi.
Plant infection and penetration assays
Plant infection assays were performed on two-week-old susceptible rice seedlings (O. sativa) CO-39 by spraying 4 ml of the conidial suspensions (5 × 104 conidia/ml in 0.2% gelatin) with a sprayer. Inoculated plants were placed in a moist chamber at 28 °C for the first 24 h in darkness, and then transferred back to another moist chamber with a photoperiod of 12 h under fluorescent lights. The disease severity was assessed at 7 days after inoculation. Approximately 6 cm long diseased rice blades were photographed to evaluate the virulence of the mutants (Chen et al., 2014). Similar spray inoculation with conidia (1 × 105 conidia/ml in 0.2% gelatin) was used for injection inoculation. For microscopic observation of penetration and infectious hyphae expansion on rice and barley tissue, rice was inoculated with 100 μl of conidial suspension (5 × 104 spores/ml) on the inner leaf sheath cuticle cells, barley was inoculated with 30 μl of conidial suspension (5 × 104 spores/ml) on the under side of the barley leaves, after 48 h (rice) and 24 h (barley) incubation under humid conditions at 28 °C, the leaf sheaths and barley leaves were observed under a microscope (Chen et al., 2015).
Additional files
Figure S1. The MoPER1 gene rescued the defect of the ∆Scper1 mutant. The ∆Scper1 mutant was transformed with the pYES2::MoPER1 construct encoding MoPer1. Serial dilutions of cultures were grown overnight on SD (galactose) and SD (galactose+ 20 μg/ml CFW) plates, and grown at 30 °C for 4 days and photographed. The experiment was repeated three times and representative results were obtained. (TIF 2001 kb)
Figure S2. Targeted gene replacement and complementation. (A) A 1384-bp fragment of the MoPER1 coding region were replaced by a 1.4-kb fragment containing the hygromycin B resistance cassette to create MoPER1 deletion mutant. (B) Southern hybridization analysis was used to validate the deletion of MoPER1 gene and the addition of a single copy integration of the HPH gene. (C) Semiquantitative RT-PCR was carried out to confirm the deletion and reintroduction of MoPER1 gene. (TIF 247 kb)
Table S1. Primers used in this study. (DOC 34 kb)
Acknowledgments
This research was supported by the Natural Science Foundation of China (Grant No. 31501599 to YC and Grant No. 31471755 to CL) and the National Key Research and Development Program of China (Project No. 2016YFD0200809 to DZ). We thank Dr. Yun Chen of Zhejiang University for providing ∆Scper1 (YCR044C) strain.
Funding
Funding for this study was provided by the Natural Science Foundation of China (Grant No. 31501599 to YC and Grant No. 31471755 to CL) and the National Key Research and Development Program of China (Project No. 2016YFD0200809 to DZ).
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files.
Authors’ contributions
YC and YL designed the research plan; YC, XW, CL, YZ and XT performed the research and analyzed the data; YC wrote the paper; DZ and YL revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Bernard M, Latge JP. Aspergillus fumigatus cell wall: composition and biosynthesis. Med Mycol. 2001;39(1):9–17. doi: 10.1080/mmy.39.1.9.17. [DOI] [PubMed] [Google Scholar]
- Bowman SM, Free SJ. The structure and synthesis of the fungal cell wall. Bioessays. 2006;28(8):799–808. doi: 10.1002/bies.20441. [DOI] [PubMed] [Google Scholar]
- Bruno KS, Tenjo F, Li L, Hamer JE, Xu JR. Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea. Eukaryot Cell. 2004;3:1525–1532. doi: 10.1128/EC.3.6.1525-1532.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhai S, Sun Y, Li M, Dong Y, Wang X, et al. MoTup1 is required for growth, conidiogenesis and pathogenicity of Magnaporthe oryzae. Mol Plant Pathol. 2015;16(8):799–810. doi: 10.1111/mpp.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zuo R, Zhu Q, Sun Y, Li M, Dong Y, et al. MoLys2 is necessary for growth, conidiogenesis, lysine biosynthesis, and pathogenicity in Magnaporthe oryzae. Fungal Genet Biol. 2014;67:51–57. doi: 10.1016/j.fgb.2014.04.001. [DOI] [PubMed] [Google Scholar]
- Chung D, Thammahong A, Shepardson KM, Blosser SJ, Cramer RA. Endoplasmic reticulum localized PerA is required for cell wall integrity, azole drug resistance, and virulence in Aspergillus fumigatus. Mol Microbiol. 2014;92(6):1279–1298. doi: 10.1111/mmi.12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R, Van Kan JA, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, et al. The top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol. 2012;13(4):414–430. doi: 10.1111/j.1364-3703.2011.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Free SJ. Fungal cell wall organization and biosynthesis. Adv Genet. 2013;81:33–82. doi: 10.1016/B978-0-12-407677-8.00002-6. [DOI] [PubMed] [Google Scholar]
- Fujita M, Jigami Y. Lipid remodeling of GPI-anchored proteins and its function. Biochim Biophys Acta. 2008;1780(3):410–420. doi: 10.1016/j.bbagen.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Fujita M, Kinoshita T. GPI-anchor remodeling: potential functions of GPI-anchors in intracellular trafficking and membrane dynamics. Biochim Biophys Acta. 2012;1821(8):1050–1058. doi: 10.1016/j.bbalip.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Fujita M, Umemura M, Yoko-o T, Jigami Y. PER1 is required for GPI-phospholipase A2 activity and involved in lipid remodeling of GPI-anchored proteins. Mol Biol Cell. 2006;17(12):5253–5264. doi: 10.1091/mbc.E06-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Yoko OT, Jigami Y. Inositol deacylation by Bst1p is required for the quality control of glycosylphosphatidylinositol-anchored proteins. Mol Biol Cell. 2006;17(2):834–850. doi: 10.1091/mbc.E05-05-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Inoue N, Takeda J. Defective Glycosyl phosphatidylinositol anchor synthesis and paroxysmal nocturnal hemoglobinuria. Adv Immunol. 1995;60:57–103. doi: 10.1016/s0065-2776(08)60584-2. [DOI] [PubMed] [Google Scholar]
- Klis FM, Mol P, Hellingwerf K, Brul S. Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS Microbiol Rev. 2002;26(3):239–256. doi: 10.1111/j.1574-6976.2002.tb00613.x. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Nishimura M, Fukada J, Moriwaki A, Fujikawa T, Ohashi M, Hibi T, et al. Mstu1, an APSES transcription factor, is required for appressorium-mediated infection in Magnaporthe grisea. Biosci Biotechnol Biochem. 2009;73(8):1779–1786. doi: 10.1271/bbb.90146. [DOI] [PubMed] [Google Scholar]
- Orlean P, Menon AK. Thematic review series: lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. J Lipid Res. 2007;48(5):993–1011. doi: 10.1194/jlr.R700002-JLR200. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Molecular cloning - a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Shi Z, Leung H. Genetic analysis of sporulation in Magnaporthe grisea by chemical and insertional mutagenesis. Mol Plant-Microbe Interact. 1995;8(6):949–959. doi: 10.1094/MPMI-8-0949. [DOI] [Google Scholar]
- Sipos G, Reggiori F, Vionnet C, Conzelmann A. Alternative lipid remodelling pathways for glycosylphosphatidylinositol membrane anchors in Saccharomyces cerevisiae. EMBO J. 1997;16:3494–3505. doi: 10.1093/emboj/16.12.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigard JA, Chumley FG, Valent B (1992) Disruption of a Magnaporthe grisea cutinase gene. Mol Gen Genet, 232(2):183–90 [PubMed]
- Talbot, NJ (2003) On the trail of a cereal killer: exploring the biology of Magnaporthe grisea. Annu Rev Microbiol 57:177–202. 10.1146/annurev.micro.57.030502.090957 [DOI] [PubMed]
- Talbot NJ, Ebbole DJ, Hamer JE. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell. 1993;5(11):1575–1590. doi: 10.1105/tpc.5.11.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot NJ, Foster AJ. Genetics and genomics of the rice blast fungus Magnaporthe grisea: developing an experimental model for understanding fungal diseases of cereals. Adv Botanical Res Incorp Adv Plant Pathol. 2001;34:263–287. [Google Scholar]
- Tanaka S, Maeda Y, Tashima Y, Kinoshita T. Inositol Deacylation of glycosylphosphatidylinositol-anchored proteins is mediated by mammalian PGAP1 and yeast Bst1p. J Biol Chem. 2004;279(14):14256–14263. doi: 10.1074/jbc.M313755200. [DOI] [PubMed] [Google Scholar]
- Wang J, Du Y, Zhang H, Zhou C, Qi Z, Zheng X, et al. The actin-regulating kinase homologue MoArk1 plays a pleiotropic function in Magnaporthe oryzae. Mol Plant Pathol. 2013;14(5):470–482. doi: 10.1111/mpp.12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhao XY, Sun J, Kang ZS, Ding SL, Xu JR, et al. A novel protein Com1 is required for normal conidium morphology and full virulence in Magnaporthe oryzae. Mol Plant-Mic Int. 2010;23(1):112. doi: 10.1094/MPMI-23-1-0112. [DOI] [PubMed] [Google Scholar]
- Zhang H, Tang W, Liu K, Huang Q, Zhang X, Yan X, et al. Eight RGS and RGS-like proteins orchestrate growth, differentiation, and pathogenicity of Magnaporthe oryzae. PLoS Pathog. 2011;7(12):e1002450. doi: 10.1371/journal.ppat.1002450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhao Q, Guo X, Guo M, Qi Z, Tang W, et al. Pleiotropic function of the putative zinc-finger protein MoMsn2 in Magnaporthe oryzae. Mol Plant-Microbe Interact. 2014;27(5):446–460. doi: 10.1094/MPMI-09-13-0271-R. [DOI] [PubMed] [Google Scholar]
- Zhang HF, Zhao Q, Liu KY, Zhang ZG, Wang YC, Zheng XB. MgCRZ1, a transcription factor of Magnaporthe grisea, controls growth, development and is involved in full virulence. FEMS Microbiol Lett. 2009;293(2):160–169. doi: 10.1111/j.1574-6968.2009.01524.x. [DOI] [PubMed] [Google Scholar]
- Zhao XH, Xue C, Kim Y, Xu JR. A ligation-PCR approach for generating gene replacement constructs in Magnaporthe grisea. Fungal Genet Newsl. 2004;51:17–18. [Google Scholar]
- Zhou X, Liu W, Wang C, Xu Q, Wang Y, Ding S, et al. A MADS-box transcription factor MoMcm1 is required for male fertility, microconidium production and virulence in Magnaporthe oryzae. Mol Microbiol. 2011;80(1):33–53. doi: 10.1111/j.1365-2958.2011.07556.x. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Li GL, Chunhua, He C. Conidiophore stalk-less1 encodes a putative zinc-finger protein involved in the early stage of Conidiation and mycelial infection in Magnaporthe oryzae. Mol Plant-Microbe Interact. 2009;22(4):402–410. doi: 10.1094/MPMI-22-4-0402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The MoPER1 gene rescued the defect of the ∆Scper1 mutant. The ∆Scper1 mutant was transformed with the pYES2::MoPER1 construct encoding MoPer1. Serial dilutions of cultures were grown overnight on SD (galactose) and SD (galactose+ 20 μg/ml CFW) plates, and grown at 30 °C for 4 days and photographed. The experiment was repeated three times and representative results were obtained. (TIF 2001 kb)
Figure S2. Targeted gene replacement and complementation. (A) A 1384-bp fragment of the MoPER1 coding region were replaced by a 1.4-kb fragment containing the hygromycin B resistance cassette to create MoPER1 deletion mutant. (B) Southern hybridization analysis was used to validate the deletion of MoPER1 gene and the addition of a single copy integration of the HPH gene. (C) Semiquantitative RT-PCR was carried out to confirm the deletion and reintroduction of MoPER1 gene. (TIF 247 kb)
Table S1. Primers used in this study. (DOC 34 kb)
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files.