Abstract
Age-related macular degeneration (AMD) is the leading cause of blindness in industrialised countries, and thereby a major individual but also a socio-economic burden. Y chromosome loss in nucleated blood cells has been implicated in age-related diseases such as Alzheimer disease and was shown to be caused by increasing age, smoking and genetic factors. Mosaic loss of Y chromosome (mLOY) in peripheral blood was estimated from normalised dosages of genotyping chip data covering the male-specific region of the Y chromosome. After quality control, we assessed the association of mLOY on AMD risk in 5772 male cases and 6732 male controls. In controls the prevalence of mLOY increased significantly with age, which is consistent with previous reports. Importantly, mLOY was associated with late-stage AMD with genome-wide significance (OR: 1.332 [95% CI: 1.206; 1.472], P = 1.60e-08), independent of age, the AMD genetic risk score and the first two principle components of ancestry. Additionally conditioning on smoking behaviour had no influence on the observed association strength. mLOY was strongest associated in individuals aged between 65 and 75 years. Taken together, mLOY is significantly associated with risk for AMD, independent of known and potential confounding factors.
Subject terms: Predictive markers, Genome-wide association studies
Introduction
Age-related macular degeneration (AMD) is the most common cause of blindness in Western societies with age identified as the major risk factor for the disease [1]. The prevalence of AMD rises significantly with age; hence demographic changes towards an increased average life expectancy are anticipated to cause a growing individual and socioeconomic burden [2]. Apart from age, environmental/modifiable as well as genetic factors have firmly been established to be strongly associated with disease risk [1, 3, 4]. Previous findings revealed ambiguous results for an association of gender with AMD and its modulation by genetic variants [2, 5–7].
The latest genome-wide association study by the International AMD Genomics Consortium (IAMDGC) [8–10] has identified numerous regions in the genome involved in AMD pathogenesis. In total, at least 34 regions with currently more than 52 independent associated lead variants have been implicated in disease risk, explaining over 50% of the heritability and more than 30% of disease variance. Although the majority of the heritability can be explained by known genetic markers, no cellular pathway besides the complement cascade as part of the innate immune system was consistently found to be involved in AMD [11].
Mosaic loss of Y chromosome (mLOY) in peripheral blood is the most common acquired mutation in the process of normal aging in men, affecting about 1.8% of the genetic material in the human genome [12]. The prevalence of mLOY increases with age and can exceed 20% in male populations older than 80 years [13]. Furthermore, the occurrence of mLOY is strongly correlated with smoking behaviour [14]. Current smokers have a more than fourfold increased risk for mLOY [13], although this effect seems to be transient as smoking cessation can result in normal mLOY levels after several years [14, 15].
Recent reports suggested a role of Y chromosome loss in risk for all-cause mortality and common age-related disease such as cancer, Alzheimer disease as well as severe atherosclerosis [12–20]. Building on such reports, we aimed to evaluate the contribution of male Y chromosome mosaicism to the risk for late-stage AMD.
Methods
Description of dataset
Initially, data from 6529 males with late-stage AMD (NV, GA, or both, GA/NV AMD) and 7815 male control individuals without fundus changes associated with AMD, all probands unrelated and of European ancestry were provided by the IAMDGC [9]. Inclusion and exclusion criteria of the IAMDGC samples as well as detailed information on ophthalmological grading, quality control of genetic and phenotypic data as well as imputation are given in detail elsewhere [9]. The age of patients and controls reported by the individual studies was usually the date the blood was drawn and the diagnosis was assessed and/or confirmed in the recruitment centre or clinic [9]. For the present study, we excluded 1046 samples, since their DNA was not retrieved from whole blood. Smoking data were available for the samples from the Regensburg, EUGENDA (Nijmegen, the Netherlands and Cologne, Germany), UCLA/Pittsburgh, Southampton and Cambridge (UK) study groups, which are part of the IAMDGC dataset. In addition, smoking status was retrieved from dbGAP for the ARED study samples [21] (accession: phs000001.v3.p1). In total, we had smoking information for 2159 samples (ever smoked/never smoked), 1210 samples (current smoker status) and 956 samples (pack-years, Table 1). The study adhered to the tenets of the declaration of Helsinki and is HIPAA compliant.
Table 1.
Baseline characteristics of the study population
| Cases | Controls | All | OR [95%CI]a | P a | |
|---|---|---|---|---|---|
| All ages [N/mLOY] | 5772/1145 | 6732/918 | 12504/2063 | 1.332 [1.206; 1.472] | 1.60 × 10−08 |
| <65 years [N/mLOY] | 538/69 | 1735/172 | 2273/241 | 1.316 [0.969; 1.771] | 0.0738 |
| 65–70 years [N/mLOY] | 675/108 | 1228/122 | 1903/230 | 1.748 [1.321; 2.311] | 9.02 × 10−05 |
| 70–75 years [N/mLOY] | 1131/209 | 1376/190 | 2507/399 | 1.422 [1.147; 1.764] | 0.0013 |
| 75–80 years [N/mLOY] | 1478/268 | 1191/175 | 2669/443 | 1.282 [1.042; 1.580] | 0.0195 |
| >80 years [N/mLOY] | 1856/482 | 1161/255 | 3017/737 | 1.229 [1.034; 1.465] | 0.0201 |
| Ever smoked [%/N] | 74.58/1137 | 63.6/1022 | 69.38/2159 | 1.688 [1.397; 2.041] | 6.11 × 10−08 |
| Current smoker [%/N] | 16.57/519 | 8.39/691 | 11.9/1210 | 2.638 [1.804; 3.886] | 6.92 × 10−07 |
| Average pack-years (SD) | 27.39 (26.2) | 19.81 (23.88) | 25.09 (25.75) | 1.012 [1.006; 1.018] | 2.47 × 10−04 |
N number of samples, mLOY number of samples with mLOY, OR [95%] odds ratio and 95% confidence interval of association between cases and controls (mLOY or smoking status), P P value of association between cases and controls, SD standard deviation
aLogistic regression models adjusted for age and the first two principal components of ancestry as well as the AMD genetic risk score
Calculation of mLOY
All samples were genotyped on the same genotyping chip (Illumina HumanCoreExome—Goncalo 15038949). A continuous variable, the mean of the log R ratio (LRR), was used to estimate the degree of LOY for each subject and calculated as the mean LRR value of 608 SNP array probes that passed quality control (i.e. call rate >95% and not monomorphic) positioned within the male-specific region of the Y chromosome (mLRRY, hg19: 2,694,521 bp—59,034,049 bp) as described elsewhere [12, 15, 20]. To exclude samples with excessive noise, we computed the standard deviation (SD) of all LRR values of 30,789 SNP array probes on chromosome 1 that passed QC (mLRR1) and excluded 974 samples with a SD of mLRR1 greater than 0.28, as suggested by the manufacturer of the array. This allowed us to assess mLOY in a total of 12,504 individuals (Table 1). We defined males to have a significant level of mLOY in blood, if their mLRRY value was smaller than (1) the mean, (2) one or (3) two SDs from the mean mLRRY in controls. In subsequent analyses, we focused on mLOY individuals with mLRRY values smaller than one SD from the mean mLRRY in controls (2), since the fraction of mLOY individuals in controls was comparable to the frequency observed in other studies.
Statistical analysis
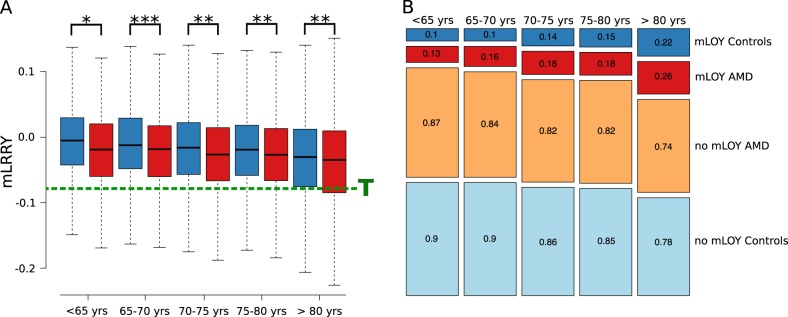
We fit multivariable linear and logistic regression models to assess the association of mLRRY and mLOY with late-stage AMD, respectively, as implemented in R [22]. The analyses were adjusted for the first two principle components of ancestry, which were calculated from the imputed genotypes. In addition, where appropriate we accounted for the age at blood collection. To exclude a potential confounding effect of known and strong AMD associated variants, a genetic risk score (GRS) was computed for AMD with 52 independent variants from 34 loci, as previously reported [10, 23]. Briefly, we calculated the sum of risk increasing alleles from 52 AMD associated variants, weighted by the respective odds ratio of the variant. To account for smoking status in the sensitivity analysis, we considered individuals who ever smoked more than one pack-year as smokers and also adjusted for current smokers as well as the number of pack-years smoked, if available. We report the odds ratio of association and the slope of correlation as well as the respective 95% confidence intervals. The frequency of mLOY in cases and controls was plotted as a mosaicplot implemented in R and the mLRRY values across different age strata are shown as a boxplot, as implemented in R.
Data availability
The genetic data of the IAMDGC can be accessed through dbGAP (accession: phs001039.v1.p1).
Results
We investigated the association of mLOY in blood cells with late-stage AMD by using the currently largest collection of late-stage AMD patients and controls [9]. In total, 12,504 male individuals from 26 studies were included in the analysis (Table 1). All samples were genotyped simultaneously on the same chip in the same genotyping centre, resulting in little heterogeneity of mLOY across studies. Previous reports indicated a strong correlation between mLOY and age [12, 13]. As expected, the mean chromosomal dosage of the Y chromosome, as measured by the average LRR of all probes on the Y chromosome (mLRRY) decreased (P < 5.15 x 10−64) with age in AMD patients and AMD free controls (Fig. 1). We also found that current smokers (slope: −0.02 [95% CI: −0.03; 0.00], P < 0.05), as well as previous smokers (slope: −0.01 [95% CI: −0.02; 0.00], P < 0.05) had reduced levels of mLRRY. In addition, the number of pack-years smoked was inversely correlated to mLRRY (slope per ten pack-years: −0.002 [95% CI: 0.004; 0.000], P < 0.05).
Fig. 1.
Boxplot and mosaicplot of mLOY in AMD patients and controls across different age groups. a The distribution of the mean LRR value across 608 SNP probes on the Y chromosome (mLRRY) for cases (red) and controls (blue) are depicted as a boxplot across different age strata. The threshold (T) for mLOY (mLRRY < −0.08) is depicted as a dashed green line. Significant differences between the mean mLRRY of cases and controls, as assessed by multivariate linear regression models adjusted for age, two principal components and AMD genetic risk score are depicted with asterisks: *P < 0.05; **P < 0.01; ***P < 0.001. b Patients with significant loss of Y chromosome (mLOY) were defined by a mLRRY smaller than one standard deviation from the mean mLRRY values of controls (mLRRY < −0.08). The occurrence of mLOY increased with age in controls (blue) as well as AMD patients (red). Conversely, cases (orange) as well as controls (light blue) with no mLOY were less frequently observed with increasing age. Absolute number of cases and the statistical assessment are given in Table 1
Importantly, we observed a genome-wide significant association of mLRRY (OR per SD increase in mLRRY: 0.867 [95% CI: 0.833; 0.899], P = 8.75 × 10−14) with late-stage AMD, independent of age, ancestry and known AMD risk variants (summarised in a single GRS per person). Next, we defined mLOY as a binary variable in cases and controls by different agnostic mLRRY thresholds. We found that mLOY was statistically significant for association with AMD, either defined as (1) mLRRY smaller than the mean mLRRY in controls (OR: 1.249 [95% CI: 1.159; 1.345], P = 4.54 × 10−09), (2) mLRRY smaller than one SD from the mean of controls (OR: 1.332 [95% CI: 1.206; 1.472], P = 1.60 × 10−08) and (3) mLRRY smaller than two SDs from the mean of controls (OR: 1. 711 [95% CI: 1.406; 2.089], P = 1.03 × 10−08). The observed association was also independent of the above-mentioned confounders. In subsequent analyses, we focused on mLOY as defined by mLRRY smaller than one SD from the mean of mLRRY of controls (2), since this treshhold resulted in a similar fraction of mLOY in controls as previously described [15, 16].
When testing the association of mLOY with AMD in different age groups (Fig. 1), we observed the strongest association when restricting the analysis to individuals aged 65 to 75. The association strength (i.e. the odds ratio) decreased in older and younger individuals (Fig. 1). In addition, we investigated the effect of smoking status on the association strength with AMD. The observed association with late-stage AMD were not diminished when adjusting for ever smoking (OR per SD increase in mLRRY: 0.854 [95% CI: 0.762; 0.932]), current smoking status (OR per SD increase in mLRRY: 0.805 [95% CI: 0.701; 0.922]) or pack-years (OR per SD increase in mLRRY: 0.877 [95% CI: 0.735; 1.044]). Similarly, the association of mLOY did not decrease when adjusting for smoking status (data not shown).
Discussion
Here, we report the association of mLOY with AMD risk, particularly in individuals between 65 and 75 years of age. We defined mLOY with agnostic thresholds from the mean and SD of the mLRRY values in controls, as previously published thresholds of mLRRY (such as −0.4 [13] or −0.15 [15]) resulted in fewer samples with mLOY than expected in this age range. This observation can be explained by the different genotyping chip architecture used in this study, which included a large exonic part to capture the variation of rare variants. However, our definition of mLOY based on one SD from the mean resulted in a similar fraction of mLOY in controls as expected in the respective age groups.
In line with independent results reported elsewhere [12, 13, 15], we also found that mLOY increased significantly with age in our study. Since age is the major risk factor for AMD, we adjusted the analysis by age, ensuring that the observed association is not driven by the increased age of our AMD samples compared to controls.
Previously, the strongest genetic associations for AMD were observed in or near genes of the complement system [9] and an increased complement activation has been shown to play a major role in AMD [11, 24, 25]. Enhanced and prolonged complement activity and thus immune activation might accelerate loss of Y chromosome and could be an important confounder in our analysis. However, the observed association of mLOY with AMD was independent of known AMD risk variants, demonstrated by adjusting the analysis for an AMD GRS, which revealed no influence on the observed association. Of note, genome-wide association studies aiming to uncover the genetic basis of mLOY have not reported any associated loci harbouring complement genes [20].
We observed that the association of mLOY with AMD is reduced in individuals over 75 years of age. This is in full agreement with a similar effect observed in the association of mLOY with Alzheimer disease [13], with decreased hazard ratios reported for older compared to younger individuals. It is known that in AMD genetic effects can be modulated by age [5, 7, 23]. It has been hypothesised that younger diseased individuals require more disease associated variants to explain the early onset of disease pathology. Similarly, mLOY at younger age may suggest an increased impairment and senescence of the immune system [12–14, 16, 17, 26], a process that is assumed to play a role in AMD and other immune related diseases [27, 28] as well as cancer [29, 30].
Taken together, we report a genome-wide significant association of mLOY with late-stage AMD independent of known and potential confounders. The mLOY marginally improved the AUC of the logistic regression model from 0.824 (GRS + smoking + age) to 0.827, which is consistent with the degree of added risk from other known, low penetrance genetic factors.
Electronic supplementary material
Acknowledgements
The authors thank Sudha K. Iyengar, Rob Igo and Alberto H. Santana (Case Western Reserve University, Cleveland, Ohio, USA) as well as the IAMDGC (http://eaglep.case.edu/iamdgc_web/) for providing the raw probe intensities. We are also grateful to valuable comments to the manuscript from Michael Gorin (Jules Stein Eye Institute, Los Angeles, CA, USA). The list of consortium members reflects the author list of the previous publication by Fritsche et al. 2016 [9], which is listed in the Supplement.
Funding
The work was funded, in parts by a grant from the German Research Foundation (GR 5065/1-1 to FG) and institutional funds of the Institute of Human Genetics, University of Regensburg (TG77 to BHFW). Genotyping was conducted as part of the IAMDGC exome-chip project supported by CIDR (contract number HHSN268201200008I) and funded by EY022310 (to Jonathan L. Haines, Case Western Reserve University, Cleveland) and 1 × 01HG006934-01 (to Gonçalo R. Abecasis, University of Michigan, Department of Biostatistics). The funding bodies had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author contributions
FG and BHFW made substantial contributions to conception and design of the study; CK and IAMDGC provided the genetic data; FG and CK devised the methodology and analyzed the clinical and genetic data; FG, CK, and BHFW wrote the manuscript. AIDH, DEW, AL and VC contributed phenotypes to the final analyses. All authors have given approval of the final version of the manuscript to be published.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41431-018-0238-8) contains supplementary material, which is available to authorized users.
References
- 1.Chakravarthy U, Wong TY, Fletcher A, et al. Clinical risk factors for age-related macular degeneration: a systematic review and meta-analysis. BMC Ophthalmol. 2010;10:31. doi: 10.1186/1471-2415-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong WL, Su X, Li X, et al. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Heal. 2014;2:e106–16. doi: 10.1016/S2214-109X(13)70145-1. [DOI] [PubMed] [Google Scholar]
- 3.Grassmann F, Ach T, Brandl C, Heid IM, Weber BHF. What does genetics tell us about age-related macular degeneration? Annu Rev Vis Sci. 2015;1:73–96. doi: 10.1146/annurev-vision-082114-035609. [DOI] [PubMed] [Google Scholar]
- 4.Grassmann F, Fauser S, Weber BHF. The genetics of age-related macular degeneration (AMD) – novel targets for designing treatment options? Eur J Pharm Biopharm. 2015;95:194–202. doi: 10.1016/j.ejpb.2015.04.039. [DOI] [PubMed] [Google Scholar]
- 5.Winkler TW, Brandl C, Grassmann F, et al. Investigating the modulation of genetic effects on late AMD by age and sex: lessons learned and two additional loci. PLoS One. 2018;13:e0194321. doi: 10.1371/journal.pone.0194321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grassmann F, Friedrich U, Fauser S, et al. A candidate gene association study identifies DAPL1 as a female-specific susceptibility locus for age-related macular degeneration (AMD) Neuromolecular Med. 2015;17:111–20. doi: 10.1007/s12017-015-8342-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grassmann F, Cantsilieris S, Schulz-Kuhnt AS, et al. Multiallelic copy number variation in the complement component 4A (C4A) gene is associated with late-stage age-related macular degeneration (AMD) J Neuroinflamm. 2016;13:81. doi: 10.1186/s12974-016-0548-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grassmann F, Heid IM, Weber BHF. Recombinant haplotypes narrow the ARMS2/HTRA1 association signal for age-related macular degeneration. Genetics. 2017;205:919–24. doi: 10.1534/genetics.116.195966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fritsche LG, Igl W, Bailey JNC, et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48:134–43. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grassmann F, Kiel C, Zimmermann ME, et al. Genetic pleiotropy between age-related macular degeneration (AMD) and sixteencomplex diseases and traits. Genome Med. 2017;9:29. doi: 10.1186/s13073-017-0418-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber BHF, Charbel Issa P, Pauly D, Herrmann P, Grassmann F, Holz FG. The role of the complement system in age-related macular degeneration. Dtsch Arztebl Int. 2014;111:133–8. doi: 10.3238/arztebl.2014.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forsberg LA. Loss of chromosome Y (LOY) in blood cells is associated with increased risk for disease and mortality in aging men. Hum Genet. 2017;136:657–63. doi: 10.1007/s00439-017-1799-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dumanski JP, Lambert JC, Rasi C, et al. Mosaic loss of chromosome Y in blood is associated with Alzheimer disease. Am J Hum Genet. 2016;98:1208–19. doi: 10.1016/j.ajhg.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dumanski JP, Rasi C, Lönn M, et al. Mutagenesis. Smoking is associated with mosaic loss of chromosome Y. Science. 2015;347:81–3. doi: 10.1126/science.1262092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou W, Machiela MJ, Freedman ND, et al. Mosaic loss of chromosome Y is associated with common variation near TCL1A. Nat Genet. 2016;48:563–8. doi: 10.1038/ng.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forsberg LA, Rasi C, Malmqvist N, et al. Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat Genet. 2014;46:624–8. doi: 10.1038/ng.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forsberg LA, Gisselsson D, Dumanski JP. Mosaicism in health and disease - clones picking up speed. Nat Rev Genet. 2017;18:128–42. doi: 10.1038/nrg.2016.145. [DOI] [PubMed] [Google Scholar]
- 18.Noveski P, Madjunkova S, Sukarova Stefanovska E, et al. Loss of Y chromosome in peripheral blood of colorectal and prostate cancer patients. PLoS One. 2016;11:e0146264. doi: 10.1371/journal.pone.0146264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haitjema S, Kofink D, van Setten J, et al. Loss of Y chromosome in blood is associated with major cardiovascular events during follow-up in men after carotid endarterectomy. Circ Cardiovasc Genet. 2017;10:e001544. doi: 10.1161/CIRCGENETICS.116.001544. [DOI] [PubMed] [Google Scholar]
- 20.Wright DJ, Day FR, Kerrison ND, et al. Genetic variants associated with mosaic Y chromosome loss highlight cell cycle genes and overlap with cancer susceptibility. Nat Genet. 2017;49:674–9. doi: 10.1038/ng.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grassmann F, Mengelkamp J, Brandl C, Harsch S, Zimmermann ME, Linkohr B, et al. A deep learning algorithm for prediction of age-related eye disease study (AREDS) severity scale for age-related macular degeneration from color fundus photography. Ophthalmology. 2018 Apr 10 [Epub ahead of print, PMID:29653860] [DOI] [PubMed]
- 22.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: the R Foundation for Statistical Computing. 2011. ISBN: 3-900051-07-0. Available online at https://www.R-project.org/.
- 23.Grassmann F, Fritsche LG, Keilhauer CN, Heid IM, Weber BHF. Modelling the genetic risk in age-related macular degeneration. PLoS One. 2012;7:e37979. doi: 10.1371/journal.pone.0037979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zipfel PF, Lauer N, Skerka C. The role of complement in AMD. Adv Exp Med Biol. 2010;703:9–24. doi: 10.1007/978-1-4419-5635-4_2. [DOI] [PubMed] [Google Scholar]
- 25.Lorés-Motta Laura, Paun Constantin C., Corominas Jordi, Pauper Marc, Geerlings Maartje J., Altay Lebriz, Schick Tina, Daha Mohamed R., Fauser Sascha, Hoyng Carel B., den Hollander Anneke I., de Jong Eiko K. Genome-Wide Association Study Reveals Variants in CFH and CFHR4 Associated with Systemic Complement Activation. Ophthalmology. 2018;125(7):1064–1074. doi: 10.1016/j.ophtha.2017.12.023. [DOI] [PubMed] [Google Scholar]
- 26.Williams GC. Pleiotropy, natural selection, and the evolution of senescence. Sci Aging Knowl Environ. 2001;2001:cp13. [Google Scholar]
- 27.Tsoukas C. Immunosenescence and aging in HIV. Curr Opin HIV AIDS. 2014;9:398–404. doi: 10.1097/COH.0000000000000077. [DOI] [PubMed] [Google Scholar]
- 28.Deeks SG. HIV infection, inflammation, immunosenescence, and aging. Annu Rev Med. 2011;62:141–55. doi: 10.1146/annurev-med-042909-093756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ershler WB. The influence of an aging immune system on cancer incidence and progression. J Gerontol. 1993;48:B3–B7. doi: 10.1093/geronj/48.1.B3. [DOI] [PubMed] [Google Scholar]
- 30.Campisi J, Kim S, Lim CS, Rubio M. Cellular senescence, cancer and aging: the telomere connection. Exp Gerontol. 2001;36:1619–37. doi: 10.1016/S0531-5565(01)00160-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genetic data of the IAMDGC can be accessed through dbGAP (accession: phs001039.v1.p1).