Abstract
Vertebrate kidneys contain nephron functional units where specialized epithelial cell types are organized into segments with discrete physiological roles. Many gaps remain in our understanding of how segment regions develop. Here, we report that the transcription factor empty spiracles homeobox gene 1 (emx1) is a novel nephron segment regulator during embryonic kidney development in zebrafish. emx1 loss of function altered the domains of distal segments without changes in cell turnover or traits like size and morphology, indicating that emx1 directs distal segment fates during nephrogenesis. In exploring how emx1 influences nephron patterning, we found that retinoic acid (RA), a morphogen that induces proximal and represses distal segments, negatively regulates emx1 expression. Next, through a series of genetic studies, we found that emx1 acts downstream of a cascade involving mecom and tbx2b, which encode essential distal segment transcription factors. Finally, we determined that emx1 regulates the expression domains of irx3b and irx1a to control distal segmentation, and sim1a to control corpuscle of Stannius formation. Taken together, our work reveals for the first time that emx1 is a key component of the pronephros segmentation network, which has implications for understanding the genetic regulatory cascades that orchestrate vertebrate nephron patterning.
Introduction
Ontogeny of the vertebrate kidney is a dynamic process involving the sequential formation and regression of multiple structures from the intermediate mesoderm (IM) during embryogenesis1. These developmental events can encompass up to three renal forms, termed the pronephros, mesonephros, and metanephros, that share a composition of nephron subunits in various anatomical arrangements1. In mammals, for example, the first two kidneys are transient structures where only the second functions during gestation, and the third form becomes the adult organ2,3. In lower vertebrates, such as fish, the pronephros is both transient as well as functional, and adults retain a mesonephros that develops during juvenile stages2,3. Nephron units serve to filter the blood, and then modify this excretory fluid to retain essential nutrients and regulate water balance. Each nephron has discrete segments of epithelial cells specialized for particular secretory and absorptive tasks. While segment organization is broadly conserved across vertebrate kidney forms, differences enable functionalities such as enhanced water retention in terrestrial species4. The pathways that direct segment regionalization in developing nephrons across kidney forms are beginning to be understood, and recent advances have revealed insights into the mechanisms of segment pattern formation, growth, and terminal differentiation in several vertebrate species5–12.
The zebrafish is a useful genetic model to study organogenesis13, and is particularly amenable to renal development and disease studies because the embryonic pronephros has a simple anatomy of two nephrons that have a segment composition like other vertebrates14–16. The pronephros develops rapidly, forming segments by 24 hours post fertilization (hpf), and the nephrons are also linear at this time, facilitating the precise assessment of segmentation events17 and subsequent physiological functions18. The nephrons share a single blood-filtering glomerulus, and their epithelial tubules have two proximal segments (proximal convoluted tubule, PCT; proximal straight tubule, PST) and two distal segments (distal early, DE; distal late, DL) that are closely associated with a gland known as the corpuscle of Stannius (CS), followed by a pronephric duct (PD)19.
To date, several studies have identified essential signaling molecules and transcription factors that mitigate pronephros segmentation, which has been speculated to offer a blueprint for conserved segmentation mechanisms across kidney forms5. During distal pronephros development, the Iroquois homeodomain transcription factor Iroquois homeobox 3b (irx3b)/Irx3 specifies the DE segment, an aspect shared between zebrafish and Xenopus, respectively20,21. In zebrafish, patterning of the DL segment is reliant on the zinc finger transcription factor MDS1 and EVI1 complex locus (mecom)22 as well as the T-box transcription factors T-box 2a (tbx2a) and T-box 2b (tbx2b)23, though the relationships between these key factors has not yet been established. A crucial upstream effector of proximal and distal cell fate decisions during the process of zebrafish pronephros segmentation is retinoic acid (RA) signaling19. RA promotes proximal segment identities and inhibits distal ones through patterning events that transpire during pre-gastrula and gastrula stages19,20,24 and lead to alterations in the expression domains of key transcription factors that include irx3b20, mecom22, tbx2a/b23, sim1a25, and etv5a26. The spatial expression domains of several other transcription factors in the renal progenitors are also altered due to the abrogation of RA biosynthesis or RA receptor signaling20, though their functional contribution(s) to nephrogenesis have yet to be assigned.
emx1 encodes a homeodomain transcription factor that is expressed in renal progenitors during early stages of zebrafish pronephros development27, and numbers among those genes that have been implicated to act downstream of RA signaling during nephron segmentation20. emx1 is related to the fruitfly gene empty spiracles (ems)28, which controls the formation of anterior head segments and the development of the olfactory system29,30. In zebrafish and rodents, emx1/Emx1 is expressed in the developing brain27,31–33, and Emx1 deficient mice evince developmental defects in the forebrain including disruption of corpus callosum formation34,35. Interestingly, microarray data curated on the Genitourinary Development Molecular Anatomy Project (GUDMAP) localized murine Emx1 transcripts in the metanephros to the renal vesicle at stage E12.5, then to S-shaped body structures, proximal tubules and loop of Henle anlage at E15.5, which may indicate roles in nephrogenesis36. Also, Emx1 expression was detected in the ureteric bud at E11.5 and ureteric tips at E15.5, suggesting involvement with collecting system formation36. Despite their intriguing expression patterns during vertebrate nephrogenesis, however, the function of emx1/Emx1 during nephron ontogeny has not been examined.
Here, we establish several roles for emx1 during nephron distal segment patterning in the zebrafish embryo kidney. Using loss of function techniques, we found that emx1 is necessary to pattern the neighboring DE and DL segment domains as well as to repress CS fate. We ascertained that RA signaling negatively regulates the domain of emx1 expression in the pronephros. Through genetic studies, we then determined that emx1 acts downstream of the distal patterning factors mecom and tbx2b, which we show here to comprise a cascade that promotes DL formation. Finally, we show that emx1 is necessary to restrict the expression domains of irx3b, irx1a and sim1a, which are known to mitigate DE and CS development, respectively. Taken together, these results provide new evidence that emx1 is a critical component of regulating segmentation in the pronephros, and suggest for the first time that emx1 has key roles in vertebrate mesoderm development. These findings have implications for understanding the basis of congenital anomalies of the kidney and urinary tract (CAKUT) and current approaches in regenerative medicine such as renal organoids.
Results
emx1 expression is dynamic in renal progenitors and then localizes to the DL segment
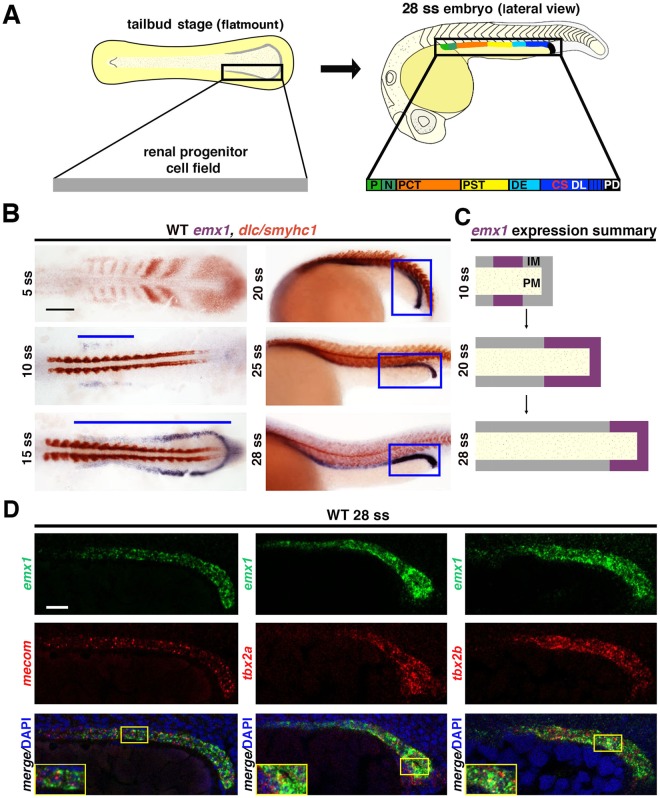
The zebrafish pronephros arises from bilateral stripes of IM precursors that develop into a pair of parallel nephrons by the 28 somite stage (ss) (Fig. 1A)17. This process involves a mesenchymal to epithelial transition of the renal progenitors37,38 along with their pattern formation into a series of at least 8 discrete segment domains, wherein the segments contain populations of like transporter cells that in some regions are intermingled with a discrete subset of multiciliated cells39. Prior studies have reported the localization of emx1 transcripts between the 15 ss and 28 ss in a spatial arrangement consistent with the IM and pronephros territories20,27.
Figure 1.
emx1 is dynamically expressed during zebrafish pronephros ontogeny. (A) The renal progenitor field is patterned into a segmented pronephros with the following 8 regions: P, podocytes; N, neck; PCT, proximal convoluted tubule; PST, proximal straight tubule; DE, distal early; CS, corpuscle of Stannius; DL, distal late; PD, pronephric duct. (B) WISH in WT embryos for emx1 (purple) and smyhc1 (red) between 5 ss and 28 ss. Blue lines and boxes indicate domains of pronephros expression. Scale bar = 100 μm. (C) Summary of the emx1 expression domain during nephrogenesis, where IM designates the intermediate mesoderm renal progenitors, and PM designates paraxial mesoderm. (D) FISH analysis of emx1, mecom, tbx2a, and tbx2b in the distal pronephros. Yellow boxes demarcate areas shown in insets. Scale bar = 50 μm.
To more closely examine the spatiotemporal location of emx1 transcripts during mesoderm development, we performed a series of double whole mount in situ hybridization (WISH) experiments over a developmental time course between the tailbud stage and the 28 ss. We used a combination of riboprobes to detect emx1 in the IM relative to the adjacent paraxial mesoderm (PM) territory using the marker deltaC (dlc), which enables visualization of the early somites, or slow myosin heavy chain 1 (smyhc1), which marks somites at later stages. emx1 expression was first detected adjacent to the somites at the 10 ss in a subset of the renal progenitor field, while transcripts were present throughout the entire renal progenitor field by the 15 ss (Fig. 1B). Between the 20–28 ss, emx1 transcripts localized to the presumptive distal segment territories and became restricted to the putative DL domain by the 28 ss (Fig. 1B). These data indicate that the emx1 expression domain is dynamic within the developing pronephros (schematized in Fig. 1C), similar to many other genes20.
To define further how the domain of emx1 expression aligned with the DL segment, we performed double fluorescent in situ hybridization (FISH) expression studies with confocal imaging at the 28 ss with several markers of this nephron population. Prior studies have localized mecom transcripts to the DL segment22 and demonstrated the co-localization of tbx2a and tbx2b with the DL specific marker solute carrier family 12 (sodium/chloride transporter), member 3 (slc12a3)23. We found that emx1 and mecom transcripts co-localized in the DL segment, and that emx1 transcripts also co-localized with tbx2a and tbx2b (Fig. 1D). These results support the conclusion that the DL segment contains a common populace of emx1, mecom, and tbx2a/b expressing cells. This suggested that emx1 may have roles in regulating DL development, and potentially other pronephros segments as well in light of its dynamic expression pattern in renal progenitors.
emx1 deficient embryos have altered distal tubule segments and CS development
We conducted loss of function studies to assess the role of emx1 in pronephros development. To create models of emx1 deficiency, we first performed knockdown studies with a morpholino combination that was designed to interfere with the splice donor site of exon 1 and the splice acceptor site of exon 2 (Fig. S1A). Following microinjection of the splice targeting combination in fertilized wild-type (WT) embryos at the 1-cell stage, we collected the embryos at the 28 ss and performed RNA extraction. Through RT-PCR analysis we determined that the splice morpholino combination led to production of an incorrectly processed emx1 transcript, with an in-frame premature stop codon due to the inclusion of intronic sequence (Fig. S1A,B). As the predicted Emx1 protein encoded by this transcript is truncated and entirely lacks the homeodomain sequence, we concluded that transcriptional activity is likely abrogated in proteins produced from these altered transcripts.
To explore the roles of emx1 in renal development, we assessed the expression domains of segment-specific markers using WISH in in emx1 deficient embryos compared to WT controls at the 28 ss. emx1 deficiency was associated with alterations in formation of both the DE and DL segment domains (Fig. 2A–C). The DE segment, marked by solute carrier family 12 (sodium/potassium/chloride transporter), member 1 (slc12a1) was expanded in emx1 deficient embryos (Fig. 2A) and measurement of absolute segment length revealed a statistically significant increase in the DE domain compared to WT control embryos (Fig. 2B). In contrast, the DL segment, marked by slc12a3 was reduced in emx1 deficient embryos (Fig. 2A) and measurement of absolute segment length revealed a statistically significant decrease in the DL compared to WT control embryos (Fig. 2C). Evaluation of the other tubule nephron segments with WISH revealed normal spatial expression domains of PCT, PST, and PD markers along with normal domains of both pan-proximal (PCT, PST) and pan-distal markers (DE, DL) compared to WT sibling control embryos (Figs S1C–J, S2). Given the alterations in the distal tubule segments, we also examined development of the CS lineage, which emerges from the IM in the vicinity occupied by DE and DL precursors19,40,41. Interestingly, emx1 deficient embryos had an increased expression domain of the CS specific marker stanniocalcin 1 (stc1) (Fig. 2D). Quantification of stc1+ expressing cells revealed that there was a significant increase in the number of CS cells associated with each nephron (Fig. 2E), while the area of individual cells was unchanged (Fig. 2F).
Figure 2.
emx1 deficient embryos exhibit a DE expansion, DL reduction and an expansion of the associated CS gland. (A) WISH analysis (lateral view) of 28 ss WT, emx1 deficient, and emx1 deficient embryos co-injected with emx1 mRNA (emx1 rescue) for the distal segment markers (purple): slc12a1 for the DE, and slc12a3 for the DL. Blue boxes demarcate areas of pronephros gene expression. Scale bar = 100 μm. (B,C) Average segment lengths of the DE and DL for WT, emx1 deficient, and emx1 rescue embryos. (D) WISH panel (dorsal view) of 28 ss WT, emx1 deficient, and emx1 rescue embryos for the CS marker stc1 (purple). Blue boxes demarcate a single CS territory while the contralateral side is unlabeled. Scale bar = 20 μm. Quantification of stc1+ cell number (E) and size (F) of WT, emx1 deficient, and rescue embryos. ***p < 0.001; N.S. = not significant. Error bars indicate standard error. (G) Summary of pronephros phenotype in emx1 deficient embryos with respect to the axial location of somites.
Next, we obtained a published emx1 morpholino that blocks the translational start site42, and examined nephron segmentation and CS development at the 28 ss (Figs S3, S4). Analysis of these emx1 deficient embryos showed similar phenotypes, with of an expanded DE segment, shortened DL segment and expanded CS populace (Figs S3, S4). The similarities in pronephros and CS phenotypes in these distinct models of emx1 deficiency strengthened our conclusion that Emx1 is necessary for formation of the DE and DL segments as well as the CS.
To further discern whether these phenotypes were caused by the loss of Emx1 activity or possibly influenced by off-target effects, we performed rescue studies in emx1 deficient embryos to test whether the expression of emx1 transcripts that encoded the full-length mature mRNA sequence would be sufficient to restore pronephros and CS development. Following in vitro mRNA synthesis and purification, we co-injected emx1 transcripts with the splice morpholino combination at the 1 cell stage and fixed embryos at the 28 ss to examine segment domains using WISH. We observed a normal DE, DL and CS in emx1 deficient embryos that were co-injected with emx1 transcripts (Fig. 2A–F). Taken together, these results led us to conclude that the loss of Emx1 specifically alters the balance of distal segment fates, leading to increases in the DE segment and CS lineages along with a concomitant reduction in the DL segment (Fig. 2G).
emx1 deficiency does not alter nephron segment traits of cell turnover, size or arrangement
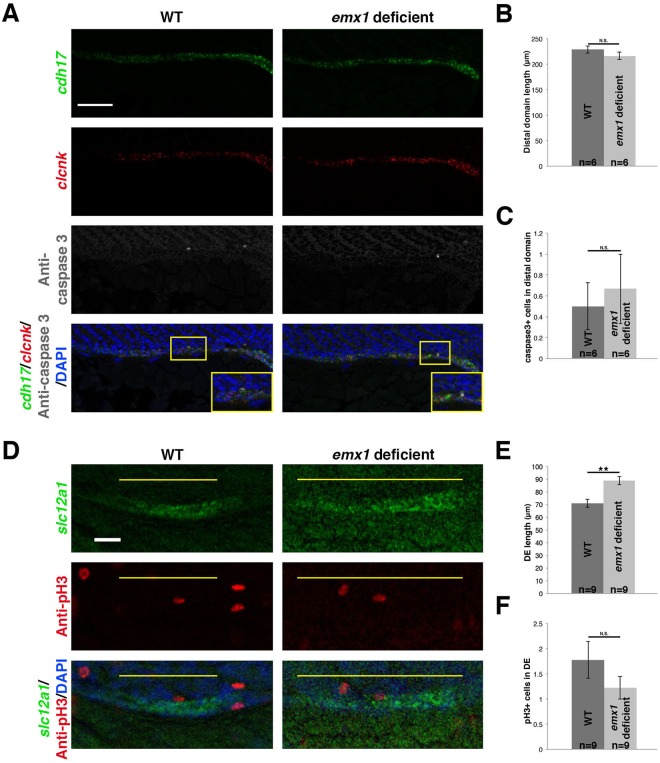
We next determined if emx1 affected the balance of distal segment fates by influencing the cellular dynamics of populations in this developing region, as early segment size in the zebrafish pronephros is affected by proliferation43. As changes in the distal segment domains of emx1 deficient embryos were first observed between the 26–28 ss (Figs 2 and S2–S4, data not shown), we examined cell dynamics at this time. Using acridine orange staining, we observed no difference in the number of cells undergoing apoptosis within the distal tubule or its neighboring tissues at the 28 ss in emx1 deficient embryos compared to WT controls (Fig. S5). Elevated numbers of acridine orange+ cells were detected within the central nervous system of emx1 deficient embryos compared to WT controls (Fig. S5), which may reflect emx1 function(s) in neuronal development. To further assess cell death, we performed whole mount immunofluorescence to detect activated endogenous Caspase-3 in combination with FISH to examine the distal pronephros based on expression of the pan-tubule marker cadherin 17 (cdh17) and the pan-distal segment/CS marker chloride channel K (clcnk). As in our prior WISH studies, the absolute length of the pan-distal segment domain was unchanged in emx1 deficient embryos, and within this domain, there was no significant difference in the number of Caspase-3+ cells compared to WT control embryos (Fig. 3A–C). These results indicate that the reduced DL segment population in emx1 deficient embryos does not originate with enhanced cell death within this region.
Figure 3.
emx1 deficiency is not associated with changes in cell death or proliferation within the pronephros. (A) Confocal imaging of double FISH analysis (lateral view) of 28 ss WT and emx1 deficient embryos for the renal epithelial cell marker cdh17 (green), the pan-distal segment marker clcnk (red), combined with protein localization of active caspase-3 (grey) and DAPI staining (blue) through IHC. Yellow boxes demarcate areas shown in insets. Scale bar = 50 μm. (B,C) Quantification of caspase-3+ cell number and clcnk domain. (D) Confocal imaging of FISH analysis (lateral view) of 28 ss WT and emx1 deficient embryos for the DE marker slc12a1 (green), with protein localization of pH3 (red) and DAPI (blue) using IF. Yellow lines demarcate the DE domain. Scale bar = 20 μm. (E,F) Quantification of pH3+ cells in the DE and average DE length. **p < 0.01; N.S. = not significant. Error bars indicate standard error.
Subsequently, we examined cell proliferation by whole mount immunofluorescence (IF). For this, we detected phospho-Histone H3 (pH3) staining in combination with FISH to identify the DE segment populace based on their expression of slc12a1 transcripts. emx1 deficient embryos formed an expanded slc12a1+ domain, as observed previously, and quantification of the pH3+ cell number within the DE showed no significant difference to WT control embryos at the 28 ss (Fig. 3D–F). This indicates that an alteration in cell proliferation within the DE segment does not underlie the significant expansion of this domain in emx1 deficient embryos. Further, alterations in pH3+ number were not observed in the neighboring regions either (data not shown), consistent with the conclusion that the CS lineage was also not proliferating at an increased rate in emx1 deficient embryos. Thus, our results indicate that Emx1 does not influence cellular turnover in the developing pronephros or CS that leads to segmentation changes.
A remaining possibility that could explain the expanded DE and reduced DL segment sizes would be alterations in the size or arrangement of individual cells in these populations. Therefore, we performed whole mount FISH and confocal imaging to examine these features. The DE and DL segment populations were again visualized based on their expression of slc12a1 and slc12a3, respectively, and cell nuclei were labeled using DAPI. First, we measured the internuclear distance (IND) of cells in each segment. Within the expanded DE of emx1 deficient embryos (Fig. 4A), there was no change in the IND compared to WT embryos (Fig. 4B). Likewise, within the reduced DL segment of emx1 deficient embryos (Fig. 4C), there was no change in the IND compared to WT embryos (Fig. 4D). We next measured segment volume in emx1 deficient embryos and WT controls. Consistent with the alterations in segment length, the DE was significantly higher in volume, and the DL was significantly reduced in volume, in emx1 deficient embryos (Fig. 4E). However, the segment volume to length ratio or the number of nuclei in a given area was not significantly different between emx1 deficient and WT control embryos in the DE or DL (Fig. 4F,G). These analyses indicate that the expanded DE and reduced DL are not related to changes in cell dimension or arrangement differences. As changes in cellular turnover were also not detected in emx1 deficient embryos, we concluded that the alterations in the DE and DL segments reflect changes in cell fates of the renal progenitors.
Figure 4.
Spatial arrangement and size of renal cells in the DE and DL are unaffected by emx1 deficiency. (A,C) Confocal imaging of FISH analysis (lateral view) of 28 ss WT and emx1 deficient embryos for the DE and DL segment makers slc12a1 and slc12a3 (red), respectively, with DAPI staining (grey). Yellow lines demarcate segment domains. Scale bar = 20 μm. (B,D) Quantification of internuclear distance of DE and DL segment cells in WT and emx1 deficient embryos. Quantification of tubule volume (E), segment volume to segment length ratio (F), and the number of nuclei per 30 μm of segment in WT and emx1 deficient DE and DL segments (G). *<0.05; N.S. = not significant. Error bars represent standard error.
RA signaling negatively regulates emx1 expression within the developing pronephros
Since our results indicated that Emx1 is required to regulate distal segment patterning by influencing cell fate of the nephron precursors, we next sought to determine the relationship of emx1 with known patterning factors that regulate fate choice in the IM. RA signaling acts to regionalize the renal progenitors into proximal and distal domains along the embryonic trunk19. Elevating bioactive retinoid levels by treatment with exogenous all-trans RA expands proximal segment domains and reduces distal segments; conversely, genetic mutations in the RA biosynthetic enzyme encoded by aldehyde dehydrogenase 1 family, member A2 (aldh1a2) or blocking RA production with the Aldh inhibitor 4-diethylaminobenzaldehyde (DEAB) expands distal segments and reduces proximal segments19,20. Alterations in the levels of RA have dramatic consequences for nephron patterning due to changes in the spatiotemporal expression of essential transcription factors. Thus, we hypothesized that RA was likely to regulate emx1 expression in the zebrafish pronephros.
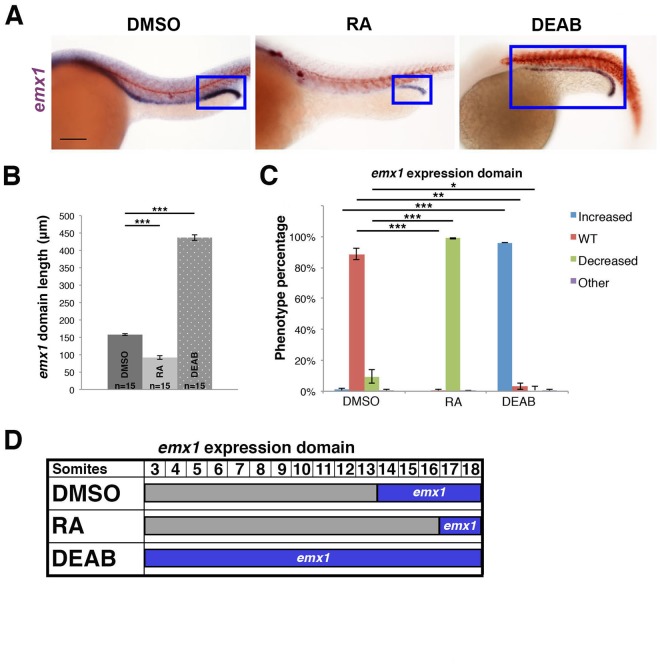
To test this, WT embryos were treated with all-trans RA, DEAB, or dimethyl sulfoxide (DMSO) as a vehicle control between 60% epiboly until 16.5 hpf when the solutions were removed and replaced with E3 media. The embryos were fixed when they reached the 28 ss, and double WISH was performed to assess the emx1 expression domain within the pronephros with respect to the axial location of somites along the trunk, where the latter was visualized based on smyhc1 expression. Exogenous RA reduced the emx1 expression domain within the pronephros (Fig. 5). Conversely, DEAB treatment expanded the emx1 domain in the nephrons (Fig. 5). These alterations in emx1 domain parallel alterations in the domain of slc12a3 expression, which marks the DL (Fig. S6). These results are consistent with the known effects of RA to promote proximal segment fates and inhibit distal segment fates, and suggest that RA signaling in renal progenitors negatively regulates the spatial domain of emx1 expression.
Figure 5.
emx1 expression within the pronephros is negatively regulated by RA signaling. (A) WISH panel (lateral view) of emx1 expression in 28 ss embryos treated with DMSO vehicle, all-trans RA, or DEAB. Blue boxes demarcate domains of emx1 expression in the pronephros. Scale bar = 100 μm. (B) Quantification of the emx1 expression domain in DMSO, RA, and DEAB treated embryos. (C) Quantification of phenotypes, categorized as increased, WT, decreased, or other, where the latter include those developmentally delayed or too dysmorphic to measure. (D) Summary of the emx1 expression domain in treatment groups. *p < 0.05; **p < 0.01; ***p < 0.001. N.S. = not significant. Error bars represent standard error.
RA signaling works by RA binding to the RA receptor (RAR), which interacts with DNA as a heterodimer with the retinoid X receptor (RXR) on regions known as Retinoic Acid Response Elements (RAREs). The consensus sequence for these RAREs is a direct repeat of the following motif: 5′- PuG(G/T)TCA spaced by 1, 2 or 5 base pairs44. Thus, we examined the promoter region of emx1 for the presence of RAREs to explore the possibility that RA directly regulates emx1. Interestingly, no RAREs were found within the emx1 promoter (data not shown), suggesting that RA has an indirect relationship with emx1.
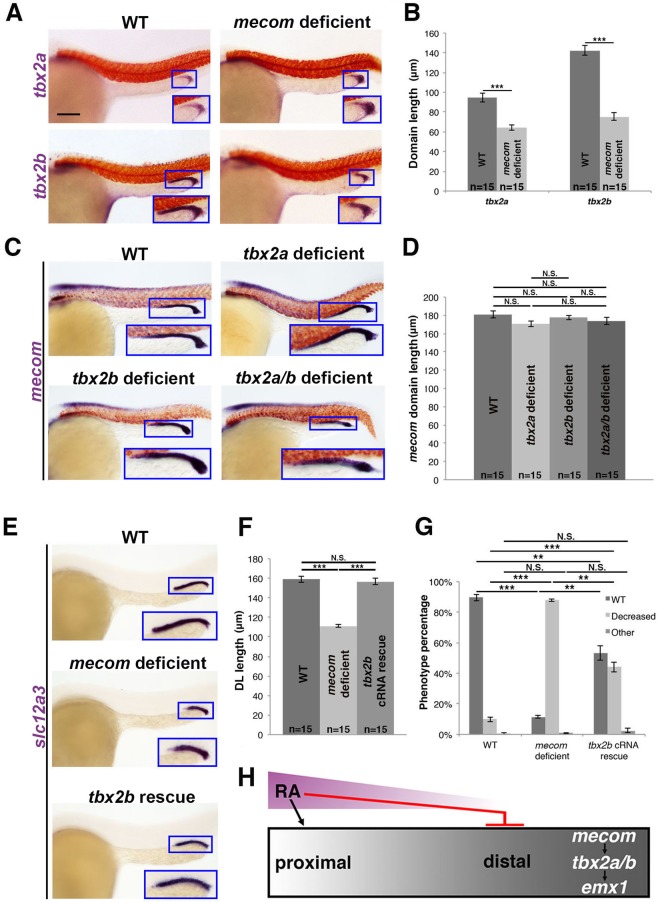
A hierarchy of the transcription factors encoded by mecom, tbx2b, and emx1 controls pattern formation of the DL nephron segment
In the zebrafish pronephros, DL segment development is reliant on activity of the transcription factors encoded by mecom and tbx2a/b, where loss of either mecom or tbx2a/b function results in a shorter DL segment22,23. Further, these genes act downstream of RA and are both negatively regulated by RA during nephron ontogeny22,23, similar to emx1. Given our results that emx1 is necessary for DL development, and causes a nearly identical DL phenotype compared to mecom or tbx2a/b loss of function, we next sought to delineate the relationships between these transcription factors and emx1.
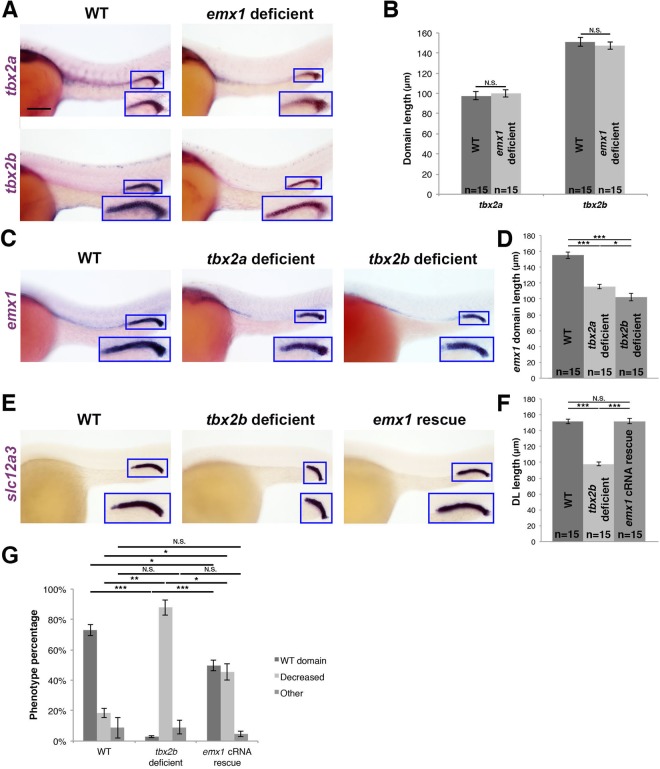
First, we assessed the relationship between emx1 and mecom. We performed WISH in emx1 deficient embryos to examine mecom expression, and observed no change compared to WT controls (Fig. 6A,B). Conversely, mecom deficient embryos had a significantly shorter domain of emx1 expression in the distal pronephros (Fig. 6C,D), suggesting that mecom is upstream of emx1. To test this, rescue studies were performed to determine if emx1 expression could restore DL segment length in the context of mecom loss of function. Provision of emx1 capped mRNA (cRNA) was sufficient to rescue DL segment length in approximately 50% of mecom deficient embryos (Fig. 6E–G). Conversely, provision of mecom cRNA was not sufficient to rescue the DL in emx1 morphants (Fig. S7). These results lead us to conclude that mecom acts upstream of emx1, and suggest that emx1 is a critical effector of Mecom, either directly or indirectly, during pronephros development.
Figure 6.
mecom acts upstream of emx1 to regulate DL segment development. (A) WISH (lateral view) of 28 ss WT and emx1 deficient embryos for mecom (purple) and smyhc1 (red) expression. (B) Quantification of mecom domain length. (C) WISH of 28 ss WT and mecom deficient embryos for emx1 (purple) and smyhc1 (red). (D) Quantification of emx1 domain length. (E) WISH (lateral view) of 28 ss WT, mecom deficient, and mecom deficient embryos co-injected with emx1 capped mRNA (emx1 cRNA rescue) for the DL segment marker slc12a3 (purple). (A, C, E) Blue boxes demarcate domains of pronephros expression. Scale bar = 100 μm. (F) Quantification of DL length for WT, mecom deficient, and emx1 cRNA rescue embryos. (G) Quantification of phenotype percentages for (F), categorized as WT, decreased, or other, where the latter include those developmentally delayed or too dysmorphic to measure. **p < 0.01; ***p < 0.001; N.S. = not significant. Error bars represent standard error.
Next, we explored the relationship between tbx2a/b and emx1. Using WISH, we found that there was no change in the spatial expression domain of either tbx2a or tbx2b in emx1 deficient embryos (Fig. 7A,B). Thus, these tbx2 genes are expressed ectopically in the expanded DE of emx1 morphants, but to no phenotypic consequence, consistent with the absence of effects on DE fate in previous tbx2a/b gain or loss of function studies23. Conversely, the emx1 expression domain was shortened in tbx2a and tbx2b deficient embryos (Fig. 7C,D). These results suggested that emx1 acts downstream of these tbx2 genes. As tbx2a acts upstream of tbx2b during DL development and tbx2a overexpression is associated with a greater incidence of dysmorphic gastrulation23, we therefore tested whether emx1 cRNA could rescue the DL in the absence of tbx2b function. Interestingly, emx1 cRNA was sufficient to rescue DL fate in approximately 50% of tbx2b deficient embryos (Fig. 7E–G). Conversely, provision of tbx2b cRNA was not sufficient to rescue the DL in emx1 morphants (Fig. S7). In sum, we concluded that emx1 acts downstream of tbx2b during DL development.
Figure 7.
tbx2b acts upstream of emx1 to regulate DL segment development. (A) WISH (lateral view) of 28 ss WT and emx1 deficient embryos for tbx2a or tbx2b (purple). (B) Quantification of tbx2a and tbx2b domain length. (C) WISH (lateral view) of 28 ss WT, tbx2a deficient, and tbx2b deficient embryos for emx1 (purple). (D) Quantification of emx1 domain length. (E) WISH (lateral view) of 28 ss WT, tbx2b deficient, and tbx2b deficient embryos co-injected with emx1 capped mRNA (emx1 cRNA rescue) for the DL segment marker slc12a3 (purple). (A,C,E) Blue boxes demarcate domains of pronephros expression. Scale bar = 100 μm. (F) Quantification of DL length in WT, tbx2b deficient, and emx1 cRNA rescue embryos. (G) Quantification of phenotype percentages for (F), categorized as WT, decreased, or other, where the latter include those developmentally delayed or too dysmorphic to measure. *p < 0.05; **p < 0.01; ***p < 0.001; N.S. = not significant. Error bars represent standard error.
In light of our findings that positioned both mecom and tbx2b as upstream of emx1, we examined the mecom-tbx2b relationship. mecom deficient embryos had reduced expression of tbx2b in the developing nephron, as well as tbx2a (Fig. 8A,B). Reciprocally, the mecom expression domain was unchanged in tbx2a or tbx2b deficient embryos (Fig. 8C,D), consistent with its pan-distal expression domain during late somitogenesis. Based on these results, we hypothesized that mecom is upstream relative to the tbx2 genes in distal pronephros ontogeny. Given the limitations in tbx2a overexpression, we tested the ability of tbx2b cRNA to rescue DL segment fate the context of mecom loss of function. Consistent with the notion that mecom acts upstream of tbx2b either directly or indirectly during DL development, tbx2b cRNA was sufficient to rescue DL length in just over 50% of mecom deficient embryos (Fig. 8E,F,G). Taken together, we concluded that a genetic cascade of mecom, followed by tbx2a/2b, regulates emx1 expression to control DL formation (Fig. 8H). Additionally, mecom may also regulate emx1 through targets other than tbx2b, and all of these genetic interactions may be direct or indirect.
Figure 8.
mecom functions upstream of tbx2b during DL segment formation (A) WISH (lateral view) of 28 ss WT and mecom deficient embryos for tbx2a or tbx2b (purple). (B) Quantification of tbx2a and tbx2b domain length. (C) WISH (lateral view) of 28 ss WT, tbx2a deficient, tbx2b deficient, and tbx2a/b deficient embryos for mecom (purple). (D) Quantification of mecom domain length. (E) WISH (lateral view) of 28 ss WT, mecom deficient, and mecom deficient injected with tbx2b capped mRNA (tbx2b cRNA rescue) for slc12a3 (purple). (A,C,E) Blue boxes demarcate domains of pronephros expression. Scale bar = 100 μm. (F) Quantification of DL length in WT, mecom deficient, and tbx2b cRNA rescue embryos. (G) Quantification of phenotype percentages for (F), categorized as WT, decreased, or other, where the latter include those developmentally delayed or too dysmorphic to measure. **p < 0.01; ***p < 0.001; N.S. = not significant. (H) RA promotes proximal segment development while inhibiting distal segments. mecom promotes distal segment fates as well as expression of tbx2a/b, which in turn promotes expression of emx1. mecom may also promote emx1 through other targets than the tbx2 genes (not shown). Error bars represent standard error.
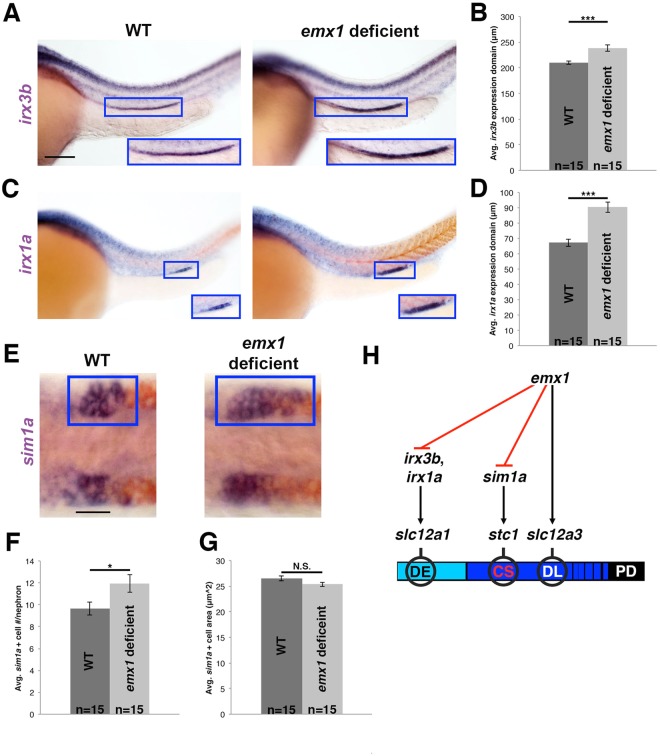
emx1 deficient nephrons show alterations in distal expression patterns of the irx3b, irx1a, and sim1a transcription factors
Following our studies analyzing the relationships between mecom, tbx2a/b and emx1 that provided insight into the molecular changes underlying the DL phenotype in emx1 deficient embryos, we turned our attention to elucidating the basis of the DE and CS phenotypes caused by emx1 loss of function. Several factors have been associated with development of the DE. In the frog pronephros, Irx3 is requisite for DE fate and abrogation of Irx3 leads to loss of Irx1 as well, leading to the hypothesis that an Irx3-mediated gene network controls the DE lineage21. Conservation of this pathway has been suggested by analysis of irx3b function in the zebrafish pronephros, where irx3b deficiency similarly caused loss of the DE lineage20,45. While irx3b transcripts are expressed in both the PST and DE segments20, expression of irx1a has been attributed to the approximate location of only the DE segment in zebrafish46.
To investigate the relationship between emx1 with these irx genes, WISH studies were performed in WT and emx1 deficient embryos. Interestingly, we observed that the expression domains of both irx3b and irx1a were distally expanded in length in emx1 deficient embryos, and that these changes were significant compared to WT controls (Fig. 9A–D). These changes correlated with the expanded DE segment size that occurs in emx1 loss of function, and suggest the intriguing hypothesis that emx1 negatively regulates DE fate by repressing irx3b and irx1a, either directly or indirectly, in renal progenitors. Alternatively, the changes in irx expression domains may simply reflect the changes in segment fates in emx1 deficient embryos.
Figure 9.
emx1 deficiency results in increased irx3b, irx1a, and sim1a expression domains. (A) WISH (lateral view) of 28 ss WT and emx1 deficient embryos for irx3b (purple). (B) Quantification of irx3b domain length. (C) WISH (lateral view) of 28 ss WT and emx1 deficient embryos for irx1a (purple) and smyhc1 (red). (A,C) Blue boxes demarcate domains of pronephros expression. Scale bar = 100 μm. (D) Quantification of irx1a domain length. (E) WISH (dorsal view) of 28 ss WT and emx1 deficient embryos for sim1a (purple) and slc12a3 (red). Blue boxes demarcate a single CS territory while the contralateral side is unlabeled. Scale bar = 20 μm. (F,G) Quantification of sim1a+ cell number and size. *p < 0.05; ***p < 0.001; N.S. = not significant. Error bars represent standard error. (H) emx1 restricts formation of the DE by inhibiting expression of irx3b and irx1a, restricts the CS anlage by inhibiting sim1a expression, and promotes DL fate (slc12a3+) by regulating unknown targets.
With respect to the CS, expression of the transcription factor single-minded family bHLH transcription factor 1a (sim1a) is necessary and sufficient for CS fate choice25,47. Thus, we next examined the pattern of sim1a expression in emx1 deficient embryos at the 28 ss, when CS anlage precursors can be visualized as bilateral clusters of cells located just dorsal to the pronephros. emx1 deficient embryos had significantly increased numbers of sim1a+ cells, which had similar dimensions as WT cells (Fig. 9E–G). These results may indicate that emx1 regulates the CS by repressing sim1a expression, either directly or indirectly, or simply that the changes in sim1a expression reflect the altered CS lineage in emx1 deficient embryos. Taken together, we propose a testable working model where emx1 is responsible for regulating the DE by controlling irx3b and irx1a, and for regulating the CS by controlling sim1a expression, which can be investigated further in future studies (Fig. 9H).
Discussion
Nephron segmentation is an important developmental process that patterns the renal functional units into structures with spatially distinct functional domains. The precise order of segments, along with their scale and cell type composition, is essential for proper kidney function. Disruptions of segment formation, as well as damage to segment populations, are associated with various forms of kidney disease48. There have been exciting advances in our knowledge about nephrogenesis49, including the expression profiles of nephron segments during development, but there is still an incomplete understanding of the mechanisms that elaborate segmental fate decisions. Further advancements in the cultivation of renal cell types from pluripotent cell sources can be facilitated by the continued discovery of nephrogenesis processes50.
Here, we have shown for the first time that emx1 is important for proper development of distal nephron segments in the zebrafish embryonic kidney. Our results indicate that emx1 is necessary for promoting DL fate and inhibiting DE fate, where emx1 acts in a genetic cascade downstream of mecom and tbx2b. We have also ascertained the relationship between emx1 and several known segmentation regulators in the pronephros. This led us to assemble an integrated hierarchy of the upstream factors that regulate emx1 (Fig. 8H), and to identify several components situated downstream of emx1 (Fig. 9H). Additional studies are needed to delineate whether these relationships involve direct or indirect interactions. The identification of a putative EMX1 binding site or consensus sequence (TAATTANNTAATTA)51 will be useful in identifying direct downstream target candidates of emx1.
Further work is also needed to situate emx1 activity with respect to other known segmentation pathways, such as Notch signaling and prostaglandin signaling. Notch signaling mitigates the choice between transporter fate and multiciliated cell (MCC) fate in several segment domains of the pronephros52,53. Because emx1 deficiency expands the DE, which is one of the regions where MCCs are found, it is possible that emx1 activity articulates with Notch to influence MCC genesis. With regard to segmentation, prostaglandin signaling is essential to control the balance of DE and DL transporter cell fates in the developing zebrafish pronephros54. Interestingly, the abrogation of prostaglandin biosynthesis or inhibition of receptor expression leads to an expanded DE and reduced DL54, which closely mimic how the emx1 loss of function impacts distal pronephros segmentation. Given these similarities, is reasonable to speculate that emx1 mitigates DE and DL fate wholly or in part through an interaction with prostaglandin signaling. Some immediate possibilities to consider are that Emx1 regulates the expression of prostaglandin synthesis enzymes or receptors within the renal progenitors or the neighboring tissues. Alternatively, emx1 may act as a key transcriptional target of prostaglandin signaling. Further studies to assess prostaglandin signaling activity in emx1 deficient embryos could be employed to ascertain the relationships between these pronephros development components.
Additionally, there may be currently unknown molecular components relevant to emx1 activity during nephron segmentation. For example, during mammalian brain development, Hedgehog signaling has been placed upstream of Emx155. Specifically, Gli3, a transcriptional regulator that is induced by Hedgehog signaling in the murine brain, is essential for Emx1 expression55. Despite the need for more studies to appreciate the relationships between emx1 and other pathways within the kidney, the genetic network maps assembled in this work provide a substantive initial framework for continued segmentation studies in the zebrafish pronephros.
The present study is the first report to attribute roles for emx1 during mesoderm development. In contrast, ems/Emx1 have long been known for their associations with the formation of the nervous system in the fly and mammal, respectively30. ems/Emx1 control specification of the neuroectoderm early in embryogenesis, and they play subsequent roles during assembly of the olfactory sensory system and visual system where they control neuron development30,56. Exciting recent proteomics approaches have begun to elucidate the molecular features associated with Emx1 loss of functionin neurogenesis57, which may provide insights into EMX1 activities that will be relevant in the context of other tissues as well. Among EMX1 interacting partners is WDR11, a WD domain-containing gene, mutations of which have been linked to Idopathic Hypogonadotropic Hypogonadism and Kallmann Syndrome (IHH/KS) in humans58. Male patients with KS display unilateral renal agenesis with a ~30% frequency59–61. The EMX1/WDR11 association warrants further investigation both in light of our findings, and given the expression of Emx1 in the developing mammalian kidney.
In contrast to Emx1, its family member Emx2 is well-established as essential for mammalian kidney development. In the developing murine kidney, for example, Emx2 is expressed in the pronephros primordium, mesonephros tubules and later the metanephros62. Within the latter, Emx2 is expressed in the invading ureteric bud and branches, along with the renal vesicles and comma shaped bodies that represent early nephron structures62. GUDMAP microarray data from E15 murine kidney have similarly shown Emx2 expression in the renal vesicles, as well as the proximal tubules, loop of Henle anlage, and collecting ducts at E15, which also mirrors expression of Emx136. Knockout of Emx2 causes severe disruptions of urogenital system formation, where the metanephric kidneys fail to develop63. Most recently, expression of Emx2 along with Hnf1b, Hnf4a and Pax8 was found to be sufficient to convert both mouse and human fibroblasts into induced renal tubular epithelial cells64. Interestingly, the same study reported the expression domains of both emx2 and emx1 in the Xenopus pronephros, where these factors initially co-localize in the nephron tubule region and then become restricted to the tubule and duct, respectively64. Similarly in the zebrafish pronephros, emx2 is expressed in the proximal renal progenitors27, while emx1 is expressed proximally and then becomes restricted to the distal renal progenitor domain, as illustrated in the present work and previous studies27. Taken together, there are clear differences across vertebrates with regard to the expression of emx1/Emx1 in developing nephrons, where lower vertebrates exclude emx1 expression from the proximal tubule during pronephros ontogeny, which is appears to be distinct compared to nephrogenesis in the mammalian metanephros. Analysis with dual in situ hybridization or immunofluorescence would be useful to further discern the spatiotemporal dynamics and respective expression domains of Emx1/Emx2 during nephrogenesis in the metanephros. Functional studies are also needed to ascertain the role(s) of emx2 in pronephros development, and the relationship between emx1 and emx2.
Here, our current study has established a framework of the transcriptional network within which emx1 functions to regulate zebrafish nephron segment development. As the conserved segment features of zebrafish and mammalian nephrons65 lend credence to the notion that insights about segmentation mechanisms in the teleost pronephros can be used as an entrée to help further our understanding of nephrogenesis in other vertebrates, the newly discovered functions of emx1 provide a novel rationale for testing the roles of its ortholog during mammalian nephrogenesis. Continuing to elucidate how nephron segmentation occurs is relevant to deciphering the basis of congenital kidney defects, and will be instrumental in improving approaches to combat renal disease with regenerative medicine.
Methods
Zebrafish husbandry and ethics statement
Zebrafish were housed and cared for in the Center for Zebrafish Research at the University of Notre Dame Freimann Life Science Center, where the Institutional Animal Care and Use Committee (IACUC) approved the experiments documented here under protocol numbers 13–021 and 16–025. All methods were carried out in accordance with relevant guidelines and regulations. Tübingen strain WT zebrafish were used and staged as described66. Embryos were incubated at 28 °C in E3 medium67.
Morpholinos
Morpholinos were purchased from Gene Tools, LLC (Philomath, OR), solubilized with DNase/RNase free water, and stored at −20 °C. The emx1 ATG-MO 5′-CCGTTGCCGAGAACATTGTCCGTGA-3′ targets the start site42. The combination of emx1 splice-blocking MOs (emx1 SB-MO), emx1 SD 5′-AATACTTACCTTGAAACCGATGTCC-3′ and emx1 SA 5′-GAGACATCATTACCTAAGATATAAC-3′ were designed to bind the splice donor and acceptor sites of exon 1 and exon 2, respectively. WT embryos were injected at the 1-cell stage with 1 nL of 133 μM emx1 ATB-MO or 400 μM emx1 SB-MO. The tbx2 morpholinos were23: MO1-tbx2a, which targets tbx2a 5′-ATCGGTGCATCCAAAAAGCCAGAT-3′; MO1-tbx2b, which targets tbx2b 5′-CCTGTAAAAACTGGATCTCTCATCG-3′; MO1-tbx2, which targets both tbx2a/b, 5′-AAAACTGGATCTCTCATCGGTGCAT-3′. The mecom morpholinos were22: MO3-mecom, 5′-CTGAGTGACTTACATATGAAGGGCT-3′ and MO4-mecom, 5′-TTGTGGCAGACCTCACCTAGAGACA-3′, which target splicing of mecom.
Reverse transcription PCR
RNA was extracted from WT embryos and emx1 SB-MO morphants at 24 hpf with Trizol reagent (Invitrogen) according to manufacturer instructions. RT-PCR was performed using the SuperScript First-Strand Synthesis System (Invitrogen)26. To confirm aberrant splicing of emx1 intron 1 in emx1 SB-MO morphant embryos, PCR primers were specifically designed to amplify a region corresponding to 620 bp of sequence that contains both the 3′-end of emx1 exon 1 and the 5′-end of emx1 intron 1 (forward: 5′-AACATCGTGAACCACTGAATTTCTACCCTTGGGTCCTAAGGAACAGGTTTTTTGGACATC-3′ and reverse 5′- CAATTGACTTCCATAATAAGAAAACAATAGCCTGCTGCTGGAAGTCATTGGTTCCAGGTTTCTAGC-3′).
cRNA synthesis and rescue experiments
An emx1 pCS2 plasmid was first constructed by obtaining an emx1 clone containing the emx1 ORF flanked by an EcoRI restriction site at the 5′ end and an XhoI restriction site at 3′ (Clone ID: ODa00797C) (GenScript). The emx1 ORF sequenced was then excised from the emx1 clone and directionally cloned into the pCS2 vector to be used for cRNA synthesis. Synthetic emx1 cDNA was synthesized from the emx1 pCS2 plasmid using an sp6 mMessage Machine kit (Ambion) after linearization of the plasmid template. mecom and tbx2b cRNA was synthesized as described22,23. To perform rescue experiments, 1-cell stage embryos were injected with 12–15 pg emx1 cRNA42, 25 pg mecom cRNA22 or 400 pg tbx2b cRNA23 along with the appropriate morpholino(s). For all rescue studies, replicate group sizes were a minimum of 30 embryos, and typically ranged between 40–60 embryos for each cohort.
WISH, FISH, IF and acridine orange staining
For gene and protein expression analyses, embryos were fixed in 4% paraformaldehyde/1X PBST and stored in 100% methanol at −20 °C. Biological triplicates of all genetic conditions were analyzed for expression of each marker, where replicates consisted of 30–40 embryos. WISH68,69,70 and FISH71 were performed as described. Antisense probes for smyhc1, deltaC, cdh17, slc4a4a, clcnk, slc20a1a, trmp7, slc12a1, stc1, slc12a3, gata3, emx1, irx3b, sim1a, and mecom were generated using IMAGE clone template plasmids20,25. Antisense probes for emx1 and irx1a were generated using IMAGE clones 9001097 and 9039039, respectively. Probe for emx1 was transcribed from PCR-generated DNA templates amplified with primers (forward: 5′-TTTATGGACACCACTGGATCG-3′ and reverse: 5′-AATTAACCCTCACTAAAGGGTTCTTCTTTTGTGTGCACTCCGGG-3′). Probe for irx1a was transcribed from the PCR-generated DNA templates with primers (forward: 5′-ATGTCTTTCCCCCAGCTGGGCTACCCG-3′ and reverse: 5′-AATTAACCCTCACTAAAGGGTCAAGCGGAGGA-3′. Immunofluorescence studies to detect pH3 and Caspase-3, and acridine orange treatment to detect apoptotic cells were performed as described71,72,73.
Chemical treatments
RA and DEAB (Sigma-Aldrich) were dissolved in 100% DMSO to make a 1 M stock solution and aliquots were stored at −80 °C in the dark74. For drug treatments, embryos were incubated in the dark from 60% epiboly to the 15 ss in either 1 × 10−7 M RA/DMSO, 1.6 × 10−5 M DEAB/DMSO, or 1.6 × 10−5 M DMSO in E3 media75. Embryos were then washed 5 times with E3 to remove exogenous chemicals, left to develop at 28 °C until 24 hpf, after which they were euthanized and fixed. These chemical treatments were fully penetrant and produced consistent results over three replicates with n = 40–60 embryos per replicate analyzed for these studies.
Measurements and statistics
For WISH and acridine orange-stained embryos, 5 embryos from both the WT and experimental group of each experiment were selected, photographed from a lateral view using a Nikon eclipse Ni with a DS-Fi2 camera, and had either the length of their pronephric stain measured in μm or the number of AO-positive cells counted using either the Nikon Elements software or FIJI. For stc1 and sim1a-stained embryos dorsal photographs were taken and cell number and cell area we measured in μm2. For FISH and IF, 3 embryos from the WT and experimental groups were selected, from which lateral view z-stacks were acquired using a Nikon A1 confocal microscope. From the z-stack data sets, the parameters of segment lengths, IND, nuclei number, and tubule volume were measured using FIJI. For statistical analysis, t-tests were performed. For phenotype percentage comparisons, the proportion data were normalized via arcsine transformation before being compared using t-tests.
Electronic supplementary material
Acknowledgements
NIH Grant R01DK100237 to RAW, and a University of Notre Dame Kinesis Fernández Richards Graduate Fellowship to EEM, supported this work. We are grateful to Elizabeth and Michael Gallagher for a generous gift to the University of Notre Dame on behalf of their family for the support of stem cell research. The funders had no role in the study design, data collection and analysis, decision to publish, or manuscript preparation. This study discussed publicly posted microarray results from the GUDMAP database36, http://www.gudmap.org, obtained 07/2018 and 09/2018 for Emx1 (Microarray Data 13796; RID: Q-48TP) and Emx2 (Microarray Data 13797; RID: Q-48TR). We thank the staffs of the Department of Biological Sciences and the Center for Zebrafish Research at the University of Notre Dame for their dedication and care of our aquarium. Finally, we thank the members of our lab for support, discussions, and insights about this work.
Author Contributions
E.E.M., N.H., B.E.D. and R.A.W. designed the experiments. E.E.M., N.H., B.E.D., J.M.C., A.N.M., A.A. and R.A.W. performed experiments. E.E.M., N.H., B.E.D. and R.W. collected and analyzed the data. E.E.M. and R.A.W. prepared the figures and wrote the manuscript.
Data Availability
The data associated with this report are provided in the figures and supplemental figures.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-36061-4.
References
- 1.Saxen, L. Organogenesis of the kidney (Cambridge University Press, 1987).
- 2.Dressler GR. The cellular basis of kidney development. Annu. Rev. Cell Dev. Biol. 2006;22:509–529. doi: 10.1146/annurev.cellbio.22.010305.104340. [DOI] [PubMed] [Google Scholar]
- 3.Romagnani P, Lasagni L, Remuzzi G. Renal progenitors: an evolutionary conserved strategy for kidney regeneration. Nat. Rev. Nephrol. 2013;9:137–146. doi: 10.1038/nrneph.2012.290. [DOI] [PubMed] [Google Scholar]
- 4.Wingert RA, Davidson AJ. The zebrafish pronephros: a model to study nephron segmentation. Kidney Int. 2008;73:1120–1127. doi: 10.1038/ki.2008.37. [DOI] [PubMed] [Google Scholar]
- 5.Desgrange A, Cereghini S. Nephron patterning: lessons from Xenopus, zebrafish and mouse studies. Cells. 2015;4:483–499. doi: 10.3390/cells4030483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindström NO, et al. 2015. Integrated Β-Catenin, BMP, PTEN, and Notch signalling patterns the nephron. Elife. 2015;3:e04000. doi: 10.7554/eLife.04000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rieger A, et al. Missense mutation of POU domain class 3 transcription factor 3 in Pou3f3L423P mice causes reduced nephron number and impaired development of the thick ascending limb of the loop of Henle. PLoS One. 2016;11:e0158977. doi: 10.1371/journal.pone.0158977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basta JM, Robbins L, Denner DR, Kolar GR, Rauchman MA. Sall1-NuRD interaction regulates multipotent nephron progenitors and is required for loop of Henle formation. Development. 2017;144:3080–3094. doi: 10.1242/dev.148692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung E, Deacon P, Park JS. Notch is required for the formation of all nephron segments and primes nephron progenitors for differentiation. Development. 2017;144:4530–4539. doi: 10.1242/dev.156661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drake KA, Adam M, Mahoney R, Potter SS. Disruption of Hox9,10,11 function results in cellular level lineage infidelity in the kidney. Sci. Rep. 2018;8:6306. doi: 10.1038/s41598-018-24782-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magella B, Mahoney R, Adam M, Potter SS. Reduced Abd-B Hox function during kidney development results in lineage infidelity. Dev. Biol. 2018;438:84–93. doi: 10.1016/j.ydbio.2018.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindström NO, et al. Conserved and divergent features of human and mouse kidney organogenesis. J. Am. Soc. Nephrol. 2018;29:785–805. doi: 10.1681/ASN.2017080887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pickart MA, Klee EW. Zebrafish approaches enhance the translational research tackle box. Transl. Res. 2014;163:65–78. doi: 10.1016/j.trsl.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Poureetezadi SJ, Wingert RA. Little fish, big catch: zebrafish as a model for kidney disease. Kidney Int. 2016;89:1204–1210. doi: 10.1016/j.kint.2016.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naylor RW, Qubisi SS, Davidson AJ. Zebrafish pronephros development. Results Probl. Cell Differ. 2017;60:27–53. doi: 10.1007/978-3-319-51436-9_2,. [DOI] [PubMed] [Google Scholar]
- 16.Morales EE, Wingert RA. Zebrafish as a model of kidney disease. Results Probl. Cell Differ. 2017;60:55–75. doi: 10.1007/978-3-319-51436-9_3. [DOI] [PubMed] [Google Scholar]
- 17.Gerlach GF, Wingert RA. Kidney organogenesis in the zebrafish: insights into vertebrate nephrogenesis and regeneration. Wiley Interdiscip. Rev. Dev. Biol. 2013;2:559–85. doi: 10.1002/wdev.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ebarasi L, Oddsson A, Hultenby K, Betsholtz C, Tryggvason K. Zebrafish: a model system for the study of vertebrate renal development, function, and pathophysiology. Curr. Opin. Nephrol. Hypertens. 2011;20:416–424. doi: 10.1097/MNH.0b013e3283477797. [DOI] [PubMed] [Google Scholar]
- 19.Wingert RA, et al. The cdx genes and retinoic acid control the positioning and segmentation of the zebrafish pronephros. PLoS Genet. 2007;3:1922–1938. doi: 10.1371/journal.pgen.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wingert RA, Davidson AJ. 2011. Zebrafish nephrogenesis involves dynamic spatiotemporal expression changes in renal progenitors and essential signals from retinoic acid and irx3b. Dev. Dyn. 2011;240:2011–2027. doi: 10.1002/dvdy.22691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reggiani L, Raciti D, Airik R, Kispert A, Brändli AW. The prepattern transcription factor Irx3 directs nephron segment identity. Genes Dev. 2007;21:2358–2370. doi: 10.1101/gad.450707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, et al. Zebrafish nephrogenesis is regulated by interactions between retinoic acid, mecom, and Notch signaling. Dev. Biol. 2014;386:111–122. doi: 10.1016/j.ydbio.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drummond BE, et al. The tbx2a/b transcription factors direct pronephros segmentation and corpuscle of Stannius formation in zebrafish. Dev. Biol. 2017;421:52–66. doi: 10.1016/j.ydbio.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naylor RW, et al. BMP and retinoic acid regulate anterior-posterior patterning of the non-axial mesoderm across the dorsal-ventral axis. Nat. Commun. 2016;7:12197. doi: 10.1038/ncomms12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng CN, Wingert RA. Nephron proximal tubule patterning and corpuscles of Stannius formation are regulated by the sim1a transcription factor and retinoic acid in zebrafish. Dev. Biol. 2015;399:100–116. doi: 10.1016/j.ydbio.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marra AN, Wingert RA. Epithelial cell fate in the nephron tubule is mediated by the ETS transcription factors etv5a and etv4 during zebrafish kidney development. Dev. Biol. 2016;411:231–245. doi: 10.1016/j.ydbio.2016.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawahara A, Dawid IB. Development expression of zebrafishemx1 during early embryogenesis. Gene Expr. Patterns. 2002;2:201–206. doi: 10.1016/S1567-133X(02)00062-5. [DOI] [PubMed] [Google Scholar]
- 28.Walldorf U, Gehring WJ. Empty spiracles, a gap gene containing a homeobox involved in Drosophila head development. EMBO J. 1992;11:2247–2259. doi: 10.1002/j.1460-2075.1992.tb05284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen SM, Jürgens G. Mediation of Drosophila head development by gap-like segmentation genes. Nature. 1990;346:482–485. doi: 10.1038/346482a0. [DOI] [PubMed] [Google Scholar]
- 30.Sen S, Reichert H, Vijay Raghavan K. Conserved roles of ems/Emx and otd/Otx genes in olfactory and visual system development in Drosophila and mouse. Open Biol. 2013;3:120177. doi: 10.1098/rsob.120177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morita T, Nitta H, Kiyama Y, Mori H, Mishina M. Differential expression of two zebrafish emx homeoprotein mRNAs in the developing brain. Neurosci. Lett. 1995;198:131–134. doi: 10.1016/0304-3940(95)11988-9. [DOI] [PubMed] [Google Scholar]
- 32.Gulisano, et al. Emx1 and Emx2 show different patterns of expression during proliferation and differentiation of the developing cerebral cortex in the mouse. Eur. J. Neurosci. 1996;8:1037–1050. doi: 10.1111/j.1460-9568.1996.tb01590.x. [DOI] [PubMed] [Google Scholar]
- 33.Chan CH, et al. Emx1 is a marker for pyramidal neurons of the cerebral cortex. Cereb. Cortex. 2001;11:1191–1198. doi: 10.1093/cercor/11.12.1191. [DOI] [PubMed] [Google Scholar]
- 34.Qiu M, et al. Mutation of the Emx-1 homeobox gene disrupts the corpus callosum. Dev. Biol. 1996;178:174–178. doi: 10.1006/dbio.1996.0207. [DOI] [PubMed] [Google Scholar]
- 35.Yoshida M, et al. Emx1 and Emx2 functions in development of dorsal telencephalon. Development. 1997;124:101–111. doi: 10.1242/dev.124.1.101. [DOI] [PubMed] [Google Scholar]
- 36.McMahon AP, et al. 2008. GUDMAP: the genitourinary developmental molecular anatomy project. J. Am. Soc. Nephrol. 2008;19:667–671. doi: 10.1681/ASN.2007101078. [DOI] [PubMed] [Google Scholar]
- 37.Gerlach GF, Wingert RA. Zebrafish pronephros tubulogenesis and epithelial identity maintenance are reliant on the polarity proteins Prkc iota and zeta. Dev. Biol. 2014;396:183–200. doi: 10.1016/j.ydbio.2014.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKee R, Gerlach GF, Jou J, Cheng CN, Wingert RA. Temporal and spatial expression of tight junction genes during zebrafish pronephros development. Gene Expr. Patterns. 2014;16:104–113. doi: 10.1016/j.gep.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marra AN, Li Y, Wingert RA. Antennas of organ morphogenesis: the roles of cilia in vertebrate kidney development. Genesis. 2016;54:457–469. doi: 10.1002/dvg.22957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Camp E, Hope R, Kortschak RD, Cox TC, Lardelli M. Expression of three spalt (sal) gene homologues in zebrafish embryos. Dev. Genes. Evol. 2003;213:35–43. doi: 10.1007/s00427-002-0284-6. [DOI] [PubMed] [Google Scholar]
- 41.Elizondo MR, et al. Defective skeletogenesis with kidney stone formation in dwarf zebrafish mutant for trpm7. Curr. Biol. 2005;15:667–671. doi: 10.1016/j.cub.2005.02.050. [DOI] [PubMed] [Google Scholar]
- 42.Viktorin G, Chiuchitu C, Rissler M, Varga ZM, Westerfield M. emx3 is required for the differentiation of dorsal telencephalic neurons. Dev. Dyn. 2009;238:1984–1998. doi: 10.1002/dvdy.22031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naylor RW, Dodd RC, Davidson AJ. Caudal migration and proliferation of renal progenitors regulates early nephron segment size in zebrafish. Sci. Rep. 2016;6:35647. doi: 10.1038/srep35647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bastien J, Rochette-Egly C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene. 2004;328:1–16. doi: 10.1016/j.gene.2003.12.005,. [DOI] [PubMed] [Google Scholar]
- 45.Marra AN, Wingert RA. Roles of Iroquois transcription factors in kidney development. Cell Dev. Biol. 2014;3:1000131. doi: 10.4172/2168-9296.1000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng CW, Hui C, Strähle U, Cheng SH. Identification and expression of zebrafish Iroquois homeobox gene irx1. Dev. Genes. Evol. 2001;211:442–444. doi: 10.1007/s004270100168. [DOI] [PubMed] [Google Scholar]
- 47.Cheng CN, Verdun V, Wingert RA. Recent advances in elucidating the genetic mechanisms of nephrogenesis using zebrafish. Cells. 2015;4:218–233. doi: 10.3390/cells4020218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schedl A. Renal abnormalities and their developmental origin. Nat. Rev. Genet. 2007;8:791–802. doi: 10.1038/nrg2205. [DOI] [PubMed] [Google Scholar]
- 49.Chambers BE, Wingert RA. Renal progenitors: roles in kidney disease and regeneration. World J. Stem Cells. 2016;8:367–375. doi: 10.4252/wjsc.v8.i11.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morales EE, Wingert RA. Renal stem cell reprogramming: prospects in regenerative medicine. World J. Stem Cells. 2014;6:458–66. doi: 10.4252/wjsc.v6.i4.458.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lim JW, et al. EMX1 Regulates NRP1-mediated wiring of the mouse anterior cingulate cortex. Development. 2015;142:3746–3757. doi: 10.1242/dev.119909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y, Narendra P, Kramer-Zucker A, Drummond IA. Notch signaling controls the differentiation of transporting epithelia and multiciliated cells in the zebrafish pronephros. Development. 2007;134:1111–1122. doi: 10.1242/dev.02806. [DOI] [PubMed] [Google Scholar]
- 53.Ma M, Jiang YJ. Jagged2a-Notch signaling mediates cell fate choice in the zebrafish pronephric duct. PLoS. Genet. 2007;3:e18. doi: 10.1371/journal.pgen.0030018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poureetezadi SJ, et al. Prostaglandin signaling regulates nephron segment patterning of renal progenitors during zebrafish kidney development. Elife. 2016;5:piie17551. doi: 10.7554/eLife.17551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Theil T, Alvarez-Bolado G, Walter A, Rüther U. Gli3 is required for Emx gene expression during dorsal telencephalon development. Development. 1999;126:3561–3571. doi: 10.1242/dev.126.16.3561. [DOI] [PubMed] [Google Scholar]
- 56.Cecchi C, Boncinelli E. Emx homeogenes and mouse brain development. Trends Neurosci. 2000;23:347–352. doi: 10.1016/S0166-2236(00)01608-8. [DOI] [PubMed] [Google Scholar]
- 57.Kobeissy FH, et al. Deciphering the role of Emx1 in neurogenesis: a neuroproteomics approach. Front. Mol. Neurosci. 2016;9:98. doi: 10.3389/fnmol.2016.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim HG, et al. WDR11, a WD protein that interacts with transcription factor EMX1, is mutated in idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am. J. Hum. Genet. 2010;87:465–479. doi: 10.1016/j.ajhg.2010.08.018.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quinton R, et al. Idiopathic gonadotropin deficiency: genetic questions addressed through phenotypic characterization. Clin. Endocrinol. 2001;55:163–174. doi: 10.1046/j.1365-2265.2001.01277.x. [DOI] [PubMed] [Google Scholar]
- 60.Massin N, et al. X chromosome-linked Kallmann syndrome: clinical heterogeneity in three siblings carrying an intragenic deletion of the KAL-1 gene. J. Clin. Endocrinol. Metab. 2003;88:2003–2008. doi: 10.1210/jc.2002-021981. [DOI] [PubMed] [Google Scholar]
- 61.Costa-Barbosa FA, et al. Prioritizing genetic testing in patients with Kallmann Syndrome using clinical phenotypes. J Clin Endocrinol Metab. 2013;98:E943–953. doi: 10.1210/jc.2012-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pellegrini M, Pantano S, Lucchini F, Fumi M, Forabosco A. Emx2 developmental expression in the primordia of the reproductive and excretory systems. Anat Embryol (Berl). 1997;196:427–433. doi: 10.1007/s004290050110. [DOI] [PubMed] [Google Scholar]
- 63.Miyamoto N, Yoshida M, Kuratani S, Matsuo I, Aizawa S. Defects of urogenital development in mice lacking Emx2. Development. 1997;124:1653–1664. doi: 10.1242/dev.124.9.1653. [DOI] [PubMed] [Google Scholar]
- 64.Kaminski MM, et al. Direct reprogramming of fibroblasts into renal tubular epithelial cells by defined transcription factors. Nat. Cell Biol. 2016;18:1269–1280. doi: 10.1038/ncb3437. [DOI] [PubMed] [Google Scholar]
- 65.Kroeger PT, Jr., Wingert RA. Using zebrafish to study podocyte genesis during kidney development and regeneration. Genesis. 2014;52:771–792. doi: 10.1002/dvg.22798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 67.Westerfield, M. The Zebrafish Book. (University of Oregon Press, Eugene, 1993).
- 68.Galloway JL, et al. Combinatorial regulation of novel erythroid gene expression in zebrafish. Exp. Hematol. 2008;36:424–432. doi: 10.1016/j.exphem.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng CN, et al. Flat mount preparation for observation and analysis of zebrafish embryo specimens stained by whole mount in situ hybridization. J. Vis. Exp. 2014;89:51604. doi: 10.3791/51604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McCampbell KK, Springer KN, Wingert RA. Analysis of nephron composition and function in the adult zebrafish kidney. J. Vis. Exp. 2014;90:e51644. doi: 10.3791/51644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marra AN, et al. Visualizing multiciliated cells in the zebrafish through a combined protocol of whole mount fluorescent in situ hybridization and immunofluorescence. J. Vis. Exp. 2017;129:e56261. doi: 10.3791/56261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McCampbell KK, Springer KN, Wingert RA. Atlas of cellular dynamics during zebrafish adult kidney regeneration. Stem Cells Int. 2015;2015:547636. doi: 10.1155/2015/547636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kroeger PT, Jr., et al. The zebrafish kidney mutant zeppelin reveals that brca2/fancd1 is essential for pronephros development. Dev. Biol. 2017;428:148–163. doi: 10.1016/j.ydbio.2017.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lengerke C, et al. 2011. Interactions between Cdx genes and retinoic acid modulate early cardiogenesis. Dev. Biol. 2011;163:134–142. doi: 10.1016/j.ydbio.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Poureetezadi SJ, Donahue EK, Wingert RA. A manual small molecule screen approaching high-throughput using zebrafish embryos. J. Vis. Exp. 2014;93:e52063. doi: 10.3791/52063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data associated with this report are provided in the figures and supplemental figures.