Highlights
-
•
High LncRNA expression was significantly correlated with the poorer OS prognosis in osteosarcoma patients. However, the cut-off values may be the source of heterogeneity.
-
•
The expression level of LncRNA was positively correlated with alkaline phosphatase, tumor size, metastasis, distant metastasis and Enneking stage.
-
•
This meta-analysis demonstrated that abnormal LncRNAs expression was correlated with advanced clinicopathological features and poor prognosis as a novel predictive biomarker in osteosarcoma.
Keywords: LncRNAs, Osteosarcoma, Prognosis, Meta-analysis
Abstract
Background
Numerous studies have reported the relationship between Long non-coding RNAs (LncRNAs) expression and prognosis of osteosarcoma, but less consensus has been reached. Our meta-analysis was conducted to quantitatively assess the relationship between the expression of LncRNAs, prognosis and clinical pathology in osteosarcoma development.
Methods
PubMed,Embase,Web of Science,The Cochrane Library,SionMed,CNKI and WanFang databases were carefully searched to identify eligible studies. The pooled hazard ratios (HRs) and 95% confidence intervals (CIs) were calculated to evaluate the prognostic significance of LncRNAs expression in osteosarcoma. Moreover, meta-regression analysis and subgroup analysis were carried out to explore the potential sources of heterogeneity.
Results
A total of 20 studies comprising 1749 patients were included in present meta-analysis. The results showed that the over-expression of LncRNA had a significant correlation with overall survival (OS) (HR = 2.16, 95% CI:1.68–2.79), and was not related to disease free survival (DFS) (HR = 0.71, 95% CI:0.05–9.53). Subgroup analysis further indicated that LncRNA transcription level was significantly associated with alkaline phosphatase (HR = 2.13, 95% CI:1.58–2.88) , tumor size (< 8/ ≥ 8:HR = 1.97, 95% CI: 1.55–2.62) , metastasis (yes/no: HR = 2.14,95% CI:1.15–3.97) , distant metastasis(presence/absence: HR = 4.02, 95% CI:3.05–5.23) and Enneking stage(IIA /IIB-III:HR = 3.2, 95% CI:2.48–4.14), but not associated with age (≤ 25/ > 25:HR = 1.01, 95% CI:0.78–1.3), gender(female/male: HR = 1.15, 95% CI: 0.96–1.37), tumor site (femur,tibia/elsewhere:HR = 1.15, 95% CI:0.94–1.4) and chemotherapy (yes/no: HR = 1.45, 95% CI:0.46–4.63).
Conclusions
This study demonstrated that abnormal LncRNAs expression might be potential prognostic markers to predict worse overall survival in osteosarcoma patients. However, the cut-off values may be the source of heterogeneity.
1. Introduction
Osteosarcoma (OS) is a highly malignant tumor of bone in children and adolescents [1], which accounts for about 2.4% of malignant tumors in children, and the incidence of OS is about 1–5 cases per million people per year [2,3]. Most patients are diagnosed as OS under the age of 25 years, and there are more men than women among the OS patients [4,5]. Both the metastasis and mortality rates of OS are high in clinic practice. About 20% OS patients were diagnosed with lung metastases at the time of the first diagnosis, and 80% OS metastases occur in the lung [6]. At present, the treatment of OS is mainly based on the combination of surgical resection and multiple chemotherapeutic drugs [7]. The average treatment rate of OS was 65% [8]. The average five-year survival rate of OS patients without metastasis was about 80% [9,10]. 90% OS patients died of recurrence or metastasis due to the presence of tumor resistance, drug side effects and other causes, and the five-year survival rate of OS patients was only 20–30% [11], [12], [13]. So far, the molecular mechanism of OS remains unclear. Therefore, finding new molecular markers for early diagnosis and prognosis and therapeutic targets of OS is very important for improving the survival rate of OS patients. Encouragingly, some LncRNAs have recently been reported to play a key role in OS pathogenesis.
LncRNA is a class of non-coding RNAs that are longer than 200 nucleotides in length, with little or no protein coding capacity [14]. Recent studies showed that LncRNA is widely transcribed in mammalian genome and plays an important role in gene regulation, which is involved in biological processes such as tumor proliferation, invasion, metastasis, apoptosis and drug resistance [15,16]. At present, the most important role of LncRNA may be associated with occurrence of cancers, and some studies have shown significant changes in LncRNA expression levels in colon, liver and lung cancers. Significantly high expressed LncRNA DLEU7-AS1 in patients with colon cancer promotes tumor invasion and metastasis through Wnt/β-catenin signaling pathway [17]. LncRNA HOTTIP can act as oncogenes to enhance the expression of anti-apoptotic factor Bcl-2 and promote chemotherapeutic resistance in small cell lung cancers by sponging miR-216a [18].LncRNA MEG3 inhibits the malignant progression of hepatocellular carcinoma by inhibiting PKM2 activity and β-catenin signaling pathway [19]. Recently, more and more literatures showed that LncRNAs are potential biomarkers for diagnosis and prognosis of OS.
So far, lots of researches have proved that LncRNA such as TUG1 [20], PVT1 [21] and ODRUL [22] are apparently highly expressed in OS patients. Jiang indicated that LncRNA DANCR can upregulate AXL expression by competitively sponging miR-33a-5p, enhance the function of cancer stem cells and promote the invasion and metastasis of OS [23]. Han found that LncRNA ATB is highly expressed in serum of OS patients, and its sensitivity and specificity are 83.33% and 90%, respectively [24]. Although many studies have assessed the correlation between LncRNA expression and OS prognosis, they are limited due to small sample sizes and different study qualities, and the prognostic value of LncRNA has not been agreed upon. Therefore, we conducted a systematic review and meta-analysis to quantitatively assess the relationship of LncRNAs expression with the prognosis and clinicopathology of OS patients.
2. Material and methods
21. Search strategy
PubMed,EMbase,Web of Science,The Cochrane Library,SionMed,CNKI and WanFang databases were searched to identify eligible studies up to November 11, 2017. The search strategy used both MeSH terms and free-text words to increase the sensitivity of the search. The search terms included: (“Long non-coding RNA”, “lncRNA”, “LincRNA”, “Long ncRNA”, “Long intergenic non-coding RNA”) AND (“Osteosarcoma”, “Osteogenic Sarcoma”, “Osteosarcoma Tumor”) with the limit to human, detailed search strategy is attached to appendix.
2.2. Inclusion and exclusion criteria
The eligible studies met the following criteria: (1) The clinical study of the expression of LncRNA in osteosarcoma; (2) Patients were confirmed osteosarcoma by pathological or histological examination; (3) qRT-PCR(Quantitative reverse transcription-polymerase chain reaction) was used to detect LncRNA expression in tissues or circulating blood of patients with osteosarcoma; (4) Studies provided sufficient information for extraction or calculation of the individual HR and 95%CI; (5) The association of LncRNAs with survivals was performed. Studies were excluded based on the following criteria: basic research of cell or animal experiment, duplicate articles, conference abstracts, case reports, review articles and letters.
2.3. Data collection and quality assessment
Two reviewers (Delong Chen and Meng Zhang) independently reviewed the eligible publications. The following data was extracted: surname of the first author, year of publication, the type of LncRNAs and expression, country, case number, cut-off value, sample type, outcome, clinicopathological features, outcome of study, HRs with their 95% CIs for OS or DFS and quality score. If the data was unavailable, we contacted study authors to request missing data. The information was extracted and recorded by using a standardized form. Two reviewers (Shan Jiang and Chi Zhou) independently assessed the quality of included studies by using Newcastle-Ottawa Quality Assessment Scale (NOS). If there are differences between the two reviewers, they can be resolved through discussion or finding another reviewer (Peng Chen).
2.4. Statistical analysis
Review Manager 5.2 (The Cochrane Collaboration, Software Update, Oxford, UK) and stata12.0 (STATA Corporation, College Station, Texas, USA) were used for meta-analysis. The HRs and 95% CI were used to evaluate the association between LncRNAs and prognosis and clinicopathological features. HR > 1 indicated that patients with upregulated LncRNA expression had poor prognosis. On the contrary, HR < 1 meant that patients with decreased LncRNA expression had better prognosis. We directly extracted if HRs and 95% CIs were reported directly in the articles. Otherwise, HRs and 95% CIs were estimated from existing data or Kaplan–Meier curve using methods previously reported by Tierney et al. Heterogeneity among the eligible studies was assessed with I2 statistics and chi-square Q test. P < 0.05 indicated statistically significant heterogeneity among studies, low heterogeneity: I2 ≥ 25%; moderate heterogeneity: I2 ≥ 50%; high heterogeneity: I2 ≥ 75%. Fixed-effects model was used when I2 < 50%. Otherwise, the random-effects model was used.
We conducted meta-regression analysis and subgroup analysis according to race, number of patients, HR availability, cut-off values and NOS scores. In order to verify the stability of the pooled results, we carried out sensitivity analysis. The publication bias was identified by Begg test and Egger test. P < 0.05 was considered statistically significant.
3. Results
3.1. Study inclusion and characteristics
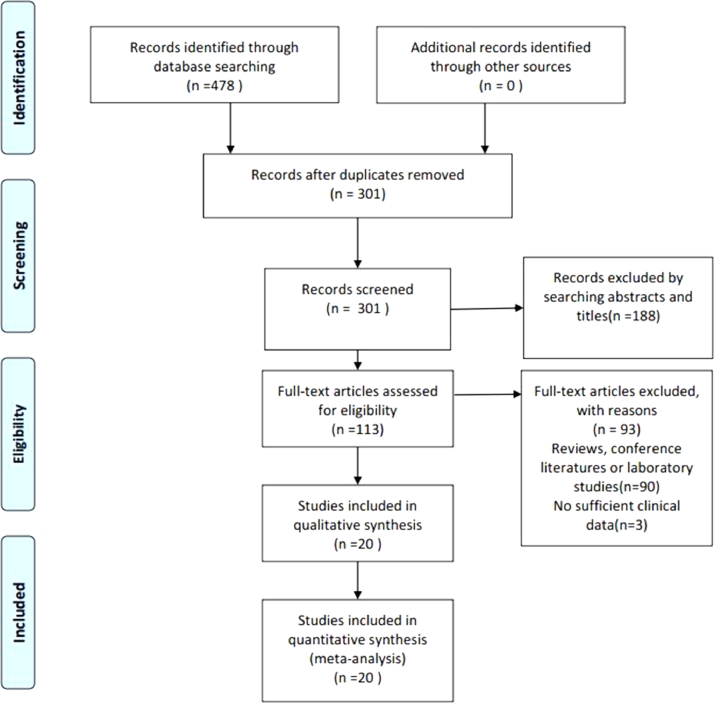
As shown in the flow diagram (Fig. 1), 478 articles were initially retrieved from PubMed,Embase, Web of Science, The Cochrane Library, SionMed, CNKI and WanFang databases. 177 duplicated articles were excluded. After the titles and abstracts were scanned, 188 irrelevant articles were removed. According to the inclusion and exclusion criteria, the full texts of remaining 113 articles were read for further evaluation and then 93 articles were excluded. Finally, 20 studies were included in the meta-analysis [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44].
Fig. 1.
Flow diagram of study selection process.
The correlations between the expression levels of LncRNA and differences in clinicopathological features in osteosarcoma patients were described in Table 1, including gender, age, tumor site, tumor size, tumor stage, metastasis, ALP and chemotherapy. The characteristics of 20 included studies were summarized in Table 2. Most of these studies were performed in China (18/20), another two studies were performed respectively in Germany and Brazil. The number of patients in 20 studies ranged from 33 to 168. Specimens were composed of osteosarcoma tissue (n = 18) and serum (n = 2). Among these 20 articles, 20 provide data on correlation between LncRNA expression and overall survival (OS), 2 on correlation between LncRNA expression and disease free survival (DFS) or progression free survival (PFS), 1 on correlation between LncRNA expression and metastasis free survival (MFS), recurrence free survival (RFS) or event free survival (EFS).
Table 1.
Comparison of p values of relationships between lncRNAs and clinicopathological features in osteosarcoma.
| Author | Year | LncRNAs | Country | Case number | Cut-off | Expression | Gender | Age | Tumor site | Tumor size | Tumor stage | Metastasis | ALP | Chemotherapy |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Jiang | 2017 | DANCR | China | 34 | NA | up-regulated | NA | NA | NA | <0.05 | NA | <0.05 | NA | NA |
| Wen | 2017 | UCA1 | China | 151 | NA | up-regulated | 0.572 | 0.199 | 0.804 | 0.907 | 0.001 | 0.007 | NA | NA |
| Cai | 2017 | HNF1A-AS1 | China | 72 | median | up-regulated | 0.215 | 0.534 | 0.143 | 0.311 | 0.019 | 0.009 | 0.128 | 0.031 |
| O'Leary | 2017 | PARTICLE | Germany | 40 | NA | up-regulated | 0.030 | NA | NA | NA | NA | 0.01 | NA | NA |
| Huo | 2017 | MALAT1 | China | 46 | median | up-regulated | 0.759 | 0.473 | NA | 0.008 | 0.058 | 0.000 | NA | NA |
| Li | 2017 | XIST | China | 145 | NA | up-regulated | 0.827 | 0.102 | 0.886 | 0.009 | 0.001 | 0.009 | 0.704 | NA |
| Wang | 2017 | SOX2-OT | China | 138 | median | up-regulated | 0.723 | 0.115 | 0.191 | 0.036 | 0.008 | 0.001 | NA | NA |
| Zhou | 2016 | CCAL | China | 46 | median | up-regulated | 0.555 | 0.200 | 0.502 | 0.134 | 0.017 | 0.006 | NA | NA |
| Peng | 2016 | BANCR | China | 84 | median | up-regulated | 0.509 | 0.505 | 0.814 | 0.008 | 0.004 | 0.020 | 0.366 | NA |
| Ju | 2016 | BCAR4 | China | 168 | median | up-regulated | 0.381 | 0.494 | 0.982 | 0.810 | 0.002 | 0.001 | 0.191 | 0.841 |
| Chen | 2016 | BCAR4 | China | 60 | median | up-regulated | 0.795 | 0.436 | 0.754 | 0.037 | 0.041 | 0.028 | NA | NA |
| Ma | 2016 | TUG1 | China | 76 | fold-change | up-regulated | 0.835 | 0.701 | 0.093 | 0.011 | 0.002 | 0.802 | 0.235 | 0.020 |
| Gao | 2016 | MALAT1 | China | 162 | median | up-regulated | 0.335 | 0.202 | 0.193 | 0.344 | 0.000 | 0.001 | NA | NA |
| Cong | 2016 | TUSC7 | China | 82 | fold-change | dowm-regulated | 0.65 | 0.473 | 0.627 | NA | 0.294 | 0.087 | NA | NA |
| Uzan | 2016 | HULC | Brazil | 33 | ROC | up-regulated | 0.999 | 0.065 | 0.274 | 0.67 | NA | 0.999 | NA | NA |
| Xia | 2016 | 91H | China | 67 | median | up-regulated | 0.497 | 0.927 | 0.114 | <0.001 | 0.015 | 0.936 | NA | 0.023 |
| Li | 2016 | UCA1 | China | 135 | median | up-regulated | 0.573 | 0.339 | 0.512 | 0.005 | <0.001 | 0.002 | NA | NA |
| Tian | 2015 | MEG3 | China | 64 | median | dowm-regulated | 0.614 | 0.302 | 0.281 | 0.076 | 0.006 | 0.011 | NA | NA |
| Li | 2015 | HOTTIP | China | 68 | median | up-regulated | 0.465 | 0.215 | 0.161 | 0.120 | 0.003 | 0.016 | NA | NA |
| Sun | 2015 | HULC | China | 78 | median | up-regulated | 0.492 | 0.352 | 0.624 | 0.496 | 0.003 | 0.005 | NA | NA |
Notes: LncRNA, long non-coding RNA;ALP, alkaline phosphatase;NA, not available.
Table 2.
Characteristics of studies included in this meta-analysis.
| Studies | LncRNAs | Country | Sample Type | Case number (High/Low) | Method | Cut-off | Tumor stage | outcome | HR availability | NOS |
|---|---|---|---|---|---|---|---|---|---|---|
| Jiang 2017 | DANCR | China | Tissue | 34(17/17) | qRT-PCR | NA | NA | OS,DFS | Directly | 7 |
| Wen 2017 | UCA1 | China | Tissue | 151 (75/76) | qRT-PCR | NA | IIA-III | OS,DFS | Directly | 7 |
| Cai 2017 | HNF1A-AS1 | China | Tissue | 72 (36/36) | qRT-PCR | median | IIA-III | OS | Directly | 6 |
| O'Leary 2017 | PARTICLE | Germany | Tissue | 40 (23/17) | qRT-PCR | NA | NA | OS,MFS | Indirectly | 7 |
| Huo 2017 | MALAT1 | China | serum | 46 (18/26) | qRT-PCR | median | I-IV | OS,PFS | Indirectly | 6 |
| Li 2017 | XIST | China | Tissue | 145 (75/70) | qRT-PCR | NA | I-IV | OS | Directly | 7 |
| Wang 2017 | SOX2-OT | China | Tissue | 138 (69/69) | qRT-PCR | median | I-III | OS | Directly | 7 |
| Zhou 2016 | CCAL | China | Tissue | 46 (23/23) | qRT-PCR | median | I-IV | OS | Directly | 7 |
| Peng 2016 | BANCR | China | Tissue | 84 (42/42) | qRT-PCR | median | IIA-III | OS | Directly | 7 |
| Ju 2016 | BCAR4 | China | Tissue | 168 (87/81) | qRT-PCR | median | IIA-III | OS | Directly | 7 |
| Chen 2016 | BCAR4 | China | Tissue | 60 (30/30) | qRT-PCR | median | I-III | OS,RFS | Directly | 7 |
| Ma 2016 | TUG1 | China | Tissue | 76 (41/35) | qRT-PCR | fold-change | I-III | OS,PFS | Directly | 8 |
| Gao 2016 | MALAT1 | China | Tissue | 162 (80/82) | qRT-PCR | median | IIA-III | OS | Directly | 7 |
| Cong 2016 | TUSC7 | China | Tissue | 82 (13/69) | qRT-PCR | fold-change | early-advanced | OS | Directly | 7 |
| Uzan 2016 | HULC | Brazil | Tissue | 33 (12/21) | qRT-PCR | ROC | NA | OS,EFS | Indirectly | 8 |
| Xia 2016 | 91H | China | serum | 67 (34/33) | qRT-PCR | median | I-III | OS | Directly | 8 |
| Li 2016 | UCA1 | China | Tissue | 135 (68/67) | qRT-PCR | median | I-III | OS | Directly | 7 |
| Tian 2015 | MEG3 | China | Tissue | 64 (32/32) | qRT-PCR | median | I-III | OS | Directly | 7 |
| Li 2015 | HOTTIP | China | Tissue | 68 (34/34) | qRT-PCR | median | IIA-III | OS | Directly | 7 |
| Sun 2015 | HULC | China | Tissue | 78 (39/39) | qRT-PCR | median | IIA-III | OS | Directly | 7 |
Notes: LncRNA, long non-coding RNA; qRT-PCR, quantities reverse transcription polymerase chain reaction; OS, overall survival; DFS, disease free survival; MFS, metastasis free survival; PFS, progression free survival; RFS, recurrence free survival; EFS, event free survival; HR, hazard ratio; NA, not available.
3.2. Prognostic value
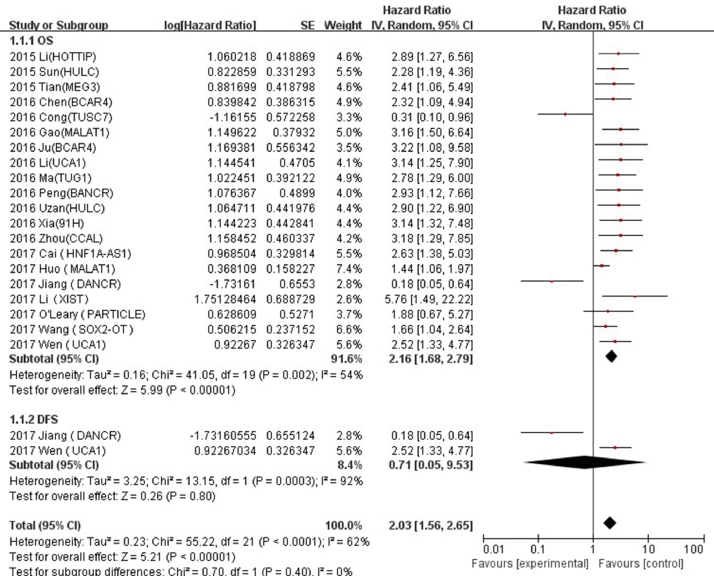
Twenty studies comprising 1749 patients were included in final analysis to assess the effect of LncRNA expression on OS. As shown in Fig. 2, the heterogeneity test indicated the existence of medium heterogeneity in OS (I2 = 54%, P < 0.00001), the pooled HR was 2.16 (95% CI: 1.68–2.79), which was calculated using random-effects model, the results suggested that a high LncRNA expression was significantly correlated with the poorer OS prognosis. The increased expressions of LncRNA UCA1 [25,38], XIST [26], SOX2-OT [28], MALAT1 [29,36], HNF1A-AS1 [30], PARTICLE [31], CCAL [32], 91H [33], HULC [34,43], BCAR4 [35,39], TUG1 [40], BANCR [41], MEG3 [42] and HOTTIP [44] indicated poorer prognoses of OS, on the contrary, the decreased expressions LncRNA DANCR [27] and TUSC7 [37] indicated better prognoses of OS. A total of 2 studies, including 185 patients, were included in the assessment of the effect of LncRNA expression on DFS (Fig. 2). There was no statistically significant correlation between low expression of LncRNA and prognosis of OS (HR = 0.71, 95%CI: 0.05–9.53, P = 0.0003, I2 = 92%).
Fig. 2.
Forest plot for the association between LncRNA expression levels with overall survival and disease-free survival in osteosarcoma.
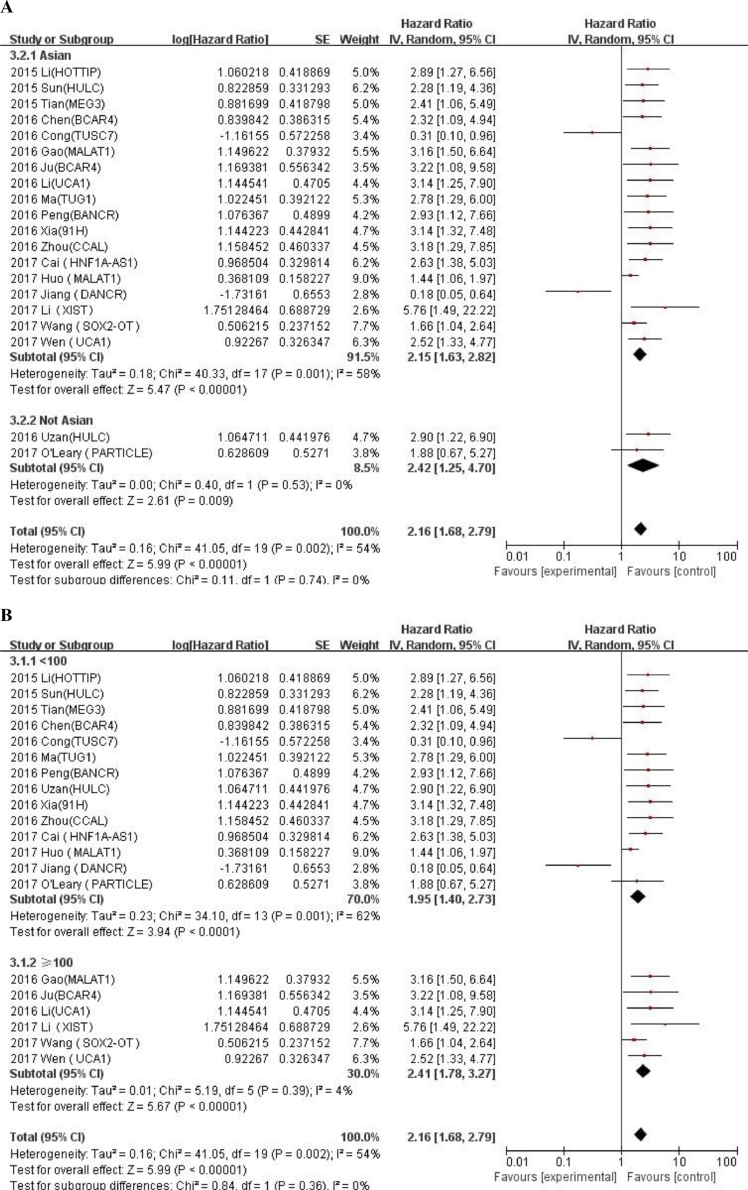
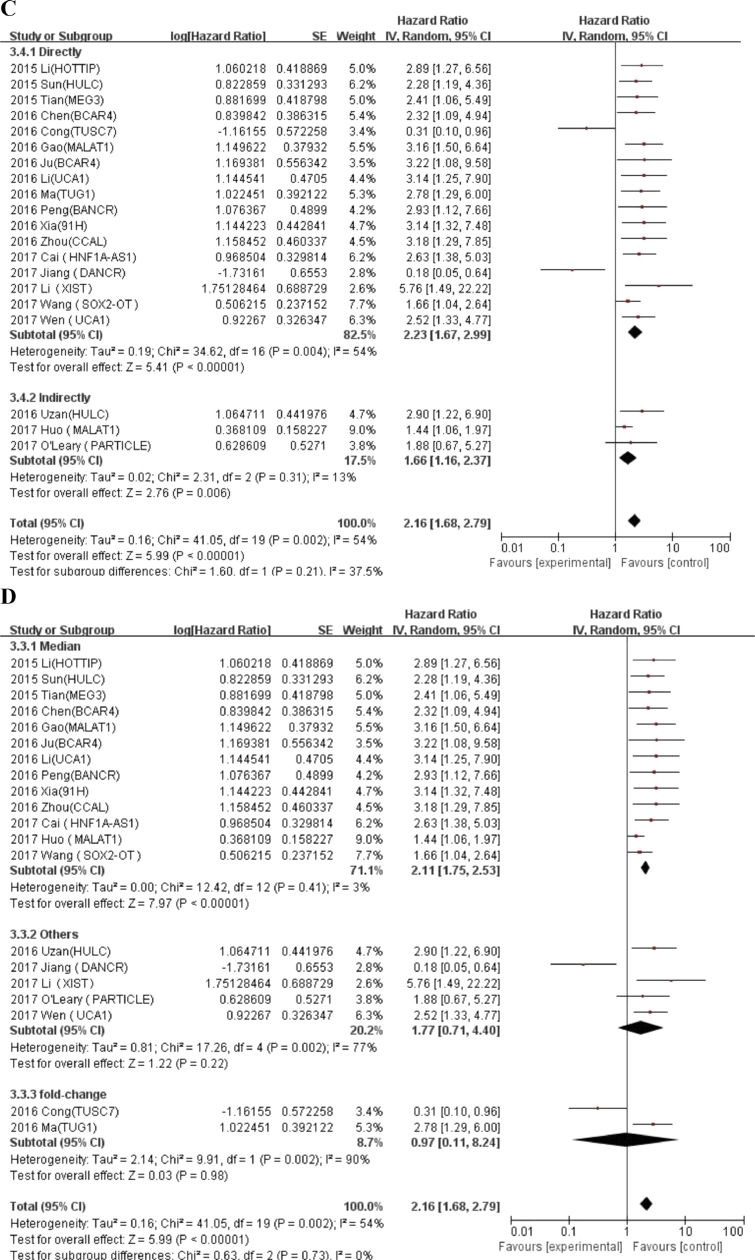
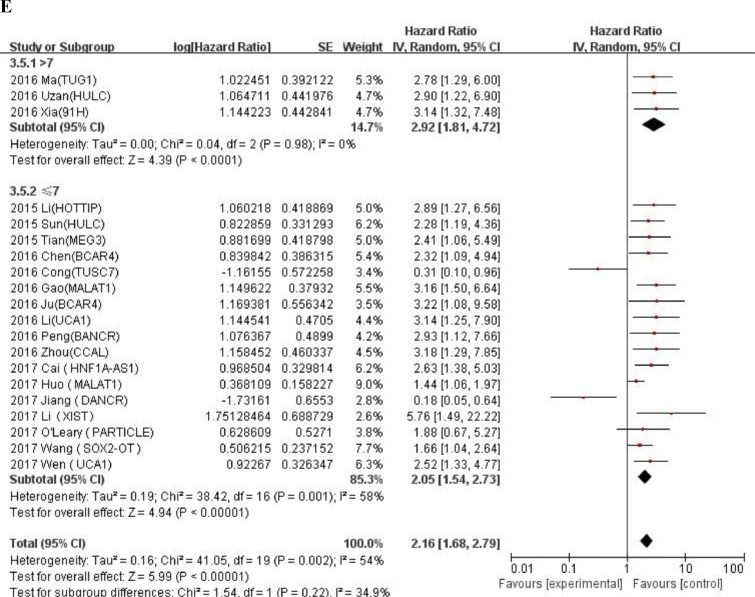
We conducted meta-regression analysis and subgroup analysis according to race, number of patients, and calculation method of HR value, cut-off values and NOS scores (Table 3). An additional file shows forest plot of HRs for subgroup analysis in more detail (Fig. A.1). In the subgroup analysis, the fold change (HR = 0.97, 95% CI: 0.11–8.24) in cut-off value was not statistically significant, other results showed that expression of LncRNA was correlated with a worse OS prognosis. Meta regression analysis revealed that there were no significant correlations between all relevant stratified factors and the heterogeneity among studies (P value ranged from 0.08 to 0.957).
Table 3.
General and subgroup analysis of the correlation between LncRNAs expression and overall survival.
| Categories | No. of studies | No. of patients | HR(95%CI)for OS | Meta-regression P-value | Heterogeneity |
|
|---|---|---|---|---|---|---|
| I-squared(%) | Chi-squared(P) | |||||
| OS | 20 | 1749 | 2.16 (1.68–2.79) | 54% | 0.0003 | |
| Race | 0.294 | |||||
| Asian | 18 | 1676 | 2.15 (1.63–2.82) | 58% | 0.001 | |
| Not Asian | 2 | 73 | 2.42 (1.25–4.7) | 0% | 0.53 | |
| No. of patients | 0.121 | |||||
| ≥100 | 6 | 899 | 2.41 (1.78–3.27) | 4% | 0.39 | |
| <100 | 14 | 777 | 1.95 (1.4–2.73) | 62% | 0.001 | |
| HR availability | 0.957 | |||||
| Directly | 17 | 1630 | 2.23 (1.67–2.99) | 54% | 0.004 | |
| Indirectly | 3 | 119 | 1.66 (1.16–2.37) | 13% | 0.31 | |
| Cut-off values | 0.08 | |||||
| Median | 13 | 1188 | 2.11 (1.75–2.53) | 3% | 0.41 | |
| Fold-change | 2 | 158 | 0.97 (0.11–8.24) | 90% | 0.002 | |
| Others | 5 | 403 | 1.77 (0.71–4.4) | 77% | 0.002 | |
| NOS scores | 0.147 | |||||
| >7 | 3 | 176 | 2.92 (1.81–4.72) | 0% | 0.98 | |
| ≤7 | 17 | 1573 | 2.05 (1.54–2.73) | 58% | 0.001 | |
Fig. A.1.
Forest plot of HRs for subgroup analysis of the correlation between LncRNAs expression and overall survival. (A) Race; (B) Number of patients; (C) HR availability; (D) Cut-off values; (E) NOS scores.
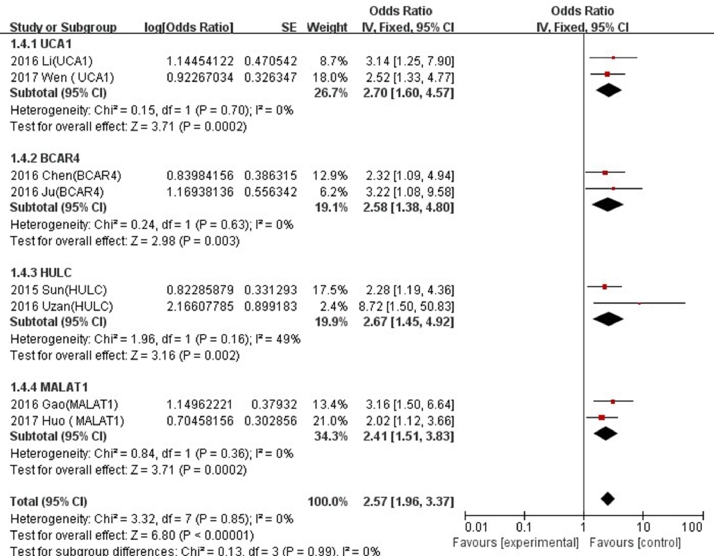
Of the total 20 LncRNAs, 4 (UCA1, BCAR4, HULC and MALAT1) were investigated in two studies. We found that the higher expressions of LncRNA UCA1 (HR = 2.70, 95% CI: 1.60–4.57), BCAR4 (HR = 2.58, 95% CI: 1.38–4.80), HULC (HR = 2.67, 95% CI: 1.45–4.92) and MALAT1 (HR = 2.41, 95% CI: 1.51–3.83), the poorer prognoses in osteosarcoma (Fig. 3).
Fig. 3.
Forest plots of studies evaluating hazard ratios of LncRNA UCAI,BCAR4,HULC and MALAT1 and overall survival of osteosarcoma patients.
3.3. The correlation between LncRNAs and clinicopathological features
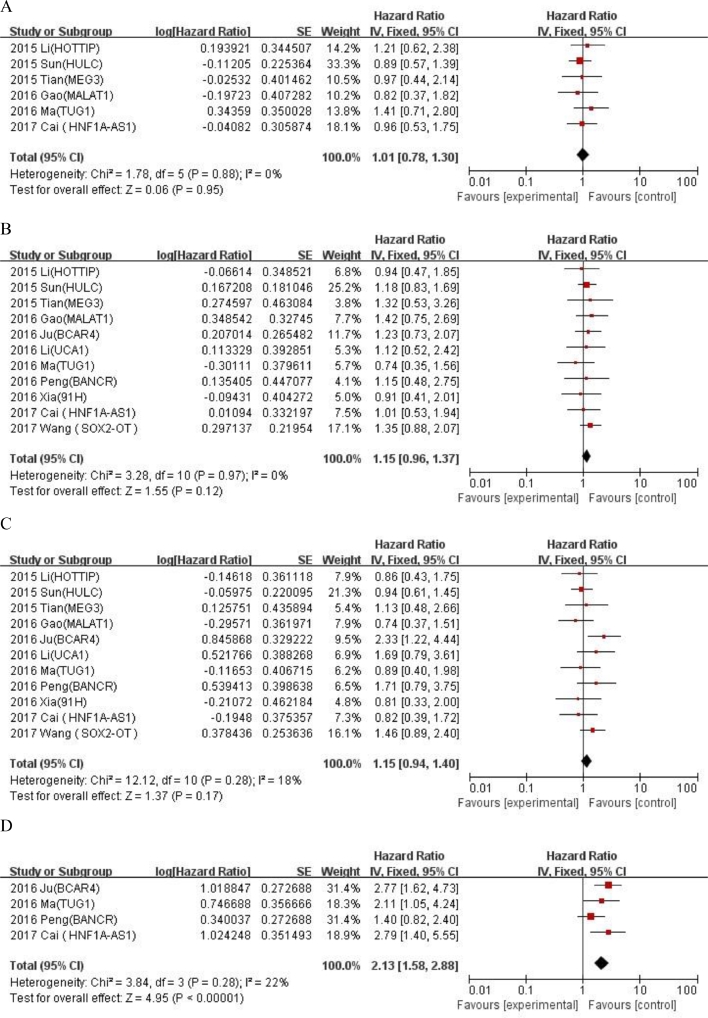
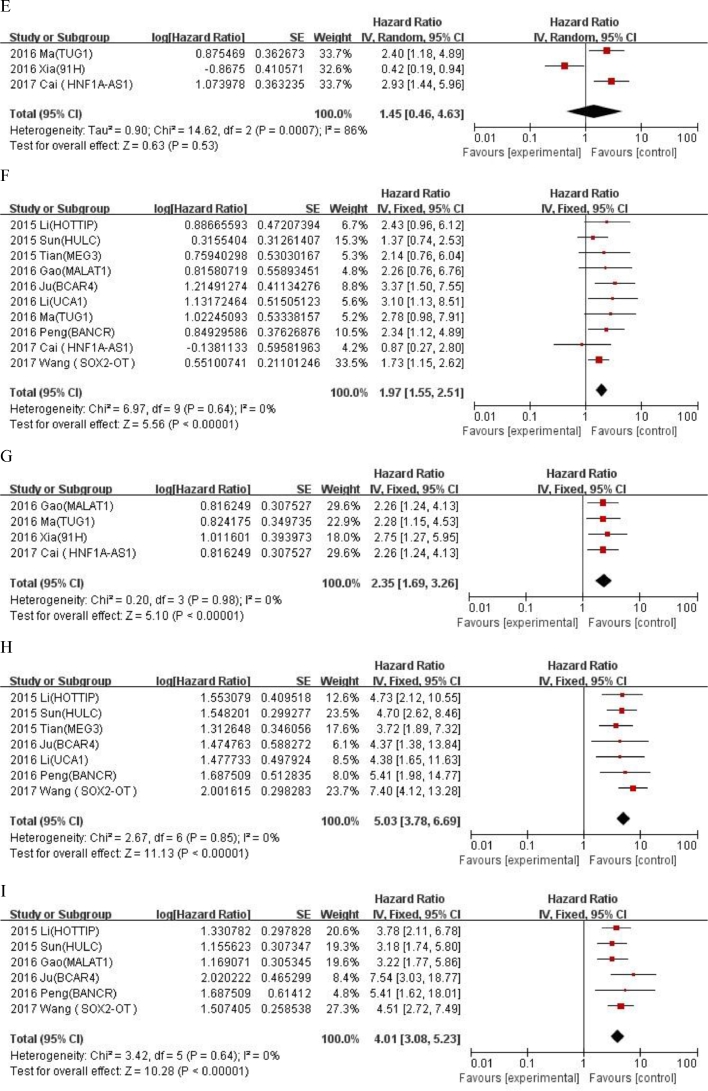
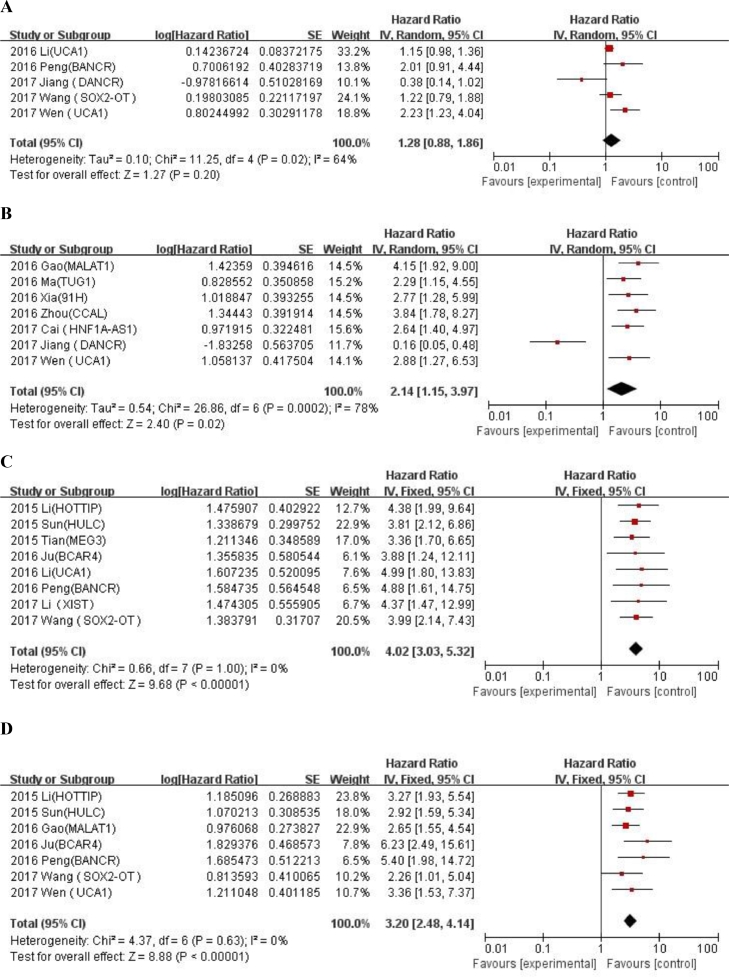
We evaluated the correlation between LncRNA expression and clinicopathological features of OS (Table 4). LncRNA transcription level was significantly correlated with alkaline phosphatase (univariate analysis: HR = 2.13, 95% CI: 1.58–2.88, tumor size (univariate analysis: < 8 / ≥ 8: HR = 1.97, 95% CI: 1.55–2.62) , metastasis (univariate analysis: yes/no: HR = 2.35, 95% CI: 1.69–3.26; multivariate analysis: yes/no: HR = 2.14, 95% CI:1.15–3.97) , distant metastasis(univariate analysis: presence/absence: HR = 5.03, 95% CI:3.78–6.69; multivariate analysis: presence/absence: HR = 4.02, 95% CI: 3.05–5.23) and Enneking stage (univariate analysis: yes/no: HR = 4.01, 95% CI: 3.08–5.23; multivariate analysis: IIA /IIB-III: HR = 3.2, 95% CI: 2.48–4.14), but not correlated with age (univariate analysis: ≤ 25 / > 25: HR = 1.01, 95% CI: 0.78–1.3), gender (univariate analysis: female/male: HR = 1.15, 95% CI: 0.96–1.37),tumor site(univariate analysis: femur,tibia/elsewhere: HR = 1.15, 95% CI: 0.94–1.4),chemotherapy (univariate analysis: yes/no: HR = 1.45, 95% CI: 0.46–4.63) and tumor size (multivariate analysis: < 8 / ≥ 8: HR = 1.28, 95% CI: 0.88–1.86) . An additional file shows forest plot for the association between overall survival time and clinicopathological features of patients in more detail (Fig. A.2 and A.3). The results showed that the high expression of LncRNA was significantly correlated with the poorer OS prognosis (univariate analysis: HR = 2.95, 95% CI: 2.37–3.66; multivariate analysis: HR = 2.16, 95% CI: 1.68–2.79).
Table 4.
Association between overall survival time and clinicopathological features of patients with osteosarcoma.
| clinicopathological Parameters | studies | Univariate analysis |
studies | Multivariate analysis |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| pooled HR(95%CI) | P-value | I-squared(%) | Chi-squared(P) | Pooled HR(95%CI) | P-value | I-squared(%) | Chi-squared(P) | |||
| Age(≤ 25 vs. > 25) | 6 | 1.01(0.78–1.3) | 0.95 | 0% | 0.88 | |||||
| Gender(Female vs. Male) | 11 | 1.15(0.96–1.37) | 0.12 | 0% | 0.97 | |||||
| Tumor site (femur,tibia vs. elsewhere) | 11 | 1.15(0.94–1.4) | 0.17 | 18% | 0.28 | |||||
| ALP | 4 | 2.13(1.58–2.88) | <0.00001 | 22% | 0.28 | |||||
| Chemotherapy(yes vs. no) | 3 | 1.45(0.46–4.63) | 0.53 | 86% | 0.0007 | |||||
| Tumor size(< 8 vs. ≥ 8) | 10 | 1.97(1.55–2.62) | <0.00001 | 0% | 0.64 | 5 | 1.28 (0.88–1.86) | 0.2 | 64% | 0.02 |
| Metastasis(yes vs. no) | 4 | 2.35 (1.69–3.26) | <0.00001 | 0% | 0.98 | 7 | 2.14 (1.15–3.97) | 0.02 | 78% | 0.0002 |
| Distant metastasis(presence vs. absence) | 7 | 5.03(3.78–6.69) | <0.00001 | 0% | 0.85 | 8 | 4.02 (3.05–5.23) | <0.0001 | 0% | 1 |
| Enneking stage(IIA vs. IIB–III) | 6 | 4.01 (3.08–5.23) | <0.00001 | 0% | 0.64 | 7 | 3.2 (2.48–4.14) | <0.0001 | 0% | 0.63 |
| LncRNA expression | 11 | 2.95 (2.37–3.66) | <0.00001 | 0% | 0.93 | 20 | 2.16 (1.68–2.79) | <0.00001 | 54% | 0.002 |
Fig. A.2.
Forest plot for the association between overall survival time and clinicopathological features of patients with osteosarcoma in univariate analysis. (A) Age (≤ 25 vs. 25); (B) Gender (Female vs. Male); (C) Tumor site (femur,tibia vs elsewhere); (D) ALP; (E) Chemotherapy (yes vs. no); (F) Tumor size (< 8 vs. ≥ 8); (G) Metastasis (yes vs. no); (H) Distant metastasis (presence vs. absence); (I) Enneking stage (IIA vs. IIB-III).
Fig. A.3.
Forest plot for the association between overall survival time and clinicopathological features of patients with osteosarcoma in multivariate analysis. (A) Tumor size (< 8 vs. ≥ 8) ; (B) Metastasis (yes vs. no); (C) Distant metastasis (presence vs. absence); (D) Enneking stage (IIA vs. IIB–III).
In addition to significant heterogeneity among studies on correlation between high expression of LncRNA and chemotherapy (univariate analysis: yes/no: I2 = 92%, P = 0.0007) , tumor size (multivariate analysis: < 8 / ≥ 8: I2 = 64%, P = 0.02) or metastasis (multivariate analysis: yes/no: I2 = 78%, P = 0.0002) no significant heterogeneity was observed among the studies on correlation between high expression of LncRNA and other clinicopathological factors.
3.4. Publication bias and sensitivity analysis
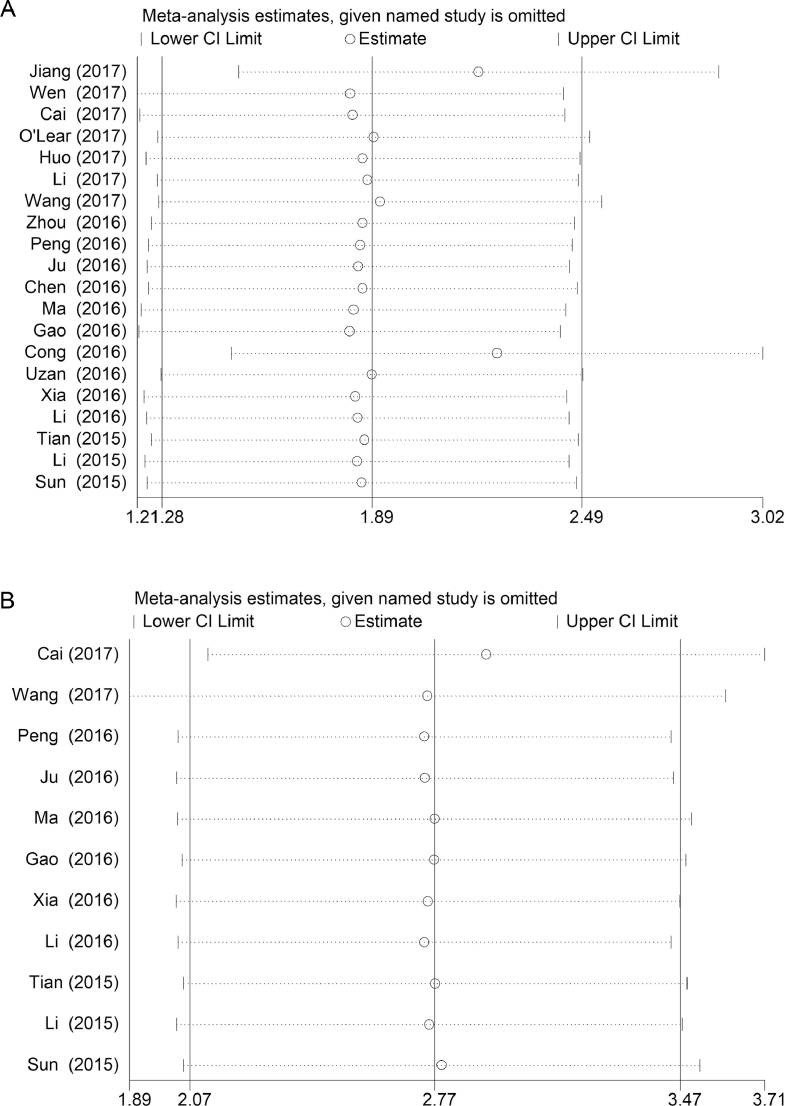
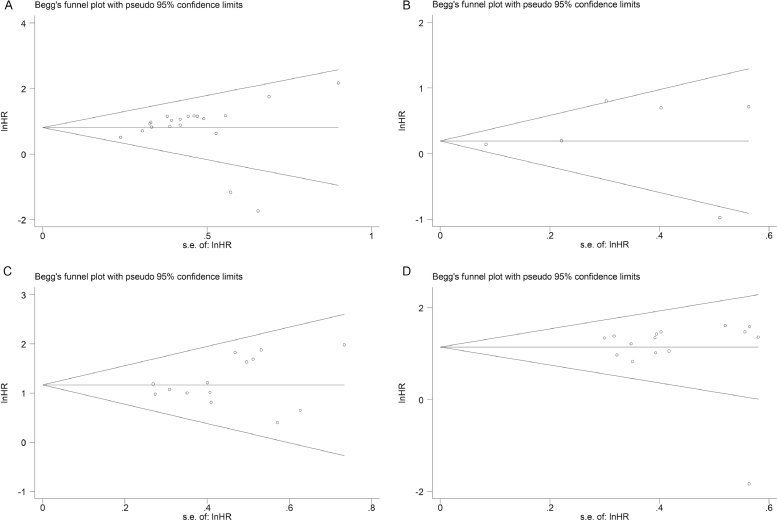
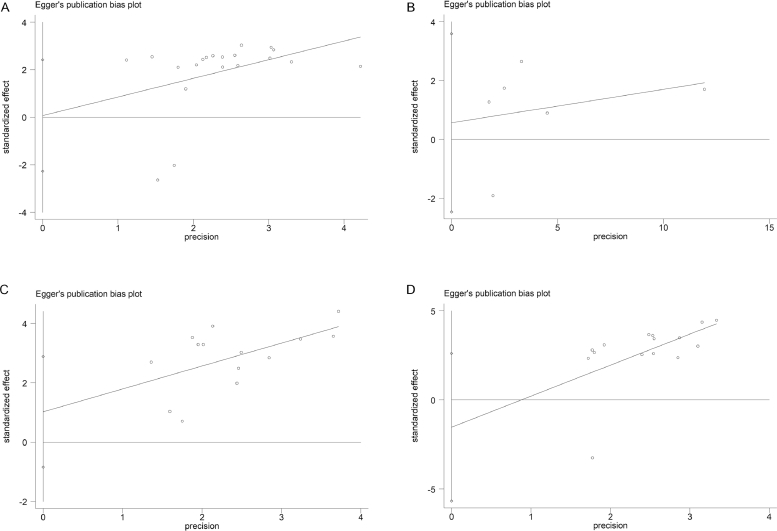
The univariate and multivariate sensitivity analyses of OS showed that the individual study had no obvious influence on the whole study results, and the overall effect had a good stability (Fig. 4.A and B). Both Begg's funnel plot and Egger's test were used to assess publication bias in the meta-analysis. The Begg's funnel plot of the pooled analysis in Fig. 5.A–D was quite symmetric, and no publication bias was detected by Egger's test due to all P-values > 0.05 (Fig. 6.A–D).
Fig. 4.
A–B Sensitivity analyses of the studies. (A) Multivariate analysis of overall survival; (B) Univariate analysis of overall survival.
Fig. 5.
A–D Begg's test for publication bias. (A) Multivariate analysis of overall survival (P = 0.127); (B) Tumor size (P = 1.000); (C) Tumor stage(P = 0.511); (D) Metastasis(P = 0.767).
Fig. 6.
A–D Egger's test for publication bias. (A) Multivariate analysis of overall survival (P = 0.951); (B) Tumor size(P = 0.631); (C) Tumor stage(P = 0.255); (D) Metastasis(P = 0.438).
4. Discussion
Osteosarcoma has become a cancer with the highest mortality rate in children and adolescents, which has characteristics such as local corrosion and systemic metastasis. Although the five-year survival rate of OS patients without metastasis is 80%, the measures to treat the metastatic OS are very limited. LncRNA is widely involved in biological processes such as tumor proliferation, invasion, metastasis, apoptosis and drug resistance. Investigating the expression and clinical significance of LncRNA in OS patients can provide a basis for prevention, diagnosis and treatment of OS in clinic. Therefore, we conducted a meta-analysis to assess the relationship of LncRNAs expression with the prognosis and clinicopathology of OS patients.
Recent studies have shown that LncRNA is closely correlated with the OS prognosis. LncRNA FOXC2-AS1 can increase the expression of FOXC2 transcription factors and promote the drug resistance of Adriamycin and multidrug resistance of OS cells [45]. High LncRNA HOST2 expression is a biomarker of poor prognosis in OS patients, it affects the proliferation, migration, invasion and apoptosis of OS cells [46]. In this meta-analysis, a total of 20 studies comprising 1749 patients were included into the final analysis. Our results showed that high LncRNA expression was significantly correlated with a poorer prognosis in OS patients, which suggested that LncRNA plays an important role in the prognosis of OS. The same results were also found in the meta-analysis of other cancers such as colon cancer [47], cervical cancer [48] and prostate cancer [49]. However, there was moderate heterogeneity in multivariate analysis of overall survival (I2 = 54%, P < 0.00001), so we performed meta-regression analysis and subgroup analysis according to race, number of patients, HR availability, cut-off values and NOS scores. The results of Meta regression analysis showed that there were no significant correlations between these factors and the heterogeneity in this study, but there was still a significant heterogeneity in the subgroup analysis. In subgroup analysis, the median cut-off value (HR = 2.11, 95% CI: 1.75–2.53; heterogeneity: P = 0.41, I2 = 3%) was significantly correlated with worse OS, but had no significant correlation with fold-change (HR = 0.97, 95% CI: 0.11–8.24; heterogeneity: P = 0.002, I2 = 90%). Significant heterogeneity existed among studies of cut-off value, fold-change and OS, and the reason maybe that fewer cases were included in studies.
In the result of our study, high expressions of LncRNA UCA1, BCAR4, HULC and MALAT1 were reported in two literatures, which predicted a worse overall survival rate in OS patients. Li found that HIF-1α enhances the expression of LncRNA UCA1 and promotes the proliferation of OS cells by inhibiting the PTEN/Akt signaling pathway [50]. The study by Kong et al. showed that LncRNA HULC exerts a sponge effect on miR-122. Overexpression of miR-122 promotes PI3K/AKT, JAK/STAT and Notch signaling pathways, down-regulates HNF4G expression and enhances tumor invasion and metastasis [51]. LncRNA MALAT1 plays an important role in the OS progression through modulating the enhancers of zeste homolog 2 (EZH2). MALAT1 provides a new target for the treatment of OS by inhibiting E-cadherin expression and enhancing β-catenin expression [52]. Therefore, LncRNA UCA1, BCAR4, HULC, and MALAT1 are independent risk factors for poor prognosis in OS patients.
We also evaluated the relationship between LncRNA expression and the clinicopathological features of OS. An univariate analysis showed that LncRNA transcription levels were not associated with age (≤ 25/ > 25), gender (male/female), tumor site (femur, tibia/elsewhere) and chemotherapy (yes/no), but were significantly correlated with ALP, tumor size (< 8/ ≥ 8), tumor metastasis (yes/no), tumor distant metastasis (presence/absence) and Ennking stage (IIA /IIB-III). However, a multivariate analysis showed that there was no significant association between LncRNA transcription level and tumor size (< 8/ ≥ 8), which may be one of the sources of heterogeneity.
In addition, there are still some deficiencies in this meta-analysis: (1) In order to reduce the bias caused by different methods of detecting LncRNA expression, we only included the studies using qRT-PCR method to detect LncRNA expression, which may bias the results due to different primers. (2) Different methods of HR extraction may also lead to bias. There were certain differences in HR and 95% CI between indirect and direct extractions. (3) The majority of populations were Asians included in this study, which may lead to biased results due to geographical differences. (4) Other factors such as different follow-up time and cut-off values will also result in bias.
In conclusion, this study shows that there is a significant correlation between the expression of LncRNA and the overall survival rate of OS patients, and it affects the prognosis of OS patients, suggesting that LncRNA may play an important role in the occurrence and development of OS. To further confirm our conclusion, a prospective high-quality study with a large sample size is needed to verify the role of LncRNA expression in the OS prognosis.
Acknowledgments
Acknowledgments
We owe our thanks to Yaolong Chen of Lanzhou University for his work on revising this manuscript. This work was financially supported by the National Natural Science Foundation of China (NSFC) (No. 81603641) and Guangdong Provincial Science and Technology Project (No. 2017A020213030).
Author contributions
Peng Chen designed the study. Delong Chen and Peng Chen wrote the manuscript. Delong Chen, Meng Zhang, Shan Jiang and Chi Zhou collected and extracted data. Haibin Wang and Bing Fang revised the manuscript. All authors read and approved the final manuscript.
Conflict of interest
The authors have no conflict of interest to declare.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jbo.2018.09.005.
Appendices
Appendix B. Supplementary materials
References
- 1.Sampson V.B., Yoo S., Kumar A., Vetter N.S., Kolb E.A. MicroRNAs and potential targets in osteosarcoma: review. Front. Pediatr. 2015;3:69. doi: 10.3389/fped.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Savage S.A., Mirabello L. Using epidemiology and genomics to understand osteosarcoma etiology. Sarcoma. 2011;2011 doi: 10.1155/2011/548151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Isakoff M.S., Bielack S.S., Meltzer P., Gorlick R. Osteosarcoma: current treatment and a collaborative pathway to success. J. Clin. Oncol. 2015;33:3029–3035. doi: 10.1200/JCO.2014.59.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu H.L., Shao L., Wang Q., Jia T., Li M., Yang D.P. A systematic review of p53 as a biomarker of survival in patients with osteosarcoma. Tumour Biol. 2013;34:3817–3821. doi: 10.1007/s13277-013-0966-x. [DOI] [PubMed] [Google Scholar]
- 5.Mirabello L., Troisi R.J., Savage S.A. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the surveillance, epidemiology, and end results program. Cancer-Am Cancer Soc. 2009;115:1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munajat I., Zulmi W., Norazman M.Z., Wan F.W. Tumour volume and lung metastasis in patients with osteosarcoma. J. Orthop. Surg. 2008;16:182–185. doi: 10.1177/230949900801600211. (Hong Kong) [DOI] [PubMed] [Google Scholar]
- 7.Ottaviani G., Jaffe N. The epidemiology of osteosarcoma. Cancer Treat. Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 8.Hagleitner M.M., Coenen M.J., Gelderblom H., Makkinje R.R., Vos H.I., de Bont E.S., van der Graaf W.T., Schreuder H.W., Flucke U., van Leeuwen F.N., Hoogerbrugge P.M., Guchelaar H.J., Te L.D. A first step toward personalized medicine in osteosarcoma: pharmacogenetics as predictive marker of outcome after chemotherapy-based treatment. Clin Cancer Res. 2015;21:3436–3441. doi: 10.1158/1078-0432.CCR-14-2638. [DOI] [PubMed] [Google Scholar]
- 9.Mirabello L., Troisi R.J., Savage S.A. International osteosarcoma incidence patterns in children and adolescents, middle ages and elderly persons. Int. J. Cancer. 2009;125:229–234. doi: 10.1002/ijc.24320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allison D.C., Carney S.C., Ahlmann E.R., Hendifar A., Chawla S., Fedenko A., Angeles C., Menendez L.R. A meta-analysis of osteosarcoma outcomes in the modern medical era. Sarcoma. 2012;2012 doi: 10.1155/2012/704872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bielack S.S., Carrle D., Hardes J., Schuck A., Paulussen M. Bone tumors in adolescents and young adults. Curr. Treat. Options Oncol. 2008;9:67–80. doi: 10.1007/s11864-008-0057-1. [DOI] [PubMed] [Google Scholar]
- 12.Guo J., Reddick W.E., Glass J.O., Ji Q., Billups C.A., Wu J., Hoffer F.A., Kaste S.C., Jenkins J.J., Ortega F.X., Quintana J., Villarroel M., Daw N.C. Dynamic contrast-enhanced magnetic resonance imaging as a prognostic factor in predicting event-free and overall survival in pediatric patients with osteosarcoma. Cancer-Am Cancer Soc. 2012;118:3776–3785. doi: 10.1002/cncr.26701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duchman K.R., Gao Y., Miller B.J. Prognostic factors for survival in patients with high-grade osteosarcoma using the surveillance, epidemiology, and end results (SEER) program database. Cancer Epidemiol. 2015;39:593–599. doi: 10.1016/j.canep.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 14.Qi P., Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod. Pathol. 2013;26:155–165. doi: 10.1038/modpathol.2012.160. [DOI] [PubMed] [Google Scholar]
- 15.Zhao W., Geng D., Li S., Chen Z., Sun M. LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer Med. 2018;7:842–855. doi: 10.1002/cam4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Zhao B., Lu Y.L., Yang Y., Hu L.B., Bai Y., Li R.Q., Zhang G.Y., Li J., Bi C.W., Yang L.B., Hu C., Lei Y.H., Wang Q.L., Liu Z.M. Overexpression of lncRNA ANRIL promoted the proliferation and migration of prostate cancer cells via regulating let-7a/TGF-beta1/ Smad signaling pathway. Cancer Biomark. 2018;21:613–620. doi: 10.3233/CBM-170683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X.B., Han C., Sun C.Z. Long non-coding RNA DLEU7-AS1 promotes the occurrence and development of colorectal cancer via Wnt/beta-catenin pathway. Eur. Rev. Med. Pharmacol. Sci. 2018;22:110–117. doi: 10.26355/eurrev_201801_14107. [DOI] [PubMed] [Google Scholar]
- 18.Sun Y., Hu B., Wang Q., Ye M., Qiu Q., Zhou Y., Zeng F., Zhang X., Guo Y., Guo L. Long non-coding RNA HOTTIP promotes BCL-2 expression and induces chemoresistance in small cell lung cancer by sponging miR-216a. Cell Death Dis. 2018;9:85. doi: 10.1038/s41419-017-0113-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng Q., Lin Z., Xu J., Lu Y., Meng Q., Wang C., Yang Y., Xin X., Li X., Pu H., Gui X., Li T., Xiong W., Lu D. Long noncoding RNA MEG3 suppresses liver cancer cells growth through inhibiting beta-catenin by activating PKM2 and inactivating PTEN. Cell Death Dis. 2018;9:253. doi: 10.1038/s41419-018-0305-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bo F.Y., Po L.X., Li L.X., Long C.G., Pei Z., Ming T.F. LncRNA TUG1 is upregulated and promotes cell proliferation in osteosarcoma. Open Med. (Wars) 2016;11:163–167. doi: 10.1515/med-2016-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Q., Chen F., Zhao J., Li B., Liang Y., Pan W., Zhang S., Wang X., Zheng D. Long non-coding RNA PVT1 promotes osteosarcoma development by acting as a molecular sponge to regulate miR-195. Oncotarget. 2016;7:82620–82633. doi: 10.18632/oncotarget.13012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu K.P., Ma X.L., Zhang C.L. LncRNA ODRUL contributes to osteosarcoma progression through the miR-3182/MMP2 Axis. Mol. Ther. 2017;25:2383–2393. doi: 10.1016/j.ymthe.2017.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Jiang N., Wang X., Xie X., Liao Y., Liu N., Liu J., Miao N., Shen J., Peng T. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 2017;405:46–55. doi: 10.1016/j.canlet.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 24.Han F., Wang C., Wang Y., Zhang L. Long noncoding RNA ATB promotes osteosarcoma cell proliferation, migration and invasion by suppressing miR-200 s. Am. J. Cancer Res. 2017;7:770–783. [PMC free article] [PubMed] [Google Scholar]
- 25.Wen J.J., Ma Y.D., Yang G.S., Wang G.M. Analysis of circulating long non-coding RNA UCA1 as potential biomarkers for diagnosis and prognosis of osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 2017;21:498–503. [PubMed] [Google Scholar]
- 26.Li G.L., Wu Y.X., Li Y.M., Li J. High expression of long non-coding RNA XIST in osteosarcoma is associated with cell proliferation and poor prognosis. Eur. Rev. Med. Pharmacol. Sci. 2017;21:2829–2834. [PubMed] [Google Scholar]
- 27.Jiang N., Wang X., Xie X., Liao Y., Liu N., Liu J., Miao N., Shen J., Peng T. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 2017;405:46–55. doi: 10.1016/j.canlet.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Wang Z., Tan M., Chen G., Li Z., Lu X. LncRNA SOX2-OT is a novel prognostic biomarker for osteosarcoma patients and regulates osteosarcoma cells proliferation and motility through modulating SOX2. IUBMB Life. 2017;69:867–876. doi: 10.1002/iub.1681. [DOI] [PubMed] [Google Scholar]
- 29.Huo Y., Li Q., Wang X., Jiao X., Zheng J., Li Z., Pan X. MALAT1 predicts poor survival in osteosarcoma patients and promotes cell metastasis through associating with EZH2. Oncotarget. 2017;8:46993–47006. doi: 10.18632/oncotarget.16551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai L., Lv J., Zhang Y., Li J., Wang Y., Yang H. The lncRNA HNF1A-AS1 is a negative prognostic factor and promotes tumorigenesis in osteosarcoma. J. Cell Mol. Med. 2017;21:2654–2662. doi: 10.1111/jcmm.12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Leary V.B., Maugg D., Smida J., Baumhoer D., Nathrath M., Ovsepian S.V., Atkinson M.J. The long non-coding RNA PARTICLE is associated with WWOX and the absence of FRA16D breakage in osteosarcoma patients. Oncotarget. 2017;8:87431–87441. doi: 10.18632/oncotarget.21086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou D.K., Yang X.W., Li H., Yang Y., Zhu Z.J., Wu N. Up-regulation of long noncoding RNA CCAL predicts poor patient prognosis and promotes tumor metastasis in osteosarcoma. Int. J. Biol. Mark. 2017;32:e108–e112. doi: 10.5301/jbm.5000240. [DOI] [PubMed] [Google Scholar]
- 33.Xia W.K., Lin Q.F., Shen D., Liu Z.L., Su J., Mao W.D. Clinical implication of long noncoding RNA 91H expression profile in osteosarcoma patients. Onco Targets Ther. 2016;9:4645–4652. doi: 10.2147/OTT.S103376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uzan V.R., Lengert A., Boldrini E., Penna V., Scapulatempo-Neto C., Scrideli C.A., Filho A.P., Cavalcante C.E., de Oliveira C.Z., Lopes L.F., Vidal D.O. High expression of HULC is associated with poor prognosis in osteosarcoma patients. Plos One. 2016;11 doi: 10.1371/journal.pone.0156774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen F., Mo J., Zhang L. Long noncoding RNA BCAR4 promotes osteosarcoma progression through activating GLI2-dependent gene transcription. Tumour Biol. 2016;37:13403–13412. doi: 10.1007/s13277-016-5256-y. [DOI] [PubMed] [Google Scholar]
- 36.Gao K.T., Lian D. Long non-coding RNA MALAT1 is an independent prognostic factor of osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 2016;20:3561–3565. [PubMed] [Google Scholar]
- 37.Cong M., Li J., Jing R., Li Z. Long non-coding RNA tumor suppressor candidate 7 functions as a tumor suppressor and inhibits proliferation in osteosarcoma. Tumour Biol. 2016;37:9441–9450. doi: 10.1007/s13277-015-4414-y. [DOI] [PubMed] [Google Scholar]
- 38.Li W., Xie P., Ruan W.H. Overexpression of lncRNA UCA1 promotes osteosarcoma progression and correlates with poor prognosis. J. Bone Oncol. 2016;5:80–85. doi: 10.1016/j.jbo.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ju L., Zhou Y.M., Yang G.S. Up-regulation of long non-coding RNA BCAR4 predicts a poor prognosis in patients with osteosarcoma, and promotes cell invasion and metastasis. Eur. Rev. Med. Pharmacol. Sci. 2016;20:4445–4451. [PubMed] [Google Scholar]
- 40.Ma B., Li M., Zhang L., Huang M., Lei J.B., Fu G.H., Liu C.X., Lai Q.W., Chen Q.Q., Wang Y.L. Upregulation of long non-coding RNA TUG1 correlates with poor prognosis and disease status in osteosarcoma. Tumour Biol. 2016;37:4445–4455. doi: 10.1007/s13277-015-4301-6. [DOI] [PubMed] [Google Scholar]
- 41.Peng Z.A., Lu R.B., Xiao D.M., Xiao Z.M. Increased expression of the lncRNA BANCR and its prognostic significance in human osteosarcoma. Genet. Mol. Res. 2016;15 doi: 10.4238/gmr.15017480. [DOI] [PubMed] [Google Scholar]
- 42.Tian Z.Z., Guo X.J., Zhao Y.M., Fang Y. Decreased expression of long non-coding RNA MEG3 acts as a potential predictor biomarker in progression and poor prognosis of osteosarcoma. Int. J. Clin. Exp. Pathol. 2015;8:15138–15142. [PMC free article] [PubMed] [Google Scholar]
- 43.Sun X.H., Yang L.B., Geng X.L., Wang R., Zhang Z.C. Increased expression of lncRNA HULC indicates a poor prognosis and promotes cell metastasis in osteosarcoma. Int. J. Clin. Exp. Pathol. 2015;8:2994–3000. [PMC free article] [PubMed] [Google Scholar]
- 44.Li F., Cao L., Hang D., Wang F., Wang Q. Long non-coding RNA HOTTIP is up-regulated and associated with poor prognosis in patients with osteosarcoma. Int. J. Clin. Exp. Pathol. 2015;8:11414–11420. [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang C.L., Zhu K.P., Ma X.L. Antisense lncRNA FOXC2-AS1 promotes doxorubicin resistance in osteosarcoma by increasing the expression of FOXC2. Cancer Lett. 2017;396:66–75. doi: 10.1016/j.canlet.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 46.Wang W., Li X., Meng F.B., Wang Z.X., Zhao R.T., Yang C.Y. Effects of the long non-coding RNA HOST2 on the proliferation, migration, invasion and apoptosis of human osteosarcoma cells. Cell Physiol. Biochem. 2017;43:320–330. doi: 10.1159/000480412. [DOI] [PubMed] [Google Scholar]
- 47.Wang J., Du S, Wang J., Fan W., Wang P., Zhang Z., Xu P., Tang S., Deng Q., Yang W., Yu M. The prognostic value of abnormally expressed lncRNAs in colorectal cancer: a meta-analysis. Plos One. 2017;12 doi: 10.1371/journal.pone.0179670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chi S., Shen L., Hua T., Liu S., Zhuang G., Wang X., Zhou X., Wang G., Wang H. Prognostic and diagnostic significance of lncRNAs expression in cervical cancer: a systematic review and meta-analysis. Oncotarget. 2017;8:79061–79072. doi: 10.18632/oncotarget.18323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma W., Chen X., Ding L., Ma J., Jing W., Lan T., Sattar H., Wei Y., Zhou F., Yuan Y. The prognostic value of long noncoding RNAs in prostate cancer: a systematic review and meta-analysis. Oncotarget. 2017;8:57755–57765. doi: 10.18632/oncotarget.17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li T., Xiao Y., Huang T. HIF1alphainduced upregulation of lncRNA UCA1 promotes cell growth in osteosarcoma by inactivating the PTEN/AKT signaling pathway. Oncol. Rep. 2018;39:1072–1080. doi: 10.3892/or.2018.6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kong D., Wang Y. Knockdown of lncRNA HULC inhibits proliferation, migration, invasion, and promotes apoptosis by sponging miR-122 in osteosarcoma. J. Cell Biochem. 2018;119:1050–1061. doi: 10.1002/jcb.26273. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Z.C., Tang C., Dong Y., Zhang J., Yuan T., Li X.L. Targeting LncRNA-MALAT1 suppresses the progression of osteosarcoma by altering the expression and localization of beta-catenin. J. Cancer. 2018;9:71–80. doi: 10.7150/jca.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.