Abstract
Multiple myeloma (MM) is the most frequent cancer to involve the skeleton with patients developing osteolytic bone lesions due to hyperactivation of osteoclasts and suppression of BMSCs differentiation into functional osteoblasts. Although new therapies for MM have greatly improved survival, MM remains incurable for most patients. Despite the major advances in current anti-MM and anti-resorptive treatments that can significantly improve osteolytic bone lysis, many bone lesions can persist even after therapeutic remission of active disease. Bone marrow mesenchymal stem cells (BMSCs) from MM patients are phenotypically distinct from their healthy counterparts and the mechanisms associated with the long-term osteogenic suppression are largely unknown. In this review we will highlight recent results of transcriptomic profiling studies that provide new insights into the establishment and maintenance of the persistent pathological alterations in MM-BMSCs that occur in MM. We will we discuss the role of genomic instabilities and senescence in propagating the chronically suppressed state and pro-inflammatory phenotype associated with MM-BMSCs. Lastly we describe the role of epigenetic-based mechanisms in regulating osteogenic gene expression to establish and maintain the pro-longed suppression of MM-BMSC differentiation into functional OBs.
Keywords: Multiple myeloma bone disease, Bone microenvironment, BMSCs, Osteoblast, Genomic, Senescence, Epigenetic
1. Introduction
Multiple Myeloma (MM) is a heterogeneous malignancy characterized by abnormal clonal plasma cell infiltration in the bone marrow [1]. Unlike solid tumors, which typically metastasize to distinct sites from the primary tumor, MM is usually widespread in multiple sites within the bone marrow. The bone marrow provides a specialized and highly supportive microenvironment in which interactions between MM and host cells, as well as components of the extracellular matrix (ECM) drive tumor development, progression and drug resistance [2]. Bone marrow mesenchymal stem cells (BMSCs) provide a special class of multi-potent cells, which can give rise to a variety of cell types, including osteoblasts (OB), osteocytes, adipocytes, chondrocytes, muscle cells and reticular fibroblasts [3]. These specialized cell types contribute to the well-organized architecture of the bone marrow, support the survival and differentiation of diverse lineages of hematopoietic cells and regulate bone formation and resorption. In addition to maintenance of bone homeostasis, immunomodulatory features of BMSCs control multiple aspects of MM pathophysiology and its associated bone disease [4]. Infiltrating MM cells co-opt a variety of mechanisms to suppress BMSC differentiation into functional OBs, which in turn enhances their support of MM cell growth, survival, and drug resistance [5], [6]. Even with the advancement of proteasome inhibitors and therapy regimens that combine chemotherapy, autologous stem cell transplantation, and maintenance strategies, which were shown to significantly improve bone formation [7] and enhance remineralization of pelvic lesions in MM patients [8], many bone lesions persist even after MM eradication due to uncoupled bone remodeling [9]. This further increases morbidity, mortality and medical costs. Research studies of MM bone disease (MMBD) have revealed a multiplicity of deregulated signaling molecules and receptor pathways associated with anti-osteogenic, pro-osteolytic and growth-supporting properties of the MM bone microenvironment (BME). However, the mechanisms associated with prolonged suppression of OB differentiation of MM-MSCs (MM-BMSCs) that occur even in the absence of signals from MM cells are still unclear. In this review, we highlight some of the phenotypic characteristics associated with the senescence and tumor-promoting features of MM-BMSCs. We discuss genome-wide transcriptional changes, chromosomal alterations as well as chromatin-based epigenetic mechanisms that contribute to osteogenic suppression of BMSC progenitors in MM bone disease.
1.1. Phenotypic alterations in BMSCs in mM
BMSCs are multipotent progenitor cells with self-renewal capacity and tri-lineage potential to differentiate into various cell types of the mesoderm lineage, including OBs, adipocytes and chondrocytes [10], [11]. The multi-lineage differentiation potential of BMSCs is highly dependent on their developmental state and interactions with their microenvironment in healthy and disease states [12]. For example, during aging and in various inflammatory and malignant conditions, the normal epigenetic reprograming and differentiation capacities of BMSCs into functional OBs are compromised, resulting in suppression of osteogenesis and increased adipogenesis. Since MM is predominantly a disease of the elderly with a median age at diagnosis of approximately 70 years [13], it is important to note the striking similarities between the changes that occur in BMSCs of older patients and those detected in BMSCs in pathologic inflammatory conditions that affect the marrow. It is important to consider whether age-related changes in BMSCs contribute to development of MM and/or progression from its precursory state, monoclonal gammopathy of undetermined significance (MGUS) to MM, or if the observed senescence-like and anti-osteogenic phenotype of MM-BMSC only results from their exposure to MM cells. Distinguishing between the relative contributions of aging per se and MM cell exposure on BMSCs has been very difficult to dissect, in part because most of the current in vivo MM models have significant limitations [14]. However, a recent metagenomic analysis of the C57BL/KaLwRij (KaLwRij) murine MM model [15], which shares many phenotypic similarities and clinical features associated with human MGUS progression to MM, demonstrated that genetic alterations in both pre-malignant B-cells and the cells of the host microenvironment, such as BMSCs and macrophages, contribute to the development of MM. This study suggests that development of malignant plasma cells together with potentially age-related alterations of multiple cell types within the BM may contribute equally to creating a permissive environment in the initial stages and progression of MM [15].
Although multiple studies have reported significant differences between normal and MM-BMSCs, there is no consensus among investigators on what these changes are. Several reports indicated that BMSCs derived from normal donors, MGUS and MM patients have similar proliferation capacities [16], [17] and that the proliferation rate of the MM-BMSCs did not correlate with the number of osteolytic lesions in MM patients [18]. These investigators found that MM-BMSCs produced abnormally high levels of cytokines that altered hematopoietic cell support and impaired osteogenesis. In contrast, Garderet et al. [19] reported that patient derived MM-BMSCs displayed impaired proliferation due to decreased levels of growth factor receptors and elevated DKK1 and IL6 expression as compared to HD-BMSCs. Furthermore, MM-BMSC expansion rate was worse in patients with advanced disease and lytic lesions [19]. Discrepancies in MM-BMSCs research studies such as these may arise from differences associated with the methodologies used in the characterization, isolation, expansion, and study design of MM-BMSCs as well as intrinsic patient-specific variability. In many cases, in vitro expansion of BMSCs is often necessary, due to the very low frequency (0.001–0.01%) of BMSCs, especially in bone marrows of elderly MM patients [20]. Typically, bulk bone marrow cell populations are subjected to plastic adherence and ex vivo expansion in proliferating media for several weeks. Increasing evidence suggests that long-term cultures of isolated BMSCs changes their phenotypic properties due to DNA methylation and epigenetic transformations [21]. However, what has been clearly shown is that the phenotypic alterations in BMSCs in MM that contribute to the drug resistance and tumor growth of MM cells result from exposure of BMSCs to MM cells in patients and/or MSC-lineage cell lines exposed to MM cells. These phenotypic features of MM-BMSCs include that they have suppressed osteogenic potential, express inflammatory cytokines, increased adipogenic differentiation and support MM growth.
Multiple mechanisms appear to contribute to changing the phenotype of normal BMSCs to tumor-promoting BMSCs in MM. For example, BMSCs isolated from MM patients can secrete protective soluble factors that confer resistance to Apo2 ligand/TRAIL-mediated apoptosis of MM cells [22]. Grigorieva et al. [23] reported that MM patient-derived BMSCs provided sufficient amounts of interleukin 6 (IL6) to effectively protect immature CD38+CD45+ MM cells from the cytotoxic effects of dexamethasone. In another report, the IL6-independent U266, ARH-77, HS-SUI-tan, and IM-9 cell lines exhibited decreased DNA synthesis when co-cultured with healthy donor BMSCs, which did not occur when they were co-cultured with MM-BMSCs [24]. MM patient-derived BMSCs also protected the IL6–dependent MM cell line, INA-6, and primary CD138+ MM cells from the cytotoxic effects of the IL6 receptor antagonist Sant7 and/or apoptosis induced by anti-gp130 monoclonal antibody treatment in vitro [25]. Hao at al. [26] showed that the U266 and H929 MM cell lines co-cultured with MM-BMSCs were protected to a greater extent from bortezomib-induced growth inhibition and apoptosis than MM cells co-cultured with normal donor (HD) BMSCs. They found that MM-BMSCs expressed higher levels of the adhesion molecules, ICAM-1 and VCAM-1, and secreted higher levels of the tumor-promoting factors, IL6 and VEGF, compared to HD-BMSCs [26]. Similar results were reported by Gupta et al. [27]. Consistent with these observations, Nefedova et al. [28] showed that media conditioned by HD-BMSCs did not protect MM cells from mitoxantrone-induced apoptosis, but after co-culture with MM cells, the BMSCs secreted soluble tumor-promoting anti-apoptotic factors. Further, co-culture of MM cells with BMSCs in direct contact provided significantly more protection from mitoxantrone treatment than separating the cells in transwell co-cultures [28]. In addition, exosomes have been implicated in exchanging proteins, DNA, mRNA, as well as non-coding RNAs between adjacent tumor cells and BMSCs in the local microenvironment [29]. Exosomes derived from MM patient bone marrow serum can prime HD-BMSCs to support the survival and growth of MM plasma cells [30]. Roccaro et al. [31] found that the amounts of the MM-inhibitory microRNA (miRNA) miR-15a were decreased in MM-BMSCS exosomes, and that the levels of oncogenic proteins, cytokines, protein kinases and adhesion molecules in exosomes differed between MM and normal BMSCs. Additionally, while exosomes from normal BMSCs inhibited MM cell proliferation, MM-BMSCs-derived exosomes promoted dissemination of MM cells in the bone marrow and enhanced tumor growth in vivo [32]. However, none of these mechanisms can explain the persistent suppression of OB differentiation that can occur in MM patients.
Therefore, in the following sections we highlight recent findings and provide insights into phenotypic alteration of myeloma exposed BMSCs. We will focus on genomic and epigenetic-based mechanisms in the context of establishment and maintenance of osteogenic suppression. We will discuss the role that epigenetic control of MSC-lineage switching from osteogenic to adipogenic commitment plays in MMBD. Lastly, we will address the role of chromatin remodeling enzymes in MMBD and the potential of employing small molecule epigenetic inhibitors currently used in the MM research as therapeutics and bone anabolic agents in the prevention or repair of osteolytic lesions in MM.
1.2. Transcriptomic characterization of the MM-BMSCs
Development of microarrays and next-generation sequencing (NGS) technologies has provided new approaches for assessing the genomic and epigenetic changes associated with the origins, progression and evolution of malignant diseases [33]. A search of the Gene Expression Omnibus (GEO) repository and available published NGS data sets demonstrate that MM plasma cells have been the primary focus of comprehensive genomic, transcriptomic and epigenetic studies in MM. Much has been learned about the changes in transcriptome signatures driven by genetic abnormalities and oncogenic transformations that occur in MM cells [34]. New appreciation for a global picture of 3-dimensional (3D) topological architecture and chromatin landscapes in the context of deregulated MM transcriptome signatures opens a new frontier of myeloma biology interrogation and drug targeting [35]. Gene expression profiling (GEP) is becoming a new tool in precision medicine for assessing myeloma subtypes and survival outcomes with a focus on drug treatment responses in high-risk MM patients [36]. The study by Botta at al. [37], found that inflammatory and cytokine/chemokine-mediated pathways are among the highest deregulated gene sets in myeloma evolution. Out of 20 selected candidate genes, differential expression of 8 genes encoding immunomodulators IL8, IL10, IL17A, CCL3, VEGEFA, EBI3, and NOS2 most accurately predicted clinical outcome and correlated with MM disease stage progression [37]. In addition to prognostic features, transcriptomic analyses have provided important insights into deregulated oncogenic and proliferative pathways in MM cells exposed to combination treatment with EZH2 and pan-HDAC inhibitors [38], as well as tofacitinib-mediated reversal of microenvironmental support for MM survival [39]. Recent comprehensive analysis of 255 MM patient samples demonstrated the importance of aberrant expression of fusion transcripts in MM progression [40]. Interestingly, alternative intronic polyadenylation known to generate various isoforms of protein coding genes has been recently defined in MM cells. In this report, global loss of intronic polyadenylation isoforms contributed to lenalidomide resistance and correlated with decreased progression-free survival of MM patients [41]. Furthermore whole transcriptome sequencing study by Samur and colleagues [42] reported that 869 long intergenic non-coding (lincRNA) were differentially expressed in MM as compared to normal plasma cells. More importantly, expression of 14 lincRNAs correlated with low- and high-risk disease stratification, which suggests their potential use in disease detection as well as prognostic factors in clinical outcome of myeloma disease [42]. Advancements in NGS sequencing at the single cell level enabled myeloma researchers to dissect and characterize cell sub-populations from bulk MM tumors. Comparative single cell genomic and global whole-exome sequencing analyses has led to identification of phylogenic divergence of myeloma cell subclones giving raise to the heterogeneity of myeloma tumors [43]. Using NOD/SCID-IL2RγNULL mouse xenografts Melchor et al. [43] further confirmed that selective pressures and the occurrence of new mutations contribute to clonal diversity during MM tumorigenesis. Newly emergent clones exhibit more resistant phenotypes and better survival properties during drug treatment or mouse engraftment [43]. Consistent with this, study by Mitra et al. [44] employed targeted transcriptome analyses of drug pre-exposed cell lines and naïve primary MM cells, to determine the proteasome inhibitor drug response GEP signature of distinct MM sub-populations. Collectively NGS technology greatly enhanced our understanding of deregulated gene expression profiles associated with MM heterogeneity and treatment resistance. The use of transcriptomic signatures for classification and diagnostic purposes in determining clinical outcome and tailored patient specific therapy are an exciting frontier of myeloma research. However, herein we present an additional layer of complexity in predicting disease evolution by accounting for the important changes and contributions of the bone marrow microenvironment to molecular pathogenesis of MM progression and drug resistance. The major clinical importance is to characterize the molecular changes that occur in the bone marrow in order to reverse its tumor fostering properties and enhance bone formation. Recent studies employing whole-transcriptome analyses of BMSC interactions with MM cells have verified previously known genes that are involved in these interactions and identified several new deregulated pathways and gene targets that play a role in MM cell homing to bone (Fig. 1).
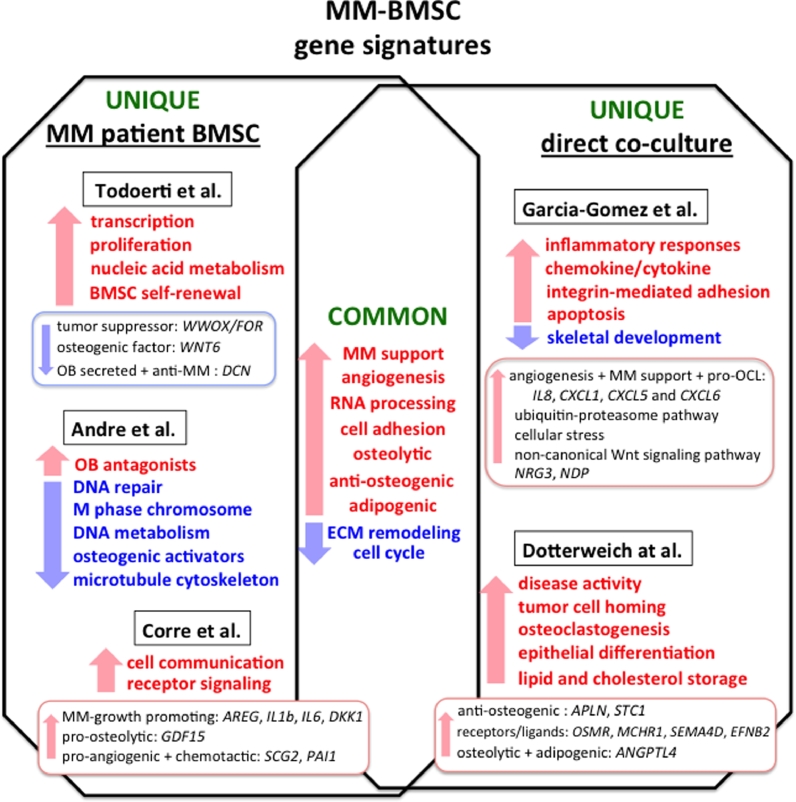
Fig. 1.
Gene expression signatures in MM-BMSCs. Depicted are transcriptional signatures of patient derived BMSCs and BMSCs co-cultured in a direct contact with MM cells, which were identified in studies using gene expression microarrays [17], [18], [57], [60], [61]. Shared gene categories reported in the studies are shown in the middle of the diagram and represent upregulated (red arrow) and downregulated (blue arrow) gene signatures, which are common to MM-BMSCs. Each side of the diagram represents unique functional categories identified in the gene expression sets for both MM-BMSCs and MM co-cultured BMSCs. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
One of the original MM-MSCs transcriptomic studies by Corre et al. [17] identified 127 annotated genes that were differentially expressed between normal and MM patient BMSCs. Almost 50% of these deregulated transcripts were associated with MM cell-BME crosstalk. These transcripts included genes encoding factors involved in cell communication, receptor signaling and extracellular matrix activity. These authors identified several upregulated genes, known to play a role in the pathogenesis of MM, including MM-growth promoting molecules such as Amphiregulin (AREG) [45], [46], the pro-inflammatory cytokines IL1b and IL6 [47] as well as DKK1, a suppressor of OB differentiation. Among the newly identified genes they reported were pro-osteolytic protein growth differentiation factor 15 (GDF15) [48], pro-angiogenic and chemotactic factor secretogranin II (SCG2), [17] and plasminogen activator inhibitor 1 (PAI-1), which were associated with disease progression of MM patients [49]. Giuliani and co-workers analyzed the transcriptional profiles of primary patient-derived BMSCs and OBs from normals (N), MGUS and MM patients [18]. This study then correlated the observed gene expression differences with the occurrence of lytic lesions among the MM patient samples. The molecular signatures of the three sample sets (N, MGUS and MM) identified genes predominantly involved in cell adhesion, cell cycle, transcriptional regulation and cell proliferation [18]. While there was not a clear distinction between MGUS and MM samples, 78 genes were differentially expressed between N and MM patient samples (in both isolated BMSCs and OBs) including HOXB2, 6 and 7, which are known to regulate mesenchymal cell lineage self-renewal and adipogenic commitment [50]. Genes predominately related to nucleic acid metabolism, RNA processing and transcription regulation were upregulated in MM-BMSCs from patients with lytic lesions, while extra cellular matrix (ECM) structural constituents, the WWOX/FOR tumor suppressor gene and WNT6, an osteogenic factor, were downregulated, consistent with their enhanced adipocytic phenotype [51]. The WWOX/FOR gene is frequently mutated in several solid tumors [52] and WWOX/FOR loss of function has been implicated in osteosarcomas [53]. However, its role in MMBD has not yet been defined. DCN, a gene encoding a small leucine-rich proteoglycan decorin, was downregulated to a greater extent in MM-BMSC/OBs from osteolytic than nonosteolytic MM patient samples [18]. Ida et al. also showed that decorin levels were downregulated in bone marrow plasma from both MGUS and MM patients as compared to healthy donors [54]. Decorin is primarily released by BMSCs and OBs [55] and exerts direct anti-MM effects by inducing apoptosis via activation of p21WAF1 [56] and blocking hepatocyte growth factor-induced migration and viability of MM cells [54]. Indirect effects of decorin on MM survival were also reported by Li et al. [56], who found that decorin decreased the capacity of osteoclasts to support primary MM cell survival.
More recently, Garcia-Gomez et al. [57] compared the transcriptional profiles of HD-BMSCs or MM patient BMSCs co-cultured for 24 hours in a direct-transwell culture system with the MM1.S human MM cell line. They found that normal and MM-derived BMSCs expressed a common upregulated functional gene signature after co-culture with MM1.S cells that included bone-ME regulatory genes involved in chemokine/cytokine and inflammatory responses, angiogenesis, proliferation, apoptosis, skeletal development, integrin-mediated adhesion and ECM remodeling. One of the highest expressed genes in MM and HD-BMSCs following MM-co-culture was IL8, which promotes angiogenesis, proliferation and chemotaxis of MM cells and stimulates osteoclastogenesis, that have important roles in the pathogenesis of MMBD (26). In support of these results, elevated IL8 levels were detected in serum of MM patients. CXCL1, CXCL5 and CXCL6, which are C-X-C chemokine/cytokine that enhance MM growth and angiogenesis, were also upregulated. Interestingly, these authors suggested that interactions of MM cells with BMSCs reduced the growth and induced early senescence of BMSCs. BMSC senescence is now emerging as an important underlying contributor to the prolonged suppression of OB differentiation in MM-BMSCs [29], [58], [59], [60]. Several gene sets were found to be specifically upregulated in MM-BMSCs. The upregulated genes were involved in RNA processing and splicing, activation of the ubiquitin-proteasome pathway, cell cycle regulation, cellular stress response, and the non-canonical Wnt signaling pathway. Neuregulin-3 (NRG3) and Norrie disease protein (NDP) were identified as putative candidates contributing to the progression of MMBD. NRG3 promoted MM cell proliferation and NDP increased RANKL-independent osteoclast formation [57]. Most likely, the gene changes induced by MM cells in HD-BMSCs may reflect genes involved in early changes in BMSCs during MM initiation, while genes uniquely deregulated in MM-patient BMSCs more likely participate in the prolonged OB suppression and enhanced adipogenic features of the later stages of lytic bone disease.
Global analysis of direct MM contact-induced gene expression changes in the BMSCs by Dotterweich at al. [61] provides new insights into deregulated osteogenic and angiogenic pathways at the bone-tumor interface. In this study, the BMSC-dependent plasmacytoma cell line, INA-6, was cultured in a direct contact with primary older donor BMSCs (average age 63) or BMSCs pre-differentiated in osteogenic media for 14-days (OP-MSC). They found that after contact with INA-6 cells, 991 and 552 probe sets were differentially expressed in BMSC and OP-MSC respectively. Pre-differentiated OP-MSCs had fewer gene expression changes after MM contact, consistent with previous reports [18] and studies suggesting that mature OBs may exhibit anti-MM properties [54], [55], [56], [62]. Gene expression profiles common to both BMSCs and OP-MSCs exposed to MM, were enriched in tumor-promoting genes involved in disease activity, tumor cell homing, angiogenesis, osteoclastogenesis, epithelial differentiation, cell-cell adhesion, and lipid and cholesterol storage. Consistent with the abnormal bone remodeling in MM, several genes known to interfere with bone metabolism including genes coding for anti-osteogenic factors APLN, STC1 and receptors/ligands OSMR, MCHR1, SEMA4D, EFNB2 were exclusively upregulated in undifferentiated BMSC. Of specific interest, adipose tissue-derived factor Angiopoietin-like 4 (ANGPTL4) was among the highest upregulated genes induced by MM [61]. Originally discovered as an endocrine regulator of lipid metabolism, ANGPTL4 affects osteoclast-mediated bone resorption, cartilage degradation and angiogenesis and has been implicated in regulating the critical balance between bone marrow adiposity and fracture risk in osteolytic disorders [63]. Importantly, the authors showed that ANGPTL4 was specifically elevated in BMSCs by INA-6 cell contact and not by contact with healthy donor CD19+ B cells [61]. These studies suggest that understanding the changes in gene networks in BMSCs induced by cross-talk with MM cells should provide a mechanistic basis for development of new therapeutic approaches for MMBD.
The transcriptomic studies described above have yielded significant insights into deregulated gene expression profiles and signaling pathways associated with phenotypic changes of BMSCs that create a permissive microenvironment for homing and growth of MM cells in the bone marrow. However, although the observed gene expression changes in skeletal precursors are striking, they occur within hours of physical contact with MM cells. It would also be very beneficial to compare these gene profiles with those obtained after extended MM co-culture, and after co-culture with distinct genotypic subtypes of primary CD138+ MM cells isolated from patients with low and high-risk MM and different levels of bone disease. Further, these studies did not determine which of the deregulated gene expression profiles they found persists after MM-cell removal. This could provide key insights into the mechanisms responsible for the prolonged osteogenic suppression, which continues in the absence of ongoing signals from MM cells.
1.3. Genomic alterations and senescence of MM-BMSCs
Genomic aberrations in patient-derived MM-BMSCs have been identified in several studies [59], [64], [65]. An array-based comparative genomic hybridization (array-CGH) analysis detected the presence of several non-recurrent chromosomal gains and losses (41Mb size) together with a discrete pattern of “hot spot” of genomic alterations in BMSCs derived from MM patients [64]. In most studies of genetically altered MM-BMSCs, the authors found deregulated expression of genes implicated in MM-BMSCs interactions, OB differentiation and MM-cell survival. Some of the significantly deregulated genes included bone morphogenetic protein 10 (BMP10), ephrin type-B receptor 1 (EPHB1), fibulin 5 EGF-like protein (FBLN5), receptors expressed in lymphoid tissues e.g. (RELL1) and ADAM metallopeptidase with thrombospondin type 1 motif 17 (ADAMTS17). However, this study did not correlate these distinct gene expression profiles with specific phenotypic changes in MM-BMSCs. While the authors do point out the possibility that in vitro culture conditions may have favored selection of genomically unstable MM-BMSC clones, these genomic imbalances were absent in identically expanded HD-BMSCs. These results suggest that the genomic instability was associated with MM exposure, rather than being an in vitro expansion artifact, and may contribute to the prolonged osteogenic-defect in MM-BMSCs [64].
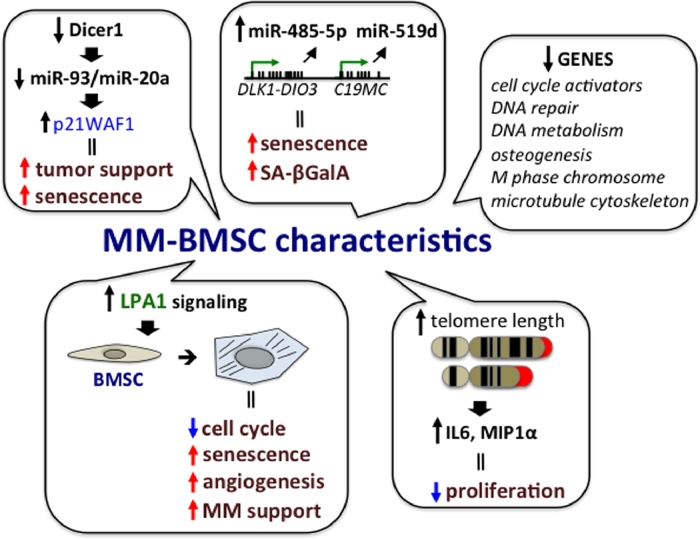
MM-induced early senescence of BMSCs also appears to contribute to the abnormalities present in MM-BMSCs (Fig 2). Senescence is a form of the cellular stress response often associated with degenerative and hyperplastic pathologies of aging. Due to dysfunctional telomeres, genotoxic stresses cause accumulation of DNA damage, perturbations to chromatin organization and strong mitogenic signals, so that senescent cells stop proliferating and undergo cell cycle arrest [66]. A causal role for senescent BMSCs in bone loss with aging has been well documented, and clinical targeting of these cells elicits both anti-resorptive and bone anabolic effects [67]. Li et al. reported that increased telomere length in BMSCs isolated from MM patients correlated with elevated expression of pro-inflammatory cytokines IL6 and MIP1α as compared to healthy donors [65]. However, consistent with other reports of possible early senescence of MM-BMSCs, these BMSCs exhibited decreased proliferative capacity but the osteogenic potential of these cells needs to be further evaluated [65]. While these results are intriguing, further mechanistic studies together with using BMSCs that are expanded less in vitro are needed to better link the changes in telomere length with functional abnormalities in MM-BMSCs. Berenstein et al. [59], showed that copy number variation and hypomethylation of the DLK1-DIO3 and C19MC genomic clusters were implicated in the senescence-associated abnormalities in MM-BMSCs isolated from patients. These genomic alterations correlated with overexpression of miR-485-5p and miR-519d, which increased senescence associated β-galactosidase activity (SA-βGalA) and S-phase cell cycle arrest. This report is consistent with previous observations that implicated involvement of miRNAs in MM pathology with potential roles in inflammation-induced cellular senescence [68], [69]. Interestingly, the senescent phenotype of MM-BMSCs was reversed after direct and transwell co-cultures with the human myeloma KMS12-PE cells [59]. Interactions with KMS12-PE MM-cells caused downregulation of the senescence-stimulating miR-485-5p and increased the proliferative capacity of the BMSCs. A recent microarray study revealed that the gene expression profile of patient-derived MM-BMSCs closely resembles that of replicative senescent human BMSCs [60]. Comparison of age matched healthy donor BMSCs with MM-BMSCs from patients that had not received anti-MM therapy revealed 646 differentially expressed probe sets between them. The downregulated genes (348) were primary associated with important biological functions pertaining to cell cycle, DNA repair, M phase chromosome, DNA metabolism and microtubule cytoskeleton regulation. The decreased expression of cell cycle activators (cyclins A, B, E, D, H; cyclin dependent kinases (CDKs) 1, 2, 4, 6; or CDC25A, B and C) was consistent with the increased expression of senescence-associated β-galactosidase, enlarged cell size and reduced proliferative capacity of MM-BMSC, due to increased accumulation of cells at S-phase. The MM-BMSCs also had decreased expression of osteogenic activators and up-regulation of OB antagonists together with an impaired mineralization potential. It is important to note that senescent abnormalities were less apparent in BMSCs from patients that had previously received anti-MM therapy [60].
Fig. 2.
Mechanisms underlying senescent phenotype of MM-BMSCs. Genomic alteration, miRNA and altered receptor LPA1 receptor signaling are contributing features to the senescent and tumor promoting phenotype of MM-BMSCs. Co-cultures of BMSCs with MM cells were shown to decrease expression of Dicer1 and subsequent processing of miR-93/miR-20a. This resulted in elevated expression of the cell cycle inhibitor p21WAF1 and initiated senescence and tumor-supporting phenotype of the MM-BMSCs [73]. Copy number variation and hypomethylation of the DLK1-DIO3 and C19MC genomic clusters caused overexpression of miR-485-5p and miR-519d, which increased senescence associated β-galactosidase activity (SA-βGalA) and S-phase cell cycle arrest [59]. The microarray study by Andre et al. [60], identified 348 downregulated genes with important biological functions in MM-BMCSs as compared to HD-BMSCs. Together with downregulated osteogenic factors, the identified gene signatures were associated with replicative senescence of MM-BMCSs. Increased telomere length has been correlated with elevated expression of pro-inflammatory cytokines IL6 and MIP1α and reduced cell proliferation of MM-BMSCs [65]. Lysophosphatidic acid receptor 1 (LPA1) was shown to modulate the senescent state and pro-inflammatory properties of MM-BMSCs. LPA1 signaling induced cell cycle arrest, promoted cellular senescence and enhanced the angiogenic and tumor-supporting properties of MM-BMSCs [58].
While senescent cells exhibit loss of proliferative capacity and osteogenic potential [70], they remain viable and metabolically active, with abnormal secretion of pro-inflammatory senescence-associated secretory profile (SASP) molecules [71]. Ozcan et al. applied oxidative stress, DNA damage and replicative exhaustion to trigger senescence in BMSCs to evaluate the effects of acute and chronic senescent BMSC SASP on MM cell survival [29]. Conditioned media derived from both acute and chronically induced senescent BMSC exhibited anti-MM effects and stimulated apoptosis and/or senescence of ARH-77 MM cells. Mass spectrometry analysis demonstrated that the content of the senescent BMSC secretome was profoundly changed when senescent BMSCs were subjected to co-culture with MM cells for 24 h. The BMSC secretome of BMSC co-cultured with MM cells lacked pro-senescent molecules and was enriched for anti-apoptotic, cancer growth promoting and pro-metastatic factors. Further, conditioned media from MM primed senescent BMSCs supported the growth of ARH-77 cells [29]. This suggests that while similar phenotypic mechanisms may be at play, MM-BMSCs are not equivalent to senescent HD-BMSCs. Lysophosphatidic acid (LPA) receptors 1 and 3 signaling has recently been implicated in modulating the senescent state and MM tumor promoting properties of MM-BMSCs [58]. LPA1 and LPA3 transduce opposite signals to tumor-surrounding BMSCs and determine if these cells enter a pro-senescent or anti-senescent state, respectively. Kanehira et al. found that LPA1 mRNA was increased in MM patient BMSCs, which correlated with their senescent phenotype. They showed that LPA1 silencing stimulated cell cycle progression, inhibited cellular senescence and trans-differentiation of BMSCs into tumor-associated fibroblasts (TAFs). Further, when IM-9 MM cells were co-transplanted with BMSCs in which LPA1 was inhibited using siRNA or the small molecule inhibitor Ki16425, tumor growth and vascularization in a mouse MM xenograft model was significantly reduced. In contrast, BM-MCSs with LPA3 knockdown and treated with IM-9 MM conditioned media readily transdifferentiated into senescent-like TAFs, and promoted tumor growth in vivo [58].
Dicer1 has also been shown to be involved in MM-induced BMSC senescence. Dicer1 is part of a multiprotein RNA-induced silencing complex (RISC), which facilitates microRNA-guided post-transcriptional mRNA regulation [72]. Knockdown of Dicer1 expression in BMSCs from healthy controls induced growth arrest, promoted cellular senescence and increased the tumor-supporting capacity of BMSCs. Similarly, co-culture of BMSC with MM cells decreased expression of Dicer1, reduced expression of miR-93/miR-20a and elevated expression of the cell cycle inhibitor p21WAF1 in BMSCs. This resulted in senescence and increased the tumor-supporting capacity of the MM-BMSCs [73]. Collectively, these studies suggest that enhanced senescence is a contributing factor to the functional changes induced in MM-BMSC by MM cells. It is still unclear if senescence of MM patient BMSCs contributes to OB suppression in the MM microenvironment in vivo, or if senescence of MM-BMSC is a result of removal of BMSCs from their BM niche with in vitro culturing that occurs in the absence of MM stimuli ex vivo. The unexpected findings reported by Berenstein et al. [59] that exposure of MM-BMSCs to MM cells reversed senescence of MM-BMSCs, suggests that a better understanding of the cause of MM-BMSC senescence is needed before therapy aimed at reversing senescence to relieve the impaired osteogenesis of MM-BMSCs is undertaken.
1.4. Chromatin regulation in MM-exposed BMSCs
Chromatin remodeling enzymes play an integral part in epigenome regulation by detecting (readers), adding (writers) and removing (erasers) active and repressive histone marks that allow for local chromatin alterations that control gene expression [74]. Key examples of epigenome flexibility are “bivalent” chromatin domains, which contain a combination of activating histone H3 lysine 4 tri-methylation (H3K4me3) and repressive H3K27me3 chromatin marks that maintain them in a quiescent state. Bivalent domains were initially identified as a key property of pluripotent embryonic stem cells and are associated with chromatin-modifying complexes and pre-loaded transcription machinery at promoters or enhancers that maintain gene expression at repressed or low levels. This “poised state,” allows for rapid gene activation by appropriate stimuli such as differentiation factors [75], [76]. Increasing evidence demonstrates that disturbances in the methylated states of bivalent chromatin and deregulation of the corresponding genes encoding developmental regulators and transcription factors are a prominent feature in cancer [77].
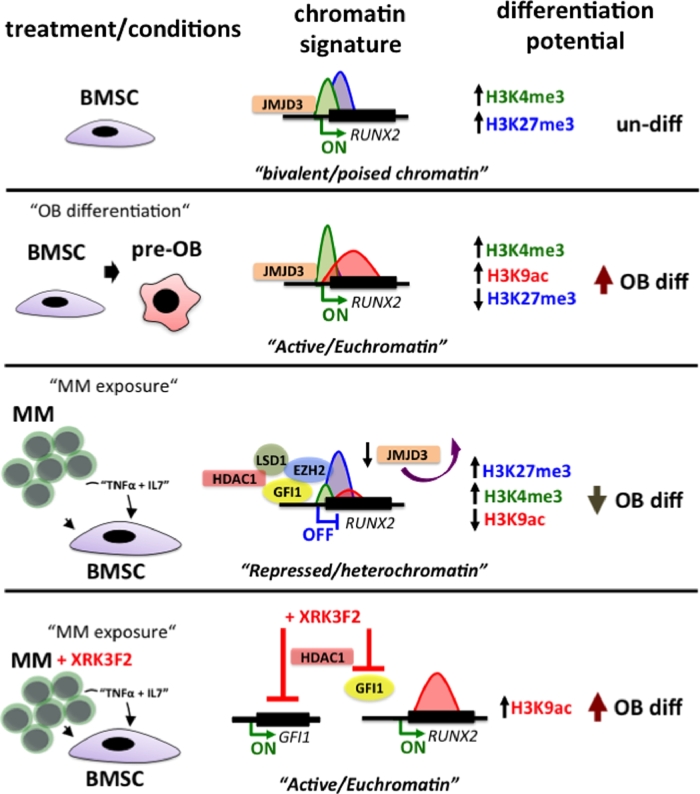
We recently showed that epigenetic-based mechanisms contribute to the initiation and persistence of osteogenic suppression of MM-BMSCs (Fig. 3)[78], [79], [80]. We found that upregulation and enhanced binding of GFI1, a transcriptional repressor, to a novel GFI1-response element within the RUNX2 promoter mediated the sustained suppression of osteogenesis [78], [80]. The role of RUNX2/CBFA1 during OB differentiation is well-established [81], and inhibition of its transcription is associated with impaired differentiation of MM-exposed human pre-osteoblasts (preOBs) and decreased bone formation of osteogenic progenitor cells from BM biopsies of MM patients with osteolytic lesions [82]. BMSCs after coculture with MM cells, BMSC from MM-bearing mice or MM-BMSCs isolated from MM patients had elevated levels of GFI1 compared to normal controls [57], [80], [83]. Both direct cell contact or exposure of BMSCs to TNFα and IL7, which are released by MM cells, increased GFI1 levels and repressed RUNX2 mRNA and protein levels in MC4 pre-OBs or naïve primary BMSCs [80]. BMSCs isolated from GFI1-knockout mice were significantly resistant to MM-induced OB suppression. Further, siRNA knockdown of GFI1 in MM-exposed MC4 pre-OBs and BMSCs from MM patients significantly restored expression of RUNX2 and OB differentiation markers. In support of GFI1’s repressive role in the BMSC cell lineage, Wang et al. [84] showed that downregulation of GFI1 in response to AMPK activation in MC4 pre-OBs upregulated Osteopontin (OPN) gene expression, which promoted osteogenesis. In addition to RUNX2, OPN is the second known direct target of GFI1 in pre-OBs. While additional GFI1 targets in MM-BMSCs/pre-OBs remain to be determined, it is interesting to note that GFI1 was among the highest represented transcription factors in a bioinformatic analysis of putative transcription factor binding sites in the promoters of deregulated genes in BMSCs cocultured with MM cells [57]. Further, GFI1 expression was also elevated in patient-derived BMSCs after MM co-culture in that study [57]. GFI1 can interact with several chromatin modifiers to repress gene expression [85], [86] In MM-exposed BMSC cells, GFI1 acts as a platform for formation of a repressive complex containing EZH2, histone deacetylase 1 (HDAC1) and lysine-specific demethylase 1 (LSD1) [78] that silences expression of RUNX2 (Fig. 3). Recruitment of the histone-modifying complex correlated with the presence of repressive chromatin architecture at RUNX2, with decreased H3K4me3, H3K36me3 and acetylated H3K9ac marks. The transcriptionally permissive bivalent nature of the RUNX2 promoter in MC4 pre-OBs co-cultured directly with MM cells changed to a “monovalent” repressive H3K27me3 architecture (Fig. 3) [78]. Kinetic Chromatin Immunoprecipitation (ChIP) analyses demonstrated that GFI1 recruitment and the RUNX2 epigenetic repression in pre-OBs occurred 36-48-hours after exposure to MM cells. Importantly, this suppression of RUNX2 by changes that were mediated by increased H3K27me3 marks on the chromatin was maintained for 4 days after MM cell removal from pre-OB co-cultures and was also present in isolated BMSCs from MM patients expanded for up to 3 weeks in the absence of MM cells [78]. Additional study by Wang and colleagues [84], suggests the possibility that upregulation of GFI1 and its co-repressors HDAC1, LSD1 and EZH2 in MM-exposed BMSCs may have more widespread epigenetic effects beyond regulation of the RUNX2 gene. The chromatin-based mechanisms responsible for the prolonged suppression of osteogenic differentiation may also contribute to the pathologic switch of BMSC differentiation toward adipocytes in the context of a pro-inflammatory MM bone marrow environment. Recent studies reported that in addition to being a potent transcriptional suppressor of osteogenic differentiation [78], [79], GFI1 plays a role downstream of AMPKα in regulating adipogenesis [84]. Overexpression of wild-type GFI1 increased adipogenesis and intracellular fat droplet content of AMPKα activated 3T3-L1 cells [84]. This argues that GFI1 with co-factors may both suppress and shift osteogenesis of MM-BMSCs to increase adipogenesis, which can further contribute to maintaining the prolonged suppression and altered phenotype of MM-BMSCs. Therapeutic significance of GFI1 targeting in MM bone disease has been demonstrated in a study by Adamik et al. [87], in which blockade of the p62-ZZ-domain signaling axis resulted in inhibition of GFI1 activity and rescued osteogenesis of myeloma-exposed preOBs. Treatment of MM-preOBs and patient-derived BMSCs with a novel small molecule inhibitor of the p62-ZZ domain, XRK3F2, prevented GFI1 upregulation and its subsequent binding and recruitment of HDAC1 to suppress Runx2. While XRK3F2 is not a bona fide epigenetic inhibitor, it prevented deacetylation of the Runx2 promoter chromatin and alleviated MM-suppressed osteogenesis in vitro [87] as well as in the syngeneic in vivo KaLwRij murine MM model [88] (Fig. 3).
Fig. 3.
Epigenetic suppression of RUNX2 promoter in MM-BMSCs. RUNX2 promoter in undifferentiated BMSCs exhibits bivalent promoter architecture having both H3K4me3 and H3K27me3 chromatin marks. During OB differentiation RUNX2 is in a transcriptionally permissive state with an open/euchromatic promoter architecture with enhanced active chromatin marks H3K3me3 and H3K9ac and reduced levels of repressive modification H3K27me3. MM exposure induces binding of transcriptional repressor Gfi1 with chromatin modifiers EZH2, HDAC1 and LSD1 to the RUNX2 promoter [78]. These modifiers deposit repressive chromatin marks on RUNX2 promoter core histones and epigenetically block its transcription. The active chromatin signature of RUNX2 changes into repressive H3K27me3-prevalent state. Treatment of preOBs with p62-ZZ-domian inhibitor XRK3F2 prevents upregulation and binding of GFI1 to the RUNX2 gene. This decreases recruitment of HDAC1 and subsequent MM-induced deacetylation of RUNX2, which leads to enhanced osteogenic differentiation [87].
Over the past years epigenetic mechanisms have been quite extensively studied in MM cells [89], but we are only beginning to understand epigenetic changes of the altered bone marrow compartments such as BMSCs in the context MM exposure. The transcriptomic analyses of the MM-BMSCs has already yielded a wealth of information, but the reported gene expression signatures are often quite variable and there is a lack of unified integration of the data from descriptive analyses into potential clinical applications. Furthermore, there is a great need for genome-wide mapping of chromatin landscapes that can be linked with the transcription profiling from MM-BMSCs in the context of both MM-preOB cell lines and MM patient BMSCs from different stages of disease progression. Correlating the changes in epigenetic and transcriptional signatures in BMSCs with changes in MM cells during MGUS to MM progression may be very informative in dissecting the nature of tumor initiation and fostering properties of MM-BMSCs. Furthermore, the field should consider the fact that just as is the case in the heterogeneity and evolution of MM tumors cells, clonal divergence and expansion may be also at play in favoring tumor-initiating and drug resistant sub-populations of BMSCs during MM disease progression. Since MM is a malignancy of both MM cells and the tumor-promoting bone microenvironment, a strong understanding of the epigenomic bases for trans-differentiation of MM-BMSCs into an altered adipogenic, senescent-like and tumor-promoting phenotype is instrumental for future studies. Therefore, epigenetic targeting should be evaluated more consciously in the context of the bone environment, to ensure that the use of epigenetic inhibitors does not adversely affect the differentiation properties and health of bone marrow cell compartments.
2. Concluding remarks
The underlying pathogenic abnormalities responsible for MM are multifactorial, and the abnormal bone metabolism seen in MMBD results from complex networks and numerous interactions among different bone cell types and their tumorigenic environment. While bone loss diseases such as MMBD may be initiated by age-related shifts in anabolic and catabolic responses that control bone homeostasis, it is becoming increasingly clear that mutual co-evolution of genetically altered plasma cells with the surrounding (aged) bone marrow cell compartments are driving factors that orchestrate the initiation and progression as well as the heterogenic abnormalities of MM cells [2]. The uncoupled bone remodeling is largely due to persistent impairment of BMSCs to differentiate into functional OBs even in the absence of active disease [9]. Studies over the past decade demonstrated that MM exposed BMSCs are phenotypically distinct from their healthy counterparts. Collectively, the evidence suggests that the prolonged suppression of BMSCs differentiation to OBs results from a combination of age-related changes and an increased inflammatory environment. These chronic changes driven by malignant plasma cells may lead to increased genomic instability, disturb epigenetic maintenance of gene expression and induce the senescent-like phenotype of BMSCs in MM.
Transcriptomic analyses demonstrate that MM cell homing to bone induces vast transcriptional changes in the interacting BMSCs, which are associated with enhanced MM-growth and drug resistance and induction of an anti-osteogenic and pro-lipogenic phenotype in BMSCs. Cross-talk between MM cells and surrounding BMSCs results from receptor-guided direct adhesive interactions as well as soluble factors and exosome-mediated exchange of genetic and protein regulatory cargo. Chromatin organization in BMSCs is based upon specific patterns of histone modifications and bivalent domains, which poises the entire genome to enter into several developmental outcomes [90]. MM cell exposure hijacks this epigenetic plasticity inducing corresponding changes in chromatin modifying enzymes such as EZH2, HDAC1 and LSD1 that shift the osteogenic differentiation potential of BMSCs toward adipogenesis. MM cells appear to act primarily on multi-potential BMSCs, as more mature OBs are largely refractory to the effects of MM cells and can exhibit anti-MM characteristics [62]. Several classes of epigenetic inhibitors have been developed, with several in pre-clinical trial and/or approved for clinical use. These epigenetic inhibitors can target the oncogenesis of MM cells [89] and their therapeutic potential for enhancing OB differentiation as a means to repair osteolytic bone lesions in MM has a promising future but needs further evaluations. Treatment modalities for multiple MM (MM) patients suffering from refractory or high-risk disease still have limited efficacy, and there are no effective, safe bone anabolic treatment approved for MM patients. Increased understanding of the underlying epigenetic and transcriptional programs responsible for multi-potential differentiation and tumorigenic support of MM-BMSCs is of high clinical significance if we are to develop new therapeutic strategies for MMBD.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jbo.2018.09.001.
Contributor Information
Deborah L Galson, Email: galson@pitt.edu.
G David Roodman, Email: groodman@iu.edu.
Appendix. Supplementary materials
References
- 1.Morgan G.J., Walker B.A., Davies F.E. The genetic architecture of multiple myeloma. Nat. Rev. Cancer. 2012;12(5):335–348. doi: 10.1038/nrc3257. [DOI] [PubMed] [Google Scholar]
- 2.Bianchi G., Munshi N.C. Pathogenesis beyond the cancer clone(s) in multiple myeloma. Blood. 2015;125(20):3049–3058. doi: 10.1182/blood-2014-11-568881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindner U., Kramer J., Rohwedel J., Schlenke P. Mesenchymal stem or stromal cells: toward a better understanding of their biology? Transfus. Med. Hemother. 2010;37(2):75–83. doi: 10.1159/000290897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olechnowicz S.W., Edwards C.M. Contributions of the host microenvironment to cancer-induced bone disease. Cancer Res. 2014;74(6):1625–1631. doi: 10.1158/0008-5472.CAN-13-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reagan M.R., Ghobrial I.M. Multiple myeloma mesenchymal stem cells: characterization, origin, and tumor-promoting effects. Clin. Cancer Res. Offi. J. Am. Assoc. Cancer Res. 2012;18(2):342–349. doi: 10.1158/1078-0432.CCR-11-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taube T., Beneton M.N., McCloskey E.V., Rogers S., Greaves M., Kanis J.A. Abnormal bone remodelling in patients with myelomatosis and normal biochemical indices of bone resorption. Eur. J. Haematol. 1992;49(4):192–198. doi: 10.1111/j.1600-0609.1992.tb00046.x. [DOI] [PubMed] [Google Scholar]
- 7.Pennisi A., Li X., Ling W., Khan S., Zangari M., Yaccoby S. The proteasome inhibitor, bortezomib suppresses primary myeloma and stimulates bone formation in myelomatous and nonmyelomatous bones in vivo. Am. J. Hematol. 2009;84(1):6–14. doi: 10.1002/ajh.21310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohan M., Samant R.S., Yoon D., Buros A.F., Branca A., Montgomery C.O., Nicholas R., Suva L.J., Morello R., Thanendrarajan S., Schinke C., Yaccoby S., van Rhee F., Davies F.E., Morgan G.J., Zangari M. Extensive remineralization of large pelvic lytic lesions following total therapy treatment in patients with multiple myeloma. J. Bone Miner. Res. 2017;32(6):1261–1266. doi: 10.1002/jbmr.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silbermann R., Roodman G.D. Current controversies in the management of myeloma bone disease. J. Cell. Physiol. 2016;231(11):2374–2379. doi: 10.1002/jcp.25351. [DOI] [PubMed] [Google Scholar]
- 10.Kim M., Kim C., Choi Y.S., Kim M., Park C., Suh Y. Age-related alterations in mesenchymal stem cells related to shift in differentiation from osteogenic to adipogenic potential: implication to age-associated bone diseases and defects. Mech. Age. Dev. 2012;133(5):215–225. doi: 10.1016/j.mad.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 11.Elahi K.C., Klein G., Avci-Adali M., Sievert K.D., MacNeil S., Aicher W.K. Human mesenchymal stromal cells from different sources diverge in their expression of cell surface proteins and display distinct differentiation patterns. Stem Cells Int. 2016;2016 doi: 10.1155/2016/5646384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei X., Yang X., Han Z.P., Qu F.F., Shao L., Shi Y.F. Mesenchymal stem cells: a new trend for cell therapy. Acta Pharmacol. Sin. 2013;34(6):747–754. doi: 10.1038/aps.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominguez L.J., Di Bella G., Belvedere M., Barbagallo M. Physiology of the aging bone and mechanisms of action of bisphosphonates. Biogerontology. 2011;12(5):397–408. doi: 10.1007/s10522-011-9344-5. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez E., Chen H., Berenson J.R. In vivo models of multiple myeloma (MM) Biochem. Pharmacol. 2014;89(3):313–320. doi: 10.1016/j.bcp.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 15.Amend S.R., Wilson W.C., Chu L., Lu L., Liu P., Serie D., Su X., Xu Y., Wang D., Gramolini A., Wen X.Y., O'Neal J., Hurchla M., Vachon C.M., Colditz G., Vij R., Weilbaecher K.N., Tomasson M.H. Whole Genome sequence of multiple myeloma-prone c57bl/kalwrij mouse strain suggests the origin of disease involves multiple cell types. PLoS One. 2015;10(5) doi: 10.1371/journal.pone.0127828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arnulf B., Lecourt S., Soulier J., Ternaux B., Lacassagne M.N., Crinquette A., Dessoly J., Sciaini A.K., Benbunan M., Chomienne C., Fermand J.P., Marolleau J.P., Larghero J. Phenotypic and functional characterization of bone marrow mesenchymal stem cells derived from patients with multiple myeloma. Leukemia. 2007;21(1):158–163. doi: 10.1038/sj.leu.2404466. [DOI] [PubMed] [Google Scholar]
- 17.Corre J., Mahtouk K., Attal M., Gadelorge M., Huynh A., Fleury-Cappellesso S., Danho C., Laharrague P., Klein B., Reme T., Bourin P. Bone marrow mesenchymal stem cells are abnormal in multiple myeloma. Leukemia. 2007;21(5):1079–1088. doi: 10.1038/sj.leu.2404621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Todoerti K., Lisignoli G., Storti P., Agnelli L., Novara F., Manferdini C., Codeluppi K., Colla S., Crugnola M., Abeltino M., Bolzoni M., Sgobba V., Facchini A., Lambertenghi-Deliliers G., Zuffardi O., Rizzoli V., Neri A., Giuliani N. Distinct transcriptional profiles characterize bone microenvironment mesenchymal cells rather than osteoblasts in relationship with multiple myeloma bone disease. Exp. Hematol. 2010;38(2):141–153. doi: 10.1016/j.exphem.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 19.Garderet L., Mazurier C., Chapel A., Ernou I., Boutin L., Holy X., Gorin N.C., Lopez M., Doucet C., Lataillade J.J. Mesenchymal stem cell abnormalities in patients with multiple myeloma. Leuk. Lymphoma. 2007;48(10):2032–2041. doi: 10.1080/10428190701593644. [DOI] [PubMed] [Google Scholar]
- 20.Marquez-Curtis L.A., Janowska-Wieczorek A., McGann L.E., Elliott J.A. Mesenchymal stromal cells derived from various tissues: biological, clinical and cryopreservation aspects. Cryobiology. 2015;71(2):181–197. doi: 10.1016/j.cryobiol.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 21.Bentivegna A., Roversi G., Riva G., Paoletta L., Redaelli S., Miloso M., Tredici G., Dalpra L. The effect of culture on human bone marrow mesenchymal stem cells: focus on dna methylation profiles. Stem Cells Int. 2016;2016 doi: 10.1155/2016/5656701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez L.E., Parquet N., Shain K., Nimmanapalli R., Alsina M., Anasetti C., Dalton W. Bone marrow stroma confers resistance to Apo2 ligand/TRAIL in multiple myeloma in part by regulating c-FLIP. J. Immunol. 2008;180(3):1545–1555. doi: 10.4049/jimmunol.180.3.1545. [DOI] [PubMed] [Google Scholar]
- 23.Grigorieva I., Thomas X., Epstein J. The bone marrow stromal environment is a major factor in myeloma cell resistance to dexamethasone. Exp. Hematol. 1998;26(7):597–603. [PubMed] [Google Scholar]
- 24.Uchiyama H., Barut B.A., Mohrbacher A.F., Chauhan D., Anderson K.C. Adhesion of human myeloma-derived cell lines to bone marrow stromal cells stimulates interleukin-6 secretion. Blood. 1993;82(12):3712–3720. [PubMed] [Google Scholar]
- 25.Chatterjee M., Honemann D., Lentzsch S., Bommert K., Sers C., Herrmann P., Mathas S., Dorken B., Bargou R.C. In the presence of bone marrow stromal cells human multiple myeloma cells become independent of the IL-6/gp130/STAT3 pathway. Blood. 2002;100(9):3311–3318. doi: 10.1182/blood-2002-01-0102. [DOI] [PubMed] [Google Scholar]
- 26.Hao M., Zhang L., An G., Meng H., Han Y., Xie Z., Xu Y., Li C., Yu Z., Chang H., Qiu L. Bone marrow stromal cells protect myeloma cells from bortezomib induced apoptosis by suppressing microRNA-15a expression. Leuk. Lymphoma. 2011;52(9):1787–1794. doi: 10.3109/10428194.2011.576791. [DOI] [PubMed] [Google Scholar]
- 27.Gupta D., Treon S.P., Shima Y., Hideshima T., Podar K., Tai Y.T., Lin B., Lentzsch S., Davies F.E., Chauhan D., Schlossman R.L., Richardson P., Ralph P., Wu L., Payvandi F., Muller G., Stirling D.I., Anderson K.C. Adherence of multiple myeloma cells to bone marrow stromal cells upregulates vascular endothelial growth factor secretion: therapeutic applications. Leukemia. 2001;15(12):1950–1961. doi: 10.1038/sj.leu.2402295. [DOI] [PubMed] [Google Scholar]
- 28.Nefedova Y., Landowski T.H., Dalton W.S. Bone marrow stromal-derived soluble factors and direct cell contact contribute to de novo drug resistance of myeloma cells by distinct mechanisms. Leukemia. 2003;17(6):1175–1182. doi: 10.1038/sj.leu.2402924. [DOI] [PubMed] [Google Scholar]
- 29.Ozcan S., Alessio N., Acar M.B., Toprak G., Gonen Z.B., Peluso G., Galderisi U. Myeloma cells can corrupt senescent mesenchymal stromal cells and impair their anti-tumor activity. Oncotarget. 2015;6(37):39482–39492. doi: 10.18632/oncotarget.5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo W., Gao Y., Li N., Shao F., Wang C., Wang P., Yang Z., Li R., He J. Exosomes: new players in cancer (Review) Oncol. Rep. 2017;38(2):665–675. doi: 10.3892/or.2017.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roccaro A.M., Sacco A., Maiso P., Azab A.K., Tai Y.T., Reagan M., Azab F., Flores L.M., Campigotto F., Weller E., Anderson K.C., Scadden D.T., Ghobrial I.M. BM mesenchymal stromal cell-derived exosomes facilitate multiple myeloma progression. J. Clin. Invest. 2013;123(4):1542–1555. doi: 10.1172/JCI66517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen B.F., Khan S., Laska B., Morgan G., Yaccoby S., Epstein J. Myeloma exosomes prime the microenvironment to support survival and growth of myeloma cells. Blood. 2016;128(22):2067. [Google Scholar]
- 33.LeBlanc V.G., Marra M.A. Next-generation sequencing approaches in cancer: where have they brought us and where will they take us? Cancers (Basel) 2015;7(3):1925–1958. doi: 10.3390/cancers7030869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szalat R., Avet-Loiseau H., Munshi N.C. Gene expression profiles in myeloma: ready for the real world? Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016;22(22):5434–5442. doi: 10.1158/1078-0432.CCR-16-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu P., Li T., Li R., Jia L., Zhu P., Liu Y., Chen Q., Tang D., Yu Y., Li C. 3D genome of multiple myeloma reveals spatial genome disorganization associated with copy number variations. Nat. Commun. 2017;8(1):1937. doi: 10.1038/s41467-017-01793-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vangsted A.J., Helm-Petersen S., Cowland J.B., Jensen P.B., Gimsing P., Barlogie B., Knudsen S. Drug response prediction in high-risk multiple myeloma. Gene. 2018;644:80–86. doi: 10.1016/j.gene.2017.10.071. [DOI] [PubMed] [Google Scholar]
- 37.Botta C., Di Martino M.T., Ciliberto D., Cuce M., Correale P., Rossi M., Tagliaferri P., Tassone P. A gene expression inflammatory signature specifically predicts multiple myeloma evolution and patients survival. Blood Cancer J. 2016;6(12):e511. doi: 10.1038/bcj.2016.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harding T., Swanson J., Van Ness B. EZH2 inhibitors sensitize myeloma cell lines to panobinostat resulting in unique combinatorial transcriptomic changes. Oncotarget. 2018;9(31):21930–21942. doi: 10.18632/oncotarget.25128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lam C., Ferguson I.D., Mariano M.C., Lin Y.T., Murnane M., Liu H., Smith G.A., Wong S.W., Taunton J., Liu J.O., Mitsiades C.S., Hann B.C., Aftab B.T., Wiita A.P. Repurposing tofacitinib as an anti-myeloma therapeutic to reverse growth-promoting effects of the bone marrow microenvironment. Haematologica. 2018;103(7):1218–1228. doi: 10.3324/haematol.2017.174482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cleynen A., Szalat R., Kemal Samur M., Robiou du Pont S., Buisson L., Boyle E., Chretien M.L., Anderson K., Minvielle S., Moreau P., Attal M., Parmigiani G., Corre J., Munshi N., Avet-Loiseau H. Expressed fusion gene landscape and its impact in multiple myeloma. Nat. Commun. 2017;8(1):1893. doi: 10.1038/s41467-017-00638-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee S.H., Singh I., Tisdale S., Abdel-Wahab O., Leslie C.S., Mayr C. Widespread intronic polyadenylation inactivates tumour suppressor genes in leukaemia. Nature. 2018;562(7721):127–131. doi: 10.1038/s41586-018-0465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samur M.K., Minvielle S., Gulla A., Fulciniti M., Cleynen A., Aktas Samur A., Szalat R., Shammas M., Magrangeas F., Tai Y.T., Auclair D., Keats J., Richardson P., Attal M., Moreau P., Anderson K.C., Parmigiani G., Avet-Loiseau H., Munshi N.C. Long intergenic non-coding RNAs have an independent impact on survival in multiple myeloma. Leukemia. 2018 doi: 10.1038/s41375-018-0116-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melchor L., Brioli A., Wardell C.P., Murison A., Potter N.E., Kaiser M.F., Fryer R.A., Johnson D.C., Begum D.B., Hulkki Wilson S., Vijayaraghavan G., Titley I., Cavo M., Davies F.E., Walker B.A., Morgan G.J. Single-cell genetic analysis reveals the composition of initiating clones and phylogenetic patterns of branching and parallel evolution in myeloma. Leukemia. 2014;28(8):1705–1715. doi: 10.1038/leu.2014.13. [DOI] [PubMed] [Google Scholar]
- 44.Mitra A.K., Mukherjee U.K., Harding T., Jang J.S., Stessman H., Li Y., Abyzov A., Jen J., Kumar S., Rajkumar V., Van Ness B. Single-cell analysis of targeted transcriptome predicts drug sensitivity of single cells within human myeloma tumors. Leukemia. 2016;30(5):1094–1102. doi: 10.1038/leu.2015.361. [DOI] [PubMed] [Google Scholar]
- 45.Mahtouk K., Hose D., Reme T., De Vos J., Jourdan M., Moreaux J., Fiol G., Raab M., Jourdan E., Grau V., Moos M., Goldschmidt H., Baudard M., Rossi J.F., Cremer F.W., Klein B. Expression of EGF-family receptors and amphiregulin in multiple myeloma. Amphiregulin is a growth factor for myeloma cells. Oncogene. 2005;24(21):3512–3524. doi: 10.1038/sj.onc.1208536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gunn W.G., Conley A., Deininger L., Olson S.D., Prockop D.J., Gregory C.A. A crosstalk between myeloma cells and marrow stromal cells stimulates production of DKK1 and interleukin-6: a potential role in the development of lytic bone disease and tumor progression in multiple myeloma. Stem Cells. 2006;24(4):986–991. doi: 10.1634/stemcells.2005-0220. [DOI] [PubMed] [Google Scholar]
- 47.Arranz L., Arriero M.D.M., Villatoro A. Interleukin-1beta as emerging therapeutic target in hematological malignancies and potentially in their complications. Blood Rev. 2017;31(5):306–317. doi: 10.1016/j.blre.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 48.Westhrin M., Moen S.H., Holien T., Mylin A.K., Heickendorff L., Olsen O.E., Sundan A., Turesson I., Gimsing P., Waage A., Standal T. Growth differentiation factor 15 (GDF15) promotes osteoclast differentiation and inhibits osteoblast differentiation and high serum GDF15 levels are associated with multiple myeloma bone disease. Haematologica. 2015;100(12):e511–e514. doi: 10.3324/haematol.2015.124511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shou L.H., Cao D., Dong X.H., Fang Q., Xu B.L., Fei J.P. Bone marrow urokinase plasminogen activator receptor levels are associated with the progress of multiple myeloma. Chin. Med. Sci. J. 2016;31(3):155–160. doi: 10.1016/s1001-9294(16)30044-x. [DOI] [PubMed] [Google Scholar]
- 50.Seifert A., Werheid D.F., Knapp S.M., Tobiasch E. Role of Hox genes in stem cell differentiation. World J. Stem Cells. 2015;7(3):583–595. doi: 10.4252/wjsc.v7.i3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cawthorn W.P., Bree A.J., Yao Y., Du B., Hemati N., Martinez-Santibanez G., MacDougald O.A. Wnt6, Wnt10a and Wnt10b inhibit adipogenesis and stimulate osteoblastogenesis through a beta-catenin-dependent mechanism. Bone. 2012;50(2):477–489. doi: 10.1016/j.bone.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pratt G. Molecular aspects of multiple myeloma. Mol. Pathol. 2002;55(5):273–283. doi: 10.1136/mp.55.5.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gordon J.A., Montecino M.A., Aqeilan R.I., Stein J.L., Stein G.S., Lian J.B. Epigenetic pathways regulating bone homeostasis: potential targeting for intervention of skeletal disorders. Curr. Osteoporos. Rep. 2014;12(4):496–506. doi: 10.1007/s11914-014-0240-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kristensen I.B., Pedersen L., Ro T.B., Christensen J.H., Lyng M.B., Rasmussen L.M., Ditzel H.J., Borset M., Abildgaard N. Decorin is down-regulated in multiple myeloma and MGUS bone marrow plasma and inhibits HGF-induced myeloma plasma cell viability and migration. Eur. J. Haematol. 2013;91(3):196–200. doi: 10.1111/ejh.12125. [DOI] [PubMed] [Google Scholar]
- 55.Nemani N., Santo L., Eda H., Cirstea D., Mishima Y., Patel C., O'Donnell E., Yee A., Raje N. Role of decorin in multiple myeloma (MM) bone marrow microenvironment. J. Bone Miner. Res. 2015;30(3):465–470. doi: 10.1002/jbmr.2371. [DOI] [PubMed] [Google Scholar]
- 56.Li X., Pennisi A., Yaccoby S. Role of decorin in the antimyeloma effects of osteoblasts. Blood. 2008;112(1):159–168. doi: 10.1182/blood-2007-11-124164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garcia-Gomez A., De Las Rivas J., Ocio E.M., Diaz-Rodriguez E., Montero J.C., Martin M., Blanco J.F., Sanchez-Guijo F.M., Pandiella A., San Miguel J.F., Garayoa M. Transcriptomic profile induced in bone marrow mesenchymal stromal cells after interaction with multiple myeloma cells: implications in myeloma progression and myeloma bone disease. Oncotarget. 2014;5(18):8284–8305. doi: 10.18632/oncotarget.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kanehira M., Fujiwara T., Nakajima S., Okitsu Y., Onishi Y., Fukuhara N., Ichinohasama R., Okada Y., Harigae H. An lysophosphatidic acid receptors 1 and 3 axis governs cellular senescence of mesenchymal stromal cells and promotes growth and vascularization of multiple myeloma. Stem Cells. 2017;35(3):739–753. doi: 10.1002/stem.2499. [DOI] [PubMed] [Google Scholar]
- 59.Berenstein R., Blau O., Nogai A., Waechter M., Slonova E., Schmidt-Hieber M., Kunitz A., Pezzutto A., Doerken B., Blau I.W. Multiple myeloma cells alter the senescence phenotype of bone marrow mesenchymal stromal cells under participation of the DLK1-DIO3 genomic region. BMC Cancer. 2015;15:68. doi: 10.1186/s12885-015-1078-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andre T., Meuleman N., Stamatopoulos B., De Bruyn C., Pieters K., Bron D., Lagneaux L. Evidences of early senescence in multiple myeloma bone marrow mesenchymal stromal cells. PLoS One. 2013;8(3):e59756. doi: 10.1371/journal.pone.0059756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dotterweich J., Schlegelmilch K., Keller A., Geyer B., Schneider D., Zeck S., Tower R.J., Ebert R., Jakob F., Schutze N. Contact of myeloma cells induces a characteristic transcriptome signature in skeletal precursor cells -Implications for myeloma bone disease. Bone. 2016;93:155–166. doi: 10.1016/j.bone.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 62.Yaccoby S., Wezeman M.J., Zangari M., Walker R., Cottler-Fox M., Gaddy D., Ling W., Saha R., Barlogie B., Tricot G., Epstein J. Inhibitory effects of osteoblasts and increased bone formation on myeloma in novel culture systems and a myelomatous mouse model. Haematologica. 2006;91(2):192–199. [PMC free article] [PubMed] [Google Scholar]
- 63.Knowles H.J. Multiple roles of angiopoietin-like 4 in osteolytic disease. Front. Endocrinol. 2017;8:80. doi: 10.3389/fendo.2017.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garayoa M., Garcia J.L., Santamaria C., Garcia-Gomez A., Blanco J.F., Pandiella A., Hernandez J.M., Sanchez-Guijo F.M., del Canizo M.C., Gutierrez N.C., San Miguel J.F. Mesenchymal stem cells from multiple myeloma patients display distinct genomic profile as compared with those from normal donors. Leukemia. 2009;23(8):1515–1527. doi: 10.1038/leu.2009.65. [DOI] [PubMed] [Google Scholar]
- 65.Li S., Jiang Y., Li A., Liu X., Xing X., Guo Y., Xu Y., Hao Y., Zheng C. Telomere length is positively associated with the expression of IL6 and MIP1alpha in bone marrow mesenchymal stem cells of multiple myeloma. Mol. Med. Rep. 2017;16(3):2497–2504. doi: 10.3892/mmr.2017.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tower J. Stress and stem cells. Wiley Interdiscip. Rev. Dev. Biol. 2012;1(6):789–802. doi: 10.1002/wdev.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Farr J.N., Xu M., Weivoda M.M., Monroe D.G., Fraser D.G., Onken J.L., Negley B.A., Sfeir J.G., Ogrodnik M.B., Hachfeld C.M., LeBrasseur N.K., Drake M.T., Pignolo R.J., Pirtskhalava T., Tchkonia T., Oursler M.J., Kirkland J.L., Khosla S. Targeting cellular senescence prevents age-related bone loss in mice. Nat. Med. 2017;23(9):1072–1079. doi: 10.1038/nm.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marasa B.S., Srikantan S., Martindale J.L., Kim M.M., Lee E.K., Gorospe M., Abdelmohsen K. MicroRNA profiling in human diploid fibroblasts uncovers miR-519 role in replicative senescence. Aging (Albany NY) 2010;2(6):333–343. doi: 10.18632/aging.100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Berenstein R., Blau I.W., Nogai A., Wächter M., Pezzutto A., Dörken B., Blau O. Aberrant expression Of miRNA and mRNA of cell cycle and adhesion-related genes in bone marrow stroma cells derived from patients with multiple myeloma. Blood. 2013;122(21):3149. [Google Scholar]
- 70.Sui B.D., Hu C.H., Zheng C.X., Jin Y. Microenvironmental views on mesenchymal stem cell differentiation in aging. J. Dent. Res. 2016;95(12):1333–1340. doi: 10.1177/0022034516653589. [DOI] [PubMed] [Google Scholar]
- 71.Campisi J. Aging, cellular senescence, and cancer. Ann. Rev. Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tijsterman M., Plasterk R.H. Dicers at RISC; the mechanism of RNAi. Cell. 2004;117(1):1–3. doi: 10.1016/s0092-8674(04)00293-4. [DOI] [PubMed] [Google Scholar]
- 73.Guo J., Li X., Zhao Y., Fei C., Chang C., Zhao S., Zheng Q. Reduction of dicer1 by multiple myeloma cells in mesenchymal stem cells promotes cellular senescence and tumor-supporting effect and decreases the differentiation. Blood. 2017;130(Suppl 1):4399. [Google Scholar]
- 74.Gardner K.E., Allis C.D., Strahl B.D. Operating on chromatin, a colorful language where context matters. J. Mol. Biol. 2011;409(1):36–46. doi: 10.1016/j.jmb.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S.L., Lander E.S. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 76.Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., Lee W., Mendenhall E., O'Donovan A., Presser A., Russ C., Xie X., Meissner A., Wernig M., Jaenisch R., Nusbaum C., Lander E.S., Bernstein B.E. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bernhart S.H., Kretzmer H., Holdt L.M., Juhling F., Ammerpohl O., Bergmann A.K., Northoff B.H., Doose G., Siebert R., Stadler P.F., Hoffmann S. Changes of bivalent chromatin coincide with increased expression of developmental genes in cancer. Sci. Rep. 2016;6:37393. doi: 10.1038/srep37393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Adamik J., Jin S., Sun Q., Zhang P., Weiss K.R., Anderson J.L., Silbermann R., Roodman G.D., Galson D.L. EZH2 or HDAC1 inhibition reverses multiple myeloma-induced epigenetic suppression of osteoblast differentiation. Mol. Cancer Res. 2018;15(4):405–417. doi: 10.1158/1541-7786.MCR-16-0242-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Adamik J., Sun Q., Roodman G.D., Galson D.L. Gfi1 inhibits osteoblast differentiation in multiple myeloma by inducing epigenetic repression of Runx2 in bone marrow stromal cells. J. Bone Miner. Res. 2014;29(Suppl 1):S54. [Google Scholar]
- 80.D'Souza S., del Prete D., Jin S., Sun Q., Huston A.J., Kostov F.E., Sammut B., Hong C.S., Anderson J.L., Patrene K.D., Yu S., Velu C.S., Xiao G., Grimes H.L., Roodman G.D., Galson D.L. Gfi1 expressed in bone marrow stromal cells is a novel osteoblast suppressor in patients with multiple myeloma bone disease. Blood. 2011;118(26):6871–6880. doi: 10.1182/blood-2011-04-346775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Komori T. Regulation of osteoblast differentiation by Runx2. Adv. Exp. Med. Biol. 2010;658:43–49. doi: 10.1007/978-1-4419-1050-9_5. [DOI] [PubMed] [Google Scholar]
- 82.Giuliani N., Colla S., Morandi F., Lazzaretti M., Sala R., Bonomini S., Grano M., Colucci S., Svaldi M., Rizzoli V. Myeloma cells block RUNX2/CBFA1 activity in human bone marrow osteoblast progenitors and inhibit osteoblast formation and differentiation. Blood. 2005;106(7):2472–2483. doi: 10.1182/blood-2004-12-4986. [DOI] [PubMed] [Google Scholar]
- 83.Roodman G.D. Genes associate with abnormal bone cell activity in bone metastasis. Cancer Metast. Rev. 2012;31(3–4):569–578. doi: 10.1007/s10555-012-9372-x. [DOI] [PubMed] [Google Scholar]
- 84.Wang Y.G., Qu X.H., Yang Y., Han X.G., Wang L., Qiao H., Fan Q.M., Tang T.T., Dai K.R. AMPK promotes osteogenesis and inhibits adipogenesis through AMPK-Gfi1-OPN axis. Cell Sig. 2016;28(9):1270–1282. doi: 10.1016/j.cellsig.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 85.Velinder M., Singer J., Bareyan D., Meznarich J., Tracy C.M., Fulcher J.M., McClellan D., Lucente H., Franklin S., Sharma S., Engel M.E. GFI1 functions in transcriptional control and cell fate determination require SNAG domain methylation to recruit LSD1. Biochem. J. 2017;473(19):3355–3369. doi: 10.1042/BCJ20160558. [DOI] [PubMed] [Google Scholar]
- 86.Duan Z., Zarebski A., Montoya-Durango D., Grimes H.L., Horwitz M. Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol. Cell Biol. 2005;25(23):10338–10351. doi: 10.1128/MCB.25.23.10338-10351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Adamik J., Silbermann R., Marino S., Sun Q., Anderson J.L., Zhou D., Xie X.Q., Roodman G.D., Galson D.L. XRK3F2 Inhibition of p62-ZZ domain signaling rescues myeloma-induced GFI1-driven epigenetic repression of the Runx2 gene in pre-osteoblasts to overcome differentiation suppression. Front. Endocrinol. 2018;9:344. doi: 10.3389/fendo.2018.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Teramachi J., Silbermann R., Yang P., Zhao W., Mohammad K.S., Guo J., Anderson J.L., Zhou D., Feng R., Myint K.Z., Maertz N., Beumer J.H., Eiseman J.L., Windle J.J., Xie X.Q., Roodman G.D., Kurihara N. Blocking the ZZ domain of sequestosome1/p62 suppresses myeloma growth and osteoclast formation in vitro and induces dramatic bone formation in myeloma-bearing bones in vivo. Leukemia. 2016;30(2):390–398. doi: 10.1038/leu.2015.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Alzrigat M., Parraga A.A., Jernberg-Wiklund H. Epigenetics in multiple myeloma: from mechanisms to therapy. Semin. Cancer Biol. 2017;51:101–115. doi: 10.1016/j.semcancer.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 90.Armstrong L. Epigenetic control of embryonic stem cell differentiation. Stem Cell Rev. 2012;8(1):67–77. doi: 10.1007/s12015-011-9300-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.