Abstract
Voltage-gated sodium channels undergo transitions between open, closed, and inactivated states, enabling regulation of the translocation of sodium ions across membranes. A recently published crystal structure of the full-length prokaryotic NavMs crystal structure in the activated open conformation has revealed the presence of a novel motif consisting of an extensive network of salt bridges involving residues in the voltage sensor, S4-S5 linker, pore, and C-terminal domains. This motif has been proposed to be responsible for maintaining an open conformation that enables ion translocation through the channel. In this study, we have used long-time molecular dynamics calculations without artificial restraints to demonstrate that the interaction network of full-length NavMs indeed prevents a rapid collapse and closure of the gate, in marked difference to earlier studies of the pore-only construct in which the gate had to be restrained to remain open. Interestingly, a frequently discussed “hydrophobic gating” mechanism at nanoscopic level is also observed in our simulations, in which the discontinuous water wire close to the gate region leads to an energetic barrier for ion conduction. In addition, we demonstrate the effects of in silico mutations of several of the key residues in the motif on the open channel’s stability and functioning, correlating them with existing functional studies on this channel and homologous disease-associated mutations in human sodium channels; we also examine the effects of truncating/removing the voltage sensor and C-terminal domains in maintaining an open gate.
Introduction
The opening of voltage-gated sodium channels (Navs) initiates the action potential in excitable cells. This is characterized by selective, rapid, and transient ion conduction, accompanied by a gating cycle involving conformational channels between open, closed, and inactivated states. In humans, mutations in different sodium-channel isoforms can cause a wide range of diseases, including epilepsy, cardiac arrhythmia, and chronic pain syndrome; hence, these channels have been the subject of extensive structural and functional studies and are key targets for drug development (1). The structure of a eukaryotic sodium channel from cockroach (determined by cryo electron-microscopy at relatively low resolution) has been determined, but it has not been shown to be functional (2). However, prokaryotic sodium channels have been shown to provide good structural and functional models for human sodium channels. They have high sequence identities to human Navs (25–30%) and common folds consisting of four regulatory voltage sensor domains surrounding an eight-helix pore bundle, which is responsible for ion translocation (3). Functionally, a number of these prokaryotic channels have been shown to exhibit similar ion flux characteristics and selectivities (3, 4, 5, 6, 7) and, in one case, very closely related sensitivity to human sodium-channel-blocking compounds (8). Different crystal structures of prokaryotic sodium channels exhibit features associated with different functional states—including activated and preactivated voltage sensors, and open and closed pore gates (5, 6, 8, 9, 10, 11, 12, 13, 14, 15). In addition, the sodium ion locations within the selectivity filter have been defined crystallographically in one of these prokaryotic channels, NavMs (16). An important difference between eukaryotic Navs (which are monomeric four-domain structures) and prokaryotic Navs (which are tetramers with each monomer corresponding to a domain of a eukaryotic Nav) is that prokaryotic Navs also possess a C-terminal domain (CTD) that extends from the end of the S6 pore helix into the cytoplasmic domain. This CTD consists of a membrane-proximal negatively charged region termed the “neck” region and a “coiled-coil” region at the end of the polypeptide chain and apparently plays a role in stabilizing the tetramers (17, 18).
Until recently, nearly all Nav crystal structures have exhibited closed conformations of the pore gate (2, 5, 10, 11, 13, 15) and hence did not represent ion-conducting forms of the channels. The NavMs pore-only construct was the first structure that exhibited an open gate (6, 9) and hence was the subject of molecular dynamics (MD) studies investigating the process of ion translocation in an open channel (7, 19). However, because that structure included neither the voltage sensor nor a visible C-terminal domain, under simulation conditions the gate tended to close unless specifically constrained, suggesting that one or both of the missing structural elements in the crystal structure might play an important role in the gating process. An “open” prokaryotic Nav structure (15)—that of the NavAb ortholog which includes a voltage sensor (VS) but in which the CTD has been deleted—has been shown to have a very similar structure to the earlier NavMs open pore structures (6, 9). There have been numerous MD studies in recent years, most of which have utilized closed forms of the pore (20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33). In addition, MD simulations were extensively adopted in addressing small-molecule channel interactions (34, 35, 36, 37, 38).
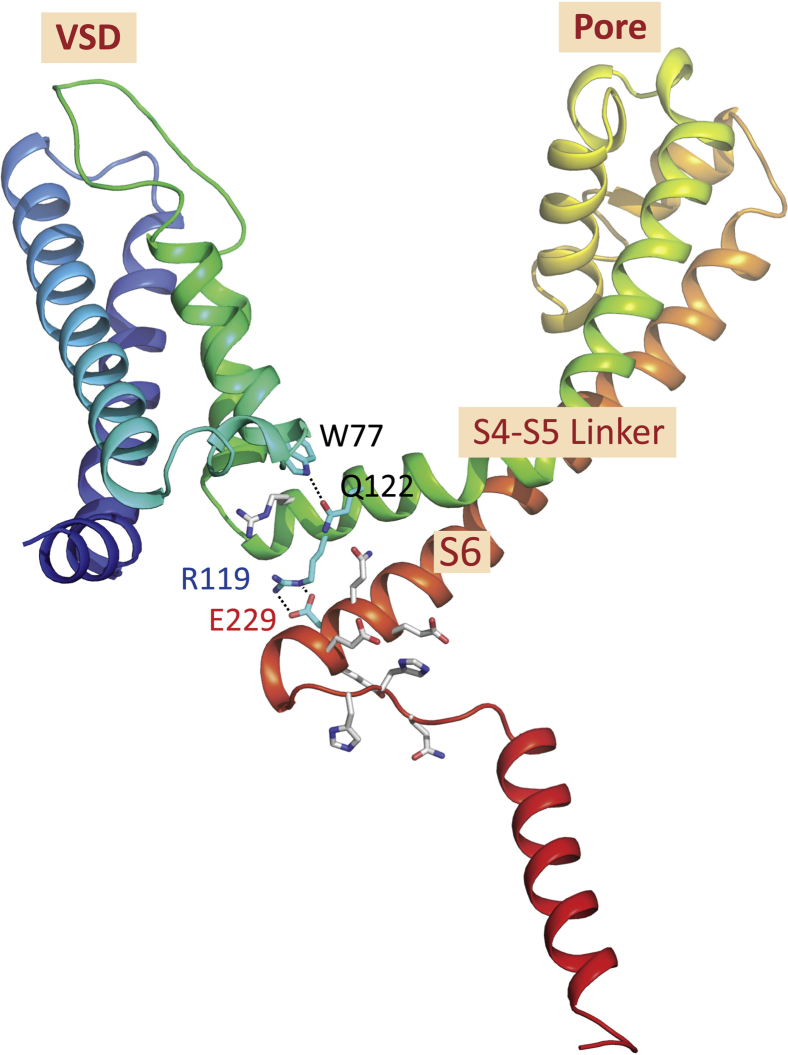
Recently, the first full-length sodium-channel structure (NavMs) has been determined, which not only exhibits an open pore but includes both an activated voltage sensor, a selectivity filter with sodium ions visible, an open pore gate, and a visible C-terminus (14). This structure reveals a novel “interaction” motif between the domains—an extensive network of hydrogen bonds and ion pairings, which appear to stabilize and be responsible for its open gate. This motif includes two regions that are conserved across prokaryotic and eukaryotic sodium channels: the W77 of the voltage-sensor S3 helix, which forms interactions with Q122 in the S4-S5 linker helix, and E229 near the end of the S6 helix, which forms a hydrogen-bonded interaction with R119, also in the S4-S5 linker. As the VS is in the activated conformation, this structure provides the first view of an activated open channel and the opportunity to utilize computational methods to investigate the translocation of sodium ions through a complete channel. In this study, equilibrium MD simulations have elucidated key interplays between the S4-S5 linker and hydrogen-bonded residues in the S6 and S3 helixes, the VS, and the CTD, shedding light on their mechanistic roles during channel gating.
Methods
MD simulations
The symmetric homotetrameric full-length open-conformation crystal structure of NavMs from Magnetococcus sp. (strain MC-1) (14) (Protein Data Bank (PDB): 5HVX) was used as the starting model in these simulations. The full-length model included residues 1–262, encompassing transmembrane helices S1–S6 and the C-terminal domain. Crystallographically disordered residues missing at the extramembrane end of helix 4 (92–99) were reconstructed using MODELER 9.15 (39) (using MODELER’s automodel module), and partial side-chain information for residues 6, 12, 74, 100, 233, 250, 254, 256, 257, 258, 261, and 262 was restored by SWISSPDB viewer automatically using a library of allowed rotamers taken from a database of structures in the PDB (40). All charged residues were treated to keep their charge states at physiological pH 7.4. The same protocol was adopted for the noVSD construct (consisting of residues 132–262 included in the structure, with terminal residues neutrally capped) and the noCTD construct (which included residues 1–222, with terminal residues neutrally capped). The protein was inserted into a pre-equilibrated membrane composed of 374 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine lipids using the g_membed method (41) in the presence of 0.5 M NaCl salt and equilibrated for 10 ns. MD simulations were performed using the supercomputers in Shanghai Jiao Tong University (Pi) and The Maryland Advanced Research Computing Center with CHARMM27 protein (42), CHARMM36 lipid force fields (43), Amber99sb-ildn force field (44), and TIP3P water models (45). Simulations were generated by Gromacs version 5.0.5 (46). All covalent bonds were constrained using the LINCS algorithm (47), allowing for an integration time step of 2 fs when updating potential energy function during simulation. A 10-Å cutoff was adopted for calculating short-range electrostatic interactions and Van der Waals interactions; particle mesh Ewald summation (48) was used for calculating long-range electrostatic interactions. Protein, hydrated ions, and lipids were each coupled separately to a heat bath with a time constant of τT = 0.1 ps using weak temperature coupling (49). An atmospheric pressure of one bar was maintained using weak semi-isotropic pressure coupling with compressibility κz = κxy = 4.6 × 10−5 bar–1 and time constant τP = 1 ps (50). Structures were displayed using VMD software (51) and demonstrations of the accessible area in the pore were done using the program HOLE (52). The simulation data were analyzed and presented by Python-based analyzing tools MDTraj (53) and Matplotlib (54).
Results
Factors involved in the stability of the open pore domain
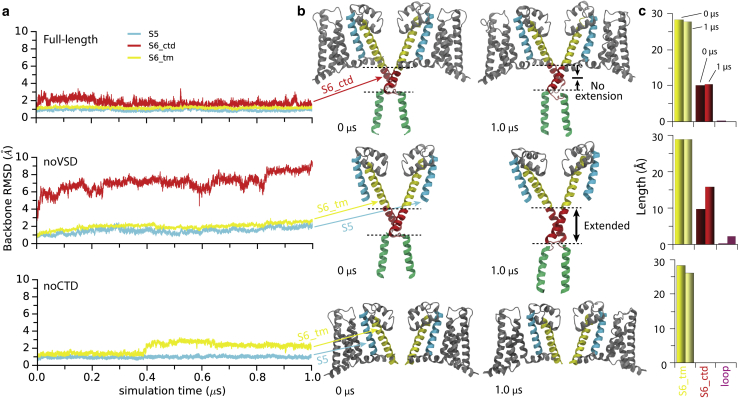
The new, full-length open NavMs structure (PDB: 5HVX) (14) has been characterized in the present MD simulation study, examining its ability to maintain an open gate without artificial constraints being required. Of particular interest was the role of the interaction motif in maintaining an open gate: when this motif was present in the structure, the gate remained open, but when it was subjected to a number of modifications, the gate closed during the simulations, an important demonstration of its essential role in gating. To examine the roles of different segments of the novel interaction motif in stabilizing the gate in the open state, three separate 1-μs simulations were performed: 1) with the full-length open NavMs structure, 2) the structure without the voltage sensor domain (noVSD), and 3) the structure without the C-terminal domain (noCTD).
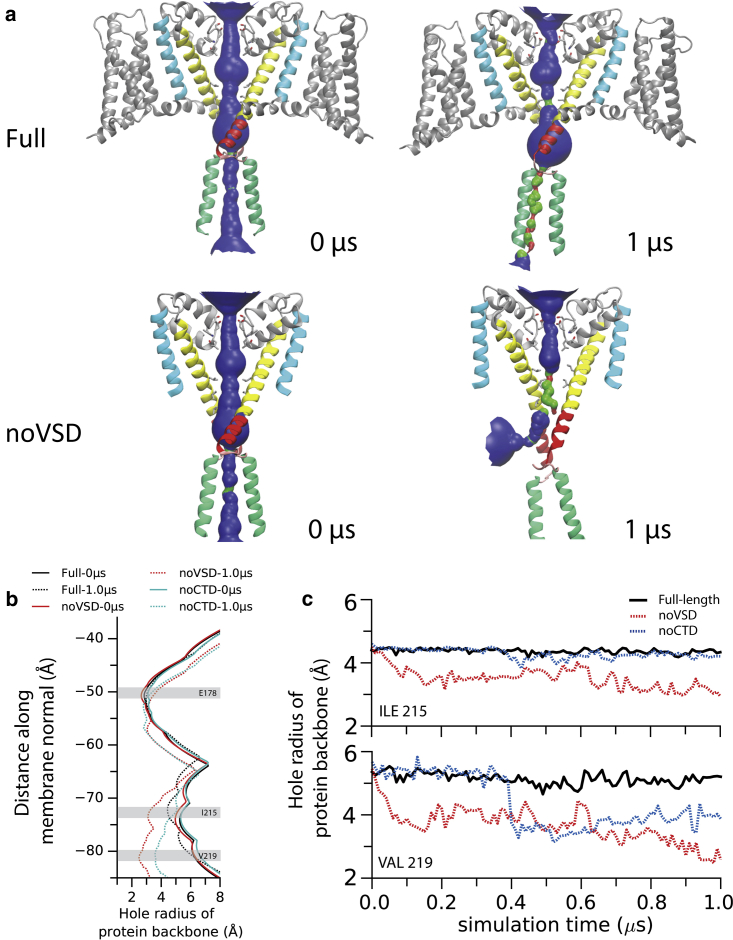
In the simulation of the full-length wild-type structure, the pore structure remained stable with an RMSD (root mean square distance) below 3 Å over the entire simulation (Fig. 1 a, top). The trajectory shows that the RMSDs plateau early in the simulation and then slightly decrease, demonstrating that convergence has been achieved. The interaction motif structure remained intact and stable during the full simulation. In contrast, when the VS and S4-S5 linker were removed (noVSD), making a construct that was essentially pore- only with a CTD (similar to the structure in the previous simulation (7) of the NavMs structure of the open pore (6), except with the CTD present), the lower part of the S6 helix (Fig. 1 a, middle, red) deviated markedly from its original conformation during the simulation. The removal of the VSD resulted in the CTD neck region becoming mobile and unpinned from the gate region. These structural changes led to the extension of the length of the S6 helix along the membrane normal (Fig. 1 b, middle). Additionally, the transmembrane part of S6 (Fig. 1 a, bottom, yellow) in the noCTD simulation (reminiscent of previous resolved transmembrane-only structures (9, 10, 11)) revealed a larger destabilizing shift when compared to full-length simulation (Fig. 1 a, top, yellow).
Figure 1.
Comparison of the simulations of the open full-length NavMs structure (PDB: 5HVX) (14) (top), the structure without the voltage sensor (noVSD, middle), and the structure without the C-terminal domain (noCTD, bottom). (a) RMSD comparisons of the trajectories of different segments in the pore domain. The plots are color-coded to match the portions of the structures shown in (b). (b) Snapshots of structures of the full-length (top), noVSD (middle), and noCTD (bottom) constructs at the beginning (0 μs) (left) and end (1 μs) (right) of the simulations with the different regions colored as follows: cyan, S5; yellow, the transmembrane part of S6; red, the C-terminal part of S6; pink, the loop between the C-terminal part of S6 and the coiled-coil; green, coiled-coil. Only two opposite subunits of the tetramer are shown in each case for clarity. (c) Comparisons of the lengths of different segments of S6 at the beginning and end of the simulations of the three different constructs. To see this figure in color, go online.
Key interactions of the full-length structures remain stable during the simulations
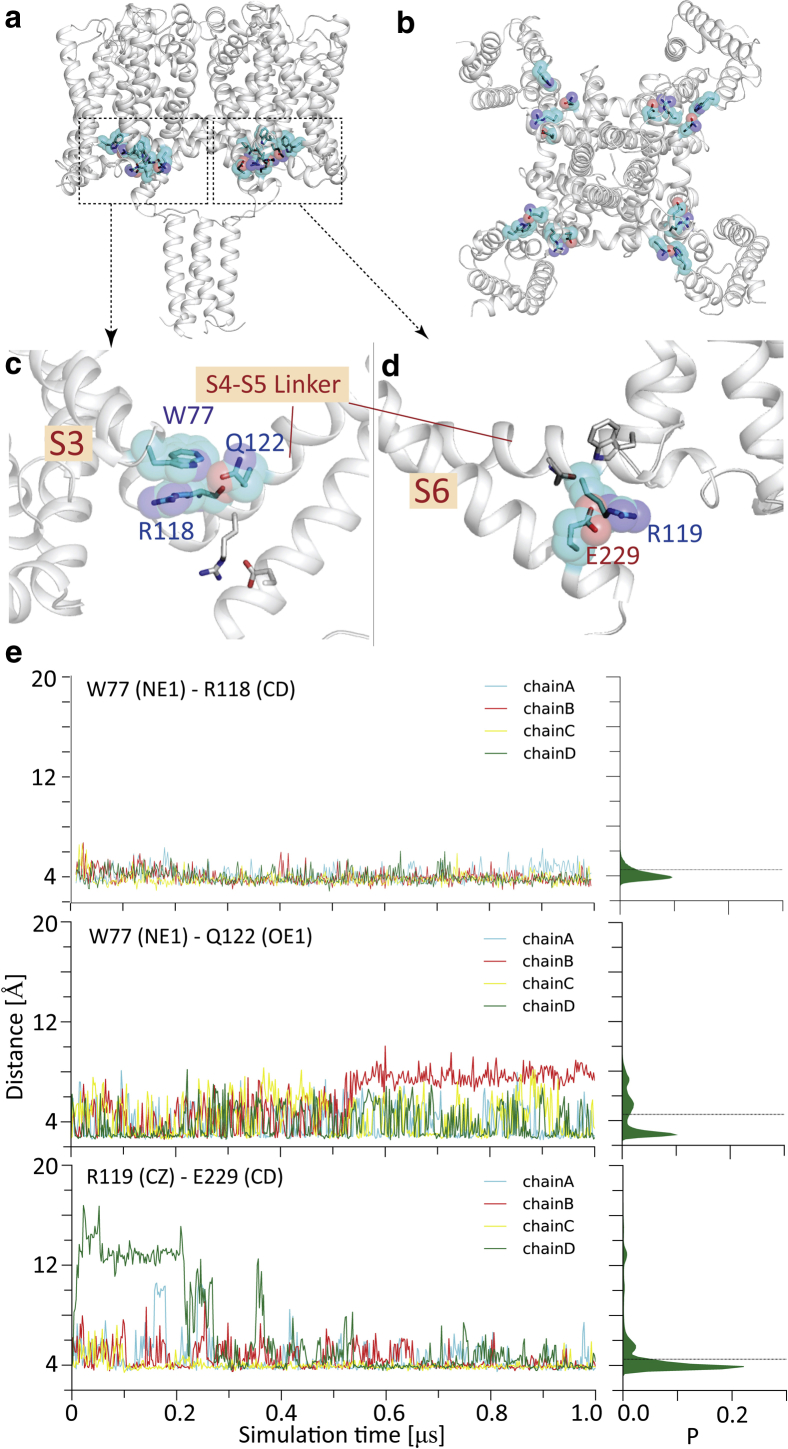
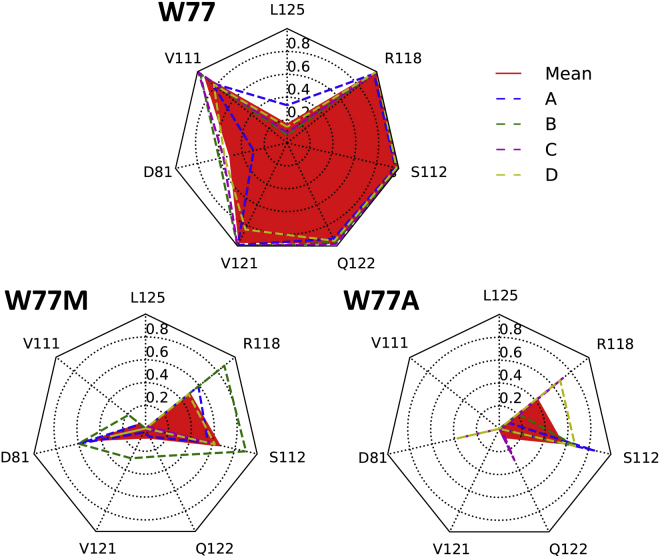
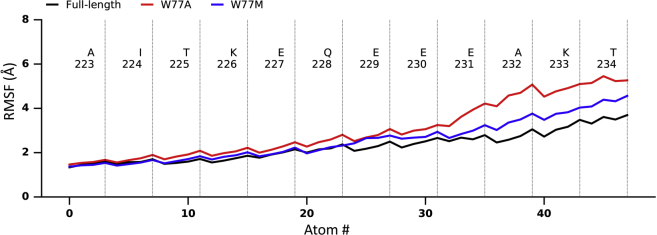
Several key interactions appear to be responsible for the differences in stability encountered (Fig. 2) between the full-length structure and the modified structures. Firstly, the conformational stability of the network of interactions involving the conserved W77 residue (present in the sequences of all prokaryotic and human sodium channels) was investigated. The important role of this residue in channel gating has been identified (14), and its potential role in human channels has been postulated based on single-site mutations of the residue that lead to disease states in humans (55). To aid in our understanding of the nature of its involvement in the dynamics of the system has been a major focus of this study. In the simulation of the full-length wild-type structure, in which all the components of the novel interaction motif exist, W77 is sandwiched by R118 (hydrophobic packing) and Q122 (H-bond interaction) (Fig. 3 top and center); as a result, all W77 side chains from four subunits remained largely stable. Selected substitutions (W77A and W77M) at position 77, which abolished channel conductance as reported by Sula et al. (14), were simulated for 1.8 μs under 200 mV membrane potential. The higher driving force enhanced the sampling and aided in distinguishing mutants from wild-type channels. These mutants, which bear smaller side chains, led to less intense interactions between S3 and the S4-S5 linker (i.e., Q122 and R118 in Fig. 4); these impaired interactions further destabilized the S6_ctd, as shown in Fig. 5, in which all gating residues started to become mobile in mutant simulations. Nonetheless, mutants at position 77 only had mild effects on other segments of the transmembrane part of the channel.
Figure 2.
The key residues of the novel interaction motif (after (14)). The intrasubunit hydrogen bond between W77 and Q122 and the salt bridge between R119 and E229 are depicted. To see this figure in color, go online.
Figure 3.
Formation and breaking of key residue-residue contacts during the simulation of the full-length construct. (a) A side view and (b) a top view of the structure. (c) An enlarged region showing the intrasubunit hydrogen bond between W77 and Q122, with cyan sticks and spheres shown for the side chains; the other salt bridge, R119-E229, is shown as white sticks. (d) The salt bridge between R119 and E229 is depicted (cyan sticks and spheres are shown for the side chains, and the other H-bond, W77-Q122, is shown with white sticks). (e) (top) Distances of the W77 and R118 side chains in each of the four different subunits during the course of the simulation; (center) lengths of the W77-Q122 H-bond in each of the four different subunits during the course of the simulation and (bottom) lengths of the E119-R229 salt bridge of four different subunits over the course of the simulation. The distribution (over all four chains) is shown on the right. To see this figure in color, go online.
Figure 4.
Contact maps for residue 77 from 1.8 μs simulations between full-length wild-type, W77A, and W77M simulations. All residues in contact with W77 in more than 5% of the simulation time are shown in a radar chart. Each vertex represents one interacting residue; the radial value is the probability (calculated as number of contacts divided by total frame numbers) of this residue to be in contact with residue 77 over 1.8 μs simulation time (every 100 ps were saved for analysis). Dotted lines represent individual monomers; red line and fill represent the mean over four monomers. To see this figure in color, go online.
Figure 5.
Comparison of S6_ctd segments’ backbone fluctuations between full-length wild-type, W77A, and W77M simulations in 1.8 μs simulations. This plot summarizes the averaged backbone root mean square fluctuation plots of each residue in the S6-ctd segment. Compared to full-length simulation (black), the lower part of the S6_ctd segment became less stable in both W77A (red) and W77M (blue) simulations. To see this figure in color, go online.
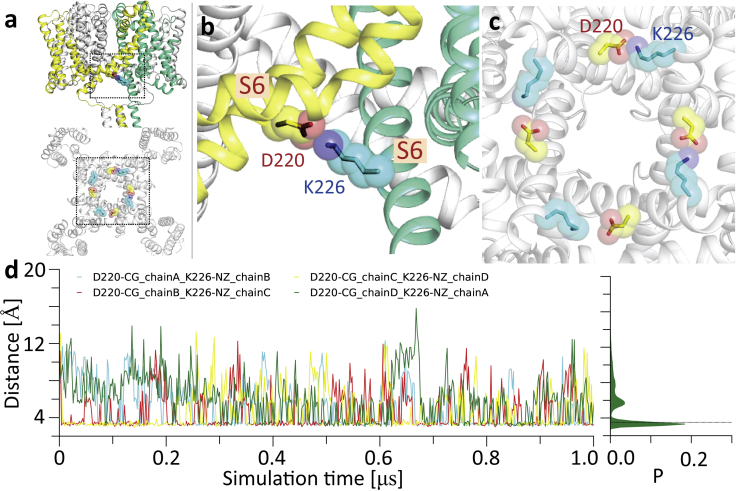
The second motif that was examined was that involving the EEE sequence of the extended S6 helix (neck region of the CTD). This is a sequence that had previously been identified by electrophysiology as having a potential functional role when all of the residues were changed to QQQ in studies of the pore-only structure (6). More recent sequence comparisons (55) suggested that the E229 of this motif might be the responsible residue because it formed a strong salt bridge with the conserved R119 in the linker. In the simulations in this study, it stayed predominantly intact in each of the four subunits (Fig. 3). This residue (but not the other Es) has also been recently identified as being mutated in several human mutations (55). The final site that was identified in these simulations was an intersubunit salt bridge formed between D220 and K226 of the adjacent subunit (Fig. 6). These interactions (which were not previously identified in the structure) also appear to be stable and may aid in enabling the pore gate to remain continuously open.
Figure 6.
The intersubunit salt bridge between D220 and K226. (a–c) A view of the intersubunit hydrogen bond between D220 (yellow sticks and spheres) and K226 (cyan sticks and spheres) in the adjacent subunit. (d) Distances of the four D220-K226 salt bridges over the course of the simulation, with a histogram over all four pairs on the right. To see this figure in color, go online.
Features of the gate in simulations of the different constructs
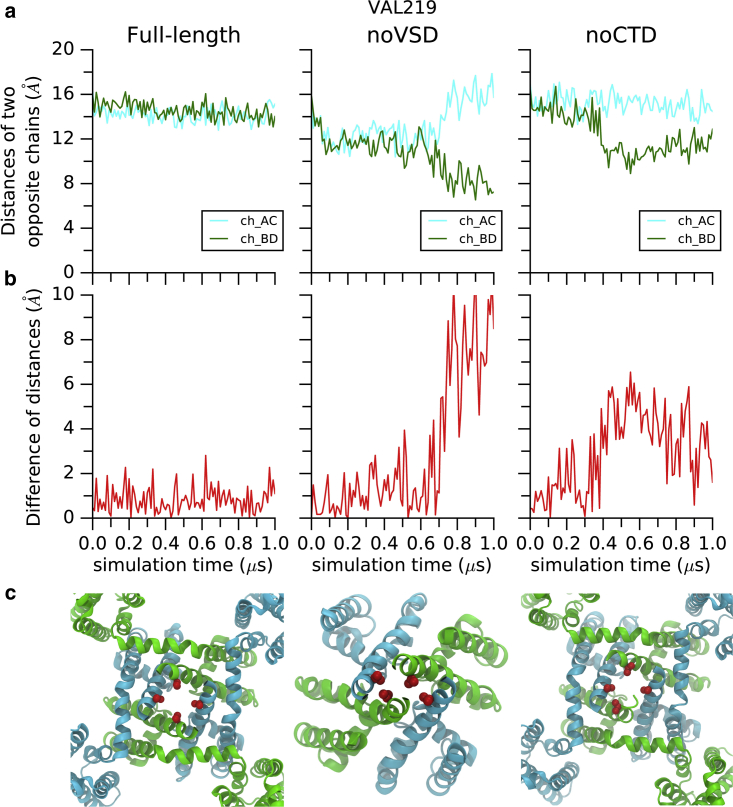
The full-length structure (but not the mutant or truncated structures) retained a fully open gate throughout the simulations: the backbone radius of pore-lining residues was almost unchanged over the 1-μs simulation time (Fig. 7, a–c, black), except for a small deviation at a series of hydrophobic residues (I215 and V219). However, when the novel interaction motif was not intact (by removing either the VSD or the CTD), the gate collapsed during the simulation (Fig. 7, red and blue). In the noVSD simulation, the removal of the VSD initiated a rapid narrowing of the pore-lining radius (I215 and V219 (Fig. 7, b and c, red)), which resulted from equivalent residues from two opposite subunits moving toward each other while the two orthogonal residues departed from each other. As a result, the dihedral angles of F214 of two closing subunits were shifted (Fig. S1). At the end of the 1-μs simulation, the gating residues encompassed an even smaller cylinder when compared to the selectivity filter (SF), indicating a nonconductive collapsed gate. This configurational change is facilitated by the formation of new intersubunit salt bridges in the neck region (E227 interacts with K226 and K233 from the subsequent subunit) (Fig. S2). Instead of a fourfold symmetry when the gate is open, this collapsed conformation was more likely to be rendered in a twofold configuration (Fig. 8, middle). Similarly, unlike the full-length simulation, the pore radius at the position of V219 was also largely reduced at the end of the 1-μs noCTD simulation, resulting in a collapsed gating configuration (Fig. 7 blue and Fig. 9, right). Taken together, the two negative control simulations suggest the important role of the novel interaction motif in stabilizing the pore in the open state.
Figure 7.
(a) View of the pore at the start (0 μs) and end (1 μs) of the simulations for the full and noVSD structures. The solvent-accessible volume (by side chain) of the interior of the pore is depicted by HOLE (52) calculations (color code for the radius: red, hole radius < 1.15 Å; green, hole radius < 2.30 Å; blue, hole radius > 2.30 Å). The small thinning of the pore at the hydrophobic region above the gate, leading to the hydrophobic gating, can be seen in the full structure. The collapse of the gate is visible in the noVSD simulation. (b) The interior radius of the pore for protein backbones between the beginnings and ends of the simulations. The shaded area depicts key residues in the selectivity filter (E178) and the gate region (I215 and V219). (c) A comparison of residues’ backbone radii (I215 and V219) over time between full-length, noVSD, and noCTD construct simulations. To see this figure in color, go online.
Figure 8.
Comparisons of pore gate symmetries. (a) Comparison of V219 Cα atom distances of two opposite chains between full-length, noVSD, and noCTD simulations; (b) comparison of the differences of orthogonal distances between full-length, noVSD, and noCTD simulations; and (c) bottom view snapshots after 1-μs simulations for full-length, noVSD, and noCTD constructs, respectively (only the transmembrane regions were shown for clarity). To see this figure in color, go online.
Figure 9.
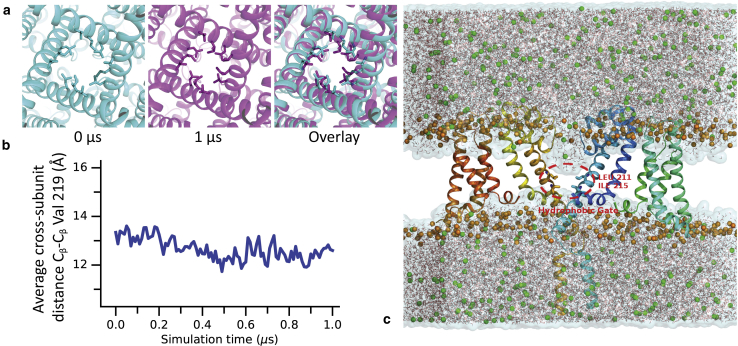
Illustration of the origin of the hydrophobic gating phenomenon at 1 μs of the full-length simulation. (a) The view down the axis of the pore. The channel slightly closes at the narrow hydrophobic region (snapshots at 0, 1 μs, and overlay) during the 1 μs simulation time. (b) This narrowing is clearly visible when plotting cross-subunit distances at this location. (c) Although enough space remains even after the narrowing, the hydrophobic side chains lead to a dewetting vaccum. Two opposite subunits are shown as a rainbow cartoon for clarity. Water surfaces are transparently shown, and water models are shown with thin lines. Orange spheres depict the phosphate atoms of lipid head groups, and green spheres depict sodium ions. To see this figure in color, go online.
Experimentally consistent conduction values in simulations of an L211A and I215A double mutant
Using previous channel constructs in which not all key domains were resolved (9, 10), it was difficult to capture stable conducting events by MD simulations without any harmonic restraints (7). Here, we attempted to investigate this mechanism in the current full-length open structure (14) using equilibrium simulations (Table S1). In all of the full-length simulations without restraints, the channel gate opening was stable, as described above, and sodium ions were preferably bound to each of the binding sites previously identified inside the selectivity filter using MD (7), which correspond well with the experimentally determined sites (6, 16).
However, the conducting sodium ion passage was occluded by a dewetting vacuum at the open gate (Fig. 9). Closer inspection revealed that the origin of this dewetting is a tiny conformational relaxation from the crystal coordinates over the 1-μs time scale. This leads to a reduction of the volume available for water molecules at this position (Figs. 7 and 9). Although the gate remains wide enough to accommodate water molecules, the water molecules in the simulation nevertheless avoided this hydrophobic region because of its hydrophobicity. This “hydrophobic gating” has previously been observed in many other ion-channel and transporter simulations, as reviewed by Aryal et al. (56). The hydrophobic gate of these membrane proteins tends to be dewetted or nonconductive even when the diameter of the gate is more than three times that of a typical water model (9 vs. 3 Å). This area was only hydrated when two long-chain hydrophobic pore-lining residues (I215 and L211) side chains were replaced by shorter chain alanines; this simulation then gave an experimentally consistent conductance rate (Fig. 10) as we observed previously in the simulation of the pore-only construct (7). Again, the mutant simulations used no restraints of any type, and the gate did not collapse and remained open. Other simulations of truncated closed pores did not observe such an effect because they did not include these residues that are at the exit of the pore.
Figure 10.
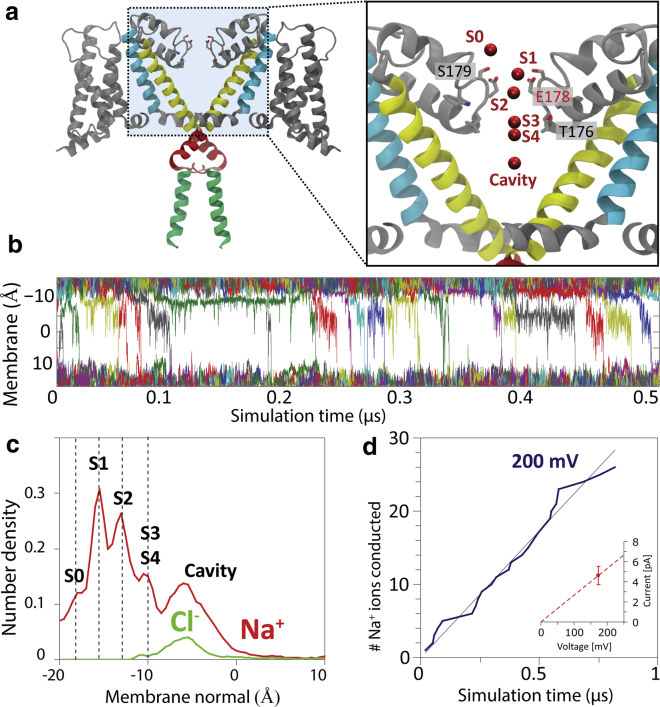
Conductance of sodium ions through the channel at a membrane voltage of 200 mV. (a) An overview of the key ion binding sites in the selectivity filter, as described previously (7). (b) Individual positions of all permeating Na+ (varying colors) along the membrane normal (x axis) versus simulation times, showing the ions hopping between the binding sites S0 and S4 in the SF and in the cavity. (c) The multi-ion number density histogram over the entire simulation. Because the simulations of the whole channel are not restrained, the histogram is less sharp than in our previous studies in which NavMs was restrained, because of channel thermal motion along the membrane normal smearing out the distribution. Binding locations are shifted up by 5 Å, as the full-size channel sits higher in the membrane than the truncated channel simulated previously. Sodium ions (red) also loiter in the cavity, where they are joined by chloride anions diffusing in through the open gate (green). (d) The overall conductance of ions through the unrestrained full-length channel is calculated as 24 pS, close to the previously determined value 34 pS for the pore-only model (7). To see this figure in color, go online.
Discussion
This MD simulation study of the recently published crystal structure of the full-length prokaryotic NavMs crystal structure in the activated open conformation (14) has revealed the critical importance of the extensive network of salt bridges involving residues in the voltage sensor, S4-S5 linker, pore, and C-terminal domains first revealed in that structure. Because these domains were absent in earlier crystal structures, the main focus of prior MD simulations was the role of the selectivity filter and its mode of action in distinguishing between different cations. Instead, the focus here is on the structurally much more complex gate region and its link with the VSD. The simulations have revealed that numerous residue pairs are critical: the interactions of W77 and R118 and Q122, R119 and E229, and D220 and K226 on the adjacent subunit. Our MD studies have shown that these residue contacts are important for maintaining an open conformation. This is clearly demonstrated if these interactions are removed: omitting or truncating domains results in rapid loss of structure at the gate region, which leads to collapse and closure of the gate.
Of particular interest is the hydrophobic dewetting observed in our simulations. This “hydrophobic gating” was first observed in simulations of model nanopores, which exhibit liquid-vapor oscillations below a critical radius, switching stochastically between wet and dry states (57, 58, 59). These liquid-vapor phase transitions are highly sensitive to the chemical environment, temperature, and pressure and involve a large change in heat capacity. Tiny changes in radius and/or polarity can drastically alter the hydration and thus the energetic barrier not only to water but also to ions. Since those early model studies, hydrophobic gating has been reported in a plethora of other MD simulations with a range of different ion channels (56, 60, 61, 62, 63, 64). This intriguing observation raises the question of how physiological stimuli may affect the conformations of the gate and subsequently how the structural dynamics can influence the mechanism of the channel gate. The consensus MD simulations data allow us to speculate that a subtle hydrophobic gating mechanism may also exist here.
Although we observe hydrophobic dewetting, it is unclear if this a genuine effect for sodium channels. Another possibility is that this is a convergence issue. Because of the system being very large, we could only achieve timescales of 1–1.8 μs. The small conformational deviation from the crystal coordinates at the hydrophobic region (Fig. 9) that causes the dewetting could well be a long-time equilibration issue that is dissolved on timescales of 10–100 μs. All simulations of membrane protein crystal structures placed in artificial membranes show an initial rapid RMSD rise with respect to the starting conformation. Some of these changes occur because of improper lateral pressure by choosing too few or too many lipid molecules in the two respective leaflets, exerting dissimilar lateral pressure on the pore domain that would not occur otherwise. When harmonic restraints to the crystal coordinates are employed, no dewetting is observed, and the channel retains its continuous water wire and conduction properties (Fig. 7, HOLE plot a). However, the main goal of our work was to not bias results via use of any restraints, so we discuss only unrestrained simulation results.
In addition, side-chain positioning could be significant: if the hydrophobic side chains of these residues are experimentally predicted to reach too much into the pore, a subtle rearrangement could render this location less hydrophobic. For example, the rotation of a single side chain promotes hydration of the central pore in TRPV1, in which hydrophobic gating is directly observed (65).
Another possibility is that the hydrophobic dewetting is caused by deficiencies in the simulation model. To check the influence of force fields, we repeated the simulations using two different protein force fields (Table S1), but the dewetting was also observed for these. As MD timescales have become much longer, there is now a large amount of new evidence that protein-protein interactions could in general be modeled as too sticky and protein-water interactions underestimated. For example, Petrov and Zagrovic have reported on the difficulties of current force fields in reproducing the precise hydration properties in a hydrophobic environment (66). We are currently investigating the hydrophobic gating using a much larger combination of protein and water force fields. The chief difficulty is that the phenomena we observe only occur during very long MD simulations in the multi-μs range. Thus, these conformational changes cannot reliably be studied by alternative short nonequilibrium potential-of-mean-force calculations because the phenomenon would not be observable. Further functional, structural, and computational approaches will be collectively required to address these issues.
The simulations presented here can be considered a first glimpse, to our knowledge, into the role of the observed interaction networks. In earlier MD simulations of ions in or near the SF, only side-chain movements of the SF-lining residues needed to be sampled, allowing for converged statistics in ∼500 ns. In contrast, the study of the interaction network at the gate and linker regions involves timescales on the order of multi-μs, with long-lived residue-residue contacts, salt bridges. and hydrogen bonds triggering gate movements and conformational transitions. Our simulations convincingly show that the full system, the pore domain, linker, VSDs, and the C-terminal domain need to be simulated to faithfully represent this interaction network. This results in system sizes of ∼200,000 atoms, making multi-μs simulations challenging. The simulations here took a substantial time to compute and are thus limited to 1–1.8 μs. The ultimate goal is to capture the long-scale transitions in these networks during gating, which will require timescales of 100 μs or more.
Regarding mutant simulations, we hypothesize that W77 is stabilized by position 118 and acts like a stationary hydrogen bond donor with different residues in the linker, i.e., Q122 during gating; therefore, the W77-Q122 interaction may not be very specific because S4-S5 is largely flexible during gating. Although Q122 and R118 may not be very well conserved across different species, similar aliphatic side chains were found in equivalent positions in both eukaryotic and prokaryotic analogs (2, 5, 6, 9, 10, 11, 12, 13, 14), which on the one side stabilize W77 and on the other provide the flexible sliding/screwing interactions mechanisms during gating. Replacement of W77 resulted in a significant altering of the contact interactions of that residue. Here, also, the 1.8 μs timescale revealed only the initial loss of contacts, with much longer simulations needed to show the more substantial conformational changes such mutations entail. With the key interactions identified here, future simulations are underway, including the EEE-QQQ mutants, which will greatly complement and enhance structural and functional studies that contribute to our understanding of the gating sodium channels.
Author Contributions
B.A.W., M.B.U., and J.P.U. designed the research. S.K. performed the simulations. S.K. and J.P.U. performed MD analyses. J.P.U. developed software and analyzed the data. S.K., J.P.U., and B.A.W. wrote the manuscript with input from the other authors.
Acknowledgments
This research was supported by a China 1000 Plan's Program for Young Talents (#13Z127060001), by the Interdisciplinary Program of Shanghai Jiao Tong University (No. YG2016MS82), and a Shanghai Jiao Tong University incentive grant (No. AF4190002), to J.P.U. Simulation resources were supported by Center for High Performance Computing, Shanghai Jiao Tong University and the Maryland Advanced Research Computing Center supercomputer facility at Johns Hopkins University. It was also supported by Biotechnology and Biological Sciences Research Council project grants L006790 and R001294 and a Biotechnology and Biological Sciences Research Council UK-USA partnering grant to B.A.W.
Editor: Chris Chipot.
Footnotes
Two figures and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)31113-5.
Contributor Information
B.A. Wallace, Email: b.wallace@mail.cryst.bbk.ac.uk.
Jakob P. Ulmschneider, Email: jakob@sjtu.edu.cn.
Supporting Material
References
- 1.Andavan G.S., Lemmens-Gruber R. Voltage-gated sodium channels: mutations, channelopathies and targets. Curr. Med. Chem. 2011;18:377–397. doi: 10.2174/092986711794839133. [DOI] [PubMed] [Google Scholar]
- 2.Shen H., Zhou Q., Yan N. Structure of a eukaryotic voltage-gated sodium channel at near-atomic resolution. Science. 2017;355:eaal4326. doi: 10.1126/science.aal4326. [DOI] [PubMed] [Google Scholar]
- 3.Ren D., Navarro B., Clapham D.E. A prokaryotic voltage-gated sodium channel. Science. 2001;294:2372–2375. doi: 10.1126/science.1065635. [DOI] [PubMed] [Google Scholar]
- 4.Yue L., Navarro B., Clapham D.E. The cation selectivity filter of the bacterial sodium channel, NaChBac. J. Gen. Physiol. 2002;120:845–853. doi: 10.1085/jgp.20028699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Payandeh J., Scheuer T., Catterall W.A. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bagnéris C., Decaen P.G., Wallace B.A. Role of the C-terminal domain in the structure and function of tetrameric sodium channels. Nat. Commun. 2013;4:2465. doi: 10.1038/ncomms3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ulmschneider M.B., Bagnéris C., Wallace B.A. Molecular dynamics of ion transport through the open conformation of a bacterial voltage-gated sodium channel. Proc. Natl. Acad. Sci. USA. 2013;110:6364–6369. doi: 10.1073/pnas.1214667110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bagnéris C., DeCaen P.G., Wallace B.A. Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism. Proc. Natl. Acad. Sci. USA. 2014;111:8428–8433. doi: 10.1073/pnas.1406855111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCusker E.C., Bagnéris C., Wallace B.A. Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nat. Commun. 2012;3:1102. doi: 10.1038/ncomms2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Payandeh J., Gamal El-Din T.M., Catterall W.A. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature. 2012;486:135–139. doi: 10.1038/nature11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X., Ren W., Yan N. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 2012;486:130–134. doi: 10.1038/nature11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaya D., Findeisen F., Minor D.L., Jr. Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels. J. Mol. Biol. 2014;426:467–483. doi: 10.1016/j.jmb.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arrigoni C., Rohaim A., Minor D.L., Jr. Unfolding of a temperature-sensitive domain controls voltage-gated channel activation. Cell. 2016;164:922–936. doi: 10.1016/j.cell.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sula A., Booker J., Wallace B.A. The complete structure of an activated open sodium channel. Nat. Commun. 2017;8:14205. doi: 10.1038/ncomms14205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lenaeus M.J., Gamal El-Din T.M., Catterall W.A. Structures of closed and open states of a voltage-gated sodium channel. Proc. Natl. Acad. Sci. USA. 2017;114:E3051–E3060. doi: 10.1073/pnas.1700761114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naylor C.E., Bagnéris C., Wallace B.A. Molecular basis of ion permeability in a voltage-gated sodium channel. EMBO J. 2016;35:820–830. doi: 10.15252/embj.201593285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Powl A.M., O’Reilly A.O., Wallace B.A. Synchrotron radiation circular dichroism spectroscopy-defined structure of the C-terminal domain of NaChBac and its role in channel assembly. Proc. Natl. Acad. Sci. USA. 2010;107:14064–14069. doi: 10.1073/pnas.1001793107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mio K., Mio M., Sato C. The C-terminal coiled-coil of the bacterial voltage-gated sodium channel NaChBac is not essential for tetramer formation, but stabilizes subunit-to-subunit interactions. Prog. Biophys. Mol. Biol. 2010;103:111–121. doi: 10.1016/j.pbiomolbio.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Ke S., Timin E.N., Stary-Weinzinger A. Different inward and outward conduction mechanisms in NavMs suggested by molecular dynamics simulations. PLoS Comput. Biol. 2014;10:e1003746. doi: 10.1371/journal.pcbi.1003746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corry B., Thomas M. Mechanism of ion permeation and selectivity in a voltage gated sodium channel. J. Am. Chem. Soc. 2012;134:1840–1846. doi: 10.1021/ja210020h. [DOI] [PubMed] [Google Scholar]
- 21.Amaral C., Carnevale V., Treptow W. Exploring conformational states of the bacterial voltage-gated sodium channel NavAb via molecular dynamics simulations. Proc. Natl. Acad. Sci. USA. 2012;109:21336–21341. doi: 10.1073/pnas.1218087109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furini S., Domene C. On conduction in a bacterial sodium channel. PLoS Comput. Biol. 2012;8:e1002476. doi: 10.1371/journal.pcbi.1002476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chakrabarti N., Ing C., Pomès R. Catalysis of Na+ permeation in the bacterial sodium channel Na(V)Ab. Proc. Natl. Acad. Sci. USA. 2013;110:11331–11336. doi: 10.1073/pnas.1309452110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boiteux C., Vorobyov I., Allen T.W. Ion conduction and conformational flexibility of a bacterial voltage-gated sodium channel. Proc. Natl. Acad. Sci. USA. 2014;111:3454–3459. doi: 10.1073/pnas.1320907111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furini S., Barbini P., Domene C. Effects of the protonation state of the EEEE motif of a bacterial Na(+)-channel on conduction and pore structure. Biophys. J. 2014;106:2175–2183. doi: 10.1016/j.bpj.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y., Liu H., Gong H. Lysine and the Na+/K+ selectivity in mammalian voltage-gated sodium channels. PLoS One. 2016;11:e0162413. doi: 10.1371/journal.pone.0162413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y., Gong H. Theoretical and simulation studies on voltage-gated sodium channels. Protein Cell. 2015;6:413–422. doi: 10.1007/s13238-015-0152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y., Sun R., Gong H. Molecular dynamics study of ion transport through an open model of voltage-gated sodium channel. Biochim. Biophys. Acta. 2017;1859:879–887. doi: 10.1016/j.bbamem.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 29.Ngo V., Wang Y., Farley R.A. K+ block is the mechanism of functional asymmetry in bacterial Na(v) channels. PLoS Comput. Biol. 2016;12:e1004482. doi: 10.1371/journal.pcbi.1004482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corry B. Na(+)/Ca(2+) selectivity in the bacterial voltage-gated sodium channel NavAb. PeerJ. 2013;1:e16. doi: 10.7717/peerj.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stock L., Delemotte L., Klein M.L. Conduction in a biological sodium selective channel. J. Phys. Chem. B. 2013;117:3782–3789. doi: 10.1021/jp401403b. [DOI] [PubMed] [Google Scholar]
- 32.Ing C., Pomès R. Simulation studies of ion permeation and selectivity in voltage-gated sodium channels. Curr. Top. Membr. 2016;78:215–260. doi: 10.1016/bs.ctm.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 33.Furini S., Domene C. Computational studies of transport in ion channels using metadynamics. Biochim. Biophys. Acta. 2016;1858:1733–1740. doi: 10.1016/j.bbamem.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 34.Boiteux C., Vorobyov I., Allen T.W. Local anesthetic and antiepileptic drug access and binding to a bacterial voltage-gated sodium channel. Proc. Natl. Acad. Sci. USA. 2014;111:13057–13062. doi: 10.1073/pnas.1408710111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaczmarski J.A., Corry B. Investigating the size and dynamics of voltage-gated sodium channel fenestrations. Channels (Austin) 2014;8:264–277. doi: 10.4161/chan.28136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin L.J., Corry B. Locating the route of entry and binding sites of benzocaine and phenytoin in a bacterial voltage gated sodium channel. PLoS Comput. Biol. 2014;10:e1003688. doi: 10.1371/journal.pcbi.1003688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin L.J., Chao R., Corry B. Molecular dynamics simulation of the partitioning of benzocaine and phenytoin into a lipid bilayer. Biophys. Chem. 2014;185:98–107. doi: 10.1016/j.bpc.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 38.Patel D., Mahdavi S., Kuyucak S. Computational study of binding of μ-conotoxin GIIIA to bacterial sodium channels NaVAb and NaVRh. Biochemistry. 2016;55:1929–1938. doi: 10.1021/acs.biochem.5b01324. [DOI] [PubMed] [Google Scholar]
- 39.Šali A., Potterton L., Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- 40.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 41.Wolf M.G., Hoefling M., Groenhof G. g_membed: efficient insertion of a membrane protein into an equilibrated lipid bilayer with minimal perturbation. J. Comput. Chem. 2010;31:2169–2174. doi: 10.1002/jcc.21507. [DOI] [PubMed] [Google Scholar]
- 42.MacKerell A.D., Bashford D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 43.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindorff-Larsen K., Piana S., Shaw D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 46.Abraham M.J., Murtola T., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 47.Hess B., Bekker J., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 48.Darden T., York D., Pedersen L. Particle mesh Ewald: an N log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 49.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 50.Berendsen H.J.C., Postma J.P.M., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 51.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38, 27–28.. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 52.Smart O.S., Neduvelil J.G., Sansom M.S.P. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 1996;14:354–360. doi: 10.1016/s0263-7855(97)00009-x. 376. [DOI] [PubMed] [Google Scholar]
- 53.McGibbon R.T., Beauchamp K.A., Pande V.S. MDTraj: a modern open library for the analysis of molecular dynamics trajectories. Biophys. J. 2015;109:1528–1532. doi: 10.1016/j.bpj.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Michael, D., H. John, …, G. Matt. 2016. matplotlib: matplotlib v1.5.1. https://matplotlib.org/1.5.1/.
- 55.Sula A., Wallace B.A. Interpreting the functional role of a novel interaction motif in prokaryotic sodium channels. J. Gen. Physiol. 2017;149:613–622. doi: 10.1085/jgp.201611740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aryal P., Sansom M.S., Tucker S.J. Hydrophobic gating in ion channels. J. Mol. Biol. 2015;427:121–130. doi: 10.1016/j.jmb.2014.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hummer G., Rasaiah J.C., Noworyta J.P. Water conduction through the hydrophobic channel of a carbon nanotube. Nature. 2001;414:188–190. doi: 10.1038/35102535. [DOI] [PubMed] [Google Scholar]
- 58.Beckstein O., Tai K., Sansom M.S. Not ions alone: barriers to ion permeation in nanopores and channels. J. Am. Chem. Soc. 2004;126:14694–14695. doi: 10.1021/ja045271e. [DOI] [PubMed] [Google Scholar]
- 59.Beckstein O., Sansom M.S. Liquid-vapor oscillations of water in hydrophobic nanopores. Proc. Natl. Acad. Sci. USA. 2003;100:7063–7068. doi: 10.1073/pnas.1136844100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dong H., Fiorin G., Klein M.L. Pore waters regulate ion permeation in a calcium release-activated calcium channel. Proc. Natl. Acad. Sci. USA. 2013;110:17332–17337. doi: 10.1073/pnas.1316969110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aryal P., Abd-Wahab F., Tucker S.J. A hydrophobic barrier deep within the inner pore of the TWIK-1 K2P potassium channel. Nat. Commun. 2014;5:4377. doi: 10.1038/ncomms5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu F., Hummer G. Drying transition in the hydrophobic gate of the GLIC channel blocks ion conduction. Biophys. J. 2012;103:219–227. doi: 10.1016/j.bpj.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu F., Hummer G. Pore opening and closing of a pentameric ligand-gated ion channel. Proc. Natl. Acad. Sci. USA. 2010;107:19814–19819. doi: 10.1073/pnas.1009313107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Birkner J.P., Poolman B., Koçer A. Hydrophobic gating of mechanosensitive channel of large conductance evidenced by single-subunit resolution. Proc. Natl. Acad. Sci. USA. 2012;109:12944–12949. doi: 10.1073/pnas.1205270109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kasimova M.A., Yazici A., Carnevale V. TRPV1 activation relies on hydration/dehydration of nonpolar cavities. bioRxiv. 2017 [Google Scholar]
- 66.Petrov D., Zagrovic B. Are current atomistic force fields accurate enough to study proteins in crowded environments? PLoS Comput. Biol. 2014;10:e1003638. doi: 10.1371/journal.pcbi.1003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.