Abstract
Background
Novel biological insights have led to consensus classification of medulloblastoma into 4 distinct molecular subgroups—wingless (WNT), sonic hedgehog (SHH), Group 3, and Group 4. We aimed to predict molecular subgrouping in medulloblastoma based on preoperative multiparametric magnetic resonance imaging (MRI) characteristics.
Methods
A set of 19 MRI features were evaluated in 111 patients with histologic diagnosis of medulloblastoma for prediction of molecular subgrouping. MRI characteristics were correlated with molecular subgroups derived from tissue samples in 111 patients (WNT = 17, SHH = 44, Group 3 = 27, and Group 4 = 23). Multinomial logistic regression of imaging parameters was performed on a training cohort (TC) of 76 patients, representing two-thirds of randomly selected patients from each of 4 molecular subgroups, to generate binary nomograms. Validation of these nomograms was performed on the remaining 35 patients as the validation cohort (VC).
Results
Medulloblastoma subgroups could be accurately predicted by preoperative MRI features in 74% of cases. Predictive accuracy was excellent for SHH (95%), acceptably high for Group 4 (78%), modest for Group 3 (56%) and worst for WNT (41%) subgroup medulloblastoma. SHH-specific nomogram was associated with excellent correlation between TC and VC, with area under the curve (AUC) of 0.939 and 0.991, respectively. AUC for Group 4 was acceptable at 0.851 and 0.788 in TC and VC, respectively; however, these values were consistently suboptimal in WNT and Group 3 medulloblastoma.
Conclusion
The predictive accuracy of MRI-based nomograms was excellent for SHH and encouraging for Group 4 medulloblastoma. Further work is needed for Group 3 and WNT-pathway medulloblastoma.
Keywords: medulloblastoma, molecular, MRI, nomogram, radiogenomics
Importance of the Study
In this observational study comprising 111 patients with medulloblastoma, preoperative MRI features accurately predicted molecular subgrouping in 82 (74%) patients, leading to development and validation of subgroup-specific nomograms. Predictive accuracy was excellent for SHH (95%), acceptably high for Group 4 (78%), modest for Group 3 (56%), and worst for WNT (41%) subgroup medulloblastoma. Subgroup-specific nomograms were developed using a training cohort (n = 76) and tested in a validation cohort (n = 35) for reliability. Nomogram-based preoperative multiparametric MRIs were able to predict molecular subgrouping in SHH and group 4 medulloblastoma reliably but had suboptimal accuracy in Group 3 and WNT-pathway tumors. A large multi-institutional cohort study has recently shown that extent of resection impacts survival only in Group 4 medulloblastoma. In this context, the use of preoperative imaging for prediction of molecular subgrouping could guide neurosurgical decision making by avoiding extensive resections in other subgroups.
Medulloblastoma (MB) is the most common pediatric malignant neoplasm of the central nervous system (CNS), comprising 20%–25% of all childhood brain tumors.1,2 Over the past decade, novel biological insights have led to the identification of various molecular subgroups in MB with distinct developmental origins, unique transcriptional profiles, diverse phenotypes, and variable clinical outcomes.3–5 The consensus classification6 comprising 4 distinct molecular subgroups—wingless (WNT), sonic hedgehog (SHH), Group 3, and Group 4 MB—has recently been incorporated into the 2016 update of the World Health Organization (WHO) classification of CNS tumors.7 For patients with clinical suspicion of brain tumors, magnetic resonance imaging (MRI) remains the investigation of choice to arrive at a presumptive diagnosis and guide further therapeutic decision making. Imaging features of MB have been well characterized and described in the literature.8 Traditionally, radiology has predominantly focused on correlating imaging with histopathological findings. However, as is now widely believed, images are more than just pictures and reflect underlying morphology and dynamics of complex biological processes including gene expression, tumor-cell proliferation, and angiogenesis. Radiogenomics or imaging genomics9 is an exciting and emerging field of research that aims to define relationships between non-invasive imaging features (radio-phenotypes) and genomic data/molecular markers (molecular phenotypes). Recently, Brisse et al have shown imaging features to correlate with tumor origin, genomic profile, and clinical outcomes in pediatric tumors.10 Some groups have tried to correlate imaging features of MB with histological subtyping and molecular subgrouping in relatively small patient cohorts.11–16 The current study represents the largest series (N = 111 patients) incorporating radiogenomic data in MB. It is based on an earlier pilot study17 correlating a set of conventional MRI features in 19 MB patients with known molecular subgroup affiliation.18 To the best of our knowledge, this is the first attempt at developing nomograms based on multi-parametric MRI to aid in the preoperative prediction of molecular subgrouping in MB.
Methods and Materials
Our study was a combined retrospective and prospective study. It was initiated in April 2014, and patient accrual was completed in August 2016. Written informed consent and assent (as appropriate) was obtained from patients and/or caregivers in the prospective cohort while waiver of consent was granted for the retrospective cohort. Previously treated MB patients coming for routine periodic follow-up during the study period were considered eligible for inclusion if preoperative MRI and tumor tissue blocks were available. Database was locked in March 2017 for final analysis after molecular profiling on tissue samples was completed for all included patients. The study was duly approved by the institutional ethics committee and partially supported through a competitive intramural research grant.
Patient Selection
All patients with histological diagnosis of MB having preoperative MRI scans and tumor tissues available for molecular profiling were considered eligible for the study. Histology was reviewed in all patients by a dedicated institutional neuropathologist for confirmation of diagnosis. Being a tertiary-care cancer center, the majority of patients had undergone surgical resection outside and were subsequently referred to our institution for adjuvant therapy.
Molecular Subgroup Affiliation
Formalin-fixed paraffin-embedded or fresh-frozen tissue samples were used for molecular profiling. Molecular subgroup affiliation (WNT, SHH, Group 3, and Group 4) was based on differential expression of 12 protein coding genes and 9 microRNAs using real-time reverse transcriptase polymerase chain reaction, which has been described in detail and validated previously.18,19
Radiological Features
Preoperative multiparametric MRI scans of all included patients were discussed in the multidisciplinary joint neuro-oncology meeting comprising specialists from neuroradiology, radiation oncology, pediatric oncology, and neurosurgical oncology. Discussants were blinded to the results of molecular profiling, although clinico-demographic data and histological features were accessible from electronic medical records. A set of unique conventional MRI features (including subcategories) were documented for all patients. Imaging features were initially extracted by a single observer, but were discussed subsequently in the multidisciplinary joint clinic and finalized by consensus. Some of the important features that were extracted included tumor location, maximum tumor size, contrast enhancement characteristics, T2-weighted characteristics, diffusion characteristics, tumor margin, peritumoral edema, intratumoral hemorrhage, necrosis, calcification, associated cysts, presence of metastases, and hydrocephalus (Supplementary Table S1). Based on the results of the earlier pilot study,17 discussants were required to predict the possible molecular subgroup on imaging by consensus.
Statistical Analysis
Data analysis was done using IBM Statistical Package for Social Sciences v20 and STATA software v14. A set of 13 unique MRI features (with some features having subcategories totaling 19) were defined. Considering 10 samples per independent variable (×13 unique features) to arrive at any meaningful analysis through multinomial logistic regression, a sample size of 130 patients was calculated for the study. Correlation of individual MRI features with molecular subgrouping was tested using the Pearson chi-square test and Fisher’s exact test as appropriate. Cohen’s kappa statistics were used for assessing the agreement between the predicted (on preoperative MRI) and actual molecular subgroup (on tumor tissues).
For construction of nomograms, the entire study sample was divided into 2 cohorts: the training cohort (TC) and the validation cohort (VC). From each of the 4 individual subgroups, two-thirds of the patients were chosen randomly to constitute the TC (n = 76), while the remaining 35 constituted the VC. Using several combinations of MRI parameters that were significant, binary logistic regression was performed in the TC for the 4 subgroups individually, to differentiate the concerned subgroup from other subgroups. Independent binary nomograms were developed for each of the 4 subgroups. Receiver operating characteristics (ROC) curves were generated following application of individual subgroup-specific nomograms to assign scores for patients in the TC. For purpose of validation, similar methodology was undertaken for VC. Optimal cutoff of total scores for individual subgroup-specific nomograms was generated from the ROC curves from the TC for acceptable range of specificity and sensitivity, and their applicability was subsequently tested in the VC. Area under the curve (AUC) with 95% CI was used for interpretation and reporting.
Results
Patient Characteristics and Molecular Subgrouping
A total of 130 patients were considered for inclusion in the study. However, the quality of extracted DNA/RNA was suboptimal in 19 patients precluding definite subgroup assignment, leaving 111 patients available for inclusion in the final analysis. Patient and disease characteristics of the study cohort are described in Supplementary Table S2. The median age of the study cohort (N = 111) was 9 years (range 2–48 y), with 27 (24%) patients aged ≥18 years at initial diagnosis. Of the 16 children aged ≤3 years at initial diagnosis (infantile MB), 8 and 7 patients belonged to Group 3 and SHH, respectively, with the remaining 1 patient belonging to Group 4. For adults (≥18 y), the most common molecular subgroup was SHH (n = 21), followed by WNT (n = 4), Group 3 (n = 1), and Group 4 (n = 1). The median interval between diagnostic MRI and tumor resection was 16 days (interquartile range, 6‒33 days).
Radiological Features of Molecular Subgroups
The distribution of MRI features across the 4 molecular subgroups is briefly described in Table 1. Eleven features were found to be statistically significantly correlated with molecular subgrouping (2-sided P ≤ 0.05). Some of the characteristic MRI features of individual molecular subgroups are depicted in Figure 1A–D and are discussed in more detail below.
Table 1.
Distribution of MRI features across the 4 molecular subgroups of medulloblastoma
| MRI Features |
WNT
(n = 17) |
SHH
(n = 44) |
Group 3
(n = 27) |
Group 4
(n = 23) |
P-value*
(2-sided) |
|---|---|---|---|---|---|
| Tumor—horizontal | |||||
| Midline | 77% | 34% | 96% | 100% | <0.001 |
| Lateral | 23% | 66% | 4% | 0% | |
| Tumor—vertical | |||||
| Superior | 6% | 48% | 0% | 0% | |
| Central | 71% | 41% | 70% | 61% | <0.001 |
| Inferior | 23% | 11% | 30% | 39% | |
| Tumor size | 0.196 | ||||
| <2 cm | 0% | 0% | 0% | 0% | |
| 2–4 cm | 35% | 18% | 11% | 13% | |
| >4 cm | 65% | 82% | 89% | 87% | |
| Brainstem involvement | |||||
| Displaced | 77% | 36% | 70% | 87% | |
| Infiltrated | 12% | 9% | 7% | 4% | 0.001 |
| Uninvolved | 11% | 55% | 23% | 9% | |
| Contrast enhancement | 0.065 | ||||
| <20% | 6% | 9% | 7% | 30% | |
| 20–80% | 35% | 47% | 44% | 57% | |
| >80% | 59% | 44% | 48% | 13% | |
| Contrast heterogeneity | |||||
| Homogeneous | 53% | 39% | 22% | 4% | |
| Heterogeneous | 47% | 55% | 74% | 83% | 0.015 |
| No uptake/not available | 0% | 6% | 4% | 17% | |
| T2-weighted intensity | |||||
| Hypointense | 35% | 20% | 15% | 30% | |
| Isointense | 65% | 68% | 74% | 39% | 0.047 |
| Hyperintense | 0% | 12% | 11% | 31% | |
| T2-weighted homogeneity | |||||
| Homogeneous | 71% | 32% | 52% | 57% | 0.030 |
| Heterogeneous | 29% | 68% | 48% | 43% | |
| Diffusion characteristics | 0.985 | ||||
| Restricted | 92% | 95% | 95% | 95% | |
| Not restricted | 8% | 5% | 5% | 5% | |
| Tumor margin | 0.630 | ||||
| Smooth | 29% | 25% | 33% | 35% | |
| Infiltrative | 65% | 64% | 67% | 61% | |
| Lobulated | 6% | 11% | 0% | 4% | |
| Peritumoral edema | |||||
| Nil/none | 59% | 9% | 74% | 70% | |
| <1.5 cm | 35% | 52% | 26% | 30% | <0.001 |
| ≥1.5 cm | 6% | 39% | 0% | 0% | |
| Hemorrhage | 0.595 | ||||
| Present | 18% | 7% | 7% | 9% | |
| Absent | 82% | 93% | 93% | 91% | |
| Necrosis | 0.067 | ||||
| Present | 0% | 14% | 0% | 18% | |
| Absent | 100% | 86% | 100% | 82% | |
| Calcification | |||||
| Present | 29% | 11% | 30% | 39% | |
| Absent | 47% | 86% | 63% | 48% | 0.009 |
| Unknown | 24% | 3% | 7% | 13% | |
| Number of cyst(s) | |||||
| Nil/none | 12% | 18% | 19% | 17% | |
| 1–3 | 0% | 0% | 22% | 0% | 0.002 |
| >3 | 88% | 82% | 59% | 83% | |
| Size of cyst(s) | |||||
| Microcyst | 71% | 50% | 33% | 57% | |
| Macrocyst | 0% | 0% | 19% | 0% | 0.017 |
| Mixed | 17% | 32% | 29% | 26% | |
| Not applicable (no cysts) | 12% | 18% | 19% | 17% | |
| Location of cyst(s) | |||||
| Central | 77% | 55% | 41% | 57% | |
| Peripheral | 0% | 0% | 11% | 0% | 0.123 |
| Mixed | 11% | 27% | 29% | 26% | |
| Not applicable (no cysts) | 12% | 18% | 19% | 17% | |
| Hydrocephalus | |||||
| Absent | 18% | 5% | 0% | 0% | |
| Mild | 41% | 52% | 11% | 22% | 0.001 |
| Moderate | 41% | 32% | 59% | 52% | |
| Severe | 0% | 11% | 30% | 26% | |
| Leptomeningeal metastases | 0.135 | ||||
| Present | 6% | 9% | 26% | 22% | |
| Absent | 94% | 91% | 74% | 78% | |
*All statistically significant P-values (≤0.05) are highlighted in bold.
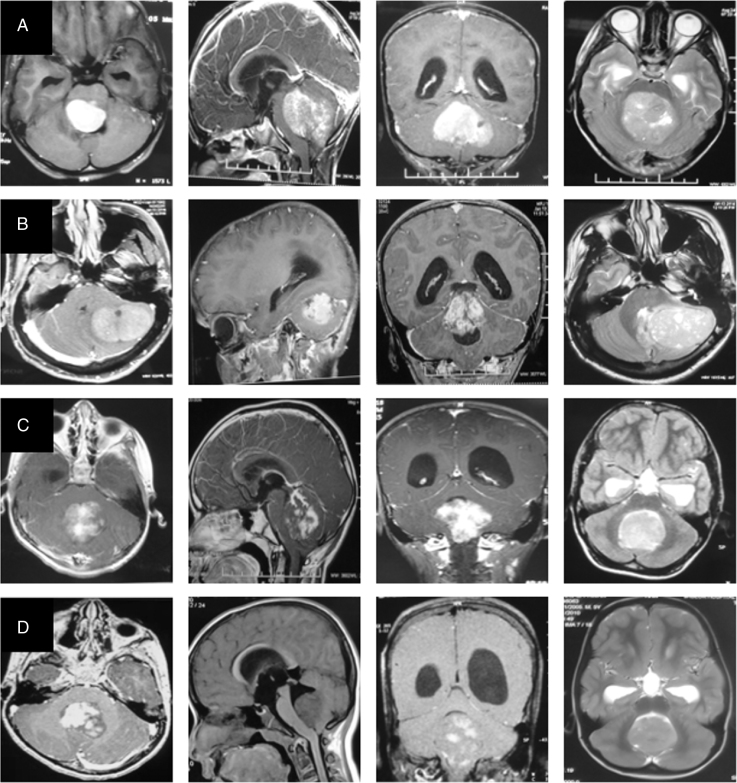
Fig. 1.
MRI image panel of the four molecular subgroups representing WNT (row A), SHH (row B), Group 3 (row C), and Group 4 (row D) medulloblastoma respectively. Post-contrast axial, sagittal, coronal T1-weighted images and axial T2-weighted images are presented from left to right, respectively, in each row.
WNT-pathway MBs were generally located in the midline (77%), but sometimes also extended laterally into the cerebellopontine angle (23%). The vast majority (94%) of WNT-subgroup patients showed significant contrast enhancement, with 59% of patients having uptake in >80% of the tumor. Homogeneous solid contrast enhancement was seen in 53% of patients with WNT-pathway MB. Although not statistically significant, hemorrhage was more commonly seen in WNT (18%) compared with other subgroups. SHH-pathway MBs were radiologically distinct from the other 3 subgroups. Overall, SHH-pathway MB was commonly located laterally over the cerebellar hemispheres (66%). However, the location of SHH-pathway MB varied significantly with the age. In infants (≤3 y) and children (>3 but <18 y), SHH-pathway MB was more commonly located in the midline (71% and 50%, respectively) as opposed to only 13% in adults (P = 0.003), where it was seen predominantly as a lateralized tumor. Almost 50% of SHH-pathway MB had a superior location abutting the tentorium, which was seen in only a single patient in other subgroups. The vast majority (91%) of SHH-pathway tumors were associated with peritumoral edema. Moderate to severe edema (>1.5 cm) was seen in 39% of patients with SHH-pathway MB compared with only 1 patient from other subgroups. Group 3 and Group 4 tumors shared some radiological features. They generally arose in the midline and tended to be located inferiorly with dilatation of superior recess of the IVth ventricle seen commonly compared with other subgroups. Group 3 tumors were more frequently isointense to uninvolved cerebellum than Group 4 MB (74% vs 39%, P = 0.001). Both subgroups showed variable and heterogeneous contrast enhancement; Group 3 having a “fluffy” type of enhancement, while Group 4 had a “patchy” enhancement due to intervening non-enhancing areas. Although not statistically significant, nearly 30% of Group 4 tumors showed either minimal or no contrast enhancement at all compared with <10% of non-enhancing tumors in other subgroups. Group 3 tumors had isolated macrocysts (19%) or multiple microcysts (22%), which were not seen in the other 3 subgroups.
Prediction of Molecular Subgrouping
The consensus prediction of molecular subgroup based on preoperative multiparametric MRI was accurate in 74% of patients included in the study. The highest predictive accuracy was for SHH-pathway MB (95%), followed by Group 4 MB (78%). Predictive accuracy was suboptimal and unacceptably low for Group 3 and WNT-pathway MB at 56% and 41%, respectively. The matrix for predicted subgroup against the actual molecular subgroup is summarized in Table 2. Overall agreement between the predicted and actual molecular subgroup was moderate (Cohen’s kappa = 0.63) for the entire cohort. Agreement was best for SHH-pathway MB (Cohen’s kappa = 0.78) followed by Group 4 (kappa = 0.64) and Group 3 MB (kappa = 0.59), with worst agreement for WNT-pathway MB (kappa = 0.36).
Table 2.
Matrix of predicted molecular subgroup on MRI (represented in rows) compared with the true molecular subgroup affiliation based on tumor-tissue profiling (represented in columns)
| True/Predicted Subgroup | WNT | SHH | Group 3 | Group 4 |
|---|---|---|---|---|
| WNT (n = 17) | 7 | 4 | 1 | 5 |
| SHH (n = 44) | 1 | 42 | 0 | 1 |
| Group 3 (n = 27) | 5 | 4 | 15 | 3 |
| Group 4 (n = 23) | 1 | 2 | 2 | 18 |
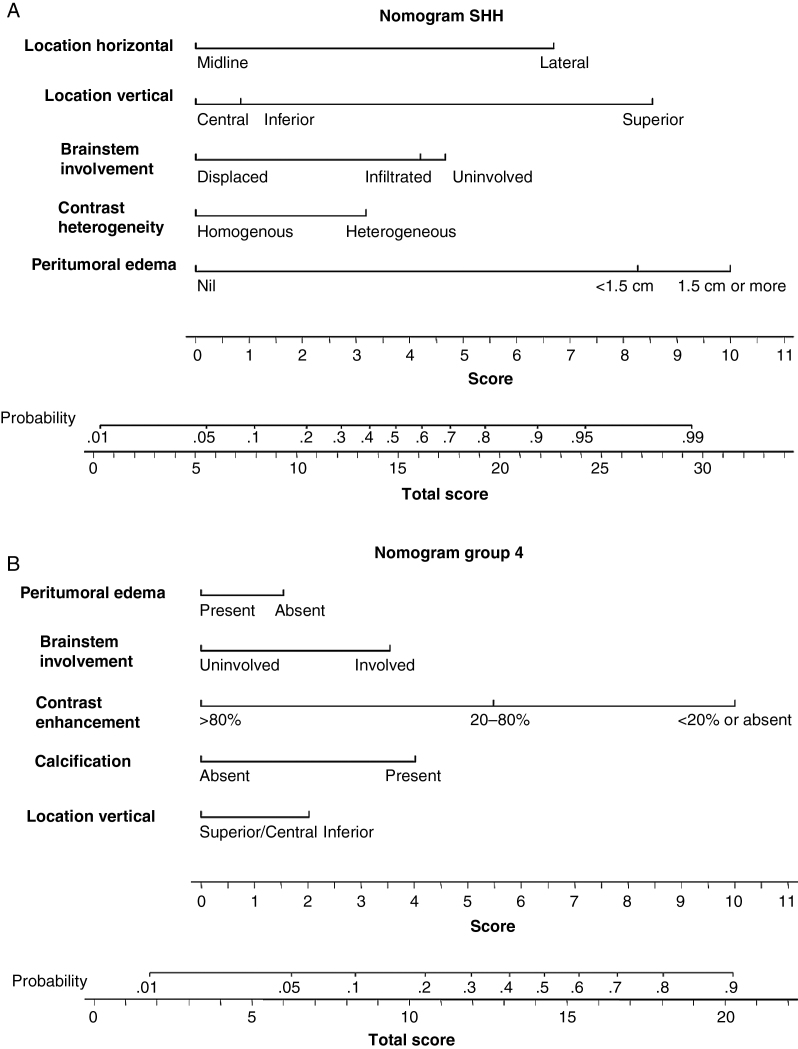
Subgroup-Specific Nomograms
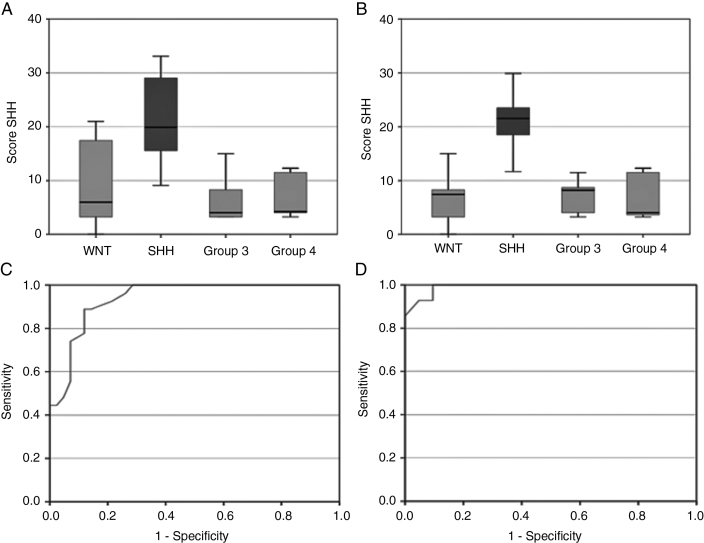
Subgroup-specific nomograms for SHH-pathway and Group 4 MB are presented in Figure 2A, B, while those for Group 3 and WNT-pathway MB are presented as online supplementary files (Supplementary Figure S1). AUCs (95% CI) from ROC curves for different molecular subgroups using subgroup-specific nomograms in the TC and VC are summarized in Table 3. Using SHH-specific nomogram, the mean “total score” for SHH-pathway MB in the TC was 21.8 (range 9.1–33.1) compared with 7.2 (range 0–21) for non-SHH subgroup tumors (Figure 3A). This was replicated in the VC as well, wherein the total score for SHH-pathway MB remained significantly higher than other molecular subgroups (Figure 3B). For a sensitivity and specificity >85% each, a cutoff total score of 13.3 was obtained from the SHH-specific nomogram in the TC. When this was tested in the VC, 93% of SHH-pathway MBs still had total score above this threshold (13.3) as opposed to only 5% of non-SHH tumors for a sensitivity of 93% and specificity of 95%. The nomogram identified even midline SHH tumors with acceptable accuracy. AUC for the SHH subgroup was excellent, with a value of 0.939 in TC (Figure 3C) and 0.991 in VC (Figure 3D), respectively. AUC for Group 4 MB was also good and acceptable at 0.851 in the TC and 0.788 in the VC. Using a Group 4–specific nomogram, a cutoff total score of 10.7 was associated with a sensitivity and specificity of >75% in the TC. Application of this threshold in the VC for Group 4–specific nomogram yielded an AUC of 0.788 at a sensitivity of 57% and specificity of 81%. AUCs were suboptimal for WNT (0.754) and Group 3 (0.726)–specific nomograms in the TC and further worsened when tested in the VC (AUC of 0.693 and 0.600, respectively).
Fig. 2.
Nomograms to predict SHH-pathway (A) and Group 4 (B) medulloblastoma based on selected MRI features. Score for individual MRI feature is obtained by dropping a perpendicular line from that feature onto the horizontal score line. Score of all individual features are summed to provide the total score. The total score (bottom line) is then used to read the predicted probability of that particular subgroup in the nomogram. For illustration, a total score of 22 in the SHH-pathway nomogram predicts a >90% probability of being SHH-pathway medulloblastoma. Similarly, a total score of 18 in Group 4 nomogram predicts >80% probability of being Group 4 medulloblastoma.
Table 3.
Area under the curve (AUC) with 95% CI) of receiver operating characteristics (ROC) curves for subgroup-specific nomograms in the training cohort and validation cohort
| Molecular Subgroup |
AUC (95% CI) in Training Cohort (n = 76) |
AUC (95% CI) in Validation Cohort (n = 35) |
|---|---|---|
| WNT | 0.754 (0.624–0.885) | 0.693 (0.416–0.970) |
| SHH | 0.939 (0.887–0.991) | 0.991 (0.971–1.000) |
| Group 3 | 0.726 (0.582–0.870) | 0.600 (0.380–0.820) |
| Group 4 | 0.851 (0.733–0.969) | 0.788 (0.632–0.945) |
Fig. 3.
Box-plots of total scores for each molecular subgroup in the SHH-specific nomogram. Note the significant difference in the mean scores for SHH-pathway medulloblastoma versus other subgroups in training cohort (A) as well as validation cohort (B). Receiver operating characteristic (ROC) curves derived using SHH-specific nomograms demonstrate excellent accuracy of predicting SHH-pathway medulloblastoma in both the training cohort (C) and the validation cohort (D).
Discussion
Novel biological insights have led to consensus classification of MB6 into 4 distinct molecular subgroups (WNT, SHH, Group 3, and Group 4), which has now been incorporated into the 2016 update of WHO classification of CNS tumors.7 The hitherto prevalent traditional risk-stratification system based entirely on clinico-radiological features20 has also been supplanted by a more contemporary risk-classification schema (incorporating molecular biology) into low-risk, standard-risk, high-risk, and very high-risk categories with expected long-term overall survival >90%, >75%–90%, >50%–75%, and <50%, respectively.21
Although molecular profiling of resected tumor tissue remains the gold standard for subgroup assignment in MB, pre-operative prediction of molecular subgroup based on imaging features can potentially guide neurosurgical decision making. Although maximum safe resection should remain the guiding principle, aggressive surgical removal of small residual tumor is not recommended, particularly, when the likelihood of precipitating neurological deficits is high. The prognostic benefit of increased extent of resection for patients with MB gets significantly attenuated after adjustment for molecular subgrouping, as recently demonstrated by Thompson and colleagues22 in a large consortium study.
The putative cells of origin for different molecular subgroups of MB are known to be different.4,5 WNT-pathway tumors arise from the nuclei in the dorsal brainstem, while SHH-pathway MBs arise from the granule neuron precursor cells,23 which explains the lateralized location of SHH-subgroup tumors. In our series, two-thirds of all SHH-pathway MBs were located laterally (over the cerebellar hemisphere), which was proportionately significantly higher in adults. Driving genetic alterations are variable across different age groups (infantile, childhood, and adult) in SHH-pathway MB,24 which may partially explain its differential location across age groups. In a study correlating anatomic location with molecular subgrouping in 60 children with MB, Teo et al11 reported that 9 of 17 (52%) patients with SHH-pathway MB had a lateralized location. In another study,12 involving 71 patients, cerebellar hemispheric location was seen in 62% of SHH-pathway tumors compared with no patient with lateralized Group 3 or Group 4 MB. Patay et al13 studied the spatial distribution in 16 patients with WNT-pathway MB and concluded that these tumors are close to the midline but are lateralized, originating from the brainstem and cerebellum around the foramen of Luschka, in accordance with the current understanding of their embryologic origins. Perreault et al14 also reported that 3 of 4 (75%) WNT-pathway tumors had extension into cerebellar peduncle and/or cerebellopontine angle cistern. Interestingly, we observed superior location of tumor to be a strong predictor for SHH-pathway MB, which was extremely rarely seen in other subgroups. Similar observation was reported by Wefers et al,12 where rostral location was more commonly seen in SHH tumors. Caudal location of Group 3 and Group 4 tumors seen in our cohort is also in accordance with previously published data.12 Inferior location with extension to (sometimes even across) the foramen magnum with dilatation of superior recess of the IVth ventricle was helpful in identifying Group 4 tumors. The relation of tumor with respect to the dorsal aspect of the brainstem serves as an important feature to discriminate between subgroups. Wefers et al12 observed 100% of non-SHH tumors to always have some contact with the brainstem, as opposed to 48% of SHH-pathway MB. Infiltration of tumor into the cuneate or dentate nucleus favors the WNT subgroup.12,13
The 4 subgroups differed among themselves with regard to contrast enhancement. WNT-pathway MB lies at one end of the spectrum with homogeneous enhancement involving almost the entire tumor, while Group 4 MB lies at the other end, with a large proportion of non-enhancing or very faintly enhancing tumor. Interestingly, Yeom et al15 had reported ring enhancement within the tumor to correlate with necrosis and large-cell/anaplastic histology; a fair proportion of these were likely Group 3 MB. In the only radiogenomic study of adult patients with MB, Keil et al16 reported WNT-pathway MB to have small tumor volumes, contact with IVth ventricle, and absence of hydrocephalus. However, imaging biomarkers for the same genetic entities were very discordant from those identified in the pediatric cohort. Recent pioneering work by Phoenix et al25 has shown presence of aberrant fenestrated vasculature driven by mutant β-catenin in WNT-pathway MB having higher association with hemorrhage compared with other subgroups. The authors also reported that 90% of WNT-pathway MB were associated with frank hemorrhages as observed during surgery from 2 independent patient cohorts. Patay et al13 had reported intratumoral hemorrhage in 31% of patients with WNT subgroup MB. We suggest that gradient-echo or susceptibility-weighted imaging be performed routinely for identifying hemorrhage in patients with suspected MB. Presence of moderate to severe edema (>1.5 cm) was quite significantly associated with SHH-pathway MB, something that has not been reported previously. Similarly, differential incidence of cyst characteristics, calcification, and hydrocephalus may have an important role in discriminating between subgroups.
Strengths and Limitations
Use of conventional imaging features easily identified on a routine diagnostic MRI scan for correlation with molecular subgrouping is a strength of our study. Clinicians and diagnosticians were blinded to the molecular subgroup at the time of extraction of imaging features and prediction of molecular subgrouping. All imaging features were initially evaluated by a single investigator to reduce interobserver bias and subsequently discussed in the multidisciplinary neuro-oncology joint clinic for consensus. The inclusion of 111 patients makes it the largest study of radiogenomics in MB. Despite the aforesaid strengths, several caveats and limitations remain. In our study, the applicability of nomograms for preoperative diagnosis was excellent for SHH-subgroup medulloblastoma. However, it had somewhat less accuracy for Group 4 and was not reliable for use in WNT-pathway and Group 3 MB. A uniform MRI protocol was not used in the study, as preoperative MRI scans were done at various centers across the country using different institutional protocols, with potential differences in interpretation and reporting. We did not attempt to correlate magnetic resonance spectroscopy (MRS) findings with molecular subgrouping, as that information was lacking in the vast majority of scans. A 5-metabolite subgroup classifier (creatine, myoinositol, taurine, aspartate, and lipid) based on MRS26 can distinguish between Group 3/Group 4 tumors and SHH-pathway MB with excellent accuracy. Data for quantifying apparent diffusion coefficient (ADC) values was also not routinely available. It is quite likely that integration of MRS and ADC values may help to further refine the nomograms and increase their predictive accuracy. Recent advances in computational technology has enabled further dissection of imaging features well beyond the potential of human capability. The use of texture analysis, artificial neural networks, and support vector machine-learning techniques may lead to better radiomic/radiogenomic classification of MB subgroups. Some non-imaging variables, such as age, gender, and histologic subtype, have higher predilection for particular molecular subgroups. For example, adults are much more likely to have SHH-pathway MB compared with children, male sex is overrepresented in Group 4 MB, and desmoplastic histology is often associated with SHH-pathway MB. Since we focused primarily on imaging features in our study, we did not include clinico-demographic characteristics that could have provided incremental value and further improved the predictive accuracy of our nomograms.
Conclusion
Conventional imaging features extracted from preoperative multiparametric MRI correlates with molecular subgrouping in MB enabling construction of subgroup-specific nomograms with variable predictive accuracy. An SHH-subgroup nomogram can be used in routine clinical practice due to its excellent predictive accuracy. The reliability of a Group 4 nomogram, though somewhat less, is still reasonable, mandating judicious use. Caution is warranted against the use of Group 3 and WNT-subgroup nomograms due to their unacceptably low and suboptimal accuracy. Further collaborative efforts are needed to improve the predictive accuracy of imaging features in Group 3 and WNT-pathway MB.
Funding
This work was supported by an intramural grant from Tata Memorial Centre, Mumbai; the Indian Council of Medical Research (ICMR), New Delhi; and the Brain Tumor Foundation (BTF) of India, Mumbai.
Supplementary Material
Acknowledgments
We are grateful to Tata Memorial Centre, Indian Council of Medical Research and Brain Tumor foundation of India for the financial support. We express our thanks to our colleagues of Radiation Oncology and Radiodiagnosis of Tata Memorial Centre and all the neurosurgery and pediatric oncology colleagues from various institutes for assistance, patient referral, and management. Special acknowledgment to efforts of Nazia Bano and Shraddha Churi for logistic and secretarial assistance during the study. Part of the data from the study were presented at the 17th International Symposium on Pediatric Neuro-Oncology (ISPNO), Liverpool, UK in 2016 (through The Children’s Hospital at Westmead International Travelling Fellowship to Archya Dasgupta) and at the 9th Annual Conference of the Indian Society of Neuro-Oncology (ISNO), Bangalore, India in 2017.
Conflict of interest statement. None of the authors have any conflicts of interest to declare.
References
- 1. Leece R, Xu J, Ostrom QT, Chen Y, Kruchko C, Barnholtz-Sloan JS. Global incidence of malignant brain and other central nervous system tumors by histology, 2003–2007. Neuro Oncol. 2017;19(11):1553–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Khanna V, Achey RL, Ostrom QT, et al. . Incidence and survival trends for medulloblastoma in the United States from 2001–2013. J Neurooncol 2017;135(3):433–441. [DOI] [PubMed] [Google Scholar]
- 3. Northcott PA, Korshunov A, Witt H, et al. . Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29(11):1408–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gajjar A, Bowers DC, Karajannis MA, Leary S, Witt H, Gottardo NG. Pediatric brain tumors: innovative genomic information is transforming the diagnostic and clinical landscape. J Clin Oncol. 2015;33(27):2986–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gupta T, Shirsat N, Jalali R. Molecular subgrouping of medulloblastoma: impact upon research and clinical practice. Curr Pediatr Rev. 2015;11(2):106–119. [DOI] [PubMed] [Google Scholar]
- 6. Taylor MD, Northcott PA, Korshunov A, et al. . Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123(4):465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Louis DN, Perry A, Reifenberger G, et al. . The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 8. Koeller KK, Rushing EJ. From the archives of the AFIP: medulloblastoma: a comprehensive review with radiologic-pathologic correlation. Radiographics. 2003;23(6):1613–1637. [DOI] [PubMed] [Google Scholar]
- 9. Kuo MD, Jamshidi N. Behind the numbers: Decoding molecular phenotypes with radiogenomics–guiding principles and technical considerations. Radiology. 2014;270(2):320–325. [DOI] [PubMed] [Google Scholar]
- 10. Brisse HJ, Blanc T, Schleiermacher G, et al. . Radiogenomics of neuroblastomas: relationships between imaging phenotypes, tumor genomic profile and survival. PLoS One. 2017;12(9):e0185190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Teo WY, Shen J, Su JM, et al. . Implications of tumor location on subtypes of medulloblastoma. Pediatr Blood Cancer. 2013;60(9):1408–1410. [DOI] [PubMed] [Google Scholar]
- 12. Wefers AK, Warmuth-Metz M, Pöschl J, et al. . Subgroup-specific localization of human medulloblastoma based on pre-operative MRI. Acta Neuropathol. 2014;127(6):931–933. [DOI] [PubMed] [Google Scholar]
- 13. Patay Z, DeSain LA, Hwang SN, Coan A, Li Y, Ellison DW. MR imaging characteristics of wingless-type-subgroup pediatric medulloblastoma. AJNR Am J Neuroradiol. 2015;36(12):2386–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Perreault S, Ramaswamy V, Achrol AS, et al. . MRI surrogates for molecular subgroups of medulloblastoma. AJNR Am J Neuroradiol. 2014;35(7):1263–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yeom KW, Mobley BC, Lober RM, et al. . Distinctive MRI features of pediatric medulloblastoma subtypes. AJR Am J Roentgenol. 2013;200(4):895–903. [DOI] [PubMed] [Google Scholar]
- 16. Keil VC, Warmuth-Metz M, Reh C, et al. . Imaging biomarkers for adult medulloblastomas: genetic entities may be identified by their mr imaging radiophenotype. AJNR Am J Neuroradiol. 2017;38(10): 1892–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Krishnatry R, Pungavkar S, Shirsat N, et al. . Radiological markers on MR imaging to identify molecular subgroups in medulloblastoma. J Cancer Res Ther 2013;9(Suppl 1):S31. [Google Scholar]
- 18. Gokhale A, Kunder R, Goel A, et al. . Distinctive microRNA signature of medulloblastomas associated with the WNT signaling pathway. J Cancer Res Ther. 2010;6(4):521–529. [DOI] [PubMed] [Google Scholar]
- 19. Kunder R, Jalali R, Sridhar E, et al. . Real-time PCR assay based on the differential expression of microRNAs and protein-coding genes for molecular classification of formalin-fixed paraffin embedded medulloblastomas. Neuro Oncol. 2013;15(12):1644–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zeltzer PM, Boyett JM, Finlay JL, et al. . Metastasis stage, adjuvant treatment, and residual tumor are prognostic factors for medulloblastoma in children: conclusions from the Children’s Cancer Group 921 randomized phase III study. J Clin Oncol. 1999;17(3):832–845. [DOI] [PubMed] [Google Scholar]
- 21. Ramaswamy V, Remke M, Bouffet E, et al. . Risk stratification of childhood medulloblastoma in the molecular era: the current consensus. Acta Neuropathol. 2016;131(6):821–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thompson EM, Hielscher T, Bouffet E, et al. . Prognostic value of medulloblastoma extent of resection after accounting for molecular subgroup: a retrospective integrated clinical and molecular analysis. Lancet Oncol. 2016;17(4):484–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gibson P, Tong Y, Robinson G, et al. . Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468(7327):1095–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kool M, Jones DT, Jäger N, et al. ; ICGC PedBrain Tumor Project Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell. 2014;25(3):393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Phoenix TN, Patmore DM, Boop S, et al. . Medulloblastoma genotype dictates blood brain barrier phenotype. Cancer Cell. 2016;29(4):508–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blüml S, Margol AS, Sposto R, et al. . Molecular subgroups of medulloblastoma identification using noninvasive magnetic resonance spectroscopy. Neuro Oncol. 2016;18(1):126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.