Abstract
Background
To date, genome-wide association studies (GWAS) have identified 25 risk variants for glioma, explaining 30% of heritable risk. Most histologies occur with significantly higher incidence in males, and this difference is not explained by currently known risk factors. A previous GWAS identified sex-specific glioma risk variants, and this analysis aims to further elucidate risk variation by sex using gene- and pathway-based approaches.
Methods
Results from the Glioma International Case-Control Study were used as a testing set, and results from 3 GWAS were combined via meta-analysis and used as a validation set. Using summary statistics for nominally significant autosomal SNPs (P < 0.01 in a previous meta-analysis) and nominally significant X-chromosome SNPs (P < 0.01), 3 algorithms (Pascal, BimBam, and GATES) were used to generate gene scores, and Pascal was used to generate pathway scores. Results were considered statistically significant in the discovery set when P < 3.3 × 10−6 and in the validation set when P < 0.001 in 2 of 3 algorithms.
Results
Twenty-five genes within 5 regions and 19 genes within 6 regions reached statistical significance in at least 2 of 3 algorithms in males and females, respectively. EGFR was significantly associated with all glioma and glioblastoma in males only and a female-specific association in TERT, all of which remained nominally significant after conditioning on known risk loci. There were nominal associations with the BioCarta telomeres pathway in both males and females.
Conclusions
These results provide additional evidence that there may be differences by sex in genetic risk for glioma. Additional analyses may further elucidate the biological processes through which this risk is conferred.
Key Points
1. EGFR was significantly associated with all glioma and glioblastoma in males only.
2. TERT was significantly associated with all glioma in females only.
3. The telomere pathway was a risk factor for glioma that varied by sex.
Importance of the Study
Glioma, like most other cancer histologies, occurs at greater frequency in males than in females, and this difference is not explained by currently known risk factors. A previous sex-stratified analysis of these datasets identified three risk loci that varied in association between males and females. In this analysis, we attempted to leverage the summary statistics generated by this previous analysis for additional discovery using gene- and pathway-based approaches. After conditioning on previously identified genetic risk loci, EGFR was significantly associated with all glioma and glioblastoma in males only and a female-specific association in TERT. There were also nominal associations with the Telomeres, Telomerase, Cellular Aging, and Immortality pathway (in BioCarta) in both males and females. These results provide additional evidence that there may be biologically relevant significant differences by sex in genetic risk for glioma.
Glioma is the most common type of primary malignant brain tumor in the United States, with an average annual age-adjusted incidence rate of 6.0 per 100000 population.1 Glioma can be broadly classified into glioblastoma (GBM, 61.9% of gliomas in adults 18+ in the US) and lower-grade glioma (non-GBM glioma, 24.2% of adult gliomas). These tumors occur more commonly in people of European ancestry, in males, and in older adults. Most glioma histologies occur with a 30–50% higher incidence in males, and this male preponderance of glial tumors increases with age (Supplementary Figure 1).1
Many environmental exposures have been investigated as sources of glioma risk, but the only validated risk factors for these tumors are ionizing radiation (which increases risk) and history of allergies or other atopic disease (which decreases risk).2,3 A minority of glioma risk is thought to arise from heritable genetic risk factors, and the contribution of common low-penetrance single nucleotide polymorphisms (SNPs) to the heritability of glioma is estimated to be ~25%.4 A recent glioma genome-wide association study (GWAS) meta-analysis validated 12 previously reported risk loci and identified 13 new risk loci, and these 25 loci in total are estimated to account for ~30% of heritable glioma risk.5 This suggests that there are both undiscovered environmental risk factors (which account for ~75% of disease incidence variance) and genetic risk factors (accounting for ~70% of heritable risk).4,5
Each individual GWAS results in regression estimates for hundreds of thousands of SNPs, only several hundred of which may meet the criteria for statistical significance to be prioritized for further investigation. While this process is appropriate for identifying individual loci that contribute to the development of disease, there is likely additional information about disease risk within these results that do not meet the stringent statistical significance thresholds used in GWAS (usually P < 5 × 10−8). Gliomas are known to be biologically complex, and as a result additional single-SNP analyses may not be appropriate to discover additional sources of genetic risk for these tumors. Multi-SNP methods—such as gene- or pathway-based approaches—can allow for additional discovery in a manner that complements single-SNP approaches, while substantially reducing the multiple testing burden associated with GWAS.6
While it is not likely that autosomal genomic sequence varies significantly by sex in the population, previous research has suggested that sex-related genetic variation may occur at the transcriptional and regulatory level.7–9 One of the primary ways that SNPs are thought to affect phenotype is through variation in gene regulation and expression.10 Sex-specific variation in regulatory processes may also affect the relationship between SNPs and phenotype; as a result it may be possible that allele frequencies in risk SNPs vary between affected males and females. A recent sex-stratified GWAS identified 3 glioma risk loci that differ in effect by sex.11 These 3 SNPs explain 1.4% of phenotypic variance in a pooled glioma sample (1.3% in males and 2.2% in females), and 0.6% of variance in GBM (0.9% in males and 0.7% in females). Other analyses have also identified sex-specific sources of risk for glioma, including an association study focused on the cAMP (cyclic adenosine monophosphate) pathway that identified SNPs in adenylate cyclase 8 as a sex-specific modifier of risk for low-grade astrocytoma in neurofibromatosis type 1.12 Genetic risk for complex traits is increasingly understood to be polygenic, and variations in risk may be the result of variation at hundreds of locations across the genome. Each individual SNP may only provide a very small contribution to genetic risk for a trait, and the mechanistic relationship between these individual, low-effect SNPs and phenotype is hard to estimate. Additional analyses using gene- and pathway-based approaches may further elucidate sex differences in genetic risk for glioma.
There is no consensus on the best method for generating gene- and pathway-based test statistics from GWAS summary statistics, and many different approaches have been developed. These pathway approaches have been utilized in other cancers with some success, but have not been widely used in glioma. The primary aim of this analysis was to contrast multiple gene-based approaches for leveraging currently existing sex-specific glioma summary statistics, as well as to assess whether these approaches may identify additional sources of genetic risk for glioma that may vary by sex.
Methods
Summary statistics generated as part of a prior sex-specific GWAS11 were used to estimate sex-specific gene and pathway scores. Data from 4 studies were divided into a testing set and a validation set. Results from the Glioma International Case-Control Study (GICC)13,14 were used as a testing set (Fig. 1A), and results from 3 prior glioma GWAS (San Francisco Adult Glioma Study GWAS,15 MD Anderson Glioma GWAS,16 and National Cancer Institute’s GliomaScan17) were combined via inverse-variance weighted fixed effects meta-analysis in META18 and used as a validation set for any statistically significant genes and pathways (Fig. 1A). See Supplementary Table 1 for an overview of characteristics for individuals included in these datasets, and Fig. 1 for an overview of the study schematic. Details of case ascertainment, genotyping, quality control, imputation, and primary analysis of these datasets are available in Melin et al (GICC), Wrensch et al (San Francisco Adult Glioma Study GWAS), Shete et al (MD Anderson Glioma GWAS), and Rajaraman et al (GliomaScan).13–17
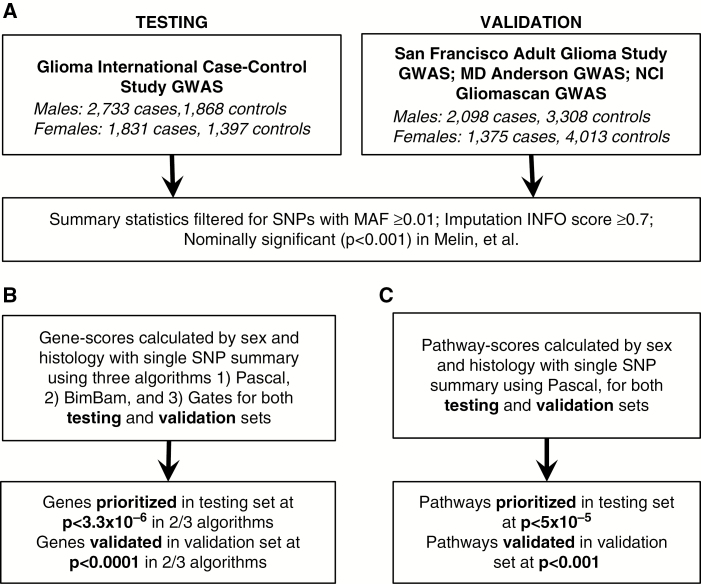
Fig. 1.
Study schematic for (A) generation of discovery and validation summary statistic sets, (B) generation, prioritization, and validation of gene scores, (C) generation, prioritization, and validation of pathway scores.
Sex-specific summary statistics studies for autosomal markers were previously generated11 using sex-stratified logistic regression models in SNPTEST19 to estimate sex-specific betas (βM and βF), standard errors (SEM and SEF), and P-values (pM and pF) (Fig. 1A). Summary statistics for the 3 studies used as a validation set were combined using META.18 Only SNPs with minor allele frequency (MAF) ≥ 0.01, imputation INFO score ≥ 0.7 and P < 0.01 in a previous 8-study pooled-sex meta-analysis,14 which included the 4 datasets used in this analysis, were used to generate gene and pathway scores. X-chromosome data were available from the GICC set only, and analyzed using the logistic regression model in the SNPTEST module “newml” assuming complete inactivation of one allele in females, and males are treated as homozygous females. X-chromosome SNPs with MAF ≥ 0.01, imputation INFO score ≥ 0.7, and single SNP association P < 0.01 were used for generation of gene scores. Linkage disequilibrium (LD) information was based on structure within the European cases from the 1000 Genomes project phase III dataset.20 All analyses were performed separately for males and females to identify genes and pathways with germline variation between cases and controls. Genes were prioritized that were identified by at least 2 of the 3 selected algorithms (Fig. 1B). Analyses were conducted for glioma overall and for glioblastoma only by sex within each dataset.
Three algorithms (Pascal,21 BimBam,22 and GATES23) were used to generate gene scores. Gene-based effects were assessed using SNPs within 50 kb of each gene (using 5ʹ and 3ʹ untranslated regions) as defined using the UCSC hg19 assembly. Pascal21 calculates gene scores using the VEGAS24 scoring algorithm and generates a gene-based test statistic using sum-of-chi-squares (SOCS) correcting for LD structure (based on a reference set). Genes that are in LD are considered to be “fusion genes” and have only one gene score calculated. BimBam22 (as implemented in the Functional Magnetic Resonance Imaging of the Brain Automated Segmentation Tool [FAST] using summary statistics25) is a Bayesian regression approach. This method calculates an average Bayes factor for all K possible models within a gene, where K is the number of SNPs. The model then uses a Laplace method to estimate posterior distributions of the model’s parameters, and distribution models are obtained using the Fletcher–Reeves conjugate gradient algorithm. GATES23 (as implemented in FAST25) uses a modified Simes test that combines SNP-based P-values, using the P-value correlation matrix to estimate the number of independent SNPs within the gene. The resulting gene-based P-values approximate a uniform distribution. For all methods implemented within FAST, SNPs were excluded if they were in complete LD (r2 = 1) with another SNP in the gene, which limited the amount of SNPs evaluated within each gene.
Pathway scores were generated using Pascal,21 using gene and fusion-gene scores generated by the Pascal algorithm (Figure 1C). The pathway score was then calculated using both independent and fusion genes. A parameter-free enrichment strategy was used to calculate pathway scores using either a chi-squared method (gene score P-values were ranked and transformed to a uniform distribution; these values were then transformed by a chi-square quantile function and summed) or an empirical sampling method (gene scores are transformed with chi-square quantile function and summed, then Monte Carlo estimate of the P-values were obtained by sampling random sets of the same size). Results from each gene and pathway algorithm were compared within each sex as well as between sexes. Pathway information was obtained from KEGG,26 Reactome,27 and BioCarta28 (as defined in MSigDB29,30).
For genes within regions that contain SNPs previously identified as significant by GWAS, conditional analyses were run for all SNPs within those regions using SNPTEST, and adjusted gene scores were calculated. All figures were generated using R v3.3.2, ggplot2, graphite, network, Intergraph, ggnetwork, igraph, gridExtra, and LocusZoom.31–37
Results
Included in gene-based analyses were 159706 SNPs from the testing set and 163115 SNPs from the validation set. Gene scores were generated for ~16000 genes and were considered significant at P < 3.3 × 10−6 (based on a Bonferroni correction for 15000 tests). P-values in the validation set were considered significant at P < 0.001 (based on a Bonferroni correction for 50 tests, for 25 total genes tested in each sex).
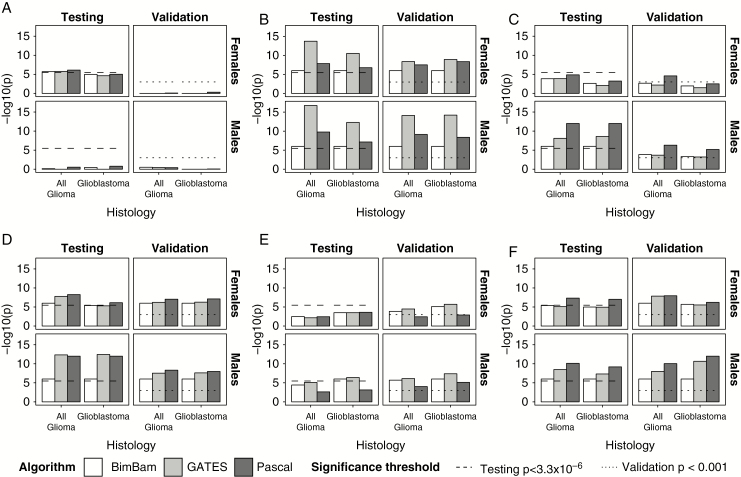
Among males, 25 genes within 5 regions had scores that reached the set significance threshold (P < 3.3 × 10−6) in at least 2 of 3 evaluated algorithms in all glioma or glioblastoma (see Fig. 2 and Supplementary Table 2 for the strongest associations within each of the 6 regions where genes met the set significance threshold). Among females, 19 genes within 6 regions had scores that reached the set significance threshold (P < 3.3 × 10−6) in at least 2 of 3 evaluated algorithms in all glioma or glioblastoma (see Fig. 2 and Supplementary Table 3 for the strongest associations within each of the 6 regions where genes met the set significance threshold). Solute carrier family 6, member 18 (SLC6A18), telomerase reverse transcriptase (TERT), cyclin-dependent kinase inhibitor 2B (CDKN2B), and stathmin 3 (STMN3) reached the set significance threshold in both males and females in glioblastoma, while SLC6A18, TERT, and STMN3 reached the set significance threshold in both sexes in all glioma. All shared associations validated.
Fig. 2.
Gene scores for prioritized genes by algorithm, histology, and sex for (A) BPESC1 (3q23), (B) TERT (5p15.33), (C) EGFR (7p11.2), (D) CDKN2B (9p21.3), (E) DNAH2 (17p13.1), and (F) RTEL1-TNFRSF6B (20q13.33).
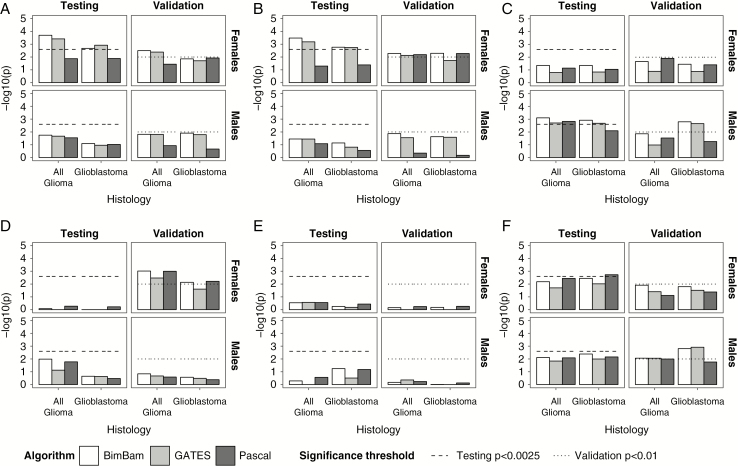
Epidermal growth factor receptor (EGFR), dynein axonemal heavy chain 2 (DNAH2), and several genes surrounding regulator of telomere elongation helicase 1 (RTEL1) on chromosome 20 (with the strongest association in the RTEL1–tumor necrosis factor [TNF] receptor superfamily member 6b [RTEL1-TNFRSF6B]) reached the significance threshold in males only (Fig. 2, Supplementary Table 2). In all glioma, CDKN2A reached the set significance threshold in males only. All genes validated in males. Blepharophimosis, epicanthus inversus and ptosis, candidate 1 (non-protein coding) (BPESC1) reached the significance threshold in all glioma in females only (Fig. 2, Supplementary Table 3), but this association was not confirmed in the validation set. The association in EGFR was nominally significant in males after conditioning on 3 SNPs previously identified by GWAS within this gene (rs75061358, rs723527, and rs11979158), including one (rs11979158) that has previously been identified as having a sex-specific effect (Supplementary Tables 4–5). When conditional single-SNP associations were examined by sex and histology in EGFR, there was a nominally significant peak apparent in both males and females, with no single SNP that approached genome-wide significance (Supplementary Fig. 3). The association at TERT was nominally significant for females in glioblastoma only after conditioning on the previous identified SNP (Fig. 3, Supplementary Table 5). When conditional single-SNP associations were examined by sex and histology in TERT, a single SNP of nominal significance (rs7705526) was identified upstream of the previously identified SNP (rs10069690; Supplementary Fig. 3). This region was apparent in males and females. Associations in STMN3 and RTEL1-TNFRSF6B remained nominally significant after conditioning in both males and females (Fig. 3, Supplementary Tables 4–5). When conditional single-SNP associations were examined by sex and histology in RTEL1-TNFRSF6B, nominally significant SNPs were identified across the gene in both males and females, with no apparent additional signal (Supplementary Fig. 4). There was no substantial difference in effect size by sex in the most significant SNP in either gene (Supplementary Fig. 5).
Fig. 3.
Conditional gene scores for prioritized genes by algorithm, histology, and sex for (A) TERT (5p15.33), (B) EGFR (7p11.2), (C) CDKN2B (9p21.3), (D) DNAH2 (17p13.1), and (E) RTEL1-TNFRSF6B (20q13.33).
There were 202886 X-chromosome SNPs with MAF ≥ 0.01 and INFO score ≥ 0.7 in the GICC dataset. Gene scores were calculated for 56 X-chromosome genes with at least 5 SNPs, and associations were considered significant at P < 8.3 × 10−4 (based on a Bonferroni correction for 60 tests). There were 12 genes within 4 chromosomal regions that reached the significance threshold in at least 2 of 3 algorithms (results from the strongest association in each region are shown in Table 1). Shroom family member 2 (SHROOM2) (Xp22.2) and armadillo repeat containing, X-linked 2 (ARMCX2) (Xq22.1) were significantly associated with both all glioma and glioblastoma, while dystrophin (DMD) (Xq21.2-p21.1) was significantly associated with all glioma only, and zinc finger protein 185 with LIM domain (ZNF185) was significantly associated with glioblastoma only.
Table 1.
Gene scores for prioritized X chromosome genes by histology
| Gene (location) | Histology | Pascal | BimBam | GATES | Algorithms P < 8.3 × 10−4 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNPsa | P-value | SNPsa | Testsb | P-value | SNPsa | Testsb | P-value | |||
| SHROOM2 (Xp22.2) | All glioma | 7 | 1.20 × 10−4 | 6 | 5.09 | 7.68 × 10−4 | 6 | 5.09 | 0.0020 | 2/3 |
| Glioblastoma | 9 | 1.45 × 10−5 | 8 | 7.06 | 5.02 × 10−4 | 8 | 7.06 | 0.0012 | 2/3 | |
| DMD (Xp21.2-p21.1) | All glioma | 88 | 3.22 × 10−5 | 79 | 59.92 | 3.13 × 10−4 | 79 | 59.92 | 0.0026 | 2/3 |
| Glioblastoma | 39 | 6.53 × 10−6 | 37 | 31.23 | 0.0047 | 37 | 31.23 | 0.0097 | 1/3 | |
| ARMCX2 (Xq22.1) | All glioma | 49 | 1.07 × 10−4 | 44 | 33.78 | 1.89 × 10−4 | 44 | 33.78 | 4.79 × 10−4 | 3/3 |
| Glioblastoma | 63 | 5.82 × 10−5 | 58 | 45.41 | 2.13 × 10−4 | 58 | 45.41 | 0.0011 | 2/3 | |
| ZNF185 (Xq28) | All glioma | 40 | 0.0018 | 33 | 24.26 | 0.0026 | 33 | 24.26 | 0.0061 | 0/3 |
| Glioblastoma | 49 | 6.19 × 10−5 | 42 | 33.52 | 3.04 × 10−4 | 42 | 33.52 | 9.22 × 10−4 | 2/3 | |
Abbreviations: SHROOM2: shroom family member 2; DMD: dystrophin; ARMCX6: armadillo repeat containing, X-linked 6; ARMCX2: armadillo repeat containing, X-linked 2; ZNF185: zinc finger protein 185 with LIM domain.
aNominally significant (P < 0.01) SNPs used in calculating gene score.
bNumber of independent SNPs after filtered for linkage disequilibrium.
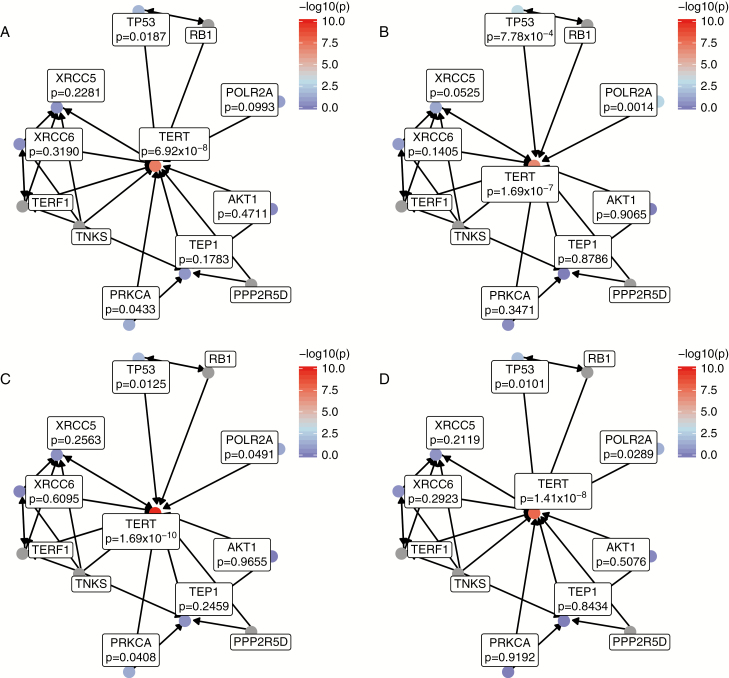
There were 1077 pathways in the combined KEGG, BioCarta, and Reactome sets, and associations were considered statistically significant in the discovery set at P < 5 × 10−5 (based on a Bonferroni correction for 1000 tests) and significant in the discovery set at P < 0.00883 (based on a Bonferroni correction for 6 tests). No pathways reached the set significance threshold, but there were several nominally significant associations. The Telomeres, Telomerase, Cellular Aging, and Immortality pathway from the BioCarta dataset reached nominal significance in both males and females in all glioma and glioblastoma (Table 2). When the gene scores for the genes contained within this pathway were examined, the association with this pathway was driven primarily by strong associations in TERT and TP53 (Fig. 4). There were nominally significant associations in POLR2A (in both males and females) and PRKCA (in males only), both genes that have not been significantly associated with glioma to date. Further interrogation of the single-SNP results for these genes found no associations significant at the P < 5 × 10−4 level in either sex or histology group.
Table 2.
Significant pathways (P < 0.001 in any testing group) by sex and histology
| Pathway (database) | Histology | Overall | Conditioned on Previous GWAS Hits | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Males | Females | Males | Females | ||||||
| Discovery | Validation | Discovery | Validation | Discovery | Validation | Discovery | Validation | ||
| Telomeres, Telomerase, Cellular Aging, and Immortality (BioCarta) | All glioma | 5.32 × 10−5 | 6.50 × 10−4 | 2.61 × 10−4 | 0.0018 | 0.3651 | 0.1046 | 0.2733 | 0.1050 |
| Glioblastoma | 5.90 × 10−5 | 8.60 × 10−4 | 8.30 × 10−4 | 0.0041 | 0.5315 | 0.4286 | 0.6885 | 0.1804 | |
| Bladder cancer (KEGG) | All glioma | 9.00 × 10−5 | 0.0013 | 0.0306 | 0.0038 | 0.5775 | 0.0514 | 0.8096 | 0.0548 |
| Glioblastoma | 1.27 × 10−4 | 5.50 × 10−4 | 0.0045 | 0.0030 | 0.4097 | 0.0679 | 0.7187 | 0.0394 | |
| Glioma (KEGG) | All glioma | 5.80 × 10−4 | 0.0057 | 0.0361 | 5.00 × 10−4 | 0.5595 | 0.1241 | 0.5701 | 0.0045 |
| Glioblastoma | 0.0011 | 0.0061 | 0.0048 | 0.0018 | 0.4818 | 0.2616 | 0.3783 | 0.0132 | |
| Melanoma (KEGG) | All glioma | 7.60 × 10−4 | 0.0032 | 0.0219 | 1.70 × 10−4 | 0.7131 | 0.0650 | 0.5803 | 0.0016 |
| Glioblastoma | 8.70 × 10−4 | 0.0020 | 0.0013 | 7.60 × 10−4 | 0.5468 | 0.0934 | 0.2391 | 0.0054 | |
| Non–small cell lung cancer (KEGG) | All glioma | 7.30 × 10−4 | 0.0171 | 0.0290 | 7.40 × 10−4 | 0.7866 | 0.4067 | 0.6097 | 0.0067 |
| Glioblastoma | 0.0013 | 0.0455 | 0.0011 | 0.0019 | 0.6823 | 0.8232 | 0.2093 | 0.0174 | |
| Pancreatic cancer (KEGG) | All glioma | 2.65 × 10−4 | 0.0132 | 0.0021 | 0.0016 | 0.3903 | 0.2530 | 0.0957 | 0.0133 |
| Glioblastoma | 0.0021 | 0.1124 | 2.01 × 10−4 | 9.10 × 10−4 | 0.6711 | 0.9026 | 0.0413 | 0.0060 | |
Fig. 4.
Gene scores for genes in the BioCarta telomere pathway for all glioma in (A) males and (B) females, and for glioblastoma in (C) males and (D) females.
Nominally significant associations were identified in 5 cancer-specific KEGG pathways: bladder cancer, glioma (Supplementary Fig. 6), melanoma (Supplementary Fig. 7), non–small cell lung cancer, and pancreatic cancer (Table 2). There is significant overlap between these gene sets (Supplementary Fig. 8), and when the gene scores used to build each pathway were examined, all the associations appear to be driven largely by strong associations in EGFR and CDKN2A, which are members of all KEGG cancer pathways found to be nominally associated with glioma in this analysis. Pathway analyses were run using single-SNP results, including conditional analyses for all SNPs within a 2 mega base window around the previously identified SNPs near TERT, EGFR, CDKN2B, TP53, and RTEL1. No pathway association reached the significance threshold when analysis included conditioned results (Table 2).
Discussion
This represents the first genome-wide sex-specific gene- or pathway-based analysis for germline risk variants in glioma. Gene-based tests are an efficient way to increase power to detect associations of low effect size, where multiple variants within a region may contribute to increased risk. Multimarker tests, such as gene- or pathway-based tests, allow investigators to leverage previously existing GWAS summary statistics for discovery as well as to increase power when strength of association for single-SNP associations may be low. Incidence of glioma is significantly higher in males compared with females, and currently identified environmental risk factors do not explain this variation in incidence.1 A previous sex-specific GWAS analysis identified 3 loci with sex-specific effects, including a previously identified SNP in 7p11.2 (rs11979158, proximate to EGFR).11 As a primary goal, this analysis aimed to compare existing gene- and pathway-based methods in the context of heritable genetic risk for glioma, and additionally to explore additional potential sources of genetic risk that may contribute to sex differences in genetic risk for glioma. All autosomal genes identified by this analysis were proximate to previously identified GWAS hits. After conditioning on previously identified SNPs, associations at TERT, EGFR, and RTEL1 remained nominally significant. The results of this conditional analysis suggest that there are remaining sources of genetic risk for glioma within these regions, including one apparent region in TERT (Supplementary Fig. 3), with a single-SNP association that approaches genome-wide significance in both sexes (Supplementary Fig. 5). There were no differences by sex in effect size and direction, which suggests that while there may be remaining genetic risk associations to be detected within these genes, they do not have sex specificity.
Four regions on the X chromosome (Xp22.2, Xp21.2-p21.1, Xq22.1, and Xq28) contained genes that reached the significance threshold in at least 2 of 3 algorithms (Table 1). These genes have not been previously associated with glioma. SNPs near SHROOM2 (Xp22.2) were previously associated with prostate and colon cancer.38–40 There are no known associations with inherited variants in the other 3 regions and increased risk for cancer, though all contain genes that have been shown to be dysregulated in some cancer cells (eg, DMD, ARMCX2, ZNF185).41–45 Without a validation set, it is not possible to know if these are true associations or the result of type 1 error. Further exploration of these genes is necessary to determine their true relationship with glioma risk.
The BioCarta Telomeres, Telomerase, Cellular Aging, and Immortality pathway reached nominal significance in both males and females in all glioma and glioblastoma (Table 2). This pathway contains EGFR, TERT, and TP53, all of which contain SNPs identified by glioma GWAS. Inherited variants affecting telomere length have been associated with many complex diseases, including glioma.46–48 Both age and sex are known to affect telomere length, and previous research has suggested that males have shorter telomeres and higher rates of telomere attrition with aging.49,50 An analysis comparing a weighted genetic score based on 8 SNPs associated with leukocyte telomere length found that telomere length was ~5% longer in glioma cases versus controls.51 The significance of the telomere maintenance pathway may explain the remaining significant association in the regions surrounding TERT, EGFR, and RTEL1, as any variants affecting telomere length could contribute to glioma risk. In addition to the strong associations in genes associated with SNPs previously identified by GWAS, there were nominally significant associations in POLR2A (in both males and females) and PRKCA (in males only).
The numerous KEGG cancer pathways found to be significant in this analysis are likely due to the strength of association in genes (CDKN2A and EGFR) that are members of all identified cancer-specific KEGG pathways. While these associations are driven by strong associations in these specific genes, they may also be evidence of shared sources of genetic risk between these cancers and glioma. Both the glioma and melanoma pathways, driven by strong associations in CDKN2A, were significantly associated with all glioma in males (Supplementary Figs. 6–7). Previous analyses suggested an association between genetic risk for glioma and melanoma, in terms of known cancer syndromes (most notably melanoma-neural system tumor syndrome, caused by inherited variants in CDKN2A3), familial glioma, and sporadic disease.52–54 Persons with a previous diagnosis of melanoma are estimated to have incidence of glioma that is 1.42 times that of the general population, while relatives of glioma patients are diagnosed with melanoma approximately 2–4 times as frequently as the general population.52–54 Melanoma GWAS to date have identified 21 genetic risk loci,55,56 including SNPs near CDKN2A and TERT, genes that have also been associated with glioma.6 The identified SNPs in these genes do not account for a large proportion of risk in either melanoma or glioma, but there is evidence that innate telomere length and variation in telomere maintenance pathways may contribute to risk in both diseases.57 When pathway analyses were re-run using single-SNP results conditioned on known GWAS hits, pathway associations no longer reached the significance threshold. Gene-specific P-values for TERT, EGFR, and RTEL1 were lowest for conditional analyses performed in Pascal, the algorithm used to calculate pathway scores compared with the other 2 algorithms. The SOCS approach used by Pascal may be more conservative than others if there are many genes with null association and few genes with significant associations. Other pathway scoring algorithms that are more sensitive to a smaller set of strong associations may be more sensitive in identifying pathway associations.
All genetic association tests require consideration of the implicit assumptions about the genetic architecture of the disease and population of interest. GWAS approaches have attempted to identify single variants that have a causal relationship with a phenotype, which requires that this variant occur repeatedly within the study population. Gene- and pathway-based tests assume that the aggregate effect of variants within a gene or pathway affect disease risk, but do not require that all individuals possess the same variant. These approaches are most appropriate for complex diseases where risk for disease is polygenic. In contrast to the logistic regression methods utilized by GWAS, the 3 analytic approaches used here do not generate measures of the magnitude of association. These methods test for enrichment of associations at single SNPs within genes, without consideration of the magnitude or direction of association. Further analysis of the identified regions is necessary to estimate the level of association with glioma.
While multimarker tests can increase power to detect associations compared with single-SNP tests, different methods may be better suited to particular types of genetic architecture. Methods vary in their performance based on whether a gene has one strong signal versus multiple signals of lower significance. One of the genes (EGFR) identified by this analysis is known to have at least 2 independent GWAS signals,5 and as a result, its identification in some methods may be affected by this bias.
There is a well-known bias in GWAS toward large genes,58 which are often enriched for tag SNPs, and this bias may influence the results of this analysis. All of the algorithms used for this analysis can be affected by gene size. Large genes with many SNPs of minimal significance and few SNPs of large effect may “dilute” the gene score in methods based on summed scores, such as Pascal. All 3 of the algorithms used for this analysis “prune” SNPs based on linkage disequilibrium statistics in attempts to obtain a set of independent SNPs. For large genes that contain multiple haplotype blocks, results may still be biased toward large genes. This analysis used a relatively large window surrounding the defined genes (±50 kb) which may further bias analyses toward large genes. While estimates of average gene size range 10–15 kb, the average size for genes included in the annotation file for this analysis was 43.32 kb. The 3 major genes identified by this analysis range in size, but all are larger than the estimated average: TERT (29.84 kb), EGFR (137.92 kb), and RTEL-TNFRSF6B (39.8 kb). Genes that did not remain significant after conditioning tended to be smaller: BPESC1 (20.98 kb), SLC6A18 (20.84 kb), CDKN2B (6.4 kb), DNAH2 (26.14 kb), and STMN3 (13.06 kb). As a result, it is possible that these methods may be biased toward identifying smaller amounts of remaining signal in larger genes compared with smaller genes. This analysis utilized a 50 kb window surrounding a gene, and it is likely that changing this window may change the identified associations. These methods will also fail to identify any associations in intergenic regions, particularly the region at 8q24.21 that has previously been identified as having a sex-specific association.11
Patterns of linkage disequilibrium within the study population may also significantly affect the performance of a method. Results for methods that use LD information, including all algorithms evaluated in this analysis, may also be significantly altered by the reference populations to estimate LD. All of the included methods attempt to adjust for potential score inflation due to LD, using the 1000 EUR super population as a reference set. FAST does this by pruning SNPs that are in complete linkage (r2 = 1), while Pascal does this by generating “fusion” gene scores for genes that are in linkage with each other. These “fusion” genes are used along with single-gene scores to generate pathway scores to decrease inflation of P-values due to the physical proximity of genes.21 Due to variations in adjustment for LD used in the 2 programs, the number of included SNPs by each gene varied slightly. Both methods require that the identifier for each variant in the summary statistics be present in the LD reference file, and as a result these methods are not able to incorporate variants that do not have a standard reference SNP cluster ID. FAST additionally limits the dataset by requiring that all markers be biallelic SNPs, and does not accept indels.
Different multimarker approaches may also perform better than others based on the computational resources available for an analysis. Permutation-based tests are more computationally intensive compared with parametric tests, especially when gene scores are calculated genome-wide. Neither Pascal nor GATES relies on permutations for estimating P-values, which significantly decreases analysis time. BimBam uses permutations to calculate exact P-values; as a result, these analyses require more time to complete. The number of permutations used to calculate determines the boundaries for an exact P-value (ranging from 1 to 1/n, where n is the number of permutations), which may result in increasing permutations for increased P-value specificity. For more stringent P-value cutoffs, such as when testing multiple phenotypes in multiple groups, the number of permutations required may substantially increase analysis time. While multimarker tests do substantially decrease the multiple testing burden compared with genome-wide single SNP approaches, it is still important to consider multiple testing when conducting these tests. In cases such as this analysis, where multiple phenotypes are being tested within population strata, multiple testing correction strategies should be used. The P-value threshold used for this analysis is adjusted only for the number of genes/pathways within the testing phase. Use of more stringent testing cutoffs may result in prioritization of fewer genes. Use of a more stringent threshold in the testing phase of P < 7.8 × 10−7 (based on a Bonferroni correction for 64000 tests, for 16000 genes in 2 phenotypes and 2 sexes) would result in DNAH2 not reaching the set significance threshold in males, and BPESC1 and STMN3 not reaching the significance threshold in females (Supplementary Tables 2 and 3). A Bonferroni correction is known to be conservative, and use of these very stringent cutoffs may result in rejection of “real” associations. Use of 2-stage testing and a validation stage provides an additional safeguard against type 1 error.
In addition to the technical limitations of the 3 algorithms utilized for this analysis, there are several limitations. All glioma cases from the included 4 GWAS datasets were recruited at time of first diagnosis, and the assigned diagnoses represent the primary tumor type according to the prevailing histologic criteria at that time. There may also be variation in the histologies contained within each set by sex. The proportion of each dataset that is composed of glioblastoma compared with lower-grade gliomas varies by both study and sex (Supplementary Table 1). Less than 50% of female glioma cases in the testing set are glioblastoma, whereas over 50% of female cases are glioblastoma in the validation sets. Glioma is a heterogonous disease, and due to all of these factors, it is likely that heterogeneity exists between the utilized datasets.
Conclusions
Multimarker tests, such as gene- or pathway-based tests, allow investigators to leverage previously existing summary statistics and increase power when strength of single-SNP associations may be low. This analysis aimed to explore additional potential sources of genetic risk that may contribute to sex differences in genetic risk for glioma. There was a nominally significant association between germline variants in RTEL1 in both males and females after conditioning on previously identified SNPs. A significant association was detected between germline variants in the telomere maintenance pathway in both males and females, which builds on previous evidence of the relationship between inherited variants related to increased telomere length and increased risk for glioma. There was also a male-specific association in EGFR, and a female-specific association in TERT that remained nominally significant after conditioning on previous GWAS hits. The results of this analysis confirm previously known information about inherited glioma risk and provide potential mechanistic explanations for how these variants may affect the process of gliomagenesis.
Funding
Q.T.O. is supported by a Research Training Grant from the Cancer Prevention and Research Institute of Texas (CPRIT; RP160097T). J.W.C. was supported by the Grant S. Roth Memorial Fund. W.H. was supported by the Young Scientist Summer Research Program. The GICC was supported by grants from the National Institutes of Health, Bethesda, Maryland (R01CA139020, R01CA52689, P50097257, and P30CA125123). Additional support was provided by the McNair Medical Institute and the Population Sciences Biorepository at Baylor College of Medicine.
In Sweden work was additionally supported by Acta Oncologica through the Royal Swedish Academy of Science (BM salary) and the Swedish Research Council and Swedish Cancer Foundation.
The UCSF Adult Glioma Study was supported by the National Institutes of Health (grant numbers R01CA52689, P50CA097257, R01CA126831, and R01CA139020), the Loglio Collective, the National Brain Tumor Foundation, the Stanley D. Lewis and Virginia S. Lewis Endowed Chair in Brain Tumor Research, the Robert Magnin Newman Endowed Chair in Neuro-oncology, and by donations from families and friends of John Berardi, Helen Glaser, Elvera Olsen, Raymond E. Cooper, and William Martinusen. This project also was supported by the National center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through UCSF-CTSI grant number UL1 RR024131. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. The collection of cancer incidence data used in this study was supported by the California Department of Public Health as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885; the National Cancer Institute’s Surveillance, Epidemiology and End Results Program under contract HHSN261201000140C awarded to the Cancer Prevention Institute of California, contract HHSN261201000035C awarded to the University of Southern California, and contract HHSN261201000034C awarded to the Public Health Institute; and the Centers for Disease Control and Prevention’s National Program of Cancer Registries, under agreement #U58DP003862-01 awarded to the California Department of Public Health.
Supplementary Material
Acknowledgments
The results of this study were previously presented at the 2017 Annual Meeting of the Society for Neuro-Oncology. A pre-publication version of this manuscript was previously made available on bioRxiv (doi: https://doi.org/10.1101/235408).
Other significant contributors for the UCSF Adult Glioma Study include: M Berger, P Bracci, S Chang, J Clarke, A Molinaro, A Perry, M Pezmecki, M Prados, I Smirnov, T Tihan, K Walsh, J Wiemels, S Zheng. GliomaScan group comprised: Laura E. Beane Freeman, Stella Koutros, Demetrius Albanes, Kala Visvanathan, Victoria L. Stevens, Roger Henriksson, Dominique S. Michaud, Maria Feychting, Anders Ahlbom, Graham G. Giles Roger Milne, Roberta McKean-Cowdin, Loic Le Marchand, Meir Stampfer, Avima M. Ruder, Tania Carreon, Goran Hallmans, Anne Zeleniuch-Jacquotte, J. Michael Gaziano, Howard D. Sesso, Mark P. Purdue, Emily White, Ulrike Peters, Julie Buring.
We are grateful to all the patients and individuals for their participation and we would also like to thank the clinicians and other hospital staff, cancer registries, and study staff in respective centers who contributed to the blood sample and data collection.
The ideas and opinions expressed herein are those of the author(s) Endorsement by the State of California Department of Public Health, the National Cancer Institute, and the Centers for Disease Control and Prevention or their contractors and subcontractors is not intended nor should be inferred. UK10K data generation and access was organized by the UK10K consortium and funded by the Wellcome Trust.
Conflict of interest statement. None declared.
Contributor Information
GliomaScan consortium:
M Berger, P Bracci, S Chang, J Clarke, A Molinaro, A Perry, M Pezmecki, M Prados, I Smirnov, T Tihan, K Walsh, J Wiemels, S Zheng, Laura E Beane Freeman, Stella Koutros, Demetrius Albanes, Kala Visvanathan, Victoria L Stevens, Roger Henriksson, Dominique S Michaud, Maria Feychting, Anders Ahlbom, Graham G Giles Roger Milne, Roberta McKean-Cowdin, Loic Le Marchand, Meir Stampfer, Avima M Ruder, Tania Carreon, Goran Hallmans, Anne Zeleniuch-Jacquotte, J Michael Gaziano, Howard D Sesso, Mark P Purdue, Emily White, Ulrike Peters, and Julie Buring
References
- 1. Ostrom QT, Gittleman H, Liao P, et al. . CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro Oncol. 2017; 19(suppl 5):v1–v88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amirian ES, Zhou R, Wrensch MR, et al. . Approaching a scientific consensus on the association between allergies and glioma risk: a report from the Glioma International Case-Control study. Cancer Epidemiol Biomarkers Prev. 2016;25(2):282–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ostrom QT, Bauchet L, Davis FG, et al. . The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. 2014;16(7):896–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kinnersley B, Mitchell JS, Gousias K, et al. . Quantifying the heritability of glioma using genome-wide complex trait analysis. Sci Rep. 2015;5:17267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Melin BS, Barnholtz-Sloan JS, Wrensch MR, et al. ; GliomaScan Consortium Genome-wide association study of glioma subtypes identifies specific differences in genetic susceptibility to glioblastoma and non-glioblastoma tumors. Nat Genet. 2017;49(5):789–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang K, Li M, Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am J Hum Genet. 2007;81(6):1278–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reinius B, Saetre P, Leonard JA, et al. . An evolutionarily conserved sexual signature in the primate brain. PLoS Genet. 2008;4(6):e1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rinn JL, Snyder M. Sexual dimorphism in mammalian gene expression. Trends Genet. 2005;21(5):298–305. [DOI] [PubMed] [Google Scholar]
- 9. Ellegren H, Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet. 2007;8(9):689–698. [DOI] [PubMed] [Google Scholar]
- 10. Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nat Rev Genet. 2015;16(4):197–212. [DOI] [PubMed] [Google Scholar]
- 11. Ostrom QT, Kinnersley B, Wrensch MR, et al. ; GliomaScan consortium Sex-specific glioma genome-wide association study identifies new risk locus at 3p21.31 in females, and finds sex-differences in risk at 8q24.21. Sci Rep. 2018;8(1):7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Warrington NM, Sun T, Luo J, et al. . The cyclic AMP pathway is a sex-specific modifier of glioma risk in type I neurofibromatosis patients. Cancer Res. 2015;75(1):16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Amirian ES, Armstrong GN, Zhou R, et al. . The Glioma International Case-Control study: a report from the genetic epidemiology of Glioma International Consortium. Am J Epidemiol. 2016;183(2):85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Melin BS, Barnholtz-Sloan JS, Wrensch MR, et al. ; GliomaScan Consortium Genome-wide association study of glioma subtypes identifies specific differences in genetic susceptibility to glioblastoma and non-glioblastoma tumors. Nat Genet. 2017;49(5):789–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wrensch M, Jenkins RB, Chang JS, et al. . Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41(8):905–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shete S, Hosking FJ, Robertson LB, et al. . Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41(8):899–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rajaraman P, Melin BS, Wang Z, et al. . Genome-wide association study of glioma and meta-analysis. Hum Genet. 2012;131(12):1877–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu JZ, Tozzi F, Waterworth DM, et al. ; Wellcome Trust Case Control Consortium Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42(5):436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39(7):906–913. [DOI] [PubMed] [Google Scholar]
- 20. Auton A, Brooks LD, Durbin RM, et al. ; 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lamparter D, Marbach D, Rueedi R, Kutalik Z, Bergmann S. Fast and rigorous computation of gene and pathway scores from SNP-based summary statistics. PLoS Comput Biol. 2016;12(1):e1004714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 2007;3(7):e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li MX, Gui HS, Kwan JS, Sham PC. GATES: a rapid and powerful gene-based association test using extended Simes procedure. Am J Hum Genet. 2011;88(3):283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu JZ, McRae AF, Nyholt DR, et al. ; AMFS Investigators A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87(1):139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chanda P, Huang H, Arking DE, Bader JS. Fast association tests for genes with FAST. PLoS One. 2013;8(7):e68585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. D’Eustachio P. Reactome knowledgebase of human biological pathways and processes. Methods Mol Biol. 2011; 694:49–61. [DOI] [PubMed] [Google Scholar]
- 28. Nishimura D. BioCarta. Biotech Software & Internet Report. 2001; 2(3):117–120. [Google Scholar]
- 29. Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Subramanian A, Tamayo P, Mootha VK, et al. . Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005; 102(43): 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. R Core Team. R: A language and environment for statistical computing. 2017; http://www.R-project.org/. [Google Scholar]
- 32. Wickham H. ggplot2: elegant graphics for data analysis. 2009; http://had.co.nz/ggplot2/book. [Google Scholar]
- 33. Csardi G, Nupusz T. The igraph software package for complex network research. InterJournal. 2006; Complex Systems:1695. [Google Scholar]
- 34. Briatte F. ggnetwork: Geometries to Plot Networks with ‘ggplot2’. R package version 0.5.1. 2016; https://CRAN.R-project.org/package=ggnetwork. [Google Scholar]
- 35. Sales G, Calura E, Romualdi C.. graphite: GRAPH Interaction from pathway Topological Environment. R package version 1.16.0. 2015; http://www.bioconductor.org/packages/release/bioc/html/graphite.html. [Google Scholar]
- 36. Auguie B. gridExtra: Miscellaneous Functions for “Grid” Graphics. R package version 2.3. 2017; https://CRAN.R-project.org/package=gridExtra. [Google Scholar]
- 37. Pruim RJ, Welch RP, Sanna S, et al. . LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Eeles RA, Olama AA, Benlloch S, et al. . Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet. 2013; 45(4):385–391, 391e381-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dunlop MG, Dobbins SE, Farrington SM, et al. ; Colorectal Tumour Gene Identification (CORGI) Consortium; Swedish Low-Risk Colorectal Cancer Study Group; COIN Collaborative Group Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk. Nat Genet. 2012;44(7):770–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Closa A, Cordero D, Sanz-Pamplona R, et al. . Identification of candidate susceptibility genes for colorectal cancer through eQTL analysis. Carcinogenesis. 2014;35(9):2039–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Darras BT, Miller DT, Urion DK.. Dystrophinopathies. In: Adam MP, Ardinger HH, Pagon RA, et al. , eds. Seattle (WA): GeneReviews(R); 1993. [Google Scholar]
- 42. Pantaleo MA, Astolfi A, Urbini M, et al. . Dystrophin deregulation is associated with tumor progression in KIT/PDGFRA mutant gastrointestinal stromal tumors. Clin Sarcoma Res. 2014;4:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang Y, Marino-Enriquez A, Bennett RR, et al. . Dystrophin is a tumor suppressor in human cancers with myogenic programs. Nat Genet. 2014;46(6):601–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hatzfeld M. The armadillo family of structural proteins. Int Rev Cytol. 1999;186:179–224. [DOI] [PubMed] [Google Scholar]
- 45. Kurochkin IV, Yonemitsu N, Funahashi SI, Nomura H. ALEX1, a novel human armadillo repeat protein that is expressed differentially in normal tissues and carcinomas. Biochem Biophys Res Commun. 2001;280(1):340–347. [DOI] [PubMed] [Google Scholar]
- 46. Codd V, Nelson CP, Albrecht E, et al. . Identification of seven loci affecting mean telomere length and their association with disease. Nat Genet. 2013; 45(4):422–427, 427e421-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Walsh KM, Codd V, Smirnov IV, et al. ; ENGAGE Consortium Telomere Group Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nat Genet. 2014;46(7):731–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Walsh KM, Wiencke JK, Lachance DH, et al. . Telomere maintenance and the etiology of adult glioma. Neuro Oncol. 2015;17(11):1445–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mayer S, Brüderlein S, Perner S, et al. . Sex-specific telomere length profiles and age-dependent erosion dynamics of individual chromosome arms in humans. Cytogenet Genome Res. 2006;112(3-4):194–201. [DOI] [PubMed] [Google Scholar]
- 50. Barrett JH, Iles MM, Dunning AM, Pooley KA. Telomere length and common disease: study design and analytical challenges. Hum Genet. 2015;134(7):679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Walsh KM, Codd V, Rice T, et al. ; ENGAGE Consortium Telomere Group Longer genotypically-estimated leukocyte telomere length is associated with increased adult glioma risk. Oncotarget. 2015;6(40):42468–42477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Scarbrough PM, Akushevich I, Wrensch M, Il’yasova D. Exploring the association between melanoma and glioma risks. Ann Epidemiol. 2014;24(6):469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Scheurer ME, Etzel CJ, Liu M, et al. ; GLIOGENE Consortium Familial aggregation of glioma: a pooled analysis. Am J Epidemiol. 2010;172(10):1099–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Paunu N, Pukkala E, Laippala P, et al. . Cancer incidence in families with multiple glioma patients. Int J Cancer. 2002;97(6):819–822. [DOI] [PubMed] [Google Scholar]
- 55. Ransohoff KJ, Wu W, Cho HG, et al. . Two-stage genome-wide association study identifies a novel susceptibility locus associated with melanoma. Oncotarget. 2017;8(11):17586–17592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kocarnik JM, Park SL, Han J, et al. . Replication of associations between GWAS SNPs and melanoma risk in the Population Architecture Using Genomics and Epidemiology (PAGE) study. J Invest Dermatol. 2014;134(7):2049–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Endicott AA, Taylor JW, Walsh KM. Telomere length connects melanoma and glioma predispositions. Aging (Albany NY). 2016;8(3):423–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mirina A, Atzmon G, Ye K, Bergman A. Gene size matters. PLoS One. 2012;7(11):e49093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.