Abstract
Background
A high heterogeneity and activation of multiple oncogenic pathways have been implicated in failure of targeted therapies in glioblastoma (GBM).
Methods
Using The Cancer Genome Atlas data, we identified subtype-specific prognostic core genes by a combined approach of genome-wide Cox regression and Gene Set Enrichment Analysis. The results were validated with 8 combined public datasets containing 608 GBMs. We further examined prognostic chromosome aberrations and mutations.
Results
In classical and mesenchymal subtypes, 2 receptor tyrosine kinases (RTKs) (MET and IGF1R), and the genes in RTK downstream pathways such as phosphatidylinositol-3 kinase (PI3K)/Akt/mammalian target of rapamycin (mTOR), and nuclear factor-kappaB (NF-kB), were commonly detected as prognostic core genes. Classical subtype-specific prognostic core genes included those in cell cycle, DNA repair, and the Janus kinase/signal transducers and activators of transcription (JAK-STAT) pathway. Immune-related genes were enriched in the prognostic genes showing negative promoter cytosine-phosphate-guanine (CpG) methylation/expression correlations. Mesenchymal subtype-specific prognostic genes were those related to mesenchymal cell movement, PI3K/Akt, mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK), Wnt/β-catenin, and Wnt/Ca2+ pathways. In copy number alterations and mutations, 6p loss and TP53 mutation were associated with poor and good survival, respectively, in the classical subtype. In the mesenchymal subtype, patients with PIK3R1 or PCLO mutations showed poor prognosis. In the glioma CpG island methylator phenotype (G-CIMP) subtype, patients harboring 10q loss, 12p gain, or 14q loss exhibited poor survival. Furthermore, 10q loss was significantly associated with the recently recognized G-CIMP subclass showing relatively low CpG methylation and poor prognosis.
Conclusion
These subtype-specific alterations have promising potentials as new prognostic biomarkers and therapeutic targets combined with surrogate markers of GBM subtypes. However, considering the small number of events, the results of copy number alterations and mutations require further validations.
Keywords: gene set, glioblastoma, heterogeneity, prognosis, subtype
Key Points
1. Cell cycle/repair/JAK-STAT pathways and TP53 mutation are prognostic in classical GBM.
2. PI3K/MAPK/Wnt pathways and PIK3R1/PCLO mutations are prognostic in mesenchymal GBM.
3. Losses of 6p and 10q are prognostic in classical and G-CIMP GBM, respectively.
Importance of the study
Despite recent advances in understanding molecular pathogenesis, GBM remains a dismal brain tumor with a high recurrence rate and remarkable resistance to current treatments. Targeted therapies have not proven substantial clinical benefit yet, and a high degree of genetic heterogeneity and activation of alternative pathways have been implicated in the failure. Hence, we identified subtype-specific prognostic signaling pathways, core genes, and genomic aberrations which have great potentials as specific molecular targets and biomarkers for risk stratification in each subtype of GBM. Our subtype-specific approach will provide insights into new strategies to overcome the heterogeneous nature of GBM and to eventually develop successful therapeutic interventions toward precision oncology.
Glioblastoma (GBM) is the most aggressive primary brain tumor in adults. Despite multimodal therapy, the overall prognosis remains dismal, largely due to almost universal recurrence. The high degree of heterogeneity and deregulation of multiple signaling pathways have been implicated in the aggressiveness of GBM and treatment failures.1,2 The aberrant signaling pathways underlying GBM pathogenesis include the receptor tyrosine kinase (RTK) pathway involving amplification or mutations of EGFR, ERBB2, PDGFRA, and MET, and other pathways, including p53/retinoblastoma (RB), phosphatidylinositol-3 kinase (PI3K)/Akt/mammalian target of rapamycin (mTOR), Ras/mitogen-activated protein kinase (MAPK), signal transducer and activator of transcription 3, Notch, Wnt, and nuclear factor-kappaB (NF-κB).2–12 Genomic and epigenomic studies have revealed that GBMs have pronounced genetic and molecular heterogeneity.13 Philips et al identified 3 transcriptional groups of high-grade glioma: proneural, proliferative, and mesenchymal.14 Similarly, comprehensive analysis of The Cancer Genome Atlas (TCGA) dataset revealed 4 subtypes: proneural, classical, mesenchymal, and neural. More recently, single-cell RNA-sequencing from primary GBMs revealed considerable intratumoral subtype heterogeneity.15 Furthermore, promoter DNA methylation profiling of TCGA GBM revealed that the proneural subtype included a distinct subset with promoter-associated cytosine-phosphate-guanine (CpG) island hypermethylation designated as a glioma-CpG island methylator phenotype (G-CIMP).16 The G-CIMP GBMs are more frequently found in secondary GBMs derived from low-grade gliomas or recurrent GBMs and are characterized by frequent mutations in IDH1, favorable prognosis, and younger age at diagnosis.
Considering the high degree of heterogeneity, complex dysregulated signaling pathways, and failure of targeted therapies in this dreadful disease,2,5,17 an approach starting with more homogeneous subsets will be a more powerful strategy to find new therapeutic possibilities and to evaluate therapeutic response efficiently. Accordingly, here we identified subtype-specific gene sets and pathways with prognostic implications. We further investigated prognostic chromosomal aberrations and mutations in each subtype of GBM.
Materials and Methods
Data Collection and Determination of Subtype
GBM expression profiles (Affymetrix HG-U133A), copy number data (Gistic2 Copy Number all thresholded by genes), and mutation data (somatic gene-level nonsilent mutation [broad]) were downloaded from TCGA (https://tcga.xenahubs.net/; Accessed 08 June 2018). DNA methylation data were downloaded from the Genomic Data Commons portal (https://portal.gdc.cancer.gov/; Accessed 08 June 2018). Sample information was obtained from 2 sources: clinical data from TCGA and a recent study.18 A total of 523 expression profiles from primary tumors with available overall survival (OS) were used. Gene symbols were updated according to the HUGO gene nomenclature committee (http://www.genenames.org/; Accessed 08 June 2018), and the final expression dataset included 11910 genes. Using 4 subtype variables (“G-CIMP Status,” “Original Subtype,” “Transcriptome Subtype,” and “Pan-glioma RNA Expression Cluster”),18,19 we selected only the samples with consistent subtype information (n = 395) (Supplementary Table 1). For validation, we collected 608 expression profiles with OS information from 8 public datasets generated by 2 platforms, Affymetrix HG-U133A or U133 plus2: GSE4412,20 GSE13041,21 GSE4271,14 Rich et al,22 GSE16011,23 GSE7696,24 E-TABM-898,25 and Repository of Molecular Brain Neoplasia Data (REMBRANDT) (Supplementary Table 2).26 The survival information of 3 datasets, GSE16011, E-TABM-898, and REMBRANDT, was obtained from GSE55981.27 Raw cel files with the same platform were preprocessed together using the RMA (robust multi-array average) algorithm.28 When multiple probe sets existed for a gene, the probe set with the largest interquartile range was selected. Using a similar procedure applied by Sandmann et al,29 the subtype of each validation sample was determined by correlation with TCGA samples based on 11634 gene expressions commonly available in both datasets. More specifically, 11634 average expression values across the samples of each TCGA subtype were compared with the expression values of each validation sample, then the subtype was defined as the same TCGA subtype showing the highest value of Pearson’s correlation coefficient (Supplementary Table 3).
Identification of Subtype-Specific Prognostic Gene Sets and Core Genes
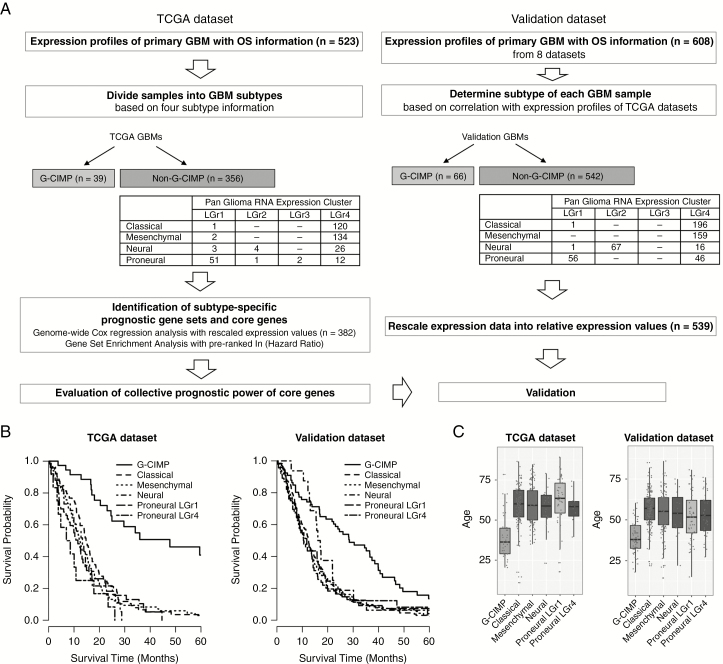
To identify subtype-specific prognostic gene sets, the prognostic power of 11910 individual genes was assessed in 6 major subtypes based on univariable Cox regression using relative expression values and OS (Fig. 1A). The TCGA GBMs included 382 samples in “Classical LGr4” (classical) (n = 120), “Mesenchymal LGr4” (mesenchymal) (n = 134), “Neural LGr4” (neural) (n = 26), “Proneural LGr4” (n = 12), “Proneural LGr1” (n = 51), and “G-CIMP” (n = 39). The relative expression values were calculated by rescaling the expression values of each gene to range from 0 to 1 across the 382 samples. Subsequently, using the pre-ranked coefficients from Cox regression, overrepresented prognostic gene sets and core genes were identified by Gene Set Enrichment Analysis (GSEA) with default settings.30 According to the positive normalized enrichment score (NES), the top 5 overrepresented gene sets were selected in 5 terms of Gene Ontology Biological Process (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), Hallmark, Pathway Interaction Database (PID), and BioCarta. A positive NES indicates that high expression of the genes is associated with poor prognosis.30 The collective prognostic power was assessed by Cox regression using the average relative expression values of the core genes in each gene set. For validation, 2 separately preprocessed expression profiles in 6 subtypes (n = 539) (Fig. 1A) were rescaled into relative expression values respectively, then combined into one dataset. The final validation dataset comprised 13093 genes common in 2 platforms. Using Cox regression, the collective prognostic power of each gene set was evaluated in the validation dataset.
Fig. 1.
Overview of the identification of subtype-specific prognostic gene sets and core genes. (A) Flowchart of data analysis. (B) Kaplan–Meier plots for overall survival. (C) Distribution of age at diagnosis.
Identification of Subtype-Specific Prognostic Chromosome Aberrations and Mutations
The prognostic chromosome aberrations and mutations were identified in 3 subtypes: classical, mesenchymal, and G-CIMP. Gain or loss of each chromosome arm was determined using TCGA gene-level copy number data including classical (n = 117), mesenchymal (n = 133), and G-CIMP (n = 39) (Supplementary Table 1). The copy number statuses of 24776 genes were thresholded into 5 categories: −2, −1, 0, 1, and 2.31 If the proportion of the genes with a negative or a positive threshold was greater than 50% in a specific chromosome arm, the copy number status of the arm was defined as a loss or a gain, respectively. Subsequently, log-rank tests were performed to assess the associations between OS and recurrent chromosome aberrations found in more than 5% of patients in each subtype. To detect prognostic mutations, the associations between OS and recurrent mutations found in more than 10% of patients in each subtype were estimated with log-rank tests in the classical (n = 55), mesenchymal (n = 60), and G-CIMP (n = 12) subtypes (Supplementary Table 1). All data analyses were performed using R statistical software (http://www. R-project.org/; Accessed 08 June 2018).
Results
The final sample numbers in 6 major subtypes were 382 and 539 in TCGA and the validation datasets, respectively (Fig. 1A). As expected, G-CIMP patients showed favorable prognoses (Fig. 1B) and younger ages at diagnosis (Fig. 1C).
Classical Subtype
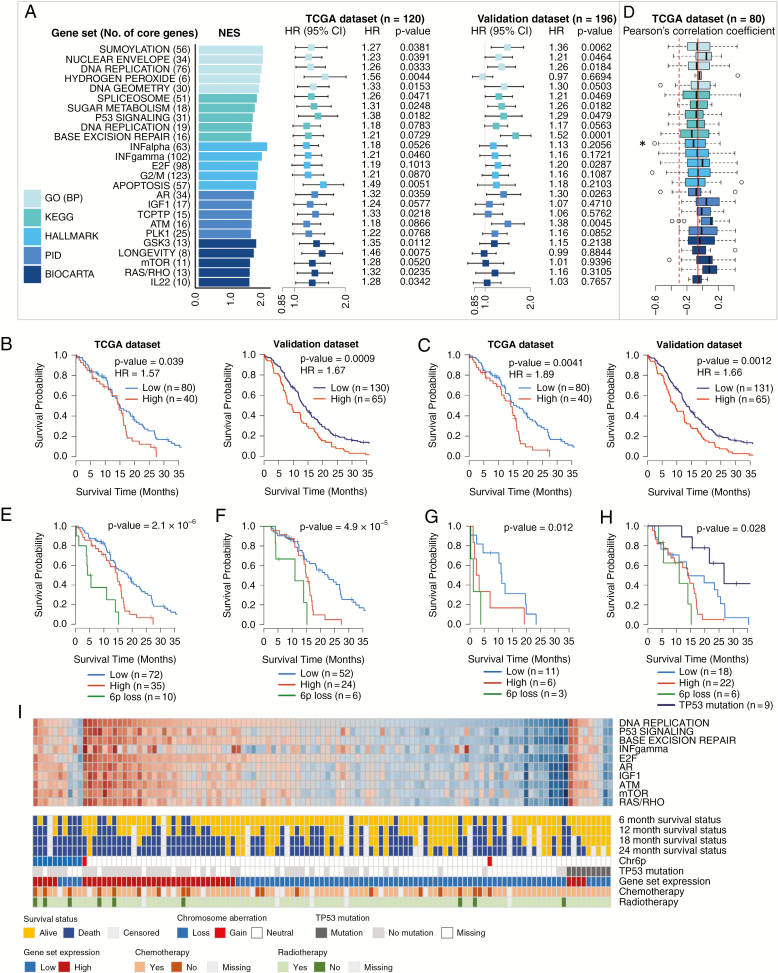
Among the top 25 prognostic gene sets, the prognostic power of core genes was validated in 10 gene sets (P < 0.05) (Fig. 2A and Supplementary Table 4), including 3 GO terms: “Sumoylation,” “Nuclear envelope,” and “DNA replication”; 4 KEGG pathways: “Spliceosome,” “Sugar metabolism,” “P53 signaling,” and “Base excision repair”; 1 Hallmark gene set: “E2F”; and 2 PID gene sets: “Androgen receptor” and “ATM.” Kaplan–Meier plots with log-rank tests using the average expression of 148 core genes in the top 3 GO gene sets revealed that the GBM patients with high average expression had poor survival (Fig. 2B). When the patients were divided into 2 or 3 groups according to the average expression of 76 core genes in the “DNA replication” gene set, the group with high expression showed significantly poor OS (Fig. 2C, Supplementary Fig. 1). Moreover, the 76 core gene expression remained a significant independent prognostic factor (adjusted hazard ratio [HR] = 1.80, P = 0.049) after adjustment for age at diagnosis, treatment (chemotherapy and radiotherapy [chemoRT]), and O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation status (Supplementary Table 5).32
Fig. 2.
Identification and validation of prognostic gene sets, chromosome aberrations, and mutations in the classical subtype. (A) Top 5 prognostic gene sets in 5 terms and forest plots of HRs per 0.1 increment of average relative expression with 95% CIs from Cox regression. (B) Kaplan–Meier plots with log-rank tests of high/low expression groups divided by the upper tertile cutoff (66.7%) of the average expression of 148 core genes in the top 3 GO gene sets. (C) Kaplan–Meier plots with log-rank tests of high/low expression groups divided by the upper tertile cutoff of the average expression of 76 core genes in the “DNA replication” gene set. (D) Boxplots of Pearson’s correlation coefficients calculated with expression profiles and β-values of CpG methylation probe of core genes. (E) Kaplan–Meier plots with log-rank tests of 3 groups: the 6p loss group and 2 expression groups defined in (C) without 6p loss. (F) Kaplan–Meier plots with log-rank tests of 3 groups defined in (E) but confined to the patients with chemotherapy and radiotherapy. (G) Kaplan–Meier plots with log-rank tests of 3 groups defined in (E) but confined to the patients without either chemotherapy or radiotherapy. (H) Kaplan–Meier plots with log-rank tests of 4 groups, a group with TP53 mutation and 3 groups defined in (E). (I) Heatmap of the average expression of core genes in 10 representative gene sets with 6- to 24-month survival statuses, 6p status (Chr6p), TP53 mutations, expression categories defined in (C), and treatment information. *P < 0.05.
To investigate whether expression changes of the prognostic core genes were associated with promoter CpG methylation, Pearson’s correlation coefficients were calculated with expression profiles and β-values of methylation probes in gene promoters (TSS1500, TSS200, 5ʹ untranslated region, and first exon). The methylation data were measured on either Illumina Infinium 27K or 450K; therefore, we analyzed only the CpG probes present on both platforms. For a given gene, only the probe with the highest negative correlation coefficient was selected. Expressions of the genes in the “INFalpha” gene set were associated with promoter CpG methylation features showing significantly different distribution of correlation coefficients toward negative relationships compared with those of the total 521 genes detected in the classical subtype (Fig. 2D, Supplementary Table 6). The genes involved in immune response (OAS2, OASL, DHX58, BST2, IFIT3, and IFITM3), DNA damage (ATRX, BRCA1, MGMT, and SMUG1), and cell cycle/cancer pathways (TGFB1, MNAT1, SMC4, and TFDP1) showed Pearson’s correlation coefficient ≤ −0.3 and changes in β-values across samples (|Δβ|) ≥ 0.25 (Supplementary Table 6).
In copy number data analysis, losses of 6p, 11p, or 12q were significantly associated with poor prognosis in classical GBM (Supplementary Table 7). However, 11p loss and 12q loss were no longer statistically significant when 6p loss was adjusted in multivariable Cox regressions. Additionally, the patients with 11p or 12q loss but not accompanied by 6p loss did not show significantly different survival from those without 6p, 11p, and 12q loss (data not shown). Accordingly, we considered that 6p loss was the main prognostic chromosomal aberration in the classical subtype. The 6p loss was detected in 10 (8.5%) of 117 patients. When the classical GBMs were divided into 3 groups according to 6p loss and the average expression level of 76 core genes in the “DNA replication” gene set, the survival curves were distinctly separated (Fig. 2E). To examine the influence of treatment, the analysis was repeated in 2 separated groups: one received chemoRT and the other did not receive either or all of chemoRT. The survival curves in both groups were similarly separated, although more favorable outcomes were generally observed in the chemoRT group (Fig. 2F and G). Finally, we investigated the association between OS and nonsilent somatic mutations. The most recurrent mutation was in EGFR; however, the EGFR mutation was not significantly associated with prognosis (Supplementary Table 8). TP53 mutation was significantly associated with prognosis (P = 0.0033, log-rank test) in a protective direction (HR = 0.24), indicating that the patients with mutated TP53 had better prognosis (Fig. 2H). Nine of 55 patients (16%) harbored TP53 mutations, including missense (n = 8) or splice site (n = 1) mutations. In Fig. 2I, the heatmap of average expressions of core genes in 10 representative prognostic gene sets was presented with 6- to 24-month survival statuses, copy number status of 6p, TP53 mutation, and high/low level of average expression of 76 core genes in the “DNA replication” gene set. The heatmap clearly showed that the average expressions of the 10 gene sets were highly correlated, which was similarly observed in the validation dataset (Supplementary Fig. 2A).
Mesenchymal Subtype
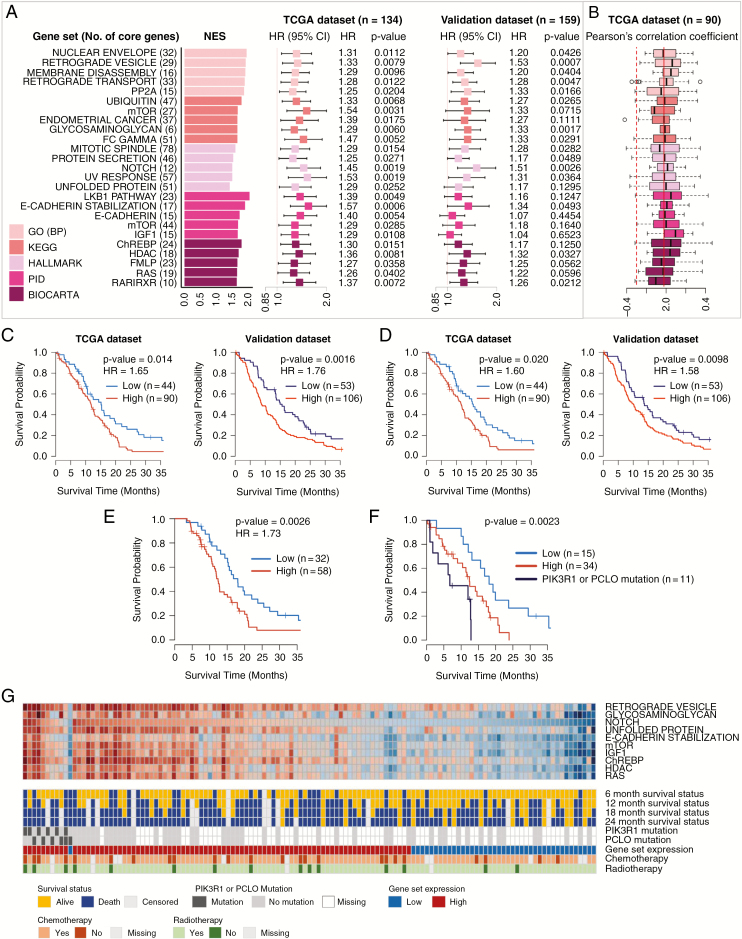
In the mesenchymal subtype, the prognostic power of 15 of the top 25 gene sets was confirmed in the validation dataset, including gene sets of GO “Retrograde vesicle,” KEGG “Glycosaminoglycan,” Hallmark “Notch,” PID “E-cadherin stabilization,” and BioCarta “HDAC” (Fig. 3A and Supplementary Table 4). In CpG methylation analysis, no particular gene set showed a significantly negative distribution of correlation coefficients (Fig. 3B). However, 2 cell cycle–related genes, EGF and PPP2R2B, showed a negative correlation with |Δβ| ≥ 0.25 (Supplementary Table 6). High average expressions of 61 core genes in the top 3 GO gene sets were significantly associated with poor OS in both TCGA and validation datasets (Fig. 3C). Only 12 core genes in the “Notch” gene set were also prognostic (Fig. 3D, Supplementary Fig. 3), which was also obvious in the patients with chemoRT (Fig. 3E). The “Notch” core genes remained an independent prognostic factor (adjusted HR = 2.15, P = 0.011) after adjustment for age at diagnosis, chemoRT, and MGMT promoter methylation status (Supplementary Table 5). No recurrent chromosome aberration was prognostic in the mesenchymal subtype; however, 2 nonsilent somatic mutations, PIK3R1 and PCLO, were associated with poor outcome (Fig. 3F and Supplementary Table 8 ). The average expressions of the core genes were highly correlated with one another in both TCGA and validation datasets (Fig. 3G and Supplementary Fig. 2B).
Fig. 3.
Identification and validation of prognostic gene sets, chromosome aberrations, and mutations in the mesenchymal subtype. (A) Top 5 prognostic gene sets in 5 terms and forest plots of HRs per 0.1 increment of average relative expression with 95% CIs from Cox regression. (B) Boxplots of Pearson’s correlation coefficients calculated expression profiles and β-values of CpG methylation probe of the core genes. (C) Kaplan–Meier plots with log-rank tests of high/low expression groups divided by the lower tertile cutoff (33.3%) of the average expression of 61 core genes in the top 3 GO gene sets. (D) Kaplan–Meier plots with log-rank tests of high/low expression groups divided by the lower tertile cutoff of the average expression of 12 core genes in the “Notch” gene set. (E) Kaplan–Meier plots with log-rank tests of 3 groups defined in (D) but confined to the patients with chemotherapy and radiotherapy. (E) Kaplan–Meier plots with log-rank tests of 3 groups, a group with PIK3R1 or PCLO mutation and 2 groups defined in (D). (G) Heatmap of the average expression of core genes in 10 representative gene sets with 6- to 24-month survival statuses, PIK3R1 and PCLO mutations, expression categories defined in (D), and treatment information.
G-CIMP Subtype
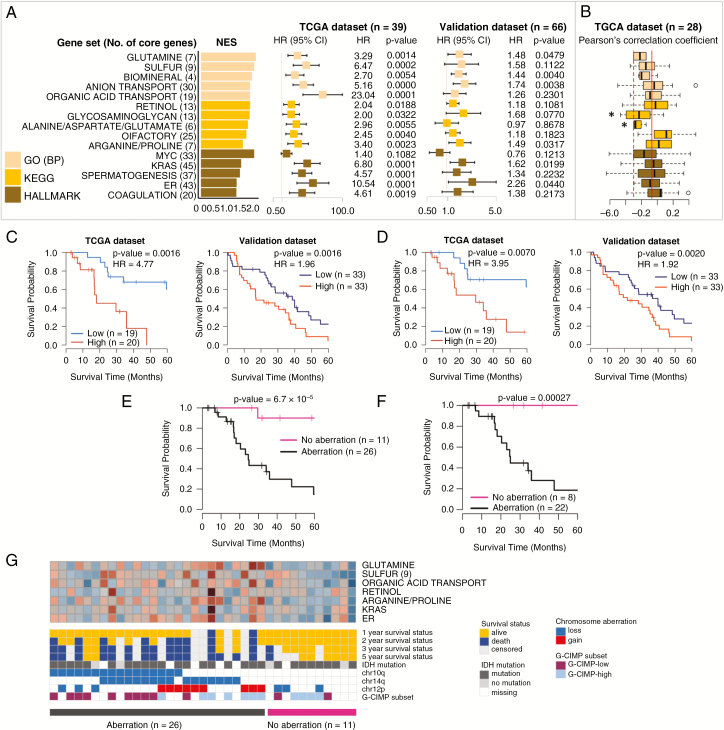
Many of the top 15 overrepresented prognostic gene sets in the G-CIMP subtype were related to cellular metabolism or transport (Fig. 4A and Supplementary Table 4). No significantly overrepresented gene set was found in PID or BioCarta. Six of 15 gene sets were confirmed in the validation dataset. In the G-CIMP subtype, the genes involved in “Glycosaminoglycan and Alanine/aspartate/glutamate” showed a significantly negative distribution of correlation coefficients between expression and CpG methylation (Fig. 4B). High average expression of 20 core genes from the top 3 GO gene sets was associated with poor prognosis (Fig. 4C), which was also substantiated with 9 core genes in the “Sulfur” gene set (Fig. 4D). Twelve chromosomal aberrations were prognostic toward an increase of risk (Supplementary Table 7). However, simultaneous aberrations were found in many patients. Additionally, 26 of 37 patients harbored at least one of the 12 risk chromosomal aberrations. The 26 patients possessed at least one of 3 of the most recurrent aberrations, 10q loss, 14q loss, and 12p gain. Furthermore, the 3 aberrations were independent prognostic factors when all 12 aberrations were added to multivariable Cox regression (P < 0.05). The G-CIMP GBMs showed differential survival curves according to presence or absence of the 3 aberrations (Fig. 4E), which was also observed in the patients with chemoRT (Fig. 4F). Fig. 4G also shows that the patients without 10q loss, 14q loss, and 12p gain had favorable outcomes. The most recurrently mutated genes in G-CIMP were TP53, IDH1, and ATRX (Supplementary Table 8). However, none was associated with prognosis.
Fig. 4.
Identification and validation of prognostic gene sets and chromosome aberrations in the G-CIMP subtype. (A) Top 5 prognostic gene sets in 3 terms and forest plots of HRs per 0.1 increment of average relative expression with 95% CIs from Cox regression. (B) Boxplots of Pearson’s correlation coefficients calculated between expression profiles and β-values of the CpG methylation probe of the core genes. (C) Kaplan–Meier plots with log-rank tests of high/low expression groups divided by the median average expression of 20 core genes in the top 3 GO gene sets. (D) Kaplan–Meier plots with log-rank tests of high/low expression groups divided by the median average expression of 9 core genes in the “Sulfur” gene set. (E) Kaplan–Meier plots with log-rank tests of 2 groups divided by the presence or absence of 10q loss, 14q loss, and 12p gain. (F) Kaplan–Meier plots with log-rank tests of 2 groups defined in (E) but confined to the patients with chemotherapy and radiotherapy. (G) Heatmap of the average expression of core genes in 7 representative gene sets with 1–3 and 5-year survival statuses, chromosome aberrations, IDH1 mutation, and G-CIMP subsets divided by methylation status. *P < 0.05.
Proneural and Neural Subtypes
Proneural GBM had been known to be associated with a favorable prognosis.14,19 However, this was attributed to a subset of G-CIMP.16 Congruently, after excluding G-CIMP, the proneural subtype did not show better OS (Fig. 1B). In TCGA pan-glioma analysis, proneural GBMs without IDH1 mutation (non–G-CIMP proneural) were clustered into 2 transcriptomic groups, LGr1 and LGr4, while most GBMs in other subtypes were clustered into one group, LGr4.18 This suggests that proneural GBM is heterogeneous even after excluding G-CIMP. Accordingly, we identified prognostic pathways in 2 groups, proneural LGr1 and LGr4, separately. Several prognostic gene sets were identified as common for the 2 groups, such as Hallmark “Genes defining epithelial-mesenchymal transition,” indicating that proneural GBMs with more mesenchymal-like features had poor prognosis. However, most of the results were not confirmed in the validation dataset (Supplementary Table 4). When the analysis was repeated with the combined proneural GBMs (LGr1 and LGr4), the results were not validated. Similarly, in the neural subtype, most of the prognostic gene sets were not validated (Supplementary Table 4). Moreover, the neural subtype was recently suspected to be normal brain contamination.33 Considering the poor validation, heterogeneity, and possible normal contamination, we did not perform further analysis for these 2 subtypes.
Discussion
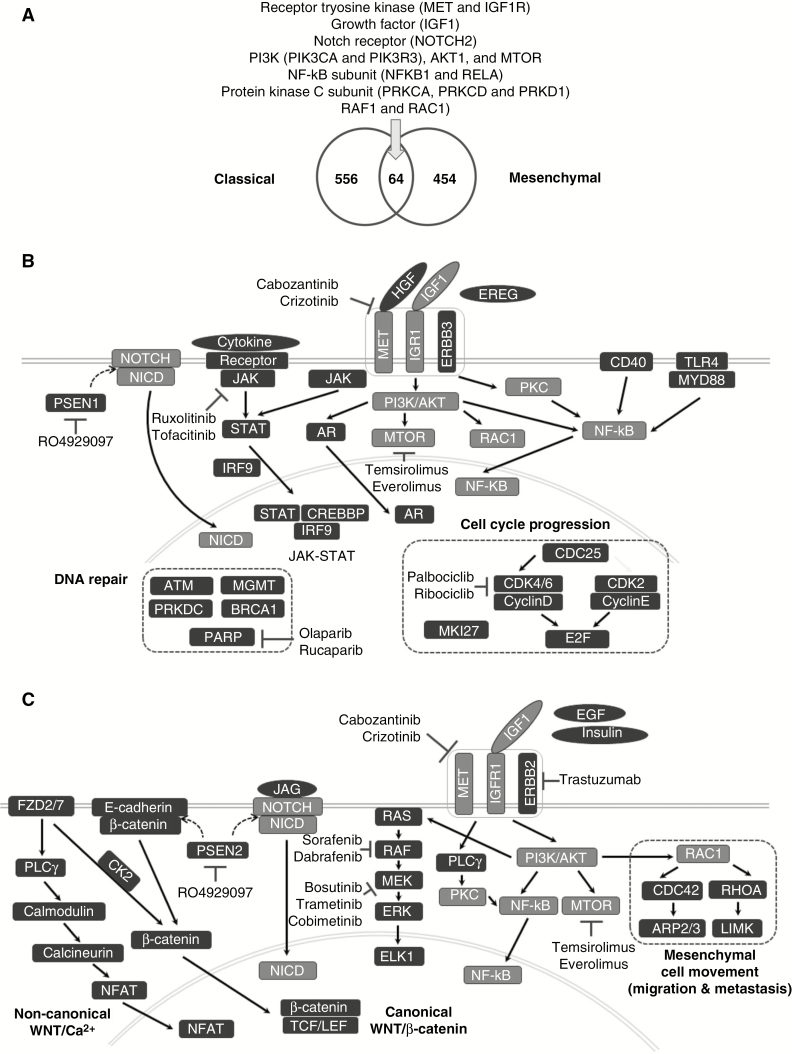
In 2 major GBM subtypes, classical and mesenchymal, many signaling pathways previously reported to be deregulated in GBM were identified as prognostic gene sets, including the p53, DNA replication, insulin-like growth factor 1 (IGF-1), mTOR, Ras, and Notch pathways (Figures 2A and 3A).8,14,19,34,35 The mTOR and IGF-1 pathways were commonly detected in both subtypes. Specifically, 64 genes were commonly identified as prognostic core genes, including 2 RTKs (MET and IGF1R), a growth factor (IGF1), a Notch receptor (NOTCH2), and many genes in RTK downstream, including PI3Ks, AKT1, MTOR, NF-κB subunits, protein kinase C subunits, RAF1, and RAC1 (Fig. 5A). Schematic diagrams of the prognostic signaling pathways with representative core genes and inhibitors are presented in Fig. 5B and C. In the classical subtype, 2 growth factors, hepatocyte growth factor (HGF) and Epiregulin (EREG), an ErbB receptor (ERBB3), and androgen receptor (AR) were the prognostic core genes. Additionally, MKI67 encoding the cell proliferation marker Ki-67, and many genes involved in the p53/RB pathway or cell cycle progression such as PLK1, CDC25A, and CDC25B, cyclins (CCND1, CCNE2, and CCNG2), cyclin-dependent kinases (CDK2/4/6/9), and E2Fs (E2F1/4) were detected as prognostic core genes, suggesting that classical GBM patients might benefit most from cancer therapy targeting cell cycle and proliferation. High expressions of many DNA repair genes (eg, ATM, PARP, BRCA1, PRKDC, MGMT) were also associated with poor prognosis in the classical subtype. In previous reports, GBM patients with MGMT silencing by promoter hypermethylation were more sensitive to treatment of temozolomide.36 A recent study reported that MGMT methylation status was predictive of temozolomide response only in the classical subtype.37 This supported our result that MGMT is a prognostic core gene only in the classical subtype. Moreover, 2 membrane receptors related to inflammatory response (CD40 and TLR4) and many genes in the JAK-STAT pathway were prognostic in the classical subtype. The JAK-STAT pathway–related prognostic genes included cytokines (IL7 and IL15), cytokine receptors (IFNAR2, IL15RA, IL22RA1, IL4R, CSF2RB, and GHR), Janus kinases (JAK1/2), STAT transcription factors (STAT1/2/3/4/5A/6), and cAMP response element binding protein (CREBBP). Potential targeted therapies inhibiting JAK-STAT or NF-κB pathways have been suggested in GBM in the context of regulating inflammatory response.4,17 In copy number and mutation data analyses, 6p loss and TP53 mutations were associated with poor and good survival, respectively (Fig. 2E and H). The association between favorable prognosis and TP53 mutations was previously reported in GBM patients without IDH1 mutation,38 but to the best of our knowledge, this is the first study reporting the association between 6p loss and poor prognosis. We found that low expression of 7 genes on 6p were significantly associated with poor prognosis (Supplementary Table 9). Among the 7 genes, NOL7 was reported as a tumor suppressor.39 However, due to the relatively small number of cases with 6p loss (10 of 117) and TP53 mutations (9 of 55), further investigations are warranted to confirm the associations.
Fig. 5.
Diagrammatic illustration of prognostic core genes and pathways. (A) Venn diagram of prognostic core genes identified in the classical and mesenchymal subtypes. (B) Schematic diagram of representative prognostic core genes and inhibitors in the classical subtype. (C) Schematic diagram of representative prognostic core genes and inhibitors in the mesenchymal subtype. Genes in light gray boxes indicate the genes commonly detected in the classical and mesenchymal subtypes.
Mesenchymal subtype-specific prognostic core genes included those involved in mesenchymal cell movement (CDC42 and RHOA), PI3K/Akt pathway genes (PIK3CD, PIK3CG, PIK3R2/5, and AKT3), and MAPK/extracellular signal-regulated kinase (ERK) pathway genes (HRAS, KRAS, NRAS, ARAF, BRAF, MAP2K1/2, MAPK1/3, and ELK1) (Fig. 5C). Many genes in canonical Wnt/β-catenin and non-canonical Wnt/Ca2+ pathways were also detected, including 2 frizzled receptors (FZD2/7), CTNNB1 (β-catenin), T-cell factor–lymphoid enhancer factor (TCF/LEF) transcription factors (TCF7L1/2 and LEF1), CDH1 (E-cadherin), phospholipase C gamma (PLCG1), calmodulins (CALM1/2/3), calcineurin (PPP3CA, PPP3CB, and PPP3CC), and nuclear factor of activated T cells (NFAT) (NFATC4). Therefore, targeting Wnt signaling might be more effective in mesenchymal patients. In mutation analysis of the mesenchymal subtype, 11 of 60 patients with mutations in PIK3R1 (n = 6) or PCLO (n = 6) showed poor prognosis. In the xenograft model, human normal astrocytes introduced by PIK3R1 somatic mutations could cause oncogenic events.40PCLO had recurrent mutations in diffuse large B-cell lymphoma.41
Recently, glioma-intrinsic transcriptional subtypes have been newly defined after excluding tumor microenvironment-related genes.42 We investigated whether subtype-specific prognostic markers could be retained in the intrinsic GBM subtypes. Many of the gene sets remained to be prognostic in the new classical subtype (11 of 25), intermediately in the new proneural subtype (8 of 25), but mostly not in the new mesenchymal subtype (Supplementary Table 10). The little congruence in the mesenchymal subtype implies that the mesenchymal prognostic core genes may originate from the tumor microenvironment and/or tumor microenvironment-associated tumor features. Indeed, several major prognostic pathways detected in the mesenchymal subtype (Fig. 5C) have been described to be linked to the tumor immune microenvironment: a possible role of Wnt signaling in the interaction between GBM and brain immune cells to create a more favorable microenvironment for invasion,43 a potential impact on immunotherapy response and regulation of immune cell infiltration by the Wnt/β-catenin pathway,44 a relationship between immune-silent tumor phenotype and the MAPK signaling pathway,45 and activation of the Notch signaling pathway by tumor-infiltrating myeloid cells.46 In addition, mesenchymal prognostic gene sets included the “Glycosaminoglycan” gene set (Fig. 3A), which is involved in biosynthesis of chondroitin sulfate, an extracellular proteoglycan that plays a crucial role in promoting a favorable tumor microenvironment.47
In the G-CIMP subtype, chromosomal aberrations were more prognostic than gene expression profiles (Fig. 4C and E), and 10q loss, 12p gain, and 14q loss were detected as the main prognostic factors. A recent study reported that G-CIMPs can be further divided into 2 different survival groups based on low/high methylation status, designated “G-CIMP-low” (poor prognosis) and “G-CIMP-high” (good prognosis).18 We found that the “G-CIMP-low” group was significantly associated with G-CIMP with 10q loss (Fig. 4G) (Fisher’s exact test P = 0.0028). Loss of chromosome 10 and phosphatase and tensin homolog (PTEN) is one of the most common events in non–G-CIMP GBMs.16 Therefore, the significant association between “G-CIMP-low” and 10q loss implies that G-CIMP with chromosomal aberrations similar to non–G-CIMP might have worse prognosis and that 10q loss might be a major prognostic marker in G-CIMP.
On the other hand, prognostic core genes showing negative methylation/expression correlations were enriched with immune function–related genes (Supplementary Table 11), suggesting that DNA methylation might influence the prognosis of GBM, probably by controlling the tumor immune microenvironment. Congruently, modulation of methylation in immune genes was proposed to play a role in GBM progression in comparative analysis of initial and recurrent primary GBM pairs.48 Moreover, genome-wide methylation analyses revealed that the top pathways enriched in hypomethylated genes in GBMs compared with normal brain tissues were all related to immune functions.49
In current practice, the G-CIMP GBMs are identified on a routine basis by detection of IDH1 mutation as a surrogate marker. However, classification of other GBM subtypes is not currently performed in clinical practice. Thus, there are many challenges in practical application of subtype-based approaches to everyday clinical management toward precision oncology. Incorporation of expression profiles into routine practice requires new molecular assays, with an increase of medical expenses. Utilization of surrogate markers of GBM subtypes will greatly facilitate subtype-specific approaches. For instance, the classical subtype is characterized by high expression of EGFR,19 which is consistently observed in both TCGA and validation datasets (Supplementary Fig. 4A). Likewise, most of the TP53 mutations and 6p losses are concurrently observed with high EGFR expressions in the classical subtype (Supplementary Fig. 4B), which sheds light on the possible use of EGFR expression as a potential surrogate marker for the classical subtype. Further identification of surrogate markers of GBM subtypes that can be easily incorporated into clinical practice will enable the application of subtype-specific approaches in risk stratification, investigation of therapeutic strategies, and clinical trial designs.
Funding
This work was supported by Sunchon National University Research Fund (A.K.P), the National R&D Program for Cancer Control, Ministry for Health and Welfare, Republic of Korea (1420020) (A.K.P), the National Research Foundation of Korea (NRF) grant funded by the Ministry of Science, ICT & Future Planning, Republic of Korea (2015R1A4A1041219) (A.K.P), and NIH grant R01LM012806 (Z.Z.). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank the Data Science and Informatics Core for Cancer Research supported by CPRIT (RP170668).
Supplementary Material
Acknowledgments
We are thankful for the reviewers’ valuable comments, which helped us improve the quality of the manuscript.
Conflict of interest statement. None declared.
References
- 1. Karsy M, Huang T, Kleinman G, Karpel-Massler G. Molecular, histopathological, and genomic variants of glioblastoma. Front Biosci (Landmark Ed). 2014;19:1065–1087. [DOI] [PubMed] [Google Scholar]
- 2. Li X, Wu C, Chen N, et al. PI3K/Akt/mTOR signaling pathway and targeted therapy for glioblastoma. Oncotarget. 2016;7(22):33440–33450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mao H, Lebrun DG, Yang J, Zhu VF, Li M. Deregulated signaling pathways in glioblastoma multiforme: molecular mechanisms and therapeutic targets. Cancer Invest. 2012;30(1):48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Atkinson GP, Nozell SE, Benveniste ET. NF-kappaB and STAT3 signaling in glioma: targets for future therapies. Expert Rev Neurother. 2010;10(4):575–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Akhavan D, Cloughesy TF, Mischel PS. mTOR signaling in glioblastoma: lessons learned from bench to bedside. Neuro Oncol. 2010;12(8):882–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lino MM, Merlo A, Boulay JL. Notch signaling in glioblastoma: a developmental drug target?BMC Med. 2010;8:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee Y, Lee JK, Ahn SH, Lee J, Nam DH. WNT signaling in glioblastoma and therapeutic opportunities. Lab Invest. 2016;96(2):137–150. [DOI] [PubMed] [Google Scholar]
- 8. Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chakravarti A, Zhai G, Suzuki Y, et al. The prognostic significance of phosphatidylinositol 3-kinase pathway activation in human gliomas. J Clin Oncol. 2004;22(10):1926–1933. [DOI] [PubMed] [Google Scholar]
- 10. Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21(21):2683–2710. [DOI] [PubMed] [Google Scholar]
- 11. Cerami E, Demir E, Schultz N, Taylor BS, Sander C. Automated network analysis identifies core pathways in glioblastoma. PLoS One. 2010;5(2):e8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sun J, Gong X, Purow B, Zhao Z. Uncovering MicroRNA and transcription factor mediated regulatory networks in glioblastoma. PLoS Comput Biol. 2012;8(7):e1002488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eder K, Kalman B. Molecular heterogeneity of glioblastoma and its clinical relevance. Pathol Oncol Res. 2014;20(4):777–787. [DOI] [PubMed] [Google Scholar]
- 14. Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. [DOI] [PubMed] [Google Scholar]
- 15. Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Noushmehr H, Weisenberger DJ, Diefes K, et al. ; Cancer Genome Atlas Research Network Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cahill KE, Morshed RA, Yamini B. Nuclear factor-κB in glioblastoma: insights into regulators and targeted therapy. Neuro Oncol. 2016;18(3):329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ceccarelli M, Barthel FP, Malta TM, et al. ; TCGA Research Network Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Verhaak RG, Hoadley KA, Purdom E, et al. ; Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Freije WA, Castro-Vargas FE, Fang Z, et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64(18):6503–6510. [DOI] [PubMed] [Google Scholar]
- 21. Lee Y, Scheck AC, Cloughesy TF, et al. Gene expression analysis of glioblastomas identifies the major molecular basis for the prognostic benefit of younger age. BMC Med Genomics. 2008;1:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rich JN, Hans C, Jones B, et al. Gene expression profiling and genetic markers in glioblastoma survival. Cancer Res. 2005;65(10):4051–4058. [DOI] [PubMed] [Google Scholar]
- 23. Gravendeel LA, Kouwenhoven MC, Gevaert O, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69(23):9065–9072. [DOI] [PubMed] [Google Scholar]
- 24. Murat A, Migliavacca E, Gorlia T, et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26(18):3015–3024. [DOI] [PubMed] [Google Scholar]
- 25. Ducray F, de Reyniès A, Chinot O, et al. An ANOCEF genomic and transcriptomic microarray study of the response to radiotherapy or to alkylating first-line chemotherapy in glioblastoma patients. Mol Cancer. 2010;9:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Madhavan S, Zenklusen JC, Kotliarov Y, Sahni H, Fine HA, Buetow K. Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res. 2009;7(2):157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee S, Piccolo SR, Allen-Brady K. Robust meta-analysis shows that glioma transcriptional subtyping complements traditional approaches. Cell Oncol (Dordr). 2014;37(5):317–329. [DOI] [PubMed] [Google Scholar]
- 28. Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. [DOI] [PubMed] [Google Scholar]
- 29. Sandmann T, Bourgon R, Garcia J, et al. Patients with proneural glioblastoma may derive overall survival benefit from the addition of bevacizumab to first-line radiotherapy and temozolomide: retrospective analysis of the AVAglio trial. J Clin Oncol. 2015;33(25): 2735–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43): 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gerson SL. MGMT: its role in cancer aetiology and cancer therapeutics. Nat Rev Cancer. 2004;4(4):296–307. [DOI] [PubMed] [Google Scholar]
- 33. Cruickshanks N, Zhang Y, Yuan F, Pahuski M, Gibert M, Abounader R. Role and therapeutic targeting of the HGF/MET pathway in glioblastoma. Cancers. 2017;9(7):doi: 10.3390/cancers9070087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kanamori M, Kawaguchi T, Nigro JM, et al. Contribution of Notch signaling activation to human glioblastoma multiforme. J Neurosurg. 2007;106(3):417–427. [DOI] [PubMed] [Google Scholar]
- 35. Trojan J, Cloix JF, Ardourel MY, Chatel M, Anthony DD. Insulin-like growth factor type I biology and targeting in malignant gliomas. Neuroscience. 2007;145(3):795–811. [DOI] [PubMed] [Google Scholar]
- 36. Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 37. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang K, Wang YY, Ma J, et al. Prognostic value of MGMT promoter methylation and TP53 mutation in glioblastomas depends on IDH1 mutation. Asian Pac J Cancer Prev. 2014;15(24):10893–10898. [DOI] [PubMed] [Google Scholar]
- 39. Doçi CL, Zhou G, Lingen MW. The novel tumor suppressor NOL7 post-transcriptionally regulates thrombospondin-1 expression. Oncogene. 2013;32(37):4377–4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Quayle SN, Lee JY, Cheung LW, et al. Somatic mutations of PIK3R1 promote gliomagenesis. PLoS One. 2012;7(11):e49466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lohr JG, Stojanov P, Lawrence MS, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109(10):3879–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang Q, Hu B, Hu X, et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell. 2018;33(1):152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Matias D, Predes D, Niemeyer Filho P, et al. Microglia-glioblastoma interactions: new role for Wnt signaling. Biochim Biophys Acta. 2017;1868(1):333–340. [DOI] [PubMed] [Google Scholar]
- 44. Pai SG, Carneiro BA, Mota JM, et al. Wnt/beta-catenin pathway: modulating anticancer immune response. J Hematol Oncol. 2017;10(1):101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hendrickx W, Simeone I, Anjum S, et al. Identification of genetic determinants of breast cancer immune phenotypes by integrative genome-scale analysis. Oncoimmunology. 2017;6(2):e1253654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ohnuki H, Jiang K, Wang D, et al. Tumor-infiltrating myeloid cells activate Dll4/Notch/TGF-β signaling to drive malignant progression. Cancer Res. 2014;74(7):2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wade A, Robinson AE, Engler JR, Petritsch C, James CD, Phillips JJ. Proteoglycans and their roles in brain cancer. FEBS J. 2013;280(10):2399–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Alentorn A, Durán-Peña A, Malousi A, et al. Differential gene methylation in paired glioblastomas suggests a role of immune response pathways in tumor progression. J Neurooncol. 2015;124(3):385–392. [DOI] [PubMed] [Google Scholar]
- 49. Lai RK, Chen Y, Guan X, et al. Genome-wide methylation analyses in glioblastoma multiforme. PLoS One. 2014;9(2):e89376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.