Abstract
Background
Patients with glioblastoma without O6-methylguanine-DNA methyltransferase (MGMT) promoter hypermethylation are unlikely to benefit from alkylating chemotherapy with temozolomide (TMZ). Trials aiming at replacing TMZ with targeted agents in unselected patient populations have failed to demonstrate any improvement of survival. Advances in molecular understanding and diagnostic precision enable identification of key genetic alterations in a timely manner and in principle allow treatments with targeted compounds based on molecular markers.
Methods
The NCT Neuro Master Match (N2M2) trial is an open-label, multicenter, phase I/IIa umbrella trial for patients with newly diagnosed isocitrate dehydrogenase (IDH) wildtype glioblastoma without MGMT promoter hypermethylation to show safety, feasibility, and preliminary efficacy of treatment with targeted compounds in addition to standard radiotherapy based on molecular characterization. N2M2 is formally divided into a Discovery and a Treatment part. Discovery includes broad molecular neuropathological diagnostics to detect predefined biomarkers for targeted treatments. Molecular diagnostics and bioinformatic evaluation are performed within 4 weeks, allowing a timely initiation of postoperative treatment. Stratification for Treatment takes place in 5 subtrials, including alectinib, idasanutlin, palbociclib, vismodegib, and temsirolimus as targeted therapies, according to the best matching molecular alteration. Patients without matching alterations are randomized between subtrials without strong biomarkers using atezolizumab and asinercept (APG101) and the standard of care, TMZ. For the phase I parts, a Bayesian criterion is used for continuous monitoring of toxicity. In the phase II trials, progression-free survival at 6 months is used as endpoint for efficacy.
Results
Molecular diagnostics and bioinformatic evaluation are performed within 4 weeks, allowing a timely initiation of postoperative treatment. Stratification for Treatment takes place in 5 subtrials, including alectinib, idasanutlin, palbociclib, vismodegib, and temsirolimus as targeted therapies, according to the best matching molecular alteration. Patients without matching alterations are randomized between subtrials without strong biomarkers using atezolizumab and asinercept (APG101) and the standard of care, TMZ. For the phase I parts, a Bayesian criterion is used for continuous monitoring of toxicity. In the phase II trials, progression-free survival at 6 months is used as endpoint for efficacy.
Discussion
Molecularly informed trials may provide the basis for the development of predictive biomarkers and help to understand and select patient subgroups who will benefit.
Keywords: CD95 ligand, MDM2, MGMT, mTOR, radiotherapy
Importance of the study
It is conceivable that targeted precision treatments work in situations of a defined molecular background. The present study addresses this topic by focusing on newly diagnosed glioblastoma, in which the analyzed tumor material best reflects the molecular specifics; further, the trial uses multiple agents in an umbrella design with postsurgical standard radiotherapy as a backbone and deferral of TMZ outside a control arm by restriction to patients with glioblastoma harboring an unmethylated MGMT promoter. The study spearheads the concept in newly diagnosed glioblastoma and leaves room for future improvement by integrating novel compounds, combinations thereof, and molecular analyses better reflecting the heterogeneity of the disease. Each subtrial may evolve into a controlled phase II trial further strengthening the therapy and the attached molecular biomarker.
The understanding of glioblastoma at the molecular level has improved dramatically in recent years.1–5 For the first time a limited defined set of molecular markers is implemented in the updated World Health Organization classification.6 These and additional markers and some others are increasingly used to support clinical decisions.7 Isocitrate dehydrogenase 1 and 2 (IDH1/2) mutations8 and 1p/19q codeletion7 are already routinely tested in glioma patients to guide diagnostics, and O6-methylguanine DNA methyltransferase (MGMT) promoter methylation9 is used to support treatment decisions.
Despite these advances, prognosis and treatment success in glioblastoma patients have only slowly been improving over the past decades, with an increase in median survival reflecting improved supportive measures and patient selection.10–12 The current standard therapy for glioblastoma patients consists of maximal safe resection followed by radiochemotherapy with temozolomide (TMZ) and 6 maintenance TMZ cycles.10MGMT methylation status was shown to be a predictive biomarker with methylation indicating a response to alkylating chemotherapy such as TMZ or lomustine. The European Organisation for Research and Treatment of Cancer (EORTC) 26981/22981 National Cancer Institute of Canada (NCIC) CE.3 trial led to the practical use of MGMT testing in daily clinical routine13—and after final confirmation by the NOA-0814 and NORDIC15 trials, it was integrated as a predictive biomarker into the current European guidelines for diagnosis and treatment of glioblastoma at least in elderly patients.7,9 Therefore, in clinical routine, treatment decisions are mainly MGMT based in elderly patients, if combined radiochemotherapy is not applicable due to age or comorbidities.7 Most other patients are treated with combined radiochemotherapy despite the unlikely benefit from an alkylating chemotherapy with a non-hypermethylated MGMT promoter. MGMT promoter methylation status does not define a molecularly distinct glioblastoma subpopulation,16 which means that other molecular lesions occur with the same frequency and there is no reason to believe that MGMT unmethylated tumors harbor further distinct molecular resistance features. However, clinical trials replacing TMZ by, for instance, temsirolimus, bevacizumab, or enzastaurin have failed to improve survival so far in molecular-wise unselected glioblastoma patients with unmethylated MGMT status.17–19
Recent developments of new targeted therapies increasingly allow subset-specific treatment for patients with expression of respective molecular markers. IDH mutations represent prognostic biomarkers and additionally are targetable by IDH inhibitors20,21 or an immunotherapeutic approach with vaccination targeting the IDH1 R132H mutation.22 Other examples for targetable alterations include variant III of epidermal growth factor receptor (EGFRvIII) mutation,23 BRAF mutations24 (although present in rare cases of adult gliomas), and CD95L.25 Lower levels of methylation of carboxypeptidase G2 (CpG2) in the promoter of cluster of differentiation ligand (CD95L) may be predictive of an improved overall survival (OS) with the CD95 inhibitory treatment with asinercept (APG101) in glioblastoma patients.25 In addition, the proneural subtype of glioblastoma according to expression analysis26 might be predictive for response to bevacizumab treatment27 and mismatch-repair deficiency or polymerase epsilon gene (POLE) mutations resulting in a hypermutator phenotype may predict response to checkpoint inhibition.28 Furthermore, improved molecular diagnostics increasingly enable individual treatments based on molecular alterations in representative tissue,29,30 building the basis for clinical trials.31
Growing evidence proposes a relevant genetic heterogeneity within one and the same disease manifestation, particularly in spatially or temporally separated tumors (ie, multifocal tumors or tumor recurrences). Whereas data from a recent study do not support uniformity within spatially heterogeneous tumors, they support the present concept of using new tissue information for informed decisions.32,33 Since the NCT Neuro Master Match (N2M2) trial relies on tissue from the surgery immediately prior to trial inclusion, the restrictions may be less relevant. In a series of dry runs, we have demonstrated feasibility of the timely molecular analysis and application of an algorithm for decision making.31
The N2M2 trial intends to translate complex molecular diagnostics in glioblastoma into clinical decision making by prospectively allocating patients with molecular profiles that match with the mode of action of a targeted therapy and might thereby indicate a higher likelihood for a response to this treatment. Glioblastoma patients harboring an unmethylated MGMT promoter status most likely benefit from alternative treatment approaches to TMZ and therefore are chosen as the study population in this trial.
Study Design
This study is designed as an open-label, parallel group, nonrandomized phase I/IIa multicenter trial of molecularly matched targeted therapies plus radiotherapy in patients with newly diagnosed glioblastoma without MGMT promoter methylation. A “match” is defined as detection of one of the predefined biomarkers of the available targeted drugs. The study is formally divided into a Discovery and a Treatment part. Discovery consists of complex molecular diagnostics including whole exome, low coverage whole-genome and transcriptome sequencing, methylome analysis using methylation arrays, and gene expression arrays to identify defined biomarkers as well as new targets and to get a more comprehensive view of affected pathways. Importantly, data from Discovery are to be confirmed with established immunohistochemical and (Sanger) sequencing techniques. The detection of predefined biomarkers for the different arms, which are considered to indicate a response to a specific available targeted therapy, forms the basis for a “match”/“no match” decision in the Treatment part of this study. Matching patients receive the respective targeted therapy in combination with radiotherapy as first-line treatment in different subtrials which are subdivided in a phase I part for determination of safety and appropriate dose by dose-escalation and a phase IIa part evaluating preliminary efficacy. The warehouse of targeted therapies in this trial consists of asinercept, alectinib, idasanutlin, atezolizumab, vismodegib, palbociclib, and temsirolimus. For asinercept and atezolizumab, biomarkers have not been considered strong enough at the present, and the “non-matching” patients will be equally allocated to receive asinercept, atezolizumab, or the current standard of care: radiotherapy with TMZ. In the latter, patients will serve as a nonrandomized but contemporary control group to the molecular informed subtrials and a randomized control for the no-match subtrials.
Objectives and Endpoints
The main objective of the N2M2 study is to demonstrate the improvement of OS of glioblastoma patients with an unmethylated MGMT promoter based on molecular characterization and use of targeted compounds in a modern trial design. Further aims are the assessment of safety and feasibility of treatment with these targeted compounds in addition to radiotherapy. Subtrials that satisfy the safety and efficacy criteria will be considered as candidates for further investigation in randomized phase II/III trials independent of the current protocol.
The phase I part evaluates safety and tolerability of the systemic molecularly defined therapy and the proof of the proposed optimal monocompound dose in conjunction with radiotherapy. The primary safety endpoint is dose-limiting toxicity (DLT), defined as all adverse events (AEs) of grade ≥3 according to Common Terminology Criteria for Adverse Events (CTCAE) v4.03 that are related to the administration of the investigational agents. The secondary objective of the phase I part is the evaluation of efficacy by determination of progression-free survival (PFS) at 6 months (PFS-6), which also defines the primary objective of the phase IIa part. Secondary objectives of the phase IIa part consist of (i) safety and tolerability of experimental therapies, (ii) PFS, (iii) OS, and (iv) biomarker development.
Trial Population
The trial population is molecularly defined by glioblastoma patients harboring an unmethylated MGMT promoter status and an IDH wildtype status.
The inclusion criteria include: (i) written informed consent; (ii) open biopsy or resection to obtain enough tumor material; (iii) availability of fresh-frozen tissue, formalin-fixed paraffin embedded tissue, and blood; (iv) histologically confirmed, newly diagnosed IDH-wildtype glioblastoma with unmethylated MGMT promoter determined by one of the accepted methods (quantitative PCR, pyrosequencing, methylation array);32,33 (v) standard MRI ≤48 (+ 6) hours postsurgery according to the present guidelines; (vi) Karnofsky performance score (KPS) ≥70%; (vii) life expectancy >6 months; (viii) age ≥18 years; (ix) no stable or decreasing steroid levels below 4 mg/day of dexamethasone during the last 3 days prior to enrollment (for a complete list, refer to the Supplementary Material). After inclusion, unmethylated MGMT status needs to be reconfirmed prior to initiation of specific treatment, otherwise the patient will be excluded from this study.
The exclusion criteria include: (i) abnormal (grade ≥2 CTCAE v4.03) laboratory values for hematology, liver, and renal function; (ii) HIV, active hepatitis B or C infection, or active infection requiring antibiotics; (iii) immunosuppression; (iv) history of other malignancies within the last 5 years; (v) prior therapy for glioma (except surgery and steroids); (vi) insufficient tumor material for molecular diagnostics; (vii) pregnancy or breastfeeding; (viii) history of hypersensitivity to the investigational medicinal product; (ix) any clinically significant condition that could interfere with the conduct of the study or absorption of oral medication or that would pose an unacceptable risk to the patient (for a complete list, refer to the Supplementary Material).
Enrollment
Patients will be enrolled in 13 Neuro-oncology Working Group trial sites of the German Cancer Society (NOA) in Germany. Based on molecular findings (“match”/“no match”), patients will be allocated in 7 different subtrials or the control group.
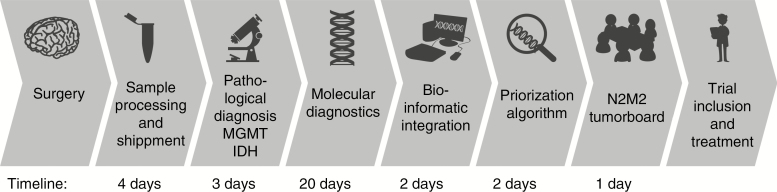
For the “match”/“no match” decision, fresh tumor tissue and blood from glioblastoma patients with an unmethylated MGMT promoter will be widely examined by neuropathological analysis. Results will be available within a maximum of 3 weeks postoperatively allowing a dedicated bioinformatics evaluation which forms the basis for the final treatment decision by the molecular tumor board and afterward a timely initiation (≤4–6 wk) of postoperative treatments. The workflow and timelines of molecular diagnostics and treatment decisions are summarized in Fig. 1.
Fig. 1.
Workflow and timelines of molecular diagnostics and treatment decision. Of note, patient tissue analyses in Heidelberg and with a start in an outside study center follow the same time intervals.
Discovery, Sequencing, and Data Processing
Molecular analysis consists of an epigenome-wide array, panel sequencing, whole exome, low-coverage whole genome, and transcriptome sequencing as well as expression array detecting somatic single nucleotide variants, small inserts/deletions, copy number variants, focal amplifications, or overexpression of affected genes and pathways.
The detected somatic mutations are assigned information from databases such as known cancer genes and the catalogue of somatic mutations in cancer, as well as custom lists of cancer-associated genes, drug targets and biomarkers (with special respect to the warehouse drugs), resistance mechanism, indirect druggability, and contraindications. The lists will continuously be updated and expanded during the project by external data and feedback from the study arms.
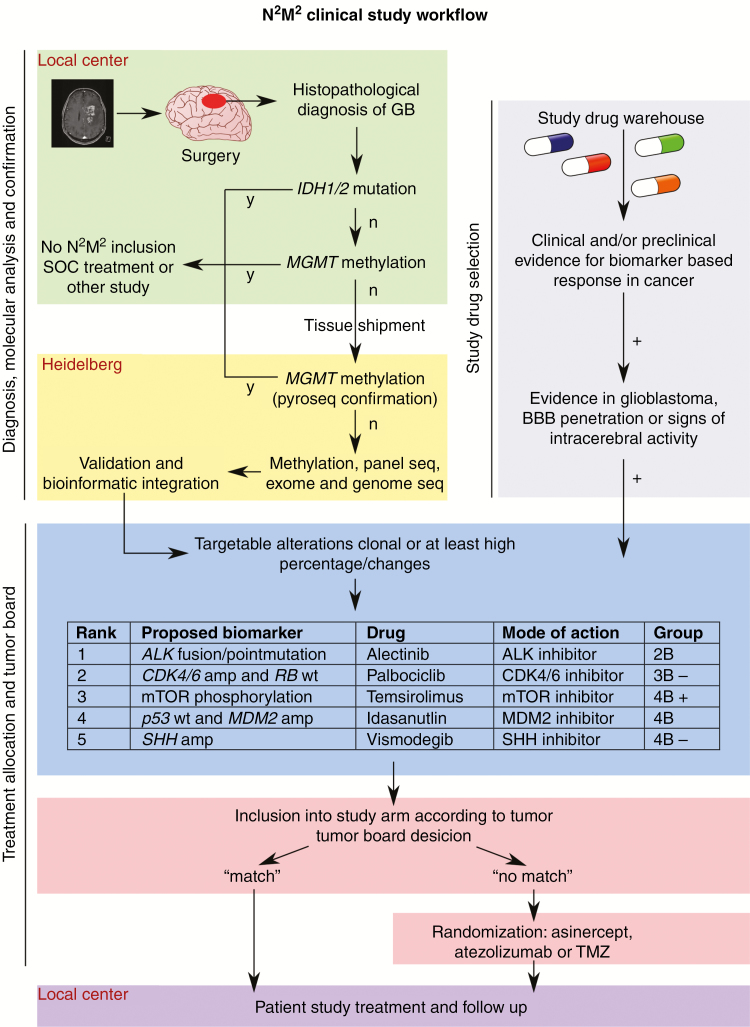
For cases with detection of several targetable mutations, a previously described ranking algorithm will be used.31 A schematic study overview, including the ranking algorithm, is depicted in Fig. 2. If more than one mutation obtains the highest rank, the match will be randomly allocated to specific subtrials or assigned for the best-performing subtrial, if already known. This process does not introduce a bias into the final evaluation, but allows for more rapid detection of a positive subtrial. All experimental test results will be confirmed by an accepted genetic test (eg, Sanger sequencing) or immunohistochemistry.
Fig. 2.
Schematic study overview. Figure shows the workflow of the N2M2 study with emphasis on molecular analysis and treatment allocation process. GB: glioblastoma; SOC: standard of care; seq: sequencing, BBB: blood–brain barrier.
Treatment Decision
The final decision about specific treatments is made by the molecular tumor board (MTB), consisting of members of the steering committee in Heidelberg, members of the participating site with patients under discussion and optionally all other participating sites invited via video conference. The aims of the MTB are to ensure reliable and consistent decisions and to provide final recommendations regarding the enrollment of patients in specific subtrials. The molecular basis for the decisions will be based on the accepted tests, not on the experimental procedures. The algorithm for decisions about patient allocation is demonstrated in Fig. 2. At complete availability of molecular information and open slots in each subtrial, the data are pre-assessed by the study chair, a molecular neuropathologist, the study coordinator, and a bioinformatician to allow suggestion of a potential match or a non-match resulting in randomization. At the MTB, the patient’s case plus the raw molecular information as well as the recommendation are intensively discussed and a decision on the allocation is rendered by consensus.
Trial Oversight Committees
An external Data Safety Monitoring Committee (DSMC) will consist of clinical and biostatistical experts to
□ meet periodically (quarterly in the phase I part of the subtrials and twice per year in the phase II parts as well as via written approval on a mailing at the end of each phase I subtrial, prior to moving to phase II) to review summarized and individual patients’ data related to safety, data integrity, and overall conduct of the trial
□ re-review specific interim analyses for safety and/or efficacy, as appropriate
□ provide recommendations to continue as originally planned, change or terminate the trial depending on these analyses
□ communicate other recommendations or concerns as appropriate
The management of the complexity and innovation of N2M2 will be facilitated by the formal implementation of a Steering Committee in addition to the DSMC. The Steering Committee will comprise representatives from all involved subspecialties to ensure input and counseling for the formal study leadership. Of note, decisions on the patient-relevant changes are made by the DSMC and the Coordinating Investigator (W.W.). The Steering Committee has advisory function; a formal role in the decision process would complicate, not improve, the study management.
Treatment, Intervention
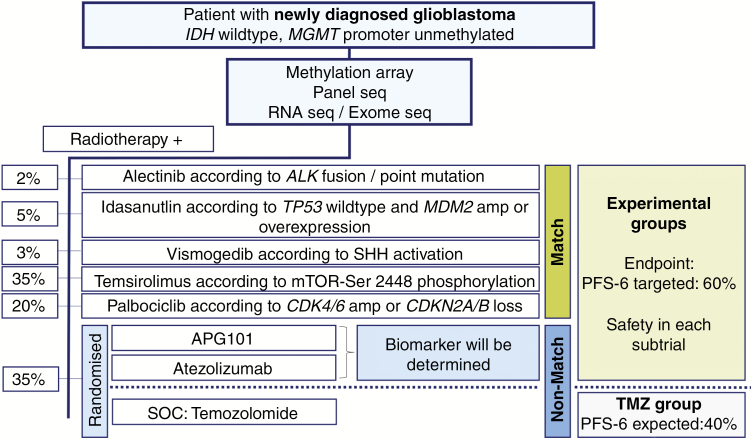
Based on the decision of the MTB, patients will be enrolled in 5 different subtrials (“match”) or randomized between asinercept, atezolizumab, and the control subtrial (“no match”) (Fig. 3). A complete randomized allocation of patients to the subtrials is not feasible due to the fact that the subtrials differ in molecular targets. As radiotherapy is considered standard of care, it is not a study procedure and builds the backbone for each subtrial with radiotherapy at 60 Gy in 2-Gy fractions in working-daily radiotherapy sessions over a period of 6 weeks.7 Experimental treatments start with the initiation of radiotherapy at the maximum tolerated dose (MTD), which is predefined or determined in phase I parts of the subtrials, and continue until progression, undue toxicity, death, or patient’s decision, whichever comes first. As a control intervention, patients without any of the defined molecular alterations receive concomitant TMZ chemotherapy (75 mg/m2 body surface area) plus radiotherapy followed by 6 cycles of TMZ maintenance therapy (150/200 mg/m2 body surface) according to the standard of care. Safety endpoints of phase I parts will be determined until the end of combined modality treatment, and efficacy data will be collected until end of study or death, whichever comes first.
Fig. 3.
Patient allocation and targeted therapies according to matching biomarkers. Percentages indicate proposed proportions of patients allocated to each subtrial. As for asinercept and atezolizumab, no specific biomarker is available so far; it will be assessed exploratively during the trial and patients will be equally allocated to each “non-match” subtrial including asinercept, atezolizumab, standard of care (SOC). RT: radiotherapy.
Subtrials, Targeted Therapies
The warehouse of targeted therapies for the different subtrials consists of alectinib, idasanutlin, vismodegib, palbociclib, and temsirolimus for the match subtrials as well as asinercept, atezolizumab, and temozolomide for the non-match randomized subtrials. Targets and biomarkers of therapies, methods for their molecular detection, as well as the prevalence of the alterations in glioblastoma patients are summarized in Table 1. Of note, no prognostic value is so far attributed to these markers.31
Table 1.
Prioritization algorithm for biomarker-based targeted treatment
| Group | Criterion |
|---|---|
| 1 | Biomarker with approved biomarker specific treatment in glioblastoma + with strong survival benefit − with moderate survival benefit or inconsistent |
| 2A | Biomarker with approved biomarker specific treatment in another cancer indication with compelling clinical evidence in glioblastoma |
| 2B | Biomarker with approved biomarker specific treatment in another cancer indication not tested in glioblastoma in a clinical setting |
| 3A | Clinical evidence in glioblastoma, but not approved in glioblastoma or any other cancer indication + mutation − amp/ expression |
| 3B | Clinical evidence in another cancer indication, makes biological sense in glioblastoma, but no clinical evidence in glioblastoma + mutation − amp/expression |
Alectinib is a second-generation inhibitor of anaplastic lymphoma kinase (ALK), which showed clinical efficacy in ALK positive non–small cell lung cancer administered orally at 600 mg twice daily.34 ALK fusions and mutations represent proven biomarkers for alectinib treatment.
The mouse double minute 2 homolog (MDM2) inhibitor idasanutlin activates the p53 pathway by blocking the inhibitory MDM2-p53 interaction in p53 wildtype tumors.3,35 Preclinical studies demonstrated a higher sensitivity toward the drug for p53 wildtype tumors with MDM2 amplification and a primary resistance of tumors harboring p53 mutations.36 Idasanutlin was effective and well tolerated in first-in-human studies in patients with acute myeloid leukemia and solid tumors. It is administered orally on 5 consecutive days of a 28-day cycle. Optimal dose will be determined in the phase I part by dose escalation from 100 mg daily until MTD.
Vismodegib, a small-molecule inhibitor of the sonic hedgehog (SHH) signaling pathway, has been approved for therapy of basal cell carcinoma in doses of 150 mg daily. Activation of the SHH pathway leads to cell proliferation, upregulation, of anti-apoptotic proteins, production of vascular endothelial growth factor, and angiopoietins37 and is considered a biomarker for a response to vismodegib treatment.
Palbociclib, an oral inhibitor of cyclin-dependent kinases (CDKs) 4 and 6, has been approved for treatment of estrogen receptor–positive, human epidermal growth factor receptor 2–negative breast cancer in combination with aromatase inhibitors or fulvestrant.38 Amplification of CDK4 and CDK6 results in dysregulation of the retinoblastoma pathway, a major regulator of cell cycle progression and proliferation. CDKN2A is an inhibitor of CDKs such as CDK4 and CDK6, and CDKN2B interacts with CDK4. Therefore, activation of CDK4 or CDK6 or CDKN2A/B codeletion serves as biomarkers for palbociclib treatment. Palbociclib will be administered initially at 75 mg with dose escalation steps to 100 and 125 mg during combination with radiotherapy and at 125 mg in adjuvant monotherapy on 21 consecutive days of a 28-day cycle.
Activation of the mechanistic target of rapamycin (mTOR) pathway is associated with reduced survival in glioma patients39,40 and leads to increased cell growth.40 Temsirolimus represents an inhibitor of the mTOR pathway, which is administered intravenously at 25 mg/week, and was evaluated as a first-line treatment in glioblastoma patients in the EORTC-26082 trial. Although primary endpoints were not reached in an unselected patient population, phosphorylation of mTOR-serine2448 (p-mTORSer2448) was retrospectively found to be predictive for response to temsirolimus.17 This association is worth prospective confirmation, which is attempted in the present subtrial. As the EORTC 26082 trial showed feasibility and safety of temsirolimus in the exact same patient population and treatment schedule, a formal phase I trial is not foreseen for this subtrial.
Asinercept (APG101), a CD95-fusion protein, has been shown to be effective and well tolerated in combination with second radiotherapy in progressive glioblastoma.25 It blocks the interaction of CD95 and its ligand CD95L and thereby inhibits the CD95 pathway, resulting in reduced proliferation and invasion of glioblastoma cells.41 Retrospective analysis suggested low methylation levels in CpG2 of the CD95L promoter as predictive for response to asinercept treatment.25 Determination of the safe combination dose of asinercept i.v. started with 600 mg/week with 3 de-/escalation steps of 200 mg (ie, D0 = 400 mg, D1 = 600 mg, D2 = 800 mg) in conjunction with radiotherapy. Atezolizumab is a monoclonal antibody targeting programmed death ligand 1 (PD-L1). PD-L1 is an inhibitory cell surface molecule which is expressed on immune and tumor cells, suppresses T-cell migration, proliferation, and secretion of cytotoxic mediators, and restricts tumor cell killing by binding the inhibitory programmed death 1 (PD-1) receptor on T cells. Predictive biomarkers for atezolizumab are currently not yet defined, but high expression of PD-L142,43 or high numbers of nonsynonymous mutations driven by mismatch-repair deficiency44 are potential candidates. Atezolizumab will be administered intravenously at 1200 mg every 3 weeks. Recent studies in colon cancer revealed that patients with mismatch repair deficiency respond better to anti-programmed death (PD)-1 therapy.44,45 Additional studies indicate that other solid tumors with mismatch repair deficiency, including glioblastoma, are sensitive to anti–PD-1 therapy.46
Temozolomide is an alkylating chemotherapy used as standard of care for patients with glioblastoma irrespective of MGMT status.7
Withdrawal of Patients
Patients must be withdrawn from trial at any time at their own request, in case of serious adverse events caused by the investigational medicinal product except for manageable abnormal laboratory values or other general safety issues by the investigator. All ongoing AEs and serious AEs of withdrawn patients will be followed up until stabilization or resolution.
Outcome Measures
An overview about diagnostic and therapeutic measures, timing of disease assessment, and study visits of participating patients is displayed in Supplementary Table 2. AEs, DLTs, concomitant medication, and safety hematological laboratory values will be recorded weekly during combined radiotherapy and medical treatment. Clinical chemistry laboratory values and physical examination will be performed every 4 weeks. MRIs are carried out twice-monthly starting 4 weeks after completion of radiotherapy. Six months after start of therapy, PFS-6 is assessed. After end of study (EOS)—that is, 6 months after start of study for the individual patient—patients will be routinely followed up until death every 3 months by phone. After EOS, patients will be routinely followed up and will be treated regarding standard of care according to the discretion of the treating physician. Patients who would still benefit from the experimental intervention after EOS might continue as part of an individual treatment or as an off-label use after consulting the coordinating physician, if medication is still available then.
Assessment of Endpoints and Statistical Analysis
Assessment of Safety
All AEs that occur during the trial after the first experimental treatment are recorded, graded according to the CTCAE v4.03 at every study visit, and followed up until resolution or stabilization. Safety endpoints will be assessed by frequency of AEs and the number of laboratory values that fall outside of predetermined ranges. AEs will be described by event, duration, seriousness, intensity, and relationship to the investigational medicinal product, actions taken, and clinical outcome and reported as tables of frequencies at Preferred Term (PT) and MedDRA System Organ Class.
Assessment of Efficacy
For the primary efficacy endpoint, PFS-6 (defined as proportion of patients with PFS 6 months after treatment start) is determined and presented in summary tables, along with Pearson–Clopper 95% CIs. Radiographic progression will be evaluated according to Response Assessment in Neuro-Oncology (RANO)47 or immunotherapy RANO for atezolizumab48 by the central neuroradiology and clinical progression by deterioration of KPS. Most importantly, the protocol contains detailed instructions to avoid too early cessation of study drug in case of presumed pseudoprogression and mandates a confirmatory scan whenever clinically possible.
For secondary efficacy endpoints, PFS and OS (defined as the time from treatment start until progression or death) will be determined and analyzed using the Kaplan–Meier method for survival curves and Greenwood’s formula for estimating the standard error of event rates. Given the low number of patients in each subtrial and the multiplicity of the analyses, all statistical tests are of strictly exploratory nature.
Efficacy will be evaluated in each subtrial separately, based on a one-sided binomial test of the null hypothesis set as PFS-6 at 40%, the rate observed in a retrospective analysis of available data in patients undergoing standard treatment,10,13 and an alternative hypothesis of 60% at the final analysis. No formal statistical comparisons between the subtrials are planned. However, results obtained for the control group and different subtrials may be used for considerations of changes regarding efficacy or recommendation for further phase II/III trials.
Interim Analysis and Stopping Rules
Two interim analyses per subtrial will be carried out once the PFS-6 endpoint has been determined for 15 and 25 patients, respectively. Tests for futility based on predictive power49 and for decisions regarding acceptance of the DLT rate of experimental treatment for a phase IIa trial are performed. For that, the posterior distribution of the DLT rate is calculated with a binomial-beta model with a non-informative prior, and a Bayesian criterion is used for continuous monitoring of toxicity.50 Recruitment will be suspended if the predictive power is lower than 10% or if the posteriori probability that the true toxicity rate (at the given dose level of dose-escalation in the phase I part of indicated subtrials) is 30% or higher exceeds 95%. In both scenarios, the DSMC will advise the coordinating investigator if patient accrual should be stopped.
Sample Size Estimation
In each of the 7 experimental subtrials, between 2 and 18 patients will be enrolled for phase I parts, depending on observed toxicities. In phase IIa parts, a maximum of 40 patients in each subtrial will be accrued, wherein 9 patients of the corresponding dose of the phase I part will be included. The exact number depends on early stopping for toxicity or futility. The “non-matching” group is anticipated to include approximately 35% of all screened study patients. Therefore, 12% of all screened patients are expected to be enrolled in the control group receiving TMZ.
Accordingly, about 450 patients with newly diagnosed glioblastoma harboring an unmethylated MGMT promoter will need to be screened, requiring approximately 5 years for recruitment and 84 months for overall duration of the trial with expected wide variability in the subtrials depending on frequency of the molecular alteration providing a match.
Ethical and Legal Aspects
The trial is conducted in accordance with the standards of Good Clinical Practice, the applicable version of the Declaration of Helsinki and local legal and regulatory requirements. The study protocol has been approved by the independent Ethics Committee (AFmu-207/2017) and the competent federal authority (Vorlagen Nummer 3051/01, Paul-Ehrlich-Institute in Langen, Germany).
For this trial, the EudraCT number 2015-002752-27 has been obtained. Monitoring and pharmacovigilance is performed by the Coordination Center for Clinical Trials (KKS) Heidelberg.
Patients are enrolled in a two-step consenting process. Oral and written explanation of the molecular testing, including interpretation and conduct of the MTB, is provided after surgery, and any trial-specific measure is started only after written informed consent. Consenting for the treatment step in the respective subtrial is done after the MTB decision prior to any subtrial-specific process.
The Discovery phase of the trial is funded by DKFZ/NCT Heidelberg. Study drugs will be provided free of charge by Hoffmann-La Roche Ltd., Apogenix AG, and Pfizer Pharma GmbH. The clinical phase is supported by funding of the German Cancer Aid (DKH, funding number 70111980) and by structural support via the German Ministry of Education and Research (BMBF) funded German Cancer Consortium (DKTK) as well as the Heidelberg Center for Personalized Oncology (HIPO 2-K25 and 2-K32R).
Discussion
The aim of this study is the development of a complex molecular and bioinformatics workup to prospectively identify patient subgroups with a potential higher likelihood for a response to a specific treatment based on predefined molecular profiles with the final objective of an improvement of OS for these patients.
At present, molecular markers are increasingly used to allocate patients for individual treatments. Some of these markers already represent a prerequisite for specific treatments, such as the IDH mutation for IDH inhibitors20,21 or vaccinations22 and the detection of EGFRvIII23 or BRAF V600E24 for respective inhibitor treatments. At least conceptually comprehensive diagnostics enable precision treatment concepts for patients based on molecular alterations and drugs with published mode of action if available.29–31 Until now these treatments have not been approved for patients with glioblastoma, and molecular markers are not validated in this patient cohort. As studies evaluating the diagnostic pipeline and consecutive prospective patient allocation are lacking, its investigation is one of the aims in the N2M2 trial. In order to translate molecular diagnostics into treatment decisions, diagnostic workup has to be performed within a maximum of 3–4 weeks to allow a timely initiation of postoperative treatment. For N2M2, dry runs already demonstrated the feasibility of this timely diagnostic process.31
Heterogeneity is observed in the mutational profile changes during the natural course of the disease, among different patients, within the tumor and through selection pressure resulting from treatment.32,33 Decisions for salvage treatments that are based on biomarker information from tissue acquired prior to any other treatment may therefore underestimate the molecular variability and result in incorrect conclusions.29,30,32,33 For that reason, patients will receive treatment with respective targeted therapies as first-line therapy in N2M2, which further enables the investigation and comprehensive molecular understanding of causes for treatment failure whenever tissue can be obtained at recurrence.
MGMT promoter status predicts the response to alkylating chemotherapies but does not define a fundamentally different subgroup of glioblastoma.16 Prior studies replacing TMZ for patients with unmethylated MGMT status failed to demonstrate a survival benefit in unselected patient populations,17–19 but retrospective analysis revealed potential predictive biomarkers for treatment response.17,19 Therefore, prospective patient allocation to respective treatments based on molecular biomarkers represents a promising approach to improve OS and establish rational alternatives to TMZ for glioblastoma patients with unmethylated MGMT promoter status unlikely benefiting from TMZ treatment.
N2M2 would be deemed successful if at least one arm made it to a full controlled phase II/III trial and if we considerably deepened our understanding of the disease and accepted molecular decisions to be integrated into primary patient care.
Funding
The Discovery phase of the trial is funded by DKFZ/NCT Heidelberg. Study drugs will be provided free of charge by Hoffmann-La Roche Ltd., Apogenix AG, and Pfizer Pharma GmbH. The clinical phase is supported by funding of the German Cancer Aid (DKH, funding number 70111980) and by structural support via the German Ministry of Education and Research (BMBF) funded German Cancer Consortium (DKTK) as well as the Heidelberg Center for Personalized Oncology (HIPO 2-K25 and 2-K32R).
Conflict of interest statement. No conflicts.
Authorship statement. Conceptualization: WW, MP, TK, AW, IKS, MB, SMP, AvD, FS
Manuscript Writing: WW, AB, TK, SD
Trial conduct: WW, SD, AB, TK, IKS, AW, FW, EP, BB, JD, AU. MB, CHM, AE, AvD, DTWJ, SMP, FS, MP
Final editing and approval of the manuscript: WW, SD, AB, TK, IKS, AW, FW, EP, BB, JD, AU. MB, CHM, AE, AvD, DTWJ, SMP, FS, MP
Supplementary Material
References
- 1. Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sturm D, Witt H, Hovestadt V, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. [DOI] [PubMed] [Google Scholar]
- 3. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Osswald M, Jung E, Sahm F, et al. Brain tumour cells interconnect to a functional and resistant network. Nature. 2015;528(7580):93–98. [DOI] [PubMed] [Google Scholar]
- 6. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 7. Weller M, van den Bent M, Tonn JC, et al. European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol. 2017;18(6):e315–e329. [DOI] [PubMed] [Google Scholar]
- 8. Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wick W, Weller M, van den Bent M, et al. MGMT testing—the challenges for biomarker-based glioma treatment. Nat Rev Neurol. 2014;10(7):372–385. [DOI] [PubMed] [Google Scholar]
- 10. Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. [DOI] [PubMed] [Google Scholar]
- 11. Stupp R, Taillibert S, Kanner A, et al. Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: a randomized clinical trial. JAMA. 2017;318(23):2306–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Herrlinger U, Tzaridis T, Mack F, et al. Phase III trial of CCNU/temozolomide (TMZ) combination therapy vs. standard TMZ therapy for newly diagnosed MGMT-methylated glioblastoma patients: the CeTeG/NOA-09 trial. Neurooncol. 2017;19(suppl. 6):vii13. [Google Scholar]
- 13. Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 14. Wick W, Platten M, Meisner C, et al. ; NOA-08 Study Group of Neuro-oncology Working Group (NOA) of German Cancer Society Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial. Lancet Oncol. 2012;13(7):707–715. [DOI] [PubMed] [Google Scholar]
- 15. Malmström A, Grønberg BH, Marosi C, et al. ; Nordic Clinical Brain Tumour Study Group (NCBTSG) Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: the Nordic randomised, phase 3 trial. Lancet Oncol. 2012;13(9):916–926. [DOI] [PubMed] [Google Scholar]
- 16. Kessler T, Sahm F, Sadik A, et al. Molecular differences in IDH wildtype glioblastoma according to MGMT promoter methylation. Neuro Oncol. 2018;20(3):367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wick W, Gorlia T, Bady P, et al. Phase II study of radiotherapy and temsirolimus versus radiochemotherapy with temozolomide in patients with newly diagnosed glioblastoma without MGMT promoter hypermethylation (EORTC 26082). Clin Cancer Res. 2016;22(19):4797–4806. [DOI] [PubMed] [Google Scholar]
- 18. Wick W, Steinbach JP, Platten M, et al. Enzastaurin before and concomitant with radiation therapy, followed by enzastaurin maintenance therapy, in patients with newly diagnosed glioblastoma without MGMT promoter hypermethylation. Neuro Oncol. 2013;15(10):1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Herrlinger U, Schäfer N, Steinbach JP, et al. Bevacizumab plus irinotecan versus temozolomide in newly diagnosed O6-methylguanine-DNA methyltransferase nonmethylated glioblastoma: the randomized GLARIUS trial. J Clin Oncol. 2016;34(14):1611–1619. [DOI] [PubMed] [Google Scholar]
- 20. Rohle D, Popovici-Muller J, Palaskas N, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340(6132):626–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pusch S, Krausert S, Fischer V, et al. Pan-mutant IDH1 inhibitor BAY 1436032 for effective treatment of IDH1 mutant astrocytoma in vivo. Acta Neuropathol. 2017;133(4):629–644. [DOI] [PubMed] [Google Scholar]
- 22. Schumacher T, Bunse L, Pusch S, et al. A vaccine targeting mutant IDH1 induces antitumour immunity. Nature. 2014;512(7514):324–327. [DOI] [PubMed] [Google Scholar]
- 23. Weller M, Butowski N, Tran DD, et al. Rindopepimut with temozolomide for patients with newly diagnosed, EGFRvIII-expressing glioblastoma (ACT IV): a randomised, double-blind, international phase 3 trial. Lancet Oncol. 2017;18(10):1373–1385. [DOI] [PubMed] [Google Scholar]
- 24. Takahashi Y, Akahane T, Sawada T, et al. Adult classical glioblastoma with a BRAF V600E mutation. World J Surg Oncol. 2015;13:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wick W, Fricke H, Junge K, et al. A phase II, randomized, study of weekly APG101+reirradiation versus reirradiation in progressive glioblastoma. Clin Cancer Res. 2014;20(24):6304–6313. [DOI] [PubMed] [Google Scholar]
- 26. Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. [DOI] [PubMed] [Google Scholar]
- 27. Sandmann T, Bourgon R, Garcia J, et al. Patients with proneural glioblastoma may derive overall survival benefit from the addition of bevacizumab to first-line radiotherapy and temozolomide: retrospective analysis of the AVAglio trial. J Clin Oncol. 2015;33(25):2735–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Johanns TM, Miller CA, Dorward IG, et al. Immunogenomics of hypermutated glioblastoma: a patient with Germline POLE deficiency treated with checkpoint blockade immunotherapy. Cancer Discov. 2016;6(11):1230–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Byron SA, Tran NL, Halperin RF, et al. Prospective feasibility trial for genomics-informed treatment in recurrent and progressive glioblastoma. Clin Cancer Res. 2018;24(2):295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wick W, Kessler T. Drug repositioning meets precision in glioblastoma. Clin Cancer Res. 2018;24(2):256–258. [DOI] [PubMed] [Google Scholar]
- 31. Pfaff E, Kessler T, Balasubramanian GP, et al. Feasibility of real-time molecular profiling for patients with newly diagnosed glioblastoma without MGMT promoter hypermethylation-the NCT Neuro Master Match (N2M2) pilot study. Neuro Oncol. 2018;20(6):826–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee JK, Wang J, Sa JK, et al. Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat Genet. 2017;49(4):594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bady P, Sciuscio D, Diserens AC, et al. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol. 2012;124(4):547–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shaw AT, Gandhi L, Gadgeel S, et al. ; study investigators Alectinib in ALK-positive, crizotinib-resistant, non-small-cell lung cancer: a single-group, multicentre, phase 2 trial. Lancet Oncol. 2016;17(2):234–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–848. [DOI] [PubMed] [Google Scholar]
- 36. Verreault M, Schmitt C, Goldwirt L, et al. Preclinical efficacy of the MDM2 inhibitor RG7112 in MDM2-amplified and TP53 wild-type glioblastomas. Clin Cancer Res. 2016;22(5):1185–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Filbin MG, Dabral SK, Pazyra-Murphy MF, et al. Coordinate activation of Shh and PI3K signaling in PTEN-deficient glioblastoma: new therapeutic opportunities. Nat Med. 2013;19(11):1518–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Finn RS, Martin M, Rugo HS, et al. Palbociclib and letrozole in advanced breast cancer. N Engl J Med. 2016;375(20):1925–1936. [DOI] [PubMed] [Google Scholar]
- 39. Chakravarti A, Zhai G, Suzuki Y, et al. The prognostic significance of phosphatidylinositol 3-kinase pathway activation in human gliomas. J Clin Oncol. 2004;22(10):1926–1933. [DOI] [PubMed] [Google Scholar]
- 40. McBride SM, Perez DA, Polley MY, et al. Activation of PI3K/mTOR pathway occurs in most adult low-grade gliomas and predicts patient survival. J Neurooncol. 2010;97(1):33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Blaes J, Thomé CM, Pfenning PN, et al. Inhibition of CD95/CD95L (FAS/FASLG) signaling with APG101 prevents invasion and enhances radiation therapy for glioblastoma. Mol Cancer Res. 2018;16(5):767–776. [DOI] [PubMed] [Google Scholar]
- 42. Herbst RS, Soria JC, Kowanetz M, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Powles T, Eder JP, Fine GD, et al. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515(7528):558–562. [DOI] [PubMed] [Google Scholar]
- 44. Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bouffet E, Larouche V, Campbell BB, et al. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J Clin Oncol. 2016;34(19):2206–2211. [DOI] [PubMed] [Google Scholar]
- 47. Wen PY, Macdonald DR, Reardon DA, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol. 2010;28(11):1963–1972. [DOI] [PubMed] [Google Scholar]
- 48. Okada H, Weller M, Huang R, et al. Immunotherapy response assessment in neuro-oncology: a report of the RANO working group. Lancet Oncol. 2015;16(15):e534–e542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Saville BR, Connor JT, Ayers GD, Alvarez J. The utility of Bayesian predictive probabilities for interim monitoring of clinical trials. Clin Trials. 2014;11(4):485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Aamot H, Gigic B, Abel U, Karapanagiotou-Schenkel I. Continuous monitoring of toxicity in clinical trials—simulating the risk of stopping prematurely. Int J Clin Pharmacol Ther. 2010;48(7):476–477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.