Abstract
Conversion of natural habitats into urban landscapes can expose wildlife to novel pathogens and alter pathogen transmission pathways. Because transmission is difficult to quantify for many wildlife pathogens, mathematical models paired with field observations can help select among competing transmission pathways that might operate in urban landscapes. Here we develop a mathematical model for the enteric bacteria Salmonella enterica in urban-foraging white ibis (Eudocimus albus) in south Florida as a case study to determine (i) the relative importance of contact-based versus environmental transmission among ibis and (ii) whether transmission can be supported by ibis alone or requires external sources of infection. We use biannual field prevalence data to restrict model outputs generated from a Latin hypercube sample of parameter space and select among competing transmission scenarios. We find the most support for transmission from environmental uptake rather than between-host contact and that ibis–ibis transmission alone could maintain low infection prevalence. Our analysis provides the first parameter estimates for Salmonella shedding and uptake in a wild bird and provides a key starting point for predicting how ibis response to urbanization alters their exposure to a multi-host zoonotic enteric pathogen. More broadly, our study provides an analytical roadmap to assess transmission pathways of multi-host wildlife pathogens in the face of scarce infection data.
Keywords: epidemiology, environmental transmission, mathematical modelling, resource provisioning, sensitivity analysis, urbanization
1. Introduction
The conversion of natural habitats to urban landscapes has complex effects on wildlife populations and communities, including consequences for pathogen transmission [1–3]. Changes in pathogen dynamics can result from shifts in wildlife behaviour, body condition, immunity, demography and trophic interactions in response to urbanization [4–6]. For example, banded mongooses (Mungos mungo) in Botswana that forage on garbage in urban habitats showed a higher incidence of tuberculosis, likely from higher contact rates and within-troop aggression [7]. Similarly, house sparrows (Passer domesticus) in urban regions of France had a higher prevalence of avian malaria and greater lead concentrations than natural populations, suggesting impaired immunity from urban contaminants [8]. Because urbanization can also produce novel species assemblages that influence interspecific contact rates [9–11], a detailed understanding of host–pathogen interactions in urban landscapes can help mitigate pathogen spillover risks.
Many wildlife pathogens, including multi-host pathogens in urban landscapes, have complex transmission processes [12–14]. Avian influenza virus and Salmonella enterica are examples of multi-host zoonotic pathogens where transmission among wildlife hosts can depend on multiple exposure routes [12,15,16]. Within birds, these pathogens are spread through faecal–oral transmission, and thus infection could be associated with both intraspecific and interspecific contacts or with exposure to environmental stages in soil or water that can persist for weeks and even months [17,18]. Resolving the roles of multiple routes of infection is challenging [18–23] and is further complicated with Salmonella, for which exposure can also be derived from human sources such as contaminated food and treated or untreated wastewater [17,24]. Investigating these transmission processes in urban wildlife is needed to better predict exposure for humans and vulnerable wildlife species and to develop management strategies to reduce infection risk.
Mathematical models offer powerful tools for understanding infectious disease dynamics when there is uncertainty or limited empirical knowledge about the underlying transmission mechanisms, as is true for many wildlife pathogens [25,26]. In one recent study, mechanistic models applied to longitudinal field data indicated that frequent inter-colony contact and immunizing exposures were necessary to explain observed seroprevalence of rabies virus in vampire bats (Desmodus rotundus) [27]. Mechanistic models of plague similarly suggest that human ectoparasites were more likely than rats to have sustained large historical epidemics in Europe [28]. Mathematical models that include different transmission routes can therefore act as virtual experiments to show which combinations of exposure routes are capable of maintaining multi-host pathogens, which can inform future field studies and intervention strategies [26,29].
In this study, we develop a mathematical model to similarly examine the transmission dynamics of Salmonella in urbanized American white ibis (Eudocimus albus) populations. White ibis are colonial wading birds that naturally reside in wetlands and forage on aquatic prey [30]. In south Florida, which includes both breeding and non-breeding ibis populations, some ibis have shifted their behaviour to forage in urban parks in recent years [31,32]. White ibis are now abundant in neighbourhood parks, golf courses and other artificial wetlands, where they show site fidelity, are fed by people and regularly interact with other urbanized birds such as gulls and ducks [33,34]. Recent work showed that Salmonella infection prevalence is greatest in urban ibis [34], possibly due to increased contact when ibis aggregate around supplemental food [4,35]. Ibis can carry Salmonella without developing clinical disease, allowing them to intermittently shed bacteria [36]. The diversity of Salmonella serotypes is also high in urban ibis, suggesting frequent environmental exposure and contributions from various sources [34,36]. As some Salmonella isolates from ibis match those from human cases [34,37], ibis could contribute to human exposure in shared urban environments. Ibises might also acquire some infections from human sources (e.g. contaminated wastewater) or other reservoir hosts (e.g. wild birds, pets).
Given uncertainties in the mechanisms by which urban habituation influences Salmonella transmission in ibis, we build a novel mathematical model of ibis–pathogen dynamics in urban habitats to determine (i) the relative importance of intraspecific contact versus environmental transmission and (ii) if sustained transmission can be supported by ibis alone or requires non-ibis sources of pathogen input. To differentiate between potential mechanisms causing exposure in urban ibis, we perform a global sensitivity analysis of our model and match predictions against temporal infection prevalence data from ibis and environmental samples in the non-breeding season. We validate our model with biannual field samples from an independent non-breeding season and simplify the model to focus on the most parsimonious and predictive assumptions. This workflow could provide a generalizable analytic roadmap for assessing transmission pathways of multi-host wildlife pathogens in the face of often scarce and sparse infection data.
2. Material and methods
2.1. Model development
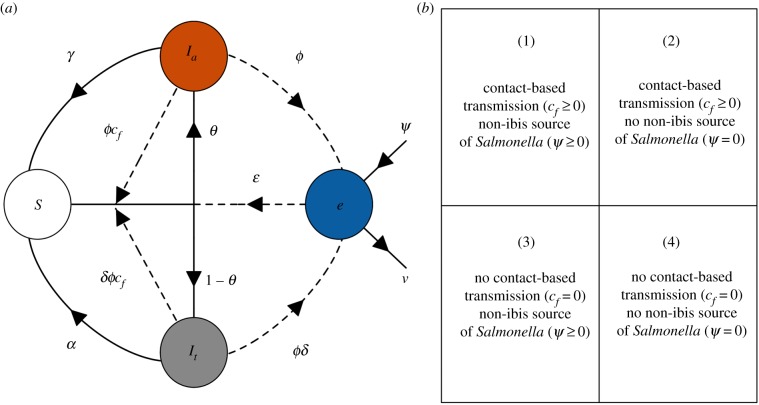
To explore Salmonella dynamics in an urban ibis flock, we use a compartmental model based on a susceptible–infected–susceptible paradigm with an environmental source of infection (figure 1a) [38]. Our model considers infection dynamics within a single non-breeding season (October to March) [39]. During the non-breeding season, ibis forage throughout the day in urban parks but roost at night in trees above water, often in wetlands and golf courses [40]. Because ibis have high winter survival, and as Salmonella is only known to cause mortality in nestlings [41,42], we ignore natural and disease-induced mortality and assume a constant flock size, N. We assume no immunity after recovery, such that re-exposure leads to new infection. As Salmonella either colonizes the gastrointestinal (GI) tract or results in transitory shedding [36], we divide infected ibis into transiently shedding () and actively colonized () hosts, where shed for a shorter duration (i.e. passage through the GI tract) relative to . Ibis either enter a transient shedding state or become actively colonized rather than an advance from transiently shedding to actively colonized (see below). As our field data from bird faecal analyses do not discriminate between and , we quantify infection prevalence () as all infected ibis divided by constant flock size (i.e. ). We track the environmental prevalence of Salmonella (e) as E/R, where E is the area of an urban park contaminated with Salmonella and R is the total park area used by ibis. Higher e can be interpreted as the probability with which ibis encounter an infectious dose (e.g. our model assumes that an environmental detection rate of 33% versus 66% doubles encounter).
Figure 1.
(a) Conceptual schematic of the mechanistic model for Salmonella dynamics in urbanized white ibis. Circles represent state variables (S = susceptible, It = transiently shedding, Ia = actively colonized, e = environmental prevalence), solid lines represent transitions and dashed lines represent exposure pathways or routes of pathogen shedding into the environmental pool. (b) The four model scenarios for varying assumptions about contact-based transmission and the contribution of non-ibis sources. Parameter definitions are provided in table 1. (Online version in colour.)
We consider two transmission routes for susceptible ibis (S): contact with infected ibis at a rate c and uptake of environmental Salmonella at a rate . To select the appropriate term for contact-based transmission, we analysed field data on ibis infection prevalence and flock size (below). We assume a positive flock size–prevalence relationship supports density-dependent transmission () and no relationship supports frequency-dependent transmission (). However, because flock size is here modelled as constant, differentiating between these contact-based transmission processes only affects the magnitude of this model parameter. A proportion of ibis become actively colonized following exposure, shed Salmonella into the environmental at rate and clear infection at rate . The remaining proportion () develop a transient infection, shed at rate δϕ (where ) and clear infection at rate (). Thus, transiently shedding and colonized ibis contribute to the environmental Salmonella pool () at rates ϕ and . Non-ibis sources, including alternative reservoir hosts or anthropogenic sources, contribute to e at a constant rate . Because e relates to the total area contaminated by Salmonella, these shedding rates represent the fraction of the foraging area contaminated per day by ibis and non-ibis sources. Only uninfected areas of the park become contaminated, and thus the rate of increase in environmental prevalence is . This specific assumption is justified given that our model only considers the park area used by foraging ibis (from which our environmental data were also derived; see below and electronic supplementary material), that ibis spend their daily foraging time well-mixed within these parks, and that the location of roost trees above water is unlikely to facilitate environmental exposure to a build-up of Salmonella [40,43]. We lastly assume that Salmonella decays from the environment at a constant rate . Salmonella infection dynamics within an urban ibis flock and the environment are illustrated in figure 1 and are provided by the following system of three ordinary differential equations:
2.2. Initial parametrization
Because many parameters relating to Salmonella dynamics in ibis are unknown, we used values primarily from published studies of poultry experimentally infected with Salmonella (given a lack of ibis-specific estimates) and performed sensitivity analyses within biologically realistic ranges (table 1). We set the probability of Salmonella colonization given exposure () from 0 to 50%, given higher odds of wild birds developing transient shedding and relatively low infection probabilities in poultry [34,44]. We assume that the minimum shedding duration by transiently shedding birds () is 2 h, based on a captive study of gut passage time for food in ibis [45]. Based on observations of experimentally inoculated chickens shedding Salmonella [46,47], we set the maximum value of to 9 days. For the shedding duration by actively colonized ibis (), we set our minimum value as the median shedding duration in orally infected chickens (13 days) [48]. Because Salmonella has been detected in experimentally inoculated chickens for up to 12 weeks [49], we set the maximum duration of shedding by colonized ibis to be 180 days.
Table 1.
Model parameters, units and input ranges used in LHS simulations. Parameters shaded in grey were varied over a log10 scale within the LHS routine.
| parameter | description | units | value | reference |
|---|---|---|---|---|
| probability of Salmonella colonization | proportion | 0–0.5 | [34,44] | |
| GI tract passage time (It) | hours | 2–216 | [45–47] | |
| duration of colonization (Ia) | days | 13–180 | [48,49] | |
| ϕ | per cent area contaminated by 1 ibis | per day | 10−6–0.1 | n.a. |
| proportional shedding of It versus Ia | proportion | 0–1 | n.a. | |
| daily effective contact rate | per day | 10−6–2 | n.a. | |
| daily environmental uptake rate | per day | 10−6–2 | n.a. | |
| duration of Salmonella persistence in environment | days | 14–180 | [17,50] | |
| per cent area contaminated by non-ibis | per day | 10−6–1 | n.a. |
As the relative shedding rates of transiently shedding and actively colonized birds are unknown, we allowed the scaling parameter for transient shedding ibis () to range from 0 to 1. We set shedding of actively colonized ibis () to range from 0 to only 0.1, the latter representing the extreme case where one ibis contaminates up to 10% of the total flock foraging area within a single day. Similarly, we allowed non-ibis sources of infection () to range from 0 to 1, in which non-ibis sources could contaminate up to the entire foraging area daily. Both the parameters for effective close contact () and environmental uptake of Salmonella () ranged from 0 to 2. Lastly, for the persistence time of Salmonella in the environment (), we considered a minimum of two weeks and a maximum of 180 days [17,50], the duration of our simulations.
2.3. Analysis of field data to inform model structure
We recorded Salmonella infection prevalence from 98 white ibis from six urban sites in south Florida (Palm Beach and Martin Counties) during four sampling periods between October and March of 2015–2016 and 2016–2017; all sites were sampled in at least three of these four time points (electronic supplementary material, figure S1). Site descriptions and methods of quantifying urbanization are given elsewhere [33,34], and details of ibis capture, environmental sample collection, flock size determination and laboratory analyses to quantify Salmonella positivity are provided in the electronic supplementary material.
Using these data, we derived the average flock size for each urban site at each sampling time point. We calculated the prevalence of Salmonella (i.e. the proportion of positive samples) (i) per site and time point and (ii) pooled across sites, using the Wilson interval to derive 95% confidence intervals as suggested for small sample sizes [51]. To assess our assumption of constant flock size over time, we used the lme4 package in R to fit a generalized linear mixed model (GLMM) with mean flock size per site and time point as the dependent variable, Gaussian errors, a fixed effect of the four sampling events, and a random intercept of the site [52,53]. To examine modelling ibis–ibis transmission as density- or frequency-dependent, we fit another GLMM to the positive and negative counts of Salmonella infection status per site and sampling event, binomial errors, a logit link, a fixed effect of log flock size and a random intercept of site.
2.4. Model analysis
We analysed model variants across a factorial design of epidemiological scenarios under which (i) non-ibis sources do or do not contribute to the environmental pool ( and ψ = 0) and (ii) contact-based transmission does or does not occur ( and ). These four model scenarios (figure 1b) include (1) contact-based transmission and non-ibis sources (, ), (2) contact-based transmission and no non-ibis sources (, ), (3) no contact-based transmission and non-ibis sources (, ) and (4) no contact-based transmission and no non-ibis sources (, ); for this latter scenario, transmission can only occur through environmental uptake (). We used Latin hypercube sampling (LHS) with the lhs and deSolve packages to efficiently sample the biological ranges of parameter values (table 1) [54–57]. We sampled 1000 unique parameter combinations, assuming prior uniform distributions of most model parameters [58]. Because we did not have a priori order-of-magnitude estimates for four model parameters (, , , ), we uniformly sampled these across a log10 scale (table 1).
In the four model scenarios, we set initial conditions to match ibis and environmental infection prevalence pooled across sites at the start of the study in autumn 2015 (figure 2), and simulations were run for the duration of the non-breeding season (tmax = 180). We then trimmed the 1000 parameter combinations to the subset that resulted in a simulated endpoint prevalence within the 95% confidence interval for the pooled proportion of positive ibis and environmental samples in spring 2016. We repeated this procedure by restricting LHS parameter combinations with only ibis prevalence (ignoring prevalence based on environmental samples) to quantify how inferences about plausible parameter sets would differ if ibis and environmental prevalence were decoupled. To assess the sensitivity of this approach to initial conditions for ibis state variables, we repeated our LHS analysis using the upper and lower bounds of the 95% confidence intervals for estimated mean urban ibis flock size and pooled ibis infection prevalence for autumn 2015. We used Salmonella data pooled across sites for these model analyses, as sample sizes per site and sampling time point were small (ibis: n = 1–8, x̄ ≈ 4; environment: n = 6 (n = 3 soil and n = 3 water)).
Figure 2.
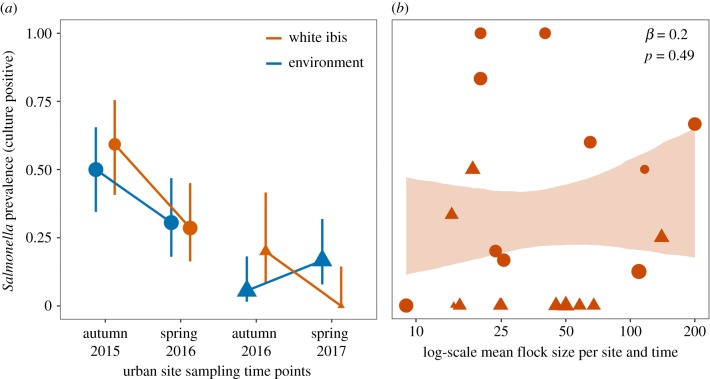
(a) Observed patterns of Salmonella prevalence pooled across six urban sites in southern Florida for white ibis (orange) and environmental samples (blue) during the 2015–2016 and 2016–2017 non-breeding seasons. Segments denote 95% confidence intervals (Wilson method), and points are scaled by sample size. (b) Test for density-dependent transmission of Salmonella as the relationship between log flock size and infection prevalence in white ibis per each site and time point. The band shows the 95% confidence interval from the GLMM (1000 bootstraps). Data from 2015 to 2016 are shown as circles, and data from 2016 to 2017 are shown as triangles. (Online version in colour.)
We visualized the distribution and median values of plausible parameter sets (i.e. those that resulted in simulated endpoint prevalence within the 95% confidence intervals of the field prevalence data in spring 2016) with violin plots across model scenarios. We then used GLMs with a Tweedie distribution or beta regressions with whether non-ibis sources were included ( and ), whether the contact-based transmission was included ( and ), and their interaction as fixed effects to quantify how treatments affected plausible parameter values (as the dependent variables) [59–61]. To assess performance across all four scenarios (figure 1b), we used the median plausible parameters and ran simulations using mean flock size and initial conditions from autumn 2016; we then qualitatively assessed how well these simulations matched our field data for end-of-season prevalence in spring 2017. Based on the relative magnitudes of estimated parameters and performance of the four model scenarios, we lastly derive a simplified model that minimized the number of parameters needed to capture observed infection dynamics.
3. Results
3.1. Analysis of field data to inform model structure
Across sites and time points, the prevalence of Salmonella in ibis was 30.6% (95% CI = 22.4–40.3), and the pooled prevalence in environmental samples was 26.7% (95% CI = 19.3–33.4). When stratified by season, Salmonella prevalence in ibis declined during the non-breeding season across both years of field data (figure 2a); this was mostly consistent within sites (electronic supplementary material, figure S1). Salmonella prevalence in ibis dropped from 59.26% to 28.57% in the 2015–2016 non-breeding season and from 20% to 0% in the 2016–2017 non-breeding season. Confidence intervals suggest either a modest decline or no change in prevalence for ibis. For environmental samples, Salmonella prevalence declined from 50% to 30.56% during the 2015–2016 non-breeding season but increased from 5.56% to 16.67% during the 2016–2017 non-breeding season (figure 2a). The confidence intervals suggest a modest increase or no change in environmental prevalence.
The mean size of urban flocks ranged from 9 to 200 ibis (figure 2b and electronic supplementary material, figure S2). The first GLMM found that flock sizes did not vary significantly across sampling time points (χ2 = 5.6, p = 0.13; electronic supplementary material, figure S2), justifying a constant flock size in our model. The binomial GLMM found no significant association between log flock size and site-level Salmonella prevalence in urban ibis during the non-breeding seasons (OR = 1.22, 95% CI = 0.72–2.37, p = 0.49; figure 2b). Results were similar for a GLMM that included the interaction between log flock size and year (χ2 = 1.08, p = 0.30). Owing to the lack of association between flock size and observed infection prevalence, we modelled ibis–ibis transmission using a frequency-dependent contact term ().
3.2. Model parametrization and restriction
We initialized models using field data from the start of the non-breeding season in 2015 (Pstart = 59%, estart = 50%, N = 68), resulting in 40 actively colonized () ibis. Given the 95% confidence intervals for Salmonella prevalence by the end of the 2015–2016 non-breeding season (figure 2a), we retained LHS parameter combinations if they produced endpoint prevalence between 16% and 45% in urban ibis and between 18% and 47% for environmental samples.
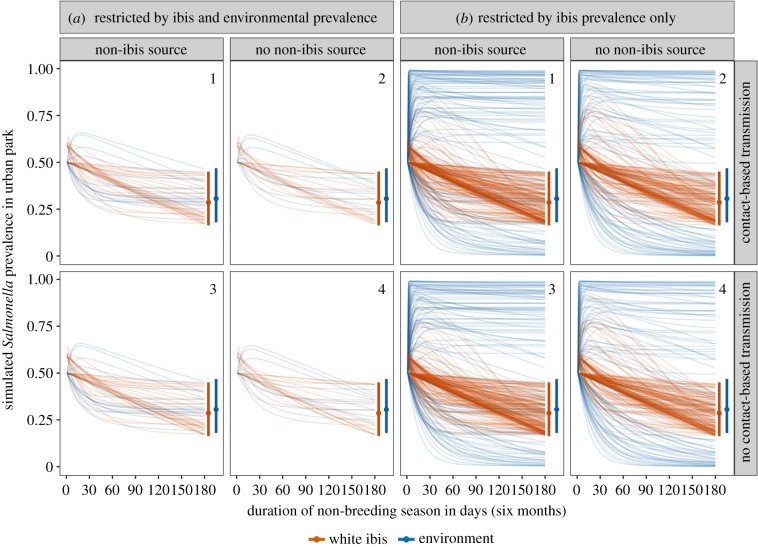
The simulated Salmonella time series in urban ibis and the environment for the plausible parameter combinations for all four model scenarios are presented in figure 3. Our parameter restriction process retained a very small number of LHS parameter combinations (1.25%, n = 50), suggesting that only a narrow region of parameter space could reproduce observed Salmonella dynamics in urban ibis and the environment (figure 3a; electronic supplementary material, table S1). Within these parameter sets, a small fraction generated time series that rapidly departed from initial conditions before returning within the observed prevalences by the end of the non-breeding season. The number of retained parameter combinations did not differ when non-ibis sources were included (; scenarios 1 and 3) nor when the contact-based transmission was included (; scenarios 2 and 4), suggesting both terms to be uninformative (figure 3a; electronic supplementary material, table S1). These results were robust to using the lower and upper 95% confidence interval for mean flock size and ibis prevalence for initial conditions (electronic supplementary material, table S1). Results were also similar when we initialized the model to have all infected ibis be transiently shedding or to evenly divide these between and (electronic supplementary material, table S2). Across all models, transiently shedding ibis () rarely contributed to overall ibis infection prevalence (electronic supplementary material, figure S3).
Figure 3.
Simulated Salmonella prevalence in ibis and the environment during the non-breeding season for parameter combinations retained from our LHS procedure. Lines show the simulated infection prevalence time series for ibis (orange) and the environment (blue) using plausible parameter combinations per model scenarios (indicated by 1–4). Rows show simulations where contact-based transmission was included (; top) or excluded (cf = 0; bottom). Columns show simulations where non-ibis sources of Salmonella were included (left in each panel; ψ ≥ 0) or excluded (right in each panel; ). Left pairs of columns show parameter combinations retained using field data on ibis and environmental prevalence (a), while right columns show those retained when only using ibis prevalence (b). Segments and points show observed prevalence and 95% confidence interval at the end of the 2015–2016 non-breeding season. (Online version in colour.)
When we ignored data on Salmonella prevalence in environmental samples for this LHS restriction process, far more parameter combinations were retained as plausible (electronic supplementary material, table S3; 9.45%, n = 378). However, the environmental prevalence in these simulations often reached 100%, which is unrealistic given the range of field data observed for these urban parks (figure 3b).
3.3. Statistical analysis of plausible parameters
While slightly more parameter combinations could reproduce observed dynamics of Salmonella in ibis and the environment when our model allowed non-ibis sources of infection (; scenarios 1 and 3; electronic supplementary material, table S1), most of these parameter values were not strongly affected by this model treatment (electronic supplementary material, table S4 and figure S4). Only the shedding rate (ϕ) varied considerably when including or excluding non-ibis sources (χ2 = 15.91); model scenarios excluding non-ibis sources had significantly higher shedding rates (predicted mean = 0.18%) than models that included non-ibis sources (predicted mean = 0.07%). No parameters were affected by including or excluding contact-based transmission (electronic supplementary material, table S4 and figure S4). Other parameters showed less variation (table 2), which suggests general estimates: the probability of colonization upon exposure (), duration of transient shedding ( hours), duration of colonization ( days), proportional shedding of transiently shedding ibis (), effective contact (), rate of environmental uptake (), duration of environmental persistence ( days) and fraction of the foraging area contaminated by non-ibis sources (ψ=0.69%). Plausible estimates of environmental uptake rates (ɛ) were consistently higher than effective contact rates (), providing support for environmental transmission as a key route of ibis exposure to Salmonella.
Table 2.
Plausible parameters (medians and range) derived from matching LHS combinations to Salmonella prevalence of urban ibis and environmental samples during the non-breeding season.
| non-ibis source () |
no non-ibis source () |
|||
|---|---|---|---|---|
| parameter | ||||
| θ (%) | 0.27 (0.01–0.47) | 0.272 (0.01–0.47) | 0.261 (0.058–0.47) | 0.261 (0.058–0.47) |
| (1/α) (h) | 6.54 (2.12–150) | 6.54 (2.12–150) | 5.64 (2.12–21.31) | 5.64 (2.12–21.31) |
| (days) | 25.64 (14.71–166.67) | 25.64 (14.71–166.67) | 22.72 (14.71–166.67) | 22.72 (14.71–166.67) |
| (%) | 0 (0–0.003) | 0 (0–0.003) | 0.002 (0–0.003) | 0.002 (0–0.003) |
| (%) | 0.59 (0.065–0.967) | 0.591 (0.065–0.967) | 0.62 (0.221–0.967) | 0.623 (0.221–0.967) |
| (per day) | 0 (0–0.009) | n.a. | 0 (0–0.009) | n.a. |
| (per day) | 0.229 (0–0.747) | 0.229 (0–0.747) | 0.266 (0–0.788) | 0.266 (0–0.788) |
| (days) | 22.72 (14.49–66.67) | 22.72 (14.49–66.67]) | 20.0 (14.49–43.48) | 20.0 (14.49–43.48) |
| ψ (%) | 0.007 (0–0.027) | 0.007 (0–0.027) | n.a. | n.a. |
3.4. Model validation
To validate parameter estimates from the initial LHS restriction procedure, we used the median parameter values and initial conditions from autumn 2016 to simulate Salmonella infection prevalence in the 2016–2017 non-breeding season. While these simulations produced moderate variability in Salmonella prevalence, all four model scenarios predicted the observed increase in Salmonella prevalence in the environment for spring 2017 (figure 4). Yet only the model scenarios excluding non-ibis sources (; scenarios 2 and 4) provided simulated outputs that fell within the 95% confidence interval observed for end-of-season ibis infectious prevalence.
Figure 4.
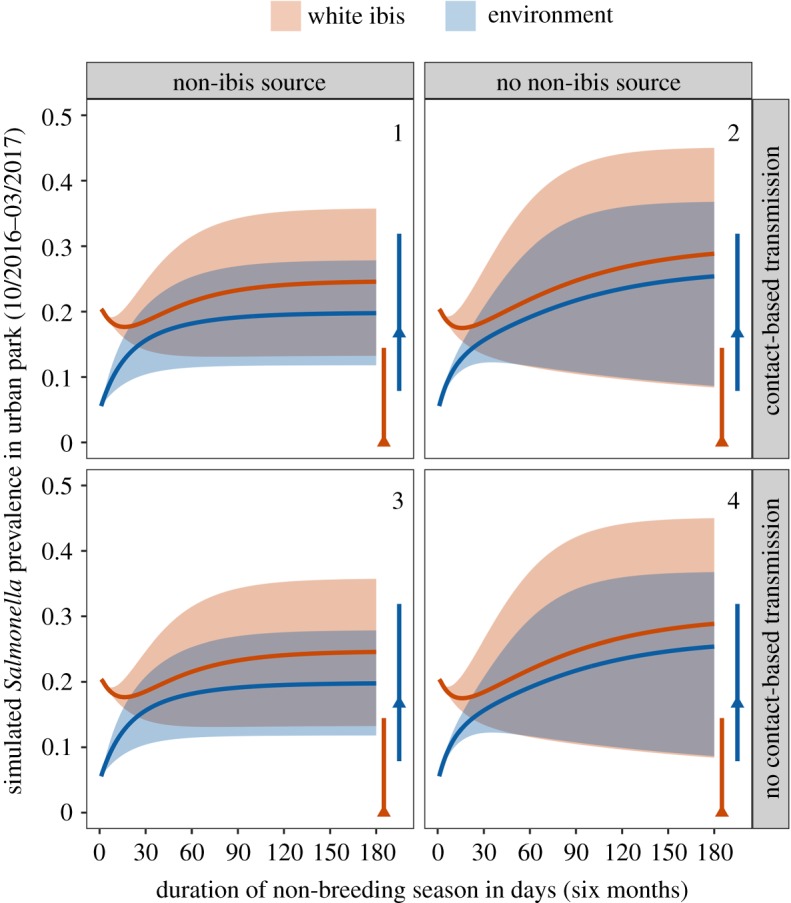
Validation of plausible parameters for each model scenario (indicated by 1–4) using mean flock size and initial conditions for autumn 2016. Lines show simulated infection prevalence time series for ibis (orange) and the environment (blue) using median values, while bands show the predictions generated using the standard error of these estimates. Rows show simulations where contact-based transmission was included (; top) or excluded (; bottom). Columns show simulations where non-ibis sources of Salmonella were included (left in each panel; ) or excluded (right in each panel; ). Segments and points show observed prevalence and 95% confidence interval at the end of the 2016–2017 non-breeding season. (Online version in colour.)
3.5. Model simplification
The above parameter restriction process and independent model validation showed that contact-based transmission, non-ibis sources and transiently shedding ibis were not necessary to capture observed temporal patterns of Salmonella infection prevalence. We thus repeated our parameter restriction procedure on a simplified model that excluded uninformative parameters (ψ, cf, δ, α). This simplified model is captured through the following two differential equations and includes actively colonized ibis, environmental exposure and Salmonella shedding into the environment:
and
Here is an aggregate parameter that combines the probability of colonization given exposure () and environmental uptake rate (). We repeated our LHS sensitivity analysis using the same starting conditions for autumn 2015, but we here used the ranges of our plausible parameter values (table 2) to set uniform distributions. The derived median parameter estimates were similar within under an order of magnitude to those from the full model ( days; days; ) and fell well within the 95% confidence intervals for the spring Salmonella prevalence data for ibis and the environment in both non-breeding seasons (figure 5).
Figure 5.
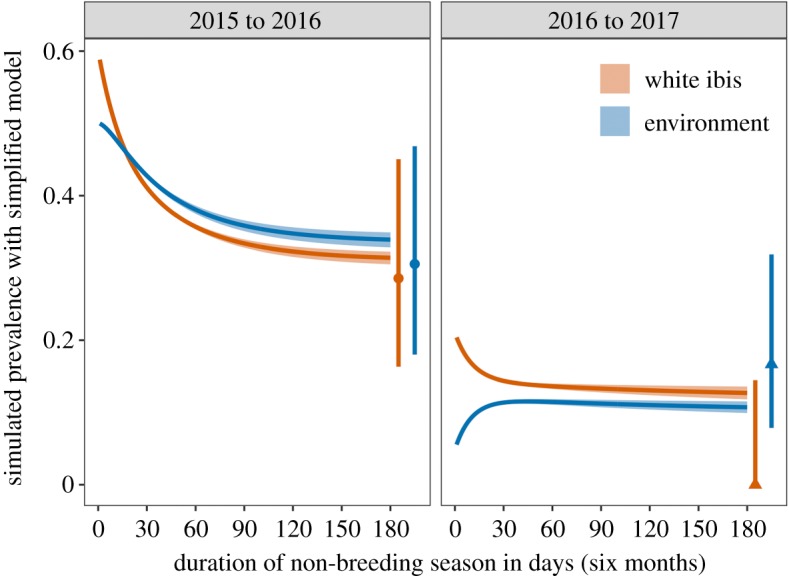
Predictions from the simplified model of ibis–Salmonella interactions (no contact-based transmission, no non-ibis sources, no transiently shedding hosts). Lines show simulated prevalence in ibis (orange) and the environment (blue) during the non-breeding season for median plausible parameter values and using mean flock size and initial conditions for autumn of each year. Bands show the range of predictions generated using the standard error of plausible parameters. Segments and points on the right side of each panel show the observed Salmonella prevalence and 95% confidence interval at the end of each non-breeding season. (Online version in colour.)
4. Discussion
Field data on infection prevalence combined with global sensitivity analyses of a compartmental model provided support for critical mechanisms for Salmonella transmission in an urban avian host. Of the four different model scenarios, we found most support for environmental uptake of Salmonella from an environmental pool (with input from ibis shedding but not external sources) as the dominant transmission route in this host–pathogen system, without requiring contact-based transmission between ibis. Sensitivity analyses also suggested that transiently shedding ibis likely contribute little to observed transmission. These findings help generate a parsimonious model of ibis–Salmonella interactions that could provide a starting point for predicting how ibis response to urbanization alters exposure to a multi-host enteric pathogen and spillover risks.
Because ibis are social and aggregate in flocks in urban parks [38], we expected that foraging in close proximity to actively shedding ibis might represent an elevated transmission risk, as has been suggested for avian influenza transmission in wild waterfowl [62]. Model analysis showed little support for the importance of this transmission route, as the fraction of plausible parameter sets did not vary between model scenarios excluding and including contact-based transmission (and the estimates for environmental uptake rates were consistently higher than effective contact rates). These findings suggest environmental shedding and persistence of infectious stages is necessary for sustained Salmonella transmission in urban ibis. Plausible estimates of Salmonella persistence times from the sensitivity analysis were of approximately 20 days, which match independent survival estimates from other non-avian systems [63–65].
A notable outcome of our model exploration was that non-ibis sources of environmental Salmonella were not necessary to produce sustained transmission and infection among ibis. Salmonella is present in abiotic reservoirs such as sewage [17,24] and can be harboured by many wildlife including bird species that frequent urban habitats [36,66–68]. However, our results suggest Salmonella could persist in urban ibis under the assumption of ibis shedding alone. When we validated the initially parametrized model with an independent year of field sampling (2016–2017) and used a simplified model that excluded non-ibis sources, we could still capture observed changes in prevalence for both ibis and the environment. This simplified model closely matched Salmonella prevalence by the end of the non-breeding season in 2015–2016, but the predictions for the end of the 2016–2017 non-breeding season tended to be higher for ibis and lower for the environment than observed in our data, suggesting that additions of Salmonella to the environment pool by non-ibis are occurring. Non-ibis sources could arise from human inputs, especially if high precipitation washes Salmonella-contaminated sewage or other substrate into parks [69,70]. While ibis may be able to sustain local transmission of Salmonella in some years, additional years of prevalence data and abiotic variables (e.g. rainfall and temperature) are needed to better understand the long-term contributions of ibis to the environmental pool [71].
The parameter restriction criteria used here identified a much wider range of plausible parameter sets when we used Salmonella prevalence from only ibis compared to using both ibis and the environment. Limiting the model by only ibis data represents contexts where prevalence is decoupled between this host species and the environment, such as if urban birds instead mostly obtain Salmonella outside of urban parks. However, the parameter retention process using all environmental prevalence data rejected more parameter combinations because simulated environmental prevalence expanded far beyond the range observed in our data. As noted above, more LHS parameter sets may have better approximated the field data if we incorporated other mechanisms of Salmonella loss, such as seasonal washout of environmental infection under heavy rain [69,70]. However, this discrepancy in regions of plausible parameter space when using both ibis and environmental prevalence more generally underscores the importance of sampling both reservoir hosts and environmental sources of infection to understand transmission.
Model simulations under plausible parameters suggested that environmental prevalence can increase rapidly when ibis arrive in urban parks at the start of the non-breeding season (e.g. figures 3a and 5). This initial increase in environmental inputs is consistent with empirical studies of other migratory and nomadic wildlife. For example, migratory saiga antelopes (Saiga tatarica) can carry and transmit nematodes to northern sheep populations, facilitating pulses of infection that coincide with annual saiga migrations [72]. Similarly, mass aggregations of ruddy turnstones (Arenaria interpres) at Delaware Bay during their spring migration fuel an ecological ‘hot spot’ for the transmission of avian influenza virus [73]. With relatively few sampling time points, we were unable to confirm this dynamical pattern in environmental prevalence. Future work could explore potential increases in prevalence early in the non-breeding season by sampling both urban soil and water more intensively over time within and before the ibis non-breeding season.
Understanding the transmission pathways of multi-host pathogens is important for better interpreting field studies and mitigating spillover risks [21,25,29]. In the case of urbanized white ibis, our model suggests that Salmonella is likely transmitted primarily through environmental exposures and may not require alternative sources of pathogen input. While the simplified model and parameter estimates derived from this analysis could be further refined by quantifying values through laboratory challenge studies and by repeating this procedure on site-specific data given larger sample sizes, our study highlights that Salmonella could be maintained through a cycle of faecal–oral environmental exposures and shedding possibly restricted to only this avian host. Ongoing sampling efforts in this system do suggest that waterfowl and other avian species also carry and shed Salmonella (S.M.H. et al., unpublished) and could therefore also contribute to the environmental pool. Although our model analysis suggests such inputs are not necessary to explain shedding patterns in ibis, future spatial and temporal surveillance of ibis and other avian species could be paired with explicitly multi-host models to understand their relative contribution as reservoir hosts of zoonotic Salmonella or whether these species instead facilitate transmission from exogenous sources of infection [34,36,37]. Furthermore, by generating the first parameter estimates for Salmonella shedding and uptake in a wild bird and elucidating a parsimonious set of model assumptions, this analysis also provides a starting point for predicting how urbanization affects Salmonella dynamics across landscapes and pathogen spillover risks [3,11,74]. Future applications of this model could critically address how urbanization modifies each transmission processes within ibis. For example, declines in infection prevalence could be driven by improved access of urban hosts to supplemental food resources that reduce susceptibility or increase pathogen clearance, as observed in other host systems [75,76]. Improved surveillance alongside quantification of how urbanization influences within-host factors, faecal–oral exposure pathways and interactions with the breeding season will be central to predict Salmonella spillover risks.
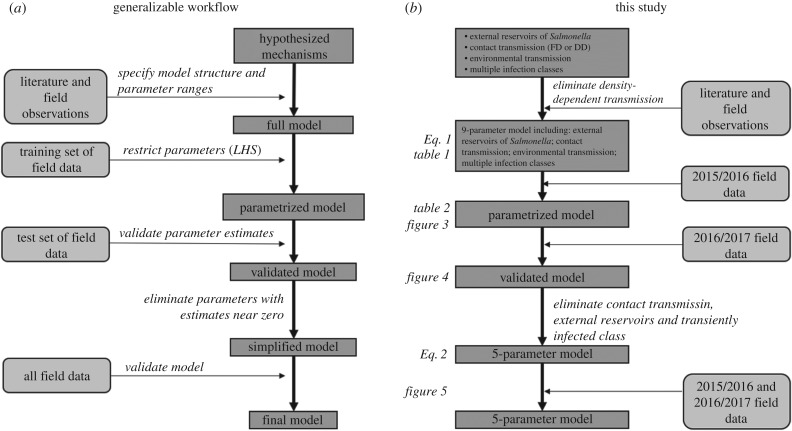
Uncertainties in transmission routes and parameter estimates are often critical limiting steps in modelling wildlife disease and using such models for generating management strategies [26,77]. Identifying a parsimonious model can often be achieved by applying likelihood-based inference to a time series of infection data [25,29], yet this can present logistical challenges for wildlife that display migratory or nomadic behaviour and can be restricted by limited sampling efforts across space and time [78–80]. Our analysis instead used an alternative workflow that generated a novel model with multiple exposure routes, infection classes, and Salmonella inputs; used previous studies and field data to inform model structure; applied LHS to cover a relevant region of parameter space; restricted model outputs by field prevalence data; validated plausible parameters against an independent dataset; and eliminated uninformative parameters to simplify our model to a more parsimonious and predictive set of assumptions (figure 6). Ultimately, the likelihood-based approaches could provide more robust parameter estimates and quantitatively select between alternative model structures. However, as our more qualitative procedure relied on only biannual sampling across two non-breeding seasons, this alternative workflow may have greater utility for often-sparse wildlife infection data. Owing to these biannual data, we note that our approach was prone to selecting a small fraction of parameter sets whose time series rapidly departed from the initial conditions before returning to within the observed end-of-season prevalences (e.g. figure 3), suggesting generally poor fits to the data. Our approach could therefore be extended to restrict parameter space with high-resolution temporal data (e.g. many sampling events within the non-breeding season), annual data over multiple years rather than biannual within-season data (e.g. for many non-migratory species) and with infection prevalence data from multiple host species. Our pooling data across sites are another limitation, as some variability in prevalence among parks (e.g. electronic supplementary material, figure S1) suggests stochasticity. Owing to small sample sizes per site, we here used an aggregate and deterministic model to capture the central tendencies in dynamics. Yet a stochastic framework could help generate such site-level variation while further refining plausible parameter selection (e.g. stochastic mortality of infected ibis could make even fewer sets plausible). Such methodological advances would further provide a more generalizable analytic roadmap to assess support for transmission pathways of multi-host wildlife pathogens.
Figure 6.
(a) Proposed generalizable analytic roadmap for inferring likely transmission processes for multi-host pathogens. (b) A summary of our study-specific modelling process.
Supplementary Material
Acknowledgements
We thank Kristen Navara, Jeffrey Hepinstall-Cymerman, Michael Yabsley and Anjelika Kidd for assistance with study design and data collection. We also thank Elizabeth Hammond, Marybeth Morrison, and government biologists who provided access to urban ibis capture sites. We lastly thank members of the Altizer and Hall groups at the University of Georgia and two anonymous reviewers for helpful comments on earlier versions of the manuscript.
Ethics
All capture and handling procedures were reviewed and approved by the University of Georgia's Institutional Animal Care and Use Committee (A2016 11-019-Y1-A0), a Florida Wildlife Conservation Commission permit (LSSC-11-00119F), and a USFWS permit (MB779238-0).
Data accessibility
R code and estimates of Salmonella prevalence are available in the Dryad Digital Repository: https://dx.doi.org/10.5061/dryad.982dd1k [81].
Authors' contributions
D.J.B. participated in model development, analysed the data and model and wrote the manuscript; C.S.T. participated in model development, contributed to model and data analysis and helped write the manuscript; M.H.M., S.E.C. and C.N.W. participated in field study design and collected data; T.E. and H.C.A. collected data; R.S.R. performed Salmonella analyses; E.K.L. and S.M.H. designed the field study, coordinated Salmonella analyses and contributed to model and data analysis; S.A. and R.J.H. participated in field study design, model development and data and model analysis. All authors provided critical comments on the manuscript and gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
D.J.B., C.S.T., M.H.M., S.E.C., C.N.W., T.E., H.C.A., R.S.R., E.K.L., S.M.H., S.A. and R.J.H. were supported by the National Science Foundation (DEB-1518611). C.S.T. was also supported by a University of Georgia Presidential Fellowship; R.J.H. was also supported by funding from the Office of Research at the University of Georgia; and D.J.B. was also supported by an appointment to the Intelligence Community Postdoctoral Research Fellowship Program at Indiana University, administered by Oak Ridge Institute for Science and Education through an interagency agreement between the US Department of Energy and the Office of the Director of National Intelligence.
References
- 1.McKinney ML. 2006. Urbanization as a major cause of biotic homogenization. Biol. Conserv. 127, 247–260. ( 10.1016/j.biocon.2005.09.005) [DOI] [Google Scholar]
- 2.Bradley CA, Altizer S. 2007. Urbanization and the ecology of wildlife diseases. Trends Ecol. Evol. 22, 95–102. ( 10.1016/j.tree.2006.11.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hassell JM, Begon M, Ward MJ, Fèvre EM. 2017. Urbanization and disease emergence: dynamics at the wildlife–livestock–human interface. Trends Ecol. Evol. 32, 55–67. ( 10.1016/j.tree.2016.09.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker DJ, Streicker DG, Altizer S. 2015. Linking anthropogenic resources to wildlife–pathogen dynamics: a review and meta-analysis. Ecol. Lett. 18, 483–495. ( 10.1111/ele.12428) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galbraith JA, Stanley MC, Jones DN, Beggs JR. 2017. Experimental feeding regime influences urban bird disease dynamics. J. Avian Biol. 48, 700–713. ( 10.1111/jav.01076) [DOI] [Google Scholar]
- 6.Plowright RK, Foley P, Field HE, Dobson AP, Foley JE, Eby P, Daszak P. 2011. Urban habituation, ecological connectivity and epidemic dampening: the emergence of Hendra virus from flying foxes (Pteropus spp.). Proc. R. Soc. B 278, 3703–3712. ( 10.1098/rspb.2011.0522) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flint BF, Hawley DM, Alexander KA. 2016. Do not feed the wildlife: associations between garbage use, aggression, and disease in banded mongooses (Mungos mungo). Ecol. Evol. 6, 5932–5939. ( 10.1002/ece3.2343) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bichet C, Scheifler R, Cœurdassier M, Julliard R, Sorci G, Loiseau C. 2013. Urbanization, trace metal pollution, and malaria prevalence in the house sparrow. PLoS ONE 8, e53866 ( 10.1371/journal.pone.0053866) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawson B, et al. 2014. Epidemiological evidence that garden birds are a source of human salmonellosis in England and Wales. PLoS ONE 9, e88968 ( 10.1371/journal.pone.0088968) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mackenstedt U, Jenkins D, Romig T. 2015. The role of wildlife in the transmission of parasitic zoonoses in peri-urban and urban areas. Int. J. Parasitol. Parasites Wildl. 4, 71–79. ( 10.1016/j.ijppaw.2015.01.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faust CL, McCallum HI, Bloomfield LS, Gottdenker NL, Gillespie TR, Torney CJ, Dobson AP, Plowright RK. 2018. Pathogen spillover during land conversion. Ecol. Lett. 21, 471–483. ( 10.1111/ele.12904) [DOI] [PubMed] [Google Scholar]
- 12.Woolhouse ME, Taylor LH, Haydon DT. 2001. Population biology of multihost pathogens. Science 292, 1109–1112. ( 10.1126/science.1059026) [DOI] [PubMed] [Google Scholar]
- 13.Haydon DT, Cleaveland S, Taylor LH, Laurenson MK. 2002. Identifying reservoirs of infection: a conceptual and practical challenge. Emerg. Infect. Dis. 8, 1468–1473. ( 10.3201/eid0812.010317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garchitorena A, et al. 2017. Disease ecology, health and the environment: a framework to account for ecological and socio-economic drivers in the control of neglected tropical diseases. Phil. Trans. R. Soc. B 372, 20160128 ( 10.1098/rstb.2016.0128) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hilbert F, Smulders FJM, Chopra-Dewasthaly R, Paulsen P. 2012. Salmonella in the wildlife-human interface. Food Res. Int. 45, 603–608. ( 10.1016/j.foodres.2011.08.015) [DOI] [Google Scholar]
- 16.Wille M, Bröjer C, Lundkvist Å, Järhult JD. 2018. Alternate routes of influenza A virus infection in mallard (Anas platyrhynchos). Vet. Res. 49, 110 ( 10.1186/s13567-018-0604-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winfield MD, Groisman EA. 2003. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl. Environ. Microbiol. 69, 3687–3694. ( 10.1128/AEM.69.7.3687-3694.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown JD, Swayne DE, Cooper RJ, Burns RE, Stallknecht DE. 2007. Persistence of H5 and H7 avian influenza viruses in water. Avian Dis. 51, 285–289. ( 10.1637/7636-042806R.1) [DOI] [PubMed] [Google Scholar]
- 19.Roche B, Lebarbenchon C, Gauthier-Clerc M, Chang C-M, Thomas F, Renaud F, van der Werf S, Guégan J-F. 2009. Water-borne transmission drives avian influenza dynamics in wild birds: the case of the 2005–2006 epidemics in the Camargue area. Infect. Genet. Evol. 9, 800–805. ( 10.1016/j.meegid.2009.04.009) [DOI] [PubMed] [Google Scholar]
- 20.Tellier R. 2006. Review of aerosol transmission of influenza A virus. Emerg. Infect. Dis. 12, 1657–1662. ( 10.3201/eid1211.060426) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown VL, Drake JM, Stallknecht DE, Brown JD, Pedersen K, Rohani P. 2013. Dissecting a wildlife disease hotspot: the impact of multiple host species, environmental transmission and seasonality in migration, breeding and mortality. J. R. Soc. Interface 10, 20120804 ( 10.1098/rsif.2012.0804) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holt PS. 1995. Horizontal transmission of Salmonella enteritidis in molted and unmolted laying chickens. Avian Dis. 37, 239–249. ( 10.2307/1591865) [DOI] [PubMed] [Google Scholar]
- 23.Hoelzer K, Moreno Switt AI, Wiedmann M. 2011. Animal contact as a source of human non-typhoidal salmonellosis. Vet. Res. 42, 34 ( 10.1186/1297-9716-42-34) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinde H, et al. 1997. Prevalence of Salmonella in municipal sewage treatment plant effluents in southern California. Avian Dis. 41, 392–398. ( 10.2307/1592195) [DOI] [PubMed] [Google Scholar]
- 25.Plowright RK, Peel AJ, Streicker DG, Gilbert AT, McCallum H, Wood J, Baker ML, Restif O. 2016. Transmission or within-host dynamics driving pulses of zoonotic viruses in reservoir–host populations. PLoS Neglect. Trop. Dis. 10, e0004796 ( 10.1371/journal.pntd.0004796) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCallum H. 2016. Models for managing wildlife disease. Parasitology 143, 805–820. ( 10.1017/S0031182015000980) [DOI] [PubMed] [Google Scholar]
- 27.Blackwood JC, Streicker DG, Altizer S, Rohani P. 2013. Resolving the roles of immunity, pathogenesis, and immigration for rabies persistence in vampire bats. Proc. Natl Acad. Sci. USA 110, 20 837–20 842. ( 10.1073/pnas.1308817110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dean KR, Krauer F, Walløe L, Lingjærde OC, Bramanti B, Stenseth NC, Schmid BV. 2018. Human ectoparasites and the spread of plague in Europe during the Second Pandemic. Proc. Natl Acad. Sci. USA 115, 1304–1309. ( 10.1073/pnas.1715640115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Restif O, et al. 2012. Model-guided fieldwork: practical guidelines for multidisciplinary research on wildlife ecological and epidemiological dynamics. Ecol. Lett. 15, 1083–1094. ( 10.1111/j.1461-0248.2012.01836.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kushlan JA. 1979. Feeding ecology and prey selection in the white ibis. Condor 81, 376–389. ( 10.2307/1366963) [DOI] [Google Scholar]
- 31.Dorn NJ, Cook MI, Herring G, Boyle RA, Nelson J, Gawlik DE. 2011. Aquatic prey switching and urban foraging by the white ibis Eudocimus albus are determined by wetland hydrological conditions. Ibis 153, 323–335. ( 10.1111/j.1474-919X.2011.01101.x) [DOI] [Google Scholar]
- 32.Traut AH, Hostetler ME. 2004. Urban lakes and waterbirds: effects of shoreline development on avian distribution. Landsc. Urban Plan. 69, 69–85. ( 10.1016/j.landurbplan.2003.08.009) [DOI] [Google Scholar]
- 33.Murray MH, Kidd AD, Curry SE, Hepinstall-Cymerman J, Yabsley MJ, Adams HC, Ellison T, Welch CN, Hernandez SM. 2018. From wetland specialist to hand-fed generalist: shifts in diet and condition with provisioning for a recently urbanized wading bird. Phil. Trans. R. Soc. B 373, 20170100 ( 10.1098/rstb.2017.0100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hernandez SM, et al. 2016. Urbanized white ibises (Eudocimus albus) as carriers of Salmonella enterica of significance to public health and wildlife. PLoS ONE 11, e0164402 ( 10.1371/journal.pone.0164402) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Becker DJ, Hall RJ. 2014. Too much of a good thing: resource provisioning alters infectious disease dynamics in wildlife. Biol. Lett. 10, 20140309 ( 10.1098/rsbl.2014.0309) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tizard I. 2004. Salmonellosis in wild birds. Semin. Avian Exotic Pet Med.13, 50–66. ( 10.1053/j.saep.2004.01.008) [DOI] [Google Scholar]
- 37.Epstein JH, McKee J, Shaw P, Hicks V, Micalizzi G, Daszak P, Kilpatrick AM, Kaufman G. 2006. The Australian white ibis (Threskiornis molucca) as a reservoir of zoonotic and livestock pathogens. EcoHealth 3, 290–298. ( 10.1007/s10393-006-0064-2) [DOI] [Google Scholar]
- 38.Keeling MJ, Rohani P. 2011. Modeling infectious diseases in humans and animals. Princeton, NJ: Princeton University Press. [Google Scholar]
- 39.Bildstein KL, Post W, Johnston J, Frederick P. 1990. Freshwater wetlands, rainfall, and the breeding ecology of white ibises in coastal South Carolina. Wilson Bull. 102, 84–98. [Google Scholar]
- 40.Welch CN. 2016. Urban land use and movements of white ibises (Eudocimus albus) in South Florida. PhD thesis, University of Georgia, Athens, USA. [Google Scholar]
- 41.Bildstein KL. 1993. White ibis. Washington, DC: Smithsonian Institution Press. [Google Scholar]
- 42.Phalen DN, Drew ML, Simpson B, Roset K, Dubose K, Mora M. 2010. Salmonella enterica subsp. enterica in cattle egret (Bubulcus ibis) chicks from central Texas: prevalence, serotypes, pathogenicity, and epizootic potential. J. Wildl Dis. 46, 379–389. ( 10.7589/0090-3558-46.2.379) [DOI] [PubMed] [Google Scholar]
- 43.Kushlan JA. 1977. Foraging behavior of the white ibis. Wilson Bull. 89, 342–345. [Google Scholar]
- 44.Smith HW, Tucker JF. 1980. The virulence of Salmonella strains for chickens: their excretion by infected chickens. Epidemiol. Infect. 84, 479–488. ( 10.1017/S0022172400027017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams EM, Frederick PC, Larkin IL, Guillette LJ. 2009. Sublethal effects of methylmercury on fecal metabolites of testosterone, estradiol, and corticosterone in captive juvenile white ibises (Eudocimus albus). Environ. Toxicol. Chem. 28, 982–989. ( 10.1897/08-253.1) [DOI] [PubMed] [Google Scholar]
- 46.Desmidt M, Ducatelle R, Mast J, Goddeeris BM, Kaspers B, Haesebrouck F. 1998. Role of the humoral immune system in Salmonella enteritidis phage type four infection in chickens. Vet. Immunol. Immunopathol. 63, 355–367. ( 10.1016/S0165-2427(98)00112-3) [DOI] [PubMed] [Google Scholar]
- 47.Barrow PA, Bumstead N, Marston K, Lovell MA, Wigley P. 2003. Faecal shedding and intestinal colonization of Salmonella enterica in in-bred chickens: the effect of host-genetic background. Epidemiol. Infect. 132, 117–126. ( 10.1017/S0950268803001274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shivaprasad HL, Timoney JF, Morales S, Lucio B, Baker RC. 1990. Pathogenesis of Salmonella enteritidis infection in laying chickens. I. Studies on egg transmission, clinical signs, fecal shedding, and serologic responses. Avian Dis. 34, 548–557. ( 10.2307/1591243) [DOI] [PubMed] [Google Scholar]
- 49.Beal RK, Wigley P, Powers C, Hulme SD, Barrow PA, Smith AL. 2004. Age at primary infection with Salmonella enterica serovar typhimurium in the chicken influences persistence of infection and subsequent immunity to re-challenge. Vet. Immunol. Immunopathol. 100, 151–164. ( 10.1016/j.vetimm.2004.04.005) [DOI] [PubMed] [Google Scholar]
- 50.Rhodes MW, Kator H. 1988. Survival of Escherichia coli and Salmonella spp. in estuarine environments. Appl. Environ. Microbiol. 54, 2902–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown LD, Cai TT, DasGupta A. 2001. Interval estimation for a binomial proportion. Stat. Sci. 16, 101–117. ( 10.1214/ss/1009213286) [DOI] [Google Scholar]
- 52.R Core Team. 2013. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See http://www.R-project.org. [Google Scholar]
- 53.Bates D, Mächler M, Bolker B, Walker S. 2015. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48. ( 10.18637/jss.v067.i01) [DOI] [Google Scholar]
- 54.Stein M. 1987. Large sample properties of simulations using Latin hypercube sampling. Technometrics 29, 143–151. ( 10.1080/00401706.1987.10488205) [DOI] [Google Scholar]
- 55.Carnell R. 2009. lhs: latin hypercube samples. R package version 0.5, 100.
- 56.Soetaert KER, Petzoldt T, Setzer RW. 2010. Solving differential equations in R: package deSolve. J. Stat. Softw. 33, 1–25. ( 10.18637/jss.v033.i09)20808728 [DOI] [Google Scholar]
- 57.Blower SM, Dowlatabadi H. 1994. Sensitivity and uncertainty analysis of complex models of disease transmission: an HIV model, as an example. Int. Stat. Rev. 62, 229–243. ( 10.2307/1403510) [DOI] [Google Scholar]
- 58.Samuel MD, Hobbelen PHF, DeCastro F, Ahumada JA, LaPointe DA, Atkinson CT, Woodworth BL, Hart PJ, Duffy DC. 2011. The dynamics, transmission, and population impacts of avian malaria in native Hawaiian birds: a modeling approach. Ecol. Appl. 21, 2960–2973. ( 10.1890/10-1311.1) [DOI] [Google Scholar]
- 59.Ferrari S, Cribari-Neto F. 2004. Beta regression for modelling rates and proportions. J. Appl. Stat. 31, 799–815. ( 10.1080/0266476042000214501) [DOI] [Google Scholar]
- 60.Smyth GK, Verbyla AP. 1999. Adjusted likelihood methods for modelling dispersion in generalized linear models. Environmetrics 10, 695–709. ( 10.1002/(SICI)1099-095X(199911/12)10:6%3C695::AID-ENV385%3E3.0.CO;2-M) [DOI] [Google Scholar]
- 61.White JW, Rassweiler A, Samhouri JF, Stier AC, White C. 2014. Ecologists should not use statistical significance tests to interpret simulation model results. Oikos 123, 385–388. ( 10.1111/j.1600-0706.2013.01073.x) [DOI] [Google Scholar]
- 62.Breban R, Drake JM, Stallknecht DE, Rohani P. 2009. The role of environmental transmission in recurrent avian influenza epidemics. PLoS Comput. Biol. 5, e1000346 ( 10.1371/journal.pcbi.1000346) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baloda SB, Christensen L, Trajcevska S. 2001. Persistence of a Salmonella enterica serovar typhimurium DT12 clone in a piggery and in agricultural soil amended with Salmonella-contaminated slurry. Appl. Environ. Microbiol. 67, 2859–2862. ( 10.1128/AEM.67.6.2859-2862.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parker WF, Mee BJ. 1982. Survival of Salmonella adelaide and fecal coliforms in coarse sands of the swan costal plain, Western Australia. Appl. Environ. Microbiol. 43, 981–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jensen AN, Dalsgaard A, Stockmarr A, Nielsen EM, Baggesen DL. 2006. Survival and transmission of Salmonella enterica serovar typhimurium in an outdoor organic pig farming environment. Appl. Environ. Microbiol. 72, 1833–1842. ( 10.1128/AEM.72.3.1833-1842.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krawiec M, Kuczkowski M, Kruszewicz AG, Wieliczko A. 2015. Prevalence and genetic characteristics of Salmonella in free-living birds in Poland. BMC Vet. Res. 11, 15 ( 10.1186/s12917-015-0332-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lillehaug A, Jonassen CM, Bergsjø B, Hofshagen M, Tharaldsen J, Nesse L, Handeland K. 2005. Screening of feral pigeon (Colomba livia), mallard (Anas platyrhynchos) and graylag goose (Anser anser) populations for Campylobacter spp., Salmonella spp., avian influenza virus and avian paramyxovirus. Acta Vet. Scand. 46, 193 ( 10.1186/1751-0147-46-193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Sousa E, Júnior AB, Pinto AA, Machado RZ, Carrasco AOT, Marciano JA, Werther K. 2010. Prevalence of Salmonella spp. antibodies to Toxoplasma gondii, and Newcastle disease virus in feral pigeons (Columba livia) in the City of Jaboticabal, Brazil. J. Zoo Wildl. Med. 41, 603–607. ( 10.1638/2008-0166.1) [DOI] [PubMed] [Google Scholar]
- 69.Schmidt N, Lipp EK, Rose JB, Luther ME. 2001. ENSO influences on seasonal rainfall and river discharge in Florida. J. Climate 14, 615–628. ( 10.1175/1520-0442(2001)014%3C0615:EIOSRA%3E2.0.CO;2) [DOI] [Google Scholar]
- 70.Vereen E, Lowrance RR, Jenkins MB, Adams P, Rajeev S, Lipp EK. 2013. Landscape and seasonal factors influence Salmonella and Campylobacter prevalence in a rural mixed use watershed. Water Res. 47, 6075–6085. ( 10.1016/j.watres.2013.07.028) [DOI] [PubMed] [Google Scholar]
- 71.Jiang C, Shaw KS, Upperman CR, Blythe D, Mitchell C, Murtugudde R, Sapkota AR, Sapkota A. 2015. Climate change, extreme events and increased risk of salmonellosis in Maryland, USA: evidence for coastal vulnerability. Environ. Int. 83, 58–62. ( 10.1016/j.envint.2015.06.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morgan ER, Medley GF, Torgerson PR, Shaikenov BS, Milner-Gulland EJ. 2007. Parasite transmission in a migratory multiple host system. Ecol. Modell. 200, 511–520. ( 10.1016/j.ecolmodel.2006.09.002) [DOI] [Google Scholar]
- 73.Krauss S, Stallknecht DE, Negovetich NJ, Niles LJ, Webby RJ, Webster RG. 2010. Coincident ruddy turnstone migration and horseshoe crab spawning creates an ecological ‘hot spot’ for influenza viruses. Proc. R. Soc. B 277, 3373–3379. ( 10.1098/rspb.2010.1090) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Becker DJ, Snedden CE, Altizer S, Hall RJ. 2018. Host dispersal responses to resource supplementation determine pathogen spread in wildlife metapopulations. Am. Nat. 192, 503–517. ( 10.1086/699477) [DOI] [PubMed] [Google Scholar]
- 75.Hwang J, Kim Y, Lee S-W, Kim N-Y, Chun M-S, Lee H, Gottdenker N. 2018. Anthropogenic food provisioning and immune phenotype: association among supplemental food, body condition, and immunological parameters in urban environments. Ecol. Evol. 8, 3037–3046. ( 10.1002/ece3.3814) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Becker DJ, et al. 2018. Livestock abundance predicts vampire bat demography, immune profiles and bacterial infection risk. Phil. Trans. R. Soc. B 373, 20170089 ( 10.1098/rstb.2017.0089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Russell RE, Katz RA, Richgels KLD, Walsh DP, Grant EHC. 2017. A framework for modeling emerging diseases to inform management. Emerg. Infect. Dis. 23, 1–6. ( 10.3201/eid2301.161452) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hoye BJ, Munster VJ, Nishiura H, Klaassen M, Fouchier RAM. 2010. Surveillance of wild birds for avian influenza virus. Emerg. Infect. Dis. 16, 1827–1834. ( 10.3201/eid1612.100589) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kuiken T, Leighton FA, Fouchier RAM, LeDuc JW, Peiris JSM, Schudel A, Stöhr K, Osterhaus ADME. 2005. Pathogen surveillance in animals. Science 309, 1680–1681. ( 10.1126/science.1113310) [DOI] [PubMed] [Google Scholar]
- 80.Stallknecht DE. 2007. Impediments to wildlife disease surveillance, research, and diagnostics. In Wildlife and emerging zoonotic diseases: the biology, circumstances and consequences of cross-species transmission (eds JE Childs, JS Mackenzie, JA Richt), pp. 445–461. Berlin, Germany: Springer. ( 10.1007/978-3-540-70962-6_17) [DOI] [PubMed] [Google Scholar]
- 81.Becker DJ, et al. 2018. Data from: Assessing the contributions of intraspecific and environmental sources of infection in urban wildlife: Salmonella enterica and white ibis as a case study Dryad Digital Repository. ( 10.5061/dryad.982dd1k) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Becker DJ, et al. 2018. Data from: Assessing the contributions of intraspecific and environmental sources of infection in urban wildlife: Salmonella enterica and white ibis as a case study Dryad Digital Repository. ( 10.5061/dryad.982dd1k) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
R code and estimates of Salmonella prevalence are available in the Dryad Digital Repository: https://dx.doi.org/10.5061/dryad.982dd1k [81].