Abstract
Background
Acute myeloid leukemia (AML) is a clinically and biologically heterogeneous disease. The survival of older patients is generally poor. In the current study, we sought to investigate the differences in molecular gene mutations between younger and older AML patients, and to identify those newly diagnosed AML patients who are more likely to respond to standard cytarabine and daunorubicin induction chemotherapy.
Methods
We retrospectively evaluated 179 patients who were newly diagnosed with non-M3 AML. A next-generation sequencing assay covering 34 genes was used to investigate recurrently mutated genes. The mutational status of fusion genes was determined by real time PCR.
Results
The median age at diagnosis was 53 years (range 18–88 years). Sixty-eight patients were 60 years or older with a median age of 67 years (range 60–88 years). Eighteen patients (10.1%) carried t(8;21)(q22;q22.1) or RUNX1–RUNX1T1 gene fusion, and there was a significantly higher incidence in younger patients (p = 0.019). At least one non-synonymous gene mutation was detected in 159 patients (88.8%). The median number of gene mutations was two (range 0–6). The mean number of molecular gene mutations at diagnosis was higher in older patients than younger patients (2.5 vs 1.83, p = 0.003). Older patients had significantly higher incidences of ASXL1 (22.1% vs 13.5%, p = 0.025) and TP53 mutations (13.2% vs 3.6%, p = 0.034). In total, 78 patients received DA60 (daunorubicin 60 mg/m2 per day on days 1–3 and cytarabine 100 mg/m2 twice per day on days 1–7) as the induction therapy, and information was available on their response to induction treatment. Patients with RUNX1–RUNX1T1 gene fusion were significantly more likely to achieve complete remission (CR) after DA60 induction therapy (p = 0.026), as were patients without the ASXL1 mutation (p = 0.007).
Conclusion
Older AML patients had a lower incidence of favorable cytogenetics and higher frequencies and burdens of molecular mutations that are associated with poor prognosis compared to younger patients. Patients with RUNX1–RUNX1T1 gene fusion or without the ASXL1 gene mutation had a better chance of achieving CR when treated with cytarabine and daunorubicin induction chemotherapy.
Keywords: Acute myeloid leukemia, Induction chemotherapy, Next-generation sequencing, Older patients
Background
Acute myeloid leukemia (AML) is a clinically and biologically heterogeneous disease characterized by the clonal expansion of undifferentiated myeloid precursors. Although induction chemotherapy with cytarabine and daunorubicin has been shown over the past 40 years to improve clinical outcomes in younger patients, survival for older patients remains very poor [1–3]. In addition to patient factors, such as old age, poor performance status and concomitant comorbidity, genetic factors are also related to outcomes in older AML patients [4–6].
The AML genome is one of the simplest cancer genomes, which makes it amenable to the clinical use of targeted next-generation sequencing (NGS). Recently, a combination of cytogenetic analysis and mutation testing has been integrated into the classification and risk assessment of AML patients [7–9]. The European LeukemiaNet revised the prognostic model for AML in 2017 to add mutations in RUNX1 and ASXL1 to the previously identified molecular risk categories defined by mutations in NPM1, CEBPA, FLT3–ITD and TP53. This model stratifies AML patients into three prognostic groups (good, intermediate and poor risk). However, recent advances in the understanding of the molecular alterations in AML are mostly derived from younger patients [9]. Some studies have demonstrated that the chromosome abnormalities and gene mutation patterns are different among older AML patients [7, 8, 10]. Less is known about the differences in gene mutations between younger and older patients with AML, especially Chinese patients.
The aim of this study was to comprehensively investigate the differences in clinicobiological features and molecular gene mutations between younger and older AML patients, and to identify those newly diagnosed AML patients who are more likely to respond to standard cytarabine and daunorubicin induction chemotherapy.
Methods
Patients
All 179 adult patients who were newly diagnosed with non-M3 AML according to the FAB criteria [11] at Peking Union Medical College Hospital (Peking, China) between July 2015 and April 2018, and for whom complete clinical, cytogenetic and laboratory data before treatment were available, were included in this study. Bone marrow samples from all patients underwent mutational analysis by NGS. Informed consent was obtained from all patients and the protocol was approved by the Peking Union Medical College Hospital Ethics Committee. The present study was performed in accordance with the ethical standards of the 1964 Declaration of Helsinki and its later amendments.
Cytogenetics and fusion genes analysis
The bone marrow samples were investigated using G-banding analysis and karyotyped according to the International System for Human Cytogenetic Nomenclature.
The mutational status of fusion genes such as RUNX1–RUNX1T1 and CBFβ–MYH11 was determined by real time PCR (RT-PCR). In addition, RT-PCR was used to detect 11 MLL-related common fusion genes, including MLL-AF1q, MLL-AF1p, MLL-AF4, MLL-AF9, MLL-AF10, MLL-AF6, MLL-ELL, MLL-ENL, MLL-AFX, MLL-SEPT6 and MLL-AF17. We used Multiplex RT-PCR Fusion Gene Kits provided by Rightongene.
Next-generation sequencing
Analyses were conducted of the relevant mutations of 34 genes, including ASXL1, BCOR, BCORL1, CALR, CBL, CEBPA, CSF3R, DNMT3A, ETV6, EZH2, FLT3, GATA2, IDH1, IDH2, JAK2, KIT, KMT2A, KRAS, MPL, NPM1, NRAS, PDGFRA, PHF6, PIGA, RUNX1, SETBP1, SF3B1, SH2B3, SRSF2, TET2, TP53, U2AF1, WT1 and ZRSR2.
Read pairs were aligned to Refseq hg19 (downloaded from the UCSC Genome Browser, URLs) by the Burrows–Wheeler Aligner version 0.7.13-r1126. Samtools version 1.3 was used to generate chromosomal coordinate-sorted BAM files. We used targeted next-generation sequencing with the Rightongene AML/MDS/MPN Sequencing Panel (Rightongene). The NGS libraries were paired-end sequenced (2 × 150 bp) on an Illumina MiSeq System (Illumina, San Diego, CA). The mean depth of each sample was 2500×, with an average 5% of the target sequence being covered sufficiently deeply for variant calling. Samtools mpileup was applied for SNV/indel calling and filter workflow.
Gene mutations were assigned to functional groups similar to those previously described as follows [12, 13]: DNA methylation and hydroxymethylation-related—DNMT3A, TET2 and IDH1/2; RNA spliceosome—SF3B1, SRSF2, ZRSR2 and U2AF1; chromatin remodeling—ASXL1, EZH2, BCOR and KMT2A; transcriptional deregulation—CEBPA, RUNX1 and WT1; activated signaling—NRAS, KRAS, CBL, KIT, JAK2 and FLT3.
Statistical analysis
Complete remission (CR) was defined according to the criteria of the International Working Group [14]. The discrete variables of patients with and without specific molecular alteration were compared using the Fisher exact test. The Fisher exact test was used to compare categorical variables, whereas the Mann–Whitney test was used to compare continuous variables between groups. We performed all statistical analyses using SPSS version 21 software (IBM Corp., Armonk, NY, USA), and considered p-values of less than 0.05 to be statistically significant.
Results
Clinical characteristics
Of 179 newly diagnosed AML patients, 116 were males and 63 were females (Table 1). The median age at diagnosis was 53 years (range 18–88 years). Sixty-eight patients were 60 years or older with a median age of 67 years (range 60–88 years). Five patients were diagnosed as treatment-related AML (one was older patient), and 21 patients were diagnosed as secondary AML (ten with myelodysplastic syndrome, three with chronic myeloid leukemia, three with chronic myelomonocytic leukemia, two with myelofibrosis, two with essential thrombocythaemia and one with myeloproliferative neoplasm). Among the 21 secondary AML patients, nine were older patients. There were no differences in gender, white blood cells (WBC) or blasts in the bone marrow between younger patients and older patients.
Table 1.
Clinical manifestations and cytogenetic abnormalities of AML patients stratified by age
| Total (n = 179) | Younger patients (n = 111) | Older patients (n = 68) | p value | |
|---|---|---|---|---|
| Age, years (median, range) | 53 (18–88) | 44 (18–59) | 67 (60–88) | |
| Male (n, %) | 116 (64.8) | 71 (64.0) | 45 (66.2) | 0.872 |
| WBC (× 109/L) (median, range) | 7.82 (0.4–362.6) | 8.48 (0.4–362.6) | 7.42 (0.6–271.4) | 0.380 |
| Blast cells in BM (%, range) | 53 (10–95) | 57 (10–94.5) | 53 (21–95) | 0.824 |
| Complex karyotype | 16/130 (12.3) | 9/81 (11.1) | 7/49 (14.3) | 0.784 |
| RUNX1–RUNX1T1 | 18 (10.1) | 16 (14.4) | 2 (2.9) | 0.019 |
| CBFβ–MYH11 | 10 (5.6) | 8 (7.2) | 2 (2.9) | 0.322 |
WBC white blood cell, BM bone marrow
Molecular gene mutations of patients
Chromosome data were available for 130 patients at diagnosis, including 63 patients (48.5%) with a normal karyotype and 16 patients (12.3%) with a complex karyotype. Eighteen patients (10.1%) carried t(8;21)(q22;q22.1) or RUNX1–RUNX1T1 gene fusion, while ten patients (5.6%) carried inv(16)(p13.1q22) or t(16;16)(p13.1;q22) or CBFβ–MYH11 gene fusion. Younger patients had a significantly higher incidence of t(8;21)(q22;q22.1) or RUNX1–RUNX1T1 gene fusion (p = 0.019) (Table 1). We also found two patients had MLL-AF9, one patient had MLL-ELL, one patient had MLL-ENL and one patient had MLL-SEPT6 gene fusion.
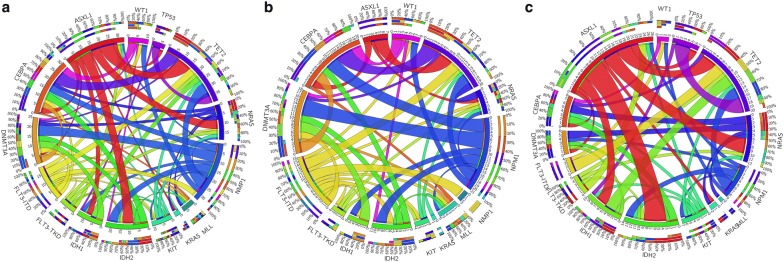
At least one non-synonymous gene mutation was detected in 159 patients (88.8%). The median number of gene mutations was two (range 0–6). The distributions of molecular gene mutations are shown in Fig. 1. The most common molecular event in the total cohort was a CEBPA mutation (17.9%), followed by TET2 (16.8%), ASXL1 (14.0%), NRAS (11.7%), NPM1 (11.2%), IDH2 (10.1%) and FLT3–ITD mutations (10.1%) (Table 2). Both the distribution of gene mutations and the pattern of mutation co-occurrence appear to be distinct between older and younger AML patients (Fig. 1). The mean number of molecular gene mutations at diagnosis was higher in older patients than younger patients (2.5 vs 1.83, p = 0.003). Older patients had significantly higher incidences of ASXL1 (22.1% vs 13.5%, p = 0.025) and TP53 (13.2% vs 3.6%, p = 0.034) mutations. The older patient group also showed a trend of more DNMT3A (14.7% vs 5.4%, p = 0.056) and RUNX1 (11.8% vs 4.5%, p = 0.081) mutations than the younger patient group. Moreover, younger patients showed a trend of more KIT (10.8% vs 2.5%, p = 0.083) and NRAS (15.3% vs 5.9%, p = 0.092) mutations. Among the different gene functional groups, the older patient group had a significantly higher incidence of DNA methylation- and hydroxymethylation-related genes (48.5% vs 27.0%, p = 0.004), RNA spliceosome (30.9% vs 14.4%, p = 0.013) and chromatin remodeling (29.4% vs 10.8%, p = 0.002) gene mutations, while the younger patient group had a significantly higher incidence of activated signaling gene mutations (37.8% vs 19.1%, p = 0.012).
Fig. 1.
Circos plots depicting the relative frequencies and pairwise co-occurrences of selected common genetic alterations: in all AML patients (a), separately in patients 60 years or older (b) and in patients younger than 60 years (c). The length of the arc corresponds to the frequency of the first gene mutation, and the width of the ribbon corresponds to the proportion of co-occurrence with the second gene mutation. Both the distribution of gene mutations and the pattern of mutation co-occurrences appear to be distinct between older and younger AML patients
Table 2.
Association of age with molecular genetic alterations
| Total (n = 179) | Younger patients (n = 111) | Older patients (n = 68) | p value | |
|---|---|---|---|---|
| Mutation number (mean) | 2.08 | 1.83 | 2.50 | 0.003 |
| CEBPA | 32 (17.9) | 22 (19.8) | 10 (14.7) | 0.428 |
| TET2 | 30 (16.8) | 15 (13.5) | 15 (22.1) | 0.153 |
| ASXL1 | 25 (14.0) | 10 (9.0) | 15 (22.1) | 0.025 |
| NRAS | 21 (11.7) | 17 (15.3) | 4 (5.9) | 0.092 |
| NPM1 | 20 (11.2) | 12 (10.8) | 8 (11.8) | 1 |
| IDH2 | 18 (10.1) | 10 (9.0) | 8 (11.8) | 0.768 |
| FLT3–ITD | 18 (10.1) | 12 (10.8) | 6 (8.8) | 0.800 |
| DNMT3A | 16 (8.9) | 6 (5.4) | 10 (14.7) | 0.056 |
| KIT | 14 (7.8) | 12 (10.8) | 2 (2.9) | 0.083 |
| TP53 | 13 (7.3) | 4 (3.6) | 9 (13.2) | 0.034 |
| RUNX1 | 13 (7.3) | 5 (4.5) | 8 (11.8) | 0.081 |
| IDH1 | 12 (6.7) | 5 (4.5) | 7 (10.3) | 0.216 |
| WT1 | 9 (5.0) | 7 (6.3) | 2 (2.9) | 0.486 |
| GATA2 | 5 (2.8) | 2 (1.8) | 3 (4.4) | 0.370 |
| FLT3–TKD | 5 (2.8) | 3 (2.7) | 2 (2.9) | 1 |
| KRAS | 4 (2.2) | 3 (2.7) | 1 (1.5) | 1 |
| DNA methylation | 63 (35.2) | 30 (27.0) | 33 (48.5) | 0.004 |
| RNA splicing | 37 (20.7) | 16 (14.4) | 21 (30.9) | 0.013 |
| Chromatin architecture | 32 (17.9) | 12 (10.8) | 20 (29.4) | 0.002 |
| Transcriptional deregulation | 50 (27.9) | 31 (27.9) | 19 (27.9) | 1 |
| Activated signaling | 55 (30.7) | 42 (37.8) | 13 (19.1) | 0.012 |
Patients with t(8;21)(q22;q22.1) or RUNX1–RUNX1T1 gene fusion had a median one gene mutation (range 0–3). Seven (38.9%) also had a KIT mutation (p < 0.001), and none had a CEBPA mutation (p = 0.046).
CEBPA mutations were detected in 32 patients, and 11 of those patients (6.1%) were bi-allelic. Five patients with a CEBPA mutation also had a GATA2 mutation, while none of the CEBPA wild-type patients had a GATA2 mutation (p < 0.001) (Table 3). Among the 25 patients who had an ASXL1 mutation, eight patients also had a TET2 mutation (p = 0.041), six also had an IDH2 mutation (p = 0.023) and four had a SETBP1 mutation (p = 0.008). Among the 20 subjects with an NPM1 mutation, six also had a DNMT3A mutation (p = 0.004) and five also had an IDH2 mutation (p = 0.034). We also noted that IDH2 and TET2 mutations were mutually exclusive. None of the 30 subjects who had a TET2 mutation had an IDH2 mutation (p = 0.046). None of the other co-mutated genes reached statistically significant incidence.
Table 3.
Correlations among different mutations
| CEBPA Mut | CEBPA WT | p value | |
|---|---|---|---|
| GATA2 Mut | 5 | 0 | |
| GATA2 WT | 27 | 147 | < 0.001 |
| ASXL1 Mut | ASXL1 WT | p value | |
|---|---|---|---|
| TET2 Mut | 8 | 22 | |
| TET2 WT | 17 | 132 | 0.041 |
| IDH2 mut | 6 | 12 | |
| IDH2 WT | 19 | 142 | 0.023 |
| SETBP1 mut | 4 | 3 | |
| SETBP` WT | 21 | 151 | 0.008 |
| NPM1 Mut | NPM1 WT | p value | |
|---|---|---|---|
| DNMT3A Mut | 6 | 10 | |
| DNMT3A WT | 14 | 149 | 0.004 |
| IDH2 mut | 5 | 13 | |
| IDH2 WT | 15 | 146 | 0.034 |
| TET2 Mut | TET2 WT | p value | |
|---|---|---|---|
| IDH2 Mut | 0 | 18 | |
| IDH2 WT | 30 | 131 | 0.046 |
Mut mutant, WT wild type
Correlations between mutations and clinical outcomes
Thirty-three patients went back to their local hospital for further chemotherapy after diagnosis in our center. Eight patients only received best support therapy. In total, 138 patients received induction chemotherapy. Among them, 78 patients received DA60 (daunorubicin 60 mg/m2 per day on days 1–3 and cytarabine 100 mg/m2 twice per day on days 1–7) as the induction therapy and information was available on their response to induction treatment. Fifteen of those patients were 60 years or older, and the overall median age was 46 years (range 18–73 years). Forty-five patients (57.7%) achieved a CR following one course of induction therapy. We found patients with RUNX1–RUNX1T1 gene fusion were significantly more likely to achieve a CR after DA60 induction therapy (p = 0.026) as well as patients without the ASXL1 mutation (p = 0.007).
Discussion
Genetic mutations in AML patients that escape cytogenetic detection are increasingly being discovered, and these mutations may serve as potential markers to extend the prognostic parameters in AML. An understanding of the frequencies of gene mutations in AML patients can facilitate the selection of targeted therapies and help us understand the potential pathways or resistance mechanisms. In the current study, we found 88.8% of AML patients had at least one mutation detected by targeted NGS. This figure is higher than what has previously been reported in the literature [15], which may be due to the larger gene panel we used. This high incidence of gene mutation detection further justifies the performance of NGS on AML patients as it can yield, at high frequency, genetic information that may be clinically actionable.
CEBPA was found to have the highest mutation rate in our cohort, which was higher than that reported in the literature [16, 17] but similar to previous reports on Chinese AML patients [18, 19]. We detected NPM1 and FLT3 mutations at frequencies of 11.2% and 12.9%, respectively, which is similar to the results reported by Hussaini et al. [20]. However, other groups have reported frequencies ranging from 20 to 33% [5–10, 19, 21]. This difference may be due to the different patient populations, but we cannot exclude technical differences as different laboratories use different mutation detection techniques and algorithms for calling mutations.
In our study we confirmed that patients with RUNX1–RUNX1T1 gene fusion had a significantly higher incidence of KIT mutation. We also found some gene mutations were correlated (e.g. CEBPA and GATA2; ASXL1 and TET2, IDH2, SETBP1; NPM1 and DNMT3A, IDH2). On the other hand, some gene mutations appeared to be potentially mutually exclusive (e.g. TET2 and IDH2). However, the clinical and biological significances of these correlations or exclusions of gene mutations are unknown.
Although much effort has been made to clarify the correlation between molecular changes and clinical outcomes of AML patients, most of the data pertain to younger patients. In our study, we consecutively recruited newly diagnosed AML patients for whom sufficient samples were available for mutation analysis without restriction of age, so we could compare the genetic alterations between younger and older AML patients. First, we showed that younger patients had more RUNX1–RUNX1T1 gene fusion than older patients. Second, older patients had more gene mutations than younger patients. Furthermore, older patients had higher incidences of ASXL1, TP53, DNA methylation- and hydroxymethylation-related, RNA spliceosome and chromatin remodeling gene mutations, and also exhibited a trend of higher incidences of DNMT3A and RUNX1 mutations. Younger patients had higher incidences of activated signaling gene mutations. Several genetic alterations were found to have prognostic significance and have been incorporated into risk stratification of AML [8]. Previously, RUNX1–RUNX1T1 gene fusion was considered a good prognostic factor and TP53 and ASXL1 mutations were poor prognostic factors [8, 22]. Our study confirmed TP53 and ASXL1 mutations are prevalent in older patients. Taken together, the data suggest the pathogenesis of AML among younger patients vs older patients may be different. In addition to a lower incidence of favorable cytogenetics, the higher frequencies and burdens of molecular mutations that are associated with poor prognosis in older patients might explain the dismal outcome in this patient group.
The use of targeted NGS testing is useful for the identification of the population of patients who have an excellent chance of achieving a CR when treated with cytarabine and daunorubicin induction chemotherapy. We have shown that patients who have a RUNX1–RUNX1T1 gene fusion or do not have an ASXL1 gene mutation constitute a group whose chance of achieving a CR is higher. An alternative induction therapy or use of a novel agent may need to be considered in patients with ASXL1 gene mutation.
Conclusion
In conclusion, our data indicate clinically targeted NGS sequencing frequently detects gene mutations in AML patients (more than 85%). Older AML patients had a lower incidence of favorable cytogenetics and higher frequencies and burdens of molecular mutations that are associated with poor prognosis than younger patients. Patients with a RUNX1–RUNX1T1 gene fusion or without an ASXL1 gene mutation have a better chance of achieving a CR when treated with cytarabine and daunorubicin induction chemotherapy.
Authors’ contributions
XC, DZ and JL contributed to the conception and design of the study; YM, QW and LZ contributed to data collection; XC wrote the paper; HC performed the next-generation sequencing and fusion genes analysis and all authors revised the paper. All authors read and approved the final manuscript.
Acknowledgements
The authors thank the patients and their families.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Informed consent was obtained from all patients and the protocol was approved by the Peking Union Medical College Hospital Ethics Committee.
Funding
The Young Scientific Research Fund of PUMC (Grant No. 2017320004).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- AML
acute myeloid leukemia
- CR
complete remission
- DA60
daunorubicin 60 mg/m2 per day on days 1–3 and cytarabine 100 mg/m2 twice per day on days 1–7
- NGS
next-generation sequencing
- RT-PCR
real time PCR
Contributor Information
Xin-xin Cao, Email: caoxinxin@pumch.cn.
Hao Cai, Email: caihao@pumch.cn.
Yue-ying Mao, Email: maoyueying@pumch.cn.
Qi Wu, Email: wuqi_15@163.com.
Lu Zhang, Email: zhanglu7@pumch.cn.
Dao-bin Zhou, Email: zhoudb@pumch.cn.
Jian Li, Email: lijian@pumch.cn.
References
- 1.Howlader N, Noone AM, Krapcho M, et al., editors. SEER cancer statistics review, 1975–2010. Bethesda: National Cancer Institute. 2013. http://seer.cancer.gov/csr/1975_2010/.
- 2.Burnett A, Wetzler M, Lowenberg B. Therapeutic advances in acute myeloid leukemia. J Clin Oncol. 2011;29:487–494. doi: 10.1200/JCO.2010.30.1820. [DOI] [PubMed] [Google Scholar]
- 3.Dӧhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373:1136–1152. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 4.Appelbaum FR, Gundacker H, Head DR, et al. Age and acute myeloid leukemia. Blood. 2006;107:3481–3485. doi: 10.1182/blood-2005-09-3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai CH, Hou HA, Tang JL, et al. Genetic alterations and their clinical implications in older patients with acute myeloid leukemia. Leukemia. 2016;30:1485–1492. doi: 10.1038/leu.2016.65. [DOI] [PubMed] [Google Scholar]
- 6.Eisfeld AK, Kohlschmidt J, Mrózek K, et al. Mutation patterns identify adult patients with de novo acute myeloid leukemia aged 60 years or older who respond favorably to standard chemotherapy: an analysis of Alliance studies. Leukemia. 2018;32:1338–1348. doi: 10.1038/s41375-018-0068-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Metzeler KH, Herold T, Rothenberg-Thurley M, et al. Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood. 2016;128:686–698. doi: 10.1182/blood-2016-01-693879. [DOI] [PubMed] [Google Scholar]
- 8.Döhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grimwade D, Ivey A, Huntly BJP. Molecular landscape of acute myeloid leukemia in younger adults and its clinical significance. Blood. 2016;127:29–41. doi: 10.1182/blood-2015-07-604496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prassek VV, Rothenberg-Thurley M, Sauerland MC, et al. Genetics of acute myeloid leukemia in the elderly: mutation spectrum and clinical impact in intensively treated patients aged ≥ 75 years. Haematologica. 2018 doi: 10.3324/haematol.2018.191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bennett JM, Catovsky D, Daniel MT, et al. Proposed revised criteria for the classification of acute myeloid leukemia. A report of the French–American–British Cooperative Group. Ann Intern Med. 1985;103:620–625. doi: 10.7326/0003-4819-103-4-620. [DOI] [PubMed] [Google Scholar]
- 12.Shlush LI, Zandi S, Mitchell A, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukemia. Nature. 2014;506:328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindsley RC, Mar BG, Mazzola E, et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood. 2015;125:1367–1376. doi: 10.1182/blood-2014-11-610543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheson BD, Bennett JM, Kopecky KJ, et al. Revised recommendations of the international working group for diagnosis, standardization of response criteria, treatment outcomes, and reporting standards for therapeutic trials in acute myeloid leukemia. J Clin Oncol. 2003;21:4642–4649. doi: 10.1200/JCO.2003.04.036. [DOI] [PubMed] [Google Scholar]
- 15.Ohgami RS, Ma L, Merker JD, et al. Next-generation sequencing of acute myeloid leukemia identifies the significance of TP53, U2AF1, ASXL1, and TET2 mutations. Mod Pathol. 2015;28:706–714. doi: 10.1038/modpathol.2014.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pabst T, Eyholzer M, Fos J, et al. Heterogeneity within AML with CEBPA mutations; only CEBPA double mutations, but not single CEBPA mutations are associated with favourable prognosis. Br J Cancer. 2009;100:1343–1346. doi: 10.1038/sj.bjc.6604977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollink IH, van den Heuvel-Eibrink MM, Arentsen-Peters ST, et al. Characterization of CEBPA mutations and promoter hypermethylation in pediatric acute myeloid leukemia. Haematologica. 2011;96:384–392. doi: 10.3324/haematol.2010.031336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen Y, Zhu YM, Fan X, et al. Gene mutation patterns and their prognostic impact in a cohort of 1185 patients with acute myeloid leukemia. Blood. 2011;118:5593–5603. doi: 10.1182/blood-2011-03-343988. [DOI] [PubMed] [Google Scholar]
- 19.Zhang M, Yin J, He Q, et al. Chinese and Europeans with acute myeloid leukemia have discordant mutation topographies. Leuk Res. 2018;70:8–12. doi: 10.1016/j.leukres.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 20.Hussaini MO, Mirza AS, Komrokji R, et al. Genetic landscape of acute myeloid leukemia interrogated by next-generation sequencing: a large cancer center experience. Cancer Genom Proteom. 2018;15:121–126. doi: 10.21873/cgp.20070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metzeler KH, Becker H, Maharry K, et al. ASXL1 mutations identify a high-risk subgroup of older patients with primary cytogenetically normal AML within the ELN favorable genetic category. Blood. 2011;118:6920–6929. doi: 10.1182/blood-2011-08-368225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.