Abstract
Background
Urban rodents and house shrews are closely correlated in terms of location with humans and can transmit many pathogens to them. Hepatitis E has been confirmed to be a zoonotic disease. However, the zoonotic potential of rat HEV is still unclear. The aim of this study was to determine the prevalence and genomic characteristics of hepatitis E virus (HEV) in rodents and house shrews.
Results
We collected a total of 788 animals from four provinces in China. From the 614 collected murine rodents, 20.19% of the liver tissue samples and 45.76% of the fecal samples were positive for HEV. From the 174 house shrews (Suncus murinus), 5.17% fecal samples and 0.57% liver tissue samples were positive for HEV. All of the HEV sequences obtained in this study belonged to Orthohepevirus C1. However, we observed a lower percentage of identity in the ORF3 region upon comparing the amino acid sequences between Rattus norvegicus and Rattus losea. HEV derived from house shrews shared a high percentage of identity with rat HEV. Notably, the first near full-length of the HEV genome from Rattus losea is described in our study, and we also report the first near full-length rat HEV genomes in Rattus norvegicus from China.
Conclusion
HEV is prevalent among the three common species of murine rodents (Rattus. norvegicus, Rattus. tanezumi, and Rattus. losea) in China. HEV sequences detected from house shrews were similar to rat HEV sequences. The high identity of HEV from murine rodents and house shrews suggested that HEV can spread among different animal species.
Electronic supplementary material
The online version of this article (10.1186/s12917-018-1746-z) contains supplementary material, which is available to authorized users.
Keywords: Genomic characteristic, Hepatitis E virus, House shrew, Murine rodent, Prevalence
Background
Hepatitis E virus (HEV) is the causative agent of hepatitis E in humans worldwide. According to 2018 data from the World Health Organization (WHO), there are 20 million HEV infections each year, leading to about 3.3 million symptomatic cases, with approximately one-third of the world’s population having been exposed to HEV [1]. HEV can cause hepatitis outbreaks and sporadic hepatitis, which is usually a self-limiting disease [2]. However, in immunosuppressed patients, HEV infection can cause rapidly progressive cirrhosis [3]. In the general population, the mortality rates of HEV infection range from 0.2–1%; however, the mortality rate is higher during pregnancy, especially in developing countries [4]. Therefore, HEV infection is a major global public health concern.
HEV is a positive-sense, single-stranded, non-enveloped RNA virus with a size of 27–34 nm. The genome of HEV contains three overlapping open reading frames (ORF), specifically ORF1 that encodes nonstructural proteins; ORF2 that encodes viral capsid protein, which is responsible for self-assembly of virus particles [5]; and ORF3 that encodes proteins involved in virus morphogenesis and release [6]. ORF4 has been identified in rat and ferret HEV strains; however, its function is still unknown [7–9].
HEV is classified in the genus Orthohepevirus in the family Hepeviridae [10]. Orthohepevirus is divided into four species, as follows: Orthohepevirus A, B, C, and D. All of the HEV strains isolated from humans belong to the species Orthohepevirus A with four genotypes, genotypes 1 and 2 can only infect humans, while genotypes 3 and 4 can infect animals and are considered zoonotic pathogens [11]. Orthohepevirus B includes avian HEV strains. Orthohepevirus C is divided into two species, one of which is Orthohepevirus C1, derived from rats, and the other of which is Orthohepevirus C2, derived from ferrets. Bat HEV is classified in Orthohepevirus D [2].
The origin of HEV is still unknown. HEV was first considered to be restricted to presentation in humans [12]. The most common transmission route of HEV is the fecal–oral route. However, in the recent years, HEV had been detected in a variety of animal species, such as pigs, wild boars, cattle, cats, rabbits, rodents, and dogs [13]. The consumption of meat or meat products and direct or indirect contact with pigs can be important modes of zoonotic HEV transmission [2, 11]. Hepatitis E has been confirmed to be a zoonotic disease [14]. In Japan, HEV strains isolated from Rattus norvegicus (R. norvegicus) that had been trapped near a pig farm where HEV was prevalent were genetically identical to the HEV strains derived from pigs [15], indicating that rats may be a reservoir for the HEV that causes infection in pigs.
Rat HEV was first detected in R. norvegicus from Germany [8]. Many studies since have reported the detection of HEV in rats. However, it is still inconclusive whether rat HEV can cross transmit to other species. In recent years, HEV has been detected in house shrews (Suncus murinus) trapped in China with a high similarity to the rat HEV strains, indicating that rat HEV might be transmitted between rats and shrews [16].
Murine rodents and house shrews are reservoirs of some human pathogens, such as mammarenavirus and hantavirus, owing to their close contact with humans [16]. However, the zoonotic potential of rat HEV is still unclear. Nonetheless, some studies have pointed out the possibility of HEV transmission between humans and rats. For example, recent investigations have shown that HEV genotype 3 has been detected in wild rats; rat HEV can successfully replicate in three human hepatoma cell lines (PLC/PRF/5, HuH-7, and HepG2 cells); and anti-rat HEV antibodies were detected in forest workers [15, 17–19].
It is important to understand the possibility of interspecies transmission of HEV between humans, rodents, and shrews. In China, the first HEV in rats was reported in 2013 [20]. So far, the presence of HEV in rats and shrews has only been reported in the Guangdong and Yunnan Provinces of China [16, 21]. Separately, the detection of full-length genomes of HEV in the Chevrier’s field mouse (Apodemus chevrieri) and Père David’s vole (Eothenomys melanogaster) have been reported in China [21]. However, the genomic characteristics of HEV in commensal rodents in China remain unclear.
In the present study, we investigated the prevalence of HEV in murine rodents and house shrews in five regions of four provinces in China. Besides, we analyzed the characteristics of the nucleic acid sequences of HEV obtained from the murine rodents and house shrews.
Results
Prevalence of HEV in murine rodents and shrews
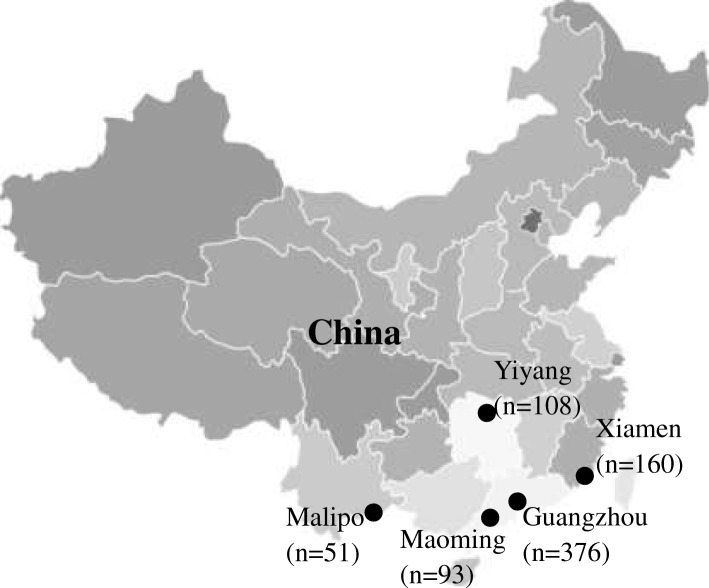
From 2014 to 2017, a total of 788 animals were trapped. Liver and fecal specimens were collected from 456 R. norvegicus, 64 Rattus tanezumi (R. tanezumi), 93 Rattus losea (R. losea), 1 Bandicota indica (B. indica), and 174 Suncus murinus (S. murinus) (Fig. 1 and Table 1).
Fig. 1.
Location of the trapping sites of the murine rodents and house shrews in China. Number of animals trapped are indicated in brackets. Map source: http://image.so.com/v?src=360pic_normal&z=1&i=0&cmg=9923e970972c38717e105d62081f0181&q=%E4%B8%AD%E5%9B%BD%E5%9C%B0%E5%9B%BE&correct=%E4%B8%AD%E5%9B%BD%E5%9C%B0%E5%9B%BE&cmsid=4372c66d63a3bc1a0c3a77413cee395a&cmran=0&cmras=0&cn=0&gn=0&kn=11#multiple=0&gsrc=1&dataindex=44&id=8c7daffc8205d4592f8e4a6e8c0e25e2&currsn=0&jdx=44&fsn=71
Table 1.
Prevalence of HEV in liver and fecal samples (%, n)
| Sample | Family | Species | Guangzhou | Yiyang | Xiamen | Malipo | Maoming | Total |
|---|---|---|---|---|---|---|---|---|
| Liver | Muridae | Rattus norvegicus | 14.57 (29/199) | 30.34 (27/89) | 12.50 (4/32) | 14.00 (7/50) | 29.07 (25/86) | 20.18 (92/456) |
| Rattus tanezumi | 0 (0/13) | 10.53 (2/19) | 16.67 (4/24) | 0 (0/1) | 0 (0/7) | 9.48 (6/64) | ||
| Rattus losea | - | - | 27.96 (26/93) | - | - | 27.96 (26/93) | ||
| Bandicota indica | - | - | 0 (0/1) | - | - | 0 (0/1) | ||
| Subtotal | 13.68 (29/212) | 26.85 (29/108) | 22.67 (34/150) | 13.72 (7/51) | 26.88 (25/93) | 20.19 (124/614) | ||
| Soricidae | Suncus murinus | 0 (0/164) | - | 10.00 (1/10) | - | - | 0.57 (1/174) | |
| Total | 7.71 (29/376) | 26.85 (29/108) | 21.88 (35/160) | 13.72 (7/51) | 26.88 (25/93) | 15.86 (125/788) | ||
| Fecal | Muridae | Rattus norvegicus | 42.21 (84/199) | 65.17 (58/89) | 28.12 (9/32) | 66.00 (33/50) | 59.30 (51/86) | 51.54 (235/456) |
| Rattus tanezumi | 15.38 (2/13) | 68.42 (13/19) | 12.50 (3/24) | 100 (1/1) | 28.57 (2/7) | 32.81 (21/64) | ||
| Rattus losea | - | - | 26.88 (25/93) | - | - | 26.88 (25/93) | ||
| Bandicota indica | - | - | 0 (0/1) | - | - | 0 (0/1) | ||
| Subtotal | 40.57 (86/212) | 65.74 (71/108) | 24.66 (37/150) | 66.67 (34/51) | 56.99 (53/93) | 45.76 (281/614) | ||
| Soricidae | Suncus murinus | 3.05 (5/164) | - | 40.00 (4/10) | - | - | 5.17 (9/174) | |
| Total | 24.20 (91/376) | 65.74 (71/108) | 25.62 (41/160) | 66.67 (34/51) | 56.99 (53/93) | 36.80 (290/788) | ||
According to nested broad-spectrum RT-PCR and nested PCR, 20.19% (124/614) of the liver tissue samples and 45.76% (281/614) of the fecal samples from murine rodents were positive for HEV. Of the 281 fecal-HEV-positive murine rodents, 84 (29.89%) also tested positive for the virus in their liver tissue. Nine (5.17%) fecal samples from house shrews were positive for HEV. Among these nine fecal-HEV-positive house shrews, one of them also was positive for HEV in the liver (Table 1).
Among the five regions of sampling, the prevalence of HEV in liver tissue samples ranged from 7.71–26.88%, while that in fecal samples ranged from 24.20–66.67%. Detection rates of HEV in liver samples from different species of animals ranged from 0 to 27.96%. With regard to fecal samples, the positive rates of HEV in different species of animals ranged from 0 to 51.54%. The positive rate of HEV in liver tissue samples from R. losea was significantly higher than in other species (χ2 = 4.399, P < 0.05). For fecal samples, the positive rate for HEV was significantly higher in R. norvegicus than in other species (χ2 = 86.309, P < 0.001) (Table 1).
Phylogenetic analysis
Partial ORF1 sequences and ORF1–ORF2 sequences were detected from liver tissue samples and fecal samples.
The identity among the representative partial ORF1 sequences obtained in our study ranged from 77.2–100% (Additional file 1: Table S1). Among these sequences, two sequences detected in R. losea from Xiamen City were totally identical (XM16 and XM27). Additionally, one sequence detected in house shrew from Xiamen City was identical to another sequence detected in R. norvegicus from Yunnan Province (XM54 and MLP51). The identity between the representative sequences obtained in this study and that reported in a previous study were also compared (GQ504010) [8] (Table 2). HEV detected from R. losea had a lower percentage of identity than that detected from other species. In this region, the percentage of identity at the amino acid level is greater than the percentage of identity at the nucleotide level.
Table 2.
Nucleotide (nt) and amino acid sequence identity for the region spanning nt positions 4139–4393 or amino acid positions 1379–1462, based on strain GQ504010
| Species | Nt identity (%) | Amino acid identity (%) |
|---|---|---|
| Rattus norvegicus (13a) | 81.5–86.6 | 88.2–96.4 |
| Rattus tanezumi (2a) | 84.3–86.2 | 95.2 |
| Rattus losea (3a) | 78.4–79.6 | 94.1 |
| Suncus murinus (1a) | 83.9 | 95.2 |
anumber of the sequences
GQ504010, GenBank accession number of a HEV strain isolated from R. norvegicus from Germany
Phylogenetic trees constructed by using two different methods were similar. Here, we only showed the phylogenetic trees constructed with MrBayes (version 3.2) (Figs. 2, 3, 4, 5, 6, 7 and 8). The phylogenetic trees obtained based on the neighbor-joining method are shown in the supplementary information file (Additional file 1: Figures S1-S7).
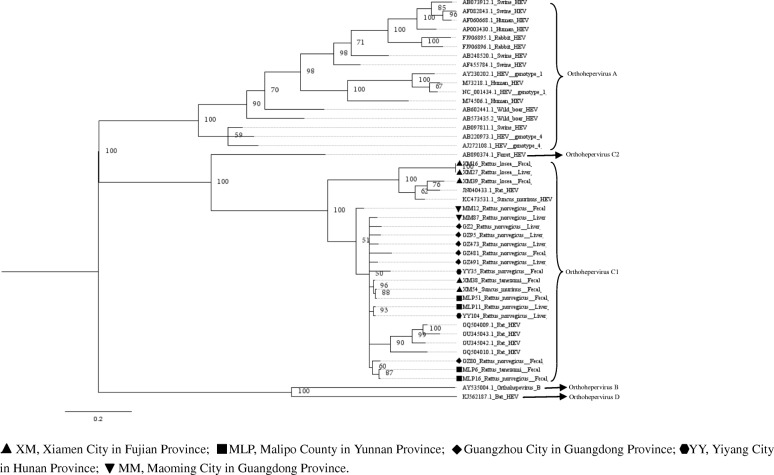
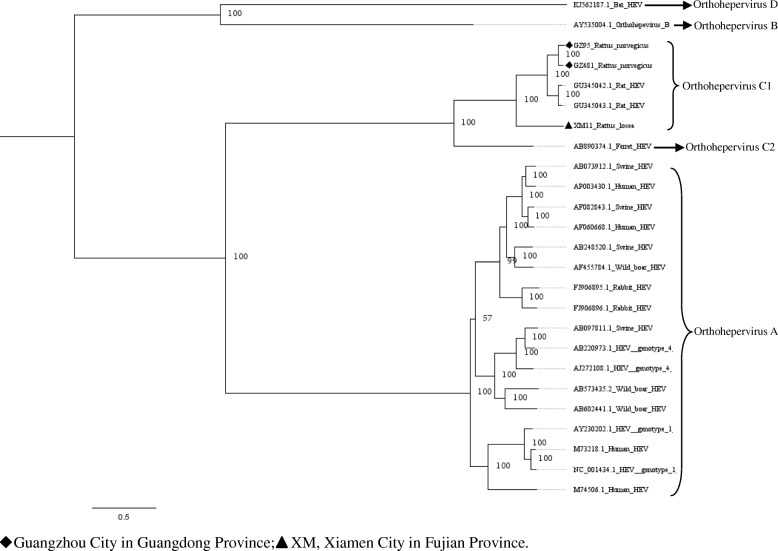
Fig. 2.
Phylogenetic tree constructed based on partial nucleotide sequences of partial ORF1 regions (255-nt) of 19 HEV strains (MrBayes, GTR + G + I nucleotide substitution model). Twenty five representative HEV isolates derived from rats, swine, humans, rabbits, wild boars, bat, and ferret are included for comparison. Percentages of the posterior probability (PP) values are indicated
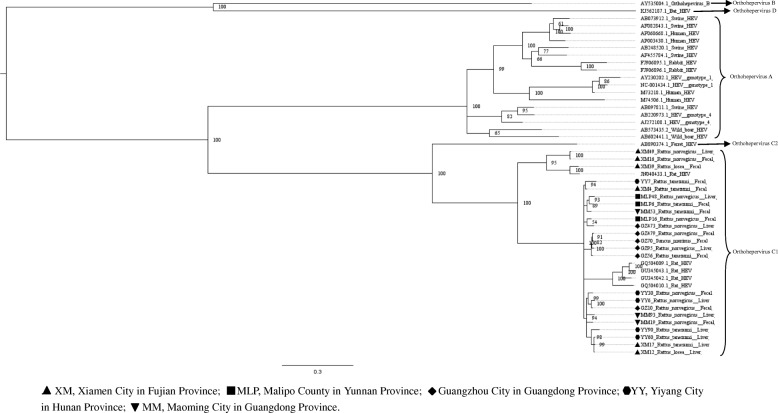
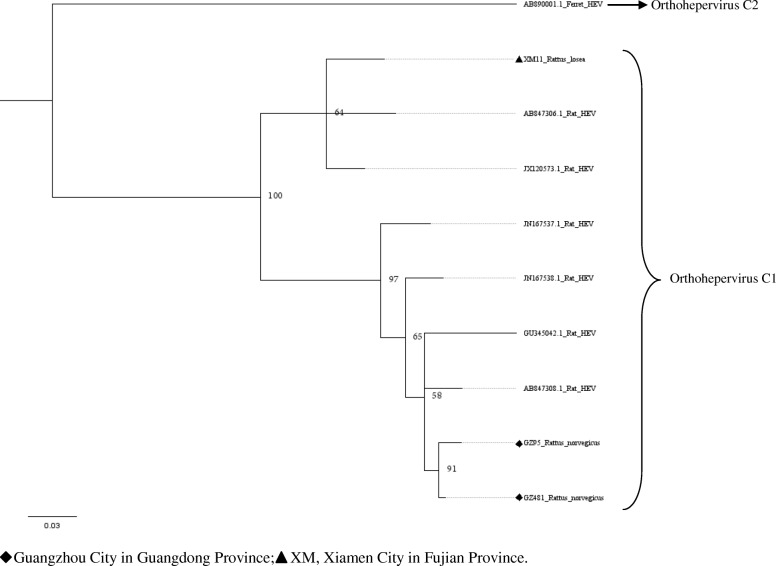
Fig. 3.
Phylogenetic tree constructed based on partial nucleotide sequences of partial ORF1–ORF2 regions (769-nt) of 23 HEV isolates (MrBayes, GTR + G + I nucleotide substitution model). Five rat HEV isolates and 20 HEV isolates derived from swine, humans, rabbits, wild boars, bat, and ferret are included for comparison. Percentages of the PP values are indicated
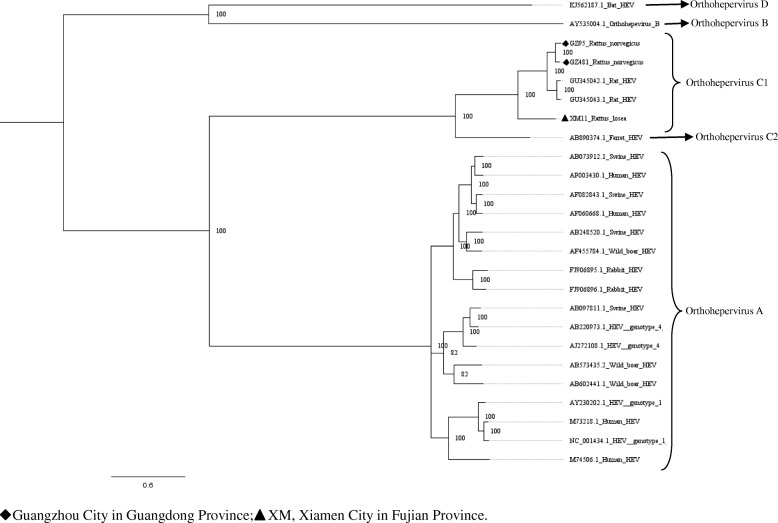
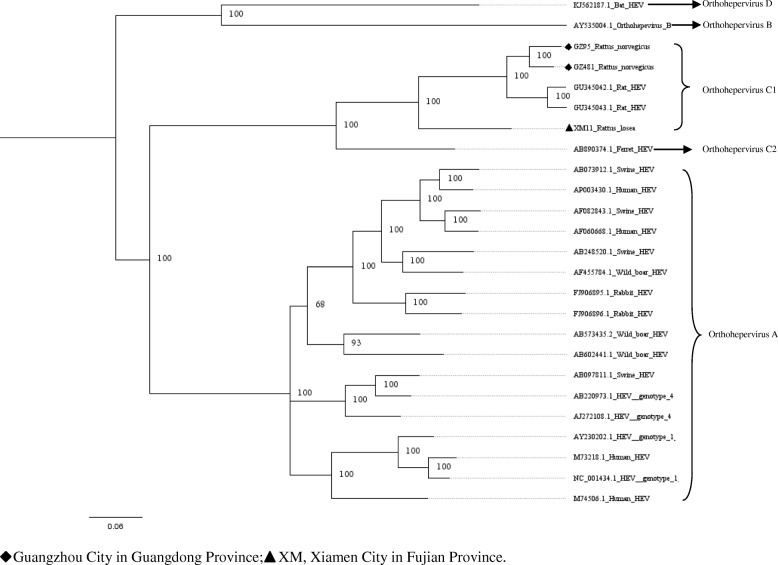
Fig. 4.
Phylogenetic tree constructed based on near full-length genomes of HEV (MrBayes, GTR + G + I nucleotide substitution model). Three rat HEV isolates and 22 HEV isolates derived from swine, humans, rabbits, wild boars, bat, and ferret are included for comparison. Percentages of the PP values are indicated
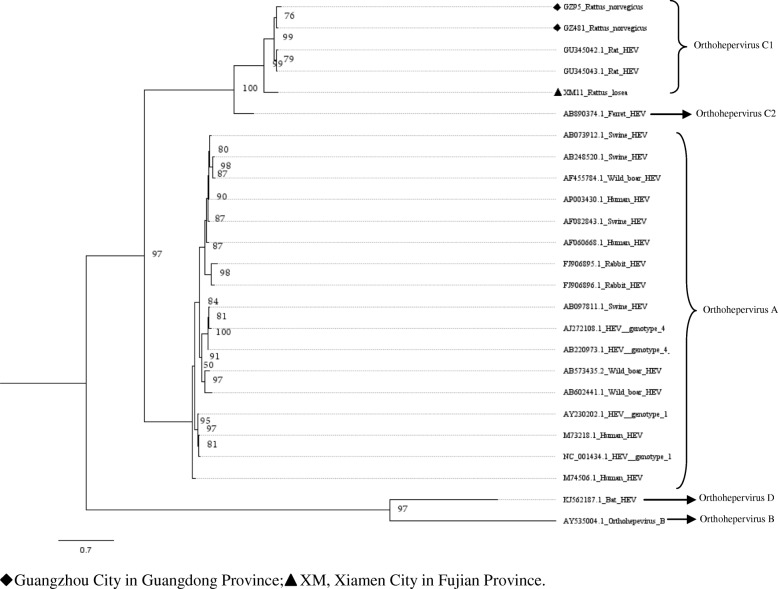
Fig. 5.
Phylogenetic tree constructed based on partial ORF1 of the near full-length genomes of HEV (MrBayes, GTR + G + I nucleotide substitution model). Three rat HEV isolates and 22 HEV isolates derived from swine, humans, rabbits, wild boars, bat, and ferret are included for comparison. Percentages of the PP values are indicated
Fig. 6.
Phylogenetic tree constructed based on partial ORF4 of the near full-length genomes of HEV (MrBayes, GTR + G + I nucleotide substitution model). Three rat HEV isolates and seven HEV isolates derived from rats and ferret are included for comparison. Percentages of the PP values are indicated
Fig. 7.
Phylogenetic tree constructed based on partial ORF2 of the near full-length genomes of HEV (MrBayes, GTR + G + I nucleotide substitution model). Three rat HEV isolates and 22 HEV isolates derived from swine, humans, rabbits, wild boars, bat, and ferret are included for comparison. Percentages of the PP values are indicated
Fig. 8.
Phylogenetic tree constructed based on ORF3 of the near full-length genomes of HEV (MrBayes, GTR + G + I nucleotide substitution model). Three rat HEV isolates and 22 HEV isolates derived from swine, humans, rabbits, wild boars, bat, and ferret are included for comparison. Percentages of the PP values are indicated
Phylogenetic trees were generated for the region spanning the nucleotides (nt) 4139 to 4393 (numbering based on a HEV sequence from R. norvegicus, GenBank accession number: JN167538), with the selected representative fragment sequences from our study and nucleotide sequences of HEV from GenBank (Fig. 2). From the result of the phylogenetic analysis, all of the novel sequences were found to belong to the species Orthohepevirus C1. The HEV sequence detected in house shrew clustered with rat HEV sequences from murine rodents, indicating that shrew HEV was similar to rat HEV. Excluding three sequences detected in R. losea from Xiamen City, all selected sequences were clustered with the HEV strains detected in Germany.
Upon analyzing the representative nested PCR products of rat HEV, the similarity ranged from 77.1–99.6% (Additional file 1: Table S2). As compared with the sequence reported in a previous study (GQ504010) [8], the HEV sequences detected in R. losea have a lower degree of identity than the sequences detected in other species (Table 3). The amino acid sequences of this region also have a higher percentage of identity than the nucleotide sequences, similar to our findings in the analysis of the partial ORF1 sequences. Phylogenetic tree was generated for the region spanning the nt 4157 to 4925 (numbering based on HEV sequence from R. norvegicus, GenBank accession number: JN167538), with the representative fragment sequences from this study and HEV sequences from GenBank. Phylogenetic analysis showed that all of the sequences belonged to the species Orthohepevirus C1. Two sequences from R. norvegicus and one sequence from R. losea clustered together with a rat HEV sequence from Asia, while the other HEV strains clustered with rat sequences from Europe (Fig. 3).
Table 3.
Nucleotide (nt) and amino acid sequence identity for the region spanning nt positions 4157–4925 or amino acid positions 1385–1636, based on strain GQ504010
| Species | Nt identity (%) | Amino acid identity (%) |
|---|---|---|
| Rattus norvegicus (12a) | 77.7–85.3 | 93.3–97.2 |
| Rattus tanezumi (8a) | 84.2–85.1 | 96.4–96.8 |
| Rattus losea (2a) | 77.5–84.5 | 93.7–96.4 |
| Suncus murinus (1a) | 84.9 | 95.6 |
anumber of the sequences
GQ504010, GenBank accession number of a HEV strain isolated from R. norvegicus from Germany
Characterization of the HEV genome
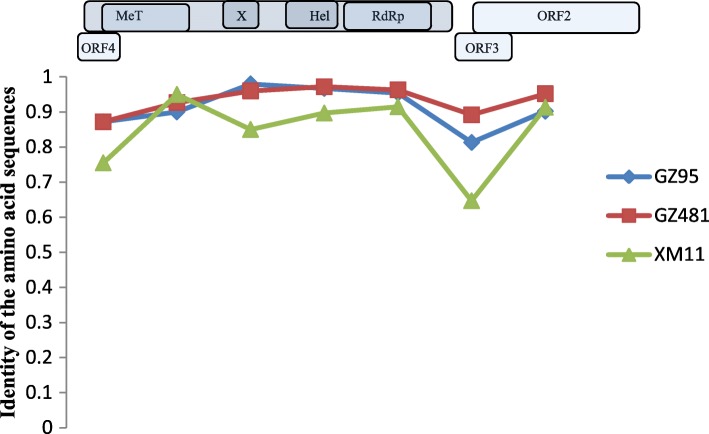
In murine rodents, three near full-length HEV genomes (GenBank accession numbers: MH729810–MH729812) detected in R. norvegicus (GZ95 and GZ481) and R. losea (XM11) were obtained. The length of these sequences ranged between the nt 6692 and 6790. These sequences were compared with the rat HEV sequence reported in a previous study (GU345042) [22]. The XM11 nucleotide and amino acid sequences had a lower percentage of identity than other HEV sequences. Additionally, the identity of the amino acid sequences of three near full-length HEV genomes in the ORF3 region were lower than other regions (Table 4 and Fig. 9). Phylogenetic trees were constructed based on these near full-length sequences and some representative HEV sequences from humans and other species of animals (Figs. 4, 5, 6, 7 and 8). The results showed that the near full-length HEV genomes obtained in this study clustered with rat HEV strains (Fig. 4). Two sequences from R. norvegicus in Guangzhou city clustered together in one branch, while the other HEV sequence from R. losea was located in another branch of the phylogenetic trees (Figs. 4, 5, 6, 7 and 8).
Table 4.
Nucleotide (nt) and amino acid identity for different regions of the near-full-length HEV genomes, based on strain GU345042
| Nt identity | Amino acid identity | |||||||
|---|---|---|---|---|---|---|---|---|
| Partial ORF1 | Partial OFR2 | ORF3 | Partial OFR4 | Partial ORF1 | Partial OFR2 | ORF3 | Partial OFR4 | |
| GZ95 | 0.836 | 0.833 | 0.899 | 0.945 | 0.907 | 0.902 | 0.813 | 0.872 |
| GZ481 | 0.846 | 0.884 | 0.935 | 0.941 | 0.919 | 0.952 | 0.892 | 0.872 |
| XM11 | 0.744 | 0.783 | 0.805 | 0.893 | 0.864 | 0.913 | 0.647 | 0.755 |
GU345042, GenBank accession number of a HEV strain isolated from R. norvegicus from Germany
Fig. 9.
The identity of the different regions of the amino acid sequences (XM11, GZ95, and GZ481), based on rat HEV (GenBank accession no. GU345042)
Five partial HEV nucleic acid sequences from house shrews, whose length ranged from 778 bp to 825 bp, were obtained (GenBank accession numbers: MH729813–MH729817). The selected representative house shrew sequence was clustered with rat HEV sequences (Figs. 2 and 3).
Discussion
In this study, we investigated HEV in four species of murine rodents and house shrews from four provinces in China via nested broad-spectrum RT-PCR assay and nested PCR method for rat HEV [8, 23]. During the conduct of this study, care was taken to limit contamination and a strict operating standard was followed during each experimental step, with a negative control established for each experiment. Our study evaluated the geographical distribution of HEV in murine rodents and house shrews in China.
HEV-positivity was detected in all of the cities investigated in this study, indicating that HEV is widely distributed among the three common species of murine rodents (R. norvegicus, R. tanezumi, and R. losea) in China. Specifically, 20.19% (124/614) of the liver tissue samples and 45.76% (281/614) of the fecal samples from the murine rodents were positive for HEV. In comparison, HEV-positivity in R. norvegicus was found to be only 2.87% in a previous study conducted in China [16]. The reason for the discrepancy in the detection rates of HEV in the present study versus the previous study is unclear. One explanation may be variations in the PCR methods used, which may affect the results obtained. The primers of two PCR methods we selected were designed based on the well-conserved regions of all types of HEV and the rat HEV genome, which may have contributed to the high detection rates of HEV in this study.
Positive liver samples were detected in all of the murine rodents, except B. indica. The positive liver samples indicated that HEV was capable of infecting these animals and subsequently replicating in them. The high prevalence and effective replication of HEV furthermore suggested that these animals are hosts of HEV.
In our study, the detection rate of HEV in house shrews was lower than in other species. This result was similar to that of the previous study conducted in China, in which only one of the 196 house shrews was positive for HEV [16]. Another study conducted in Nepal also reported the low prevalence of anti-HEV Ig-G in house shrews [24]. These indicate that house shrews might not be a natural host of HEV.
HEV has been detected in several species of murine rodents, such as R. norvegicus, R. flavipectus, and R. losea. In our study, the detection rate for HEV in R. losea in liver samples was higher than in other species of animals; this result was consistent with the findings of a previous study, indicating that R. losea is an important natural host for HEV [25]. R. norvegicus had the highest prevalence of HEV in fecal samples (Table 1). This result might be explained by the rats’ habitats. R. norvegicus mainly inhabits sewers and garbage dumps. Poor sanitation is conducive to the spread of HEV, which might explain the high HEV detection rate in the fecal samples from R. norvegicus. Further study is needed to explain why the detection rate in the liver samples from R. losea was higher than that in R. norvegicus, while the positive rate in fecal samples from R. losea was lower than that in R. norvegicus.
To our knowledge, we report the first near full-length of HEV genome from R. losea in our study. We also present the first near full-length rat HEV genomes in R. norvegicus from China. According to the phylogenetic analysis, all of the HEV sequences detected in the trapped murine rodents belonged to rat HEV. This result was in agreement with those of most previous studies [16, 26, 27], indicating that murine rodents are exclusively susceptible to rat HEV. However, some previous studies reported that HEV genotype 3 was detected in R. norvegicus [18]. Follow-up studies are necessary to confirm whether other genotypes of HEV can infect murine rodents.
Near full-length genome analysis showed that rat HEV sequences obtained in our study clustered with other rat HEV strains that were reported previously (Fig. 4). When analyzing the identity of the different regions between amino acid sequences, the partial ORF2 region seems to be a relatively conserved region (Fig. 9). The ORF2 region is known to be associated with capsid assembly and has many immune epitopes; thus, the ORF2 protein can induce strong immune responses [28]. In recent years, anti-rat HEV antibodies have been detected in forest workers and febrile patients [17, 29]. This suggests that rat HEV might infect humans and lead to liver diseases. However, further research is necessary to investigate the transmission of HEV between humans and rats.
Partial ORF1 and the ORF1–ORF2 sequences obtained in animals from different regions were clustered together, and we found that two sequences detected in R. losea from Xiamen City were totally identical, indicating that the HEV infecting the same species of animals were similar (Figs. 2 and 3 and Additional file 1: Table S1). In our study, HEV detected in R. losea showed a lower percentage of identity versus in other animal species (Tables 2 and 3). When comparing the three near full-length HEV sequences obtained in our study with the rat HEV sequence reported in a previous study (GU345042) [8], the near-full-length sequence from R. losea in Xiamen City also showed a lower percentage of identity than the sequences from R. norvegicus in Guangzhou City (Table 4). The result of the phylogenetic analysis showed that two sequences from R. norvegicus in Guangzhou City clustered together, while the other HEV strains from R. losea were in another branch of the phylogenetic trees (Figs. 5, 6, 7 and 8). This might be due to the difference in HEV infection as a result of host-specificity.
The percentages of identity at the amino acid level in the partial ORF1 and ORF1-ORF2 regions were greater than the percentages of identity at the nucleotide level (Tables 2 and 3). This might be explained by the presence of degenerate codons.
A 281 bp fragment sequence of HEV from house shrews was reported in a previous study [30]. In this investigation, 778 bp to 825 bp partial nucleic acid sequences of HEV from house shrews were obtained. One partial ORF1 sequence from house shrew obtained in our study was identical to another sequence detected in R. norvegicus. The result of phylogenetic analysis based on partial sequences of ORF1 region showed that HEV sequence detected in house shrew clustered with rat HEV sequences instead of the HEV sequence detected from house shrew reported in previous studies (Fig. 2). This may be explained by the interspecies transmission or spillover infection of HEV between murine rodents and house shrews [30].
Conclusions
There was a high prevalence of HEV in fecal samples of R. norvegicus, while a high prevalence of HEV was observed in the liver samples of R. losea. Murine rodents and house shrews shared a high identity genome of HEV, suggesting that HEV might cross transmit among different animal species.
Methods
Samples
Between 2014 and 2017, rodents and shrews were captured near human residences with cage traps. These animals were captured in five regions of four provinces in China, as follows: Yiyang City in Hunan Province, Xiamen City in Fujian Province, Maoming City and Guangzhou City in Guangdong Province, and Malipo County in Yunan Province (Fig. 1). Inhalational anesthesia was performed with diethyl ether, and the dosage of diethyl ether was adjusted according to the heart rate, respiratory frequency, corneal reflection and extremity muscle tension of the animal. Following anesthesia, the rodents and shrews were executed via cervical dislocation by trained personnel. Liver tissue samples were collected by intraperitoneal surgery in the laboratory and stored in RNAlater (Invitrogen, California, United States). The fecal samples were soaked in phosphate-buffered saline (PBS). The liver tissue and fecal samples were stored at − 80 °C and thawed at 4 °C prior to processing. The species of the trapped animals were identified by morphological identification and sequencing of the cytochrome B (cytB) gene [31].
Extraction of nucleic acid and detection of HEV
Total RNA and DNA were extracted from ~ 20 mg of liver tissue samples or 200 μL aliquot of fecal samples by using the MiniBEST Viral RNA/DNA Extraction Kit (TaKaRa, Kusatsu, Japan). We simultaneously used two polymerase chain reaction (PCR) methods for detecting HEV, one of which was a nested broad-spectrum PCR to amplify a 334 bp ORF1 fragment of all known HEV strains [8], while the other was a nested PCR to amplify the ORF1–ORF2 region of rat HEV [23]. The amplified products were separated on a 1.5% agarose gel, and the positive samples were sent to the Beijing Genomics Institute (Shenzhen, China) for sequencing.
Genome sequencing
Based on six HEV genome sequences from GenBank (GenBank accession numbers: JX120573, AB847307, GU345042, JN167538, KM516906, and LC225389), we designed eight pairs of primers to amplify the near full-length genomes of HEV (Table 5). After sequencing all of the fragments, the Lasergene SeqMan software (DNASTAR; Madison, WI, USA) was used to assemble the sequences.
Table 5.
HEV near-full-length PCR primers
| Primer | Orientation | Sequence (5'-3') | Target fragment (base pairs) |
|---|---|---|---|
| HEV-F15 | Sense (first/second round) | AGACCCATCARTATGTCG | 921 (this study) |
| HEV-R935 | Antisense (second round) | GTDGAYCKRGMCTTCTCACA | |
| HEV-R977 | Antisense (first round) | CATRAGYCKRTCCCADAT | |
| HEV-F827 | Sense (first/second round) | CATCTATGTGCGCAGCCTGT | 1340 (this study) |
| HEV-R2166 | Antisense (second round) | GTGGACAAACTGGGTGCGATC | |
| HEV-R2228 | Antisense (first round) | GTGCCACAGCGTGTATTATAG | |
| HEV-F2084 | Sense (first/second round) | GCNGTNTATGARGGRGAY | 955 (this study) |
| HEV-R3038 | Antisense (second round) | CAAAATCWATNGCNGGGATCTG | |
| HEV-R3053 | Antisense (first round) | ATVAGVCCCTTGCTYTCAAAATC | |
| HEV-F2868 | Sense (first/second round) | TKAARGCNCARTGGMGDG | 841 (this study) |
| HEV-R3708 | Antisense (second round) | ARBAYDGTCACCTGVTCHC | |
| HEV-R3740 | Antisense (first round) | ATRCGRCARTGCACDGT | |
| HEV-F3539 | Sense (first/second round) | ACCAACTTGCAGGATATAG | 699 (this study) |
| HEV-R4237 | Antisense (second round) | AAACTCGCTAAAATCATTCTCAA | |
| HEV-R4253 | Antisense (first round) | ATTCTGGGTGCTGTCAAACTCG | |
| HEV-F4980 | Sense (first/second round) | TCGTGCTCGTGYTTTTGCT | 1029 (this study) |
| HEV-R6008 | Antisense (second round) | CCTATRTCRCCYACMCCRTT | |
| HEV-R6014 | Antisense (first round) | CCCTTRCCTATRTCRCCYAC | |
| HEV-F5552 | Sense (first/second round) | GTRTCAATGTCRTTYTGG | 1153 (this study) |
| HEV-R6704 | Antisense (second round) | RTTAACAGGYCCAGYACC | |
| HEV-R6836 | Antisense (first round) | ATWGCATCAGCMACGAGGCA | |
| HEV-F6300 | Sense (first/second round) | CAACTGGCGGTCTGGTGATGTC | 535 (this study) |
| HEV-R6881 | Antisense (second round) | AGACACTGTCGGCTGCTGC | |
| HEV-R6834 | Antisense (first round) | GCATCAGCCACGAGGCAGG | |
| HE607 | Sense (first round) | CTTGGTTYAGGGCCATAGAG | 880 [23] |
| HE604 | Antisense (first round) | CAGCAGCGGCACGAACAGCA | |
| HE608 | Sense (second round) | TTYAGGGCCATAGAGAAGGC | |
| HE606 | Antisense (second round) | ACAGCAAAAGCACGAGCACG |
Phylogenetic analysis
The selected representative ORF1 fragments and ORF1–ORF2 fragments and all of the near full-length sequences obtained in this study were aligned with HEV sequences from humans and different species of animals obtained via GenBank by using the ClustalW multiple sequence alignment program in MEGA (version 7.0; Oxford Molecular Ltd., Cambridge, UK), respectively. The phylogenetic trees were constructed based on the ORF1 fragments, ORF1–ORF2 fragments, and the near full-length sequences. We constructed the phylogenetic trees via MrBayes (version 3.2) using a GTR + G + I nucleotide substitution matrix; two million Markov chain Monte Carlo (MCMC) iterations were sampled every 100 steps in order to obtain 20,000 trees, and burn-in was generally 25% of tree replicates [32, 33]. The phylogenetic trees were also constructed based on the neighbor-joining method in MEGA, with 1000 bootstrap replicates.
The identity of the different regions of the amino acid sequences and nucleotide sequences were estimated by use of the Sequence Identity Matrix program in BioEdit (version 7.2.5).
Statistical analysis
Data were analyzed using the Statistical Product and Service Solutions software (SPSS, version 13.0; IBM Corp., Armonk, NY, USA). The positive rates of HEV among different species of animals were analyzed using chi-square tests. A p-value of 0.05 was considered to be statistically significant.
Ethics guidelines
The protocol of this study was approved by the Ethics Committee of the Institutional Animal Care and Use Committee of Southern Medical University in Guangzhou, China.
Additional files
Table S1. Identity of the representative nucleotide (nt) sequences (nt positions 4139–4393) obtained in this study. Table S2. Identity of the representative nucleotide (nt) sequences (nt positions 4157–4925) obtained in this study. Figure S1. Phylogenetic tree constructed by the neighbor-joining method based on partial nucleotide sequences of ORF1 regions (255 nt) of 19 HEV strains. Twenty five representative HEV isolates derived from rats, swine, humans, rabbits, wild boars, bat and ferret are included for comparison. Bootstrap support of branches (1000 replication) is indicated. Figure S2. Phylogenetic tree constructed by the neighbor-joining method based on partial nucleotide sequences of ORF1 -ORF2 regions (769 nt) of 23 HEV isolates. Five rat HEV isolates and 20 HEV isolates derived from swine, humans, rabbits, wild boars, bat and ferret are included for comparison. Bootstrap support of branches (1000 replication) is indicated. Figure S3. Phylogenetic tree constructed by the neighbor-joining method based on near full-length genomes of HEV. Three rat HEV isolates and 22 HEV isolates derived from swine, humans, rabbits, wild boars, bat and ferret are included for comparison. Bootstrap support of branches (1000 replication) is indicated. Figure S4. Phylogenetic tree constructed by the neighbor-joining method based on ORF1 of the near full-length genomes of HEV. Three rat HEV isolates and 22 HEV isolates derived from swine, humans, rabbits, wild boars, bat and ferret are included for comparison. Bootstrap support of branches (1000 replication) is indicated. Figure S5. Phylogenetic tree constructed by the neighbor-joining method based on ORF4 of the near full-length genomes of HEV. Three rat HEV isolates and 7 HEV isolates derived from rats and ferret are included for comparison. Bootstrap support of branches (1000 replication) is indicated. Figure S6. Phylogenetic tree constructed by the neighbor-joining method based on ORF2 of the near full-length genomes of HEV. Three rat HEV isolates and 22 HEV isolates derived from swine, humans, rabbits, wild boars, bat and ferret are included for comparison. Bootstrap support of branches (1000 replication) is indicated. Figure S7. Phylogenetic tree constructed by the neighbor-joining method based on ORF3 of the near full-length genomes of HEV. Three rat HEV isolates and 22 HEV isolates derived from swine, humans, rabbits, wild boars, bat and ferret are included for comparison. Bootstrap support of branches (1000 replication) is indicated. (DOCX 309 kb)
Acknowledgements
We are grateful to Xueshan Zhong, Shaowei Chen, Xueyan Zheng, Shujuan Ma, Lina Jiang, Jin Ge, Ming Qiu, Shuting Huo, Xuemei Ke, Wen Zhou, Xing Li, Yun Mo, Xuejiao Chen, Yanxia Chen, Yongzhi Li, and Fangfei You for their participation in the collection of samples.
We would like to thank LetPub (www.LetPub.com) for providing linguistic assistance during the preparation of this manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (grant no. 81373051). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
All of the data generated or analyzed during this study are included in this manuscript and the supplementary information file.
Abbreviation
- HEV
Hepatitis E virus
Authors’ contributions
WH and QC conceived of the project and QC obtained the funding. WH and QC contributed to the writing of the paper. WH, YW, and MZ led virus detection. WH, YX, and MC performed the statistical analysis. WH, YW, YX, MZ, and MC performed the sample collection. All of the authors have read and approved the manuscript for publication.
Ethics approval and consent to participate
The protocol of this study was approved by the Ethics Committee of the Institutional Animal Care and Use Committee of Southern Medical University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Wenqiao He, Email: 1457650753@qq.com.
Yuqi Wen, Email: 498163422@qq.com.
Yiquan Xiong, Email: 747756389@qq.com.
Minyi Zhang, Email: 505133884@qq.com.
Mingji Cheng, Email: 276131617@qq.com.
Qing Chen, Email: 18002270308@163.com.
References
- 1.von Wulffen M, Westholter D, Lutgehetmann M, Pischke S, Hepatitis E. Still Waters Run Deep. J Clin Trans Hepatol. 2018;6(1):40–47. doi: 10.14218/JCTH.2017.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khuroo MS, Khuroo MS. Hepatitis E: an emerging global disease - from discovery towards control and cure. J Viral Hepat. 2016;23(2):68–79. doi: 10.1111/jvh.12445. [DOI] [PubMed] [Google Scholar]
- 3.Kamar N, Abravanel F, Lhomme S, Rostaing L, Izopet J. Hepatitis E virus: chronic infection, extra-hepatic manifestations, and treatment. Clin Res Hepatol Gastroenterol. 2015;39(1):20–27. doi: 10.1016/j.clinre.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Jaiswal SP, Jain AK, Naik G, Soni N, Chitnis DS. Viral hepatitis during pregnancy. Int J Gynaecol Obstetr. 2001;72(2):103–108. doi: 10.1016/S0020-7292(00)00264-2. [DOI] [PubMed] [Google Scholar]
- 5.Li TC, Yamakawa Y, Suzuki K, Tatsumi M, Razak MA, Uchida T, Takeda N, Miyamura T. Expression and self-assembly of empty virus-like particles of hepatitis E virus. J Virol. 1997;71(10):7207–7213. doi: 10.1128/jvi.71.10.7207-7213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emerson SU, Nguyen HT, Torian U, Burke D, Engle R, Purcell RH. Release of genotype 1 hepatitis E virus from cultured hepatoma and polarized intestinal cells depends on open reading frame 3 protein and requires an intact PXXP motif. J Virol. 2010;84(18):9059–9069. doi: 10.1128/JVI.00593-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanggis KT, Takahashi M, Jirintai S, Nishizawa T, Nagashima S, Nishiyama T, Kunita S, Hayama E, Tanaka T, et al. An analysis of two open reading frames (ORF3 and ORF4) of rat hepatitis E virus genome using its infectious cDNA clones with mutations in ORF3 or ORF4. Virus Res. 2018;249:16–30. doi: 10.1016/j.virusres.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 8.Johne R, Plenge-Bonig A, Hess M, Ulrich RG, Reetz J, Schielke A. Detection of a novel hepatitis E-like virus in faeces of wild rats using a nested broad-spectrum RT-PCR. J Gen Virol. 2010;91(3):750–758. doi: 10.1099/vir.0.016584-0. [DOI] [PubMed] [Google Scholar]
- 9.Mulyanto SJB, Andayani IG, Khalid TM, Ohnishi H, Jirintai S, Nagashima S, Nishizawa T, Okamoto H. Marked genomic heterogeneity of rat hepatitis E virus strains in Indonesia demonstrated on a full-length genome analysis. Virus Res. 2014;179:102–112. doi: 10.1016/j.virusres.2013.10.029. [DOI] [PubMed] [Google Scholar]
- 10.Purdy MA, Harrison TJ, Jameel S, Meng XJ, Okamoto H, Van der Poel WHM, Smith DB, Ictv Report C. ICTV virus taxonomy profile: Hepeviridae. J Gen Virol. 2017;98(11):2645–2646. doi: 10.1099/jgv.0.000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pavio N, Meng XJ, Doceul V. Zoonotic origin of hepatitis E. Curr Opinion Virol. 2015;10:34–41. doi: 10.1016/j.coviro.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 12.Sridhar S, Lau SK, Woo PC, Hepatitis E. A disease of reemerging importance. J Form Med Assoc Taiwan yi zhi. 2015;114(8):681–690. doi: 10.1016/j.jfma.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith DB, Simmonds P, Izopet J, Oliveira-Filho EF, Ulrich RG, Johne R, Koenig M, Jameel S, Harrison TJ, Meng XJ, et al. Proposed reference sequences for hepatitis E virus subtypes. J Gen Virol. 2016;97(3):537–542. doi: 10.1099/jgv.0.000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Syed SF, Zhao Q, Umer M, Alagawany M, Ujjan IA, Soomro F, Bangulzai N, Baloch AH, Abd El-Hack M, Zhou EM, et al. Past, present and future of hepatitis E virus infection: zoonotic perspectives. Microb Pathog. 2018;119:103–108. doi: 10.1016/j.micpath.2018.03.051. [DOI] [PubMed] [Google Scholar]
- 15.Kanai Y, Miyasaka S, Uyama S, Kawami S, Kato-Mori Y, Tsujikawa M, Yunoki M, Nishiyama S, Ikuta K, Hagiwara K. Hepatitis E virus in Norway rats (Rattus norvegicus) captured around a pig farm. BMC Res notes. 2012;5:4. doi: 10.1186/1756-0500-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang B, Cai CL, Li B, Zhang W, Zhu Y, Chen WH, Zhuo F, Shi ZL, Yang XL. Detection and characterization of three zoonotic viruses in wild rodents and shrews from Shenzhen city, China. Virol Sin. 2017;32(4):290–297. doi: 10.1007/s12250-017-3973-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dremsek P, Wenzel JJ, Johne R, Ziller M, Hofmann J, Groschup MH, Werdermann S, Mohn U, Dorn S, Motz M, et al. Seroprevalence study in forestry workers from eastern Germany using novel genotype 3- and rat hepatitis E virus-specific immunoglobulin G ELISAs. Med Microbiol Immunol. 2012;201(2):189–200. doi: 10.1007/s00430-011-0221-2. [DOI] [PubMed] [Google Scholar]
- 18.Lack JB, Volk K, Van Den Bussche RA. Hepatitis E virus genotype 3 in wild rats, United States. Emerg Infect Dis. 2012;18(8):1268–1273. doi: 10.3201/eid1808.120070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jirintai S, Tanggis M, Suparyatmo JB, Takahashi M, Kobayashi T, Nagashima S, Nishizawa T, Okamoto H. Rat hepatitis E virus derived from wild rats (Rattus rattus) propagates efficiently in human hepatoma cell lines. Virus Res. 2014;185:92–102. doi: 10.1016/j.virusres.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Li W, Guan D, Su J, Takeda N, Wakita T, Li TC, Ke CW. High prevalence of rat hepatitis E virus in wild rats in China. Vet Microbiol. 2013;165(3–4):275–280. doi: 10.1016/j.vetmic.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 21.Wang B, Li W, Zhou JH, Li B, Zhang W, Yang WH, Pan H, Wang LX, Bock CT, Shi ZL, et al. Chevrier’s Field Mouse (Apodemus chevrieri) and Pere David's Vole (Eothenomys melanogaster) in China Carry Orthohepeviruses that form Two Putative Novel Genotypes Within the Species Orthohepevirus C. Virol Sin. 2018;33(1):44–58. doi: 10.1007/s12250-018-0011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johne R, Heckel G, Plenge-Bonig A, Kindler E, Maresch C, Reetz J, Schielke A, Ulrich RG. Novel hepatitis E virus genotype in Norway rats, Germany. Emerg Infect Dis. 2010;16(9):1452–1455. doi: 10.3201/eid1609.100444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mulyanto DSN, Sriasih M, Takahashi M, Nagashima S, Jirintai S, Nishizawa T, Okamoto H. Frequent detection and characterization of hepatitis E virus variants in wild rats (Rattus rattus) in Indonesia. Arch Virol. 2013;158(1):87–96. doi: 10.1007/s00705-012-1462-0. [DOI] [PubMed] [Google Scholar]
- 24.He J, Innis BL, Shrestha MP, Clayson ET, Scott RM, Linthicum KJ, Musser GG, Gigliotti SC, Binn LN, Kuschner RA, et al. Evidence that rodents are a reservoir of hepatitis E virus for humans in Nepal. J Clin Microbiol. 2006;44(3):1208. doi: 10.1128/JCM.44.3.1208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Nguyen D, Van Nguyen C, Bonsall D, Ngo TT, Carrique-Mas J, Pham AH, Bryant JE, Thwaites G, Baker S, Woolhouse M, et al. Detection and characterization of homologues of human Hepatitis viruses and Pegiviruses in rodents and bats in Vietnam. Viruses. 2018;10(3):102. doi: 10.3390/v10030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simanavicius M, Juskaite K, Verbickaite A, Jasiulionis M, Tamosiunas PL, Petraityte-Burneikiene R, Zvirbliene A, Ulrich RG, Kucinskaite-Kodze I. Detection of rat hepatitis E virus, but not human pathogenic hepatitis E virus genotype 1-4 infections in wild rats from Lithuania. Vet Microbiol. 2018;221:129–133. doi: 10.1016/j.vetmic.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 27.Li TC, Yoshizaki S, Ami Y, Suzaki Y, Yasuda SP, Yoshimatsu K, Arikawa J, Takeda N, Wakita T. Susceptibility of laboratory rats against genotypes 1, 3, 4, and rat hepatitis E viruses. Vet Microbiol. 2013;163(1–2):54–61. doi: 10.1016/j.vetmic.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 28.Zhou Y, Zhao C, Tian Y, Xu N, Wang Y. Characteristics and functions of HEV proteins. Adv Exp Med Biol. 2016;948:17–38. doi: 10.1007/978-94-024-0942-0_2. [DOI] [PubMed] [Google Scholar]
- 29.Shimizu K, Hamaguchi S, Ngo CC, Li TC, Ando S, Yoshimatsu K, Yasuda SP, Koma T, Isozumi R, Tsuda Y, et al. Serological evidence of infection with rodent-borne hepatitis E virus HEV-C1 or antigenically related virus in humans. J Vet Med Sci. 2016;78(11):1677–1681. doi: 10.1292/jvms.16-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guan D, Li W, Su J, Fang L, Takeda N, Wakita T, Li TC, Ke C. Asian musk shrew as a reservoir of rat hepatitis E virus, China. Emerg Infect Dis. 2013;19(8):1341–1343. doi: 10.3201/eid1908.130069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arai S, Bennett SN, Sumibcay L, Cook JA, Song JW, Hope A, Parmenter C, Nerurkar VR, Yates TL, Yanagihara R. Phylogenetically distinct hantaviruses in the masked shrew (Sorex cinereus) and dusky shrew (Sorex monticolus) in the United States. Am J Trop Med Hyg. 2008;78(2):348–351. doi: 10.4269/ajtmh.2008.78.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drexler JF, Corman VM, Lukashev AN, van den Brand JM, Gmyl AP, Brunink S, Rasche A, Seggewibeta N, Feng H, Leijten LM, et al. Evolutionary origins of hepatitis a virus in small mammals. Proc Natl Acad Sci U S A. 2015;112(49):15190–15195. doi: 10.1073/pnas.1516992112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Identity of the representative nucleotide (nt) sequences (nt positions 4139–4393) obtained in this study. Table S2. Identity of the representative nucleotide (nt) sequences (nt positions 4157–4925) obtained in this study. Figure S1. Phylogenetic tree constructed by the neighbor-joining method based on partial nucleotide sequences of ORF1 regions (255 nt) of 19 HEV strains. Twenty five representative HEV isolates derived from rats, swine, humans, rabbits, wild boars, bat and ferret are included for comparison. Bootstrap support of branches (1000 replication) is indicated. Figure S2. Phylogenetic tree constructed by the neighbor-joining method based on partial nucleotide sequences of ORF1 -ORF2 regions (769 nt) of 23 HEV isolates. Five rat HEV isolates and 20 HEV isolates derived from swine, humans, rabbits, wild boars, bat and ferret are included for comparison. Bootstrap support of branches (1000 replication) is indicated. Figure S3. Phylogenetic tree constructed by the neighbor-joining method based on near full-length genomes of HEV. Three rat HEV isolates and 22 HEV isolates derived from swine, humans, rabbits, wild boars, bat and ferret are included for comparison. Bootstrap support of branches (1000 replication) is indicated. Figure S4. Phylogenetic tree constructed by the neighbor-joining method based on ORF1 of the near full-length genomes of HEV. Three rat HEV isolates and 22 HEV isolates derived from swine, humans, rabbits, wild boars, bat and ferret are included for comparison. Bootstrap support of branches (1000 replication) is indicated. Figure S5. Phylogenetic tree constructed by the neighbor-joining method based on ORF4 of the near full-length genomes of HEV. Three rat HEV isolates and 7 HEV isolates derived from rats and ferret are included for comparison. Bootstrap support of branches (1000 replication) is indicated. Figure S6. Phylogenetic tree constructed by the neighbor-joining method based on ORF2 of the near full-length genomes of HEV. Three rat HEV isolates and 22 HEV isolates derived from swine, humans, rabbits, wild boars, bat and ferret are included for comparison. Bootstrap support of branches (1000 replication) is indicated. Figure S7. Phylogenetic tree constructed by the neighbor-joining method based on ORF3 of the near full-length genomes of HEV. Three rat HEV isolates and 22 HEV isolates derived from swine, humans, rabbits, wild boars, bat and ferret are included for comparison. Bootstrap support of branches (1000 replication) is indicated. (DOCX 309 kb)
Data Availability Statement
All of the data generated or analyzed during this study are included in this manuscript and the supplementary information file.