Abstract
Why diversification rates vary so extensively across the tree of life remains an important yet unresolved issue in biology. Two prominent and potentially independent factors proposed to explain these trends reflect the capacity of lineages to expand into new areas of (i) geographical or (ii) ecological space. Here, we present the first global assessment of how diversification rates vary as a consequence of geographical and ecological expansion, studying these trends among 15 speciose passerine families (together approximately 750 species) using phylogenetic path analysis. We find that relative slowdowns in diversification rates characterize families that have accumulated large numbers of co-occurring species (at the 1° scale) within restricted geographical areas. Conversely, more constant diversification through time is prevalent among families in which species show limited range overlap. Relative co-occurrence is itself also a strong predictor of ecological divergence (here approximated by morphological divergence among species); however, once the relationship between co-occurrence and diversification rates have been accounted for, increased ecological divergence is an additional explanatory factor accounting for why some lineages continue to diversify towards the present. We conclude that opportunities for prolonged diversification are predominantly determined by continued geographical range expansion and to a lesser degree by ecological divergence among lineages.
Keywords: adaptive radiation, allopatric speciation, non-adaptive radiation, secondary sympatry, phenotypic evolution
1. Introduction
Why some clades continue to diversify towards the present, yet others have apparently exhausted their capacity to generate further species represent central questions in macroevolution [1,2]. The degree to which diversification rates decline through time has been hypothesized to reflect the capacity of lineages to undergo continued expansion into new areas of geographical and/or ecological space, as both processes can facilitate the production of genetically distinct and reproductively isolated taxa [1–10]. Although these hypotheses are well developed theoretically [2,11,12] and have received some empirical support (e.g. [7,9]), the relative influence of ecological and geographical constraints upon diversification have yet to be assessed in a global context. We address this issue by testing if geographical and eco-morphological expansion can predict variation in diversification rates among a speciose global radiation of passerine birds. We perform this analysis to understand whether geographical and ecological divergences represent significant influences upon the diversification dynamics of different passerine families.
Range expansion into new areas can result in the formation of geographically isolated populations, or lead to secondary contact among those that were recently isolated from one another [1,4,10,13–16]. Thus, an important distinction that defines the geographical dynamics of radiation among different clades is whether range expansion (following speciation) results in the sympatric build-up of closely related species (e.g. members of the same family/genus) or not [17]. At one end of this continuum lie clades that have accumulated many co-occurring species within restricted geographical settings [18,19], while at the other are those that have radiated over much larger areas, accumulating few co-occurring species, and many more geographically separated taxa [20–24]. These two extremes reflect both ecological opportunities within the region where the diversification process unfolds, and the accessibility of other regions as a consequence of colonization and range expansion [2,18]. However, whether the accumulation of co-occurring species or ecological differences predict longer-term diversification dynamics across multiple independent radiations remains largely untested (but see Harmon et al. [25]).
Although a lack of available geographical or ecological opportunity may eventually inhibit diversification [5,26], these limits are likely to differ among clades as a consequence of their ability to evolve new physiological and morphological traits that facilitate their respective lineages to exploit novel resources [2,27]. Niche evolution and character displacement allow species to co-occur with one another, because this enables the exploitation of different parts of the available resource spectrum [4,28–30]. Differences in niche space occupation reflect variation in species morphology [31], i.e. locomotive abilities, resource acquisition, and modes of foraging in the case of passerine birds [28–30,32,33]. Therefore, range expansion and co-occurrence can conceivably influence, and be influenced by, the evolution of morphological traits that are representative of key ecological differences among lineages [30].
In this study, we evaluate the effects of geographical and ecological constraints on diversification. We do so using phylogenetic path analyses [34], which enable us to directly test hypothesized causal associations between morphological divergence, species co-occurrence, and diversification rates. We study diversification rate variation among 15 species-rich families of corvoid birds (together comprising approximately 750 species) to understand whether this can be predicted by patterns of range overlap (reflecting the capacity of families to expand into new geographical space) or morphological differentiation (reflecting the capacity of families to expand into new ecological space). Corvoid passerines represent a good study system to address these questions for several reasons; (1) multiple lineages have undergone biogeographical expansion from the Australasian ancestral area, such that the overall clade has radiated across the majority of the world's insular and continental landmasses, thus encompassing a near global distribution [10,35,36], (2) corvoid lineages are extremely diverse in their morphological forms, both within and among groups (e.g. body masses of all corvoid species range from less than 10 g to greater than 1000 g [37]), and (3) the degree of geographical range overlap varies extensively among clades and across geographical space.
2. Material and methods
(a). Phylogenetic, spatial, and morphological data
We performed our comparative analyses on 15 families of corvoid passerines (table 1), which represent a subset of those considered in Kennedy et al. [41]. Together, the combined species richness of these families amounts to approximately 95% of the overall species diversity of the Corvides (741/789 species [38]). Given our aim of assessing the relationship between patterns of co-occurrence and diversification, we excluded the 16 most species-poor corvoid families from our analysis, which together represent only 48 species.
Table 1.
Species diversity, diversification rates, range size, area, morphological disparity, and average grid cell richness among 15 corvoid families. These families were delimited in accordance with the taxonomy of the IOC 2.7 Gill & Donsker [38].
| family | overall species richness | proportion of missing species from Jønsson et al. [39] | gamma (γ) estimated from Jønsson et al. [39] | gamma (γ) estimated from Kennedy et al. [40] | log (mean range size) | log (area) | FDis | mean grid cell richness |
|---|---|---|---|---|---|---|---|---|
| Artamidae | 23 | 0.17 | 0.48 ± 0.24 | 1.14 | 4.84 | 7.53 | 1.13 | 3.35 |
| Campephagidae | 92 | 0.18 | −1.16 ± 0.27 | 0.61 | 3.75 | 8.26 | 0.76 | 3.07 |
| Cinclosomatidae | 10 | 0.1 | −1.14 ± 0.32 | −0.79 | 4.03 | 6.08 | 0.35 | 1.5 |
| Corvidae | 127 | 0.11 | −1.78 ± 0.23 | −1.56 | 4.33 | 9.65 | 0.83 | 3.44 |
| Dicruridae | 24 | 0.21 | −1.3 ± 0.25 | −0.9 | 4.41 | 8.1 | 0.45 | 2.19 |
| Laniidae | 34 | 0.12 | −0.11 ± 0.35 | 0.05 | 5.48 | 9.18 | 0.47 | 1.77 |
| Malaconotidae | 50 | 0.4 | −2.21 ± 0.22 | −0.93 | 4.2 | 7.51 | 0.45 | 5.59 |
| Monarchidae | 94 | 0.07 | −1.79 ± 0.29 | −1.53 | 2.81 | 8.29 | 0.44 | 2.07 |
| Oriolidae | 35 | 0 | −1.29 ± 0.26 | −1.35 | 3.94 | 8.42 | 0.37 | 1.36 |
| Pachycephalidae | 51 | 0.1 | −0.68 ± 0.3 | −0.12 | 3.21 | 7.27 | 0.44 | 2.41 |
| Paradisaeidae | 41 | 0.02 | −1.69 ± 0.26 | −1.77 | 2.77 | 5.06 | 0.91 | 6.18 |
| Platysteiridae | 30 | 0.33 | −1.21 ± 0.44 | 0.03 | 3.97 | 7.37 | 0.4 | 2.2 |
| Rhipiduridae | 46 | 0.28 | −2.57 ± 0.23 | −1.33 | 3.27 | 7.66 | 0.37 | 1.92 |
| Vangidae | 21 | 0 | −3.38 ± 0.19 | −3.42 | 3.13 | 4.28 | 1.03 | 8.57 |
| Vireonidae | 63 | 0.18 | −3.02 ± 0.22 | −2.28 | 4.07 | 8.22 | 0.41 | 3.13 |
Estimates of the phylogenetic relationships among the 741 species which are members of the 15 families were obtained from the maximum clade credibility tree analysed in Kennedy et al. ([41]; http://dx.doi.org/10.5061/dryad.128249). Distributional data representing the breeding range were also collated for these same species from an expert-validated database [42], in which the ranges are recorded at a resolution of 1° × 1° (ca 110 km × 110 km). The range maps were determined from museum specimens, published sightings, and more than 1000 original references [42]. In addition, for 734/741 species (99% of all analysed species) PZM measured museum study skins to determine 10 different aspects of their external morphology (reflecting measurements of the wings, tail, tarsus, bill, and claws; full details of the measurement techniques can be found in electronic supplementary material, appendix A). Similar morphological measurements have been used extensively in previous comparative analyses of passerine birds (e.g. [33,43,44]), as these traits have been shown to predict important ecological differences among species [31,32,45,46]. A total of 4091 museum study skins were measured (electronic supplementary material, table S1) for a mean of 5.57 ± 1.22 specimens per species. We attempted to measure male specimens when possible, except in the relatively small number of instances where these were poorly represented in the collections, in which case we supplemented them with those from females or unsexed specimens (20/4091 measured specimens).
(b). Geographical analyses
For each of the 15 families, we determined their species richness gradients and thus the degree to which they have accumulated co-occurring forms by overlaying their range maps and summing the number of species found in each 1° grid cell. Subsequently, we computed the overall area occupied by each family (the number of unique 1° grid cells in which the family was present), the mean range size of their constituent species (the average number of 1° grid cells occupied by each species), and the mean value of species richness across all 1° grid cells. Clade area and mean range size (both log-transformed) were included in our analyses as covariates, as these variables may influence diversification rates independently from the extent to which co-occurring species accumulate [3,47].
(c). Morphological analyses
We performed a principal component analysis (PCA) on the log-transformed morphological traits. Log-transformation was necessary, because the distribution of our measured traits were all right-skewed, reflecting the evolution of large body sizes in some corvoid lineages [37]. Loadings of the log-transformed traits upon the individual principal component axes (PC1–10) can be found in electronic supplementary material, table S2. We used the species scores on the 10 principal component axes to estimate the Functional Dispersion (FDis) of each family [48] which served as a measure of the relative morphological disparity among their constituent species. These analyses were implemented in the R package metricTester [49]. The FDis of each family reflects the unweighted mean distance to the centroid of morphological space of all species [48], here represented by the principal component scores. Although other measures of morphological disparity have been proposed (e.g. functional volume, functional divergence), simulations have shown that FDis is by far the least sensitive to variation in overall species richness among groups, while also reliably capturing the range and standard deviation of the original traits [50].
(d). Diversification analyses
For each family, we assessed whether rates of diversification were either constant or slowing through time by computing the γ-statistic [51]. Values of γ provide an estimate of whether the nodes within a phylogeny are disproportionately distributed towards the root or tips of the tree. The more negative the values of γ, the greater the proportion of branching events that have occurred towards the root of the tree, indicating a decline in rates of net diversification towards the present [51]. These interpretations are drawn from the expectations of γ under the pure birth model, which is known to follow a normal distribution [51]. Although multiple alternative metrics for estimating diversification rates exist [52], we chose to analyse γ because this statistic allows us to directly quantify the relative degree to which diversification is continuing towards the present among the different families, and thus test our key hypotheses.
We computed γ for each corvoid family using the phylogeny of Kennedy et al. [41], which added 124 species to the phylogeny of Jønsson et al. [39] (who sampled 629/741 corvoid species) as polytomies based on current taxonomic information [53]. These 124 species did not have DNA sequence data available at the time of the original tree generation. The birth–death model was used to randomly resolve these polytomies and assign branch lengths for the respective diversification events, following the methods of Kuhn et al. [54]. For the vast majority of the analysed families, species sampling in the phylogeny of Jønsson et al. [39] was greater than 80% (table 1). However, we tested the robustness of our results to the exclusion of the 124 taxonomically placed species, by repeating our analyses using the mean estimates of γ across 1000 post-burnin samples of the Jønsson et al. [39] trees (electronic supplementary material, appendix B).
(e). Phylogenetic comparative analyses
While causal relationships cannot be determined using standard correlative methods, path analysis enables the relative importance of alternate causal models that propose direct and indirect relationships among variables to be compared [34,55]. We determined the causal relationships among our predictor variables (clade area, mean range size, mean grid cell richness, and FDis), and how they may influence diversification rates, using phylogenetic confirmatory path analysis [34] which combines the methodologies of phylogenetic least squares (PGLS) and d-separation (herein d-sep) [55]. We evaluated the relative support of 20 path models each proposing a biologically plausible causal structure for the variation in γ. The directed acyclic graphs (DAGs) for all models are shown in electronic supplementary material, figure S1. Models were fitted by evaluating the minimal set of conditional independencies among the predictor variables necessary for each path model to be true. We controlled for the non-independence of the data points due to the shared ancestry of the taxa [34] using a family-level phylogeny of the Corvides (our species-level phylogeny pruned to contain a single member of each family). We first tested the conditional independencies of each model by performing the relevant PGLS regressions. Subsequently, we assessed whether the proposed independencies of each model are fulfilled in the empirical relationships using Fisher's C statistic. Finally, we used the C statistic information criterion (CICc) proposed by von Hardenberg & Gonzalez-Voyer [34] to assess the relative fit of all models. These analyses were performed in the R package phylopath [56].
3. Results
(a). The distribution of γ and FDis among corvoid families
The estimated values of γ for the 15 corvoid families are shown in table 1. Eleven out of 15 families recovered negative values of γ, implying that the nodes within the respective phylogenies tend to be distributed towards the roots of the trees. Although γ is known to become negatively biased when estimated on phylogenies with incomplete species-level sampling [51], we found no evidence for a significant correlation between γ and the proportion of unsampled species per family within the Jønsson et al. [39] tree. This was the case either when γ was computed on the phylogeny of Jønsson et al. [39] (PGLS slope = 0.38, R2 = 0.01<, p = 0.88; electronic supplementary material, figure S2a), or Kennedy et al. [41] (PGLS slope = 4.2, R2 = 0.17, p = 0.13; electronic supplementary material, figure S2b). There was also no significant relationships between γ and overall family richness (γ estimated on Jønsson et al. [39]; PGLS slope = −0.006, R2 = 0.04, p = 0.46, electronic supplementary material, figure S2c. γ estimated on Kennedy et al. [41]; PGLS slope = −0.002, R2 = 0.01<, p = 0.81, electronic supplementary material, figure S2d). Given these findings, we present the results from phylogenetic path analysis of the complete species-level tree (Kennedy et al. [41]) in the main text, and those from Jønsson et al. [39] in electronic supplementary material, appendix B. Our results remained consistent in both analyses.
Estimates of FDis were also highly variable among corvoid families (table 1). Families that consist of morphologically disparate species (e.g. Artamidae, Paradisaeidae, and Vangidae) scored high values of FDis. Conversely, families containing species that possess similar morphologies (e.g. Laniidae, Oriolidae, Rhipiduridae, and Vireonidae) recovered low values of FDis. As with γ, the values of FDis were unrelated to the overall family species richness (PGLS slope = 0.001<, R2 = 0.006, p = 0.78; electronic supplementary material, figure S3).
(b). Phylogenetic comparative analyses
The results from the d-sep test for the models proposed in electronic supplementary material, figure S1 are shown in electronic supplementary material, table S3. The ΔCICc values of these models imply that model 13 was our best fitting model (relative weighting 0.50, electronic supplementary material, table S3). Two other models, model 11 and model 1 received the majority of the remaining model weighting (0.36 and 0.11, respectively). p-values from the d-sep test imply that these three models represent plausible sets of causal relationships among the variables. Despite receiving less overall weight, the ΔCICc value for model 11 (0.68) suggests that this cannot be considered a significantly poorer fit than model 13; however, the ΔCICc for model 1 (3.07) suggests that model 13 provides a significantly better explanation of the causal relationships among our variables. We present the standardized path coefficients (SPC) for the average of models 11 and 13 in figure 1, and electronic supplementary material, table S4, following the approach of von Hardenberg & Gonzalez-Voyer [34]. The SPC for the individual models is shown in electronic supplementary material, figure S4. The averaged model results show that grid cell richness is a significant negative predictor of γ (SPC = −0.71, figure 1, and electronic supplementary material, figure S5a) while FDis is a significantly positive predictor (SPC = 0.73, figure 1, and electronic supplementary material, figure S5b,c), albeit with this relationship only being proposed in model 11. Mean grid cell richness was also supported to positively determine FDis (SPC = 0.65, figure 1 and electronic supplementary material, figure S5d), meaning that families with a higher average number of co-occurring species tend to comprise groups of species that are more morphologically divergent from one another. In addition, we found significant support for a positive relationship between range size and area (SPC = 0.56, figure 1 and electronic supplementary material, figure S5e), and a negative relationship between area and mean grid cell richness (SPC = −0.67; figure 1 and electronic supplementary material, figure S5f).
Figure 1.
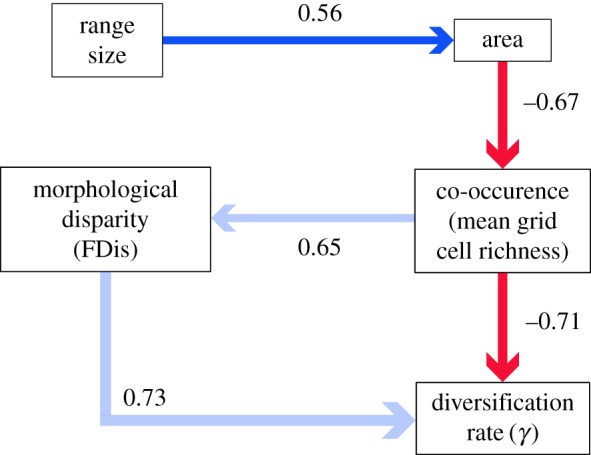
Directed acyclic graph showing the standardized path coefficients averaged among the best fitting phylogenetic path models (with ΔCICc less than 2) presented in electronic supplementary material, table S3. Bold lines indicate paths supported in both of the best fitting models, whereas faded lines illustrate paths supported only in single models. Positive path coefficients are shown in blue, while negative correlations are shown in red. All models predicted variation in gamma (γ) among 15 corvoid passerines as a consequence of the relationships between mean range size, area, morphological disparity (FDis), and mean grid cell richness.
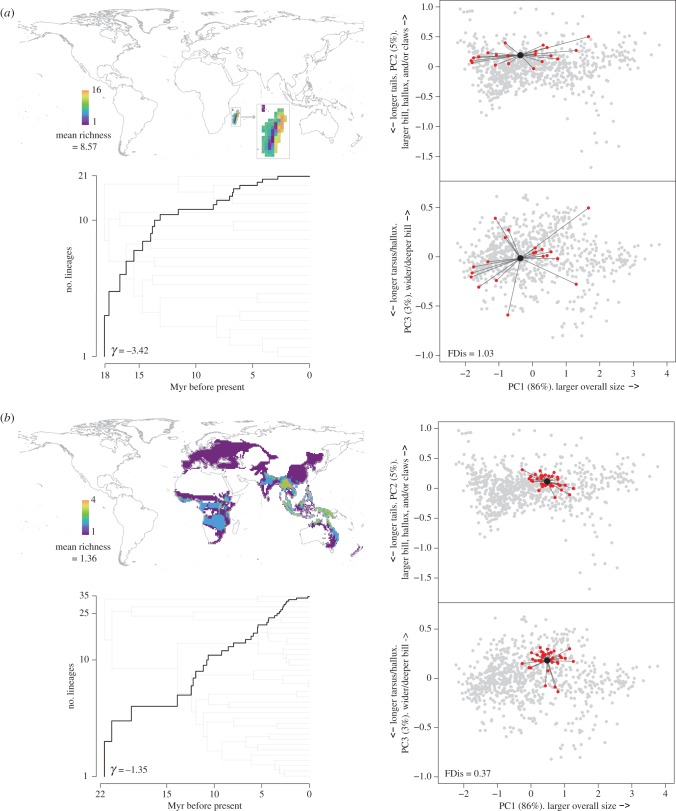
The results of these analyses indicate that corvoid families with wide-ranging species generally occur over larger areas, and that groups occurring over larger areas tend to have low average co-occurrence within 1° grid cells. Corvoid families that have more co-occurring species, or those that are relatively conserved in their morphology, are more likely to have slowed down in their rates of diversification towards the present (figure 1). In addition, higher levels of co-occurrence lead to the evolution of greater morphological disparity within families (figure 1). These trends can be evidenced by contrasting the different diversification trajectories, species richness gradients, and morphological variability of the individual corvoid families (figure 2 and electronic supplementary material, figure S6). Furthermore, these findings are generally consistent when removing species placed in the phylogeny by taxonomy alone, or when pruning taxa that have formed within the last 1 million years (electronic supplementary material, appendix B).
Figure 2.
Lineage through time plots, maps of species richness and species scores on PC axes 1–3 for two corvoid families; Vangidae ((a), N = 21) and Oriolidae ((b), N = 35). These families highlight different associations between lineage diversification, co-occurrence, and morphological disparity. In the case of the Vangidae, a strong slowdown in diversification is associated with extensive morphological differentiation and high numbers of co-occurring species that are only distributed within Madagascar. Conversely, among the Oriolidae, more continual diversification is associated with limited variability in their morphology and low numbers of co-occurring species that are distributed across an extensive geographical area. For the plots representing morphological disparity, red points show the species within the respective families and their divergence from the centroid of the families' morphospace. Grey points represent the morphospace occupation of the remaining corvoid species.
4. Discussion
Opportunities for speciation by ecological and geographical expansion have long been considered important in determining rates of lineage diversification [1,4,7,25,57]. Here, we provide the first comparative quantification of these relationships for a global radiation of birds. Our results reveal that diversification rates tend to decline both as relative range overlap increases within families [9,40], and as a result of a limited evolutionary change in eco-morphological traits (figure 1). Furthermore, we find that heightened species co-occurrence is itself a determinant of increased morphological differentiation (figure 1). Families that have expanded over large geographical areas and consist of mainly allopatric species continue to diversify at constant rates towards the present (figure 2 and electronic supplementary material, figure S5). Conversely, clades confined to restricted geographical areas and that have high levels of grid cell co-occurrence tend to exhibit diversification that is more consistent with a slowdown in rates towards the present (figure 1). These same groups also tend to show extensive morphological differentiation (figure 2 and electronic supplementary material, figure S6). However, once the influence of species co-occurrence has been accounted for, in one of our two best supported path models (electronic supplementary material, figure S5), we find evidence suggesting that the evolution of morphological variability also promotes continued diversification (figure 1 and electronic supplementary material, figure S5). Overall, our results imply that geographical opportunity and allopatric differentiation is the primary axis that enables continued radiation, while eco-morphological divergence generally evolves extensively among families that do not expand further geographically.
The accumulation of species within a clade can reflect the generation of both co-occurring and geographically isolated forms, however, in the majority of families geographical and morphological expansion have occurred largely independently among the Corvides. In other words, clades with high numbers of allopatric species tend not to be those that have diverged extensively in their morphology (electronic supplementary material, figure S6). Radiation as a consequence of predominantly ecological or geographical speciation might therefore represent extremes along a continuum of diversification trajectories for different groups. Specifically, the different diversification dynamics among corvoid clades reflect the degree to which species co-occur with one another (and thus potentially compete for the same resources), as heightened co-occurrence can lead to divergent natural selection pressures, and ultimately ecological speciation [4,17]. Intermediate scenarios (diversification that has involved both ecological and geographical speciation) are also prevalent, given that some corvoid families which occur over relatively large areas have morphologically disparate species, while also maintaining relatively high numbers of co-occurring species within certain parts of their range (electronic supplementary material, figure S6). The most obvious example of the combined influences of these different processes is the family Artamidae (electronic supplementary material, figure S6a). Here, geographical expansion and many of the most recent allopatric speciation events (located in the dark purple areas of the maps in electronic supplementary material, figure S6a) occurred among morphologically conserved Artamus woodswallows in Southeast Asia and the Pacific. Conversely, the Australian grid cells, which maintain much higher species richness, support many larger bodied species (e.g. currawongs and butcherbirds) in addition to the woodswallows, which themselves are represented by more distinct eco-morphs in this area. Thus comparatively, the Australian assemblages (and to a lesser degree those in New Guinea) have diverged more extensively from one another in their ecologies, particularly in terms of the occupation of different strata and foraging strategies, reflecting their morphological differences. Although the coupling of morphological divergence, geographical expansion, and continued diversification may also be evidenced in the families Corvidae and Campephagidae (electronic supplementary material, figure S6b,d), a more general pattern across corvoid families, is that the repeated formation of predominantly allopatric species has resulted in the proliferation of morphologically conserved forms (e.g. Dicruridae, Laniidae, Oriolidae, Pachycephalidae, Rhipiduridae, and Vireonidae; electronic supplementary material, figure S6e, f, i, j, l, and o). Conversely, lineages that diversified in more restricted geographical areas, such as the large tropical islands of New Guinea or Madagascar, underwent more extensive eco-morphological differentiation that seems to have facilitated species build-up and stable co-occurrence [58] (e.g. Paradisaeidae and Vangidae; figure 1 and electronic supplementary material, figure S6 k).
Although the hypothesis that eco-morphological divergence promotes diversification is supported in one of our best path models (electronic supplementary material, figure S5), it is also possible that morphological variability is itself determined by the degree to which co-occurring forms build-up within an area (figure 1 and electronic supplementary material, figure S5). Grid cells supporting an average higher number of species contain more morphologically differentiated forms, likely due to the increased ecological diversity of these assemblages (figure 2 and electronic supplementary material, figure S6). These trends have been generated by repeated cycles of allopatric speciation and eco-morphological divergence following secondary contact (character displacement) [59], or because ecological divergence in allopatry has enabled stable co-occurrence (ecological filtering). Despite our path models favouring the hypothesis that co-occurrence drives ecological divergence rather than the reverse scenario (evidenced by model 13 being a significantly better fit than model 1, figure 1 and electronic supplementary material, table S3), we note that we may have insufficient statistical power to differentiate between these scenarios. It is also possible that both character displacement and ecological filtering are involved in generating eco-morphological divergence and function in a synergistic manner; however, the path analysis framework is unable to test for such feedback loops [60].
The finding that geographical expansion is a major driver of continued diversification among corvoid families is in line with previous studies of the overall group, supporting the idea that dispersal and diversification throughout geographically fragmented landscapes promote lineage diversification [10,36,61]. Specifically, the heightened formation of allopatric populations on Indo-Pacific islands increases speciation rates [10,61] with the results of this analysis suggesting that these same conclusions likely apply to radiations that have taken place across more extensive continental settings. More broadly, the extent of range overlaps was shown to be an important predictor of diversification rate slowdowns among mammals [9], implying that geographical expansion and the accumulation of co-occurring forms may represent a general control upon the diversification process in vertebrates. Our other main finding that the evolution of morphological disparity can lead to faster diversification (figure 1) is also consistent with previous findings [62]. However, as with furnariid passerines [63] and plethodontid salamanders [64], we note that the capacity to explain variation in diversification rates based on morphological divergence alone is relatively low (electronic supplementary material, figure S5b).
Overall, our findings suggest the potential for a general explanation of diversification rate variation among corvoid passerine families. However, our interpretations partly rest on the assumption that our analysed traits represent a good approximation of the ecological variability among clades. While this has generally been shown to be the case for passerines [31,32,45,46], it is also notable that in some instances morphological variation could be driven by other selective forces, for example, sexual selection in the case of several species of the birds-of-paradise (Paradisaeidae; notably the genera Astrapia, Epimachus, and Parotia). Furthermore, our conclusions only apply to clades containing more than 10 species. We did not attempt to explain the diversification dynamics of the most species-poor corvoid families, for the reason that they have accumulated relatively few co-occurring forms due to their low overall species richness. Combined, these 16 families represent only 5% of the species diversity of the Corvides and are those with the lowest overall rates of diversification. These clades tend to be restricted to limited geographical areas, mostly located within the presumed Australasian ancestral area of the group [35], and particularly the central highlands of New Guinea [41]. It remains plausible that trends of eco-morphological diversification have been an important influence on their diversification dynamics. Specifically, small passerine clades have been proposed to be peripheral in ecological and morphological space compared to more speciose groups, such that the niches they occupy have provided limited opportunities for continued lineage diversification ([43], but see [65]). Alternatively, stable environmental and climatic conditions in the areas within which these taxa currently occur may enable the maintenance and preservation of these lineages over longer timescales [66,67].
The interpretation of our results comes with a number of caveats with respect to our phylogenetic hypothesis. We assessed the extent of diversification rate slowdowns among corvoid families using the γ statistic [51] which has been shown to become negatively biased when assessed on phylogenies that do not sample all known species within a clade [51,68]. We attempted to assess this issue, by repeating our analyses using estimates of γ on a phylogeny that sampled ca 85% of all species using DNA sequence alone and not taxonomic inference [39]. However, regardless of the phylogeny used to estimate γ our main results remained consistent (electronic supplementary material, appendix B). Furthermore, we could not find any evidence of a systematically negative bias in γ in the respective family phylogenies as a result of the proportion of missing species (electronic supplementary material, figure S2a,b). This latter finding may reflect that the proportion of species sampled with DNA sequence was generally high (above 80%) for most families (table 1). As our phylogeny only contained DNA sequences from extant species, we consider our values of γ are most likely to reflect the influence of recent speciation events, as opposed to extinction, or diversification over deeper timescales. However, extinction may have had a significant and variable influence upon the diversification trajectories and geographical distributions of different corvoid clades (e.g. the comparatively faster turnover of lineages that diversified throughout island archipelagoes [61]). Yet, as the fossil record of the Corvides (and passerines more broadly) is extremely poor and the validity of estimating extinction from molecular phylogenies remains questionable [69], the relative impact of extinction upon our results is difficult to assess at the current time.
It is also important to consider the potential influence of the species-level taxonomy used upon our results, due to the potential for (i) currently unrecognized corvoid species to exist, and (ii) because some recognized species at the tips of the tree may not represent good species. For the most part, these issues reflect different preferred species concepts among taxonomists, and the fact that the historical population structure of many corvoid species is poorly known; a common issue among global radiations in which many taxa occur in tropical and/or remote locations. To assess the potential influence of over-splitting, we performed sensitivity analyses, collapsing corvoid lineages that diverged within the last 1 Myr before repeating the path analyses. These analyses also continue to support our main findings (electronic supplementary material, appendix B). While the potential biases associated with unrecognized species are difficult to assess analytically, in almost all cases, these are highly likely to be young allopatric splits within already known species complexes. The existence of such taxa is thus likely to increase the number of morphologically similar and geographically separated species, strengthening the main conclusions of this study.
In summary, our findings highlight how simultaneous consideration of geographical and eco-morphological differentiation can explain variation in lineage diversification among comparable radiations. Differences in diversification rates predominantly reflect historical opportunities for geographical expansion, and the interplay between the colonization of new areas, eco-morphological divergence, and the build-up of closely related species at small spatial scales (figure 2). Importantly, eco-morphological divergence appears driven by the accumulation of co-occurring species. These findings apply to a species-rich and globally distributed radiation of passerine birds and provide insight into the processes that underlie the vast taxonomic and geographical differences in the distribution of species diversity. We conclude that the capacity for geographical expansion and the accumulation of co-occurring species is likely to be an important control upon diversification rates across the tree of life.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Louis Hansen for help in compiling the distributional datasets, and the museum collections and associated staff that made possible collection of the morphological data. In this regard, we would particularly like to thank Mark Adams, Hein van Grouw, and Robert Prys-Jones at the British Museum of Natural History, Lydia Garetano, Joel Cracraft, and Paul Sweet at the American Museum of Natural History, and Pepijn Kamminga and Steven van der Mije at the Naturalis Biodiversity Center (Netherlands). Wouter van der Bijl provided valuable advice on the phylogenetic path analyses.
Data accessibility
The morphological data are provided in electronic supplementary material, table S1 and the species-level phylogeny is available from the Dryad Digital Repository: https://dx.doi.org/10.5061/dryad.80n42 [70].
Authors' contributions
J.D.K. conceived the study, P.Z.M. collected the morphological data, J.D.K. performed the analyses and J.D.K., M.K.B., P.Z.M., A.M., J.F., and C.R. interpreted the analyses and wrote the manuscript.
Competing interests
We declare we have no competing interests.
Funding
J.D.K. was supported by an Internationalisation Fellowship (CF17-0239) from the Carlsberg Foundation, and M.K.B. by an Individual Fellowship from Marie Sklodowska-Curie actions (IDEA-707968). All authors wish to thank the Danish National Research Foundation for its support of the Center for Macroecology, Evolution and Climate (DNRF96).
References
- 1.Mayr E. 1947. Ecological factors in speciation. Evolution 1, 263–288. ( 10.1111/j.1558-5646.1947.tb02723.x) [DOI] [Google Scholar]
- 2.Simpson GG. 1953. The major features of evolution. New York, NY: Columbia University Press. [Google Scholar]
- 3.Rosenzweig ML. 1995. Species diversity in space and time. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 4.Price TD. 2008. Speciation in birds. Boulder, CO: Roberts and Co. [Google Scholar]
- 5.Phillimore AB, Price TD. 2009. Ecological influences on the temporal pattern of speciation. In Speciation and patterns of diversity, pp. 240–256. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 6.Pigot AL, Phillimore AB, Owens IP, Orme CDL. 2010. The shape and temporal dynamics of phylogenetic trees arising from geographic speciation. Syst. Biol. 59, 660–673. ( 10.1093/sysbio/syq058) [DOI] [PubMed] [Google Scholar]
- 7.Fritz SA, Jønsson KA, Fjeldså J, Rahbek C. 2012. Diversification and biogeographic patterns in four island radiations of passerine birds. Evolution 66, 179–190. ( 10.1111/j.1558-5646.2011.01430.x) [DOI] [PubMed] [Google Scholar]
- 8.Machac A, Storch D, Wiens JJ. 2013. Ecological causes of decelerating diversification in carnivoran mammals. Evolution 67, 2423–2433. ( 10.1111/evo.12126) [DOI] [PubMed] [Google Scholar]
- 9.Machac A, Graham CH, Storch D. 2018. Ecological controls of mammalian diversification vary with phylogenetic scale. Global. Ecol. Biogeogr. 27, 32–46. ( 10.1111/geb.12642) [DOI] [Google Scholar]
- 10.Kennedy JD, Borregaard MK, Jønsson KA, Marki PZ, Fjeldså J, Rahbek C. 2016. The influence of wing morphology upon the dispersal, geographical distributions and diversification of the Corvides (Aves; Passeriformes). Proc. R. Soc. B 283, 20161922 ( 10.1098/rspb.2016.1922) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gavrilets S, Losos JB. 2009. Adaptive radiation: contrasting theory with data. Science, 323, 732–737. ( 10.1126/science.1157966) [DOI] [PubMed] [Google Scholar]
- 12.Gavrilets S, Vose A. 2005. Dynamic patterns of adaptive radiation. Proc. Natl Acad. Sci. USA 102, 18 040–18 045. ( 10.1073/pnas.0506330102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayr E. 1942. Systematics and the origin of species, from the viewpoint of a zoologist. Cambridge, MA: Harvard University Press. [Google Scholar]
- 14.Barraclough TG, Vogler AP. 2000. Detecting the geographical pattern of speciation from species-level phylogenies. Am. Nat. 155, 419–434. [DOI] [PubMed] [Google Scholar]
- 15.Zink RM, Blackwell-Rago RC, Ronquist F. 2000. The shifting roles of dispersal and vicariance in biogeography. Proc. R. Soc. B 267, 497–503. ( 10.1098/rspb.2000.1028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phillimore AB, Orme CDL, Thomas GH, Blackburn TM, Bennett PM, Gaston KJ, Owens IP. 2008. Sympatric speciation in birds is rare: insights from range data and simulations. Am. Nat. 171, 646–657. ( 10.1086/587074) [DOI] [PubMed] [Google Scholar]
- 17.Huxley J. 1942. Evolution the modern synthesis. London, UK: George Allen and Unwin. [Google Scholar]
- 18.Schluter D. 2000. The ecology of adaptive radiation. Oxford, UK: Oxford University Press. [Google Scholar]
- 19.Losos JB, Mahler DL. 2010. Adaptive radiation: the interaction of ecological opportunity, adaptation, and speciation. In Evolution since Darwin: the first 150 years (eds MA Bell, DJ Futuyma, WF Eanes, JS Levinton), pp. 381–420. Oxford, UK: Oxford University Press. [Google Scholar]
- 20.Gittenberger E. 1991. What about non-adaptive radiation? Biol. J. Linn. Soc. 43, 263–272. ( 10.1111/j.1095-8312.1991.tb00598.x) [DOI] [Google Scholar]
- 21.Mayr E, Diamond J. 2001. The birds of Northern Melanesia. New York, NY: Oxford University Press, Inc. [Google Scholar]
- 22.Kozak KH, Weisrock DW, Larson A. 2006. Rapid lineage accumulation in a non-adaptive radiation: phylogenetic analysis of diversification rates in eastern North American woodland salamanders (Plethodontidae: Plethodon). Proc. R. Soc. B 273, 539–546. ( 10.1098/rspb.2005.3326) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rundell RJ, Price TD. 2009. Adaptive radiation, nonadaptive radiation, ecological speciation and nonecological speciation. Trends Ecol. Evol. 24, 394–399. ( 10.1016/j.tree.2009.02.007) [DOI] [PubMed] [Google Scholar]
- 24.Price TD. 2010. The roles of time and ecology in the continental radiation of the Old World leaf warblers (Phylloscopus and Seicercus). Phil. Trans. R. Soc. B 365, 1749–1762. ( 10.1098/rstb.2009.0269) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harmon LJ, Schulte JA, Larson A, Losos JB. 2003. Tempo and mode of evolutionary radiation in iguanian lizards. Science 301, 961–964. ( 10.1126/science.1084786) [DOI] [PubMed] [Google Scholar]
- 26.Rabosky DL. 2009. Ecological limits and diversification rate: alternative paradigms to explain the variation in species richness among clades and regions. Ecol. Lett. 12, 735–743. ( 10.1111/j.1461-0248.2009.01333.x) [DOI] [PubMed] [Google Scholar]
- 27.Cooney CR, Bright JA, Capp EJ, Chira AM, Hughes EC, Moody CJ, Nouri LO, Varley ZK, Thomas GH. 2017. Mega-evolutionary dynamics of the adaptive radiation of birds. Nature 542, 344–347. ( 10.1038/nature21074) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schluter D, Grant PR. 1984. Determinants of morphological patterns in communities of Darwin's finches. Am. Nat. 123, 175–196. ( 10.1086/284196) [DOI] [Google Scholar]
- 29.Schluter D, Price TD, Grant PR. 1985. Ecological character displacement in Darwin's finches. Science 227, 1056–1059. ( 10.1126/science.227.4690.1056) [DOI] [PubMed] [Google Scholar]
- 30.Pigot AL, Tobias JA. 2013. Species interactions constrain geographic range expansion over evolutionary time. Ecol. Lett. 16, 330–338. ( 10.1111/ele.12043) [DOI] [PubMed] [Google Scholar]
- 31.Miles DB, Ricklefs RE. 1984. The correlation between ecology and morphology in deciduous forest passerine birds. Ecology 65, 1629–1640. ( 10.2307/1939141) [DOI] [Google Scholar]
- 32.Tobias JA, Cornwallis CK, Derryberry EP, Claramunt S, Brumfield RT, Seddon N. 2014. Species coexistence and the dynamics of phenotypic evolution in adaptive radiation. Nature 506, 359–363. ( 10.1038/nature12874) [DOI] [PubMed] [Google Scholar]
- 33.Ricklefs RE. 2012. Species richness and morphological diversity of passerine birds. Proc. Natl Acad. Sci. USA 109, 14482–14487. ( 10.1073/pnas.1212079109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Hardenberg A, Gonzalez-Voyer A. 2013. Disentangling evolutionary cause-effect relationships with phylogenetic confirmatory path analysis. Evolution, 67, 378–387. ( 10.1111/j.1558-5646.2012.01790.x) [DOI] [PubMed] [Google Scholar]
- 35.Jønsson KA, Fabre PH, Ricklefs RE, Fjeldså J. 2011. Major global radiation of corvoid birds originated in the proto-Papuan archipelago. Proc. Natl Acad. Sci. USA 108, 2328–2333. ( 10.1073/pnas.1018956108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jønsson KA, Borregaard MK, Carstensen DW, Hansen LA, Kennedy JD, Machac A, Marki PZ, Fjeldså J, Rahbek C. 2017. Biogeography and biotic assembly of indo-pacific corvoid passerine birds. Annu. Rev. Ecol. Syst., 48, 231–253. ( 10.1146/annurev-ecolsys-110316-022813) [DOI] [Google Scholar]
- 37.Kennedy JD, Weir JT, Hooper DM, Tietze DT, Martens J, Price TD. 2012. Ecological limits on diversification of the Himalayan core Corvoidea. Evolution, 66, 2599–2613. ( 10.1111/j.1558-5646.2012.01618.x) [DOI] [PubMed] [Google Scholar]
- 38.Gill F, Donsker D.. 2010. IOC world bird names (v 2.7.). See http://www.worldbirdnames.org.
- 39.Jønsson KA, Fabre PH, Kennedy JD, Holt BG, Borregaard MK, Rahbek C, Fjeldså J. 2016. A supermatrix phylogeny of corvoid passerine birds (Aves: Corvides). Mol. Phylogenet. Evol. 94, 87–94. ( 10.1016/j.ympev.2015.08.020) [DOI] [PubMed] [Google Scholar]
- 40.Kennedy JD, Price TD, Fjeldså J, Rahbek C. 2017. Historical limits on species co-occurrence determine variation in clade richness among New World passerine birds. J. Biogeogr. 44, 736–747. ( 10.1111/jbi.12834) [DOI] [Google Scholar]
- 41.Kennedy JD, Borregaard MK, Jønsson KA, Holt B, Fjeldså J, Rahbek C. 2017. Does the colonization of new biogeographic regions influence the diversification and accumulation of clade richness among the Corvides (Aves: Passeriformes)? Evolution, 71, 38–50. ( 10.1111/evo.13080) [DOI] [PubMed] [Google Scholar]
- 42.Rahbek C, Hansen LA, Fjeldså J. 2012. One degree resolution database of the global distribution of birds. Copenhagen, Denmark: Natural History Museum of Denmark, University of Copenhagen. [Google Scholar]
- 43.Ricklefs RE. 2005. Small clades at the periphery of passerine morphological space. Am. Nat. 165, 651–659. ( 10.1086/429676) [DOI] [PubMed] [Google Scholar]
- 44.Claramunt S, Derryberry EP, Remsen JV, Brumfield R. 2011. High dispersal ability inhibits speciation in a continental radiation of passerine birds. Proc. R. Soc. B 279, 1567–1574. ( 10.1098/rspb.2011.1922) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miles DB, Ricklefs RE, Travis J. 1987. Concordance of ecomorphological relationships in three assemblages of passerine birds. Am. Nat. 129, 347–364. ( 10.1086/284641) [DOI] [Google Scholar]
- 46.Pigot AL, Trisos CH, Tobias JA. 2016. Functional traits reveal the expansion and packing of ecological niche space underlying an elevational diversity gradient in passerine birds. Proc. R. Soc. B 283, 20152013 ( 10.1098/rspb.2015.2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Losos J.B., Schluter D. 2000. Analysis of an evolutionary species–area relationship. Nature, 408, 847–850. ( 10.1038/35048558) [DOI] [PubMed] [Google Scholar]
- 48.Laliberté E, Legendre P. 2010. A distance-based framework for measuring functional diversity from multiple traits. Ecology 91, 299–305. ( 10.1890/08-2244.1) [DOI] [PubMed] [Google Scholar]
- 49.Miller E, Trisos C, Farine D.. 2017. metricTester: test metric and null model statistical performance. See http://CRAN.R-project.org/package=metricTester.
- 50.Oliveira BF, Machac A, Costa GC, Brooks TM, Davidson AD, Rondinini C, Graham CH. 2016. Species and functional diversity accumulate differently in mammals. Global. Ecol. Biogeogr. 25, 1119–1130. ( 10.1111/geb.12471) [DOI] [Google Scholar]
- 51.Pybus OG, Harvey PH. 2000. Testing macro–evolutionary models using incomplete molecular phylogenies. Proc. R. Soc. B 267, 2267–2272. ( 10.1098/rspb.2000.1278) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morlon H. 2014. Phylogenetic approaches for studying diversification. Ecol. Lett. 17, 508–525. ( 10.1111/ele.12251) [DOI] [PubMed] [Google Scholar]
- 53.del Hoyo J, Elliot A, Christie DA. 2009. Handbook of the birds of the world, vol. 10–14 Barcelona, Spain: Lynx Edicions. [Google Scholar]
- 54.Kuhn TS, Mooers AØ, Thomas GH. 2011. A simple polytomy resolver for dated phylogenies. Methods. Ecol. Evol. 2, 427–436. ( 10.1111/j.2041-210X.2011.00103.x) [DOI] [Google Scholar]
- 55.Shipley B. 2000. A new inferential test for path models based on directed acyclic graphs. Struct. Equ. Modeling 7, 206–218. ( 10.1207/S15328007SEM0702_4) [DOI] [Google Scholar]
- 56.van der Bijl W. 2017. phylopath: easy phylogenetic path analysis in R. bioRxiv. ( 10.1101/212068). R package version 1.0.0. [DOI] [PMC free article] [PubMed]
- 57.Simpson GG. 1944. Tempo and mode in evolution. New York, NY: Columbia University Press. [Google Scholar]
- 58.Lovette IJ, Hochachka WM. 2006. Simultaneous effects of phylogenetic niche conservatism and competition on avian community structure. Ecology 87, s14–s48. ( 10.1890/0012-9658(2006)87%5B14:SEOPNC%5D2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 59.Fjeldså J. 1983. Ecological character displacement and character release in grebes Podicipedidae. Ibis 125, 463–481. ( 10.1111/j.1474-919X.1983.tb03142.x) [DOI] [Google Scholar]
- 60.Shipley B. 2000. Cause and correlation in biology: a user's guide to path analysis, structural equations and causal inference. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 61.Marki PZ, Fabre PH, Jønsson KA, Rahbek C, Fjeldså J, Kennedy JD. 2015. Breeding system evolution influenced the geographic expansion and diversification of the core Corvoidea (Aves: Passeriformes). Evolution, 69, 1874–1924. ( 10.1111/evo.12695) [DOI] [PubMed] [Google Scholar]
- 62.Rabosky DL, Santini F, Eastman J, Smith SA, Sidlauskas B, Chang J, Alfaro ME. 2013. Rates of speciation and morphological evolution are correlated across the largest vertebrate radiation. Nat. Commun. 4, 1958 ( 10.1038/ncomms2958) [DOI] [PubMed] [Google Scholar]
- 63.Derryberry EP, et al. 2011. Lineage diversification and morphological evolution in a large-scale continental radiation: the neotropical ovenbirds and woodcreepers (Aves: Furnariidae). Evolution 65, 2973–2986. ( 10.1111/j.1558-5646.2011.01374.x) [DOI] [PubMed] [Google Scholar]
- 64.Adams DC, Berns CM, Kozak KH, Wiens JJ. 2009. Are rates of species diversification correlated with rates of morphological evolution? Proc. R. Soc. B 276, 2729–2738. ( 10.1098/rspb.2009.0543) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chira AM, Cooney CR, Bright JA, Capp EJR, Hughes EC, Moody CJA, Nouri LO, Varley ZK, Thomas GH. 2018. Correlates of rate heterogeneity in avian ecomorphological traits. Ecol. Lett. 21, 1505–1514. ( 10.1111/ele.13131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fjeldså J, Bowie RC, Rahbek C. 2012. The role of mountain ranges in the diversification of birds. Annu. Rev. Ecol. Syst. 43, 249–265. ( 10.1146/annurev-ecolsys-102710-145113) [DOI] [Google Scholar]
- 67.Fjeldså J. 2018. Mountains and the diversity of birds. In Mountains, climate and biodiversity (eds Hoorns C, Perrigo A, Antonelli A), pp. 245–256. John Wiley & Sons Ltd. [Google Scholar]
- 68.Cusimano N, Renner SS. 2010. Slowdowns in diversification rates from real phylogenies may not be real. Syst. Biol. 59, 458–464. ( 10.1093/sysbio/syq032) [DOI] [PubMed] [Google Scholar]
- 69.Rabosky DL. 2010. Extinction rates should not be estimated from molecular phylogenies. Evolution 64, 1816–1824. ( 10.1111/j.1558-5646.2009.00926.x) [DOI] [PubMed] [Google Scholar]
- 70.Kennedy JD, Borregaard MK, Marki PZ, Machac A, Fjeldså J, Rahbek C. 2018. Data from: Expansion in geographical and morphological space drives continued lineage diversification in a global passerine radiation Dryad Digital Repository. ( 10.5061/dryad.80n42) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Kennedy JD, Borregaard MK, Marki PZ, Machac A, Fjeldså J, Rahbek C. 2018. Data from: Expansion in geographical and morphological space drives continued lineage diversification in a global passerine radiation Dryad Digital Repository. ( 10.5061/dryad.80n42) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
The morphological data are provided in electronic supplementary material, table S1 and the species-level phylogeny is available from the Dryad Digital Repository: https://dx.doi.org/10.5061/dryad.80n42 [70].