Abstract
Ecological opportunity arising in the aftermath of mass extinction events is thought to be a powerful driver of evolutionary radiations. Here, we assessed how the wake of the Cretaceous–Palaeogene (K-Pg) mass extinction shaped diversification dynamics in a clade of mostly marine fishes (Carangaria), which comprises a disparate array of benthic and pelagic dwellers including some of the most astonishing fish forms (e.g. flatfishes, billfishes, remoras, archerfishes). Analyses of lineage diversification show time-heterogeneous rates of lineage diversification in carangarians, with highest rates reached during the Palaeocene. Likewise, a remarkable proportion of Carangaria's morphological variation originated early in the history of the group and in tandem with a marked incidence of habitat shifts. Taken together, these results suggest that all major lineages and body plans in Carangaria originated in an early burst shortly after the K-Pg mass extinction, which ultimately allowed the occupation of newly released niches along the benthic-pelagic habitat axis.
Keywords: mass extinctions, ecological opportunity, macroevolution, benthic-pelagic axis, diversification
1. Introduction
Patterns of initial bursts of diversification in the origin and propagation of high-level taxa are usually explained by Simpsonian theory on adaptive radiation—one where a rapidly proliferating lineage evolves into great ecological diversity and morphological disparity as a result of increased availability of resources and limited competition [1,2]. These ecological opportunities may occur when previously inaccessible resources become available via acquisition of key evolutionary innovations, colonization of new areas, or removal of competitors owing to external mechanisms of habitat depauperation [1].
The formation of vacant niches in the wake of mass extinction events is a major source of ecological opportunity [3]. Among the five major mass extinctions in the history of life on Earth, the Cretaceous–Palaeogene (K-Pg) event (ca 65 Ma) is thought to have triggered parallel rapid radiations in numerous tetrapod clades including amphibians and placental mammals [4,5]. In Actinopterygii (ray-finned fishes), the proportion of incumbent diversity that became extinct by the end of the Mesozoic is high at the family level (19%) [6] and probably represented an important source of ecological opportunity that modulated diversification dynamics in surviving acanthomorph lineages (spiny-rayed teleost fishes; a subclade of ray-finned fishes) [6–8]. Indeed, the stratigraphic distribution of acanthomorph fossils suggests significant restructuring of marine fish communities in the aftermath of the K-Pg [9], which further coincides with the expansion of the group's morphological disparity in areas of the ecospace emptied by the extinction of their (non-acanthomorph) teleost counterparts [7]. In the wake of the K-Pg, fishes also experienced a pronounced increase in abundance relative to sharks, which was presumably spurred by ecological release and ultimately prompted what has come to be known as the ‘new age of fishes’ [10].
In agreement with the fossil evidence, time-calibrated phylogenetic trees also reveal patterns in which the origin of several major acanthomorph subclades chronologically overlap with the K-Pg boundary [11–14]. One such clade featuring an explosive pulse of diversification near the K-Pg is the Carangaria [13], a diverse group with over a thousand species that includes a disparate array of benthic (e.g. flatfishes, threadfins) and pelagic (e.g. billfishes, remoras, barracudas) fish dwellers. The Carangaria also encompasses some of the most extreme morphological and ecological adaptations in vertebrates, including the asymmetric body plan of flatfishes, the endothermic body heat regulation in marlins, billfishes and swordfishes, and the hunting behaviour of archerfishes, which generate bullets of water to feed on terrestrial prey. It thus appears that the greatest phenotypic diversity in the Carangaria is associated with the early evolution of disparate morphologies that prompted adaptation to a broad array of habitats, including clear instances of adaptive peaks lying at the extremes of the benthic-pelagic spectrum in fishes (e.g. open-water billfishes and substrate-burrowing flatfishes).
Here, we investigate the dynamics of diversification, phenotypic evolution and habitat transitions in Carangaria. Based on the above observations, we hypothesize that lineage diversification varies as a function of time, with high rates reached near the clade's origin (at the Mesozoic–Cenozoic boundary) followed by a rapid drop as the carrying capacity of species diversity is reached. Furthermore, in agreement with Simpsonian predictions on adaptive radiation, we also expect that an initial expansion of morphological disparity would be subsequently replaced by a period of morphospace packing as niches become filled. To test these ideas, we estimated a multi-locus time tree that includes all major lineages of carangarians and used a suite of recently developed phylogenetic comparative approaches to assesses how rates of lineage diversification, multivariate phenotypic evolution and habitat transitions vary throughout the clade's history.
2. Material and methods
(a). Taxonomic sampling and phylogenetic inference
Carangaria's diversity is represented by a sample of 125 (out of ca 1100) species, including representatives from 26 valid families out of 28 (only Lactariidae and Paralichthodidae were not examined) and over half of the genus-level diversity (95 out of 187) in the group. This taxonomic sampling strategy was designed to maximize both phylogenetic and eco-morphological diversity within Carangaria, under the assumption that missing lineages are phylogenetically and eco-morphologically nested within the sampled ones.
The molecular dataset is based on a recent study that generated multi-locus sequences from 20 nuclear loci (19 461 sites) [15]. We expanded the molecular matrix to incorporate 10 additional outgroup species that represent major acanthomorph lineages. We used BEAST v. 1.8.4 [16] to simultaneously estimate topology and divergence times using a set of 16 fossil-based calibration points (modified from Harrington et al. [14]). Lower bounds of clade ages were defined via minimum age of earliest fossil representatives; 95% soft upper bounds were estimated based on maximum ages of the oldest fossils assigned to successive outgroups for each clade. Convergence of analyses was assessed after conducting two independent runs of 300 million generations each. To account for phylogenetic uncertainty, one hundred trees were evenly sampled from the posterior distribution, providing a robust framework for downstream macroevolutionary analyses. Detailed taxonomic sampling (electronic supplementary material, table S1), phylogenetic analyses (electronic supplementary material, figure S1) and fossil calibration information is provided in the electronic supplementary material. To further address potential phylogenetic uncertainties, we repeated some analyses using alternative trees for Carangaria estimated by other recent studies [14,17] (see below).
(b). Body-shape data
The laterally compressed body plan of most carangarians makes this group well-suited for the summarization of multivariate morphological axes using two-dimensional geometric morphometric approaches. We carefully assembled a specimen imagery database for 116 out of the 125 carangarian species in our tree. The database consists of digitized specimens from museum collections or curated images retrieved from online repositories (electronic supplementary material). We selected 15 landmarks that are extensively used to summarize general body shape variation in percomorphs [7], including both type I and type II points (electronic supplementary material, figure S2). Whereas type I landmarks are strictly homologous points, type II landmarks include points whose homology is supported by geometric evidence rather than histological data and are frequently used to describe inflexion points such as the sharpest curvature of a tooth or tips of caudal fin lobes [18]. We used tpsDig2 [19] to place the landmarks and summarized the extant species' body shape diversity using Procrustes superimposition and principal component analyses (PCA), as implemented in the R package geomorph [20]. Next, we subjected the morphological data to a phylogenetically corrected principal component analysis (pPCA) to account for possible distortions of the PCA arising from phylogenetic non-independence.
(c). Tempo and mode of lineage diversification
We assessed time variation in lineage diversification rates in Carangaria using CoMET, a Bayesian statistical model implemented in the R package TESS [21]. CoMET estimates the number of lineage diversification rate shifts along with their timing and rate parameters (i.e. speciation and extinction rates). We used TESS’ Bayes factor model selection to explicitly test the relative fit of the following series of alternative branching models to our comparative dataset: (i) time-homogeneous birth–death, (ii) continuously-decreasing rate birth–death, and (iii) episodically-varying rate that incorporates the diversification parameters (i.e. number and timing of episodic rate shifts) obtained with CoMET. Model comparison analyses were applied for both the maximum clade credibility (MCC) tree and 100 trees sampled from the posterior distribution. To accommodate biases inherent to incomplete taxonomic sampling, we applied a diversified sampling strategy correction (for both CoMET and model fitting), which is appropriate in cases where taxonomic sampling is designed to maximize phylogenetic diversity [22]. To further account for potential biases associated with incomplete taxonomic sampling and phylogenetic uncertainties, we conducted posterior-predictive simulation tests using our MCC tree and a set of independently estimated time trees that incorporate different taxonomic sampling schemes (ranging from approx. 5% of the clades diversity [14] to a nearly complete taxonomic sampling [17]; electronic supplementary material).
(d). Tempo and mode of morphological evolution
To assess the tempo and mode of morphological evolution in Carangaria, we initially estimated how multivariate morphological disparity accumulated through time using ancestral state reconstructions derived from rate-heterogeneous models of continuous trait evolution [23,24]. We estimated ancestral state values using the maximum-likelihood (ML) fastAnc function implemented in the R package phytools [24]. To account for unequal rates of evolution among the different shape axes, we inferred ancestral values for each pPC separately using rate-transformed trees in which branch lengths depict the rate of morphological change. Rate-transformed trees were estimated in BayesTraits (available from http://www.evolution.rdg.ac.uk/) using default priors. Multivariate morphological disparity was then calculated as the sum of the variances across the different pPC axes in time-slices of one million year (Myr). We compared the observed disparity against 500 curves of disparity accumulation simulated under a constant rate Brownian motion (BM) null model of continuous trait evolution. Because of the non-directional nature of trait change simulated using this BM model, we expect the underlying balance between morphospace expansion and packing to be effectively equal and constant over time. Thus, any period of time that shows substantial deviations in the observed patterns of morphological disparity accumulation (compared to the simulated null trajectories) would indicate that one of the processes (either morphospace expansion or packing) dominated over the other.
We also explicitly assessed the fit of alternative evolutionary models of body shape evolution in an ML framework using the R package mvMORPH [25]. We first fitted three alternative models of single-mode continuous-trait evolution: (i) a single rate BM model, (ii) a single regime Orstein–Uhlenbeck (OU) model, and (iii) an early burst (EB) of morphological evolution. Given that shifts in the mode of evolution may provide a more realistic explanation for the processes generating morphological disparity [26], we further considered three additional models in which processes generating disparity vary episodically: (iv) EB to independent rates OU shift, (v) BM to independent rates OU shift, and (vi) EB to independent rates BM shift. We used the function mvSHIFT, which can fit models of trait change within a mode of evolution after a fixed point. We modelled post-shift independent rates OU and BM models by allowing the drift parameter to vary after a fixed point. A temporal shift window of 46 Ma was selected based on the time of transition between stages of morphospace expansion and packing, as obtained by comparing the observed and simulated trajectories of morphological disparity (see Results).
It has been suggested that limiting macroevolutionary analyses to a narrow subset of shape dimensions (i.e. first few principal component (PC) axes) may produce erroneously strong support to more complex models, such as EB [27]. More recently, mvMORPH has been shown to produce misleading results when the N : p ratio is sufficiently low (where N is the number of species and p the number of traits). It should be noted, however, that an adequate N : p ratio level required to confidently assess the fit of alternative models using mvMORPH is still elusive [28]. To account for possible biases regarding the use of multivariate shape data, we performed analyses using two different trait subsets selected according to the proportion of body-shape variance summarized (based on 5% and 1% thresholds; see Results). The 5% subset was run using both the MCC tree and a set of 100 trees drawn from the posterior distribution; owing to computational limitations, the 1% subset was run using the MCC tree only.
(e). Tempo and mode of ecological diversification
To evaluate whether the rate of habitat transitions varied as a function of time, we first assigned species in Carangaria into three major habitat categories: benthic (bottom-dwellers), pelagic (open water dwellers), and benthopelagic (intermediate habitat states; species that swim just above the bottom) [29]. The habitat occupancy dataset (electronic supplementary material, table S1) was compiled by aggregating information from a wide range of sources, including FishBase [30], Catalog of Fishes [31] and the primary literature. We then used SIMMAP stochastic mapping, as implemented in the R package phytools [24], to reconstruct the history of trait changes in our MCC tree and to estimate the rate of habitat transitions from root through to present (i.e. number of transitions divided by the total edge length in 5 Myr time slices). Three hierarchical transition models—equal rates, symmetrical rates or all rates different—were assessed by ML with results averaged across all runs; the best-fitting model for SIMMAP was identified using likelihood ratio tests. We also explicitly tested the relative fit of models of discrete character evolution to identify the best explanation concerning the distribution of events of habitat transition throughout the evolutionary history of carangarians. We used fitDiscrete as implemented in the R package Geiger [32] to assess the fit of two contrasting models of discrete trait evolution: (i) a constant-rate model and (ii) an EB model of discrete trait diversification. For this approach, we used the set of 100 trees sampled from the Bayesian posterior distribution (see above), as well as the set of independently estimated time trees that incorporate different taxonomic sampling schemes [14,17].
3. Results
(a). Phylogenetic reconstruction and divergence times
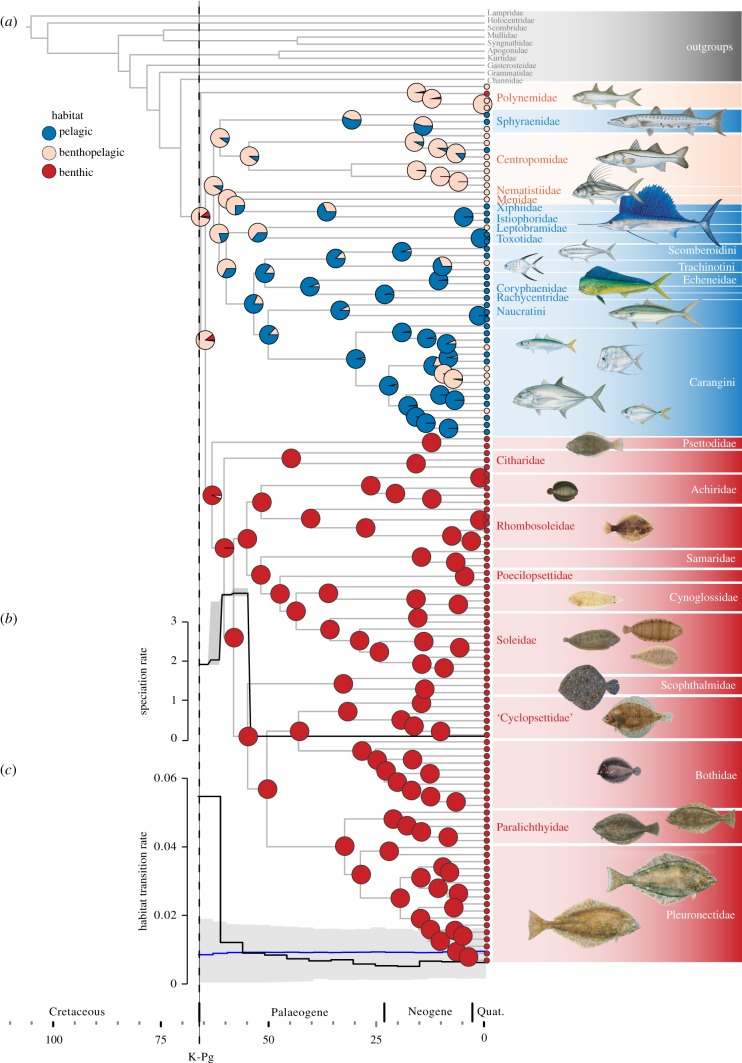
Trees, datasets and R code used for comparative analyses are available from the Dryad Digital Repository (doi:10.5061/dryad.9bq5n41) [33]. The inferred tree (figure 1; electronic supplementary material, figure S1) is largely congruent with previous multi-locus analyses of Carangaria [14,15], although placement for some lineages (e.g. barracudas and threadfins) along the backbone often varies owing to the succession of rapid speciation events at the onset of the group's evolution. Divergence-time estimates are likewise concordant with the age intervals derived from a recent study based on a phylogenomic analysis for 45 species in Carangaria [14], as well as previous estimates based on multi-locus datasets and multiple calibration points across the fish diversity [11,12,34,35] (see the electronic supplementary material, table S5 for a comparison). The evolutionary timescale inferred suggests that the origin of major carangarian lineages took place close to the Cretaceous–Palaeogene boundary, with a mean clade age ranging from 71 Ma (total group; 95% highest posterior density (HPD) 78–64 Ma) to 66 Ma (crown group; 95% HPD 72–61 Ma) [11,14].
Figure 1.
(a) Maximum clade credibility (MCC) time tree estimated for Carangaria, including pie charts for ancestral habitat reconstructions. (b) Rates of speciation through time estimated from the MCC tree using the CoMET function in TESS. (c) Rates of habitat transitions through time, estimated as number of transitions divided by the total edge length in 5 Myr time slices; blue line indicates constant rate Brownian motion model. Dashed line indicates the time of the Cretaceous–Palaeogene (K-Pg) mass extinction. Most fish illustrations are reproduced with permission from Diane Rome Peebles©.
(b). Body-shape data
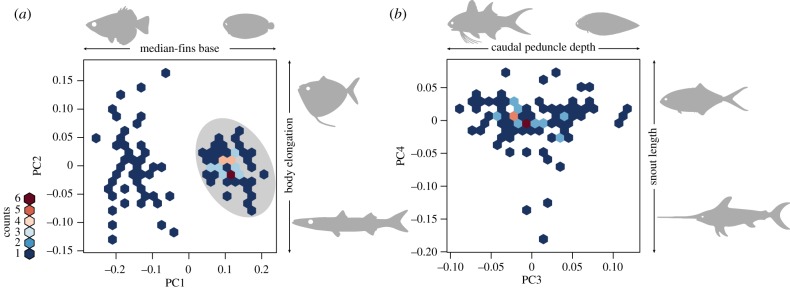
The first four PC axes accounted for more than 85% of the total shape variance and are presented as morphospace scatter plots in figure 2. The PC1 (67% of overall shape variation) describes head morphology and the length of the dorsal and anal fin bases, features that have been identified as one of the major axes of evolution in acanthomorphs [7,36]. In flatfishes, in particular, those characters are linked to some of the most extreme morphological adaptations experienced by vertebrates; i.e. the partial loss of bilateral symmetry arising from eye migration, and the dorsal advancement of median fins towards the cranium. Indeed, this is represented as a bimodal distribution of PC1 values that distinguish flatfishes from all non-pleuronectiform carangarians in our analyses (figure 2a). The PC2 (7% of total variation) summarizes differences in body elongation, a major axis of shape variation in several fish clades [36]. The PC3 and PC4 (6% and 5% of overall variation, respectively) also encompass ecologically relevant aspects of fish morphology that are frequently represented in traditional morphometric measurements (caudal peduncle depth and snout length, respectively; figure 2b). Subsequent PC axes explain lower proportions of body-shape variance. As noted in the Methods section, we selected two different trait subsets of pPC axes for downstream analyses [27,28]. The 5% and 1% subsets comprised the first 4 pPC (69% of total shape variation) and first 12 pPC (95% of total shape variation) axes, respectively.
Figure 2.
Body-shape morphospace in Carangaria. Fish silhouettes represent extreme values for each axis: (a) PC1 versus PC2 (shaded elliptical area represents the morphospace occupied by pleuronectifoms); (b) PC3 versus PC4. Colour scale indicates the number of species per hexagon.
(c). Tempo and mode of lineage diversification
These analyses show a burst of lineage diversification rate at the onset of carangarian history, with post-Cretaceous rates decreasing abruptly after the Palaeocene–Eocene boundary (56 Ma). CoMET results were rather inconsistent about the clade's evolutionary dynamics and highly sensitive to the choice of hyper-priors. However, one recurrent scenario—high initial speciation followed by a decline in speciation rates around 55 Ma (figure 1b)—demonstrates the existence of a strong signal supporting a change in the diversification regime, a result that seems robust to analytical artefacts (electronic supplementary material, figure S3).
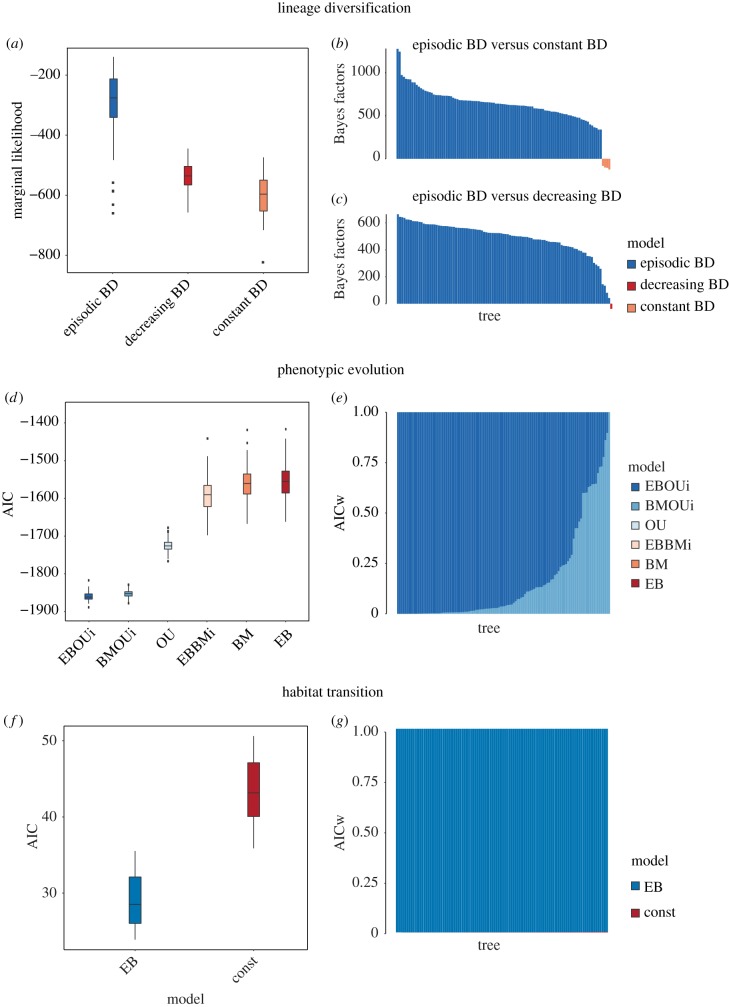
TESS’ marginal likelihood model comparison showed a preference (Bayes factors (BF) > 100) for variable-rates models (continuously-decreasing and episodically-varying rate birth-death models) over a time-homogeneous birth–death mode for most pruned resampled trees, confirming our expectation that that time-homogeneous processes cannot explain lineage diversification dynamics in the group (figure 3a-c; see the electronic supplementary material, table S2 for the MCC tree model comparison results). Moreover, comparisons between the two variable-rates models reveal that 99% of the trees provide decisive support (BF > 100) for the episodically varying rate model that incorporates one diversification rate-shift at 55 Ma. We obtained similar results for model-fit comparisons using the original set of resampled trees (electronic supplementary material, figure S4). Finally, our results on lineage diversification appear to be robust to the use of alternative phylogenetic trees incorporating a broad array of taxonomic sampling schemes as well as to the implementation of posterior-predictive simulation tests (electronic supplementary material, figures S5 and S6) or other simpler statistics (electronic supplementary material, figures S7).
Figure 3.
Model-fit comparisons based on a set of 100 trees evenly sampled from the posterior distribution. (a–c) Comparisons for alternative models of lineage diversification (using pruned versions of our 100 empirical that excludes recent cladogenetic events): (a) distribution of the marginal likelihood for the three alternative branching models; (b) Bayes factors comparing episodic birth-death (BD) and constant BD models for the 100 resampled trees; and (c) Bayes factors comparing episodic BD and decreasing BD for the 100 resampled trees. (d,e) Comparisons of alternative models of morphological evolution using the 5% threshold trait subset: (d) distribution of the Akaike information criterion (AIC) values for the six alternative models of continuous trait evolution (EBOUi, shift from a single rate EB into a multiple independent rates OU; BMOUi, shift from a single rate BM into a multiple independent rates OU; OU, Ornstein-Uhlenbeck; EBBMi, shift from a single rate EB into a multiple independent rates BM; BM, Brownian motion; EB, early burst); (e) AIC weights of each alternative model based on each resampled tree. (f,g) Comparisons of alternative models of ecological evolution: (f) distribution of the AIC values for the two models of discrete trait evolution; (e) AIC weights of each alternative model based on each resampled tree.
(d). Tempo and mode of morphological evolution
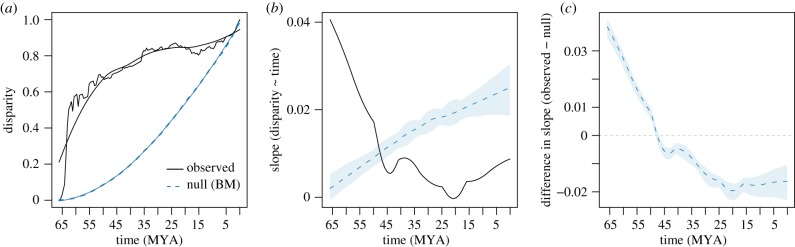
The multivariate disparity-through-time analyses reveal a remarkable proportion of Carangaria's morphological variation originating early in the clade's history. Both trait subsets analysed (4 and 12 pPCs) revealed similar patterns of morphological disparity accumulation, with 60% of the total variance arising before the Palaeocene–Eocene boundary within a time interval of just 10 Myr (figure 4a; see the electronic supplementary material, figure S8 for results using the 4 pPC subset). This proportion is particularly relevant when compared to the total variance expected to be accumulated at 55 Ma under a BM null model of trait evolution (only 5%). Comparisons against a BM also indicate a dominance of morphospace expansion early in carangarian history (figure 4c). This initial stage of accelerated morphological evolution lasted for about 20 Myr (until around 46 Ma) and was subsequently replaced by a period of morphospace packing, presumably reflecting a Simpsonian process of vacant niche filling (electronic supplementary material, figure S9).
Figure 4.
Disparity-through time plots showing the evolution of morphospace filling in Carangaria using the 1% threshold trait dataset (the highest 12 pPC axes). (a) Accumulation of multivariate disparity through time in 1 Myr time slices (thick black line, observed data; thin black line, after locally estimated scatterplot smoothing (LOESS) smoothing; blue line, constant rate Brownian motion (BM) null model). (b) Comparison of slopes for the two competing models; shaded areas represent 95% confidence intervals. (c) Differences in slope for the observed data and the BM null model; values above and below zero indicate the dominance of morphospace expansion versus morphospace packing, respectively.
We also obtained congruent results for model fitting based on both pPC trait subsets (4 and 12), indicating that the results are robust to the number of shape axes included. A simple EB model of morphological evolution—rates slowing down exponentially through time—presented the worst fit among all the competing models (Akaike weights or wA < 0.01). However, we found strong support (wA > 0.50 for approx. 90% of the trees) for a time-heterogeneous model in which body-shape evolution switches from an initial EB into a random walk with multiple and independent stationary peaks (figure 3d,e; see the electronic supplementary material, tables S3 and S4 for details on model comparisons). These independent peaks appear to represent different adaptive zones corresponding to major body-plans in Carangaria, with the random walks showing the exploration of niche space after the transition from morphospace expansion to morphospace packing ca 46 Ma.
(e). Tempo and mode of ecological diversification
In agreement with results obtained for lineage diversification and morphological evolution, plots of habitat transition rates through time show that the distribution of ecological shifts in the group is notably uneven, with initial high rates that drop slightly before the Palaeocene–Eocene boundary (figure 1c; electronic supplementary material, figure S10). Moreover, comparisons of models of discrete character evolution indicate a decisive support for an EB model in all resampled trees (wA > 0.99), suggesting that the skewed distribution of events of habitat transition would be plausibly explained by a model in which the rate of evolution decreases exponentially through time (figure 3f,g). Similar results are obtained using taxonomically-denser trees for Carangaria (electronic supplementary material, figure S10).
4. Discussion
By implementing complex models of lineage, morphological and ecological evolution, our study supports post-Cretaceous bursts of diversification as a probable explanation concerning the evolutionary trajectory of carangarians, aligning with observations from mammals, frogs and other tetrapod groups. It also highlights the apparent role of ecological release stemming from the extinction/absence of competitors in triggering ancient radiations along the benthic-pelagic axis—a well-characterized mode of diversification in recent groups of temperate fishes (e.g. sticklebacks, whitefishes [37]) whose significance is otherwise poorly understood from a macroevolutionary perspective.
Initial assessments of time variation in the rates of lineage accumulation indicated a stage of high diversification during the Palaeocene followed by a period of relative stasis towards the present. Recent efforts to explicitly assess time variation in a comparative framework across major acanthomorph groups (as well as specific subclades) have failed to detect signatures of the K-Pg mass extinction in diversification rates [12,38] (but see Price et al. [39]). By contrast, our results reveal strong support for an uneven origination of species richness in Carangaria, with high rates of lineage diversification reached in the aftermath of the end-Cretaceous mass extinction.
Early cladogenetic events giving rise to all major lineages in Carangaria (e.g. flatfishes, billfishes, robalos, moonfishes, threadfins, remoras, jacks) are entirely restricted to the Palaeocene, supporting the hypothesis that ecological opportunity arising in the wake of the K-Pg mass extinction enabled rapid radiation. Similar cladogenetic patterns are found in other specious acanthomorph fish clades. For instance, pelagiarians—a group comprising open-ocean fishes such as tunas, mackerels and cutlassfishes—appear to have radiated in the aftermath of the K-Pg mass extinction, with most of its major lineages arising in the early-Palaeogene [40]. Additionally, there are signs of rapid radiations in many species-rich reef-fish families and their early divergences in most cases also date back to this time (e.g. wrasses, grunts, surgeonfishes and blennies) [13]. The episodic decline in Carangaria's lineage diversification appears to coincide with global climatic changes during the Palaeocene–Eocene thermal maximum (55.8 ± 0.2 Ma; figure 1b), a brief interval of extreme perturbation in the global carbon cycle that resulted in record-high levels of global warming (5° to 10°C) [41]. Recent work hints that such severe environmental conditions affected reef-fish diversification dynamics [39] and are potentially linked to clade-wide extinctions in the acanthomorph order Tetraodontiformes (ocean sunfishes, pufferfishes and allies) [38]. A mode of classic niche filling, however, cannot be rejected as a plausible explanation for the decline in Carangaria's lineage diversification rates during the Palaeocene–Eocene boundary.
Plots of multivariate disparity accumulation through time revealed a remarkable proportion of Carangaria's morphological variation originating early in the clade's history, with 60% of the total clade disparity being reached before the Palaeocene–Eocene boundary. The notable dominance of morphospace expansion followed by a period of morphospace packing fits Simpsonian predictions on morphospace filling and reinforces the evolutionary consequence of the K-Pg mass extinction. This result is in line with the stratigraphic distribution of acanthomorph fossils, which reveals an early-Cenozoic expansion of the acanthomorph body-shape disparity in areas of the morphospace emptied after the mass extinction event [7]. Some of Carangaria's modern body-plans were already represented during the early Cenezoic. For instance, the late-Palaeocene †Mene purdyi resembles the contemporary morphology of its congener, the moonfish [42]. Other modern taxonomic groups, such as jacks, robalos and stem flatfishes also have been documented from the Eocene (49 Ma) deposits of Monte Bolca in northern Italy [43].
The palaeontological record is rich in evidence supporting the macroevolutionary trend of animal clades reaching high morphological disparity early in their evolutionary history [44]. However, phylogenetic comparative studies have challenged the relevance of early bursts in explaining the morphological evolution in well-established examples (though mostly younger) of adaptive radiations [45–47]. While a simple EB model also proved to be a poor explanation for the dynamics of morphological evolution in Carangaria, the apparent incompatibility between the patterns of morphospace filling and the results of model-fit comparisons was reconciled by accounting for more realistic models that incorporate variation in the processes generating morphological evolution. We found unequivocal support for a model that incorporates a shift in the mode of evolution from EB into a multiple selective peak random walk at the inferred time of transition between morphospace expansion and packing (ca 46 Ma). Although bursts of morphological evolution may be a common macroevolutionary feature, it has been demonstrated that our ability to detect them would be affected by factors such as the ecological relevance of analysed traits [23], the phylogenetic scale [46] and the use of overly simplistic evolutionary models [26]. This latter factor is probably the source of conflict in our analyses.
An important prediction of the adaptive radiation theory is that both speciation and morphological adaptations must be significantly associated with the occupation of divergent environments [2]. Habitat transitions along the benthic-pelagic axis have had important outcomes in the diversification dynamics of relatively recent freshwater fish groups such as sticklebacks, whitefishes, cichlids, minnows and perches [48,49], as well as grunts of the marine family Haemulidae [50]. However, the effects of the adoption of divergent ecological regimes remain largely unexplored at deeper macroevolutionary scales. Ancestral state reconstructions revealed that carangarians experienced higher rates of habitat transitions along the benthic-pelagic axis during the Palaeocene, notably overlapping with the early lineage diversification and morphological evolution in major clades. A notable example is the loss of the bilateral symmetry experienced by flatfishes, with laterally compressed bodies featuring both eyes on the same side of the head. Although dorsally flattened (depressed) body plans are recurrent among benthic dwellers (e.g. rays, skates, suckermouth-armoured catfishes and batfishes), flatfishes' laterally compressed plan is unusual among benthic-living species. Flatfishes further evolved other key adaptations to facilitate their burrowing into the substrate (e.g. the recessus orbitalis, a muscular sac that enables eye protrusion). The Carangaria also comprises several clades that have invaded in parallel the pelagic realm, such as istiophoriforms (swordfish, marlins and billfishes), sphyraenids (barracudas) and many carangiforms (e.g. dolphinfishes, amberjacks and the rainbow runner). Those open-water, fast-moving predators have convergently developed streamlined bodies, forked tail fins and slender tail bases (caudal peduncle). In agreement with our observations, reconstructions of the trajectory of morphological evolution in the fossil record of acanthomorphs have shown that a major component of the early-Cenozoic morphospace expansion reflected a process of ecological replacement of the pelagic non-acanthomorph fauna that became extinct by the end of the Mesozoic [7,8].
Our results also show that the uneven distribution of habitat transitions events is temporally associated with the asymmetric accumulation of species richness, with an initial stage of high rates of ecological transitions that subsequently slows down, aligning with expectations under a BM model during the Palaeocene–Eocene boundary. In agreement with this pattern, comparisons of the fit of alternative models of discrete trait evolution strongly support an exponential decrease in the rates of habitat transitions. Taken together, our results highlight the adoption of divergent ecological regimes in the origin and recovery of marine fish clades in the wake of mass extinction.
While we find bursts of evolution in carangarians to be chronologically associated with post-Cretaceous ecological release, we recognize that other possible sources of ecological opportunity have probably played an important role in their radiation during the early Cenozoic. For instance, the loss of bilateral symmetry in flatfishes and the elongation of the premaxilla bone in billfishes rank among the most extreme phenotypic adaptations in vertebrates and are candidates for key innovations that may have created additional sources of ecological opportunity in these subclades. Colonization of novel habitat regimes is another source of adaptive radiation that we here show operated in synchrony with newly released niches in the wake of the K-Pg, allowing the diversification of carangarians along the benthic-pelagic axis. Although decoupling the relative importance of these different sources of ecological opportunity may be difficult, the chronological order of events—extinction of Mesozoic marine fish fauna in the K-Pg followed by high rates of habitat transitions and the origin of singular morphologies (figure 1)—suggests that post-Cretaceous niche vacancy was the main factor behind Carangaria's evolutionary success.
Comparisons between rates of taxonomic and morphological diversity provide important insights into the dynamics of origination and diversification of higher-level taxa [51]. The results presented herein for Carangaria reveal variable dynamics during the clade's history, with high levels of lineage, morphological and ecological diversity being reached within a relatively short period in the aftermath of the K-Pg mass extinction. By and large, temporal associations of the initially accelerated rates for the three metrics investigated herein fit Simpsonian predictions on adaptive radiation. They also ultimately underscore the importance of increased ecological opportunity arising in the wake of mass extinctions by providing vacant space that prompted niche divergence along the benthic-pelagic axis and the rapid evolution of major clades.
Supplementary Material
Supplementary Material
Acknowledgements
We are grateful to Ariel Stein for assistance in collecting geometric morphometric data. We thank Sandra Raredon for providing logistic support for the examination of museum material at Smithsonian. We thank Dahiana Arcila and members of the Fish Evolution Laboratory for comments on the manuscript. We thank Shreeram Senthivasan for valuable help adapting scripts to implement job parallelization. We thank Christopher R. Cooney for sharing code for multivariate disparity analyses. We are grateful to Diane Rome Peebles for providing many of the fish illustrations used. We thank Jose Carlos Bonilla and Humberto Ortiz for providing support with bioinformatic analyses and access to the High Performance Computing facility of UPR-RP (funded by INBRE grant no. P20GM103475 from the National Institute of General Medical Sciences and the National Institutes of Health).
Ethics
The study procedures and data collection did not involve animal experimentation and do not present any ethical concerns, conservation issues, or potential risk of misuse or maltreatment of animals.
Data accessibility
Trees, datasets and R code used are available from the Dryad Digital Repository (doi:10.5061/dryad.9bq5n41) [33].
Authors' contributions
R.B.-R., E.R., A.D. and G.O. designed the study. R.B.-R. and E.R. collected data. E.R. and R.B.-R. conducted analyses. E.R. and R.A.R.-V. made the figures. E.R., R.B.-R., R.A.R.-V., A.D. and G.O. wrote the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work was financially supported by NSF awards DEB-1457184 and DEB-1541491 to R.B.-R. and DEB-1457426 to G.O. Additional support was provided by the Smithsonian P. Buck Fellowship to R.B.-R. and the Queensland-Smithsonian Fellowship to A.M.D
References
- 1.Simpson GG. 1953. Major features of evolution. New York, NY: Columbia University Press. [Google Scholar]
- 2.Schluter D. 2000. The ecology of adaptive radiation. Oxford, UK: Oxford University Press. [Google Scholar]
- 3.Erwin DH. 2001. Lessons from the past: biotic recoveries from mass extinctions. Proc. Natl Acad. Sci. USA 98, 5399–5403. ( 10.1073/pnas.091092698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pollux BJA, Meredith RW, Springer MS, Reznick DN. 2014. The evolution of the placenta drives a shift in sexual selection in livebearing fish. Nature 513, 233–236. ( 10.1038/nature13451) [DOI] [PubMed] [Google Scholar]
- 5.Feng Y-J, Blackburn DC, Liang D, Hillis DM, Wake DB, Cannatella DC, Zhang P. 2017. Phylogenomics reveals rapid, simultaneous diversification of three major clades of Gondwanan frogs at the Cretaceous-Palaeogene boundary. Proc. Natl Acad. Sci. USA 114, E5864–E5870. ( 10.1073/pnas.1704632114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavin L. 2002. Effects of the Cretaceous-Tertiary boundary event on bony fishes. In Geological and biological effects of impact events (eds Buffetaut E, Koeberl C), pp. 141–158. Berlin, Germany: Springer. [Google Scholar]
- 7.Friedman M. 2010. Explosive morphological diversification of spiny-finned teleost fishes in the aftermath of the end-Cretaceous extinction. Proc. R. Soc. B 277, 1675–1683. ( 10.1098/rspb.2009.2177) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman M. 2009. Ecomorphological selectivity among marine teleost fishes during the end-Cretaceous extinction. Proc. Natl Acad. Sci. USA 106, 5218–5223. ( 10.1073/pnas.0808468106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patterson C. 1993. Osteichthyes: teleostei. In The fossil record (ed. Benton MJ.), pp. 621–656. London, UK: Chapman and Hall. [Google Scholar]
- 10.Sibert EC, Norris RD. 2015. New age of fishes initiated by the cretaceous-paleogene mass extinction. Proc. Natl Acad. Sci. USA 2015, 8537–8542. ( 10.1073/pnas.1504985112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Betancur RR, et al. 2013. The tree of life and a new classification of bony fishes. PLoS Curr. Tree Life 1, 1–45. ( 10.1371/currents.tol.53ba26640df0ccaee75bb165c8c26288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Near TJ, et al. 2013. Phylogeny and tempo of diversification in the superradiation of spiny-rayed fishes. Proc. Natl Acad. Sci. USA 110, 12 738–12 743. ( 10.5061/dryad.d3mb4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alfaro ME, Faircloth BC, Harrington RC, Sorenson L, Friedman M, Thacker CE, Oliveros CH, Černý D, Near TJ. 2018. Explosive diversification of marine fishes at the Cretaceous–Palaeogene boundary. Nat. Ecol. Evol. 2, 688–696. ( 10.1038/s41559-018-0494-6) [DOI] [PubMed] [Google Scholar]
- 14.Harrington RC, Faircloth BC, Eytan RI, Smith WL, Near TJ, Alfaro ME, Friedman MA. 2016. Phylogenomic analysis of carangimorph fishes reveals flatfish asymmetry arose in a blink of the evolutionary eye. BMC Evol. Biol. 16, 224 ( 10.1186/s12862-016-0786-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Betancur R, Li C, Munroe TA, Ballesteros JA, Ortí G. 2013. Addressing gene tree discordance and non-stationarity to resolve a multi-locus phylogeny of the flatfishes (Teleostei: Pleuronectiformes). Syst. Biol. 62, 763–785. ( 10.1093/sysbio/syt039) [DOI] [PubMed] [Google Scholar]
- 16.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. ( 10.1093/molbev/mss075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rabosky DL, et al. 2018. An inverse latitudinal gradient in speciation rate for marine fishes. Nature 559, 392–395. ( 10.1038/s41586-018-0273-1) [DOI] [PubMed] [Google Scholar]
- 18.Bookstein FL. 1997. Morphometric tools for landmark data: geometry and biology. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 19.Rohlf FJ. 2001. TPS Dig 2.0. See http://life.bio.sunysb.edu/morph/.
- 20.Adams DC, Otárola-Castillo E. 2013. Geomorph: an r package for the collection and analysis of geometric morphometric shape data. Methods Ecol. Evol. 4, 393–399. ( 10.1111/2041-210X.12035) [DOI] [Google Scholar]
- 21.Höhna S, May MR, Moore BR. 2015. TESS: an R package for efficiently simulating phylogenetic trees and performing Bayesian inference of lineage diversification rates. Bioinformatics 32, 789–791. ( 10.1093/bioinformatics/btv651) [DOI] [PubMed] [Google Scholar]
- 22.Hohna S. 2014. Likelihood inference of non-constant diversification rates with incomplete taxon sampling. 9, 17–20. ( 10.1371/Citation) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooney CR, Bright JA, Capp EJR, Chira AM, Hughes EC, Moody CJA, Nouri LO, Varley ZK, Thomas GH. 2017. Mega-evolutionary dynamics of the adaptive radiation of birds. Nature 542, 344–347. ( 10.1038/nature21074) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223. ( 10.1111/j.2041-210X.2011.00169.x) [DOI] [Google Scholar]
- 25.Clavel J, Escarguel G, Merceron G. 2015. mv morph: an R package for fitting multivariate evolutionary models to morphometric data. Methods Ecol. Evol. 6, 1311–1319. ( 10.1111/2041-210X.12420) [DOI] [Google Scholar]
- 26.Slater GJ. 2013. Phylogenetic evidence for a shift in the mode of mammalian body size evolution at the Cretaceous-Palaeogene boundary. Methods Ecol. Evol. 4, 734–744. ( 10.1111/2041-210X.12084) [DOI] [Google Scholar]
- 27.Uyeda JC, Caetano DS, Pennell MW. 2015. Comparative analysis of principal components can be misleading. Syst. Biol. 64, 677–689. ( 10.1093/sysbio/syv019) [DOI] [PubMed] [Google Scholar]
- 28.Adams DC, Collyer ML. 2018. Multivariate phylogenetic comparative methods: evaluations, comparisons, and recommendations. Syst. Biol. 67, 14–31. ( 10.1093/sysbio/syx055) [DOI] [PubMed] [Google Scholar]
- 29.Helfman GS, Collette BB, Facey DE, Bowen BW. 2009. The diversity of fishes: biology, evolution, and ecology. Chichester, UK: Wiley-Blackwell. [Google Scholar]
- 30.Froese R, Pauly D. 2016. FishBase: World Wide Web electronic publication. See www.fishbase.org (accessed 10 February 2016).
- 31.Eschmeyer WN, Fricke R, van der Laan R. 2016. Catalog of fishes: genera, species, references. See https://www.calacademy.org/scientists/projects/catalog-of-fishes. [DOI] [PubMed]
- 32.Harmon LJ, Weir JT, Brock CD, Glor RE, Challenger W. 2008. GEIGER: investigating evolutionary radiations. Bioinformatics 24, 129–131. ( 10.1093/bioinformatics/btm538) [DOI] [PubMed] [Google Scholar]
- 33.Ribeiro E, Davis A, Rivero-Vega RA, Ortí G, Betancur-R R. 2018. Data from: Post-Cretaceous bursts of evolution along the benthic-pelagic axis in marine fishes Dryad Digital Repository. ( 10.5061/dryad.9bq5n41) [DOI] [PMC free article] [PubMed]
- 34.Near TJ, Eytan RI, Dornburg A, Kuhn KL, Moore JA, Davis MP, Wainwright PC, Friedman M, Smith WL. 2012. Resolution of ray-finned fish phylogeny and timing of diversification. Proc. Natl Acad. Sci. USA 109, 13 698–13 703. ( 10.1073/pnas.1206625109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Betancur-RR Wiley EO, Arratia G, Acero A, Bailly N, Miya M, Lecointre G, Ortí G. 2017. Phylogenetic classification of bony fishes. BMC Evol. Biol. 17, 162 ( 10.1186/s12862-017-0958-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Claverie T, Wainwright PC. 2014. A morphospace for reef fishes: elongation is the dominant axis of body shape evolution. PLoS ONE 9, e112732 ( 10.1371/journal.pone.0112732) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hollingsworth PR, Simons AM, Fordyce JA, Hulsey CD. 2013. Explosive diversification following a benthic to pelagic shift in freshwater fishes. BMC Evol. Biol. 13, 272 ( 10.1186/1471-2148-13-272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arcila D, Tyler JC. 2017. Mass extinction in tetraodontiform fishes linked to the Palaeocene–Eocene thermal maximum. Proc. R. Soc. B 284, 20171771 ( 10.1098/rspb.2017.1771) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price SA, Schmitz L, Oufiero CE, Eytan RI, Dornburg A, Smith WL, Friedman M, Near TJ, Wainwright PC. 2014. Two waves of colonization straddling the K-Pg boundary formed the modern reef fish fauna. Proc. R. Soc. B 281, 20140321 ( 10.1098/rspb.2014.0321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miya M, et al. 2013. Evolutionary origin of the scombridae (tunas and mackerels): members of a paleogene adaptive radiation with 14 other pelagic fish families. PLoS ONE 8, e73535 ( 10.1371/journal.pone.0073535) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Röhl U, Bralower TJ, Norris RD, Wefer G. 2000. New chronology for the late Palaeocene thermal maximum and its environmental implications. Geology 28, 927 ( 10.1130/0091-7613(2000)28%3C927:NCFTLP%3E2.0.CO;2) [DOI] [Google Scholar]
- 42.Friedman M, Johnson GD. 2005. A new species of mene (Perciformes: Menidae) from the Palaeocene of South America, with notes on paleoenvironment and a brief review of menid fishes. J. Vertebr. Paleontol. 25, 770–783. ( 10.1671/0272-4634(2005)025%5B0770:ANSOMP%5D2.0.CO;2) [DOI] [Google Scholar]
- 43.Bellwood DR. 1996. The Eocene fishes of Monte Bolca: the earliest coral reef fish assemblage. Coral Reefs 15, 11–19. ( 10.1007/s003380050025) [DOI] [Google Scholar]
- 44.Hughes M, Gerber S, Wills MA. 2013. Clades reach highest morphological disparity early in their evolution. Proc. Natl Acad. Sci. USA 2013, 36–38. ( 10.1073/pnas.1302642110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harmon LJ, et al. 2010. Early bursts of body size and shape evolution are rare in comparative data. Evolution 64, 2385–2396. ( 10.1111/j.1558-5646.2010.01025.x) [DOI] [PubMed] [Google Scholar]
- 46.Hopkins MJ, Smith AB. 2015. Dynamic evolutionary change in post-Paleozoic echinoids and the importance of scale when interpreting changes in rates of evolution. Proc. Natl Acad. Sci. USA 112, 3758–3763. ( 10.1073/pnas.1418153112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Slater GJ. 2015. Iterative adaptive radiations of fossil canids show no evidence for diversity-dependent trait evolution. Proc. Natl Acad. Sci. USA 112, 4897–4902. ( 10.1073/pnas.1403666111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hulsey CD, Roberts RJ, Loh YHE, Rupp MF, Streelman JT. 2013. Lake Malawi cichlid evolution along a benthic/limnetic axis. Ecol. Evol. 3, 2262–2272. ( 10.1002/ece3.633) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burress ED, Holcomb JM, Tan M, Armbruster JW. 2017. Ecological diversification associated with the benthic-to-pelagic transition by North American minnows. J. Evol. Biol. 30, 549–560. ( 10.1111/jeb.13024) [DOI] [PubMed] [Google Scholar]
- 50.Tavera J, Acero PA, Wainwright PC. 2018. Multilocus phylogeny, divergence times, and a major role for the benthic-to-pelagic axis in the diversification of grunts (Haemulidae). Mol. Phylogenet. Evol. 121, 212–223. ( 10.1016/J.YMPEV.2017.12.032) [DOI] [PubMed] [Google Scholar]
- 51.Foote M. 1993. Discordance and concordance between morphological and taxonomic diversity. Paleobiology 19, 185–204. ( 10.1017/S0094837300015864) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Ribeiro E, Davis A, Rivero-Vega RA, Ortí G, Betancur-R R. 2018. Data from: Post-Cretaceous bursts of evolution along the benthic-pelagic axis in marine fishes Dryad Digital Repository. ( 10.5061/dryad.9bq5n41) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Trees, datasets and R code used are available from the Dryad Digital Repository (doi:10.5061/dryad.9bq5n41) [33].