Abstract
Metritis is a major disease in dairy cows causing animal death, decrease of birth rate, milk production, and economic loss. Antibiotic treatment is generally used to treat such disease but has a high failure rate of 23–35%. The reason for the treatment failure remains unclear, although antibiotic resistance is postulated as one of factors. Our study investigated the prevalence of extended spectrum β-lactamase (ESBL) producing bacteria in uterine samples of cows with metritis and characterized the isolated intrauterine pathogenic Escherichia coli (IUPEC) strains using whole genome sequencing. We found that the cows with metritis we examined had a high percentage of ESBL producing IUPEC with multi-drug resistance including ceftiofur which is commonly used for metritis treatment. The ESBL producing IUPEC strains harbored versatile antibiotic resistance genes conferring resistance against 29 antibiotic classes, suggesting that transmission of these bacteria to other animals and humans may lead to antibiotic treatment failure. Furthermore, these strains had strong adhesion and invasion activity, along with critical virulence factors, indicating that they may cause infectious diseases in not only the uterus, but also in other organs and hosts.
Keywords: intrauterine pathogens, Escherichia coli, dairy cows, extended spectrum β-lactamase, metritis
Introduction
Metritis is one of the major global bacterial diseases of dairy cattle and is a prevalent reproductive disease characterized by an abnormally enlarged uterus, a watery red-brownish uterine discharge and fever within 21 days after parturition (Sheldon et al., 2006). It mainly affects dairy cows in the early postpartum period. Metritis decreases milk production, decreases conception rate, and increases culling and death (Drillich et al., 2001). Metritis affects approximately 20% of lactating dairy cattle with a range from 8% to more than 40% (Goshen and Shpigel, 2006; LeBlanc, 2008). The direct economic cost to the US dairy industry associated with this disease is approximately $600 million annually (Overton and Fetrow, 2008). The pathogens associated with metritis include Escherichia coli, Trueperella pyogenes, Fusobacterium necrophorum, Fusobacterium nucleatum, Bacteroides pyogenes, and Porphyromonas levii (Sheldon et al., 2009; Jeon et al., 2015; Cunha et al., 2018). The pathogens need essential pathogenicity traits including adhesion (Torres et al., 2005) and invasion (Sheldon et al., 2010) to host endometrial cells, motility mediated by flagella (Lane et al., 2007), and toxins like lipopolysaccharide (LPS) (Wolf, 1997) to manifest the disease. E. coli has been described as the main pathogen initiating postpartum uterine infection and disease (Bicalho et al., 2010; Sheldon et al., 2010).
Antibiotic treatment is used to treat animals with metritis, and the most common antibiotic is ceftiofur, a third generation cephalosporin that does not require milk withdrawal because the residue in milk is below the tolerance level for human consumption (Chenault et al., 2004). Ceftiofur is effective against both Gram-negative and Gram–positive pathogens (Collatz et al., 1990). However, approximately 25% of the cows fail to cure after being treated with ceftiofur (Chenault et al., 2004; McLaughlin et al., 2012). Although other antibiotics including ampicillin and tetracycline have been tested to treat metritis but they have not enhanced efficiency of curing metritis (Goshen and Shpigel, 2006; Lima et al., 2014). Multiple hypotheses have been proposed for the low cure rate of systematic administration of antibiotics, such as immunosuppression, periparturient disorders, and intrauterine placement of chemical compounds depressing the defense mechanism of cows (Smith et al., 1998). In addition, it was postulated that the continuous use of antibiotics in dairy farms attributed to the rise of antimicrobial resistance, especially to the commonly used antibiotic therapies, thereby leading to the ineffectiveness of antibiotics (Santos et al., 2010). However, the reason why a large proportion of animals do not respond to antibiotic treatment remains unclear.
Resistance to cephalosporins has been associated with bacteria carrying β-lactamases encoding genes, such as blaTEM, blaSHV, blaCTX-M, blaCMY -2, and blaampC (Zhao et al., 2001; Li et al., 2007). The plasmid-mediated CMY-2 type genes have been found in animal isolates extensively (Zhao et al., 2001; Donaldson et al., 2006; Chander et al., 2011). Isolates harboring TEM type β-lactamases usually have resistance to penicillin, but the resistance to expanded-spectrum cephalosporins was also reported (Poirel et al., 2004). In addition, CTX-M type extended spectrum β-lactamases (ESBLs) have been increasingly detected and confer resistance to expanded-spectrum cephalosporins, including cefotaxime, ceftazidime, and cefepime (Bonnet, 2004). In the UK, it was reported that in a dairy farm, 31/48 calves and 2/60 cows were positive for E. coli encoding the CTX-M gene in their fecal samples (Liebana et al., 2006). In Germany, one study found 86.7% of farms, including fecal and environmental samples, were positive for ESBL producing E. coli, most of which harbored CTX-M genes (Schmid et al., 2013). In addition, an extra-intestinal pathogenic E. coli (ExPEC) isolated from the uterus of a cow with metritis, MS499, had resistance to β-lactams, tetracyclines, and macrolides (Goldstone et al., 2014). Other pathogens resistant to β-lactam antibiotics, including Listeria monocytogenes, Staphylococcus aureus, and Enterococcus spp., have been also isolated from dairy cattle (Srinivasan et al., 2005; Tenhagen et al., 2006). Therefore, although there are multiple factors that may lead to treatment failure, we hypothesized that antimicrobial resistance is one of the key factors of this unfavored result. However, the prevalence of ESBL producing E. coli in the uterus of cows with metritis has not been reported. In this study, we focused our attention on the frequency of ESBL-producing E. coli in the uteri of cows with metritis and further characterized their antimicrobial resistance and virulence factors by whole genome sequencing and comparative genomics.
Materials and Methods
Animal Management
All animal procedures were approved by the University of Florida Institutional Animal Care and Use Committee (IACUC Protocol #: 201207405). Dairy Holstein cows were housed in freestall barns. The cows were fed twice daily with a mixed ration formulated to meet the nutrient requirements of a lactating cow weighing 650 Kg and producing 45 Kg of 3.5% fat-corrected milk per day. The animals were housed in the same farm, except one cow, the source of KCJ852.
Uterine Sample Collection
Metritis was diagnosed by the presence of red-brownish fetid uterine discharge postpartum. Uterine swab samples were collected for individual animals (n = 24) after clinical diagnosis of metritis using the procedure previously described (Jeon et al., 2016). Briefly, after disinfection of the perineum area of cows with 70% ethanol, a sterile pipette containing a sterile cotton swab was introduced to the cranial vagina. To avoid vaginal contamination of the swab, the plastic sheath containing the pipette was directed into the cervix and then to the uterus. The plastic pipette was then protruded through the plastic sheath, then the sterile swab was exposed and rolled against the uterine wall to collect sample. The swab was pulled inside the pipette then the pipette pulled inside the plastic sheath and removed from the cow. Swabs were transferred to a 15 mL tube on ice and delivered to the laboratory within 4 h. The samples were collected from the cows ranging from day 2–7 after parturition upon diagnosis of metritis.
Identification of Cefotaxime Resistant Pathogens
The swab samples were enriched in tryptic soy broth overnight in the presence of cefotaxime to select ESBL positive bacteria. Next, the samples were plated on MacConkey agar with cefotaxime (4 mg/L) and incubated at 37°C to select cefotaxime resistant colonies. Five colonies from each uterine sample grown were purified on the same media and confirmed if they were E. coli by striking them on ChromAgar E. coli (CHROMagar, France). The presence of ESBL genes (blaTEM, blaCTX-M, blaCMY) was tested by PCR using primers previously described (Mir et al., 2016). Fifteen isolates purified from 3 swab samples carried blaCTX-M gene and one isolate without ESBL genes were confirmed as E. coli; these isolates were then further characterized. One ESBL negative sample was plated on ChromAgar E. coli directly to isolate a non-ESBL endometrial pathogenic E. coli.
Adherence and Invasion Assay
Four cell lines were used in this study. Caco-2, HEK293T and Hep2 cells were cultured in Dulbecco’s modified Eagle medium (DMEM, Corning Incorporated, Corning, NY, United States) supplemented with 10% fetal bovine serum (FBS, Hyclone, Logan, UT, United States). Bovine endometrial epithelial cells (Sigma-Aldrich, St. Louis, MO, United States) were cultured in Bovine Endometrial Growth Media (Sigma-Aldrich, St. Louis, MO, United States). The cells were cultured to confluency. Then approximately 105 cells were seeded into 24-well plates and incubated overnight at 37°C and 5% CO2. The adherence assay was conducted following previously described protocol with modifications (Amigo et al., 2015). Gentamicin protection assay was used for invasion test (Falkow et al., 1987). Bacteria (KCJ4393 [DH5α], KCJ696 [the eae deletion mutant of E. coli EDL933], and KCJ698 [the tir deletion mutant of E. coli EDL933] as negative controls, E. coli EDL933 as a positive control, KCJ852, KCJ3819, KCJ3823, and KCJ3859) were cultured overnight at 37°C in Luria Bertani (LB) broth and washed with sterile phosphate buffered saline (PBS) three times. The final bacterial pellet was diluted to 106 CFU/mL and resuspended in cell culture media and added to each well. For adherence assay, the mixture of cell and bacterial culture was incubated at 37°C and 5% CO2 for 3 h. After the first incubation, the media was replaced by DMEM and the cell culture was incubated for another 3 h. Then the media was removed and cells were washed three times with PBS, and lysed by 1 mL of 0.1% Triton X-100 in PBS. The culture was serially diluted and plated on LB agar plates to enumerate the bacteria. For the invasion assay, the cultures were incubated at 37°C and 5% CO2 for 4 h. Next, the supernatant was removed and the cells were washed with PBS three times. Media containing 50 μg/mL gentamicin was added to each well and the culture was incubated for another 1.5 h. After incubation, the cells were washed and the bacteria were enumerated as described in adherence assay.
Minimal Inhibitory Concentration Test and Antimicrobial Susceptibility Test
MIC test of cefotaxime was conducted with the 15 strains carrying blaCTX-M genes and one ESBL negative strain (KCJ852). MIC was determined using broth microdilution method according to CLSI guidelines (CLSI, 2018). KCJ1409 (Mir et al., 2016), an ESBL-producing E. coli isolated from human was included as a positive control and DH5α, a non-pathogenic E. coli was included as a negative control.
Antimicrobial susceptibility test (AST) was further performed on each representative strain (KCJ3819, KCJ3823, and KCJ3859) from CTX-M positive samples (n = 3) and one ESBL negative strain (KCJ852). The standard Kirby Bauer disk diffusion method on Mueller Hinton agar was used. The control strains used for the AST were E. coli (ATCC 35401), S. aureus (ATCC 25923) and Pseudomonas aeruginosa (ATCC 27853). The antimicrobial disks used are listed below: Amikacin (K; 30 μg), Ampicillin (A; 10 μg), Amoxycillin/Clavulanic acid (X; 30 μg), Sulfisoxazole (Z; 0.25 mg), Ceftiofur (R; 30 μg), Chloramphenicol (C; 30 μg), Cephalothin (F; 30 μg), Gentamicin (G; 10 μg), Nalidixic acid (N; 30 μg), Streptomycin (S; 10 μg), Sulfamethoxazole/trimethoprim (M; 23.75 μg/1.25 μg), and Tetracycline (T; 30 μg) (BD, United States).
Whole Genome Sequencing and Phylogenetic Tree Analysis
Representative strains (n = 13) from CTX-M positive samples and the non-ESBL strain KCJ852 were sequenced by Illumina MiSeq. Pure genomic DNA was extracted from each isolate using the DNeasy blood and tissue kit (Qiagen, Valencia, CA, United States). High sensitivity double stranded DNA quantification was performed using the Qubit® fluorometer assay and the DNA was diluted to 0.2 ng/μl and used as input DNA for tagmentation with Illumina’s Nextera®XT DNA library sample preparation kit. Sequencing was performed using an Illumina MiSeq with cartridges providing 2 × 250 paired-end read coverage. De novo assemblies were performed using SPAdes (Bankevich et al., 2012) and fastq files were trimmed for quality and length using Sickle (Joshi and Fass, 2011) prior to assembly. Alignment of the de novo assembles was performed using progressiveMauve 2.4.0 (Darling et al., 2010). Single nucleotide polymorphisms (SNPs) were extracted from the whole genome alignment using MEGA 6.0 (Tamura et al., 2013) and a maximum likelihood tree was constructed using 1000 bootstrap iterations and automatic model selection with IQ-TREE (Trifinopoulos et al., 2016). To determine the best fitting roots for maximum likelihood phylogenies, TempEst was used and final tree annotations were made using FigTree (Rambaut et al., 2016). Whole genome alignments among strains in the same cluster were performed using progressiveMauve. Prophages were identified by PHAST (Zhou et al., 2011).
Virulence and Antibiotic Resistance Gene Identification
Genomes were annotated using the Virulence Factor Database (VFDB) to identify virulence and antibiotic resistance genes (Chen et al., 2005) through PATRIC (Wattam et al., 2014). The Center for Genomic Epidemiology (CGE) database was used to identify the multi-locus sequence type (MLST), serotype, and plasmid replicons for each isolate (Larsen et al., 2012; Carattoli et al., 2014; Joensen et al., 2015). The Comprehensive Antibiotic Resistance Database (CARD) was also employed for discovery of additional antibiotic resistance genes (McArthur et al., 2013).
Statistical Analysis
All experiments were carried out in triplicate. For MIC, the data were reported as mean ± SEM. For adherence and invasion assays, data were analyzed using the GLM procedure of SAS version 9.1 (SAS Institute Inc., Cary, NC, United States). Average values with standard error of means were reported. P ≤ 0.05 were considered as statistically significant.
Results
High Frequency of Cephalosporin Resistance in Cows With Metritis in the Farm Sampled
Dairy cows with metritis are usually treated with ceftiofur but the failure rate of such treatment is 23–35% (Chenault et al., 2004). To investigate if cephalosporin resistance is one of the main reasons affecting the treatment failure rate, we first evaluated the prevalence of cephalosporin resistance by plating uterine swab samples from cows with metritis on media containing cefotaxime, which is a third generation cephalosporin used to select ESBLs. Out of 24 uterine swab samples tested, 17 samples had colonies growing on media in the presence of cefotaxime, thus the percentage of cefotaxime resistance was 70.8%. PCR was conducted using total microbiome DNA to identify if the microbiota in these samples carry extended-spectrum β-lactamase (ESBL) genes. We found that CMY-2 β-lactamase gene (blaCMY) was detected in 9 samples, while ESBLs (blaCTX) was detected in 3 samples. One samples carried both blaCMY and blaTEM, and another sample carried a combination of blaCMY and blaCTX. By purifying colonies on ChromAgar E. coli, all the colonies presented blue color, suggesting all the ESBL-positive isolates were E. coli.
As CTX-M family have become the most common type of ESBLs with high clinical importance, we further purified 5 E. coli isolates from each CTX-M-positive samples and one ESBL-negative sample for comparison purpose from the cow uterus (Table 1). KCJ852 was from an ESBL negative cow, while KCJ3815 - 3819, KCJ3820 - 3824, and KCJ3855 - 3859 were from ESBL-positive cows, respectively.
Table 1.
Information of strains isolated from uterine samples.
| Strain name | Source | Animal | Disease | Lactation |
|---|---|---|---|---|
| KCJ852 | Uterine mucus | Cow 1 | Metritis | Not available |
| KCJ3815 | Uterine mucus | Cow 2 | Metritis | 3 |
| KCJ3816 | Uterine mucus | Cow 2 | Metritis | 3 |
| KCJ3817 | Uterine mucus | Cow 2 | Metritis | 3 |
| KCJ3818 | Uterine mucus | Cow 2 | Metritis | 3 |
| KCJ3819 | Uterine mucus | Cow 2 | Metritis | 3 |
| KCJ3820 | Uterine mucus | Cow 3 | Metritis | 2 |
| KCJ3821 | Uterine mucus | Cow 3 | Metritis | 2 |
| KCJ3822 | Uterine mucus | Cow 3 | Metritis | 2 |
| KCJ3823 | Uterine mucus | Cow 3 | Metritis | 2 |
| KCJ3824 | Uterine mucus | Cow 3 | Metritis | 2 |
| KCJ3855 | Uterine mucus | Cow 4 | Metritis | 5 |
| KCJ3856 | Uterine mucus | Cow 4 | Metritis | 5 |
| KCJ3857 | Uterine mucus | Cow 4 | Metritis | 5 |
| KCJ3858 | Uterine mucus | Cow 4 | Metritis | 5 |
| KCJ3859 | Uterine mucus | Cow 4 | Metritis | 5 |
Intrauterine Pathogenic E. coli Had Distinct Clonal Groups
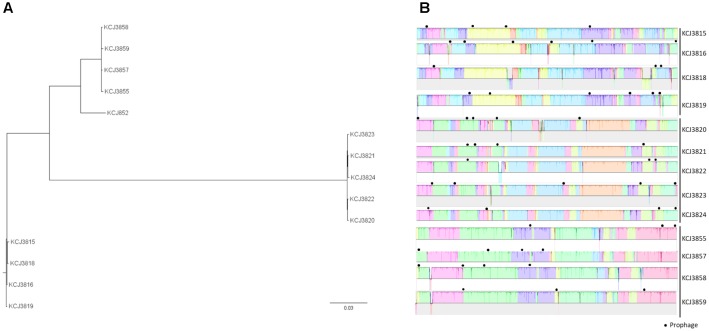
To evaluate the genomic relationship among the intrauterine pathogenic E. coli (IUPEC) isolates from uterine samples, phylogenic analysis was conducted after whole genomic sequencing. The genome size of 13 strains varied from 4.7 to 5.0 Mb, with KCJ852 being the shortest and KCJ3820-3825 being the longest. It was observed that the isolates collected from each of the cow uterine samples were clustered into the same clade (Figure 1A), suggesting that there was no transmission of isolates between animals although they were housed together. The isolates from cow #3 (KCJ3820-3824) showed longer phylogenetic distance to other isolates, demonstrating substantial genomic diversity from the other isolates. Interestingly, the ESBL-negative strain, KCJ852 (Ginn et al., 2016), was clustered closely to one clade of CTX-M-positive isolates, even though they were not collected from the same farm. To support the findings in Figure 1A, isolates in each clade were aligned by progressiveMauve (Figure 1B) to compare genome rearrangement and architecture. Within each clade, the chromosomal DNA architectures of the assembled isolates were similar to each other, suggesting they were clonal variants. However, variation in the number and content of the prophages acquired by each isolate was present. None of the isolates had the same set of prophages.
FIGURE 1.
Comparison of intrauterine pathogenic E. coli from different cows. (A) Phylogeny of intrauterine pathogenic E. coli isolated from this study. (B) Genome alignment of CTX-M positive intrauterine pathogenic E. coli. The chromosomal sequences of IUPECs collected from one animal were aligned by progressiveMauve. The background color gradient indicates homologous regions across strains. Prophages predicted by PHAST were labeled by black solid circles.
Multi-Drug Resistance of the Uterine Isolates
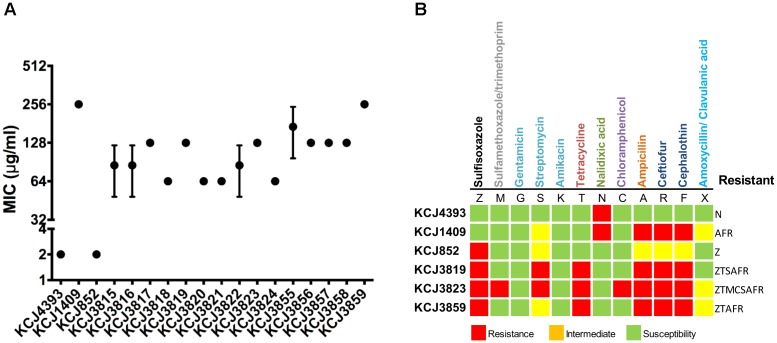
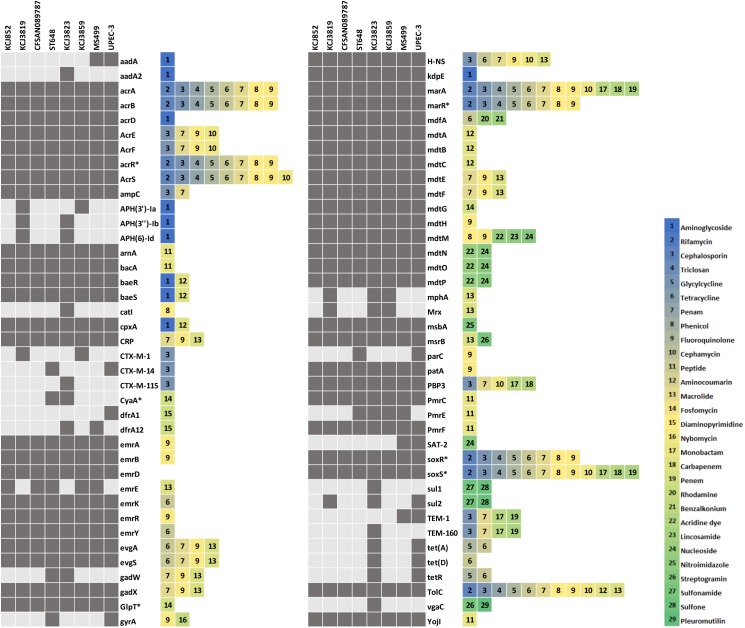
In order to address the question of if these IUPEC isolates were associated with treatment failure, we assessed multidrug resistance by MIC and AST. For MIC of cefotaxime (Figure 2A), if the level is equal or above 64 μg/mL, the strain is considered clinically relevant in human medicine. According to CLSI, if the MIC is above 4 μg/mL, the isolate is resistant to cefotaxime. The CTX-M negative isolates, E. coli DH5α and KCJ852, did not show resistance (MIC = 2 μg/mL), whereas the MIC against the CTX-M gene carriers, including the positive control KCJ1409, were higher than 64 μg/mL. The highest resistance was observed for the strains isolated from cow #4 (KCJ3855 – 3859), as the MIC ranging from 128 to 256 μg/mL, confirming the CTX-M positive isolates were highly resistant to cefotaxime. The AST of 12 different types of antibiotics, belonging to 9 classes, against representative strains were summarized in Figure 2B. The non-pathogenic E. coli DH5α had resistance to only one antibiotic, nalidixic acid, which inhibits a subunit of DNA gyrase and topoisomerase IV. The CTX-M negative strain KCJ852 showed resistance to one antibiotic, sulfisoxazole. The positive control KCJ1409 showed resistance to four antibiotics. All the IUPECs carrying CTX-M had resistance to at least five antimicrobials across multiple drug classes. KCJ3823 had resistance against eight antibiotics. The CTX-M positive strains were highly resistant to all the β-lactam antibiotics tested (ampicillin, ceftiofur, and cephalothin), whereas none of them were resistant to gentamicin, amikacin, or nalidixic acid. All the IUPEC strains showed multidrug resistance, shown by the AST results (Figure 2B). Notably, all of these ESBL-producing E. coli harbored resistance against ceftiofur, the antibiotics commonly used for the treatment of metritis. Taken together, these data indicate that it is possible that these cefotaxime resistant IUPEC isolates are associated with treatment failure of metritis using ceftiofur. Therefore, our data indicate that strains isolated from cows with uterine diseases confer multidrug resistance against 5–8 different antibiotics while carrying potential drug resistance genes which may provide resistance against a wide range of antibiotics. Furthermore, 78 antibiotic resistance genes (ARGs) responsible for resistance of 29 antibiotic classes were identified through the Comprehensive Antibiotic Resistance Database (CARD) (McArthur et al., 2013), while 71 genes were identified in the IUPEC strains (Figure 3). The majority of the resistance genes had functions related to efflux pump (acrABDEFRS, emrABDEKRY, evgAS, gadWX, marAR, mdtABCEFGHMNOP, msbA, msrB, soxRS, tetADR, and yojI) followed by antibiotic inactivation enzyme such as β-lactam resistance genes (ampC, CTX-M, TEM). KCJ3819 and 3859 had CTX-M-1 while KCJ3823 had CTX-M-115. All the IUPEC strains isolated had ampC. KCJ852 harbored less ARGs compared with other strains. In general, our IUPEC strains had very similar ARG profiles compared with the clinically important E. coli strains.
FIGURE 2.
The antimicrobial resistant profile of intrauterine pathogenic E. coli from this study and reference human clinical isolates. (A) Minimal inhibitory concentration test. Bars represent the mean ± SEM of three experiments. (B) Antimicrobial susceptibility test. The antibiotics in different colors indicate they belong to different antibiotic classes. Red squares indicate the strain was resistant to the antimicrobial; yellow squares indicate the strain was intermedium resistant to the antimicrobial; green squares indicate the strain was susceptible to the antimicrobial. KCJ4393: E. coli DH5α.
FIGURE 3.
The antibiotic resistance gene profile of intrauterine pathogenic E. coli from this study and reference human clinical isolates. Dark grey boxes indicate that the gene of the isolate has > 70% similarity to the gene in the Comprehensive Antibiotic Resistance Database.
Genomic Characterization of the Uterine Isolates
To evaluate the importance of these IUPEC strains, the genomic characteristics were compared with other representative clinical pathogenic E. coli isolates (ST648, UPEC-3, and CFSAN029787) and another IUPEC strain (MS499). Genome information of these strains were obtained from NCBI. These strains are listed in Table 2. E. coli ST648, serotype O51:H4, was isolated from pleural effusion of a patient with empyema thoracis in Beijing, China. UPEC-3, an E. coli O9:H9, was isolated from a patient with an urinary tract infection in Washington, DC, United States. CFSAN029787 was an enteroinvasive E. coli belonging to serotype O96:H19 isolated from stool samples in Milan, Italy (Table 3). The IUPECs from the same cow had the same serotype and multilocus sequence type (MLST), and the same set of plasmids (Table 3), but isolates from different samples had different serotype and MLST profiles. KCJ852 had the same serotype as KCJ3855 – 3859, but not MLST type. KCJ852 had only one plasmid replicon, IncII, whereas the other IUPEC strains shared two common plasmid replicons, IncFII and IncFIB. Based on the analysis using Center for Genomic Epidemiology (CGE) (Larsen et al., 2012; Carattoli et al., 2014; Joensen et al., 2015), KCJ3815 – 3819 and 3855 – 3859 had CTX-M-1, whereas KCJ3820 – 3824 had CTX-M-124 and TEM-1B. None of these IUPEC isolates showed a similar genomic profile compared with the reference strains. In all respects, the unique profile of isolates from each sample indicated no transmission of IUPEC strains among the cows had occurred; however, the possibility exists that the plasmids were transmitted among the strains.
Table 2.
Strains used in this study.
| Strain name | NCBI accession | Description of isolation sourceb | Reference |
|---|---|---|---|
| DH5α | N/Aa | NEB@5-alpha competent E. coli (High Efficiency) | |
| KCJ1409 | SAMN05789710 | Mir et al., 2016 | |
| ST648 | SAMN02800875 | ExPEC | Genbank |
| EDL933 | SAMN02604092 | EHEC | Genbank |
| MS499 | SAMN02839404 | IUPEC | Genbank |
| UPEC-3 | SAMN02802119 | UPEC | Genbank |
| CFSAN029787 | SAMN03612246 | EIEC | Genbank |
| KCJ852 | SAMN04396104 | IUPEC | This Study |
| KCJ3815 | SAMN04396076 | IUPEC | This Study |
| KCJ3816 | SAMN04396077 | IUPEC | This Study |
| KCJ3817 | N/A | IUPEC | This Study |
| KCJ3818 | SAMN04396078 | IUPEC | This Study |
| KCJ3819 | SAMN04396079 | IUPEC | This Study |
| KCJ3820 | SAMN04396080 | IUPEC | This Study |
| KCJ3821 | SAMN04396081 | IUPEC | This Study |
| KCJ3822 | SAMN04396082 | IUPEC | This Study |
| KCJ3823 | SAMN04396083 | IUPEC | This Study |
| KCJ3824 | SAMN04396084 | IUPEC | This Study |
| KCJ3855 | SAMN04396085 | IUPEC | This Study |
| KCJ3856 | N/A | IUPEC | This Study |
| KCJ3857 | SAMN04396086 | IUPEC | This Study |
| KCJ3858 | SAMN04396087 | IUPEC | This Study |
| KCJ3859 | SAMN04396088 | IUPEC | This Study |
aN/A, not available. bExPEC, extraintestinal pathogenic E. coli; EHEC, enterohemorrhagic E. coli; IUPEC, intrauterine pathogenic E. coli; UPEC, uropathogenic E. coli; EIEC, enteroinvasive E. coli.
Table 3.
Characterization of intrauterine pathogenic E. coli isolates.
| Strain name | Serotype | MLSTa | Plasmid replicons | Beta-lactam resistance genes |
|---|---|---|---|---|
| ST648 | O51:H4 | 648 | IncFIA, IncFIB, IncFII, Col | TEM-1B |
| MS499 | H16:O23 | 453 | IncFII, IncFIB | TEM-1C |
| UPEC-3 | O9:H9 | 162 | IncFIB, IncFIC, IncB/O/K/Z | CTX-M-14, TEM-1B |
| CFSAN029787 | O96:H19 | 99 | IncFII, IncFIB, IncQ1, IncFII | - |
| KCJ852 | O8:H19 | 708 | Incl1 | - |
| KCJ3815 | O54:H2 | 2328 | IncFII, IncFIB, IncN, ColRNAI | CTX-M-1 |
| KCJ3816 | O54:H2 | 2328 | IncFII, IncFIB, IncN, ColRNAI | CTX-M-1 |
| KCJ3818 | O54:H2 | 2328 | IncFII, IncFIB, IncN, ColRNAI | CTX-M-1 |
| KCJ3819 | O54:H2 | 2328 | IncFII, IncFIB, IncN, ColRNAI | CTX-M-1 |
| KCJ3820 | :H42 | 648 | IncFIA, IncFIB, IncFII, IncQ1 | CTX-M-124, TEM-1B |
| KCJ3821 | :H42 | 648 | IncFIA, IncFIB, IncFII, IncQ1 | CTX-M-124, TEM-1B |
| KCJ3822 | :H42 | 648 | IncFIA, IncFIB, IncFII, IncQ1 | CTX-M-124, TEM-1B |
| KCJ3823 | :H42 | 648 | IncFIA, IncFIB, IncFII, IncQ1 | CTX-M-124, TEM-1B |
| KCJ3824 | :H42 | 648 | IncFIA, IncFIB, IncFII, IncQ1 | CTX-M-124, TEM-1B |
| KCJ3855 | O8:H19 | 162 | IncFIB, IncFII, IncN | CTX-M-1 |
| KCJ3857 | O8:H19 | 162 | IncFIB, IncFII, IncN | CTX-M-1 |
| KCJ3858 | O8:H19 | 162 | IncFIB, IncFII, IncN | CTX-M-1 |
| KCJ3859 | O8:H19 | 162 | IncFIB, IncFII, IncN | CTX-M-1 |
aMLST, multilocus sequence typing.
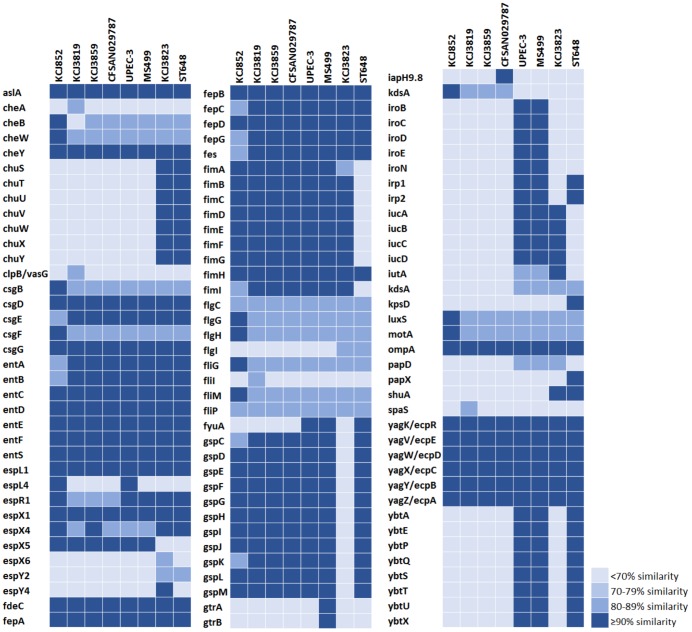
The virulence factors were identified for the same set of strains by Virulence Factor Database (VFDB) (Chen et al., 2005). The number of virulence genes IUPEC strains harbored varied from 65 to 70 (Figure 4). The IUPEC strains harbored important virulence factors including iron uptake (ent, ybt, and fep family), adherence (fim, fli, yag/ecp, and flg family), and invasion (aslA and ompA). The most invasive strain in bovine endometrial cell line, KCJ3819 (see later section of the results), harbored unique genes cheA, clpB/vasG, fliI, and spaS. Particularly, clpB/vasG, fliI, and spaS which are related to Type 6 Secretion System (Shrivastava and Mande, 2008), flagella (Dreyfus et al., 1993), and Type 3 Secretion System (Zarivach et al., 2008), respectively. Again, the IUPEC strains showed similar virulence factor profiles compared with the clinical isolates, suggesting they could be transmitted to humans and cause human infectious disease.
FIGURE 4.
Virulence factor profile of intrauterine pathogenic E. coli from this study and reference human clinical isolates.
Intrauterine Pathogenic E. coli Had High Adhesive and Invasion Ability
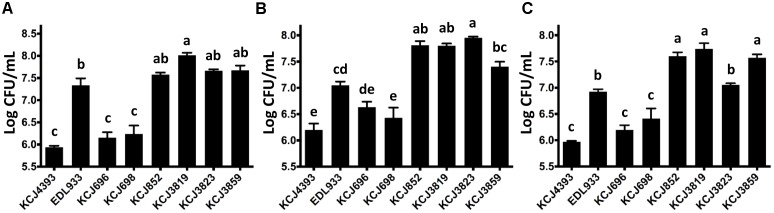
As adhesion is the first step for bacteria to colonize and cause pathogenicity, we tested the adherence ability of IUPEC strains in three human cell lines (Figure 5). The commensal E. coli strain, DH5α (KCJ4393) is known to have low adherence ability compared to pathogenic strains, thus it was included as a negative control. Oppositely, E. coli EDL933 (positive control) was reported to have high adherence activity. eae and tir are attaching and effacing virulence genes of E. coli O157:H7 EDL933 and play a key role in the adhesion to the host. Therefore, we included the deletion mutants of eae (KCJ696) and tir (KCJ698) as negative controls. In Caco-2 cells, the IUPEC strains, regardless if they carried ESBLs, adhered to the cells as efficiently as EDL933 (Figure 5A), indicating the strains were able to colonize in the intestinal tract. HEK293T is a human embryonic kidney cell line and HEP2 is a human cervix carcinoma cell line. The IUPEC strains were able to adhere to these two types of cells equal to or even more significantly than EDL933 (Figures 5B,C), showing the possibility of these strains to cause infection in different sites once invaded to the host.
FIGURE 5.
Adherence of intrauterine pathogenic E. coli to different cell lines. (A) Caco-2, (B) HEK293, (C) HEP2 cells. KCJ4393: E. coli DH5α; KCJ696: eae deletion mutant of EDL933; KCJ698: tir deletion mutant of EDL933; Bars represent the mean ± SEM of three experiments. Means with different letters differ (P < 0.05).
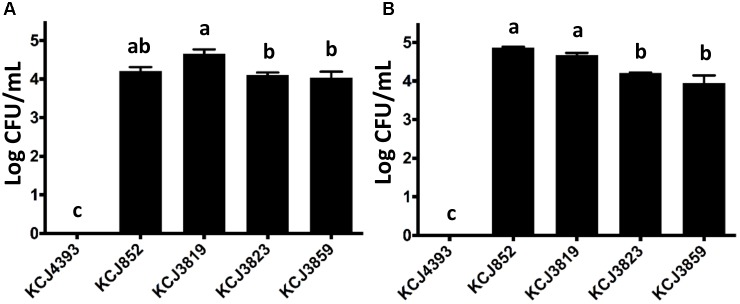
As the pathogenic E. coli associated with pelvic inflammatory disease have the ability to invade the endometrial epithelial cells, we conducted an invasion assay using a bovine endometrial epithelial cell line with representative isolates from each CTX-M-positive animal sample to confirm that the cephalosporin resistant isolates were IUPEC (Figure 6A). Also, a negative control, the non-pathogenic E. coli DH5α (KCJ4393), and KCJ852 were included. It was observed that E. coli DH5α did not have any invasive ability, while for the other four strains, significant numbers of bacteria invaded into the cells (>4 log CFU/mL). Among the pathogenic strains, KCJ3819 showed the greatest ability of invasion. These results indicate that these strains isolated from the uterus were etiological agents of metritis. Another invasion assay was further conducted with Caco-2 cells, which is a human intestinal epithelial cell line (Figure 6B). Similar results were observed that all the IUPEC strains showed high invasion abilities to the cells, suggesting that these strains may also cause infectious disease in the gastrointestinal tract.
FIGURE 6.
Invasion of intrauterine pathogenic E. coli to different cell lines. (A) Bovine endometrial epithelial and (B) Caco-2 cells. KCJ4393: E. coli DH5α; Bars represent the mean ± SEM of three experiments. Means with different letters differ (P < 0.05).
Discussion
It has frequently been reported that farm animals are reservoirs for ESBL producing Enterobacteriaceae. The prevalence of CTX-M-carrying E. coli on a dairy commercial farm using cephalosporin was reported as 39% (Dolejska et al., 2011). Also, dairy herds with ceftiofur use had an approximately 25 times higher chance to have ESBL producing E. coli compared with the herds that did not use ceftiofur (Tragesser, 2003). The prevalence of ESBL producing E. coli in the uterus of cows with metritis has not been reported. In our study, 58% of animals had ESBL producing E. coli in their uterine samples. This high percentage is likely to be associated with the continuous use of ceftiofur in the herd. A limitation of this study was that mainly one dairy farm was recruited. Therefore, the result may not represent the prevalence of ESBL producing E. coli in dairy farms overall. Further studies will be needed to compare the prevalence of ESBL carriers among the cattle population in different geographical regions. Moreover, it would be desirable to correlate the prevalence with a history of antibiotic use on individual farms.
The IUPEC isolates from each cow belonged to different MLST types. Isolates from cow #3 (KCJ3820 – KCJ3824) belong to sequencing type (ST) 648 (Table 3). Pathogenic E. coli ST648 strains have been isolated from human, companion animals and farm animals, and some of them carried ESBL genes including CTX-M-1, -3, -14, -15, and -61 (Peirano et al., 2012; Ewers et al., 2014). They have caused urinary and respiratory tract infections in human hosts (Ewers et al., 2014). They shared common virulence genes like other ExPECs harboring genes, including papC, iutA, kpsMTII, sfa/foc, and afa/dra (Johnson et al., 2003). Isolates from the uterus of cow #4 belonged to ST162. This ST type E. coli have also been found in humans, companion animals and livestock. ST162 has been frequently associated with urinary tract infections (Dierikx et al., 2012; Yuan et al., 2012). They were reported to carry ESBL type CTX-M-1 and TEM-1 (Dierikx et al., 2012). However, no information about ST708 and ST2328, which IUPEC isolates from cow #1 (KCJ852) and #2 (KCJ3815 -KCJ3819) belonged to respectively, can be found in the literature. In addition, the IUPEC isolates from different cows belong to different serotypes and MLST types, suggesting that there was low pathogen transmission among animals although the strains were isolated from the same farm at the same time. These data are consistent with the previous findings that IUPEC isolates did not cluster significantly by farms by random amplified polymorphic DNA-PCR cluster analysis (Bicalho et al., 2010). Sheldon et al. (2010) also reported that IUPEC strains were from various clusters by MLST analysis. This indicates that the strains gained antibiotic resistance either during parallel evolution or by plasmid acquisition. However, further studies with more sample numbers are needed to understand this phenomenon.
Interestingly, however, these isolates harbored two identical plasmid groups, IncFIB and FII (Table 3). IncFIB plasmid was frequently observed in avian pathogenic E. coli (Johnson et al., 2007) and IncFII plasmid was usually associated with blaCTX-M gene (Hopkins et al., 2006). IncN plasmid was carried by isolates from both cow #2 and #4, which harbored CTX-M-1. IncN plasmid was reported to carry blaCTX-M-1 and the E. coli strains which carried IncN plasmid were isolated from a dairy farm (Dolejska et al., 2011). Thus, the resistance genes may disseminate among animals via bacterial conjugation system (de Been et al., 2014). The IUPEC strains in this study carried CTX-M-1 (KCJ3815-3819 and KCJ3855-3859) and CTX-M-115 (KCJ3820-3824, Figure 3). CTX-M-1 type β-lactamase was frequently observed in both food producing animal isolates and human clinical isolates (Eckert et al., 2004; Meunier et al., 2006; Cormier et al., 2016). CTX-M-115 type was detected less frequently, but it was also found in both clinical isolates (Munoz-Price et al., 2013; Pfeifer et al., 2016) and farm animals (Cormier et al., 2016). These data further support the possible transmission of the ARGs among animals and humans, especially through plasmid acquisition.
The IUPEC strains isolated in this study harbored essential virulence genes including adherence. For example, the fimbriae are encoded by the fim gene cluster, which are necessary for adhesion to host cell surfaces (Hultgren et al., 1991). The presence of fimH positive E. coli in uteri increases the chance of establishing and persistence of uterine infection by other pathogenic Gram-negative bacteria and reduces the reproductive performance (Bicalho et al., 2012). In addition, E. coli carrying type 1 fimbrial fimH detected at 1-3 days in milk was reported to be significantly associated with development of metritis and endometritis (Bicalho et al., 2012). In this study, all the IUPEC strains isolated were fimH positive, showing that they increase the risk of uterine disease. Although some virulence factors such as siderophores (iroBCDE/iucABCD) and iron uptakes (chuSTUVWXY) were missing compared to the previously reported IUPEC strain, MS499 (Goldstone et al., 2014), all the IUPEC strains harbored adherence genes (Figure 4) and were able to adhere and invade into epithelial cells (Figures 5, 6) which is consistent with findings of other studies (Sheldon et al., 2010; Poole et al., 2017).
Conclusion
The present study showed the high frequency of ESBL producing E. coli in the uteri of cows having metritis we sampled. The IUPEC isolates were adhesive and invasive to different cell lines; this provided a clear demonstration that they can cause disease at different sites of the host. These isolates had resistance to the common antibiotic treatment of metritis (ceftiofur) and, therefore, the high treatment failure of metritis is very likely to be related with these ESBL producing pathogenic E. coli. In addition, these isolates had similar antibiotic resistance and virulence factor profiles compared with human clinical isolates, showing that they may transmit between animals and humans to cause infectious diseases.
Author Contributions
ZM, KG, and KJ designed the study. ZM, MK, and KG collected the samples. ZM, AG, and KJ analyzed the data. ZM and KJ drafted the manuscript. ZM, AG, KG, and KJ finalized this manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This material was based upon work that was supported by the National Institute of Food and Agriculture, U.S. Department of Agriculture, under award number 2015-68003-22971 to KJ.
References
- Amigo N., Mercado E., Bentancor A., Singh P., Vilte D., Gerhardt E., et al. (2015). Clade 8 and clade 6 strains of Escherichia coli O157:H7 from cattle in Argentina have hypervirulent-like phenotypes. PLoS One 10:e0127710. 10.1371/journal.pone.0127710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicalho M. L. S., Machado V. S., Oikonomou G., Gilbert R. O., Bicalho R. C. (2012). Association between virulence factors of Escherichia coli, Fusobacterium necrophorum, and Arcanobacterium pyogenes and uterine diseases of dairy cows. Vet. Microbiol. 157 125–131. 10.1016/j.vetmic.2011.11.034 [DOI] [PubMed] [Google Scholar]
- Bicalho R. C., Machado V. S., Bicalho M. L. S., Gilbert R. O., Teixeira A. G. V., Caixeta L. S., et al. (2010). Molecular and epidemiological characterization of bovine intrauterine Escherichia coli. J. Dairy Sci. 93 5818–5830. 10.3168/jds.2010-3550 [DOI] [PubMed] [Google Scholar]
- Bonnet R. (2004). Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Zankari E., Garcia-Fernandez A., Larsen M. V., Lund O., Villa L., et al. (2014). In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58 3895–3903. 10.1128/Aac.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chander Y., Oliveira S., Goyal S. M. (2011). Characterisation of ceftiofur resistance in swine bacterial pathogens. Vet. J. 187 139–141. 10.1016/j.tvjl.2009.10.013 [DOI] [PubMed] [Google Scholar]
- Chen L. H., Yang J., Yu J., Ya Z. J., Sun L. L., Shen Y., et al. (2005). VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33 D325–D328. 10.1093/nar/gki008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenault J. R., McAllister J. F., Chester S. T., Jr., Dame K. J., Kausche F. M., Robb E. J. (2004). Efficacy of ceftiofur hydrochloride sterile suspension administered parenterally for the treatment of acute postpartum metritis in dairy cows. J. Am. Vet. Med. Assoc. 224 1634–1639. [DOI] [PubMed] [Google Scholar]
- CLSI (2018). Methods for Dilution Antimicrobial Susceptibility Testes for Bacteria that Grow Aerobically. Wayne, PA: CLSI. [Google Scholar]
- Collatz E., Labia R., Gutmann L. (1990). Molecular evolution of ubiquitous beta-lactamases towards extended-spectrum enzymes active against newer beta-lactam antibiotics. Mol. Microbiol. 4 1615–1620. 10.1111/j.1365-2958.1990.tb00537.x [DOI] [PubMed] [Google Scholar]
- Cormier A. C., Chalmers G., McAllister T. A., Cook S., Zaheer R., Scott H. M., et al. (2016). Extended-spectrum-cephalosporin resistance genes in Escherichia coli from beef cattle. Antimicrob. Agents Chemother. 60 1162–1163. 10.1128/Aac.02516-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha F., Jeon S. J., Daetz R., Vieira-Neto A., Laporta J., Jeong K. C., et al. (2018). Quantifying known and emerging uterine pathogens, and evaluating their association with metritis and fever in dairy cows. Theriogenology 114 25–33. 10.1016/j.theriogenology.2018.03.016 [DOI] [PubMed] [Google Scholar]
- Darling A. E., Mau B., Perna N. T. (2010). Progressive mauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Been M., Lanza V. F., de Toro M., Scharringa J., Dohmen W., Du Y., et al. (2014). Dissemination of cephalosporin resistance genes between Escherichia coli strains from farm animals and humans by specific plasmid lineages. PLoS Genet. 10:e1004776. 10.1371/journal.pgen.1004776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierikx C. M., van Duijkeren E., Schoormans A. H. W., van Essen-Zandbergen A., Veldman K., Kant A., et al. (2012). Occurrence and characteristics of extended-spectrum-lactamase- and AmpC-producing clinical isolates derived from companion animals and horses. J. Antimicrob. Chemother. 67 1368–1374. 10.1093/jac/dks049 [DOI] [PubMed] [Google Scholar]
- Dolejska M., Jurcickova Z., Literak I., Pokludova L., Bures J., Hera A., et al. (2011). IncN plasmids carrying bla CTX-M-1 in Escherichia coli isolates on a dairy farm. Vet. Microbiol. 149 513–516. 10.1016/j.vetmic.2010.11.032 [DOI] [PubMed] [Google Scholar]
- Donaldson S. C., Straley B. A., Hegde N. V., Sawant A. A., DebRoy C., Jayarao B. M. (2006). Molecular epidemiology of ceftiofur-resistant Escherichia coli isolates from dairy calves. Appl. Environ. Microbiol. 72 3940–3948. 10.1128/Aem.02770-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus G., Williams A. W., Kawagishi I., Macnab R. M. (1993). Genetic and biochemical-analysis of Salmonella typhimurium fliI, a flagellar protein related to the catalytic subunit of the F0F1 atpase and to virulence proteins of mammalian and plant-pathogens. J. Bacteriol 175 3131–3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drillich M., Beetz O., Pfutzner A., Sabin M., Sabin H. J., Kutzer P., et al. (2001). Evaluation of a systemic antibiotic treatment of toxic puerperal metritis in dairy cows. J. Dairy Sci. 84 2010–2017. 10.3168/jds.S0022-0302(01)74644-9 [DOI] [PubMed] [Google Scholar]
- Eckert C., Gautier V., Saladin-Allard M., Hidri N., Verdet C., Ould-Hocine Z., et al. (2004). Dissemination of CTX-M-type beta-lactamases among clinical isolates of Enterobacteriaceae in Paris. France. Antimicrob. Agents Chemother. 48 1249–1255. 10.1128/Aac.48.4.1249-1255.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewers C., Bethe A., Stamm I., Grobbel M., Kopp P. A., Guerra B., et al. (2014). CTX-M-15-D-ST648 Escherichia coli from companion animals and horses: another pandemic clone combining multiresistance and extraintestinal virulence? J. Antimicrob. Chemother. 69 1224–1230. 10.1093/jac/dkt516 [DOI] [PubMed] [Google Scholar]
- Falkow S., Small P., Isberg R., Hayes S. F., Corwin D. (1987). A molecular strategy for the study of bacterial invasion. Rev. Infect. Dis. 9 S450–S455. [DOI] [PubMed] [Google Scholar]
- Ginn A., Ma Z., Galvao K. N., Jeong K. C. (2016). Draft genome sequence of an Escherichia coli O8:H19 sequence type 708 strain isolated from a Holstein dairy cow with metritis. Genome Announc. 4:e00261-16. 10.1128/genomeA.00261-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstone R. J., Popat R., Schuberth H. J., Sandra O., Sheldon I. M., Smith D. G. (2014). Genomic characterisation of an endometrial pathogenic Escherichia coli strain reveals the acquisition of genetic elements associated with extra-intestinal pathogenicity. BMC Genomics 15:1075. 10.1186/1471-2164-15-1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshen T., Shpigel N. Y. (2006). Evaluation of intrauterine antibiotic treatment of clinical metritis and retained fetal membranes in dairy cows. Theriogenology 66 2210–2218. 10.1016/j.theriogenology.2006.07.017 [DOI] [PubMed] [Google Scholar]
- Hopkins K. L., Liebana E., Villa L., Batchelor M., Threlfall E. J., Carattoli A. (2006). Replicon typing of plasmids carrying CTX-M or CMY beta-lactamases circulating among Salmonella and Escherichia coli isolates. Antimicrob. Agents Chemother. 50 3203–3206. 10.1128/Aac.00149-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultgren S. J., Normark S., Abraham S. N. (1991). Chaperone-assisted assembly and molecular architecture of adhesive pili. Annu. Rev. Microbiol. 45 383–415. [DOI] [PubMed] [Google Scholar]
- Jeon S. J., Ma Z., Kang M., Galvao K. N., Jeong K. C. (2016). Application of chitosan microparticles for treatment of metritis and in vivo evaluation of broad spectrum antimicrobial activity in cow uteri. Biomaterials 110 71–80. 10.1016/j.biomaterials.2016.09.016 [DOI] [PubMed] [Google Scholar]
- Jeon S. J., Vieira-Neto A., Gobikrushanth M., Daetz R., Mingoti R. D., Parize A. C. B., et al. (2015). Uterine microbiota progression from calving until establishment of metritis in dairy cows. Appl. Environ. Microbiology 81 6324–6332. 10.1128/Aem.0175315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joensen K. G., Tetzschner A. M. M., Iguchi A., Aarestrup F. M., Scheutz F. (2015). Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 53 2410–2426. 10.1128/Jcm.00008-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. R., Murray A. C., Gajewski A., Sullivan M., Snippes P., Kuskowski M. A., et al. (2003). Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob. Agents Chemother. 47 2161–2168. 10.1128/Aac.47.7.2161-2168.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T. J., Wannemuehler Y. M., Johnson S. J., Logue C. M., White D. G., Doetkott C., et al. (2007). Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl. Environ. Microbiol. 73 1976–1983. 10.1128/Aem.02171-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N., Fass J. N. (2011). Sickle a Sliding-Window, Adaptive, Quality-Based Trimming Tool for FastQ Files (Version 1.33). [Google Scholar]
- Lane M. C., Alteri C. J., Smith S. N., Mobley H. L. T. (2007). Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc. Natl. Acad. Sci. U.S.A. 104 16669–16674. 10.1073/pnas.0607898104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M. V., Cosentino S., Rasmussen S., Friis C., Hasman H., Marvig R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50 1355–1361. 10.1128/JCM.06094-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc S. J. (2008). Postpartum uterine disease and dairy herd reproductive performance: a review. Vet. J. 176 102–114. 10.1016/j.tvjl.2007.12.019 [DOI] [PubMed] [Google Scholar]
- Li X. Z., Mehrotra M., Ghimire S., Adewoye L. (2007). Beta-Lactam resistance and beta-lactamases in bacteria of animal origin. Vet. Microbiol. 121 197–214. 10.1016/j.vetmic.2007.01.015 [DOI] [PubMed] [Google Scholar]
- Liebana E., Batchelor M., Hopkins K. L., Clifton-Hadley F. A., Teale C. J., Foster A., et al. (2006). Longitudinal farm study of extended-spectrum beta-lactamase-mediated resistance. J. Clin. Microbiol. 44 1630–1634. 10.1128/Jcm.44.5.1630-1634.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima F. S., Vieira-Neto A., Vasconcellos G. S., Mingoti R. D., Karakaya E., Sole E., et al. (2014). Efficacy of ampicillin trihydrate or ceftiofur hydrochloride for treatment of metritis and subsequent fertility in dairy cows. J. Dairy Sci. 97 5401–5414. 10.3168/jds.2013-7569 [DOI] [PubMed] [Google Scholar]
- McArthur A. G., Waglechner N., Nizam F., Yan A., Azad M. A., Baylay A. J., et al. (2013). The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 57 3348–3357. 10.1128/Aac.00419-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin C. L., Stanisiewski E., Lucas M. J., Cornell C. P., Watkins J., Bryson L., et al. (2012). Evaluation of two doses of ceftiofur crystalline free acid sterile suspension for treatment of metritis in lactating dairy cows. J. Dairy Sci. 95 4363–4371. 10.3168/jds.2011-5111 [DOI] [PubMed] [Google Scholar]
- Meunier D., Jouy E., Lazizzera C., Kobisch M., Madec J. Y. (2006). CTX-M-1- and CTX-M-15-type beta-lactamases in clinical Escherichia coli isolates recovered from food-producing animals in France. Int. J. Antimicrob. Agents 28 402–407. 10.1016/j.ijantimicag.2006.08.016 [DOI] [PubMed] [Google Scholar]
- Mir R. A., Weppelmann T. A., Johnson J. A., Archer D., Morris J. G., Jr., Jeong K. C. (2016). Identification and characterization of cefotaxime resistant bacteria in beef cattle. PLoS One 11:e0163279. 10.1371/journal.pone.0163279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Price L. S., Arheart K., Nordmann P., Boulanger A. E., Cleary T., Alvarez R., et al. (2013). Eighteen years of experience with Acinetobacter baumannii in a tertiary care hospital. Crit. Care Med. 41 2733–2742. 10.1097/CCM.0b013e318298a541 [DOI] [PubMed] [Google Scholar]
- Overton M., Fetrow J. (2008). “Economics of postpartum uterine health,” In Proceedings of the Dairy Cattle Reproduction Council Convention, Omaha, NE, USA (Hartland, WI: DCRC; ), 39–43. [Google Scholar]
- Peirano G., van der Bij A. K., Gregson D. B., Pitout J. D. D. (2012). Molecular epidemiology over an 11-year period (2000 to 2010) of extended-spectrum beta-lactamase-producing Escherichia coli causing bacteremia in a centralized Canadian region. J. Clin. Microbiol. 50 294–299. 10.1128/Jcm.06025-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer Y., Hunfeld K. P., Borgmann S., Maneg D., Blobner W., Werner G., et al. (2016). Carbapenem-resistant Acinetobacter baumannii ST78 with OXA-72 carbapenemase and ESBL gene blaCTX-M-115. J. Antimicrob. Chemother. 71 1426–1428. 10.1093/jac/dkv462 [DOI] [PubMed] [Google Scholar]
- Poirel L., Mammeri H., Nordmann P. (2004). TEM-121, a novel complex mutant of TEM-type beta-lactamase from Enterobacter aerogenes. Antimicrob. Agents Chemother. 48 4528–4531. 10.1128/Aac.48.12.4528-4531.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole N. M., Green S. I., Rajan A., Vela L. E., Zeng X. L., Estes M. K., et al. (2017). Role for FimH in extraintestinal pathogenic Escherichia coli invasion and translocation through the intestinal epithelium. Infect. Immun. 85: e00581-17. 10.1128/IAI.00581-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Lam T. T., Max Carvalho L., Pybus O. G. (2016). Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2:vew007. 10.1093/ve/vew007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos T. M. A., Caixeta L. S., Machado V. S., Rauf A. K., Gilbert R. O., Bicalho R. C. (2010). Antimicrobial resistance and presence of virulence factor genes in Arcanobacterium pyogenes isolated from the uterus of postpartum dairy cows. Vet. Microbiol. 145 84–89. 10.1016/j.vetmic.2010.03.001 [DOI] [PubMed] [Google Scholar]
- Schmid A., Hormansdorfer S., Messelhausser U., Kasbohrer A., Sauter-Louis C., Mansfeld R. (2013). Prevalence of extended-spectrum beta-lactamase-producing Escherichia coli on bavarian dairy and beef cattle farms. Appl. Environ. Microbiol. 79 3027–3032. 10.1128/Aem.00204-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon I. M., Cronin J., Goetze L., Donofrio G., Schuberth H. J. (2009). Defining postpartum uterine disease and the mechanisms of infection and immunity in the female reproductive tract in cattle. Biol. Reprod. 81 1025–1032. 10.1095/biolreprod.109.077370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon I. M., Lewis G. S., LeBlanc S., Gilbert R. O. (2006). Defining postpartum uterine disease in cattle. Theriogenology 65 1516–1530. 10.1016/j.theriogenology.2005.08.021 [DOI] [PubMed] [Google Scholar]
- Sheldon I. M., Rycroft A. N., Dogan B., Craven M., Bromfield J. J., Chandler A., et al. (2010). Specific strains of Escherichia coli are pathogenic for the endometrium of cattle and cause pelvic inflammatory disease in cattle and mice. PLoS One 5:e9192. 10.1371/journal.pone.0009192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava S., Mande S. S. (2008). Identification and functional characterization of gene components of Type VI Secretion System in bacterial genomes. PLoS One 3:e2955. 10.1371/journal.pone.0002955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B. I., Donovan G. A., Risco C., Littell R., Young C., Stanker L. H., et al. (1998). Comparison of various antibiotic treatments for cows diagnosed with toxic puerperal metritis. J. Dairy Sci. 81 1555–1562. 10.3168/jds.S0022-0302(98)75721-2 [DOI] [PubMed] [Google Scholar]
- Srinivasan V., Nam H. M., Nguyen L. T., Tamilselvam B., Murinda S. E., Oliver S. P. (2005). Prevalence of antimicrobial resistance genes in Listeria monocytogenes isolated from dairy farms. Foodborne Pathog. Dis. 2 201–211. 10.1089/fpd.2005.2.201 [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis Version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenhagen B. A., Koster G., Wallmann J., Heuwieser W. (2006). Prevalence of mastitis pathogens and their resistance against antimicrobial agents in dairy cows in Brandenburg. Germany. J. Dairy Sci. 89 2542–2551. [DOI] [PubMed] [Google Scholar]
- Torres A. G., Zhou X., Kaper J. B. (2005). Adherence of diarrheagenic Escherichia coli strains to epithelial cells. Infect. Immun. 73 18–29. 10.1128/IAI.73.1.18-29.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tragesser L. A. (2003). Ceftiofur Use and its Association With the Recovery of Expanded-Spectrum Beta-Lactamase Producing Escherichia coli in Food Animal Populations. Columbus, OH: The Ohio State University. [Google Scholar]
- Trifinopoulos J., Nguyen L. T., von Haeseler A., Minh B. Q. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44 W232–W235. 10.1093/nar/gkw256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattam A. R., Abraham D., Dalay O., Disz T. L., Driscoll T., Gabbard J. L., et al. (2014). PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 42 D581–D591. 10.1093/nar/gkt1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf M. K. (1997). Occurence, distribution, and associations of O and H serogroups, colonizatio factor antigens, and toxins of enterotoxigenic Escherichia coli. Clin. Microbiol. Rev. 10 569–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J. Y., Xu X. G., Guo Q. L., Zhao X., Ye X. Y., Guo Y., et al. (2012). Prevalence of the oqxAB gene complex in Klebsiella pneumoniae and Escherichia coli clinical isolates. J. Antimicrob. Chemother. 67 1655–1659. 10.1093/jac/dks086 [DOI] [PubMed] [Google Scholar]
- Zarivach R., Deng W. Y., Vuckovic M., Felise H. B., Nguyen H. V., Miller S. I., et al. (2008). Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS. Nature 453 124–127. 10.1038/nature06832 [DOI] [PubMed] [Google Scholar]
- Zhao S. H., White D. G., McDermott P. F., Friedman S., English L., Ayers S., et al. (2001). Identification and expression of cephamycinase blaCMY genes in Escherichia coli and Salmonella isolates from food animals and ground meat. Antimicrob. Agents Chemother. 45 3647–3650. 10.1128/Aac.45.12.3647-3650.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Liang Y. J., Lynch K. H., Dennis J. J., Wishart D. S. (2011). PHAST: a fast phage search tool. Nucleic Acids Res. 39 W347–W352. 10.1093/nar/gkr485 [DOI] [PMC free article] [PubMed] [Google Scholar]