Abstract
Acinetobacter baumannii (Ab) is one of the most important pathogens associated with nosocomial infections, especially pneumonia. Interest in the Quorum network, i.e., Quorum Sensing (QS)/Quorum Quenching (QQ), in this pathogen has grown in recent years. The Quorum network plays an important role in regulating diverse virulence factors such as surface motility and bacterial competition through the type VI secretion system (T6SS), which is associated with bacterial invasiveness. In the present study, we investigated 30 clinical strains of A. baumannii isolated in the “II Spanish Study of A. baumannii GEIH-REIPI 2000-2010” (Genbank Umbrella Bioproject PRJNA422585), a multicentre study describing the relationship between the Quorum network in A. baumannii and the development of pneumonia and associated bacteraemia. Expression of the aidA gene (encoding the AidA protein, QQ enzyme) was lower (P < 0.001) in strains of A. baumannii isolated from patients with bacteraemic pneumonia than in strains isolated from patients with non-bacteraemic pneumonia. Moreover, aidA expression in the first type of strain was not regulated in the presence of environmental stress factors such as the 3-oxo-C12-HSL molecule (substrate of AidA protein, QQ activation) or H2O2 (inhibitor of AidA protein, QS activation). However, in the A. baumannii strains isolated from patients with non-bacteraemic pneumonia, aidA gene expression was regulated by stressors such as 3-oxo-C12-HSL and H2O2. In an in vivo Galleria mellonella model of A. baumannii infection, the A. baumannii ATCC 17978 strain was associated with higher mortality (100% at 24 h) than the mutant, abaI-deficient, strain (carrying a synthetase enzyme of Acyl homoserine lactone molecules) (70% at 24 h). These data suggest that the QS (abaR and abaI genes)/QQ (aidA gene) network affects the development of secondary bacteraemia in pneumonia patients and also the virulence of A. baumannii.
Keywords: quorum, sensing/quenching, pneumonia, bacteraemia, Acinetobacter
Introduction
Acinetobacter baumannii is a major cause of hospital-acquired infections associated with high mortality rates (Fuchs, 2016),s usually affecting patients in Intensive Care Units (ICU) (del Mar Tomas et al., 2005; Lee et al., 2017). In these patients, A. baumannii causes infections such as pneumonia or, to a lesser extent, serious infections of the bloodstream (around 10% of clinical isolates of A. baumannii cause bacteraemia) (Cisneros and Rodríguez-Baño, 2002; El Kettani et al., 2017).
The success of this bacterium as a nosocomial pathogen, has been attributed to the following factors, amongst others: (i) high genetic versatility, facilitating rapid adaptation to stressful or unfavorable situations (Gayoso et al., 2014; Trastoy et al., 2018); (ii) ability to acquire new genes horizontally by the acquisition of plasmids and phages (López et al., 2018); (iii) ability to persist for a long time on animate and inanimate surfaces (resistance to desiccation) (Gayoso et al., 2014), which is generally attributed to biofilm formation; (iv) resistance to antimicrobial agents, including broad-spectrum antibiotics such as carbapenems, colistin, and tigecycline (Fernández-Cuenca et al., 2015), as well as to disinfectants and biocides (Fernández-García et al., 2018); and (v) high virulence (colonization, invasiveness, and cytotoxicity) (Rumbo et al., 2014; Wong et al., 2017). These characteristics contribute to the fact that nosocomial outbreaks caused by A. baumannii are difficult to control and that therapeutic options to treat infections are scarce or non-existent (Fernández-Cuenca et al., 2013). In February, 2017, the World Health Organization (WHO) published a list of “priority pathogens.” The list includes antibiotic resistant bacteria, considered a serious threat to human health and for which new antibiotics are urgently needed, and is headed by carbapenem-resistant A. baumannii (Tacconelli et al., 2018).
The Quorum Sensing (QS) network is generally used by Gram-negative bacterial pathogens to regulate biological processes such as virulence, conjugation, resistance, biofilm formation (which also depends on other factors such as the lytic enzymes responsible for peptidoglycan recycling: Vijayakumar et al., 2016), motility and bacterial competition, via secretion systems (T6SS), which are associated with greater invasiveness (LaSarre and Federle, 2013; López et al., 2017a,b). Two proteins (AbaI /AbaR) identified in A. baumannii have been described as homologs of the LuxI/LuxR system found in Vibrio fischeri. This system comprises a signal or autoinducer molecule (acyl-homoserine lactone, AHL), an enzyme that synthesizes signaling molecules (AbaI) and a receptor protein activator of the QS (AbaR), which forms a complex with N-(3-hydroxydodecanoil)-L-homoserine lactone (3-OH-C12-HSL) to regulate virulence factors, biofilm formation, surface motility, and bacterial competence (T6SS) (Stacy et al., 2012). When a threshold concentration is reached, the AHL molecules present inside the cell are transported to its receptor (AbaR), putatively joining the lux-box, which is located 67 bp upstream of the ATG of AbaI, resulting in the synthesis of more AHL molecules (López et al., 2017b). The QS mechanism, on the other hand, acts naturally under environmental stress conditions such as the presence of bile salts in the gastrointestinal tract and H2O2 (ROS response) in the respiratory tract (López et al., 2017b).
A new enzyme (AidA) has recently been cloned in E. coli BL21 (DE3) and functionally characterized in clinical strains of A. baumannii capable of inhibiting their own QS (by Quorum Quenching) (López et al., 2017b). This enzyme acts by degrading signaling molecules such as N-(3-Oxo-dodecanoyl), L-homoserine lactone (3-Oxo-C12-HSL), and N-dodecanoyl-L-homoserine lactone (C12-HSL), as confirmed by observation of inhibition of motility, biofilm formation and other virulence factors associated with activation of the Quorum Sensing system (López et al., 2017b; Mayer et al., 2018). Other QQ enzymes have also recently been described in A. baumannii ATCC17978 (A1S_0383, A1S_2662, A1S_1876) (Mayer et al., 2018). Multiple QQ enzymes have been analyzed in diverse pathogens such as Pseudomonas aeruginosa (Zhang et al., 2011), Deinococcus radiodurans, Hyphomonas neptunium, Photorhabdus luminicencens, and Rhizobium spp. (Kalia et al., 2011; Krysciak et al., 2011).
Based on these findings, in the present study, we examined the relationship between the global Quorum regulatory network (QS/QQ) mediated by the abaR (QS) and aidA (QQ) genes and the development of pneumonia and bacteraemia in clinical strains of A. baumannii isolated in the “II Spanish Study of A. baumannii GEIH-REIPI 2000-2010,” a multicentre study involving 45 Spanish hospitals and 246 patients. In addition, we used an in vivo infection model consisting of larvae of the wax moth Galleria mellonella to examine the relationship between the global QS/QQ and the development of mortality by a mutant abaI (QS)-deficient strain of A. baumannii (A. baumannii ATCC17978ΔabaI) relative to that of the wild-type A. baumannii ATCC17978 strain.
Materials and Methods
Bacteria and Samples
To carry out this study, we analyzed 30 clinical strains of A. baumannii from the 465 strains isolated in the “II Spanish Study of A. baumannii GEIH-REIPI 2000-2010” multicentre study (Genbank Umbrella Bioproject PRJNA422585). The multicentre study included 45 hospitals in Spain, in which new cases of colonization or infection by A. baumannii were analyzed between February and March 2010 (Villar et al., 2014). The 30 A. baumannii strains were all isolated from respiratory samples from patients with nosocomial pneumonia (n = 13: 6 with and 7 without bacteraemia) or A. baumannii colonization of the lower respiratory tract (n = 17) (Sánchez-Encinales et al., 2017). Molecular typing was performed by Multilocus Sequence Typing (MLST) (Mosqueda et al., 2014). In addition, we used a killing assay with the Galleria mellonella infection model and an A. baumannii ATCC17978ΔabaI mutant strain (identified by Castañeda-Tamez et al., 2018).
The main clinical study variables included demographics, underlying diseases, mechanical ventilation, tracheostomy, colonization of lower respiratory airways, bacteraemic pneumonia (Pn-B), non-bacteraemic pneumonia (Pn-NB) (Horan et al., 2008) and any cause of death during hospitalization.
To design the primers and probes of the QS genes and QQ enzymes, we analyzed the presence of QS genes (abaR and abaI) and the QQ enzyme (aidA) in A. baumannii ATCC 17978 (Genbank genome accession numbers CP000521.1 [CP018664.1]) and in 1000 A. baumannii genomes by consulting the “Integrated Microbial Genomes and Microbiomes” web page (https://img.jgi.doe.gov) and using nucleotide BLAST. The gene sequences used in the search were selected from the Acinetobacter baumannii ATCC 17978 genome. A threshold of 1e-50 was used as the limit for analysis of the nucleotide sequence, where the e-value was defined as the probability of random alignments with the same score. We also calculated the percentage presence of these genes in the genomes (Figure S1).
RNA Extraction
RNA Extraction to Analyze the Quorum Regulatory Network (QS/QQ)
All clinical strains of A. baumannii were cultured on solid Luria-Bertani (LB) plates and incubated at 37° C for 24 h. One colony was removed and inoculated in liquid LB medium and incubated overnight at 37° C under stirring at 180 rpm. The inoculum was diluted (1:100) and allowed to grow until an optical density (OD600 nm) of 0.4–0.6 (corresponding to the logarithmic growth phase) was reached. The RNA was then extracted using the High Pure RNA Isolation kit (Roche, Germany) and the extract was treated with Dnase (Roche, Germany). The extracted RNA was subsequently quantified in a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies), and the concentration was adjusted to 50 ng/μl in order to yield efficiencies of 90-110% (Rumbo et al., 2013). All extractions were carried out in duplicate.
RNA Extraction to Analyze the Quorum Regulatory Network (QS/QQ) Under Stress Conditions (3-Oxo-C12-HSL and H2O2)
The 13 strains of A. baumannii, isolated from patients with pneumonia, were cultured on solid Luria-Bertani (LB) plates and incubated at 37°C for 24 h. One colony was then removed, inoculated in liquid LB medium and incubated overnight at 37°C under stirring at 180 rpm. The preinoculum was diluted (1:100) and allowed to grow until an optical density (OD600 nm) of 0.3 was reached. Aliquots of 10 μM of 3-Oxo-C12-HSL (QS-inactivating molecule by expression of the AidA protein) (Stacy et al., 2012; López et al., 2017b) and (10 μl) H2O2 were then added for 5 min (QS-activator by ROS response) (López et al., 2018). All controls were prepared by adding the same volumes of DMSO (dimethyl sulfoxide), 3-Oxo-C12-HSL and of sample, but with no H2O2. After incubation of the samples for 4 and 5 h in the presence of 3-Oxo-C12-HSL, to study the regulatory QS/QQ genes (abaR and aidA), as well as 5 min under H2O2 in static at 37°C, RNA was extracted using the High Pure RNA Isolation kit (Roche, Germany) and treated with Dnase. The extracted RNA was subsequently quantified as described above (Rumbo et al., 2013).
RT-qPCR
The studies were carried out with a Lightcycler 480 RNA MasterHydrolysis Probe (Roche, Germany), under the following conditions: reverse transcription at 63°C for 3 min, denaturation at 95°C for 30 s, followed by 45 cycles of 15 s at 95°C and 45 s at 60°C and, finally, cooling at 40°C for 30 s. The UPL primers and probes from conserved DNA regions identified by PCR (Universal Probe Library-Roche, Germany) used in the analysis are shown in Table 1.
Table 1.
Primers and Probes used in this study.
| Sequence (5′-3′) | Probe | Reference | ||
|---|---|---|---|---|
| QUORUM SENSING | ||||
| abaR | Forw | TGGCAAGAAGATTTATTATCAGCA | 119/TTGGTGGT | This study |
| Rev | TGCGGTAGATTTAACGATCTCA | |||
| Forw | AGAGGCGTTACGTTGGACTG | 155/GAAGGCAA | This study | |
| Rev | CCAAGAATCTGAGCTATTGC | |||
| QUORUM QUENCHING | ||||
| aidA | Forw | GGGAACTTCTTTCGGTGGAG | 145/CAGCGACC | López et al., 2017b |
| Rev | AACAGCAGCAAGTCGATTATCA | |||
| Forw | CCTAACCTTGCATTAGGGCTATTA | 53/TGGCAGAG | López et al., 2017b | |
| Rev | CGGTAAACCACAGGTCGGTA | |||
| HOUSEKEEPING | ||||
| rpoB | Forw | CGTGTATCTGCGCTTGG | 131/CTGGTGGT | Rumbo et al., 2014 |
| Rev | CGTACTTCGAAGCCTGCAC | |||
All of the experiments were carried out in a final volume of 20 μl per well (18 μl of master mix and 2 μl of RNA). Each experiment was carried out in duplicate with two RNA extracts (50 ng/μl). For each strain, the expression of all genes, primers, and probes was normalized relative to the reference or housekeeping gene, rpoB, for RT-qPCR studies of Quorum sensing Primer sequences (5′-3′) with Taqman probes (Rumbo et al., 2013; López et al., 2017b). Analysis of the controls without reverse transcriptase confirmed the absence of DNA contamination.
Galleria mellonella Infection Model
The Galleria mellonella model was an adapted version of that developed by Peleg et al. (2009), Yang et al. (2015). The procedure was as follows: twelve G. mellonella larvae, acquired from TruLarvTM (Biosystems Technology, Exeter, Devon, UK), were each injected with 10 μl of a suspension of A. baumannii ATCC17978, or its isogenic deficient mutant A. baumannii ATCC17978ΔabaI, diluted in sterile phosphate buffer saline (PBS) and containing 8 × 104 CFU (± 0. 5 log). The injection was performed with a Hamilton syringe (volume 100 μl) (Hamilton, Shanghai, China). In addition, a control group of twelve larvae were injected with 10 μl of sterile PBS. After being injected, the groups of larvae were placed in Petri dishes and incubated in darkness at 37°C. The number of dead larvae was recorded twice a day (morning and afternoon) for 6 days. The larvae were considered dead when they showed no movement in response to touch (Peleg et al., 2009).
Statistical Analysis
The gene expression studies were carried out in duplicate, and the data obtained were analyzed by Student's t-test, implemented with GraphPad Prism v.6 software (GraphPad Software Inc. San Diego, CA). The graphs were constructed using the GraphPad program, and the results were represented as means and their respective standard deviations.
The mortality curves corresponding to the in vivo Galleria mellonella infection model were constructed using GraphPad Prism v.6 and the data were analyzed using the Log-rank test (Mantel-Cox). In both cases, p-values < 0.05 were considered statistically significant, and the data were expressed as mean values.
The statistical analyses were applied to the following categorical variables: age, sex, immunosuppressive treatment, surgery, ICU stay, mechanical ventilation, tracheostomy, severe sepsis, septic shock, and expression of the Quorum genes in A. baumannii clinical strains (Bone et al., 1992). In addition, the severity of co-morbidities was assessed using the Charlson score (Charlson et al., 1987) and the McCabe score (McCabe and Jackson, 1962). Chi-square and Fisher tests were used in the univariate analysis of categorical variables. Continuous variables were analyzed using two-sample t-test or Mann Whitney, as appropriate. A logistic regression analysis was performed to identify factors independently associated with pneumonia and bacteraemia. Differences were considered significant at p < 0.05. All statistical analyses were performed using SPSS v.16.0 (SPSS Inc., Chicago, IL).
Results
Study of the Gene Expression of the abaR and aidA Genes of the Quorum Network (QS/QQ)
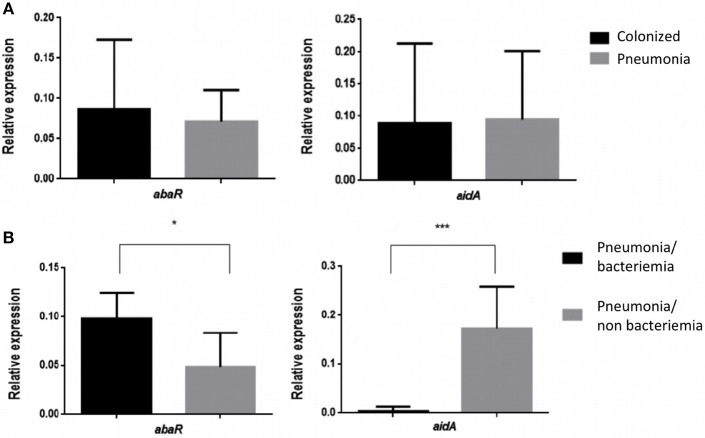
The Relative Expression (RE) of the abaR and aidA genes of the Quorum network (QS/QQ) was quantified by RT-qPCR analysis of the 17 isolates of A. baumannii from colonized patients and of the 13 isolates of A. baumannii from patients with pneumonia (Figure 1A). The mean values (of two biological replicates) are presented in Tables 2, 3. These values were first used to determine any significant differences between the two types of strains in terms of gene expression in the Quorum network.
Figure 1.
(A) Relative Expression of the abaR and aidA genes in strains of A. baumannii from patients colonized with A. baumannii and patients with pneumonia caused by A. baumannii. No significant differences (p > 0.05) were detected in either case. (B) Relative expression of the abaR and aidA genes in isolates of A. baumannii from patients with bacteraemic and non-bacteraemic pneumonia. *p-value < 0.05 and ***p-value < 0.001.
Table 2.
Results of RT-qPCR analysis of the Relative Expression (RE) of the abaR and aidA genes (Quorum network genes) in the A. baumannii isolates from colonized patients.
| Strain (MLSTa) | abaR (RE) | aidA (RE) |
|---|---|---|
| STRAINS OF A. baumannii ISOLATED FROM COLONIZED PATIENTS | ||
| Ab 22_GEIH-2010 (ST-52) | 0.043 | 0.042 |
| Ab 38_GEIH-2010 (ST-2) | 0.078 | 0.146 |
| Ab 59_GEIH-2010 (ST-269) | 0.081 | 0.003 |
| Ab 64_GEIH-2010 (ST-2) | 0.076 | 0.213 |
| Ab 77_GEIH-2010 (ST-261) | 0.018 | 0.112 |
| Ab 112_GEIH-2010 (ST-263) | 0.112 | 0.038 |
| Ab 141_GEIH-2010 (ST-264) | 0.001 | 0.010 |
| Ab 177_GEIH-2010 (ST-2) | 0.001 | 0.067 |
| Ab 205_GEIH-2010 (ST-2) | 0.131 | 0.126 |
| Ab 288_GEIH-2010 (ST-263) | 0.067 | 0.199 |
| Ab 290_GEIH-2010 (ST-264) | 0.141 | 0.006 |
| Ab 294_GEIH-2010 (ST-2) | 0.123 | 0.481 |
| Ab 326_GEIH-2010 (ST-2) | 0.123 | 0.015 |
| Ab 354_GEIH-2010 (ST79) | 0.052 | 0.020 |
| Ab 364_GEIH-2010 (ST-79) | 0.001 | 0.081 |
| Ab 399_GEIH-2010 (ST-79) | 0.061 | 0.050 |
| Ab 456_GEIH-2010 (ST-269) | 0.368 | 0.001 |
The results are expressed as the mean value of the two biological replicates.
MLST (Mutilocus Sequence Typing by Pasteur database, https://pubmlst.org/) (Villar et al., 2014). In bold, RE ≤ 0.001 not detected by RT-PCR.
Table 3.
Results of RT-qPCR analysis of the Relative Expression (RE) of the abaR and aidA genes (Quorum network genes) in the A. baumannii isolates from patients with bacteraemic pneumonia (Pn-B) or non-bacteraemic pneumonia (Pn-NB).
| A. baumannii strains from Pn-NB patients | A. baumannii strains from Pn-B patients | ||||
|---|---|---|---|---|---|
| Strain (MLSTa) | abaR (RE) | aidA (RE) | Strain (MLSTa) | abaR (RE) | aidA (RE) |
| Ab 8_GEIH-2010 (ST-2) | 0.035 | 0.108 | Ab 148_GEIH-2010 (ST-2) | 0.094 | 0.022 |
| 0.592* | 1.545* | 1.032* | 1.204* | ||
| 0.940** | 0.873** | 1.407** | 0.598** | ||
| Ab 73_GEIH-2010 (ST-2) | 0.036 | 0.361 | Ab 215_GEIH-2010 (ST-2) | 0.085 | 0.001 |
| 0.564* | 1.695* | 0.883* | 0.001* | ||
| 0.763** | 0.450** | 1.376** | 0.001** | ||
| Ab 125_GEIH-2010 (ST-257) | 0.047 | 0.156 | Ab 232_GEIH-2010 (ST-2) | 0.139 | 0.001 |
| 0.431* | 1.582* | 0.692* | 0.001* | ||
| 1.366** | 1.076** | 0.845** | 0.001** | ||
| Ab 157_GEIH-2010 (ST-2) | 0.001 | 0.172 | Ab 275_GEIH-2010 (ST-181) | 0.110 | 0.001 |
| 0.329* | 2.308* | 0.638* | 0.001* | ||
| 1.272** | 0.685** | 1.430** | 0.001** | ||
| Ab 240_GEIH-2010 (ST-2) | 0.078 | 0.150 | Ab 371_GEIH-2010 (ST-79) | 0.059 | 0.001 |
| 0.561* | 0.858* | 0.503* | 0.001* | ||
| 0.553** | 0.703** | 1.233** | 0.001** | ||
| Ab 268_GEIH-2010 (ST-181) | 0.034 | 0.139 | Ab 461_GEIH-2010 (ST-2) | 0.099 | 0.001 |
| 0.683* | 1.530* | 1.221* | 0.001* | ||
| 0.845** | 0.717** | 2.713** | 0.001** | ||
| Ab 276_GEIH-2010 (ST-181) | 0.108 | 0.126 | |||
| 0.553* | 1.007* | ||||
| 0.586** | 0.722** | ||||
The results are expressed as the mean values of the two biological replicates.
MLST (Mutilocus Sequence Typing by Pasteur database, https://pubmlst.org/) (Villar et al., 2014). In bold, RE ≤ 0.001, not detected by RT-PCR.
Results of RT-qPCR analysis of the Relative Expression (RE) of the abaR/aidA genes (Quorum network genes) in the presence of 3-Oxo-C12-HSL in strains of A. baumannii from patients with bacteraemic pneumonia (Pn-B) or non-bacteraemic pneumonia (Pn-NB).
Results of RT-qPCR analysis of the Relative Expression (RE) of the abaR/aidA genes (Quorum network genes) in the presence of H2O2 in strains of A. baumannii from patients with bacteraemic pneumonia (Pn-B) or non-bacteraemic pneumonia (Pn-NB).
The results did not reveal any significant differences in the RE of the Quorum network genes (abaR, aidA) between clinical strains of A. baumannii isolated from colonized patients and strains of A. baumannii isolated from patients with pneumonia (0.086/0.094 vs. 0.071/0.095, p > 0.05).
We then proceeded to study the RE of the abaR and aidA genes in strains of A. baumannii from patients with pneumonia, differentiating the strains isolated from patients with bacteraemia (Pn-B) from those isolated from patients without bacteraemia (Pn-NB). The resulting graphs are shown below (Figure 1B). The findings reveal significant differences in the expression of the abaR and aidA genes between clinical strains of A. baumannii from patients with bacteraemic pneumonia (Pn-B) and those with non-bacteraemic pneumonia (Pn-NB). We observed that abaR gene was overexpressed in A. baumannii isolates from Pn-B patients relative to Pn-NB patients (0.047 vs. 0.097, p < 0.05). By contrast, the aidA gene was overexpressed in A. baumannii clinical strains in Pn-NB patients relative to Pn-B patients (0.173 vs. 0.0045, p < 0.001) (Figure 1B). Only one strain, Ab 148_GEIH-2010 (ST-2), isolated from Pn-B patients, showed an aidA gene profile different from the other isolates of this group, although the RE of this gene was lower (0.022) than that of isolates from Pn-NB patients.
Study of abaR/aidA Genes (QS/QQ) Under Stress Conditions (3-Oxo-C12-HSL and H2O2)
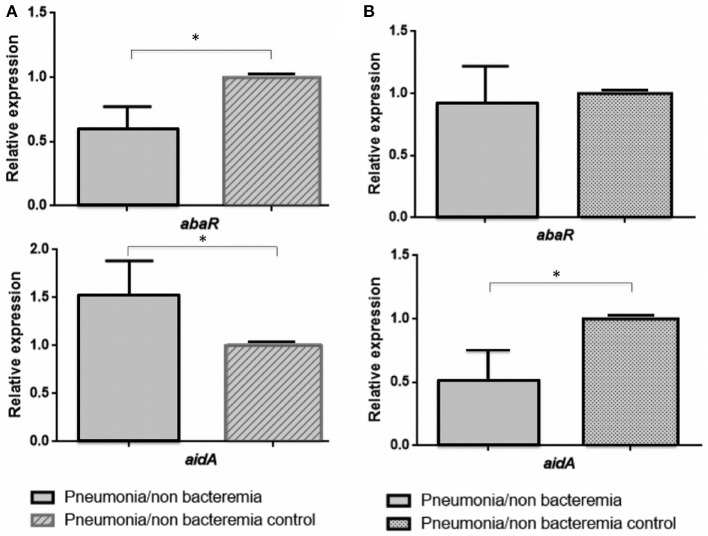
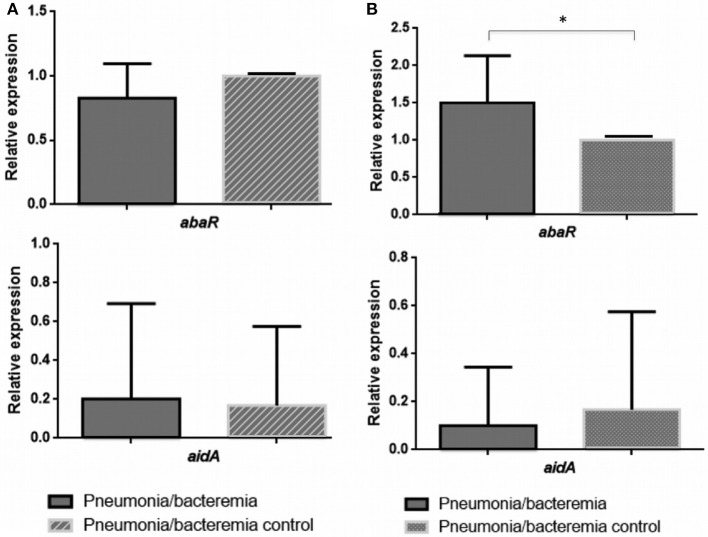
The values of the RE of the abaR and aidA genes (Quorum network) in the presence of 3-Oxo-C12-HSL (Inhibition of the QS) and H2O2 (Activation of the QS), obtained by RT-qPCR of the 13 isolates of A. baumannii from patients with pneumonia (differentiated from Pn-NB) are shown in Table 3, expressed as the mean value of the two biological replicates. These values were then analyzed to determine any significant differences in the RE of the abaR/aidA (QS/QQ) genes between the different clinical isolates (Figures 2, 3).
Figure 2.
Relative expression of the abaR and aidA genes under 3-oxo-C12-HSL (A) and H2O2 (B) in isolates of A. baumannii from patients with non-bacteraemic pneumonia (Pn-NB). *p-value < 0.05.
Figure 3.
Relative expression of the abaR and aidA genes under 3-oxo-C12-HSL (A) and H2O2 (B) in isolates of A. baumannii from patients with bacteraemic pneumonia (Pn-B). *p-value < 0.05.
In the clinical strains of A. baumannii isolated from Pn-NB (Figure 2), we observed regulation of expression of the aidA gene in the presence of 3-Oxo-C12-HSL (overexpression, RE ≥ 1.5) (Figure 2A) and of H2O2 [underexpression, RE ≤ 0.5 (Figure 2B)]. Expression of the abaR gene decreased significantly in the presence of the 3-Oxo-C12-HSL molecule (RE ≤ 0.5, Figure 2A).
In the clinical strains A. baumannii isolated from Pn-B (Figure 3), expression of the aidA gene was not regulated in the presence of 3-Oxo-C12-HSL or H2O2. However, the abaR gene was overexpressed in the presence of H2O2 (RE ≥ 1.5, Figure 3B).
These results indicate that the isolates of A. baumannii from Pn-NB may harbor a functional AidA protein (QQ enzyme), in contrast to the isolates of A. baumannii from Pn-B, which did not have this functional protein. Therefore, in the A. baumannii strains isolated from Pn-B, overexpression of the abaR gene (activation of the QS) in the presence of H2O2 (ROS response) would enable the development of the virulence factors favoring invasiveness, such as type VI secretion system (T6SS) and motility.
Quorum Network (QS/QQ) Genes and Clinical Variables
Analysis of the risk factors associated with the development of pneumonia vs. colonization by clinical strains of A. baumannii revealed only one statistically significant variable, i.e., diabetes mellitus (Table 4).
Table 4.
Univariate analysis of risk factors associated with development of pneumonia relative to colonization by clinical strains of A. baumannii.
| Variable | Colonized patients (n = 17) | Patients with pneumonia (n = 13) | P-value |
|---|---|---|---|
| Age, Med ± SEM | 55.06 ± 5.12 | 59.31 ± 5.90 | 0.590 |
| Female sex | 5 (29.41) | 6 (46.15) | 0.287 |
| Charlson score, Med ± SEM | 2.12 ± 0.67 | 2.46 ± 0.69 | 0.729 |
| Comorbity condition, no. (%) | |||
| McCabe score, ultimately or rapidly | 8 (47.06) | 5 (38.46) | 0.840 |
| Cancer | 1 (5.88) | 2 (15.38) | 0.397 |
| Diabetes | 3 (17.65) | 7 (53.85) | 0.045 |
| Cirrhosis | 0 (0) | 0 (0) | NA |
| AIDS | 0 (0) | 1 (7.69) | 0.433 |
| Chronic lung disease | 2 (11.67) | 4 (30.78) | 0.204 |
| CRF | 0 (0) | 1 (7.69) | 0.433 |
| Immunosuppression | 1 (5.88) | 2 (15.38) | 0.397 |
| Surgery, No. (%) | 5 (29.41) | 4 (30.78) | 0.623 |
| ICU stay, No. (%) | 15 (88.23) | 11 (84.61) | 0.591 |
| Tracheostomy | 4 (23.53) | 2 (15.38) | 0.469 |
| Mechanical ventilation | 10 (58.82) | 7 (53.85) | 0.538 |
| Death | 3 (17.65) | 3 (23.08) | 0.531 |
| abaR | 0.09 ± 0.02 | 0.07 ± 0.04 | 0.547 |
| aidA | 0.09 ± 0.03 | 0.09 ± 0.03 | 0.919 |
In bold and highlighted the variables that showed a p < 0.05.
However, analysis of the risk factors associated with the development of bacteraemia in pneumonia caused by A. baumannii revealed underexpression of the aidA gene as the only statistically significant variable (p < 0.05) (Table 5).
Table 5.
Univariate analysis of risk factors associated with the development of bacteraemia in pneumonia caused by A. baumannii relative to the non-bacteraemic pneumonia control.
| Variable | Pn-NB patients (N = 7) | Pn-B patients (N = 6) | P-value |
|---|---|---|---|
| Age, Med ± SEM | 58.43 ± 10.13 | 60.33 ± 6.08 | 0.880 |
| Female sex | 4 (57.14) | 2 (33.33) | 0.383 |
| Charlson score, Med ± SEM | 2.00 ± 0.79 | 3.20 ± 1.24 | 0.497 |
| Comorbity condition, no. (%) | |||
| McCabe score, ultimately or rapidly | 3 (42.86) | 2 (33.33) | 0.380 |
| Cancer | 1 (14.28) | 1 (20) | 0.731 |
| Diabetes | 2 (28.57) | 5 (83.33) | 0.078 |
| Cirrhosis | 0 (0) | 0 (0) | NA |
| AIDS | 0 (0) | 1 (20) | 0.462 |
| Chronic lung disease | 3 (42.86) | 1 (20) | 0.343 |
| CRF | 1 (14.28) | 0 (0) | 0.538 |
| Immunosuppression | 2 (28.57) | 0 (0) | 0.269 |
| Surgery, No. (%) | 3 (42.86) | 1 (20) | 0.343 |
| ICU stay, No. (%) | 5 (71.43) | 6 (100) | 0.269 |
| Tracheostomy | 0 (0) | 2 (33.33) | 0.192 |
| Mechanical ventilation | 3 (42.86) | 4 (66.67) | 0.383 |
| Severe sepsis and septic shock, No. (%) | 2 (28.57) | 3 (50) | 0.565 |
| Death | 3 (42.86) | 0 (0) | 0.122 |
| abaR | 0.78 ± 0.015 | 0.06 ± 0.02 | 0.522 |
| aidA | 0.15 ± 0.04 | 0.03 ± 0.03 | 0.045 |
In bold and highlighted the variables that showed a p < 0.05.
Mortality in the in vivo Galleria mellonella Model
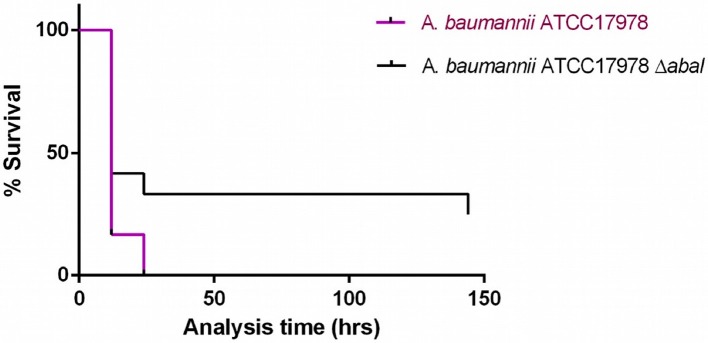
Injection of G. mellonella larvae with A. baumannii ATCC17978 at a concentration of 8 x 104 CFU/larva (± 0.5 log) caused 100% mortality after 24 h, whereas injection of the larvae with the same concentration of A. baumannii ATCC17978ΔabaI resulted in 70% mortality after 24 h (Figure 4; p < 0.05, Mantel-Cox analysis).
Figure 4.
Survival curves for G. mellonella larvae injected with A. baumannii ATCC17978 reference strain and its isogenic derivative A. baumannii ATCC17978 ΔabaI. Data from a single representative assay. For simplicity, the control group is not included in this figure.
Discussion
In this study, we analyzed the expression of Quorum network (QS/QQ) genes that differed between genomes of clinical isolates of A. baumannii, abaR and abaI (QS system) and aidA (QQ mechanism) in relation to clinical features of pneumonia and bacteraemia. Although other QQ enzymes have been described in A. baumannii ATCC 17978 (Mayer et al., 2018), these were not analyzed in the present study due to the lack of any differences between A. baumannii genomes.
In clinical strains of A. baumannii isolated from patients with bacteraemic pneumonia (Pn-B), the abaR gene was overexpressed (p < 0.05). The AbaR protein was the receptor activator of the Quorum Sensing system (QS), and the aidA gene was not expressed. Moreover, we observed regulation of aidA gene expression in clinical strains of pneumonia-causing A. baumannii (non-bacteraemic pneumonia, Pn-NB) by the 3-Oxo-C12-HSL molecule (which is an AidA enzyme substrate in QQ activity) and H2O2 (an activator of the QS system). However, there was no difference in the expression of Quorum network genes between colonized and pneumonia patients, as previously described (Stones and Krachler, 2016).
On the other hand, clinical analysis of the risk factors associated with pneumonia caused by A. baumannii revealed diabetes mellitus as only statistically significant risk factor (Kim et al., 2014). In relation to bacteraemia in A. baumannii pneumonia (P < 0.05), underexpression of the aidA gene was also the only statistically significant variable (P < 0.05).
In several pathogens, such as Yersinia pseudotuberculosis, Proteus mirabilis, and Vibrio cholerae, the QS system is the main regulatory mechanism of bacterial competence via T6SS, which is involved in the invasiveness and motility that favor the development of bacteraemia (Zhang et al., 2011; Debnath et al., 2018; Jaskólska et al., 2018; Trastoy et al., 2018). Moreover, in 86% of ICU patients, gastrointestinal tract colonization by a clinical strain of A. baumannii led to development of bacteraemia caused by genetically similar strains (Thom et al., 2010). This implies that clinical isolates of A. baumannii most capable of surviving under stress conditions (such as the presence of bile salts in the gastrointestinal tract or H2O2 in the respiratory tract) (Zheng et al., 2018) may have a higher invasive capacity due to virulence factors, such as the type VI secretion system (T6SS), previously activated under stressful conditions. Motility is also a crucial virulence factor, allowing penetration of the bacteria into the host's body and subsequent colonization (Gellatly and Hancock, 2013). Previous studies have demonstrated the existence of a relationship between motility and the origin of the isolates. Indeed, blood isolates of A. baumannii have been found to be more mobile than sputum isolates (Vijayakumar et al., 2016). Interestingly, 67% of the clinical isolates of A. baumannii were non-mobile and all of them had the AidA protein and were of respiratory origin (López et al., 2017a,b). In addition, the aidA gene was not located in the genome of the only mobile strain (clone ST79/PFGE-HUI-1) isolated from blood and which was the origin of a bacteraemic outbreak (López et al., 2017a,b).
Finally, multiple studies carried out with the abaI mutant of the M2 strain of Acinetobacter nosocomialis have analyzed the role of the abaI gene (responsible for the synthesis of quorum sensing synthesizing molecules) in various virulence factors such as biofilm formation and motility. In both cases, abaI deficiency led to a decrease in biofilm production and motility (Niu et al., 2008; Bhargava et al., 2012). The mutant lacking abaI is believed to be less virulent than the wild strain. This result was confirmed in our study in which injection of G. mellonella larvae with the reference A. baumannii ATCC17978 strain caused higher mortality than injection with the mutant A. baumannii ATCC17978ΔabaI. Regarding the mortality of the reference strain (A. baumannii ATCC17978), similar effects have been observed in other studies, in which injection of G. mellonella larvae with the reference strain A. baumannii ATCC17978 resulted in rapid death. Mortality was significantly dependent on the number of cells injected. More than 75% of the larvae died in the first 48 h of injection with at least 3.7 × 105 CFU / larva, while very few of the larvae died after being injected with a concentration equal to or lower than 3.7 × 104 CFU/larva (p < 0.01) (Clemmer et al., 2011). The results regarding the mutant A. baumannii ATCC17978ΔabaI are consistent with those obtained in a study of Pseudomonas aeruginosa (Steindler et al., 2009) in which a mutant ΔrhLI ΔlasI (QS systems homologous to abaI) was obtained, demonstrating that inactivation of both QS systems leads to a significant reduction in pathogenicity (p < 0.01) when virulence factors are not activated, such as the type VI secretion system (T6SS) and motility (Jaskólska et al., 2018).
In conclusion, our findings suggest that the QS (abaR and abaI genes)/QQ (aidA gene) network plays a role in the development of bacteraemia in patients with pneumonia caused by A. baumannii. This is the first study reporting a relationship between reduced expression of this bacterial QQ enzyme gene (AidA protein) and bacteraemia. Further studies of this relationship in the same and other bacterial QQ enzymes would be of great interest.
Author Contributions
LF-G, AA, LB, IB, ML and RA-M developed the experiments. FF-C, LM-M, JV, JR-B, JG-M, JMC, AP, JP, GB, and YS wrote the manuscript and provided the strains. MT led the experiments and manuscript redaction.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was funded by grant PI16/01163 awarded to MT within the State Plan for R+D+I 2013-2016 (National Plan for Scientific Research, Technological Development and Innovation 2008-2011) and co-financed by the ISCIII-Deputy General Directorate for Evaluation and Promotion of Research—European Regional Development Fund A way of Making Europe and Instituto de Salud Carlos III FEDER, Spanish Network for the Research in Infectious Diseases (REIPI, RD16/0016/0001, RD16/0016/0006, RD16/0016/0008, RD16/0016/0009, and RD16/0016/0010) and by the Study Group on Mechanisms of Action and Resistance to Antimicrobials, GEMARA (SEIMC, http://www.seimc.org/). MT was financially supported by the Miguel Servet Research Programme (SERGAS and ISCIII). LF-G was financially supported by a predoctoral fellowship from the Xunta de Galicia (GAIN, Axencia de Innovación).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.03105/full#supplementary-material
References
- Bhargava N., Sharma P., Capalash N. (2012). N-acyl homoserine lactone mediated interspecies interactions between A. baumannii and P. aeruginosa. Biofouling. 28, 813–822. 10.1080/08927014.2012.714372 [DOI] [PubMed] [Google Scholar]
- Bone R. C., Balk R. A., Cerra F. B., Dellinger R. P., Fein A. M., Knaus W. A., et al. (1992). Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest H101, 1644–1655. 10.1378/chest.101.6.1644 [DOI] [PubMed] [Google Scholar]
- Castañeda-Tamez P., Ramírez-Peris J., Pérez-Velázquez J., Kuttler C., Jalalimanesh A., Saucedo-Mora M. Á., et al. (2018). Pyocyanin restricts social cheating in Pseudomonas aeruginosa. Front. Microbiol. 9:1348. 10.3389/fmicb.2018.01348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlson M. E., Pompei P., Ales K. L., MacKenzie C. R. (1987). A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J. Chronic Dis. 40, 373–383. 10.1016/0021-9681(87)90171-8 [DOI] [PubMed] [Google Scholar]
- Cisneros J. M., Rodríguez-Baño J. (2002). Nosocomial bacteremia due to Acinetobacter baumannii: epidemiology, clinical features and treatment. Clin. Microbiol. Infect. 8, 687–693. 10.1046/j.1469-0691.2002.00487.x [DOI] [PubMed] [Google Scholar]
- Clemmer K. M., Bonomo R. A., Rather P. N. (2011). Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology 157, 2534–2544. 10.1099/mic.0.049791-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath I., Stringer A. M., Smith S. N., Bae E., Mobley H. L. T., Wade J. T., et al. (2018). MrpJ directly regulates proteus mirabilis virulence factors, including fimbriae and type VI secretion, during urinary tract infection. Infect Immun. 86, e00388–18. 10.1128/IAI.00388-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Mar Tomas M., Cartelle M., Pertega S., Beceiro A., Llinares P., Canle D., et al. (2005). Hospital outbreak caused by a carbapenem-resistant strain of Acinetobacter baumannii: patient prognosis and risk-factors for colonisation and infection. Clin. Microbiol. Infect. 11, 540–546. 10.1111/j.1469-0691.2005.01184.x [DOI] [PubMed] [Google Scholar]
- El Kettani A., Maaloum F., Diawara I., Katfy K., Harrar N., Zerouali K., et al. (2017). Prevalence of Acinetobacter baumannii bacteremia in intensive care units of Ibn Rochd University Hospital, Casablanca. Iran J Microbiol. 9, 318–323. [PMC free article] [PubMed] [Google Scholar]
- Fernández-Cuenca F., Tomás M., Caballero-Moyano F. J., Bou G., Martínez-Martínez L., Vila J., et al. (2015). Reduced susceptibility to biocides in Acinetobacter baumannii: association with resistance to antimicrobials, epidemiological behaviour, biological cost and effect on the expression of genes encoding porins and efflux pumps. J. Antimicrob. Chemother. 70, 3222–3229. 10.1093/jac/dkv262 [DOI] [PubMed] [Google Scholar]
- Fernández-Cuenca F., Tomás-Carmona M., Caballero-Moyano F., Bou G., Martínez-Martínez L., Vila J., et al. (2013). [In vitro activity of 18 antimicrobial agents against clinical isolates of Acinetobacter spp.: multicenter national study GEIH-REIPI-Ab 2010]. Enferm. Infecc. Microbiol. Clin. 31, 4–9. 10.1016/j.eimc.2012.06.010 [DOI] [PubMed] [Google Scholar]
- Fernández-García L., Fernandez-Cuenca F., Blasco L., Lopez-Rojas R., Ambroa A., Lopez M., et al. (2018). Relationship between tolerance and persistence mechanisms in Acinetobacter baumannii strains with AbkAB Toxin-Antitoxin system. Antimicrob Agents Chemother. 62, e00250–18. 10.1128/AAC.00250-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs R. P. (2016). Tolerance of lesions in E. coli: Chronological competition between Translesion Synthesis and Damage Avoidance. DNA Repair 44, 51–58. 10.1016/j.dnarep.2016.05.006 [DOI] [PubMed] [Google Scholar]
- Gayoso C. M., Mateos J., Méndez J. A., Fernández-Puente P., Rumbo C., Tomás M., et al. (2014). Molecular mechanisms involved in the response to desiccation stress and persistence in Acinetobacter baumannii. J. Proteome Res. 13, 460–476. 10.1021/pr400603f [DOI] [PubMed] [Google Scholar]
- Gellatly S. L., Hancock R. E. (2013) Pseudomonas aeruginosa: new insights into pathogenesis host defenses. Pathog. Dis. 67, 1590–173. 10.1111/2049-632x.12033 [DOI] [PubMed] [Google Scholar]
- Horan T. C., Andrus M., Dudeck M. A. (2008). CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am. J. Infect. Control. 36, 309–332. 10.1016/j.ajic.2008.03.002 [DOI] [PubMed] [Google Scholar]
- Jaskólska M., Stutzmann S., Stoudmann C., Blokesch M. (2018). QstR-dependent regulation of natural competence and type VI secretion in Vibrio cholerae. Nucleic Acids Res. 46, 10619–10634. 10.1093/nar/gky717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia V. C., Raju S. C., Purohit H. J. (2011). Genomic analysis reveals versatile organisms for quorum quenching enzymes: acyl-homoserine lactone-acylase and -lactonase. Open Microbiol. J. 5, 1–13. 10.2174/1874285801105010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T., Chong Y. P., Park S. Y., Jeon M. H., Choo E. J., Chung J. W., et al. (2014). Risk factors for hospital-acquired pneumonia caused by carbapenem-resistant Gram-negative bacteria in critically ill patients: a multicenter study in Korea. Diagn. Microbiol. Infect. Dis. 78, 457–461. 10.1016/j.diagmicrobio.2013.08.011 [DOI] [PubMed] [Google Scholar]
- Krysciak D., Schmeisser C., Preuss S., Riethausen J., Quitschau M., Grond S., et al. (2011). Involvement of multiple loci in quorum quenching of autoinducer I molecules in the nitrogen-fixing symbiont Rhizobium (Sinorhizobium) sp. strain NGR234. Appl. Environ. Microbiol. 77, 5089–5099. 10.1128/AEM.00112-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSarre B., Federle M. J. (2013). Exploiting quorum sensing to confuse bacterial pathogens. Microbiol. Mol. Biol. Rev. 77, 73–111. 10.1128/MMBR.00046-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. R., Lee J. H., Park M., Park K. S., Bae I. K., Kim Y. B., et al. (2017). Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front. Cell. Infect. Microbiol. 7:55. 10.3389/fcimb.2017.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López M., Blasco L., Gato E., Perez A., Fernandez-Garcia L., Martinez-Martinez L., et al. (2017a). Response to bile salts in clinical strains of Acinetobacter baumannii lacking the AdeABC efflux pump: virulence associated with quorum sensing. Front. Cell. Infect. Microbiol. 7:143. 10.3389/fcimb.2017.00143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López M., Mayer C., Fernandez-Garcia L., Blasco L., Muras A., Ruiz F. M., et al. (2017b). Quorum sensing network in clinical strains of A. baumannii: AidA is a new quorum quenching enzyme. PLoS ONE 12:e0174454. 10.1371/journal.pone.0174454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López M., Rueda A., Florido J. P., Blasco L., Fernandez-Garcia L., Trastoy R., et al. (2018). Evolution of the Quorum network and the mobilome (plasmids and bacteriophages) in clinical strains of Acinetobacter baumannii during a decade. Sci. Rep. 8:2523. 10.1038/s41598-018-20847-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C., Muras A, Romero M., López M., Tomás M., Otero A. (2018). Multiple quorum quenching enzymes are active in the nosocomial pathogen Acinetobacter baumannii ATCC17978. Front. Cell Infect. Microbiol. 8:310. 10.3389/fcimb.2018.00310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe W. R., Jackson G. G. (1962). Gram-negative bacteremia. I. Etiology and ecology. Arch Int Med. 110, 847–855. [Google Scholar]
- Mosqueda N., Gato E., Roca I., Lopez M., de Alegria C. R., Fernandez Cuenca F., et al. (2014). Characterization of plasmids carrying the blaOXA-24/40 carbapenemase gene and the genes encoding the AbkA/AbkB proteins of a toxin/antitoxin system. J. Antimicrob. Chemother. 69, 2629–2633. 10.1093/jac/dku179 [DOI] [PubMed] [Google Scholar]
- Niu C., Clemmer K. M., Bonomo R. A., Rather P. N. (2008). Isolation and characterization of an autoinducer synthase from Acinetobacter baumannii. J. Bacteriol. 190, 3386–3392. 10.1128/JB.01929-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg A. Y., Jara S., Monga D., Eliopoulos G. M., Moellering R. C., Mylonakis E. (2009). Galleria mellonella as a model system to study Acinetobacter baumannii pathogenesis and therapeutics. Antimicrob. Agents Chemother. 53, 2605–2609. 10.1128/AAC.01533-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbo C., Gato E., López M., Ruiz de Alegría C., Fernández-Cuenca F., Martínez-Martínez L., et al. (2013). Contribution of efflux pumps, porins, and β-lactamases to multidrug resistance in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 57, 5247–5257. 10.1128/AAC.00730-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbo C., Tomás M., Fernández Moreira E., Soares N. C., Carvajal M., Santillana E., et al. (2014). The Acinetobacter baumannii Omp33-36 porin is a virulence factor that induces apoptosis and modulates autophagy in human cells. Infect. Immun. 82, 4666–4680. 10.1128/IAI.02034-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Encinales V., Álvarez-Marín R., Pachón-Ibá-ez M. E., Fernández-Cuenca F., Pascual A., Garnacho-Montero J., et al. (2017). Overproduction of Outer Membrane Protein A by Acinetobacter baumannii as a risk factor for nosocomial pneumonia, bacteremia, and mortality rate increase. J. Infect. Dis. 215, 966–974. 10.1093/infdis/jix010 [DOI] [PubMed] [Google Scholar]
- Stacy D. M., Welsh M. A., Rather P. N., Blackwell H. E. (2012). Attenuation of quorum sensing in the pathogen Acinetobacter baumannii using non-native N-Acyl homoserine lactones. ACS Chem. Biol.. 7, 1719–1728. 10.1021/cb300351x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steindler L., Bertani I., De Sordi L., Schwager S., Eberl L., Venturi V. (2009). LasI/R and RhlI/R quorum sensing in a strain of Pseudomonas aeruginosa beneficial to plants. Appl. Environ. Microbiol.. 75, 5131–5140. 10.1128/AEM.02914-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stones D. H., Krachler A. M. (2016). Against the tide: the role of bacterial adhesion in host colonization. Biochem. Soc. Trans. 44, 1571–1580. 10.1042/BST20160186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis.. 18, 318–327. 10.1016/S1473-3099(17)30753-3 [DOI] [PubMed] [Google Scholar]
- Thom K. A., Hsiao W. W., Harris A. D., Stine O. C., Rasko D. A., Johnson J. K. (2010). Patients with Acinetobacter baumannii bloodstream infections are colonized in the gastrointestinal tract with identical strains. Am. J. Infect. Control. 38, 751–753. 10.1016/j.ajic.2010.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trastoy R., Manso T., Fernández-García L., Blasco L., Ambroa A., Pérez Del Molino M. L., et al. (2018). Mechanisms of bacterial tolerance and persistence in the gastrointestinal and respiratory environments. Clin. Microbiol. Rev. 31:e00023–18. 10.1128/CMR.00023-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayakumar S., Rajenderan S., Laishram S., Anandan S., Balaji V., Biswas I. (2016). Biofilm formation and motility depend on the nature of the Acinetobacter baumannii clinical isolates. Front Public Health. 4:105. 10.3389/fpubh.2016.00105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar M., Cano M. E., Gato E., Garnacho-Montero J., Miguel Cisneros J., Ruíz de Alegría C., et al. (2014). Epidemiologic and clinical impact of Acinetobacter baumannii colonization and infection: a reappraisal. Medicine 93, 202–210. 10.1097/MD.0000000000000036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong D., Nielsen T. B., Bonomo R. A., Pantapalangkoor P., Luna B., Spellberg B. (2017). Clinical and pathophysiological overview of acinetobacter infections: a century of challenges. Clin. Microbiol. Rev. 30, 409–447. 10.1128/CMR.00058-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Chen G., Hu L., Liu Y., Cheng J., Li H., et al. (2015). In vivo activity of daptomycin/colistin combination therapy in a Galleria mellonella model of Acinetobacter baumannii infection. Int. J. Antimicrob. Agents 45, 188–191. 10.1016/j.ijantimicag.2014.10.012 [DOI] [PubMed] [Google Scholar]
- Zhang W., Xu S., Li J., Shen X., Wang Y., Yuan Z. (2011). Modulation of a thermoregulated type VI secretion system by AHL-dependent quorum sensing in Yersinia pseudotuberculosis. Arch. Microbiol. 193, 351–363. 10.1007/s00203-011-0680-2 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Li Y., Long H., Zhao X., Jia K., Li J., et al. (2018). bifA regulates biofilm development of Pseudomonas putida MnB1 as a primary response to H2O2 and Mn2. Front. Microbiol. 9:1490. 10.3389/fmicb.2018.01490 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.