Abstract
The differential solvent extraction and further purification of fractions of endophytic Alternaria sp. isolated from Pinus ponderosa led to the isolation of further two perylenequinone compounds as 3,6,6a,9,10-pentahydroxy-7,8-epoxy-4-oxo-4,5,6,6a,6b,7,8,9-octahydroperylene (1) and 3,6,6a,7,10-pentahydroxy-4,9-dioxo-4,5,6,6a,6b,7,8,9-octahydroperylene (2). Structure of compounds 1–2 was determined on the basis of detailed spectroscopic analysis, as well as by comparison with literature reports. The antimicrobial, antileismanial, antimalarial and cytotoxic activities of compound 1 and 2 were evaluated.
Keyword: Natural product chemistry
1. Introduction
Perylenequinone natural products are generally dark coloured pigments isolated from diverse sources and have been used in different traditional herbal medicine through out the world. They comprise a family of natural products characterized by an oxidized pentacyclic core represented by parent perylenequinone. These vibrantly coloured natural products have the unique structural, biological and photochemical properties and have been extensively studied [1, 2]. In this article, we report further isolation of two perylenequinone compounds as 3,6,6a,9,10-pentahydroxy-7,8-epoxy-4-oxo-4,5,6,6a,6b,7,8,9-octahydroperylene (1) and 3,6,6a,7,10-pentahydroxy-4,9-dioxo-4,5,6,6a,6b,7,8,9-octahydroperylene (2) from endophytic Alternaria sp. isolated from Pinus ponderosa.
The compounds 1 (7.5 mg) and 2 (6.5 mg) were isolated from dichloromethane and ethylacetate extracts of the endophytic fungus (Alternaria sp.) respectively. Dichloromethane (200 mg) and ethylacetate (300 mg) residues were fractionated on open column chromatography [CHCl3, CHCl3-Methanol and MeOH, (silica gel 60–120, 70–230 mesh, Merck)], a solid phase extraction (SPE) (cartridge C-18 under vacuum), preparative TLC [CHCl3-MeOH (98:02)] and on preparative HPLC (Waters® Atlantis® 5μ, C18 T3, 250 × 19 mm column at a flow rate of 1 ml/min). Isolation of compounds was scaled up on preparative HPLC using a flow rate of 15 ml/min. The solvent system was first developed at analytical HPLC and the same was adopted on preparative HPLC. The isolated compounds 1 and 2 were evaluated for antileismanial, antimicrobial, antimalarial and cytotoxic activities.
2. Material and methods
2.1. General
Optical rotations were recorded using a Rudolph Research Analytical Autopol V polarimeter. UV was obtained using a Perkin-Elmer Lambda 3B UV/Vis spectrophotometer. 1H and 13C spectra were recorded on Bruker model AMX 500 NMR spectrometer with standard plus sequences, operating at 500 MHz in 1H- and 125 MHz in 13C NMR respectively. The chemical shift (δ) values were reported in parts per million units (ppm) from trimethylsilane (TMS) using known NMR solvent chemical shifts and coupling constants (J) were recorded in Hertz (Hz). Standard pulse sequences were used for COSY, DEPT, HMQC, and HMBC. A Micromass Q-Tof Micro mass spectrometer with a lock spray source was used to measure high resolution mass spectra. The experimental CD data was measured at 5 °C by Olis CD Spectrophotometer. Sephadex LH-20 (Mitsubishi Kagaku, Tokyo, Japan) and silica gel (70–230 mesh, Merck) were used for column chromatography. Fractions from column chromatography were monitored using TLC, silica gel F254, preparative TLC was carried out on silica gel 60 PF254 + 60366 plates (20 × 20 cm, 1 mm thick). Visualization of the TLC plates was achieved with a UV lamp (λ = 254 and 365 nm) and p-anisaldehyde/acid spray reagent (methanol-acetic acid-anisaldehyde-sulfuric acid, 85:9:1:5). Chemicals used were from Sigma-Aldrich (Poole, Dorset, U.K.), HPLC solvents were HPLC grades and were purchased from Sigma-Aldrich.
2.2. Preparation of endophytic fungus and its cultivation
DC401 (Alternaria sp.) was inoculated onto potato dextrose agar (PDA) at room temperature (∼25–30 °C) for two weeks with daily growth monitoring, followed by inoculation onto potato dextrose broth with shaking at 160 rpm for 14 days at 30 °C. After 14th day, the cultures were first filtered through sterile cotton using vacuum filtration; activated Dianion HP-20 was added to each flask and was returned to the shakers for another day. HP-20 was collected and washed with methanol then acetone. Methanol and acetone were combined and evaporated to dryness using a Buchi Rotavapor R-200. After evaporating all organic solvent, the remaining residue contained some portion of water (liquid-liquid extraction). The fungi strain (DC401) was grown on Potato Dextrose Agar (PDA) plates at a room temperature ∼28 °C for 14 days with daily growth monitoring. Plates were kept in refrigerator and used when needed. For small scale extractions, the fungi were grown on small scale using Potato Dextrose Broth (PDB, Sigma-Aldrich Chemie). PDB media was prepared by dissolving 24 g of PDB in 1L distilled water and autoclaved. 50 ml of PDB was placed in 125 ml conical flasks and incubated with small pieces of actively growing mycelium. The cultures were incubated at 30 °C under orbital agitation (160 rpm) for 14 days. After incubation, the contents of the flasks were filtered through sterile cotton using vacuum filtration and then the filtrates were extracted exhaustively with ethylacetate. The organic phase was vacuum concentrated to obtain the extracts. The extracts were sent to biological assays. For large scale extractions, 1L of PDB placed in 2800 ml Fernbach flasks were autoclaved and incubated with small pieces of actively growing mycelium. The cultures were incubated at 30 °C under orbital agitation (160 rpm) for 14 days. After incubation, the contents of the flasks were filtered through sterile cotton using vacuum filtration. After filtration 100 g activated ion exchange resin (Diaion-HP20) was added to each 1L of filtrates. It was returned to shakers and left overnight. After that it was filtrated and HP20 was washed with water, methanol, and then acetone. Methanol and acetone were combined and dried under vacuum. The residue was dissolved in water and extracted with hexane, dichloromethane, and ethylacetate respectively.
2.3. Extraction and isolation
Compounds 1 (7.5 mg) and 2 (6.5 mg) were isolated from dichloromethane and ethylacetate fractions respectively. Dichloromethane (200 mg) fraction was fractioned using a solid phase extraction (SPE) cartridge C-18 under vacuum. Four fractions were obtained (F1-F4). Fraction 4 (F4) was subjected to open column chromatography using chloroform, CHCl3-Methanol and MeOH, (silica gel 60, 70–230 mesh, Merck), which yielded several sub0fractions (F4a-d). Fraction F4d was chromatographed using preparative TLC with CHCl3-MeOH (90:10) three runs, affording compounds 1 (7.5 mg). The ethylacetate (300 mg) fraction was chromatographed using C-18 SPE cartridge under negative pressure, giving twelve fractions (F1-F12). Fractions 8, 9, and 10 were combined and purified by HPLC to obtain compound 2 (6.5 mg) again using a Waters® Atlantis® 5μ, C18 T3, 250 × 4.6 mm column an isocratic for 5 min 100% water + 0.1% formic acid (FA), then a gradient from 100% water + 0.1% FA to 100% MeCN + 0.1% FA at a flow rate of 1.5 ml/min over another 75 min time. It was scaled up on preparative HPLC using a Waters® Atlantis® 5μ, C18 T3, 250 × 19 mm column at a flow rate of 25.5 ml/min with the same solvent system that was used on analytical HPLC method.
2.3.1. 3,6,6a,9,10-pentahydroxy-7,8-epoxy-4-oxo-4,5,6,6a,6b,7,8,9-octahydroperylene (1)
Dark pigment (7.5 mg); [α]D20 - 22.7 (c 0.10, CHCl3); IR νmax KBr: 3415 (-OH), 2913 (Ar-H), 1718 (-C=O), 1688 (C=C), 1350 (-C-O) cm−1; HRESIMS: m/z 369.0954 [M + H]+, (calcd. for C20H10O7); 1H- (CDCl3-d1, 500 MHz) and 13C NMR (CDCl3-d1, 125 MHz) data, see Table 1.
Table 1.
1H- (500 MHz, CDCl3-d1), 13C-NMR (125 MHz, CDCl3-d1) of 1 and 2.
| Position |
1 |
2 |
||
|---|---|---|---|---|
| δC | δH | δC | δH | |
| 1 | 134.1 d | 7.80 (d, 8.8) | 134.8 d | 8.04 (d, 8.8) |
| 2 | 120.3 d | 7.09 (d, 8.8) | 122.7 d | 7.01 (d, 8.8) |
| 3 | 163.1 s | 162.1 s | ||
| 3a | 115.3 s | 114.2 s | ||
| 3b | 136.8 s | 135.4 s | ||
| 4 | 208.8 s | 206.8 s | ||
| 5 | 37.2 t | 3.43 (dd, 16.2, 1.9) | 34.3 t | 3.43 (dd, 16.0, 1.8) |
| 3.13 (dd, 16.2, 1.9) | 3.13 (dd, 16.0, 1.8) | |||
| 6 | 79.1 d | 4.65 (m) | 79.2 d | 4.74 (m) |
| 6a | 71.4 s | 69.2 s | ||
| 6b | 48.2 d | 3.50 (d, 9.5) | 46.2 d | 3.51 (d, 9.5) |
| 7 | 51.2 d | 3.86 (d, 3.5) | 69.8 d | 4.33 (d, 3.5) |
| 8 | 56.1 d | 3.56 (d, 3.5) | 42.1 t | 3.13 (dd, 16.0, 1.2) |
| 3.37 (dd, 16.0, 1.2) | ||||
| 9 | 62.8 d | 5.27 (d, 3.2) | 203.0 s | |
| 9a | 117.6 s | 119.3 s | ||
| 9b | 139.0 s | 139.4 s | ||
| 10 | 157.9 s | 162.3 s | ||
| 11 | 117.9 d | 6.86 (d, 8.8) | 112.3 d | 6.89 (d, 8.5) |
| 12 | 125.8 d | 7.80 (d, 8.8) | 132.6 d | 7.79 (d, 8.5) |
| 12a | 125.0 s | 125.8 s | ||
| 12b | 124.2 s | 125.6 s | ||
Chemical shifts are in ppm, J in parentheses are in Hertz.
2.3.2. 3,6,6a,7,10-pentahydroxy-4,9-dioxo-4,5,6,6a,6b,7,8,9-octahydroperylene (2)
Dark pigment (6.5 mg); [α]D20 - 19.3 (c 0.10, CHCl3); IR νmax KBr: 3425 (-OH), 1722 (-C=O), 2924 (Ar-H) cm−1; HRESIMS: m/z 369.0878 [M + H]+, (calcd. for C20H10O7); 1H- (CDCl3-d1, 500 MHz) and 13C NMR (CDCl3-d1, 125 MHz) data, see Table 1.
2.4. Biological assay
2.4.1. Antimicrobial assay
All organisms used for the antimicrobial evaluations of the metabolites from endophytic fungi were obtained from the American Type Culture Collection (Manassas, VA) and include the fungi Candida albicans ATCC 90028, Candida glabrata ATCC 90030, Candida krusei ATCC 6258, Cryptococcus neoformans ATCC 90133, and Aspergillus fumigates ATCC 204305 and the bacteria Staphyococcus aureus ATCC 29213, methicillin-resistant S. aureus ATCC 33591 (MRS), Escherichia coli ATCC 35218, Pseudomonas aeruginosa ATCC 27853, and Mycobacterium intracellulare ATCC 23068. Susceptibility testing is carried out using a modified version of National Committee for Clinical Laboratory Standards (NCCLS) methods. M. intracellulare was tested using a modified method [3]. Samples tested were serially-diluted in 20% DMSO/saline and transferred in duplicate to 96 well microplates. The microbial subcultures were prepared and diluted in broth to reach the desired colony forming unites (CFU) per milliliter. The samples were added to the microbial inocula to achieve a final volume of 200 μl and test concentration of 50 μg/ml for crude extracts and 20 μg/ml for pure compounds. Controls, growth (saline only), solvent, and blank (media only), were included on each test plate. Amphotericin B (ICN Biomedicals, Ohio) for fungi and ciprofloxacin (ICN Biomedicals, Ohio) for bacteria were used as positive controls in each assay. Plates are read using either optical density at 630 nm using the EL-340 Biokinetics Reader (Bio-Tek Instruments, Vermont) or fluorescence at 544ex/590em (M. intracellulare, A. fumigates) using the Polarstar Galaxy Plate Reader (BMG LabTechnologies, Germany). Percent of growth was calculated and plotted versus the test concentration to produce the IC50 which is the concentration of the sample that inhibits 50% growth of the organism. The minimum inhibitory concentration (MIC) is the lowest test concentration that does not allow any detectable growth. It is determined only for pure compounds.
2.4.2. Antileishmanial assay
The antileishmanial activity of the crude extracts/fractions/or pure compounds was measured in-vitro against a culture of Leishmania donovani promastigotes. Compounds tested were added to the Leishmania promastigotes or the axenic amastigotes cultures containing 2 × 106 cells/ml. The plates were incubated at 26 °C for 72 hours and the growth of Leishmania promastigotes/axenic amastigotes was determined using AlamarBlue® Assay. Pentamidine and amphotericin B were used as the positive controls. IC50 and IC90 values were computed from the dose-response curves.
2.4.3. Antimalarial assay
Antimalarial activity was determined in-vitro against chloroquine sensitive (D6, Sierra Leone) and resistant (W2, Indo China) strains of Plasmodium falciparum. The assay is based on evaluation of the effect of the test compounds on the growth of cultures of Plasmodium falciparum and determined by the assay of plasmodial lactate dehydrogenase (LDH) activity. The test compounds were dissolved in DMSO (2 mg/ml). A 200 μl suspension of Plasmodium falciparumculture (2% parasitemia and 2% hematocrit in RPMI 1640 medium supplemented with 10% human serum and 60 μg/mL amikacin) was added to the wells of a 96-well plate containing 10 μL of serially diluted samples. The plates were put in a humidified chamber and flushed with a gas mixture of 90% N2, 5% O2, and 5% CO2 and incubated at 37 °C for 72 hours. Plasmodial LDH activity was determined by using Malstat™ reagent (Flow Inc., Portland, OR). Briefly, 100 μL of the Malstat reagent was added to 20 μL of the incubation mixture and incubated for 30 min. After that, 20 μL of a 1:1 mixture of NBT/PES (Sigma, St. Louis, MO) was added and the plate was further incubated for 1 h in dark. The reaction was stopped by adding 100 μL of a 5% acetic acid solution. The plate was measured at 650 nm using the EL-340 Biokinetics Reader (Bio-Tek Instruments, Vermont). The primary screening of the crude extracts/fractions was run at a single concentration 15 μg/ml only on chloroquine sensitive (D6) strain of P. falciparum. The extracts showing ≥50% inhibition in the growth of the parasite were tested at three concentrations in the secondary screening. Chloroquine and artemisinin were included in each assay as the positive controls while DMSO (0.25%) was used as the negative control. IC50 values were obtained from the dose-response curves generated by plotting percent growth versus concentrations of tested compounds.
2.4.4. In-vitro cytotoxicity assay
The cytotoxicity of compounds was determined against mammalian kidney fibroblasts (VERO cells) obtained from American Type Culture Collection (ATCC, Rockville, MD). The assay was carried out in 96-well tissue culture-treated microplates. Cells were seeded in the wells of a 96-well plate at a density of 25,000 cells/well and incubated for 24 h. Samples were added at different concentrations to the wells and plates were again incubated for 48 h. The number of viable cells was measured using Neutral Red according to a modification of the procedure of Borenfreund and Babich. IC50 values were measured from dose response curves. Doxorubicin was chosen as a positive control and DMSO was used as a negative control.
3. Results and discussion
3.1. Structure identification
As a part of our programme to work out phytochemically on endophytic fungi, for a continuous chase for bioactive as well as structurally different molecules, we further fractionated the fungus (Alternaria sp.) and isolated two new perylenequinones 1 and 2.
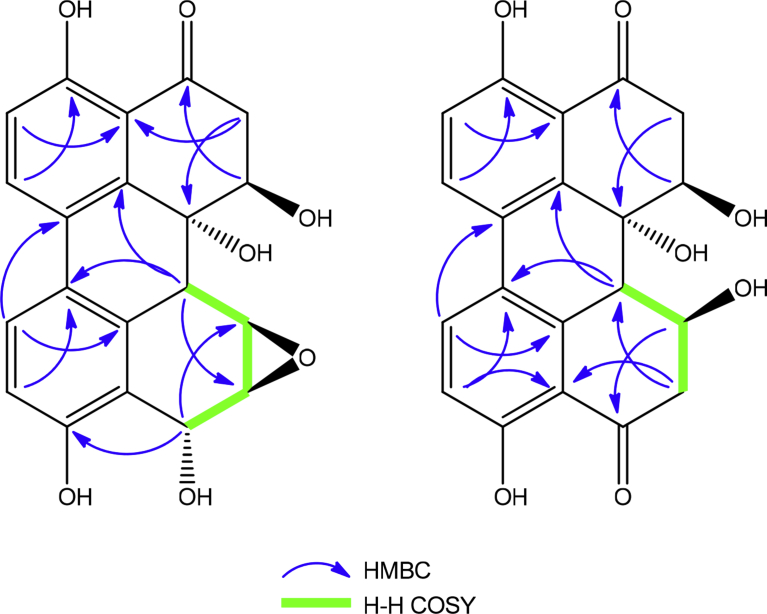
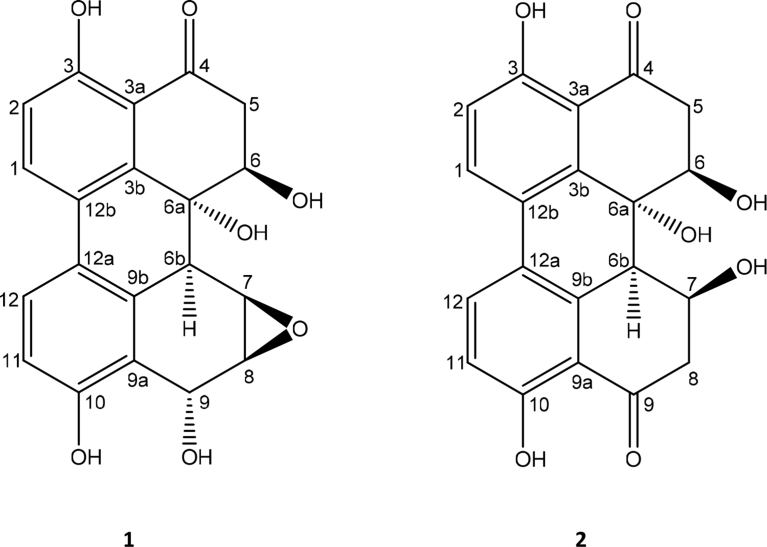
The compound 1 depicted molecular ion peak at 369.0954 [M + H]+ in the positive ESI-MS. The molecule was again analyzed as through the fragments A - D illustrated in our already published article [4]. The 13C NMR showed 20 resonances corresponding to the 20 carbons, which in DEPT spectra further revealed the presence of one methylene at δC 37.2. Both 1H and 13C NMR showed the peaks in shielded and deshielded region of the NMR spectra, revealed the presence of aliphatic and aromatic protons. The signals at δH/δC 3.43, 3.12/37.2 were assigned to methylene group protons flanked by carbonyl and carbinolic carbons. The signals at δH/δC 4.65/79.1, 71.4 and 5.27/62.8 were assigned to carbinolic carbon protons of H-6, H-6a and H-9 respectively. The resonances at δH/δC 3.86/51.2, 3.56/56.1 were assigned to epoxy protons. The signals at δC 163.1 and 157.9 were assigned to phenolic carbons. The 1H NMR doublets at δH 7.80, 7.09, 6.86 and 7.80 were assigned to aromatic protons at H-1, H-2, H-11 and H-12 respectively. Based on the above discussion structure 1 was assigned to compound 1. The positions of carbonyl, hydroxyl and epoxide were supported by HMBC, HMQC and H-H COSY 2D NMR experiments (Fig. 2). The compound 1 was finally elucidated to be 3,6,6a,9,10-pentahydroxy-7,8-epoxy-4-oxo-4,5,6,6a,6b,7,8,9-octahydroperylene (Fig. 1).
Fig. 2.
H-H COSY and HMBC of Compounds 1–2 (For better interpretation, the reader is referred to web version of the figure).
Fig. 1.
Structure of Compounds 1–2.
The compound 2 showed the molecular ion peak at 369.0878 [M + H]+ in agreement with the molecular formula C20H10O7. The 13C NMR spectra depicted C-20 carbon skeleton, which is almost similar with that of compound 1, except the presence of additional carbonyl signal at δC 203.0, absence of epoxide, presence of two methylene (CH2) in the DEPT spectra. The absence of epoxide and the presence of additional methylene with that of 1, indicates opening of epoxide ring, which lead to the conversion/formation of –C-OH and –CH2, which showed the resonances at δC/δH 69.8/4.33 and 42.1/3.13, 3.37 respectively. On an whole the northern part of compound 2 is exactly same as that of 1, but in southern part C-7/C-8 epoxide is reduced to -C-OH (C-7) and –CH2 (C-8) and an oxidized carbonyl at C-9 (δC 203.0). These assignments were further supported by HMBC and HMBC NMR experiments (Fig. 2). Based on this compound 2 was elucidated to be 3,6,6a,7,10-pentahydroxy-4,9-dioxo-4,5,6,6a,6b,7,8,9-octahydroperylene (Fig. 1).
3.2. Biological evaluation
Compound 1 showed antileishmanial activity against Leishmania donovani with IC50 = 2.55 μg/ml and IC90 = 8.28 μg/ml. Compound 2 showed antileishmanial activity against Leishmania donovani with IC50 = 4.40 μg/ml and IC90 = 9.54 μg/ml and antimalarial activity against both the chloroquine sensitive (D6) and chloroquine resistant (W2) clones of P. falciparum with IC50 = 4.24 μg/ml and 3.65 μg/ml respectively and cytotoxicity activity against mammalian kidney fibroblasts (VERO cells) with IC50 = 3.59 μg/ml.
3.3. Plausible biosynthesis of perylenequinones and subsequent isomerisation
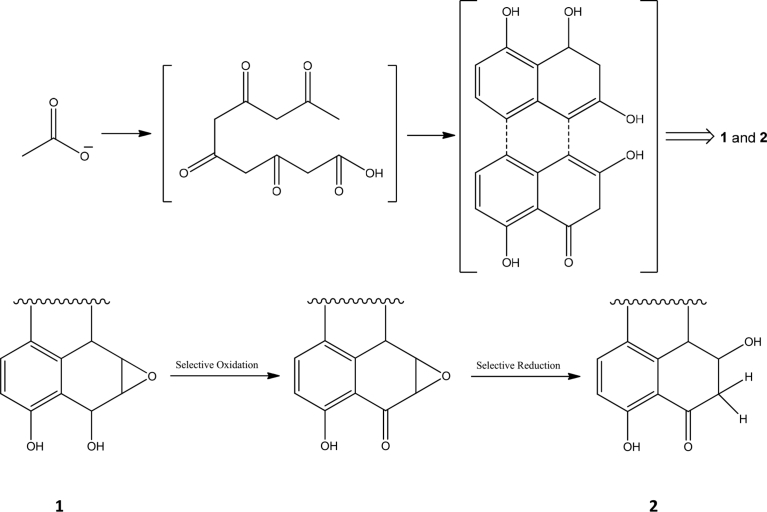
Compounds 1 and 2 are examples of reduced perylenequinones so far identified in fungi. The biosynthesis of these compounds occurs most probably via oxidative coupling of two molecules of a tetralone derivative synthesized from a pentaketide derivative by head to head coupling, followed by reduction and hydroxylation in different positions. The plausible biosynthetic pathway of reduced perylenequinones 1 and 2 and possible isomerization is shown in figure (Fig. 3).
Fig. 3.
Figure depicting plausible biosynthetic pathway of perylenequinones 1 and 2 and probable biogenetic isomerization of compound 1 to 2.
Declarations
Author contribution statement
Mudasir Tantry, Ahmed Idris, John S. Williamson, Tasfi Shafi, Jehangir S. Dar, Tauseef A. Malik, Bashir A. Ganai, Abdul S. Shawl: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
Mudasir Tantry was supported by CSIR-New Delhi (grant IA-27401).
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- 1.Mulrooney C.A., O'Brien E.M., Morgan B.J., Kozlowski M.C. Perylenequinones: isolation, synthesis and biological activity. Eur. J. Org. Chem. 2012;21:3887–3904. doi: 10.1002/ejoc.201200184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mulrooney C.A., Morgan B.J., Li X., Kozlowski M.C. Perylenequinone natural products: enantioselective synthesis of the oxidized pentacyclic core. J. Org. Chem. 2010;75:16–29. doi: 10.1021/jo9013832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franzblau S.G., Witzig R.S., McLaughlin J.C., Torres P., Madico G., Hernandez A., Degnan M.T., Ferguson R.M., Gilman R.H. Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis isolates by using the microplate alamar blue assay. J. Clin. Microbiol. 1998;36:362–366. doi: 10.1128/jcm.36.2.362-366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Idris A., Tantry M.A., Ganai B.A., Kamili A.N., Williamson J.S. Reduced perylenequinone derivatives from an endophytic Alternaria sp. isolated from Pinus ponderosa. Phytochem. Lett. 2015;11:264–269. [Google Scholar]