Abstract
Background
Genomic aberrations have been identified in metastatic castration-resistant prostate cancer (mCRPC), but molecular predictors of resistance to abiraterone acetate/prednisone (AA/P) treatment are not known.
Patients and methods
In a prospective clinical trial, mCRPC patients underwent whole-exome sequencing (n = 82) and RNA sequencing (n = 75) of metastatic biopsies before initiating AA/P with the objective of identifying genomic alterations associated with resistance to AA/P. Primary resistance was determined at 12 weeks of treatment using criteria for progression that included serum prostate-specific antigen measurement, bone and computerized tomography imaging and symptom assessments. Acquired resistance was determined using the end point of time to treatment change (TTTC), defined as time from enrollment until change in treatment from progressive disease. Associations of genomic and transcriptomic alterations with primary resistance were determined using logistic regression, Fisher‘s exact test, single and multivariate analyses. Cox regression models were utilized for determining association of genomic and transcriptomic alterations with TTTC.
Results
At 12 weeks, 32 patients in the cohort had progressed (nonresponders). Median study follow-up was 32.1 months by which time 58 patients had switched treatments due to progression. Median TTTC was 10.1 months (interquartile range: 4.4–24.1). Genes in the Wnt/β-catenin pathway were more frequently mutated and negative regulators of Wnt/β-catenin signaling were more frequently deleted or displayed reduced mRNA expression in nonresponders. Additionally, mRNA expression of cell cycle regulatory genes was increased in nonresponders. In multivariate models, increased cell cycle proliferation scores (≥ 50) were associated with shorter TTTC (hazard ratio = 2.11, 95% confidence interval: 1.17–3.80; P = 0.01).
Conclusions
Wnt/β-catenin pathway activation and increased cell cycle progression scores can serve as molecular markers for predicting resistance to AA/P therapy.
Keywords: castrate-resistance prostate cancer, abiraterone acetate, Wnt/β-catenin signaling
Key Message
Genome- and transcriptome-wide analyses of metastases from castration-resistance prostate cancer (CRPC) patients revealed molecular markers of resistance to abiraterone acetate/prednisone. These observations enhance the development of predictive biomarker-based strategies for managing patients with advanced CRPC stage.
Introduction
Progression to lethal metastatic castration-resistant prostate cancer (mCPRC) is observed in almost all hormone sensitive prostate cancer patients treated with androgen deprivation therapy (ADT). The emergence of mCRPC has been attributed to several mechanisms including intracrine biosynthesis of androgens, expression of androgen receptor (AR) variants, activation of AR target genes by alternative steroid receptors, and evolution to an AR-null neuroendocrine phenotype [1–4]. Intracrine biosynthesis of androgens can be inhibited by blocking cytochrome P450 17A1 (CYP17A1) activity with abiraterone acetate/prednisone (AA/P). AA/P slows disease progression, but many patients fail to respond suggesting primary resistance [5]. Although prospective and retrospective whole exome and transcriptome analyses have identified a number of recurrent genomic and gene expression alterations in primary and metastatic CRPC stage [2, 6–9], associations with primary or acquired AA/P resistance is unknown. We conducted a prospective study (https://clinicaltrials.gov/identifier NCT #01953640) with the aim to identify genomic alterations in metastases associated with primary and acquired resistance to AA/P.
Methods
The ‘PROMOTE’ (Prostate Cancer Medically Optimized Genome-Enhanced Therapy) study was initiated in May 2013 after approval by the Mayo Clinic Institutional Review Board (IRB). All enrolled patients provided written informed consent to undergo two serial metastatic tissue biopsies with the first carried out before initiation of AA/P and the second after 12 weeks of treatment. Patients had to experience progression on continuous ADT at the time of study enrollment. Full eligibility criteria are available in an abbreviated study protocol document under supplementary attachments, ‘Promote Study Protocol’. The primary goal of the study was to determine genomic alterations associated with prechemotherapy AA/P treatment resistance.
Sequencing and genomic aberration analysis methods
Whole-exome sequencing (WES) was carried out on Illumina HiSeq 2500 and whole transcriptome sequencing (RNA-seq) was carried out on Illumina HiSeq 2000. Details of sequencing methods, copy number variation analysis, somatic mutation, and RNA-seq analysis along with gene pathway scores and analyses are provided under supplementary Methods, available at Annals of Oncology online.
Statistical methods
Patients showing progression after 12 weeks of AA/P treatments were categorized as ‘nonresponders’. Definitions of responders and nonresponders and statistical tests used to compare the two groups are provided in supplementary Methods, available at Annals of Oncology online. Fisher‘s exact test was used to evaluate the association between somatic aberrations and 12-week treatment response. Cox regression models were used to assess association of gene-based pathways of AR, cell cycle progression (CCP), neuroendocrine prostate cancer (NEPC), Wnt inhibitor (25-gene panel), Wnt inhibitor (4-gene panel), and Wnt activator scores (analyzed as continuous variables) with lack of response at 12 weeks for primary resistance and with time to treatment change (TTTC) for acquired resistance.
Results
Patient and sequencing results
Between May 2013 and September 2015, 92 of 110 patients targeted for enrollment were accrued in this prospective trial of patients initiating prechemotherapy AA/P. Patient accrual was halted in September 2015, as a result of competing drug options in this stage approved after the trial began which slowed accrual. Metastatic biopsy sites included bone, lymph nodes, and soft tissues (Figure 1A). Tumor nucleic acid yield and purity of lymph nodal versus skeletal biopsies are shown in supplementary Figure S1, available at Annals of Oncology online. Of the 92 patients, 86 had analyzable RNA-seq or WES data. The biopsy sites and 12-week outcomes for these patients with analyzable RNA-seq or WES data are shown in Figure 1B. Clinical and demographic characteristics of the cohort are listed in ‘Study Cohort Demographics Table’ under supplementary Results, available at Annals of Oncology online. Clinical and sequencing statistics for WES and RNA-seq data with response assessment at 12 weeks are described in supplementary Tables S1 and S2, available at Annals of Oncology online, respectively.
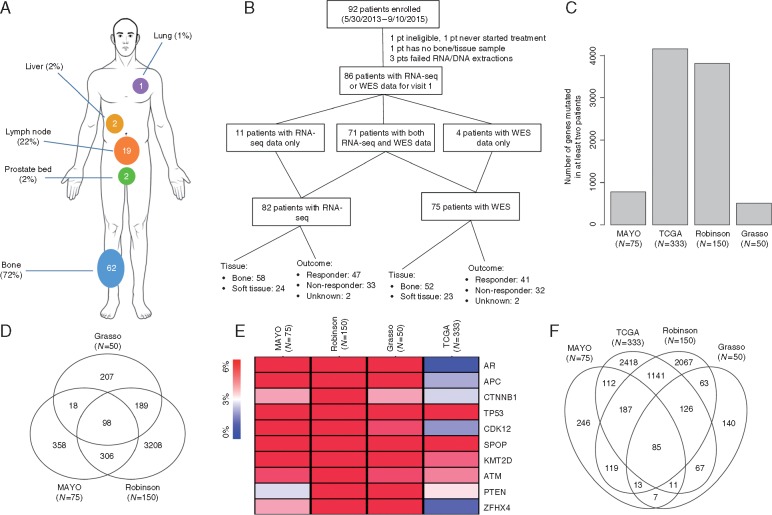
Figure 1.
(A) Biopsy sites of 86 patients participating this study. (B) CONSORT flow diagram of patients involved in this study. Of the 11 patients who only had RNA-seq available, 8 failed exome extraction or sequencing library preparation, 1 had exome sample contamination, and 2 did not have data available at the time of data freeze. Of the four patients who only had WES data available, two failed RNA extraction or sequencing library preparation and two had no RNA bone/tissue sample available. (C) Histogram showing the number of genes mutated in at least two patients in each of the four cohorts including Mayo’s CRPC cohort (N = 75), Robinson’s CPRC cohort (N = 150), Grasso’s CPRC cohort (N = 50), and TCGA’s localized, hormone-naive prostate cancer cohort (N = 333 specimens). (D) Venn diagram showing the overlap in genes mutated in at least two patients between the three CRPC cohorts including Mayo’s CRPC cohort (N = 75), Robinson’s CPRC cohort (N = 150), Grasso’s CPRC cohort (N = 50). (E) Heat map comparing gene level mutation frequencies between the three CPRC cohorts and the TCGA cohort. (F) Venn diagram showing the overlap in genes mutated in at least two patients between all four datasets. CPRC, castration-resistant prostate cancer.
Somatic mutations and primary resistance to AA/P
In total, 5390 nonsilent somatic mutations were identified in this cohort, which corresponds to a mutation burden of 55 mutations per tumor genome [median, interquartile range (IQR): 40–67)] (supplementary Table S3, available at Annals of Oncology online). The mutation burden between responders and nonresponders was not different (P = 0.51), although two nonresponders exhibited hypermutated genomes with over 500 nonsilent mutations (supplementary Figure S2A, available at Annals of Oncology online). The mutation burden detected in our cohort is 2.9 times higher (P < 2.2 × 10−16, two-tailed Wilcoxon rank sum test) than that reported in the TCGA study (median = 19, IQR: 13–25) of 333 localized, hormone-naive prostate cancer specimens [8] but 23% lower (P = 9.8 × 10−7) than that detected in the AACR/PCF Stand-Up-To-Cancer (SU2C) WES study (median = 71.5, IQR: 55–120) of 150 CRPC metastases [6] (supplementary Figure S2B and D, available at Annals of Oncology online). An average of 68.9 genes (median = 54, IQR: 38.5–65.5) were mutated in each tumor genome. A total of 3971 genes were detected with nonsilent mutations, of which 780 genes were recurrently mutated in two or more specimens. Consistent with previous reports, the most frequently mutated genes were TP53 (24%), MLL3 (14.7%), AR (10.7%), FOXA1 (10.7%), APC (10.7%), and SPOP (6.7%) (supplementary Table S4, available at Annals of Oncology online)[6, 9]. Additional genes with recurrent mutations not previously reported in CRPC included TDG (12%), PSPH (10.67%), and FGFR3 (8%) (supplementary Table S5, available at Annals of Oncology online).
We compared the 780 genes displaying recurrent mutations in our cohort (N = 75) with two previously published whole-exome studies carried out in CRPC metastases [Grasso et al. [9] (N = 50) and Robinson et al. (N = 150) [6]]. A set of 98 genes were mutated in all three CRPC datasets in two or more tumor specimens (supplementary Figure S3, available at Annals of Oncology online). We then calculated MutSig-CV q-values to identify CRPC-associated significantly mutated genes (SMGs) and found 17 SMGs including TP53 and PTEN (supplementary Figure S3 and Table S15, available at Annals of Oncology online). Of the 98 genes, 10 were CRPC-specific SMGs in at least one of three CRPC cohort-based studies (Figure 1E;supplementary Results, available at Annals of Oncology online). Pathway analysis of the 10 genes identified ‘regulation of nuclear β-catenin signaling and target gene transcription’ (q-value = 3.97 × 10−8) suggesting a high frequency of mutations in the Wnt/β-catenin signaling pathway associated genes in CRPC. Compared with the TCGA study of hormone-naive localized prostate cancer (N = 333 specimens); 85 of these 98 genes were observed to be mutated in both stages of the disease (Figure 1F) although the mutation frequencies were different between the three CRPC cohorts versus the TCGA cohort (supplementary Figure S3, available at Annals of Oncology online). We detected higher mutation frequencies for AR, CDK12, CACNA1C, TNIK, DMD, VCAN, SLIT3, and NOTCH1 in CRPC patients (supplementary Figure S3 and Table S5, available at Annals of Oncology online).
Associations between somatic mutations and 12-week primary resistance to treatment were evaluated at the single gene level and the gene pathway/network level. Associations for each of the 744 gene mutated in two or more specimens with complete outcome data (N=73) are provided in supplementary Table S6, available at Annals of Oncology online. Using the risk ratio (RR) of 2 as a threshold, the 744 genes were divided into three nonoverlapping categories: 113 genes that were more frequently mutated in nonresponders (i.e. associated with primary resistance; RR > 2); 292 genes that were more frequently mutated in responders (i.e. associated with primary response; RR < 0.5); and 339 genes that were mutated at similar frequencies in both responders and nonresponders (0.5 ≤ RR ≤ 2) (supplementary Table S6, available at Annals of Oncology online).We carried out interaction network analysis for these three groups of genes to identify ‘hub’ genes with hyperconnectivity to other genes. From the set of genes having similar mutational frequencies in responders and nonresponders, we can identify networks with high mutational burden in this cohort. We identified several networks with ‘hub’ genes (TP53, ATM, APC, MYC, and PTEN) involved in ATM-dependent DNA damage response (supplementary Figure S4A and B, available at Annals of Oncology online), and one hub gene (CSNK2A1) involved in CCP (supplementary Figure S4, available at Annals of Oncology online). Although the genes used for this last network analysis are not limited to CRPC-specific genes, the mutation burden in these networks may contribute to CRPC stage. For genes with higher mutation frequencies in responders, we identified AR- and AKT1-centered networks. The mutation frequency of AR centered gene network in responders (24/41 = 58.5%) was significantly higher compared with nonresponders (3/32 = 9.4%) (P = 1.9 × 10−5, two-tailed Fisher’s exact test), suggesting mutations in the AR regulated gene network is associated with decreased response to AA/P (supplementary Figure S5 and Table S7, available at Annals of Oncology online). For genes with higher mutation frequencies in nonresponders, we identified a CTNNB1 (β-catenin-1) centered gene network. The mutation frequency of CTNNB1-centered gene network in non-responders (18/32 = 56.25%) was higher compared with responders (7/41 = 17.1%) (P = 0.001, two-tailed Fisher's exact test), suggesting dysregulated Wnt/β-catenin signaling in non-responders (supplementary Figure S6 and Table S7, available at Annals of Oncology online).
Copy number alteration and primary resistance to abiraterone acetate
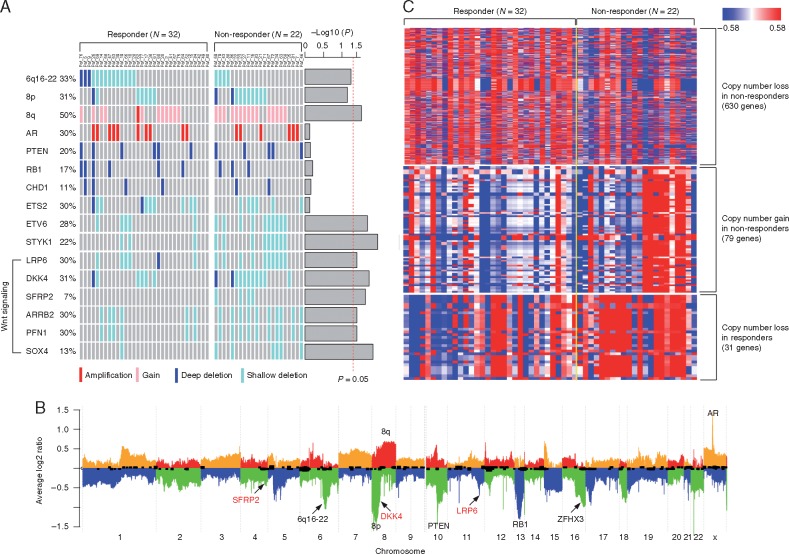
We evaluated somatic copy number alterations using WES data after removing samples with tumor purity lower than 15%. This yielded samples from 22 nonresponders and 32 responders. Consistent with previous genomic analyses of CRPC, we observed frequent amplification of AR (30%) [10], gain of 8q (50%) [11], loss of 8p (31%), as well as deletion of RB1 (17%), CHD1 (11%), and PTEN (20%) [12] (Figure 2A and B). The copy number alteration frequencies of AR, RB1, CHD1, and PTEN were not significantly different between responders and nonresponders (Figure 2A, supplementary Table S7, available at Annals of Oncology online). The 6q16-22 region has been reported as the second most common genomic deletion in prostate cancer, occurring in 22%–62% of cases [13, 14]. In our cohort, 6q16-22 was deleted in 14 out of 32 (44%) responders and in 4 out of 22 (18%) nonresponders suggesting that deletion of this region is associated with response to AA/P (P = 0.027) (Figure 2A and B, supplementary Table S7, available at Annals of Oncology online).
Figure 2.
(A) Comparing the arm-level (6q16-22, 8p, and 8q) and gene level copy number alteration frequencies between responders (N = 32) and nonresponders (N = 22). P-values were calculated using logistic regression and indicated as barplot on the right. The vertical dash line indicated P = 0.05. (B) Heat map showing genes with different copy number alterations (P < 0.05) between responders (N = 32) and nonresponders (N = 22). (C) Average copy number profiles of the 54 tumor genomes. Individual chromosomes are separated by alternating colors with ‘orange’ and ‘red’ indicating copy number gain and ‘blue’ and ‘green’ indicating copy number loss. Key aberrant genes and segments are indicated. Wnt pathway inhibitors (SFRP2, DKK4, and LRP6) are indicated in red.
We identified 630 and 31 genes with copy number loss occurring more frequently (P < 0.05) in nonresponders and responders, respectively (Figure 2C, supplementary Table S8, available at Annals of Oncology online). Similarly, we detected 79 genes with copy number gain occurring more frequently (P < 0.05) in nonresponders, but no genes were detected with copy number gain occurring more frequently in responders (Figure 2C, supplementary Table S8, available at Annals of Oncology online) suggesting increased genomic instability in nonresponders. Functional annotation analysis suggested that genes involved in ‘negative regulation of TCF-dependent signaling by WNT ligand antagonists’ (DKK4, LRP6, and SFRP2) were more frequently lost in nonresponders (q-value < 0.001) (Figure 2A and B, supplementary Table S9, available at Annals of Oncology online).
Gene expression signatures and primary resistance to AA/P
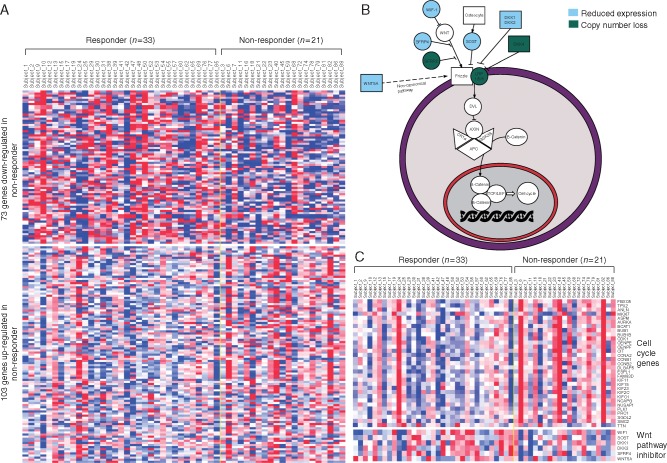
Next, analysis of RNA-sequencing data was carried out to identify gene expression signatures associated with primary resistance to AA/P. This analysis was restricted to the 54 patients with skeletal metastases as this was the most abundant site of metastases. In these 54 patients, there were 33 responders and 21 nonresponders. Using a false discovery rate (FDR) threshold 0.01, we identified 103 genes that were up-regulated and 73 genes that were down-regulated in nonresponders (Figure 3A, supplementary Table S10, available at Annals of Oncology online). Pathway analysis using ConsensusPathDB [15] and Ingenuity Pathway Analysis (IPA) revealed that down-regulated genes in nonresponders were enriched for negative regulators of WNT signaling (Figure 3B, supplementary Table S11, available at Annals of Oncology online). For example, WIF1 (WNT Inhibitory Factor 1), an inhibitor of the canonical WNT signaling pathway, was significantly down-regulated in nonresponders (FDR = 1.0 × 10−4). Expression of other Wnt antagonists, SOST (Sclerostin, FDR = 2.9 × 10−8), DKK1 (Dickkopf WNT Signaling Pathway Inhibitor 1, FDR = 8.9 × 10−4), and DKK3 (FDR = 1.9 × 10−3) was also significantly decreased in nonresponders. Additional Wnt antagonists including SFRP4 (Soluble frizzled-related protein 4, P = 8.9 × 10−3, FDR = 0.21) and WNT5A (P = 8.3 × 10−3, FDR = 0.20) were down-regulated in nonresponders, but these did not reach FDR thresholds (Figure 3C). On the other hand, 31 of 103 (30.0%) genes that were up-regulated in nonresponders were involved in CCP (adjusted P = 9.2 × 10−17). Both ConsensusPathDB and IPA identified CCP pathways as the most highly enriched pathways in nonresponders. These pathways included ‘Polo-like kinase 1 (PLK1) signaling’ and ‘G2/M DNA damage checkpoint regulation’ (Figure 3C, supplementary Table S11, available at Annals of Oncology online).
Figure 3.
(A) Heat map showing genes differentially expressed between 33 responders and 21 nonresponders with skeletal metastases. (B) WNT signaling pathway diagram. Genes with reduced expression in nonresponders were indicated with blue background, and genes with copy number loss in nonresponders were indicated with green background. (C) Heat map showing differentially expressed cell cycle genes and WNT pathway inhibitors.
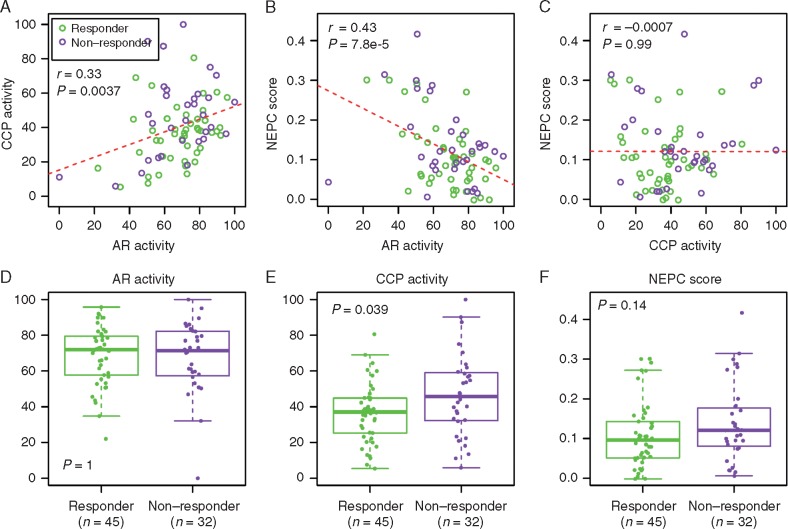
We estimated gene panel expression scores to determine correlations between biological pathways and primary resistance. These previously reported scores included an AR activity score, CCP activity score [16], and NEPC score [17] (supplementary Methods, available at Annals of Oncology online). We detected AR activity displayed a positive albeit weak correlation with CCP (r = 0.33, P = 3.7 × 10−3) and a negative correlation with NEPC (r = −0.43, P = 7.8 × 10−5) (Figure 4A and B). No association was observed between CCP and NEPC (r = −0.0007, P = 0.99) (Figure 4C). The positive correlation observed between AR activity and CCP is inconsistent with a recent report indicating these signatures were negatively correlated [2]. The negative correlation between AR activity and NEPC score is consistent with the observation that NEPC tumors often lose the expression of AR and AR-regulated genes such as KLK3 [18]. We found no difference in AR activity score between the responders and nonresponders (P = 1, two-tailed Wilcoxon rank sum test) (Figure 4D). Expectedly, high CCP scores were significantly associated with nonresponders (P = 0.039) (Figure 4E). We also noted high NEPC scores for nonresponders although with insignificant P = 0.14 (Figure 4F). We developed a Wnt inhibitor activity (WIA) score and Wnt activator activity (WAA) score to assess the role of Wnt pathway activation in CRPC patients. WIA was calculated from four genes including WIF1, SOST, DKK1, and DKK3 that were significantly down-regulated in nonresponders (Figure 3C). WIA was also calculated from previously published 25 Wnt negative regulator genes [19]. The WAA score was calculated from eight genes including RSPO1/2/3/4, NDP, DACT1/2, and FRAT1. In multivariate analysis, only the CCP and WIA scores were significantly associated with primary drug resistance (Table 1).
Figure 4.
Upper panel: associations between AR activity and CCP activity (A), AR activity and NEPC score (B), and CCP activity and NEPC score (C). Lower panel: Comparing AR activity (D), CCP activity (E), NEPC score (F) between responders (green) and nonresponders (purple). r, Pearson correlation coefficient; CCP, cell cycle progression; NEPC, neuroendocrine prostate cancer.
Table 1.
Univariate and multivariate analyses of pathway scores’ association with abiraterone acetate response
| Pathway score | N | Hazard ratio | 95% CI | P-value |
|---|---|---|---|---|
| CCP | 77 | 1.02 | 1.00–1.03 | 0.02 |
| CCP | ||||
| <50 | 57 | 1.0 | ||
| ≥50 | 20 | 1.93 | 1.10–3.35 | 0.02 |
| AR | 77 | 1.00 | 0.99–1.02 | 0.95 |
| NEPC | 79 | 4.20 | 0.32–55.9 | 0.28 |
| NEPC | ||||
| <0.05 | 16 | 1.0 | ||
| ≥0.05 | 63 | 1.57 | 0.79–3.10 | 0.20 |
| Wnt inhibitor activity (25 genes) | 79 | 0.92 | 0.76–1.11 | 0.38 |
| Wnt inhibitor activity (4 genes) | 77 | 1.01 | 1.00–1.01 | 0.36 |
| Wnt inhibitor activity (4 genes) | ||||
| <20 | 19 | 1.0 | ||
| ≥20 | 58 | 1.61 | 0.87–2.98 | 0.13 |
| Wnt activator activity | 79 | 0.94 | 0.81–1.10 | 0.45 |
| Multivariate analysis of pathway scores | ||||
| CCP | ||||
| <50 | 57 | 1 | ||
| ≥50 | 20 | 2.42 | 1.34–4.37 | 0.004 |
| Wnt inhibitor activity (4 genes) | ||||
| <20 | 19 | 1 | ||
| ≥20 | 58 | 2.07 | 1.08–3.97 | 0.03 |
CCP, cell cycle progression score; AR, androgen receptor; NEPC, neuroendocrine prostate cancer score.
Gene fusions and splicing variants associated with primary resistance
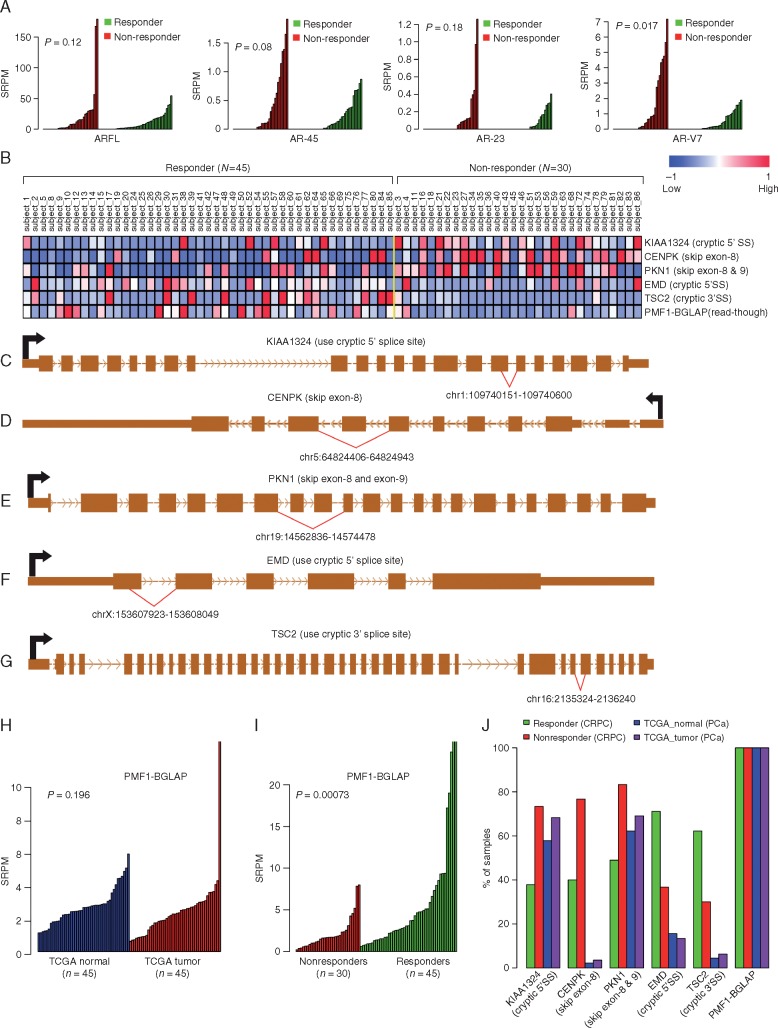
We identified 27 gene fusions (supplementary Tables S2 and S12, available at Annals of Oncology online) from the 82 samples with RNA-seq data. Detailed results of association are included under supplementary Results, available at Annals of Oncology online, including AR splicing variants (AR-Vs) with clinical outcomes (Figure 5A). Results of additional alternative mRNA splicing events and primary AA/P resistance (Figure 5B–J, supplementary Table S13, available at Annals of Oncology online) are also detailed further under supplementary Results, available at Annals of Oncology online.
Figure 5.
Comparing the expression of ARFL, AR45, AR23, and AR-V7 between responders and nonresponders. Overexpression of AR-V7 lacking the ligand-binding domain (LBD) was significantly (P = 8.7 × 10−3, Wilcoxon rank sum test) associated with primary resistance to AA/P. In contrast, the expression levels of other AR-Vs with LBD were not associated with primary resistance. (A) Comparing the expression levels of AR splicing variants between responders (green) and nonresponders (red). P-values were calculated using Wilcoxon rank sum test. (B) Heat map showing six aberrant splicing isoforms differentially expressed between 45 responders and 30 nonresponders. Aberrant splicing of KIAA1324, CENPK, PKN1, EMD, and TSC2 are illustrated in (C)–(G). (H) PMF1-BGLAP expression in 45 TCGA tumor and matched normal samples. (I) PMF1-BGLAP expression in 45 responders and 30 nonresponders. (J) Aberrant splicing frequency (y-axis) in CPRC patients who are responding to AA/P treatment (green), CPRC patients who are not responding to AA/P treatment (red), TCGA primary prostate cancer patients (purple), and normal prostate tissues (blue). SRPM, spliced reads per million. AA/P, abiraterone acetate/prednisone; CPRC, castration-resistant prostate cancer.
For determining acquired resistance at the time of disease progression on AA/P, the time to event variable used was defined as TTTC are presented under supplementary Results, available at Annals of Oncology online. Briefly, CCP gene pathway scores (≥ 50) were associated with shorter TTTC [hazard ratio (HR) = 1.94, 95% confidence interval [CI]: 1.09–3.43; P = 0.02).
Efficacy of wnt and cell cycle inhibition in abiraterone-resistant organoids
Based on the clinical observations, we hypothesized that activated Wnt signaling and enhanced cell cycle activities are associated with primary resistance to AA/P. To test this hypothesis, we developed three organoid lines (MC-PRX01, MC-PRX04, and MC-PRX05). MC-PRX01 and MCPRX05 were derived from patients who were AA/P nonresponders and MCPRX-04 was derived from a patient responding to AA/P. The pretreatment tissue biopsies were grown and expanded for several generations in patient-derived xenograft tumors, and organoids were grown from these models. All the organoids displayed strong AR staining compared with LNCaP cells cultured under the same organoid conditions (supplementary Figure S7A, available at Annals of Oncology online). Genome mutational status of the organoids is summarized under supplementary Results and in supplementary Table S14, available at Annals of Oncology online.
The organoids were treated with the CDK4/6 inhibitor palbociclib [20, 21] or the Wnt inhibitor XAV939 [22] alone or in combination with AA/P (supplementary Figure S7B, available at Annals of Oncology online). Organoids were all pretreated with 100 nM pregnenolone, a precursor of DHEA and downstream androstenedione. This reaction is catalyzed by CYP17, the target of abiraterone acetate. As expected, pregnenolone significantly enhanced the growth of organoids (supplementary Figure S7B, available at Annals of Oncology online). Abiraterone had no effect on pregnenolone-stimulated growth indicating that the MCPRX-01 and MCPRX-05 organoids retained abiraterone resistance (supplementary Figure S7B, available at Annals of Oncology online). However, the MCPRX-04 organoid was growth-suppressed by abiraterone. Treatment with palbociclib or XAV939 as single agents or in combination with abiraterone significantly inhibited AA/P-resistant organoid growth, although the combination with either drug showed more profound growth inhibition. In contrast, palbociclib or XAV939 treatment alone or combined with abiraterone showed similar response to abiraterone alone in the MCPRX-04 organoid (supplementary Figure S7B, available at Annals of Oncology online). Collectively, these results provide a rationale for clinical trials with inhibitors of these pathways to achieve better outcomes.
Discussion
A genome-wide detailed analysis of the genomic heterogeneity of metastases carried out in this study revealed specific candidates associated with resistance to AA/P. Genes in the Wnt/β-pathway were more frequently mutated and altered expression of Wnt/β-pathway components occurred more frequently in patients exhibiting primary resistance. Increased expression of CCP pathway related genes was also associated with primary resistance, as well as a shorter TTTC (acquired resistance). Mutations of the canonical Wnt/β-catenin signaling pathway have been linked to tumorigenesis and implicated in metastasis of many cancers including colon, breast, and pancreas. Links between Wnt/β-catenin signaling and therapeutic resistance have also been reported in ovarian, colon, and pancreatic cancer [23–25]. A recent study detected a higher CTNNB1 mutational frequency in CRPC patients who experienced rapid disease progression on enzalutamide therapy [26], and another study found that noncanonical Wnt signaling is associated with enzalutamide resistance [27]. A previous study also found that a CCP score had prognostic significance for disease-specific mortality [28]. Our pathway analysis indicates that cell cycle regulation–based markers may be useful for predicting resistance to AA/P in the future. We did observe that in this metastatic CRPC cohort the detection rates of common genomic alterations (such as TP53 mutation and ETS fusions) were lower when compared with Robinson et al. [6], but the results of genomic associations relative to disease progression do not appear to be affected by this (see detailed analysis in the supplementary Results, available at Annals of Oncology online),
Conclusions
CRPC patients with mutations or altered expression of genes in the Wnt pathway and increased CCP scores are likely to fail AA/P. Future studies will be needed to assess the feasibility of testing combination drug treatments or novel drug approaches to overcome drug resistance for such patient populations.
Data availability
Whole exome and RNA-seq data have been deposited into the database of Genotypes and Phenotypes (dbGaP) with accession number phs001141.v1.p1.
Supplementary Material
Acknowledgements
We are grateful for the administrative assistance provided by Bobbi-Ann Jebens for this submission. We also would like to thank all patients who participated in this study for their selfless contribution in bringing precision medicine to future advanced prostate cancer patients. We highly appreciate the support of family members as well. We gratefully acknowledge the helpful recruitment efforts of the following physicians who made patient referrals to the ‘PROMOTE’ program; Sandeep Basu (Mayo Clinic Health Systems), Daniel Burns (Mayo Clinic Health Systems), Kevin Cockerill (Mayo Clinic Health Systems), Sarah Kratz (Mayo Clinic Health Systems), Mohammad Ranginwala (Mayo Clinic Health Systems), Amrit Singh (Mayo Clinic Health Systems), Gautam Jha (University of Minnesota), Badrinath Konety (University of Minnesota), Mir Ali Khan (CGH Medical Center), Ferdinand Addo (Prairie Lakes Healthcare System), Kevin Panico (Altru Health System) and Laura Joque (Essentia Health Brainerd Clinic).
Funding
This study was funded in part by the Mayo Clinic Center for Individualized Medicine (MC1351 to MK and LW); Minnesota Partnership for Biotechnology and Medical Genomics (MNP#14.37 to MK and SMD); Department of Defense (W81XWH-15-1-0634 to SMD, MK); Prostate Cancer Foundation (2015 Prostate Cancer Foundation Challenge Award to HH, MK, and SMD); National Institute of Health-National Cancer Institute, R01 CA174777 to SMD; AT Suharya and Ghan DH.; Joseph and Gail Gassner; and Mayo Clinic Schulze Cancer for Novel Therapeutics in Cancer Research (to MK and LW). Other contributing groups include the Mayo Clinic Cancer Center (no grant numbers apply), the Pharmacogenomics Research Network (PGRN) (no grant numbers apply), and the Janssen Research & Development, LLC (no grant numbers apply), which provided partial drug support on study from April 2014 onwards for study patient number 45–92, and funding support for collaborative bioinformatics analysis).
Disclosure
The authors have declared no conflicts of interest.
References
- 1. Chen Y, Clegg NJ, Scher HI.. Anti-androgens and androgen-depleting therapies in prostate cancer: new agents for an established target. Lancet Oncol 2009; 10(10): 981–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kumar A, Coleman I, Morrissey C. et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med 2016; 22(4): 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Knudsen KE, Scher HI.. Starving the addiction: new opportunities for durable suppression of AR signaling in prostate cancer. Clin Cancer Res 2009; 15(15): 4792–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Watson PA, Arora VK, Sawyers CL.. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer 2015; 15(12): 701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ryan CJ, Smith MR, de Bono JS. et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med 2013; 368(2): 138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Robinson D, Van Allen EM, Wu YM. et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015; 161(5): 1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pritchard CC, Mateo J, Walsh MF. et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med 2016; 375(5): 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cancer Genome Atlas Research N. The molecular taxonomy of primary prostate cancer. Cell 2015; 163: 1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grasso CS, Wu YM, Robinson DR. et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012; 487(7406): 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Attar RM, Takimoto CH, Gottardis MM.. Castration-resistant prostate cancer: locking up the molecular escape routes. Clin Cancer Res 2009; 15(10): 3251–3255. [DOI] [PubMed] [Google Scholar]
- 11. El Gammal AT, Bruchmann M, Zustin J. et al. Chromosome 8p deletions and 8q gains are associated with tumor progression and poor prognosis in prostate cancer. Clin Cancer Res 2010; 16(1): 56–64. [DOI] [PubMed] [Google Scholar]
- 12. Choucair K, Ejdelman J, Brimo F. et al. PTEN genomic deletion predicts prostate cancer recurrence and is associated with low AR expression and transcriptional activity. BMC Cancer 2012; 12(1): 543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sun J, Liu W, Adams TS. et al. DNA copy number alterations in prostate cancers: a combined analysis of published CGH studies. Prostate 2007; 67(7): 692–700. [DOI] [PubMed] [Google Scholar]
- 14. Taylor BS, Schultz N, Hieronymus H. et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010; 18(1): 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kamburov A, Wierling C, Lehrach H, Herwig R.. ConsensusPathDB—a database for integrating human functional interaction networks. Nucleic Acids Res 2009; 37(Suppl 1): D623–D628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cuzick J, Swanson GP, Fisher G. et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol 2011; 12(3): 245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Beltran H, Prandi D, Mosquera JM. et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 2016; 22(3): 298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aggarwal R, Zhang T, Small EJ, Armstrong AJ.. Neuroendocrine prostate cancer: subtypes, biology, and clinical outcomes. J Natl Compr Canc Netw 2014; 12(5): 719–726. [DOI] [PubMed] [Google Scholar]
- 19. Cruciat CM, Niehrs C.. Secreted and transmembrane wnt inhibitors and activators. Cold Spring Harb Perspect Biol 2013; 5(3): a015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. O'Leary B, Finn RS, Turner NC.. Treating cancer with selective CDK4/6 inhibitors. Nat Rev Clin Oncol 2016; 13: 417–430. [DOI] [PubMed] [Google Scholar]
- 21. Ehab M, Elbaz M.. Profile of palbociclib in the treatment of metastatic breast cancer. Breast Cancer 2016; 8: 83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ma L, Wang X, Jia T. et al. Tankyrase inhibitors attenuate WNT/beta-catenin signaling and inhibit growth of hepatocellular carcinoma cells. Oncotarget 2015; 6(28): 25390–25401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Varma RR, Hector SM, Clark K. et al. Gene expression profiling of a clonal isolate of oxaliplatin-resistant ovarian carcinoma cell line A2780/C10. Oncol Rep 2005; 14: 925–932. [PubMed] [Google Scholar]
- 24. Anastas JN, Kulikauskas RM, Tamir T. et al. WNT5A enhances resistance of melanoma cells to targeted BRAF inhibitors. J Clin Invest 2014; 124(7): 2877–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peng C, Zhang X, Yu H. et al. Wnt5a as a predictor in poor clinical outcome of patients and a mediator in chemoresistance of ovarian cancer. Int J Gynecol Cancer 2011; 21(2): 280–288. [DOI] [PubMed] [Google Scholar]
- 26. Wyatt AW, Azad AA, Volik SV. et al. Genomic alterations in cell-free DNA and enzalutamide resistance in castration-resistant prostate cancer. JAMA Oncol 2016; 2(12): 1598–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miyamoto DT, Zheng Y, Wittner BS. et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science 2015; 349(6254): 1351–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Crawford ED, Scholz MC, Kar AJ. et al. Cell cycle progression score and treatment decisions in prostate cancer: results from an ongoing registry. Curr Med Res Opin 2014; 30(6): 1025–1031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole exome and RNA-seq data have been deposited into the database of Genotypes and Phenotypes (dbGaP) with accession number phs001141.v1.p1.