Acinetobacter baumannii is an opportunistic pathogen and is a leading cause of hospital-acquired infections. A. baumannii is difficult to eradicate and to manage, because this bacterium is known to robustly survive desiccation and to quickly gain antibiotic resistance. We sought to investigate biofilm formation in A. baumannii, since much remains unknown about biofilm formation in this bacterium. Biofilms, which are multicellular communities of bacteria, are surface attached and difficult to eliminate from hospital equipment and implanted devices. Our research identifies multifaceted physiological roles for the conserved bacterial protease Lon in A. baumannii. These roles include biofilm formation, motility, and viability. This work broadly affects and expands understanding of the biology of A. baumannii, which will permit us to find effective ways to eliminate the bacterium.
KEYWORDS: Acinetobacter, Lon protease, biofilms
ABSTRACT
Acinetobacter baumannii is a Gram-negative opportunistic pathogen that is known to survive harsh environmental conditions and is a leading cause of hospital-acquired infections. Specifically, multicellular communities (known as biofilms) of A. baumannii can withstand desiccation and survive on hospital surfaces and equipment. Biofilms are bacteria embedded in a self-produced extracellular matrix composed of proteins, sugars, and/or DNA. Bacteria in a biofilm are protected from environmental stresses, including antibiotics, which provides the bacteria with selective advantage for survival. Although some gene products are known to play roles in this developmental process in A. baumannii, mechanisms and signaling remain mostly unknown. Here, we find that Lon protease in A. baumannii affects biofilm development and has other important physiological roles, including motility and the cell envelope. Lon proteases are found in all domains of life, participating in regulatory processes and maintaining cellular homeostasis. These data reveal the importance of Lon protease in influencing key A. baumannii processes to survive stress and to maintain viability.
IMPORTANCE Acinetobacter baumannii is an opportunistic pathogen and is a leading cause of hospital-acquired infections. A. baumannii is difficult to eradicate and to manage, because this bacterium is known to robustly survive desiccation and to quickly gain antibiotic resistance. We sought to investigate biofilm formation in A. baumannii, since much remains unknown about biofilm formation in this bacterium. Biofilms, which are multicellular communities of bacteria, are surface attached and difficult to eliminate from hospital equipment and implanted devices. Our research identifies multifaceted physiological roles for the conserved bacterial protease Lon in A. baumannii. These roles include biofilm formation, motility, and viability. This work broadly affects and expands understanding of the biology of A. baumannii, which will permit us to find effective ways to eliminate the bacterium.
INTRODUCTION
Acinetobacter baumannii is a Gram-negative opportunistic pathogen that is a major cause of hospital-acquired infections. Making it especially problematic is the propensity for A. baumannii to survive desiccation and to rapidly gain antibiotic resistance (1–3). Specifically, it has been found that clinical A. baumannii strains form desiccation-resistant biofilms on both abiotic and biotic surfaces (1, 4). Biofilms are surface-attached bacterial multicellular communities embedded in a self-produced extracellular matrix (5, 6). This matrix may be composed of polysaccharides, proteins, and/or extracellular DNA (7). Biofilms are often studied in the laboratory as colonies and as pellicles, which are biofilms formed at the air-liquid interface (8). Biofilms are especially important in the context of opportunistic pathogens, because they provide a mechanism for cells to robustly survive stress through both physical resilience and differential gene expression (9). In addition, biofilms may increase a cell’s capacity for variability through genetic exchange (10). Because of the clinical implications, such as A. baumannii biofilm development on implanted devices, it is critical to further elucidate mechanisms of biofilm development in this organism.
Regulation of biofilm development is sophisticated, involving environmental and internal signals to initiate biofilm formation; this includes initiating the production of the extracellular matrix and pili or fimbriae for surface attachment (5). In Bacillus subtilis, a model organism for biofilm formation, there are many well-characterized regulatory networks, which highlight the intricate regulation underlying biofilm development (11). Interestingly, in a recent collaborative work, we found that the DNA damage response (DDR) influences biofilm formation in B. subtilis (12). While some research has been performed on A. baumannii biofilm development (13), much remains unknown. Currently, there are some identified biofilm genes, including the pilus csu gene and the biofilm-associated protein bap gene (14–16). However, much work has yet to be done to characterize key regulatory pathways in A. baumannii for biofilm development.
Here, we expand the knowledge of biofilm regulation and study the physiological impact of the Lon protease in A. baumannii. The Lon family of proteases are evolutionarily conserved intracellular ATP-dependent enzymes responsible for targeted degradation of misfolded proteins and of certain regulatory proteins (17–20). It was shown previously that, in Pseudomonas aeruginosa and Vibrio cholerae, Lon proteases influence biofilm formation (21, 22). Through a random transposon mutagenesis screen for biofilm development and cell envelope mutants, we expanded the role of the Lon protease in biofilm formation to the opportunistic pathogen A. baumannii. We found that a Lon-protease-deficient transposon mutant formed a weak pellicle biofilm and had decreased adherence to polystyrene surfaces. This phenotype was conserved in a clean Lon deletion mutant. Unexpectedly, we found that, compared to the parental strain and despite decreased pellicle biofilms, Lon protease mutant colony biofilms had increased architecture and Congo red staining. Congo red binds to secreted polysaccharides and surface proteins (23–25). As this staining suggests, we found that the Lon-deficient mutant had an altered cell morphology and cell envelope, including surface protein and capsule production. We also observed decreased motility for the Lon-deficient mutant. Altogether, these data may provide an explanation for the discrepancy between colony and pellicle biofilms. Additionally, we found that Lon has an important role in UV survival. We further identified a surface antigen, encoded by surA1 (A1S_1383), that was strongly upregulated in a Lon-deficient mutant. We found that the increased upregulation of this surface antigen was independent of biofilm and UV sensitivity phenotypes but that Lon mediated cell viability in a surA1-dependent manner and had a role in its regulation.
RESULTS
Lon protease is necessary for biofilm formation in A. baumannii.
We performed a transposon mutagenesis screen and generated a library of single-insertion mutants. We sought to isolate mutants of A. baumannii ATCC 17978 with altered staining phenotypes, compared to the parental strain, on brain heart infusion (BHI) agar supplemented with Congo red and Coomassie brilliant blue (referred to as Congo red plates), dyes that detect changes in components of a typical biofilm matrix (23, 24). Congo red also differentially stains cell envelope proteins. Due to our focus on the regulation of the DDR in the opportunistic pathogen A. baumannii and previous findings on the connection between the DDR and biofilm development in B. subtilis (12), the parental strain we used in the transposon experiment bears a PrecA–gfp chromosomal reporter. The DDR in A. baumannii is dependent on RecA (26). Mutants with staining different from that of the parental strain were isolated and further studied. We screened ∼2,000 transposon mutants and obtained mutants with either decreased or increased staining at a frequency of approximately 3 × 10−3. To demonstrate the variation of Congo red staining we observed, three mutants and their biofilm phenotypes are shown in Fig. S1A in the supplemental material.
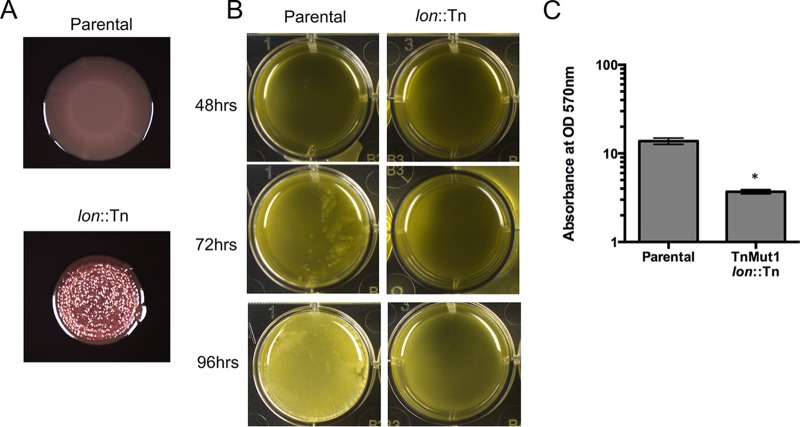
Using whole-genome sequencing (WGS), we mapped the locations of the transposon insertions. WGS also allowed us to confirm that there were no spontaneous mutations that might have been responsible for the observed phenotype. One of our transposon mutants with strong Congo red staining (VGC3) (Table S2) has an insertion in the likely promoter of A1S_1031 (position 1191130 in the genome; GenBank accession no. NC_009085.1), which encodes a Lon protease. It should be noted that this gene was previously misannotated as two genes (A1S_1031 and A1S_1030), due to an erroneous frameshift mutation (27). The lon gene is in fact a 2,331-bp-long gene (positions 1191086 to 1188755 in the genome). Because the colony of the mutant strain had increased Congo red staining and wrinkling, compared to the smooth and less stained colony of the parental strain (Fig. 1A), we hypothesized that it would have a strong biofilm pellicle phenotype. We tracked pellicle formation over time and, surprisingly, the lon::Tn mutant formed no observable pellicle in BHI liquid medium, compared to the parental strain, even after 96 h (Fig. 1B). This finding suggested that the Lon mutant was unable to form pellicle biofilms but might overproduce some extracellular polysaccharide or have an altered cell envelope composition that is stained by Congo red.
FIG 1.
Transposon mutagenesis screen identifies biofilm mutants with altered colony and pellicle biofilm formation. (A) Colony biofilms of parental and lon::Tn strains on BHI Congo red plates. (B) Biofilm pellicles formed in BHI medium in a 12-well untreated polystyrene dish and imaged at 48, 72, and 96 h. Panels A and B show representative images, at 96 h, of experiments performed in at least triplicate. (C) Absorbance at 570 nm for crystal violet staining, at 96 h, of polystyrene-adhered cells from pellicle biofilms formed in BHI medium. The data are representative data for experiments performed in triplicate. Error bars represent standard deviations. An unpaired two-tailed t test was used for statistical analysis, relative to the parental strain. *, P < 0.05.
Although the Lon mutant was incapable of forming pellicle biofilms at the air-liquid interface, it was possible that the lon::Tn cells were still adhering to the surface of the wells, i.e., forming a surface-attached biofilm. To quantify this growth, we performed crystal violet staining for cells adhering to the polystyrene wells (28). Crystal violet staining confirmed that the lon::Tn mutant had significantly decreased polystyrene adherence (Fig. 1C). To test whether this result was medium specific, we grew pellicle biofilms in yeast-tryptone (YT) medium, with a similar result (Fig. S1B and C). Therefore, lon::Tn cells were unable to develop biofilms under these conditions.
Because the insertion in the transposon mutant was in the likely promoter of the lon gene (Fig. S1D), we needed to determine whether the transposon insertion was for loss or gain of expression in lon. Therefore, we performed quantitative PCR (qPCR) on the lon open reading frame (ORF). Transcription of lon was strongly downregulated in the transposon mutant, demonstrating that the transposon insertion was indeed in the lon promoter and disrupted gene expression and that the lon::Tn mutant was lon deficient (Fig. S1E). These data, along with the biofilm phenotypes, suggest that loss of the Lon protease represses biofilm formation in A. baumannii.
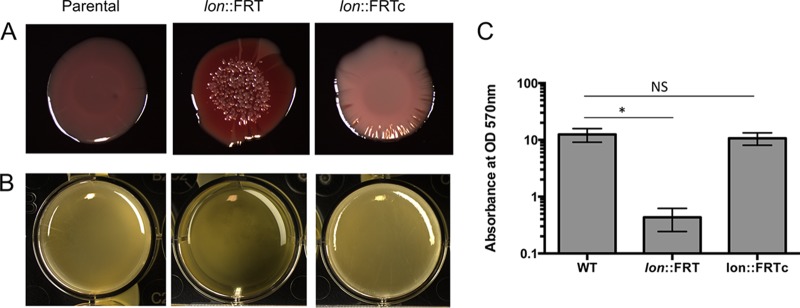
To confirm this result, we wanted to test a clean lon deletion. Tucker et al. previously constructed a clean markerless lon::FRT deletion strain, replacing the full ORF with the flippase recognition target (FRT) cassette, as a test for a novel recombineering system in A. baumannii (27), and we were able to use that strain in further experiments. Indeed, the lon::FRT strain had increased Congo red staining (Fig. 2A) and decreased surface adherence (Fig. 2B and C). To confirm that the biofilm phenotype was due solely to inactivation of lon, we complemented the lon::FRT mutant with a low-copy-number plasmid, PNLAC1 (29), bearing the native promoter and the lon ORF (A1S_1031 and A1S_1030) (referred to as lon::FRTc). We found that the extrachromosomal copy of lon could rescue the pellicle biofilm formation defect of the lon::FRT strain (Fig. 2B and C). The lon::FRTc strain colonies lacked the deep red coloring and appeared to have an intermediate phenotype, with light coloring and a mixture of smooth and wrinkled edges, perhaps due to sensitivity to gene dosage (Fig. 2A).
FIG 2.
Lon protease negatively regulates biofilm formation in A. baumannii. (A) Colony biofilms of WT, lon::FRT, and lon::FRTc strains on BHI Congo red plates. (B) Pellicle biofilms of the same strains as in panel A in BHI medium. Panels A and B show representative images, at 96 h, of experiments performed in at least triplicate. (C) Absorbance at 570 nm for crystal violet staining, at 96 h, of polystyrene-adhered cells from pellicle biofilms formed in BHI medium. The data are representative data for experiments performed in triplicate. Error bars represent standard deviations. An unpaired two-tailed t test was used for statistical analysis, relative to the WT strain. *, P < 0.05; NS, not significant.
The lon-deficient mutant is UV sensitive but does not have an altered response to DNA damage.
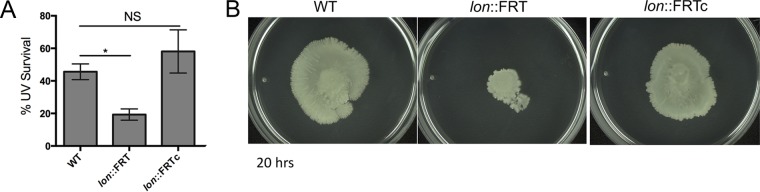
In many bacteria, including Escherichia coli, Lon-defective mutant strains are hypersensitive to UV light, because Lon degrades the cell division inhibitor SulA (30, 31). To test whether this was also the case in A. baumannii, we measured the UV sensitivity of the lon::FRT strain, compared to the isogenic wild-type (WT) strain. We found that the lon::FRT mutant was ∼2-fold more sensitive to UV light (27 J/m2), compared to WT cells (Fig. 3A). A. baumannii lacks a known SulA homologue (32), suggesting that the UV sensitivity pathway in A. baumannii is different from that of E. coli, whose lon-deficient UV sensitivity is >10-fold greater (30). We did not see a significant difference in UV sensitivity between the WT strain and the complemented strain (Fig. 3A).
FIG 3.
Lon-deficient mutant has increased UV sensitivity and decreased motility. (A) UV (27 J/m2) survival of WT, lon::FRT, and lon::FRTc strains. Data shown are the means for experiments performed in biological duplicate, and error bars represent standard deviations. An unpaired two-tailed t test was used for statistical analysis, relative to the WT strain. *, P < 0.05; NS, not significant. (B) WT, lon::FRT, and lon::FRTc strains were spotted on modified 0.3% YT-agarose to test motility. Representative images for experiments performed in at least triplicate are shown.
Because the original transposon screen was performed in a strain bearing a reporter for the DDR (PrecA-gfp), we investigated whether the lon::Tn strain had altered responses to DNA damage. We found that fluorescence expression was similar for the two strains upon induction with ciprofloxacin, a DNA-damaging agent (Fig. S2A to C). Consistent with our previous findings, we observed both bright and dim subpopulations in response to DNA damage (33). We also observed an ∼2-fold decrease in UV sensitivity between the lon::Tn strain and the parental strain (Fig. S2D). This evidence suggested to us that the altered biofilm formation pathway involving Lon does not affect the DDR. It also suggests that the observed UV sensitivity is due to processes independent of the recA-mediated DDR pathway. Indeed, RecA-independent DDR pathways have been observed in other bacteria, including Mycobacterium tuberculosis and Caulobacter crescentus (34, 35). Such examples suggest that there may be redundant pathways for DNA damage repair in bacteria.
The Lon protease mutant has decreased motility.
A. baumannii lacks flagella but displays twitching motility (36). Motility has been shown to be important for biofilm formation in other bacteria (37). To test for motility defects, the lon::FRT strain and the parental WT strain were spotted on an optimized soft agarose medium (P. Muller and V. G. Godoy, unpublished data). After 20 h, the parental strain had migrated outward, while the lon::FRT mutant had little motility outside the initial inoculum (Fig. 3B). This difference in motility might contribute to the inability to form pellicle or submerged biofilms and might account for the different colony biofilm architecture observed for the mutant. The lon::FRTc strain was as motile as the parental strain (Fig. 3B). These data suggest that Lon influences motility, which in turn may influence biofilm development in A. baumannii; however, a mechanistic relationship between these phenotypes remains to be established and elucidated.
The lon-deficient mutant shows differential protein expression and strongly upregulates a surface antigen gene, surA1 (A1S_1383).
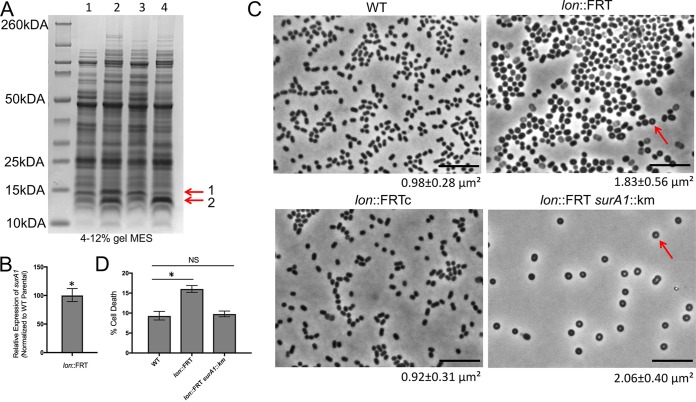
Due to the role of Lon in protein degradation (17), we hypothesized that the lon::Tn strain might have an altered proteomic profile. Therefore, we separated cell-free lysates of the mutant and parental strains by SDS-PAGE (Fig. S3). We found a strong difference for two small proteins in the lon::Tn lysate, compared to the parental strain (Fig. S3, red arrows). These bands were excised and identified using mass spectrophotometry. The larger of the bands (band 1; size estimate of ∼12 kDa, based on a standard curve for the ladder) was strongly enriched for a surface antigen (A1S_1383) of 12.44 kDa (Fig. 4A). This surface antigen was purified previously from an inner membrane fractionation (38). Recently, this surface antigen was described as SurA1 in a virulent A. baumannii isolate and was shown to be important for virulence and bacterial fitness (39). It should be noted that the protein described as SurA1 has an amino acid change in the last residue (M105I), compared to the A. baumannii ATCC 17978 protein. We further separated cell-free lysates from the WT and lon::FRT strains and observed the same extra banding, as expected (Fig. 4A, lanes 1 and 2). These two bands were not seen in the complemented strain (Fig. 4A, lane 3). The same banding pattern was observed in the exponential phase for all of the strains (data not shown); therefore, this is not dependent on the growth phase. Full mass spectrophotometry data are included in Table S1.
FIG 4.
Lon-deficient mutant upregulates a surface antigen and has unique cell morphology. (A) Separation of cell-free lysates from the WT (lane 1), lon::FRT (lane 2), lon::FRTc (lane 3), and lon::FRT A1S_1383::Km (lane 4) strains by SDS-PAGE on a 4 to 12% polyacrylamide gel in MES buffer. The same protein concentration of each lysate was loaded in the wells, to allow direct comparison between strains. Arrows point to two strongly upregulated bands. Band 1 and band 2 were identified by mass spectrometry to be enriched with A1S_1383 and A1S_2230, respectively. (B) Real-time qPCR performed in biological duplicate to determine expression of the A1S_1383 gene transcript from the lon::FRT mutant, relative to the WT strain. Expression was standardized to 16S rRNA expression, and the value for the WT strain was set to 1. Error bars represent standard deviations. An unpaired two-tailed t test was used for statistical analysis, compared to A1S_1383 expression of the WT strain. *, P < 0.05. (C) Single-cell phase microscopy of saturated cultures. Arrows point to the phase bright center. The sizes of cells were measured using MicrobeJ (62). The mean cell areas and standard deviations (n > 1,000) are provided below the images. Representative microscopic images of the strains are shown. Scale bars represent 10 μm. (D) Percentage of dead cells, among total cells, quantified at 24 h over multiple fields of view (>8 fields), with at least 2,700 cells counted. The error bars represent the 95% confidence intervals, and a chi-square test was used for statistical analysis. *, P < 0.05; NS, not significant.
Since we observed an increase in the surface antigen protein, there were two likely possibilities; there was either an alteration in the surA1 transcript or direct posttranslational regulation by Lon itself. To distinguish between these two possibilities, we performed qPCR of the surA1 gene in the lon insertion mutant and the parental strain. Strikingly, the lon mutant had ∼100 times more transcript of the gene encoding the surface antigen than did the parental strain (Fig. 4B). This finding suggests that the surA1 transcript is indirectly regulated through Lon protease depletion, perhaps through stabilization of an otherwise Lon-sensitive transcriptional regulator.
Lon protease influences the cell envelope.
We found a striking difference in cell morphology between the lon-defective (lon::FRT) and WT strains upon microscopic observation of cells from saturated cultures, perhaps indicating an altered cell envelope. The lon-defective (lon::FRT) cells were misshapen, rounder, and visibly larger (∼87% increase) than the WT cells (areas of 1.83 and 0.98 μm2, respectively) (Fig. 4C). Notably, the lon-defective (lon::FRT) cells often had a phase bright spot in the center (Fig. 4C, arrows). The lon-complemented strain (lon::FRTc) had cell morphology similar to that of the parental strain (areas of 0.98 and 0.92 μm2, respectively) (Fig. 4C). These data suggest that Lon has an influence on cell shape.
We hypothesized that the altered Congo red staining of the lon-defective (lon::FRT) strain, compared to the WT strain, might be due to an altered cell envelope. In E. coli, Lon protease affects capsule production through degradation of an activator for capsular polysaccharide biosynthesis (40). To determine whether surA1 had a direct role in the observed phenotypes, we constructed a double deletion mutant (lon::FRT and surA1::Km). We verified that the extra band 1 in Fig. 4A, lane 2, was indeed due to the surA1 protein, because we saw it disappear in the double mutant (Fig. 4A, lane 4). However, we found no significant change in biofilm phenotypes, motility, or UV sensitivity (Fig. S4A and B and data not shown), indicating that these phenotypes are independent of surA1 and dependent on other Lon processes. Interestingly, we noticed that, similar to some lon::FRT cells, the double mutant cells were almost always round, with distinct phase bright spots in the middle; however, we noted that there appeared to be less cell death (detected as light rather than dark cells) in the phase images (Fig. 4C). To quantify this phenomenon, we performed LIVE/DEAD staining of saturated cultures for these strains at 24 h, and we found that the percentage of cell death in the sampled population increased significantly, from ∼9% for WT cells to ∼16% for lon::FRT cells (Fig. 4D; also see Fig. S4C). The lon::FRT surA1::Km cells, however, had a cell death rate of ∼9%, similar to that for WT cells. This finding suggests that there may be a physiological role of Lon in regulating the virulence factor surA1, which may lead to cell death when it is highly upregulated.
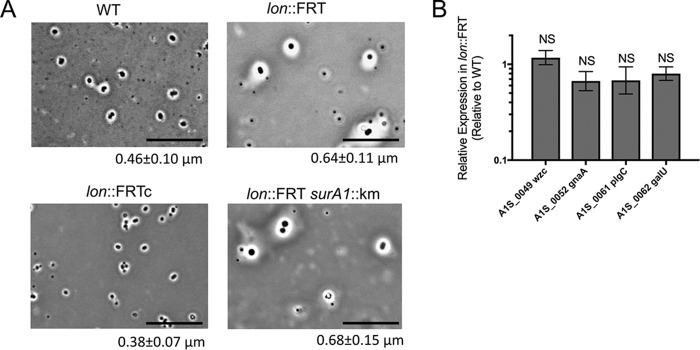
To directly visualize the capsule, we performed a negative staining procedure, with crystal violet and copper sulfate for the capsule (41), on stationary-phase cells of the WT, lon::FRT, lon::FRTc, and lon::FRT surA1::Km strains. The lon::FRT mutant showed increased capsule (39% increase), compared to the WT cells (measured thickness of 0.64 μm, compared with 0.46 μm for WT cells) (Fig. 5A, white area around the cells). The lon-complemented strain had a capsule thickness similar to that of the WT strain (0.38 μm, compared to 0.46 μm), while the lon::FRT surA1::Km capsule thickness matched that of the lon::FRT cells (0.64 μm, compared to 0.68 μm). These data suggest that Lon negatively regulates capsule production.
FIG 5.
Lon protease influences the cellular envelope. (A) Negative capsule staining (41) was performed on WT, lon::FRT, lon::FRTc, and lon::FRT surA1::Km strains. The white space surrounding cells is indicative of capsule. Representative microscopic images of the strains are shown. The thickness of the capsule was measured with ImageJ (NIH) (n = 100), and the mean thickness and standard deviation are provided below each image. Scale bars represent 10 μm. Staining was performed in three independent experiments. (B) Real-time qPCR was performed in biological triplicate to determine expression of the K locus in the lon::FRT and WT strains. Expression was standardized to that of 16S rRNA. Relative quantification of the K locus gene transcripts is shown, with the value for the WT strain being set to 1. Error bars represent standard deviations. An unpaired two-tailed t test was used for statistical analysis, compared to expression of the parental strain. NS, not significant (P > 0.05).
In A. baumannii, the K locus is the main capsular polysaccharide operon involved in capsule formation (42). To investigate whether there were changes in K locus gene expression that would account for the difference in polysaccharide profiles, we performed qPCR for four genes of the K locus (wzc, gnaA, plcC, and galU) under the control of independent promoters. Interestingly, we did not see a significant change in the expression of any of these genes in the lon::FRT strain, compared to the WT strain (Fig. 5B). This evidence suggests that the altered cell envelope is not significantly due to K locus gene expression.
To test for modifications that may not change transcript levels but perhaps may alter polysaccharide contents, we performed analysis of surface polysaccharides of standardized amounts of cells, as performed before for A. baumannii (43). We found that most major polysaccharide component bands (described in the Fig. S5 legend) remained in the lon::FRT strain, albeit with some differences. In an effort to quantify the differences, we measured whether bands had shifted with respect to each other, as well as measuring the width of each band. From quantification of the profile of each band, band *2, which represents the K-locus-dependent capsule polysaccharide components (43), shifted minimally in the lon::FRT strain and in the lon::FRT surA1::Km strain, compared to the WT strain (Fig. S5A; also see Fig. S5B for the density profile of each pattern). There was also a shift in the band pattern for the lon::FRTc strain, although the width of band *2 was rescued to that of the WT strain (Fig. S5A, lane 3; width numbers are listed below each lane). This band was smaller for the lon::FRT (Fig. S5A, lane 2) and lon::FRTsurA1::Km (Fig. S5A, lane 4) strains. In addition, another low-molecular-weight band (band *3) was less intense in both the WT and lon::FRTc strains (Fig. S5A, compare lanes 1 and 3 with lanes 2 and 4). These data indicate that there may be alterations of polysaccharide components that are specific to lon-defective genotypes, although the exact nature of those alterations is unclear. These data also suggest that the larger capsule in the lon::FRT strain, compared to the WT strain (Fig. 5A), is apparently not due to the K locus. Alternatively, it is possible that the Lon mutant retains a tighter capsule than WT cells, and thus more polysaccharide remains cell associated. This would explain the increase in capsule but the lack of significant changes in K locus expression and total polysaccharide extraction.
DISCUSSION
Here, we find a relationship between Lon protease and biofilm development in A. baumannii, providing insights into the regulation of biofilm development and the maintenance of the cell envelope in this opportunistic pathogen. We found that a lon-deficient mutant had decreased surface adherence and motility (Fig. 1 to 3). Interestingly, we found that the lon-deficient mutant had different cell morphology and an altered cell envelope (Fig. 4 and 5). We found that a surface antigen gene, A1S_1383 (surA1), was strongly upregulated, in an indirect manner, in the absence of Lon and may be regulated by Lon to mediate fitness (Fig. 4).
An altered cell envelope may hinder the ability of cells to adhere to surfaces and to grow. This finding could explain the observations that the lon::FRT cells were unable to form pellicle biofilms but had increased Congo red staining (Fig.1). Evidence for this kind of interplay between an altered cell envelope and biofilm development in bacteria was observed previously (44–46). For example, increased capsular polysaccharide production in Vibrio vulnificus inhibits biofilm formation by restricting continual biofilm growth (63). In A. baumannii, we observed increased negative staining for capsule and no significant changes in K locus genes at the transcript level in the lon-deficient mutant, indicating that there may be posttranscriptional regulation or additional components in the cell envelope that were not described previously (47). Another possibility is that Lon affects the amount of K locus capsular polysaccharides retained at the cell surface.
Additionally, impaired motility has been known to contribute to decreased biofilm development in some bacteria (48–50). Type IV pili in A. baumannii are involved in twitching motility, and inhibitors of type IV pilus biogenesis have decreased biofilm formation (51). Previously, much work on A. baumannii biofilms was performed with strains besides ATCC 17978. Recent work has shown that, as in other reported strains (specifically, A. baumannii ATCC 19606 [15]), the Csu pilus system influences biofilm formation and motility in A. baumannii ATCC 17978 (52). A hypothetical histidine kinase/response regulator (A1S_2811) is involved in positive regulation of the csuA/ABCDE operon (52). This response is independent of the type IV twitching motility pili. Interestingly, transcriptional data on a strain with deletion of the putative histidine kinase/response regulator A1S_2811 showed somewhat increased expression of Lon protease (A1S_1031) and decreased expression of surA1 (52). The lon-deficient mutant also appeared to be overexpressing the gene A1S_2230 (enrichment of band 2 in Fig. 4A; also see Table S1 in the supplemental material). This encodes a protein of unknown function (DUF2171) that is downregulated (2-fold) in the A1S_2811 mutant (52). There are many unknowns in the Lon-dependent pathways in A. baumannii, but these data suggest that they are independent of the csuA/ABCDE operon. Thus, the motility defect could be due to defects of type IV pili or could involve purely physical limitations for functional pili due to the altered cell envelope. In Pseudomonas aeruginosa, Lon protease is also a negative regulator of quorum-sensing systems (53). Notably, there are signal peptides that appear in the mass spectrophotometry results for the proteomic profile of the lon-deficient mutant (see Table S1 in the supplemental material), which could indicate a role for quorum sensing as well. However, more work needs to be done to elucidate the specific mechanisms underlying the motility and biofilm defects.
Interestingly, we observed that Lon has a role in regulating the surface antigen gene surA1. We observed a significant increase in expression of the surA1 gene in the lon-deficient mutant, which does not appear to have a direct role in biofilm formation or UV sensitivity but plays a role in cell death. Intriguingly, this surface antigen gene was one of 40 genes whose products are phosphorylated in the phosphoproteome of A. baumannii, suggesting that its protein is posttranslationally regulated (54). Previous work also showed downregulation of surA1 in the antibiotic-resistant A. baumannii strain ATCC 19606, with the possible role being linked to evasion of the host defense mechanisms (38). Another group showed that this gene was upregulated upon stimulation with ethanol (55). SurA1 was first characterized and named in a study using a virulent strain isolated from chickens (A. baumannii strain CCGGD201101), which had increased expression of surA1. SurA1 was found to be cytotoxic to host cells and had a role in mediating A. baumannii survival in human serum and virulence in a moth model (39). In the lon-deficient mutant, dramatic upregulation of this protein may be an energetic burden (phosphorylation sink) or may physically hinder functions of the cell, thus having a role in inducing cell death. Interestingly, transposon insertions in Lon led to significant attenuation in the persistence of A. baumannii in an in vivo lung infection (56). The decreased viability and biofilm formation may contribute to this observation. The full molecular mechanisms through which surA1 influences virulence in A. baumannii remain to be elucidated. Here, we identify indirect regulation of surA1 by Lon.
Overall, our data reveal a novel relationship between Lon protease and the cell shape, cell envelope, motility, and biofilm development in A. baumannii. It is interesting that A. baumannii is able to survive under very harsh conditions outside clinical settings, such as the dry environment of the desert (57). This indicates that A. baumannii has evolved strategies to survive and to deal with extreme stressors. Here, we show that an altered cell envelope surface may be an example of an alternative to biofilm development as a protective strategy, which may correspond to A. baumannii having bimodal responses to stress (33); this would serve as a bet-hedging strategy for population protection. The multifaceted role of Lon is an intriguing finding, suggesting that Lon may be key to A. baumannii biology.
MATERIALS AND METHODS
Strains and culture conditions.
The strains and plasmids used are listed in Table S2 in the supplemental material. All bacterial cultures were routinely grown in LB medium, unless otherwise noted, and were incubated at 37°C, with shaking at 225 rpm, for liquid cultures. Kanamycin (35 μg/ml), tetracycline (12 μg/ml), gentamicin (10 μg/ml), and ciprofloxacin (A. baumannii MIC, 0.6 μg/ml) were added to the medium as indicated below and in Table S2. YT medium is composed of 2.5 g/liter NaCl, 10 g/liter tryptone, and 1 g/liter yeast extract. BHI medium was prepared from BBL BHI broth, modified (product no. 299070; BD).
Strain construction.
To construct the PrecA-gfp transcriptional reporter (CC444) (Table S2), the promoter of A1S_1962 fused to gfp was amplified from a PrecA-gfp reporter previously constructed on a different plasmid backbone (33), using the primers RecAprF and GFPR, which contained SalI and NotI (New England Biolabs) restriction sites, respectively (Table S3). This fragment and the vector pBJ4648 (58), which does not contain an origin of replication for A. baumannii, were digested with the same enzymes. The recombinant plasmid was introduced into A. baumannii by electroporation, and colonies were selected for gentamicin resistance. Candidate colonies were tested by PCR and confirmed by sequencing.
To construct the lon::FRT A1S_1383::Km strain (CC737), flanking regions of A1S_1383 were amplified from genomic DNA using the primer pairs 1383upF/1383upR and 1383downF/1383downR, along with the kanamycin resistance cassette from a previous plasmid, puAC66 (33), using the primers kanF and kanR, containing restriction sites needed for a three-fragment ligation into pUC19 (New England Biolabs). The ligated fragment was amplified using 1383upF and 1383downR and ligated into pBJ4648 (58). The recombinant plasmid was introduced into A. baumannii strain AT08 (27) (Table S2) by electroporation and was recombined onto the chromosome using selection for kanamycin resistance and loss of the backbone. Candidate colonies were checked by PCR and confirmed by sequencing.
To construct the Plon-lon plasmid for Lon complementation (pCC1) (Table S2), a fragment containing the A. baumannii A1S_1031 putative promoter region to the end of the A1S_1030 ORF was amplified using primers P1031F and 1030R (Table S3). The PCR amplicons and the plasmid pNLAC1 (29) (Table S2) were digested with PstI (New England Biolabs). The insert and vector were then ligated with T4 ligase (Promega). After transformation into chemically competent E. coli and selection for tetracycline resistance, all plasmids were confirmed by sequencing using the primers PNLACF and PNLACR, to verify correct assembly. All plasmids were introduced into A. baumannii cells by electroporation, as described previously (33).
All accession numbers are from the genome with GenBank accession no. NC_009085.1. Corresponding GenBank accession numbers for the A. baumannii ATCC 17978-mff genome (GenBank accession no. NZ_CP012004.1) are WP_001292274.1 for Lon protease (A1S_1031 and A1S_1030), WP_000132045.1 for A1S_1383, and WP_001094391.1 for A1S_2230.
Transposon mutagenesis screen.
To perform transposon mutagenesis, electrocompetent cells were made as described previously (33). The transposon containing plasmid pDL1073, which does not replicate in A. baumannii, was introduced by electroporation. Cells were allowed to recover for 2 h and were selected for kanamycin resistance on LB plates. pDL1073 harbors a mini-Tn10 transposon, which transfers a kanamycin resistance marker within the minitransposon onto the chromosome at 37°C. The annotated sequence of pDL1073 is available upon request. LB plates containing transposon mutants were replica plated onto BHI plates containing 200 μg/ml Congo red and 100 μg/ml Coomassie brilliant blue (Congo red plates).
Biofilm and adherence assays.
For colony biofilm assays, cells were grown to exponential phase and 2 μl of the culture was spotted onto Congo red plates. The plates were incubated at 30°C. For pellicle biofilm assays, cells were grown to exponential phase and inoculated at a 1:1,000 dilution of the culture into YT or BHI liquid medium in untreated 12-well polystyrene microtiter plates, which were then incubated statically at 25°C. Images of colony and pellicle biofilms were taken using a Leica MSV269 dissecting microscope and a Leica DMC2900 camera with the same settings, to permit direct comparison. For the adherence assays, standard crystal violet staining was carried out as performed previously for Staphylococcus aureus (59).
Whole-genome sequencing.
Genomic DNA was prepared as suggested by the manufacturer for the Qiagen DNeasy Blood and Tissue kit (Qiagen). Genomic DNA was prepared using the Nextera DNA XT Library preparation kit (Illumina). MiSeq v2 paired-end 250-bp sequencing was performed at Tufts University Core Facility Genomics Services (Boston, MA). Data were analyzed using CLC Genomics Workbench software (Qiagen).
Quantitative real-time PCR.
Total RNA from A. baumannii cells grown to exponential phase was extracted using the Zymo Direct-zol RNA kit, as directed (Zymo). The total RNA concentration was measured with a NanoDrop 2000 spectrophotometer (Thermo Scientific). RNA was then treated with RNase-free DNase I according to the manufacturer’s protocol (Thermo Scientific). RNA was converted to cDNA using the cloned avian myeloblastosis virus (AMV) first-strand cDNA synthesis kit, as suggested by the manufacturer (Thermo Scientific). qPCR was performed with serially diluted cDNA, using primers 16SRNAF and 16SRNAR and qPCR primer pairs listed in Table S3, following the protocol for Luna qPCR Mastermix (Thermo Scientific) with the StepOnePlus real-time PCR system (Applied Biosystems). Absence of genomic DNA was verified by carrying out qPCR on a no-reverse transcriptase control. The comparative ΔΔCT was calculated for each strain in biological duplicate and technical triplicate. The relative expression was standardized using the endogenous 16S rRNA as a control. An unpaired two-tailed t test was used for statistical analysis (P < 0.05).
Ciprofloxacin treatment, UV survival, and motility assays.
Ciprofloxacin treatment and UV survival assays were carried out as described previously (60). An unpaired two-tailed t test was used for statistical analysis (P < 0.05). Motility was assessed by spotting 2 μl of a saturated culture on optimized 0.3% YT-agarose medium (P. Muller and V. G. Godoy, unpublished data). These plates were incubated at 30°C.
Polysaccharide analysis, protein lysate extraction, SDS-PAGE analysis, and mass spectrometry analysis.
Polysaccharide analysis was performed as described previously (43), using stationary-phase cells directly from culture, which represented both cell-associated and secreted polysaccharides. Separation was performed on a 4 to 12% polyacrylamide gel with 2-(N-morpholino)ethanesulfonic acid (MES) buffer (Invitrogen). Analysis was done using ImageJ (NIH) (61).
Whole-protein lysate was extracted from diluted cultures and saturated cultures using Bugbuster reagent, according to the manufacturer’s protocol (EMD Millipore). Lysate concentrations were standardized, lysates were separated on a 4 to 12% or 12% polyacrylamide gel with MES buffer (Invitrogen), and proteins were stained with Coomassie brilliant blue (Bio-Rad). Bands containing the proteins of interest were excised and sent for mass spectrometry analysis at the Taplin Mass Spectrometry Facility at Harvard Medical School (Boston, MA).
Microscopy, capsule staining, and viability staining.
Cells (1 ml) were collected in a microcentrifuge at 14,000 rpm for 1 min, resuspended in 100 μl of SMO buffer (100 mM NaCl and 20 mM Tris, pH 7.5), and imaged as described previously (60). For negative staining, 3 μl of cells directly from a saturated culture was added to a glass slide and air dried. Cells were covered with 0.1% crystal violet solution and stained for 5 min. The slide was then washed twice with 1% copper sulfate solution and dried (41). For LIVE/DEAD staining, cells were washed and resuspended in 0.85% NaCl solution and were stained with the LIVE/DEAD BacLight bacterial viability kit (Thermo Fisher), according to the manufacturer’s instructions. Imaging across strains was performed with the same exposure settings, using a Leica MicroStation5000 with a Leica DM3000G camera. Fluorescence and cell sizes were quantified using ImageJ and MicrobeJ (61, 62). To measure the thickness of the capsule, the diameter of the cell body length measurement was subtracted from the diameter of the whole encapsulated cell, and the result was divided by 2.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Bryan Davies (University of Texas at Austin) for his gracious gift of the lon::FRT strain and Eddie Geisinger (Northeastern University) for technical assistance and for providing us with plasmids. We thank Merlin Brychcy, a visiting scholar funded by the DAAD, and Breanna Isley, a biology undergraduate student, for experimental assistance.
This work was funded by an NIH grant supplement (grant GM088230) to V.G.G., and A.C. was supported by an NIH grant (grant AI055058). C.C. was supported by Northeastern University, and B.Y. and C.O. were supported by Northeastern University Undergraduate Provost Awards.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00536-18.
REFERENCES
- 1.Jawad A, Seifert H, Snelling AM, Heritage J, Hawkey PM. 1998. Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J Clin Microbiol 36:29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valencia R, Arroyo LA, Conde M, Aldana JM, Torres M-J, Fernández-Cuenca F, Garnacho-Montero J, Cisneros JM, Ortiz C, Pachón J, Aznar J. 2009. Nosocomial outbreak of infection with pan–drug‐resistant Acinetobacter baumannii in a tertiary care university hospital. Infect Control Hosp Epidemiol 30:257–263. doi: 10.1086/595977. [DOI] [PubMed] [Google Scholar]
- 3.Custovic A, Smajlovic J, Tihic N, Hadzic S, Ahmetagic S, Hadzagic H. 2014. Epidemiological monitoring of nosocomial infections caused by Acinetobacter baumannii. Med Arch 68:402–406. doi: 10.5455/medarh.2014.68.402-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wendt C, Dietze B, Dietz E, Ruden H. 1997. Survival of Acinetobacter baumannii on dry surfaces. J Clin Microbiol 35:1394–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toole GO, Kaplan HB, Kolter R. 2000. Biofilm formation as microbial development. Annu Rev Microbiol 54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 6.Kolter R, Greenberg EP. 2006. Microbial sciences: the superficial life of microbes. Nature 441:300–302. doi: 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- 7.Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 8.Branda SS, González-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. 2001. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A 98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 10.Molin S, Tolker-Nielsen T. 2003. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr Opin Biotechnol 14:255–261. doi: 10.1016/S0958-1669(03)00036-3. [DOI] [PubMed] [Google Scholar]
- 11.Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. 2013. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol 11:157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gozzi K, Ching C, Paruthiyil S, Zhao Y, Godoy VG, Chai Y. 2017. Bacillus subtilis utilizes the DNA damage response to manage multicellular development. NPJ Biofilms Microbiomes 3:8. doi: 10.1038/s41522-017-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaddy JA, Actis LA. 2009. Regulation of Acinetobacter baumannii biofilm formation. Future Microbiol 4:273–278. doi: 10.2217/fmb.09.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loehfelm TW, Luke NR, Campagnari AA. 2008. Identification and characterization of an Acinetobacter baumannii biofilm-associated protein. J Bacteriol 190:1036–1044. doi: 10.1128/JB.01416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomaras AP, Dorsey CW, Edelmann RE, Actis LA. 2003. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149:3473–3484. doi: 10.1099/mic.0.26541-0. [DOI] [PubMed] [Google Scholar]
- 16.Luo L, Wu L, Xiao Y, Zhao D, Chen Z, Kang M, Zhang Q, Xie Y. 2015. Enhancing pili assembly and biofilm formation in Acinetobacter baumannii ATCC19606 using non-native acyl-homoserine lactones. BMC Microbiol 15:62. doi: 10.1186/s12866-015-0397-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu KG, Smith JM, Markovitz DM. 1997. Bacterial protease Lon is a site-specific DNA-binding protein. J Biol Chem 272:534–538. doi: 10.1074/jbc.272.1.534. [DOI] [PubMed] [Google Scholar]
- 18.Gottesman S. 1989. Genetics of proteolysis in Escherichia coli. Annu Rev Genet 23:163–198. doi: 10.1146/annurev.ge.23.120189.001115. [DOI] [PubMed] [Google Scholar]
- 19.Charette MF, Henderson GW, Markovitz A. 1981. ATP hydrolysis-dependent protease activity of the lon (capR) protein of Escherichia coli K-12. Proc Natl Acad Sci U S A 78:4728–4732. doi: 10.1073/pnas.78.8.4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dierksen KP, Marks J, Chen DD, Trempy JE. 1994. Evidence for structural conservation of Lon and RcsA. J Bacteriol 176:5126–5130. doi: 10.1128/jb.176.16.5126-5130.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marr AK, Overhage J, Bains M, Hancock REW. 2007. The Lon protease of Pseudomonas aeruginosa is induced by aminoglycosides and is involved in biofilm formation and motility. Microbiology 153:474–482. doi: 10.1099/mic.0.2006/002519-0. [DOI] [PubMed] [Google Scholar]
- 22.Rogers A, Townsley L, Gallego-Hernandez AL, Beyhan S, Kwuan L, Yildiz FH. 2016. The LonA protease regulates biofilm formation, motility, virulence, and the type VI secretion system in Vibrio cholerae. J Bacteriol 198:973–985. doi: 10.1128/JB.00741-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Surgalla MJ, Beesley ED. 1969. Congo red-agar plating medium for detecting pigmentation in Pasteurella pestis. Appl Microbiol 18:834–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romero D, Aguilar C, Losick R, Kolter R. 2010. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci U S A 107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishiguro EE, Ainsworth T, Trust TJ, Kay WW. 1985. Congo red agar, a differential medium for Aeromonas salmonicida, detects the presence of the cell surface protein array involved in virulence. J Bacteriol 164:1233–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norton MD, Spilkia AJ, Godoy VG. 2013. Antibiotic resistance acquired through a DNA damage-inducible response in Acinetobacter baumannii. J Bacteriol 195:1335–1345. doi: 10.1128/JB.02176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tucker AT, Nowicki EM, Boll JM, Knauf GA, Burdis NC, Trent MS, Davies BW. 2014. Defining gene-phenotype relationships in Acinetobacter baumannii through one-step chromosomal gene inactivation. mBio 5:e01313-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merritt JH, Kadouri DE, O’Toole GA. 2011. Growing and analyzing static biofilms. Curr Protoc Microbiol Chapter 1:Unit 1B.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luke NR, Sauberan SL, Russo TA, Beanan JM, Olson R, Loehfelm TW, Cox AD, Michael FS, Vinogradov EV, Campagnari AA. 2010. Identification and characterization of a glycosyltransferase involved in Acinetobacter baumannii lipopolysaccharide core biosynthesis. Infect Immun 78:2017–2023. doi: 10.1128/IAI.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howard-Flanders P, Simson E, Theriot L. 1964. A locus that controls filament formation and sensitivity to radiation. Genetics 49:237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizusawa S, Gottesman S. 1983. Protein degradation in Escherichia coli: the lon gene controls the stability of sulA protein. Proc Natl Acad Sci U S A 80:358–362. doi: 10.1073/pnas.80.2.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson A, Brzoska AJ, Turner KM, Withers R, Harry EJ, Lewis PJ, Dixon NE. 2010. Essential biological processes of an emerging pathogen: DNA replication, transcription, and cell division in Acinetobacter spp. Microbiol Mol Biol Rev 74:273–297. doi: 10.1128/MMBR.00048-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacGuire AE, Ching MC, Diamond BH, Kazakov A, Novichkov P, Godoy VG. 2014. Activation of phenotypic subpopulations in response to ciprofloxacin treatment in Acinetobacter baumannii. Mol Microbiol 92:138–152. doi: 10.1111/mmi.12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller AU, Imkamp F, Andreas UM, Weber-Ban E. 2018. The mycobacterial LexA/RecA-independent DNA damage response is controlled by PafBC and the Pup-proteasome system. Cell Rep 23:3551–3564. doi: 10.1016/j.celrep.2018.05.073. [DOI] [PubMed] [Google Scholar]
- 35.Modell JW, Kambara TK, Perchuk BS, Laub MT. 2014. A DNA damage-induced, SOS-independent checkpoint regulates cell division in Caulobacter crescentus. PLoS Biol 12:e1001977. doi: 10.1371/journal.pbio.1001977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clemmer KM, Bonomo RA, Rather PN. 2011. Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology 157:2534–2544. doi: 10.1099/mic.0.049791-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guttenplan SB, Kearns DB. 2013. Regulation of flagellar motility during biofilm formation. FEMS Microbiol Rev 37:849–871. doi: 10.1111/1574-6976.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tiwari V, Vashistt J, Kapil A, Moganty RR. 2012. Comparative proteomics of inner membrane fraction from carbapenem-resistant Acinetobacter baumannii with a reference strain. PLoS One 7:e39451. doi: 10.1371/journal.pone.0039451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu D, Liu Z, Hu P, Cai L, Fu B, Li Y, Lu S, Liu N, Ma X, Chi D, Chang J, Shui Y, Li Z, Ahmad W, Zhou Y, Ren H. 2016. Characterization of surface antigen protein 1 (SurA1) from Acinetobacter baumannii and its role in virulence and fitness. Vet Microbiol 186:126–138. doi: 10.1016/j.vetmic.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 40.Ebel W, Skinner MM, Dierksen KP, Scott JM, Trempy JE. 1999. A conserved domain in Escherichia coli Lon protease is involved in substrate discriminator activity. J Bacteriol 181:2236–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jasmin AM. 1945. An improved staining method for demonstrating bacterial capsules, with particular reference to Pasteurella. J Bacteriol 50:361–363. [PubMed] [Google Scholar]
- 42.Kenyon JJ, Hall RM. 2013. Variation in the complex carbohydrate biosynthesis loci of Acinetobacter baumannii genomes. PLoS One 8:e62160. doi: 10.1371/journal.pone.0062160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geisinger E, Isberg RR. 2015. Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS Pathog 11:e1004691. doi: 10.1371/journal.ppat.1004691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davey ME, Duncan MJ. 2006. Enhanced biofilm formation and loss of capsule synthesis: deletion of a putative glycosyltransferase in Porphyromonas gingivalis. J Bacteriol 188:5510–5523. doi: 10.1128/JB.01685-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yi H, Yuan B, Liu J, Zhu D, Wu Y, Wang M, Jia R, Sun K, Yang Q, Chen S, Liu M, Chen X, Cheng A. 2017. Identification of a wza-like gene involved in capsule biosynthesis, pathogenicity and biofilm formation in Riemerella anatipestifer. Microb Pathog 107:442–450. doi: 10.1016/j.micpath.2017.04.023. [DOI] [PubMed] [Google Scholar]
- 46.Petruzzi B, Briggs RE, Tatum FM, Swords WE, De Castro C, Molinaro A, Inzana TJ. 2017. Capsular polysaccharide interferes with biofilm formation by Pasteurella multocida serogroup A. mBio 8:e01843-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weber BS, Harding CM, Feldman MF. 2015. Pathogenic Acinetobacter: from the cell surface to infinity and beyond. J Bacteriol 198:880–887. doi: 10.1128/JB.00906-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 49.Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jørgensen A, Molin S, Tolker-Nielsen T. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol 48:1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x. [DOI] [PubMed] [Google Scholar]
- 50.Yildiz FH, Liu XS, Heydorn A, Schoolnik GK. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol Microbiol 53:497–515. doi: 10.1111/j.1365-2958.2004.04154.x. [DOI] [PubMed] [Google Scholar]
- 51.Chabane YN, Mlouka M, Alexandre S, Nicol M, Marti S, Pestel-Caron M, Vila J, Jouenne T, Dé E. 2014. Virstatin inhibits biofilm formation and motility of Acinetobacter baumannii. BMC Microbiol 14:62. doi: 10.1186/1471-2180-14-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen R, Lv R, Xiao L, Wang M, Du Z, Tan Y, Cui Y, Yan Y, Luo Y, Yang R, Song Y. 2017. A1S_2811, a CheA/Y-like hybrid two-component regulator from Acinetobacter baumannii ATCC17978, is involved in surface motility and biofilm formation in this bacterium. Microbiologyopen 6:e00510. doi: 10.1002/mbo3.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takaya A, Tabuchi F, Tsuchiya H, Isogai E, Yamamoto T. 2008. Negative regulation of quorum-sensing systems in Pseudomonas aeruginosa by ATP-dependent Lon protease. J Bacteriol 190:4181–4188. doi: 10.1128/JB.01873-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soares NC, Spät P, Méndez JA, Nakedi K, Aranda J, Bou G. 2014. Ser/Thr/Tyr phosphoproteome characterization of Acinetobacter baumannii: comparison between a reference strain and a highly invasive multidrug-resistant clinical isolate. J Proteomics 102:113–124. doi: 10.1016/j.jprot.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 55.Nwugo CC, Arivett BA, Zimbler DL, Gaddy JA, Richards AM, Actis LA. 2012. Effect of ethanol on differential protein production and expression of potential virulence functions in the opportunistic pathogen Acinetobacter baumannii. PLoS One 7:e51936. doi: 10.1371/journal.pone.0051936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang N, Ozer EA, Mandel MJ, Hauser AR. 2014. Genome-wide identification of Acinetobacter baumannii genes necessary for persistence in the lung. mBio 5:e01163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Repizo GD, Viale AM, Borges V, Cameranesi MM, Taib N, Espariz M, Brochier-Armanet C, Gomes JP, Salcedo SP. 2017. The environmental Acinetobacter baumannii isolate DSM30011 reveals clues into the preantibiotic era genome diversity, virulence potential, and niche range of a predominant nosocomial pathogen. Genome Biol Evol 9:2292–2307. doi: 10.1093/gbe/evx162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jung JY, Lee DH, Wang EW, Nason R, Sinnwell TM, Vogel JP, Chole RA. 2011. P. aeruginosa infection increases morbidity in experimental cholesteatomas. Laryngoscope 121:2449–2454. doi: 10.1002/lary.22189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen Y, Gozzi K, Yan F, Chai Y. 2015. Acetic acid acts as a volatile signal to stimulate bacterial biofilm formation. mBio 6:e00392-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ching C, Gozzi K, Heinemann B, Chai Y, Godoy VG. 2017. RNA-mediated cis regulation in Acinetobacter baumannii modulates stress-induced phenotypic variation. J Bacteriol 199:e00799-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ducret A, Quardokus E, Brun Y. 2016. MicrobeJ, a high throughput tool for quantitative bacterial cell detection and analysis. Nat Microbiol 1:16077. doi: 10.1038/nmicrobiol.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Joseph LA, Wright AC. 2004. Expression of Vibrio vulnificus capsular polysaccharide inhibits biofilm formation. J Bacteriol 186:889–893. doi: 10.1128/JB.186.3.889-893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.