All organisms use a variety of mechanisms to allocate limited resources to match their needs in their current environment. Here, we explore how halophilic microbes use a novel mechanism to allow efficient production of rhodopsin, a complex of an opsin protein and a retinal prosthetic group. We previously demonstrated that Halobacterium salinarum bacterioopsin directs available resources toward retinal by inhibiting synthesis of bacterioruberin, a molecule that shares precursors with retinal. In this work, we show that this mechanism can be carried out by proteins from halophilic Archaea that are not closely related to H. salinarum and those in at least one species of Bacteria. Therefore, opsin-mediated inhibition of bacterioruberin synthesis may be a highly conserved, ancient regulatory mechanism.
KEYWORDS: cofactor biosynthesis, carotenoid biosynthesis, C50 carotenoid, membrane protein biogenesis, microbial rhodopsin, proteorhodopsin, UbiA prenyltransferase
ABSTRACT
Halophilic Archaea are a distinctive pink color due to a carotenoid pigment called bacterioruberin. To sense or utilize light, many halophilic Archaea also produce rhodopsins, complexes of opsin proteins with a retinal prosthetic group. Both bacterioruberin and retinal are synthesized from isoprenoid precursors, with lycopene as the last shared intermediate. We previously described a regulatory mechanism by which Halobacterium salinarum bacterioopsin and Haloarcula vallismortis cruxopsin inhibit bacterioruberin synthesis catalyzed by lycopene elongase. In this work, we found that opsins in all three major Halobacteria clades inhibit bacterioruberin synthesis, suggesting that this regulatory mechanism existed in the common Halobacteria ancestor. Halophilic Archaea, which are generally heterotrophic and aerobic, likely evolved from an autotrophic, anaerobic methanogenic ancestor by acquiring many genes from Bacteria via lateral gene transfer. These bacterial “imports” include genes encoding opsins and lycopene elongases. To determine if opsins from Bacteria inhibit bacterioruberin synthesis, we tested bacterial opsins and found that an opsin from Curtobacterium, in the Actinobacteria phylum, inhibits bacterioruberin synthesis catalyzed by its own lycopene elongase, as well as that catalyzed by several archaeal enzymes. We also determined that the lycopene elongase from Halococcus salifodinae, a species from a family of Halobacteria lacking opsin homologs, retained the capacity to be inhibited by opsins. Together, our results indicate that opsin-mediated inhibition of bacterioruberin biosynthesis is a widely distributed mechanism found in both Archaea and Bacteria, possibly predating the divergence of the two domains. Further analysis may provide insight into the acquisition and evolution of the genes and their host species.
IMPORTANCE All organisms use a variety of mechanisms to allocate limited resources to match their needs in their current environment. Here, we explore how halophilic microbes use a novel mechanism to allow efficient production of rhodopsin, a complex of an opsin protein and a retinal prosthetic group. We previously demonstrated that Halobacterium salinarum bacterioopsin directs available resources toward retinal by inhibiting synthesis of bacterioruberin, a molecule that shares precursors with retinal. In this work, we show that this mechanism can be carried out by proteins from halophilic Archaea that are not closely related to H. salinarum and those in at least one species of Bacteria. Therefore, opsin-mediated inhibition of bacterioruberin synthesis may be a highly conserved, ancient regulatory mechanism.
INTRODUCTION
Halophilic archaea that make up the class Halobacteria grow to such high densities that they provide the brilliant pink, red, and orange colors characteristic of salt lakes and ponds. These colors are primarily produced by 50-carbon carotenoid pigments called bacterioruberins. Although their primary function has not been fully characterized in vivo, bacterioruberins are proposed to protect cells from osmotic stress (1) or damage from UV light (2). In Archaea, both carotenoids and the hydrophobic tails of membrane lipids are produced from isoprenoid precursors (3). Universally, isoprenoids are derived from the 5-carbon precursor molecules isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), which themselves are generated from acetyl coenzyme A (acetyl-CoA) and ATP. Therefore, IPP and DMAPP represent a substantial cellular investment of reduced carbon and energy. IPP and DMAPP are then consumed in consecutive condensation reactions to generate the 20-carbon geranylgeranyl pyrophosphate (GGPP), the last shared intermediate of carotenoid and polar lipid biosynthesis in Archaea (3). In the committed step of carotenoid biosynthesis, phytoene synthase catalyzes the condensation of two GGPP molecules to form the 40-carbon phytoene (4). Phytoene is then desaturated by phytoene dehydrogenase (phytoene desaturase) to generate lycopene (5). For bacterioruberin synthesis, lycopene elongase (Lye) catalyzes the addition of two C5 isoprenoids to generate the 50-carbon bacterioruberin, which may then be modified by further desaturation and hydroxylation (Fig. 1A) (6, 7).
FIG 1.
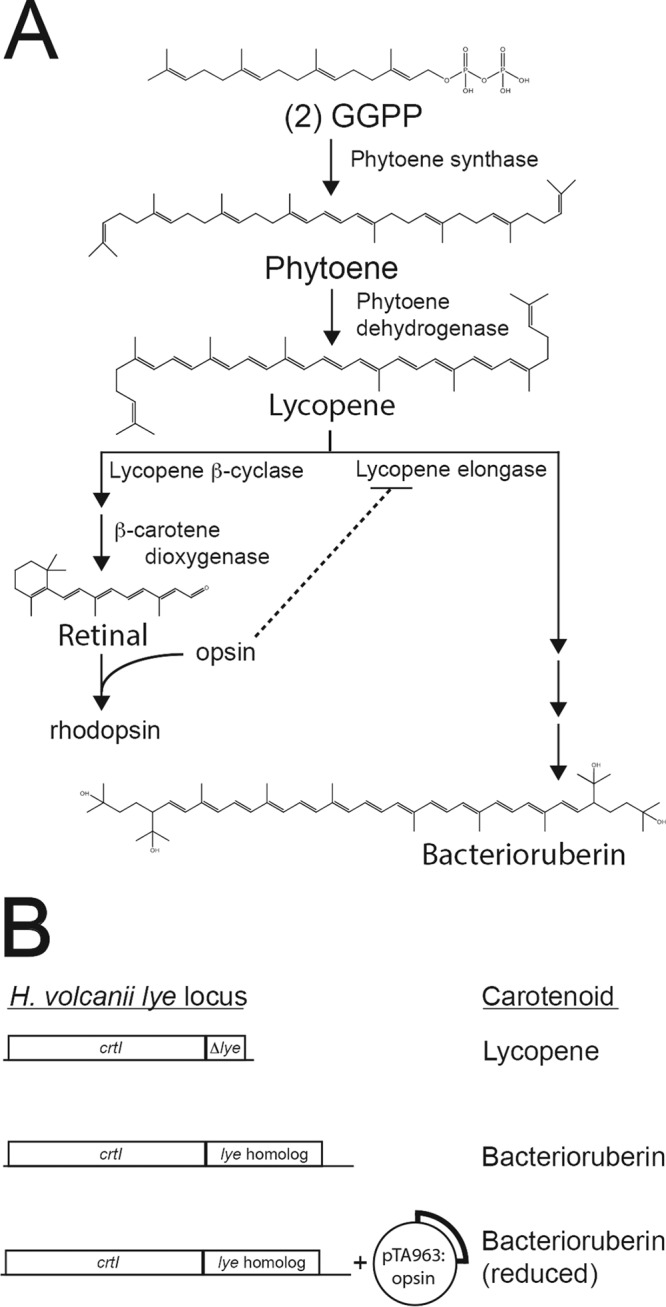
(A) Carotenoid biosynthesis in halophilic Archaea. Enzyme names are indicated next to reactions catalyzed. A broken line indicates the regulatory mechanism of an opsin inhibiting the committed step in bacterioruberin synthesis. GGPP, geranylgeranyl pyrophosphate. (B) Experimental approach for testing opsin-mediated inhibition of Lye in H. volcanii. The H. volcanii lye locus is shown approximately to scale on the left and the carotenoid produced is indicated on the right. The lye gene encoding lycopene elongase is directly downstream and in a putative operon with crtI, the gene that encodes phytoene dehydrogenase. No bacterioruberin is produced in the H. volcanii Δlye strain (Fig. 2). lye homologs are incorporated into the lye locus, maintaining the integrity of crtI, and bacterioruberin synthesis is restored (Fig. 2). Opsins are then expressed by introducing pTA963 (34), harboring the gene encoding the opsin, and bacterioruberin production is quantified to determine if the opsin inhibits Lye activity.
For the many halophilic Archaea that produce rhodopsin complexes for energy conversion, lycopene is the final shared intermediate for two end products, namely, bacterioruberin and retinal, the prosthetic group (sometimes called “cofactor”) of rhodopsin. To generate retinal, lycopene is cyclized to form β-carotene, which then undergoes oxidative cleavage to form retinal (8–10). Given the energy input required for carotenoid biosynthesis and the requirement for retinal to form functional rhodopsins, these organisms must have regulatory processes to optimally allocate lycopene for bacterioruberin or retinal synthesis. We previously described a mechanism of regulation in which Halobacterium salinarum bacterioopsin (BO), when not bound by retinal, inhibits bacterioruberin synthesis catalyzed by Lye (6). We subsequently identified a similar inhibition by the opsin of Haloarcula vallismortis (11). This inhibition likely ensures that lycopene is conserved for retinal biosynthesis when opsins are available. In the work described here, we examined opsins and Lyes from multiple halophilic Archaea. Our findings demonstrate that opsin-mediated inhibition of bacterioruberin synthesis is distributed throughout the Halobacteria and indicate that this mechanism may have existed in the last common Halobacteria ancestor.
We next explored the possibility that opsin-mediated inhibition of bacterioruberin synthesis existed at the origin of halophilic Archaea. Substantial evidence indicates that halophilic Archaea arose from a methanogenic species (12, 13). The transition from a presumably anaerobic, autotrophic metabolism to the generally aerobic, heterotrophic metabolism of halophilic Archaea has been proposed to be driven by acquisition of bacterial genes (14, 15). A recent study examining this proposed acquisition predicted Lye and bacterioopsin of Halobacteria to be among these laterally transferred genes from Bacteria (15). To gain insight into the evolution of this mechanism, we examined the possibility that opsins from Bacteria inhibit Lye homologs. Of two bacterial opsins analyzed, one, an opsin homolog from a member of the Actinobacteria, Curtobacterium sp. strain ER1/6, inhibited its own Lye. In addition, the Curtobacterium sp. ER1/6 opsin also inhibited the Lye from several Archaea species, suggesting some level of functional and structural conservation even across domains. To analyze if Lyes from a species lacking opsins could be subject to opsin-mediated inhibition, we used the Lye from Halococcus salifodinae, a species isolated from Permian halite deposits (16, 17) that lacks opsin homologs (18). We found that the H. salinarum bacterioopsin could inhibit bacterioruberin synthesis catalyzed by H. salifodinae Lye, suggesting that this enzyme retains structural features allowing inhibition in this organism devoid of opsin homologs.
RESULTS AND DISCUSSION
Bacterioruberin and retinal synthesis is common and widely distributed in Halobacteria species.
To assess if the need to coordinate bacterioruberin and retinal production is common in halophilic Archaea, we analyzed Halobacteria genome sequences to determine the prevalence of organisms with the necessary genes for synthesis of these molecules (Table 1; homolog accession numbers listed in Table S1 in the supplemental material). We conducted a preliminary examination of all 348 Halobacteria genome sequences available on GenBank. To allow consistent analysis, we selected one representative genome for each species and excluded genomes with too many contigs and those missing highly conserved, essential genes. Among the 220 genomes that remained after this selection, we found that carotenoid synthesis is nearly universally conserved, as 218 genomes harbor the necessary genes, encoding phytoene synthase and phytoene dehydrogenase, needed to catalyze the conversion of GGPP to lycopene. Of these genomes that have genes for lycopene production, only 3 lack a gene encoding Lye, needed for bacterioruberin synthesis. All 43 completely assembled genomes contain all genes necessary for bacterioruberin production, suggesting that incomplete sequences may account for genomes that lack these genes (Table 1). Overall, our analysis indicates that the ability to generate bacterioruberin from isoprenoid precursors is a nearly universal characteristic of halophilic Archaea.
TABLE 1.
Number of Halobacteria genomesa harboring genes allowing synthesis of lycopene, bacterioruberin, retinal, and/or opsins
| Product(s) (enzyme[s] encoded) | Complete genomes (no.) | Incomplete genomesb (no.) | Total (no.) |
|---|---|---|---|
| Lycopene (phytoene synthase, phytoene dehydrogenase) | 43 | 175 | 218 |
| Bacterioruberin (lycopene elongase) | 43 | 172 | 215 |
| Retinal (lycopene cyclase, β-carotene dioxygenase) | 25 | 114 | 139 |
| Opsin | 29 | 123 | 152 |
| Retinal and opsin | 24 | 112 | 136 |
| Retinal, opsin, and bacterioruberin | 24 | 111 | 135 |
Halobacteria genome sequences were obtained from the National Center for Biotechnology Information (NCBI; www.ncbi.nlm.nih.gov) in August 2018, and homologs were identified as described in Materials and Methods. All homologs identified are listed in Table S1 in the supplemental material. In total, 220 genomes were analyzed, 43 complete and 177 incomplete.
Genome assembly levels “contig” and “scaffold,” as labeled in the NCBI database, were considered to be incomplete.
In contrast, the capacity to produce rhodopsin complexes is widespread but not universal in Halobacteria species. We found that 152 genomes out of 220 harbor a gene for at least one opsin, and 136 of these also have genes encoding enzymes necessary for generating retinal from lycopene, lycopene β-cyclase (9) and β-carotene dioxygenase (8, 10). Comparing these values, 16 species, including 5 completely assembled genomes, have genes encoding opsins but apparently do not have the ability to generate retinal. These proteins may perform functions that do not require retinal binding, and a role in responding to stimuli other than light has been previously proposed for these putatively “blind” opsins (19). Interestingly, three genomes that have genes for retinal synthesis, including the completely assembled Halopenitus persicus genome, lack identifiable opsin homologs, suggesting that retinal functions in other roles aside from serving as an opsin prosthetic group. In total, 135 genomes out of the 220 analyzed contained genes for bacterioruberin, retinal, and rhodopsin synthesis (Table 1), indicating that approximately half of the known Halobacteria species require a mechanism to direct available lycopene toward retinal or bacterioruberin.
Although the phylogeny of the Halobacteria is not fully characterized, a combination of 16S rRNA and RNA polymerase subunit B′ sequence comparisons defined two clades, with other organisms placed in a third group (20). With many more genome sequences available, a subsequent study using 40 concatenated conserved marker genes broadly supported the earlier phylogeny and clearly defined three clades within Halobacteria (18). We had previously found that opsins from two organisms in clade 3, H. salinarum and H. vallismortis, inhibit their respective Lyes. To examine if this mechanism occurs throughout Halobacteria, we selected nine organisms distributed throughout the class and included at least two members from each of the three clades (Table 2). Since our previous work indicated that opsins from proton pumps mediated this activity, we chose opsin homologs that had residues equivalent to aspartates at positions 85 and 96, demonstrated to be required for proton translocation in H. salinarum bacteriorhodopsin (21). Very few halophilic Archaea have been developed to be genetically tractable, so our approach to study potential interaction between these proteins was heterologous expression and assessment of bacterioruberin production in Haloferax volcanii (Fig. 1B).
TABLE 2.
Source species of Lye and opsin homologs used in this study
| Species or strain | Domain | Archaeal cladeb | UniProtKB accession no. for: |
Genome reference or source | |
|---|---|---|---|---|---|
| Lye homolog | Opsin homolog(s) | ||||
| Halobacterium salinarum | Archaea | 3 | Q9HPD9 | P02945 | 38 |
| Haloarcula marismortui ATCC 43049 | Archaea | 3 | Q5V532 | Q5UXY6, Q5V0R5 | 39 |
| Halorhabdus utahensis DSM 12940 | Archaea | 3 | C7NVL7 | C7NQP3 | 40 |
| Natronomonas moolapensis DSM 18674 | Archaea | 3 | M1XS73 | M1Y5I7 | 41 |
| Halomicrobium mukohataei DSM 12286 | Archaea | 3 | C7P310 | C7P2T2 | 42 |
| Haloarcula vallismortis ATCC 29715 | Archaea | 3 | A0A1H2XMF1 | P94854 | Unpublished |
| Haloquadratum walsbyi DSM 16854 | Archaea | 2 | G0LJV2 | G0LFX8, G0LFY1 | 43 |
| Halorubrum distributum JCM 10118 | Archaea | 2 | M0F851 | M0F390a | 18 |
| Halorubrum sp. A07HR67 | Archaea | 2 | V4XXE3 | V4ZQH6 | 44 |
| Natrinema pellirubrum DSM 15624 | Archaea | 1 | L0JK84 | L0JG40 | 18 |
| Haloterrigena mahli H13 | Archaea | 1 | A0A179EP62 | A0A179EFG5 | 45 |
| Haloferax volcanii DS2 | Archaea | 1 | D4GTV9 | None | 46 |
| Halococcus salifodinae DSM 8989 | Archaea | NA | M0N6J9 | None | 18 |
| Rubrobacter xylanophilus DSM 9941 | Bacteria | NA | Q1ATY0 | Q1AUE6 | Unpublished |
| Curtobacterium sp. ER1/6 | Bacteria | NA | A0A1E5MGI3 | A0A1E5MMK0 | 26 |
Lycopene elongases are functionally expressed in H. volcanii.
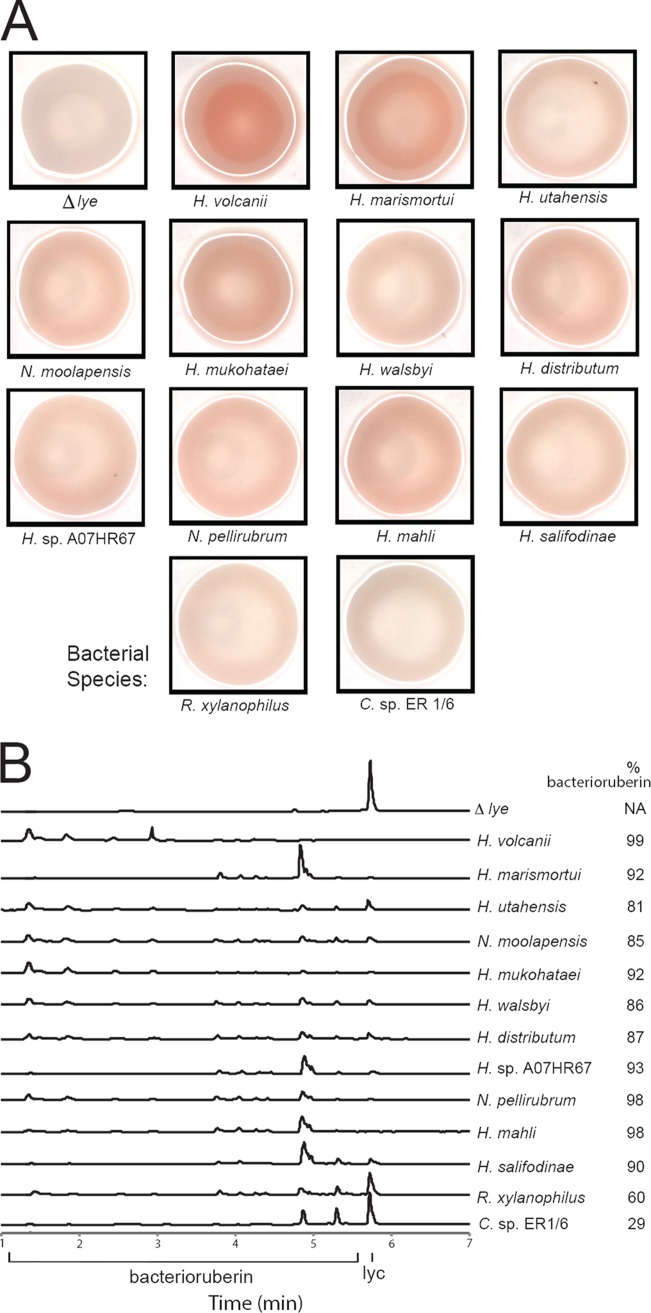
As a first step, we obtained codon-optimized synthetic genes encoding Lye from the nine Halobacteria species. These genes were integrated into the chromosome of H. volcanii, exactly replacing the open reading frame of the native lye gene (Fig. 1B) to allow similar expression to the native Lye protein. All resulted in a shift in colony color to pink, compared to the off-white of the lye deletion strain (Fig. 2A), indicating a rescue of bacterioruberin synthesis. To confirm bacterioruberin production, carotenoids were extracted from liquid cultures and analyzed by high-performance liquid chromatography (HPLC). In wild-type H. volcanii cells, bacterioruberin constitutes ∼99% of the total lycopene-derived carotenoid. While exhibiting slightly reduced activity compared to that of native Lye, all Lyes from Halobacteria restored at least 86% of bacterioruberin synthesis in H. volcanii (Fig. 2B). Thus, heterologous expression in H. volcanii is a suitable technique to assess Lye activity.
FIG 2.
Expression of Lye homologs in H. volcanii restores bacterioruberin production. (A) Photographs of representative colonies of H. volcanii strains with the native lye deleted (Δlye) or replaced with lye homologs from the indicated species. Colonies were prepared from cultures, grown for 4 days, and photographed as described in Materials and Methods. (B) Reverse-phase ultra-high-performance liquid chromatography (RP-UHPLC) traces of carotenoid extracts from H. volcanii Δlye, H. volcanii H1209 (parental strain), and strains expressing Lye homologs from the indicated species. Positions of bacterioruberin and lycopene (lyc) standards are noted. Traces were normalized for total carotenoid concentration, corrected for slight differences in retention time using an internal standard, and offset along the vertical axis for clarity. Traces are representative of at least 2 replicates for each strain, except for Halorubrum sp. A07HR67, where only 1 trace was available. Values indicate bacterioruberin as a molar percentage of total lycopene and bacterioruberin.
Expression of opsins often inhibits the Lyes of their respective species.
We had previously determined that expression of H. salinarum BO in H. volcanii cells allowed effective inhibition of bacterioruberin synthesis catalyzed by H. salinarum Lye (11). To assess potential opsin-mediated inhibition of bacterioruberin synthesis across the Halobacteria class, we expressed opsins in the H. volcanii strains harboring the lye genes from their respective organisms. Lyes from five organisms out of the nine tested exhibited a reduction in bacterioruberin production upon expression of an opsin from their genome (Fig. 3). The most active opsin tested was Bop2 from Haloquadratum walsbyi, a clade 2 species, which resulted in an 82% reduction in bacterioruberin synthesis. H. walsbyi Bop1 also reduced bacterioruberin, but only by 30%. A similar difference in activity was observed in the two opsins tested for Haloarcula marismortui, a clade 3 species, as Bop1 inhibited bacterioruberin production by 33% (Fig. 3), compared to only 19% inhibition by Bop2. Expression of opsins from the clade 1 species Natrinema pellirubrum, clade 2 species Halorubrum sp. strain A07HR67, and clade 3 species Halorhabdus utahensis also significantly inhibited bacterioruberin production from their respective Lye enzymes. When the H. volcanii strains expressing these opsins were grown in the presence of retinal to convert the available opsin to the rhodopsin complex, bacterioruberin synthesis was partially, but significantly, restored in nearly all strains, with only the H. marismortui Bop2-expressing strain lacking a statistically significant increase in bacterioruberin (Fig. 3). Since H. volcanii lack any known proteins that interact with retinal, this increase in bacterioruberin is strong evidence that inhibition of Lye is caused by opsin. Moreover, this reduction in inhibition with retinal indicates that the opsins folded into structures that allowed at least a partial formation of the rhodopsin complex.
FIG 3.
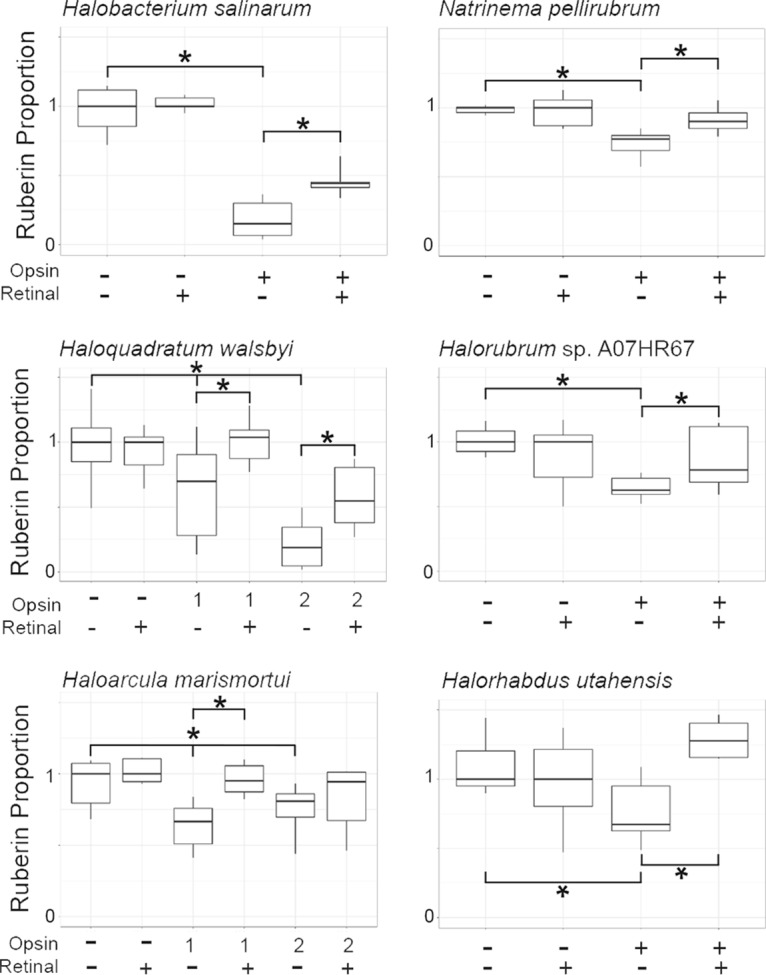
Expression of opsins inhibits Lye-catalyzed bacterioruberin production in Halobacteria species. Box plot (Tukey’s [37]) indicating proportionate bacterioruberin levels compared to those of the empty vector control in H. volcanii strains expressing Lye homologs from the indicated species. Bacterioruberin levels were determined by colony pigment analysis, as described previously (11). Heavy horizontal bars indicate the median value, and boxes demarcate the upper and lower quartiles. Whiskers extend to the smaller value of 1.5 times the interquartile range or to the most extreme value. Asterisks indicate a Bonferroni adjusted P value of <0.05 (n ≥ 6).
To assess if inhibition was correlated to opsin abundance, we constructed opsin genes with C-terminal hexahistidine (His6) tags and analyzed expression via immunoblotting. We had previously used a His6-tagged H. salinarum BO to assess expression (11), and this BO also inhibited bacterioruberin synthesis catalyzed by H. salinarum Lye (data not shown). Among the opsins that exhibited inhibitory activity, BO homologs from N. pellirubrum, Halorubrum sp. A07HR67, and H. utahensis all had abundances at 50% or greater relative to that of H. salinarum BO (Fig. 4). Intriguingly, for organisms with two BO homologs, abundance was inversely correlated to inhibitory activity; that is, H. marismortui Bop1 was much less abundant than H. marismortui Bop2. For H. walsbyi, the less active Bop1 was measured at 17% of H. salinarum BO abundance, but the H. walsbyi Bop2, highly active in inhibiting bacterioruberin, was not detected in our immunoblots (Fig. 4). Although data relating to the native physiological abundance of opsins and Lye in these species are not available, our results suggest that even a small amount of opsin can inhibit bacterioruberin production.
FIG 4.
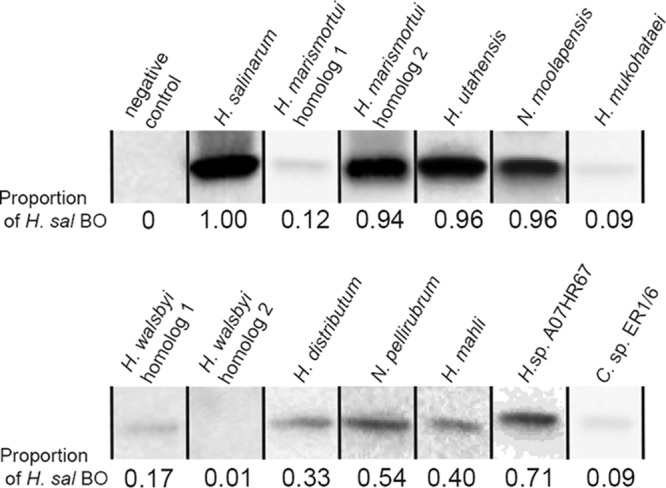
Expression of opsins in H. volcanii. H. volcanii strains harboring expression plasmids for indicated opsins with C-terminal His6 tags were grown in cultures using the same procedures as that for carotenoid analysis. Cell lysates were normalized by total protein concentration and separated by polyacrylamide gel electrophoresis. Blots were probed with anti-His6 primary antibody. For standardization across multiple blots, cell lysates from H. volcanii expressing H. salinarum BO were included on all blots. Subsequently, each immunoblot image was adjusted (as an entire image) so that the BO bands from all blots had identical net (band subtracting background) mean gray values. The proportion indicated on the image was determined by dividing the net mean gray value of each opsin band into the net mean gray value of H. salinarum BO. Bands shown are from different blots, as indicated by black lines, and each is representative of at least 3 biological replicates. All full immunoblots are shown in Fig. S1 in the supplemental materials.
The expression of opsins from four species, Haloterrigena mahli, Halorubrum distributum, Halomicrobium mukohataei, and Natronomonas moolapensis, did not result in a significant decrease in bacterioruberin synthesis (data not shown), but expression of all proteins was detected at various levels. Given that the expression of these opsins was generally reduced compared to that of opsins that exhibited inhibition of bacterioruberin synthesis, it may be that these proteins have structural differences when expressed in H. volcanii that prevent normal inhibition of Lye activity. Alternatively, this regulatory mechanism may not occur in these species.
Taken together, our results indicate that opsin-mediated inhibition of bacterioruberin production is not confined to clade 3 species and is likely common in Halobacteria. Overall, we have evidence that this mechanism occurs in 7 out of 11 total species tested. Opsin-mediated inhibition is nearly as likely to occur in clade 1 species, (1 of 2 tested) and clade 2 species (2 of 3 tested) as it is in clade 3 species (2 of 4 tested in this report and 4 out of 6 when including the previously reported species H. salinarum and H. vallismortis). Additionally, due to differences in environmental conditions, some of the opsin-Lye pairs tested by heterologous expression in H. volcanii are likely false negatives. From our results, we think the simplest explanation is that this regulatory mechanism existed in the last common ancestor of the Halobacteria and may have been present when the halophilic Archaea arose from an influx of bacterial genes into a methanogen.
A bacterial opsin is capable of inhibiting bacterioruberin production.
To determine if opsin-mediated inhibition of Lye could occur in Bacteria, we conducted similarity (BLAST) searches of the UniProt microbial proteomes database to identify genomes in Bacteria with homologs to H. salinarum bacterioopsin and Lye. To focus our search on organisms that likely produced the retinal cofactor of opsin from a lycopene precursor, searches of the same database were conducted using enzymes that catalyze the conversion of lycopene to β-carotene, namely, H. salinarum lycopene cyclase (9) (UniProtKB accession number Q9HNE5), Solanum lycopersicum chloroplastic lycopene cyclase (22) (UniProtKB accession number Q43503), and Synechococcus sp. lycopene cyclase (23) (UniProtKB accession number A5A545). Similarity searches were then conducted with β-carotene dioxygenases from H. salinarum (10) (UniProtKB accession number Q9HNE6) and Salinibacter ruber (24) (UniProtKB Q2S6A1), which both catalyze the oxidative cleavage of β-carotene to retinal but are not homologous. From this analysis, 13 bacterial genomes were identified that encode homologs of bacterioopsin, enzymes that allow retinal synthesis from lycopene, and homologs of Lye. The opsin sequences were analyzed to identify those with the required residues to allow light-driven proton translocation. The region of the genomes containing the lye homologs was explored to identify genes potentially involved in carotenoid synthesis. Lastly, we conducted a literature search to determine if the identified species or close relatives produced a red or pink pigment that could indicate a modification of lycopene.
The best matches for potential opsin-mediated inhibition of Lye activity from this screen were genes from two species (Table 1), both in the Actinobacteria phylum, namely, Rubrobacter xylanophilus, a thermophilic freshwater organism (25), and Curtobacterium sp. ER1/6, a citrus tree endophyte (26). The R. xylanophilus opsin has demonstrated proton-pumping activity (27). The Curtobacterium sp. ER1/6 protein is not annotated as an opsin but includes a lysine residue at position 241 for likely retinal binding and acidic residues at positions 101 and 112 involved in proton translocation. Both R. xylanophilus and Curtobacterium sp. ER1/6 lye homolog open reading frames (ORFs) are upstream and overlap genes homologous to Haloarcula japonica crtD, a gene demonstrated to encode a carotenoid 3,4-desaturase, which catalyzes the processing of the first bacterioruberin intermediate to mature bacterioruberin (7). Interestingly, this potential operon mirrors that observed in Halobacteria genomes, except that the orientation is reversed, with the crtD homolog upstream of lye homologs in Halobacteria.
To examine potential opsin-mediated inhibition, we obtained genes encoding Lye and opsin homologs codon optimized for expression in halophilic Archaea. Incorporation of the R. xylanophilus or Curtobacterium sp. ER1/6 lye homolog into the H. volcanii lye locus resulted in partial recovery of bacterioruberin production (Fig. 2) despite the dramatically increased salinity compared to the environments in the native organisms. Expression of the R. xylanophilus opsin in the H. volcanii strain with the R. xylanophilus lye homolog resulted in no reduction in bacterioruberin production (data not shown). In contrast, expression of the Curtobacterium sp. ER1/6 opsin homolog resulted in a small, but significant, reduction in bacterioruberin synthesis catalyzed by the Curtobacterium sp. ER1/6 Lye homolog (Fig. 5). To examine potential cross-domain interactions, we tested the ability of the Curtobacterium sp. ER1/6 opsin to inhibit Lye from halophilic Archaea. While expression of Curtobacterium sp. ER1/6 opsin had no significant effect on H. volcanii Lye, it significantly reduced bacterioruberin production catalyzed by H. salinarum, H. vallismortis, and Halorubrum sp. A07HR67 Lye (Fig. 5). This result raises the possibility that the opsin-Lye interaction existed in the ancestor that gave rise to both bacterial and archaeal proteins.
FIG 5.
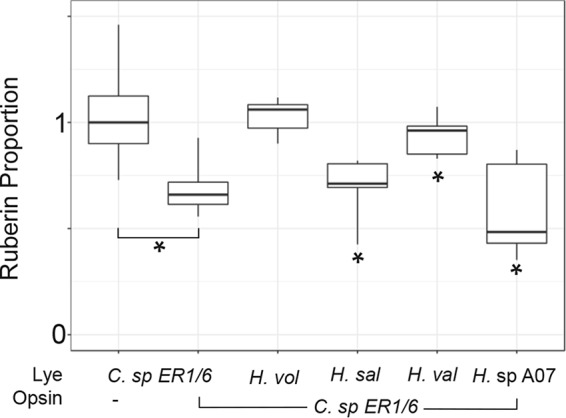
A bacterial opsin inhibits its own Lye and several archaeal Lyes. Box plot indicating proportionate bacterioruberin levels of H. volcanii strains expressing the indicated Lye and, if noted, the Curtobacterium opsin homolog. Relative bacterioruberin levels were determined by colony color analysis, and data were plotted as described for Fig. 3. Asterisks indicate a Bonferroni adjusted P value of <0.05 compared to empty vector controls (n ≥ 6).
A Lye from a deeply rooted branch of Halobacteria that natively lack opsins is capable of being inhibited by an opsin.
Halococcus is the only genus within the Halococcaceae family, and a multimarker phylogeny places the halococci as a deep-rooted branch outside the three major Halobacteria clades (18). There are seven Halococcus genome sequences available, and all of them encode enzymes that allow bacterioruberin production from GGPP (see Table S1 in the supplemental material). None of the available genomes encode detectable homologs to opsins or retinal biosynthetic enzymes. The Halococcus common ancestor may have lost genes for rhodopsins and retinal biosynthesis before radiation. An alternative explanation is that opsin and retinal biosynthetic genes were acquired by other Halobacteria species after the Halococcus lineage diverged. We chose to study the Lye of Halococcus salifodinae, an organism cultured from halite deposited in the Permian period (16), suggesting that the species has been isolated for approximately 250 million years. A gene encoding H. salifodinae Lye was integrated into the H. volcanii lye locus, and bacterioruberin synthesis was restored to 90% of wild-type levels (Fig. 2B). Expression of BO from H. salinarum, a clade 3 species, sharply reduced bacterioruberin synthesis by 74%. To determine if H. salifodinae Lye could be inhibited by other opsins, we tested active opsins from Halobacteria clade 1, N. pellirubrum bacterioopsin, and from clade 2, H. walsbyi bacterioopsin 2. Neither opsin significantly inhibited H. salifodinae Lye. In addition, we tested the bacterial opsin from Curtobacterium sp. ER1/6, and no inhibition was observed. Therefore, a halobacterial Lye that has not been exposed to selective pressure from opsin expression for a long period may retain features that allow specific opsin-mediated inhibition.
Conclusion. We previously demonstrated that opsin-mediated inhibition of bacterioruberin biosynthesis occurs in two relatively closely related species, H. salinarum and H. vallismortis, from clade 3 of the Halobacteria. In this work, we demonstrate that opsins throughout the Halobacteria have the ability to inhibit the activity of their Lye enzymes. We also demonstrate that a bacterial opsin inhibits its Lye and those of least three Archaea species. In addition, a Lye homolog from an early diverged species outside the 3 major clades, H. salifodinae, can be inhibited by a modern opsin. Together, our results demonstrate that that opsin-mediated inhibition of Lye is widely conserved in Halobacteria species and may occur between similar proteins in Bacteria species.
Halophilic Archaea likely arose from a methanogenic species (12, 13), and this emergence is correlated to lateral acquisition of many bacterial genes (14, 15). One explanation for the widespread distribution of opsin-mediated inhibition of Lye in Halobacteria is that this regulatory mechanism existed when both opsin and Lye were acquired together in the common ancestor of Halobacteria. A recent paper explored the genomes of two members of Methanonatronarchaeia, newly isolated halophilic methanogens that are proposed to form a class most closely related to Halobacteria (28). This study suggests that both opsins and Lye were acquired by the common ancestor of Halobacteria. In this scenario, some lineages of Halobacteria lost genes encoding opsins, while, as we found in our analysis, lye and other genes allowing bacterioruberin synthesis were nearly universally maintained. This interpretation is supported by the finding that the Lye from H. salifodinae, a species from a genus that lacks any known opsins, retains the ability to be inhibited by opsin (Fig. 6). We also have evidence that this mechanism is found in Bacteria, as we observed that expression of the Curtobacterium sp. ER1/6 opsin inhibits its Lye activity. However, we have determined that not all Lyes are inhibited by all or any opsins, a phenomenon that indicates that other selective pressures on the genes may have resulted in the loss of this inhibitory interaction.
FIG 6.
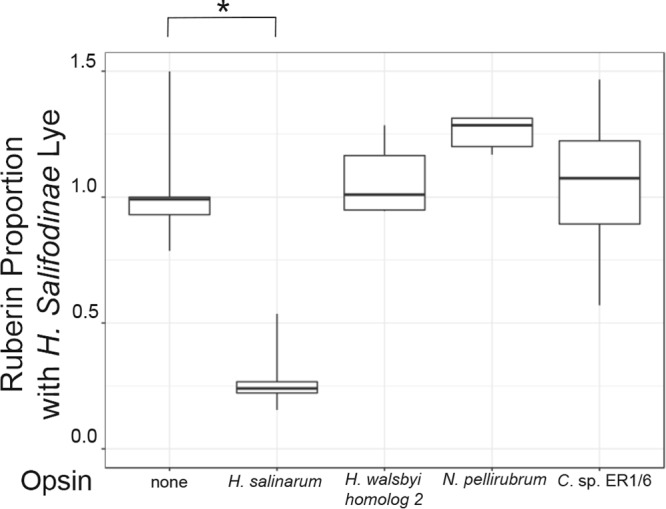
Lye from a Halobacteria species that lacks opsins is inhibited by H. salinarum BO. Box plot indicating proportionate bacterioruberin levels of H. volcanii strains expressing H. salifodinae Lye and the indicated opsin homologs. Relative bacterioruberin levels were determined by colony color analysis, and data were plotted as described for Fig. 3. The asterisk indicates a Bonferroni adjusted P value of <0.05 compared to empty vector controls (n ≥ 6).
An alternative hypothesis is that this inhibition arose after radiation of the Halobacteria and spread by lateral gene transfer. To examine the possibility that genes encoding opsin or Lye exist on horizontally transferred regions, we examined predicted genomic islands (29) for the species listed in Table 1. Although many genomic islands were predicted, none of the archaeal Lye or opsin homologs used in our study were in these regions. In the bacterial genes used in our study, only the R. xylanophilus opsin, which notably did not show inhibition activity, was predicted to be in the region of the genome inherited by lateral transfer. We also think that inheritance by lateral transfer of smaller genome regions is unlikely, as the lye and bop homologs are not closely associated in any genome we examined. Our data also do not exclude the possibility that this mechanism is so favorable that it arose independently in multiple lineages. We plan to continue exploring required structural motifs in both opsin and Lye proteins to provide additional insight into the evolution of this regulatory interaction.
MATERIALS AND METHODS
Genomic analysis.
As of August 2018, 348 genomes were classified as Halobacteria in the GenBank database (30). For analysis, we eliminated genomes assembled to more than 200 contigs or scaffolds and chose one genome from each available species. If genomes from multiple strains in a species were available, we chose the type strain genome if available but otherwise chose the genome sequence with the most complete assembly. BLAST (31) databases for each genome were constructed from FASTA amino acid (.faa) files for proteins and FASTA nucleotide files (.fna) for nucleotides, using makeblastdb version 2.6.0 with default options. BLASTP (version 2.6.0) searches to determine the presence of proteins in each genome were conducted using default options and an E value cutoff of 1 × 10−10. To check for missed annotations, genomes were subjected to TBLASTN (version 2.6.0) searches with default options of nucleotide sequences with an E value cutoff of 1 × 10−10. Since our initial analyses revealed that many incompletely assembled genomes were likely missing significant sections of DNA sequence, we assessed genome completeness by performing BLAST and TBLASTN searches for essential, highly conserved genes, as reported in reference 32. Out of the 93 genes identified as highly conserved in both Archaea and Eukarya (see Tables S2 and S3 in reference 32), we chose a subset of 10 genes that represented a variety of cellular processes. The H. salinarum homolog of the following genes were used for these searches (protein name [NCBI gi number, UniProtKB accession number for H. salinarum homolog]): aspartate-tRNA ligase (6323011, O07683), RIO-type serine/threonine protein kinase Rio1 (6324693, B0R7D7), RNase L inhibitor (6320296, Q9HMC1), RNA polymerase subunit A′ (6324690, P0CX02), ribosomal protein S3 (6324151, P15009), rRNA small subunit methyltransferase A (6324989, Q9HQH1), guanine nucleotide exchange factor SDO1 (6323050, Q9HQ87), protein translocase subunit SecY (6323411, Q9HPB1), signal recognition particle 54-kDa protein (6325345, Q9HMN5), and TATA box-binding protein E (6320996, Q9HN56). Notably, all 43 genomes labeled with an assembly level of “complete” harbored homologs for all 10 of these genes, indicating that genomes with assembly levels of “contig” or “scaffold” may be missing substantial portions of the actual genome sequence.
After eliminating genomes lacking one or more of these essential genes, we assessed the ability to produce carotenoids and opsins by conducting BLAST searches using the following genes: to assess the ability to generate lycopene from GGPP, H. salinarum phytoene synthase 2 (UniProtKB accession number Q9HPE1) and H. salinarum phytoene dehydrogenase 1 (Q9HPD8); to identify species with the capability to synthesize bacterioruberin, H. salinarum Lye (Q9HPD9); and to determine the ability to synthesize retinal, H. salinarum lycopene β-cyclase (Q9HNE5) and β-carotene dioxygenase (Q9HNE6). Genes encoding opsins were identified using H. salinarum bacterioopsin (P02945), haloopsin (P0DMH7), sensory opsin-1 (P0DMH8), and sensory opsin-2 (P71411).
H. volcanii strain construction.
H. volcanii strains are listed in Table S2 in the supplemental material. Synthetic gene sequences and details of plasmid construction are given in Tables S3 and S4, respectively, in the supplemental material. Synthetic genes encoding Lyes and opsins were designed using codon optimization for H. marismortui, using the IDTDNA Codon Optimization Tool (https://www.idtdna.com/CodonOpt), and ordered from GenScript (Piscataway, NJ). Plasmids all harbored an ampicillin resistance marker and were propagated in Escherichia coli DH5α cells (New England Biolabs, Ipswich, MA) in LB media with 100 μg/ml ampicillin (Gold Biotechnology, St. Louis, MO). For integration into the H. volcanii lye locus, synthetic genes encoding Lye were amplified with appropriate primers (Table S5 in the supplemental material) and ligated into pRFP128 (11), an integrative plasmid derived from pTA131 (33), using NdeI and BglII sites.
For expression of opsins, codon-optimized synthetic genes (Table S3) were inserted into pTA963 (34), using NdeI and EcoRI sites, to allow expression from the p.tnaA promoter. As in our previous studies (11), we relied on the moderate level of expression of this promoter when H. volcanii strains with pTNA963-based plasmids are grown in H. volcanii-yeast extract-peptone-Casamino Acids (Hv-YPC) medium (35). Genes encoding opsins with C-terminal His6 tags were amplified with primer RP268 (anneals to pTA963-based plasmids) and appropriate primers (Table S5) to encode His6, using the previously constructed opsin expression plasmids as the template. These PCR products were inserted into pTA963 using NdeI and EcoRI sites. All plasmid modifications were confirmed by sequencing. Genetic modification of H. volcanii was carried out in derivatives of strain H1209, as previously described (34). Proper integration of lye homologs into the lye locus was confirmed by PCRs that included one primer from the lye homolog and one primer, RP558 (Table S5), with a binding site on the chromosome but not in the integrative plasmid. The Lye-encoding region, and plasmid inserts in all H. volcanii strains, were sequenced to confirm accurate genotypes.
Carotenoid analysis.
For carotenoid analysis from liquid cultures, H. volcanii cultures were grown in Hv-YPC medium (33) supplemented with 40 μg/ml thymidine. The carotenoids were extracted and analyzed by reverse-phase ultra-high-performance liquid chromatography, as previously described (11).
For calculation of bacterioruberin as a proportion of total bacterioruberin and lycopene, colonies were grown on Hv-YPC solid medium supplemented with thymidine (40 μg/ml) and supplemented with retinal (50 μM) where noted. Images were obtained, and a Ruberin metric was calculated as described previously (11). Briefly, colonies were grown from cultures normalized by turbidity for 4 days and photographed with a Moticam 5.0-megapixel (MP) camera (Motic, Hong Kong, China) mounted on a dissection microscope (VistaVision; VWR, Radnor, PA). Color quantification was obtained by sampling approximately 15% of the total colony area with the Adobe Photoshop CS6 (Adobe, San Jose, CA) eyedropper tool to determine values for hue and saturation. These values were normalized to those of H. volcanii H1209 colonies grown under identical conditions on the same plates. The ruberin metric was calculated using the inverse-logit function previously determined to accurately predict bacterioruberin content, as follows:
Since Lye enzymes from different organisms had various levels of activity when expressed in H. volcanii, ruberin proportions were calculated for each H. volcanii strain by dividing the ruberin metric for strains expressing an opsin by the ruberin metric from the corresponding control strain with the empty vector. For statistical comparisons between strains, we used the permutation test as implemented by the R package perm (36).
Quantification of opsin abundance in H. volcanii.
Cultures of H. volcanii strains harboring expression plasmids for opsins with C-terminal His6 tags were grown, cell lysates prepared, and total protein quantified as described previously (11). Cell lysates were diluted to 2 mg of protein per ml using Laemmli sample buffer (Bio-Rad, Hercules, CA) with 2-mercaptoethanol (355 mM) as a reducing agent. Protein samples (30 µl) were separated by polyacrylamide gel electrophoresis and transferred to nitrocellulose membrane. Opsin levels were quantified by immunoblotting with THE His tag mouse antibody (Genscript) diluted to a concentration of 0.2 µg/ml. The blot was subsequently probed using the One-Hour Western advanced kit (Genscript) according to the manufacturer’s protocol. Chemiluminescence for visualization was developed using LumiSensor horseradish peroxidase substrate (Genscript) following the manufacturer’s instructions, and images were obtained with an Azure c600 imaging system (Azure Biosystems, Dublin, CA). Bands were quantified by densitometry using the “mean gray value” determination in Photoshop CS6 (Adobe).
Supplementary Material
ACKNOWLEDGMENTS
R.F.P. designed the research, directed the biological interpretation of the data, and wrote the majority of the manuscript. S.M.G. assisted in the experimental design, performed much of the strain construction, and directed data collection and analysis. A.M.G. performed strain construction and data collection and assisted in biological interpretation. All authors read and approved of the paper.
We thank Kang Mo Ku and Moo Jung Kim for conducting the RP-UHPLC analysis. David Angelini was an invaluable resource for assistance with statistical analysis. Frank Fekete provided insightful comments on the manuscript. Samantha Lee and Emily Shaw provided important technical assistance, and Erika Smith and Margot Miranda-Katz contributed thoughtful advice.
This project was supported by an Institutional Development Award from the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health under grant P20GM103423. Additional funding was provided by the Colby College Natural Science Division.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00576-18.
REFERENCES
- 1.Lazrak T, Wolff G, Albrecht A-M, Nakatani Y, Ourisson G, Kates M. 1988. Bacterioruberins reinforce reconstituted Halobacterium lipid membranes. Biochim Biophys Acta 939:160–162. doi: 10.1016/0005-2736(88)90057-0. [DOI] [Google Scholar]
- 2.Shahmohammadi HR, Asgarani E, Terato H, Saito T, Ohyama Y, Gekko K, Yamamoto O, Ide H. 1998. Protective roles of bacterioruberin and intracellular KCl in the resistance of Halobacterium salinarium against DNA-damaging agents. J Radiat Res 39:251–262. doi: 10.1269/jrr.39.251. [DOI] [PubMed] [Google Scholar]
- 3.Matsumi R, Atomi H, Driessen AJM, van der Oost J. 2011. Isoprenoid biosynthesis in Archaea—biochemical and evolutionary implications. Res Microbiol 162:39–52. doi: 10.1016/j.resmic.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Sieiro C, Poza M, De Miguel T, Villa TG. 2003. Genetic basis of microbial carotenogenesis. Int Microbiol 6:11–16. doi: 10.1007/s10123-003-0097-0. [DOI] [PubMed] [Google Scholar]
- 5.Sandmann G. 2009. Evolution of carotene desaturation: the complication of a simple pathway. Arch Biochem Biophys 483:169–174. doi: 10.1016/j.abb.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 6.Dummer AM, Bonsall JC, Cihla JB, Lawry SM, Johnson GC, Peck RF. 2011. Bacterioopsin-mediated regulation of bacterioruberin biosynthesis in Halobacterium salinarum. J Bacteriol 193:5658–5667. doi: 10.1128/JB.05376-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y, Yatsunami R, Ando A, Miyoko N, Fukui T, Takaichi S, Nakamura S. 2015. Complete biosynthetic pathway of the C50 carotenoid bacterioruberin from lycopene in the extremely halophilic archaeon Haloarcula japonica. J Bacteriol 197:1614–1623. doi: 10.1128/JB.02523-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim YS, Kim NH, Yeom SJ, Kim SW, Oh DK. 2009. In vitro characterization of a recombinant Blh protein from an uncultured marine bacterium as a β-carotene 15,15′-dioxygenase. J Biol Chem 284:15781–15793. doi: 10.1074/jbc.M109.002618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peck RF, Johnson EA, Krebs MP. 2002. Identification of a lycopene β-cyclase required for bacteriorhodopsin biogenesis in the archaeon Halobacterium salinarum. J Bacteriol 184:2889–2897. doi: 10.1128/JB.184.11.2889-2897.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peck RF, Echavarri-Erasun C, Johnson EA, Ng WV, Kennedy SP, Hood L, DasSarma S, Krebs MP. 2001. brp and blh are required for synthesis of the retinal cofactor of bacteriorhodopsin in Halobacterium salinarum. J Biol Chem 276:5739–5744. doi: 10.1074/jbc.M009492200. [DOI] [PubMed] [Google Scholar]
- 11.Peck RF, Pleşa AM, Graham SM, Angelini DR, Shaw EL. 2017. Opsin-mediated inhibition of bacterioruberin synthesis in halophilic archaea. J Bacteriol 199:e00303–e00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forterre P, Brochier C, Philippe H. 2002. Evolution of the Archaea. Theor Popul Biol 61:409–422. doi: 10.1006/tpbi.2002.1592. [DOI] [PubMed] [Google Scholar]
- 13.Wolf YI, Makarova KS, Yutin N, Koonin EV. 2012. Updated clusters of orthologous genes for Archaea: a complex ancestor of the Archaea and the byways of horizontal gene transfer. Biol Direct 7:46. doi: 10.1186/1745-6150-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson-Sathi S, Dagan T, Landan G, Janssen A, Steel M, McInerney JO, Deppenmeier U, Martin WF. 2012. Acquisition of 1,000 eubacterial genes physiologically transformed a methanogen at the origin of Haloarchaea. Proc Natl Acad Sci U S A 109:20537–20542. doi: 10.1073/pnas.1209119109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson-Sathi S, Sousa FL, Roettger M, Lozada-Chávez N, Thiergart T, Janssen A, Bryant D, Landan G, Schönheit P, Siebers B, McInerney JO, Martin WF. 2015. Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature 517:77–80. doi: 10.1038/nature13805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denner EBM, McGenity TJ, Busse H-J, Grant WD, Wanner G, Stan-Lotter H. 1994. Halococcus salifodinae sp. nov., an archaeal isolate from an Austrian salt mine. Int J Syst Bacteriol 44:774–780. doi: 10.1099/00207713-44-4-774. [DOI] [Google Scholar]
- 17.Legat A, Denner E, Dornmayr-Pfaffenhuemer M, Pfeiffer P, Knopf B, Claus H, Gruber C, König H, Wanner G, Stan-Lotter H. 2013. Properties of Halococcus salifodinae, an isolate from Permian rock salt deposits, compared with halococci from surface waters. Life (Basel) 3:244–259. doi: 10.3390/life3010244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker EA, Seitzer PM, Tritt A, Larsen D, Krusor M, Yao AI, Wu D, Madern D, Eisen JA, Darling AE, Facciotti MT. 2014. Phylogenetically driven sequencing of extremely halophilic archaea reveals strategies for static and dynamic osmo-response. PLoS Genet 10:e1004784. doi: 10.1371/journal.pgen.1004784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becker EA, Yao AI, Seitzer PM, Kind T, Wang T, Eigenheer R, Shao KSY, Yarov-Yarovoy V, Facciotti MT. 2016. A large and phylogenetically diverse class of type 1 opsins lacking a canonical retinal binding site. PLoS One 11:e0156543. doi: 10.1371/journal.pone.0156543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walsh DA, Bapteste E, Kamekura M, Doolittle WF. 2004. Evolution of the RNA polymerase B′ subunit gene (rpoB′) in halobacteriales: a complementary molecular marker to the SSU rRNA gene. Mol Biol Evol 21:2340–2351. doi: 10.1093/molbev/msh248. [DOI] [PubMed] [Google Scholar]
- 21.Lanyi JK. 1997. Mechanisms of ion transport across membranes: bacteriorhodopsin as a prototype for proton pumps. J Biol Chem 272:31209–31212. doi: 10.1074/jbc.272.50.31209. [DOI] [PubMed] [Google Scholar]
- 22.Pecker I, Gabbay R, Cunningham FX, Hirschberg J. 1996. Cloning and characterization of the cDNA for lycopene β-cyclase from tomato reveals decrease in its expression during fruit ripening. Plant Mol Biol 30:807–819. doi: 10.1007/BF00019013. [DOI] [PubMed] [Google Scholar]
- 23.Maresca JA, Graham JE, Wu M, Eisen JA, Bryant DA. 2007. Identification of a fourth family of lycopene cyclases in photosynthetic bacteria. Proc Natl Acad Sci U S A 104:11784–11789. doi: 10.1073/pnas.0702984104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mongodin EF, Nelson KE, Daugherty S, DeBoy RT, Wister J, Khouri H, Weidman J, Walsh DA, Papke RT, Sanchez Perez G, Sharma AK, Nesbo CL, MacLeod D, Bapteste E, Doolittle WF, Charlebois RL, Legault B, Rodriguez-Valera F. 2005. The genome of Salinibacter ruber: convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc Natl Acad Sci U S A 102:18147–18152. doi: 10.1073/pnas.0509073102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carreto L, Moore E, Nobre MF, Wait R, Riley PW, Sharp RJ, da Costa MS. 1996. Rubrobacter xylanophilus sp. nov., a new thermophilic species isolated from a thermally polluted effluent. Int J Syst Bacteriol 46:460–465. doi: 10.1099/00207713-46-2-460. [DOI] [Google Scholar]
- 26.Garrido LM, Alves JMP, Oliveira LS, Gruber A, Padilla G, Araújo WL. 2016. Draft genome sequence of Curtobacterium sp. strain ER1/6, an endophytic strain isolated from citrus sinensis with potential to be used as a biocontrol agent. Genome Announc 4:e01264–e01216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehara K, Yoshizawa S, Tsukamoto T, Sudo Y. 2017. A phylogenetically distinctive and extremely heat stable light-driven proton pump from the eubacterium Rubrobacter xylanophilus DSM 9941 T. Sci Rep 7:44427. doi: 10.1038/srep44427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sorokin DY, Makarova KS, Abbas B, Ferrer M, Golyshin PN, Galinski EA, Ciordia S, Mena MC, Merkel AY, Wolf YI, van Loosdrecht MCM, Koonin EV. 2017. Discovery of extremely halophilic, methyl-reducing euryarchaea provides insights into the evolutionary origin of methanogenesis. Nat Microbiol 2:17081. doi: 10.1038/nmicrobiol.2017.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bertelli C, Laird MR, Williams KP, Lau BY, Hoad G, Winsor GL, Brinkman FSL. 2017. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res 45:W30–W35. doi: 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2017. GenBank. Nucleic Acids Res 45:D37–D42. doi: 10.1093/nar/gkw1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 32.Williams TA, Foster PG, Nye TMW, Cox CJ, Embley TM. 2012. A congruent phylogenomic signal places eukaryotes within the Archaea. Proc R Soc B Biol Sci 279:4870–4879. doi: 10.1098/rspb.2012.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allers T, Ngo HP, Mevarech M, Lloyd RG. 2004. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl Environ Microbiol 70:943–953. doi: 10.1128/AEM.70.2.943-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allers T, Barak S, Liddell S, Wardell K, Mevarech M. 2010. Improved strains and plasmid vectors for conditional overexpression of His-tagged proteins in Haloferax volcanii. Appl Environ Microbiol 76:1759–1769. doi: 10.1128/AEM.02670-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Large A, Stamme C, Lange C, Duan Z, Allers T, Soppa J, Lund PA. 2007. Characterization of a tightly controlled promoter of the halophilic archaeon Haloferax volcanii and its use in the analysis of the essential cct1 gene. Mol Microbiol 66:1092–1106. doi: 10.1111/j.1365-2958.2007.05980.x. [DOI] [PubMed] [Google Scholar]
- 36.Fay MP, Shaw PA. 2010. Exact and asymptotic weighted logrank tests for interval censored data: the interval R package. J Stat Softw 36:i02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGill R, Tukey JW, Larsen WA. 1978. Variations of box plots. Am Stat 32:12–16. doi: 10.2307/2683468. [DOI] [Google Scholar]
- 38.Pfeiffer F, Schuster SC, Broicher A, Falb M, Palm P, Rodewald K, Ruepp A, Soppa J, Tittor J, Oesterhelt D. 2008. Evolution in the laboratory: the genome of Halobacterium salinarum strain R1 compared to that of strain NRC-1. Genomics 91:335–346. doi: 10.1016/j.ygeno.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 39.Baliga NS, Bonneau R, Facciotti MT, Pan M, Glusman G, Deutsch EW, Shannon P, Chiu Y, Weng RS, Gan RR, Hung P, Date SV, Marcotte E, Hood L, Ng WV. 2004. Genome sequence of Haloarcula marismortui: A halophilic archaeon from the Dead Sea. Genome Res 14:2221–2234. doi: 10.1101/gr.2700304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson I, Tindall BJ, Pomrenke H, Göker M, Lapidus A, Nolan M, Copeland A, del Rio TG, Chen F, Tice H, Cheng JF, Lucas S, Chertkov O, Bruce D, Brettin T, Detter JC, Han C, Goodwin L, Land M, Hauser L, Chang YJ, Jeffries CD, Pitluck S, Pati A, Mavromatis K, Ivanova N, Ovchinnikova G, Chen A, Palaniappan K, Chain P, Rohde M, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP. 2009. Complete genome sequence of Halorhabdus utahensis type strain (AX-2 T). Stand Genomic Sci 1:218–225. doi: 10.4056/sigs.31864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dyall-Smith ML, Pfeiffer F, Oberwinkler T, Klee K, Rampp M, Palm P, Gross K, Schuster SC, Oesterhelt D. 2013. Genome of the haloarchaeon Natronomonas moolapensis, a neutrophilic member of a previously haloalkaliphilic genus. Genome Announc 1:e0009513. doi: 10.1128/genomeA.00095-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tindall BJ, Schneider S, Lapidus A, Copeland A, del Rio TG, Nolan M, Lucas S, Chen F, Tice H, Cheng JF, Saunders E, Bruce D, Goodwin L, Pitluck S, Mikhailova N, Pati A, Ivanova N, Mavrommatis K, Chen A, Palaniappan K, Chain P, Land M, Hauser L, Chang YJ, Jeffries CD, Brettin T, Han C, Rohde M, Göker M, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Klenk HP, Kyrpides NC, Detter JC. 2009. Complete genome sequence of Halomicrobium mukohataei type strain (arg-2T). Stand Genomic Sci 1:270–277. doi: 10.4056/sigs.42644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dyall-Smith ML, Pfeiffer F, Klee K, Palm P, Gross K, Schuster SC, Rampp M, Oesterhelt D. 2011. Haloquadratum walsbyi: limited diversity in a global pond. PLoS One 6 doi: 10.1371/journal.pone.0020968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Podell S, Emerson JB, Jones CM, Ugalde JA, Welch S, Heidelberg KB, Banfield JF, Allen EE. 2014. Seasonal fluctuations in ionic concentrations drive microbial succession in a hypersaline lake community. ISME J 8:979–990. doi: 10.1038/ismej.2013.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ding J-Y, Lai M-C. 2010. The biotechnological potential of the extreme halophilic archaea Haloterrigena sp. H13 in xenobiotic metabolism using a comparative genomics approach. Environ Technol 31:905–914. doi: 10.1080/09593331003734210. [DOI] [PubMed] [Google Scholar]
- 46.Hartman AL, Norais C, Badger JH, Delmas S, Haldenby S, Madupu R, Robinson J, Khouri H, Ren Q, Lowe TM, Maupin-Furlow J, Pohlschroder M, Daniels C, Pfeiffer F, Allers T, Eisen JA. 2010. The complete genome sequence of Haloferax volcanii DS2, a model archaeon. PLoS One 5:e9605. doi: 10.1371/journal.pone.0009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.