Biofilms are crucial for bacterial survival, adaptation, and dissemination in natural, industrial, and medical systems. Sessile cells embedded in the self-produced extracellular matrix of the biofilm benefit from a division of labor and are protected from environmental insults. However, as the biofilm ages, cells become stressed because of overcrowding, starvation, and accumulation of waste products. How does the sessile biofilm community sense and respond to stressful conditions? Here, we show that in Bacillus subtilis, the transcription factors SigB and SinR control whether cells remain in or leave a biofilm when metabolic conditions become unfavorable. This novel SigB-SinR regulatory circuit might be important for controlling the fitness of biofilms (either beneficial or harmful) in diverse environments.
KEYWORDS: Bacillus subtilis, biofilm aging, biofilm dispersal, sigma B, stress activation
ABSTRACT
Bacterial biofilms are important in natural settings, biotechnology, and medicine. However, regulation of biofilm development and its persistence in different niches is complex and only partially understood. One key step during the biofilm life cycle is dispersal, when motile cells abandon the mature biofilm to spread out and colonize new niches. Here, we show that in the model bacterium Bacillus subtilis the general stress transcription factor SigB is essential for halting detrimental overgrowth of mature biofilm and for triggering dispersal when nutrients become limited. Specifically, SigB-deficient biofilms were larger than wild-type biofilms but exhibited accelerated cell death, significantly greater sensitivity to different stresses, and reduced dispersal. Interestingly, the signal detected by SigB to limit biofilm growth was transduced through the RsbP-dependent metabolic arm of the SigB regulatory cascade, which in turn positively controlled expression of SinR, the master regulator of biofilm formation and cell motility. This novel SigB-SinR regulatory circuit might be important in controlling the fitness of biofilms (either beneficial or harmful) in diverse environments.
IMPORTANCE Biofilms are crucial for bacterial survival, adaptation, and dissemination in natural, industrial, and medical systems. Sessile cells embedded in the self-produced extracellular matrix of the biofilm benefit from a division of labor and are protected from environmental insults. However, as the biofilm ages, cells become stressed because of overcrowding, starvation, and accumulation of waste products. How does the sessile biofilm community sense and respond to stressful conditions? Here, we show that in Bacillus subtilis, the transcription factors SigB and SinR control whether cells remain in or leave a biofilm when metabolic conditions become unfavorable. This novel SigB-SinR regulatory circuit might be important for controlling the fitness of biofilms (either beneficial or harmful) in diverse environments.
INTRODUCTION
Bacteria form multicellular and cooperative communities known as biofilms, which are critical for bacterial survival, adaptation, and dissemination in natural, industrial, and medical systems (1, 2). Sessile cells embedded in the self-produced extracellular matrix of a biofilm benefit from a division of labor and are protected from environmental insults (3). However, as the biofilm ages, the population of cells becomes stressed due to overcrowding, nutrient restriction, and waste product accumulation. However, thus far it has not been resolved how a sessile biofilm community senses and responds to stressful conditions (1, 2).
One strategy that is believed to be beneficial for rejuvenation of a stressed biofilm is triggering the escape of motile cells. This phenomenon, known as biofilm dispersal (4, 5), is a natural step of the biofilm life cycle and is important for many pathogenic bacteria with regard to the transmission, exacerbation, and spread of infections, especially in periodontitis, cystic fibrosis, pneumonia, and catheter-associated endocarditis (1, 5, 6). Proficiency in flagellum synthesis, which is essential for swimming and swarming activities, is required for the ability to disperse from liquid (pellicle) and solid (colony) biofilms (5–8). Regardless, the molecular mechanisms that coordinate the aging and rejuvenation of biofilm through cell dispersal in Gram-positive bacteria are poorly understood (4).
Bacillus subtilis is a Gram-positive, endospore-forming bacterium that serves as a model organism for the study of different prokaryote phenomena (e.g., biofilm development) at both physiological and molecular levels (4, 9–13). In B. subtilis, SinR is considered a master regulator that controls entry into two alternative physiological states: a motile state, activated by SinR, in which cells are able to swim or swarm (on liquid or wet surfaces, respectively), and a nonmotile state, repressed by SinR, in which cells form a sessile multicellular biofilm (14, 15). Under conditions that favor planktonic growth (i.e., bacterial growth with shaking), SinR activates the genes required for flagellum synthesis (hag) and cell chain separation (lytA and lytF) and represses those required for a sessile lifestyle (i.e., eps and tasA) (10). When B. subtilis develops in media that support a sessile lifestyle (i.e., bacterial growth without shaking), a subset of the population produces and secretes the complex polysaccharides (extracellular polysaccharides, or EPSs) and proteins, mainly the amyloid-like fiber TasA and the hydrophobin BslA, that will constitute the main components of the extracellular matrix of the biofilm (4). Notably, the operons responsible for the synthesis of EPS and TasA (the 15 genes of the epsA-epsO (epsA–O) operon and the three-cistron tapA-sipW-tasA operon, respectively) are repressed by SinR (14, 15).
In B. subtilis, other bacilli, Listeria, and Staphylococcus, the transcription factor SigB controls the general stress response that comprises more than 200 genes, the products of which confer the bacterium with resistance to multiple stresses (16, 17). Activation of SigB is controlled by the partner-switching RsbV-RsbW-SigB module (16, 18, 19). Under nonstress conditions, SigB is maintained in an inactive state in a complex with the anti-sigma factor/kinase RsbW; the third partner, RsbV, is inactive because of phosphorylation by RsbW (20–23). However, under stress, release of SigB from the inactive SigB::RsbW complex is achieved via the dephosphorylated form of the anti-anti-sigma factor RsbV. In B. subtilis, activation (dephosphorylation) of RsbV, and therefore SigB activation, is achieved by alternative phosphatases that sense energy or environmental stress (RsbP or RsbU phosphatase, respectively) (20–23). SigB is also activated by cold shock (24, 25) independently of RsbP, RsbU, and RsbV activities (24).
Here, we describe a novel mechanism involving the transcription factors SigB and SinR by which B. subtilis regulates whether a cell remains in or leaves a biofilm when environmental and metabolic conditions become unfavorable.
RESULTS
SigB and its role during the biofilm life cycle.
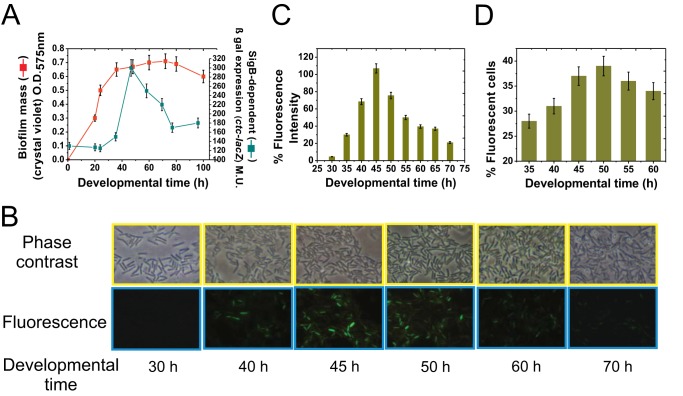
In planktonic cultures, expression of at least 200 genes (∼5% of the genome) is estimated to be under direct or indirect control of the general stress sigma factor SigB (16, 17). Induction of this SigB-dependent regulon confers the cell with general, unspecific, and preventive stress protection (16, 17). We sought to determine if SigB is activated during biofilm formation and, if so, what its role is during biofilm development. To address this issue, we monitored the activity of SigB during growth of the B. subtilis reference strain JH642 (Table 1) under biofilm-supporting conditions. This domesticated laboratory strain is able to form a substantial biofilm (i.e., floating pellicles) under developmental control (26–28) when cultivated in nutritionally enriched media such as LBY broth (Luria-Bertani broth fortified with 4% yeast extract) (29). As shown in Fig. 1A, the low β-galactosidase activity driven from the SigB-dependent ctc promoter (16, 17) during the first 20 to 30 h of cultivation indicated that SigB was not very active when the biofilm was juvenile. However, the transcription factor SigB became active at later incubation times (after 40 h), with the highest activity occurring when the biofilm reached its plateau of growth (50 h). Although SigB activity declined after reaching maximal expression (300 Miller units [MU] at 50 h of development), the level was still significantly higher (220 MU at 70 h of development) than that when the biofilm was younger (120 MU at 20 h of development) (Fig. 1A). The pattern of fluorescence protein production by a JH642-isogenic strain harboring the sigB-gfp reporter fusion (strain MR655) (Table 1) confirmed the presence of SigB activity under biofilm-supporting conditions (Fig. 1B to D). The activation pattern of SigB during the biofilm life cycle (Fig. 1A) was similar to the previously reported SigB activity pattern in planktonic cultures of B. subtilis (21, 30, 31), as shown in Fig. S1 in the supplemental material. Overall, these results suggest that for genetically equivalent (kin) cells living as a sessile biofilm (Fig. 1) and in a free planktonic state (Fig. S1), entry into the stationary phase of growth represents a stressful and threatening condition that B. subtilis might at least partially manage via activation of SigB and enhanced expression of its regulon. The other adaptive pathway that B. subtilis biofilm cells might employ for stress management at the onset of the stationary phase is sporulation (9, 11, 32). However, sporulation is largely prevented in nutrient-enriched media because it is subject to nitrogen and carbon catabolite control (9, 32). Under our experimental growth conditions in rich medium (i.e., LBY), the sporulation frequency of the wild-type strain after 50 h of biofilm development was less than 0.05% (1.8 × 108 viable cells and 1.1 × 105 spores ml−1, respectively). Therefore, the sporulation pathway does not significantly influence biofilm stress adaptation under our experimental conditions.
TABLE 1.
Strains used in this work
| Strain | Relevant phenotype and/or genotype | Comment and/or sourcea |
|---|---|---|
| JH642 | B. subtilis wild-type strain | Laboratory collection (12) |
| NCIB3610 | B. subtilis Marburg strain | Laboratory collection (47) |
| RG4365 | B. subtilis natto strain | Laboratory collection (37) |
| MR644 | JH642 ΔsigB::cat | Laboratory collection (25) |
| RG5567 | RG4365 ΔsigB::cat | MR644→RG4365 (this work) |
| RG5568 | NCIB3610 ΔsigB::cat | MR644→NCIB3610 (this work) |
| MR655 | JH642 amyE::PsigB-gfp::cat | Laboratory collection (24) |
| MR101 | JH642 amyE::Pctc-lacZ::cat | Laboratory collection (25) |
| NRS2289 | JH642 amyE::PbslA-lacZ::kan | Laboratory collection (12) |
| RG5569 | NR52289 ΔsigB::cat | NR52289→JH642 |
| RG5570 | JH642 amyE::PepsG-lacZ::kan | Laboratory collection (12) |
| RG5571 | RG5570 ΔsigB::cat | RG5570→JH642 |
| RG5572 | JH642 ΔrsbU::kan | Laboratory collection (24) |
| RG5573 | JH642 ΔrsbP::spc | Laboratory collection (24) |
| RG5574 | JH642 ΔrsbUP::kan-spc | Laboratory collection (24) |
| RG5575 | RG5570 amyE::Pctc-lacZ::cat | MR101→RG5570 (this work) |
| RG5576 | RG5571 amyE::Pctc-lacZ::cat | MR101→RG5571 (this work) |
| RG5577 | RG5572 amyE::Pctc-lacZ::cat | MR101→RG5572 (this work) |
| RG438 | JH642 amyE::PsinR-lacZ::spc | Laboratory collection (40) |
| RG432 | JH642 ΔsinR::ery | Laboratory collection (40) |
| RG4576 | NCIB3610 ΔsinR::ery | RG432→NCIB3610 (this work) |
| RG5578 | MR644 ΔsinR::ery | RG432→MR644 (this work) |
| RG5580 | RG438 ΔsigB::cat | RG438→MR644 (this work) |
| RG4500 | JH642 amyE::Pspo0A-lacZ::ery | Laboratory collection (37) |
| RG4501 | RG4500 ΔsigB::cat | MR644→RG4500 (this work) |
| RG4503 | JH642 amyE::PabrB-lacZ::ery | Laboratory collection (40) |
| RG4504 | RG4503 ΔsigB::cat | MR644→RG4503 (this work) |
| RG4505 | JH642 amyE::PsinI-lacZ::ery | Laboratory collection (40) |
| RG4506 | RG4505 ΔsigB::cat | MR644→RG4505 (this work) |
| RG4507 | JH642 amyE::PcodY-lacZ::ery | Laboratory collection (13) |
| RG4508 | RG4507 ΔsigB::cat | MR644→RG4507 (this work) |
| RG4509 | JH642 amyE::PcomA-lacZ::ery | Laboratory collection (13) |
| RG4510 | RG4509 ΔsigB::cat | MR644→RG4509 (this work) |
| RG4511 | JH642 amyE::PdegSU-lacZ::ery | Laboratory collection (13) |
| RG4512 | RG4511 ΔsigB::cat | MR644→RG4511 (this work) |
| RG4513 | JH642 amyE::Prok-lacZ::ery | Laboratory collection (13) |
| RG4514 | RG4513 ΔsigB::cat | MR644→RG4513 (this work) |
| RG4515 | JH642 amyE::PluxS-lacZ::ery | Laboratory collection (47) |
| RG4516 | RG4515 ΔsigB::cat | MR644→RG4515 (this work) |
| RG4517 | JH642 amyE::PsigH-lacZ::ery | Laboratory collection (25) |
| RG4518 | RG4517 ΔsigB::cat | MR644→RG4517 (this work) |
| RG4519 | JH642 amyE::PtnrA-lacZ::ery | Laboratory collection (13) |
| RG4520 | RG4519 ΔsigB::cat | MR644→RG518 (this work) |
| TB24 | NCIB3610 Δhag::spc | Laboratory collection (12) |
| RG4521 | RG5568 Δhag::spc | TB24→RG5568 (this work) |
| TB25 | TB24 ΔbslA::kan | Laboratory collection (12) |
| RG4522 | TB24 ΔsinR::ery | RG432→TB24 (this work) |
| RG4523 | MR101 ΔsinR::ery | RG432→MR101 (this work) |
| RG5581 | NCIB3610 ΔrsbU::kan | RG5572→NCIB3610 (this work) |
| RG5582 | NCIB3610 ΔrsbP::spc | RG5573→NCIB3610 (this work) |
| RG5583 | NCIB3610 ΔrsbUP::kan-spc | RG5581→RG5582 (this work) |
Comments are formatted as DNA from donor strain→receptor strain.
FIG 1.
SigB expression during the biofilm cell cycle. (A) Kinetics of biofilm formation and expression of the SigB-dependent ctc gene in biofilm-supporting LBY static culture. O.D.575nm, optical density at 575 nm; β-Gal, β-galactosidase. (B) Representative photographs from inner biofilm regions of GFP-expressing isogenic JH642 cells (strain MR655) at the indicated development times. Cells from exterior regions of the biofilm did not exhibit significant SigB-directed fluorescence (data not shown). Representative results from three independent experiments performed in parallel are shown. (C) Relative fluorescence intensity, indicated as a percentage of maximal (100%) SigB-directed gfp expression by the JH642 isogenic strain MR655 (amyE::PsigB-gfp::cat). Maximal fluorescence (100%) was considered to occur at 45 h of biofilm incubation. All of the other relative fluorescence levels, at the different times, were calculated as a percentage of the maximum fluorescence observed at 45 h of biofilm development. (D) Percentage of cells taken from inner regions of the biofilm that express GFP (MR655 strain). Each data point shown in panels A, C, and D is the mean ± SEM from a representative experiment performed in triplicate.
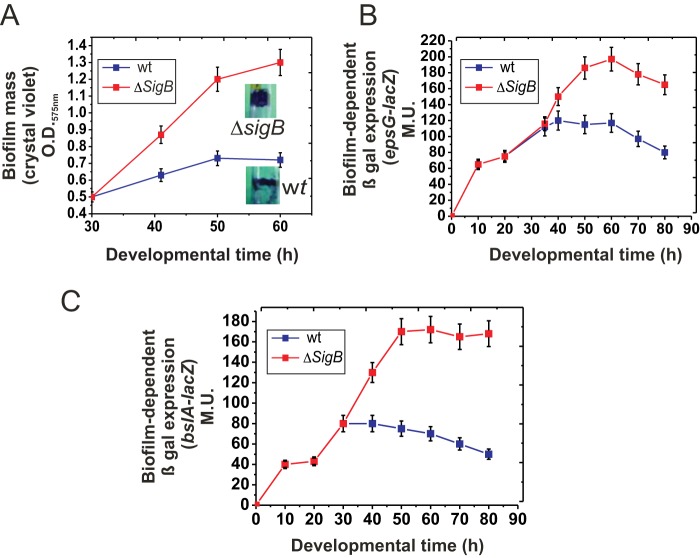
To evaluate the effect of SigB activity on biofilm development, we monitored biofilm formation in JH642 cells with and without SigB activity (JH642 and MR644 strains, respectively) (Table 1). At early times of incubation (from 0 to 30 h of development), there was no significant difference in biofilm generation between SigB-proficient and SigB-deficient cultures. However, at later times of biofilm development, the ΔsigB cells produced more biofilm than did wild-type cells, and when the wild-type biofilm had reached the stationary phase and halted growth (50 h of incubation), the ΔsigB biofilm continued to grow (Fig. 2A). Synthesis of the main biofilm matrix components (e.g., the hydrophobin BslA and exopolysaccharide) (4, 33) continued during the stationary phase of the ΔsigB biofilm but ceased in the case of the wild-type biofilm, finally resulting in a 2.5-fold higher mass for the former than the latter (Fig. 2B and C). To assess whether the biofilm phenotype observed in ΔsigB cells derived from the JH642 strain was due to its domesticated nature (29, 34–36), we analyzed the effect of SigB on biofilm formation in wild (undomesticated) strains of B. subtilis. The increased production of biofilm by SigB-deficient cells relative to the amount produced by wild-type cells was independent of the genetic pedigree (domesticated versus undomesticated) of different wild B. subtilis isolates (i.e., the Marburg- and natto-related strains NCIB3610 and RG4365, respectively) (34, 37) (Table 1 and Fig. S2A). Additionally, the observed increase in the amount of ΔsigB biofilms compared to that of wild-type biofilm was independent of the growth medium used (MSgg or LB-glycerol-manganese broth) (data not shown) as well as the supportive surface (i.e., glass or plastic; tubes, bottles, or microtiter wells) (Fig. S2B). Therefore, SigB acts as a negative regulator of biofilm formation in domesticated and undomesticated (wild) B. subtilis strains.
FIG 2.
SigB regulates biofilm growth. (A) Rate of biofilm (floating pellicle) formation by wild-type (wt) JH642 and isogenic ΔsigB cells grown in static LBY broth. Inset photographs are representative of typical biofilms developed for 50 h and stained with crystal violet. (B) β-Galactosidase activity of the biofilm-reporter fusion epsG-lacZ::amyE in wild-type and isogenic ΔsigB cells grown in static LBY broth. A typical output of three independent experiments performed in parallel is shown. (C) β-Galactosidase activity of the biofilm-reporter fusion bslA-lacZ::amyE in wild-type and isogenic ΔsigB cells grown in static LBY broth. Each data point is the mean ± SEM. A typical output of three independent experiments performed in parallel is shown.
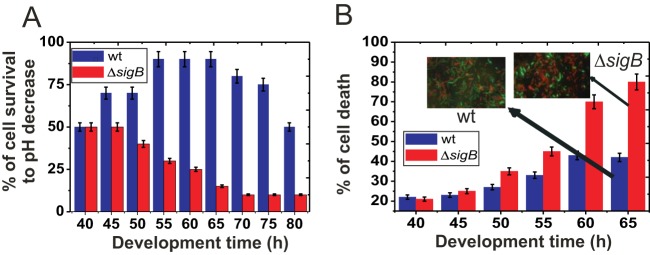
The products of SigB-controlled genes allow planktonic cells to adapt to and survive different environmental and/or metabolic stresses (21, 30, 31, 38). To examine the role of SigB in stress adaptation and the survival of embedded biofilm cells, we monitored the survival of SigB-proficient and SigB-deficient biofilms after exposure to particular stresses at different times after the start of biofilm formation. A drastic drop in pH (Fig. 3A) and heat shock, alcohol stress, and biocide treatments (Fig. S3) all resulted in higher resistance in wild-type biofilms during the stationary phase than in SigB-deficient biofilms that had reached the stationary phase. Thus, despite the fact that SigB-deficient cells produced thicker biofilms, they were more susceptible to stressful conditions. Interestingly, the cell destiny of the biofilms differed significantly. As shown in Fig. 3B, a higher proportion of SigB-deficient cells than wild-type cells displayed cell death at advanced times of biofilm development (15% and 55% of cellular survival in ΔsigB and wild-type biofilms after 60 h of biofilm development, respectively) in the absence of external stress. Therefore, SigB-deficient cells produce more biofilm than do wild-type cells but die faster.
FIG 3.
SigB regulates biofilm fitness. (A) Cell survival in wild-type JH642 and ΔsigB (strain MR644) biofilms after a drastic decrease in pH. At the indicated developmental times, wild-type and ΔsigB biofilms were exposed to a pH of 3.0 for 1 h before viable cells were counted, as indicated in Materials and Methods. A typical output of three independent experiments performed in parallel is shown. (B) Cell survival in wild-type JH642 and ΔsigB (strain MR644) biofilms of different ages were measured as the percentage of the total number of fluorescent cells observed in 10 different fields after staining with the LIVE/DEAD BacLight kit (green and red cells are alive and dead cells, respectively). Inset photographs are representative of several pictures taken at the indicated time. Each data point is the mean ± SEM from a representative experiment performed in triplicate.
The RsbP-dependent pathway controls SigB activation during biofilm development.
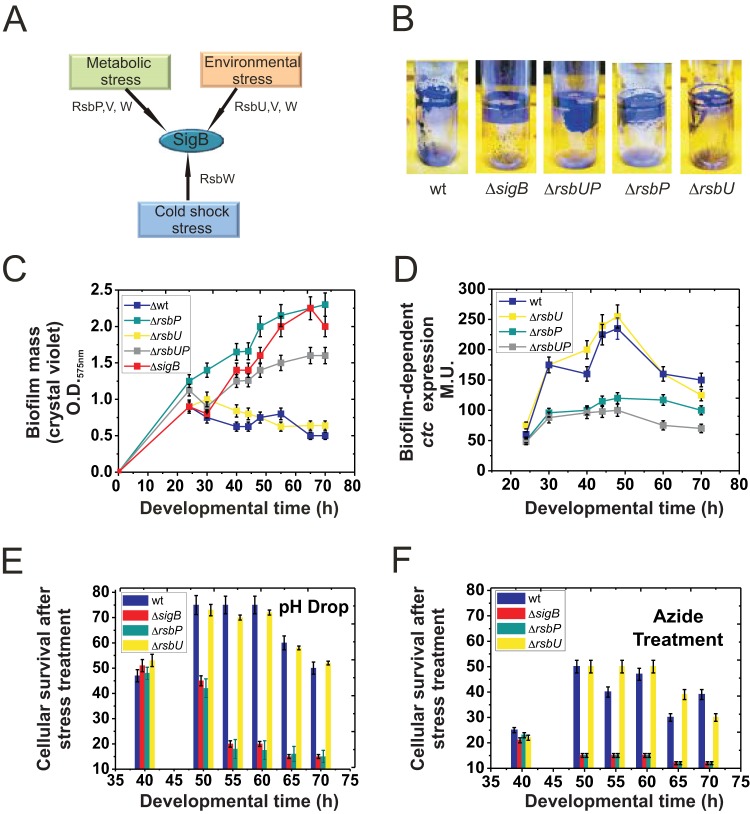
We next explored the regulatory route (Fig. 4A) that controls activation of the alternative sigma factor SigB during biofilm development. To this end, we monitored biofilm formation in isogenic JH642 strains with altered pathways of SigB activation (i.e., RG5572-ΔrsbU, RG5573-ΔrsbP, and RG5574-ΔrsbUP strains) (Table 1). For the ΔrsbU strain, the amount and kinetics of biofilm formation were very similar to those displayed by wild-type cells (Fig. 4B and C). Interestingly, the biofilm-forming capacity of ΔrsbP and ΔrsbUP strains resembled that of ΔsigB cells (Fig. 4B and C). These results strongly suggest that during biofilm development in LBY broth, the RsbP route is responsible for activation of SigB inside the biofilm. Monitoring of β-galactosidase activity from the SigB-dependent ctc-lacZ-reporter fusion in B. subtilis cells without RsbP and/or RsbU activities confirmed the proposed conclusion (Fig. 4D). The RsbP route senses energy depletion (nutrient deprivation) in planktonic cultures of B. subtilis (16, 17, 21, 23), and therefore it can be assumed that this route also senses energy (nutrient) depletion inside the biofilm. However, the possibility that RsbP senses an unknown signal that might be present exclusively in the biofilm and absent from (or weaker in) planktonic cultures cannot be excluded.
FIG 4.
SigB input during the biofilm cell cycle. (A) Cartoon summarizing the three possible routes of SigB activation (see the text for details). (B) Fifty-hour-old biofilms, as observed after crystal violet staining, of wild-type JH642 and isogenic mutants with altered routes of SigB activation: strain MR644 (ΔsigB), strain RG5572 (ΔrsbU), strain RG5573 (ΔrsbP), and strain RG5574 (ΔrsbUP) (Table 1). Representative pictures of five independent experiments are shown. (C) Rate of biofilm (pellicle) formation by the wild type and different isogenic mutants with altered SigB activity shown in panel B. A typical output of three independent experiments performed in parallel is shown. (D) β-Galactosidase activity of the SigB-dependent reporter fusion ctc-lacZ::amyE in wild-type and isogenic mutants with altered SigB activity grown in static LBY broth. (E and F) Cell survival in biofilms of wild-type JH642 and isogenic mutants with altered SigB activity after a drastic decrease in pH (E) or treatment with azide (F). The percentage of cellular survival is referenced to the number of CFU of biofilms of the same age that had not been exposed to stress. For panels B to F, typical output ± SEM from five independent experiments is shown.
The effect of SigB on biofilm formation was independent of incubation temperature and not related to particular temperatures and the edge of the growth range (24, 25) (Fig. S4). Exposure of aged biofilms lacking either the RsbP or RsbU route of environmental or energy stress (strong pH variation or acute ATP depletion, respectively) sensing confirmed that the RsbP-dependent pathway controls activation of SigB and confers general, unspecific, and preventive stress protection (16, 17) to aging biofilms (Fig. 4E and F).
To further support the current findings, we monitored biofilm development in the well-studied biofilm-forming strain NCIB3610 (34) and isogenic derivatives affected in different routes of SigB activation (Fig. S5). The data obtained confirmed that the RsbP-dependent route controls SigB activation under biofilm-supporting conditions in domesticated and undomesticated B. subtilis isolates (strains JH642 and NCIB3610, respectively) (29, 34).
SigB maintains SinR levels in sessile B. subtilis populations.
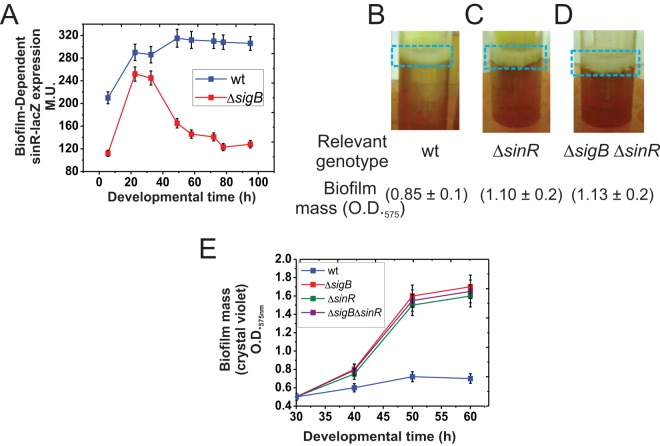
To assess how SigB prevents biofilm overgrowth, we tested the best-characterized B. subtilis biofilm regulators in SigB-proficient and SigB-deficient isogenic strains under biofilm-supporting conditions. SigB did not significantly affect expression of the genes encoding the following direct and indirect key regulators of biofilm formation: ComA (39), Spo0A (29), AbrB (26), SigH (34), SinI (40), CodY (41), DegSU (42), LuxS (37), TnrA (32), and Rok (39) (Fig. S6). However, SigB was required to maintain expression of sinR (40) during the stationary phase of sessile cultures of B. subtilis (Fig. 5A). Interestingly, SinR is a master (negative) regulator of B. subtilis biofilm formation because it represses expression of the key operons, namely, epsA–O and tapA-sigW-tasA, involved in the synthesis of exopolysaccharide and the TasA amyloid fibers of the biofilm matrix, respectively (14, 15). Therefore, in ΔsigB cells, lower levels of sinR expression would result in increased biofilm formation. To obtain further experimental evidence that supports this interpretation, we measured biofilm growth in B. subtilis cultures with and without SigB and/or SinR activities. Interestingly, the increased biofilm formation observed for the ΔsinR strain (relative to the final size of the wild-type biofilm) (Fig. 5B and C) was quite similar to that observed for ΔsinR ΔsigB cells (Fig. 5D). Thus, biofilm growth (Fig. 5E) did not increase above the level observed in SigB-deficient cells by inactivating the natural negative regulator of biofilm formation, SinR. Additionally, SinR did not affect SigB expression (Fig. S7). Overall, these results indicate that SigB operates upstream of SinR to control the growth and fate of B. subtilis biofilms. The finding that the biofilm phenotype and its size were almost identical in ΔsigB, ΔsinR, and ΔsigB ΔsinR strains strongly suggests that SinR is the main target of SigB to regulate biofilm fate.
FIG 5.
SigB regulates sinR expression. (A) β-Galactosidase activity from the sinR-lacZ::amyE reporter fusion in wild-type and isogenic ΔsigB cells grown in static LBY. Typical output from one of four independent experiments is shown. (B to D) SinR and SigB do not generate additive effects on biofilm growth. Fifty-hour-old representative biofilms developed by standing cultures of the wild type (strain JH642) and isogenic mutants with altered SinR (ΔsinR, strain RG432) and/or SigB (ΔsigB ΔsinR, strain RG5578) activity. Biofilm mass was measured using the crystal violet technique, as indicated. Similar results were observed in several independent experiments. (E) Rate of biofilm formation by the wild type and different isogenic mutants with altered SigB, SinR, and SigB-SinR activities. Each data point in panels A and E is an average ± SEM from five independent representative experiments.
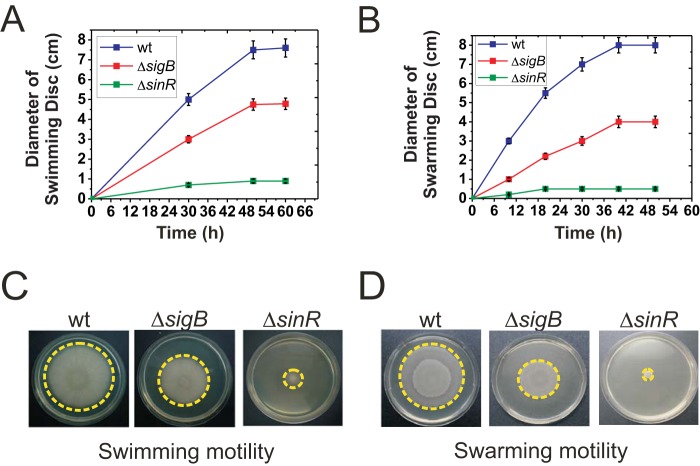
SinR is also essential for flagellum-mediated B. subtilis motility (swimming and swarming), and sinR mutant strains are completely nonmotile (12, 43, 44). Therefore, if SigB is required to maintain sinR expression, it would be expected that flagellum-dependent motility would also be affected by SigB. Accordingly, swimming (Fig. 6A and C) and swarming (Fig. 6B and D) were reduced in ΔsigB cells compared to their levels in wild-type cells. However, in contrast to biofilm thickness, motility displayed SigB-dependent, SigB-independent, and SinR-dependent components. Conversely, sliding proficiency, which is another type of social motility behavior that is flagellum independent (and therefore independent of SinR activity) (12), was not affected by inactivating SigB (Fig. S8). In summary, the results confirmed that SigB-dependent downregulation of biofilm overgrowth is mediated by controlling the activity of SinR.
FIG 6.
SigB regulates flagellum-dependent motilities in B. subtilis. (A) Swimming proficiencies of wild-type (strain JH642) and isogenic mutants with altered SigB or SinR activity (strains MR644 and RG432, respectively). (B) Swarming proficiencies of the wild type (strain NCIB3610) and isogenic mutants with altered SigB or SinR activity (strains RG5568 and RG4576, respectively). For the swarming experiments, the NCIB3610 strain was utilized instead of the JH642 strain, because the latter strain does not swarm. (C and D) Representative pictures from several independent experiments of top-viewed swimming (C) or swarming (D) cells after 48 h of incubation are shown. Each data point in panels A and B is the mean ± SEM from a representative experiment performed in triplicate.
SigB regulates biofilm dispersal.
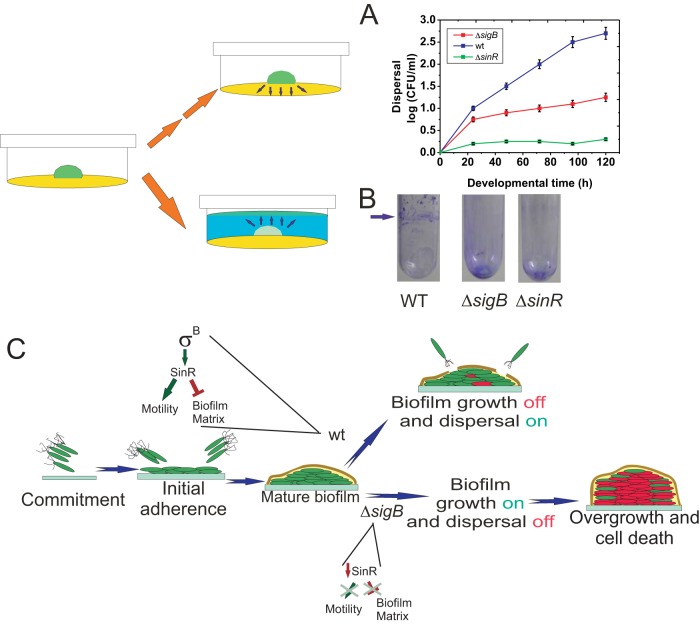
Although dispersal is likely to be crucial for bacterial survival, the gene network and signal transduction pathways controlling this phenomenon in bacilli are completely unexplored (4). In old (mature) biofilms, surface-attached cells detach and disperse into the environment to search for new food supplies and/or to colonize new sites. Studies of biofilm dispersal in Gram-negative bacteria and Gram-positive cocci have focused on identifying signals that trigger dispersal, one of which is nutrient availability (5, 7, 8, 45). Accordingly, we found that RsbP (which, in planktonic cultures, senses the nutritional status of the population) activated SigB inside the biofilm (Fig. 4), which in turn maintained the expression levels of the cell motility regulator SinR (Fig. 5). Therefore, it can be predicted that dispersal, a biofilm property that relies on motility proficiency, is also positively affected by the alternative sigma factor SigB. To test this hypothesis, we monitored the cell dispersal of wild-type and ΔsigB B. subtilis biofilms over time. Because biofilm dispersal has not been previously studied in Gram-positive bacilli, we adapted methodologies used to monitor the dispersal of other bacteria (8, 46). In one case (Fig. 7A), we quantified dispersal from a solid biofilm (colony) by counting the dispersed (swimming) cells (in CFU) present in the medium surrounding the biofilm (8); in the second case, we measured formation of a cellular ring (biofilm pellicle) located on top of a preformed biofilm from where dispersed cells produced the aforementioned ring (8) (Fig. 7B). As expected, in both cases, significantly less dispersal (2.5- to 3.5-fold) occurred for ΔsigB biofilms than for wild-type biofilms (Fig. 7A and B). The reduction in the number of cells dispersing from the biofilm might be a direct consequence of the lack of SigB activity or an indirect consequence of the reduced number of living cells observed in aging ΔsigB biofilms than in wild-type biofilms (Fig. 3B). However, dispersal also requires proficiency in cellular motility (5–8, 45, 46), and in B. subtilis this property is regulated by SinR (43, 44), whose levels are reduced in ΔsigB biofilms (Fig. 5A). Furthermore, as shown in Fig. 7A, the reduced dispersal of ΔsigB biofilm cells (compared with the dispersal of wild-type biofilm cells) is evident at early times of incubation (before 50 h) well before significant differences in cell death between both types of biofilms occur (Fig. 3B). Again, and similar to motility, dispersal displayed a SigB-dependent but also a SigB-independent but still SinR-dependent component, because dispersal was further reduced in a ΔsinR mutant compared to that of a ΔsigB mutant. In summary, the results presented allow us to hypothesize that the physiological role of SigB during the biofilm life cycle is to halt overgrowth of a mature biofilm and to prevent massive cell death through SinR-mediated dispersal activation when environmental and/or metabolic conditions become suboptimal or detrimental to maintaining the fitness of the population (Fig. 7C).
FIG 7.
SigB regulates biofilm dispersal. (A and B) Quantification of cell dispersal from wild-type and ΔsigB biofilms. (A) Colonies of JH642 and isogenic ΔsigB and ΔsinR derivatives (strains MR644 and RG432, respectively) developed on 2-cm-diameter wells poured with LBY medium (yellow in the cartoon) prepared with 0.8% agar and incubated for 30 h at 37°C, as indicated in Materials and Methods. At the indicated times, the agar (yellow in the cartoon) surrounding each solid biofilm (green in the cartoon) was carefully removed, and trapped cells were eluted and washed before appropriate serial dilutions were plated on LB agar plates as described in Materials and Methods. After 24 h of incubation at 37°C, the number of CFU ml−1 was calculated and plotted. The means ± SEM from 10 independent experiments are shown. (B) Colonies of wild-type (JH642), ΔsigB (MR644), and ΔsinR (RG432) strains were developed for 30 h at 37°C on 0.2 ml of LBY medium solidified with 2% agar at the bottom of a glass tube. The colonies were then carefully covered with 2.5 ml of LBY broth (blue in the cartoon) and incubated for another 15 h at 25°C to visualize the cellular ring (new green pellicle biofilm in the cartoon) produced at the liquid-air interface by the dispersed cells (see Materials and Methods for details). Ring cells adhered to the well walls were visualized by crystal violet staining (arrow). In the case of ΔsigB and ΔsinR biofilms, the staining at the bottom of the tubes represents the dispersal-defective SigB and SinR cells adhered to the glass bottom. A typical result from 10 independent experiments is shown. (C) Proposed model for the role of SigB during the biofilm life cycle and its regulation on sinR (see the text for details).
DISCUSSION
The idea of what we now recognize as a bacterial biofilm likely emerged in the 17th century, when Anton Van Leeuwenhoek observed and described microbial aggregates adhered to his teeth. However, it was not until forty years ago that the first reports pointing to the clinical relevance of biofilms appeared (47). Since then, accumulating evidence indicates that biofilms are important for the persistence, evolution, and dissemination of bacteria in multiple environments (2). Not all bacterial biofilms are harmful; indeed, some biofilms play critical and beneficial roles in nature. For example, plant-beneficial bacteria form biofilms in the plant rhizosphere and phyllosphere to colonize plant tissues (roots and leaves, respectively) and promote plant growth (48), and animal and human probiotics produce beneficial biofilms at the gut mucosa to improve host health and immunity (49, 50). Interestingly, it was recently demonstrated that probiotic B. subtilis can prevent S. aureus biofilm formation and colonization in both humans and mice when the bacilli were coadministered with the diet (51). In addition, other useful biofilms are used in industry, for example, to reduce steel corrosion and to explore novel compounds of biotechnological interest (52).
However, the formation of biofilms provokes stressful conditions for the cells involved (53, 54). When conditions are favorable, altruism exists between cells in the biofilm (kin cooperation), but when conditions become unfavorable, competition arises (kin rivalry). How kin cooperation and kin competition are resolved at the community level is beginning to be understood (5, 33, 55–57). For example, as a biofilm grows in size, the resident bacteria become crowded, and cells in the inner parts will experience reduced access to nutrients and electron acceptors and will accumulate waste products and toxin-like by-products (54). How do trapped biofilm cells cope with this self-generated life-threatening situation? Larger colonies produce greater detachments of individual cells than do smaller colonies, an observation that inspired the proposal that a certain maturation stage of the colony, as defined by its size, is linked to the nutritional status of the inner parts of the community to trigger dispersal events (58, 59). Supporting this interpretation, a positive relationship between dispersal proficiency and biofilm size has been reported for the pathogens Streptococcus intermedius and Pseudomonas aeruginosa (60, 61). In P. aeruginosa, swimming biofilm cells appear to initiate detachment when the size of the colony reaches a threshold diameter (60). In the early steps of infection by S. intermedius, the high-molecular-mass polysaccharide hyaluronan (HA), a major component of the extracellular matrix of connective tissue, allows bacterial adherence and biofilm formation. A common mechanism used by S. intermedius and other bacteria to allow dispersal involves the production of extracellular enzymes that degrade adhesive components of the biofilm extracellular matrix (7, 8, 45). Accordingly, hyaluronidase-defective mutant strains of S. intermedius are deficient in dispersal, forming 31% more biofilm than a wild-type strain in medium supplemented with HA (61). Importantly, and in accordance with our results, Singh et al. recently reported that the general stress-responsive transcription factor RpoS (SigS), the SigB functional homolog present in Gram-negative bacteria, is also activated by starvation to trigger biofilm dispersal in the human pathogen Vibrio cholerae (62).
Here, we show that SigB-deficient cells are unable to sense stress and maintain SinR levels as the biofilm ages (Fig. 4 and 5). As a consequence of such metabolic imbalance, the biofilm continued to grow (Fig. 2) and became less resistant to diverse stresses (Fig. 3; see also Fig. S3 in the supplemental material); dispersal was also downregulated (Fig. 7). How SigB regulates sinR expression is an unsolved question. The only known signal that regulates (activates) the expression of sinR is alcohol stress (40), which is also known as one of the best inducers of the SigB-dependent general stress response (16, 17). The simplest scenario for the influence of SigB on sinR expression would be direct binding of SigB to the sinR promoter. However, even the most comprehensive characterization of the transcriptional landscape of B. subtilis performed to date (63) did not reveal a SigB-dependent promoter directly upstream of sinR or in front of sinI. Unfortunately, all studies on regulation of SigB activity have so far been uniquely confined to the planktonic growth style of B. subtilis but not related to its sessile life style. Therefore, it is likely that SigB activates sinR expression indirectly via an unidentified pathway that operates under biofilm-supporting conditions. The likely target of this regulation is the promoter directly upstream of sinR (Fig. 5A), because no effect was observed on expression of sinI (Fig. S6C).
Because SigB and its functional homologs (i.e., SigS) are also present in pathogenic bacteria (17, 62, 64–66) and likely perform similar functions, blocking expression or activity of SigB or its homologs in pathogens might constitute a novel strategy for fighting harmful biofilms. Nonetheless, B. subtilis and other bacilli (i.e., Bacillus coagulans, B. amyloliquefaciens, and B. thuringiensis) have beneficial effects on agriculture and human health (13, 48, 51, 52, 67). In both situations, formation of a beneficial biofilm at the level of the plant rhizosphere or the human gut mucosa would be desired, and industrial/medical interventions with molecules or treatments that induce SigB might contribute to this goal.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
The B. subtilis strains used in this study are summarized in Table 1. Bacterial cultures were maintained in Luria-Bertani (LB) broth; for biofilm formation, LB was fortified with 4% yeast extract (LBY medium) as described previously (29). When appropriate, antibiotics were included at the following final concentrations: 1 μg/ml erythromycin (Ery), 5 μg/ml kanamycin (Kan), 5 μg/ml chloramphenicol (Cat), and 50 μg/ml spectinomycin (Spc). Transformation of B. subtilis to obtain isogenic derivatives of the parental strains was carried out as previously described (12). The cultures used to assess β-galactosidase activity were grown at 37°C in LBY broth without shaking (static cultures) (29). For each time point, 5 ml of LBY inoculated with 20 µl of an overnight culture of the corresponding strain was incubated in glass test tubes until the indicated developmental time. After the corresponding incubations, each pellicle biofilm was disrupted (dissolved) by vortexing during at least 3 min at room temperature and centrifuged at 4,500 rpm for 10 min, and the cellular (biofilm) pellet was resuspended in 1 ml of Z buffer for measuring of specific β-galactosidase activity (expressed in Miller units, or MU) as described previously (29). An inverted fluorescence microscope (Axiovert 25; Zeiss) was used to visualize green fluorescent protein (GFP) expression, and images were photographed, recorded, and analyzed with an image capture system (Olympus software and Fiji software). The LIVE/DEAD BacLight kit from Thermo-Fisher was used to evaluate the viability of biofilm cells according to the manufacturer’s recommendations. For the case of SigB-directed GFP expression during biofilm formation, 10 different microscope fields for each developmental time point were analyzed in triplicate, and the percentage of fluorescent cells refers to the total number of GFP-expressing and non-GFP-expressing cells counted at that time. For sporulation efficiency, B. subtilis biofilms were developed in LBY broth for 48 h, disrupted, and resuspended in sterile water. The biofilm cells were serially diluted, and appropriate dilutions were plated on LB agar plates and incubated for 24 h at 37°C to determine the total number of viable cells (spores plus vegetative cells). The same serial dilutions were treated with 10% CHCl3 for 5 min before plating to determine the number of spores (32).
Biofilm formation and dispersal.
The different B. subtilis strains used in this study were grown overnight at 37°C in LB medium with antibiotic supplementation (if appropriate); 20 µl of each culture was diluted in 5 ml of fresh LBY broth for pellicle formation in test tubes or as indicated in the figure legends when the biofilm was developed in microplates or bottles. The tubes (or microplates or bottles, as indicated) were statically incubated at 37°C for the indicated times. The amount of biofilm was measured by the crystal violet technique (29).
For dispersal experiments, a solid biofilm (colony) was developed on the center of 2-cm-diameter wells (12-well microtiter plates) containing LBY medium with 0.8% agar; this agar concentration allows swimming cells to enter the agar surrounding the solid biofilm. At the indicated times, the soft agar surrounding the solid biofilm was carefully removed and soaked (at 50 rpm) in 5.0 ml of sterile water supplemented with a sub-MIC amount of nalidixic acid (0.1 µg/ml) to avoid cellular duplication for 2 h at room temperature. After this incubation time, the undissolved soft agar was carefully removed and the solution containing the eluted cells was centrifuged at 4,500 rpm for 15 min at room temperature. The supernatant was discarded and the cellular pellet was dissolved in 1 ml of sterile water, and appropriate serial dilutions were plated on LB agar plates. After 24 h of incubation at 37°C, the number of CFU per milliliter was calculated and plotted. In another set of experiments, biofilm dispersal was detected by the formation of a second biofilm (a ring or pellicle) on top of the first biofilm (8). To this end, a solid biofilm (colony) was developed on 0.2 ml of LBY medium solidified with 2.0% agar for 30 h at 37°C, as indicated above, on the bottom of a glass tube. After this incubation time, 2.5 ml of LBY broth was carefully added through the wall of the glass tube to completely cover the preformed solid biofilm (without detaching or disturbing it), and the sample was incubated for 15 h at 25°C. Formation of a cellular ring (located at the liquid-air interface) indicated the formation of a second biofilm by cells dispersed from the first biofilm.
Stress treatments.
Isogenic JH642 and ΔsigB cells were subjected to different types of environmental and metabolic stresses, as described previously (31). For pH stress, biofilms were developed for the indicated times in 48-well microtiter plates at 37°C, and 500 µl of a pH 3.0 HCl solution was carefully added to the top of the biofilm. After 1 h of incubation at 25°C, the biofilms were disrupted and washed, and appropriate serial dilutions were plated on LB agar plates. CFU were counted after 24 h of incubation at 37°C, and the percentage of cell survival was referenced to the number of CFU of biofilms of the same age that had not been exposed to HCl. For azide treatment, 300 µl of 50 mM azide solution was carefully added to the top of biofilms developed in 48-well microtiter plates at 37°C. After 1 h of incubation at 25°C, the biofilms were disrupted and washed; appropriate serial dilutions were then plated on LB agar plates and incubated for 24 h at 37°C before CFU counting. Each data point in the figures represents the mean ± standard errors of the mean (SEM) of a representative experiment performed in triplicate.
Spreading (swimming, swarming, and sliding) experiments.
To assess surface (swarming and sliding) and swimming motilities, LB plates fortified with 0.7% or 0.3% agar, respectively, and dried for 1 h were inoculated at the center with 1 µl of an 8 × 107-cells ml−1 culture grown to mid-log phase at 37°C in LB broth. The inoculated petri dishes were then incubated at 37°C for the indicated times. The developed swimming, swarming, and sliding cellular discs were visualized using a Stemi 2000 (Zeiss) stereomicroscope with the KL1500LCD (Zeiss) illumination system. A PowerShot A80 (Canon) system was used to capture images.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by national grants from FONCyT (PICT2014 startup 3777) and CONICET to G.R. We thank the Fulbright (Washington, DC) and Pew (Philadelphia, PA) foundations for their support.
We thank David Zeigler from BGSC (Bacillus Genetic Stock Center, Columbus, OH) for providing strains and technical support. We also thank Alexander Reder for helpful discussions about the structure of the SigB regulon and Romina Coullery for her technical help at the initial stage of this work.
M.B., S.C., D.V., L.R., C.L., C.B., F.A., M.F., L.S., and J.M.V. carried out the experiments. All authors contributed to the experimental design and concepts, and all authors contributed to the manuscript. R.G. and U.V. wrote the main text with contributions from all other authors.
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00473-18.
REFERENCES
- 1.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 2.Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. 2016. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol 14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 3.Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 4.Abee T, Kovács AT, Kuipers OP, van der Veen S. 2011. Biofilm formation and dispersal in Gram-positive bacteria. Curr Opin Biotechnol 22:172–179. doi: 10.1016/j.copbio.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 5.McDougald D, Rice SA, Barraud N, Steinberg P, Kjelleberg S. 2011. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat Rev Microbiol 10:39–50. doi: 10.1038/nrmicro2695. [DOI] [PubMed] [Google Scholar]
- 6.Dalton T, Dowd SE, Wolcott R, Sun Y, Watters C, Griswold J, Rumbaugh K. 2011. An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS One 6:e27317. doi: 10.1371/journal.pone.0027317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barraud N, Kjelleberg S, Rice SA. 2015. Dispersal from microbial biofilms. Microbiol Spectrum 3:MB-0015-2014. [DOI] [PubMed] [Google Scholar]
- 8.Guilhen C, Forestier C, Balestrino D. 2017. Biofilm dispersal: multiple elaborate strategies for dissemination of bacteria with unique properties. Mol Microbiol 105:188–210. doi: 10.1111/mmi.13698. [DOI] [PubMed] [Google Scholar]
- 9.Piggot PJ, Coote JG. 1976. Genetic aspects of bacterial endospore formation. Bacteriol Rev 40:908–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cairns L, Hobley L, Stanley-Wall N. 2014. Biofilm formation by Bacillus subtilis: new insights into regulatory strategies and assembly mechanisms. Mol Microbiol 93:587–598. doi: 10.1111/mmi.12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Losick R. 2015. A love affair with Bacillus subtilis. J Biol Chem 290:2529–2538. doi: 10.1074/jbc.X114.634808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grau R, de Oña P, Kunert M, Leñini C, Gallegos-Monterrosa R, Mhatre E, Vileta D, Donato V, Hölscher T, Boland W, Kuipers OP, Kovács ÁT. 2015. A duo of potassium-responsive histidine kinases govern the multicellular destiny of Bacillus subtilis. mBio 6:e00581-15. doi: 10.1128/mBio.00581-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ayala F, Bauman C, Cogliati S, Lenini C, Bartolini M, Grau R. 2017. Microbial flora, probiotics, Bacillus subtilis and the search for a long and healthy human longevity. Microb Cell 4:133–136. doi: 10.15698/mic2017.04.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kearns DB, Chu F, Branda SS, Kolter R, Losick R. 2005. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol 55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- 15.Chu F, Kearns DB, Branda SS, Kolter R, Losick R. 2006. Targets the master regulator of biofilm formation in Bacillus subtilis. Mol Microbiol 59:1216–1228. doi: 10.1111/j.1365-2958.2005.05019.x. [DOI] [PubMed] [Google Scholar]
- 16.Price CW. 2002. General stress response, p 369–384. In Sonenshein AL, Hoch JA, Losick R (ed), Bacillus subtilis and its closest relatives. From genes to cells ASM Press, Washington, DC. [Google Scholar]
- 17.Hecker M, Pané-Farré J, Völker U. 2007. SigB-dependent general stress response in Bacillus subtilis and related Gram-positive bacteria. Annu Rev Microbiol 61:215–236. doi: 10.1146/annurev.micro.61.080706.093445. [DOI] [PubMed] [Google Scholar]
- 18.Benson AK, Haldenwang WG. 1993. Bacillus subtilis σB is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc Natl Acad Sci U S A 90:2330–2334. doi: 10.1073/pnas.90.6.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dufour A, Haldenwang WG. 1994. Interactions between a Bacillus subtilis anti-sigma factor (RsbW) and its antagonist (RsbV). J Bacteriol 176:1813–1820. doi: 10.1128/jb.176.7.1813-1820.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wise AA, Price CW. 1995. Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor σB in response to environmental signals. J Bacteriol 177:123–133. doi: 10.1128/jb.177.1.123-133.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voelker U, Voelker A, Maul B, Hecker M, Dufour A, Haldenwang WG. 1995. Separate mechanisms activate SigB of Bacillus subtilis in response to environmental and metabolic stresses. J Bacteriol 177:3771. doi: 10.1128/jb.177.13.3771-3780.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, Kang CM, Brody MS, Price CW. 1996. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev 10:2265–2275. doi: 10.1101/gad.10.18.2265. [DOI] [PubMed] [Google Scholar]
- 23.Vijay K, Brody MS, Fredlund E, Price CW. 2000. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the σB transcription factor of Bacillus subtilis. Mol Microbiol 35:180–188. doi: 10.1046/j.1365-2958.2000.01697.x. [DOI] [PubMed] [Google Scholar]
- 24.Brigulla M, Hoffmann T, Krisp A, Völker A, Bremer E, Völker U. 2003. Chill induction of the SigB-dependent general stress response in Bacillus subtilis and its contribution to low-temperature adaptation. J Bacteriol 185:4305–4314. doi: 10.1128/JB.185.15.4305-4314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Méndez MB, Orsaria LM, Philippe V, Pedrido ME, Grau R. 2004. Novel roles of the master transcription factors Spo0A and sigma B for survival and sporulation of Bacillus subtilis at low growth temperature. J Bacteriol 186:989–1000. doi: 10.1128/JB.186.4.989-1000.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamon M, Lazazzera B. 2001. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol Microbiol 42:1199–1209. [DOI] [PubMed] [Google Scholar]
- 27.Stanley N, Britton R, Grossman A, Lazazzera B. 2003. Identification of catabolite repression as a physiological regulator of biofilm formation by Bacillus subtilis by use of DNA microarrays. J Bacteriol 185:1951–1957. doi: 10.1128/JB.185.6.1951-1957.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamon M, Stanley N, Britton R, Grossman A, Lazazzera B. 2004. Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis. Mol Microbiol 52:842–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pedrido ME, de Oña P, Ramirez W, Leñini C, Goñi A, Grau R. 2013. Spo0A links de novo fatty acid synthesis to sporulation and biofilm development in Bacillus subtilis. Mol Microbiol 87:348–367. doi: 10.1111/mmi.12102. [DOI] [PubMed] [Google Scholar]
- 30.Völker U, Maul B, Hecker M. 1999. Expression of the σB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J Bacteriol 181:3942–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Höper D, Völker U, Hecker M. 2005. Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis. J Bacteriol 187:2810–2826. doi: 10.1128/JB.187.8.2810-2826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, Grau R, Perego M, Hoch JA. 1997. A novel histidine kinase inhibitor regulating development in Bacillus subtilis. Genes Dev 11:2569–2579. doi: 10.1101/gad.11.19.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. 2013. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol 11:157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Branda SS, González-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. 2001. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A 98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Branda SS, González-Pastor JE, Dervyn E, Ehrlich SD, Losick R, Kolter R. 2004. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J Bacteriol 186:3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLoon AL, Guttenplan SB, Kearns DB, Kolter R, Losick R. 2011. Tracing the domestication of a biofilm-forming bacterium. J Bacteriol 193:2027–2034. doi: 10.1128/JB.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lombardia E, Rovetto AJ, Arabolaza AL, Grau R. 2006. A LuxS-dependent cell-to-cell language regulates social behavior and development in Bacillus subtilis. J Bacteriol 188:4442–4452. doi: 10.1128/JB.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaidenko TA, Price CW. 1998. General stress transcription factor-σΒ and sporulation transcription factor-σH each contribute to survival of Bacillus subtilis under extreme growth conditions. J Bacteriol 180:3730–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kovács A, Kuipers O. 2011. Rok regulates yuaB expression during architecturally complex colony development of Bacillus subtilis 168. J Bacteriol 193:998–1002. doi: 10.1128/JB.01170-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gottig N, Pedrido ME, Méndez M, Lombardía E, Rovetto A, Philippe V, Orsaria L, Grau R. 2005. The Bacillus subtilis SinR and RapA developmental regulators are responsible for the inhibition of spore development by alcohol. J Bacteriol 187:2662–2672. doi: 10.1128/JB.187.8.2662-2672.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sonenshein AL. 2005. CodY, a global regulator of stationary phase and virulence in Gram-positive pathogens. Curr Opin Microbiol 8:203–207. doi: 10.1016/j.mib.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 42.Rodríguez Ayala F, Bauman C, Bartolini M, Saball E, Salvarrey M, Leñini C, Cogliati S, Strauch M, Grau R. 2017. Transcriptional regulation of adhesive properties of Bacillus subtilis to extracellular matrix proteins through the fibronectin binding protein YloA. Mol Microbiol 104:804–821. doi: 10.1111/mmi.13666. [DOI] [PubMed] [Google Scholar]
- 43.Fredrick K, Helmann JD. 1996. FlgM is the primary regulator of sigD activity, and its absence restores motility to a sinR mutant. J Bacteriol 178:7010–7013. doi: 10.1128/jb.178.23.7010-7013.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kearns DB, Losick R. 2003. Swarming motility in undomesticated Bacillus subtilis. Mol Microbiol 49:581–590. [DOI] [PubMed] [Google Scholar]
- 45.Kaplan JB. 2010. Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J Dent Res 89:205–2018. doi: 10.1177/0022034509359403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stacy A, Everett J, Jorth P, Trivedi U, Rumbaugh KP, Whiteley M. 2014. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc Natl Acad Sci U S A 111:7819–7824. doi: 10.1073/pnas.1400586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Costerton JW, Geesey GG, Cheng K-J. 1978. How bacteria stick. Sci Am 238:86–95. doi: 10.1038/scientificamerican0178-86. [DOI] [PubMed] [Google Scholar]
- 48.Beauregard PB, Chaib Y, Vlamakis H, Losick R, Kolter R. 2013. Bacillus subtilis biofilm induction by plant polysaccharides. Proc Natl Acad Sci USA 110:1621–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Donato V, Rodriguez Ayala F, Cogliati S, Bauman C, Costa JG, Leñini C, Grau R. 2017. Bacillus subtilis biofilm extends Caenorhabditis elegans longevity through downregulation of the insulin-like signaling pathway. Nat Commun 8:14332. doi: 10.1038/ncomms14332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smolentseva O, Gusarov I, Gautier L, Shamovsky I, De Francesco A, Losick R, Nudler E. 2017. Mechanism of biofilm-mediated stress resistance and lifespan extension in C. elegans. Sci Rep 7:7137. doi: 10.1038/s41598-017-07222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yue Zheng P, Nguyen TH, Dickey SW, Joo HS, Villaruz AE, Glose KA, Fisher EL, Hunt R, Li B, Chiou J, Pharkjaksu S, Khongthong S, Cheung G, Kiratisin P, Otto M. 2018. Pathogen elimination by probiotic Bacillus via signalling interference. Nature 562:532–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morikawa M. 2006. Beneficial biofilm formation by industrial bacteria, Bacillus subtilis and related species. J Biosci Bioeng 101:1–8. doi: 10.1263/jbb.101.1. [DOI] [PubMed] [Google Scholar]
- 53.Jefferson K. 2004. What drives bacteria to produce a biofilm? FEMS Microbiol Lett 236:163–173. doi: 10.1016/j.femsle.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 54.Liu J, Prindle A, Humphries J, Gabalda-Sagarra M, Asally M, Lee DY, Ly S, Garcia-Ojalvo J, Süel GM. 2015. Metabolic co-dependence gives rise to collective oscillations within biofilms. Nature 523:550–554. doi: 10.1038/nature14660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Le Galliard JF, Ferriere R, Dieckmann U. 2005. Adaptive evolution of social traits: origins, trajectories, and correlations of altruism and mobility. Am Nat 165:206–224. doi: 10.1086/427090. [DOI] [PubMed] [Google Scholar]
- 56.Lyons NA, Kraigher B, Stefanic P, Mandic-Mulec I, Kolter R. 2016. A combinatorial kin discrimination system in Bacillus subtilis. Curr Biol 26:733–742. doi: 10.1016/j.cub.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wall D. 2016. Kin recognition in bacteria. Annu Rev Microbiol 70:143–160. doi: 10.1146/annurev-micro-102215-095325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tolker-Nielsen T, Brinch UC, Ragas PC, Andersen JB, Jacobsen CS, Molin S. 2000. Development and dynamics of Pseudomonas sp. biofilm. J Bacteriol 182:6482–6489. doi: 10.1128/JB.182.22.6482-6489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davies DG, Marques CN. 2009. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J Bacteriol 191:1393–1403. doi: 10.1128/JB.01214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Purevdorj-Gabe B, Costerton WJ, Stoodley P. 2005. Phenotypic differentiation and seeding dispersal in non-mucoid and mucoid Pseudomonas aeruginosa biofilms. Microbiology 151:1569–1576. doi: 10.1099/mic.0.27536-0. [DOI] [PubMed] [Google Scholar]
- 61.Pecharki D, Petersen FC, Scheie A. 2008. Role of hyaluronidase in Streptotococcus intermedius biofilm. Microbiology 154:932–938. doi: 10.1099/mic.0.2007/012393-0. [DOI] [PubMed] [Google Scholar]
- 62.Singh PK, Bartalomej S, Hartmann R, Jeckel H, Vidakovic L, Nadell C, Drescher K. 2017. Vibrio cholerae combines individual and collective sensing to trigger biofilm dispersal. Curr Biol 27:3359–3366. doi: 10.1016/j.cub.2017.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nicolas P, Mäder U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, Becher D, Bisicchia P, Botella E, Delumeau O, Doherty G, Denham EL, Fogg MJ, Fromion V, Goelzer A, Hansen A, Härtig E, Harwood CR, Homuth G, Jarmer H, Jules M, Klipp E, Le Chat L, Lecointe F, Lewis P, Liebermeister W, March A, Mars RA, Nannapaneni P, Noone D, Pohl S, Rinn B, Rügheimer F, Sappa PK, Samson F, Schaffer M, Schwikowski B, Steil L, Stülke J, Wiegert T, Devine KM, Wilkinson AJ, van Dijl JM, Hecker M, Völker U, Bessières P, Noirot P. 2012. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335:1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- 64.Dangel A, Ackermann N, Abdel-Hadi O, Maier R, Önder K, Francois P, Müller CW, Pané-Farré J, Engelmann S, Schrenzel J, Heesemann J, Lindermayr C. 2013. A de-novo-designed antimicrobial peptide with activity against multiresistant Staphylococcus aureus acting on RsbW kinase. FASEB J 27:4476–4488. doi: 10.1096/fj.13-234575. [DOI] [PubMed] [Google Scholar]
- 65.Ringus DL, Gaballa A, Helmann JD, Wiedmann M, Boor KJ. 2013. Fluoro-phenyl-styrene-sulfonamide, a novel inhibitor of sigma B activity, prevents the activation of sigma B by environmental and energy stresses in Bacillus subtilis. J Bacteriol 195:2509–2517. doi: 10.1128/JB.00107-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feklístov A, Sharon BD, Darst SA, Gross CA. 2014. Bacterial sigma factors: a historical, structural, and genomic perspective. Annu Rev Microbiol 68:357–376. doi: 10.1146/annurev-micro-092412-155737. [DOI] [PubMed] [Google Scholar]
- 67.Cutting S. 2011. Bacillus probiotic. Food Microbiol 28:214–220. doi: 10.1016/j.fm.2010.03.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.