Abstract
Macropinocytosis is a prevalent and essential pathway in macrophages where it contributes to anti-microbial responses and innate immune cell functions. Cell surface ruffles give rise to phagosomes and to macropinosomes as multi-functional compartments that contribute to environmental sampling, pathogen entry, plasma membrane turnover and receptor signalling. Rapid, high resolution, lattice light sheet imaging demonstrates the dynamic nature of macrophage ruffling. Pathogen-mediated activation of surface and endosomal Toll-like receptors (TLRs) in macrophages upregulates macropinocytosis. Here, using multiple forms of imaging and microscopy, we track membrane-associated, fluorescently-tagged Rab8a expressed in live macrophages, using a variety of cell markers to demonstrate Rab8a localization and its enrichment on early macropinosomes. Production of a novel biosensor and its use for quantitative FRET analysis in live cells, pinpoints macropinosomes as the site for TLR-induced activation of Rab8a. We have previously shown that TLR signalling, cytokine outputs and macrophage programming are regulated by the GTPase Rab8a with PI3 Kγ as its effector. Finally, we highlight another effector, the phosphatase OCRL, which is located on macropinosomes and interacts with Rab8a, suggesting that Rab8a may operate on multiple levels to modulate phosphoinositides in macropinosomes. These findings extend our understanding of macropinosomes as regulatory compartments for innate immune function in macrophages.
This article is part of the Theo Murphy meeting issue ‘Macropinocytosis’.
Keywords: macropinocytosis, Rab GTPase, Toll-like receptors, ruffles, phosphoinositides, FRET
1. Introduction
(a). Macropinocytosis—a highway into macrophages
Macrophages play a multitude of roles in homeostasis and innate immunity [1]. As scavengers and professional phagocytes, they actively engulf debris and dead cells for the remodelling and maintenance of tissues and seek out invading pathogens to ingest and destroy. Macrophages constantly sample tissue environments through macropinocytosis, a pathway for indiscriminate internalization of solutes and fluids [2]. The gulping of fluid is accomplished by voluminous (greater than 0.2 µm) macropinosomes, which are internalized and rapidly shrink as they deploy membrane and cargo into other intracellular pathways [3,4]. Unlike other cells, macrophages have both constitutive macropinocytosis and induced or ‘receptor stimulated’ form of macropinocytosis, which is best known as a response to growth factor receptor stimulation and results in the formation of larger macropinosomes and increased fluid uptake [2,5,6]. In innate immune cells, macropinosomes also support antigen capture and subsequent antigen presentation. Incoming fluid phase cargo is surveilled by scavenger receptors and by pathogen recognition receptors (PRRs) which detect danger signals and pathogen signatures [3,7]. The macrophage surface undergoes constitutive ruffling, through the dynamic formation and collapse of undulating dorsal and peripheral F-actin-rich membrane ruffles. Upon ruffle closure, they give rise to macropinosomes or to phagosomes [8]. While macropinocytosis and phagocytosis provide portals for the uptake and destruction of microbial pathogens by macrophages, these pathways can also be subverted by viruses and intracellular bacteria for deliberate invasion and host colonization [9].
Using macropinocytosis, macrophages can also efficiently ingest many other things. The uptake of amino acids or protein as energy sources is a property of macropinosomes [10] and one that can be upregulated in cancer cells [11], but the use of macropinocytosis for sourcing protein may also be required by innate immune cells operating in poor nutrient environments at sites of infection or inflammatory tissue damage. Macrophages have a metabolic role in the uptake of lipids and lipid particles, which is accomplished at least partly through macropinocytosis in a process that can also be pathognomonic in atherosclerosis [12]. Finally, the ardent macropinocytosis pathway in macrophages is increasingly targeted in biotechnology for the delivery of nanoparticles, drugs, biologicals, vaccines, labels, inhibitors, viral vectors, RNA and DNA into macrophages or into the body [3]. Therefore, learning more about macropinocytosis in macrophages serves a very broad remit.
Importantly, macropinosomes are also active trafficking hubs, especially in macrophages which have a plasma membrane turnover rate far exceeding most other cell types [13]. Macropinocytic pathways offer the capacity to handle this constant membrane internalization and to recycle membrane rapidly back to the cell surface from macropinosomes via tubules or vesicles connecting with the large recycling endosome network in these cells [14]. All of the steps in macropinosome formation, beginning from surface ruffles, through closure, internalization and maturation, require extensive remodelling of the actin cytoskeleton and membrane phospholipids and involve multiple membrane trafficking events. Accordingly, a large array of molecular machinery, including families of actin-binding proteins, lipid kinases, GTPases, SNX proteins and many others are involved in these processes [3,4]. Moreover, an equally complex machinery is recruited to macropinosomes to regulate the processes occurring on macropinosomal platforms, such as cargo sorting, receptor signalling, pH regulation and protein modifications. Small GTPases, particularly of the Rab, Rho and Arf subfamilies [15–18], are required for ruffling and macropinocytosis but also contribute to cargo-related functions. The Rab small GTPases typically and ubiquitously govern membrane trafficking and receptor signalling pathways. We and others have previously characterized multiple Rabs (including Rab35, 20, 8, 5, 34 and 7) that are recruited and act discretely, but in sequence, on surface ruffles, macropinosomes or phagosomes and ensuing endosomal compartments [17,19–23]. Most recently we have shown a role for Rab13 in forming characteristic tent-pole ruffles on activated macrophages which give rise to enlarged macropinosomes [24]. As part of this sequence, Rab8a is enriched on ruffles and early macropinosome membranes, where, like Rab5 and some other Rabs, it is involved in receptor signalling [19,21].
(b). Rab8a as a Toll-like receptor signalling regulator in macropinosomes
Rab8a is a multifunctional GTPase, promiscuous in its ability to couple with multiple effectors for roles in different cell pathways, including cell ruffling and migration, neurite outgrowth and multiple steps of vesicular traffic to and from the recycling endosome, to polarized plasma membrane domains and to cilia [25–29]. Fluorescently tagged Rab8a expressed in macrophages is enriched on dorsal ruffles at the cell surface and in macropinosomes in live and fixed cells [19,21]. Previously, using live cell imaging, we identified a peak in Rab8a enrichment in early macropinosomes, during a window in time where the signalling phosphoinositide PI(3,4,5)P3 is enriched, preceding the conversion of the macropinosome to the early endosome-like, Rab5 and PI(3)P positive state [19]. This early window in maturation of the macropinosome has been termed the signalling macropinosome in other cell types [30]. Accordingly, as it transitions between ruffles and the early macropinosome, Rab8a can be found co-located with Toll-like receptor (TLR) 4 and TLR adaptor proteins [19,21].
The TLR family members are pattern recognition receptors and are activated by a wide range of pathogens to guard against infection. TLRs can be activated on the macrophage cell surface by contact with pathogens in tissue environments, while other TLRs are strategically located on intracellular membranes with the goal of detecting intracellular pathogens or pathogen-associated nucleic acids [31]. Complex TLR-induced signalling pathways produce transcription and secretion of arrays of pro- and anti-inflammatory cytokines and chemokines which shape inflammatory responses and trigger adaptive immune responses [32]. TLR signalling and cytokine outputs are varied through the actions of signalling adaptors, kinases, membrane phosphoinositides, GTPases and transcriptional regulators. There is an emerging inventory of the molecular machinery involved in regulating TLR signalling and cytokine secretion and our research has highlighted a role for macropinosomal Rab8a in TLR signalling [19,21].
Interest in macrophage Rab8a was piqued upon identifying a lipid kinase, of the class 1B PI3Kγ, as a non-canonical effector for this GTPase [21]. Evidence from genetic or CRISPR deletion of PI3Kγ or Rab8a implicated this complex in regulating Akt and mTOR signalling downstream of TLR4, and later of other TLRs, and this signalling was found to bias cytokine programmes, limiting the secretion of pro-inflammatory cytokines and enhancing anti-inflammatory cytokines [19,21]. PI3Kγ has emerged as an important and pharmacologically tractable regulator in TLR-driven inflammation where it constrains inflammation and promotes macrophage reprogramming or polarization towards an M2-like phenotype [21,33]. Several lines of evidence point to early macropinosomes as the site for Rab8a-mediated regulation of TLR signalling [19] but further exploration of Rab8a in this environment is needed.
The current study is motivated by the emerging potential for Rab8a to serve as a critical and multi-functional switch for TLR signalling and other functions in macrophage macropinosomes. A more complete spatial and functional molecular landscape of these organelles will significantly advance our understanding about how macropinosomes shape innate immune responses.
2. Material and methods
(a). Reagents, plasmids and biosensor constructs
Antibodies and reagents were sourced commercially as follows: mouse anti-HA.11 (Covance, MMS-101R); rabbit anti-OCRL1 (Sigma-Aldrich, Cat No. 07640); GFP antibody (LifeTechnologies, Australia, A6455); 10 K MW Alexa Fluor 647, 70 K MW Oregon Green-dextran and Texas Red-WGA (Molecular Probes (Invitrogen)); HRP-conjugated goat anti-mouse and anti-rabbit antibodies (81-6520) (Zymed Laboratories Inc. San Francisco, CA); 5-(N-Ethyl-N-isopropyl)amiloride (EIPA) (Sigma-Aldrich); lipopolysaccharide (LPS), purified from Salmonella enterica serotype Minnesota Re 595 (Sigma-Aldrich Australia); CpG-containing oligonucleotide ODN-1688 (Genscript); Poly(I:C) (Integrated Sciences, Chatswood, Australia). LPS, poly(I:C), and CpG DNA were used at 10 ng ml−1, 10 µg ml−1, and 0.3 µM, respectively.
Constructs containing GFP-, tdTomato-, mCherry-Rab8a, GFP-Rab8a-Tail domain, mCherry-Rab5a, PH-PLCδ–GFP, 2xFYVE-mCherry, GFP-Rab11a, GFP-Rab35, SidC(P4C)-GFP, APPL1-GFP have all been described previously [19,34–36]. peGFP-LifeAct, OCRL1-GFP and HA-OCRL1 were kindly provided by colleagues Fredric Meunier (University of Queensland), Sergio Grinstein (University of Toronto) and Christina Mitchell (Monash University) respectively.
For construction of a Rab8 biosensor, a Rab13 biosensor [37] was used as the backbone. This biosensor consists of the N-terminal Rab binding domain of MICAL-L2, linked to the mCerulean3 donor, a linker, mVenus acceptor followed by full-length Rab13. The small GTPase at the C-terminus acts as the lipid-binding anchor and has been shown to faithfully replicate Rab13 localization when expressed in cells [37]. Mouse Rab8a specific primers—forward 5′-TATAGAATTCA TGGCGAAGACCTACGATTAC-3′, reverse 5′-TATACTCGAGTCACAGGAGACTGCACCG-3′—were used to PCR the wild-type (WT), Q67 L constitutively active (CA) and T22N dominant negative (DN) open reading frames. Rab13 was replaced by restriction digestion with EcoRI and XhoI. To generate stable cell lines the whole biosensor open reading frame was transferred to the pEF6/V5-His TOPO (Invitrogen, Carlsbad, USA) vector using the GIBSON cloning method utilizing the NEB Gibson Assembly Master Mix (NEB, Massachusetts, USA) and the universal forward primer 5′-GCTTGGTACCGAGCTCGGATCCTATACCATGGGAGAGGAGCA GCA-3′ and reverse 5′- TCGAGGCTGATCAGCGGGTTTAAACTGATGG TGATGGTGGTGCTCGAG-3′.
(b). Cell culture and immunoprecipitation
The RAW 264.7 mouse macrophage-like cell line was sourced from ATCC. Cells were cultured in RPMI 1640 (Thermo Fisher Scientific, USA) supplemented with 10% heat-inactivated FCS (Thermo Fisher Scientific, USA) and 2 mM L-glutamine (Invitrogen, Carlsbad, USA) at 37°C in humidified 5% CO2. Transfections were performed using Lipofectamine 2000 (Thermo Fisher Scientific, USA) as per the manufacturer protocol. Immunoprecipitation was performed as described previously [19]. Briefly, RAW264.7 cells stably expressing GFP-Rab8a were lysed by passage through successive needles in lysis buffer (20 mM Tris pH 7.4, 150 mM NaCl, 1% NP-40 (Sigma), 5% glycerol) with addition of complete protease inhibitors (Roche Applied Science) and phosSTOP tablets (Roche Applied Science). After centrifugation at 17 000g for 15 min at 4°C, the supernatant was collected and incubated with GFP-Nanotrap beads (Protein expression facility, University of Queensland) for 1 h at 4°C. Beads were then washed 4 times with lysis buffer and bound proteins were eluted in SDS-PAGE sample buffer. 1% of lysates and 20% of eluates were loaded and separated by 10% SDS-PAGE and analysed by immunoblotting.
(c). Imaging and Förster (or fluorescence) resonance energy transfer (FRET) calculations
Immunofluorescence staining was performed on cells fixed in 4% paraformaldehyde, as previously described [38]. Ratiometric imaging of full-length Rab8a versus the Rab8a-tail domain has previously been described [21]. Live-cell imaging set-up and the use of the personal Deltavision system, Zeiss 710 confocal and Zeiss Spinning disk system has previously been described [19]. Microscopes used for imaging are designated in the figure legends. Briefly, for FRET imaging, the Zeiss Spinning Disk Confocal System (Zeiss Axiovert 200 with CSU-X1 scanhead) was used with a LCI Plan-Neofluar 63×/1.30 W Korr DIC M27 lens or Plan-Apochromat 40×/0.95 Korr M27 with 458 nm laser excitation dual 512 × 512 EMCCD cameras with a 510LP dichroic and dual 485/30 and 562/45 emission filters. As the FRET biosensor is a single chain sensor with equimolar concentrations of donor and acceptor, the FRET measurements were taken as the ratio of FRET/donor after flat field corrections were performed [39]. For flat-field correction, we used the average of 20 frames acquired using an empty dish filled with imaging medium under equivalent laser and exposure conditions to those used for image acquisition for in-cell FRET measurements. Imaging of medium alone was performed with and without laser power to generate the dark field [DF] and light field [LF] images for correction calculations. Corrections were performed as per the following equation; where DFmean is the mean intensity of the DF image and SF is the scaling factor: a scaling factor of 1000 for our imaging set-up.
A 2 × 2 mean filter was applied to the corrected FRET and corrected donor images before being ratioed and multiplied by a binary mask of the cells of interest. All statistics were calculated using Prism v. 7 software.
Scanning electron microscopy (SEM) and cryo-immuno electron microscopy were performed as previously described [36,40]. For some live cell imaging and SEM, we used Salmonella enterica serovar typhimurium (SL1344) added to cells at a multiplicity or infection (MOI) of 10 during imaging. For live cell imaging using lattice light sheet microscopy (LLSM), RAW264.7 macrophages expressing GFP-LifeAct were grown on 5 mm glass coverslips overnight. Cells were imaged in Phenol Red Free Leibovitz's L15 media (ThermoFisher), supplemented with 5% FBS, 1% L-glutamine and penicillin/streptomycin. Images were acquired on a 3i LLSM microscope (IMB) equipped with a 500 mW 488 nm Coherent Sapphire laser, and were observed with a Nikon 25× LWD 1.1 NA dipping objective, 2.5× tube lens and dual Hamamatsu ORCA Flash4.0 camera. The annular mask position used was 0.465/0.5 (inner/outer NA) with the lattice pattern generated at 488 nm, 1.049 spacing, 35 beams, 0.2 cropping. Images were acquired using 3i Slidebook with a step spacing of 0.553 µm and 120 steps per capture, with a time interval of 5 s between frames, and were deskewed (32.8°) using Microvolution. Post processing deconvolution was performed using 30 cycles of deconvolution with a generated PSF (200 nm beads Thermofisher #F8811) in Microvolution on a Dell R740 with 1TB of RAM and 2 NVidia V100 GPUs. Movies were visualized and produced using Imaris 9.2 (Bitplane) at 10 fps.
(d). Dextran uptake assay
Macrophages grown on glass coverslips were inverted over a 50 µl drop of media containing 100 µg ml−1 of fluorescent dextran and incubated for 15 min. Cells were then washed twice in ice cold PBS prior to fixation with 4% paraformaldehyde. The cell surface was then labelled with TMR-WGA before analysis. Using ImageJ thresholding and the ‘analyse particles’ function, the area of macropinosomes (thresholded dextran) and the number of cells (segmented using WGA) were counted for each condition. All statistics were calculated using Prism 7 software.
3. Results
(a). Macropinocytosis in Toll-like receptor-activated macrophages
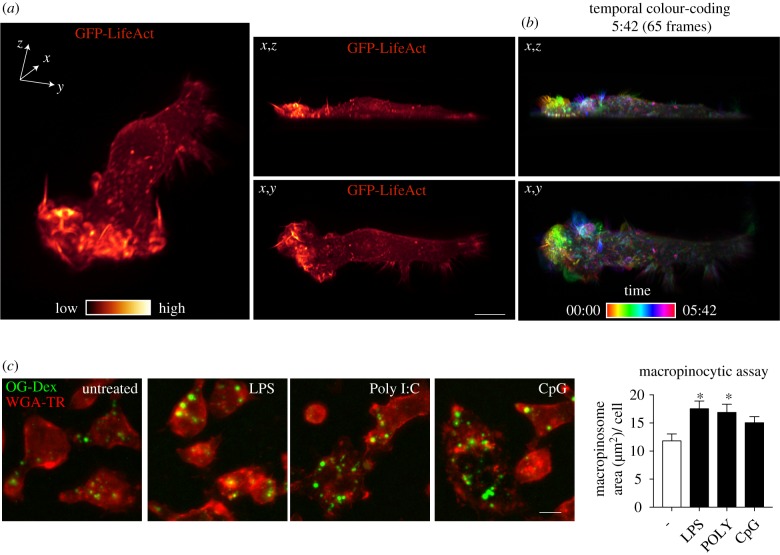
Macrophages ruffle constitutively and upon the activation of the cells after contact with pathogens or other danger signals. The active dorsal ruffling on the cell surface is revealed by lattice light sheet imaging of live RAW 264.7 macrophages stably expressing LifeAct to label F-actin-rich ruffles (figure 1a). Temporal colour coding is used to display the sequential appearance and short lifetime of individual ruffles on a cell surface that is in constant motion (figure 1b and electronic supplementary material, movie S1).
Figure 1.
Activation of macrophages enhances ruffling and macropinocytosis. (a) RAW 264.7 macrophages stably transfected with GFP-LifeAct were imaged using LLSM over a 30 min time-course. Three orientations are shown, an x,y,z perspective (left panel), x,z (upper middle) and x,y (lower middle). GFP-LifeAct is pseudocoloured with the GLOW colour map LUT from Imaris. (b) Temporal-colour coding of 65 frames over 5 min and 42 s using the thermal LUT from FIJI. (c) Macropinocytosis assay in RAW 264.7 macrophages untreated and treated with 10 ng ml−1 LPS, 10 µg ml−1 Poly I:C or 0.3 µM CpG-DNA. Cells were prestimulated for 30 min before addition of 70 K MW OG-dextran for 15 min. Quantification of total macropinosome area (μm2) per cell. Scale bar, 10 µm.
Ruffling results in the formation of macropinosomes and both ruffling and macropinocytosis can be enhanced in macrophages by contact with pathogens or by growth factors [19,41–43]. LPS activation of TLR4 has typically been used in such studies to show upregulation of macropinocytosis [24]. Here we have extended this analysis to the activation of other TLRs by quantitatively assaying macropinocytosis in cells activated by Poly I:C and CpG-DNA, the agonists for TLR3 and TLR9 respectively (figure 1c). The images and quantification in figure 1c indicate that, compared to untreated cells, treatment with LPS and Poly I:C significantly increased macropinocytic uptake, while treatment with CpG DNA showed a similar trend. This means that TLR detection of diverse pathogens, ranging from TLR4 recognition of external Gram-negative bacteria, to TLR3 and TLR9 detection of intracellular viral and bacterial DNA, all have in common the ability to upregulate macropinocytosis.
(b). Temporal association of Rab8a with macropinosomal and recycling pathways
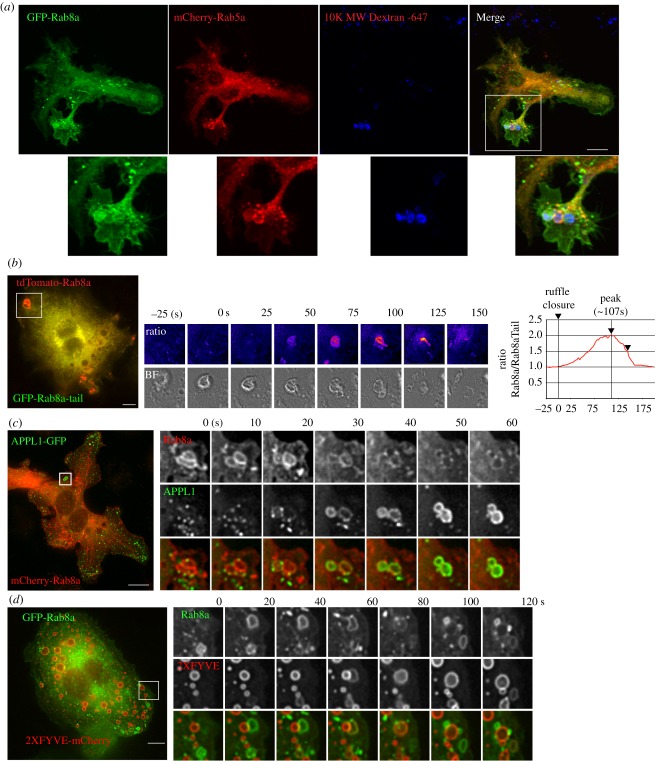
We previously reported that Rab8a is localized on macrophage membranes, particularly on ruffles and macropinosomes [19]. Here we set out to explore in more detail the molecular landscape and timing of Rab8 recruitment and activation on macropinosomes which are the purported sites for Rab8a-mediated TLR signalling. GFP-Rab8a expressed in macrophages is associated with intracellular vesicles and macropinosomes but, in addition, Rab8a is found on sections of the plasma membrane, particularly ruffling edges and dorsal ruffles on the cell surface near adjacent macropinosomes (figure 2a). Macropinosomes are co-labelled with dextran-647 and mCherry-Rab5 which overlap partially with GFP-Rab8a on the macropinosomes (figure 2a), further confirming the earlier identification of these Rab8a compartments [19,21]. The specific enrichment of Rab8a on early macropinosomes can be demonstrated by ratiometric imaging of full-length tdTomato-Rab8a and the GFP-Rab8a-tail in live cells using a previously described approach [21]. When combined with brightfield views, this live imaging can clearly demonstrate the point of enrichment of full-length Rab8a soon after ruffle circularization during closure and formation of early macropinosomal membranes (figure 2b). The macropinosomes also begin to tubulate at this point (see inset at 100 and 125) with Rab8a also enriched on the tubule membranes. We previously demonstrated that Rab8a resides on macropinosomes transiently and prior to peak recruitment of Rab5 [19]. This temporal sequence is reinforced by comparing Rab8a recruitment to that of the Rab5 effector, APPL1 (figure 2c) and the effector substrate and early endosomal lipid PI(3)P (figure 2d) which both show Rab8a depleting as these markers are enriched on the maturing macropinosomes. Thus, Rab8a is a bona fide and prominent component of the early macropinosome.
Figure 2.
Rab8a recruitment analysis during macropinosome formation. (a) RAW 264.7 cells transiently expressing GFP-Rab8a and mCherry-Rab5a. Cells were pulsed for 15 min with 647-dextran to label newly formed macropinosomes imaged on a Zeiss AxioImager. (b) Ratiometric time-lapse imaging of full-length tdTomato-Rab8a and GFP-Rab8a-Tail domain. Right shows ratio of two channels compared to formation of the macropinosome, depicted by brightfield image. Time-lapse imaging of cells co-expression of (c) APPL1-GFP and mCherry-Rab8a and (d) GFP-Rab8a and 2xFYVE-mCherry. Cells in (b) and (c) were imaged using the DeltaVision Deconvolution microscope. Scale bar, 10 µm.
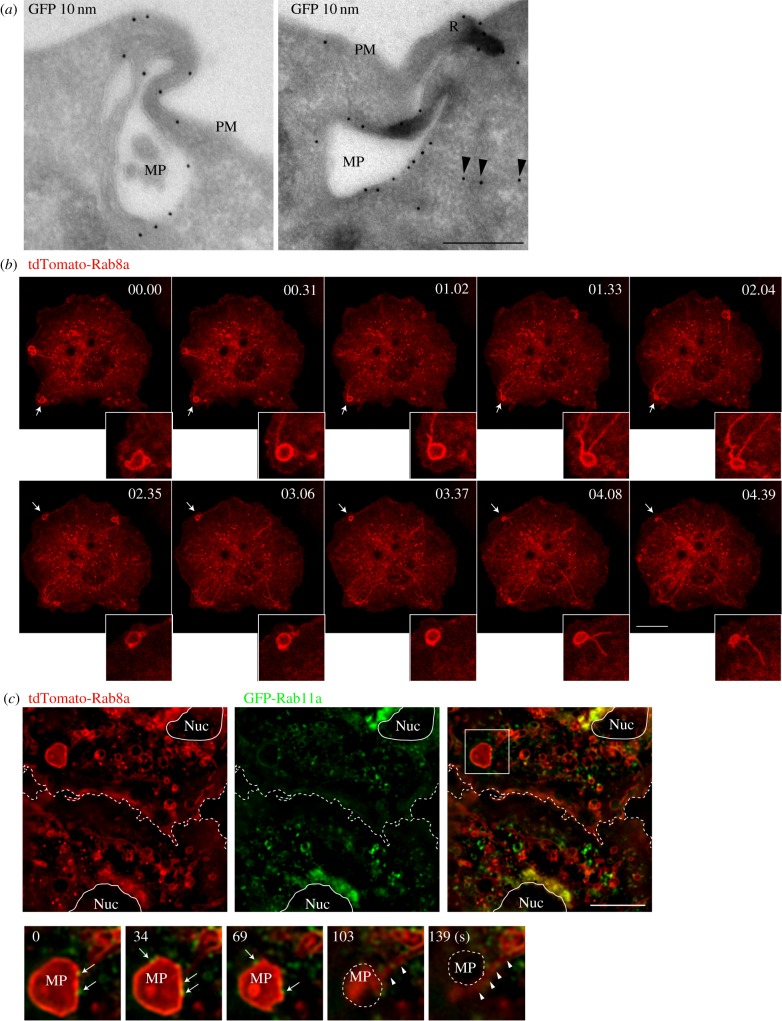
The early-stage macropinosomes are also sites for major membrane remodelling and protein sorting as the macropinosomes mature, condense and move into the cell interior. Tubulation of membranes driven by BAR domain proteins is a conspicuous feature of the macropinosomes, contributing to the sorting of membrane proteins and cargo destined for recycling endosomes, and other organelles [44]. At an ultrastructural level, immunogold labelling also depicts GFP-Rab8a on surface ruffles and resulting macropinosomes and associated tubules (figure 3a). This dynamic tubulation of macropinosomes and the appearance of Rab8a-positive tubules is also demonstrated by live cell microscopy. In figure 3b (electronic supplementary material, movie S2) multiple large Rab8a positive, peripheral macropinosomes condense and tubulate over a 12 min time course with tubules that move into the cell interior (figure 3b). Additionally, the use of live-cell structured illumination microscopy (SIM) captures the initial enrichment of GFP-Rab8a on early macropinosomes, followed by its tubulation from the macropinosomal membrane (figure 3c, arrowheads). These tubules move towards the centre of the cell, where they are often found in close proximity to the recycling endosomal Rab11a (figure 3c, arrows), suggesting a connection of Rab8a with a recycling endosome trafficking route. Rab11a also sometimes decorates the Rab8a-positive macropinosomes in the cell periphery, and then in the perinuclear region, Rab8a and Rab11a are strongly colocalized on the central recycling endosome (figure 3c). This imaging depicts macropinosomal tubulation from multiple perspectives as a process that has been infrequently studied in macrophages. It also serves to visually connect the Rab8a tubules with the recycling network, suggesting the recycling endosome as the post-macropinosomal destination of Rab8a and a proportion of macropinocytic cargo in these cells.
Figure 3.
Rab8a recruitment to macropinosome-associated tubules and recycling pathways. (a) Cryo-immuno (anti-GFP) electron microscopy of RAW 264.7 macrophages expressing GFP-Rab8a. Arrowheads indicate presence of GFP-Rab8a on a macropinosome associated with a putative tubule. PM, plasma membrane; R, ruffle; MP, macropinosome. Scale bar, 250 nm. (b) RAW 264.7 macrophages transiently expressing tdTomato-Rab8a were treated with 10 ng ml−1 LPS and imaged using a Zeiss 710 confocal microscope. (c) Macrophages transiently expressing tdTomato-Rab8a and GFP-Rab11a were treated with 10 ng ml−1 LPS and imaged using a Nikon Structured Illumination Microscope (SIM). Dotted line is the border between two cells, and the nucleus is a solid line. MP, macropinosome; inset shows a frame every ∼34 s, with arrowheads indicating Rab8 on tubules emanating from a macropinosome and arrows indicating Rab11a on Rab8-positive macropinosomes. Scale bar, 10 µm.
(c). Rab8a macropinosomes in pathogen entry and detection
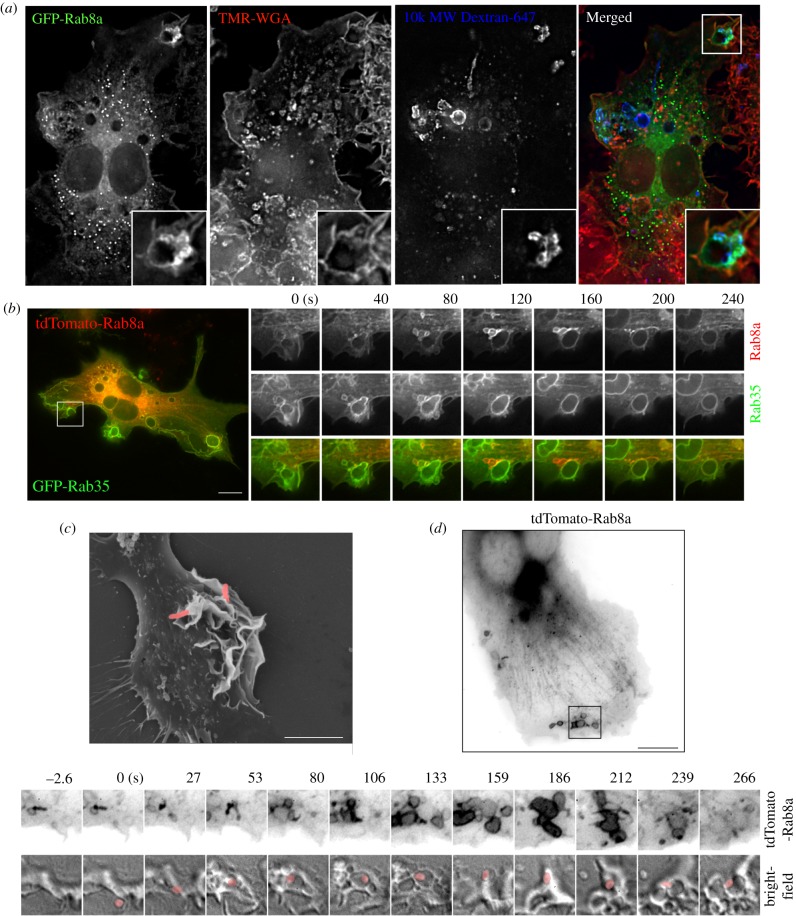
Peripheral ruffles give rise to both macropinosomes and phagosomes [14] which share some functional features and machinery. Both types of structures coexist in areas of active plasma membrane remodelling and during phagosome formation, tightening and internalization. In fixed cells, dextran-loaded macropinosomes are often seen surrounding newly forming phagosomes (figure 4a) and, while GFP-Rab8a faintly decorates the phagosomal membrane, it is significantly more enriched on the surrounding macropinosomes. In cotransfected live cells, during phagocytosis of opsonized sheep red blood cells (sRBCs), Rab8a is again clearly recruited to the surrounding macropinosomes, while the plasma membrane and phagosome are labelled more strongly and in a distinct fashion by coexpressed GFP-Rab35 (figure 4b). These images show that while ruffles may give rise to phagosomes and macropinosomes, Rabs are partitioned between these membranes, with Rab8a recruited preferentially to the macropinosomes but not to the phagosomes themselves. Finally, intracellular bacteria such as Salmonella use and indeed induce ruffling for cell entry, primarily by macropinocytosis, and these bacteria-associated and enlarged ruffles are demonstrated in the scanning electron micrograph in figure 4c. Combined fluorescence and bright-field live imaging of td-Tomato-Rab8a in macrophages shows Rab8a around the Salmonella-induced macropinosomes, including early compartments with rod-shaped bacteria and the enlarged macropinosomes that eventually lose Rab8a labelling (figure 4d). Thus, in these positions, around phagosomes and on macropinosomes, Rab8a is exposed to pathogen uptake and entry and it is in a prime position to be involved in pathogen detection and activation of the macrophages.
Figure 4.
Rab8a recruitment to phagosome-associated macropinosomes. (a) Medium containing IgG opsonized 3 µm latex beads was added to RAW 264.7 macrophages transiently expressing GFP-Rab8a for 10 min before fixation. Cells were counterstained with TMR-WGA without permeabilisation. (b) RAW 264.7 cells transiently expressing tdTomato-Rab8a and GFP-Rab35 were imaged after addition of IgG opsonized sheep red blood cells. (c) SEM of RAW 264.7 macrophages after infection with Salmonella enterica. Salmonella is pseudo-coloured pink. Scale bar, 5 µm. (d) Live-cell imaging of S. enterica infection of RAW 264.7 macrophages transiently expressing tdTomato-Rab8a. Still image and inset frames are inverted and represented in grey-scale. Salmonella is pseudo-coloured pink. Scale bar, 10 µm.
Having shown (in figure 1) that multiple TLR agonists can enhance macropinocytosis, we next performed labelling of Rab8a to determine whether Rab8a is localized on macropinosomes in TLR4-, 3- and 9-activated cells. tdTomato-labelled Rab8a was imaged on ruffles and early macropinosomes partially overlapping with a PI(4,5)P2 probe (PH-PLCδ-GFP) (electronic supplementary material, figure S1). Rab8a is thus found on peripheral macropinosomes in untreated cells and in cells treated with TLR4, TLR3 and TLR9 agonists. This is important confirmation that Rab8a is present on this compartment, poised to participate in signalling downstream of multiple TLRs. Finally, growth factors such as CSF-1 are well known stimulants for generating enlarged macropinosomes in macrophages [2] and here we also show that Rab8a is present in these growth-factor enlarged macropinosomes (electronic supplementary material, figure S1). Thus, Rab8a is habitually a transient component of ruffles and macropinosomes in macrophages responding to many pathogenic or physiological triggers.
(d). Rab8a is activated on macropinosome membranes
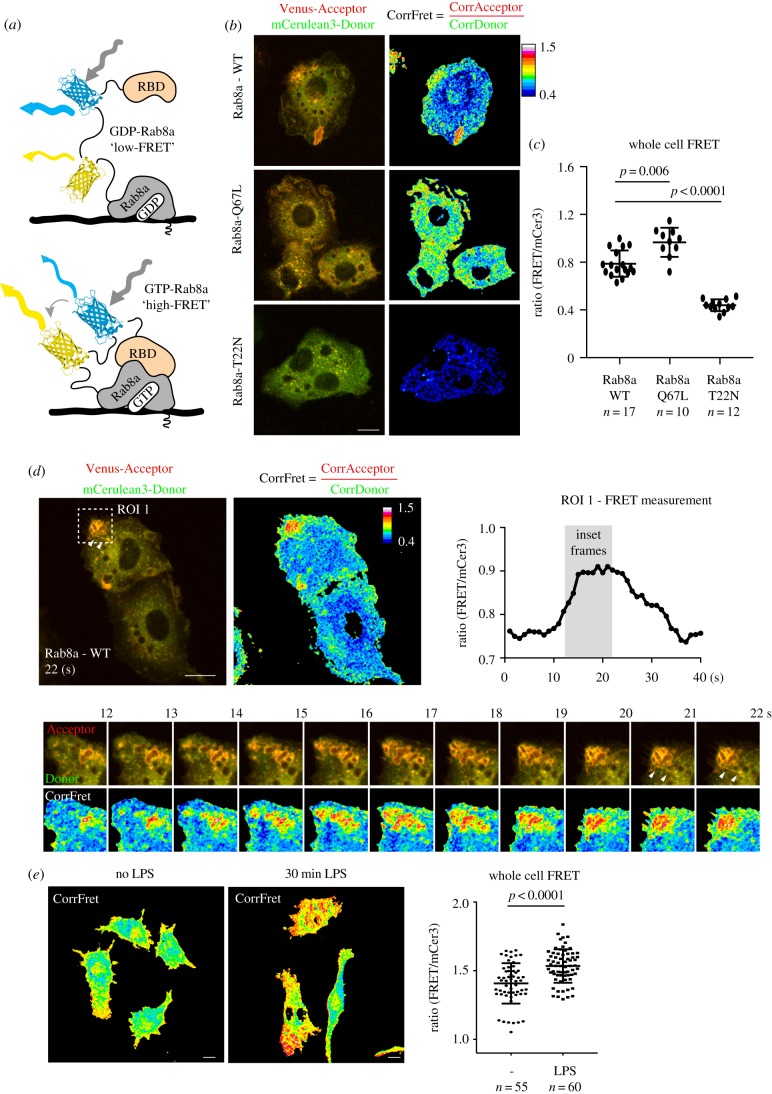
As with all GTPases, nucleotide exchange is required to generate the active form of GTP-Rab8a which recruits effectors. We have demonstrated the activation of Rab8a in cell extracts by TLR agonists [19], but the actual site for Rab8a activation on cell membranes has not been determined. To assess in-cell activation, we prepared a Rab8a biosensor, using the backbone of a single molecule FRET-based biosensor for Rab13 [37] containing the MICAL-L2 Rab binding domain (RBD) which not only binds to Rab13, but also to Rabs 8, 10 and 15 [45] (figure 5a). To test the ability of this biosensor to function, we expressed the constitutively active Rab8a (Q67 L mutant) as a high-FRET control and the dominant negative Rab8a T22N protein as a low-FRET control (figure 5b). Rab8 Q67L is mainly localized to the plasma membrane and early macropinosomes, while the T22N biosensor remains largely in the cytoplasm as well as showing association with some small endosome-like structures (figure 5b). The Rab8a WT biosensor localized in a similar fashion to tdTomato or GFP-Rab8a on the plasma membrane, macropinosomes and tubules (figure 5b). Quantification was performed on whole-cell FRET measurements and it demonstrated a biosensor dynamic range of 1.5 fold comparing Q67L versus T22N mutants (figure 5c). Then, in live LPS-activated cells, FRET measurements using the Rab8a biosensor in a region of interest (ROI) over peripheral ruffling and macropinosome formation showed the rise and fall of FRET (from approx. 0.75 rising to 0.91 before dropping back down to approx. 0.75) demonstrating the transient activation of Rab8a on these membranes (figure 5d). In the sequence of ROI movie frames, the most active Rab8a (a ratio of 1.7 at 22 s) was detected on the macropinosome membrane (figure 5d). In these frames, active Rab8 could also be detected on tubules emanating from these macropinosomes (figure 5d). Next, the biosensor was used for FRET on whole cells to demonstrate the LPS-induced activation of Rab8a (figure 5e). Thus, this Rab8a biosensor is used here to show for the first time that, despite its multiple locations in macrophages, Rab8a located on macropinosome membranes is activated by LPS, coinciding with the site we proposed for its binding to PI3Kγ and regulation of TLR4 signalling. In the future, the biosensor can be used to examine whether Rab8a is activated at the same site downstream of intracellular TLRs 3 and 9, or indeed by other macrophage receptors.
Figure 5.
Rab8a biosensor reveals enhanced Rab8a activity on macropinosomes and tubules. (a) Schematic diagram illustrating the architecture of the Rab8a biosensor. High-FRET is observed on membrane domains where GTP-Rab8a recruits the MICAL-L2 Rab-binding domain at the extreme C-terminus of the biosensor. (b,c) Whole cell FRET measurements of Rab8a WT-, Q67L- and T22N-expressing RAW 264.7 macrophages. The mCer3 ‘donor’ channel is pseudo coloured green and the Venus ‘acceptor’ channel is pseudo coloured red for visualization. (d) Live-cell imaging and quantification of the Rab8a WT biosensor. Region of Interest 1 (ROI 1) is boxed and measurements were taken every second over 48 s. Inset frames represent the rise from low to high FRET measurements. (e) Whole cell FRET measurements of cells left untreated or treated with 10 ng ml−1 LPS. Cells were imaged using the Zeiss Axiovert 200 spinning disk microscope with a 60× objective for (b–d) and 40× for (e). Data represented as mean ± s.d. Scale bar, 10 µm.
(e). Rab8a—a master of multiple macropinosome effectors?
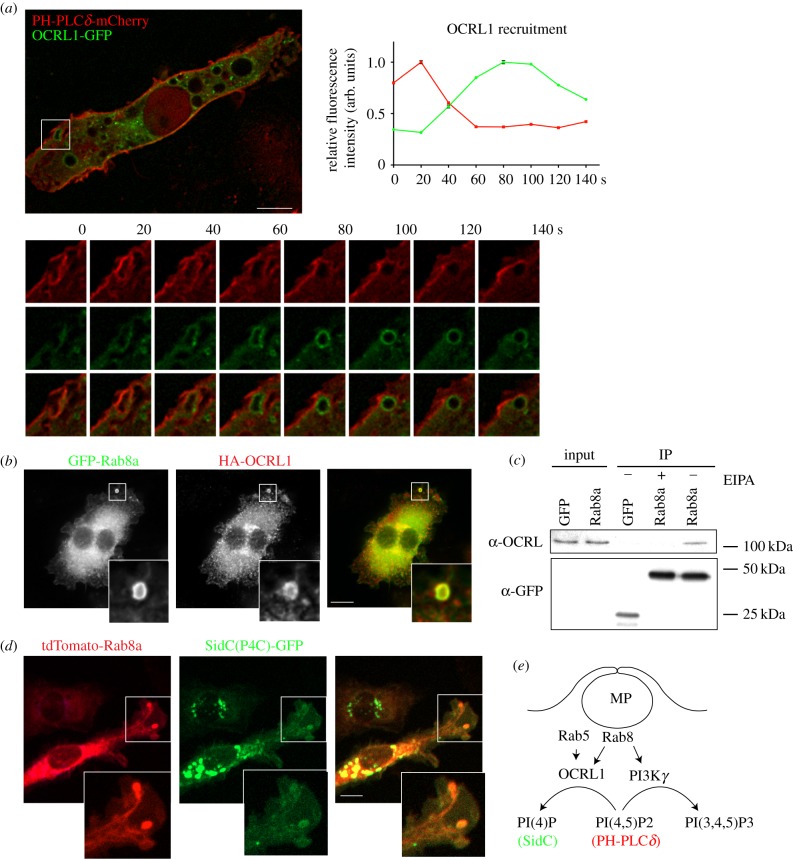
We have reported that in response to TLR activation, Rab8a recruits the class IB PI3Kγ as an effector to modulate TLR signalling [19,21]. PI3Kγ typically promotes the formation of PI(3,4,5)P3 from PI(4,5)P2 to enhance the recruitment of signalling kinases [46], and live-cell imaging with phosphoinositide probes confirmed that this lipid transition occurs in the macropinosomes of activated macrophages [19]. Interestingly, another one of Rab8a's known effectors is Lowe oculocerebrorenal syndrome protein (OCRL1 or INPP5F) [47], a 5′ phosphatase that dephosphorylates the same substrate that is phosphorylated by PI3K, converting PI(4,5)P2 to PI(4)P [48]. OCRL1 is found associated with Rab5 and APPL1-positive endosomes and it is on phagosomes in macrophages [49]. Preliminary experiments were carried out to explore the possibility that OCRL1 is on macrophage macropinosomes. Indeed when we express GFP-OCRL1 with PH-PLCδ-mCherry, we could see clear recruitment of OCRL1 during the depletion of PI(4,5)P2 on macropinosomes (figure 6a), reflecting similar timing to Rab8a recruitment during loss of PI(4,5)P2 [19]. Immunostaining here confirms that GFP-Rab8a and HA-OCRL1 are on peripheral macropinosomes together and immunoprecipitation shows that GFP-Rab8a coprecipitates HA-OCRL1 (figure 6b,c). Since there is also considerable perinuclear overlap of GFP-Rab8a and HA-OCRL1 staining, co-immunoprecipitation analysis was performed in the presence and absence of EIPA, to inhibit macropinocytosis [50] (figure 6c). This analysis reveals that the interaction of GFP-Rab8a with OCRL1 is inhibited by blocking macropinocytosis. Further evidence suggesting that Rab8a may be mediating OCRL1 function on the macropinosome is illustrated by co-expression and co-localization of Rab8a with SidC(P4C)-GFP a phosphoinositide marker for PI(4)P [51], the product of OCRL1-mediated hydrolysis of PI(4,5)P2. This preliminary data is suggestive of OCRL1 as a second Rab8a effector that is recruited during early stage macropinocytosis, opening up the possibility that Rab8a can perform two functions, one with PI3Kγ and the other with OCRL1, as two effectors that serve to remove PI(4,5)P2 to shape the macropinosome lipid environment. Through further analysis, the timing and reciprocal binding and functions of Rab8a with these effectors in macropinosomes can be pursued.
Figure 6.
OCRL1 interaction with Rab8a on macropinosomes. (a) RAW 264.7 macrophages transiently expressing PH-PLCδ-mCherry with OCRL1-GFP. Inset shows example of a ruffle transitioning to a macropinosome. OCRL1 recruitment analysis is displayed in arbitrary units and represented relative to each channels' peak intensity. (b) Co-expression of GFP-Rab8a and HA-OCRL1 in RAW 264.7 macrophages. Cells were imaged in (a,b) using the DeltaVision deconvolution microscope. (c) Co-immunoprecipitation analysis of RAW 264.7 macrophages, transiently expressing GFP-Rab8a or GFP-empty vector as a control. Cells were treated with or without 25 µM EIPA to block macropinocytosis and immunoprecipitations were performed using GFP-nanotrap agarose. Immunoblots were probed for anti-GFP and anti-OCRL. (d) Live-cell imaging of RAW 264.7 macrophages transiently expressing tdTomato-Rab8a and SidC(P4C)-GFP. Cells were imaged using Zeiss Axiovert 200 spinning disk microscope. (e) Summary diagram illustrating two possible macropinosome-associated Rab8a effectors that act on PI(4,5)P2.
4. Discussion
Macropinocytosis is a multifunctional pathway in macrophages, serving in several guises for pathogen detection, receptor activation and signalling and pathogen entry. It is a key compartment for membrane traffic, supporting the ingestion of fluid, protein, lipids and particles and it is a high capacity pathway for plasma membrane turnover and recycling. In this study we highlight the small GTPase, Rab8a, as a key component of macropinosomes in macrophages under several of these conditions. Data herein confirm and extend our previous findings, showing how Rab8a is poised to support TLR signalling in macropinosomes, and hinting at more extensive roles for this GTPase through additional effector interactions in this compartment.
Macropinocytosis is shown to be enhanced by activation of TLRs 4, 3 and to a lesser extent, TLR9, indicating that a range of extracellular and intracellular bacterial and viral pathogens induce macrophages to upregulate macropinocytic uptake. This extends our knowledge of macropinocytosis as an inducible process, one that can be tuned to suit physiological or pathophysiological conditions, ranging from pathogen challenge to growth factor stimulation, starvation or transformation in cancer [4,6,10,52]. In macrophages, it is not exactly clear how enhanced macropinocytosis supports innate immune functions, but resulting increases in fluid sampling, protein uptake and the trafficking of surface receptors are all likely to facilitate anti-microbial responses. Alternatively, or in concert, increased macropinocytosis may serve as an enhanced platform for phosphoinositide metabolism to accommodate receptor signalling [30,53]. Using various types of imaging, we show here that Rab8a is localized on macropinosomes in macrophages interacting with pathogens and those activated by different TLR agonists. We previously reported that LPS, poly I:C and CpG DNA all activate Rab8a (by increasing levels of GTP-Rab8a) in macrophages and that resulting TLR signalling and cytokine outputs are affected after CRISPR knockout of Rab8a [19]. Taken together, current and earlier evidence points to Rab8a, with its PI3Kγ effector, being recruited at the level of macropinosomes by different pathogens for regulation in multiple TLR signalling pathways. Given the emergent role of PI3Kγ as a key driver of macrophage reprogramming and in supressing inflammation, its common recruitment by Rab8a downstream of distinct TLRs sits as a distinguishing feature of these signalling pathways, compared to GPCRs and RTKs pathways which alternatively employ Ras-recruited PI3Kγ [46].
The production and use of a Rab8a biosensor in live cells provides, for the first time, compelling evidence that macropinosomes in the cell periphery (early macropinosomes) are sites where active, GTP-bound Rab8a is enriched in LPS-stimulated macrophages and that Rab8a activation temporally follows ruffling and macropinosome formation. This not only confirms the macropinosome as a site for Rab8a function, it provides further evidence that this is a key intersecting locale where Rab8a meets and can direct signalling from internalizing or endosomal TLRs, given that Rab8a is not associated directly with TLR complexes [21]. Furthermore, while the concept of endosomal sites for signalling from internalized TLR4 is frequently promulgated, the precise endosomal compartment(s) involved remain ill-defined since many prior studies are based largely on flow cytometry to detect surface versus intracellular TLRs [42,54]. The concept of the macropinosome as this site fits with the conversion of PI(4,5)P2 required for adaptor recruitment on TLR4 [55], it is congruent with internalization of CD14/MD2/TLR4 complexes independent of a receptor cytoplasmic tail signals [56] and it supports the localization of TLR4 and TRAM on these membranes [19]. Moreover, the labelling shown in this study presents scenarios where macropinosomes neighbour phagosomes and Rab11a coexists transiently on Rab8a macropinosomes, offering opportunities for the reported Rab11a involvement in TLR4 signalling [57]. We believe emerging data from other approaches provide an increasingly strong case for macropinosomes as key sites for TLR4 internalization and for TRIF/TRAM signalling from TLR4 and from intracellular TLRs. With the Rab8a biosensor now available, we will be able to examine Poly I:C- and CpG-stimulated cells to determine where Rab8a intersects with TLRs 3 and 9 for signalling given that signalling sites have not yet been pinpointed among their intracellular membrane locations [19].
We have shown that Rab8a can recruit PI3Kγ to mediate Akt signalling through the production of PI(3,4,5)P3 from PI(4,5)P2. Now we have also introduced the possibility that Rab8a is interacting with another phosphoinositide effector protein, OCRL1. During phagocytosis, Rab5 is responsible for recruitment of APPL1 and OCRL to hydrolyse PI(4,5)P2 to help remove F-actin for the final stages of particle engulfment [49]. While Rab5 is typically found associated with both phagosomes and macropinosomes, imaging here shows that Rab8a is recruited preferentially to macropinosomes and moreover this occurs before enrichment of Rab5 and APPL1. Notably, we have previously found no role for Rab8a in macropinosome formation since knockdown of Rab8 in macrophages does not alter dextran uptake [19,21]. By now showing that Rab8a can potentially bind to both PI3Kγ and to OCRL1 on macropinosomes, it will be interesting to decipher the precise spatial and temporal interactions involving Rabs 8a and 5 with OCRL1 and other effectors and accessory proteins during early macropinosome formation. It is reminiscent of Rabs being recruited in sequence or in networks to act as enzymatic cascades [58]. In the newly forming and early macropinosome, the removal of PI(4,5)P2 is important both for F-actin depolymerization and macropinosome closure [2], as well as for TLR signalling [55,59]. The Rab-mediated recruitment of a phosphatase and a kinase that mediate these actions highlights a level of spatio-temporal regulation that can support pathogen ingestion and receptor activation.
5. Conclusion
The ongoing study of macropinosomes, and specifically of Rab8a on macropinosomes in macrophages, allows us to draw conclusions that expand our understanding of the location and behaviour of this GTPase in the context of innate immune functions. Through multiple forms of imaging in macrophages, the localization of Rab8a on membranes in the dynamic dorsal ruffle–macropinosome–tubules–recycling endosome corridor is herein confirmed. Use of a biosensor revealed that LPS-induced activation of Rab8a occurs most intensely on ruffles transitioning to early macropinosomes, with lesser amounts of active Rab8a on macropinosome-derived tubules. This pinpoints the sites for LPS-induced Rab8a recruitment of a known effector, PI3Kγ, and a likely new effector in this context, OCRL1. The data provide further, much-needed evidence to define the functional sites for signalling from multiple TLRs, emerging increasingly as macropinosomes. Finally, Rab8a is a ubiquitous GTPase with many possible functions, inviting much wider examination of this GTPase in the context of macropinosomes. Broader studies will be aided by powerful and diverse experimental systems that reaffirm, pleasingly, how closely spatio-temporal organization and molecular function in macropinocytosis are recapitulated across species and cell types. Technologically, further studies will draw upon new imaging and visualization modalities to reproduce dynamic macropinosomal landscapes. Physiological and pathophysiological roles for macropinocytosis in immunity, nutrition, migration, cell growth and death have emerged and cemented this pathway as a critical and essential portal into cells.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Tatiana Khromykh, IMB and Darren Brown, IMB for valuable technical assistance. P. McPherson (McGill University, Canada) and L. Hodgson (Albert Einstein College of Medicine, USA) generously provided the Rab13 biosensor and helpful advice. We thank colleagues as noted for providing reagents. Imaging was performed in IMB's Cancer Biology Imaging and CUF Facilities funded by the Australian Cancer Research Foundation.
Data accessibility
The datasets supporting this article have been uploaded as part of the electronic supplementary material.
Authors' contributions
A.A.W. and J.L.S. designed the study; A.A.W., N.D.C. and L.L. collected and analysed the data. N.D.C. performed LLSM and post-acquisition processing. A.A.W. and J.L.S. wrote the article.
Competing interests
We declare we have no competing interests.
Funding
The work was supported by funding from the National Health and Medical Research Council of Australia (1003021, 1098710 to J.L.S.); Australian Research Council (grants LE170100206 and DP140101461 to J.L. Stow and DP180101910 to J.L. Stow and A.A.W) N.D.C. obtained PhD scholarship funding from the Australian Government and The Yulgilbar Foundation.
References
- 1.Wynn TA, Chawla A, Pollard JW. 2013. Macrophage biology in development, homeostasis and disease. Nature 496, 445–455. ( 10.1038/nature12034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canton J, Schlam D, Breuer C, Gutschow M, Glogauer M, Grinstein S. 2016. Calcium-sensing receptors signal constitutive macropinocytosis and facilitate the uptake of NOD2 ligands in macrophages. Nat. Commun. 7, 11284 ( 10.1038/ncomms11284) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kerr MC, Teasdale RD. 2009. Defining macropinocytosis. Traffic 10, 364–371. ( 10.1111/J.1600-0854.2009.00878.X) [DOI] [PubMed] [Google Scholar]
- 4.Buckley CM, King JS. 2017. Drinking problems: mechanisms of macropinosome formation and maturation. FEBS J. 284, 3778–3790. ( 10.1111/febs.14115) [DOI] [PubMed] [Google Scholar]
- 5.Bosedasgupta S, Moes S, Jenoe P, Pieters J. 2015. Cytokine-induced macropinocytosis in macrophages is regulated by 14-3-3ζ through its interaction with serine-phosphorylated coronin 1. FEBS J. 282, 1167–1181. ( 10.1111/febs.13214) [DOI] [PubMed] [Google Scholar]
- 6.Pacitto R, Gaeta I, Swanson JA, Yoshida S. 2017. CXCL12-induced macropinocytosis modulates two distinct pathways to activate mTORC1 in macrophages. J. Leukoc. Biol. 101, 683–692. ( 10.1189/jlb.2A0316-141RR) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canton J, Neculai D, Grinstein S. 2013. Scavenger receptors in homeostasis and immunity. Nat. Rev. Immunol. 13, 621–634. ( 10.1038/nri3515) [DOI] [PubMed] [Google Scholar]
- 8.Swanson JA. 2008. Shaping cups into phagosomes and macropinosomes. Nat. Rev. Mol. Cell Biol. 9, 639–649. ( 10.1038/nrm2447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mercer J, Helenius A. 2009. Virus entry by macropinocytosis. Nat. Cell Biol. 11, 510–520. ( 10.1038/ncb0509-510) [DOI] [PubMed] [Google Scholar]
- 10.Yoshida S, Pacitto R, Yao Y, Inoki K, Swanson JA. 2015. Growth factor signaling to mTORC1 by amino acid-laden macropinosomes. J. Cell Biol. 211, 159–172. ( 10.1083/jcb.201504097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Commisso C, et al. 2013. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497, 633–637. ( 10.1038/nature12138) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore KJ, Sheedy FJ, Fisher EA. 2013. Macrophages in atherosclerosis: a dynamic balance. Nat. Rev. Immunol. 13, 709–721. ( 10.1038/nri3520) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steinman RM, Brodie SE, Cohn ZA. 1976. Membrane flow during pinocytosis. A stereologic analysis . J. Cell Biol. 68, 665–687. ( 10.1083/jcb.68.3.665) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stow JL, Condon ND. 2016. The cell surface environment for pathogen recognition and entry. Clin. Transl. Immunol. 5, e71 ( 10.1038/cti.2016.15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radhakrishna H, Al-Awar O, Khachikian Z, Donaldson JG. 1999. ARF6 requirement for Rac ruffling suggests a role for membrane trafficking in cortical actin rearrangements. J. Cell Sci. 112, 855–866. [DOI] [PubMed] [Google Scholar]
- 16.Boshans RL, Szanto S, Van Aelst L, D'Souza-Schorey C. 2000. ADP-ribosylation factor 6 regulates actin cytoskeleton remodeling in coordination with Rac1 and RhoA. Mol. Cell. Biol. 20, 3685–3694 ( 10.1128/MCB.20.10.3685-3694.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schnatwinkel C, Christoforidis S, Lindsay MR, Uttenweiler-Joseph S, Wilm M, Parton RG, Zerail M. 2004. The Rab5 effector Rabankyrin-5 regulates and coordinates different endocytic mechanisms. PLoS Biol. 2, E261 ( 10.1371/journal.pbio.0020261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujii M, Kawai K, Egami Y, Araki N. 2013. Dissecting the roles of Rac1 activation and deactivation in macropinocytosis using microscopic photo-manipulation. Sci. Rep. 3, 2385 ( 10.1038/srep02385) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wall AA, Luo L, Hung Y, Tong SJ, Condon ND, Blumenthal A, Sweet MJ, Stow JL. 2017. Small GTPase Rab8a-recruited phosphatidylinositol 3-kinase gamma regulates signaling and cytokine outputs from endosomal Toll-like receptors. J. Biol. Chem. 292, 4411–4422. ( 10.1074/jbc.M116.766337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeo JC, Wall AA, Luo L, Stow JL. 2016. Sequential recruitment of Rab GTPases during early stages of phagocytosis. Cell. Logist. 6, e1140615 ( 10.1080/21592799.2016.1140615) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo L, et al. 2014. Rab8a interacts directly with PI3Kgamma to modulate TLR4-driven PI3K and mTOR signalling. Nat. Commun. 5, 4407 ( 10.1038/ncomms5407) [DOI] [PubMed] [Google Scholar]
- 22.Sun P, Yamamoto H, Suetsugu S, Miki H, Takenawa T, Endo T. 2003. Small GTPase Rah/Rab34 is associated with membrane ruffles and macropinosomes and promotes macropinosome formation. J. Biol. Chem. 278, 4063–4071. ( 10.1074/jbc.M208699200) [DOI] [PubMed] [Google Scholar]
- 23.Egami Y, Araki N. 2012. Spatiotemporal localization of Rab20 in live RAW264 macrophages during macropinocytosis. Acta Histochem. Cytochem. 45, 317–323. ( 10.1267/ahc.12014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Condon ND, Heddleston JM, Chew TL, Luo L, McPherson PS, Ioannou MS, Hodgson L, Stow JL, Wall AA. 2018. Macropinosome formation by tent pole ruffling in macrophages. J. Cell Biol. 217, 3873 ( 10.1083/jcb.201804137) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peranen J. 2011. Rab8 GTPase as a regulator of cell shape. Cytoskeleton 68, 527–539. ( 10.1002/cm.20529) [DOI] [PubMed] [Google Scholar]
- 26.Nakajo A, et al. 2016. EHBP1L1 coordinates Rab8 and Bin1 to regulate apical-directed transport in polarized epithelial cells. J. Cell Biol. 212, 297–306. ( 10.1083/jcb.201508086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi H, Etoh K, Ohbayashi N, Fukuda M. 2014. Rab35 promotes the recruitment of Rab8, Rab13 and Rab36 to recycling endosomes through MICAL-L1 during neurite outgrowth. Biol. Open 3, 803–814. ( 10.1242/bio.20148771) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Morita Y, Mazelova J, Deretic D. 2012. The Arf GAP ASAP1 provides a platform to regulate Arf4- and Rab11-Rab8-mediated ciliary receptor targeting. EMBO J. 31, 4057–4071. ( 10.1038/emboj.2012.253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng S, Knodler A, Ren J, Zhang J, Zhang X, Hong Y, Huang S, Peränen J, Guo W. 2012. A Rab8 guanine nucleotide exchange factor-effector interaction network regulates primary ciliogenesis. J. Biol. Chem. 287, 15 602–15 609. ( 10.1074/jbc.M111.333245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zoncu R, Perera RM, Balkin DM, Pirruccello M, Toomre D, De Camilli P. 2009. A phosphoinositide switch controls the maturation and signaling properties of APPL endosomes. Cell 136, 1110–1121. ( 10.1016/j.cell.2009.01.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brubaker SW, Bonham KS, Zanoni I, Kagan JC. 2015. Innate immune pattern recognition: a cell biological perspective. Annu. Rev. Immunol. 33, 257–290. ( 10.1146/annurev-immunol-032414-112240) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawasaki T, Kawai T. 2014. Toll-like receptor signaling pathways. Front. Immunol. 5, 461 ( 10.3389/fimmu.2014.00461) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaneda MM, et al. 2016. PI3Kγ is a molecular switch that controls immune suppression. Nature 539, 437–442. ( 10.1038/nature19834) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo L, et al. 2017. SCIMP is a transmembrane non-TIR TLR adaptor that promotes proinflammatory cytokine production from macrophages. Nat. Commun. 8, 14133 ( 10.1038/ncomms14133) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeo JC, Wall AA, Luo L, Condon ND, Stow JL. 2016. Distinct roles for APPL1 and APPL2 in regulating Toll-like receptor 4 signaling in macrophages. Traffic 17, 1014–1026. ( 10.1111/tra.12415) [DOI] [PubMed] [Google Scholar]
- 36.Micaroni M, et al. 2013. Rab6a/a’ are important Golgi regulators of pro-inflammatory TNF secretion in macrophages. PLoS ONE 8, e57034 ( 10.1371/journal.pone.0057034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ioannou MS, et al. 2015. DENND2B activates Rab13 at the leading edge of migrating cells and promotes metastatic behavior. J. Biol. Chem. 208, 629–648. ( 10.1083/jcb.201407068) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murray RZ, Kay JG, Sangermani DG, Stow JL. 2005. A role for the phagosome in cytokine secretion. Science 310, 1492–1495. ( 10.1126/science.1120225) [DOI] [PubMed] [Google Scholar]
- 39.Spiering D, Bravo-Cordero JJ, Moshfegh Y, Miskolci V, Hodgson L. 2013. Quantitative ratiometric imaging of FRET-biosensors in living cells. Methods Cell Biol. 114, 593–609. ( 10.1016/B978-0-12-407761-4.00025-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beaumont KA, et al. 2011. The recycling endosome protein Rab17 regulates melanocytic filopodia formation and melanosome trafficking. Traffic 12, 627–643. ( 10.1111/j.1600-0854.2011.01172.x) [DOI] [PubMed] [Google Scholar]
- 41.Patel PC, Harrison RE. 2008. Membrane ruffles capture C3bi-opsonized particles in activated macrophages. Mol. Biol. Cell 19, 4628–4639. ( 10.1091/mbc.E08-02-0223) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zanoni I, Ostuni R, Marek LR, Barresi S, Barbalat R, Barton GM, Granucci F, Kagan JC. 2011. CD14 controls the LPS-induced endocytosis of Toll-like receptor 4. Cell 147, 868–880. ( 10.1016/J.Cell.2011.09.051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Racoosin EL, Swanson JA. 1989. Macrophage colony-stimulating factor (rM-CSF) stimulates pinocytosis in bone marrow-derived macrophages. J. Exp. Med. 170, 1635–1648. ( 10.1084/jem.170.5.1635) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang JT, Kerr MC, Karunaratne S, Jeanes A, Yap AS, Teasdale RD. 2010. The SNX-PX-BAR family in macropinocytosis: the regulation of macropinosome formation by SNX-PX-BAR proteins. PLoS ONE 5, e13763 ( 10.1371/journal.pone.0013763) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fukuda M, Kanno E, Ishibashi K, Itoh T. 2008. Large scale screening for novel Rab effectors reveals unexpected broad Rab binding specificity. Mol. Cell. Proteomics 7, 1031–1042. ( 10.1074/mcp.M700569-MCP200) [DOI] [PubMed] [Google Scholar]
- 46.Schmid MC, et al. 2011. Receptor tyrosine kinases and TLR/IL1Rs unexpectedly activate myeloid cell PI3Kγ, a single convergent point promoting tumor inflammation and progression. Cancer Cell 19, 715–727. ( 10.1016/j.ccr.2011.04.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hou X, Hagemann N, Schoebel S, Blankenfeldt W, Goody RS, Erdmann KS, Itzen A. 2011. A structural basis for Lowe syndrome caused by mutations in the Rab-binding domain of OCRL1. EMBO J. 30, 1659–1670. ( 10.1038/emboj.2011.60) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levin R, Grinstein S, Schlam D. 2015. Phosphoinositides in phagocytosis and macropinocytosis. Biochim. Biophys. Acta 1851, 805–823 ( 10.1016/j.bbalip.2014.09.005) [DOI] [PubMed] [Google Scholar]
- 49.Bohdanowicz M, Balkin DM, De Camilli P, Grinstein S. 2012. Recruitment of OCRL and Inpp5B to phagosomes by Rab5 and APPL1 depletes phosphoinositides and attenuates Akt signaling. Mol. Biol. Cell 23, 176–187. ( 10.1091/mbc.E11-06-0489) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koivusalo M, Welch C, Hayashi H, Scott CC, Kim M, Alexander T, Touret N, Hahn KM, Grinstein S. 2010. Amiloride inhibits macropinocytosis by lowering submembranous pH and preventing Rac1 and Cdc42 signaling. J. Cell Biol. 188, 547–563. ( 10.1083/jcb.200908086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo X, Wasilko DJ, Liu Y, Sun J, Wu X, Luo ZQ, Mao Y. 2015. Structure of the Legionella virulence factor, SidC reveals a unique PI(4)P-specific binding domain essential for its targeting to the bacterial phagosome. PLoS Pathog. 11, e1004965 ( 10.1371/journal.ppat.1004965) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Recouvreux MV, Commisso C. 2017. Macropinocytosis: a metabolic adaptation to nutrient stress in cancer. Front. Endocrinol. 8, 261 ( 10.3389/fendo.2017.00261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Welliver TP, Swanson JA. 2012. A growth factor signaling cascade confined to circular ruffles in macrophages. Biol. Open 1, 754–760. ( 10.1242/bio.20121784) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. 2008. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat. Immunol. 9, 361–368. ( 10.1038/ni1569) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kagan JC, Medzhitov R. 2006. Phosphoinositide-mediated adaptor recruitment controls Toll-like receptor signaling. Cell 125, 943–955. ( 10.1016/J.Cell.2006.03.047) [DOI] [PubMed] [Google Scholar]
- 56.Tan Y, Zanoni I, Cullen TW, Goodman AL, Kagan JC. 2015. Mechanisms of Toll-like receptor 4 endocytosis reveal a common immune-evasion strategy used by pathogenic and commensal bacteria. Immunity 43, 909–922. ( 10.1016/j.immuni.2015.10.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Husebye H, et al. 2010. The Rab11a GTPase controls Toll-like receptor 4-induced activation of interferon regulatory factor-3 on phagosomes. Immunity 33, 583–596. ( 10.1016/j.immuni.2010.09.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Novick P. 2016. Regulation of membrane traffic by Rab GEF and GAP cascades. Small GTPases 7, 252–256. ( 10.1080/21541248.2016.1213781) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bonham KS, et al. 2014. A promiscuous lipid-binding protein diversifies the subcellular sites of Toll-like receptor signal transduction . Cell 156, 705–716. ( 10.1016/j.cell.2014.01.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article have been uploaded as part of the electronic supplementary material.