Abstract
One-fourth of the human population is estimated to have been exposed to Mycobacterium tuberculosis (Mtb) and carries the infection in its latent form. This latent infection presents a lifelong risk of developing active tuberculosis (TB) disease, and persons with latent TB infection (LTBI) are significant contributors to the pool of active TB cases. Genetic polymorphisms among hosts have been shown to contribute to the outcome of Mtb infection. The SP110 gene, which encodes an interferon-induced nuclear protein, has been shown to control host innate immunity to Mtb infection. In this study, we provide experimental data demonstrating the ability of the gene to control genetic susceptibility to latent and active TB infection. Genetic variants of the SP110 gene were investigated in the Taiwanese population (including 301 pulmonary TB patients, 68 LTBI individuals, and 278 healthy household contacts of the TB patients), and their association with susceptibility to latent and active TB infection was examined by performing an association analysis in a case-control study. We identified several SNPs (rs7580900, rs7580912, rs9061, rs11556887, and rs2241525) in the SP110 gene that are associated with susceptibility to LTBI and/or TB disease. Our studies further showed that the same SNPs may have opposite effects on the control of susceptibility to LTBI versus TB. In addition, our analyses demonstrated that the SP110 rs9061 SNP was associated with tumor necrosis factor-α (TNFα) levels in plasma in LTBI subjects. The results suggest that the polymorphisms within SP110 have a role in controlling genetic susceptibility to latent and active TB infection in humans. To the best of our knowledge, this is the first report showing that the SP110 variants are associated with susceptibility to LTBI. Our study also demonstrated that the identified SP110 SNPs displayed the potential to predict the risk of LTBI and subsequent TB progression in Taiwan.
1. Introduction
Tuberculosis (TB), caused by Mycobacterium tuberculosis (Mtb) infection, remains one of the top ten causes of death in the world [1]. Approximately one-fourth of the world population has been infected with Mtb [2], but only 10% of these infected persons develop progressive disease during their lifetimes [3]. The majority of infected individuals remain healthy and noninfectious but carry Mtb in a latent form. Latent TB infection (LTBI) is a state during which a persistent host immune response to stimulation by Mtb antigens is sustained without evidence of clinically manifested active TB [4, 5]. As many as 10% of people with LTBI will go on to develop progressive disease in the near or remote future (a process named “TB reactivation”), and the risk is significantly higher in the presence of predisposing factors, such as coinfection with human immunodeficiency virus (HIV).
Persons with LTBI can be diagnosed by skin (tuberculin skin test (TST)) and/or blood (interferon-gamma (IFNγ) release assay (IGRA)) tests [6]. A positive result from these assays indicates an immune response to stimulation by Mtb antigens in the LTBI population, despite the fact that these individuals have a negative bacteriological test. However, neither TST nor IGRA can distinguish between LTBI and active TB disease or predict who will progress to active TB [7]. Given that the LTBI individuals represent a potential reservoir for future active TB cases, preventive therapy for LTBI is as important a goal as timely anti-TB treatment to reduce the burden of TB [8]. Therefore, identifying and treating cases with LTBI will contribute to TB elimination. Fundamental research for the development of diagnostic assays with improved performance and predictive assessment for TB reactivation will have practical applications and offer a substantial benefit for LTBI management.
In both humans and experimental animal models, genetic polymorphisms among hosts have been shown to contribute to the outcome of Mtb infection [9–13]. In mice, the Ipr1 (intracellular pathogen resistance 1) gene is located within the sst1 (supersusceptibility to tuberculosis 1) locus on chromosome 1 (49-54 cM) [14] and has been identified as a genetic determinant conferring host innate immunity to Mtb infection [15]. The previous studies indicate that the Ipr1 gene may function to integrate mechanisms on controlling cell death, innate immunity, and pathogenesis during intracellular pathogen infections [15, 16]. The gene orthologous to the mouse Ipr1 in humans is SP110, located on chromosome 2q37.1. Expression of both Ipr1 and SP110 genes is intensively regulated by IFNs, suggesting that the function of both genes is related to the IFN-mediated immune response [17].
Genetic defects in the SP110 gene have been found to be responsible for hepatic veno-occlusive disease and immunodeficiency [18, 19], indicating that the gene plays important roles in immunity [20]. The SP110 gene encodes the SP110 nuclear body protein, which has at least three isoforms, including the dominantly expressed SP110a, b, and c isoforms that are believed to be the result of alternative mRNA splicing. Our recent study demonstrated that SP110b, which is most similar to mouse Ipr1 and is expressed more abundantly than SP110a and SP110c, modulates nuclear factor-κB (NF-κB) activity resulting in the downregulation of tumor necrosis factor-α (TNFα) production and concomitant upregulation of NF-κB-induced antiapoptotic gene expression, thereby suppressing IFNγ-mediated monocyte/macrophage cell death [21]. This indicates that the protein is crucial in the control of the activation of macrophages, the reservoir for Mtb persistence.
Although a number of genetic variants of the SP110 gene have been reported to be associated with susceptibility to human TB, the results of studies regarding the relationship between SP110 polymorphisms and TB susceptibility are inconsistent [22–29]. A family-based study in West Africa identified 3 SP110 polymorphisms that are associated with TB susceptibility [22]; however, no significant associations between SP110 and disease susceptibility were identified by other, larger case-control studies conducted on various populations [24–26]. After screening Taiwanese populations for polymorphisms in SP110, we identified some single-nucleotide polymorphisms (SNPs) in SP110 that are significantly associated with susceptibility to LTBI as well as TB disease. These results suggest that the SP110 variants may provide novel predictive markers for TB infection status and disease outcome.
2. Materials and Methods
2.1. Human Subject Study
The study was conducted in accordance with the terms of the informed consent that was provided to, and received from, participants prior to inclusion in the study. This study was approved by the National Taiwan University Hospital Institutional Review Board (IRB No. 200612009M and IRB No. 201512169RINA). Human blood was ethically collected from patients with culture-confirmed pulmonary TB and their household contacts as described previously [30]. Briefly, the participating contacts received chest radiography, and mycobacteriology studies were conducted for 3 sputum samples (including acid-fast smear and mycobacterial culture) to exclude the possibility of active TB disease. Because routine BCG vaccination for newborns in Taiwan could affect the accuracy of tuberculin skin test, all enrolled contacts were then tested for LTBI using a T-SPOT.TB assay (Oxford Immunotec Ltd., Abingdon, UK) or QuantiFERON-TB Gold In-Tube assay (QFT) (Qiagen, Hilden, Germany), and the assays were interpreted according to the manufacturers' criteria. Both patients and contacts were excluded if they were tested positive for HIV infection. In total, 301 pulmonary TB patients, 68 individuals with LTBI, and 278 healthy household contacts were included in the study, and genomic DNA was extracted from their peripheral blood (1-2 mL) using a kit from Qiagen according to the manufacturer's protocol. SP110 polymorphisms were identified from the National Center for Biotechnology Information dbSNP database (https://www.ncbi.nlm.nih.gov/snp). The SNPs were genotyped using the MassARRAY System (Sequenom, San Diego, CA, US), and the primer extension products were analyzed by MALDI-TOF mass spectrometry as previously described [31, 32]. Details of the primers that were used are listed in Table S1 in the Supplementary Materials.
Plasma samples were prepared from blood samples by centrifugation and then stored at −80°C until analysis. The TNFα levels in plasma samples were determined by a MAGPIX® platform (Luminex Corp., Austin, TX. US) with a MILLIPLEX MAP Human Cytokine/Chemokine Magnetic Bead Panel I kit (Merck Millipore, Billerica, MA, US) according to the manufacturer's instructions.
2.2. Statistical Analyses
The associations of gene polymorphisms with LTBI and TB disease were analyzed by SAS 9.4 software (SAS Institute, Cary, NC, US) [33]. Linkage disequilibrium and haplotype analyses were performed using Haploview (https://www.broadinstitute.org/haploview) [34]. Chi-square tests were used to compare frequencies. Odds ratios (ORs) were calculated with 95% confidence intervals (CIs). Bonferroni correction or false discovery rate (FDR) correction was applied for multiple comparison adjustments as indicated. The difference of the TNFα levels in plasma between two genotype groups of samples was calculated using a two-tailed unpaired t-test with Prism software (GraphPad, San Diego, CA, US). p values less than 0.05 were considered to indicate statistical significance, and the number of asterisks represents the degree of significance with regard to the p values.
3. Results
3.1. Association of Polymorphisms in SP110 with LTBI and TB Susceptibility
To investigate the association between the SP110 gene and control of Mtb infection, we examined polymorphisms in the gene of members of the Taiwanese population for genetic association with TB disease status. In this study, 301 pulmonary TB patients (202 males and 99 females; mean age: 63.1 ± 19.9 years), 68 individuals with LTBI (35 males and 33 females; mean age: 46.9 ± 17.7 years), and 278 healthy household contacts of the patients (92 males and 186 females; mean age: 47.1 ± 17.3 years) were included (Table 1). In total, 20 SNPs in the SP110 gene were selected for analysis. Of these, 10 SNPs that were not polymorphic or had a minor allele frequency less than 1% were not further included in the analysis, and the remaining 10 SNPs were then analyzed. We found that 3 SNPs (rs7580912, p = 0.0426, OR: 1.52, 95% CI: 1.01–2.39; rs7580900, p = 0.008, OR: 1.68, 95% CI: 1.14–2.48; and rs9061, p = 0.0026, OR: 0.39, 95% CI: 0.21–0.73) showed differential allele frequency distributions in LTBI cases vs. healthy controls (Table 2). After Bonferroni correction, rs9061 remained significant in this analysis (p < 0.05). Although allele frequencies of none of the 10 SNPs differed significantly in TB patients vs. healthy controls (Table 2), 3 SNPs (rs7580900, p = 0.0319, OR: 0.66, 95% CI: 0.45–0.97; rs9061, p = 0.0116, OR: 2.21, 95% CI: 1.18–4.15; and rs2241525, p = 0.0309, OR: 0.6, 95% CI: 0.38–0.96) exhibited differential allele frequency distributions in TB cases vs. LTBI individuals (Table 2). All of these SNPs were in accordance with the Hardy-Weinberg equilibrium (HWE).
Table 1.
Demographic characteristics of TB patients, LTBI cases, and healthy controls in this study.
| Group | Number | Age (years) | |||
|---|---|---|---|---|---|
| Total | Male (%) | Female (%) | Mean ± SD | Range | |
| Health | 278 | 92 (33) | 186 (67) | 47.1 ± 17.3 | 15.2-93.9 |
| LTBI | 68 | 35 (51) | 33 (49) | 46.9 ± 17.7 | 19.5-86.5 |
| TB | 301 | 202 (67) | 99 (33) | 63.1 ± 19.9 | 19.4-98.7 |
Table 2.
Allele frequencies of polymorphisms in SP110 in TB patients, LTBI cases, and healthy controls.
(a) Allele frequencies in LTBI cases and healthy controls and odds ratio estimates for LTBI
| SNP ID | Position1 | Location2 | HWEp | Alleles | LTBI n (%) |
Health n (%) |
OR (95% CI) |
p value |
|---|---|---|---|---|---|---|---|---|
| rs7580912 | 230216690 | Intron 2-3 | 0.1113 | A | 48 (40) | 165 (30.4) | 1.52 (1.01-2.39) | 0.0426 |
| G | 72 (60) | 377 (69.6) | ||||||
| rs7580900 | 230216669 | Intron 2-3 | 0.5446 | A | 67 (52.3) | 214 (39.5) | 1.68 (1.14-2.48) | 0.008 |
| G | 61 (47.7) | 328 (60.5) | ||||||
| rs11556887 | 230212961 | Exon 4 | 0.0593 | C | 113 (88.3) | 476 (87.8) | 0.96 (0.53-1.74) | 0.8862 |
| T | 15 (11.7) | 66 (12.2) | ||||||
| rs9061 | 230212395 | Exon 5 | 0.1272 | G | 118 (90.8) | 427 (79.4) | 0.39 (0.21-0.73) | 0.0026 ∗ |
| A | 12 (9.2) | 111 (20.6) | ||||||
| rs3820974 | 230211574 | Intron 5-6 | 0.8257 | C | 93 (71.5) | 366 (67.5) | 0.83 (0.54-1.26) | 0.3774 |
| A | 37 (28.5) | 176 (32.5) | ||||||
| rs1365776 | 230207994 | Exon 8 | 0.4341 | A | 111 (91) | 479 (89.7) | 0.86 (0.44-1.70) | 0.6708 |
| G | 11 (9) | 55 (10.3) | ||||||
| rs41309108 | 230201006 | Intron 9-10 | 0.0333 | T | 105 (80.8) | 415 (76.6) | 0.78 (0.48-1.26) | 0.3039 |
| A | 25 (19.2) | 127 (23.4) | ||||||
| rs2241525 | 230178086 | Intron 13-14 | 0.451 | G | 30 (24.6) | 93 (17.4) | 1.55 (0.97-2.47) | 0.067 |
| A | 92 (75.4) | 441 (82.6) | ||||||
| rs1135791 | 230177560 | Exon 14 | 0.4826 | T | 107 (89.2) | 472 (87.4) | 0.76 (0.41-1.43) | 0.5952 |
| C | 13 (10.8) | 68 (12.6) | ||||||
| rs10498244 | 230173117 | Intron 14-15 | 0.5064 | A | 95 (89.6) | 459 (86.6) | 0.75 (0.38-1.47) | 0.3972 |
| G | 11 (10.4) | 71 (13.4) |
1NCBI Reference Sequence: NC_000002.12. 2Based on SP110c (NCBI Reference Sequence: NM_080424.2). HWE: Hardy-Weinberg equilibrium; OR: odds ratio; CI: confidence interval; ORs are adjusted for gender. The significant ORs are shown in italic. ∗OR remains significant after Bonferroni correction.
(b) Allele frequencies in TB patients and healthy controls and odds ratio estimates for TB
| SNP ID | Position1 | Location2 | HWEp | Alleles | LTBI n (%) |
Health n (%) |
OR (95% CI) |
p value |
|---|---|---|---|---|---|---|---|---|
| rs7580912 | 230216690 | Intron 2-3 | 0.9813 | A | 201 (33.6) | 165 (30.4) | 1.16 (0.91-1.49) | 0.2524 |
| G | 397 (66.4) | 377 (69.6) | ||||||
| rs7580900 | 230216669 | Intron 2-3 | 0.8338 | A | 251 (42) | 214 (39.5) | 1.11 (0.88-1.41) | 0.393 |
| G | 347 (58) | 328 (60.5) | ||||||
| rs11556887 | 230212961 | Exon 4 | 0.7296 | C | 543 (90.8) | 476 (87.8) | 0.73 (0.50-1.07) | 0.1028 |
| T | 55 (9.2) | 66 (12.2) | ||||||
| rs9061 | 230212395 | Exon 5 | 0.0162 | G | 485 (81.6) | 427 (79.4) | 0.86 (0.64-1.16) | 0.3326 |
| A | 109 (18.4) | 111 (20.6) | ||||||
| rs3820974 | 230211574 | Intron 5-6 | 0.9602 | C | 195 (32.6) | 17 (32.5) | 1.01 (0.79-1.29) | 0.9609 |
| A | 403 (67.4) | 366 (67.5) | ||||||
| rs1365776 | 230207994 | Exon 8 | 0.0396 | A | 67 (11.2) | 55 (10.3) | 1.10 (0.75-1.59) | 0.6242 |
| G | 531 (88.8) | 479 (89.7) | ||||||
| rs41309108 | 230201006 | Intron 9-10 | 0.0393 | T | 463 (77.4) | 415 (76.6) | 0.95 (0.72-1.26) | 0.7314 |
| A | 135 (22.6) | 127 (23.4) | ||||||
| rs2241525 | 230178086 | Intron 13-14 | 0.102 | G | 495 (83.6) | 441 (82.6) | 0.93 (0.68-1.27) | 0.6447 |
| A | 97 (16.4) | 93 (17.4) | ||||||
| rs1135791 | 230177560 | Exon 14 | 0.3517 | T | 92 (15.4) | 68 (12.6) | 1.28 (0.92-1.80) | 0.176 |
| C | 506 (84.6) | 472 (87.4) | ||||||
| rs10498244 | 230173117 | Intron 14-15 | 0.5808 | A | 88 (14.7) | 71 (13.4) | 1.13 (0.81-1.59) | 0.525 |
| G | 510 (85.3) | 459 (86.6) |
1NCBI Reference Sequence: NC_000002.12. 2Based on SP110c (NCBI Reference Sequence: NM_080424.2). HWE: Hardy-Weinberg equilibrium; OR: odds ratio; CI: confidence interval; ORs are adjusted for gender.
(c) Allele frequencies in TB patients and LTBI cases and odds ratio estimates for TB
| SNP ID | Position1 | Location2 | HWEp | Alleles | LTBI n (%) |
Health n (%) |
OR (95% CI) |
p value |
|---|---|---|---|---|---|---|---|---|
| rs7580912 | 230216690 | Intron 2-3 | 0.7351 | A | 397 (66.4) | 72 (60) | 0.76 (0.51-1.14) | 0.1797 |
| G | 201 (33.6) | 48 (40) | ||||||
| rs7580900 | 230216669 | Intron 2-3 | 0.9968 | A | 347 (58) | 61 (47.7) | 0.66 (0.45-0.97) | 0.0319 |
| G | 251 (42) | 67 (52.3) | ||||||
| rs11556887 | 230212961 | Exon 4 | 0.8656 | C | 543 (90.8) | 113 (88.3) | 0.76 (0.42-1.40) | 0.3804 |
| T | 55 (9.2) | 15 (11.7) | ||||||
| rs9061 | 230212395 | Exon 5 | 0.0995 | G | 109 (18.4) | 12 (9.2) | 2.21 (1.18-4.15) | 0.0116 |
| A | 485 (81.6) | 118 (90.8) | ||||||
| rs3820974 | 230211574 | Intron 5-6 | 0.8727 | C | 195 (32.6) | 37 (28.5) | 1.22 (0.80-1.85) | 0.3577 |
| A | 403 (67.4) | 93 (71.5) | ||||||
| rs1365776 | 230207994 | Exon 8 | 0.028 | A | 67 (11.2) | 11 (9) | 1.27 (0.65-2.49) | 0.4786 |
| G | 531 (88.8) | 111 (91) | ||||||
| rs41309108 | 230201006 | Intron 9-10 | 0.2174 | T | 135 (22.6) | 25 (19.2) | 1.22 (0.76-1.97) | 0.4039 |
| A | 463 (77.4) | 105 (80.8) | ||||||
| rs2241525 | 230178086 | Intron 13-14 | 0.1275 | G | 495 (83.6) | 92 (75.4) | 0.60 (0.38-0.96) | 0.0309 |
| A | 97 (16.4) | 30 (24.6) | ||||||
| rs1135791 | 230177560 | Exon 14 | 0.3733 | T | 92 (15.4) | 13 (10.8) | 1.50 (0.81-2.77) | 0.1979 |
| C | 506 (84.6) | 107 (89.2) | ||||||
| rs10498244 | 230173117 | Intron 14-15 | 0.8897 | A | 88 (14.7) | 11 (10.4) | 1.49 (0.77-2.89) | 0.2363 |
| G | 510 (85.3) | 95 (89.6) |
1NCBI Reference Sequence: NC_000002.12. 2Based on SP110c (NCBI Reference Sequence: NM_080424.2). HWE: Hardy-Weinberg equilibrium; OR: odds ratio; CI: confidence interval; ORs are adjusted for gender. The significant ORs are shown in italic.
The associations between SP110 genotypes and susceptibility to LTBI and TB were then analyzed. In LTBI cases vs. healthy controls, we found that genotypes “GG” in rs7580912 (p = 0.025, OR: 2.451, 95% CI: 1.12–5.364) and “GG” in rs7580900 (p = 0.015, OR: 2.584, 95% CI: 1.208–5.53) were associated with LTBI risk and that genotype “GA” in rs9061 exhibited a protective effect on LTBI (p = 0.044, OR: 0.494, 95% CI: 0.239–0.981) (Table 3). We also found that genotypes “GG” in rs7580912 (p = 0.02, OR: 0.392, 95% CI: 0.179–0.86) and “GG” in rs7580900 (p = 0.017, OR: 0.392, 95% CI: 0.182–0.848) in TB patients vs. LTBI cases (Table 3), as well as “CT” in rs11556887 (p = 0.039, OR: 0.626, 95% CI: 0.401–0.976) in TB patients vs. healthy controls had a protective effect on TB (Table 3). These results indicated that several SNPs (rs7580900, rs7580912, rs9061, and rs11556887) in SP110 were associated with susceptibility to LTBI and/or TB disease.
Table 3.
Association analyses of SP110 SNP genotypes with LTBI and TB susceptibility.
(a) Association between SP110 SNP genotypes and LTBI risk in LTBI cases vs. healthy controls
| SNP ID | Genotypes | Health no. (%) | LTBI no. (%) | OR (95% CI) | p value |
|---|---|---|---|---|---|
| rs7580912 | AA | 134 (49) | 26 (43) | Ref | — |
| GA | 111 (41) | 20 (33) | 0.884 (0.464-1.686) | 0.709 | |
| GG | 27 (10) | 14 (23) | 2.451 (1.12-5.364) | 0.025 | |
| rs7580900 | AA | 100 (37) | 16 (25) | Ref | — |
| GA | 128 (47) | 29 (45) | 1.403 (0.716-2.749) | 0.324 | |
| GG | 43 (16) | 19 (30) | 2.584 (1.208-5.53) | 0.015 | |
| rs11556887 | CC | 206 (76) | 49 (77) | Ref | — |
| CT | 64 (24) | 15 (23) | 0.982 (0.513-1.882) | 0.957 | |
| TT | 1 (0.4) | 0 (0) | <0.001 (<0.001-999.999) | 0.967 | |
| rs9061 | GG | 177 (65) | 54 (82) | Ref | — |
| AA | 16 (6) | 0 (0) | <0.001 (<0.001-999.999) | 0.901 | |
| GA | 79 (29) | 12 (18) | 0.494 (0.239-0.981) | 0.044 | |
| rs3820974 | CC | 125 (46) | 34 (52) | Ref | — |
| AA | 29 (11) | 6 (9) | 0.712 (0.269-1.886) | 0.494 | |
| CA | 118 (43) | 25 (38) | 0.703 (0.391-1.264) | 0.239 | |
| rs1365776 | AA | 217 (81) | 52 (83) | Ref | — |
| GA | 47 (18) | 10 (16) | 0.757 (0.345-1.66) | 0.487 | |
| GG | 4 (1) | 1 (1) | 1.212 (0.129-11.39) | 0.867 | |
| rs41309108 | TT | 153 (56) | 41 (63) | Ref | — |
| AA | 10 (4) | 1 (1) | 0.395 (0.048-3.248) | 0.387 | |
| TA | 109 (40) | 23 (35) | 0.768 (0.433-1.362) | 0.367 | |
| rs2241525 | GG | 186 (69) | 36 (58) | Ref | — |
| AA | 10 (4) | 4 (7) | 1.984 (0.579-6.802) | 0.276 | |
| GA | 75 (28) | 22 (35) | 1.509 (0.828-2.75) | 0.179 | |
| rs1135791 | TT | 206 (75) | 48 (79) | Ref | — |
| CC | 3 (1) | 0 (0) | <0.001 (<0.001-999.999) | 0.943 | |
| CT | 64 | 13 (21) | 0.86 (0.434-1.703) | 0.667 | |
| rs10498244 | AA | 199 (75) | 44 (83) | Ref | — |
| AG | 61 (23) | 7 (13) | 0.523 (0.223-1.223) | 0.135 | |
| GG | 5 (2) | 2 (4) | 1.9 (0.354-10.203) | 0.454 |
OR: odds ratio; CI: confidence interval; ORs are adjusted for gender. The significant ORs are shown in italic.
(b) Association between SP110 SNP genotypes and TB risk in TB patients vs. LTBI cases
| SNP ID | Genotypes | Health no. (%) | LTBI No. (%) | OR (95% CI) | p value |
|---|---|---|---|---|---|
| rs7580912 | AA | 26 (43) | 129 (43) | Ref | — |
| GA | 20 (33) | 139 (47) | 1.377 (0.731-2.595) | 0.322 | |
| GG | 14 (23) | 31 (10) | 0.392 (0.179-0.86) | 0.02 | |
| rs7580900 | AA | 16 (25) | 98 (33) | Ref | — |
| GA | 29 (45) | 151 (50) | 0.863 (0.444-1.878) | 0.664 | |
| GG | 19 (30) | 50 (17) | 0.392 (0.182-0.848) | 0.017 | |
| rs11556887 | CC | 49 (77) | 248 (83) | Ref | — |
| CT | 15 (23) | 47 (16) | 0.628 (0.324-1.219) | 0.169 | |
| TT | 0 (0) | 4 (1) | >999.999 (<0.001->999.999) | 0.988 | |
| rs9061 | GG | 54 (82) | 203 (68) | Ref | — |
| AA | 0 (0) | 15 (5) | >999.999 (<0.001->999.999) | 0.974 | |
| GA | 12 (18) | 79 (27) | 1.831 (0.924-3.627) | 0.083 | |
| rs3820974 | CC | 34 (52) | 136 (45) | Ref | — |
| AA | 6 (9) | 32 (11) | 1.261 (0.483-3.293) | 0.637 | |
| CA | 25 (38) | 131 (44) | 1.31 (0.739-2.323) | 0.356 | |
| rs1365776 | AA | 52 (83) | 240 (80) | Ref | — |
| GA | 10 (16) | 51 (17) | 1.264 (0.58-2.753) | 0.556 | |
| GG | 1 (1) | 8 (3) | 2.028 (0.244-16.888) | 0.513 | |
| rs41309108 | TT | 41 (63) | 176 (59) | Ref | — |
| AA | 1 (1) | 12 (4) | 2.401 (0.3-19.219) | 0.409 | |
| TA | 23 (35) | 111 (37) | 1.096 (0.621-1.932) | 0.753 | |
| rs2241525 | GG | 36 (58) | 211 (71) | Ref | — |
| AA | 4 (7) | 12 (4) | 0.47 (0.142-1.562) | 0.218 | |
| GA | 22 (35) | 73 (25) | 0.564 (0.31-1.026) | 0.061 | |
| rs1135791 | TT | 48 (79) | 212 (71) | Ref | — |
| CC | 0 (0) | 5 (2) | >999.999 (<0.001->999.999) | 0.986 | |
| CT | 13 (21) | 82 (27) | 1.33 (0.68-2.602) | 0.404 | |
| rs10498244 | AA | 44 (83) | 215 (72) | Ref | — |
| AG | 7 (13) | 80 (27) | 2.137 (0.917-4.981) | 0.079 | |
| GG | 2 (4) | 4 (1) | 0.441 (0.074-2.62) | 0.368 |
OR: odds ratio; CI: confidence interval; ORs are adjusted for gender. The significant ORs are shown in italic.
(c) Association between SP110 SNP genotypes and TB risk in TB patients vs. healthy controls
| SNP ID | Genotypes | Health no. (%) | LTBI no. (%) | OR (95% CI) | p value |
|---|---|---|---|---|---|
| rs7580912 | AA | 134 (49) | 129 (43) | Ref | — |
| GA | 111 (41) | 139 (47) | 1.258 (0.869-1.822) | 0.224 | |
| GG | 27 (10) | 31 (10) | 0.877 (0.474-1.622) | 0.676 | |
| rs7580900 | AA | 100 (37) | 98 (33) | Ref | — |
| GA | 128 (47) | 151 (50) | 1.258 (0.851-1.86) | 0.249 | |
| GG | 43 (16) | 50 (17) | 1.023 (0.601-1.74) | 0.934 | |
| rs11556887 | CC | 206 (76) | 248 (83) | Ref | — |
| CT | 64 (23.6) | 47 (16) | 0.626 (0.401-0.976) | 0.039 | |
| TT | 1 (0.4) | 4 (1) | 2.387 (0.243-23.458) | 0.456 | |
| rs9061 | GG | 176 (65) | 203 (68) | Ref | — |
| AA | 17 (6) | 15 (5) | 0.792 (0.36-1.742) | 0.561 | |
| GA | 79 (29) | 79 (27) | 0.886 (0.597-1.313) | 0.546 | |
| rs3820974 | CC | 125 (46) | 136 (45) | Ref | — |
| AA | 29 (11) | 32 (11) | 0.871 (0.473-1.603) | 0.657 | |
| CA | 118 (43) | 131 (44) | 0.942 (0.651-1.363) | 0.749 | |
| rs1365776 | AA | 217 (81) | 240 (80) | Ref | — |
| GA | 47 (18) | 51 (17) | 0.931 (0.582-1.489) | 0.767 | |
| GG | 4 (2) | 8 (3) | 2.291 (0.631-8.322) | 0.208 | |
| rs41309108 | TT | 153 (56) | 176 (59) | Ref | — |
| AA | 9 (3) | 12 (4) | 0.982 (0.382-2.527) | 0.971 | |
| TA | 109 (40) | 111 (37) | 0.852 (0.593-1.225) | 0.387 | |
| rs2241525 | GG | 186 (69) | 211 (71) | Ref | — |
| AA | 10 (3) | 12 (4) | 1.004 (0.402-2.505) | 0.994 | |
| GA | 75 (28) | 73 (25) | 0.85 (0.569-1.268) | 0.425 | |
| rs1135791 | TT | 206 (75) | 212 (71) | Ref | — |
| CC | 3 (1) | 5 (2) | 1.681 (0.371-7.617) | 0.5 | |
| CT | 64 (24) | 82 (27) | 1.177 (0.788-1.76) | 0.426 | |
| rs10498244 | AA | 199 (75) | 215 (72) | Ref | — |
| AG | 61 (23) | 80 (27) | 1.185 (0.787-1.785) | 0.417 | |
| GG | 5 (2) | 4 (1) | 0.774 (0.191-3.133) | 0.719 |
OR: odds ratio; CI: confidence interval; ORs are adjusted for gender. The significant ORs are shown in italic.
3.2. Association Analyses in Various Inheritance Models
The minor allele of each SNP was presumed as a risk factor compared to the major allele, and the associations between SNPs and susceptibility to LTBI and TB were analyzed in various inheritance models (Table 4). We found that in LTBI cases vs. healthy controls, rs9061 showed a protective effect on LTBI in both additive (p = 0.0059, OR: 0.41, 95% CI: 0.22–0.78) and dominant (p = 0.0112, OR: 0.42, 95% CI: 0.21–0.82) models, while rs7580900 was associated with LTBI risk in both additive (p = 0.0147, OR: 1.62, 95% CI: 1.1–2.39) and recessive (p = 0.0195, OR: 2.13, 95% CI: 1.13–4.03) models. In addition to this finding, rs7580912 was also associated with LTBI risk in a recessive model (p = 0.0142, OR: 2.49, 95% CI: 1.2–5.18). In TB patients vs. LTBI cases, rs9061 was associated with TB risk in both additive (p = 0.015, OR: 2.17, 95% CI: 1.16–4.06) and dominant (p = 0.0273, OR: 2.14, 95% CI: 1.09–4.22) models, while rs7580900 exhibited a protective effect on TB in both additive (p = 0.0261, OR: 0.64, 95% CI: 0.43–0.95) and recessive (p = 0.0117, OR: 0.45, 95% CI: 0.24–0.84) models and rs2241525 was associated with protection from TB in both additive (p = 0.0407, OR: 0.62, 95% CI: 0.4–0.98) and dominant (p = 0.0389, OR: 0.55, 95% CI: 0.31–0.97) models. Additionally, rs7580912 had a protective effect on TB in a recessive model (p = 0.0043, OR: 0.35, 95% CI: 0.17–0.72). After false discovery rate (FDR) correction, rs7580900 and rs7580912 remained significant (p = 0.041 and 0.0301, respectively) in a recessive model in this analysis.
Table 4.
Association analyses of SP110 SNP genotypes with LTBI and TB susceptibility in various inheritance models.
(a) Association analyses of SP110 SNP genotypes in an additive model
| SNP ID | TB vs. health | TB vs. LTBI | LTBI vs. health | |||
|---|---|---|---|---|---|---|
| OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | p value | |
| rs9061 | 0.89 (0.66,1.2) | 0.4382 | 2.17 (1.16,4.06) | 0.015 | 0.41 (0.22,0.78) | 0.0059 |
| rs7580900 | 1.07 (0.83,1.38) | 0.5839 | 0.64 (0.43,0.95) | 0.0261 | 1.62 (1.1,2.39) | 0.0147 |
| rs3820974 | 0.94 (0.72,1.23) | 0.6661 | 1.2 (0.79,1.82) | 0.4009 | 0.79 (0.51,1.21) | 0.2761 |
| rs41309108 | 0.9 (0.66,1.22) | 0.4917 | 1.2 (0.73,1.97) | 0.4824 | 0.73 (0.44,1.23) | 0.2366 |
| rs7580912 | 1.05 (0.8,1.37) | 0.7307 | 0.74 (0.49,1.1) | 0.1387 | 1.4 (0.94,2.08) | 0.0992 |
| rs1135791 | 1.2 (0.83,1.73) | 0.3242 | 1.46 (0.77,2.77) | 0.2444 | 0.81 (0.42,1.56) | 0.525 |
| rs1365776 | 1.1 (0.75,1.62) | 0.6153 | 1.32 (0.7,2.5) | 0.3933 | 0.84 (0.43,1.65) | 0.6133 |
| rs10498244 | 1.1 (0.77,1.59) | 0.5919 | 1.4 (0.71,2.73) | 0.3303 | 0.77 (0.4,1.5) | 0.4457 |
| rs2241525 | 0.91 (0.66,1.25) | 0.5681 | 0.62 (0.4,0.98) | 0.0407 | 1.46 (0.92,2.32) | 0.1102 |
| rs11556887 | 0.72 (0.48,1.08) | 0.1155 | 0.75 (0.41,1.39) | 0.362 | 0.96 (0.5,1.81) | 0.8916 |
OR: odds ratio; CI: confidence interval; ORs are adjusted for gender. The significant ORs are shown in italic.
(b) Association analyses of SP110 SNP genotypes in a dominant model
| SNP ID | TB vs. health | TB vs. LTBI | LTBI vs. health | |||
|---|---|---|---|---|---|---|
| OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | p value | |
| rs9061 | 0.87 (0.6,1.26) | 0.4646 | 2.14 (1.09,4.22) | 0.0273 | 0.42 (0.21,0.82) | 0.0112 |
| rs7580900 | 1.22 (0.84,1.76) | 0.2929 | 0.68 (0.37,1.27) | 0.2276 | 1.74 (0.93,3.24) | 0.083 |
| rs3820974 | 0.93 (0.65,1.32) | 0.6879 | 1.3 (0.76,2.24) | 0.3378 | 0.71 (0.41,1.23) | 0.2193 |
| rs41309108 | 0.86 (0.6,1.23) | 0.4092 | 1.15 (0.66,2.01) | 0.6191 | 0.74 (0.42,1.3) | 0.2917 |
| rs7580912 | 1.18 (0.83,1.68) | 0.3571 | 0.98 (0.55,1.72) | 0.9312 | 1.2 (0.68,2.12) | 0.5388 |
| rs1135791 | 1.2 (0.81,1.78) | 0.3678 | 1.41 (0.72,2.75) | 0.3146 | 0.83 (0.42,1.64) | 0.5948 |
| rs1365776 | 1.03 (0.66,1.6) | 0.9033 | 1.34 (0.64,2.82) | 0.4408 | 0.79 (0.37,1.67) | 0.5353 |
| rs10498244 | 1.15 (0.77,1.72) | 0.4807 | 1.75 (0.81,3.78) | 0.1536 | 0.63 (0.29,1.35) | 0.2341 |
| rs2241525 | 0.87 (0.59,1.27) | 0.4667 | 0.55 (0.31,0.97) | 0.0389 | 1.57 (0.88,2.78) | 0.1239 |
| rs11556887 | 0.66 (0.43,1.02) | 0.0609 | 0.67 (0.35,1.3) | 0.2363 | 0.97 (0.51,1.86) | 0.9282 |
OR: odds ratio; CI: confidence interval; ORs are adjusted for gender. The significant ORs are shown in italic.
(c) Association analyses of SP110 SNP genotypes in a recessive model
| SNP ID | TB vs. health | TB vs. LTBI | LTBI vs. health | |||
|---|---|---|---|---|---|---|
| OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | p value | |
| rs9061 | 0.82 (0.38,1.78) | 0.6227 | — | — | — | — |
| rs7580900 | 0.92 (0.57,1.48) | 0.737 | 0.45 (0.24,0.84) | 0.0117 | 2.13 (1.13,4.03) | 0.0195 ∗ |
| rs3820974 | 0.92 (0.52,1.62) | 0.7796 | 1.13 (0.45,2.83) | 0.8018 | 0.84 (0.33,2.15) | 0.7209 |
| rs41309108 | 1.05 (0.41,2.66) | 0.9241 | 2.35 (0.3,18.52) | 0.4186 | 0.43 (0.05,3.55) | 0.437 |
| rs7580912 | 0.79 (0.44,1.42) | 0.4367 | 0.35 (0.17,0.72) | 0.0043 | 2.49 (1.2,5.18) | 0.0142 ∗ |
| rs1135791 | 1.63 (0.36,7.51) | 0.5284 | — | — | — | — |
| rs1365776 | 2.25 (0.63,8.05) | 0.2129 | 1.94 (0.23,16.11) | 0.5375 | 1.24 (0.13,11.51) | 0.8504 |
| rs10498244 | 0.74 (0.18,3.05) | 0.6804 | 0.38 (0.06,2.23) | 0.2844 | 2.27 (0.42,12.19) | 0.3385 |
| rs2241525 | 1.05 (0.42,2.6) | 0.9172 | 0.56 (0.17,1.83) | 0.3374 | 1.73 (0.52,5.81) | 0.3747 |
| rs11556887 | 2.63 (0.27,25.7) | 0.4064 | — | — | — | — |
OR: odds ratio; CI: confidence interval; ORs are adjusted for gender. The significant ORs are shown in italic. ∗ORs remain significant after false discovery rate (FDR) correction.
3.3. Linkage Disequilibrium and Haplotype Analyses
We then examined linkage disequilibrium (LD) with the SNP markers in the SP110 gene using a Haploview analysis. We found that the “ATATACGCGG” and “ATGTACGCGA” haplotypes met statistical significance (p = 0.0193, OR: 2.11, 95% CI: 1.11–3.99 and p = 0.0225, OR: 2.76, 95% CI: 1.12–6.84, respectively) for association with LTBI risk in LTBI cases vs. healthy controls (see Figure S1 in the Supplementary Materials) and that the “ATGTACGCGA” and “ATGAAAGCGA” haplotypes were statistically significant (p = 0.03, OR: 0.38, 95% CI: 0.16–0.95 and p = 0.0324, OR: 0.24, 95% CI: 0.06–0.99, respectively) with a protective effect on TB in TB cases vs. LTBI individuals (see Figure S2 in the Supplementary Materials). Interestingly, the “GCGTACGCGG” haplotype was associated with TB risk (p = 0.0169, OR: 3.81, 95% CI: 1.18–12.31) in TB cases vs. healthy controls (see Figure S3 in the Supplementary Materials), although none of the SNPs studied show a significant difference in frequency distributions in this comparison (Table 2). Noteworthily, when we compared “ATGTACGCAA” (the most frequent haplotype) with “ATGTACGCGA” (the haplotype that was associated with LTBI risk in LTBI cases vs. healthy controls and had a protective effect on TB in TB cases vs. LTBI individuals), the latter was found to be significantly affected by the rs7580900 SNP in both comparisons (p = 0.0225, OR: 2.76, 95% CI: 1.12–6.84 for LTBI cases vs. healthy controls; p = 0.03, OR: 0.38, 95% CI: 0.16–0.95 for TB cases vs. LTBI individuals) (see Figures S1 and S2 in the Supplementary Materials).
We analyzed a block that includes 3 SNPs (rs9061, rs7580900, and rs7580912) in two comparisons (LTBI cases vs. healthy controls and TB cases vs. LTBI individuals) (Figure 1). We found that the “GAA” (p = 0.0037, OR: 1.77, 95% CI: 1.20–2.60), “GGG” (p = 0.0009, OR: 2.06, 95% CI: 1.34–3.17), “GGA” (p = 0.0009, OR: 2.81, 95% CI: 1.49–5.27), and “GAG” (p = 0.0001, OR: 10, 95% CI: 2.36–42.32) haplotypes were associated with disease risk and that the “AGG” haplotype had a protective effect on LTBI (p = 0.0254, OR: 0.41, 95% CI: 0.18–0.92) for LTBI cases vs. healthy controls (Table 5). In addition, the “GAA” (p = 0.0111, OR: 0.61, 95% CI: 0.42–0.90), “GGG” (p = 0.0408, OR: 0.65, 95% CI: 0.43–0.98), “GGA” (p = 0.0001, OR: 0.3, 95% CI: 0.16–0.57), and “GAG” (p = 0.0001, OR: 0.1, 95% CI: 0.02–0.4) haplotypes showed a protective effect on TB disease for TB patients vs. LTBI individuals (Table 5). These genetic studies suggest that the SP110 gene plays a key role in modulating susceptibility to latent and active TB infection.
Figure 1.
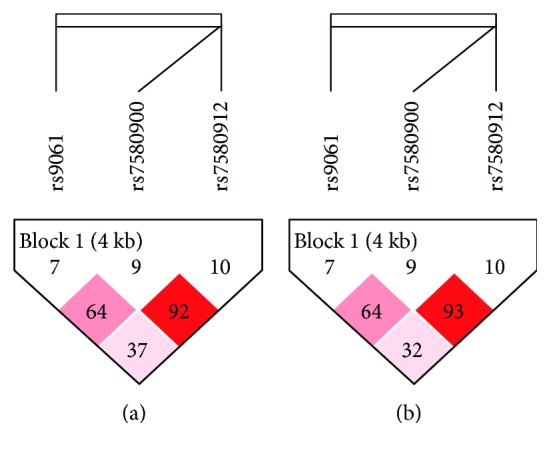
Haplotype block maps for SP110 with 3 SNPs (rs9061, rs7580900, and rs7580912). The haplotype blocks were analyzed in LTBI cases vs. healthy controls (a) and TB cases vs. LTBI individuals (b), respectively.
Table 5.
Association of the haplotype frequencies of SP110 SNPs with 3 SNPs (rs9061, rs7580900, and rs7580912) with LTBI and TB susceptibility.
(a) Association of haplotype frequencies with LTBI risk in LTBI cases and healthy controls
| Haplotypes | Frequencies | Chi-square | OR (95% CI) | p value | |
|---|---|---|---|---|---|
| LTBI | Health | ||||
| GAA | 0.417 | 0.558 | 8.417 | 1.77 (1.20-2.60) | 0.0037 |
| GGG | 0.310 | 0.179 | 11.048 | 2.06 (1.34-3.17) | 9.00E − 04 |
| AGG | 0.053 | 0.120 | 4.997 | 0.41 (0.18-0.92) | 0.0254 |
| GGA | 0.134 | 0.052 | 10.99 | 2.81 (1.49-5.27) | 9.00E − 04 |
| AGA | 0.021 | 0.043 | 1.429 | 0.47 (0.13-1.68) | 0.232 |
| AAA | 0.019 | 0.042 | 1.579 | 0.44 (0.12-1.65) | 0.209 |
| GAG | 0.047 | 0.005 | 14.5 | 10.00 (2.36-42.32) | 1.00E − 04 |
OR: odds ratio; CI: confidence interval; ORs are adjusted for gender. The significant ORs are shown in italic.
(b) Association of haplotype frequencies with TB risk in TB cases vs. LTBI individuals
| Haplotypes | Frequencies | Chi-square | OR (95% CI) | p value | |
|---|---|---|---|---|---|
| TB | LTBI | ||||
| GAA | 0.540 | 0.417 | 6.445 | 0.61 (0.42-0.90) | 0.0111 |
| GGG | 0.228 | 0.313 | 4.184 | 0.65 (0.43-0.98) | 0.0408 |
| AGG | 0.103 | 0.053 | 3.191 | 2.05 (0.91-4.62) | 0.074 |
| GGA | 0.043 | 0.130 | 14.591 | 0.30 (0.16-0.57) | 0.0001 |
| AGA | 0.045 | 0.021 | 1.564 | 2.16 (0.62-7.49) | 0.211 |
| AAA | 0.035 | 0.018 | 0.993 | 1.95 (0.51-7.53) | 0.3191 |
| GAG | 0.005 | 0.047 | 16.031 | 0.10 (0.02-0.40) | 0.0001 |
OR: odds ratio; CI: confidence interval; ORs are adjusted for gender. The significant ORs are shown in italic.
3.4. Association between the SP110 rs9061 SNP and the TNFα Production in LTBI Subjects
TNFα plays a crucial role in controlling Mtb infection and TB reactivation; however, overproduction of TNFα may cause pathology [35, 36]. Our previous studies demonstrated that the SP110b protein, which is encoded by the SP110 gene and whose expression is upregulated by IFNs, downregulates TNFα production in monocyte/macrophage cells activated by IFNγ, thereby alleviating cell death [21]. This finding indicates that the protein functions as a regulator of proinflammatory cytokines of host immunity contributing to a reduction in tissue damage caused by excessive inflammation [21]. To further investigate the potential association between the SP110 SNPs and disease status, we next analyzed clinical parameters in the studied subjects. We measured TNFα levels in plasma from LTBI individuals who carry the different genotypes of the SP110 SNPs and demonstrated that the “GA” genotype of rs9061 in LTBI individuals was associated with lower TNFα levels in plasma compared to “GG” LTBI subjects (Figure 2). The TNFα levels of healthy controls with both genotypes were undetectable (not shown). These data are in agreement with the protective role of the “GA” genotype of rs9061 in LTBI (Table 3) and further support our recent finding showing that the SP110b protein prevents cell death and tissue damage by downregulating TNFα production.
Figure 2.
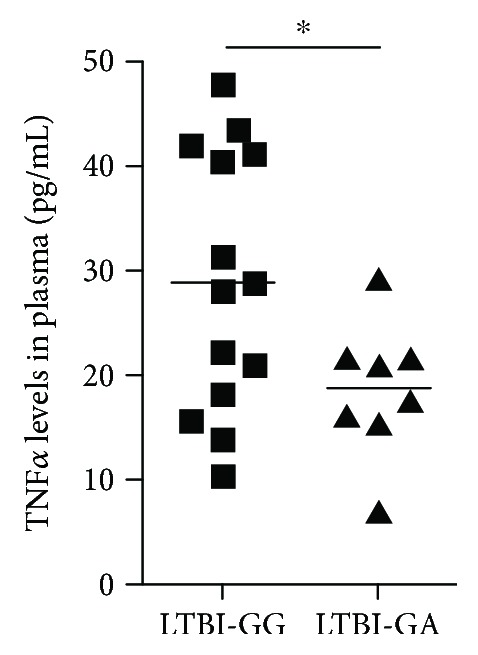
Association between the SP110 rs9061 SNP and the TNF levels in plasma of LTBI subjects. The TNF production was determined in plasma samples from LTBI cases carrying the different genotypes of the rs9061 SNP by a MAGPIX instrument with a MILLIPLEX MAP Human Cytokine/Chemokine Magnetic Bead Panel I kit. Sample numbers are 14 for “GG” and 8 for “GA” genotypes, respectively. Statistical significance was calculated using a two-tailed unpaired t-test. ∗ p < 0.05.
4. Discussion
SP110 is strongly regulated by IFNs [17], suggesting its possible role in microbial immunity. Although many groups have studied the associations between the gene and TB susceptibility in a variety of populations, these studies show inconclusive results [22–29]. In our study, we recruited healthy household contacts of TB patients as controls, as these contacts are at a high risk of exposure to Mtb. It has been reported that approximately 80-100% of the contacts may have Mtb infection, and on average, 20% of them may develop disease [37], indicating that household contacts are at high risk of LTBI and active TB disease. Therefore, examination of this group carries a considerable importance for prevention and control of TB disease. To the best of our knowledge, this is the first study to demonstrate an association between SP110 and LTBI, and it is the first study of the gene in the Taiwanese population. This work will help clarify the relationship between genetic variation in SP110 with latent and active TB infection in an Asian population.
Several SNPs (rs7580900, rs7580912, rs9061, rs11556887, and rs2241525) in SP110 showed an association with susceptibility to LTBI and/or TB disease in our study. In Table 3, we found that genotypes “GG” in rs7580912 (p = 0.025, OR: 2.451, 95% CI: 1.12–5.364) and “GG” in rs7580900 (p = 0.015, OR: 2.584, 95% CI: 1.208–5.53) were associated with LTBI susceptibility in LTBI cases vs. healthy controls, while the same SNP genotypes exhibited a protective effect on LTBI (“GG” in rs7580912, p = 0.02, OR: 0.392, 95% CI: 0.179–0.86; “GG” in rs7580900, p = 0.017, OR: 0.392, 95% CI: 0.182–0.848) in TB patients vs. LTBI individuals. In the Haploview analysis, we also found that the haplotype of multiple SNPs “ATGTACGCGA” in the SP110 gene was significantly associated with LTBI risk (p = 0.0225, OR: 2.76, 95% CI: 1.12–6.84) in LTBI cases vs. healthy controls, while the same haplotypes had a protective effect on TB disease (p = 0.03, OR: 0.38, 95% CI: 0.16–0.95) in LTBI individuals vs. TB patients (see Figures S1 and S2 in the Supplementary Materials). In addition, 3 SNPs (rs9061, rs7580900, and rs7580912) with the haplotype “GAA” (p = 0.0037, OR: 1.77, 95% CI: 1.20–2.60), “GGG” (p = 0.0009, OR: 2.06, 95% CI: 1.34–3.17), “GGA” (p = 0.0009, OR: 2.81, 95% CI: 1.49–5.27), and “GAG” (p = 0.0001, OR: 10, 95% CI: 2.36–42.32) were associated with disease risk in LTBI cases vs. healthy controls, while the same haplotypes showed a protective effect on TB disease for TB patients vs. LTBI individuals (p = 0.0111, OR: 0.61, 95% CI: 0.42–0.90 for “GAA”; p = 0.0408, OR: 0.65, 95% CI: 0.43–0.98 for “GGG”; p = 0.0001, OR: 0.3, 95% CI: 0.16–0.57 for “GGA”; and p = 0.0001, OR: 0.1, 95% CI: 0.02–0.4 for “GAG”) (Table 5). These results revealed that the same SNP genotypes or haplotypes in the SP110 gene had opposite effects on the control of susceptibility to LTBI and TB disease, suggesting that the gene may have differential roles in the control of susceptibility to LTBI and TB disease.
SNP rs9061 (G→A) introduces an amino acid change from glutamic acid to lysine at codon position 207 of the SP110 protein. Transforming an acidic amino acid to a basic amino acid may alter the protein structure or posttranslational modification of the SP110 protein leading to better downregulation of TNFα production. It has been suggested that the A allele may cause changes in α-helices and β-sheets in the secondary structure of the SP110 protein compared with the G allele [38]. In addition, we analyzed the SP110 protein using the ELM database (http://elm.eu.org/) and found that this amino acid change generates a potential binding motif for the C-terminal ubiquitin-like domain (CTD) of ubiquitin specific protease 7 (USP7), one of the most abundant deubiquitinases [39]. USP7 plays important roles in various biological activities, including cell survival, proliferation, apoptosis, and tumorigenesis [40, 41]. As shown in Figure 2, the data demonstrated an association between the “GA” genotype of rs9061 with lower TNFα levels in plasma from LTBI individuals compared to “GG” LTBI subjects. One possible explanation for this result is that SP110 with the “GA” genotype at rs9061 may interact with USP7 and thus is more stable than the SP110 with the “GG” genotype, resulting in a more efficient downregulation of TNFα production. However, further functional studies are needed to elucidate the exact effects of SNP rs9061 on the SP110 gene and disease risk.
Individuals with LTBI, based on a positive result in the TST or the IGRA, usually show no disease symptoms and acquire an effective adaptive immunity. However, a proportion of people with LTBI might reactivate and develop clinical disease. The positive results from these assays indicate an immune response to stimulation by Mtb antigens in the LTBI population; however, neither TST nor IGRA can distinguish between LTBI and active TB. In addition, these assays also cannot predict which LTBI cases will progress to active TB [7]. To control and eliminate TB, worldwide TB eradication endeavors have been focused on identifying and treating cases with LTBI. Therefore, our studies identified several SNPs in SP110 that are significantly associated with controlling susceptibility to LTBI and TB disease and thus may provide novel predictive markers for latent and active TB infection. This study may also yield an improved strategy that can identify persons at increased risk of the disease.
There were some limitations in our study. First, this study dealt with relatively small sample sizes (301, 68, and 278 for TB patients, LTBI cases, and healthy household contacts, respectively). Further large-scale studies are, therefore, needed to validate the predictive value of the SP110 SNPs identified in this study. Second, the LTBI number of our study group is small, and besides, the results on plasma TNFα levels were analyzed from groups with even smaller sample sizes; therefore, the difference in TNFα levels in plasma of LTBI cases with two genotypes may be underestimated and should be verified in a different population. In addition, the analysis of TNFα levels in plasma of active TB patients with the different genotypes is in progress. The result may help support our finding in LTBI individuals and clarify the potential association between the SP110 SNPs and disease status.
5. Conclusions
The results suggest that the SP110 variants have a role in controlling genetic susceptibility to latent and active TB infection in humans. To the best of our knowledge, this is the first report demonstrating associations of polymorphisms in SP110 with LTBI susceptibility. Additionally, we provide evidence that the SP110 rs9061 SNP is associated with plasma TNFα levels in LTBI individuals. These data suggest that the identified SP110 SNPs may serve as a biomarker for latent and active TB infection in Taiwan.
Acknowledgments
This research was funded by the Ministry of Science and Technology, Taiwan (NSC 97-2320-B-010-006-MY3, NSC 99-2321-B-002-022-, NSC 100-2321-B-002-009-, NSC 100-2320-B-002-106-, NSC 101-2321-B-002-001-, NSC 101-2320-B-002-022-, NSC 103-2320-B-002-044-MY3, and 106-2320-B-002-020- to BSY; NSC 99-2320-B-002-078-MY3, NSC 103-2314-B-002-083-, and 104-2314-B-002-075- to MLC), and Institute for Biotechnology and Medicine Industry, Taiwan. We thank Hsiang-Ting Hsu and Bo-Wen Chen for technical assistance. We also thank the expert assistance provided by Microarray Core Facility of National Research Program for Genomic Medicine of National Science Council in Taiwan.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare no conflict of interest.
Supplementary Materials
Table S1: primers for genotype analysis for SP110. Figure S1: association of SP110 SNP haplotypes with LTBI risk in LTBI cases vs. healthy controls. (a) Haplotype block map for SP110 with 10 SNPs. (b) Association of haplotype frequencies with LTBI risk in LTBI cases and healthy controls. Figure S2: association of SP110 SNP haplotypes with TB risk in TB cases vs. LTBI individuals. (a) Haplotype block map for SP110 with 10 SNPs. (b) Association of haplotype frequencies with TB risk in TB cases vs. LTBI individuals. Figure S3: association of SP110 SNP haplotypes with TB risk in TB cases vs. healthy controls. (a) Haplotype block map for SP110 with 10 SNPs. (b) Association of haplotype frequencies with TB risk in TB cases and healthy controls.
References
- 1.WHO. Tuberculosis Fact Sheet N°104. WHO; 2018. [Google Scholar]
- 2.Houben R. M. G. J., Dodd P. J. The global burden of latent tuberculosis infection: a re-estimation using mathematical modelling. PLoS Medicine. 2016;13(10) doi: 10.1371/journal.pmed.1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloom B. R., Murray C. J. L. Tuberculosis-commentary on a reemergent killer. Science. 1992;257(5073):1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 4.Dye C., Scheele S., >Dolin P., Pathania V., Raviglione M. C., for the WHO Global Surveillance and Monitoring Project Global burden of tuberculosis - estimated incidence, prevalence, and mortality by country. JAMA. 1999;282(7):677–686. doi: 10.1001/jama.282.7.677. [DOI] [PubMed] [Google Scholar]
- 5.Frieden T. R., Sterling T. R., Munsiff S. S., Watt C. J., Dye C. Tuberculosis. Lancet. 2003;362(9387):887–899. doi: 10.1016/S0140-6736(03)14333-4. [DOI] [PubMed] [Google Scholar]
- 6.Pai M., Riley L. W., Colford J. M., Jr Interferon-γ assays in the immunodiagnosis of tuberculosis: a systematic review. Lancet Infectious Diseases. 2004;4(12):761–776. doi: 10.1016/S1473-3099(04)01206-X. [DOI] [PubMed] [Google Scholar]
- 7.Barry C. E., Boshoff H. I., Dartois V., et al. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nature Reviews Microbiology. 2009;7(12):845–855. doi: 10.1038/nrmicro2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rose D. N. Benefits of screening for latent Mycobacterium tuberculosis infection. Archives of Internal Medicine. 2000;160(10):1513–1521. doi: 10.1001/archinte.160.10.1513. [DOI] [PubMed] [Google Scholar]
- 9.Apt A., Kramnik I. Man and mouse TB: contradictions and solutions. Tuberculosis. 2009;89(3):195–198. doi: 10.1016/j.tube.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casanova J. L., Abel L. Genetic dissection of immunity to mycobacteria: the human model. Annual Review of Immunology. 2002;20(1):581–620. doi: 10.1146/annurev.immunol.20.081501.125851. [DOI] [PubMed] [Google Scholar]
- 11.Fortin A., Abel L., Casanova J. L., Gros P. Host genetics of mycobacterial diseases in mice and men: forward genetic studies of BCG-osis and tuberculosis. Annual Review of Genomics and Human Genetics. 2007;8(1):163–192. doi: 10.1146/annurev.genom.8.080706.092315. [DOI] [PubMed] [Google Scholar]
- 12.Hill A. V. S. The genomics and genetics of human infectious disease susceptibility. Annual Review of Genomics and Human Genetics. 2001;2(1):373–400. doi: 10.1146/annurev.genom.2.1.373. [DOI] [PubMed] [Google Scholar]
- 13.Lynch C. J., Pierce-Chase C. H., Dubos R. A genetic study of susceptibility to experimental tuberculosis in mice infected with mammalian tubercle bacilli. The Journal of Experimental Medicine. 1965;121(6):1051–1070. doi: 10.1084/jem.121.6.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kramnik I., Dietrich W. F., Demant P., Bloom B. R. Genetic control of resistance to experimental infection with virulent Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(15):8560–8565. doi: 10.1073/pnas.150227197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan H., Yan B. S., Rojas M., et al. Ipr1 gene mediates innate immunity to tuberculosis. Nature. 2005;434(7034):767–772. doi: 10.1038/nature03419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kramnik I. Genetic dissection of host resistance to Mycobacterium tuberculosis: the sst1 locus and the Ipr1 gene. Current Topics in Microbiology and Immunology. 2008;321:123–148. doi: 10.1007/978-3-540-75203-5_6. [DOI] [PubMed] [Google Scholar]
- 17.Kadereit S., Gewert D. R., Galabru J., Hovanessian A. G., Meurs E. F. Molecular cloning of two new interferon-induced, highly related nuclear phosphoproteins. Journal of Biological Chemistry. 1993;268(32):24432–24441. [PubMed] [Google Scholar]
- 18.Cliffe S. T., Wong M., Taylor P. J., et al. The first prenatal diagnosis for veno-occlusive disease and immunodeficiency syndrome, an autosomal recessive condition associated with mutations in SP110. Prenatal Diagnosis. 2007;27(7):674–676. doi: 10.1002/pd.1759. [DOI] [PubMed] [Google Scholar]
- 19.Roscioli T., Cliffe S. T., Bloch D. B., et al. Mutations in the gene encoding the PML nuclear body protein Sp110 are associated with immunodeficiency and hepatic veno-occlusive disease. Nature Genetics. 2006;38(6):620–622. doi: 10.1038/ng1780. [DOI] [PubMed] [Google Scholar]
- 20.Cliffe S. T., Bloch D. B., Suryani S., et al. Clinical, molecular, and cellular immunologic findings in patients with SP110-associated veno-occlusive disease with immunodeficiency syndrome. Journal of Allergy and Clinical Immunology. 2012;130(3):735–742.e6. doi: 10.1016/j.jaci.2012.02.054. [DOI] [PubMed] [Google Scholar]
- 21.Leu J. S., Chen M. L., Chang S. Y., et al. SP110b controls host immunity and susceptibility to tuberculosis. American Journal of Respiratory and Critical Care Medicine. 2017;195(3):369–382. doi: 10.1164/rccm.201601-0103OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tosh K., Campbell S. J., Fielding K., et al. Variants in the SP110 gene are associated with genetic susceptibility to tuberculosis in West Africa. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(27):10364–10368. doi: 10.1073/pnas.0603340103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fox G. J., Sy D. N., Nhung N. V., et al. Polymorphisms of SP110 are associated with both pulmonary and extra-pulmonary tuberculosis among the Vietnamese. PLoS One. 2014;9(7, article e99496) doi: 10.1371/journal.pone.0099496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thye T., Browne E. N., Chinbuah M. A., et al. No associations of human pulmonary tuberculosis with Sp110 variants. Journal of Medical Genetics. 2006;43(7):p. e32. doi: 10.1136/jmg.2005.037960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Babb C., Keet E. H., van Helden P. D., Hoal E. G. SP110 polymorphisms are not associated with pulmonary tuberculosis in a South African population. Human Genetics. 2007;121(3-4):521–522. doi: 10.1007/s00439-007-0335-1. [DOI] [PubMed] [Google Scholar]
- 26.Szeszko J. S., Healy B., Stevens H., et al. Resequencing and association analysis of the SP110 gene in adult pulmonary tuberculosis. Human Genetics. 2007;121(2):155–160. doi: 10.1007/s00439-006-0293-z. [DOI] [PubMed] [Google Scholar]
- 27.Moller M., Hoal E. G. Current findings, challenges and novel approaches in human genetic susceptibility to tuberculosis. Tuberculosis. 2010;90(2):71–83. doi: 10.1016/j.tube.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Png E., Alisjahbana B., Sahiratmadja E., et al. Polymorphisms in SP110 are not associated with pulmonary tuberculosis in Indonesians. Infection, Genetics and Evolution. 2012;12(6):1319–1323. doi: 10.1016/j.meegid.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 29.Lei X., Zhu H., Zha L., Wang Y. SP110 gene polymorphisms and tuberculosis susceptibility: a systematic review and meta-analysis based on 10 624 subjects. Infection, Genetics and Evolution. 2012;12(7):1473–1480. doi: 10.1016/j.meegid.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 30.Wang J. Y., Shu C. C., Lee C. H., Yu C. J., Lee L. N., Yang P. C. Interferon-gamma release assay and rifampicin therapy for household contacts of tuberculosis. The Journal of Infection. 2012;64(3):291–298. doi: 10.1016/j.jinf.2011.11.028. [DOI] [PubMed] [Google Scholar]
- 31.Jurinke C., van den Boom D., Cantor C. R., Köster H. The use of MassARRAY technology for high throughput genotyping. Advances in Biochemical Engineering/Biotechnology. 2002;77:57–74. doi: 10.1007/3-540-45713-5_4. [DOI] [PubMed] [Google Scholar]
- 32.Jurinke C., van den Boom D., Cantor C. R., Köster H. Automated genotyping using the DNA MassArray technology. Methods in Molecular Biology. 2002;187:179–192. doi: 10.1385/1-59259-273-2:179. [DOI] [PubMed] [Google Scholar]
- 33.Tung C. C., Wong J. M., Lee W. C., et al. Combining TNFSF15 and ASCA IgA can be used as a predictor for the stenosis/perforating phenotype of Crohn’s disease. Journal of Gastroenterology and Hepatology. 2014;29(4):723–729. doi: 10.1111/jgh.12496. [DOI] [PubMed] [Google Scholar]
- 34.Barrett J. C., Fry B., Maller J., Daly M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 35.Bekker L. G., Moreira A. L., Bergtold A., Freeman S., Ryffel B., Kaplan G. Immunopathologic effects of tumor necrosis factor alpha in murine mycobacterial infection are dose dependent. Infection and Immunity. 2000;68(12):6954–6961. doi: 10.1128/IAI.68.12.6954-6961.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ehlers S., Benini J., Held H. D., Roeck C., Alber G., Uhlig S. αβ T cell receptor-positive cells and interferon-gamma, but not inducible nitric oxide synthase, are critical for granuloma necrosis in a mouse model of mycobacteria-induced pulmonary immunopathology. The Journal of Experimental Medicine. 2001;194(12):1847–1859. doi: 10.1084/jem.194.12.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Self-Study Modules on Tuberculosis. Atlanta, GA, USA: U.S. Department of Health and Human Services, CDC; 1999. Contact investigations for Tuberculosis. [Google Scholar]
- 38.Zhang S., Wang X. B., Han Y. D., Wang C., Zhou Y., Zheng F. Certain polymorphisms in SP110 gene confer susceptibility to tuberculosis: a comprehensive review and updated meta-analysis. Yonsei Medical Journal. 2017;58(1):165–173. doi: 10.3349/ymj.2017.58.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Everett R. D., Meredith M., Orr A., Cross A., Kathoria M., Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO Journal. 1997;16(7):1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim R. Q., Sixma T. K. Regulation of USP7: a high incidence of E3 complexes. Journal of Molecular Biology. 2017;429(22):3395–3408. doi: 10.1016/j.jmb.2017.05.028. [DOI] [PubMed] [Google Scholar]
- 41.Nicholson B., Suresh Kumar K. G. The multifaceted roles of USP7: new therapeutic opportunities. Cell Biochemistry and Biophysics. 2011;60(1-2):61–68. doi: 10.1007/s12013-011-9185-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: primers for genotype analysis for SP110. Figure S1: association of SP110 SNP haplotypes with LTBI risk in LTBI cases vs. healthy controls. (a) Haplotype block map for SP110 with 10 SNPs. (b) Association of haplotype frequencies with LTBI risk in LTBI cases and healthy controls. Figure S2: association of SP110 SNP haplotypes with TB risk in TB cases vs. LTBI individuals. (a) Haplotype block map for SP110 with 10 SNPs. (b) Association of haplotype frequencies with TB risk in TB cases vs. LTBI individuals. Figure S3: association of SP110 SNP haplotypes with TB risk in TB cases vs. healthy controls. (a) Haplotype block map for SP110 with 10 SNPs. (b) Association of haplotype frequencies with TB risk in TB cases and healthy controls.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


