Significance
Iron limits the growth of photosynthetic organisms, especially in marine environments. Understanding the response of photosynthetic organisms to changing iron concentrations is therefore important for agriculture and biotechnology. We have identified a protein that is essential for the correct response to changing iron concentrations in photosynthetic bacteria (cyanobacteria). This protein was previously annotated as an electron transfer component of photosynthesis, called Fed2, and contains an iron−sulfur cluster. We tested Fed2, and found that it cannot act in photosynthetic electron transport. The corresponding gene is essential, and is highly conserved between cyanobacteria, algae, and higher plants. By specifically perturbing its function, we could show that it is essential for the low-iron response at the posttranscriptional level.
Keywords: iron, ferredoxin, cyanobacteria, fed2, FdC2
Abstract
Iron chronically limits aquatic photosynthesis, especially in marine environments, and the correct perception and maintenance of iron homeostasis in photosynthetic bacteria, including cyanobacteria, is therefore of global significance. Multiple adaptive mechanisms, responsive promoters, and posttranscriptional regulators have been identified, which allow cyanobacteria to respond to changing iron concentrations. However, many factors remain unclear, in particular, how iron status is perceived within the cell. Here we describe a cyanobacterial ferredoxin (Fed2), with a unique C-terminal extension, that acts as a player in iron perception. Fed2 homologs are highly conserved in photosynthetic organisms from cyanobacteria to higher plants, and, although they belong to the plant type ferredoxin family of [2Fe-2S] photosynthetic electron carriers, they are not involved in photosynthetic electron transport. As deletion of fed2 appears lethal, we developed a C-terminal truncation system to attenuate protein function. Disturbed Fed2 function resulted in decreased chlorophyll accumulation, and this was exaggerated in iron-depleted medium, where different truncations led to either exaggerated or weaker responses to low iron. Despite this, iron concentrations remained the same, or were elevated in all truncation mutants. Further analysis established that, when Fed2 function was perturbed, the classical iron limitation marker IsiA failed to accumulate at transcript and protein levels. By contrast, abundance of IsiB, which shares an operon with isiA, was unaffected by loss of Fed2 function, pinpointing the site of Fed2 action in iron perception to the level of posttranscriptional regulation.
Oxygenic photosynthesis evolved on Earth over 2.4 billion years ago, resulting in oxygenation of the previously anaerobic atmosphere (1). As soluble Fe2+ was oxidized to Fe3+, this led to a drastic decrease in the bioavailability of iron, which now limits the productivity of many aquatic systems (2). This is a particular problem for cyanobacteria, the symbiotic ancestors of chloroplasts (3), in which multiple heme, FeS cluster, and Fe cofactors are required for both respiratory and photosynthetic electron transport chains. Limitation of cyanobacterial growth by iron is thought to occur in 40% of the ocean (4), demonstrated by the massive blooms that can result from iron fertilization of aquatic environments (5, 6). In contrast to its essential role in cofactors, iron is potentially highly toxic in its unbound form in the cellular environment, catalyzing the production of dangerous hydroxyl radicals from hydrogen peroxide (7). To survive, it is therefore essential for organisms to develop highly sensitive mechanisms for the perception of cellular iron status and transduction of such signals into acclimation responses. These mechanisms optimize iron utilization, while avoiding the toxic effects of excess iron.
In cyanobacteria, the most dramatic responses to low iron include reorganization of the photosynthetic apparatus (8): Expression of the IsiA antenna protein (also called CP43′) is induced (9), and the iron-containing electron carrier protein ferredoxin (Fd) is exchanged for the functionally equivalent, noniron-requiring flavodoxin (Fld, IsiB) (10). Both photosystems are also down-regulated, in particular, photosystem I (PSI) (11), which contains three [4Fe-4S] clusters. IsiA associates with the remaining PSI (12) where it is suggested to act either as a fluorescence quencher (13) or to accelerate electron transport through PSI (14). IsiA supercomplexes with PSI and photosystem II (PSII) have been identified after prolonged iron deprivation (15). IsiA and IsiB are expressed from the iron stress-induced (isi) operon as isiA and isiB, respectively (9, 16). Expression of the isiAB operon was thought to be repressed in iron-replete conditions by the global iron regulator FurA on binding of Fe2+ (17, 18), although a subsequent study found that the principle regulator acts upstream of this site, and has yet to be identified (19). Total abundance of the IsiA protein is controlled at the level of mRNA stability by an internal antisense RNA, IsrR (20). However, little is known about the regulators of multiple other cyanobacterial iron-responsive elements, such as idiCB operon (Z48754) (21).
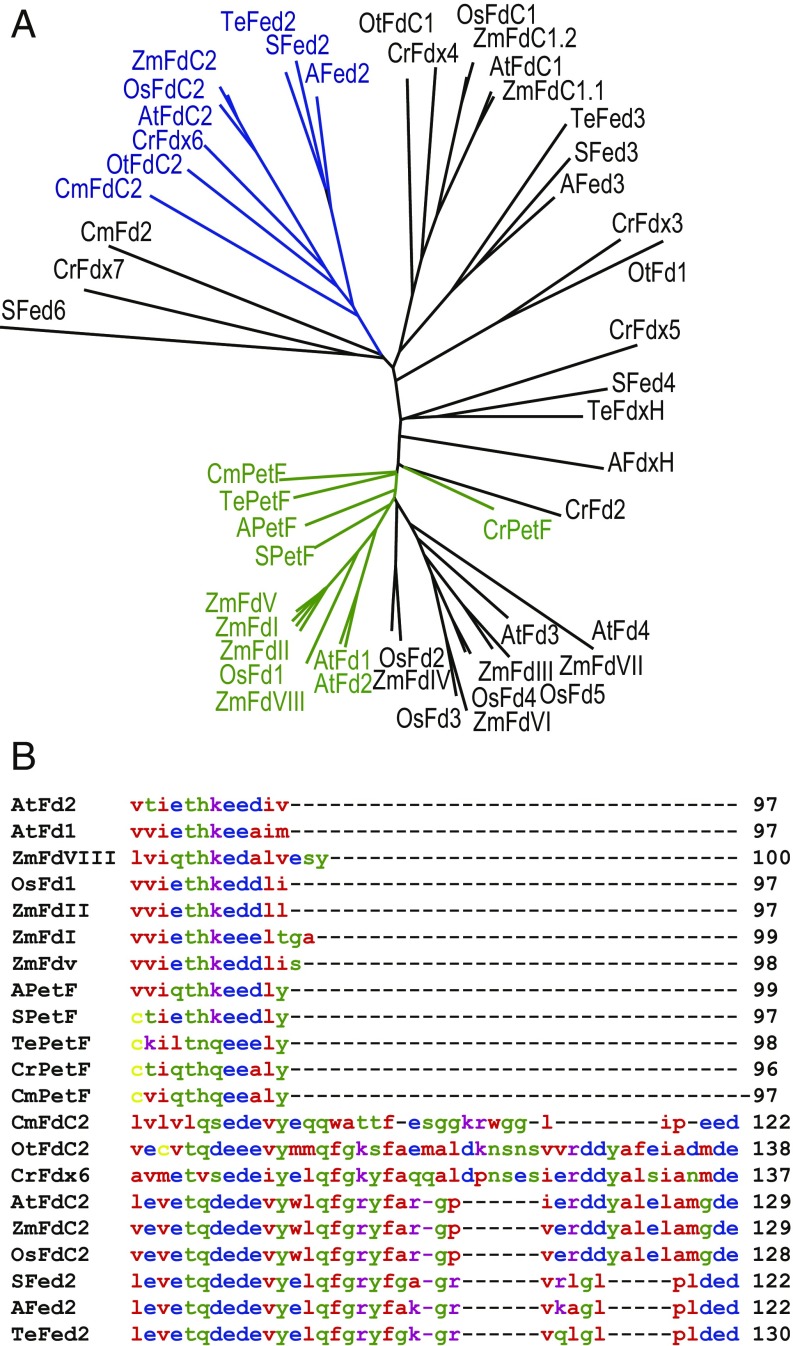
While the photosynthetic Fd (petF) gene is repressed in response to iron limitation, most cyanobacteria also contain at least three other genes encoding [2Fe-2S] Fds (22). These include a specific heterotrophic FdxH involved in heterocyst or dark metabolism, and one to two Fds with extended C-termini. Homologs of these Fd proteins with C-terminal extensions have been named FdC1 (Fd with C-terminal extension) and FdC2 in higher plants (Fig. 1A). In the model cyanobacterium Synechocystis sp. PCC 6803 (hereafter called Synechocystis), the closest homolog of higher plant FdC2 has been named Fed2 (sll1382), and has a 22-amino acid C-terminal extension relative to PetF (22) (Fig. 1B). Due to the presence of up to 10 different genes encoding [2Fe-2S] Fds in higher plant and algal genomes (23), the nomenclature of Fed2/FdC2 homologs is highly variable, and they have variously been named Fd6 or FdC2 (AT1G32550.1) in Arabidopsis thaliana (24), FdC2 (Os03g0685000) in rice (Oryza sativa) (25, 26), and Fdx6 (ABC88605.1) in the alga Chlamydomonas reinhardtii (27).
Fig. 1.
Fed2/FdC2 proteins are highly conserved in photosynthetic organisms. (A) Unrooted phylogenetic tree of Fds from different photosynthetic organisms, highlighting known photosynthetic type (in green) and Fed2/FdC2 type (in blue). The tree was generated at the Phylodendron website using an alignment generated in Clustal Omega (62). Transit peptide amino acids were removed from eukaryotic proteins for accurate comparison of mature proteins. Fd isoproteins from different species are as follows: Synechocystis PCC 6803: SFed2 sll1382, SFed3 slr1828, SPetF ssl0020, SFed6 ssl2559, SFed4 slr0150; Anabeana PCC7120: Afed2 all2919, Afed3 alr0784, APetF all4148, AFdxH all1430; T. elongatus: TeFdxH WP_011057078.1, TePetF WP_011056851.1, TeFed3 WP_011056338.1, TeFed2 WP_011057493.1; C. reinhardtii: CrFdx6 ABC88605.1, CrPetF AAA33085.1, CrFdx2 ABC88601, CrFdx3 ABC88602, CrFdx4 ABC88603.1, CrFdx7 EDP05356, CrFdX5 ABC88604.1; Ostreochoccus tauri: OtFdC2 CAL53412.1, OtFdC1 CAL53607, OtFdI CAL58543.1; Cyanidioschyzon merolae: CmPetF CMV193c, CmFd2 CMR177c, CmFdC2 CMR278c; A. thaliana: AtFdC2 AT1G32550.1, AtFdC1 AT4G14890, AtFd1 AT1G10960, AtFd2 AT1G60950, AtFd3 AT2G27510, AtFd4 AT5G10000.1; O. sativa: OsFdC2 Os03g0685000, OsFdC1 Os03g0659200, OsFd1 Os08g0104600, OsFd2 Os04g0412200, OsFd3 Os05g0443500, OsFd4 Os03g0835900, OsFd5 Os01g0860601; and Zea mays: ZmFdC2 ACG28100.1, ZmFdC1.1 ACG25123, ZmFdC1.2 AAT42183, ZmFdI AAA33459.1, ZmFdII BAA32348.1, ZmFdIII BAA19251.1, ZmFdIV NP_001150016, ZmFdV ACA34366.1, ZmFdVI BAA19249.1, ZmFdVII ACF78894, ZmFdVIII ACN29244. (B) Comparison of the C-terminal region of photosynthetic Fds with Fed2/FdC2 proteins from the same Clustal Omega alignment. Amino acid coloring is as follows: red, small and hydrophobic; blue, acidic; magenta, basic; yellow, cysteine; and green, other polar (full alignment shown in SI Appendix, Fig. S1); same protein codes as in A.
A Fed2/FdC2 homolog seems to be present in all photosynthetic organisms (23), and, intriguingly, it was found to be essential in cyanobacteria (22). While no transposable element insertion (T-DNA) knockout has been identified, to date, in higher plants, a single amino acid substitution results in slower growth and a pale green leaf phenotype in rice (25, 26). Although, in Synechocystis, expression of fed2 is up-regulated in response to oxidative and heavy metal stress (22), and expression of the homologous Fdx6 in Chlamydomonas is increased in low iron (27), its biological function remains unknown. In a previous study on recombinant, His-tagged Arabidopsis Fd6/FdC2, Kolton et al. (28) report that the protein is capable of electron transport between PSI and NADPH, and can be reduced by Fd:NADP(H) oxidoreductase (FNR), although the affinity is very low between FdC2 and FNR in both assays. Moreover, although the protein was detected bound to thylakoid membranes and mRNA, no specific functional role for the protein was identified. In this study, we aimed to understand the physiological role of Fed2 by disrupting its function in Synechocystis. Our data indicate that Fed2 is a critical component in cyanobacterial iron perception and/or homeostasis.
Results
Fed2 Shows Unique Structural Features and Lacks Activity with FNR.
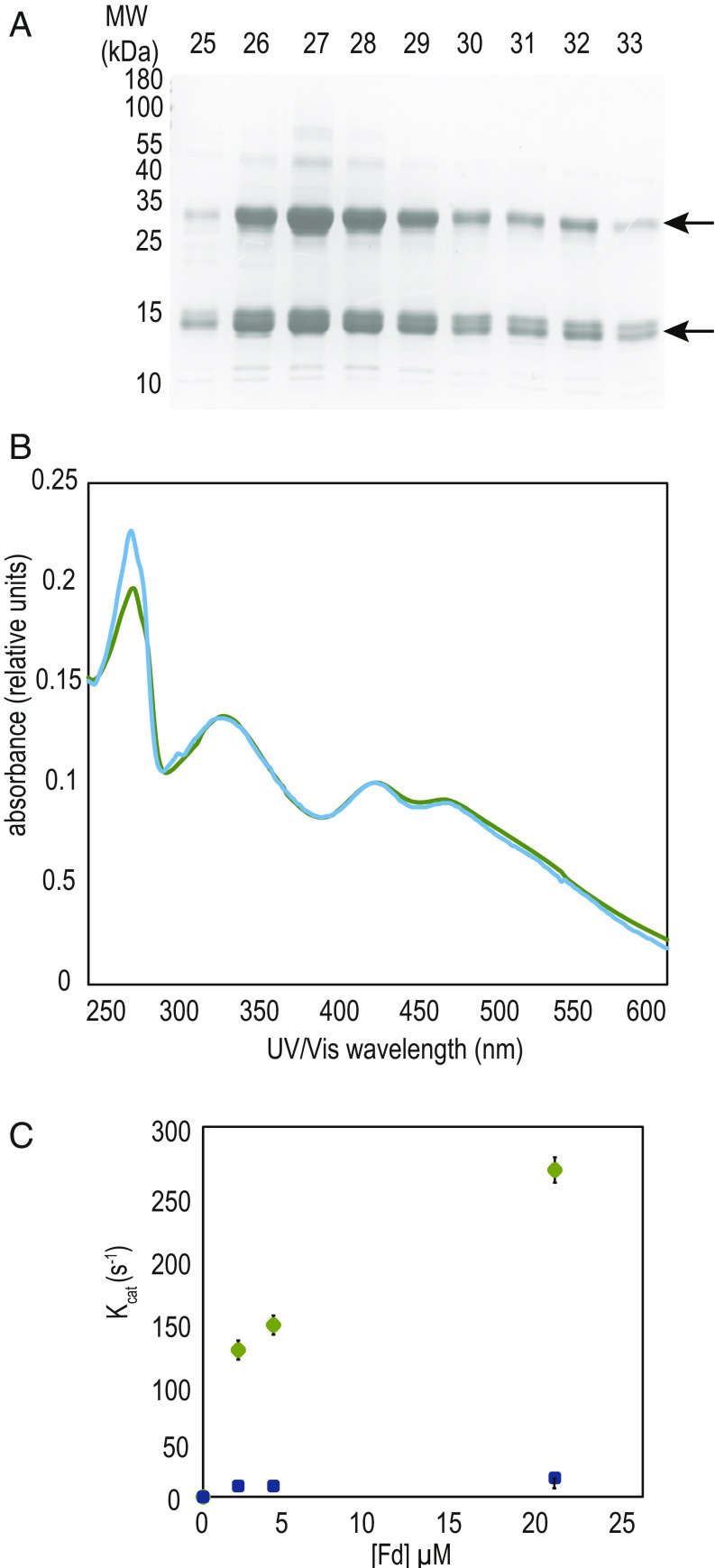
Fed2/FdC2 proteins are highly conserved (Fig. 1A) and distant from the photosynthetic PetF proteins, but present through the “green cut” of cyanobacterial genes with homologs conserved through algae to higher plants (29). The characteristically extended C terminus is well conserved, usually beginning with a Gln−Phe−Gly−Arg/Lys−Tyr−Phe motif, and ending with several acidic residues at the C terminus (Fig. 1B). These 22 to 36 extra residues at the C terminus are significant, as it is known that the C-terminal region of PetF (which lacks them) plays a critical role in interaction with PSI during PetF reduction (30, 31). Following cloning into a protein expression vector, the [2Fe-2S] cluster of purified Synechocystis Fed2 proved to be unstable, and we therefore purified recombinantly expressed Fed2 from the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1 to investigate protein function. Remarkably for an Fd, the purified T. elongatus Fed2 showed migration over a size exclusion column as an oligomer, part of which forms a stable dimer during SDS/PAGE (Fig. 2A), with purity and band identity confirmed by mass spectrometry (SI Appendix, Table S1). The protein showed a characteristic [2Fe-2S] cluster UV−visible (UV-VIS) spectrum (Fig. 2B), although it had very poor ability to transfer electrons with the classical FNR compared with cyanobacterial PetF (Fig. 2C). Our data contrast with the findings of Kolton et al. (28), who measured reasonable electron transport with FNR by the higher plant homolog, using a His-tagged higher plant (A. thaliana) FdC2. This could relate to species differences, or the noncleaved His-tag used in the study by Kolton et al., where the protein also showed an atypical UV-VIS spectrum for a [2Fe-2S] cluster. To further establish whether Fed2 could function in photosynthetic electron transport, we measured the redox midpoint potential of the recombinant protein. The spectra in SI Appendix, Fig. S2 indicate a midpoint potential of −465 ± 3 mV vs. Ag/AgCl/KCl (3 M). Corrected for normal hydrogen electrode (NHE) reference, this gives a value of −243 ± 3 mV. This is much more positive than the values of −412, −320, and −320 mV measured for Synechocystis PetF (32), cyanobacterial FNR (33), and NADP(H), respectively, making electron donation from cyanobacterial Fed2 to FNR and then NADP+ energetically unfavorable. These factors, in combination with the very low protein content reported for the algal homolog (27), indicate that it is highly unlikely that Fed2/FdC2 proteins function directly in photosynthetic electron transport or are reduced by FNR. Moreover, this redox potential also indicates that electron donation from Fed2/FdC2 proteins to many other classical Fd-dependent enzymes, such as nitrite reductase (34) or glutamate synthase (35), is also energetically less favorable.
Fig. 2.
Unusual properties of the recombinant purified Fed2 protein. (A) Coomassie-stained SDS/PAGE gel showing two bands in eluting fractions from size exclusion chromatography in the final step of recombinant T. elongatus Fed2 (TFed2) purification. (B) Comparison of UV-Vis spectra of recombinant purified PetF (green), cloned from Synechocystis sp.PCC 6803, and TeFed2 (blue). Spectra measured at 0.1 mM concentration. (C) Electron transport between FNR and Fds. Activity was measured in cytochrome c coupled assay with 20 nM FNR over a range of concentrations of Synechocystis PetF (green circles) and TeFed2 (blue squares).
Fed2 Is Essential in Synechocystis.
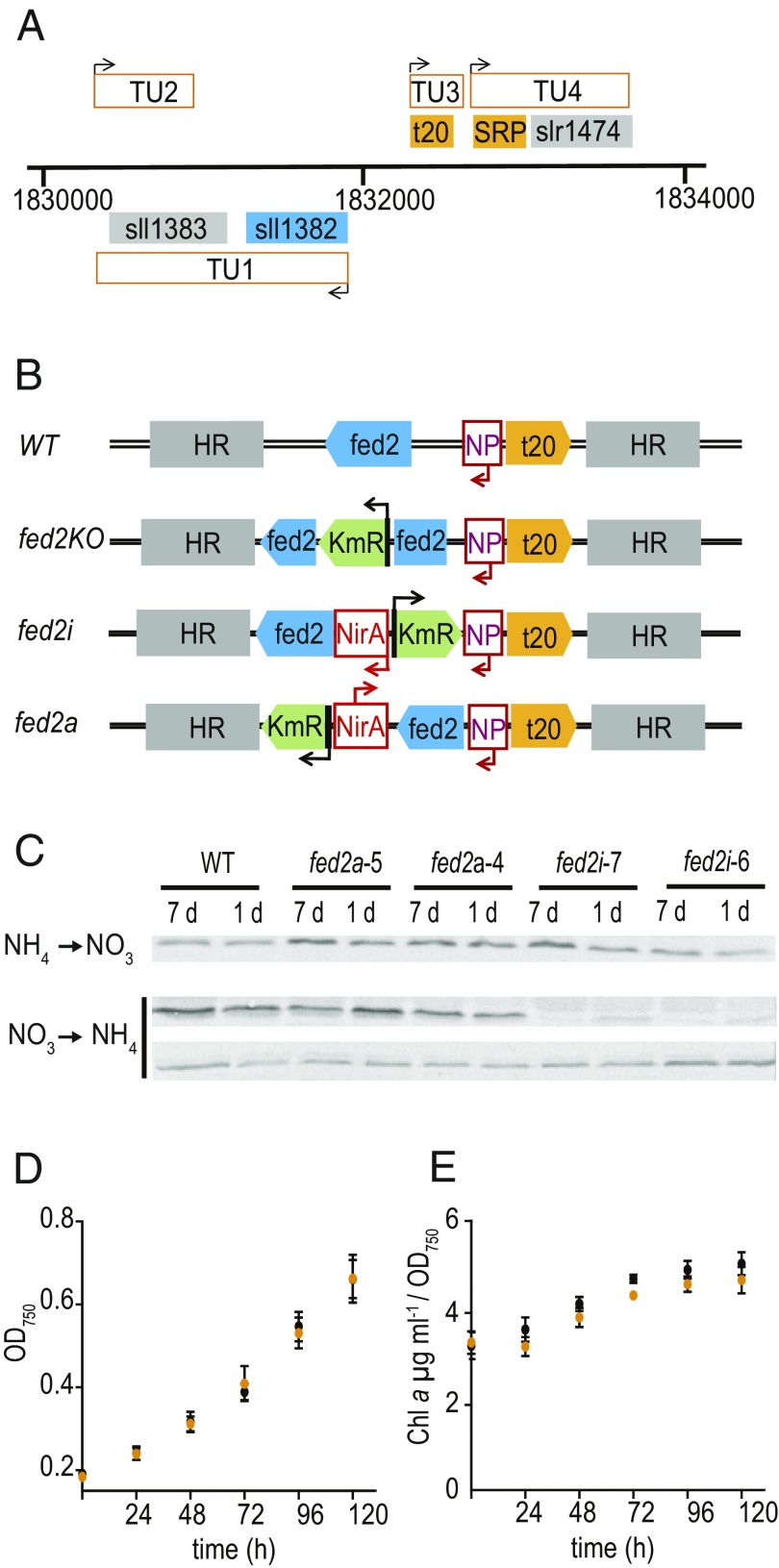
To identify the physiological role of Fed2, we aimed to knock out the gene in Synechocystis. fed2 (Sll1382) forms a transcriptional unit with suhB (inositol-1 monophosphatase) in Synechocystis (Fig. 3A adapted from ref. 36) and is part of the same operon in various other cyanobacteria (22). Despite this, we chose to insert all selectable markers downstream of fed2, as only 71 bp separate the first codon of the fed2 gene and the first nucleotide of a unique tRNA-Ser gene on the opposite strand, whose expression we considered it critical not to disturb. Moreover, we hypothesize that any impact on suhB expression will make a small contribution to the phenotype, because its knockout in Synechococcus only results in a mild phenotype related to osmotic stress (37). In contrast, disruption of the fed2 gene by insertion of a kanamycin resistance cassette in Synechocystis failed to yield a fully segregating strain (SI Appendix, Fig. S3A) even after restreaking cells multiple times in the presence of glucose, suggesting that Fed2 is essential.
Fig. 3.
Genomic environment and disruption of fed2 in Synechocystis. (A) Genomic context of fed2 in Synechocystis. The sll1382 (fed2) and sll1383 (suhB) form a single transcriptional unit (TU1). In the region 5′ from fed2 there are two more transcriptional units. One encodes a unique t-RNA-Ser (t20) and the other a predicted signal recognition particle (SRP) in combination with slr1471, a protein of unknown function. (B) Strategies for fed2 knockdown and knockout. Arrows show direction of promoter action; HR, homologous region taken for cloning; KmR, kanamycin resistance; NP, fed2 native promoter; NirA, nitrate responsive promoter. From top to bottom: WT genomic arrangement, knockout insertion of KmR, inducible knockdown by replacing the native promoter with NirA, and inducible antisense by introduction of the NirA promoter 3′ of fed2 in the anticoding direction. (C) Protein abundance of Fed2 following transfer of single colony derived cultures of inducible antisense and inducible expression lines between inducing and noninducing media, detected by Western blotting for Fed2 (see SI Appendix, Fig. S3 for segregation of strains). For nitrate to ammonium transfer, Top is Fed2 and Bottom is a loading control of a nonspecific band on the same blot. (D) Growth and (E) chlorophyll content of Synechocystis WT (black) and fed2i (orange) cells following transfer to ammonium media (suppression of fed2 expression in fed2i line). Chlorophyll a content is expressed as a function of the cell density. Values are mean ± SD of at least three independent measurements. Data from ref. 36.
We next tried to knock down fed2 using two independent inducible expression systems (Fig. 3B): firstly, with an inducible 3′ promoter running antisense to fed2 (fed2a) and, secondly, by exchanging an inducible promoter for the native fed2 promoter (fed2i). As fed2 transcript abundance responds to some metals (22), we avoided the classical Cu-inducible system (38) and used the nitrate-inducible nirA promoter from Synechococcus (39). These strains readily segregated (SI Appendix, Fig. S3B) in nitrogen regimes that facilitate fed2 expression (ammonia for antisense, nitrate for sense). On transfer to the alternative N source (nitrate for fed2a, ammonia for fed2i), to repress transcript abundance, no changes in Fed2 protein were observed in fed2a lines, but fed2i lines showed a large decrease in Fed2 abundance (Fig. 3C). Surprisingly, the fed2i strain showed no significant changes in cell growth or chlorophyll a content in these conditions (Fig. 3 D and E), indicating that even very low abundance of Fed2 is sufficient for regular cyanobacterial growth.
Attenuated Fed2 Function Results in a Growth Penalty.
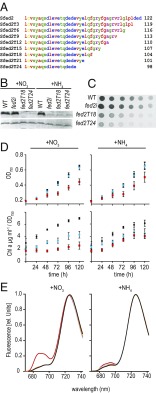
Rather than decreasing protein abundance, we then sought to disrupt, but not eliminate, Fed2 function by mutation. The most obvious conserved feature of Fed2 proteins is the C-terminus region (Fig. 1A), so we performed a sequential truncation, introducing stop codons to remove amino acids from the C terminus in groups of three (Fig. 4). Truncations of up to 18 amino acids (fed2T18) readily segregated to full mutation (SI Appendix, Fig. S3C), while deletions of more than 21 amino acids (fed2T21 and fed2T24) only segregated after extensive rounds of screening were performed (SI Appendix, Fig. S3 C and D). To compare phenotypes to the fed2i line, we also performed experiments on the truncation lines in both ammonium and nitrate growth media. Western blot analysis detected much less Fed2 protein in both the fed2T18 and fed2T24 lines, similar to that seen in the fed2i knockdown line (Fig. 4B). Because expression of fed2T18 and fed2T24 is driven by the native promoter in these lines, the decreased Fed2 protein content is probably due to structural destabilization caused by truncation, leading to faster protein turnover. In contrast to fed2i, however, the truncation mutants both showed decreased growth on agar (Fig. 4C). This phenotype is almost certainly due to perturbed Fed2 function, rather than disruption of other genomic elements or compensatory mutation, as it is seen in the two independent truncation lines (one of which segregated rapidly), and not observed in fed2a, where the kanamycin resistance gene is inserted in an identical position in the genome. For fed2T24, this translated into significantly slower growth in liquid media (Fig. 4D). Interestingly, we also observed significantly decreased chlorophyll a content relative to cell density for the truncation lines in both nitrate and ammonium liquid growth conditions (Fig. 4D), while the fed2i line did not differ significantly from the WT in ammonium growth conditions (Fig. 3E).
Fig. 4.
Truncation of Fed2 results in perturbed growth. (A) Sequential truncation of the Fed2 protein by three amino acid steps. (B) Western blot analysis of Fed2 content in fully segregated fed2T18, fed2T24, and fed2i lines (see SI Appendix, Fig. S3 for segregation analysis). Cells were cultivated for 1 wk in liquid media supplemented with either nitrate or ammonium. Following SDS/PAGE gel (20 µg of total protein per lane) and Western blotting, proteins were visualized via a secondary antibody conjugated to alkaline phosphatase. Top is Fed2, and Bottom is a loading control of a nonspecific band on the same blot. (C) Growth of Synechocystis WT, fed2i, fed2T18, and fed2T24 truncation lines. A cell suspension of 0.2 µg⋅mL−1 of chlorophyll was plated in a dilution series on regular BG11 plates followed by 1 wk of growth. (D) Growth and pigment analysis of the same lines as in C, cultivated in liquid media containing either nitrate or ammonium as a N source. WT (black circles), fed2T18 (blue circles), and fed2T24 (red squares). Precultured cells (grown in ammonium-containing media) were washed in nitrogen-free BG11 media and subsequently diluted to an OD750 of 0.2 in media containing either nitrate or ammonium. Growth rate was monitored for 5 d. Chlorophyll a content is expressed as a function of the cell density. Values are mean ± SD of at least three independent measurements. (E) The 77 K fluorescence emission spectra of Synechocystis WT (black), inducible fed2i (orange), and truncation lines fed2T18 and fed2T24 (blue and red, respectively) grown for 7 d in media containing either nitrate (Left) or ammonium (Right) as N source. Cells were adjusted to a chlorophyll a content of 3 µg⋅mL−1. The excitation wavelength was 430 nm. Each spectrum shown is the mean of five spectra, normalized to the characteristic PSI fluorescence peak emitted at 725 nm.
Response to Low Iron Is Disrupted in fed2 Truncation Lines.
To investigate how decreased chlorophyll a content impacted the photosynthetic apparatus, we monitored the 77 K emission spectra (Fig. 4E), where emission peaks at 685 and 695 nm correspond to PSII reaction centers, and the peak at 720 nm corresponds to PSI reaction centers. Fed2T24 cells grown in nitrate medium showed a dramatic increase in the characteristic PSII peaks relative to PSI (Fig. 4E, Left). In ammonium medium (Fig. 4E, Right), both truncation lines showed a much smaller increase in 685-nm fluorescence, and the fed2i only deviated very slightly from the WT spectrum. A relative increase at 685 nm, such as that seen in the fed2T24 line in nitrate growth conditions, has previously been ascribed to the IsiA antennae and is typically associated with iron depletion in cyanobacteria (12, 16).
Since previous studies detected increased fed2 expression in response to high cadmium and zinc (22) and expression of Fdx6, the Fed2/FdC2 homolog in Chlamydomonas, is up-regulated in response to low iron (27), this result prompted us to examine the sensitivity of the truncation mutants to altered metal contents (SI Appendix, Fig. S4), with a particular focus on iron (Fig. 5). Although growth of the truncation mutants at increased iron concentrations is still perturbed relative to the WT, use of media with depleted iron results in similar growth of all genotypes (Fig. 5A). Following transfer of cells to depleted iron liquid media, cell proliferation is the same for WT and the fed2T24 line, but total chlorophyll concentrations are severely affected in the truncation line (Fig. 5B). The 77 K fluorescence spectra shown in Fig. 5C demonstrate that, after growth in liquid medium lacking iron, the fluorescence peaks of PSII increase relative to PSI in both genotypes. However, this is much more dramatic in the fed2T24 line than in WT, irrespective of the N source. This contrasts to the 77 K phenotype of fed2T24 under iron-replete conditions, which is only seen in nitrate media. Although growth on solid media with elevated copper, cobalt, and zinc also eliminated the difference in growth between the WT and truncation mutants (SI Appendix, Fig. S4), only low iron induced the dramatic increase in PSII relative to PSI fluorescence. After identifying this phenotype in fed2T24, we subjected fed2T18 to iron-depleted media and measured 77 K spectra (Fig. 5D). Unexpectedly, fed2T18 showed a trend opposite to that of fed2T24. Instead of an exaggerated increase in PSII/PSI fluorescence, this genotype is less responsive than the WT. Thus, a difference of just six amino acids at the C terminus of Fed2 results in opposite responses at the level of PSII to PSI ratio.
Fig. 5.
Truncated Fed2 perturbs adaptation to low iron. (A) Twofold dilution series of Synechocystis WT, fed2i inducible knockdown, and truncated fed2 lines fed2T18 and fed2T24. Cell suspensions of 2 µg⋅mL−1 of chlorophyll were spotted onto BG11 plates with the indicated iron concentrations. Plates were scanned after 7 d of growth. (B) Growth and pigment analysis comparing WT and the Fed2 truncation line fed2T24 cultivated in liquid media containing either nitrate or ammonium as a N source; WT, black; fed2T24, red. Precultured cells (grown in ammonium-containing media) were washed in nitrogen-free BG11 media and subsequently diluted to an OD750 of 0.2 in media containing either nitrate or ammonium in the indicated iron concentrations. Growth rate was monitored for 5 d. Chlorophyll a content is expressed as a function of the cell density. Results are typical of three independent experiments. (C) The 77 K fluorescence emission spectra of Synechocystis WT (black) and truncation line fed2T24 (red) grown for 7 d in media containing either nitrate or ammonium as N source in the indicated iron concentrations. Cells were adjusted to a chlorophyll a content of 5 µg⋅mL−1. The excitation wavelength was 430 nm. Experiments were performed three times with basically the same result. (D) Comparison of 77 K spectra from WT (black), fed2T18 (blue), and fed2T24 (red) lines following 1 wk of growth in liquid media lacking iron, normalized to the PSI emission peak of 720 nm (Left) and the PSII emission peak of 685 nm (Right). Experiments were performed three times with basically the same result.
Iron Import Is Not Disturbed in Fed2 Truncation Lines.
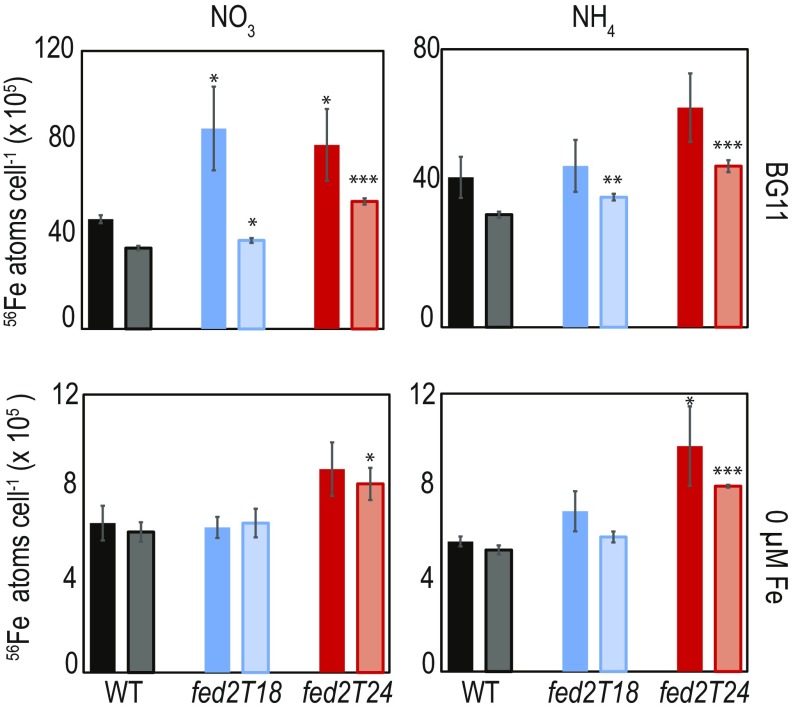
A major pathway of iron acquisition by Synechocystis relies on reduction of ferric iron (Fe3+) to ferrous iron (Fe2+) during import (40). Although some of the components of this machinery are identified (40–43), the original source of these electrons remains unclear (44). We initially hypothesized that Fed2 might be involved in providing this reducing power to the plasma membrane, and we therefore measured iron concentrations in WT and truncation mutants of Fed2 by inductively coupled plasma mass spectrometry (ICP-MS). Cellular iron contents were compared for iron-replete and iron-deprived cells (Fig. 6). Significant amounts of unimported iron may be stored in the Synechocystis periplasm (41), so we also measured iron contents of EDTA-washed cells (pale bars, Fig. 6). Interestingly, despite the apparent iron stress of fed2T24 under normal growth conditions indicated by the 77 K spectrum (Fig. 4E) and the disturbed PSI/PSII emission ratios in 77 K spectra of both mutants following iron depletion (Fig. 5), neither line showed decreased cellular iron relative to WT. In fact, contents of both total cellular iron and intracellular iron (EDTA-washed cells) increased relative to the WT (up to 1.8 times higher, depending on the growth conditions).
Fig. 6.
Iron content of Synechocystis WT (black) and Fed2 truncation lines fed2T18 (blue) and fed2T24 (red) as measured by ICP-MS. Cells were precultured in liquid BG11 before washing three times in minimal BG11 media without iron or copper and subsequently diluted to an OD750 of 0.5 in regular BG11 or BG11 lacking iron. After 1 wk of growth, 500 μL of the cell culture was harvested and washed two times with either chelex-treated water (solid colors) for total cellular Fe, or 5 mM EDTA (pH 8.0) (pale colors) to remove periplasmic iron. Cells were resuspended in 200 μL of chelex-treated water and analyzed by ICP-MS. Values are mean ± SE of three independent measurements. Values are typical of two independent experiments. Statistically significant differences from WT in a Students t test: *P < 0.05; **P < 0.01; ***P < 0.001.
Iron Is Not Correctly Perceived on Fed2 Truncation.
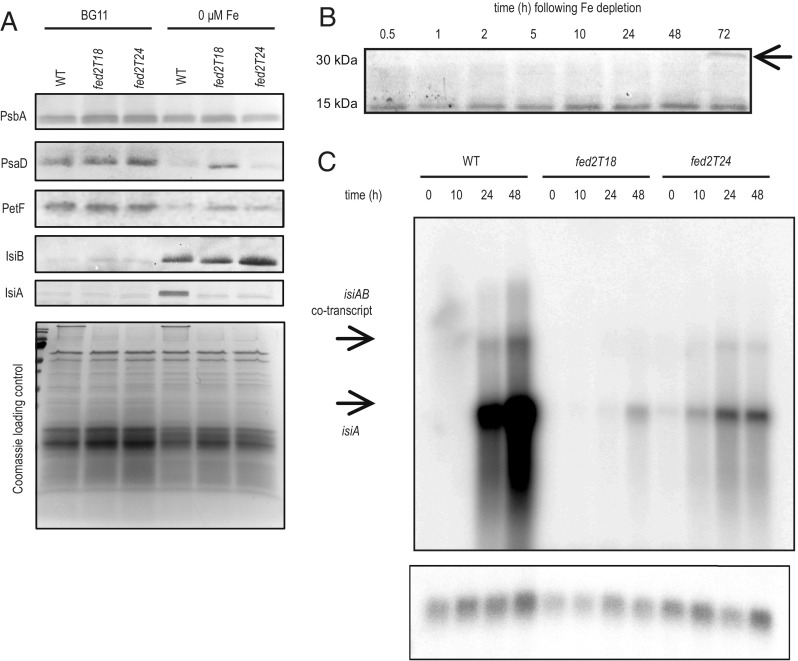
The contradiction between the photosystem response to decreased iron (Fig. 5) and actual cellular iron content (Fig. 6) prompted us to investigate whether other cellular responses to low iron were activated in the Fed2 truncation lines. We therefore checked the abundance of classical iron depletion marker proteins in cells grown with either iron-replete or iron-limited media (Fig. 7A). As this phenotype is seen in both N conditions, we limited the experiment to nitrate growth only. While iron limitation leads to a decrease in the PSI protein PsaD, this is less pronounced in the fed2T18 line, consistent with its greater PSI fluorescence than WT under these conditions (Fig. 5D). The change in PSI acceptors, with decreased Fd (PetF) and increased Fld (IsiB), occurs in all genotypes, although the increase in Fld levels is more pronounced in fed2T24 than in WT or fed2T18. Dramatically, abundance of IsiA, the classical marker for iron depletion, does not increase in either Fed2 truncation line after transfer to low-iron conditions. This is especially striking, as isiA and isiB are thought to form a single transcriptional unit, with the promoter region upstream of isiA (45). Our data indicate that processing and translation of this cotranscript is more complex than previously believed. Although it was reported that Synechocystis fed2 transcripts do not respond to iron, we examined Fed2 protein contents over time in the WT following iron depletion (Fig. 7B). Increased abundance of Fed2 is visible 10 h after iron depletion, and, by 72 h, a new band at the same predicted migration size as a Fed2 dimer (∼30 kDa) appears. This is of interest in the context of the Fed2 dimer obtained on purification of the recombinant protein (Fig. 2A).
Fig. 7.
Truncation of Fed2 results in perturbed transcriptional responses to iron concentrations. (A) Western blot analysis of photosynthetic components. WT, fed2T18, and fed2T24 lines were grown in regular BG11 and BG11 containing no iron, with NO3 as a N source. Cells were diluted to the same OD750 in fresh BG11 and washed once before harvesting. Subsequently, cell pellets were treated with SDS-loading buffer and boiled for 5 min before SDS/PAGE. After electrophoresis, proteins were either subject to Western blotting and the indicated proteins were visualized using specific primary antibodies raised against PsbA, PsaD, PetF, IsiB, and IsiA, with a secondary antibody conjugated to alkaline phosphatase (Top), or stained with Coomassie brilliant blue as a loading control (Bottom). Results are typical of three independent experiments. (B) Fed2 abundance responds to iron starvation. Western blot was performed to detect Fed2 in protein samples taken from WT Synechocystis cells following transfer to no-iron BG11. Cells were precultured in iron-containing ammonium BG11 media before washing in iron-free media and transfer to iron-free BG11. Sampling was performed at indicated time points. Results are typical of three independent experiments. (C) The isiA transcript responses detected by Northern blotting at the indicated time points following transfer of the cells to no-iron-containing media (Top). RnpB detection as control (Bottom). Results are typical of two independent experiments.
As Fed2 appears critical for IsiA accumulation, but does not negatively impact IsiB, we investigated whether misregulation occurs at the protein or transcript level, by detecting isiA transcript abundance following the same treatment (Fig. 7C). In the WT, 24 h of iron limitation is sufficient for a large induction of isiA transcript expression. However, neither truncation mutant shows a significant response, although a small amount of transcript is visible, especially in fed2T24.
As the iron depletion response is disrupted on truncation of Fed2, we checked whether this was also true when Fed2 contents are decreased in the fed2i line (SI Appendix, Fig. S5). While growth rate was not affected, the chlorophyll content was slightly decreased in low iron (SI Appendix, Fig. S5A), and the 685-nm peak in 77 K responded more than in the WT (SI Appendix, Fig. S5B). This mirrors the response in the fed2T24 line but is less severe. These observations were confirmed by Western blotting, where fed2i showed an intermediate increase in IsiA contents between WT and fed2T18 (SI Appendix, Fig. S5E). These effects were most severe in ammonium media, but also seen in nitrate media. In the fed2i line, fed2 expression is driven by the nirA promoter, which can be additionally down-regulated in iron starvation (46, 47), explaining the decreased Fed2 content in the fed2i line (SI Appendix, Fig. S5C), and the corresponding phenotypic changes in chlorophyll concentration (SI Appendix, Fig. S5A), 77 K fluorescence (SI Appendix, Fig. S5B), and protein response (SI Appendix, Fig. S5C). It is also clear from the corresponding Northern blots that, although isiA induction is perturbed in the fed2i line (SI Appendix, Fig. S5D), this is to a lesser extent than in the truncation lines (Fig. 7C).
Disrupted Thylakoid Structure in Fed2 Truncation Lines.
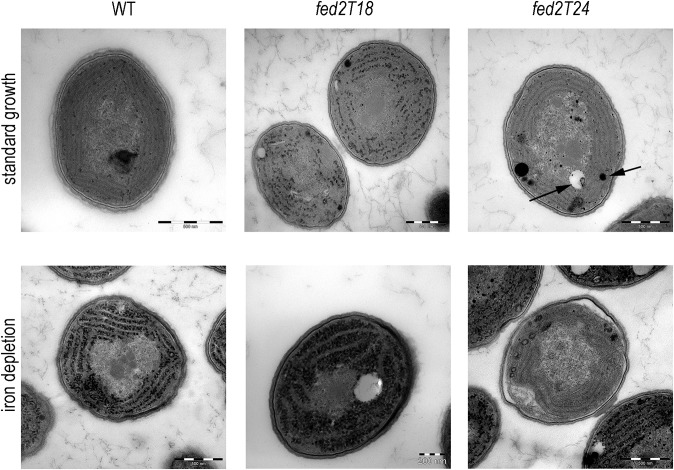
It was previously reported that thylakoid structure was disrupted in mutants of isiA (16), and we therefore checked whether this feature was consistent with the Fed2 truncation lines, in which IsiA does not accumulate. Transmission electron microscopy (TEM) was performed on Synechocystis WT, fed2T18, and fed2T24 lines that had been grown in either iron-replete or iron-deprived conditions (Fig. 8). Greater numbers of cells at lower magnification are shown in SI Appendix, Fig. S6. Under iron-deprived conditions, all genotypes show large numbers of electron-dense particles in the cytosol, principally between the thylakoids. Interestingly, the fed2T18 line also shows some accumulation of these structures under iron-replete conditions. Even in iron-replete conditions, cells of the fed2T24 line appear stressed, with large inclusion bodies present in the cytosol of most cells. On iron depletion, a significant proportion of fed2T24 cells appear to have decreased abundance of high-density cytosolic particles relative to the other genotypes. Some of these cells contain very little internal membrane, with the remainder highly disorganized. This is not a uniform phenotype but is present in a large percentage of the cells (∼21% for fed2T24), some of which are clearly dividing (SI Appendix, Fig. S6). If these cells were dead, a decreased growth rate of fed2T24 on low iron would be expected. However, cellular growth is very similar to the WT (Fig. 5), indicating that these cells are alive but lack significant thylakoid structures. Nonviable cells are easily identified by their green fluorescence following chlorophyll excitation (48), so we compared the genotypes following iron depletion using confocal microscopy (SI Appendix, Fig. S7). As seen in SI Appendix, Fig. S7B, the T18 and T24 lines have a similar, or even a lower, frequency of nonviable cells compared with the WT, indicating that the low membrane density T24 cells are viable. The fed2T18 line shows the same phenotype under iron deprivation, but, in this case, a smaller proportion of the cells (around 8%) are affected (SI Appendix, Fig. S6B). Both lines are completely segregated (SI Appendix, Fig. S3), so the binary nature of this phenotype (either WT-like or lacking organized thylakoids) must reflect either induction or absence of a specific developmental response, to which cells with perturbed Fed2 function are more prone.
Fig. 8.
Impact of Fed2 truncation on cyanobacterial cell structure. Synechocystis WT, fed2T18, and fed2T24 lines were transferred to BG11 with NO3 as a N source, either with iron (Top) or in the absence of iron (Bottom), for 7 d before fixation and treatment for TEM. Typical phenotypes are displayed. Arrows on fed2T24 cell in standard growth media indicate typical spherical inclusions of high or low density interpreted as inclusion bodies. Additional cells are shown in SI Appendix, Fig. S6.
Discussion
In this article, we show that Fed2, an Fd with a uniquely extended C terminal, which is conserved throughout photosynthetic organisms (Fig. 1), is involved in the response to low iron in cyanobacteria. Following iron depletion, the Synechocystis lines with perturbed Fed2 function, fed2T18 and fed2T24, failed to induce isiA transcript (Fig. 7C) and protein accumulation (Fig. 7A), demonstrating that a functional Fed2 is critical for the cyanobacterial response to iron depletion. IsiA forms an operon with isiB (36), but, under the same conditions, the IsiB protein accumulated to similar or even higher levels in fed2i, fed2T18, and fed2T24 than in WT (Fig. 7A and SI Appendix, Fig. S5C), and was therefore independent of Fed2 function. When we additionally probed for the isiB transcript by Northern blotting (SI Appendix, Fig. S5D), we confirmed the original finding that it is predominantly present as a cotranscript with IsiA (49). Our findings indicate that IsiB can be translated before other regulatory processes prevent translation of IsiA. This indicates that Fed2 might impact at the level of RNA stability rather than transcriptional processes. IsiA transcript abundance is known to be regulated by the small antisense RNA IsrR, forming a double-stranded RNA that is targeted for destruction (20). The latest model proposes that IsrR expression is constant and prevents short-term iron fluctuations from causing expensive induction of isiA, driven by the iron-sensing transcriptional regulator FurA (50). Our data show that a functional Fed2 also plays a critical role in accumulating isiA transcript. A previous study demonstrated that recombinant higher plant FdC2 protein is capable of binding chloroplast transcripts (28), although it is still unclear whether this interaction is part of any significant regulatory process.
In Synechocystis, Fed2 is critical for the up-regulation of IsiA, and the phenotype of the truncation mutants resembles that of isiA mutants under low-iron conditions (16), with a similar growth rate to WT, but low chlorophyll content. However, in iron-replete conditions, the isiA mutant is not distinguishable from the WT, whereas the failure of fed2 insertion mutants to segregate (SI Appendix, Fig. S3) agrees with previous reports that this protein is essential for cell growth (22). Surprisingly, large decreases in Fed2 protein abundance do not impact growth rate (Fig. 2), indicating that only a very small amount of protein is required to support growth under our laboratory conditions. The increased iron contents of Fed2 truncation mutant cells (Fig. 6) indicate that iron homeostasis itself is disrupted. In this case, loss of Fed2 could be lethal through dangerous hydroxyl radical formation in the Fenton reaction between free ferrous iron and hydrogen peroxide. The function of Fed2 could also extend beyond regulating IsiA and other components of iron homeostasis, to a more global role in cellular homeostasis. Indeed, the electron micrographs indicate that large inclusion bodies are also formed in the fed2T24 cells even under iron-replete conditions (Fig. 8), and the truncation mutants grow at similar or slightly better rates than the WT on some high heavy metal solid media (SI Appendix, Fig. S4).
There are also differences between the truncation lines fed2T18 and fed2T24, indicating that the C terminus has a critical functional role. Most strikingly, a six-amino acid difference results in opposite trends in the PSI/PSII ratio on iron depletion. In iron-deprived fed2T24 cells, which do not induce IsiA, the 685-nm peak detected in 77 K measurements is exaggerated relative to the P700 peak at 720 nm (Fig. 5). fed2i shows the same trend, but a more moderate effect. By contrast, fed2T18 shows low induction of the characteristic 685-nm peak on iron depletion (Fig. 5). The 685-nm peak is thought to reflect activity of chlorophylls bound to IsiA (16), and purified free IsiA protein shows strong emission at this wavelength (12). However, our data demonstrate that IsiA is not necessary for induction of a strong 685-nm peak. This is consistent with the findings of Wilson et al. (51), who report that isiA mutants actually exhibit a faster rise in 77 K emission spectra at 685 nm than WT on iron starvation. They attribute this to high fluorescence from disconnected phycobilisomes, which act as part of a quenching mechanism to balance the effect of the decreased PSI/PSII ratio. The variation in 685-nm signal might therefore partly reflect differences in phycobilisome dissociation between the genotypes following iron depletion. This may be related to the decrease in abundance of photosystem proteins, which is less severe than WT in fed2T18 (Fig. 7A) and fed2i (SI Appendix, Fig. S5E), but more severe in fed2T24 (Fig. 7A). As the fed2T24 line shows other signs of an exaggerated low-iron response, such as an enhanced accumulation of IsiB relative to the WT (Fig. 7A), the difference between the two truncation lines likely reflects changes in the speed or amplitude of the low-iron response at the photosystem level. This corresponds to a faster response than WT in the fed2T24 and fed2i lines, but a slower response than WT in fed2T18.
We initially conducted our growth experiments on two separate N sources, due to the use of the nitrate-inducible promoter in fed2i line (Fig. 3), and noted that, for fed2T24, the 685-nm emission phenotype is more severe when cyanobacteria are grown on nitrate than on ammonia media. It may be that perception of iron deprivation is more sensitive under nitrate growth conditions, as iron deprivation regulates expression of several genes involved specifically in nitrate assimilation (46, 47, 52). The typical induction of IsiA is also attenuated on low N (53), although differences between separate nitrogen sources were not investigated until now. In addition, two more enzymes are required for assimilation of nitrate than for assimilation of ammonium, nitrate reductase and nitrite reductase, both of which possess essential iron-containing cofactors.
There is a highly complex network of factors involved in regulating the response of cyanobacteria to low iron. These include multiple players at the level of posttranscriptional regulation, with 10 small RNAs and 62 antisense RNAs identified so far (52, 54). Our data show that Fed2 is a critical player in specific parts of this response. It will be interesting to see whether the regulatory role of Fed2 is conserved in higher plants, and, although Fed2/FdC2 appears to be the only C-terminal type Fd that is almost uniformly conserved (Fig. 1), some species contain multiple Fds with extended C termini (27) that could potentially also act as posttranslational regulators. Future studies should aim to identify the interaction partners and functional mechanism of Fed2/FdC2 proteins, which may be related to dimerization (Figs. 2A and 7B), as has been demonstrated for other iron- and redox-sensing regulators (55–57).
Materials and Methods
Strains and Culture Conditions.
A glucose-tolerant Synechocystis strain sp. PCC 6803 (58) obtained from N. Murata, National Institute for Basic Biology, Okazaki, Japan, was used as WT in this work. Cells were grown photoautotrophically on BG11_NH (NH4+ as nitrogen source) or BG11_NO (NO3− as nitrogen source) medium (59) at 30 °C under continuous illumination of 50 µE⋅m−2⋅s−1. For plate cultures, media was supplemented with 1% (wt/vol) agar. If not otherwise indicated, kanamycin was added to a final concentration of 50 µg⋅mL−1. Experiments were performed using cultures from the midlogarithmic phase (2 μg to 3 µg chlorophyll per milliliter) in BG11_NH or BG11_NO medium supplemented with the indicated amounts of CuSO4, CoCl2, ZnCl2, and ferric ammonium citrate. Escherichia coli D18 cells were grown in LB medium and supplemented with 200 µg⋅mL−1 of ampicillin or 12.5 µg⋅mL−1 of kanamycin when required.
Mutagenesis of Synechocystis Genes.
Primers used in this work are listed in SI Appendix, Table S2. If not otherwise mentioned, all restriction enzymes were purchased from Thermo Scientific. Gene disruption of fed2 was achieved by PCR amplification of a 1,293-bp fragment from the genomic DNA of Synechocystis using primers P1 and P2 (SI Appendix, Table S2). The fragment containing the fed2 gene (369 bp) and additional homologous regions (492 bp at the 5′ end and 432 bp at the 3′ end) was gel-purified and ligated into the multiple cloning site of a pJET1.2/blunt vector using the CloneJET PCR Cloning Kit (Thermo Scientific), resulting in the formation of pJET_FDC2. Following digestion with SfiI and treatment with blunting enzyme, a kanamycin resistance cassette derived from the pUK-4K vector cut with HincII was ligated into pJET_FDC2, resulting in the formation of pfed2KO (Fig. 3B), which was finally transformed into WT Synechocystis. For the nitrate responsive promoter, a 166-bp fragment upstream of the nirA operon from Synechococcus sp. strain PCC 7942 was synthesized (Eurofins Genomics) with additional SacI and AgeI restriction sites flanking the 5′ and 3′ ends, respectively. The pex-A2 vector harboring the nirA promoter was digested with KpnI and treated with blunting enzyme, followed by ligation with a kanamycin resistance cassette derived from a pUK-4K vector cut with HincII, resulting in the formation of pex-A2_NK.
In a parallel experiment, multiple cloning sites containing StuI and AgeI restriction sites were added at the 3′ (in front of the start codon) or the 5′ end (behind the stop codon) of the fed2 coding sequence via site-directed mutagenesis of pJET_FDC2 using primers P3 and P4 or P5 and P6 (SI Appendix, Table S2), respectively (Q5 Site-Directed Mutagenesis Kit; New England Biolabs). The resulting vectors pfdC2-StAge and pfdC2-StAge2 were digested with AgeI and StuI and ligated with the expression cassette containing the nirA promoter and the kanamycin selection marker derived from pex-A2_NK2 cut with the same enzymes, generating pfed2i and pfed2a (Fig. 3B), which were finally transformed into WT Synechocystis. Stepwise truncation of Fed2 was obtained by sequential site-directed mutagenesis introducing stop codons to shorten the C terminus at specific positions on the vector pfed2a with primer P7 and one of the primers P8 to P15 (SI Appendix, Table S2), resulting in the formation of plasmids pfed2trunc3 to pfed2trunc24 (Fig. 4A).
Protein Analysis.
T. elongatus fed2 was cloned from genomic DNA (kind gift of Yuichi Fujita, Nagoya University, Nagoya, Japan) into the 6XHN-tag expression vector pT3871 by in-fusion cloning, followed by purification on Talon resin (all from Clontech Laboratories). This was followed by precipitation in 25% saturation (NH4)2SO4. The His-tag was cleaved with enterokinase and removed by passing through Talon resin, followed by size-exclusion chromatography (Superose 12 10/300; GE Healthcare Bio-Sciences). Anti-cyanobacterial Fed2 was generated by injection of this protein into a rabbit (Pineda Antikörper Service). Synechocystis petF was cloned into pTRC99A, then expressed and purified basically as described previously (24).
Proteins were detected by SDS/PAGE followed by Western blot analysis with detection by alkaline phosphatase reaction. The cells were broken in 10 consecutive cycles of vortexing in the presence of 50 mM Tris Buffer (pH 7.6) and about 250 µL of glass beads (0.25- to 0.3-mm diameter) for 1 min at 4 °C, each cycle followed by 30 s resting at 4 °C. Cell debris was removed by centrifugation, and the supernatant was recovered. Primary antibodies were 1:40,000 dilutions of antisera against TeFed2, maize Fd1 (PetF), PsbA and PsaD (both raised against synthetic peptides; Agrisera), Synechocystis CP43’ (IsiA), and Synechocystis Fld (IsiB, kind gift of Néstor Carrillo, Rosario University, Rosario, Argentina). Specificity of the anti-Fed2 antiserum was confirmed by comparing detection of Fed2 with 100 times more PetF, the most abundant Fd in Synechocystis (SI Appendix, Fig. S8). The rate of electron transfer between FNR and Fds was followed by change in absorption at 550 nm in a cytochrome c-linked assay, as described previously (24).
Redox Potentiometry.
The spectroelectrochemical measurements were carried out with a “honeycomb” gold working electrode, gold counter electrode integrated on the same card (AKSTCKIT3; PINE Instruments), using a Ag/AgCl/KCl 3 M reference electrode. All solutions were in 100 mM Tris buffer with 150 mM NaCl (pH = 7.5). The electrolyte contained the following mediators at 50 mM concentration: A1, anthraquinone-2,6-disulfonic acid (Fluka; >97%), E0′ = −(419 ± 5) mV vs. Ag/AgCl; H1, 2-hydroxy-1,4-naphthoquinone (Alfa Aesar; >98%), E0′ = −(435 ± 5) mV vs. Ag/AgCl; H2, 5-hydroxy-1,4-naphthoquinone (Alfa Aesar; >99%), E0′ = −(222 ± 5) mV vs. Ag/AgCl; D1, 1,1′-dimethyl-4,4′-bipyridinium chloride (Riedel-de Haën, 99.9%), E0′ = −(653 ± 5) mV vs. Ag/AgCl; and D2, 1,1′-dibenzyl-4,4′-bipyridinium bromide (synthesized according to standard methods), E0′ = −(508 ± 5) mV vs. Ag/AgCl.
When present, Fed2 concentration was 750 µM. The potentiostat was a PAR 173/179, and the spectrometer was HP 8453 controlled by the software Agilent Chemstation. All measurements were performed under argon. Potential-dependent spectra were taken from 0 mV to −700 mV (vs. Ag/AgCl/KCl 3 M) using reasonable small steps. Before the absorbance measurement, the electrode was potentiostated at each potential for 4 min. The reversibility was examined by taking spectra with potential steps in the reverse direction. To get the precise redox potential of the protein, the spectra of the mediators without protein was subtracted from that of mediators with protein. The titration curve was calculated from the change at 437 nm (largest Fe-II/III response). The resulting midpoint value vs. Ag/AgCl (−465 ± 3 mV) was corrected for the NHE reference by addition of +222 mV to yield 243 ± 3 mV.
The 77 K Fluorescence Emission Spectra.
The 77 K fluorescence spectroscopy was performed as described by Ossenbühl et al. (60) using excitation wavelengths of 430 nm. Synechocystis cultures were concentrated to a chlorophyll a content of 2 µg⋅mL−1 to 3 µg⋅mL−1. Emission spectra were recorded with 1-nm step width. Five consecutive technical replicates were recorded and averaged. Emission spectra were normalized for the characteristic PSI peak emitted at 725 nm.
Electron Microscopy.
Synechocystis cells were fixed with 2% glutaraldehyde buffered with 0.1 M phosphate. Sample staining of cells embedded in low-melting agarose was performed for 3.5 h with 1% OsO4, followed by consecutive dehydration of the sample in serial ethanol dilutions: 5 min in 30%, 5 min in 50%, 10 min in 70%, 10 min in 80%, and 10 min in 95% ethanol. Final dehydration was obtained by sample treatment in 95% and 100% ethanol, each for 15 min. For infiltration, the sample was transferred to a 1:1 mixture of propylene oxide and ethanol for 10 min, followed by two incubation steps in propylene oxide for 5 min and one final infiltration step with a 1:3 mixture of propylene oxide and Epon [1:1 mixture A/B plus 1.5% 2,2-dimethoxypropane (DMP) 30]. For embedding, the sample was washed three times with Epon (1:1 mixture A/B plus 1.5% DMP) followed by polymerization at 65 °C for 72 h. The sample was sectioned at the Ultramicrotome (Ultracut UCT; Leica) and stained for 30 min with 2% uranyl acetate and then for 20 min with lead citrate. The TEM images were recorded with the EM 902 from Zeiss.
ICP-MS.
Cells were collected at midlogarithmic phase before rapidly washing two times in either chelex-treated water or 5 mM EDTA. Cells were then suspended in 200 μL of chelex-treated H2O and mixed with 800 μL of 65% Suprapur HNO3 (Merck Millipore) to digest, before dilution (1 in 5) with 2.5% Suprapur HNO3 for analysis. Quantitative analysis of metal content was determined using an XSERIES-2 ICP mass spectrometer (Thermo Fisher Scientific) following calibration with elemental standards that were matrix-matched to the sample. Internal standards (beryllium, silver, and indium) were used to correct for any variations in analytical performance. Mean and SD values determined from three biological replicates.
Northern Blot Analysis.
Cells grown under iron-replete conditions (time point 0) were washed two times with iron-free medium and further grown under iron depletion for 24 and 48 h (time points 24 and 48). RNA extraction, blotting, and hybridization were performed as described previously (61), and RNA was hybridized to a radioactively labeled isiA DNA probe (including the 5′ UTR region) using the Rediprime II DNA labeling system (GE Healthcare Life Sciences). As a control, the same blot was hybridized with a riboprobe against the rnpB RNA using the T7 polymerase Maxiscript kit (Ambion). Primers used to generate the isiA (P16 and P17), rnpB (P18 and P19), and isiB (P20 and P21) probes are given in SI Appendix, Table S2.
Supplementary Material
Acknowledgments
We thank Professor Conrad Mullineaux (Queen Mary University of London) for helpful discussions and insight, Giulia Mastoianni (Queen Mary University of London) for excellent technical support in the production and interpretation of TEM images, and Werner Bigott (University of Freiburg) for isolation of RNA. We thank Néstor Carrillo (Rosario University) for the kind gift of the antibody against IsiB. We acknowledge funding from the Deutsche Forschungsgemeinschaft Grant HA5921/1-1 (to G.T.H.) for some of this work.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810379115/-/DCSupplemental.
References
- 1.Lyons TW, Reinhard CT, Planavsky NJ. The rise of oxygen in Earth’s early ocean and atmosphere. Nature. 2014;506:307–315. doi: 10.1038/nature13068. [DOI] [PubMed] [Google Scholar]
- 2.Norman L, Cabanesa DJ, Blanco-Ameijeiras S, Moisset SA, Hassler CS. Iron biogeochemistry in aquatic systems: From source to bioavailability. Chimia (Aarau) 2014;68:764–771. doi: 10.2533/chimia.2014.764. [DOI] [PubMed] [Google Scholar]
- 3.Margulis L. Symbiotic theory of the origin of eukaryotic organelles; criteria for proof. Symp Soc Exp Biol, 1975:21–38. [PubMed] [Google Scholar]
- 4.Falkowski PG, Barber RT, Smetacek V. Biogeochemical controls and feedbacks on ocean primary production. Science. 1998;281:200–207. doi: 10.1126/science.281.5374.200. [DOI] [PubMed] [Google Scholar]
- 5.Coale KH, et al. A massive phytoplankton bloom induced by an ecosystem-scale iron fertilization experiment in the equatorial Pacific Ocean. Nature. 1996;383:495–501. doi: 10.1038/383495a0. [DOI] [PubMed] [Google Scholar]
- 6.Martin JH, et al. Testing the iron hypothesis in ecosystems of the equatorial Pacific Ocean. Nature. 1994;371:123–129. [Google Scholar]
- 7.Kehrer JP. The Haber-Weiss reaction and mechanisms of toxicity. Toxicology. 2000;149:43–50. doi: 10.1016/s0300-483x(00)00231-6. [DOI] [PubMed] [Google Scholar]
- 8.Fraser JM, et al. Photophysiological and photosynthetic complex changes during iron starvation in Synechocystis sp. PCC 6803 and Synechococcus elongatus PCC 7942. PLoS One. 2013;8:e59861. doi: 10.1371/journal.pone.0059861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laudenbach DE, Straus NA. Characterization of a cyanobacterial iron stress-induced gene similar to psbC. J Bacteriol. 1988;170:5018–5026. doi: 10.1128/jb.170.11.5018-5026.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hutber GN, Hutson KG, Rogers LJ. Effect of iron dificiency on levels of two ferredoxins and flavodoxin in a cyanobacterium. FEMS Microbiol Lett. 1977;1:193–196. [Google Scholar]
- 11.Öquist G. Iron deficiency in the blue-green alga Anacystis nidulans: Fluorescence and absorption spectra recorded at 77°K. Physiol Plant. 1974;31:55–58. [Google Scholar]
- 12.Bibby TS, Nield J, Barber J. Iron deficiency induces the formation of an antenna ring around trimeric photosystem I in cyanobacteria. Nature. 2001;412:743–745. doi: 10.1038/35089098. [DOI] [PubMed] [Google Scholar]
- 13.van der Weij-de Wit CD, et al. Fluorescence quenching of IsiA in early stage of iron deficiency and at cryogenic temperatures. Biochim Biophys Acta. 2007;1767:1393–1400. doi: 10.1016/j.bbabio.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 14.Sun J, Golbeck JH. The presence of the isiA-PSI supercomplex leads to enhanced photosystem I electron throughput in iron-starved cells of Synechococcus sp. PCC 7002. J Phys Chem B. 2015;119:13549–13559. doi: 10.1021/acs.jpcb.5b02176. [DOI] [PubMed] [Google Scholar]
- 15.Ma F, et al. Dynamic changes of isiA-containing complexes during long-term iron deficiency in Synechocystis sp. PCC 6803. Mol Plant. 2017;10:143–154. doi: 10.1016/j.molp.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 16.Burnap RL, Troyan T, Sherman LA. The highly abundant chlorophyll-protein complex of iron-deficient Synechococcus sp. PCC7942 (CP43′) is encoded by the isiA gene. Plant Physiol. 1993;103:893–902. doi: 10.1104/pp.103.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrews SC, Robinson AK, Rodríguez-Quiñones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 18.González A, Bes MT, Valladares A, Peleato ML, Fillat MF. FurA is the master regulator of iron homeostasis and modulates the expression of tetrapyrrole biosynthesis genes in Anabaena sp. PCC 7120. Environ Microbiol. 2012;14:3175–3187. doi: 10.1111/j.1462-2920.2012.02897.x. [DOI] [PubMed] [Google Scholar]
- 19.Kunert A, Vinnemeier J, Erdmann N, Hagemann M. Repression by Fur is not the main mechanism controlling the iron-inducible isiAB operon in the cyanobacterium Synechocystis sp. PCC 6803. FEMS Microbiol Lett. 2003;227:255–262. doi: 10.1016/S0378-1097(03)00689-X. [DOI] [PubMed] [Google Scholar]
- 20.Dühring U, Axmann IM, Hess WR, Wilde A. An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc Natl Acad Sci USA. 2006;103:7054–7058. doi: 10.1073/pnas.0600927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nodop A, et al. Transcript profiling reveals new insights into the acclimation of the mesophilic fresh-water cyanobacterium Synechococcus elongatus PCC 7942 to iron starvation. Plant Physiol. 2008;147:747–763. doi: 10.1104/pp.107.114058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cassier-Chauvat C, Chauvat F. Function and regulation of ferredoxins in the cyanobacterium, Synechocystis PCC6803: Recent advances. Life (Basel) 2014;4:666–680. doi: 10.3390/life4040666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanke G, Mulo P. Plant type ferredoxins and ferredoxin-dependent metabolism. Plant Cell Environ. 2013;36:1071–1084. doi: 10.1111/pce.12046. [DOI] [PubMed] [Google Scholar]
- 24.Hanke GT, Kimata-Ariga Y, Taniguchi I, Hase T. A post genomic characterization of Arabidopsis ferredoxins. Plant Physiol. 2004;134:255–264. doi: 10.1104/pp.103.032755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li C, et al. Mutation of FdC2 gene encoding a ferredoxin-like protein with C-terminal extension causes yellow-green leaf phenotype in rice. Plant Sci. 2015;238:127–134. doi: 10.1016/j.plantsci.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 26.Zhao J, et al. Functional inactivation of putative photosynthetic electron acceptor ferredoxin C2 (FdC2) induces delayed heading date and decreased photosynthetic rate in rice. PLoS One. 2015;10:e0143361. doi: 10.1371/journal.pone.0143361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terauchi AM, et al. Pattern of expression and substrate specificity of chloroplast ferredoxins from Chlamydomonas reinhardtii. J Biol Chem. 2009;284:25867–25878. doi: 10.1074/jbc.M109.023622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolton M, et al. Plastidic redox switches: Ferredoxins as novel RNA-binding proteins. J Endocytobiosis Cell Res. 2011;21:1–18. [Google Scholar]
- 29.Karpowicz SJ, Prochnik SE, Grossman AR, Merchant SS. The GreenCut2 resource, a phylogenomically derived inventory of proteins specific to the plant lineage. J Biol Chem. 2011;286:21427–21439. doi: 10.1074/jbc.M111.233734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gou P, et al. Higher order structure contributes to specific differences in redox potential and electron transfer efficiency of root and leaf ferredoxins. Biochemistry. 2006;45:14389–14396. doi: 10.1021/bi061779d. [DOI] [PubMed] [Google Scholar]
- 31.Lelong C, Sétif P, Lagoutte B, Bottin H. Identification of the amino acids involved in the functional interaction between photosystem I and ferredoxin from Synechocystis sp. PCC 6803 by chemical cross-linking. J Biol Chem. 1994;269:10034–10039. [PubMed] [Google Scholar]
- 32.Bottin H, Lagoutte B. Ferredoxin and flavodoxin from the cyanobacterium Synechocystis sp PCC 6803. Biochim Biophys Acta. 1992;1101:48–56. doi: 10.1016/0167-4838(92)90465-p. [DOI] [PubMed] [Google Scholar]
- 33.Sancho J, Peleato ML, Gomez-Moreno C, Edmondson DE. Purification and properties of ferredoxin-NADP+ oxidoreductase from the nitrogen-fixing cyanobacteria Anabaena variabilis. Arch Biochem Biophys. 1988;260:200–207. doi: 10.1016/0003-9861(88)90441-9. [DOI] [PubMed] [Google Scholar]
- 34.Hirasawa M, Tollin G, Salamon Z, Knaff DB. Transient kinetic and oxidation-reduction studies of spinach ferredoxin:nitrite oxidoreductase. Biochim Biophys Acta. 1994;1185:336–345. doi: 10.1016/0005-2728(94)90249-6. [DOI] [PubMed] [Google Scholar]
- 35.Ravasio S, et al. Properties of the recombinant ferredoxin-dependent glutamate synthase of Synechocystis PCC6803. Comparison with the Azospirillum brasilense NADPH-dependent enzyme and its isolated alpha subunit. Biochemistry. 2002;41:8120–8133. doi: 10.1021/bi020083r. [DOI] [PubMed] [Google Scholar]
- 36.Mitschke J, et al. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc Natl Acad Sci USA. 2011;108:2124–2129. doi: 10.1073/pnas.1015154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang FK, Latifi A, Chen WL, Zhang CC. The inositol monophosphatase All2917 (IMPA1) is involved in osmotic adaptation in Anabaena sp. PCC7120. Environ Microbiol Rep. 2012;4:622–632. doi: 10.1111/j.1758-2229.2012.00388.x. [DOI] [PubMed] [Google Scholar]
- 38.Kuchmina E, Wallner T, Kryazhov S, Zinchenko VV, Wilde A. An expression system for regulated protein production in Synechocystis sp. PCC 6803 and its application for construction of a conditional knockout of the ferrochelatase enzyme. J Biotechnol. 2012;162:75–80. doi: 10.1016/j.jbiotec.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 39.Maeda S, Kawaguchi Y, Ohe TA, Omata T. cis-acting sequences required for NtcB-dependent, nitrite-responsive positive regulation of the nitrate assimilation operon in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1998;180:4080–4088. doi: 10.1128/jb.180.16.4080-4088.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kranzler C, Lis H, Shaked Y, Keren N. The role of reduction in iron uptake processes in a unicellular, planktonic cyanobacterium. Environ Microbiol. 2011;13:2990–2999. doi: 10.1111/j.1462-2920.2011.02572.x. [DOI] [PubMed] [Google Scholar]
- 41.Badarau A, et al. FutA2 is a ferric binding protein from Synechocystis PCC 6803. J Biol Chem. 2008;283:12520–12527. doi: 10.1074/jbc.M709907200. [DOI] [PubMed] [Google Scholar]
- 42.Bradley RW, Bombelli P, Lea-Smith DJ, Howe CJ. Terminal oxidase mutants of the cyanobacterium Synechocystis sp. PCC 6803 show increased electrogenic activity in biological photo-voltaic systems. Phys Chem Chem Phys. 2013;15:13611–13618. doi: 10.1039/c3cp52438h. [DOI] [PubMed] [Google Scholar]
- 43.Katoh H, Hagino N, Ogawa T. Iron-binding activity of FutA1 subunit of an ABC-type iron transporter in the cyanobacterium Synechocystis sp. Strain PCC 6803. Plant Cell Physiol. 2001;42:823–827. doi: 10.1093/pcp/pce106. [DOI] [PubMed] [Google Scholar]
- 44.Thorne RJ, Schneider K, Hu H, Cameron PJ. Iron reduction by the cyanobacterium Synechocystis sp. PCC 6803. Bioelectrochemistry. 2015;105:103–109. doi: 10.1016/j.bioelechem.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 45.Leonhardt K, Straus NA. An iron stress operon involved in photosynthetic electron transport in the marine cyanobacterium Synechococcus sp. PCC 7002. J Gen Microbiol. 1992;138:1613–1621. doi: 10.1099/00221287-138-8-1613. [DOI] [PubMed] [Google Scholar]
- 46.Saxena RK, Pandey PK, Bisen PS. Physiological and biochemical alterations in Anabaena 7120 under iron stress. Indian J Exp Biol. 2002;40:594–599. [PubMed] [Google Scholar]
- 47.Esen M, Ozturk Urek R. Ammonium nitrate and iron nutrition effects on some nitrogen assimilation enzymes and metabolites in Spirulina platensis. Biotechnol Appl Biochem. 2015;62:275–286. doi: 10.1002/bab.1268. [DOI] [PubMed] [Google Scholar]
- 48.Schulze K, López DA, Tillich UM, Frohme M. A simple viability analysis for unicellular cyanobacteria using a new autofluorescence assay, automated microscopy, and ImageJ. BMC Biotechnol. 2011;11:118. doi: 10.1186/1472-6750-11-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vinnemeier J, Kunert A, Hagemann M. Transcriptional analysis of the isiAB operon in salt-stressed cells of the cyanobacterium Synechocystis sp. PCC 6803. FEMS Microbiol Lett. 1998;169:323–330. doi: 10.1111/j.1574-6968.1998.tb13336.x. [DOI] [PubMed] [Google Scholar]
- 50.Legewie S, Dienst D, Wilde A, Herzel H, Axmann IM. Small RNAs establish delays and temporal thresholds in gene expression. Biophys J. 2008;95:3232–3238. doi: 10.1529/biophysj.108.133819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson A, Boulay C, Wilde A, Kerfeld CA, Kirilovsky D. Light-induced energy dissipation in iron-starved cyanobacteria: Roles of OCP and IsiA proteins. Plant Cell. 2007;19:656–672. doi: 10.1105/tpc.106.045351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hernández-Prieto MA, et al. Iron deprivation in Synechocystis: Inference of pathways, non-coding RNAs, and regulatory elements from comprehensive expression profiling. G3 (Bethesda) 2012;2:1475–1495. doi: 10.1534/g3.112.003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schrader PS, Milligan AJ, Behrenfeld MJ. Surplus photosynthetic antennae complexes underlie diagnostics of iron limitation in a cyanobacterium. PLoS One. 2011;6:e18753. doi: 10.1371/journal.pone.0018753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Georg J, et al. Acclimation of oxygenic photosynthesis to iron starvation is controlled by the sRNA IsaR1. Curr Biol. 2017;27:1425–1436.e7. doi: 10.1016/j.cub.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 55.Khoroshilova N, Popescu C, Münck E, Beinert H, Kiley PJ. Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity. Proc Natl Acad Sci USA. 1997;94:6087–6092. doi: 10.1073/pnas.94.12.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pecqueur L, et al. Structural changes of Escherichia coli ferric uptake regulator during metal-dependent dimerization and activation explored by NMR and X-ray crystallography. J Biol Chem. 2006;281:21286–21295. doi: 10.1074/jbc.M601278200. [DOI] [PubMed] [Google Scholar]
- 57.Shen G, et al. SufR coordinates two [4Fe-4S]2+, 1+ clusters and functions as a transcriptional repressor of the sufBCDS operon and an autoregulator of sufR in cyanobacteria. J Biol Chem. 2007;282:31909–31919. doi: 10.1074/jbc.M705554200. [DOI] [PubMed] [Google Scholar]
- 58.Williams JGK. Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol. 1988;167:766–778. [Google Scholar]
- 59.Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology. 1979;111:1–61. [Google Scholar]
- 60.Ossenbühl F, Inaba-Sulpice M, Meurer J, Soll J, Eichacker LA. The Synechocystis sp PCC 6803 oxa1 homolog is essential for membrane integration of reaction center precursor protein pD1. Plant Cell. 2006;18:2236–2246. doi: 10.1105/tpc.106.043646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.David C, Schmid A, Adrian L, Wilde A, Bühler K. Production of 1,2-propanediol in photoautotrophic Synechocystis is linked to glycogen turn-over. Biotechnol Bioeng. 2018;115:300–311. doi: 10.1002/bit.26468. [DOI] [PubMed] [Google Scholar]
- 62.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.