Significance
Fibrous sheath interacting protein 1 (FSIP1) has been identified as a poor prognostic indicator of breast cancer. Here, we report that FSIP1 positively regulates the proliferation and invasion of triple-negative breast cancer (TNBC) cells, and its silencing promotes drug resistance by inducing autophagy, by reducing mitochondrial biogenesis, and by enhancing AMP-activated protein kinase activation. Collectively, our findings may aid in the design of therapies for advanced TNBC.
Keywords: chemoresistance, FSIP1 knockdown, AMP-activated protein kinase, ULK1 serine/threonine protein kinase
Abstract
Fibrous sheath interacting protein 1 (FSIP1) is a cancer antigen expressed in the majority of breast cancer tissues and is associated with poor prognosis. However, the role of FSIP1 in the progression and drug sensitivity of triple-negative breast cancer (TNBC) has not been explored. Here, we show that FSIP1 deficiency by shRNA-mediated knockdown or CRISPR-Cas9–mediated knockout significantly inhibits the proliferation and invasion of TNBC cells and impairs chemotherapy-induced growth inhibition in vivo. Computational modeling predicted that FSIP1 binds to ULK1, and this was established by coimmunoprecipitation. FSIP1 deficiency promoted autophagy, enhanced AMP-activated protein kinase (AMPK) signaling, and decreased mechanistic target of rapamycin (mTOR) and Wnt/β-catenin activity. In contrast, knockdown of AMPK or inhibition of autophagy restored the sensitivity to chemotherapy drugs in TNBC cells. Our findings uncover a role of FSIP1 as well as mechanisms underlying FSIP1 action in drug sensitivity and may, therefore, aid in design of TNBC therapies.
Breast cancer is the most prevalent malignancy in women and its incidence is increasing worldwide (1). Although patients with early diagnosed breast cancer can be cured, those at an advanced stage or with recurrent cancer usually depend on hormonal therapy, chemotherapy, radiation therapy, and various target therapies (2). While patients with luminal or HER2+ breast cancers can be treated with hormone and targeted therapies, patients with basal-like cancer, such as triple-negative breast cancer (TNBC), usually depend on chemotherapy drugs such as docetaxel, doxorubicin, and others (3, 4). However, these patients often develop resistance to chemotherapy with a downhill course (5, 6). Hence, discovery of the mechanisms underlying the chemoresistance of TNBC will be of significance.
Autophagy is a process through which cytoplasmic components are degraded in lysosomes to maintain cellular homeostasis (7, 8). Many stimuli activate the serine/threonine protein kinase ULK1, which interacts with autophagy-related protein 13 (ATG13), ATG101, and Rb1-inducible coiled-coil protein 1 (FIP200) in a complex to initiate the autophagic process by activating the Beclin 1–class III phosphatidylinositol 3-kinase (PI3KC3) complex (9, 10). Thereafter, microtubule-associated protein 1A/1B light chain 3 (LC3) is recruited to induce the formation of autophagosomes, in which the LC3 on the autophagosomal inner membrane is converted to LC3-II by conjugating to phosphatidylethanolamine (11). LC3-II can recruit sequestosome 1 (SQSTM1), which promotes the degradation of cargo proteins (12). Autophagy is positively regulated by AMP-activated protein kinase (AMPK), particularly in stress conditions, but is regulated negatively by the mechanistic target of rapamycin (mTOR) C1 (9). Previous studies have shown that enhanced autophagy is associated with drug resistance in several types of cancers, including TNBC (13, 14). Although enhanced autophagy supports the survival of tumor cells, particularly in a condition of hypoxia and nutrient deprivation by sustaining tumor metabolism, aberrant autophagy can also lead to apoptosis, which enhances drug sensitivity (15). However, how enhanced autophagy promotes resistance to chemotherapeutic reagents and what factors regulate autophagy in TNBC remain poorly understood.
Fibrous sheath interacting protein 1 (FSIP1) is a cancer/testis antigen (16) expressed in the majority of breast cancers, bladder cancers, and nonsmall cell lung cancer (17–19). Our previous findings and those of others show that high FSIP1 expression is associated with increased invasiveness and poor prognosis in breast cancer (19, 20). We found that FSIP1 interacts with HER2, and that FSIP1 silencing inhibits the proliferation and invasiveness of ER+ or HER2+ breast cancer cells (21). Such data suggest that FSIP1 may be a potential therapeutic target for ER+ and HER2+ breast cancers. Although FSIP1 is expressed in TNBC, the role of FSIP1 in the proliferation and migration of TNBC has not been explored. Furthermore, it is still unclear which cellular protein(s) bind to FSIP1 and how the interaction affects autophagy and sensitivity to common chemotherapeutic drugs, such as docetaxel and doxorubicin, in TNBC.
In this study, we employed TNBC MDA-MB-231 and MDA-MB-468 cells to explore the impact of FSIP1 deficiency on their proliferation, migration, and drug sensitivity. Furthermore, we investigated the potential mechanisms underlying the action of FSIP1 in regulating autophagy and drug resistance. Our data indicate that FSIP1 silencing inhibits the proliferation and migration of TNBC cells and the growth of implanted tumors but enhances drug resistance to docetaxel, doxorubicin, and other chemotherapeutic drugs by promoting autophagy.
Results
FSIP1 Deficiency Inhibits Proliferation and Migration of Breast Cancer Cells.
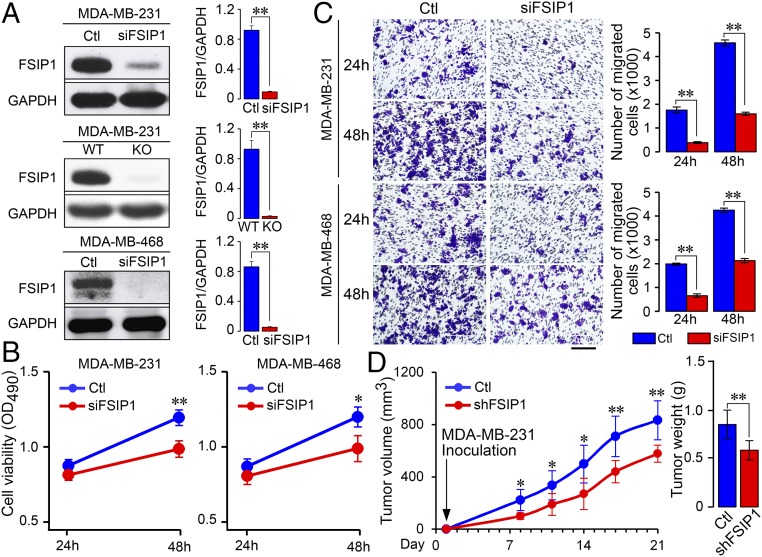
We showed previously that FSIP1 silencing inhibited the proliferation, migration, and invasion of HER2+ breast cancer cells (21). To further determine the function of FSIP1, MDA-MB-231 and MDA-MB-468 cells were transfected with FSIP1-specific (siFSIP1) or control siRNA, and their proliferation and migration were determined by 3-(4, 5-dimethylthiazol-2-yl)-5 (3-carboxymethonyphenol)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) and transwell migration assays. Transfection with siFSIP1 reduced FSIP1 expression by 90% in both cell lines (Fig. 1A). We also generated an FSIP1-knockout (KO) MDA-MB-231 cell line using CRISPR-Cas9 technology (Fig. 1A). FSIP1 silencing decreased viability of both MDA-MB-231 and MDA-MB-468 cells at 48 h posttransfection (Fig. 1B) and reduced the numbers of migrated cells at 24 and 48 h (Fig. 1C). To further understand the role of FSIP1 in the progression of TNBC in vivo, we established stable FSIP1-silenced (shFSIP1) or control MDA-MB-231 cells. FSIP1 silencing reduced the dynamic growth of MDA-MB-231 tumors in mice (Fig. 1D). Collectively, these data indicate that FSIP1 promotes the progression of TNBC.
Fig. 1.
FSIP1 deficiency inhibits proliferation and migration of TNBC cells and TNBC tumor growth in mice. TNBC MDA-MB-231 and MDA-MB-468 cells were transfected with negative control (Ctl) or siFSIP1 (n = 3 per group). Furthermore, FSIP1-KO MDA-MB-231 cells were established using CRISPR-Cas9 technology. (A) FSIP1 silencing or FSIP1 KO reduced FSIP1 expression (Western blot). FSIP1 deficiency decreased cell proliferation (B) and migration (C) compared with Ctl. (Scale bar in C: 100 µm.) (D) To study the effect of FSIP1 silencing in vivo, BALB/c nude mice were implanted with MDA-MB-231 cells that were transduced with lentivirus expressing shFSIP1 or Ctl shRNA (n = 5 to 8 per group). Tumor volumes and weights were lower in mice with FSIP1-silenced xenotransplant at day 21. Mean ± SEM; one-way ANOVA followed by post hoc Newman-Keuls test; *P < 0.05, **P < 0.01.
FSIP1 Deficiency Reduces the Sensitivity to Chemotherapeutic Drugs in TNBC.
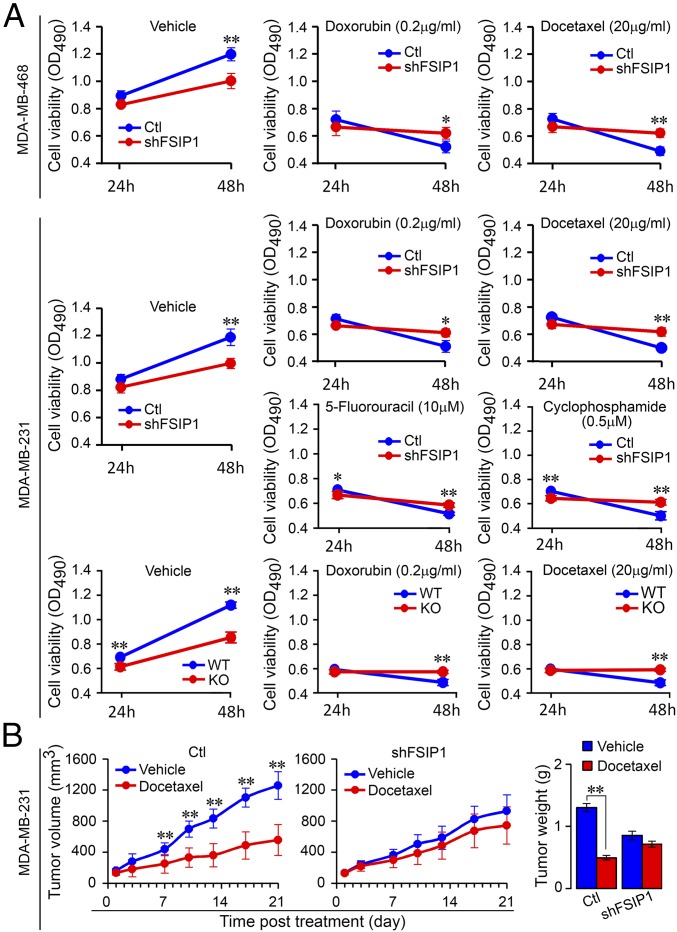
For patients with advanced TNBC, chemotherapies are widely used for treatment even after surgical removal of local tumors. However, the efficacy of chemotherapy varies due to primary resistance to chemotherapeutic reagents (22). To explore the potential role of FSIP1 in regulating drug sensitivity, FSIP1-silenced or control MDA-MB-231 and MDA-MB-468 cells were treated with vehicle, doxorubicin (0.2 µg/mL), or docetaxel (20 µg/mL), and cell viability was determined at 24 and 48 h (MTS assay). FSIP1 silencing significantly reduced cell viability at 48 h (Fig. 2A, Left; compare with Fig. 1B). Furthermore, while control cells were sensitive to chemotherapeutic reagents, FSIP1-silenced and FSIP1-KO cells were resistant, with a significantly higher viability at 48 h despite their low proliferation capacity (Fig. 2A, Middle and Right). Similarly, FSIP1-silenced MDA-MB-231 cells were resistant to 5-fluorouracil (10 µM) and cyclophosphamide (500 nM) (Fig. 2A). Consistent with the in vitro data, docetaxel reduced the size of implanted control, but not FSIP1-silenced, MDA-MB-231 tumors and decreased the tumor weights in mice (Fig. 2B).
Fig. 2.
FSIP1 deficiency promotes chemoresistance in TNBC. (A) MDA-MB-231 and MDA-MB-468 cells expressing shFSIP1 or control (Ctl) shRNA, as well as FSIP1-KO MDA-MB-231 cells, were cultured overnight in 96-well plates (1 × 103 cells per well) and treated with vehicle, doxorubicin (0.2 µg/mL), or docetaxel (20 µg/mL) (n = 3 per group). MDA-MB-231 cells expressing shFSIP1 or Ctl were also treated with 5-fluorouracil (10 µM) or cyclophosphamide (500 nM). Cell viability was determined after 24 or 48 h (MTS assay). (B) BALB/c nude mice were injected with 1 × 106 Ctl or shFSIP1 MDA-MB-231 cells into their flanks (n = 8 per group). When s.c. tumors grew to 100 to 200 mm3, the mice were randomized and injected i.p. with vehicle or docetaxel (10 mg/kg) on days 1, 7, and 14. Tumor sizes were measured twice a week. On day 21, mice were killed and tumors were dissected and weighed. Docetaxel reduced the size and weight of Ctl, but not FSIP1-silenced, MDA-MB-231 tumors. Mean ± SEM; one-way ANOVA followed by post hoc Newman-Keuls test; *P < 0.05, **P < 0.01.
FSIP1 Interacts with ULK1 in TNBC Cells.
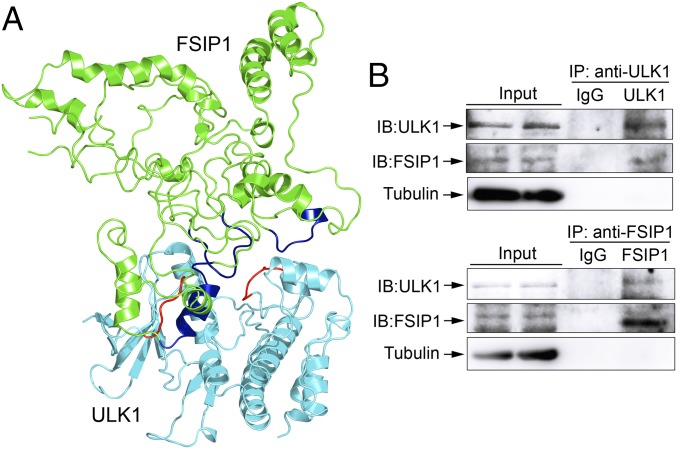
Docetaxel binds to β-tubulin through hydrophobic and van der Waals interactions to stabilize microtubules and prevent their depolymerization (23–25). The lower sensitivity of FSIP1-deficient MDA-MB-231 cells to docetaxel indicates a dynamic instability and catastrophes of microtubules. This dynamic instability, together with the reduced proliferation, suggests that FSIP1 silencing may promote autophagy (26), which is associated with drug resistance in breast cancers (27, 28). It is well known that the ULK1 (ATG1) complex is crucial for the initiation of autophagy (29). Accordingly, we hypothesized that FSIP1 may interact with ULK1 to inhibit autophagy in TNBC cells. To test the hypothesis, we employed protein–protein docking technology to evaluate the potential direct interaction of FSIP1 F2 domain (amino acids 191 to 581) with ULK1 using the ZDOCK software. Three regions on the FSIP1 F2 domain (helix: Ser479-Asn492; loop: Tyr310-Gln316; loop: Pro541-Leu549) were predicted to interact with ULK1 (loop: Ile22-Ala26; loop: Ala179-Ser184) through hydrophobic interactions (Fig. 3A). To confirm their interaction, we performed coimmunoprecipitation and Western blotting in MDA-MB-231 cells. While control IgG did not precipitate any protein of interest, the immune complex precipitated by anti-ULK1 contained FSIP1 and that precipitated by anti-FSIP1 contained ULK1 (Fig. 3B).
Fig. 3.
FSIP1 interacts with ULK1 in MDA-MB-231 cells. The potential interaction of FSIP1 F2 domain (amino acids 191 to 581) with ULK1 (Protein Data Bank ID code 4WNO) was predicted by the ZDOCK program, based on the modeled FSIP1 F2 domain using the ab initio modeling protocol implemented in the ROSETTA software. (A) Ribbon diagram of the potential interactions was depicted for FSIP1 (green) and ULK1 (cyan). Three regions on FSIP1 (helix: Ser479-Asn492; loop: Tyr310-Gln316; loop: Pro541-Leu549; in blue) were predicted to interact with ULK1 (loop: Ile22-Ala26; loop: Ala179-Ser184; in red) by hydrophobic interactions. (B) MDA-MB-231 cell lysates were incubated with anti-FSIP1 or anti-ULK1 antibody. The complexes were immunoprecipitated (IP) by protein G microbeads and resolved by SDS/PAGE, followed by immunoblotting (IB) using anti-FSIP1, anti-ULK1, or control anti-tubulin antibody. Data are representative images from two separate experiments.
FSIP1 Deficiency Promotes Autophagy in Breast Cancer.
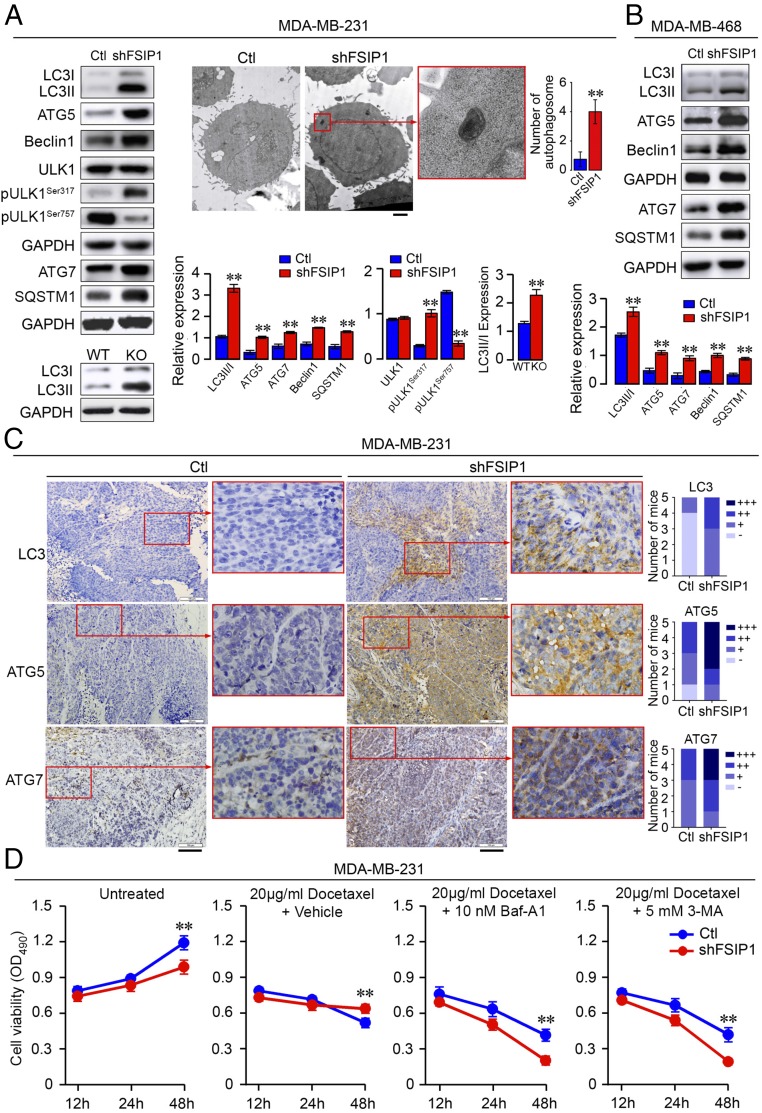
Next, we tested whether FSIP1 deficiency could promote autophagy in TNBC cells. We found that the ratios of LC3-II to LC3-I and the levels of ATG5, ATG7, Beclin 1, and SQSTM1 in FSIP1-silenced MDA-MB-231 and MDA-MB-468 cells were higher than in control cells (Fig. 4B). A similar increase in the ratio of LC3-II to LC3-I was detected in FSIP1-KO cells (Fig. 4A). FSIP1 silencing enhanced Ser317 and reduced Ser757 phosphorylation of ULK1 in MDA-MB-231 cells (Fig. 4A). Transmission electron microscopy analysis showed increased numbers of autophagosomes in FSIP1-silenced MDA-MB-231 cells (Fig. 4A). Consistent with this, LC3, ATG5, and ATG7 levels in FSIP1-silenced MDA-MB-231 tumors were higher than in control tissues (Fig. 4C).
Fig. 4.
FSIP1 deficiency enhances autophagy and drug resistance in TNBC. (A and B) Expression levels of LC3-I, LC3-II, ATG5, ATG7, Beclin 1, SQSTM1, and ULK1, as well as ULK1 Ser317 and Ser757 phosphorylation in control (Ctl) and shFSIP1 MDA-MB-231 and MDA-MB-468 cells were determined by Western blot. Shown also are LC3-I and LC3-II expression levels in Ctl and FSIP1-KO MDA-MB-231 cells. Autophagosome number was examined by transmission election microscopy (n = 12 per group). (Scale bar in A: 1 µm.) (C) Expression levels of LC3, ATG5, and ATG7 in Ctl and shFSIP1 MDA-MB-231 tumor tissues (two sections each, from Fig. 1D) were examined by immunohistochemistry and semiquantified. (Scale bars in C: 100 µm.) (D) Inhibition of autophagy sensitizes TNBC cells to docetaxel in vitro. Ctl and shFSIP1 MDA-MB-231 cells were treated with vehicle or docetaxel (20 µg/mL) in the presence or absence of Baf-A1 (10 nM) or 3-MA (5 mM). Cell viability was measured at 12, 24, and 48 h (MTS). Data are representative images or expressed as mean ± SEM; one-way ANOVA followed by post hoc Newman-Keuls test; **P < 0.01.
We further investigated how the enhanced autophagy was associated with drug resistance in FSIP1-silenced TNBC cells. FSIP1-silenced or control MDA-MB-231 cells were treated with vehicle, docetaxel (20 µg/mL), or a combination of docetaxel and either bafilomycin A1 (Baf-A1, 10 nM) or 3-methyladenine (3-MA, 5 mM). Cell viability was determined at 12, 24, and 48 h (MTS assay). While FSIP1-silenced cells were resistant to docetaxel, their sensitivity was restored by cotreatment with Baf-A1 or 3-MA (Fig. 4D). Together, these data indicated that FSIP1 deficiency promoted autophagy, which was associated with increased resistance to chemotherapy in TNBC cells.
FSIP1 Silencing Promotes AMPK Activation, Contributing to Its Drug Resistance to Chemotherapy.
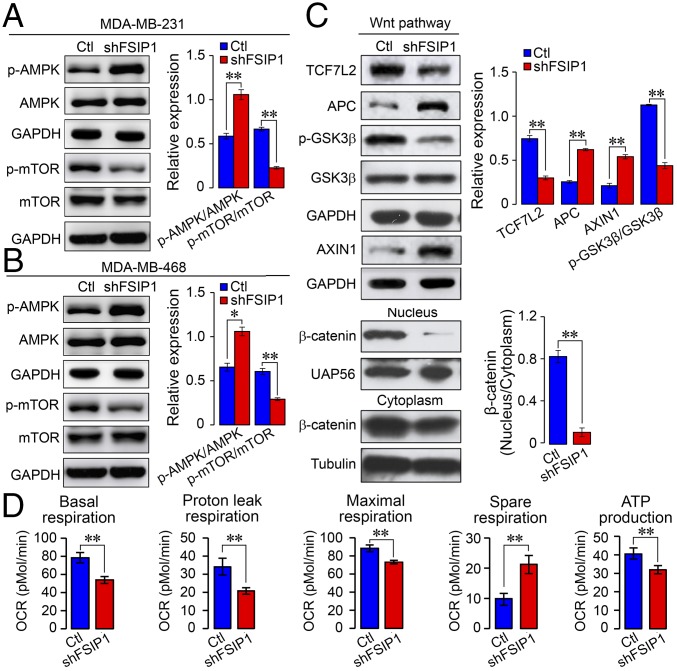
AMPK is a key sensor of stress. Its activation enhances autophagy and inhibits PI3K/protein kinase B (AKT) and mTOR signaling, which commonly cross-regulates Wnt/β-catenin and other signaling pathways (30, 31). To further understand the molecular mechanisms underlying the action of FSIP1 silencing, we studied AMPK and mTOR activation in MDA-MB-231 and MDA-MB-468 cells. While FSIP1 silencing did not change total AMPK and mTOR expression, it enhanced AMPK activation by increasing its phosphorylation and attenuated mTOR signaling by reducing its phosphorylation (Fig. 5 A and B). Furthermore, FSIP1 silencing increased expression of adenomatous polyposis coli and AXIN1, but not glycogen synthase kinase 3β (GSK-3β), and it decreased GSK-3β phosphorylation, β-catenin nuclear translocation, and transcription factor TCF7L2 expression in MDA-MB-231 cells (Fig. 5C). Thus, FSIP1 silencing enhanced AMPK activation and reduced mTOR and β-catenin activation in TNBC cells, which may contribute to drug resistance.
Fig. 5.
FSIP1 silencing enhances AMPK activation but reduces mTOR and β-catenin activation and mitochondrial biogenesis in TNBC cells. (A and B) Western blots showing total and phosphorylated AMPK and mTOR levels in control (Ctl) and shFSIP1 MDA-MB-231 and MDA-MB-468 cells. (C) Western blots showing expression of β-catenin signaling molecules in Ctl and shFSIP1 MDA-MB-231 cells. (D) Extracellular flux analysis of mitochondrial OCRs in Ctl and shFSIP1 MDA-MB-231 cells. Data are representative images or expressed as mean ± SEM; *P < 0.05, **P < 0.01.
Since activated AMPK and mTOR regulate oxygen consumption in the mitochondria, we measured the changes in oxygen consumption rates (OCRs) in control and FSIP1-silenced MDA-MB-231 cells using the Seahorse XF24 Extracellular Flux Analyzer. We found that FSIP1 silencing reduced basal, ATP-independent (proton-leaking), ((4-(trifluoromethoxy)phenyl)hydrazono)malononitrile–stimulated, and electron transport chain inhibition-related OCRs as well as ATP production but increased spare respiration capacity (Fig. 5D). Therefore, FSIP1 silencing mitigated the oxidation-related OCR in the mitochondria and reduced metabolism, which may also contribute to reduced sensitivity to chemotherapeutic drugs in TNBC cells.
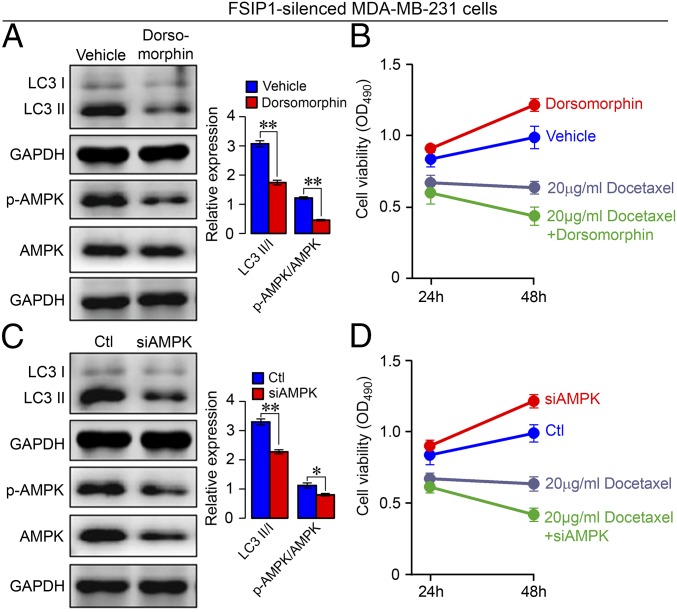
Lastly, we tested whether inhibition of AMPK activity and expression could rescue the sensitivity to chemotherapy in FSIP1-silenced MDA-MB-231 cells. We found that the AMPK inhibitor dorsomorphin decreased AMPK phosphorylation and LC3-II expression but increased the viability of FSIP1-silenced cells, indicating that inhibition of AMPK activation attenuated autophagy but promoted cell proliferation (Fig. 6 A and B). Cotreatment with docetaxel, however, reduced cell viability. Therefore, inhibition of AMPK activation attenuated autophagy and increased TNBC sensitivity to docetaxel. Similarly, transfection with AMPK-specific siRNA, but not control siRNA, reduced AMPK expression and phosphorylation as well as LC3-II expression in FSIP1-silenced MDA-MB-231 cells (Fig. 6C). AMPK knockdown alone increased cell proliferation, but cotreatment with docetaxel reduced cell viability (Fig. 6D). Collectively, these two lines of evidence demonstrate that inhibition of AMPK signaling attenuated autophagy and rescued the sensitivity to docetaxel in FSIP1-silenced MDA-MB-231 cells.
Fig. 6.
Inhibition of AMPK activity or AMPK silencing rescues docetaxel sensitivity in FSIP1-silenced MDA-MB-231 cells. shFSIP1 MDA-MB-231 cells were pretreated with vehicle or dorsomorphin (10 µM) for 2 h. For AMPK knockdown, the cells were transfected with control (Ctl) or AMPK-specific siRNA (siAMPK) for 24 h. (A and C) Western blot analysis showing levels of LC3-I, LC3-II, and AMPK activation. (B and D) MTS analysis of the sensitivity to docetaxel (20 µM) at 24 and 48 h. Data are representative images or expressed as mean ± SEM; *P < 0.05, **P < 0.01.
Discussion
FSIP1 is a cancer/testis antigen and its expression is associated with poor prognosis in breast cancer (19, 20). We have previously shown that FSIP1 promoted proliferation and invasion of ER+ and HER2+ breast cancer cells (19). In this study, we report the expression of FSIP1 in TNBC cells. FSIP1 silencing inhibited the proliferation and migration of TNBC cells in vitro and attenuated the growth of implanted tumors in vivo. These data extended previous findings (19, 21) and support the notion that FSIP1 acts as a prooncogenic molecule to promote the development and progression of breast cancers, including TNBC.
We explored the potential role and mechanisms of FSIP1 in promoting cell proliferation and invasion of TNBC cells. We found that FSIP1 deficiency enhanced autophagy in MDA-MB-231 and MDA-MB-468 cells in vitro and in implanted MDA-MB-231 tumors in vivo. These data suggest that FSIP1 may inhibit spontaneous autophagy and promote the proliferation and invasion of TNBC cells, contributing to the development and progression of TNBC. Currently, the precise role of autophagy in the growth and invasion of TNBC remains controversial. There is sufficient evidence that autophagic components such as Beclin 1 and ATG4C are tumor suppressors and that their deficiency promotes the development of malignancy (9, 32). Here, we find that the reduction of autophagy through pharmacological inhibition of AMPK or AMPK silencing promoted the proliferation of FSIP1-silenced MDA-MB-231 cells. In contrast, autophagy can recycle cytoplasmic components to maintain metabolic balance in a stress condition, which supports the survival of both healthy and tumor cells (33, 34). A previous study has shown that high levels of LC3-II expression are associated with poor prognosis in TNBC, augmenting that autophagy promotes proliferation and migration of TNBC (13). It is possible that different outcomes of autophagy-regulated proliferation of TNBC cells may depend on the degree of autophagy and the stage of TNBC development.
TNBC is very aggressive and is associated with recurrence and distant metastasis (3). Patients with recurrent and metastatic TNBC are commonly treated with chemotherapies and often develop resistance after prolonged use (5). We explored the potential role of FSIP1 in the development of resistance to docetaxel, doxorubicin, and other drugs. We found that FSIP1 silencing reduced the sensitivity to docetaxel and doxorubicin in MDA-MB-231 and MDA-MB-468 cells in vitro and to docetaxel in the implanted MDA-MB-231 tumors in vivo, which were associated with enhanced autophagy. More importantly, pharmacological inhibition of early and late stages of autophagy significantly rescued the sensitivity to docetaxel in FSIP1-silenced MDA-MB-231 cells. We surmise that the enhanced autophagy by FSIP1 silencing is crucial for the chemoresistance to docetaxel in TNBC cells. Our data were consistent with previous findings showing that inhibition of autophagy enhances the sensitivity to adriamycin and cyclophosphamide in TNBC cells (13, 14). It is well known that chemotherapies can induce autophagy to sustain metabolism and stress tolerance in cancer cells (34), and treatment with epirubicin or doxorubicin can induce autophagy by inducing HSF-1 and ATG7 expression (28, 35). Consequently, the enhanced autophagy may up-regulate permeability-glycoprotein expression to increase drug resistance in TNBC cells (36). Given that the lack of effective drugs to inhibit TNBC growth is the main cause of therapy failure and tumor relapse (4), treatment with chemotherapies together with inhibitors of autophagy may be valuable for management of patients with advanced TNBC. Therefore, our findings may aid in the design of new therapies for TNBC.
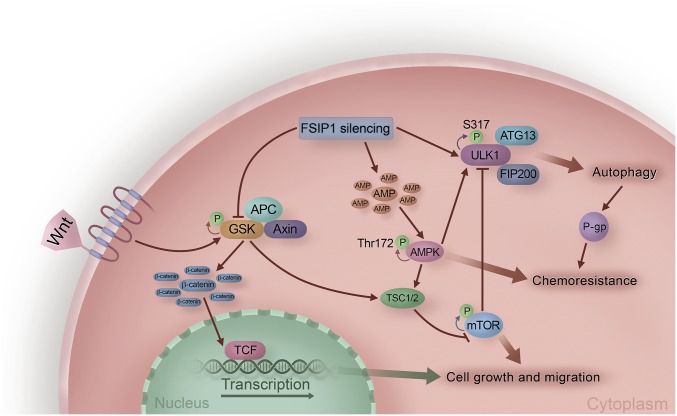
Several signaling pathways regulate autophagy. Through computational modeling, we predicted that the FSIP1 F2 domain interacts with the serine/threonine kinase ULK1, which was validated by coimmunoprecipitation experiments. ULK1, ATG13, FIP200, and ATG101 form a complex, which controls ATG9 trafficking and promotes the formation of Beclin 1–vps34 complex to drive the formation and movement of autophagosomes (37, 38). The enhanced ULK1 expression and autophagy by FSIP1 silencing indicated that the basal levels of autophagy were dependent on ULK1 expression and activity in TNBC cells. The enhanced autophagy by FSIP1 silencing suggests that FSIP1 may interfere with the ULK1 complex and downstream autophagosome formation (Fig. 7). Alternatively, this interaction may affect ULK1 phosphorylation by preventing AMPK-mediated phosphorylation (Fig. 7). Indeed, we found higher levels of ULK1 Ser317 phosphorylation in FSIP1-silenced cells. Thus, our data uncovered the molecular basis of how FSIP1 negatively regulates autophagy in TNBC.
Fig. 7.
FSIP1 silencing promotes autophagy by releasing ULK1 from the FSIP1–ULK1 complex and causes chemoresistance through AMPK activation, Wnt/β-catenin pathway inhibition, and reduction in cell proliferation and migration, thus regulating the survival of TNBC.
Autophagy can be positively regulated by AMPK activation and negatively regulated by mTOR activation (29). In this study, we found that FSIP1 silencing enhanced ULK1 Ser317 and AMPK phosphorylation, but decreased ULK1 Ser757 and mTOR phosphorylation in MDA-MB-231 cells. How does FSIP1 silencing enhance AMPK activation? Since FSIP1 silencing reduced mitochondrial OCR and ATP production, it is likely that dysregulated mitochondrial biogenesis promotes autophagy (39). The reduced ATP production should increase AMP contents in TNBC cells. As a result, the increased ratios of AMP to ATP would activate the AMPK at Thr172, thereby inhibiting PI3K/AKT/mTOR signaling (40, 41). Simultaneously, the activated AMPK can phosphorylate ULK1 that drives the autophagic process (29, 42). Therefore, it is possible that FSIP1 silencing reduces the mitochondrial biogenesis and OCR to activate AMPK, which releases ULK1 from the FSIP1–ULK1 complex and down-regulates mTOR activation, leading to enhanced autophagy in TNBC cells. Our previous study and those of others have shown that Wnt/β-catenin signaling is crucial for the proliferation and invasion of TNBC cells (43, 44). Intriguingly, knockdown of β-catenin promotes AMPK activation and survival of head and neck squamous carcinoma cells (45). Conversely, AMPK activation inhibits Wnt/β-catenin signaling by phosphorylating TSC1/2 in breast cancer cells, and treatment with metformin to induce AMPK activation inhibits Wnt/β-catenin signaling and growth of breast cancer (46, 47). In this study, we observed that FSIP1 silencing not only enhanced AMPK activation and autophagy but also decreased Wnt/β-catenin signaling in TNBC cells. Our data indicate that a cross talk exists between AMPK and Wnt/β-catenin signaling pathways, which in turn regulates autophagy and survival of TNBC.
Methods
Please see SI Appendix, Supplemental Materials and Methods for details. Briefly, transient knockdown of FSIP1 or AMPK was performed using specific siRNAs and Mission siRNA transfection reagents (Sigma). Stable shFSIP1-expressing cells were generated using lentivirus (OriGene). FSIP1-KO cells were generated with an FSIP1-targeted guide RNA using CRISPR-Cas9 technology. For cytotoxicity, 1 × 103 cells were exposed to chemotherapeutic agents for 24 or 48 h, followed by proliferation assay (Promega). To determine cell migration, transwell assays were carried out. Mitochondrial respiration parameters were measured by extracellular flux analysis (Seahorse XF24) on 2 × 104 cells per well plated overnight. Transmission electron microscopy was done using 70-nm cell sections. For xenograft, 1 × 106 tumor cells were injected into the flanks of 6-wk-old female BALB/c nude mice. The experimental protocol was approved by the Animal Research and Care Committee of China Medical University.
Supplementary Material
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grants 81572609 and 31601142) and by National Institutes of Health Grants R01 AG23176, R01 AR065932, and R01 AR067066 (to M.Z.) and DK113627 (to M.Z. and L.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809681115/-/DCSupplemental.
References
- 1.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64:52–62. doi: 10.3322/caac.21203. [DOI] [PubMed] [Google Scholar]
- 2.Goldhirsch A, et al. Panel members Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol. 2013;24:2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowerman CJ, et al. Docetaxel-loaded PLGA nanoparticles improve efficacy in taxane-resistant triple-negative breast cancer. Nano Lett. 2017;17:242–248. doi: 10.1021/acs.nanolett.6b03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 5.Gluz O, et al. Triple-negative breast cancer–Current status and future directions. Ann Oncol. 2009;20:1913–1927. doi: 10.1093/annonc/mdp492. [DOI] [PubMed] [Google Scholar]
- 6.Gómez-Miragaya J, et al. Resistance to Taxanes in triple-negative breast cancer associates with the dynamics of a CD49f+ tumor-initiating population. Stem Cell Reports. 2017;8:1392–1407. doi: 10.1016/j.stemcr.2017.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie R, Nguyen S, McKeehan WL, Liu L. Acetylated microtubules are required for fusion of autophagosomes with lysosomes. BMC Cell Biol. 2010;11:89. doi: 10.1186/1471-2121-11-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wirth M, Joachim J, Tooze SA. Autophagosome formation—The role of ULK1 and Beclin1-PI3KC3 complexes in setting the stage. Semin Cancer Biol. 2013;23:301–309. doi: 10.1016/j.semcancer.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L, et al. Discovery of a small molecule targeting ULK1-modulated cell death of triple negative breast cancer in vitro and in vivo. Chem Sci. 2017;8:2687–2701. doi: 10.1039/c6sc05368h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Q, Fan W, Zhong Q. Regulation of Beclin 1 in autophagy. Autophagy. 2009;5:713–716. doi: 10.4161/auto.5.5.8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee Y, Weihl CC. Regulation of SQSTM1/p62 via UBA domain ubiquitination and its role in disease. Autophagy. 2017;13:1615–1616. doi: 10.1080/15548627.2017.1339845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lefort S, et al. Inhibition of autophagy as a new means of improving chemotherapy efficiency in high-LC3B triple-negative breast cancers. Autophagy. 2014;10:2122–2142. doi: 10.4161/15548627.2014.981788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park JH, Kim KP, Ko JJ, Park KS. PI3K/Akt/mTOR activation by suppression of ELK3 mediates chemosensitivity of MDA-MB-231 cells to doxorubicin by inhibiting autophagy. Biochem Biophys Res Commun. 2016;477:277–282. doi: 10.1016/j.bbrc.2016.06.057. [DOI] [PubMed] [Google Scholar]
- 15.Karantza-Wadsworth V, White E. Role of autophagy in breast cancer. Autophagy. 2007;3:610–613. doi: 10.4161/auto.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cappell KM, et al. Multiple cancer testis antigens function to support tumor cell mitotic fidelity. Mol Cell Biol. 2012;32:4131–4140. doi: 10.1128/MCB.00686-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mao Y, Xu R, Liu X, Shi W, Han Y. Elevated fibrous sheath interacting protein 1 levels are associated with poor prognosis in non-small cell lung cancer patients. Oncotarget. 2017;8:12186–12193. doi: 10.18632/oncotarget.14575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun M, et al. Fibrous sheath interacting protein 1 overexpression is associated with unfavorable prognosis in bladder cancer: A potential therapeutic target. OncoTargets Ther. 2017;10:3949–3956. doi: 10.2147/OTT.S143491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, et al. Expression and clinicopathological significance of FSIP1 in breast cancer. Oncotarget. 2015;6:10658–10666. doi: 10.18632/oncotarget.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chapman KB, et al. Elevated expression of cancer/testis antigen FSIP1 in ER-positive breast tumors. Biomarkers Med. 2013;7:601–611. doi: 10.2217/bmm.13.58. [DOI] [PubMed] [Google Scholar]
- 21.Liu T, et al. FSIP1 binds HER2 directly to regulate breast cancer growth and invasiveness. Proc Natl Acad Sci USA. 2017;114:7683–7688. doi: 10.1073/pnas.1621486114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100:672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 23.Mackeh R, Perdiz D, Lorin S, Codogno P, Poüs C. Autophagy and microtubules–New story, old players. J Cell Sci. 2013;126:1071–1080. doi: 10.1242/jcs.115626. [DOI] [PubMed] [Google Scholar]
- 24.McGrogan BT, Gilmartin B, Carney DN, McCann A. Taxanes, microtubules and chemoresistant breast cancer. Biochim Biophys Acta. 2008;1785:96–132. doi: 10.1016/j.bbcan.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 25.Snyder JP, Nettles JH, Cornett B, Downing KH, Nogales E. The binding conformation of taxol in beta-tubulin: A model based on electron crystallographic density. Proc Natl Acad Sci USA. 2001;98:5312–5316. doi: 10.1073/pnas.051309398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casimiro MC, et al. Cyclin D1 restrains oncogene-induced autophagy by regulating the AMPK-LKB1 signaling axis. Cancer Res. 2017;77:3391–3405. doi: 10.1158/0008-5472.CAN-16-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen S, et al. Autophagy is a therapeutic target in anticancer drug resistance. Biochim Biophys Acta. 2010;1806:220–229. doi: 10.1016/j.bbcan.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 28.Sun WL, Chen J, Wang YP, Zheng H. Autophagy protects breast cancer cells from epirubicin-induced apoptosis and facilitates epirubicin-resistance development. Autophagy. 2011;7:1035–1044. doi: 10.4161/auto.7.9.16521. [DOI] [PubMed] [Google Scholar]
- 29.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao C, et al. Autophagy negatively regulates Wnt signalling by promoting dishevelled degradation. Nat Cell Biol. 2010;12:781–790. doi: 10.1038/ncb2082. [DOI] [PubMed] [Google Scholar]
- 31.Henry WS, et al. Aspirin suppresses growth in PI3K-mutant breast cancer by activating AMPK and inhibiting mTORC1 signaling. Cancer Res. 2017;77:790–801. doi: 10.1158/0008-5472.CAN-16-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maiuri MC, et al. Control of autophagy by oncogenes and tumor suppressor genes. Cell Death Differ. 2009;16:87–93. doi: 10.1038/cdd.2008.131. [DOI] [PubMed] [Google Scholar]
- 33.Madrigal-Matute J, Cuervo AM. Regulation of liver metabolism by autophagy. Gastroenterology. 2016;150:328–339. doi: 10.1053/j.gastro.2015.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dokladny K, et al. Regulatory coordination between two major intracellular homeostatic systems: Heat shock response and autophagy. J Biol Chem. 2013;288:14959–14972. doi: 10.1074/jbc.M113.462408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang LH, et al. Enhanced autophagy reveals vulnerability of P-gp mediated epirubicin resistance in triple negative breast cancer cells. Apoptosis. 2016;21:473–488. doi: 10.1007/s10495-016-1214-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lazarus MB, Novotny CJ, Shokat KM. Structure of the human autophagy initiating kinase ULK1 in complex with potent inhibitors. ACS Chem Biol. 2015;10:257–261. doi: 10.1021/cb500835z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma B, et al. Dapper1 promotes autophagy by enhancing the Beclin1-Vps34-Atg14L complex formation. Cell Res. 2014;24:912–924. doi: 10.1038/cr.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salem AF, Whitaker-Menezes D, Howell A, Sotgia F, Lisanti MP. Mitochondrial biogenesis in epithelial cancer cells promotes breast cancer tumor growth and confers autophagy resistance. Cell Cycle. 2012;11:4174–4180. doi: 10.4161/cc.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meijer AJ, Codogno P. AMP-activated protein kinase and autophagy. Autophagy. 2007;3:238–240. doi: 10.4161/auto.3710. [DOI] [PubMed] [Google Scholar]
- 41.Meijer AJ, Codogno P. Autophagy: Regulation by energy sensing. Curr Biol. 2011;21:R227–R229. doi: 10.1016/j.cub.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 42.Xie CM, Liu XY, Sham KW, Lai JM, Cheng CH. Silencing of EEF2K (eukaryotic elongation factor-2 kinase) reveals AMPK-ULK1-dependent autophagy in colon cancer cells. Autophagy. 2014;10:1495–1508. doi: 10.4161/auto.29164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.King TD, Suto MJ, Li Y. The Wnt/β-catenin signaling pathway: A potential therapeutic target in the treatment of triple negative breast cancer. J Cell Biochem. 2012;113:13–18. doi: 10.1002/jcb.23350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao Z, et al. Nestin positively regulates the Wnt/β-catenin pathway and the proliferation, survival and invasiveness of breast cancer stem cells. Breast Cancer Res. 2014;16:408. doi: 10.1186/s13058-014-0408-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang HW, et al. Knockdown of β-catenin controls both apoptotic and autophagic cell death through LKB1/AMPK signaling in head and neck squamous cell carcinoma cell lines. Cell Signal. 2013;25:839–847. doi: 10.1016/j.cellsig.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 46.Choo AY, Roux PP, Blenis J. Mind the GAP: Wnt steps onto the mTORC1 train. Cell. 2006;126:834–836. doi: 10.1016/j.cell.2006.08.025. [DOI] [PubMed] [Google Scholar]
- 47.Parris AB, et al. Buformin inhibits the stemness of erbB-2-overexpressing breast cancer cells and premalignant mammary tissues of MMTV-erbB-2 transgenic mice. J Exp Clin Cancer Res. 2017;36:28. doi: 10.1186/s13046-017-0498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.