Significance
The International Union for Conservation of Nature (IUCN) Red List of Threatened Species is a key tool for the conservation of biological diversity. The evaluation and addition of species to this list is a time-consuming and costly task, and as such, a large number of species are not listed. For example, only 5% of plant species housed in the Global Biodiversity Information Facility are currently listed on the IUCN Red List. The simple and integrated protocol presented here enables conservation researchers and managers to identify unassessed species most likely at risk and, thus, assists in the direction of resource allocation for conservation. Our results suggest that efforts have been highly skewed geographically, and identify conservation hotspots in need of further evaluation.
Keywords: plantae, conservation, predictive modeling, random forest, IUCN
Abstract
The conservation status of most plant species is currently unknown, despite the fundamental role of plants in ecosystem health. To facilitate the costly process of conservation assessment, we developed a predictive protocol using a machine-learning approach to predict conservation status of over 150,000 land plant species. Our study uses open-source geographic, environmental, and morphological trait data, making this the largest assessment of conservation risk to date and the only global assessment for plants. Our results indicate that a large number of unassessed species are likely at risk and identify several geographic regions with the highest need of conservation efforts, many of which are not currently recognized as regions of global concern. By providing conservation-relevant predictions at multiple spatial and taxonomic scales, predictive frameworks such as the one developed here fill a pressing need for biodiversity science.
Biodiversity is essential for ecosystem function (1, 2) yet is being lost at an unprecedented rate (3). This threat to ecosystem function has downstream economic (4) and cultural (1) consequences that affect human health and well-being (5, 6). Plants are the foundation of ecosystem architecture and agriculture, and as such, changes in plant species diversity strongly influence processes such as biomass production, decomposition, and nutrient cycling (7, 8). Plant diversity is therefore critical for diversity on other trophic levels (9, 10).
Conserving biodiversity is a complex task that includes scientific, social, and political challenges. Both species (11) and geographic areas (12) must be identified as targets for conservation while considering time, monetary costs (13), and community acceptance (14). For these reasons, the International Union for Conservation of Nature (IUCN) Red List of Threatened Species (Red List) is a key conservation tool for both policy makers and researchers. This list represents the most comprehensive and consistent listing of conservation status for animal and plant species worldwide (15). However, despite the essential ecological role of plant species, plants are not as well represented on the Red List as animals (13) and are often neglected in favor of charismatic vertebrates (14). The 2010 Convention on Biological Diversity (CBD) Global Strategy for Plant Conservation aims to protect 75% of known threatened plant species, yet only about one-tenth of plant species are on the Red List (16, 17), whereas some (1,777) are classified as Data Deficient (DD) and many unlisted species are likely to be at risk (18, 19). Consequently, there is an urgent need for more efficient methods of identifying at-risk species. To meet this need, we developed and evaluated a predictive protocol that permits a rapid initial assessment of conservation status for understudied plant taxa.
Our framework assesses risk for all land plant (hereafter, plant) species with geographic coordinates available on the Global Biodiversity Information Facility (GBIF). We use a machine-learning approach to predict plant species Red List status using open source geographic, environmental, and morphological trait data for over 150,000 species, allowing us to provide conservation-relevant predictions at multiple spatial and taxonomic scales. Random forest (RF), a technique that builds random decision trees for classification and prediction (20, 21), has recently been applied to the exploration of biodiversity and conservation (e.g., refs. 18 and 22), and we use it to establish a predictive protocol for at-risk species at continental and global scales. We calculate the probability of each unlisted or DD species as belonging to a Red List non-Least Concern (non-LC) category (i.e., likely of being at risk on some level) and identify variables that are the most important in predicting conservation risk. We then identify global conservation hot- and coldspots and provide direct tools for local and global conservation needs. Our results indicate that a large number of unassessed species have a high probability of being at risk, and these probabilities can be used to establish assessment prioritization. Further, our work identifies global regions in need of conservation efforts, some of which are not currently recognized as regions of global concern. When appropriate, these results can be readily applied to direct conservation efforts at both the species and landscape scales.
Results and Discussion
Unlisted Species with Conservation Risk.
Plants represent the base of both natural and human-modified ecosystems and are central in sustaining full food chains. However, because of the resources required to perform detailed species assessments, only a small proportion of all described plant species are currently assessed by the Red List (6.5% according to the Catalogue of Life www.catalogueoflife.org/). The low representation of these groups within the Red List appears to be at least partially the result of a focus on charismatic species, differences in resource allocation across the globe, and an unbalanced presence of collectors across the world (16, 23, 24). Here, we respond to the challenge of global assessment and predict the Red List classification of unassessed land plant species. We evaluated several downsampling and resampling schemes to overcome biases in the data, and unless stated otherwise, the results presented below are based on predictions from downsampled data with LC vs. non-LC categories, as these produced the lowest balanced error rates (see SI Appendix, Supplementary Methods and Results for all analyses). Demonstrating the need of Red List assessments in plants, an astonishing 95% (153,057; Table 1) of the taxa databased in GBIF (with parameters that pass our filters) have never been assessed under the Red List protocol. The overall accuracy of our classifiers fell within the same range of those obtained in other studies (18, 25), at 73 to 82% globally. As we found previously (22), down- and subsampling balanced the error rates across categories (SI Appendix, Tables S3–S6) and should generally be applied in RF analyses in which datasets have unequal representation across categories (response variables).
Table 1.
Number of species for each IUCN Red List category and those not listed for each continent
| Continent | Red List Category | Not listed, n | Available to build classifier, n | |||||
| LC | NT | VU | EN | CR | DD | |||
| Africa | 941 | 153 | 505 | 335 | 81 | 43 | 23,142 | 2,015 |
| Asia | 1,363 | 113 | 184 | 67 | 30 | 64 | 31,584 | 1,757 |
| Australia | 488 | 40 | 48 | 24 | 5 | 5 | 22,538 | 605 |
| Central America | 351 | 54 | 136 | 91 | 29 | 15 | 14,999 | 661 |
| Europe | 753 | 66 | 60 | 57 | 29 | 71 | 17,597 | 965 |
| North America | 1,181 | 28 | 32 | 22 | 7 | 16 | 31,074 | 1,270 |
| South America | 888 | 353 | 872 | 411 | 84 | 63 | 44,590 | 2,608 |
| Global species | 2,134 | 130 | 205 | 108 | 32 | 61 | 30,424 | 2,609 |
Using the best classifiers built for two types of datasets, those containing only spatial data (“spatial”) and those containing both spatial and morphological data (“spatial+morpho”), we predicted Red List status for 213,927 and 17,231 species, respectively, and summarized the number of species predicted as non-LC and most likely in need of some conservation action (Table 2). For the spatial dataset, on average, 7.9% (range across continents, 3.4 to 13.6%) and 29.5% (range across continents, 18.7 to 41.9%) of plant taxa were predicted as non-LC at a probability of >0.80 and >0.60, respectively. For the spatial+morpho dataset, on average, 5.1% (range across continents, 0 to 10.1%) and 21.4% (range across continents, 10.2 to 43.3%) were predicted as non-LC at a probability of >0.80 and >0.60, respectively.
Table 2.
Number of species predicted as non-LC at probabilities above 0.80 and 0.60 and total number of predicted species and error rates
| Continent | Spatial | Spatial+Morpho | ||||||
| Species predicted, n | Error rate | Non-LC >0.80 | Non-LC >0.60 | Species predicted, n | Error rate | Non-LC >0.80 | Non-LC >0.60 | |
| Africa | 23,185 | 0.1983 | 1,631 | 4,352 | 899 | 0.1737 | 81 | 193 |
| Asia | 31,648 | 0.2462 | 1,553 | 9,336 | 928 | 0.2998 | 113 | 325 |
| Australia | 22,543 | 0.2709 | 2,854 | 7,185 | 7,866 | 0.2519 | 580 | 2,074 |
| Central America | 15,014 | 0.2731 | 1,115 | 4,841 | 1,552 | 0.2495 | 158 | 672 |
| Europe | 15,336 | 0.1825 | 2,089 | 5,095 | 1,247 | 0.4824 | 0 | 170 |
| North America | 31,090 | 0.2500 | 2,676 | 13,041 | 1,125 | 0.2506 | 33 | 115 |
| South America | 44,653 | 0.2520 | 1,534 | 9,616 | 1,295 | 0.3151 | 54 | 275 |
| Global species | 30,458 | 0.2197 | 1,797 | 8,303 | 1,319 | 0.2654 | 20 | 240 |
We identified a core set of species that were consistently predicted as non-LC at a high probability (Table 3 and Dataset S1). We also found that the vast majority of species predicted with the spatial dataset have characteristics that make them good candidates for further assessment (e.g., restricted ranges, endemism, and exposure to threats), further validating the predictions made using the RF model. However, this was not true for many of the species from the spatial+morpho dataset (Table 3 and Dataset S1). We suspect that the smaller dataset used to construct the classifiers in the spatial+morpho analyses (SI Appendix, Table S1) led to low power of these classifiers in most regions, and we recommend that these results be considered carefully.
Table 3.
Number of top 30 species predicted as non-LC in the spatial analysis likely to be listed as non-LC
| Region | Non-LC support |
| Africa | 29/30 |
| Asia | 23/30 |
| Australia | 21/30 |
| Central America | 20/30 |
| Europe | 22/30 |
| North America | 28/30 |
| South America | 25/30 |
| Global | 28/30 |
Results based on information from bibliographic searches. Results are shown for all regions. For the full list of species names, see Dataset S1, part 1.
From a practical perspective, the species-centered predictions can be used to prioritize risk assessment. Species with the highest probabilities in one or both datasets represent the most critical targets for future studies (see Dataset S2 for a complete list of species probabilities across all predictive models and continents). Biases in the search and assessment of species are widespread, and this also biases resource allocations toward species that are more visually attractive (24). The predictive protocol presented here is valuable, in that it successfully exploits open-source data, providing critical information for policy and decision makers who are responsible for the allocation of resources toward the investigation of conservation risk, and has the potential to increase the efficiency of conservation efforts and amplify the impact of biodiversity data in public data repositories. Notably, the computational requirements of the analyses are relatively low, permiting the use of personal computers, even for large datasets. Thus, this protocol is an extremely efficient way to prioritize species and geographic regions for conservation assessments and enables the optimization of both human and economic resources for the conservation of biodiversity.
Traits That Predict Extinction Risk.
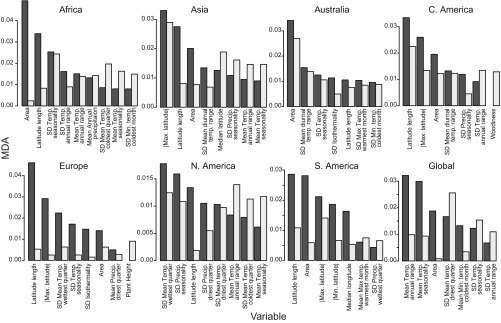
Although we did identify trends in the variables that contribute the most to at-risk classifiers across continents, there is no one single global variable that predicts conservation status. This result highlights the importance of considering local dynamics and conditions when making conservation-related decisions. In most cases, the geographic variables (area, length of latitude, and distance from the equator) were important predictors for which species are at risk (Fig. 1), in agreement with other work on conservation and biodiversity (17, 26). For example, species range size has long been considered in identifying taxa at risk [represented in Red List criterion B (27)], in part because small populations are more likely to go extinct than larger ones (represented in Red List criteria C and D; refs. 27 and 28). The fact that our analyses identify these variables as important strongly indicates the adequacy of RF to predict IUCN conservation status. Indeed, several Red List criteria relate to the spatial variables used in our analyses, and since RF is a classification algorithm, the use of these variables can represent an appropriate way of mechanizing the initial search of species at risk. Along with the geographic variables, some bioclimatic traits related to temperature [e.g., temperature seasonality (BIO4) and temperature annual range (BIO7)], ranked regularly among the top explanatory variables for all continents. Results from the spatial+morpho datasets are complementary to the spatial datasets. Even though morphological traits were used for all datasets, the Europe and Central America datasets were the only ones that identified a single top explanatory variable (woodiness and plant height, respectively). This result agrees with previous studies conducted at more restricted taxonomic/spatial scales, which indicate that plant habit can affect diversification rates (e.g., ref. 29). The full global dataset was more influenced by the bioclimatic variables pertaining to temperature than by the geographic variables.
Fig. 1.
Variable importance ranked by the MDA for all analyzed continents. Black bars indicate the spatial dataset; gray bars indicate the spatial+morpho dataset. Only the top five predictor variables for each model are included for simplicity, and they are ordered according to the spatial data.
The predictive framework developed here is an example of the capability for global analyses to complement local studies; investigations on both scales are thus complementary and important for conservation decision-making. We identify both spatial and morphological traits that are thought to influence the ability of plants to survive when facing threats. Such identification of mechanistic processes via the analysis of large datasets not only demonstrates the utility of information contained in open-source repositories but also the adequacy of the protocol presented here to identify species at risk.
Global Distribution of Extinction Risk.
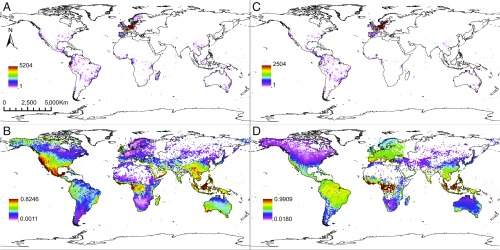
While our predictions can be used to prioritize the assessment of species, they also provide global information about the predicted distribution of expected conservation hotspots (Fig. 2). By associating each species’ GPS coordinates with their predicted probabilities (Fig. 2 B and D), we facilitate the identification of understudied regions in need of further conservation focus. Our predictions based on the spatial dataset identified several regions recognized as global biodiversity hotspots (30) and currently considered Key Biodiversity Areas (31) that are expected to harbor a large number of at-risk species (Fig. 2B). Some of these were the California Floristic Province, Mesoamerica, southeast North America, southwest Australia, parts of Sundaland and the Philippines, Madagascar, and the West African Rainforest. Other regions not recognized as hotspots (30) but recognized as Key Biodiversity Areas (31) were also recovered in our analyses, such as the Middle Eastern coast and Tasmania. Lastly, our results also recovered unrecognized regions, such as the southern coast of the Arabian Peninsula. The spatial+morpho dataset identified the West African Rainforest, including large portions of central Africa, the peri-Mediterranean, the Amazon basin, the California Floristic Province, and Sundaland as those likely to harbor non-LC species (Fig. 2D). These results agree with, and complement, other studies (30–32). An important but troubling result of our analysis is that currently well-assessed regions (e.g., Europe; Fig. 2 A and C) do not necessarily match major regions in need of conservation (Fig. 2 B and D). For instance, there is little overlap between the number of species assessed in a region and the likelihood of it harboring at-risk species. This stresses more strongly the importance of our discoveries: Our protocol can assist species assessments in regions where most species are yet to be assessed, thus translating global conservation predictions into specific regional actions. From another perspective, it is important to note that even though our methodology and datasets were substantially different from those used by Brum et al. (32), we recover similar areas likely in need of conservation. Further, because both Brum et al. (32) and our study considered all organisms belonging to high taxonomic ranks (mammals in ref. 32; land plants in our study), the results, together, should be seen as detecting consistent biodiversity patterns that are likely to be present across taxa and should therefore be considered in global, regional, and local conservation decision-making.
Fig. 2.
(A and C) Number of GPS coordinates per grid-cell that are of non-LC categories from the IUCN Red List used in the RF classifier for the spatial (A) and spatial+morpho (C) datasets. (B and D) Average per grid-cell probability of being listed as non-LC calculated by the RF classifier using the spatial (B) and spatial+morpho (D) datasets. See scales for values.
Conclusions
Plants are involved in myriad interspecific interactions [e.g., pollination, herbivory, and mycorrhizae (33, 34)], contribute to the diversification of organisms on Earth (35), can prevent natural disasters [e.g., flooding (36)], and contribute to ecosystem productivity in general (37). Given the CBD’s Global Strategy for Plant Conservation, a substantial number of species in need will go unprotected, resulting in quicker biodiversity loss than we are already experiencing (38). On a global scale, the level of threat to plants is much higher than expected. Additionally, our results indicate that several geographic regions should receive more attention from conservation biologists and/or decision-makers (Fig. 2) than they currently do. Both the IUCN Red List and other soundly defined conservation assessments can have an important impact on policy-making and conservation actions at various scales (39–41). Because it can be used for any assessment system that follows a structured protocol, our approach can be used on the region for which the system was created (e.g., province, country, continent, or biome), reflecting in each case the needs and specificities of the evaluated scale (41). Thus, when exploiting different scales, our predictive protocol provides researchers and conservation practitioners with a comprehensive list to assist the decision-making of resource allocation in conservation, regardless of the classification system used in each geographic region. Lastly, our results can be used to develop strategies that establish or sustain conservation actions to protect important geographic regions, thereby protecting key ecological systems and ecosystem services (42). For example, these data can be used in conjunction with information about how funding levels are distributed globally and where conservation measures will have the most impact (43, 44).
Materials and Methods
Data.
All georeferenced occurrence data for land plants were downloaded from the GBIF biodiversity database in March 2017 (see GitHub for links to all downloaded datasets, https://github.com/AnahiEspindola/PelletierEtAlPNAS). Many studies have pointed toward different biases and errors present in global biodiversity databases, such as taxonomic and spatial biases and coordinate inaccuracies (16, 23, 45–47). Even though some of these errors cannot be excluded, we sought to reduce them by taxonomic choice and checks and by spatial and data manipulations. It has been shown that the GBIF database harbors a globally good representation of the taxonomic diversity of land plants (16), indicating that our predictions should have a low taxonomic bias. However, it is known that GBIF can carry some taxonomic issues, in particular related to the presence of taxonomic synonyms and poorly taxonomically curated taxa (48). For this reason, we checked all species and, when needed, corrected their taxonomy using the classification function in the R package taxize (49). Lastly, we selected only those GPS coordinates with “no known coordinate issues” and that had come from human observation, observation, literature occurrence, specimen, and material sample. After removing duplicates, we retained species with five or more GPS coordinates to eliminate species that might be incorrectly assigned to a continent, and we used GPS coordinates that contained between one and six decimal values. To evaluate the bias that known georeferencing errors could bring into our results, we tested the filter proposed in ref. 46, and our results indicated that these errors did not affect our results in significant ways (SI Appendix, Figs. S4 and S5). We bounded our search locations by continent, excluding Antarctica, and using most of the regions defined in the 1:5,000,000 ESRI World Continents shapefile (50). An exception to this was that we split the original North America designation so as to include (i) Mexico, Canada, and the United States (“North America”), and (ii) all countries south of Mexico and north of South America (“Central America”). There were totals of 1,048,575 geographic coordinates from Africa; 1,821,626 from Asia; 7,329,297 from Australia; 858,234 from Central America; 48,394,349 from Europe; 4,326,043 from North America; and 2,855,312 from South America (Table 1 and SI Appendix, Fig. S1). Because some species are present in several continents, we also ran analyses using species endemic to each continent (seven “endemic” datasets) and those globally present (one “global” dataset using species that are found on at least two continents). In parallel, all plant species on the Red List were downloaded by continent from the IUCN website in March 2017. Species are listed as either DD, LC, Near Threatened (NT), Vulnerable (VU), Endangered (EN), Critically Endangered (CR), Extinct in the Wild (EW), or Extinct (EX). Red List categories were assigned to species present in our GBIF database. IUCN listings from before 2001 were recoded so that listings were consistent across time periods: Near Threatened (LR/nt) = NT, Conservation Dependent (LR/cd) = VU, and Least Concern (LR/lc) = LC; DD was recoded as Not Available (NA) and predicted with our classifiers. EW and EX taxa were excluded from all analyses. To generalize our results, we also coded all listed taxa as LC (LC and NT) vs. non-LC (CR, EN, VU), and CR vs. non-CR. This allowed us to increase the number of observations per predicted category, which had a direct impact on the quality of our classifiers and resulted in overall better predictors (SI Appendix, Tables S2–S6). Analyses were done on three sets of response categories (i.e., all IUCN categories, LC vs. non-LC, and CR vs. non-CR), but only the LC vs. non-LC category is presented in the main manuscript (see also SI Appendix, Supplementary Methods and Results). Based on preliminary analyses, and because of the very different biogeographic history of Hawaii, GPS coordinates from these islands were excluded from the North America dataset.
All analyses were run in R v.3.4.0 (R Core 2017), using custom scripts [data and scripts are available on GitHub (https://github.com/AnahiEspindola/PelletierEtAlPNAS)]. For all GPS coordinates retained from the above search, we extracted data from a series of spatial and environmental variables on a species-by-species basis using the R packages geosphere (51), raster (52), and plyr (53). First, coordinates were used to draw a polygon, from which only the area falling on land masses was used as a proxy for range area. For latitude and longitude, we extracted maxima and minima, absolute value of maxima, and median. Lastly, for each locality, we extracted values for the 19 bioclimatic WorldClim variables (54) at a resolution of 30 s (∼1 km2) and calculated their mean values and SDs on a species-by-species basis. We refer to this as the spatial dataset [available on github (URL: https://github.com/AnahiEspindola/PelletierEtAlPNAS)].
Bioclimatic variables often display a certain level of correlation. Even though this has been suggested to affect the ability of RF to accurately identify the contribution of each variable to the classifier (e.g., ref. 55), this does not influence the predictive ability of the classifier because subsets of variables are used to create each decision tree (see ref. 56 for a review). To understand the level of correlation of our variables and its potential impact, we performed pairwise correlations of all our variables. This analysis indicated that they are overall little correlated (Dataset S3) and display a level of correlation that falls within ranges demonstrated to not affect the ability of RF to correctly interpret variable importance (55).
Along with the spatial data, we obtained morphological trait data from the plant trait database TRY (ref. 57; https://www.try-db.org/). Because the TRY traits were not equally represented across taxa, and RF does not accommodate missing data, we selected between one and three traits that were best represented across taxa for each continental dataset. These traits included woodiness (global and all continents), leaf phenology (global, Africa, Asia, Europe, and North America), and plant height (global, Europe, and North America). Some continents (Africa, Europe, and North America) had species lacking values for traits that belong to families in which more than 50% of the species had that trait data available; to allow for the accommodation of missing data, these values were imputed using the missForest function in the missForest R package (58). Imputation was done within the family, with replacement, and with the parameter ntree set to 100. We refer to these as the spatial+morpho datasets.
RF with Spatial Data.
RF is an appropriate method for global biodiversity datasets because it can accommodate large amounts of data, including high numbers of both observations and predictor variables; it does not overfit; and it circumvents issues associated with variable correlation. It constructs decision trees built on a series of random samples of variables and observations and is extremely efficient in classification and prediction for complex datasets (20, 21). The RF method has been shown to perform better than other machine-learning approaches in predicting ecological status in some restricted clades (18, 25). We used the R package randomForest (59) to build classifiers based on 1,000 random trees, using the following variables as predictor variables: range area, maximum distance from the equator, minimum distance from the equator, median latitude, length of latitude (degrees the latitude extends), median longitude, and the mean values and SD of each of the 19 bioclimatic variables. RF samples the data with replacement for each decision tree in the forest, and uses the unsampled datasets (one third of the total dataset) to test the model. This information is used to build a confusion matrix for the prediction and calculate the out-of-bag (OOB) error rates.
We conducted RF analyses on six spatial datasets per continent, for both “all species” and “endemics only”. These datasets contained either (i) all species, without any manipulation to avoid Red List category imbalance, or (ii) iterated downsampling of the majority class(es) to match the value of the minority class (Table 1 and SI Appendix, Tables S1 and S2) (22, 60). For each of these, we ran RF analyses using (i) all IUCN Red List categories as the response variable, (ii) IUCN categories coded as either LC or non-LC as the response variable, or (iii) IUCN Red List categories coded as either CR or non-CR as the response variable. For RF on the downsampled datasets, we subsampled the majority class(es) 100 times to match that of the minority class, ran the RF, and then averaged results from these iterations. Lastly, we conducted two additional LC vs. non-LC RF analyses after removing species that were misclassified in the model 90% and 80% of the time, in an attempt to improve model accuracy. This was done under the assumption that some species may be inaccurately categorized by IUCN or contain abnormal characteristics for being on the Red List, and their removal should therefore increase the accuracy of the prediction.
RF with Spatial+Morpho Data.
We used the R package randomForest (59) to build classifiers based on 1,000 random trees. Here, we used all spatial variables present in the spatial dataset, but also added the morphological trait values presented in the Data section and calculated OOB error rates as above. This dataset was analyzed using approaches similar to those used for the spatial dataset; however, because the response classes were extremely imbalanced, we applied downsampling and resampling strategies only on the LC vs. non-LC coded data (22, 60): (i) downsampling of the majority class to match the value of the minority class, and (ii) resampling of classes to double the minority class count (SI Appendix, Table S1). We conducted 1,000 iterations, and the results were averaged.
RF Predictions.
For each continent and dataset type (i.e., spatial and spatial+morpho), we used the OOB error rates to identify the most accurate classifiers, considering both overall OOB error and within-class OOB error (SI Appendix, Tables S2–S6). We used the predict.randomForest function in the randomForest R package to calculate the probability of belonging to each Red List category of nonassessed species from each continent. For all downstream analyses and discussion, we refer to the LC vs. non-LC downsampled RF results for both the spatial and spatial+morpho datasets (all predictions found in Dataset S2).
To understand the level of agreement between predictions from the spatial and spatial+morpho datasets, we calculated the number of species that were predicted to belong to the same category by both methods, using a probability threshold of 80% (Dataset S1). Because we suspected the predictive power of the spatial dataset to be greater than that of the spatial+morpho dataset, we further evaluated predictions from this spatial dataset for the top 30 non-LC predicted species for each region (Table 3 and Dataset S1). We performed online bibliographic searches to identify whether or not those species already display any indication of being potentially in need of conservation actions (e.g., endemics, restricted ranges, occupying threatened regions, rare species, etc.).
Variable Importance.
The importance of each variable was determined by measuring the mean decrease in accuracy (MDA) of the prediction after the removal of each variable from the predictive function. For our downsampling schemes, we calculated the mean and range MDA for all iterations. Lastly, we compared these results across datasets and for each continent (the top five variables from each dataset are shown in Fig. 1; the values for the endemic datasets are reported in SI Appendix, Fig. S2).
Global Distribution of Non-LC.
One of our goals was to use our predictions to inform conservation not only at the species level, but also on a global scale. To achieve this goal, we associated the probability value of being non-LC for all unassessed species to each of their own georeferenced GPS coordinates. After doing so, we used the raster package (52) in R to calculate the average probability of non-LC for all GPS coordinates within each cell of a 1° × 1° grid covering the world (see SI Appendix, Fig. S3 for the endemic plots).
All data are deposited on GitHub (https://github.com/AnahiEspindola/PelletierEtAlPNAS) and further analyses are presented in SI Appendix.
Supplementary Material
Acknowledgments
We thank Michael Cummings and James Foster for discussions on RF. This work was supported by the National Science Foundation (Grants DEB-1457519 and DEB-1457726) and the Institute for Bioinformatics and Evolutionary Studies at the University of Idaho (supported by NIH Grants NCRR 1P20RR016454-01 and NCRR 1P20RR016448-01 and by the NSF Grant EPS-809935). This study has been supported by the TRY initiative on plant traits (https://www.try-db.org/). The TRY initiative and database is hosted, developed, and maintained by J. Kattge and G. Bönisch (Max Planck Institute for Biogeochemistry, Jena, Germany). TRY is currently supported by Diversitas/Future Earth and the German Centre for Integrative Biodiversity Research Halle-Jena-Leipzig.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All data and scripts used to analyze the data are available on GitHub (https://github.com/AnahiEspindola/PelletierEtAlPNAS).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804098115/-/DCSupplemental.
References
- 1.Cardinale BJ, et al. Biodiversity loss and its impact on humanity. Nature. 2012;486:59–67. doi: 10.1038/nature11148. [DOI] [PubMed] [Google Scholar]
- 2.Oliver TH, et al. Biodiversity and resilience of ecosystem functions. Trends Ecol Evol. 2015;30:673–684. doi: 10.1016/j.tree.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Tittensor DP, et al. A mid-term analysis of progress toward international biodiversity targets. Science. 2014;346:241–244. doi: 10.1126/science.1257484. [DOI] [PubMed] [Google Scholar]
- 4.Bateman IJ, et al. Bringing ecosystem services into economic decision-making: Land use in the United Kingdom. Science. 2013;341:45–50. doi: 10.1126/science.1234379. [DOI] [PubMed] [Google Scholar]
- 5.Perrings C, Folke C, Maler KG. The ecology and economics of biodiversity loss–The research agenda. Ambio. 1992;21:201–211. [Google Scholar]
- 6.Handa IT, et al. Consequences of biodiversity loss for litter decomposition across biomes. Nature. 2014;509:218–221. doi: 10.1038/nature13247. [DOI] [PubMed] [Google Scholar]
- 7.Tilman D, Wedin D, Knops J. Productivity and sustainability influenced by biodiversity in grassland ecosystems. Nature. 1996;379:718–720. [Google Scholar]
- 8.Cardinale BJ, et al. The functional role of producer diversity in ecosystems. Am J Bot. 2011;98:572–592. doi: 10.3732/ajb.1000364. [DOI] [PubMed] [Google Scholar]
- 9.Siemann E, Tilman D, Haarstad J, Ritchie M. Experimental tests of the dependence of arthropod diversity on plant diversity. Am Nat. 1998;152:738–750. doi: 10.1086/286204. [DOI] [PubMed] [Google Scholar]
- 10.Zak DR, Holmes WE, White DC, Peacock AD, Tilman D. Plant diversity, soil microbial communities, and ecosystem function: Are there any links? Ecology. 2003;84:2042–2050. [Google Scholar]
- 11.Winter M, Devictor V, Schweiger O. Phylogenetic diversity and nature conservation: Where are we? Trends Ecol Evol. 2013;28:199–204. doi: 10.1016/j.tree.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 12.Forest F, et al. Preserving the evolutionary potential of floras in biodiversity hotspots. Nature. 2007;445:757–760. doi: 10.1038/nature05587. [DOI] [PubMed] [Google Scholar]
- 13.Stuart SN, Wilson EO, McNeely JA, Mittermeier RA, Rodríguez JP. Ecology. The barometer of life. Science. 2010;328:177. doi: 10.1126/science.1188606. [DOI] [PubMed] [Google Scholar]
- 14.Balding M, Williams KJ. Plant blindness and the implications for plant conservation. Conserv Biol. 2016;30:1192–1199. doi: 10.1111/cobi.12738. [DOI] [PubMed] [Google Scholar]
- 15.Vié J-C, Hilton-Taylor C, Stuart SN. Wildlife in a Changing World—An Analysis of the 2008 IUCN Red List of Threatened Species. IUCN; Gland, Switzerland: 2009. [Google Scholar]
- 16.Troudet J, Grandcolas P, Blin A, Vignes-Lebbe R, Legendre F. Taxonomic bias in biodiversity data and societal preferences. Sci Rep. 2017;7:9132. doi: 10.1038/s41598-017-09084-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schluter D, Pennell MW. Speciation gradients and the distribution of biodiversity. Nature. 2017;546:48–55. doi: 10.1038/nature22897. [DOI] [PubMed] [Google Scholar]
- 18.Bland LM, Collen B, Orme CD, Bielby J. Predicting the conservation status of data-deficient species. Conserv Biol. 2015;29:250–259. doi: 10.1111/cobi.12372. [DOI] [PubMed] [Google Scholar]
- 19.Keith DA, et al. Detecting extinction risk from climate change by IUCN Red List criteria. Conserv Biol. 2014;28:810–819. doi: 10.1111/cobi.12234. [DOI] [PubMed] [Google Scholar]
- 20.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 21.Biau G. Analysis of a random forests model. J Mach Learn Res. 2012;13:1063–1095. [Google Scholar]
- 22.Espíndola A, et al. Identifying cryptic diversity with predictive phylogeography. Proc Biol Sci. 2016;283:20161529. doi: 10.1098/rspb.2016.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donaldson MR, et al. Taxonomic bias and international biodiversity conservation research. Facets. 2016;1:105–113. [Google Scholar]
- 24.Martín-López B, González JA, Montes C. The pitfall-trap of species conservation priority setting. Biodivers Conserv. 2011;20:663–682. [Google Scholar]
- 25.Cutler DR, et al. Random forests for classification in ecology. Ecology. 2007;88:2783–2792. doi: 10.1890/07-0539.1. [DOI] [PubMed] [Google Scholar]
- 26.Niskanen AK, Heikkinen RK, Väre H, Luoto M. Drivers of high-latitude plant diversity hotspots and their congruence. Biol Conserv. 2017;212:288–299. [Google Scholar]
- 27.Mace GM, et al. Quantification of extinction risk: IUCN’s system for classifying threatened species. Conserv Biol. 2008;22:1424–1442. doi: 10.1111/j.1523-1739.2008.01044.x. [DOI] [PubMed] [Google Scholar]
- 28.Lande R. Risks of population extinction from demographic and environmental stochasticity and random catastrophes. Am Nat. 1993;142:911–927. doi: 10.1086/285580. [DOI] [PubMed] [Google Scholar]
- 29.Soltis DE, et al. Phylogenetic relationships and character evolution analysis of Saxifragales using a supermatrix approach. Am J Bot. 2013;100:916–929. doi: 10.3732/ajb.1300044. [DOI] [PubMed] [Google Scholar]
- 30.Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GA, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 31.International Union for the Conservation of Nature . Version 1.0. 1st Ed. IUCN; Gland, Switzerland: 2016. A Global Standard for the Identification of Key Biodiversity Areas; p. 46. [Google Scholar]
- 32.Brum FT, et al. Global priorities for conservation across multiple dimensions of mammalian diversity. Proc Natl Acad Sci USA. 2017;114:7641–7646. doi: 10.1073/pnas.1706461114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farrell B, Mitter C, Futuyma DJ. Diversification at the insect-plant interface. Bioscience. 1992;42:34–42. [Google Scholar]
- 34.Brundrett MC. Coevolution of roots and mycorrhizas of land plants. New Phytol. 2002;154:275–304. doi: 10.1046/j.1469-8137.2002.00397.x. [DOI] [PubMed] [Google Scholar]
- 35.Drès M, Mallet J. Host races in plant-feeding insects and their importance in sympatric speciation. Philos Trans R Soc Lond B Biol Sci. 2002;357:471–492. doi: 10.1098/rstb.2002.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McIvor AL, Möller I, Spencer T, Spalding M. 2012. Reduction of wind and swell waves by mangroves. (The Nature Conservancy and Wetlands International, Cambridge, UK), Natural Coastal Protection Series, Report 1. Cambridge Coastal Research Unit Working Paper 40, p 27.
- 37.Loreau M, et al. Biodiversity and ecosystem functioning: Current knowledge and future challenges. Science. 2001;294:804–808. doi: 10.1126/science.1064088. [DOI] [PubMed] [Google Scholar]
- 38.Jiang L, Pu Z. Different effects of species diversity on temporal stability in single-trophic and multitrophic communities. Am Nat. 2009;174:651–659. doi: 10.1086/605961. [DOI] [PubMed] [Google Scholar]
- 39.Persson Å, et al. Editorial: Environmental Policy Integration: Taking stock of policy practice in different contexts. Environ Sci Policy. 2018;85:113–115. [Google Scholar]
- 40.Rodrigues ASL, Pilgrim JD, Lamoreux JF, Hoffmann M, Brooks TM. The value of the IUCN Red List for conservation. Trends Ecol Evol. 2006;21:71–76. doi: 10.1016/j.tree.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 41.De Grammont PC, Cuarón AD. An evaluation of threatened species categorization systems used on the American continent. Conserv Biol. 2006;20:14–27. doi: 10.1111/j.1523-1739.2006.00352.x. [DOI] [PubMed] [Google Scholar]
- 42.Harvey E, Gounand I, Ward CL, Altermatt F. Bridging ecology and conservation: From ecological networks to ecosystem function. J Appl Ecol. 2017;54:371–379. [Google Scholar]
- 43.Jenkins CN, Pimm SL, Joppa LN. Global patterns of terrestrial vertebrate diversity and conservation. Proc Natl Acad Sci USA. 2013;110:E2602–E2610. doi: 10.1073/pnas.1302251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waldron A, et al. Targeting global conservation funding to limit immediate biodiversity declines. Proc Natl Acad Sci USA. 2013;110:12144–12148. doi: 10.1073/pnas.1221370110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruete A. Displaying bias in sampling effort of data accessed from biodiversity databases using ignorance maps. Biodivers Data J. 2015:e5361. doi: 10.3897/BDJ.3.e5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maldonado C, et al. Estimating species diversity and distribution in the era of Big Data: To what extent can we trust public databases? Glob Ecol Biogeogr. 2015;24:973–984. doi: 10.1111/geb.12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.James SA, et al. Herbarium data: Global biodiversity and societal botanical needs for novel research. Appl Plant Sci. 2018;6:e1024. doi: 10.1002/aps3.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boyle B, et al. The taxonomic name resolution service: An online tool for automated standardization of plant names. BMC Bioinformatics. 2013;14:16. doi: 10.1186/1471-2105-14-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chamberlain SA, Szöcs E. taxize: Taxonomic search and retrieval in R. F1000 Res. 2013;2:191. doi: 10.12688/f1000research.2-191.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Environmental Systems Research, Inc. 2018. World continents. The World Factbook, ed Global Mapping International USCIA (ESRI, Redlands, CA)
- 51.Hijmans RJ, Williams E, Vennes C. 2016 geosphere: Spherical trigonometry for geographic applications, R Package version 1.5-5. Available at https://cran.r-project.org/web/packages/geosphere/index.html. Accessed October 30, 2018.
- 52.Hijmans RJ, et al. 2016 raster: Geographic analysis and modeling with raster data, version 2.5-8. Available at https://cran.r-project.org/web/packages/raster/index.html. Accessed October 30, 2018.
- 53.Wickham H. The split-apply-combine strategy for data analysis. J Stat Softw. 2011;40:1–29. [Google Scholar]
- 54.Fick SE, Hijmans RJ. WorldClim 2: New 1-km spatial resolution climate surfaces for global land areas. Int J Climatol. 2017;37:4302–4315. [Google Scholar]
- 55.Gregorutti B, Michel B, Saint-Pierre P. Correlation and variable importance in random forests. Stat Comput. 2017;27:659–678. [Google Scholar]
- 56.Biau G, Scornet E. A random forest guided tour. Test. 2016;25:197–227. [Google Scholar]
- 57.Kattge J, et al. TRY–A global database of plant traits. Glob Chang Biol. 2011;17:2905–2935. [Google Scholar]
- 58.Stekhoven DJ, Bühlmann P. MissForest–Non-parametric missing value imputation for mixed-type data. Bioinformatics. 2012;28:112–118. doi: 10.1093/bioinformatics/btr597. [DOI] [PubMed] [Google Scholar]
- 59.Liaw A, Wiener M. Classification and regression by randomForest. R News. 2002;2:18–22. [Google Scholar]
- 60.Chen C, Liaw A, Breiman L. 2004 Using random forest to learn imbalanced data (University of California, Berkeley, CA), pp 1–12. Available at statistics.berkeley.edu/sites/default/files/tech-reports/666.pdf. Accessed October 30, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.